Abstract
Background
Apolipoprotein (apo) C-III is involved in several processes that increase triglyceride levels, inflammation, and insulin resistance. Four of its proteoforms have been the focus of several studies and have shown differential associations with cardiovascular risk biomarkers, mostly lipids. However, there are other proteoforms of apoC-III that have not yet been investigated in detail. The aim of this study was to evaluate the associations of seven apoC-III proteoforms with a comprehensive set of biomarkers, including lipid metabolism, inflammation, and glucose homeostasis.
Methods
Seven apoC-III proteoforms (apoC-III0a, apoC-III0b, apoC-III1, apoC-III1d, apoC-III2, apoC-III2d, and apoC-III0f) were measured using a mass spectrometry immunoassay in 875 participants from the cross-sectional study of the Di@bet.es cohort. The complete lipoprotein profile was obtained via the Liposcale test, and the proton nuclear magnetic resonance (1H-NMR)-assessed glycoprotein signals were also obtained as biomarkers of inflammation.
Results
Three proteoform ratios (apoC-III2d, apoC-III2, and apoC-III0f normalized to apoC-III1) showed protective associations with most of the cardiovascular risk biomarkers in comparison with total apoC-III in linear regression models and were negatively associated with triglycerides (β=-0.173, p < 0.001; β=-0.297, p < 0.001; β=-0.223, p = 0.002), very low-density (VLDL) particle concentration (β=-0.133, p < 0.001; β=-0.265, p < 0.001; β=-0.203, p < 0.001), GlycA (β=-0.148, p < 0.001; β=-0.263, p < 0.001; β=-0.211, p < 0.001) and homeostatic model assessment of insulin resistance (HOMA-IR) (β=-0.096, p = 0.003; β=-0.199, p < 0.001; β=-0.114, p = 0.002). These associations were partly independent of total apoC-III concentrations. Participants with high levels of these proteoforms had a lower prevalence of cardiometabolic disorders, such as type 2 diabetes (p = 0.022), obesity (p = 0.001), and metabolic syndrome (p = 0.013).
Conclusions
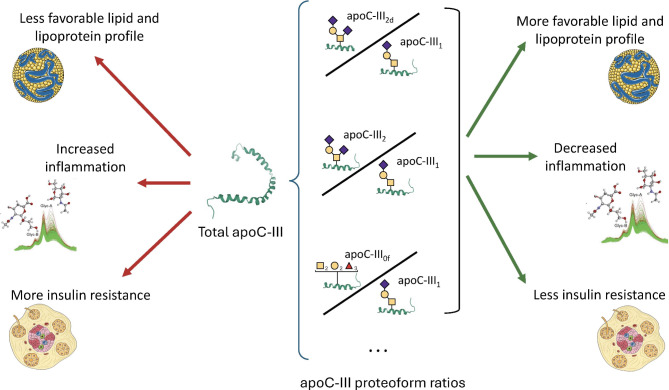
While apoC-III is positively associated with biomarkers of cardiometabolic risk, the proportions of three apoC-III proteoforms show opposite associations, independent of total apoC-III concentrations. Measuring not only apoC-III but also the proportions of apoC-III proteoforms can provide valuable information since individuals with similar levels of total apoC-III could display opposite lipid profiles depending on the proportion of apoC-III proteoforms.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12933-024-02531-5.
Keywords: Apolipoprotein C-III, Proteoforms, Cardiovascular disease, Type 2 diabetes, Dyslipidemia, Insulin resistance, Inflammation, Triglycerides, Glycoproteins, Lipoproteins
Background
Apolipoprotein (apo) C-III is a small protein (8.8 kDa) that is synthesized in the liver and intestine. It is mostly bound to lipoproteins, especially very low-density lipoproteins (VLDL) and high-density lipoproteins (HDL) [1], although it can also be detected in low-density lipoproteins (LDL) and intermediate-density lipoproteins (IDL) [2]. It has raised interest as a cardiovascular risk factor: loss-of-function mutations in the APOC3 gene are associated with a lower risk of both ischemic vascular disease and coronary heart disease in two large cohort studies [3, 4], and apoC-III concentrations in VLDL and LDL are predictors of recurrent coronary events [5].
ApoC-III has multiple functions, all of which are related to a worse cardiometabolic profile, which could explain the increased cardiovascular risk associated with apoC-III. These functions can be classified into three areas: lipid metabolism, glucose homeostasis, and inflammation. With respect to lipid metabolism, apoC-III is a major regulator of triglyceride concentrations: it decreases the uptake of triglyceride-rich lipoproteins (TRLs) by liver receptors [6], inhibits lipoprotein lipase (LPL) and hepatic lipase (HL) (although it is not clear whether LPL inhibition occurs in vivo) [7], and enhances the synthesis and secretion of VLDL [8].
In relation to glucose homeostasis, apoC-III impairs the function of pancreatic β cells, and preclinical studies suggest a role for apoC-III in the pathology of both type 1 and type 2 diabetes. In the context of the insulin resistance found in type 2 diabetes (T2DM), a local increase in the production of apoC-III within the pancreatic islets causes inflammation and apoptosis of β cells, thus impairing β-cell function [9]. In type 1 diabetes (T1DM), increased serum apoC-III causes an increase in intracellular Ca2+ and β-cell apoptosis [10].
In vitro experiments have shown that apoC-III also promotes the secretion of proinflammatory cytokines in endothelial cells [11] and induces inflammasome activation and apoptosis in monocytes [12]. Moreover, LDL particles containing apoC-III are more prone to aggregation in the endothelium [13].
This study focuses on apoC-III posttranslational modifications, which have the potential to alter, modulate or reverse the above-described functions. ApoC-III is posttranslationally modified inside the cell and is secreted as a glycosylated protein. In the circulation, four main forms of the protein were initially described: nonglycosylated apoC-III (apoC-III0a) and three proteoforms originating from O-glycosylation at threonine 74: apoC-III0b, with mucin-type core-1, Galactose (Gal)– N-acetylgalactosamine (GalNAc); and apoC-III1 and apoC-III2, with one or two sialic acid residues bound to the Gal-GalNAc core, respectively [14].
These four proteoforms have been studied because of their different behaviors in terms of cardiovascular risk and lipid metabolism. There are several population studies reporting that higher proportions of apoC-III2 and lower proportions of apoC-III1 are associated with improved lipids and lipoproteins and insulin resistance, whereas less consensus exists regarding apoC-III0a and apoC-III0b [15–17]. Mechanistic in vivo studies have revealed a different clearance pathway of apoC-III depending on its glycosylation status: some hepatic receptors such as the heparan sulfate proteoglycan preferentially clear apoC-III2, whereas others like the LDL receptor and the LDL receptor-related protein-1 are more prone to the uptake of apoC-III1 [18].
ApoC-III sialylation is also a biomarker of colorectal cancer [19] and incident cardiovascular events [20, 21] Therefore, it seems clear that not only apoC-III but also the way it is glycosylated is important in relation to the lipid profile and cardiovascular risk.
In addition to these four widely studied apoC-III proteoforms, other less abundant proteoforms have been identified by mass spectrometry: apoC-III1d and apoC-III2d, with the same glycosylation pattern as apoC-III1 and apoC-III2, respectively, but lacking the last alanine residue; and apoC-III0f, which is further fucosylated (Fuc) at the same residue (Gal2GalNAc2Fuc3). The literature on these proteoforms is scarce and has focused mostly on proteomics [17, 22, 23]; therefore, to the best of our knowledge, cohort studies assessing the associations between these proteoforms and clinical or biochemical biomarkers are lacking.
In the present study, we quantified up to 7 apoC-III proteoforms: the four main proteoforms apoC-III0a, apoC-III0b, apoC-III1, and apoC-III2 and the three less abundant proteoforms apoC-III1d, apoC-III2d and apoC-III0f. We analyzed the associations of all these proteoforms with a comprehensive set of biomarkers related to the three areas of apoC-III functions (namely, lipid metabolism, glucose homeostasis, and inflammation). Additionally, we combined all these proteoforms to define groups of people with different metabolic and cardiovascular risk profiles.
Methods
Study design and participants
The study population consisted of a subgroup of 875 participants from the cross-sectional study of the Di@bet.es cohort, which is a population-based study of the Spanish general population conducted in 2008–10 [24].
The samples were taken from a previous study aimed at exploring an advanced NMR lipoprotein and glycoprotein profile in relation to the prevalence and incidence of diabetes and cardiovascular disease. As such, the study population was enriched with people with different cardiometabolic disorders, including diabetes, prediabetes, and cardiovascular disease (CVD). People without any of these diseases and with similar sex frequencies were also included in a proportion close to 2:1.
Anthropometric data were collected and adherence to the Mediterranean diet was assessed via a 14-item questionnaire that had been previously developed and validated [25].
Biochemistry
Upon enrolment in the Di@bet.es study, serum samples were obtained, frozen and stored at -80 °C for subsequent analyses. Lipid and routine biochemistry values were obtained as previously reported [24]. The homeostatic model assessment of insulin resistance (HOMA-IR) index was calculated from glucose and insulin measurements. Apolipoproteins, lipoprotein (a) (Lp(a)) and high-sensitivity C-reactive protein (hsCRP) were measured using immunoturbidimetric assays (apoA-I, apoB: BioSystems, Spain; apoC-III, apoE: Randox, UK; hsCRP, Lp(a): Spinreact, Spain) with a Spin240 autoanalyzer (Spinreact, Spain).
Proton nuclear magnetic resonance (1H-NMR) lipoprotein and glycoprotein measurements
The particle concentrations of three subclasses (small, medium, and large) of each lipoprotein type (VLDL, LDL, and HDL) and the mean diameter of each lipoprotein type were obtained from the Liposcale® test (Biosfer Teslab SL, Spain) of serum samples, an advanced lipoprotein test based on 2D diffusion-ordered 1H-NMR spectroscopy and utilizing diffusion coefficients to quantify the number of lipoprotein particles, as previously described [26].
The glycoprotein signals GlycA, GlycB, and GlycF were obtained from analysis of the region of the 1H-NMR spectrum where the glycoproteins resonate (2.15–1.90 ppm) via several analytical functions, as previously described [27], by Biosfer Teslab SL (Spain). For each function, the total area (proportional to the concentration) was determined. The area of GlycA provides the concentration of protein-bound N-acetylneuraminic acid, and the area of GlycB provides those of N-acetylglucosamine [28]. The GlycF area is derived from the concentration of the acetyl groups of N-acetylglucosamine, N-acetylgalactosamine, and N-acetylneuraminic acid unbound to proteins (free fraction) [29].
Mass spectrometry immunoassay
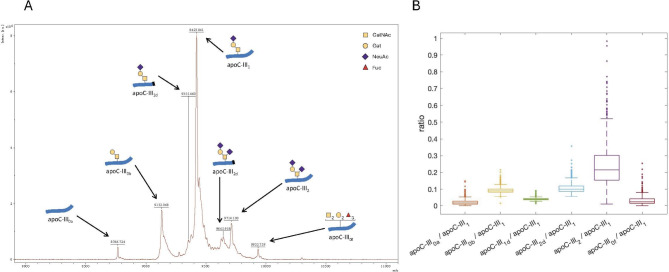
ApoC-III proteoforms were analyzed via a mass spectrometry immunoassay at the Centre for Omic Sciences (COS)– Eurecat, Reus, Spain. Briefly, serum samples were incubated with a biotinylated goat anti-apoC-III antibody (Fortis Life Sciences, MA, USA) and total apoC-III was immobilized on streptavidin AssayMAP cartridges (Agilent Technologies, CA, USA) using an automated AssayMAP BRAVO platform (Agilent Technologies, CA, USA). After elution, total apoC-III was dried and resuspended in a sinapinic acid matrix (15 mg/mL sinapinic acid in a solution of 33% acetonitrile and 0.4% trifluoroacetic acid in water), and 1 µL was loaded onto a ground steel MALDI plate. Mass spectra were acquired with an UltrafleXtrem III MALDI-TOF/TOF instrument (Bruker, Germany) operating in positive ion mode and with a range from 4 − 20 kDa. An average of 5000 laser shot mass spectra were saved for each sample spot. Mass spectra were internally calibrated using protein calibration standard-I (Bruker, Germany) and further processed using Flex Analysis 3.0 software (Bruker Daltonics). (Fig. 1A).
Fig. 1.
Detection and distribution of apoC-III proteoforms. A Representative mass spectrum of apoC-III proteoforms. B Distribution of the six apoC-III proteoform ratios in the study population. The number of observations in which each proteoform ratio could be computed was 745 (apoC-III0a/apoCIII1), 860 (apoC-III0b/apoCIII1), 739 (apoC-III1d/apoCIII1), 823 (apoC-III2d/apoCIII1), 871 (apoC-III2/apoCIII1) and 687 (apoC-III0f/apoCIII1). Apo, apolipoprotein; Fuc, fucose; Gal, galactose; GalNAc, N-acetylgalactosamine; NeuAc, N-acetylneuraminic acid (sialic acid)
The peaks were labeled, and peak intensity lists were generated. The extraction of peak intensities for each proteoform was automated within a defined confidence mass range spanning the theoretical m/z values 8765 (apoC-III0a), 9136 (apoC-III0b), 9350 (apoC-III1d), 9422 (apoC-III1), 9641 (apoC-III2d), 9713 (apoC-III2) and 9934 (apoC-III0f). Given the complexity of the spectra, the assignment of minor proteoforms was manually reviewed for each sample to ensure accuracy. For comparison, the peak intensities corresponding to each proteoform were normalized to the peak intensity of the apoC-III1 peak (the most abundant proteoform) to account for sample dilution; therefore, ratios of each apoC-III proteoform to the apoC-III1 proteoform were obtained: apoC-III0a/apoC-III1, apoC-III0b/apoC-III1, aoC-III1d/apoC-III1, apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1, as previously reported [16]. In some spectra, certain peak intensities could not be reliably quantified (especially those corresponding to the least abundant proteoforms) and were considered missing values (see Fig. 1 legend for the exact number of observations of each proteoform ratio). Batch correction was performed to address the inter-day variability in mass spectra acquisition.
Statistical analyses
The normality of the variables was assessed via histograms and qq-plots of the distributions. For linear regression analyses, triglycerides, Lp(a); the total VLDL particle concentration (VLDLP); small, medium, and large VLDL particle concentrations (S-VLDLP, M-VLDLP, and L-VLDLP, respectively); insulin; fasting glucose; HOMA-IR; GlycA; GlycB; GlycF; and hsCRP were log-transformed.
Multivariate linear regression models were built for each studied parameter (lipids, lipoproteins, and apolipoproteins; inflammatory markers and glucose metabolism variables) as dependent variables, with one proteoform ratio at a time used as a predictor. All the models were adjusted for age, sex, body mass index (BMI), lipid-lowering medication, exercise, and the Mediterranean diet score, plus specific confounders, depending on the analyses: smoking history for lipid- or inflammation-related variables and anti-inflammatory medication for inflammation-related variables. To study the independence of the observed effects from triglyceride levels and apoC-III, additional models were built and adjusted for triglycerides or apoC-III in addition to the abovementioned confounders. Both the dependent variable and the proteoform ratio were standardized.
Clustering of the data was performed via all of the proteoform ratios: apoC-III0a/apoC-III1, apoC-III0b/apoC-III1, apoC-III1d/apoC-III1, apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 (only in those cases with data of all the proteoform ratios available, n = 645). The k-means and k-medoids algorithms were tested, and the silhouette coefficient was calculated. On the basis of the silhouette values and interpretability of the data, two clusters were defined using the k-medoids algorithm. The characteristics of the two clusters were analyzed via principal component analysis (PCA) (using all the proteoform ratios) and the loadings of each proteoform.
Differences in potential confounders (age, sex, BMI, anti-inflammatory medication, oral antidiabetic medication, insulin treatment, and smoking) between clusters were assessed using the Student’s t-test for continuous, normally distributed variables; Wilcoxon’s test for continuous, nonnormally distributed variables; and the chi-square test for categorical variables.
Differences in lipid, lipoprotein, glucose, and inflammation-related parameters between clusters were assessed via ANCOVA, controlling for confounders that were found to differ between clusters and could interfere with the studied variable: age and sex for all variables, plus anti-inflammatory medication for inflammation-related parameters. To compare variables related to glucose metabolism, all participants with T1DM or previously diagnosed with T2DM were excluded. The false discovery rate of the differences between clusters was controlled via the Benjamini and Hochberg method, with a false discovery rate threshold of 5%. The prevalence of metabolic disorders was compared between clusters using the chi-square test.
Univariate differences in proteoform ratios between groups of treated and nontreated participants were assessed using Wilcoxon’s test.
All the statistical analyses were performed using MATLAB version R2024a (MathWorks Inc. USA).
Results
Characteristics of the study population
The study population was a subgroup of the Di@bet.es cohort, consisting of 875 participants with a mean age of 56.1 ± 14.6 years, 46.2% of whom were men. The mean lipid levels were within the normal range, and a high proportion of participants had diabetes: 239 (27.3%), of whom 126 (14.4% of the study population) were not previously diagnosed (without treatment) (Table 1).
Table 1.
Characteristics of the study population (n = 875)
| Age, y | 56.08 (47.00–67.00) |
| Male sex, n (%) | 404 (46.17) |
| BMI, kg/m2 | 29.28 (25.93–32.06) |
| Systolic blood pressure, mm Hg | 136.94 (123.25–150.00) |
| Diastolic blood pressure, mm Hg | 79.33 (71.62–85.50) |
| Obesity, n (%) | 338 (38.99) |
| Type 2 diabetes, n (%) | 220 (25.14) |
| Hypertension, n (%) | 515 (59.06) |
| Metabolic syndrome, n (%) | 458 (54.39) |
| Under treatment | |
| Lipid-lowering medication, n (%) | 204 (23.34) |
| Oral antidiabetic medication, n (%) | 77 (8.80) |
| Insulin treated, n (%) | 31 (3.54) |
| Anti-hypertensive medication, n (%) | 305 (34.86) |
| Anti-inflammatory medication, n (%) | 196 (22.43) |
| Current smoking, (%) | 187 (21.37) |
| Total cholesterol, mg/dL | 199.00 ± 39.32 |
| LDLc, mg/dL | 107.65 ± 29.88 |
| HDLc, mg/dL | 50.34 (41.18–57.23) |
| Triglycerides, mg/dL | 133.05 (82.15-157.88) |
| Lp(a), mg/dL | 26.61 (4.43–37.32) |
| apoA-I, mg/dL | 130.39 (105.64-150.34) |
| apoB100, mg/dL | 90.93 ± 13.95 |
| apoC-III, mg/dL | 10.07 (7.63–11.87) |
| apoE, mg/dL | 4.08 (3.51–4.55) |
| Fasting glucose, mg/dL | 108.60 (89.82-115.74) |
| hsCRP, mg/L | 4.07 (0.97–4.49) |
The number of observations and percentage are shown for categorical variables, the mean ± standard deviation for normally distributed continuous variables, and the median (quartile 1–quartile 3) for nonnormally distributed continuous variables. Apo, apolipoprotein; BMI, body mass index; HDLc, high-density lipoprotein cholesterol; hsCRP, high-sensitivity C-reactive protein; LDLc, low-density lipoprotein cholesterol; Lp(a), lipoprotein (a)
The median apoC-III concentration was 10.07 mg/dL. Among the 7 measured apoC-III proteoforms, the ratios of 6 proteoforms (apoC-III0a, apoC-III0b, apoC-III1d, apoC-III2d, apoC-III2, and apoC-III0f) to the most abundant proteoform apoC-III1 were computed. The proteoform with the highest proportion in relation to apoC-III1 was apoC-III2 (24% of the apoC-III1 peak), followed by apoC-III2d and apoC-III0b (11% and 9%, respectively). The apoC-III0a, apoC-III1d, and apoC-III0f peaks were less pronounced (2%, 4%, and 3% of the apoC-III1 peak, respectively, and not quantifiable in part of the study sample) (Fig. 1B).
ApoC-III, apoC-III proteoforms, and lipid metabolism
Triglycerides and VLDL
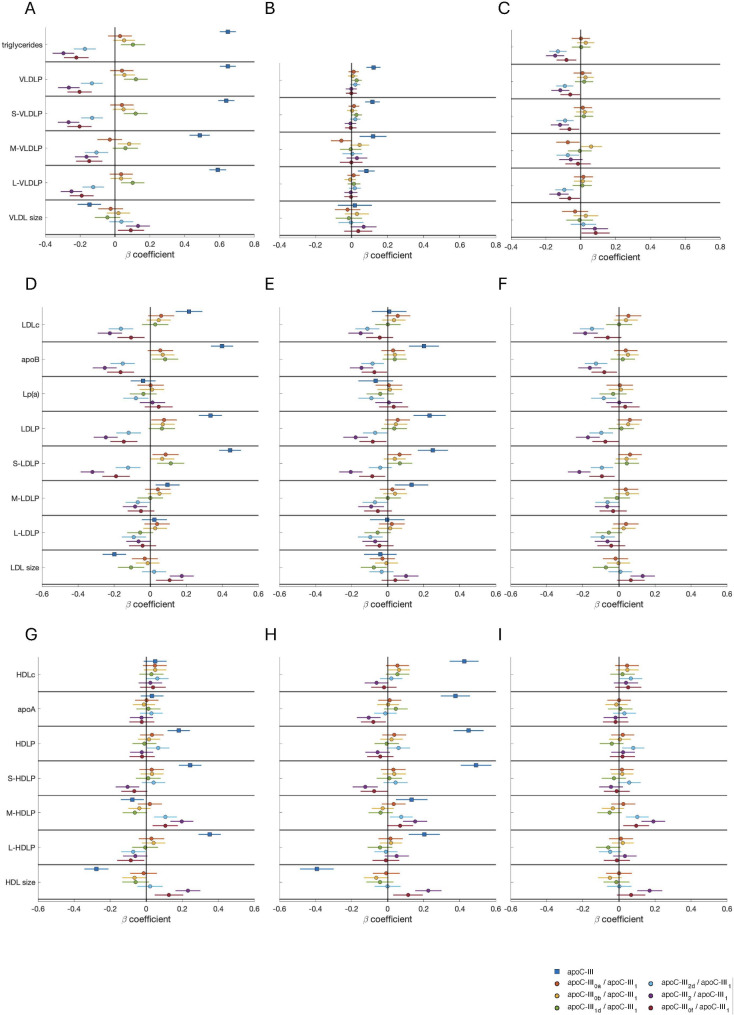
Total apoC-III was positively associated with triglyceride (β = 0.648, p < 0.001) and VLDL particle concentrations (β = 0.648, p < 0.001; β = 0.639, p < 0.001; β = 0.486, p < 0.001 and β = 0.590, p < 0.001; for total, small, medium and large VLDL particles, respectively) (Fig. 2A). The association with all VLDL particle subclasses was still significant after controlling for the triglyceride concentration, although the magnitude of the effect was markedly lower (Fig. 2B).
Fig. 2.
Associations between apoC-III proteoforms and parameters of TRL (A–C), LDL (D–F) and HDL (G–I) metabolism. Beta coefficients and 95% confidence intervals of apoC-III or proteoform ratios (predictors) for individual multivariate linear regression models on each lipid-related variable (dependent variables) are shown. In each model, both the predictor and dependent variable are standardized; therefore, the coefficients indicate the SD variation in the lipid-related parameter for a 1 SD increase in the apoC-III or apoC-III proteoform ratio. A, D, G: Models were adjusted for age, sex, BMI, lipid-lowering medication, exercise, Mediterranean diet score, and smoking history. B, E, H: Models were adjusted for confounders in the first models (A, D, G) plus triglycerides. C, F, I: Models were adjusted for confounders in the first models (A, D, G) plus apoC-III. Apo, apolipoprotein; BMI, body mass index; HDL, high-density lipoprotein; HDLc, HDL cholesterol; HDLP, HDL particle concentration; L-HDLP, large HDL particle concentration; L-LDLP, large LDL particle concentration; L-VLDLP, large VLDL particle concentration; LDL, low-density lipoprotein; LDLc, LDL cholesterol; LDLP, LDL particle concentration; Lp(a), lipoprotein (a); M-HDLP, medium HDL particle concentration; M-LDLP, medium LDL particle concentration; M-VLDLP, medium VLDL particle concentration; VLDL, very low-density lipoprotein; VLDLP, VLDL particle concentration; S-HDLP, small HDL particle concentration; S-LDLP, small LDL particle concentration; S-VLDLP, small VLDL particle concentration; SD, standard deviation; TRL, triglyceride-rich lipoprotein
Analysis of individual apoC-III proteoforms revealed that the ratio apoC-III1d/apoC-III1 was also positively associated with triglycerides, total, large, and small VLDL particles. However, the ratios of three proteoforms (apoC-III2d, apoC-III2, and apoC-III0f) to apoC-III1 were negatively associated with triglycerides and the concentrations of all VLDL particles. ApoC-III2/apoC-III1 had the greatest negative associations (β=-0.297, p < 0.001; β=-0.265, p < 0.001; β=-0.267, p < 0.001; β=-0.163, p < 0.001; β=-0.250, p < 0.001; for triglycerides; total, small, medium and large VLDL particle concentrations, respectively), but all three ratios had similar regression coefficients (Fig. 2A). These effects were significant even after controlling for total apoC-III concentrations (except with medium VLDL particles), albeit with a lower magnitude (Fig. 2C), but were lost after adjusting for triglycerides (Fig. 2B). An interesting negative association between the same three ratios (apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1) and VLDL cholesterol (VLDLc) (β=-0.135, p < 0.001; β=-0.206, p < 0.001; β=-0.160, p < 0.001; respectively) was also observed, although it was not independent of triglyceride levels either (data not shown).
An increase in total apoC-III was associated with a smaller VLDL size, whereas the ratios apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 were associated with a larger VLDL size (Fig. 2A). For the ratio apoC-III2/apoC-III1, this association was independent of total apoC-III (Fig. 2C) but was lost when controlling for triglycerides (Fig. 2B).
LDL
Concerning the metabolism of LDL cholesterol (LDLc), total apoC-III was positively associated with LDLc (β = 0.214, p < 0.001), apoB100 (β = 0.399, p < 0.001), and total, small and medium LDL particle concentrations (β = 0.333, p < 0.001; β = 0.443, p < 0.001; β = 0.096, p = 0.004; respectively) (Fig. 2D). Except for LDLc, this effect was independent of the triglyceride concentration (Fig. 2E).
The associations of apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 ratios were opposite to those of total apoC-III, with apoC-III2/apoC-III1 demonstrating the strongest negative association with LDLc (β=-0.223, p < 0.001), apoB100 (β=-0.253, p < 0.001) and LDL particles (β=-0.246, p < 0.001; β=-0.320, p < 0.001; β=-0.084, p < 0,001 for total, small and medium LDL, respectively) (Fig. 2D). These effects remained significant after controlling for triglycerides (all the associations) (Fig. 2E) or total apoC-III (all except medium LDL particles; Fig. 2F).
Interestingly, higher levels of apoC-III were related to smaller LDL particles (β=-0.199, p < 0.001), but a greater proportion of apoC-III2 and apoC-III0f over apoC-III1 was associated with larger particles (β = 0.175, p < 0.001; β = 0.108, p = 0.005), which, in the case of apoC-III2, was still significant after controlling for total triglycerides or total apoC-III.
HDL
Total apoC-III was also associated with increased HDL cholesterol (HDLc) (β = 0.424, p < 0.001), apoA-I (β = 0.376, p < 0.001) and HDL particles (β = 0.449, p < 0.001; β = 0.491, p < 0.001; β = 0.134, p = 0.003; β = 0.203, p < 0.001; for total, small, medium and large HDL particles, respectively) when regressions were adjusted for triglyceride levels (due to the strong relationship between triglycerides and HDL, both directly related to apoC-III, the magnitude and significance of some of the previous associations was only evident after adjusting for triglycerides, Fig. 2G-H).
After adjusting for triglyceride concentrations, the ratios apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 showed the opposite trend to that of total apoC-III in relation to apoA-I (β=-0.105, p = 0.002 for apoC-III2/apoC-III1; β=-0.079, p = 0.024 for apoC-III0f/apoC-III1) and small HDL particles (β=-0.124, p < 0.001 for apoC-III2/apoC-III1 and a nonsignificant trend for apoC-III0f/apoC-III1: β=-0.075, p = 0.052), although this trend was not independent of total apoC-III. These two ratios, together with ApoC-III2d/apoC-III1, were positively associated with medium HDL particles after adjusting for total apoC-III (β = 0.102, p = 0.001; β = 0.191, p < 0.001; β = 0.096, p = 0.009 for apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1, respectively. Figure 2H-I).
An increase in apoC-III concentrations was related to smaller HDL particles (β=-0.393, p < 0.001 with HDL size, controlling for triglycerides), whereas increased apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 ratios were associated with larger HDL particles (β = 0.226, p < 0.001; β = 0.113, p = 0.007, respectively, controlling for triglycerides). The association with apoC-III2/apoC-III1 remained significant after adjusting for total apoC-III (Fig. 2H-I).
ApoC-III, apoC-III proteoforms, and inflammation
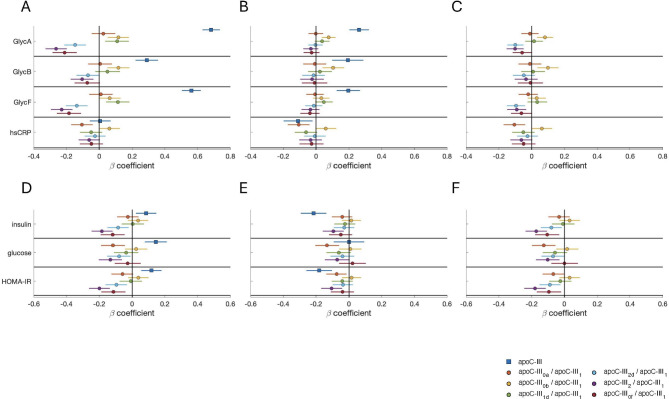
Because of the proinflammatory role of apoC-III, the associations between apoC-III and its proteoforms and distinct markers of inflammation (hsCRP and 1H-NMR-assessed glycoprotein signals GlycA, GlycB, and GlycF) were explored.
Strong and positive associations between apoC-III and GlycA (β = 0.684, p < 0.001), GlycB (β = 0.289, p < 0.001), and GlycF (β = 0.562, p < 0.001) (Fig. 3A) were observed, which remained significant after adjusting for triglycerides. After this adjustment, a weak negative association with hsCRP (β=-0.110, p = 0.015) was discovered (Fig. 3B).
Fig. 3.
Associations between apoC-III proteoforms and parameters of inflammation (A–C) and glucose metabolism (D–F). Beta coefficients and 95% confidence intervals of apoC-III or proteoform ratios (predictors) for individual multivariate linear regression models on each inflammation or glucose-related variable (dependent variables) are shown. In each model, both the predictor and dependent variable are standardized; therefore, the coefficients indicate the SD variation in the inflammation- or glucose-related parameters for a 1 SD increase in the apoC-III or apoC-III proteoform ratio. A: Models were adjusted for age, sex, BMI, lipid-lowering medication, exercise, Mediterranean diet score, smoking history, and anti-inflammatory medication. B: Models were adjusted for confounders in A plus triglycerides. C: Models were adjusted for confounders in A plus apoC-III. D: Models were adjusted for age, sex, BMI, lipid-lowering medication, exercise, and the Mediterranean diet score. E: Models were adjusted for confounders in D plus triglycerides. F: Models were adjusted for confounders in D plus apoC-III. Apo, apolipoprotein; BMI, body mass index; HOMA-IR, homeostatic model assessment of insulin resistance; hsCRP, high-sensitivity C-reactive protein; SD, standard deviation
The apoC-III0b/apoC-III1 ratio was also positively associated with GlycA (β = 0.116, p < 0.001) and GlycB (β = 0.116, p < 0.001), and this effect was independent of triglycerides or total apoC-III. Conversely, the apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 ratios exhibited the opposite trend in terms of their relationships with GlycA (β=-0.148, p < 0.001; β=-0.263, p < 0.001; β=-0.211, p < 0.001) and GlycF (β=-0.138, p < 0.001; β=-0.230, p < 0.001; β=-0.1845, p < 0.001). The trends involving ratio of apoC-III2d and apoC-III2 were independent of total apoC-III, but none of them resisted the adjustment for triglycerides. The apoC-III0a/apoC-III1 ratio was negatively associated with hsCRP levels (β=-0.106, p = 0.002) independently of triglycerides or total apoC-III, whereas the apoC-III0b/apoC-III1 ratio showed the opposite trend (Fig. 3A-C).
ApoC-III, apoC-III proteoforms, and glucose metabolism
Since apoC-III is tightly controlled by insulin and glucose and has been reported to affect the function of β cells in pancreatic islets, the relationships between apoC-III and its proteoforms and the clinical parameters of glucose metabolism were also studied. In this context, participants under treatment for type 1 or type 2 diabetes were excluded to eliminate the potential effects of drugs or insulin injection on the studied parameters.
Total apoC-III was associated with increased plasma concentrations of insulin (β = 0.086, p = 0.007) and fasting glucose (β = 0.145, p < 0.001) and, consequently, with an increased HOMA-IR index (β = 0.118, p < 0.001) (Fig. 3D). Interestingly, after adjustment for triglycerides, the association with HOMA-IR became negative (β=-0.181, p < 0.001) (Fig. 3E).
The apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 ratios showed the opposite association to that observed for total apoC-III in relation to HOMA-IR (β=-0.096, p = 0.003; β=-0.199, p < 0.001 and β=-0.114, p = 0.002, respectively) as did apoC-III0a/apoC-III1 (β=-0.060, p = 0.077). These relationships were independent of total apoC-III, but only apoC-III2/apoC-III1 and apoC-III0a/apoC-III1 were significantly associated with HOMA-IR after controlling for triglycerides (Fig. 3D-F).
Clusters of apoC-III proteoforms
The previous results were obtained from models that used only one proteoform ratio at a time. Since interesting associations were observed in more than just one proteoform ratio, we wanted to assess whether the combination of all proteoforms could characterize subgroups of people with different profiles. All 6 ratios of apoC-III proteoforms were used to cluster the data into two clusters using the k-medoids algorithm (since this number of clusters and this algorithm retrieved the best average silhouette coefficient among those tested). Only the observations with available data for all ratios were used for this analysis (n = 645).
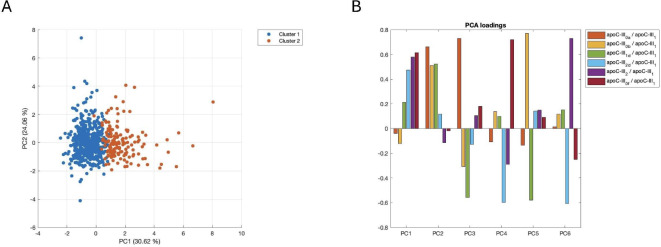
The PCA of all 6 ratios revealed that the two clusters were separated by principal component (PC) 1, with Cluster 1 (n = 494) having mostly positive values and Cluster 2 (n = 151) having negative values for PC1 (Fig. 4A). This principal component was defined by positive loadings for apoC-III1d/apoC-III1 and, with a greater magnitude, apoC-III2d/apoC-III1, apoC-III2/apoC-III1 and apoC-III0f/apoC-III1 and negative values of apoC-III0a/apoC-III1 and apoC-III0b/apoC-III1 (although less important in magnitude) (Fig. 4B). Therefore, high values of the three proteoform ratios that were associated with a more protective profile (in terms of lipids, lipoproteins, biomarkers of inflammation, and glucose metabolism, according to the results from the previous section) defined Cluster 2, whereas low values of these proteoform ratios defined Cluster 1.
Fig. 4.
Clusters of apoC-III proteoforms. A: PCA of the study sample (only observations with data available for all proteoform ratios, n = 645) generated via the 6 apoC-III proteoform ratios showing the two clusters. B: PCA loadings of the 6 principal components. Apo, apolipoprotein; PC, principal component; PCA, principal component analysis
Compared with those in Cluster 2, the participants in Cluster 1 were older (p = 0.002) and had a higher BMI (p < 0.001), as well as a higher proportion of individuals treated with anti-inflammatory drugs (p = 0.045) or insulin (p = 0.007) (Table S1); therefore, subsequent analyses were performed taking into account these potential confounders.
Lipid metabolism, inflammation, and glucose metabolism parameters were compared across the two clusters (Table 2). Compared with those in Cluster 2, participants in Cluster 1 presented higher triglyceride (p < 0.001), apoC-III (p < 0.001), VLDLc (p < 0.001), and VLDL particle (p = 0.002) concentrations. LDLc (p = 0.001), apoB100 (p = 0.003), and the concentration of LDL particles (p = 0.002) were also higher in Cluster 1. In addition, Cluster 1 displayed increased levels of small LDL particles (p < 0.001) and a smaller mean LDL size (p = 0.001). Cluster 2 had higher concentrations of medium HDL particles (p < 0.001) and larger HDL particles (p = 0.003).
Table 2.
Lipid, lipoprotein, glucose, and inflammation-related differences between cluster 1 and 2
| Cluster 1 (n = 494) | Cluster 2 (n = 151) | p value | |
|---|---|---|---|
| triglycerides, mg/dL | 130.20 (95.66-177.15) | 96.55 (71.74-137.29) | 0.004 |
| HDLc, mg/dL | 47.56 (40.22–56.07) | 51.43 (41.57–59.55) | 0.470 |
| LDLc, mg/dL | 111.78 ± 29.15 | 102.85 ± 30.43 | 0.006 |
| VLDLc, mg/dL | 41.38 (34.80-48.72) | 37.51 (31.42–45.15) | 0.004 |
| Lp(a), mg/dL | 13.04 (4.52–37.68) | 12.77 (4.55–34.61) | 0.384 |
| apoB100, mg/dL | 93.22 ± 13.83 | 89.42 ± 14.13 | 0.007 |
| apoA-I, mg/dL | 122.28 (103.77-146.66) | 130.54 (109.84–153.50) | 0.904 |
| apoE, mg/dL | 4.04 (3.58–4.63) | 4.06 (3.48–4.59) | 0.382 |
| apoC-III, mg/dL | 10.12 (8.12–12.72) | 8.99 (6.87–11.45) | 0.004 |
| VLDLP, nmol/L | 54.07 (35.66–81.43) | 37.61 (29.20-58.86) | 0.006 |
| S-VLDLP, nmol/L | 47.76 (30.99–73.52) | 32.80 (25.65–51.10) | 0.006 |
| M-VLDLP, nmol/L | 4.59 (3.36–6.09) | 3.79 (2.68–4.92) | 0.116 |
| L-VLDLP, nmol/L | 1.44 (1.07–2.02) | 1.12 (0.83–1.53) | 0.058 |
| LDLP, nmol/L | 1460.25 ± 280.25 | 1380.63 ± 265.43 | 0.007 |
| S-LDLP, nmol/L | 828.41 (733.71-937.63) | 764.43 (681.74-847.47) | < 0.001 |
| M-LDLP, nmol/L | 413.73 ± 136.88 | 416.25 ± 140.62 | 0.705 |
| L-LDLP, nmol/L | 191.89 ± 38.46 | 192.70 ± 38.49 | 0.730 |
| HDLP, µmol/L | 27.47 (24.47–30.58) | 28.28 (25.57–31.23) | 0.884 |
| S-HDLP, µmol/L | 18.63 ± 4.01 | 18.63 ± 4.08 | 0.387 |
| M-HDLP, µmol/L | 8.92 (8.06–9.75) | 9.38 (8.54–10.45) | 0.005 |
| L-HDLP, µmol/L | 0.28 (0.25–0.31) | 0.28 (0.25–0.30) | 0.528 |
| VLDL size, nm | 41.88 (41.62–42.13) | 42.01 (41.75–42.22) | 0.120 |
| LDL size, nm | 20.96 (20.70-21.15) | 21.07 (20.83–21.28) | 0.007 |
| HDL size, nm | 8.25 (8.20–8.30) | 8.27 (8.23–8.33) | 0.007 |
| fasting insulin, mIU/L | 9.16 (6.38–13.10) | 6.44 (4.96–8.19) | 0.010 |
| fasting glucose, mg/dL | 99.00 (90.36-112.45) | 94.41 (86.04-105.39) | 0.096 |
| HOMA-IR | 2.29 (1.54–3.45) | 1.51 (1.13–2.04) | 0.016 |
| GlycA, µmol/L | 734.14 (646.91-851.53) | 660.54 (596.33-783.84) | 0.002 |
| GlycB, µmol/L | 258.34 (227.31-296.65) | 250.97 (215.50-287.38) | 0.465 |
| GlycF, µmol/L | 293.12 (258.72-330.04) | 273.93 (236.92-312.41) | 0.006 |
| hsCRP, mg/L | 2.21 (1.07–4.68) | 1.76 (0.72–3.60) | 0.393 |
The mean ± standard deviation is reported for variables that are normally distributed in both clusters, and the median (quartile 1–quartile 3) is reported for variables that are nonnormally distributed in at least one cluster. P values were calculated via ANCOVA with age and BMI as covariates (for all variables except inflammation-related variables) and age, BMI, and anti-inflammatory medication for GlycA, GlycB, GlycF, and hsCRP; and further corrected for multiple comparisons via the Benjamini and Hochberg procedure with a false discovery rate threshold of 5%. Participants with T1DM or previously diagnosed T2DM were excluded from the models for glucose, insulin, and HOMA-IR. Apo, apolipoprotein; HDL, high-density lipoprotein; HDLc, HDL cholesterol; HDLP, HDL particle concentration; HOMA-IR, homeostatic model assessment of insulin resistance; hsCRP, high-sensitivity C-reactive protein; L-HDLP, large HDL particle concentration; L-LDLP, large LDL particle concentration; L-VLDLP, large VLDL particle concentration; LDL, low-density lipoprotein; LDLc, LDL cholesterol; LDLP, LDL particle concentration; Lp(a), lipoprotein (a); M-HDLP, medium HDL particle concentration; M-LDLP, medium LDL particle concentration; M-VLDLP, medium VLDL particle concentration; VLDL, very low-density lipoprotein; VLDLc, VLDL cholesterol; VLDLP, VLDL particle concentration; S-HDLP, small HDL particle concentration; S-LDLP, small LDL particle concentration; S-VLDLP, small VLDL particle concentration T1DM, T2DM,
With respect to glucose metabolism and inflammation, Cluster 1 had increased insulin resistance, as assessed by HOMA-IR (p = 0.008), and the 1H-NMR-assessed biomarkers of inflammation, GlycA, and GlycF were also higher in Cluster 1 (p < 0.001 and p = 0.003, respectively) than in Cluster 2 (Table 2).
Since the above results for lipids, glucose metabolism, and inflammation clearly revealed one cluster with a worse metabolic and inflammatory profile, we studied whether there were any differences in the prevalence of several metabolic conditions across the two clusters. We found a greater prevalence of obesity (p = 0.001), metabolic syndrome (p = 0.013), and T2DM (p = 0.022) within Cluster 1. Surprisingly, the history of cardiovascular events was higher in Cluster 2 (p = 0.009) (Table 3).
Table 3.
Prevalence of metabolic disorders across the clusters defined by apoC-III proteoforms
| Cluster 1 | Cluster 2 | p value | |
|---|---|---|---|
| Prediabetes | 116 (25.72%) | 26 (20.97%) | 0.277 |
| Type 2 diabetes | 87 (19.29%) | 13 (10.48%) | 0.022 |
| CVD | 100 (20.24%) | 46 (30.46%) | 0.009 |
| obesity | 210 (42.86%) | 42 (28.19%) | 0.001 |
| metabolic syndrome | 288 (60.63%) | 70 (48.95%) | 0.013 |
| hypertension | 304 (61.91%) | 81 (53.64%) | 0.07 |
The number of observations and percentage are reported for each outcome. P values were computed via the chi-square test. For the prevalence of prediabetes and type 2 diabetes, those participants with T1DM or previously diagnosed with T2DM were excluded. CVD, cardiovascular disease
Figure titles and legends
Discussion
In the present study, we were able to quantify seven different apoC-III proteoforms in more than 800 individuals from a subgroup of the Di@bet.es cohort and assess their relationship with a complete set of parameters related to cardiovascular risk.
Lipoprotein profile
In terms of lipid metabolism, we observed that three proteoform ratios (apoC-III2d/apoC-III1, apoC-III2/apoC-III1, and apoC-III0f/apoC-III1) displayed the opposite behavior to that of total apoC-III since they were associated with lower levels of triglycerides and LDLc, as well as their transport particles (VLDL and LDL) and apoB100. Notably, the strongest association between these three ratios and LDL particles was found with the atherogenic small LDL subtype, a hallmark of diabetic dyslipidemia [30, 31] and associated with a higher risk of CVD [32]. This feature was again opposed to those of total apoC-III. While the associations with triglyceride-related parameters were dependent on total triglyceride concentrations, the negative association with LDLc and LDL particles was, in part, independent of triglycerides. This finding is especially relevant in the case of reduced small LDL particles, a subtype that is normally a consequence of increased triglyceride concentrations [33] and highlights the importance of apoC-III glycosylation beyond its effects on triglyceride metabolism. Some of these protective proteoform ratios were negatively correlated with total apoC-III (Supplemental Figure S1). However, the observed associations were in part independent of apoC-III levels, which indicated that they were not completely driven by the lower apoC-III levels associated with these proteoforms and emphasized the importance of glycosylation.
The literature on the associations between minor apoC-III proteoforms (apoC-III1d, apoC-III2d, and apoC-III0f) and lipid or lipoprotein parameters is scarce, and most of the available studies analyze the four main proteoforms with a focus on the apoC-III2/apoC-III1 ratio. Our results on these proteoforms and triglyceride metabolism are in line with those reported from previous studies on people without diabetes [15, 34], with prediabetes or diabetes [16], with hypertriglyceridemia and receiving apoC-III-lowering therapy [18] or in the MESA cohort [20]. Yassine et al. reported that the ratios of apoC-III0a, apoC-III0b, and apoC-III1 to apoC-III2 were positively correlated with fasting triglycerides, which supports our findings that higher apoC-III2 and/or lower apoC-III1 were associated with fewer triglycerides and VLDL. We were unable to find such clear associations regarding the proportion of apoC-III0a and apoC-III0b, but in our results, we normalized these proteoforms by apoC-III1, and in the mentioned article, the ratios are computed to apoC-III2. These results suggest that apoC-III0a and apoC-III0b, as well as apoC-III1, are directly related to triglycerides, which, in our method, could mask the associations between the ratios of these proteoforms and triglycerides. Koska et al. showed that both triglycerides and LDLc are strongly and negatively correlated with the ratio of apoC-III2/apoC-III1, which is in agreement with the trends found in our study regarding these proteoforms. This protective role of a greater relative proportion of apoC-III2 that we and others have observed in relation to triglyceride metabolism could be explained by less efficient interference with TRL uptake by the liver [35] or less inhibition of LPL activity [16].
The strong relationship between apoC-III2/apoC-III1 and fewer small LDL particles (or a larger mean LDL size) that we observed was also reported in the abovementioned study, also in an apoC-III-independent fashion [16]; and, in Mendoza et al., apoC-III2 was the only proteoform that was not related to a decreased LDL size [17]. However, other studies have shown a negative association between the apoC-III2 production rate and LDL size [36]. Further research with comparable proteoform metrics (ratios, absolute concentrations, and kinetic metrics) is needed to understand the full picture of the relationship between apoC-III proteoforms and LDL size.
In relation to HDL, we observed a clear positive association of total apoC-III with HDLc and HDL particles after adjusting for triglycerides. This relationship may seem counterintuitive because of the known association of apoC-III with a more proatherogenic lipid profile, but it could be explained by the presence of apoC-III on the surface of HDL particles and, after controlling for triglyceride levels (which impact the HDL particle composition [33]), apoC-III and HDL were directly related, as previously observed in the MESA cohort [20]. ApoC-III2/apoC-III1 and apoC-III0f/apoC-III1 were associated with decreased small HDL and a larger mean HDL size. This finding is consistent with the protective lipid profile associated with these proteoform ratios that we observed regarding the LDL and VLDL parameters since small and dense HDL are a hallmark of diabetic dyslipidemia, and a larger HDL size is related to reduced cardiovascular risk [37]. However, HDL size is a controversial parameter: most of the published results on HDL size and cardiovascular risk are not independent of HDLc or other traditional risk factors, and some studies even reported opposite associations with cardiovascular mortality [38]. Here, we also observed an association of these proteoforms with medium HDL independent of the total apoC-III concentration, which should be further studied. Other studies showed that apoC-III2 was also associated with increased HDLc and that apoC-III1 was associated with decreased HDLc, which supports the suggested protective role of the apoC-III2/apoC-III1 ratio [20, 34]; however, in our study, we did not observe such associations.
Inflammation
While total apoC-III was undoubtedly associated with more inflammation when assessed by GlycA, GlycB, or GlycF, there was no association between apoC-III and hsCRP (and it was even negative after controlling for triglycerides). This finding is in line with previous data from our group, where iPCSK9 therapy resulted in a parallel decrease in apoC-III and glycoprotein signals, while no such association was observed with hsCRP [39].
The negative associations between apoC-III2d/apoC-III1, apoC-III2/apoC-III1, and apoC-III0f/apoC-III1 and 1H-NMR-glycoprotein signals are explained by plasma triglycerides, and the positive relationship between glycoprotein signals and total apoC-III was also diminished when adjusting for triglycerides (although it remained significant). This result was expected because of the pro-inflammatory nature of triglycerides and their high correlation with glycoproteins. However, the positive association of apoC-III0b/apoC-III1 with GlycA and GlycB seemed to be partly independent of triglyceride concentrations and total apoC-III. The apoC-III0a/apoC-III1 ratio was the only ratio that was associated with lower hsCRP, whereas the ratio of the other proteoform with no sialic acids, apoC-III0b/apoC-III1, showed the opposite trend, suggesting that not only sialic acids but also the GalGalNAc moiety is related to differential associations among apoC-III proteoforms.
To the best of our knowledge, we are among the first group to report associations between apoC-III proteoforms and 1H-NMR-assessed glycoprotein signals, and most importantly, these associations were partly independent of total apoC-III concentrations.
Glycoproteins are strongly related to cardiometabolic risk [40, 41] and are predictors of CVD [42] and T2DM [43]. Accordingly, the same proteoforms that are associated with a more protective lipid profile also show negative regression coefficients with GlycA, GlycF and, to a lesser extent, GlycB. However, Hiukka et al. [13] reported that, compared with other proteoforms, apoC-III2 stimulated greater secretion of proinflammatory cytokines in vitro; this finding could also be explained by the different types of inflammation that are assessed and the methodology used, although further research on the role of apoC-III glycosylation in inflammation should be performed, including a greater number of biomarkers of inflammation, to fully understand its implications.
Glucose metabolism
Few glucose homeostasis biomarkers are available in our cohort, but the same protective proteoform ratios were found to be associated with reduced insulin resistance, in agreement with the lipid profile associated with these proteoforms. ApoC-III0a/apoC-III1 also showed a similar coefficient with HOMA-IR, which was less expected because, in general, it exhibited a trend towards a more dyslipidaemic lipid profile and a significant association with the small LDL subclass; but, conversely, it was also related to less inflammation, as assessed by hsCRP. The literature provides similar results in relation to fasting glucose in the MESA cohort [20] and in people with diabetes [16]. In addition, the present results revealed that the associations between these proteoforms and insulin resistance were again independent of total apoC-III concentrations.
The analyses performed in all three areas (lipid metabolism, inflammation, and glucose homeostasis) revealed protective associations among the three proteoform ratios. Nevertheless, multivariate models including all proteoform ratios confirmed an independent effect of apoC-III2/apoC-III1 but suggested that the associations of apoC-III2d/apoC-III1 and, to a lesser extent, apoC-III0f/apoC-III2 were dependent on the apoC-III2/apoC-III1 ratio (data not shown).
ApoC-III proteoform clusters
Measuring and analyzing as many as seven apoC-III proteoforms allowed us to identify more proteoforms associated with a protective lipid profile than those previously reported. The PCA loadings showed that the three protective proteoform ratios (all three in the same direction) were the variables that contributed the most to the unsupervised clustering of the sample by the k-medoids algorithm, splitting the population into one small cluster with high values and another greater cluster with smaller values of these ratios. As expected, they differed in most of the lipid, lipoprotein, inflammation, and glucose homeostasis parameters that were already associated with these ratios and clearly showed a more proatherogenic profile of Cluster 1 versus a more protective profile of Cluster 2, which translated to a higher prevalence of metabolic disorders in Cluster 1.
Surprisingly, the history of cardiovascular events was significantly lower in the first cluster. In the literature, similar findings have been reported in the MESA cohort, where apoC-III2/apoC-III1 shows a positive association and apoC-III0b/apoC-III1 shows a negative association with the risk of PAD [21] and CVD [20], and several studies have linked the presence of sialic acid in plasma with greater inflammation and CVR [44–46]. In other studies, a lower incidence of MACE has been reported in people with a greater apoC-III2/apoC-III1 ratio in a triglyceride-dependent fashion [16]. However, given that all cardiovascular risk factors were decreased in Cluster 2, the observed higher prevalence of CVD in this cluster might be due to certain confounders, such as treatment (see below).
ApoC-III proteoforms and lipid-lowering and anti-inflammatory therapies
Lipid-lowering therapies and antidiabetic treatment can alter the proportion of apoC-III proteoforms towards a higher apoC-III2/apoC-III1 ratio [16, 47, 48], and we observed that people under either lipid-lowering or anti-inflammatory medications showed increased proportions of some of the protective proteoform ratios (Supplemental Figure S2 A-B). In our case, we cannot rule out the possibility that our finding for CVD prevalence was affected by treatment. A similar situation might occur regarding the prevalence of T2DM: Cluster 2 showed a lower prevalence of T2DM when we excluded treated diabetes from the analysis (Table 3), but we observed no significant differences when we also included people with treated diabetes (data not shown), who had increased ratios of protective proteoforms and were classified into Cluster 2 (Supplemental Figure S2 C-E).
Overall, the results of the present study provide information on proteoforms that are not usually studied (apoC-III1d, apoC-III2d, and apoC-III0f) and complete the existing knowledge on previously studied proteoforms by assessing their relationship with advanced biomarkers (the complete lipoprotein profile and 1H-NMR-measured Glyc concentrations). Owing to the study design, the associations observed throughout the study do not imply a causal role of proteoform ratios on the studied parameters. The fact that differences in proteoform ratios exist between treatment groups could suggest that, to some extent, the proteoform composition might be a consequence of the metabolic state. Conversely, other evidence from in vitro and mechanistic studies supports a direct effect of proteoform composition on some of the studied roles, as discussed above. To gain a deeper understanding of the physiological role and mechanisms of action of the different apoC-III proteoforms, further research in more controlled environments, such as in vitro systems or animal models, could complement the present findings. Another limitation of the study is the great variability of the study sample in the prevalence and duration of exposure to metabolic disorders and treatments, and despite robust control for covariates, other confounders that are not collected in our dataset may be important. From a clinical perspective, the study of the potential therapeutic implications of the observed results, i.e., different responses to triglyceride-lowering treatments depending on the proteoform distribution, would be of interest. Additionally, for future large cohort studies on apoC-III proteoforms or to consider any application in a clinical environment, a faster and simpler way to detect the most informative proteoforms would be highly useful.
Conclusions
In conclusion, our results support the importance of measuring not only apoC-III but also the relative proportions of its proteoforms, since similar levels of total apoC-III could be related to very different lipid profiles in people with different distributions of apoC-III proteoforms, not only in relation to lipoprotein metabolism but also inflammation and glucose metabolism.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The Di@bet.es project is a collaborative study with various phases and subprojects in which a large number of researchers and technicians have collaborated, to whom we are indebted. We are especially grateful to the Steering Committee of the study together with all the collaborators who have made it possible (https://www.sediabetes.org/cientifico-y%20asistencial/investigacion/proyectos-de-investigacion/estudio-dibet-es/ accessed on 21 September 2024). CIBERDEM Biorepository (IDIBAPS Biobank, Barcelona, Spain) supplied the samples. The mass spectrometry immunoassay was carried out at the Centre for Omic Sciences (Joint Unit Eurecat-Universitat Rovira i Virgili, Unique Scientific and Technical Infrastructure (ICTS)) facilities and with the guidance of its staff.
Abbreviations
- apo
Apolipoprotein
- BMI
Body mass index
- CVD
Cardiovascular disease
- Fuc
Fucose
- Gal
Galactose
- GalNAc
N-acetylgalactosamine
- 1H-NMR
Proton nuclear magnetic resonance
- HDL
High-density lipoprotein
- HDLc
HDL cholesterol
- HDLP
HDL particle concentration
- HL
Hepatic lipase
- HOMA-IR
Homeostatic model assessment of insulin resistance
- hsCRP
High-sensitivity C-reactive protein
- L-HDLP
Large HDL particle concentration
- L-LDLP
Large LDL particle concentration
- L-VLDLP
Large VLDL particle concentration
- LDL
Low-density lipoprotein
- LDLc
LDL cholesterol
- LDLP
LDL particle concentration
- Lp(a)
Lipoprotein (a)
- LPL
Lipoprotein lipase
- M-HDLP
Medium HDL particle concentration
- M-LDLP
Medium LDL particle concentration
- M-VLDLP
Medium VLDL particle concentration
- PC
Principal component
- PCA
Principal component analysis
- S-HDLP
Small HDL particle concentration
- S-LDLP
Small LDL particle concentration
- S-VLDLP
Small VLDL particle concentration
- T1DM
Type 1 diabetes
- T2DM
Type 2 diabetes
- TRL
Triglyceride-rich lipoprotein
- VLDL
Very low-density lipoprotein
- VLDLc
VLDL cholesterol
- VLDLP
VLDL particle concentration
Author contributions
MG, JR, JG, and PR were responsible for the conception and design of the work. GR, RR, YE, PR, HB, and GG-A acquired the data. PR, JG, EO, HB, GG-A, MG, and JR analyzed the data. PR, JG, MG, JR, GG-A, HB, AG-L, and NA interpreted the data. PR, JG, MG, and JR drafted the manuscript. JR, JG, MG, and GG-A revised the manuscript. All authors approved the final version of the manuscript.
Funding
This work has been funded by Instituto de Salud Carlos III (ISCIII) through the projects PI21/01294 and PI16/00507 and co-funded by the European Union, European Regional Development Fund (ERDF); CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM); and by the Cerca Programme, Generalitat de Catalunya. P. R. is a recipient of a predoctoral fellowship from the Spanish Ministerio de Universidades (FPU19/04610).
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The research was carried out in accordance with the Declaration of Helsinki (WHO 2011) of the World Medical Association. Written informed consent was obtained from all the participants. The study was approved by the Ethics and Clinical Investigation Committee of the Hospital Regional Universitario de Málaga (Malaga, Spain) in addition to other regional ethics and clinical investigation committees all over Spain.
Consent for publication
Not applicable.
Competing interests
N.A. is a stock owner of Biosfer Teslab and has a patent on the method for lipoprotein profiling described in the present manuscript.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pere Rehues and Josefa Girona have contributed equally to this work.
References
- 1.Borén J, Packard CJ, Taskinen MR. The roles of ApoC-III on the metabolism of Triglyceride-Rich Lipoproteins in humans. Front Endocrinol (Lausanne). 2020;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos H, Perlov D, Khoo C, Sacks FM. Distinct patterns of lipoproteins with apoB defined by presence of apoE or apoC-III in hypercholesterolemia and hypertriglyceridemia. J Lipid Res. 2001;42(8):1239–49. [PubMed] [Google Scholar]
- 3.Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371(1):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371(1):32–41. [DOI] [PubMed] [Google Scholar]
- 5.Sacks FM, Alaupovic P, Moye LA, Cole TG, Sussex B, Stampfer MJ, Apolipoproteins VLDL et al. B, CIII, and E, and Risk of recurrent coronary events in the cholesterol and recurrent events (CARE) Trial [Internet]. 102, Circulation. 2000. http://www.circulationaha.org [DOI] [PubMed]
- 6.Gordts PLSM, Nock R, Son NH, Ramms B, Lew I, Gonzales JC, et al. ApoC-III inhibits clearance of triglyceride-rich lipoproteins through LDL family receptors. J Clin Invest. 2016;126(8):2855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendivil CO, Zheng C, Furtado J, Lel J, Sacks FM. Metabolism of very-low-density lipoprotein and low-density lipoprotein containing apolipoprotein C-III and not other small apolipoproteins. Arterioscler Thromb Vasc Biol [Internet]. 2010 [cited 2024 Mar 21];30(2):239–45. https://pubmed.ncbi.nlm.nih.gov/19910636/ [DOI] [PMC free article] [PubMed]
- 8.Qin W, Sundaram M, Wang Y, Zhou H, Zhong S, Chang CC, et al. Missense mutation in APOC3 within the C-terminal lipid binding domain of human ApoC-III results in impaired assembly and secretion of triacylglycerol-rich very low density lipoproteins: evidence that ApoC-III plays a major role in the formation of lipid precursors within the microsomal lumen. J Biol Chem. 2011;286(31):27769–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Åvall K, Ali Y, Leibiger IB, Leibiger B, Moede T, Paschen M, et al. Apolipoprotein CIII links islet insulin resistance to β-cell failure in diabetes. Proc Natl Acad Sci USA. 2015;112(20):E2611–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juntti-Berggren L, Refai E, Appelskog I, Andersson M, Imreh G, Dekki N, et al. Apolipoprotein CIII promotes Ca2+-dependent beta cell death in type 1 diabetes. Proc Natl Acad Sci USA. 2004;101(27):10090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao Y, Xiong Y, Wang H, Chu S, Zhong R, Wang J et al. APOC3 induces endothelial dysfunction through TNF-α and JAM-1. Lipids Health Dis [Internet]. 2016;15(1):1–8. 10.1186/s12944-016-0326-0 [DOI] [PMC free article] [PubMed]
- 12.Zewinger S, Reiser J, Jankowski V, Alansary D, Hahm E, Triem S, et al. Apolipoprotein C3 induces inflammation and organ damage by alternative inflammasome activation. Nat Immunol. 2020;21(1):30–41. [DOI] [PubMed] [Google Scholar]
- 13.Hiukka A, Ståhlman M, Pettersson C, Levin M, Adiels M, Teneberg S, et al. ApoCIII-enriched LDL in type 2 diabetes displays altered lipid composition, increased susceptibility for sphingomyelinase, and increased binding to biglycan. Diabetes. 2009;58(9):2018–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaith P, Assmann G, Uhlenbruck G. Characterization of the oligosaccharide side chain of apolipoprotein C-III from human plasma very low density lipoproteins. BBA - Gen Subj. 1978;541(2):234–40. [DOI] [PubMed] [Google Scholar]
- 15.Yassine HN, Trenchevska O, Ramrakhiani A, Parekh A, Koska J, Walker RW, et al. The association of human apolipoprotein C-III sialylation proteoforms with plasma triglycerides. PLoS ONE. 2015;10(12):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koska J, Phd M, Yassine Md H, Phd OT, Phd SS, Schwenke Phd DC et al. Disialylated Apolipoprotein C-III proteoform is associated with improved lipids in prediabetes and Type 2 diabetes. J Lipid Res. 2016;57(October 2015). [DOI] [PMC free article] [PubMed]
- 17.Mendoza S, Trenchevska O, King SM, Nelson RW, Nedelkov D, Krauss RM et al. Changes in low-density lipoprotein size phenotypes associate with changes in apolipoprotein C-III glycoforms after dietary interventions. J Clin Lipidol [Internet]. 2017;11(1):224–233.e2. 10.1016/j.jacl.2016.12.009 [DOI] [PMC free article] [PubMed]
- 18.Kegulian NC, Ramms B, Horton S, Trenchevska O, Nedelkov D, Graham MJ, et al. ApoC-III glycoforms are differentially cleared by hepatic TRL (triglyceride-Rich lipoprotein) receptors. Arterioscler Thromb Vasc Biol. 2019;39(10):2145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kianičková K, Pakanová Z, Květoň F, Holazová A, Kundalia PH, Baráth P et al. O-glycoprofiling of serum apolipoprotein C-III in Colorectal Cancer. Front Bioscience - Landmark. 2024;29(1). [DOI] [PubMed]
- 20.Sinari S, Koska J, Hu Y, Furtado J, Jensen MK, Budoff MJ et al. ApoC-III Proteoforms, plasma lipids, and cardiovascular risk in MESA. Arterioscler Thromb Vasc Biol [Internet]. 2023; https://www.ahajournals.org/doi/10.1161/ATVBAHA.123.319035 [DOI] [PMC free article] [PubMed]
- 21.Koska J, Hansen S, Hu Y, Jensen MC, Billheimer D, Nedelkov D et al. Relationship of apolipoprotein C-III proteoform composition with ankle-brachial index and peripheral artery disease in the multi-ethnic study of atherosclerosis (MESA). Atherosclerosis. 2024;395. [DOI] [PMC free article] [PubMed]
- 22.Bondarenko PV, Cockrill SL, Watkins LK, Cruzado ID, Macfarlane RD. Mass spectral study of polymorphism of the apolipoproteins of very low density lipoprotein. J Lipid Res. 1999;40(3):543–55. [PubMed] [Google Scholar]
- 23.Nicolardi S, Burgt YEM, Van Der, Dragan I, Hensbergen PJ, Deelder AM. Identification of new apolipoprotein-CIII glycoforms with ultrahigh resolution MALDI-FTICR Mass Spectrometry of Human Sera. J Proteome Res. 2013. [DOI] [PubMed]
- 24.Soriguer F, Goday A, Bosch-Comas A, Bordiú E, Calle-Pascual A, Carmena R, et al. Prevalence of diabetes mellitus and impaired glucose regulation in Spain: the Di@bet.es Study. Diabetologia. 2012;55(1):88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröder H, Fitó M, Estruch R, Martínez-González MA, Corella D, Salas-Salvadó J, et al. A short screener is valid for assessing mediterranean diet adherence among older Spanish men and women. J Nutr. 2011;141(6):1140–5. [DOI] [PubMed] [Google Scholar]
- 26.Mallol R, Amigo N, Rodriguez MA, Heras M, Vinaixa M, Plana N et al. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J Lipid Res. 2015. [DOI] [PMC free article] [PubMed]
- 27.Fuertes-Martín R, Moncayo S, Insenser M, Martínez-García MÁ, Luque-Ramírez M, Grau NA, et al. Glycoprotein A and B height-to-width ratios as obesity-independent novel biomarkers of low-Grade chronic inflammation in women with polycystic ovary syndrome (PCOS). J Proteome Res. 2019;18(11):4038–45. [DOI] [PubMed] [Google Scholar]
- 28.Mallagaray A, Rudolph L, Lindloge M, Mölbitz J, Thomsen H, Schmelter F et al. Towards a precise NMR quantification of acute phase inflammation proteins from human serum. Angewandte Chemie - Int Ed. 2023;62(35). [DOI] [PubMed]
- 29.Amigó N, Fuertes-Martín R, Malo AI, Plana N, Ibarretxe D, Girona J et al. Glycoprotein Profile measured by a1 H-Nuclear magnetic resonance based on approach in patients with diabetes: a New Robust Method to assess inflammation. Life. 2021;11(12). [DOI] [PMC free article] [PubMed]
- 30.Haffner SM. Management of dyslipidemia in adults with diabetes [Internet]. http://diabetesjournals.org/care/article-pdf/21/1/160/585066/21-1-160.pdf [DOI] [PubMed]
- 31.Taskinen MR, Björnson E, Kahri J, Söderlund S, Matikainen N, Porthan K et al. Effects of evolocumab on the postprandial kinetics of apo (apolipoprotein) B100- and B48-Containing lipoproteins in subjects with type 2 diabetes. Arterioscler Thromb Vasc Biol. 2021;(February):962–75. [DOI] [PubMed]
- 32.Quesada JA, Bertomeu-González V, Orozco-Beltrán D, Cordero A, Gil-Guillén VF, López-Pineda A, et al. The benefits of measuring the size and number of lipoprotein particles for cardiovascular risk prediction: a systematic review and meta-analysis. Clin e Investigacion en Arterioscler. 2023;35(4):165–77. [DOI] [PubMed] [Google Scholar]
- 33.Reiner Ž. Hypertriglyceridaemia and risk of coronary artery disease. Nature Reviews Cardiology. Volume 14. Nature Publishing Group; 2017. pp. 401–11. [DOI] [PubMed]
- 34.Demus D, Naber A, Dotz V, Jansen BC, Bladergroen MR, Nouta J, et al. Large-scale analysis of Apolipoprotein CIII glycosylation by ultrahigh resolution mass spectrometry. Front Chem. 2021;9(May):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann CJ, Troussard AA, Yen FT, Hannouche N, Najib J, Fruchart JC, et al. Inhibitory effects of specific apolipoprotein C-III isoforms on the binding of triglyceride-rich lipoproteins to the lipolysis-stimulated receptor. J Biol Chem. 1997;272(50):31348–54. [DOI] [PubMed] [Google Scholar]
- 36.Mauger JF, Couture P, Bergeron N, Lamarche B. Apolipoprotein C-III isoforms: kinetics and relative implication in lipid metabolism. J Lipid Res. 2006;47(6):1212–8. [DOI] [PubMed] [Google Scholar]
- 37.Kontush A. HDL particle number and size as predictors of cardiovascular disease. Front Pharmacol. 2015;6(OCT). [DOI] [PMC free article] [PubMed]
- 38.Li R, Chen JX, Lu Q, Geng TT, Xia PF, Wang Y, et al. Associations of lipoprotein subclasses with risk of all-cause and cardiovascular disease mortality in individuals with type 2 diabetes: a prospective cohort study. Diabetes Obes Metab. 2023;25(11):3259–67. [DOI] [PubMed] [Google Scholar]
- 39.Rehues P, Girona J, Guardiola M, Plana N, Scicali R, Piro S et al. PCSK9 Inhibitors Have Apolipoprotein C-III-Related Anti-Inflammatory Activity, Assessed by 1H-NMR Glycoprotein Profile in Subjects at High or very High Cardiovascular Risk. Int J Mol Sci [Internet]. 2023 Feb 1 [cited 2024 Feb 24];24(3). https://pubmed.ncbi.nlm.nih.gov/36768645/ [DOI] [PMC free article] [PubMed]
- 40.Andreychuk N, Llop D, Moreno-Vedia J, Girona J, Ibarretxe D, Rodriguez-Borjabad C, et al. Glycoprotein serum concentrations assessed by 1H-NMR are increased in patients with high blood pressure. Hypertension. 2023;80(2):460–9. [DOI] [PubMed] [Google Scholar]
- 41.Moreno-Vedia J, Rosales R, Ozcariz E, Llop D, Lahuerta M, Benavent M et al. Triglyceride-Rich lipoproteins and glycoprotein A and B assessed by 1H-NMR in metabolic-associated fatty liver disease. Front Endocrinol (Lausanne). 2022;12. [DOI] [PMC free article] [PubMed]
- 42.Duprez DA, Otvos J, Sanchez OA, Mackey RH, Tracy R, Jacobs DR. Comparison of the predictive value of GlycA and other biomarkers of inflammation for total death, incident cardiovascular events, noncardiovascular and noncancer inflammatory-related events, and total cancer events. Clin Chem. 2016;62(7):1020–31. [DOI] [PubMed] [Google Scholar]
- 43.Akinkuolie AO, Pradhan AD, Buring JE, Ridker PM, Mora S. Novel protein glycan side-chain biomarker and risk of Incident Type 2 diabetes Mellitus. Arterioscler Thromb Vasc Biol. 2015;35(6):1544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawanishi K, Dhar C, Do R, Varki N, Gordts PLSM, Varki A. Human species-specific loss of CMP-N-acetylneuraminic acid hydroxylase enhances atherosclerosis via intrinsic and extrinsic mechanisms. Proc Natl Acad Sci USA. 2019;116(32):16036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reganon E, Vila V, Martínez-Sales V, Vayá A, Mira Y, Ferrando F et al. Sialic acid is an inflammation marker associated with a history of deep vein thrombosis. Thromb Res [Internet]. 2007 [cited 2023 Jul 14];119(1):73–8. https://pubmed.ncbi.nlm.nih.gov/16500696/ [DOI] [PubMed]
- 46.Zhang L, Wei TT, Li Y, Li J, Fan Y, Huang FQ, et al. Functional Metabolomics characterizes a key role for N-Acetylneuraminic acid in coronary artery diseases. Circulation. 2018;137(13):1374–90. [DOI] [PubMed] [Google Scholar]
- 47.Naber A, Demus D, Slieker RC, Nicolardi S, Beulens JWJ, Elders PJM, et al. Apolipoprotein-CIII O-Glycosylation is Associated with Micro- and macrovascular complications of type 2 diabetes. Int J Mol Sci. 2024;25(10):5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savinova OV, Fillaus K, Harris WS, Shearer GC. Effects of niacin and omega-3 fatty acids on the apolipoproteins in overweight patients with elevated triglycerides and reduced HDL cholesterol. Atherosclerosis. 2015;240(2):520–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.