Abstract
The fungi present in the breeding waters of mosquitoes have been scarcely investigated. This work explored the diversity of cultivable fungi present in the breeding sites of the South American malaria vector mosquito Anopheles darlingi. Water samples were collected from four sites located in the municipalities of Coari and São Gabriel da Cachoeira and four different culture media were used for the isolation of fungi. Two-hundred-and-six fungal strains were isolated and morphologically similar fungi were grouped into 30 morphotypes. Their taxonomic identities were assigned by macro and microscopic observations and sequencing of rDNA internal transcribed spacers (ITS1-5.8S-ITS2). Representatives of 26 morphotypes were identified at the genus level, one only at the family level, and three were not identified. The identified morphotypes belong to the phyla, Ascomycota (80.6%), Basidiomycota (11.7%), and Mucoromycota (2.4%), distributed in five classes, ten orders, 25 families, and 26 genera. This study fills a considerable knowledge gap about the fungi present in the breeding sites of An. darlingi mosquitoes.
Introduction
The Amazon basin has the largest volume of fresh water on the planet [1]. In that tropical environment biodiversity abounds, and much remains to be explored about species diversity and the ecological relationships among them. The mosquito Anopheles darlingi Root, 1926, is the main malaria vector in the Amazon region [2–4]. Despite the importance of malaria in the Amazon and the tropics, little effort has been made to study and identify the fungi associated with these vector mosquitoes [5, 6], especially compared to studies of the bacteria associated with them [7–11].
Although the complex mosquito-associated microbiota is made up of bacteria, fungi, protists, viruses, and nematodes, fungi have been largely neglected [12]. Fungi are an important part of the mosquito larval diet, providing long-chain polyunsaturated fatty acids and phytosterols [13]. Furthermore, fungi synthesize and secrete volatile molecules that attract gravid female mosquitoes and signal suitable oviposition sites [14, 15]. Concerning public health applications, entomopathogenic fungi as well as fungi-derived enzymes and toxins have been used effectively in mosquito control [16, 17], providing alternatives to conventional chemically-synthesized insecticides [16, 18, 19]. Fungi with low levels of pathogenicity can modulate the immune system of mosquitoes, interfering with the development of malaria parasites and other pathogens [20].
The fungi present in the aquatic habitats of mosquito larvae may provide the appropriate means to develop and implement biological control measures against malaria vectors [21–23], however, few mosquito-fungi interactions have been characterized. Therefore, this study aimed to explore the diversity of cultivable fungi present in the aquatic habitats of Anopheles darlingi larvae in two malaria-endemic municipalities in the state of Amazonas, and to fill a considerable gap in knowledge of the fungi present in the breeding sites of this important malaria vector.
Methods
Sampling sites and collection
Water samples were collected from permanent An. darlingi breeding sites identified by the Epidemiological Surveillance of the Municipal Health Secretariat from the municipalities of Coari and São Gabriel da Cachoeira, in the State of Amazonas—Brazil (Table 1), with official authorization (21263–1) granted by the Biodiversity Authorization and Information System (SISBIO) of the Brazilian Ministry of the Environment (MMA). Two sites were selected in each municipality and from each site water samples were collected at four equidistant points (5 m from each other) (S1 Fig). Water samples were collected on the surface of the breeding sites in 1-liter sterile glass bottles and kept at 4°C during transportation to the Laboratory of Bioassays and Microorganisms of the Amazon (LaBMicrA) of the Federal University of Amazonas (Universidade Federal do Amazonas, UFAM).
Table 1. Location and characteristics of the sites for collecting water samples from Anopheles darlingi breeding sites.
| Site | Location | Characteristics | GPS Coordinates | Date | |
|---|---|---|---|---|---|
| Latitude (S) | Longitude (W) | (Month/year) | |||
| Coari | |||||
| 1 - | Sítio do Gordo (C1) | - Permanent dam area, with fish and vegetation on the margins | 4°06’43.7" | 63°07’43.6" | 02/2017 |
| 2 - | Sítio João do Boi (C2) | - Permanent natural lake, with fish and vegetation on the margins. | 4°06’56.6" | 63°08’34.4" | 02/2017 |
| São Gabriel da Cachoeira | |||||
| 3 - | Sítio Matador (S1) | - Permanent fishpond with fish and no vegetation on the margins. | 0°6’54.873’’ | 67°5’12.859’’ | 02/2017 |
| 4 - | Sítio do Pelado (S2) | - Permanent natural lake, with vegetation on the margins and no fish. | 0°7’6.866’’ | 67°4’24.576’’ | 02/2017 |
Isolation of fungi
For the isolation of filamentous fungi, 100 μl aliquots of sample materials were transferred to Petri dishes (90x15 mm) containing one of the following culture media: AVA (10 g/l oats, 15 g/l agar, 4 g/l dextrose, 4 g/l yeast extract, and 10 g/l malt extract), PDA + L (200 g/l potato, 20 g/l dextrose, and 15 g/l of agar plus 2 g/l yeast extract) [24], ISP2 (10 g/l agar, 10 g/l starch, 4 g/l dextrose, 4 g/l yeast extract, and 10 g/l extract malt), or SDAY (15 g/l agar, 40 g/l dextrose, 10 g/l yeast extract, and 10 g/l peptone). Inoculation in each medium was in triplicate, and all media were supplemented with tetracycline and ampicillin (50 μg/ml each) to inhibit bacterial growth.
The plates were incubated at 26°C for up to 20 days and monitored daily. Beginning on the fifth day of incubation, visible fungal colonies were transferred individually to new plates with the same culture medium. Successive reinoculations were performed until pure cultures were obtained. All purified cultures were preserved at -80°C in 20% glycerol. Those with conidia or spores were also preserved in distilled water [25]. The isolated and preserved strains were deposited in the LaBMicrA/UFAM work collection and registered under the SisGen (National System for the Management of Genetic Heritage and Traditional Knowledge Associated) number AD64E07.
Morphological analysis
Morphological identifications followed taxonomic keys [26–31], according to the macro and micromorphological characteristics observed. Macroscopic characters included, color, shape, colony diameter, texture, mycelium elevation, and pigment diffusion.
For microscopic examinations, strains were inoculated in a Petri dish at three equidistant points, 1 cm from the edge. Coverslips were placed on top of two of these inocula, leaving the third as a visual control of the colonies. Whenever differentiation from the vegetative mycelium was observed, one coverslip was removed and stained with lactophenol blue to confirm the appearance of the reproductive structures. Additional incubation time was allowed before removing the second micro-cultivation coverslip and staining, when necessary. The vegetative and reproductive microstructures were examined and microphotographed using the Axio Lab. A1 trinocular microscope (Zeiss) with 400X and 1000X magnification. Fungal strains that exhibited similar morphological characteristics were grouped into morphotypes. At least 5% of the strains of each morphotype were randomly chosen to perform rDNA sequencing.
DNA extraction, rDNA amplification, and sequencing
Each fungal strain was grown in 125 ml Erlenmeyer flasks containing 50 ml of Potato Dextrose Broth (PDB) medium for 24–72 h at 26°C and 120 rpm. The mycelium was separated by vacuum filtration on Whatman paper, No. 4, and crushed with SilicaFlash Irregular Silica Gel G60 (SiliCycle) to lyse the cells. Genomic DNA was extracted with a ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, USA), according to the manufacturer’s instructions. The DNA quantity and quality were assessed by optical density measurements (NanoDrop 2000, Thermo Scientific, USA), and gel electrophoresis, respectively.
Approximately 700 bp DNA fragments including internal transcribed spacers (ITS1-5.8S-ITS2) of the rDNA were amplified using primers ITS1 and ITS4 [32]. The amplification reaction had the final volume of 25 μl containing: 0.5 μl of each primer at 10 pmol (Invitrogen), 1 μl DNA at 50–100 ng/μl, 2.5 μl 10x EasyTaq Buffer with Mg2 (TransGen Biotech Co.), 0.3 μl EasyTaq DNA Polymerase at 5 U/μl (TransGen Biotech Co.), 1 μl dNTP at 2.5 mM (TransGen Biotech Co.), and 19.2 μl of milli-Q water. PCR was performed using a BioRad S1000 thermal cycler (BioRad Laboratory, CA) with an initial incubation at 94°C for 3 min, followed by 30 cycles of [94°C for 30 s, 58°C for 30 s, and 72°C for 1 min], and a final incubation at 72°C for 5 min.
Amplicons were visualized by electrophoresis on 1.5% agarose gel stained with GelRed (Invitrogen). PCR products were treated with ExoSAP Ilustra—ExoProStar (GE Healthcare) prior to sequencing reactions using BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems) and a 3500 Genetic Analyzer (Applied Biosystems) sequencer. Sequencing reactions were performed using primers ITS1 and ITS4 [31].
Sequence analysis and taxonomy assignment
Consensus sequences were assembled using DNA Sequence Assembly BASER Software v.4.5.0 (http://www.dnabaser.com/index.html) and all sequences generated in this study were deposited in the NCBI GenBank database (accession numbers MZ781245—MZ781299). The sequences were then compared with the sequences stored in GenBank at the NCBI (National Centre for Biotechnology Information) using the BLASTn algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Alignments were performed using the MAFFT online interface [33], followed by manual adjustments using MEGA v.7 [34]. Maximum likelihood analyses were performed using RAxMLHPC2 v.8.2.8 [35] in XSEDE. Phylogenetic trees were projected in FigTree 1.4 [36].
Data statistical analysis
For statistical analysis, we used raw data detailing isolates from each breeding site and culture medium. We computed Shannon-Wiener (H’) diversity and Jaccard similarity indices to characterize species richness and community across breeding sites, with dendrograms illustrating point clustering to visualize these relationships. The normality of the data was assessed using the Shapiro-Wilk test. Due to frequent deviations from the normal distribution, we used the non-parametric Kruskal Wallis test to assess differences in fungal richness between sites, followed by Dunn post-hoc test for pairwise comparisons, with a significance level of 95% (p ≤ 0.05). Non-metric multidimensional scaling (nMDS) was performed using Bray-Curtis dissimilarity index to analyze species distribution patterns and similarities among sites. To determine the significance of differences between clusters, we performed a Similarity Analysis (ANOSIM) using R [37]. In addition, a Venn diagram was generated with the tool Interacti Venn developed by Heberle [38] to visualize the overlap and unique components of fungal communities across breeding sites.
Results
Richness, diversity, and characterization of cultivable fungi from Anopheles darlingi breeding sites
A total of 206 fungal strains have grown from An. darlingi larvae breeding waters, collected in Coari (sites C1 and C2) and São Gabriel da Cachoeira (sites S1 and S2). The culture media AVA (oats, agar, dextrose, yeast extract, and malt extract), PDA + L (potato, dextrose, and agar plus yeast extract), ISP2 (agar, starch, dextrose, yeast extract, and extract malt), and SDAY (agar, dextrose, yeast extract, and peptone) used in this work supported the growth of many fungi. Small differences were observed when comparing the number of isolates successfully grown on each medium, SDAY (n = 57), PDA+L (n = 52), ISP2 (n = 50), and AVA (n = 47)—(S1 Table), indicating their applicability in the recovery of fungi from aquatic freshwater habitats. The C1 site (n = 107) yielded the highest number of isolates, followed by the sites S2 (n = 45), C2 (n = 44), and S1 (n = 10) (Table 2).
Table 2. Identification of taxa and allocation at the collection sites of fungi isolated from the waters of Anopheles darlingi larvae breeding sites in the Brazilian Amazon.
| Class | Taxon | No of isolated strains | Distribution of isolates at collection sites | |||
|---|---|---|---|---|---|---|
| C1 | C2 | S1 | S2 | |||
| Agaricomycetes | Emmia 1 | 4 | 0 | 0 | 0 | 4 |
| Hypomontagnella 1 | 7 | 3 | 0 | 0 | 4 | |
| Peniophora 1 | 5 | 5 | 0 | 0 | 0 | |
| Trametes 1 | 8 | 3 | 2 | 1 | 2 | |
| Dothideomycetes | Cladosporium 2 | 2 | 2 | 0 | 0 | 0 |
| Cucurbitariaceae 1 | 2 | 1 | 1 | 0 | 0 | |
| Epicoccum 1 | 1 | 1 | 0 | 0 | 0 | |
| Hongkongmyces 1 | 5 | 2 | 3 | 0 | 0 | |
| Microsphaeropsis 1 | 19 | 16 | 3 | 0 | 0 | |
| Nigrograna 1 | 1 | 1 | 0 | 0 | 0 | |
| Ochronis 2 | 12 | 8 | 3 | 0 | 1 | |
| Paraconiothyrium 8 | 23 | 19 | 0 | 3 | 1 | |
| Pyrenochaetopsis 2 | 4 | 4 | 0 | 0 | 0 | |
| Eurotiomycetes | Aspergillus 5 | 12 | 5 | 2 | 2 | 3 |
| Penicillium 2 | 14 | 2 | 1 | 1 | 10 | |
| Talaromyces 4 | 8 | 2 | 4 | 1 | 1 | |
| Mucoromycetes | Gongronella 1 | 5 | 1 | 1 | 0 | 3 |
| Sordariomycetes | Albifimbria 1 | 1 | 1 | 0 | 0 | 0 |
| Chrysoporthe 1 | 4 | 2 | 1 | 0 | 1 | |
| Cytospora 3 | 3 | 0 | 3 | 0 | 0 | |
| Diaporthe 3 | 6 | 1 | 2 | 0 | 3 | |
| Eutypella 2 | 2 | 0 | 2 | 0 | 0 | |
| Fusarium 4 | 19 | 12 | 4 | 0 | 3 | |
| Hyphodermella 1 | 3 | 1 | 1 | 0 | 1 | |
| Sarocladium 2 | 5 | 1 | 1 | 0 | 3 | |
| Striaticonidium 1 | 3 | 1 | 1 | 0 | 1 | |
| Trichoderma 2 | 17 | 6 | 6 | 1 | 4 | |
| NID1 1 | 6 | 6 | 0 | 0 | 0 | |
| NID2 2 | 1 | 0 | 1 | 0 | 0 | |
| NID3 3 | 4 | 1 | 2 | 1 | 0 | |
| Total | 30 | 206 | 107 | 44 | 10 | 45 |
NID (unidentified)—refers to fungi grouped into morphotypes that could not be identified by morphological characters or rDNA sequencing. n—Number of specimens sequenced.
Macro and micromorphological characterization of the isolated strains allowed their classification into 30 morphotypes. The diversity of morphotypes grown in different culture media was different. PDA+L supported the highest diversity of morphotypes (n = 23), followed by SDAY (n = 21), AVA (n = 18), and ISP2 (n = 15). Morphological data, together with ribosomal RNA sequences, and analysis of phylogenetic trees identified 26 of the morphotypes at their genus level and one at the family level. In all cases, the molecular data confirmed the morphology-based taxonomy, therefore, for diversity and richness estimation, all strains within a given morphotype were assigned to the same taxon (S2 Table). The ideal outcome would be to assign species-level taxonomy to each queried sequence, however, the limited resolution of the sequenced locus prevented accurate classification at the species level. Three morphotypes proved difficult to cultivate under the conditions described above and their DNA was not sequenced (Table 2). Overall, one in every 6.9 isolated fungal strains was successfully classified at the genus level (S3 Table; S2 Fig).
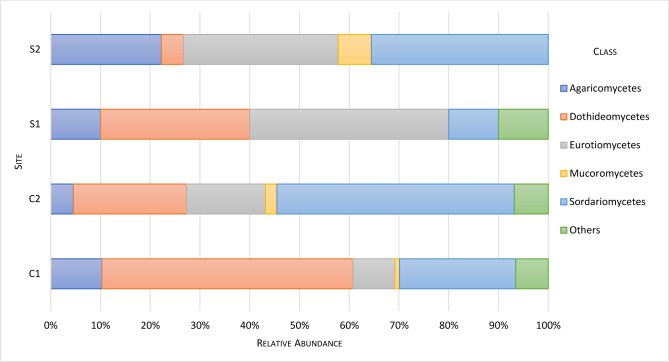
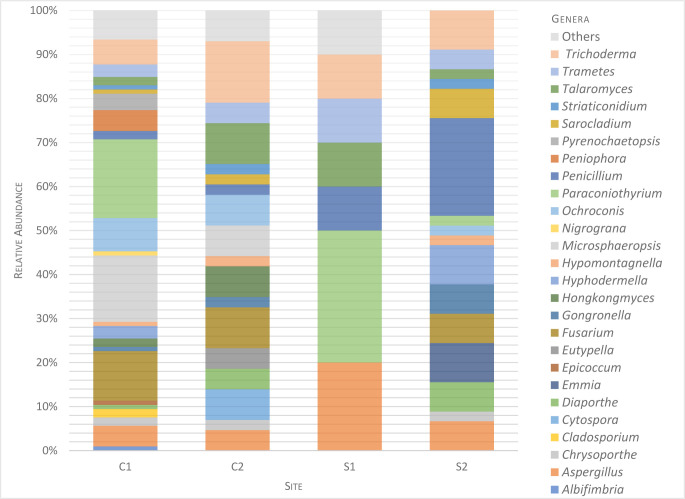
The sequenced morphotypes belong to three phyla: 80.6% Ascomycota, followed by Basidiomycota and Mucoromycota, 11.7% and 2.4%, respectively. The three most represented classes were Dothideomycetes, Sordariomycetes, and Eurotiomycetes corresponding to 33.5%, 30.6%, and 16.5%, respectively (Fig 1). Ten orders were identified, the three main ones being: Pleosporales 26.7%, Hypocreales 21.9%, and Eurotiales 16.5% (Fig 2). The sequenced morphotypes had representatives of 25 families and 26 genera (S1 Table). The genus Paraconiothyrium was the most prevalent, represented by 11.2% of all isolated fungi strains, followed by Fusarium and Microsphaeropsis, both with 9.2% (Fig 3).
Fig 1. Fungal community composition in different Anopheles darlingi breeding sites at class level.
Others—refers to fungi that could not be classified at class level using the analyses carried out.
Fig 2. Fungal community composition in different Anopheles darlingi breeding sites at the order level.
Others—refers to fungi that could not be classified at order level using the analyses carried out.
Fig 3. Composition of fungal communities at the genera level at different collection sites.
Others—refers to fungi that could not be classified at genera level using the analyses carried out.
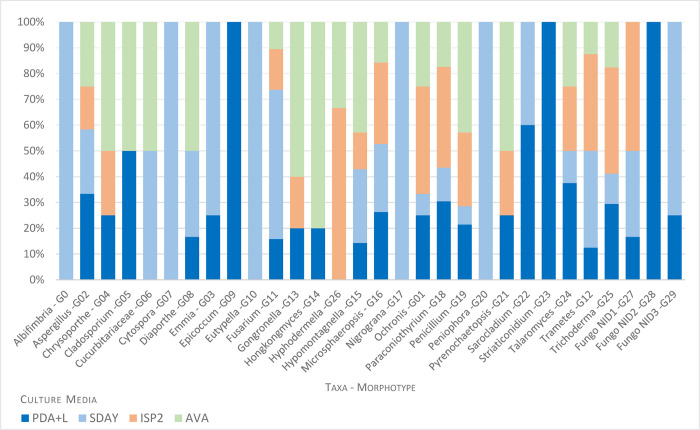
Fungi of genera Albifimbria, Cytospora, Eutypella, Nigrograna, and Peniophora only grew on the SDAY medium, while fungi of genera Epicoccum, Striaticonidium, and Fungo NID2 grew only on PDA+L medium (Fig 4).
Fig 4. Fungi isolated from An. darlingi larvae breeding sites in the different culture media AVA, ISP2, PDA + L, and SDAY.
Taxonomic assignments of 30 morphotypes (G0-G29) was based on morphological and molecular characterization. Fungo NID (unidentified)1, Fungo NID2 e Fungo NID3—refer to morphotypes that could not be identified by morphology or rDNA sequencing.
Fungi taxonomic diversity analysis
The diversity and richness of the fungi, represented by the Shannon (H’) and Chao1 indices, indicated that the samples from the C1 site showed the highest values (Chao1 = 33.5 and H’ = 2.772). Regarding the dominance (Simpson D) and equitability (Equitability_J) indices, the samples from site C2 showed the highest values (D = 0.931 and J = 0.944). In contrast, the S1 site showed the lowest values of diversity (H ’ = 1.834), richness (Chao1 = 12), and dominance (D = 0.82) (S4 Table).
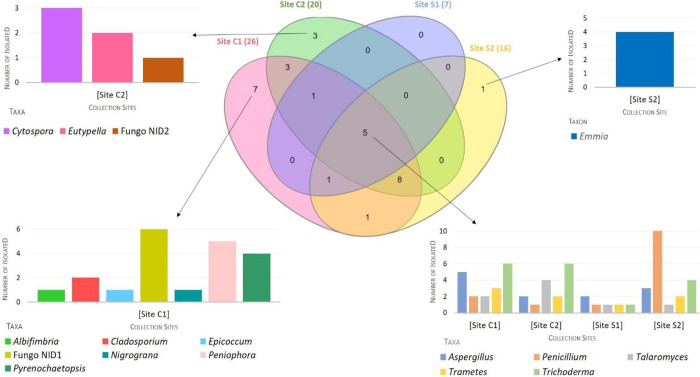
Representatives of five genera (Aspergillus, Penicillium, Talaromyces, Trametes, and Trichoderma) were shared among the four sampling sites. Seven taxa (Albifimbria, Cladosporium, Epicoccum, Nigrograna, Peniophora, and Pyrenochaetopsis genera, and NID1), were isolated only from site C1. Taxa exclusively isolated from the sites C2 (Cytospora and Eutypella genera, and ND2) and S2 (Emmia) were also observed. All taxa isolated from S1 were also present in at least one other site (Fig 5).
Fig 5. Venn diagram showing the number of fungal taxa shared or exclusive of collection sites C1, C2, S1 and S2.
Fungo NID (unidentified)1, Fungo NID2 e Fungo NID3—refer to morphotypes that C1, C2, S1 could not be identified by morphology or rDNA sequencing.
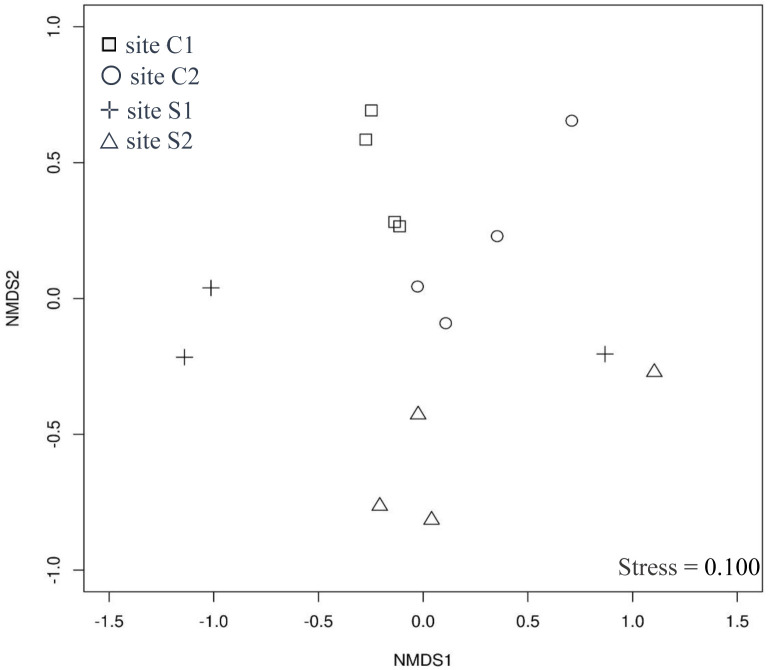
The jaccard similarity index identified three groups, with C2 and S2 showing the highest similarity, while S1 showed the greatest divergence (S3 Fig). The Kruskal-Wallis test followed by Dunn’s post hoc comparisons revealed a borderline statistical difference in richness between sites C2 and S2. In contrast to the comparisons between sites C2 and S2, all other pairwise comparisons between sites were significantly different (p <0.05) (S5 Table). Non-metric multidimensional scaling (nMDS) analysis based on all fungal isolates obtained in the samples and separated by collection site shows that the spatial distribution of the composition of the fungal community varied between the collection sites, with site S1 being less similar among the others. Two sub-sites of the C1 site were the most similar, with the same microbial composition (Fig 6).
Fig 6. Non-metric multidimensional scaling (nMDS) using the Bray-Curtis distance to show the similarities between the collection sites in relation to the distribution of fungal isolates.
In each site, four sub-sites were sampled, however, the S1 site presented isolates in only 3 sub-sites.
Discussion
Anopheles mosquitoes breed in a variety of natural and artificial water bodies, such as riverbanks, streams, lakes, ponds, dams, and fishponds, which generally contain organic matter and aquatic vegetation [39–42]. This aquatic habitat hosts a wide variety of fungi, some of which are associated with mosquitoes throughout their life cycle.
Fungi present in mosquito breeding waters are ingested during larval feeding [13, 43, 44] and attach to their cuticle and external body wall structures. Although most of the microbiota associated with mosquito larvae does not persist after metamorphosis, some fungi are transstadially transmitted across developmental stages, survive metamorphosis, and are inherited by their progeny [20]. Adult mosquitoes can also contact fungi and/or bacteria when standing on or ingesting water from the breeding site immediately after emerging from the pupal stage [40–42]. These adults can introduce or reintroduce fungi into aquatic habitats through contact, urine deposition [45], or during egg laying [46, 47], affecting the microbiota to which larvae are exposed [47].
In this study, we explored and compared the diversity of cultivable fungi in Anopheles darlingi breeding water samples from the Brazilian Amazon, collected in the municipalities of Coari (C1 and C2) and São Gabriel da Cachoeira (S1 and S2). Coari is located on the banks of the Solimões River, while São Gabriel da Cachoeira is on the banks of the Negro River, two distinct hydrological basins. Both municipalities are considered highly endemic areas for malaria [48].
The distinct characteristics of the black water of the Negro River, characterized by acidity (pH < 5.0), low productivity, low suspended sediment concentration, and low electrical conductivity, contrast with the white water of the Solimões River, which has a neutral pH (7.0), is rich in nutrients and high in suspended matter and dissolved salts, resulting in a greater diversity of microorganisms [49–52]. According to Fonseca [50] and Tadei [53], the characteristics of black water provide more suitable conditions for the breeding of the malaria vector.
The isolated strains in this study represent only a fraction of all fungi present in the sampled sites. Likely, other fungi could be isolated by collecting and exploring additional samples from the same or other An. darlingi breeding sites and investigating fungi that grow optimally in other cultivation media. Culture techniques do not capture the full spectrum of microbial diversity; in fact, approximately 99% of naturally occurring microorganisms have been suggested to remain unknown [54–56].
The frequent and heavy rainfall events in the Amazon rain forest, especially during the period when collections were carried out for this study, suggest that some of the fungi found in mosquito breeding waters are transient, carried with plant and soil residues, mainly by rainwater [57, 58]. A longitudinal investigation could reveal details of the fungal population dynamics in the studied breeding sites. Therefore, these results must be interpreted with attentiveness as a number of limitations should be borne in mind.
Ascomycota was highly predominant among the three phyla found in the breeding waters of An. darlingi, followed by Basidiomycota, while only one morphotype was from the Mucoromycota phylum. This is consistent with previous reports showing that Ascomycota is the largest phylum of fungi, encompassing approximately two-thirds of all described fungal species [59, 60], while Basidiomycota is the second richest phylum in number of species [61]. Both the Ascomycota and Basidiomycota phyla are ubiquitous in nature and are the main phyla found in freshwater environments [60, 62, 63]. The scarcer phylum Mucoromycota consists mainly of mycorrhizal fungi, root endophytes, and plant material decomposers [64]. However, as observed in this work, Mucoromycota is also found in freshwater environments [65].
Similar results were found when analyzing fungi in other mosquito breeding sites in diverse localities worldwide [66, 67]. For example, Tawidian [65], who analyzed the fungal microbiota in samples of Aedes albopictus from Manhattan, KS, USA, and water from the larval breeding sites, identified representatives of the phyla Ascomycota (59.5%), Basidiomycota (30.8%), and Mucoromycota (0.46%), among others.
The most represented classes, Dothideomycetes, Sordariomycetes, and Eurotiomycetes, belong to the most abundant Ascomycota phylum. Agaricomycetes and Mucoromycetes were less abundant and were the unique classes found for their respective phyla. Sordariomycetes contain nearly half of all known freshwater Ascomycota, corresponding to approximately 300 out of 620 taxa [68, 69]. The three main classes in this study are also the most abundant in waters from aquatic environments of the High Arctic [70] and other marine environments [57], indicating they are adapted to occupy very different niches.
The C1 site showed the highest richness index, with the highest number of isolates, seven taxa were found exclusively in C1 and 19 were shared with other sites (Fig 5). Such diversity and richness are consistent with the suitable conditions for fungal growth found at the C1 site, a permanent dam with fish and vegetation on the margins and, therefore, rich in organic particles. Water dams such as C1 have characteristics similar to natural environments and are favorable to breeding Anopheles [41].
The C2 and S2 sites, the second and third sites in richness, respectively, are permanent natural lakes, with fish and vegetation on the margins, a situation similar to that found in the site C1. The diversity in C2 was 20 taxa, while S2 had 16. The lowest diversity and richness of fungi were attributed to the site S1. This breeding site is a permanent fishpond, without vegetation on the margins and near a deforested area, most affected by anthropic actions, and probably poorer in conditions for fungal survival. In fact, anthropogenic activities affect the biogeochemical properties of breeding sites and, in turn, affect the microbiota of mosquitoes [63, 71, 72]. S1 had only seven taxa: none exclusive, two common to two other sites, and five common to all sites (Fig 5).
Five genera of fungi found in the four sites sampled in this work, Aspergillus, Penicillium, Talaromyces, Trametes, and Trichoderma, are known to be ubiquitous in the environment [57, 73–75]. Interactions between species of these five genera with mosquitoes have been studied. Penicillium species present in the midgut of Anopheles made mosquitoes more susceptible to Plasmodium infection [76, 77], Talaromyces make Aedes aegypti more susceptible to dengue virus infection [78], metabolites produced by Trametes species showed larvicidal activity against Ae. aegypti [79], and fungi belonging to the genera Penicillium, Aspergillus, and Trichoderma have larvicidal and adulticidal activities against mosquitoes [19, 80–83].
The present study explored the diversity of cultivable fungi from An. darlingi breeding sites in the state of Amazonas, Brazil, revealing rich and diverse fungal communities in more natural freshwaters and poorer diversity in the anthropic influenced one. The knowledge generated by this study could be leveraged to achieve a more holistic understanding of mosquito biology and its associated microbiome. Similar to bacterial endosymbionts, mosquito-associated fungi could harbor both beneficial and antagonistic traits. [44, 46, 84–86]. Fungi from the genera found in this work have been investigated as potential agents against Anopheles larvae. However, native fungus isolates may offer a superior alternative to the introduction of foreign biocontrol agents, as they can be better adapted to infect and kill local mosquitoes and survive in the Amazonian local environmental conditions, with high temperatures and abundant rainfall [87]. In fact, extracts of Amazonian fungi isolated during this investigation, Albifimbria 1160 and Diaporthe 1203, are active in the killing of mosquito larvae [76].
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors thank the Graduate Program in Biodiversity and Biotechnology/PPG-BIONORTE, Embrapa Western Amazon/EMBRAPA, and the technicians of the Laboratório de Malária e Dengue/INPA for their help with sample collections. We also thank the LabMicrA laboratory of the Central Analitica–CAM/UFAM, for providing space and equipment for the development of the work. This work is dedicated in memorian to the great researcher and collaborator Wanderli Pedro Tadei. This study is part of the doctoral thesis of MRO (Universidade Federal do Amazonas).
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The information and accession numbers of the sequences deposited in GenBank can be found as described in line 149: All sequences generated in this study have been deposited in the NCBI GenBank database (accession numbers MZ781245 - MZ781299), and in the table in the supporting material with the GenBank deposit number: S5 Table. Taxonomic classification of fungi isolated from An. darlingi breeding sites in the municipalities of Coari (C1 and C2) and São Gabriel da Cachoeira (S1 and S2), based on sequencing of the ITS (Internal Transcribed Spacer) region of the nuclear ribosome and comparison of the sequences with those in the NCBI database, and analysis of phylogenetic trees.
Funding Statement
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001, by the project Pró-Amazônia: Biodiversidade e Sustentabilidade (process number 23038.009442/2013-12) and FAPEAM. This study is part of the doctoral thesis of MRO (Universidade Federal do Amazonas). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ministério do Meio Ambiente M. Áreas Prioritárias para Conservação, Uso Sustentável e Repartição de Benefícios da Biodiversidade Brasileira: Atualização—Portaria MMA n°9, de 23 de janeiro de 2007. 1st ed. Brasília: Ministério do Meio Ambiente, Secretaria de Biodiversidade e Florestas; 2007. Available: https://www.mma.gov.br/estruturas/chm/_arquivos/biodiversidade31.pdf [Google Scholar]
- 2.Rocha EM, Katak R de M, Campos de Oliveira J, Araujo M da S, Carlos BC, Galizi R, et al. Vector-Focused Approaches to Curb Malaria Transmission in the Brazilian Amazon: An Overview of Current and Future Challenges and Strategies. Tropical Medicine and Infectious Disease. 2020;5: 161. doi: 10.3390/tropicalmed5040161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consoli RAGB, Oliveira RL de. Principais mosquitos de importância sanitária no Brasil. Editora FIOCRUZ; 1994. Available: http://books.scielo.org/id/th [Google Scholar]
- 4.Brasil, Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de Vigilância Epidemiológica. "Guia de vigilância em saúde. 3rd ed. Brasília–DF: Ministério da Saúde, Secretaria de Vigilância em Saúde, Coordenação-Geral de Desenvolvimento da Epidemiologia em Serviços; 2019. Available: http://bvsms.saude.gov.br/bvs/publicacoes/guia_vigilancia_saude_3ed.pdf [Google Scholar]
- 5.Pereira E da S, Sarquis MI de M, Ferreira-Keppler RL, Hamada N, Alencar YB. Filamentous fungi associated with mosquito larvae (Diptera: Culicidae) in municipalities of the Brazilian Amazon. Neotrop entomol. 2009;38: 352–359. doi: 10.1590/s1519-566x2009000300009 [DOI] [PubMed] [Google Scholar]
- 6.Hegde S, Khanipov K, Hornett EA, Nilyanimit P, Pimenova M, Saldaña MA, et al. Interkingdom interactions shape the fungal microbiome of mosquitoes. bioRxiv; 2023. p. 2023.08.11.552965. doi: 10.1101/2023.08.11.552965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrade RM, Rachou RG. Sampling of plankton organisms in various breeding places of Anopheles darlingi in southern Brazil. Revista Brasileira de Malariologia e Doenças Tropicais. 1954;6: 481–496. [PubMed] [Google Scholar]
- 8.Rocha EM, Marinotti O, Serrão DM, Correa LV, Katak R de M, de Oliveira JC, et al. Culturable bacteria associated with Anopheles darlingi and their paratransgenesis potential. Malar J. 2021;20: 40. doi: 10.1186/s12936-020-03574-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson LKJ, de Oliveira MR, Marinotti O, Rocha EM, Håkansson S, Tadei WP, et al. Characterization of Bacterial Communities in Breeding Waters of Anopheles darlingi in Manaus in the Amazon Basin Malaria-Endemic Area. Microb Ecol. 2019;78: 781–791. doi: 10.1007/s00248-019-01369-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arruda A, Ferreira GS, Lima NC da S, dos Santos Júnior A, Custódio MGF, Benevides-Matos N, et al. A simple methodology to collect culturable bacteria from feces of Anopheles darlingi (Diptera: Culicidae). Journal of Microbiological Methods. 2017;141: 115–117. doi: 10.1016/j.mimet.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 11.Terenius O, de Oliveira CD, Pinheiro WD, Tadei WP, James AA, Marinotti O. 16S rRNA Gene Sequences from Bacteria Associated with Adult Anopheles darlingi (Diptera: Culicidae) Mosquitoes. Journal of Medical Entomology. 2008;45: 172–175. doi: 10.1603/0022-2585(2008)45[172:srgsfb]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 12.Malassigné S, Valiente Moro C, Luis P. Mosquito Mycobiota: An Overview of Non-Entomopathogenic Fungal Interactions. Pathogens. 2020;9: 564. doi: 10.3390/pathogens9070564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt RW, Dadd RH, Walker ED. Feeding Behavior, Natural Food, and Nutritional Relationships of Larval Mosquitoes. Annual Review of Entomology. 1992;37: 349–374. doi: 10.1146/annurev.en.37.010192.002025 [DOI] [PubMed] [Google Scholar]
- 14.Girard M, Martin E, Vallon L, Raquin V, Bellet C, Rozier Y, et al. Microorganisms Associated With Mosquito Oviposition Sites: Implications for Habitat Selection and Insect Life Histories. 2021. [cited 4 Aug 2021]. doi: 10.3390/microorganisms9081589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eneh LK, Saijo H, Borg-Karlson A-K, Lindh JM, Rajarao GK. Cedrol, a malaria mosquito oviposition attractant is produced by fungi isolated from rhizomes of the grass Cyperus rotundus. Malaria Journal. 2016;15: 478. doi: 10.1186/s12936-016-1536-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei G, Lai Y, Wang G, Chen H, Li F, Wang S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. PNAS. 2017;114: 5994–5999. doi: 10.1073/pnas.1703546114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh G, Prakash S. New Prospective on Fungal Pathogens for Mosquitoes and Vectors Control Technology. Journal of Mosquito Research. 2014;4. Available: http://www.emtoscipublisher.com/index.php/jmr/article/view/629 [Google Scholar]
- 18.Scholte E-J, Knols BG, Takken W. Autodissemination of the entomopathogenic fungus Metarhizium anisopliae amongst adults of the malaria vector Anopheles gambiae s.s. Malar J. 2004;3: 45. doi: 10.1186/1475-2875-3-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivekanandhan P, Bedini S, Shivakumar MS. Isolation and identification of entomopathogenic fungus from Eastern Ghats of South Indian forest soil and their efficacy as biopesticide for mosquito control. Parasitology International. 2020;76: 102099. doi: 10.1016/j.parint.2020.102099 [DOI] [PubMed] [Google Scholar]
- 20.Nattoh G, Bargul JL, Magoma G, Mbaisi L, Butungi H, Mararo E, et al. The fungus Leptosphaerulina persists in Anopheles gambiae and induces melanization. PloS one. 2021;16: e0246452. doi: 10.1371/journal.pone.0246452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. Bacteria in Midguts of Field-Collected Anopheles albimanus Block Plasmodium vivax Sporogonic Development. J Med Entomol. 2003;40: 371–374. doi: 10.1603/0022-2585-40.3.371 [DOI] [PubMed] [Google Scholar]
- 22.Lindh JM, Terenius O, Faye I. 16S rRNA Gene-Based Identification of Midgut Bacteria from Field-Caught Anopheles gambiae Sensu Lato and A. funestus Mosquitoes Reveals New Species Related to Known Insect Symbionts. Appl Environ Microbiol. 2005;71: 7217–7223. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. PNAS. 2007;104: 9047–9051. doi: 10.1073/pnas.0610451104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Souza AQL, de Souza ADL, Astolfi Filho S, Pinheiro MLB, Sarquis MI de M Pereira JO. Antimicrobial activity of endophytic fungi isolated from amazonian toxic plants: Palicourea longiflora (aubl.) rich and Strychnos cogens bentham. Acta Amazonica. 2004;34: 185–195. [Google Scholar]
- 25.Castellani A. Maintenance and cultivation of common pathogenic fungi of man in sterile distilled water. Further researches. Journal of Tropical Medicine & Hygiene. 1967;70: 181–184. [Google Scholar]
- 26.Barnett HL, Hunter BB. Illustrated genera of imperfect fungi. Minnesota: American Phytopathological Society (APS Press); 1988. [Google Scholar]
- 27.Bononi VLR, Grandi R. A. P. Zigomicetos, Basidiomicetos e Deuteromicetos: noções básicas de taxonomia e aplicações biotecnológicas. São Paulo: Instituto de Botanica; 1999. [Google Scholar]
- 28.Hanlin RT. Illustrated genera of Ascomycetes. Minnesota: Aps Press; 1997. [Google Scholar]
- 29.Hawksworth DL, Kirk PM, Bc Sutton, Pegler DN. Ainsworth & Bisby’s Dictionary of the Fungi. 8th ed. Oxfordshire: Oxford University Press; 1995. [Google Scholar]
- 30.Funder S. Practical mycology. Manual for identification of fungi. Practical mycology Manual for identification of fungi. 1953. [Google Scholar]
- 31.Dugan FM, Dugan FM. The identification of fungi: an illustrated introduction with keys, glossary, and guide to literature. 2006. [Google Scholar]
- 32.White TJ, Bruns T, Lee SJWT, Taylor J Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18: 315–322. [Google Scholar]
- 33.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution. 2016;33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambaut A. FigTree v1.4: tree figure drawing tool. In: molecular evolution, phylogenetics and epidemiology [Internet]. 2007. [cited 15 Nov 2020]. Available: http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- 37.R Core Team. R: A language and environment for statistical computing. 2017. [cited 4 Sep 2020]. Available: https://www.r-project.org/index.html [Google Scholar]
- 38.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16: 169. doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu VM, Sallum MAM, Moore TE, Emerson KJ, Schlichting CD, Conn JE. Evidence for family-level variation of phenotypic traits in response to temperature of Brazilian Nyssorhynchus darlingi. Parasites & vectors. 2020;13: 55. doi: 10.1186/s13071-020-3924-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forattini OP. Culicidologia Médica: Identificação, Biologia. Epidemiologia São Paulo: Editora da Universidade de São Paulo. 2002;2: 864. [Google Scholar]
- 41.de Freitas Rodrigues A, Escobar AL, Souza-Santos R. Spatial analysis and determination of malaria control areas in the State of Rondônia. Revista da Sociedade Brasileira de Medicina Tropical. 2008;41. [DOI] [PubMed] [Google Scholar]
- 42.Arcos AN, Ferreira FA da S, da Cunha HB, Tadei WP. Characterization of artificial larval habitats of Anopheles darlingi (Diptera: Culicidae) in the Brazilian Central Amazon. Revista Brasileira de Entomologia. 2018;62: 267–274. doi: 10.1016/j.rbe.2018.07.006 [DOI] [Google Scholar]
- 43.Chukalo E, Abate D. Bacterial populations of mosquito breeding habitats in relation to maize pollen in Asendabo, south western Ethiopia. African Journal of Microbiology Research. 2017;11: 55–64. [Google Scholar]
- 44.Tawidian P, Rhodes VL, Michel K. Mosquito-fungus interactions and antifungal immunity. Insect Biochemistry and Molecular Biology. 2019;111: 103182. doi: 10.1016/j.ibmb.2019.103182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urubschurov V, Janczyk P. Biodiversity of yeasts in the gastrointestinal ecosystem with emphasis on its importance for the host. The Dynamical Processes of Biodiversity-Case Studies of Evolution and Spatial Distribution. 2011; 277–302. [Google Scholar]
- 46.Steyn A, Roets F, Botha A. Yeasts Associated with Culex pipiens and Culex theileri Mosquito Larvae and the Effect of Selected Yeast Strains on the Ontogeny of Culex pipiens. Microb Ecol. 2016;71: 747–760. doi: 10.1007/s00248-015-0709-1 [DOI] [PubMed] [Google Scholar]
- 47.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Molecular Ecology. 2016;25: 5806–5826. doi: 10.1111/mec.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.FVS-AM, Fundação de Vigilância em Saúde do Amazonas. Boletim Epidemiológico de Vigilância em Saúde. 2018. [cited 23 Aug 2020]. Available: http://www.fvs.am.gov.br/media/publicacao/boletim_Epidemiol%C3%B3gico_2018.pdf [Google Scholar]
- 49.Queiroz MMA, Horbe AMC, Seyler P, Moura CAV. Hidroquímica do rio Solimões na região entre Manacapuru e Alvarães: Amazonas—Brasil. Acta Amaz. 2009;39: 943–952. doi: 10.1590/S0044-59672009000400022 [DOI] [Google Scholar]
- 50.Fonseca F, Martinez J-M, Balieiro A, Orellana J, Santos JD, Filizola N. Relationship between the colours of the rivers in the Amazon and the incidence of malaria. Malaria Journal. 2023;22: 358. doi: 10.1186/s12936-023-04789-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfarth BR, Filizola N, Tadei WP, Durieux L. Epidemiological analysis of malaria and its relationships with hydrological variables in four municipalities of the State of Amazonas, Brazil. Hydrological Sciences Journal. 2013;58: 1495–1504. doi: 10.1080/02626667.2013.831977 [DOI] [Google Scholar]
- 52.do Nascimento Monte C, Correa Saldanha E, Costa I, Shinaigger Rocha do Nascimento T, Santos Pereira M, Farias Batista L et al. The physical-chemical characteristics of surface waters in the management of quality in clearwater rivers in the Brazilian Amazon. Water Policy. 2021;23: 1303–1313. doi: 10.2166/wp.2021.258 [DOI] [Google Scholar]
- 53.Tadei WP, Rodrigues IB, Rafael MS, Sampaio RTM, Mesquita HG, Pinheiro VCS, et al. Adaptative processes, control measures, genetic background, and resilience of malaria vectors and environmental changes in the Amazon region. Hydrobiologia. 2017;789: 179–196. doi: 10.1007/s10750-016-2960-y [DOI] [Google Scholar]
- 54.Kim M, Yoon H, You Y-H, Kim Y-E, Woo J-R, Seo Y, et al. Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. J Microbiol Biotechnol. 2013;23: 1347–1356. doi: 10.4014/jmb1306.06068 [DOI] [PubMed] [Google Scholar]
- 55.Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proc Natl Acad Sci USA. 2016;113: 5970–5975. doi: 10.1073/pnas.1521291113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vitorino LC, Bessa LA. Microbial Diversity: The Gap between the Estimated and the Known. Diversity. 2018;10: 46. doi: 10.3390/d10020046 [DOI] [Google Scholar]
- 57.Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanová L, Padgett D, et al. Fungal biodiversity in aquatic habitats. Biodivers Conserv. 2007;16: 49–67. doi: 10.1007/s10531-006-9120-z [DOI] [Google Scholar]
- 58.Voronin LV. Terrigenous micromycetes in freshwater ecosystems (review). Inland Water Biol. 2014;7: 352–356. doi: 10.1134/S1995082914040191 [DOI] [Google Scholar]
- 59.Naranjo‐Ortiz MA, Gabaldón T. Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biological Reviews. 2019;94: 2101–2137. doi: 10.1111/brv.12550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panzer K, Yilmaz P, Weiß M, Reich L, Richter M, Wiese J, et al. Identification of Habitat-Specific Biomes of Aquatic Fungal Communities Using a Comprehensive Nearly Full-Length 18S rRNA Dataset Enriched with Contextual Data. PLOS ONE. 2015;10: e0134377. doi: 10.1371/journal.pone.0134377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao R-L, Li G-J, Sánchez-Ramírez S, Stata M, Yang Z-L, Wu G, et al. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Diversity. 2017;84: 43–74. doi: 10.1007/s13225-017-0381-5 [DOI] [Google Scholar]
- 62.Lepère C, Domaizon I, Humbert J-F, Jardillier L, Hugoni M, Debroas D. Diversity, spatial distribution and activity of fungi in freshwater ecosystems. PeerJ. 2019;7: e6247. doi: 10.7717/peerj.6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grossart H-P, Van den Wyngaert S, Kagami M, Wurzbacher C, Cunliffe M, Rojas-Jimenez K. Fungi in aquatic ecosystems. Nature Reviews Microbiology. 2019;17: 339–354. doi: 10.1038/s41579-019-0175-8 [DOI] [PubMed] [Google Scholar]
- 64.Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108: 1028–1046. doi: 10.3852/16-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tawidian P, Coon KL, Jumpponen A, Cohnstaedt LW, Michel K. Host-environment interplay shapes fungal diversity in mosquitoes. Microbiology; 2020. Dec. doi: 10.1101/2020.12.01.407494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luis P, Vallon L, Tran F-H, Hugoni M, Tran-Van V, Mavingui P, et al. Aedes albopictus mosquitoes host a locally structured mycobiota with evidence of reduced fungal diversity in invasive populations. Fungal Ecology. 2019;39: 257–266. doi: 10.1016/j.funeco.2019.02.004 [DOI] [Google Scholar]
- 67.Thongsripong P, Chandler JA, Green AB, Kittayapong P, Wilcox BA, Kapan DD, et al. Mosquito vector-associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecology and Evolution. 2018;8: 1352–1368. doi: 10.1002/ece3.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai L, Hu D-M, Liu F, Hyde KD, Jones EBG. 3. The molecular phylogeny of freshwater Sordariomycetes and Discomycetes. Freshwater Fungi. De Gruyter; 2014. pp. 47–72. Available: https://www.degruyter.com/document/doi/ doi: 10.1515/9783110333480.47/html [DOI] [Google Scholar]
- 69.Shearer CA, Raja HA. Freshwater Ascomycete Database. 2010. [cited 15 Nov 2020]. Available: http://fungi.life.illinois.edu/ [Google Scholar]
- 70.Zhang T, Wang N-F, Zhang Y-Q, Liu H-Y, Yu L-Y. Diversity and Distribution of Aquatic Fungal Communities in the Ny-Ålesund Region, Svalbard (High Arctic): Aquatic Fungi in the Arctic. Microb Ecol. 2016;71: 543–554. doi: 10.1007/s00248-015-0689-1 [DOI] [PubMed] [Google Scholar]
- 71.Scolari F, Casiraghi M, Bonizzoni M. Aedes spp. and Their Microbiota: A Review. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.02036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ortiz-Vera MP, Olchanheski LR, da Silva EG, de Lima FR, Martinez LR del PR, Sato MIZ, et al. Influence of water quality on diversity and composition of fungal communities in a tropical river. Sci Rep. 2018;8: 14799. doi: 10.1038/s41598-018-33162-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Almeida VR, Szpoganicz B, Chou L, Baert K, Hubin A, Bonneville S. Equilibrium and Out-Of-Equilibrium Investigation of Proton Exchange and CuII and ZnII Complexation on Fungal Mycelium (Trametes hirsuta). J Braz Chem Soc. 2016;27: 15–29. doi: 10.5935/0103-5053.20150236 [DOI] [Google Scholar]
- 74.Samuels GJ. Trichoderma: a review of biology and systematics of the genus. Mycological research. 1996;100: 923–935. [Google Scholar]
- 75.Wong MKM, Goh T-K, Hodgkiss IJ, Hyde KD, Ranghoo VM, Tsui CKM, et al. Role of fungi in freshwater ecosystems. Biodiversity and Conservation. 1998;7: 1187–1206. doi: 10.1023/A:1008883716975 [DOI] [Google Scholar]
- 76.Angleró-Rodríguez YI, Talyuli OA, Blumberg BJ, Kang S, Demby C, Shields A, et al. An Aedes aegypti-associated fungus increases susceptibility to dengue virus by modulating gut trypsin activity. Soldati-Favre D, editor. eLife. 2017;6: e28844. doi: 10.7554/eLife.28844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balakrishnan S, Santhanam P, Srinivasan M. Larvicidal potency of marine actinobacteria isolated from mangrove environment against Aedes aegypti and Anopheles stephensi. J Parasit Dis. 2017;41: 387–394. doi: 10.1007/s12639-016-0812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angleró-Rodríguez YI, Blumberg BJ, Dong Y, Sandiford SL, Pike A, Clayton AM, et al. A natural Anopheles-associated Penicillium chrysogenum enhances mosquito susceptibility to Plasmodium infection. Scientific reports. 2016;6: 34084. doi: 10.1038/srep34084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waweru AW, Omolo JO, Cheplogoi PK, Njue AW. Mosquito larvicidal trihydroxylindene derivative from submerged cultures of Trametes species. African Journal of Biotechnology. 2017;16: 1457–1460. [Google Scholar]
- 80.de Faria MR, Wraight SP. Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biological control. 2007;43: 237–256. [Google Scholar]
- 81.Farenhorst M, Knols BG, Thomas MB, Howard AF, Takken W, Rowland M, et al. Synergy in efficacy of fungal entomopathogens and permethrin against West African insecticide-resistant Anopheles gambiae mosquitoes. PLoS One. 2010;5: e12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raj MM, Srikanth G, Rajikkannu M, Nandakumar R. Evaluation of botanicals against: Mosquito Larvae to the Extracts of Fungus Beauveria Species. World Scientific News. 2017;88: 199–210. [Google Scholar]
- 83.Vivekanandhan P, Kavitha T, Karthi S, Senthil-Nathan S, Shivakumar MS. Toxicity of Beauveria bassiana-28 Mycelial Extracts on Larvae of Culex quinquefasciatus Mosquito (Diptera: Culicidae). International Journal of Environmental Research and Public Health. 2018;15: 440. doi: 10.3390/ijerph15030440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asahina S. Food Material and Feeding Procedures for Mosquito Larvae. [PMC free article] [PubMed] [Google Scholar]
- 85.Valzania L, Martinson VG, Harrison RE, Boyd BM, Coon KL, Brown MR, et al. Both living bacteria and eukaryotes in the mosquito gut promote growth of larvae. PLOS Neglected Tropical Diseases. 2018;12: e0006638. doi: 10.1371/journal.pntd.0006638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Oliveira MR, Katak R de M, da Silva GF, Marinotti O, Terenius O, Tadei WP, et al. Extracts of Amazonian Fungi With Larvicidal Activities Against Aedes aegypti. Front Microbiol. 2021;12. doi: 10.3389/fmicb.2021.743246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bilgo E, Lovett B, St. Leger RJ, Sanon A, Dabiré RK, Diabaté A. Native entomopathogenic Metarhizium spp. from Burkina Faso and their virulence against the malaria vector Anopheles coluzzii and non-target insects. Parasites & Vectors. 2018;11: 209. doi: 10.1186/s13071-018-2796-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. The information and accession numbers of the sequences deposited in GenBank can be found as described in line 149: All sequences generated in this study have been deposited in the NCBI GenBank database (accession numbers MZ781245 - MZ781299), and in the table in the supporting material with the GenBank deposit number: S5 Table. Taxonomic classification of fungi isolated from An. darlingi breeding sites in the municipalities of Coari (C1 and C2) and São Gabriel da Cachoeira (S1 and S2), based on sequencing of the ITS (Internal Transcribed Spacer) region of the nuclear ribosome and comparison of the sequences with those in the NCBI database, and analysis of phylogenetic trees.