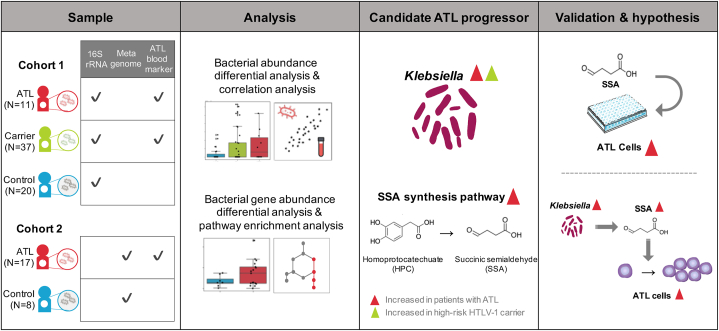
Abstract
Adult T-cell leukemia/lymphoma (ATLL) is a refractory blood cancer with severe immunodeficiency resulting from retroviral infection. ATLL develops in only 5 % of HTLV-1-infected individuals, but the entire mechanism of ATLL progression remains unknown. Since recent studies have reported that the gut microbiome influences the progression of various diseases, we hypothesized that ATLL is also related to the gut microbiome and aimed to investigate this relationship.
We analyzed the taxonomic and functional profiles of the gut microbiota of ATLL patients (n = 28) and HTLV-1-infected individuals (n = 37). We found that the succinic semialdehyde (SSA) synthesis pathway was significantly enriched in the gut microbiome of ATLL patients (P = 0.000682), and Klebsiella, whose abundance was significantly greater in ATLL patients and high-risk HTLV-1-infected individuals (P = 0.0326), was the main contributor to this pathway. Administration of SSAs to ATLL cell lines resulted in significant cell proliferation.
Herein, we propose that the gut microbiome can regulate ATLL progression via metabolites.
1. Introduction
Gut microbes produce various metabolites that influence gastrointestinal functions, including those influencing local and systemic immunity [1]. However, specific metabolites produced by the gut microbiota may have harmful effects [2] and the specific gut microbiota contributes to opportunistic infections [3]. Notably, a reduction in gut microbiota populations that release substances that activate gut barrier function and suppress immunity has been linked to leukemia development. For example, in acute myeloid leukemia (AML), the release of lipopolysaccharide (LPS) into the bloodstream, accompanied by a decrease in intestinal barrier function due to changes in the gut microbiome, may contribute to the deterioration of AML [4]. In addition, several other associations between hematologic tumors and the gut microbiome have been reported [[5], [6], [7]]. One specific type of leukemia is adult T-cell leukemia/lymphoma (ATLL), which is caused by the oncogenic retrovirus human T-cell leukemia virus type 1 (HTLV-1), and ATLL is an endemic virus that affects an estimated 5–10 million individuals worldwide [8]. HTLV-1 infects and transforms host T cells through the viral-encoded oncoprotein Tax over an extended period, followed by clonal expansion due to the presence of HTLV-1/HBZ and the accumulation of genomic and epigenomic abnormalities in HTLV-1-infected T cells, ultimately leading to leukemia-lymphoma. ATLL symptoms manifest as leukemic cell infiltration into organs and opportunistic infections, resulting in decreased survival rates [9]. While 95 % of HTLV-1-infected individuals remain asymptomatic throughout their lifetime and are called asymptomatic HTLV-1 carriers, approximately 5 % develop ATLL after decades of infection [10]. However, the specific factors contributing to ATLL initiation and progression have not been identified, suggesting the involvement of specific host factors.
Immunosuppressive states, such as those amenable to the development of opportunistic infections, may promote the development of ATLL [11]. In HTLV-1 carriers coinfected with Strongyloides stercoralis, which is known to cause opportunistic infection, type 2 helper T cells are suppressed [12,13], increasing the susceptibility of HTLV-1 carriers to ATLL. Several gut microbes are also responsible for opportunistic infections [14]. In addition, opportunistic infections such as Clostridium difficile infection have been observed in HIV-infected patients, a disease-causing virus analogous to HTLV-1 [15], suggesting that fluctuations in the gut microbiome may play a role in disease progression [16]. Moreover, since ATLL cells frequently infiltrate the gut mucosa [17], ATLL cells may interact with the gut microbiome, including metabolites. Thus, we hypothesized that the gut microbiome may play a role in the development and progression of ATLL.
To investigate the significance of the gut microbiome in the development and progression of ATLL, we characterized the gut microbiomes of asymptomatic HTLV-1 carriers and patients with ATLL. We identified significantly enriched metabolic pathways associated with ATLL progression in gut microbes through shotgun metagenomic data analysis. Additionally, we validated our findings using in vitro experiments. We revealed that metabolites that are likely enriched in the guts of patients with ATLL can promote ATLL cell proliferation. Based on these findings, we propose a mechanism involving metabolites derived from the gut microbiome in the progression of ATLL.
2. Results
2.1. Overview of gut microbial analysis in patients with ATLL and asymptomatic HTLV-1 carriers
In this study, we aimed to identify the gut microbiome and factors related to the development and progression of ATLL. We obtained fecal samples from two cohorts because fecal samples were collected from asymptomatic HTLV-1 carriers at home after the hospital visit using a preservation solution kit [18], while fresh fecal samples were collected from patients with ATLL at the same time as hospital admission. Therefore, one cohort included asymptomatic HTLV-1 carriers (n = 37) and patients with ATLL (n = 11), most of whom had less advanced disease-type, such as the smoldering type, and healthy controls (n = 20) (cohort 1, Table 1). In the asymptomatic HTLV-1 carrier group, there were eight asymptomatic HTLV-1 carriers with HTLV-1 proviral loads (PVLs) above 40 (per 1000 peripheral blood mononuclear cells (PBMCs)), and we considered that they have a high risk for developing ATLL (high-risk carriers) because it has been reported that asymptomatic HTLV-1 carriers with a PVL greater than 4 % have an increased risk of developing ATLL in a previous study [19]. The other cohort included patients with advanced-stage ATLL (n = 17), such as acute type, and healthy controls (n = 8) (cohort 2, Table 2). Most patients with ATLL in this study had a history of antibiotic use within one week of fecal collection to treat opportunistic infections because a symptom of ATLL is immunodeficiency.
Table 1.
Characteristics of the subjects in cohort 1.
| ATLL | Asymptomatic HTLV-1 Carrier | Healthy control | ||
|---|---|---|---|---|
| Number of subjects | 11 | 37 | 20 | |
| Disease type | Acute | 2 | – | |
| Lymphoma | 0 | – | – | |
| Chronic | 1 | – | – | |
| Smoldering | 8 | – | – | |
| High-risk carrier | – | 8 | – | |
| Low-risk carrier | – | 29 | – | |
| Age | 67.4 ± 5.1 | 60.6 ± 10.4 | 58.6 ± 3.0 | |
| Sex (F/M) | 4:7 | 28:9 | 0:20 | |
| PVL (/1000PBMC) | 243.0 ± 274.6 | 60.6 ± 91.3 | – | |
| sIL-2Ra(U/mL) | 2564.8 ± 4357.5 | 450.1 ± 146.3 | – | |
| Chemotherapy prior to blood sampling | 1 | 1 | – | |
| Chemotherapy prior to stool sampling | 1 | 1 | 0 | |
| Antibiotics prior to blood sampling | 2 | 4 | – | |
| Antibiotics prior to stool sampling | 2 | 4 | 4 | |
Table 2.
Characteristics of the subjects.
| ATLL | Asymptomatic HTLV-1 Carrier | Healthy control | ||
|---|---|---|---|---|
| Number of subjects | 17 | 0 | 8 | |
| Disease type | Acute | 12 | – | – |
| Lymphoma | 5 | – | – | |
| Chronic | 0 | – | – | |
| Smoldering | 0 | – | – | |
| Age | 68.5 ± 6.5 | – | 43.2 ± 9.4 | |
| Sex (F/M) | 8:9 | – | 4:4 | |
| PVL (/1000PBMC) | 478.9 ± 499.1 | – | – | |
| sIL-2Ra(U/mL) | 16284.7 ± 13830.5 | – | – | |
| Chemotherapy prior to blood sampling | 14 | – | 0 | |
| Chemotherapy prior to stool sampling | 14 | – | 0 | |
| Antibiotics prior to blood sampling | 16 | – | 0 | |
| Antibiotics prior to stool sampling | 16 | – | 0 | |
We analyzed the gut microbiomes of ATLL patients and asymptomatic HTLV-1 carriers in cohort 1. The results showed that the abundance of Klebsiella was increased in patients with ATLL and high-risk HTLV-1 carriers. In addition, metagenomic data from cohort 2 suggested that the homoprotocatechuate (HPC) degradation pathway was significantly enriched in the gut microbiome of ATLL patients. The end product of this degradation pathway is succinic semialdehyde (SSA). Thus, we tested the efficacy of ATLL cells by adding them to ATLL cell lines derived from patients with ATLL. Notably, in lymphocytes, specific concentrations of SSA promoted cell proliferation.
In conclusion, this study identified a candidate ATLL progressor in the gut via in silico analysis, and we confirmed its function via in vitro experimental validation (Fig. 1).
Fig. 1.
Overview of this study.
2.2. The community structure of the gut microbiota in patients with ATLL and asymptomatic HTLV-1 carriers
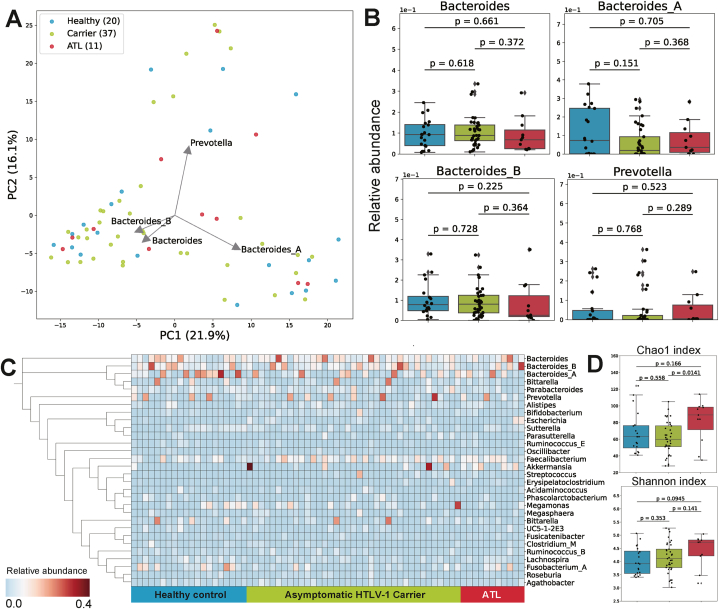
We examined the gut microbiome of patients with ATLL and asymptomatic HTLV-1 carriers in both groups. Using 16S rRNA gene amplicon sequence data from cohort 1, we first conducted a principal component analysis (PCA) to compare the genus-level profiles. There were three ATLL subtypes in cohort 1: acute (n = 2), chronic (n = 1), and smoldering (n = 8). Several criteria define these subtypes and reflect ATLL progression and the risk of ATLL development [20].
The PCA results showed variability in the composition of the gut microbiota among the subjects, and the top four principal components were related to Bacteroides and Prevotella (Fig. 2A). These bacteria are known to contribute to enterotype [21]. The median relative abundance of the four bacterial genera characterizing each cluster was approximately 10 % (Fig. 2B). Fig. 2C shows the 30 genera with the highest average abundance. The Chao1 index, which represents the taxonomic alpha diversity, was significantly greater in patients with ATLL than in asymptomatic carriers (P = 0.0141) (Fig. 2D). The results obtained from cohort 2, which consisted of only patients with ATLL and healthy controls and no asymptomatic HTLV-1 carriers (Table 2), are shown in Supplementary Fig. S3.
Fig. 2.
Comparison of the gut microbial community structure between patients with ATLL and asymptomatic HTLV-1 carriers.
Blue, healthy controls; green, asymptomatic HTLV-1 carriers; red, patients with ATLL.
(A) PCA plot of the subjects' gut microbiota taxonomic composition.
(B) Variation in the relative abundance of the four dominant genera in the subjects' guts. P values were calculated by the two-tailed Brunner–Munzel test.
(C) Relative abundance of the top 30 genera in patients with ATLL, asymptomatic HTLV-1 carriers, and healthy controls. A phylogenetic tree was constructed from 16S rRNA gene sequences.
(D) Alpha diversity of the subjects' gut microbiota. P values were calculated by the two-tailed Mann‒Whitney U test. The Shannon index did not significantly differ among all the groups.
Furthermore, to investigate which factors contribute to characterizing the gut microbiome of subjects, we performed a permutational multivariate analysis of variance (PERMANOVA) with Bray–Curtis distance (Supplementary Table 1). There were no significant differences between patients treated with antibiotics or chemotherapy and nonusers in cohort 1 (P > 0.05), although the gut microbial composition significantly differed among patients with different ATLL subtypes.
2.3. The specific gut microbial abundances differed between patients with ATLL and asymptomatic HTLV-1 carriers
To investigate individual bacterial abundance changes, we compared the abundances of each gut microbial genus in cohort 1 using 16S rRNA gene amplicon sequences. There were eight high-risk HTLV-1 carriers and 29 low-risk HTLV-1 carriers in the HTLV-1 asymptomatic carrier group in cohort 1.
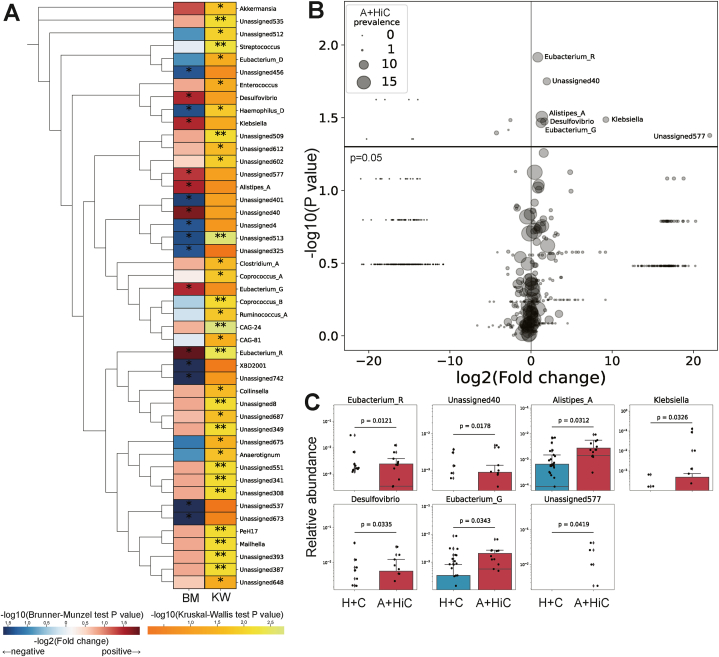
Using the Kruskal‒Wallis test, we identified 32 bacterial genera with differential abundance among the four groups (P < 0.05) (Fig. 3A, right column, “KW”). Further pairwise comparisons with the Brunner‒Munzel test were also conducted between the two groups: patients with ATLL and high-risk HTLV-1 carriers (A + HiC) and healthy controls and low-risk HTLV-1 carriers (H + C). This comparison revealed 17 bacterial genera that exhibited significant differences in abundance between the two groups (P < 0.05) (Fig. 3A, left column, “BM”). Specifically, seven genera were significantly more abundant in the A + HiC group (Fig. 3A, left column, Fig. 3B), namely, Eubacterium_R (P = 0.0121); an unassigned bacterial genus from the Ruminococcaceae family labeled “Unassigned 40” (P = 0.0178); Alistipes_A (P = 0.0312); Klebsiella (P = 0.0326); Desulfovibrio (P = 0.0335); Eubacterium_G (P = 0.0343); and an unassigned bacterial genus from the Acidaminococcaceae family labeled “Unassigned 577” (P = 0.0419) (Fig. 3C). The results of the Kruskal‒Wallis test for these seven bacteria are shown in Supplementary Fig. S4A. In addition, the distributions of these seven bacteria in cohort 2 are shown in Supplementary Fig. S4B.
Fig. 3.
Differential analysis of the abundance of each genus in the gut microbiota.
A + HiC, a combined group of patients with ATLL and high-risk HTLV-1 carriers; H + C, a combined group of healthy controls and asymptomatic HTLV-1 carriers.
(A) The distribution of P values for 46 bacterial genera with significant differences in abundance according to the two-tailed Brunner–Munzel test (left column, “BM”) and Kruskal‒Wallis test (right column, “KW”). Bacteria with significantly increased abundance compared to H + C are represented as red, and those with decreased abundance are represented as blue on the left side of the heatmap. The color intensity reflects the magnitude of the -log10 (P value). ∗, P < 0.05; ∗∗, P < 0.01. A phylogenetic tree was constructed from the V3-V4 region of the 16S rRNA gene sequences.
(B) Comparison of the abundance of each bacterial genus between the A + HiC and H + C groups. The X-axis represents the ratio of the mean relative abundance of each group (fold change) transformed by log2; the Y-axis represents the P value transformed by -log10. The horizontal line represents P = 0.05. P values were calculated by the two-tailed Brunner–Munzel test. The plot size represents the prevalence of each bacterium (number of subjects) in the A + HiC group.
(C) The relative abundance of seven genera was significantly greater in the A + HiC group (red) than in H + C group (blue). P values were calculated by the two-tailed Brunner–Munzel test.
As a definitive result, we observe clear differences in the relative abundance of specific bacterial genera among ATLL patients, high-risk and low-risk HTLV-1 carriers, and healthy controls.
2.4. The abundances of Klebsiella and Eubacterium_G were positively correlated with the ATLL-specific tumor markers
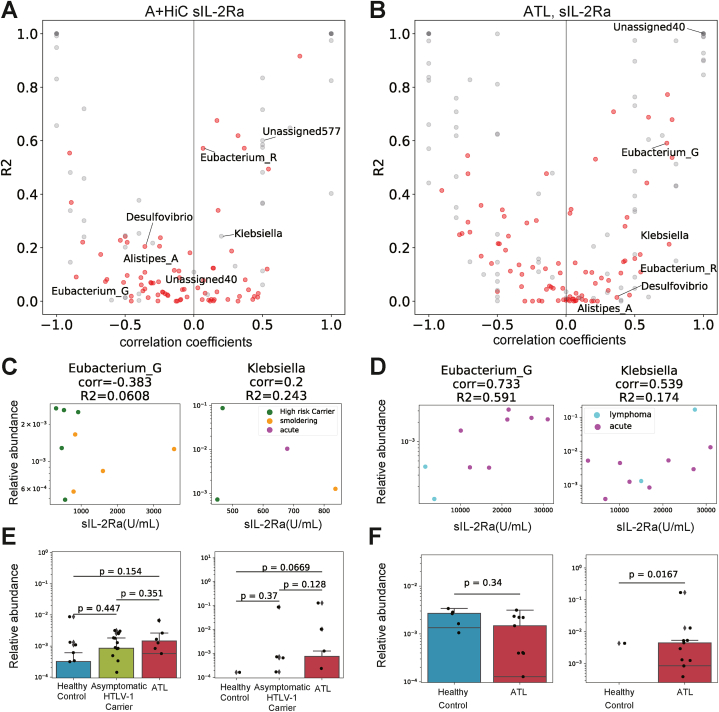
To understand the relationships between the seven significantly increased gut microbial genera and the development and progression of ATLL, we investigated the correlation between the abundance of the increased gut microbial genera and soluble interleukin 2 receptor-alpha (sIL-2Ra) protein concentration, an ATLL biomarker that reflects ATLL progression, in the peripheral blood [22]. We analyzed gut microbial taxonomic profiles determined via 16S rRNA gene amplified sequences in both cohorts.
For high-risk HTLV-1 carriers and patients with ATLL in cohort 1, we did not find a positive correlation between sIL-2Ra levels and the two microbial abundances (Eubacterium_G: ρ = −0.383, Klebsiella: ρ = 0.2) (Fig. 4A–C). However, there were positive correlations between sIL-2Ra levels and these two microbial abundances in patients with ATLL in cohort 2 (ρ = 0.733, 0.539; Fig. 4B–D). The abundance of Klebsiella was significantly greater in the patients with ATLL than in the controls in both cohorts (P = 0.0326, 0.0167; Fig. 3C; Fig. 4F). In comparison, the abundance of Eubacterium_G was not significantly greater in cohort 2 (P = 0.34; Fig. 4F). The correlation analysis results for the other five candidate microbiota and sIL-2Ra levels are presented in Supplementary Fig. S5.
Fig. 4.
Correlations between tumor markers and the relative abundance of each bacteria.
(A), (B) Distribution of Spearman correlation coefficients between sIL-2Ra levels and the relative abundance of each individual bacteria (X-axis) and the R2 score (Y-axis) for patients with ATLL and high-risk HTLV-1 carriers in cohort 1 (A) and cohort 2 (B). The bacterial names in the figure represent the bacterial genera whose abundance increased with the progression of ATLL identified in the previous section. Red plots represent bacteria that were found in five or more subjects; subjects had measurable bacterial abundance values and sIL-2Ra levels. A relative abundance of 0 was omitted before the correlation coefficient was calculation.
(C), (D) The distribution of sIL-2Ra levels (X-axis) and relative abundance (Y-axis) of Eubacterium_G (left) and Klebsiella (right). The relative abundance values were converted by log10. Corr, Spearman's correlation coefficient: R2, coefficient of determination. The plot color represents ATLL subtypes: green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type. (C) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (D) patients with ATLL in cohort 2.
(E), (F) The relative abundance of Eubacterium_G (left) and Klebsiella (right). The relative abundance values were converted by log10. P values were calculated by the two-tailed Brunner–Munzel test. (E) Cohort 1 and (F) cohort 2.
Furthermore, specific gut microbial abundances, such as those of Coprococcus_B and Ruminococcus_E, exhibited strong correlations with HTLV-1 proviral load values and sIL-2Ra levels (Supplementary Figs. S6 and S7).
Thus, several gut microbial abundances were correlated with ATLL biomarkers, and the abundances of Eubacterium_G and Klebsiella could be positively related to the progression of ATLL.
2.5. The succinic semialdehyde synthesis pathway is enriched in the gut microbiome of patients with ATLL, promoting the proliferation of ATLL cells
To explore the functional composition of the gut microbiota in patients with ATLL and the functions of the identified ATLL-associated candidate microbiota, we analyzed functional ortholog gene profiles from whole-genome shotgun sequencing data of fecal samples from patients with ATLL in cohort 2.
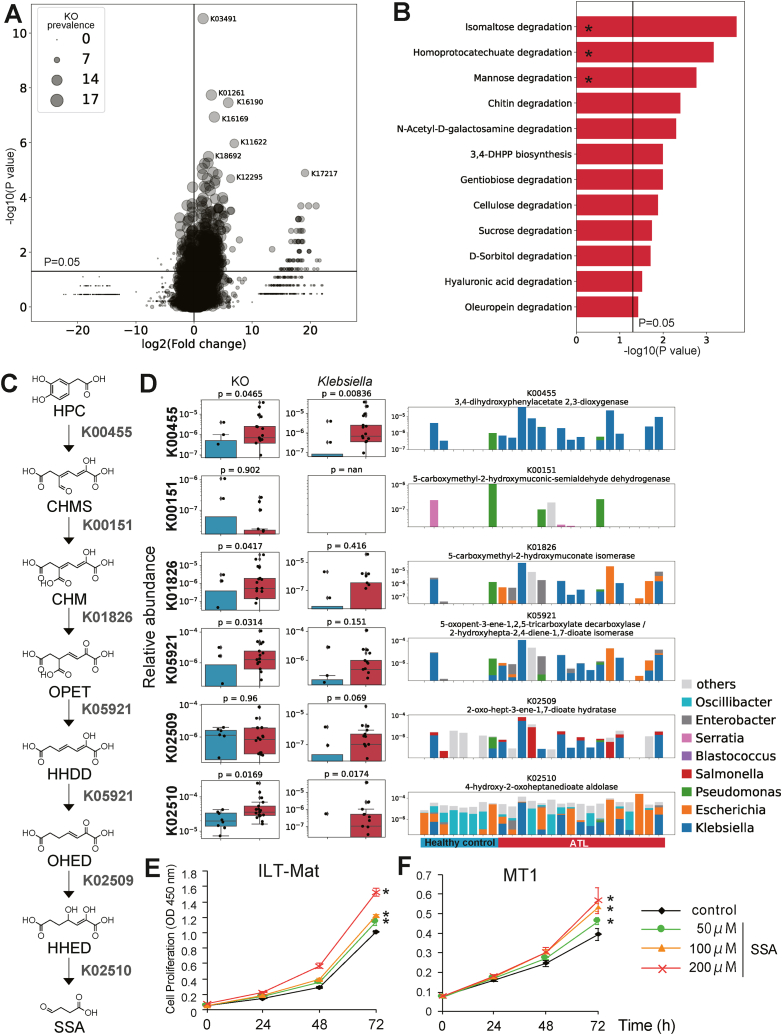
Through differential analysis of functional gene (KO) profiles, we identified 788 KOs with significantly different abundances between patients with ATLL and healthy controls (P = 0.05). Among these KOs, 463 were significantly more abundant in the ATLL patient group than in the control group. These KOs were present in more than 14 individuals in the ATLL patient group (representing approximately 80 % of the patients with ATLL) (P = 0.05; Fig. 5A). To determine which pathways are involved in these KOs, enrichment analysis was performed using Enteropathway [23], a map specific to metabolic pathways and modules in the human gut microbiome. Enrichment analysis revealed 12 upregulated metabolic modules in patients with ATLL (P < 0.05), and three modules had Q values less than 0.05 after FDR correction (Fig. 5B, Supplementary Fig. S8). We also conducted enrichment analysis for the 154 KOs, the abundances of which were significantly lower in the gut microbiome of patients with ATLL than in that of controls (Supplementary Fig. S9).
Fig. 5.
Gut microbial functional modules with altered abundance in the ATLL patient group.
(A) Comparison of KO abundances between patients with ATLL and healthy controls. X-axis, fold change (log2-transformed) in the mean relative abundance of each bacterium in each group; Y-axis, -log10-transformed P values from pairwise comparisons. The horizontal line represents P = 0.05. P values were calculated by the two-tailed Brunner–Munzel test. The plot size shows the KO prevalence (number of subjects) in the ATLL patient group.
(B) Enrichment analysis of KOs that were significantly more abundant in patients with ATLL. P values are transformed by -log10. The vertical line represents P = 0.05. ∗, q < 0.05.
(C) SSA synthesis module. Abbreviations: HPC, homoprotocatechuate; CHMS, 5-carboxymethyl-2-hydroxymuconic-semialdehyde; CHM, 5-carboxymethyl-2-hydroxymuconate; OPET, 5-oxo-pent-3-ene-1,2,5-tricarboxylate; HHDD, 2-hydroxyhepta-2,4-diene-1,7-dioate; OHED, 2-oxo-hept-3-ene-1,7-dioate; HHED, 2,4-dihydroxy-hept-2-ene-1,7-dioate; SSA, succinic semialdehyde.
(D) Association of KO abundance and genus Klebsiella with the SSA synthesis module. The box plot represents the relative abundance of the KOs (left) and ortholog genes assigned to the KO from Klebsiella (right). The bar plot represents the bacterial composition of the KOs of each subject (right). The top two genera with the highest average relative abundance in at least one KO are displayed.
(E) (F) Cell growth rate. (E) IL-2-dependent ILT-Mat cells and (F) IL-2-independent MT-1 cells treated with SSA. The values represent the means ± s.d. ∗P < 0.05; unpaired two-tailed Student's t-test.
Of the three significantly enriched modules, we focused on the homoprotocatechuate (HPC) degradation module (Fig. 5B and C) because succinic semialdehyde (SSA), the end product of the module, is considered to lead to oxidative stress, and oxidative stress is known to be involved in the development of tumors [24,25].
Klebsiella and Escherichia were the main genera associated with the module (Fig. 5D, right bar graph), which is consistent with findings from previous reports [[26], [27], [28]]. The gut microbiota associated with the other two significantly enriched modules are shown in Supplementary Figs. S10 and S11.
Finally, to demonstrate the impact of SSA on ATLL cell lines, we treated three types of leukemia cell lines (MT-1, ILT-Mat, and ATN cells) derived from patients with ATLL with various concentrations of SSA for three days (Fig. 5E and F; Supplementary Fig. S12). We measured cell proliferation based on the absorbance values. We observed that, compared with the control, 200 μM SSA significantly promoted the proliferation of ATLL cell lines, which was not dependent on IL-2. Thus, these findings suggest that SSA promotes the proliferation of the ATLL cells and that Klebsiella may contribute to this effect.
3. Discussion
In this study, we investigated the variations in the gut microbiome of patients with ATLL and asymptomatic HTLV-1 carriers using in vitro and in silico analyses. Our findings suggest a potential role for the gut microbiome and metabolites in ATLL progression. This study is one of the few to explore the link between hematological tumors and the gut microbiome, shedding light on the mechanism of tumor progression.
We observed an increased abundance of Klebsiella and other genera in high-risk HTLV-1 carriers and patients with ATLL (Fig. 3C), which was positively correlated with tumor marker levels of patients with ATLL (Fig. 4D). Notably, our metagenomic data revealed enrichment of SSA synthesis pathway in the gut microbiota, particularly Klebsiella was a main contributor of it in the guts of patients with ATLL (Fig. 5B–D). We confirmed that SSA promoted the proliferation of ATLL cells (Fig. 5E and F). Overall, our study provides insights into the potential involvement of the gut microbiota in ATLL progression.
Antibiotics increase Klebsiella abundance [3] and cause dysbiosis of the gut microbiota [29]. Indeed, most patients with ATLL in this study had a history of antibiotic use within one week of fecal collection. It is possible to hypothesize that antibiotics might induce ATLL by increasing Klebsiella abundance and worsen ATLL by disrupting the gut microbiota. However, this remains a hypothesis, and further analyses, such as tracking the gut microbiota of ATLL patients before and after antibiotic treatment over time, are necessary. In addition, a species in the genus Klebsiella is also known to cause opportunistic infections [3]. Previous reports have suggested that S. stercoralis antigens may promote ATLL via IL-2 activation [30,31]. Therefore, the increasing abundance of Klebsiella may alter the host immune system in the gut, affecting ATLL development.
SSA is known to be a reactive carbonyl and may induce oxidative stress in cells. Oxidative stress can cause tumor expansion [25]. A previous study reported that HTLV-1 Tax-expressing cells showed antiapoptotic property under oxidative stress [32]. These findings suggest that oxidative stress caused by SSA may lead to ATLL progression via ATLL cell proliferation. In other words, it is possible that SSA is a potential driver of ATLL. However, another study showed that Tax itself stimulates reactive oxidative stress (ROS) production and induces the apoptosis of T cells [33]. In addition, Tax expression is temporary, and more than half of HTLV-1-infected cells do not express Tax [32]. Thus, the mechanism of ATLL cell proliferation by oxidative stress and SSA needs to be determined.
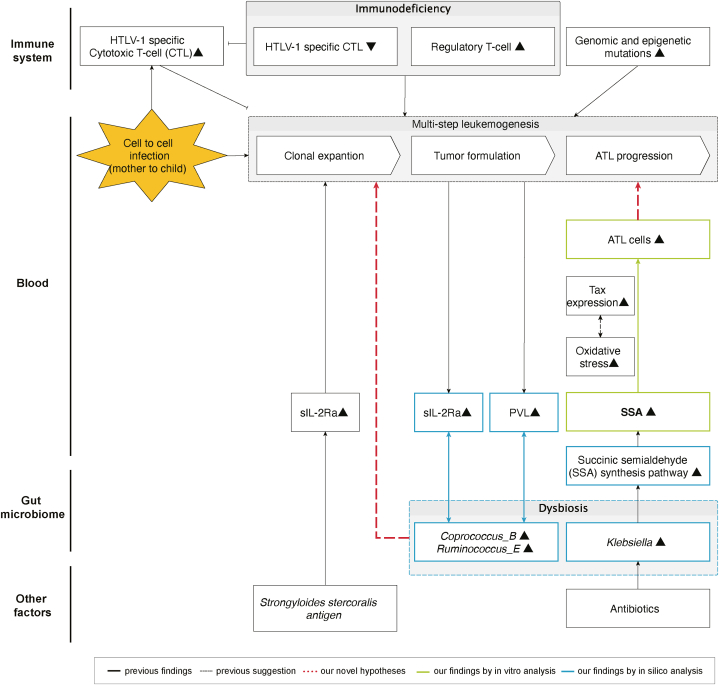
Based on our findings and those of previous studies, we propose a hypothesis for ATLL development and progression. An increase in the abundance of gut Klebsiella leads to enhanced homoprotocatechuate degradation, resulting in elevated SSA concentrations and promoting ATLL cell expansion (Fig. 6).
Fig. 6.
Hypotheses and previous findings on the mechanism of development and progression of ATLL.
We summarized our hypotheses and previous findings related to the development and progression of ATLL.
Several limitations exist in our study. First, the SSA concentration in the intestinal tract or peripheral blood due to increased homoprotocatechuate degradation by Klebsiella still needs to be better understood. Measurement of the SSA concentration in patients with ATLL and SPF ATLL model animals colonized by Klebsiella is needed. Second, several subjects had a history of illness or treatment (antibiotics or chemotherapy) before fecal sampling, potentially influencing the study outcome. Analysis of untreated, sex- and age-unbiased samples is needed, but no associations between increased Klebsiella abundance and antibiotic/chemotherapy history or age- or sex-related bias were found in this study (data not shown). Taken together, further experiments and analyses that address these limitations will provide additional evidence about the association between the gut microbiome and ATLL progression.
4. STAR methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human gut microbial DNA from fecal samples | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 medium | FUJIFILM Wako Pure Chemical Corporation | 18902025 |
| fetal bovine serum | Cosmobio | 3031 CCP-FBS-BR-500 |
| Penicillin- streptomycin | FUJIFILM Wako Pure Chemical Corporation | 168-23191 |
| nonessential amino acids | FUJIFILM Wako Pure Chemical Corporation | 13915651 |
| interleukin-2 | BioLegend | 589104 |
| succinic semialdehyde | Santa Cruz Biotechnology | sc-281158 |
| Critical commercial assays | ||
| WST-8 method with a Cell Counting Kit | Dojindo | 341-07624 |
| preservation solution kit of fecal sample | Technosuruga | |
| PowerFecal DNA Isolation Kit | QIAGEN | 12830-50 |
| QIAmp DNA Stool | QIAGEN | 51504 |
| Deposited data | ||
| Raw and analyzed gut bacterial DNA sequence data | This paper | DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA), DRA016992. |
| Experimental models: Cell lines | ||
| MT-1 | JCRB CELL BANK | JCRB |
| ILT-Mat | RIKEN BRC CELL BANK | RCB0475 |
| ANT | RIKEN BRC CELL BANK | RCB1440 |
| Oligonucleotides | ||
| 16S rRNA-Forward Primer 341F: 5′- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNCCTACGGGNGGCWGCAG-3′ |
Oral Microbiome Center, Taniguchi Dental Clinic | N/A |
| 16S rRNA-Reverse Primer −806R: 5’ – GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNGACTACHVGGGTATCTAATCC-3′ |
Oral Microbiome Center, Taniguchi Dental Clinic | N/A |
| Software and algorithms | ||
| Fastp | Chen et al. [34] | https://github.com/OpenGene/fastp |
| VSEARCH | Rognes et al. [35] | https://github.com/torognes/vsearch |
| SeqKit | Shen et al. [36] | https://github.com/shenwei356/seqkit/releases |
| Deblur | Amir et al. [37] | https://github.com/biocore/deblur |
| SortMeRNA | Kopylova et al. [38] | https://github.com/sortmerna/sortmerna |
| BLASTn | Camacho et al. [39] | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Cutadapt | Marcel [40] | https://github.com/marcelm/cutadapt |
| Bowtie2 | Langmead et al. [41] | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MEGAHIT | Li et al. [42] | https://github.com/voutcn/megahit |
| Prodigal | Hyatt et al. [43] | https://github.com/hyattpd/Prodigal |
| MMseqs2 | Steinegger et al. [44] | https://github.com/soedinglab/MMseqs2 |
| Diamond | Buchfink et al. [45] | https://github.com/bbuchfink/diamond |
| MAFFT | Katoh et al. [46] | https://mafft.cbrc.jp/alignment/software/ |
| RAxML | Stamatakis et al. [47] | https://github.com/amkozlov/raxml-ng |
| iTOL | Letunic et al. [48] | https://itol.embl.de/ |
| Enteropathway | Shiroma et al. [23] | https://enteropathway.org/#/ |
| Other | ||
| GTDB | Donovan et al. [49] | https://gtdb.ecogenomic.org/ |
| MGnify | Mitchell et al. [50] | https://github.com/EBI-Metagenomics |
| Python | Python Software Foundation | https://www.python.org/downloads/release/python-376/ |
5. Resource availability
This study did not generate new unique reagents.
6. Data availability statement
6.1. Data
The raw sequencing data reported in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA), Tokyo, Japan, under accession number DRA016992.
6.2. Code
This paper does not report the original code.
6.3. Experimental model and study participant details
6.3.1. In vitro experiments
ATLL patient-derived leukemia cell lines (MT-1, ATN-1, and ILT-Mat) were procured from the JCRB or RIKEN cell bank in 2022. The cell lines were authenticated utilizing STR-PCR profiling and tested negative for mycoplasma. MT-1 cells were cultured in RPMI-1640 medium (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10 % fetal bovine serum (Cosmobio, Tokyo, Japan), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) (complete RPMI-1640 medium). ATN-1 and ILT-Mat cells were cultured in complete RPMI-1640 medium supplemented with 1 × nonessential amino acids (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) and interleukin-2 (BioLegend, San Diego, USA), respectively. The cells were maintained at 37 °C in a humidified atmosphere with 5 % CO2.
Cells were seeded into 96-well plates at densities ranging from 50 × 103 to 200 × 103 cells in 100 μL of complete cell culture medium, and succinic semialdehyde (Santa Cruz Biotechnology, Texas, USA) was added to the medium for 72 h at 37 °C with 5 % CO2. Cell proliferation was assessed every 24 h using the WST-8 method with a Cell Counting Kit (Dojindo, Kumamoto, Japan) following the manufacturer's instructions. The experiments were performed in triplicate and repeated at least three times.
6.3.2. Study participants’ sample collection
The Miyazaki University Faculty of Medicine Ethics Committee and participating institution approved the study (approved number 2020-565). Samples and clinical information were obtained with informed consent. Patients with ATLL and asymptomatic HTLV-1 carriers aged over 20 years old with confirmed HTLV-1 infection were recruited from Miyazaki University Hospital and Imamura General Hospital in Japan. Healthy controls aged over 20 years old with no family history of ATLL were recruited. We obtained fecal samples from two cohorts because fecal samples were collected from asymptomatic HTLV-1 carriers at home after the hospital visit using a preservation solution kit [18], while fresh fecal samples were collected from patients with ATLL at the same time as hospital admission. Therefore, one cohort included asymptomatic HTLV-1 carriers (n = 37) and patients with less advanced types of ATLL (n = 11), such as the smoldering type and healthy controls (n = 20) (cohort 1, Table 1). In the asymptomatic HTLV-1 carrier group, there were eight asymptomatic HTLV-1 carriers with HTLV-1 proviral loads (PVLs) above 40 (per 1000 peripheral blood mononuclear cells (PBMCs)), and we defined them as high-risk carriers [19]. The other cohort included patients with advanced-stage ATLL, such as acute type (n = 17), and healthy controls (n = 8) (cohort 2, Table 2). In cohort 1, patients with ATLL were significantly older (Mann‒Whitney U test, two-tailed, P = 0.00164 vs. asymptomatic HTLV-1 carriers, P = 1.39 × 10−9 vs. healthy controls). There were more women in the asymptomatic HTLV-1 carrier group than in the control group, whereas the healthy controls were all men. In cohort 2, patients with ATLL were significantly older than healthy controls (P = 3.89 × 10−19).
The soluble interleukin 2 receptor-alpha (sIL-2Ra) levels, PVL levels, and clinical data were collected from the medical records (Table 1). sIL-2Ra is a blood biomarker that reflects tumor size and disease progression in patients with ATLL. sIL-2Ra also reflects lymphoma and other immune disorders, but none of the study subjects had these diseases. The samples and clinical information used in this study were obtained after providing informed consent and with the approval of the institutional review boards of each participating institute of the University of Miyazaki.
7. Method details
7.1. 16S rRNA gene sequencing
To obtain 16S rRNA gene sequencing data from cohort 1, DNA was extracted from fecal samples with preserved guanidine solution using the PowerFecal DNA Isolation Kit (QIAGEN, Hilden, Germany). To obtain 16S rRNA gene sequencing data from cohort 2, DNA was extracted from fresh fecal samples using the QIAmp DNA Stool (QIAGEN, Hilden, Germany). After extraction, the libraries were prepared following the official Illumina protocol for 16S Metagenomics Sequencing Library Preparation.
The V3-V4 region of the 16S rRNA gene was amplified using 341F-806R primers and KAPA HiFi HotStar (Nippon Genetics, Tokyo, Japan). The resulting DNA was purified with AMpureXP (Nippon Genetics, Tokyo, Japan) and subjected to two washes with 80 % ethanol, followed by elution with 10 mM Tris buffer (pH 8.5). The resulting library was denatured with 0.2 N NaOH after adjusting to 4 nM and HT1 buffer, and paired-end sequencing was performed using MiSeq with the addition of 20 % 10 pM PhiX DNA (Oral Microbiome Center, Taniguchi Dental Clinic, Kagawa, Japan). On average, 197,008 reads were obtained per sample.
7.2. Shotgun metagenomic sequencing
For whole-genome shotgun sequencing data acquisition, DNA was extracted from fecal samples stored at 4 °C using a QIAamp DNA Stool (QIAGEN, Hilden, Germany) within 24 h. One stool sample from a patient with ATLL was stored at 4 °C for five days before DNA extraction. PCR was performed according to the official Illumina protocol for the Nextera DNA Flex Library preparation kit five times after DNA fragmentation. Subsequently, 12 pM PhiX DNA was added at 5 %, and sequencing was subsequently conducted on a NovaSeq instrument. On average, 143,084,977 reads were obtained per sample (Oral Microbiome Center, Taniguchi Dental Clinic, Kagawa, Japan).
7.3. Taxonomic profiling
To estimate the phylogenetic composition, we utilized the reads obtained from 16S rRNA gene sequence data and processed them through our in-house pipeline (Supplementary Fig. S1). Fastp (version 0.19.6) [34] was used for quality filtering, which involved removing adaptor sequences, poly G sequences longer than 10 base pairs, reads shorter than 200 base pairs, reads containing one or more undetermined nucleotides (N) from the sequencer, and reads with an average quality score lower than 15.
Next, VSEARCH (version 2.7.0_linux_x86_64) [35] was used to combine forward and reverse reads, resulting in an average of 136,908.7 high-quality reads per sample. The length distribution of these reads was examined with SeqKit (version 0.5.5)36 and the reads were trimmed to 400 base pairs using the deblur trim command in Deblur (version 1.1.0-dev) [37]. Duplicate reads were counted, and singletons were removed with the deblur dereplicate command. PhiX-derived reads were eliminated with the deblur remove-artifacts command using a file of PhiX sequences registered in Deblur and converted with SortMeRNA (version 2.0)38. Multiple sequence alignments were conducted with the deblur multiple-seq-alignment command to detect sequencing errors, followed by denoising with the deblur deblur-seqs command. Finally, the uchime_denovo command with VSEARCH was used to remove chimeric sequences generated during read merging. After all these steps, a total of 6,838 reads (ASVs) were retrieved.
Phylogenetic assignments for these reads were obtained from the GTDB (https://data.gtdb.ecogenomic.org/releases/release86/86.1/bac120_ssu_r86.1.fna) [49]. BLASTn (version 2.8.1) 39was utilized for homology searches. Genus names were assigned from identity ≥ 94 % match to registered sequences, while species names were assigned with an identity ≥ 99 % match to reference sequences. Ultimately, 6,093 reads were assigned a phylogenetic name above the genus level (Supplementary Fig. S1).
7.4. Functional profiling
To estimate the functional composition, we used the reads obtained from whole-genome shotgun sequencing data (Supplementary Fig. S2). For quality filtering, we first removed adaptor sequences from the forward and reverse reads by using Cutadapt (version 1.9.1)40. The command for forward reads was as follows:
cutadapt -a CTGTCTCTTATACACATCTCCGAGCCCACGAGAC -O 33
The command for reverse reads was as follows:
cutadapt -a CTGTCT CTTATACACATCTGACGCTGCCGACGA -O 32
Fastp was subsequently used to remove poly G sequences longer than 10 base pairs, reads shorter than 200 base pairs, reads containing one or more undetermined bases (N) from the sequencer, and reads with an average quality score lower than 15. PhiX and human-derived DNA sequences were removed using Bowtie2 (version 2.2.9)41. Only paired-end reads were counted, resulting in a total of 141,279,674 high-quality reads.
MEGAHIT (version 1.1.1–2) [42] was used for assembly, in which contigs were created. Mapping of high-quality reads to the contigs was performed using Bowtie2. Gene prediction for the contigs was accomplished using Prodigal (version 2.6.3) [43] to obtain amino acid sequences. Representative sequences were obtained by clustering sequences with 100 % similarity using MMseq2 (version 61ca48883b50714be51fc35fc9b77325ffde53fb) [44]. Functional assignment was performed through a homology search with the representative sequence using DIAMOND (version 0.9.10) [45] against the Unified Human Gastrointestinal Protein (UHGP) catalog in the MGnify (version 2019.8)50. The abundance of functional genes was calculated based on sequence identity >40, bit score >70, and coverage >80, matching the list of genes assigned to the gene name of the UHGP with the mapping output. Additionally, the abundance of orthologs was calculated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) [51] linked to the UHGP of MGnify. The results revealed 4,287,107 assigned genes and 7,861 KOs (Supplementary Fig. S2).
The contribution (abundance) of each bacterium to each KO was calculated from the output of the UHGP gene profile. The gene profile included the ID of each gene, the strain name of the bacteria containing that gene, and the ID of the KO.
7.5. Phylogenetic tree
For phylogenetic tree estimation, the 16S rRNA amplicon sequences of all the strains belonging to the genera were aligned using MAFFT [46]. RAxML [47] was subsequently used to construct the phylogenetic tree, the parameters of which were set to raxmlHPC-SSE3 -m GTRGAMMA -p 12345. The visualization of the tree was performed using iTOL [48].
7.6. Statistical analysis
To calculate the fold change in the mean abundance of each microbe, 1.0 × 10−9 was added to all the bacterial relative abundance values. For the fold change in KOs, 1.0 × 10−12 was added to all the KO abundance values.
Principal component analysis (PCA) was performed on the relative abundance of each bacterial strain using the sklearn.decomposition module of scikit-learn in Python (version 3.9.7). The relative abundances were converted to z scores using scipy stats.
For two-group comparisons of bacterial and KO abundances, two-sided Brunner–Munzel tests were used. The significance level for bacterial and KO abundance comparisons was set at p = 0.05. In this analysis, the significance level was set at p = 0.05 unless otherwise mentioned. Additionally, statistical significance was determined by using the following tests:
-
1.
Two-sided Mann‒Whitney U test to determine differences in demographic information and alpha diversity between groups.
-
2.
Two-sided Brunner-Munzel tests to determine differences in the bacterial and KO abundances between groups.
-
3.
Two-sided Kruskal‒Wallis test to determine differences in bacterial abundance between the four groups.
For correlation analysis between bacterial abundance and blood biomarker values, the Spearman correlation coefficient was used. The goodness of fit of the correlation coefficient was assessed using the coefficient of determination (R2 score). Enrichment analysis was performed using Enteropathway [23]. This analysis focused on the KOs with significant differences in abundance between the ATLL patient group and healthy controls. The significance level for this analysis was set at p = 0.05.
Ethics statements
This study was reviewed and approved by Miyazaki University with the approval number: 2020-565, dated March 16, 2021.
All participants provided written informed consent to participate in the study and for their data to be published.
Grant support
This work was supported by grants from the Japan Society for the Promotion of Science (JJSPS) KAKENHI (19K07693, 22K07172 to S.Nakahata; 15K15087 to K.M.; JP16H06279 (PAGS) to T.Y.), the Japan Agency for Medical Research and Development (JP21ck0106546h0002, JP21cm0106477, JP22ama221404) (to T.Y.), Japan Science and Technology Agency AIP Acceleration Research (JPMJCR19U3) (to T.Y.), and JST SPRING (JPMJSP2106) (to N.C.).
CRediT authorship contribution statement
Nodoka Chiba: Writing – review & editing, Writing – original draft, Validation, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Shinya Suzuki: Investigation. Daniel Enriquez-Vera: Investigation. Atae Utsunomiya: Resources. Yoko Kubuki: Resources. Tomonori Hidaka: Resources. Kazuya Shimoda: Resources. Shingo Nakahata: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Investigation, Funding acquisition, Formal analysis, Conceptualization. Takuji Yamada: Writing – review & editing, Writing – original draft, Validation, Supervision, Investigation, Funding acquisition, Formal analysis, Conceptualization. Kazuhiro Morishita: Writing – review & editing, Validation, Supervision, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Takuji Yamada reports a relationship with Metagen Therapeutics Inc that includes: board membership. Takuji Yamada reports a relationship with Metagen Inc that includes: board membership. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank all the participants and their families who participated in this study. We thank Dr. Hirotsugu Shiroma (Tokyo Institute of Technology) for constructing the whole-genome shotgun metagenomic sequence pipeline. We would also like to acknowledge Dr. Chikako Tani (University of Miyazaki) and Ms. Satomi Harazono (Kagoshima University) for providing research assistance. We would like to thank Dr. Sayaka Mizutani (Tokyo Institute of Technology) for advice on the paper-writing technique.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38507.
Contributor Information
Shingo Nakahata, Email: snakahata@kufm.kagoshima-u.ac.jp.
Takuji Yamada, Email: takuji@bio.titech.ac.jp.
Kazuhiro Morishita, Email: kmorishi@med.miyazaki-u.ac.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Taxonomic profiling pipeline.
Functional profiling pipeline.
Comparison of the gut microbial community structure in cohort 2
|Blue, healthy controls; red, patients with ATLL.
(A) PCA plot of the subjects' gut microbiota taxonomic composition.
(B) Alpha diversity of the subjects' gut microbiota. P values were calculated by the two-tailed Mann‒Whitney U test.
(C) Relative abundance of the top 30 genera in patients with ATLL and healthy controls. A phylogenetic tree was constructed from 16S rRNA gene sequences.
Differential analysis of the abundance of each genus in the gut microbiota
Seven genera were significantly more abundant in the A + HiC group in cohort 1.
(A) The relative abundance of the seven genera in cohort 1. P values were calculated by the two-tailed Kruskal–Wallis test.
(B) The relative abundance of the seven genera in cohort 2. P values were calculated by the two-tailed Brunner–Munzel test.
Correlation between tumor markers (sIL-2Ra) and the relative abundance of each bacteria
Distribution of sIL-2Ra levels (X-axis) and the relative abundance of the seven genera that were significantly more abundant in the A + HiC group (Y-axis). The values of relative abundance were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type.
(A) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (B) patients with ATLL in cohort 2.
Correlations between tumor markers (sIL-2Ra) and the relative abundance of each bacteria
The distribution of sIL-2Ra levels (X-axis) and the relative abundance of the genera (Y-axis) that were positively and strongly (ρ > 0.4 and R2>0.3) correlated with sIL-2Ra levels. The relative abundance were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type.
(A) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (B) patients with ATLL in cohort 2.
Correlations between tumor makers (proviral load) and the relative abundance of each bacteria
(A), (B) Distribution of Spearman correlation coefficients between proviral load levels and the relative abundance of each bacterium (X-axis) and the R2 score (Y-axis) for patients with ATLL and high-risk HTLV-1 carriers in cohort 1 (A) and cohort 2 (B). The bacterial names in the figure represent the bacterial genera whose abundance increased with the progression of ATLL identified in the previous section. Red plots are bacteria that were found in five or more subjects; subjects had measurable bacterial abundance values and proviral load levels. A relative abundance of 0 was omitted before the correlation coefficient was calculated.
C, (D) The distributions of proviral load levels (X-axis) and the relative abundances of genera (Y-axis) that were positively and strongly (ρ > 0.4 and R2>0.3) correlated with the proviral load levels. The relative abundance values were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type. (C) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (D) patients with ATLL in cohort 2.
Results of enrichment analysis of KOs that were significantly increased in abundance in patients with ATLL in cohort 2.
Results from the enrichment analysis by Enteropathway. P values are transformed by -log10. The vertical line represents P = 0.05.
Results of enrichment analysis of KOs whose abundance significantly decreased in patients with ATLL in cohort 2
Results from the enrichment analysis by Enteropathway. P values are transformed by -log10. The vertical line represents P = 0.05.
Isomaltose degradation module
Association of KO abundance and the genus Klebsiella with the isomaltose degradation module. The box plot represents the relative abundance of the KOs (left) and ortholog gene in the KO from Klebsiella (right). The bar plot represents the bacterial composition of the KOs of each subject. Abbreviations: Isomaltose 6P, isomaltose-6-phosphate.
Mannose degradation module
Association of KO abundance and the genus Klebsiella with the mannose degradation module. The box plot represents the relative abundance of KOs (left) and ortholog gene in the KO from Klebsiella (right). The bar plot represents the bacterial composition of the KOs of each subject. Abbreviations: Mannose N-AcGln, Mannose N-acetylglucosamine; D-Man, D-Mannose; D-Man 6P, D-Mannose-6-phosphate; Beta-D-Fru 6P, Beta-D-fructose-6-phosphate.
Effects of SSA on ATL cell proliferation
Cell growth rate. IL-2-independent ATL cell line ATN1 with SSA. The values represent the means ± s.d. ∗P < 0.05; unpaired two-tailed Student's t-test.
References
- 1.Hou K., Wu Z.X., Chen X.Y., Wang J.Q., Zhang D., Xiao C., Zhu D., Koya J.B., Wei L., Li J., et al. Microbiota in health and diseases. Signal Transduct. Targeted Ther. 2022;71(7):1–28. doi: 10.1038/s41392-022-00974-4. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalmasso G., Cougnoux A., Delmas J., Darfeuille-Michaud A., Bonnet R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microb. 2014;5:675–680. doi: 10.4161/19490976.2014.969989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raffelsberger N., Hetland M.A.K., Svendsen K., Småbrekke L., Löhr I.H., Andreassen L.L.E., Brisse S., Holt K.E., Sundsfjord A., Samuelsen Ø., et al. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microb. 2021;13 doi: 10.1080/19490976.2021.1939599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R., Yang X., Liu J., Zhong F., Zhang C., Chen Y., Sun T., Ji C., Ma D. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 2022;131 13:1–18. doi: 10.1038/s41467-022-30240-8. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allegra A., Innao V., Allegra A.G., Ettari R., Pugliese M., Pulvirenti N., Musolino C. Role of the microbiota in hematologic malignancies. Neth. J. Med. 2019;77:67–80. [PubMed] [Google Scholar]
- 6.Niemann C., Coelho M., Öztürk S., Seiffert M. 2023. The Diversity of the Microbiome Impacts Chronic Lymphocytic Leukemia Development in Mice and Humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma T., Chen Y., Li L.J., Zhang L.S. Opportunities and challenges for gut microbiota in acute leukemia. Front. Oncol. 2021;11 doi: 10.3389/FONC.2021.692951/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips A.A., Harewood J.C.K. Adult T cell leukemia-lymphoma (ATL): state of the art. Curr. Hematol. Malig. Rep. 2018;13:300–307. doi: 10.1007/S11899-018-0458-6. [DOI] [PubMed] [Google Scholar]
- 9.Tsukasaki K., Marçais A., Nasr R., Kato K., Fukuda T., Hermine O., Bazarbachi A. Diagnostic approaches and established treatments for adult T cell leukemia lymphoma. Front. Microbiol. 2020;11 doi: 10.3389/FMICB.2020.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamagishi M., Suzuki Y., Watanabe T., Uchimaru K. 2022. Clonal Selection and Evolution of HTLV-1-Infected Cells Driven by Genetic and Epigenetic Alteration. Viruses 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawano N., Nagahiro Y., Yoshida S., Tahara Y., Himeji D., Kuriyama T., Tochigi T., Nakaike T., Shimokawa T., Yamashita K., et al. Clinical features and treatment outcomes of opportunistic infections among human T-lymphotrophic virus type 1 (HTLV-1) carriers and patients with adult T-cell leukemia-lymphoma (ATL) at a single institution from 2006 to 2016. J. Clin. Exp. Hematop. 2019;59:156–167. doi: 10.3960/JSLRT.18032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porto A.F., Neva F.A., Bittencourt H., Lisboa W., Thompson R., Alcântara L., Carvalho E.M. HTLV-1 decreases Th2 type of immune response in patients with strongyloidiasis. Parasite Immunol. 2001;23:503–507. doi: 10.1046/J.1365-3024.2001.00407.X. [DOI] [PubMed] [Google Scholar]
- 13.Montes M., Sanchez C., Verdonck K., Lake J.E., Gonzalez E., Lopez G., Terashima A., Nolan T., Lewis D.E., Gotuzzo E., et al. Regulatory T cell expansion in HTLV-1 and strongyloidiasis Co-infection is associated with reduced IL-5 responses to Strongyloides stercoralis antigen. PLoS Neglected Trop. Dis. 2009;3 doi: 10.1371/JOURNAL.PNTD.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens E.J., Bates K.A., King K.C. Host microbiota can facilitate pathogen infection. PLoS Pathog. 2021;17 doi: 10.1371/JOURNAL.PPAT.1009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Anazi A.R. Gastrointestinal opportunistic infections in human immunodeficiency virus disease. Saudi J. Gastroenterol. 2009;15:95. doi: 10.4103/1319-3767.48965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zevin A.S., McKinnon L., Burgener A., Klatt N.R. Microbial translocation and microbiome dsybiosis in HIV-associated immune activation. Curr. Opin. HIV AIDS. 2016;11:182. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tag-Adeen M., Hashiguchi K., Akazawa Y., Ohnita K., Yasushi S., Daisuke N., Nakao K. An unusual presentation of adult T-cell leukemia/lymphoma. Ecancermedicalscience. 2018;12 doi: 10.3332/ECANCER.2018.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimoto Y., Mizutani S., Nakajima T., Hosoda F., Watanabe H., Saito Y., Shibata T., Yachida S., Yamada T. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut. 2016;65:1574–1575. doi: 10.1136/GUTJNL-2016-311937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwanaga M., Watanabe T., Utsunomiya A., Okayama A., Uchimaru K., Koh K.R., Ogata M., Kikuchi H., Sagara Y., Uozumi K., et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: a nationwide prospective study in Japan. Blood. 2010;116:1211–1219. doi: 10.1182/BLOOD-2009-12-257410. [DOI] [PubMed] [Google Scholar]
- 20.Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. Br. J. Haematol. 1991;79:428–437. doi: 10.1111/J.1365-2141.1991.TB08051.X. [DOI] [PubMed] [Google Scholar]
- 21.Arumugam M., Raes J., Pelletier E., Paslier D. Le, Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamihira, S., Atogami, S., Sohda, H., Momita, S., Yamada, Y., and Tomonaga, M. Significance of soluble interleukin-2 receptor levels for evaluation of the progression of adult T-cell Leukemia. 10.1002/1097-0142(19940601)73:11. [DOI] [PubMed]
- 23.Shiroma H., Darzi Y., Terajima E., Nakagawa Z., Tsuchikura H., Tsukuda N., Moriya Y., Okuda S., Goto S., Yamada T. Enteropathway: the metabolic pathway database for the human gut microbiota. bioRxiv. 2023;2023:6. doi: 10.1101/2023.06.28.546710. 28.546710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niemi A.K., Brown C., Moore T., Enns G.M., Cowan T.M. Evidence of redox imbalance in a patient with succinic semialdehyde dehydrogenase deficiency. Mol. Genet. Metab. Reports. 2014;1:129. doi: 10.1016/J.YMGMR.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes J.D., Dinkova-Kostova A.T., Tew K.D. Oxidative stress in cancer. Cancer Cell. 2020;38:167–197. doi: 10.1016/J.CCELL.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A., Pande A., Sabrin A., Thapa S.S., Gioe B.W., Grove A. MarR family transcription factors from Burkholderia species: hidden clues to control of virulence-associated genes. Microbiol. Mol. Biol. Rev. 2019;83 doi: 10.1128/MMBR.00039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Méndez V., Agulló L., González M., Seeger M. The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS One. 2011;6 doi: 10.1371/JOURNAL.PONE.0017583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galán B., Kolb A., Sanz J.M., García J.L., Prieto M.A. Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res. 2003;31:6598. doi: 10.1093/NAR/GKG851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez J., Guarner F., Bustos Fernandez L., Maruy A., Sdepanian V.L., Cohen H. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 2020;10:731. doi: 10.3389/FCIMB.2020.572912/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dykie A., Wijesinghe T., Rabson A.B., Madugula K., Farinas C., Wilson S., Abraham D., Jain P. Human T-cell leukemia virus type 1 and Strongyloides stercoralis: partners in pathogenesis. Pathogens. 2020;9:1–22. doi: 10.3390/PATHOGENS9110904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh M., Toma H., Sugahara K., Etoh K.I., Shiroma Y., Kiyuna S., Takara M., Matsuoka M., Yamaguchi K., Nakada K., et al. Involvement of IL-2/IL-2R system activation by parasite antigen in polyclonal expansion of CD4+25+ HTLV-1-infected T-cells in human carriers of both HTLV-1 and S. stercoralis. Oncogene. 2002;2116 21:2466–2475. doi: 10.1038/sj.onc.1205329. 2002. [DOI] [PubMed] [Google Scholar]
- 32.Mahgoub M., Yasunaga J.I., Iwami S., Nakaoka S., Koizumi Y., Shimura K., Matsuoka M. Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E1269–E1278. doi: 10.1073/pnas.1715724115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi M., Higuchi M., Makokha G.N., Matsuki H., Yoshita M., Tanaka Y., Fujii M. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood. 2013;122:715–725. doi: 10.1182/BLOOD-2013-03-493718. [DOI] [PubMed] [Google Scholar]
- 34.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/BIOINFORMATICS/BTY560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/PEERJ.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen W., Le S., Li Y., Hu F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016;11 doi: 10.1371/JOURNAL.PONE.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., et al. vol. 2. PDF; 2017. (Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopylova E., Noé L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/BIOINFORMATICS/BTS611. [DOI] [PubMed] [Google Scholar]
- 39.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:1–9. doi: 10.1186/1471-2105-10-421/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 41.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/NMETH.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D., Luo R., Liu C.M., Leung C.M., Ting H.F., Sadakane K., Yamashita H., Lam T.W. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/J.YMETH.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:1–11. doi: 10.1186/1471-2105-11-119/TABLES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017;3511 35:1026–1028. doi: 10.1038/nbt.3988. 2017. [DOI] [PubMed] [Google Scholar]
- 45.Buchfink B., Reuter K., Drost H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;184 18:366–368. doi: 10.1038/s41592-021-01101-x. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772. doi: 10.1093/MOLBEV/MST010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312. doi: 10.1093/BIOINFORMATICS/BTU033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/NAR/GKZ239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;3610 36:996–1004. doi: 10.1038/nbt.4229. 2018. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., et al. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2020;48:D570–D578. doi: 10.1093/NAR/GKZ1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27. doi: 10.1093/NAR/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomic profiling pipeline.
Functional profiling pipeline.
Comparison of the gut microbial community structure in cohort 2
|Blue, healthy controls; red, patients with ATLL.
(A) PCA plot of the subjects' gut microbiota taxonomic composition.
(B) Alpha diversity of the subjects' gut microbiota. P values were calculated by the two-tailed Mann‒Whitney U test.
(C) Relative abundance of the top 30 genera in patients with ATLL and healthy controls. A phylogenetic tree was constructed from 16S rRNA gene sequences.
Differential analysis of the abundance of each genus in the gut microbiota
Seven genera were significantly more abundant in the A + HiC group in cohort 1.
(A) The relative abundance of the seven genera in cohort 1. P values were calculated by the two-tailed Kruskal–Wallis test.
(B) The relative abundance of the seven genera in cohort 2. P values were calculated by the two-tailed Brunner–Munzel test.
Correlation between tumor markers (sIL-2Ra) and the relative abundance of each bacteria
Distribution of sIL-2Ra levels (X-axis) and the relative abundance of the seven genera that were significantly more abundant in the A + HiC group (Y-axis). The values of relative abundance were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type.
(A) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (B) patients with ATLL in cohort 2.
Correlations between tumor markers (sIL-2Ra) and the relative abundance of each bacteria
The distribution of sIL-2Ra levels (X-axis) and the relative abundance of the genera (Y-axis) that were positively and strongly (ρ > 0.4 and R2>0.3) correlated with sIL-2Ra levels. The relative abundance were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type.
(A) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (B) patients with ATLL in cohort 2.
Correlations between tumor makers (proviral load) and the relative abundance of each bacteria
(A), (B) Distribution of Spearman correlation coefficients between proviral load levels and the relative abundance of each bacterium (X-axis) and the R2 score (Y-axis) for patients with ATLL and high-risk HTLV-1 carriers in cohort 1 (A) and cohort 2 (B). The bacterial names in the figure represent the bacterial genera whose abundance increased with the progression of ATLL identified in the previous section. Red plots are bacteria that were found in five or more subjects; subjects had measurable bacterial abundance values and proviral load levels. A relative abundance of 0 was omitted before the correlation coefficient was calculated.
C, (D) The distributions of proviral load levels (X-axis) and the relative abundances of genera (Y-axis) that were positively and strongly (ρ > 0.4 and R2>0.3) correlated with the proviral load levels. The relative abundance values were converted by log10. ρ, Spearman's correlation coefficient; R2, coefficient of determination. The plot color represents ATLL subtypes; green, high-risk HTLV-1 carrier; yellow, smoldering type; pink, acute type; and cyan, lymphoma type. (C) Patients with ATLL and high-risk HTLV-1 carriers in cohort 1 and (D) patients with ATLL in cohort 2.
Results of enrichment analysis of KOs that were significantly increased in abundance in patients with ATLL in cohort 2.
Results from the enrichment analysis by Enteropathway. P values are transformed by -log10. The vertical line represents P = 0.05.
Results of enrichment analysis of KOs whose abundance significantly decreased in patients with ATLL in cohort 2
Results from the enrichment analysis by Enteropathway. P values are transformed by -log10. The vertical line represents P = 0.05.
Isomaltose degradation module
Association of KO abundance and the genus Klebsiella with the isomaltose degradation module. The box plot represents the relative abundance of the KOs (left) and ortholog gene in the KO from Klebsiella (right). The bar plot represents the bacterial composition of the KOs of each subject. Abbreviations: Isomaltose 6P, isomaltose-6-phosphate.
Mannose degradation module
Association of KO abundance and the genus Klebsiella with the mannose degradation module. The box plot represents the relative abundance of KOs (left) and ortholog gene in the KO from Klebsiella (right). The bar plot represents the bacterial composition of the KOs of each subject. Abbreviations: Mannose N-AcGln, Mannose N-acetylglucosamine; D-Man, D-Mannose; D-Man 6P, D-Mannose-6-phosphate; Beta-D-Fru 6P, Beta-D-fructose-6-phosphate.
Effects of SSA on ATL cell proliferation
Cell growth rate. IL-2-independent ATL cell line ATN1 with SSA. The values represent the means ± s.d. ∗P < 0.05; unpaired two-tailed Student's t-test.
Data Availability Statement
6.1. Data
The raw sequencing data reported in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA), Tokyo, Japan, under accession number DRA016992.
6.2. Code
This paper does not report the original code.
6.3. Experimental model and study participant details
6.3.1. In vitro experiments
ATLL patient-derived leukemia cell lines (MT-1, ATN-1, and ILT-Mat) were procured from the JCRB or RIKEN cell bank in 2022. The cell lines were authenticated utilizing STR-PCR profiling and tested negative for mycoplasma. MT-1 cells were cultured in RPMI-1640 medium (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10 % fetal bovine serum (Cosmobio, Tokyo, Japan), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) (complete RPMI-1640 medium). ATN-1 and ILT-Mat cells were cultured in complete RPMI-1640 medium supplemented with 1 × nonessential amino acids (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) and interleukin-2 (BioLegend, San Diego, USA), respectively. The cells were maintained at 37 °C in a humidified atmosphere with 5 % CO2.
Cells were seeded into 96-well plates at densities ranging from 50 × 103 to 200 × 103 cells in 100 μL of complete cell culture medium, and succinic semialdehyde (Santa Cruz Biotechnology, Texas, USA) was added to the medium for 72 h at 37 °C with 5 % CO2. Cell proliferation was assessed every 24 h using the WST-8 method with a Cell Counting Kit (Dojindo, Kumamoto, Japan) following the manufacturer's instructions. The experiments were performed in triplicate and repeated at least three times.
6.3.2. Study participants’ sample collection
The Miyazaki University Faculty of Medicine Ethics Committee and participating institution approved the study (approved number 2020-565). Samples and clinical information were obtained with informed consent. Patients with ATLL and asymptomatic HTLV-1 carriers aged over 20 years old with confirmed HTLV-1 infection were recruited from Miyazaki University Hospital and Imamura General Hospital in Japan. Healthy controls aged over 20 years old with no family history of ATLL were recruited. We obtained fecal samples from two cohorts because fecal samples were collected from asymptomatic HTLV-1 carriers at home after the hospital visit using a preservation solution kit [18], while fresh fecal samples were collected from patients with ATLL at the same time as hospital admission. Therefore, one cohort included asymptomatic HTLV-1 carriers (n = 37) and patients with less advanced types of ATLL (n = 11), such as the smoldering type and healthy controls (n = 20) (cohort 1, Table 1). In the asymptomatic HTLV-1 carrier group, there were eight asymptomatic HTLV-1 carriers with HTLV-1 proviral loads (PVLs) above 40 (per 1000 peripheral blood mononuclear cells (PBMCs)), and we defined them as high-risk carriers [19]. The other cohort included patients with advanced-stage ATLL, such as acute type (n = 17), and healthy controls (n = 8) (cohort 2, Table 2). In cohort 1, patients with ATLL were significantly older (Mann‒Whitney U test, two-tailed, P = 0.00164 vs. asymptomatic HTLV-1 carriers, P = 1.39 × 10−9 vs. healthy controls). There were more women in the asymptomatic HTLV-1 carrier group than in the control group, whereas the healthy controls were all men. In cohort 2, patients with ATLL were significantly older than healthy controls (P = 3.89 × 10−19).
The soluble interleukin 2 receptor-alpha (sIL-2Ra) levels, PVL levels, and clinical data were collected from the medical records (Table 1). sIL-2Ra is a blood biomarker that reflects tumor size and disease progression in patients with ATLL. sIL-2Ra also reflects lymphoma and other immune disorders, but none of the study subjects had these diseases. The samples and clinical information used in this study were obtained after providing informed consent and with the approval of the institutional review boards of each participating institute of the University of Miyazaki.