Abstract
Adipose tissue is traditionally classified into two main types based on their functions: brown adipose tissue (BAT) and white adipose tissue (WAT). Each type plays a distinct role in the body's energy metabolism. Additionally, a third type, beige adipose tissue, can develop within subcutaneous WAT (including inguinal WAT, iWAT) in response to specific stimuli and exhibits characteristics of both BAT and WAT. Extracellular vesicles (EVs) are crucial for intercellular communication, carrying a diverse array of biomolecules such as proteins, lipids, and nucleic acids. While the functional diversity and endocrine roles of adipose tissues are well-documented, a comparative analysis of the functions of EVs released by different adipose tissues from mice housed at room temperature has not been thoroughly explored. MicroRNAs (miRNAs), which are highly enriched in small extracellular vesicles (sEVs), offer a promising avenue for investigating the complex functions and unique roles of various adipose tissues. In this study, we isolated sEVs from different adipose tissues under basal conditions and performed a comprehensive analysis of their miRNA content. By comparing miRNA profiles across different adipose tissues, we aim to elucidate the potential roles of sEV-derived miRNAs in mediating intercellular communication and the distinct physiological functions of adipose tissues. Understanding the molecular features of miRNAs in adipose tissue EVs could reveal new aspects of adipose tissue biology and lay the groundwork for further research into their physiological significance.
Keywords: Adipose tissues, Small extracellular vesicles, miRNAs, Functional analysis
Graphical abstract
Highlights
-
•
Analysis of miRNA expression patterns in small extracellular vesicles (sEVs) derived from different adipose tissues.
-
•
Compare and analyze the possible functions of EV-miRNAs derived from different adipose tissues.
-
•
Exploring the potential roles of adipose tissue-enriched sEV-miRNAs in regulating systemic homeostasis.
1. Introduction
Adipose tissue is traditionally classified into two main types based on their functions: brown adipose tissue (BAT) and white adipose tissue (WAT) [1]. Each type plays a distinct role in the body's energy metabolism. BAT is primarily involved in non-shivering thermogenesis, a process facilitated by uncoupling protein-1 (UCP-1) in its mitochondria, which converts chemical energy into heat [[2], [3], [4]].This thermogenic ability is crucial for BAT's role in thermoregulation and its potential use in obesity management through increased energy expenditure [5,6].On the other hand, WAT serves as the main energy reservoir, storing lipids as triglycerides within large, unilocular adipocytes [7]. Additionally, there is a third type of adipose tissue—beige adipose tissue—which can develop within subcutaneous WAT in response to specific stimuli, such as cold exposure or physical activity in both rodents and humans. Beige adipose tissue exhibits characteristics of both BAT and WAT, including thermogenic potential and lipid storage capabilities [8]. Adipocytes play a central role in maintaining metabolic homeostasis and preventing various diseases. Dysfunction in adipocytes is linked to a range of conditions, including metabolic syndrome, cardiovascular diseases, type 2 diabetes, and several cancers [[9], [10], [11], [12]].This study examines adipose tissues based on their anatomical locations and histological features in mice, distinguishing between interscapular BAT (iBAT), inguinal WAT (iWAT), and epididymal WAT (eWAT). iWAT, a type of subcutaneous adipose tissue, primarily stores energy but can also develop beige characteristics under certain conditions. eWAT, a type of visceral adipose tissue, is located around the epididymis in mice.
Extracellular vesicles (EVs) are recognized as key mediators of intercellular communication [13]. EVs carrying a wide range of biomolecules, including proteins, lipids, and nucleic acids, with microRNAs (miRNAs) being particularly prominent. EVs are secreted by virtually all cell types and are abundant in body fluids, playing significant roles in both physiological and pathological contexts [14]. Small extracellular vesicles (sEVs), which are less than 200 nm in diameter, have been extensively studied, especially in relation to pathological conditions such as obesity [15,16], type 2 diabetes [17], melanoma [18,19], and colorectal cancer [20,21].
microRNAs (miRNAs), crucial regulators of gene expression, influence various biological pathways and are involved in processes such as adipocyte differentiation, maturation, and metabolism [[22], [23], [24]]. These miRNAs, which are known to be enriched in sEVs, can be secreted by various cells to exert their functions systemically [25]. Research has demonstrated the role of adipose tissue-derived sEVs in regulating physiological homeostasis. For example, Thomou et al. found that transplantation of both white and brown adipose tissues, particularly brown adipose tissue, into ADicerKO mice restored the levels of several circulating miRNA [26]. Our previous study also showed that miR-378a-3p, which is predominantly expressed in iBAT, can be transferred to the liver via sEVs during cold exposure, thereby stimulating hepatic gluconeogenesis in male mice [27]. Additionally, emerging research on EVs from adipocyte-derived mesenchymal cells (ADSC-EVs) has highlighted their potential benefits, including anti-inflammatory effects [26]. However, a comparative analysis of the distinct functions of EVs released by iBAT, iWAT, and eWAT from mice housed at room temperature has not yet been thoroughly explored.
Characterizing and analyzing sEV-derived miRNAs can offer significant insights into the regulatory mechanisms of adipose tissue function. This study involves the isolation of sEVs from adipose tissue, characterization of miRNAs derived from these sEVs, and an analysis of their content to clarify their roles in intercellular communication and the promotion of tissue-specific functions. Gaining an understanding of the molecular characteristics of miRNAs in adipose tissue-derived further exploration of their physiological roles.
2. Methods
2.1. Animals
10-week-old male and female C57BL/6J WT mice were obtained from Gem-Pharmatech Laboratory (Nanjing, China). The experimental animals were individually housed in a specific pathogen-free (SPF) facility at Nanjing University, maintained at 25 ± 2 °C and a relative humidity of 55 ± 10 %, and provided with ad libitum access to pellet normal chow diet and water, and were kept on a 12-h light/12-h dark cycle.
Euthanasia of mice was performed by inhalation of carbon dioxide, followed by immersion of the mouse body in 75 % ethanol for 5 min. Three types of adipose tissues were carefully dissected using scissors and forceps. Further details of in vivo experiments were provided for each respective section.
2.2. Hematoxylin–eosin (H&E) staining
For hematoxylin–eosin (H&E) staining, mouse tissues were promptly preserved in 4 % formaldehyde for 24 h, rinsed with 70 % ethanol, embedded in paraffin, and serially sectioned at 5 μM. The slides were visualized using an Olympus microscope CX31. Subsequently, adipocyte diameter measurements were conducted using NIH ImageJ software (NIH, Bethesda, MD, USA) in a minimum of three fields per slide (20 × magnification).
2.3. Small extracellular vesicles (sEVs) collection
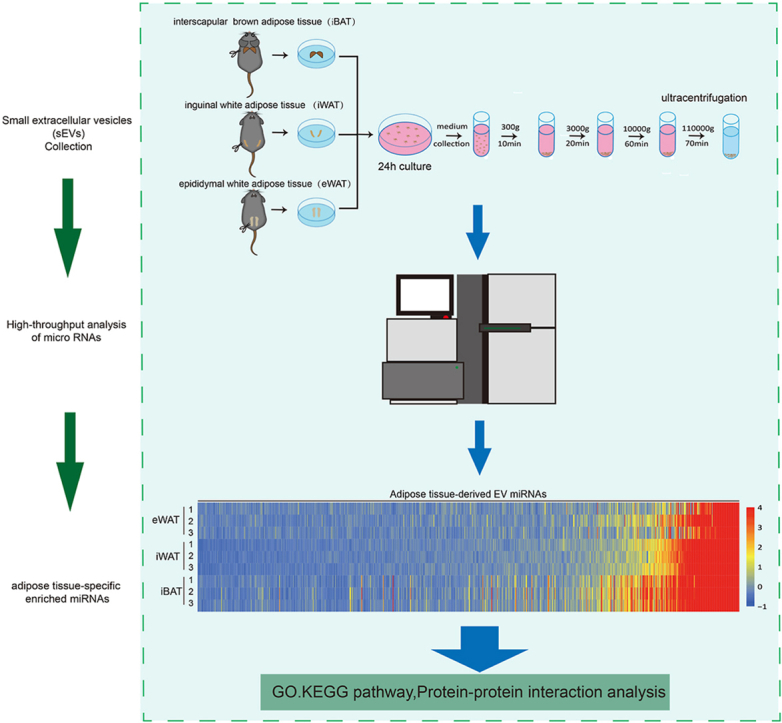
Small extracellular vesicles (EVs) derived from iBAT, iWAT, and eWAT were isolated using ultracentrifugation.The process involved excising total iBAT, iWAT, and eWAT from mice and transferring them into 2 ml DMEM with 1 % P/S, followed by gentle cutting into 2 mm3 pieces, respectively. The chopped tissues were then centrifuged for 5 min at 1000×g at room temperature, and the resulting pellets were resuspended in DMEM before being transferred into a Petri dish. After a 30-min incubation in DMEM containing 2 % EV-free FBS (YOBIBIO cat#U45-852A), 0.5 mg/ml DNase I, 0.2 mg/ml RNase A, and 1 % P/S, the culture medium was replaced with DMEM containing 2 % EV-free FBS. The iBAT, iWAT, and eWAT tissues were then incubated for 24 h at 37 °C, 5 % CO2, respectively, and the culture supernatant was collected for EV isolation using the ultracentrifugation method.
In summary, the medium underwent centrifugation at 300×g for 10 min, 3000×g for 20 min, and 10,000×g for 30 min to remove tissues, cells, debris, and medium/large vesicles. The resulting supernatant was filtered through a 0.22 μm filter and then subjected to ultracentrifugation at 110,000×g using a 70Ti fixed-angle rotor placed in a Beckman Coulter Optima LE-80K ultracentrifuge for 70 min at 4 °C to pellet EVs. The EV collections were washed once, resuspended in PBS, and then centrifuged a second time at 110,000×g for 16 h at 4 °C. Finally, the EV pellet was resuspended in DMEM or PBS.
2.4. Transmission electron microscopy (TEM)
The isolated EVs suspension was applied to a copper grid with carbon film and allowed to sit for 3–5 min. Excess liquid was then absorbed using filter paper. Subsequently, 2 % phosphotungstic acid (Servicebio, cat# G1102) was added to the copper grid to stain the EVs for 1–2 min, and excess liquid was again absorbed using filter paper. After the grids were air-dried, they were observed using a HT7800 transmission electron microscope manufactured by HITACHI in Tokyo, Japan, and operated at 120 kV.
2.5. Nanoparticle tracking analysis (NTA)
To analyze the size distribution o Evs, a dark-field microscope Nanosight NS300 manufactured by Malvern Panalytical in Amesbury, UK, was utilized. The EV samples were initially diluted with saline to achieve a concentration of 107–108 particles/mL for analysis. Each sample was measured in triplicate using the camera and then analyzed using NanoSight NTA 3.2 software.
2.6. Western blot analysis
To identify Evs markers, the Evs collected from iBAT, iWAT, and eWAT were dissolved in PBS, and the proteins were extracted for Western blot analysis. Evs,cells or tissues were lysed in ice-cold RIPA buffer (Beyotime, cat#P0013B) supplemented with a protease and phosphatase inhibitor cocktail. The protein concentration was measured using the Pierce™ BCA protein assay kit (Thermo Scientific, cat#23227). The following antibodies were used for Western blot analysis: anti-UCP1 (1:1000) antibody(Cell Signaling Technology, cat#72298),anti-Alix (1:1000) antibody (ABclonal, cat#A2215), anti-CD9 (1:1000) antibody(Cell Signaling Technology, cat#13174), anti-CD63 (1:1000) antibody(Cell Signaling Technology, cat#52090), anti-p-AKT (1:1000) antibody (Cell Signaling Technology, cat#4060), anti-AKT (1:1000) antibody(Cell Signaling Technology, cat#9272), anti-LAMIN A/C (1:10000) antibody(ABclonal, cat#A19524), anti-TGFBI (1:1000) antibody(ABclonal, cat#A21964), anti-LDHA/B (1:1000) antibody (ABclonal, cat#A21893). Following the Western blot procedure, the results were analyzed using ImageJ. The signal of each image was normalized such that the average of all control samples equaled 1.0.
2.7. Cell migration assay
The migratory capacity of the human hepatoma cell line HepG2 (ATCC, HB-8065) was assessed using a wound healing assay. For the wound healing assay, HepG2 cells were plated in 6-well plates with DMEM containing 10 % FBS and incubated at 37 °C with 5 % CO2 for 24 h until the cells reached 70–80 % confluence. A straight scratch was made on the cell monolayer using a sterile 20 μL disposable serological pipette tip. The cells were washed with 1 mL PBS to remove debris and smooth the edges of the scratch, then the medium was replaced with DMEM containing 2 % EV-Free FBS for co-culture. iBAT-sEV, iWAT-sEV, and eWAT-sEV were added to three experimental groups for 24 h of co-culture, with an sEV concentration of 10000 particles/cell. Images of cell proliferation were captured using a microscope at 0 and 24 h post-scratch. The percentage of migrated area was calculated using ImageJ software (version 1.48v, National Institutes of Health, USA).
2.8. Cell proliferation assay
Cell viability was measured using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. Briefly, cells were seeded into 96-well plates and cultured with DMEM containing 10 % FBS for 24 h. The medium was then replaced with DMEM containing 2 % EV-Free FBS, and iBAT-sEV, iWAT-sEV, and eWAT-sEV were added to the respective experimental groups for 24 h of co-culture, with an sEV concentration of 10000 particles/cell. The medium was changed to complete medium containing 10 % FBS, and CCK-8 reagent (10 μL per well) was added, followed by further incubation at 37 °C and 5 % CO2 for 2–4 h. The rate of cell proliferation was determined by measuring the absorbance at 450 nm.
2.9. Isolation of adipose SVF cells
Mice were euthanized, and the tissues of iBAT/iWAT/eWAT were harvested and soaked in 75 % ethanol for 5 min, followed by washing in PBS containing 1 % PS to remove hair. The tissues were minced into pieces of approximately 1 mm3 and digested in a mixed enzyme solution containing collagenase D (Sigma, cat#11088866001) and Dispase II (Sigma, cat#4942078001) at 37 °C with shaking at 150 rpm for 30 min. After the addition of 5 mL of complete growth medium to terminate the digestion, the mixture was centrifuged at 700g for 10 min to remove excess lipids from the supernatant. The pellet was resuspended, filtered through a 70 μm cell strainer, centrifuged again at 700g for 10 min, and the supernatant was discarded. The resulting pellet was resuspended in complete growth medium, and the cells were plated in culture dishes. After 2 h, the medium was changed to remove excess blood cells and other contaminating cells, and the cells were allowed to grow for approximately 72 h at 37 °C in a 5 % CO2 incubator.
2.10. sEV Co-culture experiment
Adipose SVF cells from iBAT, iWAT, and eWAT were plated in 6-well plates and grown at 37 °C in a 5 % CO2 incubator for 24 h. The medium was replaced with DMEM containing 2 % EV-Free FBS, and sEVs extracted from the corresponding adipose tissues were added to the cultures at a concentration of 10,000 particles per cell for 24 h of co-culture. In the context of iWAT and eWAT stromal vascular fraction (SVF) cells, an additional experimental group was warranted. Following a 12-h co-culture with sEVs, a further supplementation of 5 nM insulin was introduced for an additional 12 h of incubation. Subsequently, the cells were harvested for the assessment of relevant markers using quantitative polymerase chain reaction (q-PCR) or Western blot analysis.
2.11. Total RNA isolation
RNA extraction from cultured cell lines or tissue samples was performed using RNAiso Plus (Takara, cat#9109), according to protocols provided by the supplier. Utilizing 1 μg of the extracted RNA, reverse transcription to synthesize first-strand cDNA was conducted using HiScript III RT SuperMix (Vazyme, cat#R323). Subsequent analysis of the synthesized cDNA involved quantitative PCR, employing ChamQ Universal SYBR qPCR Master Mix (Vazyme, cat#Q711) on a LightCycler®480 system (Roche Diagnostics, Mannheim, Germany). The PCR thermal cycling parameters were set as follows: an initial denaturation at 95 °C for 5 min, succeeded by 40 amplification cycles (95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s). Quantification of relative mRNA levels was executed through normalization against β-actin expression, applying the 2 -ΔΔCT technique for calculation.
2.12. Microarray analysis for detection of microRNA
In microarray studies, samples of iBAT, iWAT, and eWAT were harvested from male C57BL/6J mice at 10 weeks of age. For each tissue type, sEV samples were collected from 6 mice, with pooled samples from 2 distinct mice, resulting in a total of three composite sample pools. Total RNA was extracted from these pooled samples using TRIzol reagent, in preparation for subsequent analysis via the Affymetrix miRNA microarray platform, a service provided by Nanjing Hepu Biotechnology Co., Ltd. Briefly, after RNA extraction, the miRNA molecules were labeled with a fluorescent dye. The labeled miRNAs were then hybridized to an Affymetrix miRNA microarray. After washing away nonspecifically bound molecules, the microarray was scanned to detect the fluorescence emitted from the labeled miRNAs. The intensity of the fluorescence signal was measured and used to determine the expression levels of specific miRNAs. Consequently, after subtracting the background signal and normalizing across different samples or experiments, the resultant fluorescence signal intensity reflects the relative expression level of each miRNA.
2.13. Functional enrichment analysis
MiRNAs identified in EVs from iBAT, iWAT, and eWAT were subjected to Gene Ontology (GO) and KEGG pathway analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8 [[28], [29], [30]]. The protein–protein interaction (PPI) network analysis was performed by applying the STRING® programme version 11 [31,32]. The resultant PPI images were then imported into Cytoscape_version 3.10.1 for a better resolution based on adjustment of the node interaction degree and scores [33,34].
2.14. Quantification and statistical analysis
Statistical analysis was conducted using GraphPad Prism 9 software. Firstly, the normality of the data was assessed using either D'Agostino-Pearson's normality test (n > 7) or Shapiro-Wilk's test (n ≤ 7). The results are presented as the mean ± SEM. GPower 3.1 was used to determine effect size and power values for data with significant differences. Specific tests used for statistical analyses are indicated in the figure legends, and further statistical details can be found in the Source Data.
3. Results
3.1. Characterization of sEVs isolated from different adipose tissues of mice
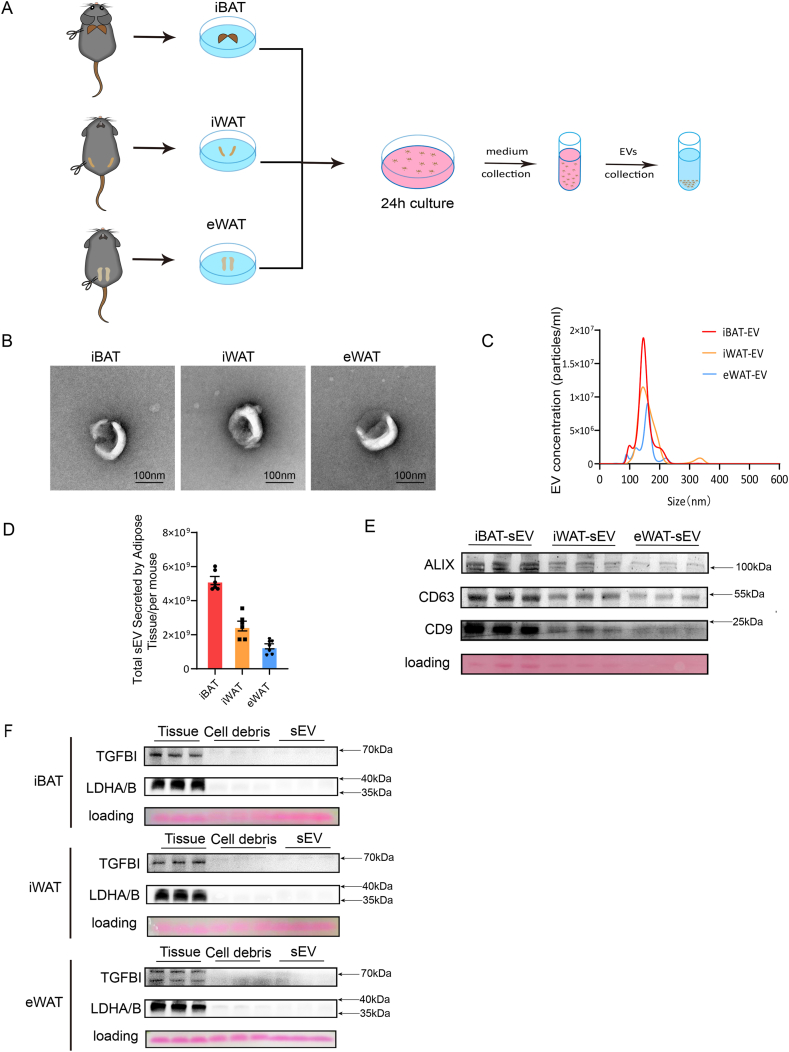
To investigate the physiological functions of iBAT, iWAT, and eWAT, adipose tissues were isolated from C57BL/6 mice housed at room temperature (25 °C). Consistent with previous reports, Hematoxylin and Eosin (H&E) staining revealed that eWAT is characterized by larger unilocular lipid droplets, iWAT contains relatively smaller lipid droplets, and iBAT features the smallest lipid droplets with a multilocular structure, as also confirmed by cell size quantification (Supplementary Figures S1A and S1B). The expression of UCP1 in iBAT was significantly higher compared to iWAT and eWAT at room temperature (Supplementary Figures S1C and S1D). The small extracellular vesicles (sEVs) were isolated from the three different adipose tissues using ultracentrifugation (Fig. 1A). Transmission electron microscopy (TEM) images demonstrated that the sEVs from all three sources maintained a typical EV morphology (Fig. 1B). Under electron microscopy, the vesicles displayed distinct membrane boundaries and appeared as cup-shaped or saucer-like vesicular structures with a diameter of approximately 100 nm, with no significant morphological or size differences among the sEVs from these different adipose tissues. Nanoparticle tracking analysis (NTA) revealed that although iBAT had the lowest mass, it secreted the highest number of sEVs, followed by iWAT, with eWAT secreting the fewest sEVs (Supplementary Figure S1E and Fig. 1C–D). The levels of EV marker proteins, including CD9, CD63, and ALIX, differed among iBAT, iWAT, and eWAT, reflecting the variations in sEV secretion (Fig. 1E), which aligns with the NTA findings. Additionally, we examined the presence of non-vesicular extracellular particles (NVEPs), such as TGFBI and LDHA/B, which were typically enriched in exomeres or supermeres, in the isolated sEV samples. Fig. 1F shows that TGFBI and LDHA/B were minimally detected in sEVs from iBAT, iWAT, or eWAT, in contrast to their abundant expression in the corresponding tissues. Moreover, the nuclear protein Lamin A/C was not detected in sEVs (Figure S1F). Therefore, based on the established characteristics of EVs and the analysis of EV markers, the isolated sEVs demonstrate a relatively high level of purity.
Fig. 1.
Isolation and characterization of sEVs from different adipose tissues of mice. (A) Schematic diagram of the experimental procedure for the isolation of sEVs. (B) Transmission electron microscopy (TEM) images of sEVs released from iBAT, iWAT, and eWAT. (C and D) Nanoparticle tracking analysis (NTA) and quantification of sEVs derived from iBAT, iWAT, and eWAT (n = 6 each group, from 2 independent experiments). (E) Western blot detection of EV markers ALIX, CD63, and CD9 in sEVs derived from iBAT, iWAT, and eWAT (n = 6 each group, from 2 independent experiments). (F) Western blot analysis of non-vesicular extracellular particles (NVEPs) TGFBI and LDHA in the tissue, cell debris, and their derived sEVs from iBAT, iWAT, and eWAT (n = 6 each group, from 2 independent experiments).
3.2. Comparative analysis of sEV-Derived miRNA expression profiles in different adipose tissues
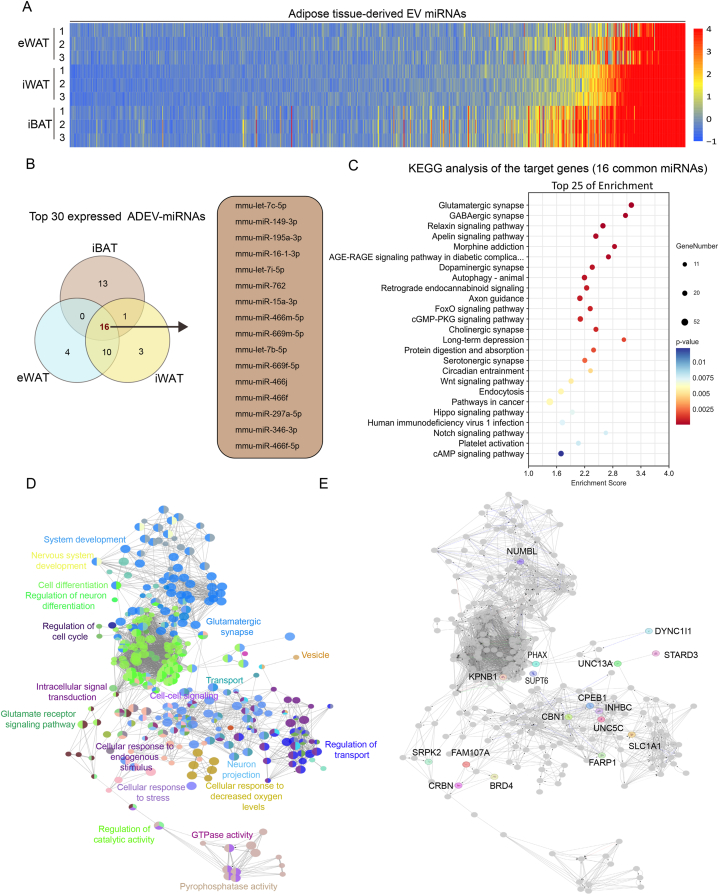
In our comparative study, we conducted a high-throughput analysis to identify miRNAs derived from sEVs in iBAT, iWAT, and eWAT. The heatmap reveals a distinct expression landscape, indicating different miRNA expression levels (Fig. 2A). For each type of adipose tissue, we selected the top 30 most abundantly expressed sEV-derived miRNAs from the 850 detected miRNAs in iBAT, iWAT, and eWAT for further analysis (Supplementary Table S1 and Table S2). The Venn diagram illustrates both shared and unique sEV-derived miRNAs among these adipose tissues (Fig. 2B). Some miRNAs were found to be commonly expressed, potentially reflecting their fundamental regulatory roles, while others were tissue-specific, suggesting specialized functions within their respective adipose depots. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis further clarified the roles of the predicted targets of 16 commonly enriched sEV-derived miRNA across all three types of adipose tissue, highlighting their involvement in essential biological pathways (Fig. 2C and Supplementary Table S3). Significant enrichment was observed in pathways related to neuronal guidance and intracellular signaling, indicating the possible impact of these miRNAs on adipose tissue function. Gene Ontology (GO) analysis was performed to categorize the genes targeted by the 16 common sEV-derived miRNAs based on functional annotations (Fig. 2D). The proximity of the dots represents the strength of the association between these target genes within specific biological processes (BP). We observed tight clusters related to 'neuron projection', 'cell-cell signaling,' and 'GTPase activity', suggesting a coordinated regulatory network that may influence physiological outcomes within adipose tissues and intercellular regulation. The molecular interaction networks mapped the potential pathways and interactions governed by the 16 common sEV-derived miRNAs (Fig. 2E). These networks highlight key regulatory genes, which could be crucial for adipose tissue signaling and homeostasis. Notable genes such as 'NUMBL' and 'KPNB1′ emerged as central nodes in the regulation of adipose tissue function through sEV-miRNA signaling, which may deserve further experimental exploration.
Fig. 2.
Comparative Analysis of sEV-Derived miRNA Expression Profiles in Different Adipose Tissues. (A) The heatmap illustrates the expression levels of EV-derived miRNAs from eWAT, iWAT, and iBAT, The color scale represents the correlation between color and expression levels, with intensity ranging from low (blue) to high (red). For each tissue type, samples were collected from 6 mice, with each row representing pooled samples from 2 distinct mice, resulting in a total of three composite sample pools. (B) Among the top 30 expressed adipose-derived extracellular vesicle-miRNAs (ADEV-miRNAs), there are 16 miRNAs common to the three types of adipose tissues. (C) The bubble chart of KEGG enrichment analysis for the target genes of the 16 common miRNAs, with p-values indicating the statistical significance of each pathway. (D) GO analysis of the pathways related to the target genes of the 16 common miRNAs, conducted using DAVID Bioinformatics Resources and categorized based on biological processes, cellular components, and molecular functions, with each color representing a different category. (E) Molecular interaction network based on the pathways related to the target genes of the 16 common miRNAs.
3.3. Analysis of iBAT-enriched sEV-derived miRNAs and their possible functions
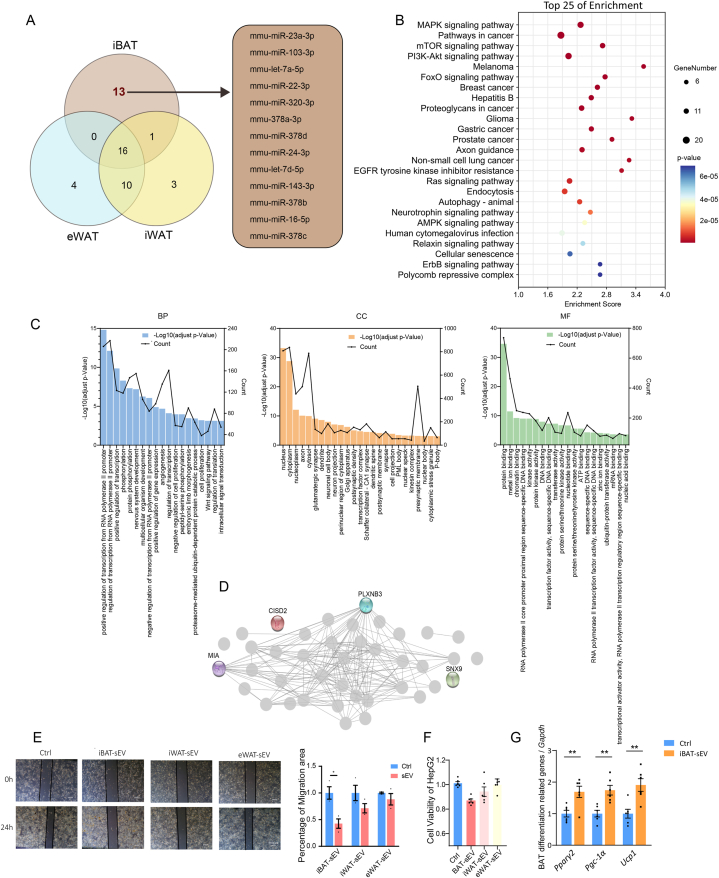
Among the top 30 miRNAs expressed in sEVs from various adipose tissues, 13 miRNAs were uniquely enriched in sEVs derived from iBAT (Fig. 3A). Notably, mmu-miR-23a and mmu-miR-378a have been shown to play specialized regulatory roles within brown adipose tissue [27,35]. KEGG pathway analysis revealed that these iBAT-specific miRNAs target genes involved in key pathways that regulate essential physiological processes, including cell growth, survival, differentiation, and metabolism (Fig. 3B and Supplementary Table S4). For instance, the MAPK signaling pathway is crucial for cell differentiation, growth, and apoptosis; the PI3K-Akt pathway regulates cell survival, growth signaling, and glucose homeostasis; and the mTOR pathway is a key regulator of cellular metabolism, growth, and proliferation. Interestingly, several of these pathways are associated with cancers such as lung cancer, melanoma, breast cancer and prostate cancer. This observation aligns with recent reports suggesting that activation of brown adipose tissue may suppress tumor growth [35,36], highlighting the potential importance of iBAT-specific miRNAs in both cancer research and broader physiological contexts. The Gene Ontology (GO) analysis, combined with STRING network prediction, revealed four key genes targeted by the 13 iBAT-enriched sEV-derived miRNAs, including plexin B3 (PLXNB3), sorting nexin 9 (SNX9), melanoma inhibitory activity (MIA), and CDGSH iron sulfur domain (CISD2) (Fig. 3C and D), suggesting they may represent crucial regulatory nodes in the network. To explore the impact of these BAT-sEV enriched miRNAs on cancer, we co-cultured iBAT-derived sEVs (iBAT-sEVs) with HepG2 cells. As shown in Fig. 3E and F, treatment with iBAT-sEVs significantly reduced the migratory capacity and viability of HepG2 cells. Additionally, treatment with iWAT-derived sEVs resulted in a modest reduction in both the migratory capacity and viability of HepG2 cells, although the effect was less pronounced compared to that seen with iBAT-derived sEVs. In contrast, eWAT-derived sEVs had minimal impact on the migratory capacity and viability of HepG2 cells (Fig. 3E and F).
Fig. 3.
Comprehensive Analysis of iBAT-Specific EV-derived miRNA and Their Unique Profiles. (A) Venn diagram illustrating a subset of 13 extracellular vesicle (EV)-derived microRNAs (miRNAs) exclusive to iBAT. (B) KEGG enrichment analysis bubble plot for the target genes of 13 iBAT-specific EV-derived miRNAs, with p-values indicating the statistical significance of each pathway. (C) GO analysis of the pathways associated with the target genes of iBAT-specific EV-derived miRNAs, conducted using DAVID Bioinformatics Resources and categorized by biological process, cellular component, and molecular function, with each color representing a different category.(D) Molecular interaction network of the target genes of 13 iBAT-specific EV-derived miRNAs, highlighting key nodes that represent genes with a high degree of connectivity. (E) Graphical representation of the quantitative analysis of HepG2 cell migration, expressed as the percentage of wound closure relative to the initial scratch area at time 0. Scale bars: 200 μm. (F) Assessment of HepG2 cell viability using the Cell Counting Kit-8 (CCK-8) assay (n = 6 each group, from 2 independent experiments). (G) Relative expression levels of genes associated with iBAT differentiation (n = 6 each group, from two independent experiments).
Moreover, we investigated the functions of iBAT-derived sEVs by co-culturing them with SVF-derived cells from iBAT. We found that iBAT-sEVs significantly enhanced the differentiation of preadipocytes in iBAT-SVF, evidenced by increased expression of genes such as Pparγ2, Pgc1α, and Ucp1 (Fig. 3G). This suggests that iBAT-sEVs also play important autocrine and paracrine roles in maintaining specific thermogenic functions in iBAT. The same phenomenon was observed for iBAT-sEV derived from female mice (Supplementary Figure S2).
3.4. Analysis of iWAT-specific enriched miRNAs and overlapping miRNAs with iBAT
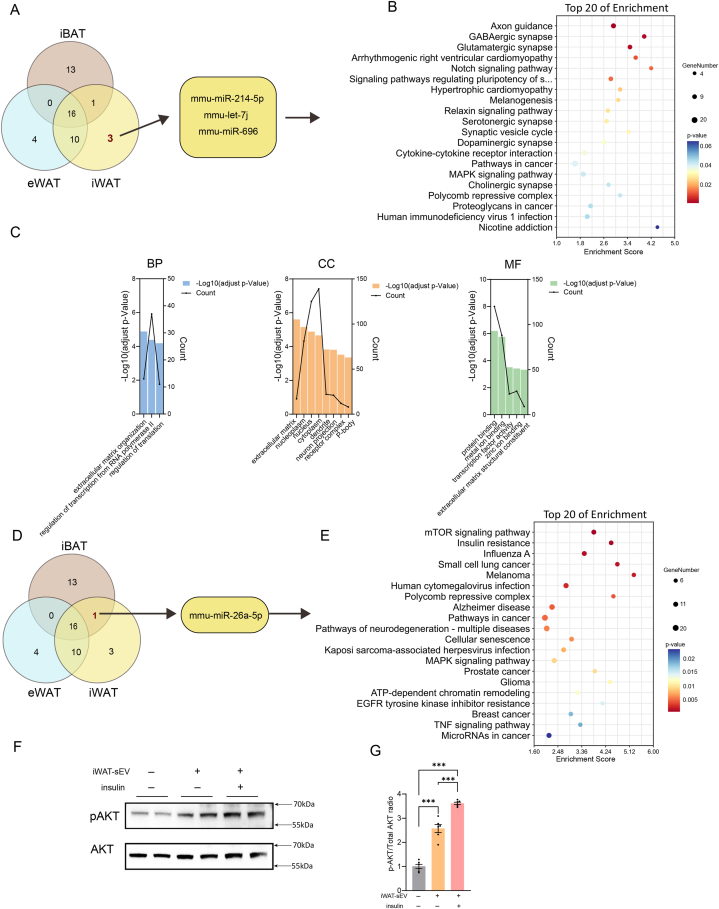
We analyzed three miRNAs specific to iWAT derived from small extracellular vesicles (sEVs): mmu-miR-214-5p, mmu-let-7j, and mmu-miR-696 (Fig. 4A). These miRNAs were first subjected to KEGG pathway analysis (Fig. 4B and Supplementary Table S5). The results revealed that their target genes are primarily involved in axon guidance, GABAergic synapses, and glutamatergic synapses. This suggested that iWAT-enriched miRNAs may regulate pathways associated with neural and synaptic functions. Next, we performed GO analysis on the target genes of iWAT-enriched miRNAs (Fig. 4C). This analysis showed that these target genes are predominantly associated with cellular processes, cellular components, and binding functions. Additionally, the Venn diagram identified one miRNA shared between iWAT and iBAT: mmu-miR-26a-5p (Fig. 4D). KEGG analysis and pathway visualizations showed that mmu-miR-26a-5p targets genes involved in key physiological processes such as cell growth, metabolism, and survival. Significant pathways affected by this miRNA include the mTOR signaling pathway, insulin resistance, and influenza A (Fig. 4E and Supplementary Figure S3 and Supplementary Table S6). We also explored the possible autocrine and paracrine functions of adipose-derived sEVs. Western blot and quantification analysis of phosphorylated AKT following co-culturing iWAT-derived sEVs with their SVF cells demonstrated that treatment with iWAT-derived sEVs increased AKT phosphorylation levels, confirming their role in regulating insulin sensitivity in adipose tissue (Fig. 4F and G). The same phenomenon was observed for iWAT-sEV derived from female mice (Supplementary Figures S2B).
Fig. 4.
Analysis of iWAT-Specific Enriched miRNAs and Shared miRNAs with iBAT. (A) Venn diagram highlighting a subset of three EV-derived miRNAs exclusively identified in iWAT. (B) KEGG enrichment analysis bubble plot for the target genes of three iWAT-specific EV-derived miRNAs, with p-values indicating the statistical significance of each pathway. (C) GO analysis of the pathways associated with the target genes of iWAT-specific EV-derived miRNAs, conducted using DAVID Bioinformatics Resources and categorized by biological process, cellular component, and molecular function, with each color representing a different category. (D) Venn diagram focusing on a subset of one EV-derived miRNA shared between iBAT and iWAT. (E) KEGG enrichment analysis bubble plot for the target genes of the shared miRNA between iBAT and iWAT, with p-values indicating the statistical significance of each pathway. (F and G) Western blot and quantification analysis of phosphorylated AKT in iWAT-derived SVF cells (n = 6 biological replicates, from three independent experiments).
3.5. Analysis of eWAT-Specific sEV-derived miRNAs and overlapping miRNAs between eWAT and iWAT
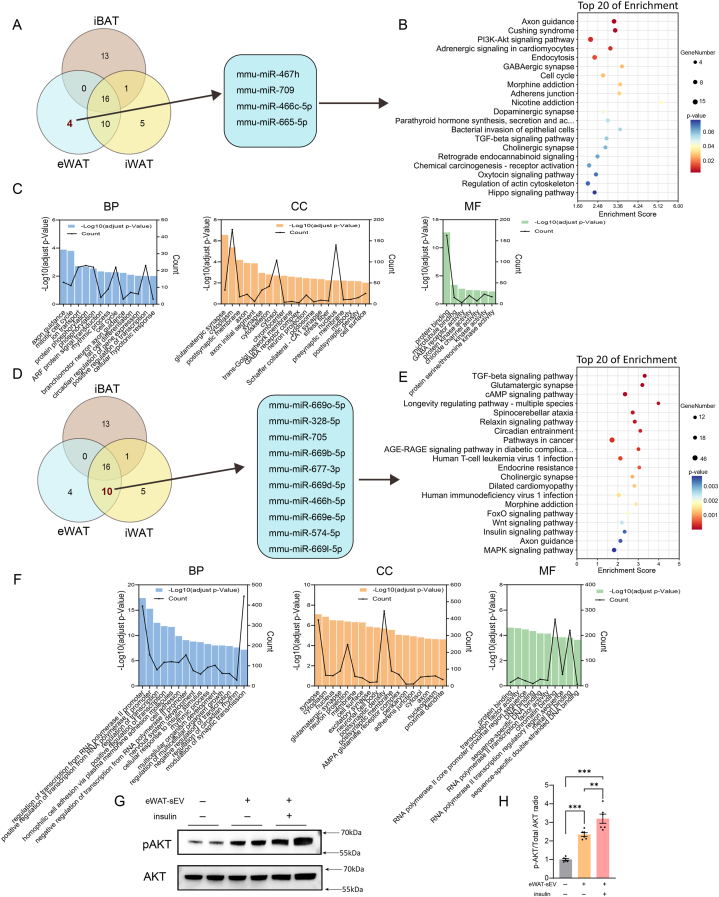
We identified four extracellular vesicle (EV)-derived microRNAs (miRNAs) uniquely associated with eWAT: mmu-miR-467h, mmu-miR-709, mmu-miR-466c-5p, and mmu-miR-665-5p (Fig. 5A). KEGG pathway linked these eWAT-specific sEV-derived miRNAs to key pathways involved in cellular function, development, and physiological responses (Fig. 5B and C and Supplementary Table S7). The enrichment scores and p-values underscore the relevance of axon guidance signaling, which directs axon growth during development, and pathways like PI3K-Akt signaling, adrenergic signaling in cardiomyocytes, and endocytosis, each contributing uniquely to cellular function and physiology. To further elucidate these findings, we employed bioinformatics to analyze protein data related to the identified pathways. GO analysis revealed that most proteins were associated with cell signaling and the regulation of cellular functions in the biological process (BP) category. In the cell component (CC) analysis, the majority of proteins were linked to neural signaling components. Molecular function (MF) classification highlighted their roles in synaptic transmission and neuronal signaling (Fig. 5C).
Fig. 5.
Analysis of eWAT-Specific EV-derived miRNAs and shared miRNAs between eWAT and iWAT. (A) Venn diagram highlighting a subset of four extracellular vesicle (EV)-derived microRNAs (miRNAs) exclusively identified in eWAT. (B) KEGG enrichment analysis bubble plot for the target genes of four eWAT-specific EV-derived miRNAs, with p-values indicating the statistical significance of each pathway. (C) Gene Ontology (GO) analysis of the pathways associated with the target genes of eWAT-specific EV-derived miRNAs, performed using DAVID Bioinformatics Resources and categorized by biological process, cellular component, and molecular function, with distinct colors representing different categories. (D) Venn diagram illustrating the overlap of 10 EV-derived miRNAs shared between eWAT and iWAT. (E). KEGG enrichment analysis bubble plot for the target genes of 10 EV-derived miRNAs common to both eWAT and iWAT, with p-values indicating the statistical significance of each pathway. (F) GO analysis of the target genes of EV-derived miRNAs shared between eWAT and iWAT, performed using DAVID Bioinformatics Resources and categorized by biological process, cellular component, and molecular function, with each color representing a different category. (G and H) Western blot and quantification analysis of phosphorylated AKT in eWAT-derived SVF cells (n = 6 biological replicates, from 3 independent experiments).
Additionally, the Venn diagram showed 10 miRNAs shared between iWAT and eWAT. KEGG pathway analysis of these shared miRNAs revealed their involvement in cellular signaling and physiological regulation, including intercellular signal transduction, neuronal communication, and intracellular signaling pathways (Fig. 5D and E and Supplementary Table S8). These processes reflect the transmission of information within and outside the cell and the regulation of cellular functions, crucial for maintaining cellular homeostasis and normal physiological state. GO analysis indicated that the proteins related to these shared miRNAs primarily function in regulating RNA polymerase II activity at specific DNA sequences, impacting gene expression and cellular functions (Fig. 5F). Cell component analysis linked most proteins to neural signaling components and the maintenance of cellular structural integrity, while molecular function classification highlighted their roles in controlling gene expression by modulating transcription factor activity and sequence-specific DNA binding.
Furthermore, we also explored the possible autocrine and paracrine effects of adipose tissue-derived sEVs. Western blot and quantification analysis of phosphorylated AKT following co-culturing sEVs from eWAT with their SVF cells showed that eWAT-derived sEVs significantly increased AKT phosphorylation levels, also indicating their role in maintaining insulin sensitivity in adipose tissue (Fig. 5G and H). The same phenomenon was observed for eWAT-sEV derived from female mice (Supplementary Figures S2C).
4. Discussion
In mammals, there are two main types of adipose tissue: WAT and BAT. Structural and functional similarities between human and mouse adipose tissues are evident, including similarities in basic tissue composition and the role of adipocytes as primary energy storage cells. However, there are also significant differences, such as the distribution of adipose tissue. For example, in rodents like mice, brown adipocytes are primarily located in the interscapular region and are characterized by multilocular lipid droplets and a high density of mitochondria, which are mainly involved in dissipating stored energy as heat. While it was once thought that brown adipocytes were limited to specific regions in infants and adults exposed to extreme cold, recent research suggests that brown adipocytes, or those with features of both brown and white adipocytes (known as 'beige' or 'brite' adipocytes), may be more common in adult humans than previously believed [37]. Nonetheless, WAT remains the dominant type in adult humans and is characterized by large adipocytes with a single lipid droplet and fewer mitochondria compared to brown adipocytes. In humans, adipose tissue is categorized into distinct subtypes: subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT). In contrast, mice have more gonadal WAT (eWAT), which shares similar functional and structural properties with human VAT. Additionally, inguinal WAT (iWAT) in mice resembles SAT, highlighting functional differences in fat tissues between species. Our current study aimed to investigate the potential physiological significance of sEVs derived from different adipose tissues in mice housed at room temperature. Specifically, we conducted a systematic analysis of miRNAs—one of the most abundant cargo RNAs in sEVs (also newly named as RNAkines [38])—from various adipose tissues under normal physiological conditions in mice.
We first focused on the top 30 most abundantly expressed miRNAs across different adipose tissue-derived sEVs. Among these, 30 miRNAs were conserved between humans and mice (Supplementary Table S2), suggesting potential similarities in the functions of corresponding adipose tissues across species.
SAT, which is found abundantly beneath the skin in both mice and humans, plays a vital role in storing body fat and regulating metabolic balance. It is involved in the secretion of endocrine hormones, immune modulation, and maintaining tissue homeostasis—key functions for preserving internal stability and overall health [39]. Insulin resistance, a condition characterized by reduced insulin sensitivity and/or responsiveness, impairs the ability of insulin to facilitate glucose uptake and utilization, leading to elevated blood glucose levels and potentially resulting in diabetes [40]. Our study suggests that miRNAs secreted by iWAT, which is the most extensively studied type of SAT in mice, can target genes associated with insulin resistance. This indicates that iWAT holds significant potential for further research and development in the treatment of diabetes and other related metabolic disorders.
eWAT, a crucial type of VAT in mice, is vital for energy storage and release and acts as a significant endocrine organ [41,42]. The specific miRNAs derived from its sEVs are involved in regulating key cellular functions, development, and physiological responses. Our analysis reveals that these miRNAs play roles in cellular signal transduction, regulation, and neural processes. These findings enhance our understanding of the regulatory mechanisms of eWAT and highlight the potential impact of sEV-derived miRNAs on adipose tissue metabolism and physiological functions. Furthermore, 10 miRNAs derived from sEVs are shared between iWAT and eWAT and are involved in intercellular signal transduction and physiological regulation. These insights offer valuable information on the regulatory mechanisms and physiological roles of sEV-derived miRNAs in adipose tissue, underscoring their potential significance in metabolic and physiological processes.
Most intriguingly, our analysis suggests that sEVs from BAT, known for its high thermogenic activity and energy dissipation, may also play a role in inhibiting tumorigenesis. Previous research by Cao et al. demonstrated that cold-induced activation of BAT can suppress tumor growth [43]. Consistent with these findings, our analysis of iBAT-specific sEV-derived miRNAs under physiological conditions provides new insights into the role of iBAT in cancer biology. We identified a nuanced regulatory network within iBAT that may impact the expression of oncogenes and tumor suppressor genes. Our GO analysis, combined with STRING prediction, highlights four key genes potentially linked to tumor suppression. For example, PLXNB3, involved in semaphorin signaling, is crucial for neural development and angiogenesis. Abnormal expression of PLXNB3 has been associated with prostate cancer and gliomas, where it contributes to tumor growth and metastasis [44]. MIA, a biomarker for melanoma, affects the extracellular matrix and influences cell adhesion, migration, and tissue structure. Elevated MIA levels in serum are correlated with aggressive melanoma, increased tumor thickness, metastatic potential, and poor prognosis [45]. SNX9 is essential for endocytosis and interacts with regulators of the actin cytoskeleton, impacting cell migration, adhesion, and shape changes. Dysregulation of SNX9 can contribute to metastatic behavior in cancers such as breast cancer [46]. CISD2, a mitochondrial protein, is important for maintaining mitochondrial integrity, iron homeostasis, and regulating reactive oxygen species (ROS) levels. Elevated CISD2 expression is linked to aggressive forms of breast cancer, hepatocellular carcinoma, and pancreatic cancer [47]. These four proteins are involved in critical cellular functions, including cell survival, adhesion, migration, and lipid metabolism. Their dysregulated expression is associated with tumor growth and metastasis, suggesting a mechanistic link between miRNA actions and potential tumorigenesis. The alignment of these miRNAs with key signaling pathways, particularly those abnormally activated in cancer, positions them as crucial molecular switches in the interplay between adipose physiology and cancer. Our findings suggest that BAT's role extends beyond energy expenditure and glucose regulation [48]. It also functions as an important endocrine organ, secreting factors that may target cancer-related pathways and inhibit tumor growth [49]. Given that our previous study showed that BAT-derived sEVs can be secreted to various organs, including the liver, kidneys, heart, and white adipose tissues, we speculate that BAT-derived sEVs may also act as potential tumor suppressors. This possibility warrants further investigation.
Furthermore, in addition to their anticipated endocrine roles, which may have systemic effects, our findings demonstrate that sEVs derived from adipose tissue significantly influence the tissues themselves. Specifically, sEVs from interscapular iBAT notably enhanced the differentiation of preadipocytes in the iBAT SVF cells, as evidenced by increased expression of Pparγ2, Pgc1α, and Ucp1. Additionally, sEVs from iWAT and eWAT improved insulin sensitivity and metabolic activity in their respective SVF cells. Thus, alongside well-known adipose-derived hormones such as leptin, resistin, and adiponectin, our study underscores the significant autocrine, paracrine, and potential endocrine roles of adipose tissue-derived sEVs, particularly due to their enriched miRNA content.
In summary, we isolated sEVs from various adipose tissues (eWAT, iWAT, and iBAT) under basal conditions and characterized the sEV-derived miRNAs using microarray and bioinformatics analysis. Our findings reveal complex regulatory networks mediated by these miRNAs and their significant impact on adipose tissue functions. This study underscores the distinct roles of adipose-derived sEVs in normal physiological processes and their potential involvement in disease onset, offering new insights into the molecular mechanisms of adipose tissue communication under physiological conditions.
5. Limitations of the study
While our study provides a comprehensive analysis and comparison of miRNAs from various adipose tissues, primarily using male mice, several limitations must be acknowledged. Firstly, although we validated some functions of sEVs using samples from female mice, most of our analyses were based on male mice. It would be beneficial to determine if these findings apply to female mice as well, considering that sex-specific differences in adipose tissue function and miRNA expression might exist. Secondly, our study utilized differential ultracentrifugation (dUC) methods to isolate sEVs, which makes it challenging to completely eliminate all contaminants from the samples. Additionally, while we investigated the functional roles of the identified miRNAs in cancer pathways, further research—including overexpression and knockout experiments—is necessary to clarify their specific effects on cancer-related gene expression and tumor cell behavior. Lastly, the physiological functions of shared miRNAs across different adipose tissues were not thoroughly explored. Further investigation into their roles in intercellular signaling and physiological regulation is needed to gain deeper insights into the regulatory mechanisms and physiological functions of these miRNAs in adipose tissues.
CRediT authorship contribution statement
Jiaqi Wang: Writing – review & editing, Writing – original draft, Visualization, Methodology, Data curation. Yuan Ji: Writing – review & editing, Writing – original draft, Validation, Investigation. Xiaoqin Cao: Writing – original draft, Validation, Methodology, Data curation. Ruixue Shi: Writing – original draft, Validation. Xiaohui Lu: Conceptualization. Ye Wang: Data curation. Chen-Yu Zhang: Supervision, Funding acquisition. Jing Li: Writing – review & editing, Writing – original draft, Formal analysis, Data curation, Conceptualization. Xiaohong Jiang: Writing – review & editing, Writing – original draft, Validation, Supervision, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Ethics declaration
All animal experimental procedures were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and received approval from the Animal Ethical Board of Nanjing University (IACUC-2204007).
Data availability
The raw data from the Affymetrix microarray has been uploaded to the GEO database (GSE 256424). We have also included the expression levels of all detected miRNAs (normalized signal intensity from the microarray) in Supplementary Table S1, along with a list of the top 30 enriched miRNAs in sEVs from various adipose tissues (iBAT, iWAT, and eWAT) in Supplementary Table S2. Additionally, the predicted target genes are detailed in Supplementary Tables S3–S8. Any other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (32371245,31972912), Natural Science Foundation of Jiangsu Province (BK20211153), the Fundamental Research Funds for the Central Universities (020814380203).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39149.
Contributor Information
Jing Li, Email: jingli220@nju.edu.cn.
Xiaohong Jiang, Email: xiaohongjiang@nju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schoettl T., Fischer I.P., Ussar S. Heterogeneity of adipose tissue in development and metabolic function. J. Exp. Biol. 2018;221:17. doi: 10.1242/jeb.162958. [DOI] [PubMed] [Google Scholar]
- 2.Echtay K.S., et al. Uncoupling proteins: martin Klingenberg's contributions for 40 years. Arch. Biochem. Biophys. 2018;657:41–55. doi: 10.1016/j.abb.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Giralt M., Villarroya F., White Brown. Beige/brite: different adipose cells for different functions? Endocrinology. 2013;154(9):2992–3000. doi: 10.1210/en.2013-1403. [DOI] [PubMed] [Google Scholar]
- 4.Li Y.G., Fromme T. Uncoupling protein 1 does not produce heat without activation. Int. J. Mol. Sci. 2022;23(5):14. doi: 10.3390/ijms23052406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamsi F., Wang C.H., Tseng Y.H. The evolving view of thermogenic adipocytes - ontogeny, niche and function. Nat. Rev. Endocrinol. 2021;17(12):726–744. doi: 10.1038/s41574-021-00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen P., Kajimura S. The cellular and functional complexity of thermogenic fat. Nat. Rev. Mol. Cell Biol. 2021;22(6):393–409. doi: 10.1038/s41580-021-00350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwok K.H.M., Lam K.S.L., Xu A.M. Heterogeneity of white adipose tissue: molecular basis and clinical implications. Exp. Mol. Med. 2016;48:12. doi: 10.1038/emm.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartelt A., Heeren J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014;10(1):24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 9.Sakers A., et al. Adipose-tissue plasticity in health and disease. Cell. 2022;185(3):419–446. doi: 10.1016/j.cell.2021.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oikonomou E.K., Antoniades C. The role of adipose tissue in cardiovascular health and disease. Nat. Rev. Cardiol. 2019;16(2):83–99. doi: 10.1038/s41569-018-0097-6. [DOI] [PubMed] [Google Scholar]
- 11.Harvey I., Boudreau A., Stephens J.M. Adipose tissue in health and disease. Open Biology. 2020;10(12):12. doi: 10.1098/rsob.200291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longo M., et al. Adipose tissue dysfunction as determinant of obesity-associated metabolic complications. Int. J. Mol. Sci. 2019;20(9) doi: 10.3390/ijms20092358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grange C., Bussolati B. Extracellular vesicles in kidney disease. Nat. Rev. Nephrol. 2022;18(8):499–513. doi: 10.1038/s41581-022-00586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanayama R. Emerging roles of extracellular vesicles in physiology and disease. J. Biochem. 2021;169(2):135–138. doi: 10.1093/jb/mvaa138. [DOI] [PubMed] [Google Scholar]
- 15.Clement E., et al. Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. Embo Journal. 2020;39(3):20. doi: 10.15252/embj.2019102525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazar I., et al. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res. 2016;76(14):4051–4057. doi: 10.1158/0008-5472.Can-16-0651. [DOI] [PubMed] [Google Scholar]
- 17.Jafari N., et al. Adipocyte-derived exosomes may promote breast cancer progression in type 2 diabetes. Sci. Signal. 2021;14(710):11. doi: 10.1126/scisignal.abj2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J.E., et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene. 2019;38(26):5158–5173. doi: 10.1038/s41388-019-0782-x. [DOI] [PubMed] [Google Scholar]
- 19.Chen G., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718) doi: 10.1038/s41586-018-0392-8. 382-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S.L., et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020;13(1):19. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooks T., et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018;9:15. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzelmann J., et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J. Urol. 2011;29(3):367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 23.Pasquinelli A.E. NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 24.Saliminejad K., et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019;234(5):5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 25.Moraes F.C., et al. miRNA delivery by nanosystems: state of the art and perspectives. Pharmaceutics. 2021;13(11):20. doi: 10.3390/pharmaceutics13111901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin K.O., et al. Exosomes from Human Adipose Tissue-Derived Mesenchymal Stem Cells Promote Epidermal Barrier Repair by Inducing de Novo Synthesis of Ceramides in Atopic Dermatitis. Cells. 2020;9(3):23. doi: 10.3390/cells9030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J.H., et al. Cold-activated brown fat-derived extracellular vesicle-miR-378a-3p stimulates hepatic gluconeogenesis in male mice. Nat. Commun. 2023;14(1):19. doi: 10.1038/s41467-023-41160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killcoyne S., et al. In: Protein Networks and Pathway Analysis. Nikolsky Y., Bryant J., editors. Humana Press Inc; 2009. Cytoscape: a community-based framework for network modeling; pp. 219–239. 999 Riverview Dr, Ste 208, Totowa, Nj 07512-1165 USA. [Google Scholar]
- 35.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;127(3):5–19. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Yang M.M., et al. The translational regulation in mTOR pathway. Biomolecules. 2022;12(6):13. doi: 10.3390/biom12060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W.S., Seale P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016;17(11):691–702. doi: 10.1038/nrm.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., et al. RNAkines are secreted messengers shaping health and disease. Trends in Endocrinology & Metabolism. 2024;35(3):201–218. doi: 10.1016/j.tem.2023.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Scheja L., Heeren J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019;15(9):507–524. doi: 10.1038/s41574-019-0230-6. [DOI] [PubMed] [Google Scholar]
- 40.Lebovitz H.E. Insulin resistance: definition and consequences. Exp. Clin. Endocrinol. Diabetes. 2001;109:S135–S148. doi: 10.1055/s-2001-18576. [DOI] [PubMed] [Google Scholar]
- 41.Joffin N., et al. Mitochondrial metabolism is a key regulator of the fibro-inflammatory and adipogenic stromal subpopulations in white adipose tissue. Cell Stem Cell. 2021;28(4) doi: 10.1016/j.stem.2021.01.002. 702-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brestoff J.R., et al. Intercellular mitochondria transfer to macrophages regulates white adipose tissue homeostasis and is impaired in obesity. Cell Metabol. 2021;33(2) doi: 10.1016/j.cmet.2020.11.008. 270-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seki T., et al. Brown-fat-mediated tumour suppression by cold-altered global metabolism. Nature. 2022;608(7922) doi: 10.1038/s41586-022-05030-3. 421-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadanandam A., et al. Identification of functional cell adhesion molecules with a potential role in metastasis by a combination ofin vivoPhage display andin silicoAnalysis. OMICS A J. Integr. Biol. 2007;11(1):41–57. doi: 10.1089/omi.2006.0004. [DOI] [PubMed] [Google Scholar]
- 45.Riechers A., Bosserhoff A.K. Melanoma inhibitory activity in melanoma diagnostics and therapy – a small protein is looming large. Exp. Dermatol. 2013;23(1):12–14. doi: 10.1111/exd.12281. [DOI] [PubMed] [Google Scholar]
- 46.Bendris N., et al. SNX9 promotes metastasis by enhancing cancer cell invasion via differential regulation of RhoGTPases. Mol. Biol. Cell. 2016;27(9):1409–1419. doi: 10.1091/mbc.E16-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim E.H., et al. CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer. Cancer Lett. 2018;432:180–190. doi: 10.1016/j.canlet.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 48.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 49.Rosina M., et al. Ejection of damaged mitochondria and their removal by macrophages ensure efficient thermogenesis in brown adipose tissue. Cell Metabol. 2022;34(4):533–548.e12. doi: 10.1016/j.cmet.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from the Affymetrix microarray has been uploaded to the GEO database (GSE 256424). We have also included the expression levels of all detected miRNAs (normalized signal intensity from the microarray) in Supplementary Table S1, along with a list of the top 30 enriched miRNAs in sEVs from various adipose tissues (iBAT, iWAT, and eWAT) in Supplementary Table S2. Additionally, the predicted target genes are detailed in Supplementary Tables S3–S8. Any other data supporting the findings of this study are available from the corresponding author upon reasonable request.