Abstract
Pre-trained chemical language models (CLMs) have attracted increasing attention within the domains of cheminformatics and bioinformatics, inspired by their remarkable success in the natural language processing (NLP) domain such as speech recognition, text analysis, translation, and other objectives associated with language. Furthermore, the vast amount of unlabeled data associated with chemical compounds or molecules has emerged as a crucial research focus, prompting the need for CLMs with reasoning capabilities over such data. Molecular graphs and molecular descriptors are the predominant approaches to representing molecules for property prediction in machine learning (ML). However, Transformer-based LMs have recently emerged as de-facto powerful tools in deep learning (DL), showcasing outstanding performance across various NLP downstream tasks, particularly in text analysis. Within the realm of pre-trained transformer-based LMs such as, BERT (and its variants) and GPT (and its variants) have been extensively explored in the chemical informatics domain. Various learning tasks in cheminformatics such as the text analysis that necessitate handling of chemical SMILES data which contains intricate relations among elements or atoms, have become increasingly prevalent. Whether the objective is predicting molecular reactions or molecular property prediction, there is a growing demand for LMs capable of learning molecular contextual information within SMILES sequences or strings from text inputs (i.e., SMILES). This review provides an overview of the current state-of-the-art of chemical language Transformer-based LMs in chemical informatics for de novo design, and analyses current limitations, challenges, and advantages. Finally, a perspective on future opportunities is provided in this evolving field.
Keywords: Transformers, Natural language processing, SMILES, Machine learning, Cheminformatics, BERT, GPT
1. Introduction
SMILES [1], which stands for simplified molecular input line entry system, is a line notation that allows a user to represent and describe a chemical structure or a chain of letters, numbers and characters that specify the atoms, their connectivity, bond order and chirality in a way that can be used by the computer for various tasks such as natural language processing (NLP) related tasks [2]. SMILES strings are composed of ASCII characters and provide a textual representation of molecular structures. Consider the example of SMILES notation for Paracetamol is CC(=O)Nc1ccc(O)cc1. In SMILES representation atoms are represented by the their atomic symbols. Metal atoms are represented with the symbols in square brackets (‘[]’), however, brackets can be omitted for B, C, N, O, P, S, and Halides. Aromatic C, O, S, and N atoms are shown in lower case ‘c’, ‘o’, ‘s’, and ‘n’. Aliphatic C, O, S, and N atoms are shown in upper case ‘C’, ‘O’, ‘S’, and ‘N’. On the other hand single, double, and triple bonds are represented by the symbols (-, =, and #) respectively. Adjacent atoms are assumed to be connected to each other by a single or aromatic bond. Branches are specified by enclosing them in parentheses, which can be nested or arranged. SMILES is widely used to represent chemical structures in text form, efficiently encoding molecular information and facilitating chemical data interoperability [3].
Driven by novel reactions and diverse building blocks, the fully enumerated REAL database has grown significantly from approximately 170 million compounds in 2017 to over 5.5 billion compounds in 2022 [4]. It now forms the majority of the popular and widely-used ZINC20 virtual screening database [5]. Chemical structure representation such as SMILES, is a critical aspect of computational chemistry or cheminformatics, aiding in the analysis and prediction of several tasks such as molecular properties, compound-protein interaction, and de novo molecular generation in the field of drug discovery. Recently, deep learning (DL) techniques have been widely adopted to accelerate and improve drug discovery and have delivered impressive results in various applications, such as molecular property prediction, drug–drug interaction (DDI) prediction, drug–target interaction (DTI) prediction, drug repositioning, and de novo drug generation [6], [7], [8], [9]. However, the problem of limited labeled data problem is still challenging and also restricts the performance of these techniques in specific tasks. Additionally, data labeling is a resource-intensive and time-consuming task to be accomplished in every stage of drug discovery pipeline [10].
Based on the probably approximately correct learning theory, language models (LMs) heavily depend on the size of their training data or in other words, larger training datasets assist the model in making highly accurate predictions. Therefore, an effective and promising approach involves leveraging both a vast number of unlabeled data or molecules and the limited available labeled data, wherein CLMs acquire insights or more informative molecular representation through pre-training (i.e., unsupervised pre-training) with unlabeled data (molecules) and fine-tuning with labeled data for specific downstream tasks. LMs leverage and customize algorithms originally designed for natural language processing (NLP) to understand the language of chemistry. This adaptation is facilitated by the use of string representations, such as SMILES strings. [3].
Transformer-based models have revolutionized numerous fields within artificial intelligence (AI), most notably in NLP, where they have demonstrated unparalleled capabilities in understanding and generating human language [11]. This same architecture, known for its ability to capture long-range dependencies and contextual information, has recently been adapted for use in computational chemistry focusing on their use with SMILES representations to enhance the prediction of molecular properties [12], [13]. Here, it facilitates the interpretation and prediction of chemical properties from SMILES strings, a text-based representation of chemical structures [14]. Suitability of transformer-based models can be seen in terms of the criteria such as: Diverse Applications: Transformer-based models are well-suited for a wide range of molecular properties analysis tasks, including drug discovery, toxicity prediction, and materials science. State-of-the-Art Performance: With appropriate fine-tuning and optimization, transformer models have demonstrated state-of-the-art performance across various molecular property prediction benchmarks. Flexibility: Transformers can accommodate different types of molecular representations beyond SMILES, such as graph-based representations, enabling their application to diverse cheminformatics tasks. Continual Improvement: As transformer architectures evolve and new training methodologies emerge, their suitability for molecular properties analysis is expected to improve further, addressing current limitations and expanding their applicability in the field.
Over the years, various methods have been employed to encode molecular structures, with recent advancements focusing on transformer and attention-based models. Consequently, this literature review aims to provide a comprehensive overview of the application of transformer and attention-based models in computational chemistry handling SMILES representations to facilitate the prediction of molecular properties. In this study, we aim to provide a thorough review of various transformer (i.e., attention) based CLMs as well as a systematic taxonomy of the applications in handling SMILES representations. To summarize, our contributions are:
-
•
We provide a detailed review over existing transformer based CLMs. We present a general design pipeline and discuss the variants of each module based on transformer architecture.
-
•
We discuss major applications and their corresponding methods contributed to various chemical properties predictions.
-
•
We propose open problems for future research and provide a thorough analysis of each problem and propose future research directions.
2. Background
This section discusses the limitations of traditional molecular representation approaches and highlights the need for more advanced models to capture intricate relationships within molecular structures.
Before the breakthrough of transformer and attention-based models, traditional molecular representation methods such as molecular descriptors, fingerprints, and graph-based representations were predominant approaches to representing molecules. These are also common methods previously employed for predicting molecular properties which often utilize machine learning (ML) models. While effective, these methods often required expert-engineered or pre-designed molecular features representation and extensive domain knowledge to enhance predictive performance [15]. Moreover, numerous research studies have incorporated explicit 3D atomic coordinates to achieve even higher performance levels [15]. Another significant research direction focuses on optimizing the model architecture, whether it operates on molecular descriptors or fingerprints, directly processes SMILES strings, or analyzes the underlying molecular graph [16].
The effectiveness of DL methods relies greatly on having large-scale labeled training data. While various research areas such as image processing often reaches millions of labeled samples, molecular property prediction faces a severely different scenario. Gathering such a vast number of labeled molecular properties through screening experiments is exceptionally costly and labor-intensive. This resembles the challenge in natural language modeling, where there's an abundance of unlabeled data but only a tiny is labeled. To address this, the prevailing approach is the pre-training and fine-tuning framework or semi-supervised learning architecture [17]. This method involves initially training the model in an unsupervised manner, then fine-tuning it with labeled data. Consequently, few unsupervised fingerprint methods have been presented previously as a solution to the challenge of limited labeled data.
The seq2seq et al. [18] and Seq3seq et al. [19] fingerprint models are the pioneered semi-supervised frameworks, leveraging large-scale unlabeled data to enhance prediction accuracy. However, the efficiency of these methods such as Seq3seq, is somewhat limited due to its recurrent neural networks (RNNs) [20] encoder-decoder structure, with the decoder primarily serving as a scaffold during pre-training without significantly contributing to the final prediction or fine-tuning. Hence, these fingerprint methods lack computational efficiency in this regard [12]. In general, these models train deep neural networks to generate robust vector representations using extensive unlabeled datasets. These vector representations are then utilized for supervised training with various classifiers. As these deep models are trained on enormous amount of dataset, the representations are anticipated to contain adequate information for effective inference. However, such methods are not directly trained on prediction tasks, leading to representations optimized solely for the recovery task of the original raw data representation. Consequently, they may not offer optimal inference performance for broader prediction tasks.
Seq2Seq / Seq3Seq fingerprint methods (RNN-based encoder-decoder)
-
•
Architecture: These fingerprint methods such as Seq2Seq typically relies on RNNs, including variations like long short-term memory (LSTM) [20] or gated recurrent units (GRUs) [21], for both the encoder and decoder.
-
•
Encoder: In this approach, the encoder processes an input sequence (e.g., a molecular representation like SMILES) one token at a time. The hidden states are updated sequentially, and the final hidden state summarizes the entire input sequence.
-
•
Decoder: The decoder also uses an RNN and generates the output sequence based on the hidden state from the encoder. In tasks like molecule generation or prediction, the decoder sequentially outputs each element of the sequence (e.g., fingerprint), using the previous output as input for the next.
-
•
Handling long-term dependencies: Since they rely on RNNs, these fingerprint methods such as Seq2Seq can suffer from issues like vanishing gradients when handling long sequences, even with advanced variants like LSTMs. The model must retain the entire sequence context through hidden states passed along each time step.
Transformers [11], by contrast, leverage the self-attention mechanism to model dependencies and interactions within sequences, making them particularly well-suited in this regard for SMILES strings, which encode complex molecular structures in a linear text format. The ability of transformers to process and generate sequences aligns perfectly with the requirements for interpreting SMILES strings, enabling the extraction of rich, contextually aware representations of molecular structures.
Transformer encoder-decoder structure
-
•
Architecture: The Transformer uses self-attention mechanisms instead of RNNs. Both the encoder and decoder are built using layers of multi-head self-attention and feed-forward networks, which operate in parallel rather than sequentially.
-
•
Encoder: The transformer encoder processes the entire input sequence at once, applying self-attention across all tokens in the sequence. This allows the model to capture long-range dependencies and relationships between tokens without the sequential bottleneck of RNNs. Each token attends to every other token in the sequence, capturing context in a global manner.
-
•
Decoder: Similarly, the decoder uses both self-attention and encoder-decoder attention. The self-attention focuses on previously generated tokens (autoregressive generation), while the encoder-decoder attention helps the decoder focus on relevant parts of the input sequence. This parallelization improves efficiency and long-range dependency handling.
-
•
Handling long-term dependencies: Transformers handle long sequences more effectively because self-attention allows every token to directly attend to every other token in the sequence. This avoids the problem of vanishing gradients and limits on sequence length that RNN-based Seq2Seq models may face.
-
•
Attention mechanism (core component): In the transformer, attention mechanisms are the core of both the encoder and decoder. The model doesn't rely on sequential hidden state propagation but instead uses attention to compute dependencies between tokens.
In summary, while both fingerprint methods and transformers follow an encoder-decoder structure, methods such as Seq2Seq or Seq3Seq relies on sequential processing via RNNs, whereas transformers use self-attention mechanisms for parallel processing, allowing them to handle long sequences more effectively and efficiently. Nonetheless, utilizing large amounts of unlabeled SMILES data along with a limited set of labeled data, transformer LMs can effectively learn and capture meaningful molecular representations from SMILES strings [3]. These models first learn from the abundant unlabeled data and are then fine-tuned using labeled data for specific downstream tasks [4]. This approach addresses the challenge of obtaining large quantities of labeled molecular-property data, which is often resource-intensive and requires significant labor through screening experiments and manual data annotation [6], [10].
3. Transformer architecture and attention mechanism
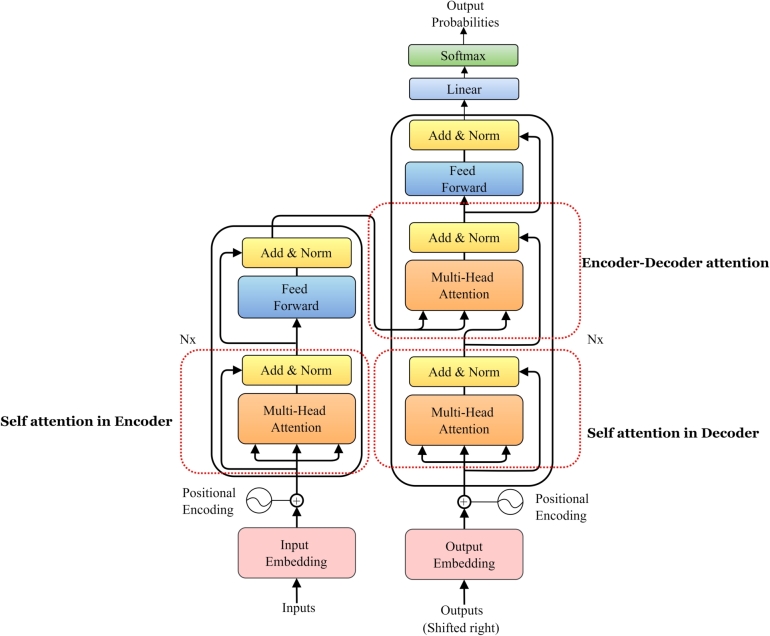
The core structure of a transformer is comprised of two fundamental components: the encoder and the decoder, as illustrated in Fig. 1. Transformer models, originally designed for NLP, have shown remarkable success in various domains, such as computational chemistry. Specific transformer-based architectures, such as bidirectional encoder representations from transformers (BERT) [22] and generative pre-trained transformer (GPT) [23], [24] in this regard can be adapted and utilized using for various tasks in cheminformatics such as virtual screening and drug discovery, molecular property prediction, and de novo molecular design. Remarkable capabilities of transformer lies in its innovative self-attention mechanism as shown in Fig. 1. This mechanism, considered the cornerstone of the transformer intelligence, revolutionizes how transformer models capture and understand relationships within sequences [25], [26]. Attention mechanisms have proven to be pivotal in capturing long-range dependencies among the elements in an input sequence. Since certain words or SMILES tokens are more impactful in defining the context and meaning of a sequence. The attention mechanism plays a crucial role in assigning the different attention weights to each input elements or tokens [25], The attention mechanism mimics this mechanism by enabling each word (or token) to consider the significance of other words or tokens, thereby grasping their relative importance. The attention weights indicate the significance or relevance of a particular input sequence element at given step, put simply elements with large weight values play a bigger role in comprehending the entire input sequence. This interactive process guarantees that tokens convey contextual information from throughout the entire sequence, which is essential for interpreting complex language structures. Therefore, this facilitates and assists Transformer LMs to concentrate on the most pertinent elements within the given input sequence, aiding in better comprehension and analysis.
Figure 1.
Transformer (attention-based) architecture. CLMs in this field typically utilize one of three core transformer structures: encoder-only models (e.g., BERT), decoder-only models (e.g., GPT), or the encoder-decoder architecture (e.g., BART, T5).
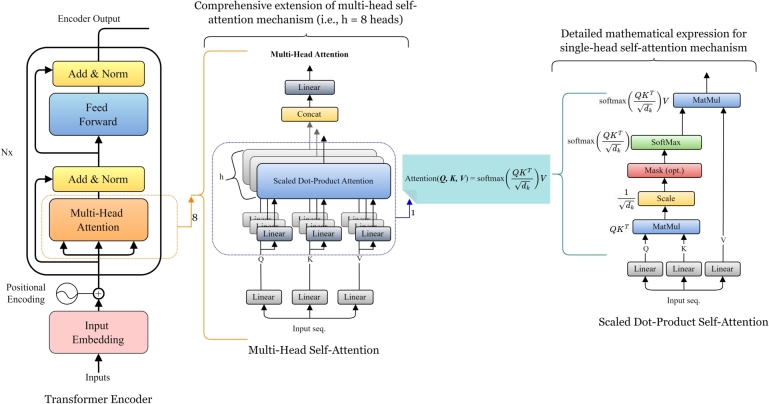
The attention mechanism used in Transformer is named scaled dot-product attention for each attention head [11]. It maps or transform the input sequence into three different parts corresponding to query (Q), key (K), and values (V) matrices as depicted in Fig. 2. The terms Q, K, and V utilized in here are inspired by information retrieval systems and databases [25]. It is important to note that, rather than relying on a single attention (single head self-attention) mechanism, multi-head attention utilizes multiple attention mechanisms operating in parallel, enabling the model to capture diverse types of relationships and patterns within the sequence as depicted in Fig. 1. Each separate attention head focuses on various aspects of the input, accelerating and enhancing the attention mechanism's potency [12]. In this context of multi-head attention, three matrices correspond to each single attention head, hence there will be sets of Q, K, and V matrices. Each part (i.e., Q, K, and V) is introduced with weight matrices that can be fit as model parameters during model training, to make self-attention mechanism more flexible and amenable during model optimization for a given input sequence [25]. The Q matrix works together with the K matrix to serve as the input of the Softmax. Then Softmax creates the attention weights, which will be applied to the V matrix to generate the output features with the attention on the whole sequence.
Figure 2.
Self-attention mechanism architecture. Case study: Encoder.
In summary, as shown in Fig. 2, multiple layers of self-attention mechanisms, known as multi-head self-attention repeats the scaled dot-product attention computation multiple times in parallel and the results are concatenated and then linearly transformed, producing a comprehensive and refined representation [26]. Thus the multi heads of attention allow the model to capture information from different parts of the input sequence similar to multiple kernels which produce multiple channels in a convolutional neural network (CNNs) [27], in which each channel capture different feature information. Multi-head attention offers several advantages such as:
-
•
Enhanced feature representation: Multiple attention heads allow the model to focus on different parts of the input sequence simultaneously. Each head can learn to capture different relationships between SMILES tokens, leading to a richer representation of the complex molecular structures.
-
•
Parallelization: Multi-head attention allows multiple attention mechanisms to operate in parallel, which improves computational efficiency compared to processing attention sequentially.
-
•
Improved learning: By using multiple attention heads, the model can better capture various linguistic patterns and dependencies at different levels of abstraction, enhancing its ability to understand both local and global relationships in the complex molecular structures data.
-
•
Reduced overfitting: The use of multiple heads helps mitigate overfitting, as it enables the model to consider diverse perspectives of the same data, rather than relying on a single point of view.
The scaled dot-product self attention is formulated in Equation (1) and shown in Fig. 2.
| (1) |
where is defined as the input feature matrix, , , and correspond to the query weight matrix, the key weight matrix, and the value weight matrix respectively. is a scaling factor preventing softmax from being pushed into a region with an extremely small gradient and Z indicates the output of the attention layer. Fig. 2 illustrates deeper the structural architecture of the attention mechanism.
Attention scores calculation: The self-attention mechanism computes attention scores for each token through the dot product of its Q vector with the K vectors of all other tokens. These scores represent the relevance of the token relative to others in the sequence.
Softmax and Weighted Values: The attention scores are passed through a softmax function, which converts them into probabilities. These probabilities indicate how much weight each token's V vector contributes to the final output.
Output via weighted sum of Vs: The final output for each token is generated by computing the weighted sum of the V vectors, with the weights determined by the softmax probabilities. This output captures both local and global contextual relationships.
Other accompanied components are as explained as follows:
-
•
Normalization layer (Layer Normalization): Stabilizes training by standardizing activations, ensuring they have a mean of zero and variance of one, which helps maintain consistent gradients and prevents exploding or vanishing gradients.
-
•
Feed-forward neural networks: Provide additional non-linearity and learning capacity within each layer, enhancing the model's ability to generalize and capture complex patterns, while the residual connections help in stabilizing this process.
-
•
Residual connections: Skip connections bypass layers, allowing gradients to flow directly through the network, thus mitigating the vanishing gradient problem and helping deeper layers learn more effectively. Each components incorporates a residual input to better utilize the original information or maintain strong information signals to avoiding vanishing gradient phenomenon and overfitting, to ensure stable training.
Together, these elements contribute to efficient training, stability, and generalization in Transformer-based CLMs.
4. Transformer-based models in computational chemistry
Transformer and attention-based models have demonstrated exceptional performance in various tasks in computational chemistry. This section provides an in-depth analysis of how these models contribute to molecular property prediction, virtual screening, and de novo molecular design using SMILES-encoded information.
Molecular Property Prediction: Transformer-based CLMs have shown significant promise in predicting various molecular properties, such as solubility, toxicity, and binding affinity. By training on large datasets of SMILES strings, these models learn to encode molecular structures into high-dimensional representations that capture relevant chemical information. Fine-tuning these models on specific prediction tasks further enhances their accuracy, often surpassing traditional machine learning methods.
Virtual Screening and Drug Discovery: In drug discovery, virtual screening is a crucial step where millions of compounds are evaluated for potential biological activity. Transformers can process large chemical libraries encoded in SMILES format, efficiently identifying promising candidates for further experimental validation. The scalability of transformers allows them to handle extensive datasets, making them invaluable in high-throughput screening applications.
De Novo Molecular Design: De novo molecular design involves generating new chemical compounds with desired properties. Transformer models, particularly those with generative capabilities like GPT can be trained to generate valid and novel SMILES strings. This generative process can be guided by desired chemical properties, enabling the design of molecules tailored for specific applications, such as drug development or materials science.
Transformer-based models generally offer several strengths in computational chemistry for molecular properties and chemical reactions analysis using SMILES representation in various criteria such as:
Learn complex relationships: Transformers excel at capturing intricate relationships between elements in sequential data, making them well-suited for analyzing the complex structural patterns present in SMILES strings.
Scalability: These models scale large datasets efficiently, enabling the analysis of extensive molecular libraries containing millions of compounds.
Contextual understanding: Transformers can capture contextual information from the surrounding atoms and bonds, allowing for a more nuanced understanding of molecular structures.
Transfer learning: Pre-trained transformer models can be fine-tuned on molecular property prediction tasks with relatively small amounts of labeled data, leveraging knowledge learned from pre-training on large unlabeled datasets.
4.1. Comparative analysis
Transformer-based models have marked a significant advancement in the field of computational chemistry, particularly in predicting molecular properties from SMILES representations. Table 1 provides a comparative analysis of various transformer-based CLM used in this field. This analysis delves into various categories of transformer models based on the architectures like encoder-based, decoder-based, and encoder-decoder-based, each with unique strength and suited for specific tasks. We further highlighting their applications, strengths, and limitations in molecular property prediction. These models differ in their architectures and specific applications, yet all leverage the core strengths of the transformer architecture as shown in Table 1. For example encoder-based models such as BERT and its variants excel in capturing detailed contextual information or extracting meaningful context from input sequences [26]. These are particularly effective for tasks requiring deep molecular understanding. Decoder-based models like GPT and its derivatives shine in creative generation tasks, enabling the creation of novel molecules with desired properties. Encoder-decoder models combine the strengths of both approaches, offering versatile solutions for complex sequence-to-sequence tasks.
Table 1.
Case studies: Transformer-Based Language Models for computational chemistry and cheminformatics.
| Backbone | Encoder-based CLMs | Decoder-based CLMs | Encoder-decoder CLMs |
|---|---|---|---|
| Transformers(Attention) | BERT [28] | Guo et al. [29] | TransAntivirus (IUPAC) [30] |
| MOLBERT [14] | GPT-2-based model [31] | Chemformer (BART-based) [32] | |
| SMILES-BERT [12] | MolXPT (GPT-2-based) [33] | MolT5 [34] | |
| ChemBERTa [13] | BioGPT [35] | Transformer-based models [36] | |
| ChemBerta-2 [37] | MolGPT (Relative Attention) [38] | X-MOL [39] | |
| GPT-MolBERTa [40] | Mol-Instructions [41] | SMILES transformer [42] | |
| RoBERTa-based model [43] | Transformer Neural Network [44] | ||
| MoLFormer [45] | Transformer-based ANN [46] | ||
| SELFormer (SELFIES) [47] | RT (SELFIES) [48] | ||
| Mol-BERT [49] | |||
| MolRoPE-BERT [50] | |||
| BET [51] |
4.1.1. Encoder-based CLMs
-
•
BERT. Herein, BERT [28] model was employed as shown in Table 1, and pre-trained on large corpora and fine-tuned for specific tasks. In cheminformatics, it has been adapted to process SMILES strings, enabling the extraction of rich, contextual molecular representations. Applications: Property prediction, molecular similarity search, and drug discovery. Strengths: BERT captures bidirectional context, enhancing the understanding of chemical environments and molecular interactions. They excel in transfer learning, requiring smaller datasets for fine-tuning. Limitations: High computational cost and memory usage due to the bidirectional nature of the model.
-
•
MOLBERT. This model is a chemistry-specific adaptation of BERT, tailored for handling SMILES strings. Applications: Predicting physicochemical properties, bioactivity, and molecular interactions. Strengths: Specialized training on chemical datasets enhances performance on cheminformatics tasks. Limitations: Requires large, labeled datasets for optimal performance.
-
•
SMILES-BERT. This is a variant of BERT explicitly designed to work with SMILES representations. Applications: Predicting molecular properties and chemical reactions. Strengths: Effective at learning molecular representations directly from SMILES strings without requiring extensive feature engineering. Limitations: Like other BERT-based models, it demands substantial computational resources.
-
•
ChemBERTa and ChemBERTa-2. These models are based on the RoBERTa [52] which is the adaptation of BERT [22], designed for chemical property prediction, optimized for handling SMILES inputs. Applications: Broad range of property predictions, including toxicity, solubility, and bioactivity. Strengths: Enhanced with domain-specific training, improving prediction accuracy. Limitations: The complexity and resource demands are significant.
-
•
RoBERTa-based Model. This model also refine BERT by training with more data and longer sequences, further adapted for cheminformatics. Applications: Property prediction and molecular classification tasks. Strengths: Improved training protocols lead to better generalization in chemical datasets. Limitations: Increased computational requirements for training and inference.
-
•
Mol-BERT and MolRoPE-BERT. These models are also based on and utilized BERT architecture to handle chemical SMILES representation for predicting molecular properties and chemical reactions. The primary distinction between these models lies in their approach to position embedding (PE) implementation. Mol-BERT [49] model employs absolute PE, which limits the length of encoding sequences, impacting the effectiveness of downstream model prediction tasks [50]. On the other hand to address the aforementioned issues, Liu et al. [50] proposed MolRoPE-BERT, which applies rotary PE.
4.1.2. Decoder-based CLMs
-
•
GPT-2-based Models (e.g., MolGPT, MolXPT). In overview these models, derived from the generative pre-trained transformer 2 (GPT-2) [24], are used for generative tasks, including SMILES string generation. Applications: De novo drug design and molecular optimization. Strengths: Excellent at generating novel, chemically valid molecules by leveraging learned distributions of SMILES strings. Limitations: Tend to focus on one-directional context, which may limit the understanding of bidirectional dependencies in molecules.
-
•
BioGPT. This model focuses on generating biomedical text and mining. It appears to outperform other related models such as BioBERT [53], PubMedBERT [54], GPT-2 model in this regard. Applications: Biomedical text generation and Mining. Strength: Tailored to handle large-scale biological data, making it suitable for biomedicine, biomedical text mining and generation. Limitation: Generative focus may not always capture the most relevant features for certain predictive tasks.
-
•
MolGPT. This proposed model employs the generative pre-trained transformer (GPT) architecture along with relative attention for de novo drug design [38]. Relative attention, a variation of the attention mechanism, enables the model to understand the relative distances and relationships between tokens in the input sequence. Unlike the standard attention mechanism, which uses fixed-PEs to encode positional information, relative attention dynamically incorporates relative positional encodings during the attention calculation. This enables the model to learn the syntax of new and unseen tokens efficiently.
4.1.3. Encoder-decoder-based CLMs
-
•
TransAntivirus (IUPAC). A model designed to predict antiviral properties by processing chemical structures in the IUPAC nomenclature format. Applications: Antiviral drug discovery. Strengths: Specifically optimized for antiviral property prediction, leveraging encoder-decoder architecture for better sequence-to-sequence learning. Limitations: Performance may be dataset-dependent, requiring high-quality training data.
-
•
Chemformer (BART-based). Based on the Bidirectional and Auto-Regressive Transformers (BART) architecture, Chemformer is tailored for generating and predicting molecular properties. Applications: Molecular property prediction and compound generation. Strengths: Combines the strengths of both encoder and decoder models, providing robust performance in property prediction and generation tasks. Limitations: Complex training process and higher computational overhead.
-
•
SMILES Transformer. This model utilizes the transformer architecture for converting SMILES strings into meaningful representations and predicting their properties. Applications: Broad applications including drug discovery, toxicity prediction, and property optimization. Strengths: Effective at handling the sequential nature of SMILES strings, capturing intricate molecular features. Limitations: Computationally intensive and requires significant preprocessing.
-
•
MolT5. An adaptation of the T5 (Text-to-Text Transfer Transformer) model for molecular tasks. Applications: Molecular property prediction, reaction prediction, and molecular optimization. Strengths: Unified framework for various tasks, leveraging transfer learning for improved performance. Limitations: Training and fine-tuning require substantial computational resources and high-quality data.
In general, transformer-based CLMs especially encoder-based models have been widely employed and contributed significantly in this field followed by encoder-decoder based models with few studies utilize molecular representations other than SMILES such as SELFIES, IUPAC, and biomedicine text data representations as shown in Table 1.
5. Materials and methods
5.1. Datasets
Dataset in this field in primarily various chemical compound representations as shown in Table 4. In this review, most of LMs primarily utilized SMILES dataset however few studies have used IUPAC and SELFIES as shown in Table 1. DeepSMILES is defined as a variant of SMILES, altering how branches and rings are represented while maintaining the same fundamental atom-level characters with SMILES [55]. SMILES is a notation that allows a user to represent a chemical structure in a way that can be used by the computer. A SMILES training dataset would typically consist of molecular structures encoded using the SMILES notation, paired with corresponding labels or information about the properties of the molecules. SMILES strings are composed of ASCII characters and provide a textual representation of molecular structures. Ensuring the accuracy or correctness of SMILES strings representations is crucial, as invalid representations can result in the misinterpretation of molecular structures by the model. Data preprocessing may involve tasks such as canonicalization of SMILES strings (ensuring a unique representation for the same molecule), handling chirality, and other relevant transformations to standardize the data. Creating a SMILES training dataset often involves leveraging existing databases of chemical compounds, experimental data, or cheminformatics resources such as ZINC [5], [56], PubChem [57], [58], and ChEMBL [59], [60] databases. Researchers and practitioners in computational chemistry or cheminformatics use such datasets to develop models for tasks such as molecular property prediction, drug discovery, and chemical informatics.
Table 4.
Molecular strings representations case study: ibuprofen.
| Type | Molecules representation |
|---|---|
| IUPAC Name | 2-[4-(2-methylpropyl)phenyl]propanoic acid |
| SMILES (Canonical or Isomeric) | CC(C)Cc1ccc(cc1)[C@@H](C)C(=O)O |
| DeepSMILES [55] | CCC)Ccccccc6))[C@@H]C)C==O)O |
| SELFIES [74] | [C][C][Branch1][C][C][C][C][=C][C][=C][Branch1][Branch1][C][=C][Ring1] |
| [=Branch1][C@@H1][Branch1][C][C][C][=Branch1][C][=O][O] | |
| InChI | 1S/C13H18O2/c1-9(2)8-11-4-6-12(7-5-11)10(3)13(14)15/ |
| h4-7,9–10H,8H2,1–3H3,(H,14,15) | |
| InChIKey | HEFNNWSXXWATRW-UHFFFAOYSA-N |
5.1.1. Pre-training dataset
The dataset serves as the input to the ML algorithm, facilitating the learning of patterns, relationships, and features by the LMs. Herein, dataset is categorized into two primary groups based on pre-training and fine-tuning strategies, aiming to fully utilize the potential of transformer-based CLMs. A pre-training dataset typically enables the model to acquire more robust patterns and enhance generalization to unseen data. Creating an effective pre-training dataset involves meticulous data collection, pre-processing, and cleaning to ensure that the model learns meaningful patterns. The quality of the pre-training dataset significantly impacts the model's performance and it's ability to generalize to new data.
5.1.2. Fine-tuning dataset
The fine-tuning serves as the foundation for adapting a pre-trained model to a specific task or domain, enabling the model to learn task-specific patterns and achieve optimal performance, leveraging transfer learning by building on the knowledge and representations learned by the pre-trained model. The fine-tuning dataset plays a crucial role in this process by providing examples that are relevant to the target task. The size of the fine-tuning dataset can vary depending on factors such as task complexity, the availability of labeled data, and computational resources. Generally, a larger and higher quality fine-tuning dataset with accurate labels and minimal errors, enhances the model's capacity to learn robust patterns and generalize effectively to unseen data. Conversely, poor-quality data may impede performance and necessitate additional pre-processing or cleaning efforts. Fine-tuning datasets are often divided into training, validation, and test sets, with the former employed to update the model's parameters during fine-tuning and the latter utilized to monitor performance and fine-tune hyperparameters.
All transformer-based CLMs in this review study are fine-tuned or evaluated on several regression and classification tasks from MoleculeNet introduced by Wu et al. [61]. MoleculeNet, a large scale benchmark for molecular machine learning (ML) that curates multiple public datasets as shown in Table 2. However most data still struggle to deal with complex tasks under data scarcity and highly imbalanced classification.
Table 2.
Datasets used in cheminformatics for pre-training and fine-tuning transformer-based CLMs.
| Pre-training | Fine-tuning | SMILES Data | Task | # Task | # Compounds | ||
|---|---|---|---|---|---|---|---|
| Databases | ZINC [5], [56] | Category | Physical Chemistry [61] | ESOL | R | 1 | 1128 |
| PubChem [57], [58] | FreeSolv | R | 1 | 642 | |||
| ChEMBL [59], [60] | Lipophilicity | R | 1 | 4200 | |||
| Biophysics [61] | PCBA | C | 128 | 437929 | |||
| MUV | C | 17 | 93087 | ||||
| HIV | C | 1 | 41127 | ||||
| PDBbind | R | 1 | 11908 | ||||
| BACE | C | 1 | 15513 | ||||
| Physiology [61] | BBBP | C | 1 | 2039 | |||
| Tox21 | C | 12 | 7831 | ||||
| ToxCast | C | 617 | 8575 | ||||
| SIDER | C | 27 | 1427 | ||||
| ClinTox | C | 2 | 1478 | ||||
| Proposed[62], [63], [64] | Antimalarial | C | 1 | 4794 | |||
| Cocrystals | C | 1 | 3282 | ||||
| Covid | C | 1 | 740 | ||||
| Genes | R | 1 | 201 |
R - Regression.
C - Classification.
5.2. SMILES tokenization and encoding
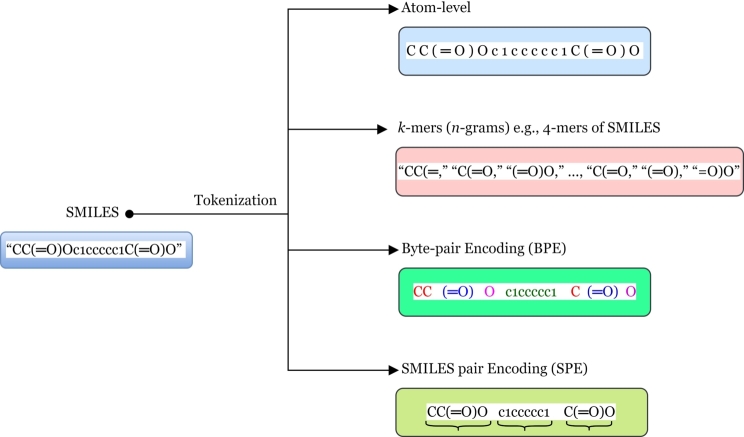
In the context of NLP especially transformer-based models in cheminformatics, tokenization refers to the process of breaking down SMILES strings data into their constituent elements or substructure tokens. Tokens are then converted into embeddings—numerical representations that capture the semantic information of the tokens. These embeddings undergo processing through a series of identical layers, with each layer comprising two sub-layers: multi-head self-attention and position-wise feedforward neural networks. SMILES strings represents chemical structures using a simplified notation system, where each character or group of characters represents specific elements, bonds, or substructures features such as functional groups. During data-driven tokenization of SMILES data, each character or group of characters that represents a specific element, bond, or structural feature is identified and treated as a separate token as shown in Fig. 3. The partitioning or tokenization alter the perception of molecules owing to the internal relationships among its atomic elements or components, and allows NLP models to process and understand the chemical structure represented by the SMILES strings data more effectively. Tokenization is a crucial preprocessing step before feeding SMILES data into a NLP models to facilitate both pretraining and fine-tuning tasks such as chemical property prediction, reaction prediction (i.e., drug-target predictions, API-coformer predictions). Tokenization can significantly influence prediction quality within the framework of text generation [65]. In this realm, tokenization of SMILES accelerates and increases the SMILES-based prediction performance [66].
Figure 3.
Tokenization mechanism.
In the feld of cheminformatics, SMILES tokenization is typically different compared to other NLP domains which involve features like white spaces within a sequence and more than one sequence of sentences. In SMILES, there is no presence of white space, and parentheses play the essential role of grammatical punctuation by indicating the start and end of a branch [28]. Therefore, square brackets are used to indicate atoms and charged atoms. During tokenization parentheses and square brackets are crucial for tokenizers to understand the substructures effectively. Additionally, there are two tokens namely <CLS> defined as the class token and <SEP> which indicates separation token, however in SMILES case it represent end of sequence as there is only single sequence input. After tokenization of SMILES strings, each token is mapped to three vectors: Q, K, and V, which are generated from the embeddings of the input tokens.
Previous studies have discussed and demonstrated various tokenization algorithms of SMILES towards SMILES-based prediction performance as shown in Table 3 and Fig. 3. In the field of Chemistry, tokenizing SMILES into atom-wise tokens is primarily method used for training chemical NLP models utilizes the character-level tokenization with certain adjustment to ensure that atoms are identified and extracted as individual tokens. Alongside this atom-wise SMILES tokenization approach, there have been recent introductions of novel molecular representations like SELFIES and DeepSMILES. Additionally, specialized tokenization methods such as byte-pair encoding (BPE), have emerged [65]. BPE is a hybrid combines elements of both character and word-level representations, enabling the management of extensive vocabularies in natural language collections. It operates on the principle that uncommon or unfamiliar words can often be broken down into recognizable subwords. BPE identifies the optimal segmentation of words by iteratively and greedily combining commonly occurring pairs of characters [13]. As observed in Table 3, most of the studies have been inspired or employed BPE as their architectural back-bone for their proposed tokenization algorithms such as a data-driven substructural tokenization algorithm namely SMILES pair encoding (SPE) [67].
Table 3.
Tokenization strategies from Previous studies.
5.3. Embedding information
Following the tokenization of the SMILES string data is the computation of embedding information. The tokens are first embedded into the feature space. Besides the token embedding, positional embedding is also included to add sequential information used in self-attention layer to utilize the temporal information of the inputs. Positional embeddings (PEs) are crucial for representing sequential data and grasping complex relationships within sequences, allowing models to capture both the presence and the order of tokens. Various PEs have been utilized such as absolute, relative key, relative key query, rotary [68], and sinusoidal positional encoding [11]
Notably, transformer-based CLMs in chemistry differs from other domains in the following perspectives:
-
•
Single Masked SMILES Recovery is employed on large scale unlabeled dataset.
-
•
Segmentation embedding is not included in models like BERT and their variants since SMILES data does not involve the continuous sentences training.
5.4. Pre-training
In the context of transformer-based language models (LMs) applied to SMILES, pre-training involves training the model on a large dataset of chemical structures represented using SMILES notation. The model architecture used for pre-training is based on transformers, a type of neural network (NN) architecture known for its self-attention mechanism which is particularly well-suited for learning dependencies. As a result, transformer-based LMs have demonstrated great effectiveness in capturing long-range dependencies and contextual information in sequences or within SMILES strings. Pre-training in transformer-based CLMs refers to the initial phase of training where a model is pre-trained on a large corpus and diverse corpus of unlabeled text data (i.e., SMILES) before being fine-tuned on a specific downstream tasks. Thus, pretraining offers several advantages, especially in chemical tasks such as:
-
•
Rich Representations: Pre-training on large datasets (e.g., molecular structures or chemical reactions) helps the model learn general patterns, such as chemical bonding, molecular interactions, and reaction mechanisms. This provides a strong foundation for downstream tasks.
-
•
Data Efficiency: Pre-training reduces the need for large, labeled datasets in specific chemical tasks, as the model can leverage knowledge from pre-training to quickly adapt to new tasks with fewer data points.
-
•
Improved Generalization: Pre-trained models generalize better, as they have been exposed to diverse chemical data. This improves their ability to handle complex tasks like molecular property prediction or reaction outcome forecasting.
Herein, the CLMs are trained on a large amount of data without any specific task labels, and this is often referred to as an unsupervised learning strategy. During unsupervised pre-training, the objective is typically a language modeling task known as masked language modeling (MLM), where the model learns to predict the next word or token in a sequence given the context of the preceding words. Pre-training allows the model to capture rich contextual information, general language patterns, semantics and linguistic nuances from diverse text sources in this case chemical contextual and topological information of SMILES compounds. The pre-trained model acts as a strong language understanding base that can be adapted to different tasks, making it more effective and efficient in which it has been successful in improving the performance of language models (LMs). The size of the pre-trained model and the amount of data are crucial factors in determining the model's performance.
5.5. Fine-tuning
Fine-tuning is the subsequent phase after pre-training, where the learned parameters of the pre-trained model can be transferred to a specific downstream task. This fine-tuning involves further training the model on a smaller, task-specific labeled datasets, adjusting the weights based on the task's objectives and more focused on the specific application or domain of interest. The pre-trained model, having learned general language features and equipped with a foundational understanding of the structural characteristics and chemical context of molecules encoded in SMILES notation, can then be fine-tuned on task-specific datasets for more specialized applications within the domain of computational chemistry and cheminformatics. Fine-tuning is faster and requires less labeled data than training a model from scratch for various tasks such as classification and regression. Therefore, fine-tuning is paramount considering various factors such as:
-
•
Task Specialization: Chemical tasks (e.g., toxicity prediction, drug discovery) often require specific knowledge. Fine-tuning adapts the general representations from pre-training to these specialized tasks, allowing the model to focus on relevant features.
-
•
Performance Boost: Fine-tuning refines the model's parameters on the specific task data, leading to improved accuracy and task-specific performance, ensuring that the pre-trained knowledge is fully leveraged and optimized for the target chemical problem.
Thus, pre-training provides a broad knowledge base for the model to learn a general and broad understanding of the relationships, patterns, and features within molecular structures encoded in the SMILES format. While fine-tuning adapts this general knowledge to specific cheminformatics downstream tasks, leading to more effective and task-specific models. This two-step process has been instrumental in the success of transformer-based CLMs in various NLP applications. In addition, to enable meaningful comparisons between CLMs in cheminformatics, a standardized set of evaluation metrics is essential. These metrics assess the performance across multiple dimensions, such as molecular representation, property prediction, and reaction modeling. These can be observed in various works in cheminformatics including Chemberta [13], SMILES-BERT [12], Mol-BERT [49].
6. Challenges and limitations
This section addresses these challenges and suggests potential avenues for future research. Although successful, transformer and attention-based models face limitations in handling large-scale chemical datasets.
Data efficiency: Despite their ability to generalize well with limited labeled data, transformer models may still require substantial amounts of training data to achieve optimal performance, particularly for complex tasks or rare molecular properties. Given that pre-training language models such as CLMs requires substantial amounts of data, this challenge can be addressed through various strategies to mitigate the demand for extensive datasets:
-
•
Transfer Learning: By pre-training models on large, general chemical datasets, then fine-tuning on specific tasks with limited data, the model leverages learned knowledge to improve performance on data-scarce tasks.
-
•
Data Augmentation: This involves creating variations of existing chemical data, such as altering molecular structures while preserving their properties, to increase the diversity of training samples without needing new data.
-
•
Synthetic Data Generation: Generating synthetic chemical data using techniques like generative models such as generative adversarial networks (GANs) [69] or variational autoencoders (VAEs) [70] can provide additional training examples, enhancing data efficiency and helping the model learn a broader set of chemical patterns.
These approaches improve data efficiency, reduce dependency on large datasets, and enhance the performance of chemical language models.
Interpretability: The inherent complexity of transformer architectures can make it challenging to interpret the learned representations and understand how specific molecular features contribute to property predictions.
Computational resources: Training transformer models, especially large-scale variants, demands significant computational resources and time, which may limit their accessibility to researchers with constrained resources. Computational resource constraints are a significant concern when training and deploying large chemical language models (CLMs). To address these limitations, various optimization techniques can be employed:
-
•
Model Pruning: This involves removing less critical neurons or layers in the model, reducing its size and computational demands while maintaining performance.
-
•
Quantization: By reducing the precision of model weights (e.g., using 8-bit instead of 32-bit floating points), quantization lowers memory and computation requirements, enabling faster inference without significant loss in accuracy.
-
•
Distributed Training: Distributing the training process across multiple GPUs or nodes accelerates the training of large models, allowing them to scale efficiently and complete in a reasonable time frame despite hardware constraints.
These strategies help optimize CLMs for efficiency, making them more accessible in resource-limited environments.
Handling rare molecules: Transformers may struggle to effectively model rare or novel molecular structures that deviate significantly from the patterns observed in the training data. Handling rare molecules in CLMs is challenging due to the limited availability of training examples for these compounds. This might be addressed through:
-
•
Domain Adaptation: Adapting a pre-trained model from a broad chemical domain to a specific subset of rare molecules helps the model better capture unique molecular features, improving performance even with limited examples.
-
•
Few-Shot Learning: This technique allows CLMs to learn from only a few examples by leveraging prior knowledge from pre-training. It enables the model to generalize to unseen or rare molecules with minimal additional data.
Both strategies help improve the model's ability to handle rare molecules by efficiently transferring or adapting knowledge from more common data.
Majority of the attention-based CLMs proposed and discussed in this review utilize SMILES particularly canonical SMILES notations, which serves as the most widely adopted string-based encoding for molecules, in both pre-training and fine-tuning phase in this field as shown in Table 1. Despite being widely used and showing potential utility in computational chemistry, SMILES has its limitations and poses several challenges concerning its validity and robustness, potentially impeding the model's ability to effectively extract the underlying knowledge within the data [47]. As discussed by Yüksel et al. [47] the limitations of SMILES-based representations include:
-
•
The potential for non-canonical representations of the same molecule, reducing string uniqueness.
-
•
In SMILES notation, many symbol combinations yield invalid outcomes that lack a valid chemical interpretation [71].
-
•
The possibility of valid SMILES strings having invalid chemical properties, such as exceeding atom valency [72].
-
•
Inadequate capture of spatial information.
-
•
Lastly insufficiency in fully representing molecular characteristics due to its lack of syntactic and semantic robustness [71], [73].
Furthermore, current benchmark datasets may lack the comprehensiveness needed to tackle emerging real-world challenges and diseases effectively. Therefore, the integration of novel benchmark datasets focusing on diseases like malaria, cancer, and COVID-19 is essential. These new datasets can significantly aid transformer-based CLMs in comprehensively exploring and evaluating their efficacy in addressing complex real-world scenarios. By incorporating datasets that specifically target these diseases, researchers can enhance the capacity of transformer-based models to address pressing healthcare challenges and contribute to the advancement of diagnostic and therapeutic solutions.
7. Future research
Novel molecular representation: While SMILES stands as the most commonly and widely used and has demonstrated potential utility in computational chemistry, it is somehow insufficient to fully represent the complexity and properties of the molecules [47] Therefore, other molecular representations as shown in Table 4 such as DeepSMILES, SELFIES, IUPAC Names, InChI could be taken into consideration and utilized during pre-training and fine-tuning to access the performance of transformer and attention-based models. For example, DeepSMILES were proposed as an improvement of SMILES, to address unbalanced parentheses and ring closure pairs which cause invalid syntax [55]. SELFIES encapsulates all molecular graph information, including atoms, bonds, branches, and rings using characters enclosed in brackets and therefore, it appears to effectively resemble the unique identification of a given molecule [71], [74]. For example, normal benzene SMILES is ‘c1ccccc1’ when converted from SMILES to SELFIES will be ‘[C][=C][C][=C][C][=C][Ring1][=Branch1]’, and lastly if converted from SELFIES back to SMILES will be ‘C1=CC=CC=C1’. Recent discussions have extensively explored potential future extensions of SELFIES aimed at addressing the limitations of current molecular string representations [3], [71]. In addition SELFIES may also serve as inspiration for potential variants of SMILES and DeepSMILES [3]. Expanding chemical languages to encompass more intricate molecular entities presents a promising avenue for advancing the capabilities of transformer based CLMs in chemistry. This expansion could involve addressing complex structures like proteins and peptides containing non-natural amino acids, as well as exploring areas such as crystals and supramolecular chemistry.
Role of SMILES token characters: Understanding the role of different SMILES token characters—such as metals, non-metals, bonds, and branches—is crucial in the development and enhancement of chemical language models (CLMs). By examining how these specific parts of the SMILES strings impact the model's performance, especially in terms of information loss, researchers can improve the focus of CLMs on essential tokens rather than relying on random masked language modeling (MLM). For instance, during the pre-training phase of MLM, researchers can strategically hide tokens representing metals, non-metals, bonds, or branches in the SMILES sequences. By doing so, they can assess how the model performs with these tokens masked, thereby gaining insights into which components are most critical for accurate molecular property prediction. This targeted masking approach helps in refining the CLMs to concentrate on significant elements of the SMILES strings, ultimately enhancing their predictive capabilities.
Few-shot learning: The approach of few-shot learning is particularly pertinent in fields like NLP and Chem-informatics, where acquiring labeled data can be challenging. Few-shot learning entails training a model with a small number of labeled examples per class, aiming for effective generalization with minimal training data. This approach becomes particularly valuable in situations where gathering extensive labeled data is impractical or costly. By combining few-shot learning approaches with large-scale pre-trained transformer based CLMs, prospective applications of CLMs are anticipated to see significant enhancements [3], [75]. Furthermore, enhancing the capability of CLMs in cheminformatics to propose synthesizable molecules holds the potential to elevate their practical relevance in both cheminformatics and bionformatics for drug discovery efforts [76].
Effective analysis in drug repurposing or repositioning: Drug repurposing, also known as drug repositioning, represents a cost-effective strategy for identifying new therapeutic indications for existing drugs within a short period of time. This approach holds promise in addressing challenges such as the emergence of drug resistance and the reemerging of viral infections [77]. In the context of drug repurposing, certain drug molecules may exhibit diverse biological activities, showing efficacy or lack thereof against specific targeted organisms across different clinical trials aimed at treating various diseases using similar drug compounds. For instance, a chemical compound that demonstrates promising efficacy in treating COVID-19 may exhibit inactive effects when used to combat malaria. Therefore, it becomes imperative for LMs such as transformer based models to discern between identical compound molecules exhibiting multiple biological or chemical activity profiles across different therapeutic domains.
Improvement of donwnstream task datasets: Recent benchmark datasets [61] in the field of NLP shown in Table 2, while valuable for assessing model performance, may not fully address the complexities and challenges encountered in real-world scenarios, particularly in the context of diseases like malaria, cancer, and COVID-19. To overcome this limitation and facilitate more comprehensive evaluations, it is imperative to incorporate new benchmark datasets that specifically target these diseases. By leveraging datasets that reflect the intricacies of these conditions, transformer-based models can be better equipped to explore and evaluate their effectiveness in addressing real-world challenges. This approach not only enhances the applicability of these models but also contributes to advancements in disease diagnosis, treatment, and prevention strategies. Therefore, in this review, below and as shown in Table 2, we propose various new datasets that we are convinced can be added and utilized during fine-tuning to address the aforementioned challenges.
-
1.
Covid dataset [78]. This dataset consist of drug chemical compounds that have shown experimental activeness against coronavirus drug targets such as SARS-CoV, MERS-CoV, SARS-CoV-2, ORF1ab - ORF1a polyprotein; ORF1ab polyprotein (Betacoronavirus England 1), surface glycoprotein (SARSCoV-2), and Replicase polyprotein 1ab (SARS-CoV-2)). Furthermore, in this dataset we incorporate the information about the chemical structure representation of the proven drug target enzyme i.e., Mpro, or 3CLpro which is a proven drug discovery target in the case of the SARS-CoV-2 that shares a genome sequence similar to that of other members of the betacoronavirus group like MERS-CoV and SARS-CoV. This target is also essential for the replication and life cycle of coronaviruses.
-
2.
Cocrystal formation dataset [62]. The dataset comprises chemical reactions between active pharmaceutical ingredients (APIs) and different coformers, with the aim of forming cocrystals. APIs have garnered significant pharmaceutical interest due to their intrinsic properties, and are consequently employed as adjunct solid dosage forms following FDA guidance and approval of pharmaceutical cocrystals as a promising avenue for drug substance development.
-
3.
Antimalarial drugs dataset [63]. This dataset encompasses drugs exhibiting antimalarial activity against Plasmodium falciparum, serving as a valuable resource for various ML models aimed at predicting and facilitating the discovery of novel antimalarial drugs. In addition to drugs, the dataset includes three potential and high-priority targets—Acetyl CoA Synthetase (PfAcAS), Bifunctional Farnesyl/Geranylgeranyl Pyrophosphate Synthase (F/GGPPS), and Monoacylglycerol Lipase (PfMAGL)—present in Plasmodium and crucial for advancing antimalarial drug discovery efforts. Despite global initiatives, malaria remains a formidable health challenge, affecting nearly half of the world's population in 2020. The emergence of parasite resistance to existing antimalarial treatments or drugs have drawn the attention and critical need for innovative therapeutic strategies capable of addressing the diverse stages of the Plasmodium life cycle.
-
4.
Cancer dataset [64]. The dataset comprises a gene expression matrix containing data from 8,046 genes across 734 cell lines, along with a drug response matrix encompassing 201 drugs tested on the same cell lines. By integrating gene expression data with biological networks, the dataset encapsulates crucial insights into the mechanisms underlying drug responses in cancer treatment. Cancer patients' genomic profiles, including gene expression patterns, have emerged as pivotal resources for predicting drug responses in the realm of personalized medicine. With the availability of extensive drug screening datasets involving cancer cell lines, numerous computational methodologies, notably ML approaches, have been employed for the prediction of drug responses.
However, when fine-tuning transformer-based CLMs, it is crucial that newly proposed datasets meet certain criteria to ensure the model learns to generalize across diverse chemical tasks and environments. Such key considerations include:
-
•
Diverse chemical structures: The dataset should encompass a wide range of molecular structures, including small molecules, polymers, peptides, and more complex organic and inorganic compounds. This ensures that the model is exposed to different bonding patterns, functional groups, and stereochemistry, enhancing its ability to understand a variety of molecular architectures.
-
•
Varying levels of complexity: Including molecules with different levels of structural and chemical complexity is essential. This means incorporating both simple, well-known compounds (e.g., methane, ethanol) and more intricate ones (e.g., natural products, pharmaceuticals). This allows the model to develop a robust understanding of both fundamental chemical principles and more nuanced molecular behaviors.
-
•
Representation of different chemical environments: The dataset should capture molecules under various conditions, such as different solvents, temperatures, pH levels, and reaction environments. This helps the model adapt to the variability in how molecules behave in different settings, which is critical for tasks like reaction prediction and property estimation.
-
•
Coverage of functional groups and reactivity: A well-rounded dataset should include molecules with a wide range of functional groups (e.g., alcohols, amines, ketones) and reactivity patterns. This diversity ensures that the model can predict and understand chemical transformations and interactions relevant to different fields, from organic synthesis to material science.
-
•
Balanced data representation: Ensuring that no single class of molecules dominates the dataset is crucial for balanced learning. Over-representation of certain types of molecules (e.g., small organic compounds) could bias the model and hinder its performance on other less-represented molecular classes (e.g., transition metal complexes).
-
•
Task-relevant annotations: For specific fine-tuning tasks, the dataset should contain relevant annotations, such as molecular properties (e.g., solubility, toxicity, reactivity), bioactivity data, or reaction outcomes. This allows the CLM to specialize in predictive tasks or property estimation by learning the associations between chemical structures and their real-world behaviors.
By ensuring that fine-tuning datasets meet these criteria, the chemical language model will be able to generalize well to a broad range of tasks, from molecular property prediction to chemical reaction modeling, and improve its ability to handle the complexities of real-world chemical environments.
The dual use risk of and potential harms of CLMs: The integration of AI such as CLMs in chemistry indeed raises significant ethical questions, particularly regarding unintended consequences and biases. For instance, Ahmad et al. [37] not only open-sourced their trained models but also emphasized the dual-use risks associated with CLMs. They argue that the challenge of synthesizing novel molecules serves as a safeguard against potential harms that could arise from the release of updated models. Furthermore, they acknowledge that the balance of risks may change as their research progresses, and they have committed to continuously evaluating the dual-use implications of future open-source releases.
-
•Unintended consequences
-
–Safety risks: CLMs may suggest novel chemical compounds or reactions that, while theoretically sound, could pose safety risks in practice. Unanticipated reactions or toxic byproducts might emerge, leading to hazardous outcomes.
-
–Environmental impact: CLM-driven processes for materials synthesis or drug discovery might inadvertently prioritize efficiency over sustainability, resulting in harmful environmental effects. This includes the potential for increased waste or the development of substances that could harm ecosystems.
-
–Over-reliance on automation: As CLMs, take on more decision-making roles, there may be a risk of diminishing critical thinking among researchers, leading to over-reliance on AI suggestions without adequate scrutiny or validation.
-
–
-
•Biases
-
–Data Bias: CLMs are trained on existing datasets, which may be biased due to underrepresentation of certain chemical structures or properties. This can lead to skewed predictions that favor more common compounds while neglecting rare or novel ones, limiting innovation and discovery in important areas.
-
–Reinforcement of existing inequalities: If the datasets used to train CLMs reflect historical biases in chemical research (such as underrepresentation of certain demographics in drug development), the CLMs may perpetuate these inequalities by favoring the needs of well-studied populations while overlooking others.
-
–Interpretation bias: The interpretation of CLM-generated results may be influenced by human biases, affecting how findings are perceived and applied in research and industry. This could lead to misinformed decisions based on CMLs outputs.
-
–
Addressing these ethical questions requires a proactive approach, including careful dataset curation, ongoing evaluation of CLMs, and fostering a culture of responsible CLMs use in the chemical sciences. Engaging with diverse stakeholders and prioritizing transparency can help mitigate risks and ensure that CLMs serves to advance ethical and equitable practices in chemistry.
8. Discussion
Dataset bias can significantly impact the performance of CLMs, potentially limiting their generalizability and effectiveness in real-world applications. When datasets are not representative of the broad spectrum of chemical diversity and complexity, models trained on them may develop skewed or incomplete understandings of chemical principles. Here are the potential impacts of dataset bias and why ensuring representative datasets is crucial:
-
•
Reduced generalization: If a dataset is biased toward specific classes of molecules (e.g., small organic compounds), the CLM may struggle to generalize to underrepresented molecular categories, such as inorganic complexes, natural products, or polymers. This leads to poor performance when the model encounters unfamiliar chemical structures or environments in real-world tasks.
-
•
Suboptimal prediction of molecular properties: A biased dataset might overemphasize certain molecular properties (e.g., small molecules with known pharmacological properties) and ignore others (e.g., materials science compounds). As a result, the model may perform well in predicting specific chemical attributes (like drug-likeness) but poorly in others (like catalytic efficiency or solubility in non-standard solvents), limiting its usefulness across different domains of chemistry.
-
•
Inaccurate reaction predictions: Biases in reaction datasets can cause CLMs to favor certain reaction mechanisms or conditions that are over-represented in the training data, leading to inaccurate or incomplete predictions for less common reactions. For example, if the dataset heavily features common organic reactions, the model may fail to predict outcomes in organometallic chemistry or biocatalysis.
-
•
Challenges in handling rare or complex molecules: Dataset bias against rare, large, or structurally complex molecules can lead to poor handling of these types in real-world scenarios. This becomes a critical issue in areas like drug discovery or materials design, where unique molecular scaffolds or highly functionalized structures are often key to innovation.
-
•
Overfitting and narrow focus: Models trained on biased data may overfit to specific molecular patterns or chemical properties, narrowing their focus. This overfitting makes the model less flexible and more prone to errors when faced with new tasks that require broader chemical knowledge or cross-domain adaptability.
Nevertheless, representative datasets for real-world challenges are of utmost importance for various reasons, such as:
-
•
Capturing chemical diversity: Real-world chemical challenges involve a wide range of molecular structures, chemical reactions, and environmental conditions. Ensuring that datasets capture this diversity—spanning small molecules, macromolecules, organometallic compounds, and biologically relevant structures—allows the CLM to better predict real-world chemical behavior and outcomes. Representative datasets help prevent models from becoming overly specialized and allow them to remain versatile across various domains.
-
•
Balanced molecular property distribution: It's essential that datasets contain molecules with a variety of properties, such as polarity, solubility, reactivity, and toxicity. This balance ensures the model can perform well in diverse tasks, from drug design to materials science, and enhances the model's robustness when predicting different chemical properties.
-
•
Addressing underrepresented domains: Representative datasets ensure that underexplored areas of chemistry, such as sustainable catalysis, green chemistry, and rare molecular scaffolds, are included. Through this, CLMs are better prepared to tackle cutting-edge research challenges and can contribute to areas where innovative solutions are needed, such as renewable energy materials or environmentally friendly chemical processes.
-
•
Improving transferability across tasks: Models trained on biased datasets may have limited ability to transfer learned knowledge to new tasks or domains. By ensuring the dataset is comprehensive and diverse, the model can more easily adapt to novel challenges, making it more useful across different branches of chemistry, from pharmaceuticals to environmental chemistry.
-
•
Preventing systematic errors: A representative dataset helps in reducing the likelihood of systematic errors. For example, if a dataset disproportionately represents reactions or molecular structures from academic literature, it may underperform in industrial contexts where reactions may follow different protocols. Broadening the dataset to include real-world industrial reactions ensures the model's robustness in a variety of applications.
To ensure that CLMs are robust, flexible, and capable of addressing real-world chemical challenges, it is crucial to mitigate dataset bias and ensure newly proposed datasets are representative of the full range of chemical diversity, environments, and molecular properties. This enhances the model's generalizability, reduces errors, and ensures it performs optimally across various domains in chemistry, from drug discovery to materials science and environmental chemistry.
Future research should also focus on optimizing these models for better data efficiency, optimizing computational requirements, and enhancing model interpretability. Developing methods to better handle rare or novel molecular structures will further expand the applicability of transformers in computational chemistry. Additionally, integrating these CLMs with other ML approaches, such as graph neural networks (GNNs) [63], CNNs [79], could further improve their capability to understand and predict complex molecular properties.
9. Conclusion
Transformer-based models have revolutionized computational chemistry, offering robust tools for analyzing and predicting molecular properties from SMILES representations. Their ability to learn complex relationships or dependencies, scale to large datasets, and leverage contextual information positions them as one of the powerful tools in drug discovery, molecular design, and property prediction. CLMs have had a transformative impact on cheminformatics by revolutionizing the way chemical data is analyzed and interpreted. They have significantly enhanced the ability to predict molecular properties, design novel compounds, and streamline drug discovery by leveraging vast chemical datasets in ways that were previously unattainable. CLMs have enabled more accurate and scalable representations of molecular structures, improving both efficiency and precision in cheminformatics tasks. Beyond cheminformatics, the broader implications of CLMs extend to related fields such as materials science, where they assist in the discovery of new materials, and synthetic biology, where they aid in the design of biological molecules. Their ability to process and model complex chemical and biological information is driving innovation across multiple scientific disciplines.
As the field continues to evolve, ongoing innovations and addressing current limitations will be crucial to further enhance the potential capabilities and applications of these CLMs in cheminformatics. In general, this literature review summarizes the evolution of molecular representation methods, focusing on the emergence of transformer and attention-based CLMs. By providing a thorough overview of their architectures, applications, and challenges, this review aims to guide researchers and practitioners in leveraging these advanced CLMs for enhanced molecular analysis and prediction.
For the potential future directions of this review, we plan to address various important issues in our forthcoming work, such as: Integration with experimental chemistry: Explore how transformer-based CLMs can be integrated with experimental workflows, such as reaction optimization or compound screening. Discuss the potential for AI to accelerate real-time experimental design and feedback. Expansion to Multimodal Models: Address the potential for combining chemical data with other modalities (e.g., spectroscopy, biological data) to create more holistic models that better represent complex chemical systems and drug discovery pipelines. Ethical and Regulatory Considerations: Emphasize the growing importance of addressing ethical issues, such as dual-use risks, data biases, and the need for regulations governing AI in chemistry. This future direction could drive more responsible development and deployment of CLMs. Scalability and Resource Efficiency: Discuss advances in model pruning, quantization, and distributed training to address computational constraints. Highlight how these techniques can make large-scale models more accessible and environmentally sustainable. Personalized Medicine and Chemical Design: Suggest how future CLMs could be tailored for applications like personalized drug discovery or the design of custom materials, where predictions are finely tuned for individual or specialized use cases.
Funding
Following are results of a study on the “Leaders in INdustry-university Cooperation 3.0” Project, supported by the Ministry of Education and National Research Foundation of Korea. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2020R1A6A1A12047945).
CRediT authorship contribution statement
Medard Edmund Mswahili: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Young-Seob Jeong: Validation, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The Data Analysis and Artificial Intelligence (DAI) Lab http://dai.chungbuk.ac.kr/.
Data availability
No additional data was used for the research described in the article.
References
- 1.Weininger D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 2.Arús-Pous J., et al. Randomized smiles strings improve the quality of molecular generative models. J. Cheminform. 2019;11:1–13. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grisoni F. Chemical language models for de novo drug design: challenges and opportunities. Curr. Opin. Struct. Biol. 2023;79 doi: 10.1016/j.sbi.2023.102527. [DOI] [PubMed] [Google Scholar]
- 4.Sadybekov A.V., Katritch V. Computational approaches streamlining drug discovery. Nature. 2023;616:673–685. doi: 10.1038/s41586-023-05905-z. [DOI] [PubMed] [Google Scholar]
- 5.Irwin J.J., et al. Zinc20—a free ultralarge-scale chemical database for ligand discovery. J. Chem. Inf. Model. 2020;60:6065–6073. doi: 10.1021/acs.jcim.0c00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L., Su Y., Liu Y., Zeng X. Review of unsupervised pretraining strategies for molecules representation. Brief. Funct. Genomics. 2021;20:323–332. doi: 10.1093/bfgp/elab036. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Wang H., Wang X., Chang H. Predicting drug-target interactions via fm-dnn learning. Curr. Bioinform. 2020;15:68–76. [Google Scholar]
- 8.Zeng X., et al. Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics. 2020;36:2805–2812. doi: 10.1093/bioinformatics/btaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters W.P., Barzilay R. Applications of deep learning in molecule generation and molecular property prediction. Acc. Chem. Res. 2020;54:263–270. doi: 10.1021/acs.accounts.0c00699. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., et al. Ai-based language models powering drug discovery and development. Drug Discov. Today. 2021;26:2593–2607. doi: 10.1016/j.drudis.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaswani A., et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017;30 [Google Scholar]
- 12.Wang S., Guo Y., Wang Y., Sun H., Huang J. Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. 2019. Smiles-bert: large scale unsupervised pre-training for molecular property prediction; pp. 429–436. [Google Scholar]
- 13.Chithrananda S., Grand G., Ramsundar B. Chemberta: large-scale self-supervised pretraining for molecular property prediction. 2020. arXiv:2010.09885 preprint.
- 14.Fabian B., et al. Molecular representation learning with language models and domain-relevant auxiliary tasks. 2020. arXiv:2011.13230 preprint.
- 15.Yang K., et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 2019;59:3370–3388. doi: 10.1021/acs.jcim.9b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr A., et al. Large-scale comparison of machine learning methods for drug target prediction on chembl. Chem. Sci. 2018;9:5441–5451. doi: 10.1039/c8sc00148k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai A.M., Le Q.V. Semi-supervised sequence learning. Adv. Neural Inf. Process. Syst. 2015;28 [Google Scholar]
- 18.Xu Z., Wang S., Zhu F., Huang J. Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. 2017. Seq2seq fingerprint: an unsupervised deep molecular embedding for drug discovery; pp. 285–294. [Google Scholar]
- 19.Zhang X., et al. Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. 2018. Seq3seq fingerprint: towards end-to-end semi-supervised deep drug discovery; pp. 404–413. [Google Scholar]
- 20.Sherstinsky A. Fundamentals of recurrent neural network (rnn) and long short-term memory (lstm) network. Phys. D: Nonlinear Phenom. 2020;404 [Google Scholar]
- 21.Dey R., Salem F.M. 2017 IEEE 60th International Midwest Symposium on Circuits and Systems (MWSCAS) IEEE; 2017. Gate-variants of gated recurrent unit (gru) neural networks; pp. 1597–1600. [Google Scholar]
- 22.Devlin J., Chang M.-W., Lee K., Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. 2018. arXiv:1810.04805 preprint.
- 23.A. Radford, K. Narasimhan, T. Salimans, I. Sutskever, et al., Improving language understanding by generative pre-training. OpenAI, 2018.
- 24.Brown T., et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020;33:1877–1901. [Google Scholar]
- 25.Raschka S., Liu Y.H., Mirjalili V., Dzhulgakov D. Packt Publishing Ltd; 2022. Machine Learning with PyTorch and Scikit-Learn: Develop Machine Learning and Deep Learning Models with Python. [Google Scholar]
- 26.A. Abaskohi, Navigating transformers: a comprehensive exploration of encoder-only and decoder-only models, right shift, and beyond (2023) Accessed: 2024-09-26.
- 27.Li Z., Liu F., Yang W., Peng S., Zhou J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE Trans. Neural Netw. Learn. Syst. 2021;33:6999–7019. doi: 10.1109/TNNLS.2021.3084827. [DOI] [PubMed] [Google Scholar]
- 28.Lee I., Nam H. Infusing linguistic knowledge of smiles into chemical language models. 2022. arXiv:2205.00084 preprint.
- 29.Guo T., et al. What can large language models do in chemistry? A comprehensive benchmark on eight tasks. Adv. Neural Inf. Process. Syst. 2024;36 [Google Scholar]
- 30.Mao J., et al. Transformer-based molecular generative model for antiviral drug design. J. Chem. Inf. Model. 2023 doi: 10.1021/acs.jcim.3c00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adilov S. Generative pre-training from molecules. ChemRxiv. 2021 [Google Scholar]
- 32.Irwin R., Dimitriadis S., He J., Bjerrum E.J. Chemformer: a pre-trained transformer for computational chemistry. Mach. Learn.: Sci. Technol. 2022;3 [Google Scholar]
- 33.Liu Z., et al. Molxpt: wrapping molecules with text for generative pre-training. 2023. arXiv:2305.10688 preprint.
- 34.Oniani D., et al. Emerging opportunities of using large language models for translation between drug molecules and indications. 2024. arXiv:2402.09588 preprint. [DOI] [PMC free article] [PubMed]
- 35.Luo R., et al. Biogpt: generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 2022;23 doi: 10.1093/bib/bbac409. [DOI] [PubMed] [Google Scholar]
- 36.Omote Y., Matsushita K., Iwakura T., Tamura A., Ninomiya T. Proceedings of the 1st Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics and the 10th International Joint Conference on Natural Language Processing. 2020. Transformer-based approach for predicting chemical compound structures; pp. 154–162. [Google Scholar]
- 37.Ahmad W., Simon E., Chithrananda S., Grand G., Ramsundar B. Chemberta-2: towards chemical foundation models. 2022. arXiv:2209.01712 preprint.
- 38.Haroon S., Hafsath C., Jereesh A. Generative pre-trained transformer (gpt) based model with relative attention for de novo drug design. Comput. Biol. Chem. 2023;106 doi: 10.1016/j.compbiolchem.2023.107911. [DOI] [PubMed] [Google Scholar]
- 39.Xue, D. et al., X-mol: large-scale pre-training for molecular understanding and diverse molecular analysis. bioRxiv 2020–12 (2020). [DOI] [PubMed]
- 40.Balaji S., Magar R., Jadhav Y., et al. Gpt-molberta: Gpt molecular features language model for molecular property prediction. 2023. arXiv:2310.03030 preprint.
- 41.Fang Y., et al. Mol-instructions: a large-scale biomolecular instruction dataset for large language models. 2023. arXiv:2306.08018 preprint.
- 42.Honda S., Shi S., Ueda H.R. Smiles transformer: pre-trained molecular fingerprint for low data drug discovery. 2019. arXiv:1911.04738 preprint.
- 43.Tran T., Ekenna C. Molecular descriptors property prediction using transformer-based approach. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241511948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris P., St. Clair R., Hahn W.E., Barenholtz E. Predicting binding from screening assays with transformer network embeddings. J. Chem. Inf. Model. 2020;60:4191–4199. doi: 10.1021/acs.jcim.9b01212. [DOI] [PubMed] [Google Scholar]
- 45.Ross J., et al. Large-scale chemical language representations capture molecular structure and properties. Nat. Mach. Intell. 2022;4:1256–1264. [Google Scholar]
- 46.Krasnov L., Khokhlov I., Fedorov M.V., Sosnin S. Transformer-based artificial neural networks for the conversion between chemical notations. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-94082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yüksel A., Ulusoy E., Ünlü A., Doğan T. Selformer: molecular representation learning via selfies language models. Mach. Learn.: Sci. Technol. 2023 [Google Scholar]
- 48.Born J., Manica M. Regression transformer enables concurrent sequence regression and generation for molecular language modelling. Nat. Mach. Intell. 2023;5:432–444. [Google Scholar]
- 49.Li J., Jiang X. Mol-bert: an effective molecular representation with bert for molecular property prediction. Wirel. Commun. Mob. Comput. 2021;2021:1–7. [Google Scholar]
- 50.Liu Y., et al. Molrope-bert: an enhanced molecular representation with rotary position embedding for molecular property prediction. J. Mol. Graph. Model. 2023;118 doi: 10.1016/j.jmgm.2022.108344. [DOI] [PubMed] [Google Scholar]
- 51.Chen D., Zheng J., Wei G.-W., Pan F. Extracting predictive representations from hundreds of millions of molecules. J. Phys. Chem. Lett. 2021;12:10793–10801. doi: 10.1021/acs.jpclett.1c03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y., et al. Roberta: a robustly optimized bert pretraining approach. 2019. arXiv:1907.11692 preprint.
- 53.Lee J., et al. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Y., et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthcare. 2021;3:1–23. [Google Scholar]
- 55.O'Boyle N., Dalke A. Deepsmiles: an adaptation of smiles for use in machine-learning of chemical structures. ChemRxiv. 2018 [Google Scholar]
- 56.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. Zinc: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim S., et al. Pubchem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S., et al. Pubchem 2023 update. Nucleic Acids Res. 2023;51:D1373–D1380. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaulton A., et al. The chembl database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaulton A., et al. Chembl: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Z., et al. Moleculenet: a benchmark for molecular machine learning. Chem. Sci. 2018;9:513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mswahili M.E., et al. Cocrystal prediction using machine learning models and descriptors. Appl. Sci. 2021;11:1323. [Google Scholar]
- 63.Mswahili M.E., Ndomba G.E., Jo K., Jeong Y.-S. Graph neural network and bert model for antimalarial drug predictions using plasmodium potential targets. Appl. Sci. 2024;14:1472. [Google Scholar]
- 64.Kim S., Bae S., Piao Y., Jo K. Graph convolutional network for drug response prediction using gene expression data. Mathematics. 2021;9:772. [Google Scholar]
- 65.Ucak U.V., Ashyrmamatov I., Lee J. Improving the quality of chemical language model outcomes with atom-in-smiles tokenization. J. Cheminform. 2023;15:55. doi: 10.1186/s13321-023-00725-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Domingo M., Garcıa-Martınez M., Helle A., Casacuberta F., Herranz M. How much does tokenization affect neural machine translation? 2018. arXiv:1812.08621 preprint.
- 67.Li X., Fourches D. Smiles pair encoding: a data-driven substructure tokenization algorithm for deep learning. J. Chem. Inf. Model. 2021;61:1560–1569. doi: 10.1021/acs.jcim.0c01127. [DOI] [PubMed] [Google Scholar]
- 68.Su J., et al. Roformer: enhanced transformer with rotary position embedding. Neurocomputing. 2024;568 [Google Scholar]
- 69.Goodfellow I., et al. Generative adversarial networks. Commun. ACM. 2020;63:139–144. [Google Scholar]
- 70.Doersch C. Tutorial on variational autoencoders. 2016. arXiv:1606.05908 preprint.
- 71.Krenn M., et al. Selfies and the future of molecular string representations. Patterns. 2022;3 doi: 10.1016/j.patter.2022.100588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wigh D.S., Goodman J.M., Lapkin A.A. A review of molecular representation in the age of machine learning. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022;12 [Google Scholar]
- 73.Li Z., Jiang M., Wang S., Zhang S. Deep learning methods for molecular representation and property prediction. Drug Discov. Today. 2022;27 doi: 10.1016/j.drudis.2022.103373. [DOI] [PubMed] [Google Scholar]
- 74.Krenn M., Häse F., Nigam A., Friederich P., Aspuru-Guzik A. Self-referencing embedded strings (selfies): a 100% robust molecular string representation. Mach. Learn.: Sci. Technol. 2020;1 [Google Scholar]
- 75.Abdel-Aty H., Gould I.R. Large-scale distributed training of transformers for chemical fingerprinting. J. Chem. Inf. Model. 2022;62:4852–4862. doi: 10.1021/acs.jcim.2c00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao W., Coley C.W. The synthesizability of molecules proposed by generative models. J. Chem. Inf. Model. 2020;60:5714–5723. doi: 10.1021/acs.jcim.0c00174. [DOI] [PubMed] [Google Scholar]
- 77.Roy K. Academic Press; 2019. In silico drug design: repurposing techniques and methodologies. [Google Scholar]
- 78.Harigua-Souiai E., et al. Deep learning algorithms achieved satisfactory predictions when trained on a novel collection of anticoronavirus molecules. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.744170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang M., et al. Nonlinear modeling of temperature-induced bearing displacement of long-span single-pier rigid frame bridge based on dcnn-lstm. Case Stud. Therm. Eng. 2024;53 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data was used for the research described in the article.