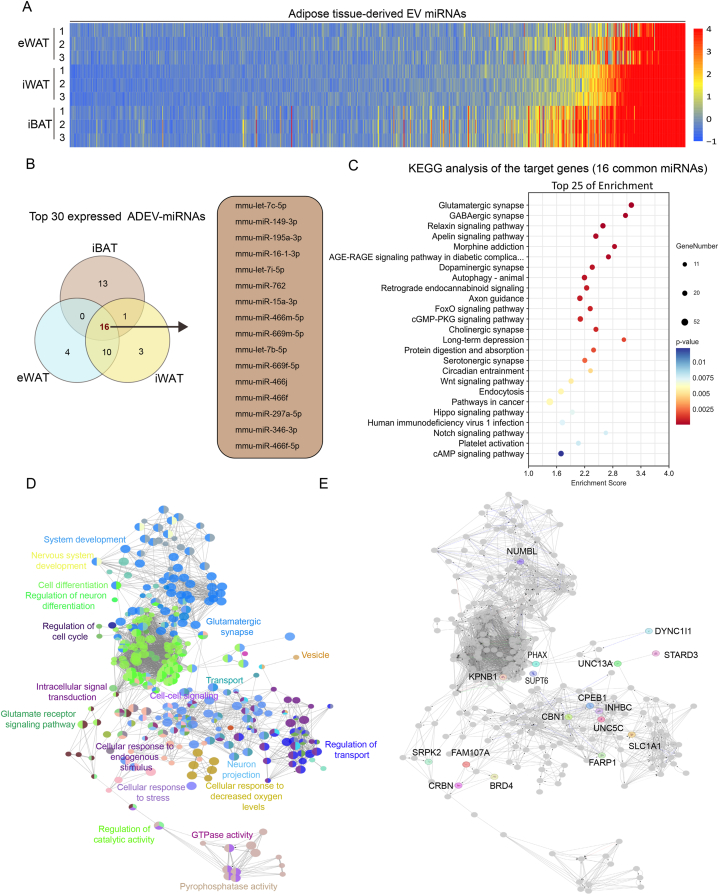
Fig. 2.
Comparative Analysis of sEV-Derived miRNA Expression Profiles in Different Adipose Tissues. (A) The heatmap illustrates the expression levels of EV-derived miRNAs from eWAT, iWAT, and iBAT, The color scale represents the correlation between color and expression levels, with intensity ranging from low (blue) to high (red). For each tissue type, samples were collected from 6 mice, with each row representing pooled samples from 2 distinct mice, resulting in a total of three composite sample pools. (B) Among the top 30 expressed adipose-derived extracellular vesicle-miRNAs (ADEV-miRNAs), there are 16 miRNAs common to the three types of adipose tissues. (C) The bubble chart of KEGG enrichment analysis for the target genes of the 16 common miRNAs, with p-values indicating the statistical significance of each pathway. (D) GO analysis of the pathways related to the target genes of the 16 common miRNAs, conducted using DAVID Bioinformatics Resources and categorized based on biological processes, cellular components, and molecular functions, with each color representing a different category. (E) Molecular interaction network based on the pathways related to the target genes of the 16 common miRNAs.