Abstract
Hearing loss is a common disease. More than 100 genes have been reported to be associated with hereditary hearing loss. However, the distribution of these genes and their variants across diverse populations remains unclear. In this study, we gathered 347 hearing-impaired patients from four language families (Sinitic, Tibeto-Burman, Kra-Dai, and Hmong-Mien) in Southwestern China, excluding cases caused by common mutations in the GJB2 gene. By using next generation sequencing, 122 genes associated with hereditary hearing loss were analyzed on these patients. Rare candidate variants were identified in 71.93 % (264/347) of patients with hearing loss. The diagnostic rate varied around 10 % across different language families. The most frequently identified causative genes in successfully diagnosed cases were SLC26A4, MYO7A and TMPRSS3. Moreover, a substantial number of variants of unknown significance (VUS) were identified in our patient cohort. This underscores the critical need for establishing ethnicity-specific genomic databases for hearing loss. It will significantly improve the clinical diagnostic rate for hearing loss in this region.
Keywords: Hearing loss, Next generation sequencing, Diagnostic rate, Ethnicity, Variants of unknown significance (VUS)
Highlights
-
•
Yunnan province, situated in southwestern China, is characterized by its diverse multi-ethnic population.
-
•
The study made a first comprehensive investigation on hearing loss genes in patients with hearing Loss of this region involving 4 Language Families of 15 Ethnicities.
-
•
The hearing loss diagnostic rate varied around 10 % across different language families of this region.
-
•
A large number of variants of unknown significance (VUS) were identified in unsolved patients.
-
•
This underscores the critical need for establishing ethnicity-specific genomic databases for hearing loss.
1. Introduction
Hearing loss is a common disease [1] with a newborn incidence of approximately 1 ‰ [2]. In developed countries, 50 %–60 % of childhood hearing loss are associated with genetic factors. Hearing loss is associated with environmental factors, genetic factors or a combination [2,3]. Non-syndromic hearing loss accounts for 70 % of cases, while 30 % of hereditary hearing loss is syndromic [[4], [5], [6]]. In non-syndromic cases, only hearing impairment is observed, and the majority (around 80 %) is characterized by autosomal recessive inheritance with prelingual severe-to-profound hearing loss. Autosomal dominant, X-linked and mitochondrial inheritance patterns are also observed in non-syndromic hearing loss, with incidences of approximately 20 %, 1 % and 1 %, respectively [5].
Hearing loss is a genetically heterogeneous disorder, with over 100 identified genes implicated in hereditary hearing loss [7,8]. Additionally, more than 140 loci are linked to non-syndromic hearing loss. Mutations in the same gene can lead to both autosomal recessive and dominant forms of hearing loss [9]. For example, in our previous study, distinct mutations in the MYO7A gene were associated with both non-syndromic hearing loss and Usher syndrome [10]. Interestingly, even the same variant can result in variable phenotypes of hearing loss, as seen in the GJB2 c.109 G > A mutation [11].
Targeted next-generation sequencing is extensively employed in the molecular diagnosis of Mendelian disorders [[12], [13], [14]]. In contrast to Sanger sequencing, it allows for the simultaneous screening of mutations across multiple genes. Hearing loss is a heterogeneous disorder associated with over 100 genes [15]. Following the guidelines established by the American College of Medical Genetics and Genomics (ACMG), molecular genetic testing plays a crucial role in ensuring the accurate diagnosis of hereditary hearing loss. Identifying specific variants associated with hearing loss significantly aids in confirming the clinical diagnosis [16,17].
The global spectrum of DNA variants in patients with hearing loss remains unclear. Yunnan, a province in Southwestern China, is characterized by its diverse ethnic composition. In our previous study involving 84 hearing-impaired patients from Southwestern China, potential pathogenic variants were identified in 34 genes associated with hearing loss [18]. However, the distribution of these genes and their variants has been minimally explored. To further clarify this point, we expanded our investigation to include 347 patients with hearing loss, representing four language families and encompassing 15 different ethnicities. Notably, the hearing loss in these patients was not caused by common mutations of GJB2 gene. In the present study, we employed targeted next-generation sequencing of 122 genes to analyze the molecular pathology in this expanded cohort.
2. Materials and methods
2.1. Statement of ethnics, patients and DNA preparation
Ethics Committee of First People's Hospital of Yunnan Province (Affiliated Hospital of Kunming University of Science and Technology) approved the present study. Peripheral blood samples of patients with hearing loss were collected from Kunming, Dehong, Baoshang, Lijiang, Puer and Tengchong cities in Yunnan province. All the patients were clinically diagnosed as hearing loss by Dr. Jiahong Pei. Each patient in this study signed informed consent. For the minors or children, one of their guardians signed the informed consent. We extracted genomic DNA from the peripheral blood leukocytes by using E.Z.N.A.® Blood DNA Kit (cat. no. D3392-02; Omega Bio-tek, Inc, USA).
2.2. Targeted sequence capture and high-throughput sequencing
Total 122 genes (supplementary material 1) of hearing loss were captured and sequenced in this study. Experimental procedures of target gene capture sequencing were referred to our previous published studies [10]. A probe capture panels (Roche NimbleGen Inc., Madison, WI) targeting 122 genes were generated, and the total size for targeted regions were about 4M. Briefly, DNA of patients were fragmented, end-repaired and ligated to adapter oligonucleotides. After that, a library preenrichment amplification were performed by using PCR. Qualified libraries were used in capture of targeted genes. The captured fragments were sequenced by Hiseq2500 Analyzers (Illumina, San Diego, USA).
2.3. Data filtering, read mapping, variant detection and analysis pipeline
Data filtering, read mapping and variant detection were referred to our previous published studies [10]. Briefly, primary data was achieved after image analysis, error estimation and base calling. Clean reads were achieved after data filtering. UCSC hg19 reference genome was used in detection of SNPs (single-nucleotide polymorphisms) and indels (insertion-deletions). BWA (Burrows-Wheeler Aligner) Multi Vision software package was used in the sequence alignment. SOAPsnp software and GATK Indel Genotyper (http://www.broadinstitute.org/gsa/wiki/index.php/, The Genome Analysis Toolkit) were used in detection of SNPs (single-nucleotide polymorphisms) and indels (insertion-deletions). Frequency of these variants were referred to the 1000 Genomes public variant databases, dbSNP and ExAC databases. Pathogenicity classification of these variants was based on ACMG guidelines [19], and further checked in ClinVar and HGMD databases. SNPs variants were evaluated using InterVar (http://wintervar.wglab.org/) [20]. For indels and splice-site variants, PP3 evidence was not used in ACMG scores. PP3 evidence referred to multiple lines of computational evidence to support a deleterious effect on the gene or its product (such as conservation, evolutionary impact, splicing effects, etc.). Bioinformatic prediction tools for indels and splice-site variants were more limited compared to those available for SNPs, and often did not generate multiple lines of evidence. For correlation between the gene variants and phenotype of patients, missense, indel, nonsense and splice-site variants were considered. The selected variants were further confirmed by Sanger sequencing. CNV (copy number variation) was analyzed by referring to the method described by by Nord et al. [21].
3. Results
3.1. Patients with hearing loss and target next-generation sequencing
A total of 347 patients with hearing loss, representing 15 different ethnicities, were recruited from Yunnan province of Southwestern China. Notably, the hearing loss in these patients was not caused by common GJB2 mutations (c.35delG, c.109 G > A, c.167delT, c.176_191del16, c.235delC and c.299_300delAT). The distribution of patients among different ethnic groups was as follows: A-Chan/5, Bai/20, Tibetan/1, Dai/37, Hani/32, Han/110, Hui/6, Jingpo/10, Lisu/16, Miao/21, Wa/1, Yao/1, Yi/83, Zhuang/3 and Lagu/1. These patients were classified into four language families, namely Sinitic (n = 116), Tibeto-Burman (n = 172), Kra-Dai (n = 37) and Hmong-Mien (n = 22). DNA was extracted from peripheral white blood cells, and a total of 122 known genes associated with hearing loss were captured and sequenced using massively parallel sequencing. The targeted region spanned approximately 4 million base pairs (4M), encompassing 122 nuclear genes associated with hearing loss. Full genomic region of CDH23, GJB2, GJB3, GJB6, LOXHD1, MYO15A, MYO7A, OTOF, OTOG, PCDH15, SLC26A4, TRIOBP and USH2A genes were captured. For other genes, all exons, splice sites and immediate flanking intron sequences of 100bp were captured.
3.2. Bioinformatic analysis of variants identified in patients of hearing loss
The average sequencing depth exceeded 400X, meeting the criteria for bioinformatic analysis. The sequencing data exhibited high quality, with coverage rates of Q20 and Q30 exceeding 96 % and 90 %, respectively. In each patient, both SNPs and Indels were detected. Variants with a minor allele frequency (MAF) > 0.01 were excluded from further analysis. Nonsynonymous and splicing site variants that were directly or potentially associated with patient phenotypes were selected for subsequent analysis. In accordance with the ACMG guidelines, these candidate variants were categorized as “pathogenic”, “likely pathogenic”, “variants of unknown significance (VUS) ", “likely benign” or “benign”.
3.3. Causative genes in patients with hearing loss of different ethnicities
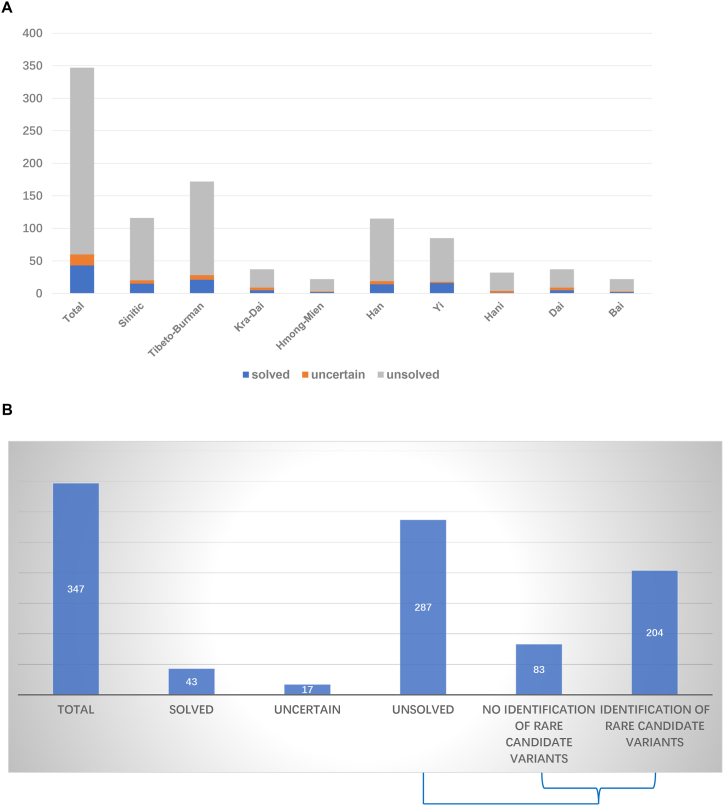
The methodology for analyzing pathogenic genes in the current study was referred to the descriptions by Yan et al. [22]. Patients representing 15 different ethnicities were divided into three groups: solved, uncertain and unsolved, as shown in Fig. 1A and B. Rare candidate variants were identified in 71.93 % (264/347) of patients with hearing loss. Among the solved patients, hearing loss was caused by variants classified as pathogenic or likely pathogenic. A total of 43 solved cases were identified among all patients with hearing loss, resulting in an etiologic diagnostic rate of 12.39 % (43/347). The diagnostic rates within the four language families were 12.93 % (15/116, Sinitic), 12.21 % (21/172, Tibeto-Burman), 13.51 % (5/37, Kra-Dai) and 9.09 % (2/22, Hmong-Mien). Additionally, diagnostic rates for different ethnicities were 12.17 % (14/115, Han), 18.82 % (16/85, Yi), 3.13 % (1/32, Hani), 13.51 % (5/37, Dai) and 9.09 % (2/22, Bai). Uncertain patients were characterized by the presence of one variant of unknown significance (VUS) and one likely pathogenic or pathogenic variant, resulting in the identification of 17 uncertain cases. The proportion of uncertain patients was 4.9 % (17/347).
Fig. 1.
Representation of solved, uncertain and unsolved subjects in total patients with hearing loss (A) categorized by language families or five main ethnicities (Han, Yi, Hani, Dai and Bai). In 287 unsolved patients, rare candidate variants were identified in 204 patients (B). Most of these rare variants were classified as VUS (variant of uncertain significance) according to ACMG guidelines.
A total of 287 patients with hearing loss were categorized as unsolved cases, constituting 82.71 % of the overall patient population (287/347). Of 287 unsolved patients, rare candidate variants were identified in 204 patients, while they were not found in other 83 patients (Fig. 1B). Most of these rare variants were classified as VUS (variant of uncertain significance) according to ACMG guidelines. The distribution of unsolved patients within the four language families was as follows: 82.76 % (96/116, Sinitic), 83.72 % (144/172, Tibeto-Burman), 75.68 % (28/37, Kra-Dai) and 86.36 % (19/22, Hmong-Mien). Among major ethnicities, the percentage of unsolved patients was 83.48 % (96/115, Han), 78.82 % (67/85, Yi), 87.50 % (28/32, Hani), 75.68 % (28/37, Dai) and 86.36 % (19/22, Bai).
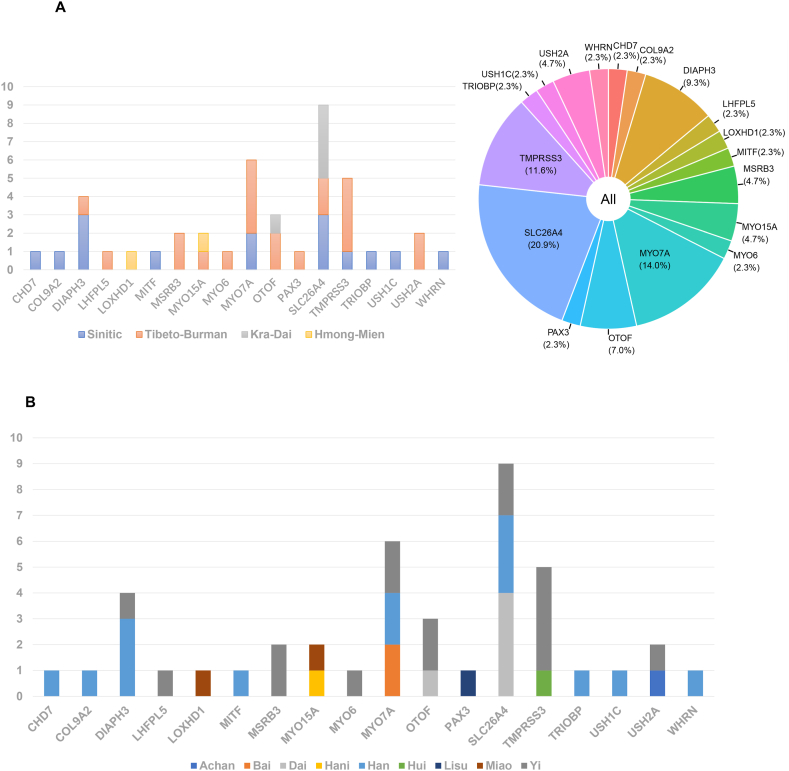
Candidate pathogenic variants were identified in a total of 69 genes, with 18 genes detected in the solved patient group. The most common genes among solved patients were SLC26A4 (9/43), MYO7A (6/43) and TMPRSS3 (5/43), as shown in Fig. 2 and Table 1. Distribution of causative DNA variants in solved patients of four language families and four main ethnicities were shown in Fig. 3A and B respectively. Among uncertain patients, 9 genes were identified, as shown in supplementary material 1. These patients harbored at least one variant of unknown significance (VUS). The most common genes among uncertain patients included GJB2 (5/17), SLC26A4 (2/17), CDH23 (2/17), MYO15A (2/17) and TRIOBP (2/17).
Fig. 2.
Number of solved patients with hearing loss for each gene in language families (A) or ethnicities(B).
Table 1.
Identified likely pathogenic and pathogenic variants in the solved patients with hearing loss.
| Subject | Ethnicity | Gene | Chromosome | Transcript | Genotype | Protein change | HGMD | ClinVar | GnomAD | Zygosity | ACMG | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PL32 | Han | SLC26A4 | chr7:107323713 | NM_000441 | c.833delC | p.T278fs | Het | Likely Pathogenic(PVS1+PM2) | AR | |||
| SLC26A4 | chr7:107323898 | NM_000441 | c.919-2A > G | Reported | Pathogenic | 0.000336 | Het | Pathogenic (PVS1+PS3+PM2) | AR | |||
| PL51 | Han | SLC26A4 | chr7:107323898 | NM_000441 | c.919-2A > G | Reported | Pathogenic | 0.000336 | Het | Pathogenic (PVS1+PS3+PM2) | AR | |
| SLC26A4 | chr7:107334920 | NM_00044 | c. 1336T > C | p.Q446X | Reported | Pathogenic | 0.00000723 | Het | Pathogenic (PVS1+PM2+PP3) | AR | ||
| PL67 | Han | SLC26A4 | chr7:107323898 | NM_000441 | c.919-2A > G | Reported | Pathogenic | 0.000336 | Hom | Pathogenic (PVS1+PS3+PM2) | AR | |
| EBH8 | Han | MYO7A | chr11:76890874 | NM_001127180 | c. 2461C > T | p.Q821X | Reported | Pathogenic | Het | Pathogenic (PVS1+PM2+PP3+ PP5) |
AR/AD | |
| MYO7A | chr11:76917153 | NM_000260 | c. 5648G > A | p.R1883Q | Reported | Pathogenic | 0.00003649 | Het | Likely pathogenic (PM1+PM2+ PP3+PP5) | AR/AD | ||
| PL15 | Han | DIAPH3 | chr13:60385059 | NM_001042517 | c.3028-2- > TAAG | Het | Likely pathogenic (PVS1+PM2) | AD | ||||
| PL23 | Han | DIAPH3 | chr13:60385059 | NM_001042517 | c.3028-2- > TAAG | Het | Likely pathogenic (PVS1+PM2) | AD | ||||
| PL29 | Han | DIAPH3 | chr13:60385059 | NM_001042517 | c.3028-2- > TAAG | Het | Likely pathogenic (PVS1+PM2) | AD | ||||
| PL50 | Han | WHRN | chr9:117168961 | NM_001173425 | c.1890_1909del | p.P630fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| PL63 | Han | COL9A2 | chr1:40768483 | NM_001852 | c.1604-2- > CTCC | Het | Likely pathogenic (PVS1+PM2) | AR/AD | ||||
| PL65 | Han | MYO7A | chr11:76905496 | NM_001127180 | c.4251delC | p.I1417fs | Het | Likely pathogenic (PVS1+PM2) | AR/AD | |||
| PL8 | Han | MITF | chr3:69990458 | NM_198177 | c.691delC | p.P231fs | Het | Likely pathogenic (PVS1+PM2) | AR/AD | |||
| DH5 | Han | CHD7 | chr8:61778038 | NM_017780 | c.8541delA | p.G2847fs | Het | Likely pathogenic (PVS1 +PM2) | AD | |||
| DH66 | Han | TRIOBP | chr22:38119391 | NM_001039141 | c.828_829 insAG |
p.P276fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| DH68 | Han | USH1C | chr11:17547893 | NM_001297764 | c.674 + 1 G > A |
Hom | Likely pathogenic (PVS1 +PM2) | AR | ||||
| h201 | Yi | MYO7A | chr11:76890874 | NM_001127180 | c. 2461 C > T | p.Q821X | Reported | Pathogenic | Het | Pathogenic (PVS1+PM2+PP3+ PP5) |
AR/AD | |
| MYO7A | chr11:76901128 | NM_001127180 | c.3695_3705del | p.R1232fs | 0.00000526 | Het | Likely pathogenic (PVS1 +PM2) | AR/AD | ||||
| h248 | Yi | OTOF | chr2:26700596 | NM_001287489 | c. 2236 C > T | p.Q746X | 0.00000407 | Hom | Pathogenic (PVS1+PM2+PP3) | AR | ||
| K1 | Yi | OTOF | chr2:26700596 | NM_001287489 | c. 2236 C > T | p.Q746X | 0.00000407 | Hom | Pathogenic (PVS1+PM2+ PP3) | AR | ||
| L23 | Yi | MYO6 | chr6:76554623 | NM_001300899 | c. 826 C > T | p.R276X | Pathogenic | 0.00001627 | Hom | Pathogenic (PVS1+PM2+PP3+ PP5) |
AR/AD | |
| h7 | Yi | TMPRSS3 | chr21:43803307 | NM_001256317 | c.616_617 insAG |
p.A206fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| h108 | Yi | MYO7A | chr11:76895766 | NM_001127179 | c.3510_3531del | p.V1170fs | Hom | Likely pathogenic (PVS1+PM2) | AR/AD | |||
| h120 | Yi | TMPRSS3 | chr21:43803307 | NM_001256317 | c.616_617 insAG |
p.A206fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| h148 | Yi | SLC26A4 | chr7:107312588 | NM_000441 | c.311_321 del |
p.A104fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| h173 | Yi | TMPRSS3 | chr21:43803307 | NM_001256317 | c.616_617 insAG |
p.A206fs | Het | Likely pathogenic (PVS1+PM2) | AR | |||
| TMPRSS3 | chr21:43803309 | NM_001256317 | c.617-2- > CT | Het | Likely pathogenic (PVS1+PM2) | AR | ||||||
| K13 | Yi | DIAPH3 | chr13:60385059 | NM_001042517 | c.3028-2- > TAAG | Het | Likely pathogenic (PVS1+PM2) | AD | ||||
| K18 | Yi | TMPRSS3 | chr21:43803307 | NM_001256317 | c.616_617 insAG |
p.A206fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| L4 | Yi | SLC26A4 | chr7:107312588 | NM_000441 | c.311_321 del |
p.A104fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| L9 | Yi | MSRB3 | chr12:65847507 | NM_198080 | c.314-1G > C | Hom | Likely pathogenic (PVS1+PM2) | AR | ||||
| L19 | Yi | MSRB3 | chr12:65847507 | NM_198080 | c.314-1G > C | Hom | Likely pathogenic (PVS1+PM2) | AR | ||||
| L38 | Yi | LHFPL5 | chr6:35773827 | NM_182548 | c. 380 A > G | p.Y127C | Reported | Pathogenic | 0.00002844 | Hom | Likely pathogenic (PM1+PM2+PP2+ PP5) |
AR |
| h127 | Yi | USH2A | chr1:216595579 | NM_206933 | c.99_100 insT |
p.R34fs | Reported | 0.00000362 | Hom | Likely pathogenic (PVS1 +PM2) | AR | |
| K64 | Dai | OTOF | chr2:26706449 | NM_001287489 | c. 1273 C > T | p.R425X | Pathogenic | 0.00001083 | Hom | Pathogenic (PVS1+PM2+PP3+ PP5) |
AR | |
| s3 | Dai | SLC26A4 | chr7:107315543 | NM_000441 | c. 754 T > C | p.S252P | Reported | Pathogenic | 0.00000406 | Het | Likely pathogenic (PM1+PM2+PP3+PP5+BP1) | AR |
| SLC26A4 | chr7:107338487 | NM_000441 | c.1546dupC | p.F515fs | Likely pathogenic | 0.00002033 | Het | Likely pathogenic (PVS1+PM2) | AR | |||
| s12 | Dai | SLC26A4 | chr7:107315543 | NM_000441 | c. 754 T > C | p.S252P | Reported | Pathogenic | 0.00000406 | Het | Likely pathogenic (PM1+PM2+PP3+PP5+BP1) | AR |
| SLC26A4 | chr7:107338487 | NM_000441 | c.1546dupC | p.F515fs | 0.00002033 | Het | Likely pathogenic (PVS1+PM2) | AR | ||||
| s13 | Dai | SLC26A4 | chr7:107315543 | NM_000441 | c. 754 T > C | p.S252P | Reported | Likely pathogenic | 0.00000406 | Hom | Likely pathogenic (PM1+PM2+PP3+PP5+BP1) | AR |
| s72 | Dai | SLC26A4 | chr7:107315543 | NM_000441 | c. 754 T > C | p.S252P | Reported | Pathogenic | 0.00000406 | Het | Likely pathogenic (PM1+PM2+PP3+PP5+BP1) | AR |
| SLC26A4 | chr7:107338487 | NM_000441 | c.1546dupC | p.F515fs | Likely pathogenic | 0.00002033 | Het | Likely pathogenic (PVS1+PM2) | AR | |||
| PL14 | Miao | LOXHD1 | chr18:44122689 | NM_144612 | c.3748 + 1 G > A |
Het | Likely pathogenic (PVS1+PM2) | AR | ||||
| LOXHD1 | chr18:44127015 | NM_144612 | c. 3357 C > G | p.Y1119X | Het | Pathogenic (PVS1+PM2+PP3) | AR | |||||
| h225 | Miao | MYO15A | chr17:18035824 | NM_016239 | c. 4264 C > T | p.Q1422X | Het | Pathogenic (PVS1+PM2+PP3) | AR | |||
| MYO15A | chr17:18049382 | NM_016239 | c.6471_6478del | p.G2157fs | Het | Likely pathogenic (PVS1+PM2) | AR | |||||
| K55 | Hani | MYO15A | chr17:18075606 | NM_016239 | c.10350 + 2 T > G |
0.00000406 | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
| V44 | Bai | MYO7A | chr11:76858866 | NM_001127180 | c.156_157 del |
p.N52fs | Hom | Likely pathogenic (PVS1+PM2) | AR/AD | |||
| V56 | Bai | MYO7A | chr11:76858866 | NM_001127180 | c.156_157 del |
p.N52fs | Hom | Likely pathogenic (PVS1+PM2) | AR/AD | |||
| s36 | Lisu | PAX3 | chr2:223086019 | NM_181461 | c.879dupG | p.F294fs | Reported | Het | Likely pathogenic (PVS1 +PM2) | AR/AD | ||
| PL9 | Hui | TMPRSS3 | chr21:43803309 | NM_001256317 | c.617-2- > CT | Hom | Likely pathogenic (PVS1+PM2) | AR | ||||
| DH45 | Achan | USH2A | chr1:215848878 | NM_206933 | c.12374_12375insAT | p.F4125fs | Hom | Likely pathogenic (PVS1+PM2) | AR | |||
AD: autosomal dominant inheritance.
AR: autosomal recessive inheritance.
ACMG: American College of Medical Genetics.
HGMD: The Human Gene Mutation Database, http://www.hgmd.cf.ac.uk.
ClinVar: http://www.ncbi.nlm.nih.gov/clinvar/.
GnomAD: The Genome Aggregation Database, http://gnomad-sg.org/.
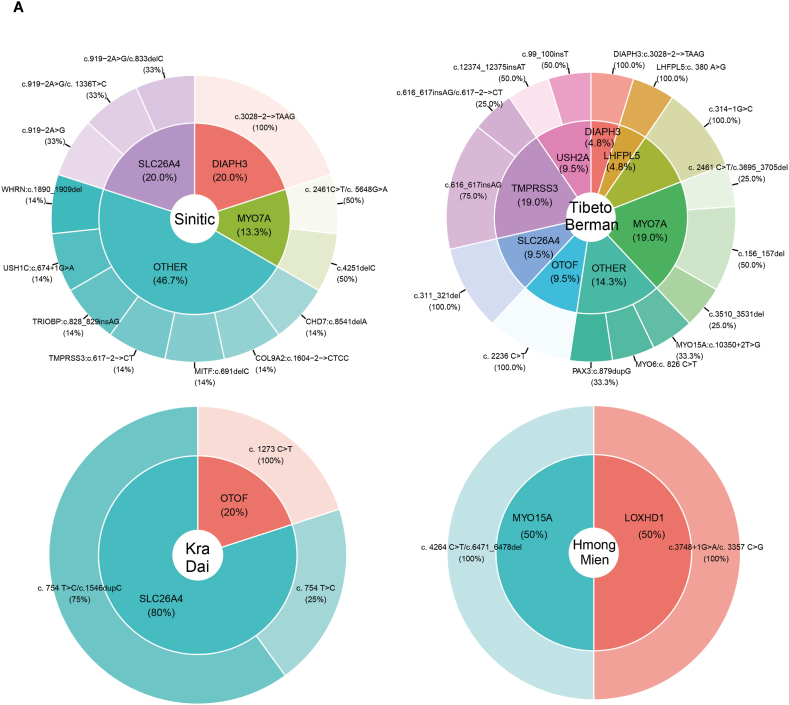
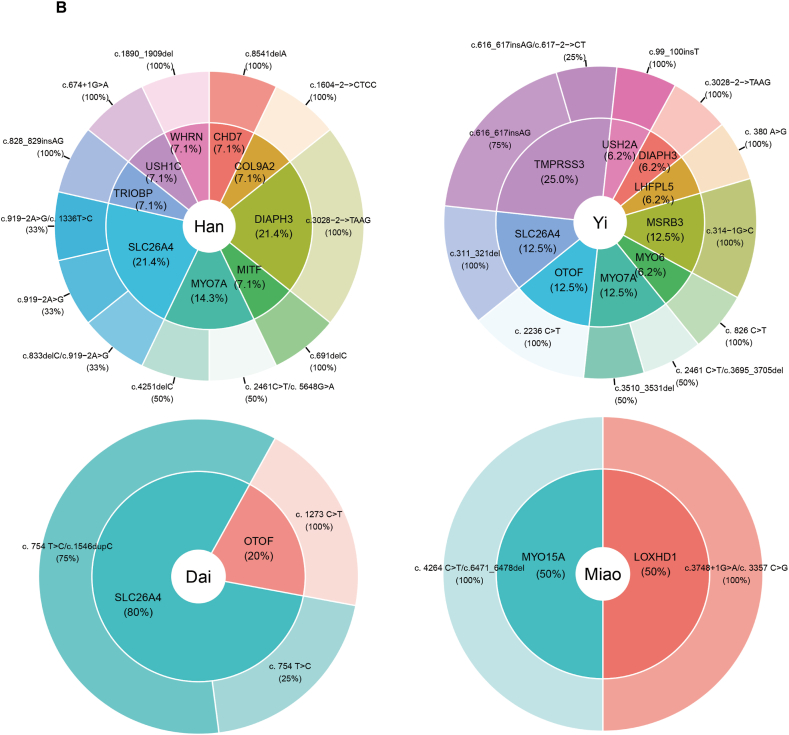
Fig. 3.
Distribution of causative DNA variants among solved patients from four language families (A) including Sinitic, Tibeto-Burman, Kra-Dai and Hmong-Mien, and four ethnicities (B) comprising Han, Yi, Dai and Miao from Southwestern China.
According to the ACMG guidelines [19], 34 pathogenic and likely pathogenic variants were identified in the solved patients, as shown in Table 1. Among these, 8 variants were documented in the HGMD databases, and 10 variants were reported in the ClinVar databases. In the solved patient group, 79.07 % (34/43) exhibited autosomal recessive inheritance, while 20.93 % (9/43) demonstrated autosomal dominant inheritance. No cases of X-linked inheritance were identified among the solved patients. Within the autosomal recessive inheritance category, 70.59 % (24/34) of patients carried homozygous variants, and 29.41 % (10/34) carried compound heterozygous variants. Interestingly, in the solved hearing-impaired individuals from ethnic minorities such as the Yi, Dai and Hani, pathogenic and likely pathogenic variants were predominantly homozygous. In contrast, among non-ethnic minority Han Chinese individuals with hearing loss, the variants appeared to be compound heterozygous. This suggests the possibility of a shared ancestral origin for these variants in ethnic minority populations.
No pathogenic or likely pathogenic copy number variations (CNVs) were detected in the solved patients. Additionally, digenic inheritance of hearing loss was not observed among our solved patient cohort.
4. Discussion
Yunnan province, situated in southwestern China, is characterized by its diverse multi-ethnic population. The distribution of hearing loss genes among individuals in this region remains unclear. In our preliminary study, we investigated hearing loss genes in a cohort of 84 patients from southwestern China. Beyond common hearing loss genes such as GJB2, SLC26A4 and MT-RNR1, we identified candidate pathogenic variants in 34 other hearing loss genes, including MYO7A, PDZD7 and OTOF [18]. To further understanding of the distribution of hearing loss genes in southwestern China, we expanded our study to encompass 347 hearing loss patients representing 15 ethnicities and four language families within the Yunnan province. Among the solved patients in the present study, the most frequently identified pathogenic genes were SLC26A4, MYO7A and TMPRSS3 (Fig. 2).
122 genes of hearing loss were sequenced and analyzed in our cohorts of patients. Notably, the diagnostic rate showed a variation of around 10 % across different language families. The etiologic diagnostic rates for individuals belonging to the Sinitic, Tibeto-Burman, Kra-Dai and Hmong-Mien language families were 12.93 %, 12.21 %, 13.51 % and 9.09 %, respectively (Fig. 1). The solved rate observed in our study differs from some previous studies. For example, in Yan et al.'s study [22], the most common genes identified were MYO15A, USH2A, MYO7A, MYO6 and TRIOBP. Their study reported a diagnostic rate of 28 % for patients from non-sub-Saharan African countries, contrasting with a 4 % rate for sub-Saharan Africa. In Sloan-Heggen et al.'s study, the diagnostic rate was 39 % in a cohort of 1119 patients with hearing loss [23], with GJB2 gene causing 21.6 % of the cases. Wu et al. [24], in a recent study involving 1027 patients with hearing loss, reported a high diagnostic rate of 57.25 %, with 20.2 % attributed to the GJB2 gene. Gu et al.'s study reported a solved rate of 13 % for strictly pre-screened sporadic patients [25]. The patients in their study were strictly pre-screened and were all sporadic patients. In a literature review by Shearer et al. [26], the diagnostic rate greatly varied (range 10 %–83 %) in different studies and different regions. It is crucial to note that the comparison among these studies has limited significance due to several factors: i) variation in the pre-screening of patients by common hearing loss genes such as GJB2 in some studies but not in others, ii) differences in the genetic backgrounds of patients across studies, and iii) inconsistency in the selection of candidate genes for hearing loss research in different studies.
Shearer et al. conducted a reassessment of reported pathogenic non-syndromic hearing loss variants [27]. Their analysis revealed that 93 variants previously classified as pathogenic were, in fact, benign. Consequently, the pathogenic and likely pathogenic variants identified in our study (Table 1) require additional verification to ensure accuracy in clinical applications. Meanwhile, standards and guidelines for the interpretation of sequence variants were continuously updated by ACMG. It is important to recognize that the interpretation of variant pathogenicity in the present study may vary based on the evidence codes of different patients (such as segregation data and functional data) and future iterations of ACMG guidelines.
CNVs have been demonstrated to be associated with hearing loss [28,29], although their contribution to the overall causation of hearing loss is relatively low [22]. In the solved cases of the present study, no pathogenic CNVs related to hearing loss were identified. It is possible that variants, rather than CNVs, are more likely to be the predominant cause of hearing loss in this region. Pathogenic CNVs often affect multiple genes, leading to syndromic disorders, while the majority of patients in our study were non-syndromic. This may explain the absence of identified pathogenic CNVs associated with hearing loss in our study.
Next generation sequencing is widely used in clinical diagnosis [17], leading to the identification of numerous newly discovered variants. According to the ACMG guidelines, most of these variants were classified as VUS, posing a significant challenge to the precise diagnosis of hereditary hearing loss and other hereditary diseases [30,31]. In the present study, the etiologic diagnosis rate in solved patients was only 12.39 %, while the percentage of unsolved patients was 82.71 % (Fig. 1). This was primarily attributed to unsolved patients harboring a large number of VUS variants (Fig. 1B and supplementary material 1), even though the clinical phenotypes of some patients were likely associated with these genes. Therefore, a thorough investigation into the pathogenicity of these variants is imperative. Functional studies, employing cellular and animal models, are needed for a comprehensive understanding.
Hearing loss is mostly managed by cochlear implants. Previous studies indicated that patients harboring pathogenic variants in genes such as GJB2, OTOF, TMPRSS3, CDH23, SLC26A4, MYO7A, MYO15A, MYTH9, ACTG1 and COCH tend to experience improved outcomes with cochlear implants [1,32]. Genetic testing plays a crucial role in aiding patients in their decision-making process regarding management options. SLC26A4, MYO7A, MYO15A, OTOF and TMPRSS3 genes were identified in our cohort of patients with hearing loss (Table 1), suggesting a potential favorable response to cochlear implantation for these patients. For cases of high-frequency sensorineural hearing loss associated with mutations in the DIAPH1 and mitochondrial MT-RNR1 genes, electro-acoustic stimulation (EAS) has shown promising outcomes [1,33]. Our previous studies on mitochondrial genes have identified MT-RNR1 variants in individuals with hearing loss from southwestern China [34], indicating the potential efficacy of EAS for these patients. It is noteworthy that gene therapy and stem cell therapy appeared to be promising for hearing loss treatment [35]. There have been multiple cases showing successful gene therapies in mice models. Recently, AAV1-hOTOF gene therapy for DFNB9 (autosomal recessive deafness9) were successful in clinical trial [36,37]. This therapy offers a novel treatment approach for children affected by DFNB9. Nonetheless, larger-scale trials with extended follow-up periods are imperative to comprehensively assess the safety and efficacy of gene therapy and stem cell therapy.
5. Conclusion
In summary, the etiological diagnostic rate among our cohorts of patients with hearing loss was relatively low by using a panel testing with 122 genes. Based on these findings, several recommendations have been proposed regarding genetic diagnostic methods and management for hearing loss. First, numerous variants of VUS were detected in our patients, encompassing 15 ethnicities from southwestern China. It is essential to validate the pathogenicity of these variants to enhance the overall diagnostic rate for patients in this region, potentially through the establishment of ethnicity-specific genomic databases for hearing loss. Second, Whole Exome Sequencing (WES) and Whole-genome sequencing (WGS) seem to be more promising in the future. However, significant challenges such as high costs, huge data analysis and the interpretation of VUS need to be carefully considered and resolved. Third, even if a genetic testing of hearing loss is negative, the possibility of a genetic etiology might still remain. This point is important to emphasize, as it may be misunderstood by clinicians, patients and their families.
Funding
The present study was supported by the National Natural Science Foundation of China (82360331,81860190), Yunnan Provincial Talents Program for Top Young Talents (YNWR-QNBJ-2018-128), the Joint Special Research Funds of Kunming Medical University (2017FE468 [-010]), the Foundation of Medical Discipline Leaders Program of Health and Family Planning Commission of Yunnan Province (D-201668) and the Personnel Training Project of Yunnan Province(2017HB043).
Data availability statements
Data will be made available on request.
CRediT authorship contribution statement
Jingyu Li: Investigation, Data curation. Shiyu Zhou: Formal analysis, Data curation. Jiahong Pei: Formal analysis, Data curation. Wanzhen Li: Formal analysis, Data curation. Rongjie Cui: Data curation. Xiaofei Ren: Data curation. Jingru Wei: Data curation. Qian Li: Conceptualization. Baosheng Zhu: Formal analysis, Data curation. Yaliang Sa: Data curation. Yunlong Li: Writing – review & editing, Writing – original draft, Formal analysis, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Ms. Suyun Li, Ms. Na Feng and Ms. Huayan Wang for collecting samples, and all the participants in the present study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38802.
Contributor Information
Yaliang Sa, Email: sayalian@126.com.
Yunlong Li, Email: yunlongli.km@qq.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sharma N., et al. A systematic review of the monogenic causes of Non-Syndromic Hearing Loss (NSHL) and discussion of Current Diagnosis and Treatment options. Clin. Genet. 2023;103(1):16–34. doi: 10.1111/cge.14228. [DOI] [PubMed] [Google Scholar]
- 2.Morton C.C., Nance W.E. Newborn hearing screening--a silent revolution. N. Engl. J. Med. 2006;354(20):2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 3.Fink D. Review of hearing loss in children. JAMA. 2021;325(12):1223–1224. doi: 10.1001/jama.2021.0387. [DOI] [PubMed] [Google Scholar]
- 4.Brewer C.C., King K.A. Genetic hearing loss: the audiologist's perspective. Hum. Genet. 2022;141(3–4):311–314. doi: 10.1007/s00439-021-02360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angeli S., Lin X., Liu X.Z. Genetics of hearing and deafness. Anat. Rec. 2012;295(11):1812–1829. doi: 10.1002/ar.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nance W.E. The genetics of deafness. Ment. Retard. Dev. Disabil. Res. Rev. 2003;9(2):109–119. doi: 10.1002/mrdd.10067. [DOI] [PubMed] [Google Scholar]
- 7.Finsterer J., Fellinger J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. Int. J. Pediatr. Otorhinolaryngol. 2005;69(5):621–647. doi: 10.1016/j.ijporl.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Duman D., Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci (Landmark Ed) 2012;17(6):2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D., Liu X.Z. Cochlear molecules and hereditary deafness. Front. Biosci. 2008;13:4972–4983. doi: 10.2741/3056. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., et al. Identification of four novel mutations in MYO7A gene and their association with nonsyndromic deafness and Usher Syndrome 1B. Int. J. Pediatr. Otorhinolaryngol. 2019;120:166–172. doi: 10.1016/j.ijporl.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 11.Huang S., et al. The relationship between the p.V37I mutation in GJB2 and hearing phenotypes in Chinese individuals. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0129662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng S.B., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bademci G., et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet. Med. 2016;18(4):364–371. doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shearer A.E., et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tekin D., et al. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear. Res. 2016;333:179–184. doi: 10.1016/j.heares.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King P.J., et al. Etiologic diagnosis of nonsyndromic genetic hearing loss in adult vs pediatric populations. Otolaryngol. Head Neck Surg. 2012;147(5):932–936. doi: 10.1177/0194599812453553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shearer A.E., et al. Advancing genetic testing for deafness with genomic technology. J. Med. Genet. 2013;50(9):627–634. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., et al. Targeted next-generation sequencing of deaf patients from Southwestern China. Mol Genet Genomic Med. 2021;9(4):e1660. doi: 10.1002/mgg3.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. 2017;100(2):267. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nord A.S., et al. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genom. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan D., et al. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum. Genet. 2016;135(8):953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sloan-Heggen C.M., et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016;135(4):441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J., et al. Molecular diagnose of a large hearing loss population from China by targeted genome sequencing. J. Hum. Genet. 2022;67(11):643–649. doi: 10.1038/s10038-022-01066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu X., et al. Genetic testing for sporadic hearing loss using targeted massively parallel sequencing identifies 10 novel mutations. Clin. Genet. 2015;87(6):588–593. doi: 10.1111/cge.12431. [DOI] [PubMed] [Google Scholar]
- 26.Shearer A.E., Smith R.J. Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol. Head Neck Surg. 2015;153(2):175–182. doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shearer A.E., et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am. J. Hum. Genet. 2014;95(4):445–453. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rentas S., Abou Tayoun A. Utility of droplet digital PCR and NGS-based CNV clinical assays in hearing loss diagnostics: current status and future prospects. Expert Rev. Mol. Diagn. 2021;21(2):213–221. doi: 10.1080/14737159.2021.1887731. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg C., et al. Genomic copy number alterations in non-syndromic hearing loss. Clin. Genet. 2016;89(4):473–477. doi: 10.1111/cge.12683. [DOI] [PubMed] [Google Scholar]
- 30.Tsai A.C., Liu X. Toward best practice in using molecular diagnosis to guide medical management, are we there yet? N. Am. J. Med. Sci. 2014;7(4):199–200. [PMC free article] [PubMed] [Google Scholar]
- 31.Aronson S.J., Rehm H.L. Building the foundation for genomics in precision medicine. Nature. 2015;526(7573):336–342. doi: 10.1038/nature15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong X., et al. Advances in cochlear implantation for hereditary deafness caused by common mutations in deafness genes. J Bio-X Res. 2019;2(2):74–80. [Google Scholar]
- 33.Incerti P.V., Ching T.Y.C., Cowan R.J.T.i.H. A systematic review of electric-acoustic stimulation. 2013;17(1) doi: 10.1177/1084713813480857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S., et al. Distribution of mitochondrial MT-RNR1, MT-TL1, MT-TS1, MT-TK and MT-TE genes variants associated with hearing loss in Southwestern China. Int. J. Pediatr. Otorhinolaryngol. 2024;181 doi: 10.1016/j.ijporl.2024.111979. [DOI] [PubMed] [Google Scholar]
- 35.Jiang L., et al. Advances in gene therapy hold promise for treating hereditary hearing loss. Mol. Ther. 2023;31(4):934–950. doi: 10.1016/j.ymthe.2023.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv J., et al. AAV1-hOTOF gene therapy for autosomal recessive deafness 9: a single-arm trial. 2024;403(10441):2317–2325. doi: 10.1016/S0140-6736(23)02874-X. [DOI] [PubMed] [Google Scholar]
- 37.Akil O., et al. Dual AAV-mediated gene therapy restores hearing in a DFNB9 mouse model. Proc. Natl. Acad. Sci. U.S.A. 2019;116(10):4496–4501. doi: 10.1073/pnas.1817537116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.