Abstract
Background
Transcription factor lark has been demonstrated to play multiple functions in Drosophila, but the function of this gene in embryonic development remains to be elucidated.
Results
In this study, the CRISPR/Cas9 gene-editing method was used to construct a Bmlark mutant strain of Bombyx mori to investigate the roles of this gene. The results showed that the homozygous mutant Bmlark−/− was lethal. The Bmlark−/− embryos showed obvious developmental defects, such as defective sclerotization and melanization of the exoskeleton. A transcriptomic comparison of Bmlark−/− and wild-type embryos showed that the differentially expressed genes were mainly enriched in the structure and metabolic processes of chitin and cuticles. While the expression levels of chitin metabolism-related enzyme genes did not significantly change, in the mutant embryos compared to the wild-type embryos, the expression levels of 63 putative cuticle protein genes showed significant differences. Among which, 35 genes were downregulated and 28 genes were upregulated. The expression levels of the transcription factor BmPOUM2 and eight wing disc cuticle protein genes (WCP) also changed. BmPOUM2, WCP5, WCP9, WCP10, WCP11 were downregulated and WCP1, WCP2, WCP3, WCP6 were upregulated in Bmlark−/− embryos. While the expression level of TH in the tyrosine-mediated pigmentation pathway was upregulated in the mutant embryos, the expression levels of the four key pigment synthesis genes DDC, aaNAT, Laccase2A, and yellow-f2 were significantly downregulated.
Conclusions
The expression levels of 63 putative cuticle protein genes, eight WCP genes, and five pigment synthesis genes significantly changed in Bmlark mutant B. mori compared to those of the wildtype. These results suggest that Bmlark is essential for normal development of cuticle and tyrosine-mediated melanization in silkworm embryos.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11107-2.
Keywords: Bmlark, Mutant, Embryonic development, Sclerotization, Melanization, Bombyx mori
Background
LARK is a vital transcription factor and plays multiple biological functions in many species. In Drosophila, the lark gene (Dmlark) was first identified from a mutant with an abnormal eclosion rhythm [1]. Dmlark has a broad spatiotemporal expression profile in many tissues, including the nervous system, ring gland, gut and Malpighian tubules, at different developmental stages, including eggs, larvae, and pupae [2]. Dmlark was found to participate in the regulation of the eclosion rhythm [3, 4], circadian rhythm [2, 5–7], eye development [6], embryonic development [1], oogenesis [8], neuronal development and physiology [1, 4, 9, 10], and gut immunocompetence [11]. In Drosophila, recessive homozygous embryonic lethality was observed in lark mutants caused by a P-element insertion, but the specific regulatory mechanism was not reported [1]. The dead embryos showed defects in morphology and structures that were almost unrecognizable, and the nervous system was severely disorganized [1].
The homologous gene of Dmlark in mammals is RBM4 (RNA-binding motif protein 4). By regulating alternative splicing of target mRNA precursors, RBM4 contributes to diverse biological functions, such as muscle cell differentiation [12], development of brown adipocytes [13], neuronal cell differentiation and neurite outgrowth [14], neuronal migration [15], migration and invasion of human colorectal cancer cells [16], and cancer suppression [17–19]. Human RBM4 was also revealed to participate in microRNA-guided gene regulation [20] and RNA translation [21]. RBM4 participated in translation suppression via Argonaute2 during muscle cell differentiation [22]. Mouse LARK was found to affect circadian rhythms, possibly by modulating the translation of mPER1 mRNA [23].
Bmlark in Bombyx mori, an economically important silk producing insect, has two splicing isoforms, Bmlark-PA and Bmlark-PB. Bmlark-PA is the longer transcript and contains two RNA recognition motif domains and one retroviral-type zinc finger domain, while Bmlark-PB lacks all these functional domains and may perform only basic functions [24]. Bmlark is consistently expressed across various tissues, including head, testis, ovary, and flight muscle, and at different developmental stages, including eggs, larvae, pupa and adults [24, 25]. Recently, it was reported that BmLARK is involved in the alternative splicing of the sex determination gene doublesex [26]. Our laboratory found that B. mori LARK is involved in regulating of the expression of B. mori acyl-CoA binding protein gene (BmACBP), and therefore lipid metabolism [27]. BmLARK was found to bind to G4 structures in the promoters of the transcription factor BmPOUM2 and many other genes that contain G4 structures in B. mori and humans to regulate their expression [28].
In this study, a Bmlark knockout strain of silkworm was generated using the CRISPR/Cas9 gene editing method. We found that embryos of the homozygous mutant Bmlark−/− died during embryogenesis, suggesting that Bmlark is essential for the embryonic development in silkworm. The expression levels of BmPOUM2, cuticle protein genes (CPs), and pigment synthesis related genes in the Bmlark−/− mutant were notably changed, and therefore may be the main factors leading to the death of the mutant embryos.
Materials and methods
Insects
Bombyx mori strain P50 used in this study was obtained from the Sericulture and Agri-Food Research Institute of Guangdong Academy of Agricultural Sciences, China. Eggs were incubated at 25 °C -27 °C and 75%-85% relative humidity, with a photoperiod cycle of 12 h light:12 h dark. Larvae were reared with fresh mulberry leaves at 25 °C -27 °C and 50%-60% relative humidity, with a photoperiod cycle of 12 h light:12 h dark.
CRISPR/Cas9-mediated gene editing
The CRISPR/Cas9 system was used to knock out the Bmlark gene (Gene ID: 692732) in silkworm. Oligonucleotide sequences (20 nt) that targeted Bmlark were designed with the online tool CRISPRdirect (http://crispr.dbcls.jp/) [29]. The complete single-guide RNA (sgRNA, 116 nt) containing a T7 promoter, a target site sequence and a guide RNA sequence (Supplementary Table S1) was synthesized using a MEGAscript™ T7 transcription kit (Thermo Scientific, Shanghai, China). All primers used for the synthesis of the sgRNA are listed in Supplementary Table S1. The experimental procedure was performed with reference to the literature [30].
Fertilized silkworm eggs were microinjected within 3 h after oviposition. The sgRNA and Cas9 protein (PNA Bio, California, USA) were co-injected at final concentrations of 600 ng/μL, in a volume of 10 nL per egg. Hatched larvae were recorded as the G0 generation. The offspring of G0 mating were recorded as G1, and so forth for subsequent generations.
With the intent to not affect survival, larval-pupal molt residues or partial wings of adult were collected to isolate genomic DNA. The target region sequences were amplified by PCR from the genomic DNA and sequenced to examine the mutations. The G0 heterozygous mutant moths (Bmlark+/-) were back-crossed with the wild-type moths to produce a G1 generation, and then Bmlark+/- mutants in the G1 generation carrying the same mutation were self-crossed to obtain homozygous mutants (Bmlark−/−). All primers used for synthesizing sgRNAs and screening mutants are listed in Supplementary Table S1.
Transmission electron microscope (TEM) examination
Bmlark−/− and wild-type (WT) embryos were collected from eggs 168 h after oviposition (about the beginning of head pigmentation) and fixed in 2.5% glutaraldehyde at 4℃ overnight. Then, the embryos were washed using 0.1 M phosphate buffer (PB, pH 7.4) three times, 15 min each time. Embryos were fixed with 1% OsO4 in 0.1 M PB for 2 h in the dark at room temperature. After removing the OsO4, the embryos were washed again in 0.1 M PB three times, 15 min each time. The embryos were dehydrated at room temperature using an ethanol gradient of 30%, 50%, 70%, 80%, 95%, 100%, and 100%, each for 20 min, and finally incubated twice in acetone for 15 min. Resin penetration and embedding were performed at 37℃ using the following reagents for the specified time periods: acetone:EMBed 812 (1:1) for 4 h; acetone:EMBed 812 (1:2) overnight; and pure EMBed 812 for 8 h. Then, pure EMBed 812 was poured into embedding molds and the embryos were immerged into the pure EMBed 812 at 37℃ overnight. The embedding molds containing the resin and embryos were placed in a 65℃ oven to polymerize for 48 h. Then, the resin blocks were removed from the embedding molds at room temperature and 70 nm thin cross sections of the embryo samples were cut using an ultramicrotome (EM UC7, Leica, Germany). The ultrathin sections were placed onto the 150-mesh cuprum grids with Formvar film (WFHM-150, Servicebio, China). The samples in the cuprum grids were immersed in a 2% uranium acetate saturated alcohol solution for 8 min for staining while avoiding light, and then rinsed in 70% ethanol three times and in ultrapure water three times. Then the samples were immersed in 2.6% lead citrate for 8 min while avoiding CO2 and rinsed with ultrapure water three times. After drying with a filter paper, the cuprum grids were placed onto a grid board and dried overnight at room temperature. The cuprum grids were observed and photographed under a transmission electron microscope (HT7800, HITACHI, Japan).
Chitin staining
Bmlark−/− and WT embryos were dissected from eggs 168 h and 180 h after oviposition and fixed in 4% paraformaldehyde for more than 24 h. The fixed samples were dehydrated with an ethanol gradient of 50%, 70%, 85%, 95%, and 100%, each for 1 h, treated with ethanol:xylene (1:1) for 30 min, and then treated with xylene twice for 30 min each time. The treated samples were infiltrated with paraffin and then embedded into paraffin blocks. Paraffin sections were prepared for subsequent chitin staining and hematoxylin–eosin (HE) staining. The sections were placed in xylene twice, each time for 5 min, to completely remove the paraffin. After being treated with xylene:ethanol (1:1) for 5 min, the sections were treated with 100%, 95%, 90%, 80%, 70%, and 50% alcohol, each for 5 min, to rehydrate. Fluorescent brightener 28 (10 μg/mL) (Sigma-Aldrich, USA) was used as a chitin staining dye. Sections were treated with the dye for 10 s, followed immediately by washing three times with distilled water, 1 min each time. Cell nuclei were stained with propidium iodide (10 μg/mL) (Sigma-Aldrich, USA) for 10 s and washed twice with distilled water, each time for 1 min. Glycerol and 1 × PBS (pH 7.4) were mixed in equal volumes as a sealing agent. The sealed sections were observed under a confocal laser microscope (FV3000, Olympus, Japan).
HE staining
Paraffin sections of Bmlark−/− and WT embryos prepared as described above were also used for HE staining analysis. After being deparaffinized and rehydrated as described above, the sections were treated with a hematoxylin staining solution for 5 min, then immediately washed with running tap water to remove the excess staining solution. The sections were treated with 1% hydrochloric acid in 70% alcohol three times, each time for 10 s, and rinsed with running tap water for 2 min. The sections were then treated with 1% ammonia to color the nuclei, washed with double-distilled water for 2 min and treated sequentially with 70%, 80%, 90%, and 95% alcohol, each for 5 min. The sections were stained with 0.5% eosin in 95% alcohol for 1 min, dehydrated with 95%, 100%, and 100% alcohol, each for 5 min; treated with a mixture of xylene and alcohol (1:1) for about 5 min; and finally treated twice with xylene for 5 min each time. The excess xylene was removed and the samples were sealed with a neutral resin. The sections were observed under a microscope (ECLIPSE Ni-U, Nikon, Japan).
Determination of chitin content
An insect chitin ELISA kit (MEIMIAN, China) was used to determine the chitin content of Bmlark−/− and WT embryos 168 h and 180 h after oviposition. Three replicates were performed on each sample. Each replicate contained 20–50 individuals. The wells of a microplate were coated with purified insect chitin antibody to produce a solid phase of antibody. Bmlark−/− and WT embryos were dissected and homogenized in 1 × PBS (9 mL of 1 × PBS per gram of embryo sample), followed by centrifugation at 3,000 rpm for 20 min. The supernatant was collected for chitin content analysis. After a five-fold dilution, 50 µL of diluted samples were added to the wells of microplate and maintained for 30 min at 37 ℃. Then, the samples were discarded, and each well was washed five times with a 1 × washing solution, each time for 30 s. Then, 50 µL of enzyme-labeled reagent was added to each well (except the blank well) and incubated for 30 min at 37 ℃. After incubation, each well was washed five times with 1 × washing solution, each time for 30 s. Then, 50 µL of color developing agent A was added to each well, followed by 50 µL of color developing agent B. The mix was gently shaken and incubated in the dark for 10 min at 37 ℃. Finally, 50 µL of a termination solution was added to each well to terminate the reaction. The absorbance spectrum of the sample in each well was measured at 450 nm using a multi-mode microplate reader (FlexStation 3, Molecular Devices, USA) within 15 min after adding termination solution. A standard curve was plotted using the standard concentrations of chitin and the corresponding OD values. The chitin concentration of the sample was calculated based on the standard curve established. The average chitin content per gram of Bmlark−/− and WT embryo samples was calculated and compared.
RNA sequencing (RNA-seq)
Wild-type and Bmlark−/− embryos 174 h post oviposition were dissected for RNA-seq. Each sample was replicated three times, and each contained 30 embryos. The isolation of total RNA, library construction, and sequencing were carried out by Novogene (Beijing, China). Gene expression levels were represented by fragments per kilobase of transcript per million fragments mapped (FPKM). Differentially expressed genes (DEGs) were identified using DESeq2 software [31] with p-value < 0.05 and |log2(FoldChange)|> 0.0 as the threshold. The clusterProfiler software was used to perform Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the DEGs.
RNA isolation and quantitative real-time PCR analysis (RT-qPCR)
Total RNA was extracted from the Bmlark−/− and WT embryos 168 h after oviposition (head pigmentation stage) using RNAiso Plus (Trizol) (Takara, Dalian, China). After precipitation with isopropanol and washing with 75% ethanol, the dried RNA pellet was dissolved in RNase-free water. The quantity and quality of the obtained RNA samples were analyzed by a Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). Two micrograms of RNA from each sample was used for reverse transcription into cDNA using the reverse transcriptase M-MLV (Takara, Dalian, China). The synthesized cDNA served as a template for RT-qPCR to analyze mRNA levels of the target genes. All primers used for RT-qPCR were designed by Primer Premier 5.0 software (Premier, Canada) and are listed in Supplementary Table S1. RT-qPCR was performed on a QuantStudio™ 6 Flex real-time PCR system (ABI, Life Technologies, USA) using Eastep® qPCR Master Mix (Promega, Shanghai, China), following the manufacturer’s instructions. The housekeeping gene ribosomal protein 49 (BmRP49) (Gene ID: 778453) was used as an internal reference to normalize the data. Relative expression levels of genes were analyzed using the 2−△△Ct method [32]. All the data are presented using the means of three independent biological replicates.
Western blotting
Proteins from the Bmlark−/− mutant and wild-type embryos 168 h after oviposition were extracted using RIPA lysis buffer (BestBio, Shanghai, China). The concentrations of the protein samples were analyzed with a BCA protein quantification kit (BestBio, Shanghai, China). A total of 20 μg of protein from each sample was denatured and then loaded onto a 12% SDS-PAGE gel for electrophoresis, followed by transferring to 0.45 μm PVDF membrane (Merck Millipore, Germany). After blocking with TBST buffer (20 mM Tris–HCl, 150 mM NaCl, 0.05% Tween-20, pH 8.0) containing 3% (w/v) bovine serum albumin (BSA) at 37 ℃ for 2 h, the membrane was incubated overnight at 4 °C with a primary antibody diluted in TBST containing 1% (w/v) BSA. The membrane was washed in TBST three times, 10 min each time and then incubated with HRP-conjugated second antibody (Dingguo, China) diluted (1:10,000) in TBST containing 1% (w/v) BSA at 30 °C for 2 h. β-actin (Dingguo, China) was used as a reference to normalize protein loading. After washing several times with TBST followed by TBS (20 mM Tris–HCl, 150 mM NaCl, pH 8.0), the membrane was immersed in a mixture of buffer A and B (1:1) from an Enhanced Chemiluminescence Detection Kit (Vazyme, Nanjing, China) for 1 min. Then, the membrane was scanned with a ChemiDoc™ imaging system (Bio-Rad, USA).
Statistical analysis
GraphPad Prism version 8.0 software was used for graphing and analyzing the data. The data are presented as mean ± standard error (SE). p < 0.05 was considered statistically significant using Student’s t-test.
Results
CRISPR/Cas9-mediated knockout of Bmlark
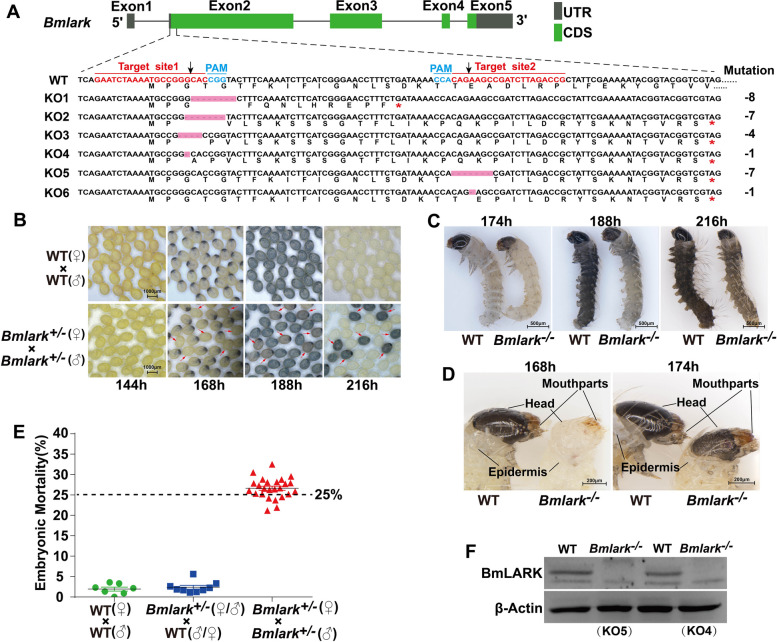
Two target sites were designed to specifically knock out Bmlark in B. mori. To ensure complete loss of gene function, the target sites were located near the ATG start codon (Fig. 1A). Six mutations (KO1 to KO6) in the Bmlark knockout (KO) that led to premature termination of protein translation were detected by mutation screening (Fig. 1A; Supplementary Table S2 and Figure S1).
Fig. 1.
CRISPR/Cas9-mediated Bmlark knockout and the phenotypes of Bmlark knockout mutants. A Schematic diagram of the Bmlark gene, target sites, and mutation types. UTR: untranslated regions; CDS: coding sequence; red asterisks represent the stop codon; pink boxes represent missing sequences; sequences in red are the target sites; sequences in cyan are the protospacer adjacent motif (PAM); black arrows indicate the positions at which Cas9 cleaved the target DNA. WT: wild-type sequence; KO1-KO6: mutant sequences. B Developmental difference of Bmlark knockout eggs of the G2 generation 144–216 h post oviposition. Red arrows indicate the Bmlark−/− eggs. WT: wild-type; time (h) is after oviposition. C Phenotypes of the WT and homozygous Bmlark−/− embryos at 174, 188 and 216 h post oviposition. D Detailed differences in the head and epidermis between WT and Bmlark−/− embryos 168 and 174 h post oviposition. E Embryo mortality of offspring of different genotype crossings. The bars show the means (in black) and standard error (in green, blue, or red) of each group. The results are presented as mean ± SE of multiple biological replicates (Supplementary Table S3). The results of the χ2-test showed that the observed value was not significantly different from the expected value (p > 0.05, Supplementary Table S3), indicating that the mortality rate was consistent with the assumed probability of 25%. F Western blot analysis of BmLARK protein in the WT and Bmlark−/− mutant (KO4 and KO5) embryos at 168 h post oviposition
Knockout of Bmlark resulted in the death of homozygous embryos
Bmlark+/- heterozygotes of the G1 generation with the same type of mutation were mated to produce homozygous individuals. However, once two Bmlark+/- were mated, some of the offspring eggs did not hatch. The embryos in those eggs showed obvious developmental delays, which occurred as early as at the beginning of the head pigmentation stage. The eggs displayed abnormal color during subsequent development and finally died (Fig. 1B and C). These mutant eggs had a lighter color than the WT eggs and eventually displayed a brown color instead of the black color of the WT eggs. Compared with the wild-type embryos, the mutant embryos showed abnormal phenotypes, such as wrinkling, delayed hardening and darkening of the head and epidermis exoskeleton, and particularly of the mouthparts in the head (Fig. 1C and D). These abnormal embryos accounted for about 25% of all the offsprings produced by self-crossing Bmlark+/- (Fig. 1E; Supplementary Table S3), implying that these abnormal embryos were homozygous Bmlark−/− mutants. Genomic DNA was extracted from some of these mutants for PCR and sequencing analysis, and the results confirmed their genotype of homozygous Bmlark−/− mutants (Supplementary Figure S1). Bmlark−/− mutant and the WT eggs at the head pigmentation stage were collected for Western blot analysis to confirm whether BmLARK protein had been eliminated. The result indicated that BmLARK protein was absent in the Bmlark−/− mutants, further indicating that these abnormal embryos were homozygous Bmlark−/− mutants (Fig. 1F). These results suggest that deletion of Bmlark resulted in abnormal development and the death of embryos, and thus Bmlark is essential for normal embryonic development in B. mori.
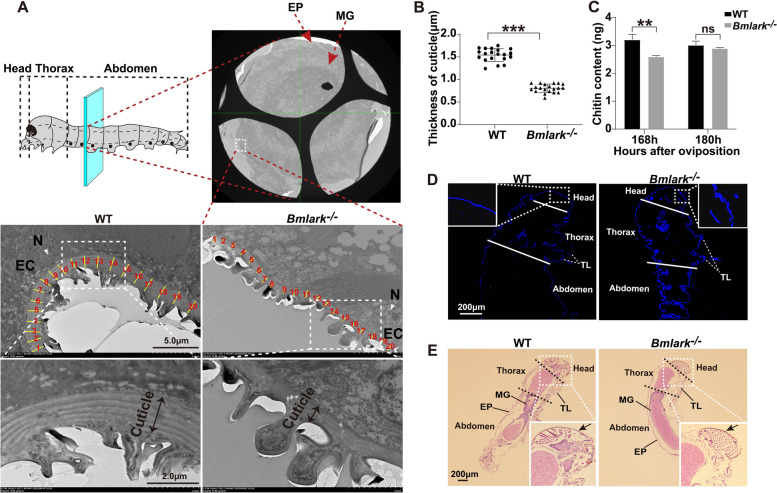
The cuticle of Bmlark−/− embryos was defective
A TEM examination comparing Bmlark−/− and WT embryos at the beginning of the head pigmentation stage showed that the cuticle of Bmlark−/− embryos had a less distinct lamellar structure (Fig. 2A, lower section). Twenty positions of the cuticle were randomly selected and their thickness was measured using a Nano Measurer 1.2 (Fig. 2A, middle section; Supplementary Table S4). The results revealed that the cuticle of the Bmlark−/− mutant embryos was significantly thinner than that of the WT embryos (Fig. 2B). The embryos were stained for chitin, the main component of cuticle, but no significant difference in chitin content was observed between the Bmlark−/− and WT embryos. However, compared to the WT embryos, the Bmlark−/− embryos had wrinkled and incompletely hardened cuticle in the head (Fig. 2D). HE staining also revealed wrinkled cuticle in the Bmlark−/− mutant embryos (Fig. 2E, indicated by arrows). To more accurately determine the chitin content, a chitin ELISA kit was used to determine the chitin content of Bmlark−/− and WT embryos. At the beginning of the head pigmentation stage (168 h after oviposition), the chitin content of the Bmlark−/− mutant embryos was lower than that of the WT embryos; however, at the later development stages (for example, at 180 h after oviposition), the chitin content was similar in the mutant and WT embryos (Fig. 2C, Supplementary Table S5). These results suggest that deletion of Bmlark did not significantly change the chitin content of the cuticle, but instead changed the cuticle architecture in the Bmlark−/− mutant embryos, leading to embryonic death.
Fig. 2.
Structural differences in the cuticle of WT and Bmlark−/− embryos at the head pigmentation stage. A TEM images of the cross sections of WT and Bmlark−/− embryos at the head pigmentation stage (168 h post oviposition). The upper part of the figure shows the crosscut position of the embryo, along with a TEM image of the entire associated transverse section (original images are presented in Supplementary Figure S2A). The cyan plane represents the cut position, which is consistent in both WT and Bmlark−/− embryos, and located roughly in the second segment of the abdomen. EP: epidermis. MG: midgut. EC: epidermal cell. N: nucleus. The yellow lines, which are perpendicular to the tangent lines along the cuticle, represent the thickness of the cuticle at 20 randomly selected regions. B Comparison of the thickness of the WT and Bmlark−/− cuticle in the epidermis of the abdomen. The length of yellow lines at 20 different regions per individual in Figure (A) were calculated using Nano Measurer 1.2 software, and represent the thickness of the cuticles in the epidermis of WT and Bmlark−/− embryos. Student’s t test was used to evaluate statistical significance. Results are presented as the mean ± SE. ***p ≤ 0.001. C Chitin content of WT and Bmlark−/− embryos 168 and 180 h post oviposition. The ordinate represents the mass of chitin per gram of embryonic tissues. Student’s t test was used to evaluate statistical significance. Results are presented as the mean ± SE of three biological replicates. **p ≤ 0.01; ns: no significance, p > 0.05. D Chitin staining of the sagittal sections of the WT and Bmlark−/− mutant embryos at 180 h post oviposition. The white lines divide different parts of the body. Chitin was stained blue with fluorescent brightener 28. TL: thoracic leg. (E) HE stained images of the sagittal sections of the WT and Bmlark−/− embryos at 180 h post oviposition. The arrows indicate the head cuticle. The black dotted linesdivided different parts of the body. MG: midgut. EP: epidermis. TL: thoracic leg. Enlarged original images of the head of WT and Bmlark−/− embryos are presented in Supplementary Figure S2B
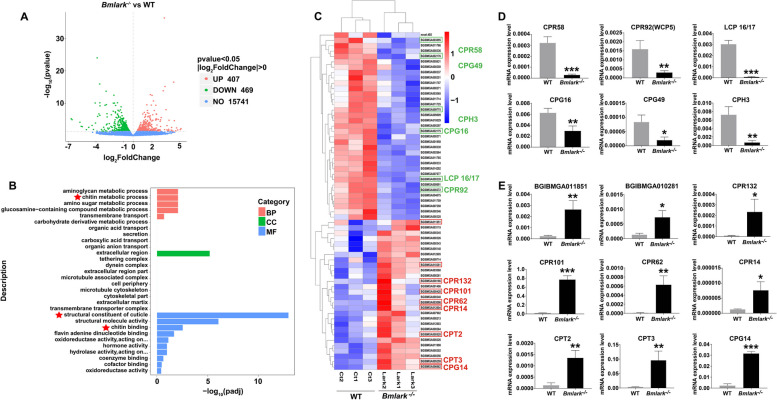
Changes in gene expression in the Bmlark−/− mutant embryo
RNA-seq was performed with Bmlark−/− and WT eggs at the head pigmentation stage to examine changes in gene expression after Bmlark was knocked out. Transcriptomic analysis identified 876 DEGs, including 407 upregulated DEGs and 469 downregulated DEGs (Fig. 3A, Supplementary Table S6). The top 30 GO terms from an enrichment analysis of these DEGs are shown in Fig. 3B. The DEGs were significantly enriched in nine functional terms (padj < 0.05) related to the metabolism, structure and components of cuticle, such as chitin metabolic processes, structural constituents of cuticle and chitin binding (Supplementary Table S7). These results suggested that chitin and cuticle proteins were most significantly impacted by the loss of Bmlark.
Fig. 3.
Analyses of the transcriptome and differentially expressed genes (DEGs) of the Bmlark−/− mutant and WT embryos at the head pigmentation stage. A Volcano plot of DEG analysis between WT and Bmlark−/− embryos. UP: upregulated genes; Down: downregulated genes. NO: not significantly changed genes. B Top 30 GO enrichment terms of the Bmlark−/− mutant embryo DEGs. Red asterisks represent the enriched cuticle-related GO terms. C Cluster analysis of 63 CPs in the Bmlark−/− mutant and WT embryos. (D and E) RT-qPCR analysis of CPs in different CP families. Student’s t-test was used to evaluate statistical significance. Results are presented as the mean ± SE of three biological replicates. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
Because chitin is one of the most foundational components of cuticle, its synthesis and degradation related enzyme genes were first analyzed. Twelve genes of chitin metabolism (two chitin synthase genes and ten chitinase genes) were examined for their expression in the transcriptomic data, and no significant difference in the expression levels of these genes was found between the WT and Bmlark−/− mutant embryos (Supplementary Figure S3A). RT-qPCR analysis of two chitin synthase genes (ChsA and ChsB) and two well-studied chitinase genes (Cht5 and Cht10) showed similar results (Supplementary Figure S3B), indicating that chitin synthesis and degradation were not significantly affected by the deletion of Bmlark. This result is consistent with the observations and measurements of the Bmlark−/− and WT embryos at the later head pigmentation stage (Fig. 2C), which showed no difference in the chitin content.
CPs are another important component of cuticle. To determine whether CPs were impacted by the deleting Bmlark, the expression of CPs in the mutant and wild type embryos was determined. A total of 218 genes annotated as CPs or CP-related were selected from the transcriptomic data (Supplementary Table S8) for further analyses, including all the enriched genes of the three most significantly enriched chitin-related GO terms (Fig. 3B, marked with red stars); the known CP genes of different CP families [33, 34], such as R&R consensus bearing CPR, glycine-rich cuticular protein (CPG), Tweedle protein (CPT), and hypothetical cuticular protein (CPH) genes, and all the chitin binding-related genes. The genes underwent differential expression analysis using p < 0.05 and |log2(FoldChange)|> 0.0 as the threshold, followed by cluster analysis. The results showed that 63 genes were differentially expressed between the WT and Bmlark−/− embryos (Fig. 3B), including 47 known CP genes and 16 chitin binding-related CP genes, according to the genome annotation (Supplementary Table S9). Some of these genes from different CP families were selected for further confirmation by analyzing their expression levels using RT-qPCR (Supplementary Table S10). The expression levels of these genes showed significant down- or upregulated changes in the Bmlark−/− mutant embryos compared with the WT embryos (Fig. 3D and E), which was consistent with the transcriptomic data. These results suggested that the deletion of Bmlark either upregulated or downregulated the expression pattern of CP genes in Bmlark−/− mutants, which might be responsible for the abnormal exoskeleton development in the head and epidermis of the mutant embryos.
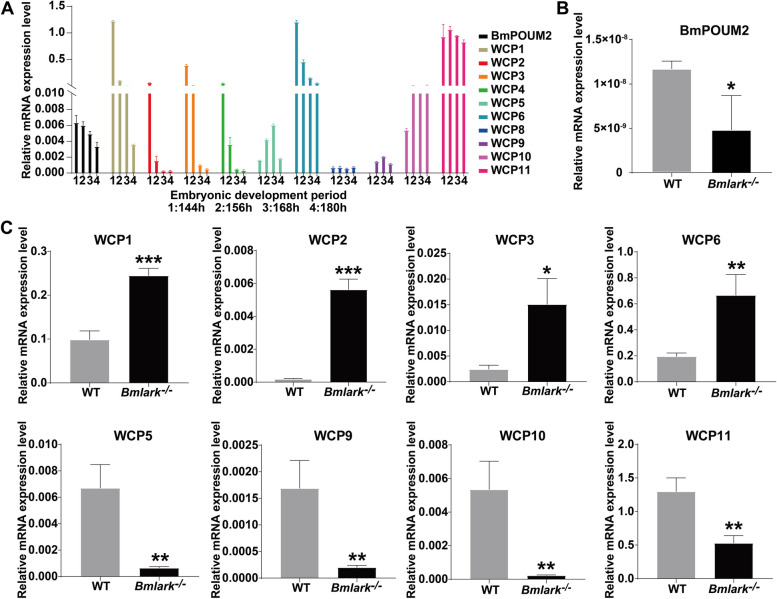
Given the importance of the transcription factor BmPOUM2 in the expression of cuticle proteins [35, 36], the expression profiles of BmPOUM2 and wing disc cuticle protein(WCP) genes (listed in Supplementary Table S11) were analyzed prior to and at the head pigmentation stage (144–180 h after oviposition) (Fig. 4A). The results showed that in the WT embryos (Fig. 4A), the expression level of BmPOUM2 gradually decreased from the beginning of the head pigmentation stage (168 h after oviposition), while WCP1, WCP2, WCP3, WCP4, and WCP6 were highly expressed before the head pigmentation stage and then gradually decreased, suggesting that these genes may play a vital role in the development of the embryos as they enter the head pigmentation stage, after which their expression declined. The expression of WCP5 and WCP9 reached their highest level at the beginning of the head pigmentation stage, while the expression of WCP10 and WCP11 began to increase at this stage and remained relatively stable afterwards, suggesting that these genes may be closely related to the sclerotization and melanization of the embryonic cuticle in the pigmentation stage of embryonic development. The expression of BmPOUM2 at the beginning of the head pigmentation stage was downregulated in the Bmlark−/− mutant embryos as compared with that in the WT embryos (Fig. 4B), while WCP1, WCP2, WCP3, and WCP6 were still at a high level in this stage due to delayed development (Fig. 4C). However, the expression levels of WCP5, WCP9, WCP10 and WCP11 were significantly lower in the Bmlark−/− mutant embryos than in the WT embryos (Fig. 4C). Among them, WCP5, WCP10 and WCP11 were reported to have high expression levels in the late embryonic development stages of Bombyx mori [37], which may be important for the sclerotization and melanization of the embryonic cuticle. These differences in gene expression explain the observed defects in the cuticle structure of the Bmlark−/− mutant embryos.
Fig. 4.
Expression analysis of BmPOUM2 and WCP genes in the WT and Bmlark−/− mutant embryos. A Expression profiles of BmPOUM2 and WCP genes in WT embryos at 144, 156, 168, and 180 h post oviposition. Hours post oviposition are indicated in the figure. B RT-qPCR analysis of BmPOUM2 expression in the Bmlark−/− mutant and WT embryos. C RT-qPCR analysis of the expression of WCP genes in the Bmlark−/− mutant and WT embryos. Student’s t-test was used to evaluate statistical significance. Results are presented as the mean ± SE of three biological replicates. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
Gene expression was altered in the tyrosine-mediated cuticle tanning pathway
Melanin is one of the many diverse and critical pigments for cuticle coloring and hardening in insects [38]. Melanin synthesis begins in epidermal cells and the produced pigment precursors are transported to the cuticle formation on the outside surface of the epidermis where they precipitate. These processes are regulated by a series of genes, including TH, DDC, ebony, tan, aaNAT, yellow, Laccase 2 and black, which covert the products of tyrosine and uracil metabolism, such as dopa, dopamine, N-β-alanyldopamine (NBAD) and N-acetyldopamine (NADA), into black, brown, yellow or colorless pigments [39, 40] (Fig. 5A). To examine whether these genes contributed to the abnormal pigmentation in Bmlark−/− mutant embryos, the expression of genes related to the tyrosine-mediated melanin pathway (listed in Supplementary Table S12) was determined in the mutant and WT embryos. At the beginning of the head pigmentation stage, several key genes for melanin synthesis were differentially expressed between the mutant and WT embryos: TH was upregulated in the Bmlark−/− mutant embryos (Fig. 5B), and DDC, aaNAT, Laccase2A and yellow-f2 were downregulated (Fig. 5C). These results suggest that the synthesis of dopa from tyrosine likely proceeded normally in the mutant, but the subsequent processes of pigment production from dopa to melanin were hindered, and was likely responsible for the abnormal melanization in the Bmlark−/− mutant.
Fig. 5.
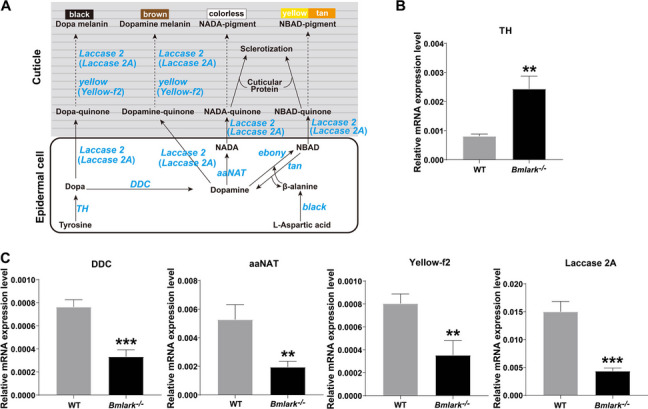
The tyrosine-mediated pigment synthesis pathway and expression analysis of key genes. A Schematic diagram of the tyrosine-mediated pigment synthesis pathway [39, 40]. Dashed lines indicate multistep biochemical reactions. The key genes that encode the enzymes involved in the tyrosine-mediated pigment synthesis pathway are shown in blue. (B and C) RT-qPCR analysis of the expression of the key genes in the Bmlark−/− mutant and WT embryos at the head pigmentation stage. Student’s t-test was used to evaluate statistical significance. Results are presented as the mean ± SE of three biological replicates. **p ≤ 0.01; ***p ≤ 0.001
Discussion
The D. melanogaster lark gene has been reported to play multiple functions in a variety of developmental processes, including the circadian rhythm [4, 5], embryonic development [1], oogenesis [8, 41], eclosion [3, 9], and physiology and development of the nervous system [10], indicating the constitutive importance of this gene. The mammalian lark homologous gene RBM4 has also been revealed to participate in RNA silencing [20], alternative splicing [42], and translation regulation [22, 23]. Our previous studies showed that silkworm BmLARK acts as a G4-binding protein to regulate the transcription of BmPOUM2 [28, 43, 44]. This study further demonstrated the diverse and essential functions of Bmlark in embryonic development in silkworm.
In this study, CRISPR/Cas9 gene editing was used to knock out Bmlark to investigate the effects on silkworm development caused by the loss of BmLARK function. We found that heterozygous Bmlark+/- individuals developed normally into adults without obvious phenotypic defects, but the homozygous Bmlark−/− mutants were embryonic lethal, indicating that Bmlark was indispensable in silkworm embryonic development. Bmlark−/− eggs could not be visibly distinguished from wild-type or heterozygous eggs until the head pigmentation stage (about 168 h after oviposition), although embryonic development might have already been delayed before this stage. The Bmlark−/− embryos were unable to hatch normally and eventually died. Compared with the WT embryos, the cuticle of the Bmlark−/− mutant embryos showed obvious defects in sclerotization and melanization. Although chitin synthesis and its levels did not significantly change, the architecture of the cuticle in the head and epidermis changed in the Bmlark−/− mutant embryos. Normal development of the cuticle is critical for the development of insect embryos to help ensure that the first instar larvae can survive in the environment outside the eggs after hatching. Cuticle sclerotization and melanization of the embryo body wall become visibly observable at the head pigmentation stage of embryonic development in silkworm (Fig. 1B-D); consistently, the negative impacts (phenotype) of knocking out Bmlark did not appear until this stage.
The formation and differentiation of the cuticles usually begin at late embryogenesis and have different compositions and functions depending on developmental stage and where they form. For example, the body cuticle of holometamorphic insect larvae is usually soft, while the head skeleton, which they use to chew food, is usually hard. Some of developmental events, such as setae formation, chitinization of the body and pigmentation of mandible begin prior to the head pigmentation stage in silkworm embryos, although they are not obvious until this stage. In the embryos of Bmlark−/− mutants, the morphological differences in these aspects from the wild type embryos became obvious at this head pigmentation stage, including abnormal pigmentation, lighter head and mandible color (Fig. 1B-D), delayed and less erect setae formation (Fig. 1C). The delay of these developmental events in the Bmlark−/− mutants was the results of defective sclerotization and melanization of the cuticles, and it can expected that this kind of soft exoskeleton in the Bmlark−/− mutant embryos may be fatal to their survival. Insect cuticle is mainly composed of chitin and structural CPs [45, 46]. The type and number of CPs contribute to the physical properties of cuticle [45, 46]. In this study, the RNA-seq results indicated that there were no significant changes in the expression of genes related to chitin synthesis and degradation, and the chitin content was not significantly changed in the mutant embryos, suggesting that the deletion of Bmlark did not affect chitin synthesis or metabolism. However, 63 predicted CPs in different CP families, such as CPR (including RR-1 and RR-2), CPG, CPT, and CPH, showed differences in expression between the Bmlark−/− and WT embryos. Thus, changes in the expression of these CPs in the Bmlark−/− mutant might alter the composition and structure of the cuticle, resulting in a dysfunctional exoskeleton in the mutant, as observed in Fig. 1D and Fig. 2.
In our previous studies, we demonstrated that BmLARK bound to the G4 structure in the promoter of BmPOUM2 and regulated its transcription [28, 43, 44]. BmPOUM2 is a transcription factor involved in the regulation of several WCPs [35, 36]. In this study, the expression of BmPOUM2 and several BmWCP genes were significantly down- or upregulated in the Bmlark−/− mutant embryos, suggesting that these genes are regulated by BmLARK and necessary for the embryonic development. Moreover, knockout of BmPOUM2 caused an embryonic lethal phenotype [47] that was quite similar to the phenotype of the Bmlark knockout in this study. Thus, BmLARK may act as an upstream regulatory factor of BmPOUM2 by interacting with its G4 structure or through other mechanisms [28, 43, 44]. This demonstrates that both BmLARK and BmPOUM2 are vital for embryonic development. Because some CP protein genes are upregulated while others are downregulated when BmLARK (this study) or BmPOUM2 [35, 47] are knocked out, how BmLARK and BmPOUM2 regulate the expression of CPs and how these CPs participate the regulation of embryonic development require further investigation.
The abnormal body color of the Bmlark−/− mutant attracted our attention to another developmental event, pigmentation or melanization. In insects, body cuticle melanization is usually achieved through the tyrosine-mediated pigment synthesis pathway (Fig. 5A). In Bmlark−/− mutants, TH was upregulated, but DDC, aaNAT, Laccase2A, and yellow-f2 were all downregulated. Although the upregulation of TH may accelerate the conversion of tyrosine to dopa, the conversion of dopa and dopamine to the final pigment melanin may be prevented due to the suppression of DDC, aaNAT, Laccase2A or yellow-f2 in the absence of Bmlark, thus producing abnormal melanization in the mutant. We noticed that various TH mutants of B. mori showed different body colors in different experiments. For example, BmTH RNAi prevented pigmentation and hatching and caused a sex-linked chocolate phenotype (sch) [48]. When Bm-iAANAT was knocked out, BmTH was also downregulated and a darker pattern in the sclerified tissues (head, thoracic legs, and anal plate) of melanism mutant (mln) was observed [49]. In this study, we found that when the lark gene was deleted, the expression of TH was increased, while the expression of DDC, aaNAT, Yellow-f2 and Laccase 2A was decreased (Fig. 5B and C), which might lead to the accumulation of dopa and dopamine and change the proportion of the pigments and cause Bmlark−/− mutant embryos to become brown, different from the black color of WT (Fig. 1C). These results indicate that the body color may be affected by different pigment synthesis pathways and pigment proportions. This needs to be further investigated in future.
In addition, NBAD-quinone and NADA-quinone generated in the process of melanin synthesis are cross-linking agents of CPs, and their incorporation into CPs is required for cuticle sclerotization [50]. The degree of cross-linking of cuticle components determines the hardness and function of cuticle [39]. Therefore, the tyrosine-mediated pigment synthesis pathway affects not only melanization, but also sclerotization of the cuticle. In this study, incomplete sclerotization of the head capsule, especially of the mouthparts, may be responsible for the failure of Bmlark−/− embryos to hatch and their subsequent death. Mouthpart defects may be one of the causations for hatching failure, as seen in the Ddc mutant of Vanessa cardui [51].
As described above, the lark gene has been reported to play multiple functions. In Drosophila, lark has been found to be involved in circadian rhythms [2, 5–7], neural development [10], eye development [6], embryonic development [1], oogenesis [8], neuronal development and physiology [1, 4, 9, 10], and gut immunocompetence [11]. In humans, the LARK homolog, RBM4 has been reported to modulate diverse biological functions by regulating alternative splicing of target mRNA, such as, in muscle cell differentiation [12], brown adipocytes development [13], neuronal cell differentiation and neurite outgrowth [14], neuronal migration [15], human colorectal cancer cells migration and invasion [16], and cancer suppression [17–19]. In B. mori, Bmlark has been reported to participate in the alternative splicing of the sex determination gene doublesex [26]. Our laboratory found that BmLARK is involved in regulating BmACBP expression, and subsequently lipid metabolism [27]. Furthermore, we found that BmLARK binds to G4 structures in the promoters of the transcription factor BmPOUM2 and many other genes that contain G4 structures in B. mori and humans, regulating their expression [28]. Therefore, we believe that as a transcription factor, LARK may have multiple regulatory functions that affect many cellular and physiological processes.
We examined Bmlark mRNA levels during embryonic development and protein levels in different tissues of larvae in the wandering stage (Supplementary Figure S14) and found that Bmlark has a broad expression profile and is expressed in many types of tissue. These results imply the functional diversity and importance of Bmlark in B. mori. In this study, by knocking out Bmlark, we found that this gene is necessary for embryonic development, probably by regulating the expression of cuticle protein genes and pigment synthesis genes via BmPOUM2. This is a novel finding that has not been reported before. While we cannot exclude other possible molecular and physiological functions of LARK in B. mori embryonic development, which need to be investigated in the future, we can conclude that Bmlark is essential for embryonic development in B. mori.
Conclusions
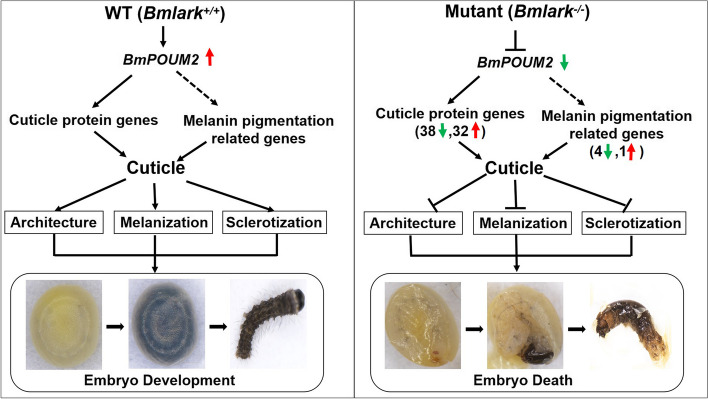
In summary, the cuticle exoskeleton of insects has various important functions, such as protecting the body from physical damage and pathogen invasion, preventing water evaporation, and providing mechanical support for body movement, eating, and sensing [52, 53]. Therefore, the cuticle is crucial for insect survival. This study found that Bmlark regulates the expression of cuticle protein genes and pigmentation genes and maintains the normal structure, sclerotization and melanization of the embryonic exoskeleton cuticle (Fig. 6, left). However, deletion of Bmlark altered the BmPOUM2-mediated expression of cuticle proteins and pigmentation genes, and in turn deleteriously altered the architecture, sclerotization and melanization of the cuticles in the body and head of the insect (Fig. 6, right), resulting in embryonic death.
Fig. 6.
Schematic diagram of embryonic death caused by the Bmlark knockout. The solid arrows represent direct regulation, and the dashed arrows indicate that regulation has not yet been determined to be direct or indirect. The red and green arrows indicate up- or downregulated expression, respectively
Supplementary Information
Acknowledgements
Not applicable.
Authors’ contributions
YP performed most of the experiments, organized the data, carried out statistical analyses and wrote the manuscript. JL helped extract parts of the genomic DNA and RNA. KN, MW, YC and CT provided helps in the experimental methods and techniques. QF designed and supervised the project and finalized the manuscript.
Funding
This work was supported by the grants of the Chinese National Natural Science Foundation (Grant no.: 32100383, 32250710148, 31930102) and the China Postdoctoral Science Foundation (Grant no.: 2021M691096).
Data availability
Data is provided within the manuscript or supplementary information files. The RNA-seq raw data have been deposited and can be found in the Sequence Read Archive (SRA) under the accession number PRJNA1119663 at NCBI. The original images are provided in the file named Supplementary Figures.
Declarations
Ethics approval and consent to participate
Ethics approval is not applicable for experimental animals in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Newby LM, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135(4):1077–90. [DOI] [PMC free article] [PubMed]
- 2.Zhang XL, Mcneil GP, Hilderbrand-Chae MJ, Franklin TM, Schroeder AJ, Jackson FR. Circadian regulation of the lark RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45(1):14–29. [DOI] [PubMed] [Google Scholar]
- 3.Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31(1):117–28. [DOI] [PubMed]
- 4.Sundram V, Ng FS, Roberts MA, Millán C, Ewer J, Jackson FR. Cellular Requirements for LARK in the Drosophila Circadian System. J Biol Rhythms. 2012;27(3):183–95. [DOI] [PMC free article] [PubMed]
- 5.McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20(2):297–303. [DOI] [PubMed] [Google Scholar]
- 6.Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL. The Drosophila FMRP and LARK RNA-Binding Proteins Function Together to Regulate Eye Development and Circadian Behavior. J Neurosci. 2008;28(41):10200–5. [DOI] [PMC free article] [PubMed]
- 7.Huang YM, McNeil GP, Jackson FR. (2014) Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation. PLoS Genet. 2014;10(9): 10.1371/journal.pgen.1004536. [DOI] [PMC free article] [PubMed]
- 8.McNeil GP, Kaur M, Purrier S, Kang R. The Drosophila RNA-binding protein Lark is required for localization of Dmoesin to the oocyte cortex during oogenesis. Dev Genes Evol. 2009;219(1):11–19. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder AJ, Genova GK, Roberts MA, Kleyner Y, Suh J, Jackson FR. Cell-specific expression of the LARK RNA-binding protein in Drosophila results in morphological and circadian behavioral phenotypes. J Neurogenet. 2003;17(2–3):139–69. [PubMed]
- 10.Huang YM, Howlett E, Stern M, Jackson FR. Altered LARK Expression Perturbs Development and Physiology of the Drosophila PDF Clock Neurons. Mol Cell Neurosci. 2009;41(2):196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bou Sleiman M, Frochaux MV, Andreani T, Osman D, Guigo R, Deplancke B. Enteric infection induces Lark-mediated intron retention at the 5′ end of Drosophila genes. Genome Biol. 2020; 21(1): 10.1186/s13059-019-1918-6. [DOI] [PMC free article] [PubMed]
- 12.Lin JC, Tarn WY. Multiple roles of RBM4 in muscle cell differentiation. Front Biosci (Schol Ed). 2012;4(1):181–9. [DOI] [PubMed]
- 13.Lin JC, Chi YL, Peng HY, Lu YH. RBM4-Nova1-SRSF6 splicing cascade modulates the development of brown adipocytes. Biochim Biophys Acta. 2016;1859(11):1368–79. [DOI] [PubMed]
- 14.Tarn WY, Kuo HC, Yu HI, Liu SW, Tseng CT, Dhananjaya D, Hung KY, Tu CC, Chang SH, Huang GJ, Chiu IM. RBM4 promotes neuronal differentiation and neurite outgrowth by modulating Numb isoform expression. Mol Biol Cell. 2016;27(10):1676–83. [DOI] [PMC free article] [PubMed]
- 15.Dhananjaya D, Hung KY, Tarn WY. (2018) RBM4 Modulates Radial Migration via Alternative Splicing of Dab1 during Cortex Development. Mol Cell Biol. 2018;38(12): 10.1128/MCB.00007-18. [DOI] [PMC free article] [PubMed]
- 16.Lin JC, Lee YC, Tan TH, Liang YC, Chuang HC, Fann YC, Johnson KR, Lin YJ. RBM4-SRSF3-MAP4K4 splicing cascade modulates the metastatic signature of colorectal cancer cell. Biochim Biophys Acta Mol Cell Res. 2018;1865(2):259–72. [DOI] [PubMed]
- 17.Wang Y, Chen D, Qian HL, Tsai YS, Shao SJ, Liu Q, Dominguez D, Wang ZF. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374–89. [DOI] [PMC free article] [PubMed]
- 18.Chen JY, Liu LP, Xu JF. Decrease of RBM4 indicates poor prognosis in patients with hepatocellular carcinoma after hepatectomy. Onco Targets Ther. 2017;10:339–45. [DOI] [PMC free article] [PubMed]
- 19.Yong HM, Zhao W, Zhou XY, Liu ZY, Tang Q, Shi HC, Cheng RH, Zhang X, Qiu ZN, Zhu J, Feng ZQ. RNA-Binding Motif 4 (RBM4) Suppresses Tumor Growth and Metastasis in Human Gastric Cancer. Med Sci Monit. 2019;25:4025–34. [DOI] [PMC free article] [PubMed]
- 20.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8(11):1052–60. [DOI] [PMC free article] [PubMed]
- 21.Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci U S A. 2007;104(7):2235–40. [DOI] [PMC free article] [PubMed]
- 22.Lin JC, Tarn WY. RNA-binding Motif Protein 4 Translocates to Cytoplasmic Granules and Suppresses Translation via Argonaute2 during Muscle Cell Differentiation. J Biol Chem. 2009;284(50):34658–65. [DOI] [PMC free article] [PubMed]
- 23.Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104(6):1859–64. [DOI] [PMC free article] [PubMed]
- 24.Wang ZL, Li J, Xia QY, Zhao P, Duan J, Zha XF, Xiang ZH. Identification and expression pattern of Bmlark, a homolog of the Drosophila gene lark in Bombyx mori. DNA Seq. 2005;16(3):224–9. [DOI] [PubMed]
- 25.Iwai S, Takeda M. Expression analysis of two types of transcripts from circadian output gene lark in Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 2007;146(4):470–6. [DOI] [PubMed]
- 26.Zheng ZZ, Sun X, Zhang B, Pu J, Jiang ZY, Li M, Fan YJ, Xu YZ. Alternative splicing regulation of doublesex gene by RNA-binding proteins in the silkworm Bombyx mori. RNA Biol. 2019;16(6):809–20. [DOI] [PMC free article] [PubMed]
- 27.Xiang LJ, Niu KK, Peng YL, Zhang XJ, Li XY, Ye RQ, Yu GX, Ye GJ, Xiang H, Song QS, Feng QL. DNA G-quadruplex structure participates in regulation of lipid metabolism through acyl-CoA binding protein. Nucleic Acids Res. 2022;50(12):6953–67. [DOI] [PMC free article] [PubMed]
- 28.Niu KK, Xiang LJ, Jin Y, Peng YL, Wu F, Tang WH, Zhang XJ, Deng HM, Xiang H, Li S, Wang J, Song QS, Feng QL. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019;47(14):7306–20. [DOI] [PMC free article] [PubMed]
- 29.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31(7):1120–3. [DOI] [PMC free article] [PubMed]
- 30.Zhu GH, Xu J, Cui Z, Dong XT, Ye ZF, Niu DJ, Huang YP, Dong SL. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem Mol Biol. 2016;75:1–9. [DOI] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12): 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- 32.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25(4):402–8. [DOI] [PubMed]
- 33.Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm. Bombyx mori Insect Biochem Mol Biol. 2008;38(12):1138–46. [DOI] [PubMed]
- 34.Dong Z, Zhang W, Zhang Y, Zhang X, Zhao P, Xia Q. Identification and Characterization of Novel Chitin-Binding Proteins from the Larval Cuticle of Silkworm. Bombyx mori J Proteome Res. 2016;15(5):1435–45. [DOI] [PubMed]
- 35.Deng HM, Zhang JL, Li Y, Zheng SC, Liu L, Huang LH, Xu WH, Palli SR, Feng QL. Homeodomain POU and Abd-A proteins regulate the transcription of pupal genes during metamorphosis of the silkworm, Bombyx mori. Proc Natl Acad Sci U S A. 2012;109(31):12598–603. [DOI] [PMC free article] [PubMed]
- 36.Deng HM, Zheng SC, Yang XH, Liu L, Feng QL. Transcription factors BmPOUM2 and BmβFTZ-F1 are involved in regulation of the expression of the wing cuticle protein gene BmWCP4 in the silkworm. Bombyx mori Insect Mol Biol. 2011;20(1):45–60. [DOI] [PubMed] [Google Scholar]
- 37.Xu GF, Gong CC, Lyu H, Deng HM, Zheng SC. Dynamic transcriptome analysis of Bombyx mori embryonic development. Insect Sci. 2022;29(2):344–62. [DOI] [PubMed]
- 38.Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;20(1):65–71. [DOI] [PubMed] [Google Scholar]
- 39.Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y. Cuticle formation and pigmentation in beetles. Curr Opin Insect Sci. 2016;17:1–9. [DOI] [PubMed] [Google Scholar]
- 40.Wu SY, Tong XL, Peng CX, Xiong G, Lu KP, Hu H, Tan D, Li CL, Han MJ, Lu C, Dai FY. Comparative analysis of the integument transcriptomes of the black dilute mutant and the wild-type silkworm Bombyx mori. Sci Rep. 2016;6: 10.1038/srep26114. [DOI] [PMC free article] [PubMed]
- 41.McNeil GP, Zhang X, Roberts M, Jackson FR. Maternal function of a retroviral-type zinc-finger protein is essential for Drosophila development. Dev Genet. 1999;25(4):387–96. [DOI] [PubMed]
- 42.Wang WY, Quan WL, Yang F, Wei YX, Chen JJ, Yu H, Xie J, Zhang Y, Li ZF. RBM4 modulates the proliferation and expression of inflammatory factors via the alternative splicing of regulatory factors in HeLa cells. Mol Genet Genomics. 2020;295(1):95–106. [DOI] [PubMed] [Google Scholar]
- 43.Niu KK, Zhang XJ, Deng HM, Wu F, Ren YD, Xiang H, Zheng SC, Liu L, Huang LH, Zeng BJ, Li S, Xia QY, Song QS, Palli SR, Feng QL. BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 2018;46(4):1710–23. [DOI] [PMC free article] [PubMed]
- 44.Peng YL, Niu KK, Yu GX, Zheng MX, Wei QL, Song QS, Feng QL. Identification of binding domains and key amino acids involved in the interaction between BmLARK and G4 structure in the BmPOUM2 promoter in Bombyx mori. Insect Sci. 2021;28(4):929–40. [DOI] [PubMed]
- 45.Togawa T, Nakato H, Izumi S. Analysis of the chitin recognition mechanism of cuticle proteins from the soft cuticle of the silkworm. Bombyx mori Insect Biochem Mol Biol. 2004;34(10):1059–67. [DOI] [PubMed]
- 46.Okamoto S, Futahashi R, Kojima T, Mita K, Fujiwara H. Catalogue of epidermal genes: genes expressed in the epidermis during larval molt of the silkworm Bombyx mori. BMC Genomics. 2008;9:10.1186/1471-2164-9-396. [DOI] [PMC free article] [PubMed]
- 47.Tong CM, Zhang K, Rong ZX, Mo WY, Peng YL, Zheng SC, Feng QL, Deng HM. Alternative splicing of POUM2 regulates embryonic cuticular formation and tanning in Bombyx mori. Insect Sci. 2023;30(5):1267–81. [DOI] [PubMed]
- 48.Liu C, Yamamoto K, Cheng TC, Kadono-Okuda K, Narukawa J, Liu SP, Han Y, Futahashi R, Kidokoro K, Noda H, Kobayashi I, Tamura T, Ohnuma A, Banno Y, Dai FY, Xiang ZH, Goldsmith MR, Mita K, Xia QY. Repression of tyrosine hydroxylase is responsible for the sex-linked chocolate mutation of the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 2010;107(29):12980–5. [DOI] [PMC free article] [PubMed]
- 49.Dai FY, Qiao L, Tong XL, Cao C, Chen P, Chen J, Lu C, Xiang ZH. Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J Biol Chem. 2010;285(25):19553–60. [DOI] [PMC free article] [PubMed]
- 50.Qiao L, Li Y, Xiong G, Liu X, He S, Tong X, Wu S, Hu H, Wang R, Hu H, Chen L, Zhang L, Wu J, Dai F, Lu C, Xiang Z. Effects of altered catecholamine metabolism on pigmentation and physical properties of sclerotized regions in the silkworm melanism mutant. PLoS One. 2012;7(8): 10.1371/journal.pone.0042968. [DOI] [PMC free article] [PubMed]
- 51.Zhang LL, Martin A, Perry MW, van der Burg KR, Matsuoka Y, Monteiro A, Reed RD. Genetic Basis of Melanin Pigmentation in Butterfly Wings. Genetics. 2017;205(4):1537–50. [DOI] [PMC free article] [PubMed]
- 52.Liang J, Zhang L, Xiang ZH, He NJ. Expression profile of cuticular genes of silkworm, Bombyx mori. BMC Genomics. 2010;11: 10.1186/1471-2164-11-173. [DOI] [PMC free article] [PubMed]
- 53.Balabanidou V, Grigoraki L, Vontas J. Insect cuticle: a critical determinant of insecticide resistance-sciencedirect. Curr Opin Insect Sci. 2018;27:68–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files. The RNA-seq raw data have been deposited and can be found in the Sequence Read Archive (SRA) under the accession number PRJNA1119663 at NCBI. The original images are provided in the file named Supplementary Figures.