Abstract
The members of genus Moniezia are the common parasites of livestock in tropical areas. The tapeworm, Moniezia expansa is commonly found in the gastrointestinal tract of the small and large ruminants. The present study focused on reporting the prevalence of M. expansa in small ruminants of southern Punjab: sheep and goats, in relation with epidemiological factors like age and gender. An overall prevalence of 27.2% was estimated for the small ruminants with higher infection rates in males (29.8%) and younger age group (<1 year; 32.9%). Moreover, the molecular characterization and phylogenetic analysis of the isolates based on partial cox1 gene indicated the placement of these sequences in the M. expansa cluster. Two distinct haplotypes, without any host tropism, were identified within the Pakistani isolates. A meta-analysis for M. expansa was run for all available global reports exhibiting an overall pooled prevalence of 21.3% (CI 95%: 13.5–29.0). Additionally, a global dataset encompassing 59 partial cox1 sequences submitted from different geographical locations was also assessed. Moderate haplotype diversity (0.760 ± 0.051) and significantly negative deviations from neutrality were estimated. The median joining haplotype network for these sequences revealed an interesting population structure indicating highly divergent sequences from China and Iraq compared to Pakistan, India, Vietnam, Senegal and Ethiopia. Given inconsistencies in genetic data there is a dire need to carry out molecular studies across the entire distributional range of M. expansa to delineate genetic diversity and population structure of the species. This will also be crucial in reevaluating the taxonomy of genus Moniezia.
Introduction
Moniezia species have cosmopolitan distribution inhabitating the intestinal tract of ruminants [1] and are classified as members of the Anoplocephalidae family within the Cyclophyllidea order [2]. Moniezia spp. have typical cestode body structure; scolex, neck followed by long chained strobila. Four large suckers on the scolex act as holdfast organs for the attachment to the host [3,4]. It is distinguished by having suckers devoid of spines and by lacking rostellar hooks and rostellum [5].
Moniezia’s life cycle is indirect, using oribatid mites as intermediate hosts that usually thrive unhindered in grass and soil [6–8]. Domestic animals ingest the oribatid mites that are infected with the eggs of Moniezia. These eggs become infectious larvae (cysticercoids) which infect domestic ruminants upon ingestion [7,9]. Larvae then migrate to the ruminant’s small intestine, attach with their suckers, and develop into adult tapeworms [10,11] and are responsible for the onset of monieziasis, the gastrointestinal disorder [2].
These tapeworms are typically thought to have little pathogenicity, particularly in adult livestock, but they can cause significant illness in calves and lambs, leading to economic losses in stockbreeding [12,13]. However, a heavy infection frequently results in unfavorable clinical manifestations like pot-belly, reduced development rate, diarrhea, anemia, intestinal disease, poor quality wool, fleshless condition, and even death of the ruminant host [3,14].
Despite the significance of these tapeworms, little is known about their ecology, evolutionary biology, and population genetics. Morphological identification of these species is not easy, so far at least 12 species of the genus Moniezia have been identified in both domestic and wild ruminants, largely characterized by a limited set of physical traits [15]. However, these traits are convergent, leading to ongoing debates over the taxonomy of this genus [2], although genetic data are only known for three species: Moniezia expansa, Moniezia benedeni, and Moniezia monardi [16]. Among these, M. expansa is commonly distributed worldwide with varying regional prevalence. For instance, Alberfkani et al. [11] reported 16% prevalence from Iraq, while Bashtar et al. [4] from Egypt observed significantly higher prevalence (74%) in sheep. Most studies on the prevalence of M. expansa have relied on faecal [17,18] and post mortem examination [11,19–21]. A few studies employed multilocus enzyme electrophoresis and isoenzyme electrophoresis method to genetically compare the Moniezia spp. [22,23]. Similarly, few studies have employed the molecular markers (SSU rDNA, ITS1and ITS2, cox1 and nad1) for the correct identification of the M. expansa and M. benedeni [2,12,24–26]. Nonetheless, due to existence of cryptic species further studies are needed to clarify the genetic diversity of Moniezia spp. around the world [2].
Pakistan is an agricultural country and livestock production significantly contributes to the sustenance of farmers by providing food, revenue and employment. Pakistan has a large livestock population and the prevalence of M. expansa is reported from both small and large ruminants (0.86–17.7%) [21,27,28]. However, nearly all the studies have reported prevalence via fecal examination and no published molecular report is available till date.
The aim of the present study was to investigate the epidemiological factors and molecular identification of the intestinal tapeworm, Moniezia expansa in small ruminants from Punjab, Pakistan, by utilizing the partial cox1 gene. In addition to this, global overview about prevalence of this parasite and its genetic diversity analysis were performed on a global dataset for the partial cox1 gene sequences.
Materials and methods
Sample collection
Present study involved the collection of tapeworm infected small intestines (n = 464) from sheep (n = 144) and goats (n = 320) from various slaughterhouses in the Muzaffargarh and Rajanpur districts of Punjab, Pakistan (Fig 1). As the tapeworm samples were collected from slaughterhouses, hence ethival approval was not required. When available, tapeworms were removed from infected small intestines by using the forceps and washed with the saline solution. Tapeworms were stored in sterile labelled bottles with 5 mL of 70% ethanol. The tapeworms were identified on the basis of morphological characters described for Moniezia spp [29]. Epidemiological risk factors like age and gender of host and parasitic burden per animal (total number of tapeworms found) were noted at the time of sample collection, along with documenting the length and width of the parasite. A total of 80 worms were selected for calculating the length and width.
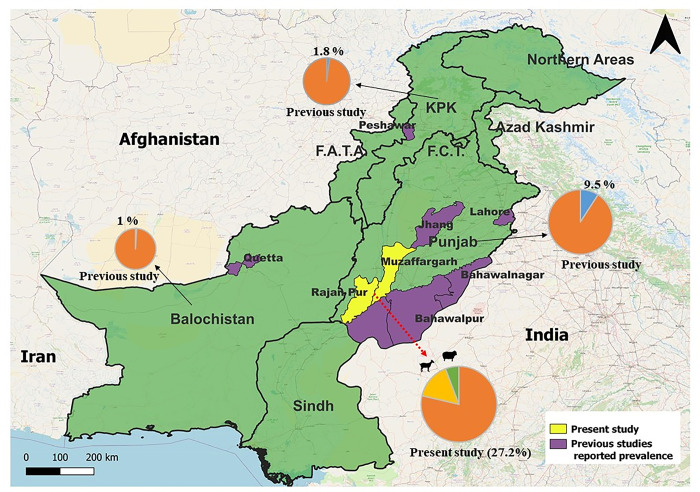
Fig 1. Map of Pakistan showing sampling location and prevalence from present study and estimated pooled prevalence from different provinces (based on previously published data).
Molecular analysis
The parasite tissue samples were rinsed thrice with phosphate buffered saline (PBS) to remove ethanol in which the tapeworms were preserved. Genomic DNA was extracted by using commercial kit (Gene JET Genomic DNA Purification kit, ThermoScientific, USA) according to the given instructions. PCR amplification targeted a partial mitochondrial gene encoding cytochrome c oxidase subunit 1 (cox1) using the primer pair JB3/JB4.5 [30] following the established protocol as reported previously by Muqaddas et al. [31]. The amplicons (450 bp) were separated on ethidium stained agarose gel (1.5%) which was later visualized and digitally captured by UV transilluminator gel documentation system (UVidoc, UK). Gel extraction kit (GeneJET, ThermoScientific, USA) was used to purify positive amplicons, which were later commercially sequenced (1st Base, Malaysia) using both forward and reverse primers as used during the PCR.
The obtained raw sequences were examined using FinchTV viewer (Geospiza, Seattle, WA, USA) to identify vague bases and check the peak quality. The obtained sequences were subsequently compared using the Basic Local Alignment Search Tool (BLAST) in the NCBI (National Centre for Biotechnology Information) database. Multiple sequence alignment of sequences obtained in the present study and those of comparable sequences downloaded from NCBI was performed in ClustalX2 software. Inter and intraspecific phylogenies have been assessed through MEGA11 software by using the Maximum Likelihood method based on the best model [32].
Analysis of genetic diversity of M. expansa
All available nucleotide sequences for M. expansa (partial cox1 gene) from various countries around the world were retrieved from NCBI. In order to analyze the global haplotypes analysis, a final dataset of 59 sequences was trimmed to equal base length of 317 bp, which included sequences generated from the present study and those from other regions of the world. To display the haplotype network, PopART software was used by keeping the statistical parsimony [33]. Furthermore, for global genetic diversity analysis, such as DNA polymorphism, variable sites and their number and population diversity, number of haplotypes (hn), haplotype diversity (hd) and nucleotide diversity (nd) were estimated. The neutrality indices like Tajima’s D and Fu’s Fs were also computed by using DnaSP 6 [34].
Meta-analysis for M. expansa
Data from 94 articles reporting prevalence of M. expansa either through fecal examination or postpartum inspection was entered in Microsoft Excel 2016, and the detailed characteristics of each study included: region, country, host species, diagnostic method, parasitic species and reported prevalence. Later, estimated pool prevalence (%) based on 95% CI and heterogeneity (I2%) was estimated through OpenMetaAnalyst software.
Statistical analysis
The descriptive statistical tool was used for estimating prevalence rates among the hosts, gender and age groups. Prevalence, estimated in percentage, was computed with the number of infected animals relative to the examined animals [35]. To establish the role of different epidemiological factors with the prevalence of monieziasis, Chi square statistic (χ2) was employed. All data were analyzed through SPSS (version 25) at significance level of 0.05.
Results
Prevalence of Moniezia spp. in South Punjab, Pakistan
The tapeworms belonging to genus Moniezia were detected in 126 out of 464 small ruminants during GI tract examination, with an overall prevalence of 27.2% (Table 1). The prevalence of Moniezia spp. was further determined in two age groups. The animals in the age group <1 year had the highest prevalence (32.9%), while those in the age group >1 year had the lowest prevalence (10.7%). With regards to gender, higher prevalence was recorded among males (29.8%) than females (20.9%). The mean burden of Moniezia spp. was three (n = 3±2.98) parasites per host with a maximum number of 12 worms retrieved from a 6 month old male goat. Moreover, the average length of worms was 167.6±2.37 cm with the mean width of 1.1± 0.27cm.
Table 1. Prevalence of M. expansa in different age group and gender of host species.
| Variables | Category | Examined | Infected | Prevalence % | Chi-square (χ2) | Significance value (p) |
|---|---|---|---|---|---|---|
| Host | Goat | 320 | 92 | 28.7 | 1.326 | 0.250 |
| Sheep | 144 | 34 | 23.6 | |||
| Age group | <1 | 343 | 113 | 32.9 | 22.28 | 0.001 *** |
| >1 | 121 | 13 | 10.7 | |||
| Gender | Male | 325 | 97 | 29.8 | 3.97 | 0.46 |
| Female | 139 | 29 | 20.9 | |||
| Total | 464 | 126 | 27.2 | |||
P > 0.05 = Non significant; P < 0.001 = Highly significant (***).
Phylogenetic analysis of present study
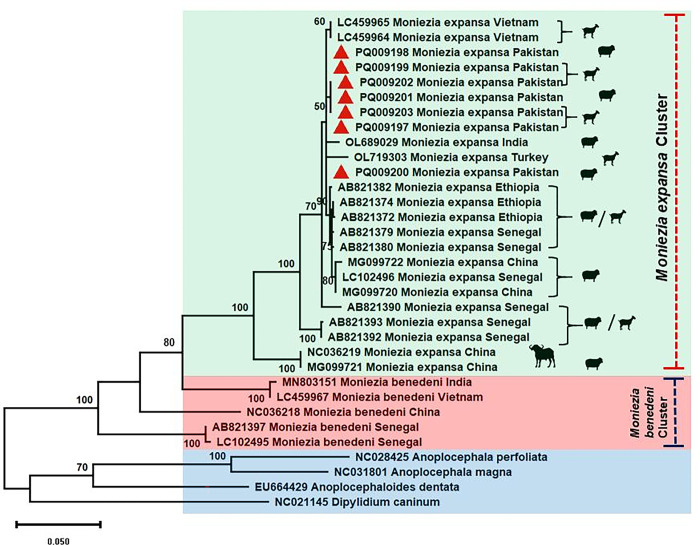
Overall, 25 Moniezia tapeworms were sequenced for the partial cox1 gene (392bp) which resembled with M. expansa after BLAST analysis. Two distinct haplotypes were found with no genetic distinction for sheep or goat. Among these, only seven isolates, based on host and geographical location, were submitted to GenBank (accession numbers PQ009197-203). The sheep sampled from Muzaffargarh only harbored a single dominant haplotype of M. expansa whereas, the goats harbored both haplotypes. For phylogenetic analysis, M. benedeni and other Anoplocephalid species were chosen along with Dipylidium caninum as an outgroup. Across phylogenetic analyses, the studied sequences fell into the clade of M. expansa (Fig 2), and showed a high similarity to M. expansa from Vietnam (LC459964-65) and India (OL689029).
Fig 2. A maximum likelihood tree of M. expansa constructed from sequences of mtDNA cox1 gene (392bp) displaying distinct M. expansa and M. benedeni clades.
Bootstrap values are shown as numbers on the nodes.
Haplotype analysis of M. expansa world population
The polymorphism analysis of global data set (n = 59; 317bp) revealed the presence of 16 haplotypes characterized by 9 singleton variable sites and 50 parsimony informative sites (Table 2). Genetic diversity estimates for M. expansa isolates revealed moderately high haplotype diversity (0.760±0.051) accompanied with low nucleotide diversity (0.0196±0.0059). A significantly negative trend was estimated for Fu’s Fs (-0.253, p < 0.05), whereas Tajima’s D exhibited negative and significant trend (-1.8996, p< 0.05).
Table 2. Diversity and neutrality indices for M. expansa population originating from different countries of the world.
| Amplified gene | Cox1 (317bp) |
|---|---|
| No of isolates | 59 |
| No. of polymorphic sites | 65 |
| Singleton variable sites | 9 |
| Parsimony informative sites | 50 |
| Parsimony informative sites (two variants) | 45 |
| Parsimony informative sites (three variants) | 4 |
| No of Haplotypes | 16 |
| K = Average number of pairwise nucleotide difference | 6.23144 |
| Haplotype diversity (Hd) ± SD | 0.760±0.051 |
| Nucleotide diversity (π) ± SD | 0.01966 ±0.00594 |
| Tajima’s D | -1.89968, p< 0.05 |
| Fu’s Fs | -0.253, p < 0.05 |
P < 0.05 = Least significant.
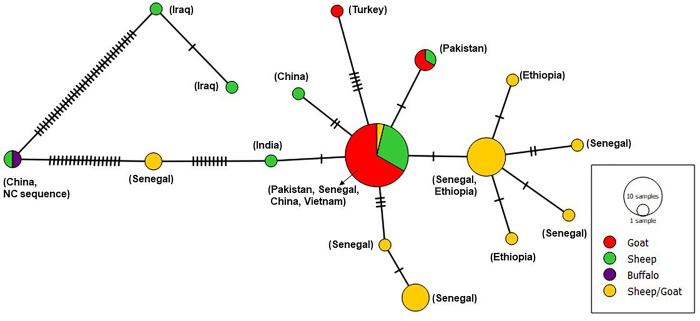
The parsimony haplotype network for M. expansa global data set showed a star shaped topology with most prevalent and centrally located haplotype from Pakistan, Senegal, China and Vietnam (Fig 3). Whereas the second most common haplotype with single mutation from centrally located haplotype is from Senegal and Ethiopia. Two haplotypes from Iraq showed large number of mutations from the central haplotype. Similarly, the reference sequence NC036219 from China is distantly related to central haplotype with large number of mutations in a small conserved region of mitochondrial gene cox1.
Fig 3. Overall haplotypic profile of M. expansa populations based on partial cox1 (317bp) gene from different countries and their associated hosts.
Hatch marks represent the number of mutations between each haplotype and the size of circle corresponds to the frequency of each haplotype in the population.
Meta-analysis
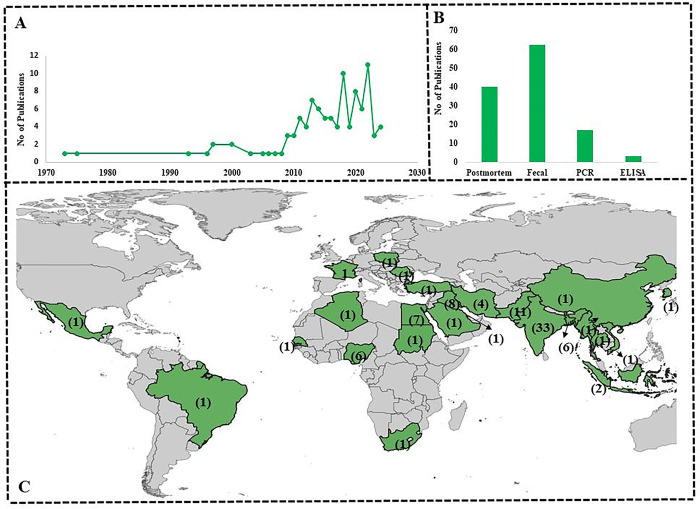
After review of the global literature, it seemed that monieziasis is quite common in ruminants of the tropical region. Prevalence reports mostly came from countries in Asia (n = 72), followed by few reports from Africa (n = 17), Europe (n = 3) and America (n = 2). There were great variations in prevalence between various countries and continents with an overall estimated pooled prevalence of 12.4% (CI: 95%, 10.9–13.8) reflecting diverse epidemiological dynamics within the region and countries. Among the countries in Asia, Vietnam and Iraq exhibited the highest estimated pooled prevalence of 21.5% (CI: 95%, 15.8–27.2) and 21.3% (CI: 95%,13.5–29.0) respectively. With regards to South Asia, highest number of studies (n = 33) are reported from India, in total, 23837 animals were tested and 1908 were found positive, giving rise to an estimated pooled prevalence of 9.2% (CI: 95%, 7.7–10.6%) (Table 3; Fig 4) followed by Pakistan with 11 studies with 257 positive animals with estimated pooled prevalence of 5.2% (CI: 95%, 3.0–7.4). Within Pakistan, a notable disparity in the prevalence rates was found across the provinces, with Punjab having the highest pooled prevalence (9.5%) and Baluchistan harboring the lowest pooled prevalence (1%) (Table 4).
Table 3. M. expansa prevalence in animals from the main continents and their countries.
| Parameter | No. data sets | No. tested | No. positive | Pooled estimate % based on 95% CI | Heterogeneity I2% |
|---|---|---|---|---|---|
| Overall | 94 | 48828 | 5551 | 12.4 (10.9–13.8) | 99.09 |
| Africa | 17 | 6368 | 2276 | 19.8 (7.8–31.8) | 99.73 |
| Egypt | 7 | 3962 | 1980 | 26.8 (-3.4–57.1) | 99.84 |
| Algeria | 1 | 116 | 60 | 51.7 (42.6–60.8) | NA |
| Nigeria | 6 | 1452 | 151 | 11.2 (4.7–17.7) | 96.52 |
| South Africa | 1 | 283 | 6 | 2.1 (0.4–3.8) | NA |
| Senegal | 1 | 510 | 74 | 14.5 (11.5–17.6) | NA |
| Sudan | 1 | 45 | 5 | 11.1 (1.9–20.3) | NA |
| Asia | 72 | 39966 | 3032 | 9.0 (8.0–10.0) | 97.77 |
| Iraq | 8 | 1675 | 176 | 21.3 (13.5–29) | 98.02 |
| Vietnam | 1 | 200 | 43 | 21.5 (15.8–27.2) | NA |
| India | 33 | 23837 | 1908 | 9.2 (7.7–10.6) | 98.44 |
| Bangladesh | 6 | 2084 | 137 | 11.6 (7.1–16.1) | 96.08 |
| Korea | 1 | 546 | 91 | 16.7 (13.5–19.8) | NA |
| Bahrain | 1 | 170 | 4 | 2.4 (0.1–4.6) | NA |
| Indonesia | 2 | 340 | 31 | 9.1 (6.1–12.2) | 0 |
| China | 1 | 1011 | 167 | 16.5 (14.2–18.8) | NA |
| Turkey | 1 | 4003 | 125 | 3.1 (2.6–3.7) | NA |
| Myanmar | 1 | 380 | 53 | 13.9 (10.5–17.4) | NA |
| Thailand | 1 | 185 | 10 | 5.4 (2.1–8.7) | NA |
| Iran | 4 | 277 | 16 | 4.8 (1.7–7.9) | 24.74 |
| Saudia Arabia | 1 | 1200 | 14 | 1.2 (0.6–1.8) | NA |
| Pakistan | 11 | 4058 | 257 | 5.2 (3.0–7.4) | 95.65 |
| Europe | 3 | 362 | 111 | 33.2 (14.7–51.6) | 93.77 |
| Poland | 1 | 158 | 39 | 24.7 (18.0–31.4) | NA |
| France | 1 | 118 | 24 | 20.3 (13.1–27.6) | NA |
| Romania | 1 | 86 | 48 | 55.8 (45.3–66.3) | NA |
| America | 2 | 2132 | 132 | 11.9 (-4.3–28.2) | 98.02 |
| Mexico | 1 | 1823 | 69 | 3.8 (2.9–4.7) | NA |
| Brazil | 1 | 309 | 63 | 20.4 (15.9–24.9) | NA |
Fig 4.
Graph illustrating the number of publications per year on M. expansa (A), techniques used for identification (B), The world map showing the number of studies conducted in various countries on the prevalence of M. expansa (C).
Table 4. Prevalence of M. expansa among different provinces of Pakistan.
| Parameter | No. data sets | No. tested | No. positive | Pooled estimate % based on 95% CI | Heterogeneity I2% |
|---|---|---|---|---|---|
| Overall | 11 | 4058 | 257 | 5.2 (3.0–7.4) | 95.65 |
| Punjab | 6 | 2220 | 224 | 9.5 (4.5–14.4) | 97.68 |
| Khyber Pakhtunkhwa | 4 | 1325 | 28 | 1.8 (0.7–2.8) | 39.92 |
| Baluchistan | 1 | 513 | 5 | 1.0 (0.1–1.8) | NA |
Discussion
In the present study, genetic diversity and other epidemiological factors contributing to M. expansa prevalence were studied. Till date, only a few studies have been carried out regarding the prevalence of monieziasis in Pakistan on domestic host species. There are gaps in knowledge about the disease and no study reporting molecular epidemiology exists in literature.
M. expansa is commonly found in sheep and goats and rarely in cattle [23,36]. The present study reported 27.2% prevalence of M. expansa in the small ruminants with goats exhibiting a higher frequency (28.7%) compared to sheep (23.6%). Nearly similar prevalence of 24% was reported from India in sheep and goats [37]. However, earlier studies in Pakistan suggested only a prevalence of 0.26–5.81% in the small ruminants from different areas of the country [38,39]. The occurrence of gastrointestinal helminths is influenced by agro-climatic factors such as pasture quality and management practices, temperature, humidity, and the grazing habits of the livestock species [40]. There is an increased parasitic burden in the host species during the rainy (monsoon) season [41] due to favorable conditions for parasite propagation and larval development [42]. In gender based analysis, a higher prevalence was observed in males (32.9%) compared to females (10.7%). Similar results were reported by Zvinorova et al. [43] where a higher number of males were infected with gastrointestinal helminths owing to differential susceptibility of males due to hormonal control and genetic predisposition. A higher chance of contact due to increased browsing time may also be attributed to high infection status [44]. In the parasitic diseases, females generally have higher infection rates due to the stress of pregnancy and parturition. However, the practice of stall-feeding females during pregnancy may reduce their exposure to contaminated grazing areas [39]. Age is one of the infection determinants for monieziasis; in current investigation animals with age bracket of less than one year were found to be more infected than age greater than one year. Increased contact with parasites and favorable conditions for ecdysis of the mite species may result in higher infection rates [45]. Moreover, it is reported that M. expansa is usually more common in animals younger than 6 to 8 months, and older animals generally exhibit reduced susceptibility. After the age of two years, the animals rarely host more than one or a small number of worms [46].
The parasite M. expansa is believed to have European origin with little variation across geographical populations on the basis of isoenzyme electrophoresis [23]. Nonetheless, taxonomic uncertainties exist for the genus Moneizia and both M. expansa and M. benedeni represent a species complex due to the presence of cryptic species [22,23]. A characteristic diagnostic morphological feature of pattern of interproglottidal glands in M. expansa is refuted in some studies and the utilization of genetic markers is therefore necessitated for correct species identification [2,22,23]. In current study, 25 isolates were sequenced for the partial cox1 gene and all the samples were identified as M. expansa after phylogenetic analysis. Three clades of M. expansa were identified from sheep and goats of Senegal and Ethiopia by Diop et al. [2], however, none of the current study sequences resembled sequences in these clades despite being placed among M. expansa cluster. The current study sequences of M. expansa also differed from those identified from China and were somewhat more similar to Indian and Vietnamese isolates (Fig 2). These results indicated that the genetic variation among all geographical populations of M. expansa must be studied based on longer genetic (mitochondrial and nuclear) markers to reevaluate this species complex enabling more reliable identification.
The global data set of M. expnasa based on partial cox1 gene (317 bp) was characterized by the presence of 16 haplotypes from a total of 59 sequences submitted from China, India, Vietnam, Pakistan, Iraq, Turkey, Senegal and Ethiopia. A moderate haplotype diversity and low nucleotide diversity were identified with negative trends for neutrality indices suggesting population expansion. The median joining haplotype network for these sequences revealed an interesting population structure placing the Pakistani isolates in the middle as a major cluster from which African isolates from Senegal and Ethiopia were branching off. The isolates from China and Iraq were highly divergent from the main cluster represented by several mutational differences within the small cox1 fragment (Fig 3). This kind of population structure reiterates the need to address the taxonomic issues within M. expansa species complex.
Conclusion
Current study reflects upon the prevalence and distribution of M. expansa across different continents with main locales present in Asia and Africa. Apart from outlining geographical presence, these studies fail to establish the population structure as most of the studies are not supported by molecular evidence. There is a dire need to carry out molecular studies across the entire distributional range of M. expansa to delineate genetic diversity and population structure of the species. Moreover, the taxonomical controversy about the existence of cryptic species can also be resolved by keeping in view the biological features, morphology and host tropism in addition to geographical distribution. There is also a need to identify relevant morphological characters which may be able to reliably distinguish different species of the genus Moniezia as most of these parasites employ similar hosts.
Data Availability
The datasets generated and/or analyzed during the current study are available in the GenBank repository, with Accession numbers PQ009197-203 for Moniezia expansa https://www.ncbi.nlm.nih.gov/nuccore/ PQ009197 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009198 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009199 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009200 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009201 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009202 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009203.
Funding Statement
The authors extend their appreciation to the Researchers supporting Project number (RSPD2024R971 to A.S.), King Saud University, Riyadh, Saudi Arabia, for funding this research.
References
- 1.Jyoti SNK, Juyal PD. Prevalence of gastro-intestinal parasites in buffalo calves from different agro-climatic zones of Punjab. J. Parasit. Dis. 2014; 38(4): 367–370. 10.1007/s12639-013-0259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diop G, Yanagida T, Hailemariam Z, Menkir S, Nakao M, Sako Y, et al. Genetic characterization of Moniezia species in Senegal and Ethiopia. Parasitol. Int. 2015; 64(5): 256–260. 10.1016/j.parint.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Zhao WJ, Zhang H, Bo X, Li Y, Fu X. Generation and analysis of expressed sequence tags from a cDNA library of Moniezia expansa. Mol Biochem Parasitol. 2009; 164(1): 80–85. 10.1016/j.molbiopara.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Bashtar AR, Hassanein M, Abdel-Ghaffar F, Al-Rasheid K, Hassan S, Mehlhorn H, et al. Studies on monieziasis of sheep I. Prevalence and antihelminthic effects of some plant extracts, a light and electron microscopic study. Parasitol. Res. 2011; 108(1): 177–186. doi: 10.1007/s00436-010-2060-2 [DOI] [PubMed] [Google Scholar]
- 5.Atiyah KM, Azzal GY. Biological study of Moniezia spp. isolated from slaughtered sheep in Basrah Provence, Southern Iraq. J. Global Sci. Res. 2022; 7(4): 2227–2233. https://www.gsjpublications.com/jgsr15920064. [Google Scholar]
- 6.Akrami MA, Mostowfizadeh-Ghalamfarsa R, Ebrahimi F, Moazeni M. Molecular detection of Moniezia spp.(Cestoda) in Pergalumna persica (Acari: Oribatida) in Iran. Syst Appl Acarol. 2014: 23(10): 1931–1939. [Google Scholar]
- 7.Schuster R, Coetzee L, Putterill JF. Oribatid mites (Acari, Oribatida) as intermediate hosts of tapeworms of the family Anoplocephalidae (Cestoda) and the transmission of Moniezia expansa cysticercoids in South Africa. Onderstepoort J. Vet. Res. 2000; 67(1):49–55 . [PubMed] [Google Scholar]
- 8.Roczen-Karczmarz M, Tomczuk K. Oribatid mites as vectors of invasive diseases. Acarologia. 2016; 56(4), 613–623. https://doi.org/ff10.1051/acarologia/20164143. [Google Scholar]
- 9.Abdelhamid M, Vorobiev VI, Lapteva ML, Dyab AK. Combined effect of monieziosis and hypomicroelementosis on some hematological, biochemical and hormonal parameters in merino sheep. Pak. Vet. J. 2021; 41(1): 107–111. 10.29261/pakvetj/2020.068. [DOI] [Google Scholar]
- 10.Guo A. Moniezia benedeni and Moniezia expansa are distinct cestode species based on complete mitochondrial genomes. Acta Trop. 2017; 166: 287–292. 10.1016/j.actatropica.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Alberfkani MI, Albarwary AJ, Jaafar GM, Zubair AI, Abdullah RY. Molecular characterization and phylogenetic analysis of cox 1 and ITS 1 gene fragments of Moniezia species isolated from sheep. Pak. Vet. J. 2022; 42(4): 566–570. 10.29261/pakvetj/2022.073. [DOI] [Google Scholar]
- 12.Al-Otaibi BO, Degheidy NS, Al-Malki JS. Prevalence, incidence and molecular characterization of tape worms in Al Taif governorate, KSA and the effectiveness of Spirulina platensis as a biological control in vitro. Saudi J. Biol. Sci. 2021; 28(11): 6272–6278. 10.1016/j.sjbs.2021.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazyad SA, el-Nemr H I. The endoparasites of sheep and goats, and shepherd in North Sinai Governorate, Egypt. J. Egypt. Soc. Parasitol. 2002; 32(1): 119–126. . [PubMed] [Google Scholar]
- 14.Yan H, Bo X, Liu Y, Lou Z, Ni X, Shi W, et al. Differential diagnosis of Moniezia benedeni and M. expansa (Anoplocephalidae) by PCR using markers in small ribosomal DNA (18S rDNA). Acta Vet. Hung. 2013; 61(4), 463–472. doi: 10.1556/AVet.2013.035 [DOI] [PubMed] [Google Scholar]
- 15.Schmidt GD, 1986. Key to the genera Anoplocephalinae. pp. 421–451. In: Handbook of Tapeworm identification. CRC Press, Boca Raton. [Google Scholar]
- 16.Ohtori M, Aoki M, Itagaki, T. Sequence differences in the internal transcribed spacer 1 and 5.8S ribosomal RNA among three Moniezia species isolated from ruminants in Japan. J. Vet. Med. Sci. 2015; 77(1): 105–107. 10.1292/jvms.14-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azrul LM, Poungpong K, Jittapalapong S, Prasanpanich S. Descriptive prevalence of gastrointestinal parasites in goats from small farms in Bangkok and vicinity and the associated risk factors. Annual Res. Rev. Biol. 2017; 16(2): 1–7. 10.9734/ARRB/2017/34932. [DOI] [Google Scholar]
- 18.Pilarczyk B, Tomza-Marciniak A, Pilarczyk R, Bombik E, Seremak B, Udała J, et al. A comparison of the prevalence of the parasites of the digestive tract in goats from organic and conventional farms. Animals 2021; 11(9): 2581. doi: 10.3390/ani11092581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Seify MA, Elshahawy IS, Ibrahim O, Ahamed ZK. An abattoir-based survey on the prevalence of some gastrointestinal helminths of camels (Camelus dromedarius) in Aswan Province, Egypt. SVU-Int. J. Vet. Sci. 2021; 4(3), 119–130. 10.21608/svu.2021.83625.1133. [DOI] [Google Scholar]
- 20.Ndom M, Diop G, Quilichini Y, Yanagida T, Ba CT, Marchand B. Prevalence and scanning electron microscopic identification of anoplocephalid cestodes among small ruminants in Senegal. J. Parasitol. Res. 2016; 3937292. doi: 10.1155/2016/3937292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaman MA, Sajid M, Sikandar A, Awais MM. Point prevalence of gastrointestinal helminths and their association with sex and ge of buffaloes in lower Punjab, Pakistan. Int. J. Agric. Biol. 2014; 16(6): 1229–1231. [Google Scholar]
- 22.Chilton NB, O’Callaghan MG, Beveridge I, Andrews RH. Genetic markers to distinguish Moniezia expansa from M. benedeni (Cestoda: Anoplocephalidae) and evidence of the existence of cryptic species in Australia. Parasitol. Res. 2007; 100(6): 1187–1192. [DOI] [PubMed] [Google Scholar]
- 23.Ba CT, Wang XQ, Renaud F, Euzet L, Marchand B, De Meeüs T. Diversity and specificity in cestodes of the genus Moniezia: genetic evidence. Int. J. Parasitol. 1993; 23(7): 853–857. doi: 10.1016/0020-7519(93)90049-5 [DOI] [PubMed] [Google Scholar]
- 24.Alfatlawi MA, Ismail YK, Ali MJ, Karawan AC, Ibadi IN. Molecular differentiation of Thysaniezia (Helictometra) giardi and Moniezia species based on 18s rRNA gene in small ruminants. Iraqi J. Vet. Sci. 2021; 35(1): 105–108. 10.33899/ijvs.2020.126407.1313. [DOI] [Google Scholar]
- 25.Tam TT, Lan NTK, Doanh PN. Morphological differences and molecular phylogenetic relationship of two tapeworm species, Moniezia expansa and Moniezia benedeni collected from domestic ruminants in northern Vietnam. Parasitol. Int. 2020; 74: 101998. 10.1016/j.parint.2019.101998. [DOI] [PubMed] [Google Scholar]
- 26.Haukisalmi V, Laaksonen S, Oksanen A, Beckmen K, Halajian A, Yanagida T, et al. Molecular taxonomy and subgeneric classification of tapeworms of the genus Moniezia Blanchard, 1891 (Cestoda, Anoplocephalidae) in northern cervids (Alces and Rangifer). Parasitol. Int. 2018; 67(2): 218–224. 10.1016/j.parint.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Shah A, Rehman N. Coprological examination of domestic livestock for intestinal parasites in Village Bahlola, District Charsaddah (Pakistan). Pak. J. Zool. 2001; 33(4). 344–346. [Google Scholar]
- 28.Ijaz M, Zaman MA, Mariam F, Farooqi SH, Aqib AI, Saleem S, et al. Prevalence, hematology and chemotherapy of gastrointestinal helminths in camels. Pak. Vet. J. 2018; 38(1): 81–85. doi: 10.29261/pakvetj/2018.016 [DOI] [Google Scholar]
- 29.Spasski AA. Essentials of cestology. Anoplocephalate tapeworms of domestic and wild animals. Vol. 1. 1961. [Google Scholar]
- 30.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992; 54(2): 165–173. doi: 10.1016/0166-6851(92)90109-W [DOI] [PubMed] [Google Scholar]
- 31.Muqaddas H, Mehmood N, Arshad M. Genetic variability and diversity of Echinococcus granulosus sensu lato in human isolates of Pakistan based on cox1 mt-DNA sequences (366bp). Acta Trop. 2020; 207: 105470. 10.1016/j.actatropica.2020.105470. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Boil. Evol. 2018; 35(6): 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Boil. Evol. 199916(1)37–48. doi: 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- 34.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Boil. Evol. 2017; 34(12): 3299–302. doi: 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 35.Mehmood N, Arshad M, Ahmed H, Simsek S, Muqaddas H. Comprehensive account on prevalence and characteristics of hydatid cysts in livestock from Pakistan.Korean J. Parasitol. 2020. Apr;58(2):121. doi: 10.3347/kjp.2020.58.2.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen TD, Le QD, Huynh VV, Nguyen ST, Nguyen TV, Vu-Khac H. The development of PCR methodology for the identification of species of the tapeworm Moniezia from cattle, goats and sheep in central Vietnam. J. Helminthol. 2012; 86(4): 426–9. doi: 10.1017/S0022149X11000629 [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Kaur H. Molecular characterization of Moniezia denticulata (Rudolphi, 1810) and its distinction from M. expansa infecting sheep and goats raised in the north and north-western regions of India. Parasitology. 2023; 150(9): 831–41. 10.1017/S003118202300063X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayaz MM, Raza MA, Murtaza S, Akhtar S. Epidemiological survey of helminths of goats in southern Punjab, Pakistan. Trop. Biomed. 2013; 30(1): 62–71. . [PubMed] [Google Scholar]
- 39.Raza MA, Arshad HM, Ayaz MM, Bachaya SM, Basit A. Gastrointestinal helminthiasis in goats at the localities of Jatoi, Punjab, Pakistan. Sci. Int. (Lahore). 2012; 24(2): 171–5. [Google Scholar]
- 40.Gupta A, Singh NK, Singh H, Rath SS. Assessment of risk factors associated with prevalence of gastrointestinal helminths in buffaloes from Punjab State, India. Buffalo Bullet. 2018; 37(3): 279–90. [Google Scholar]
- 41.Pathak AK, Pal S. Seasonal prevalence of gastrointestinal parasites in goats from Durg district of Chhattisgarh. Vet. World. 2008; 1(5):136–138. [Google Scholar]
- 42.Faizal AC, Rajapakse RP. Prevalence of coccidia and gastrointestinal nematode infections in cross bred goats in the dry areas of Sri Lanka. Small Rumin. Res. 2001; 40(3): 233–238. doi: 10.1016/s0921-4488(01)00179-1 [DOI] [PubMed] [Google Scholar]
- 43.Zvinorova PI, Halimani TE, Muchadeyi FC, Matika O, Riggio V, Dzama K. Prevalence and risk factors of gastrointestinal parasitic infections in goats in low-input low-output farming systems in Zimbabwe. Small Rumin. Res. 2016; 143: 75–83. doi: 10.1016/j.smallrumres.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouattara L, Dorchies P. Gastro-intestinal helminths of sheep and goats in subhumid and sahelian areas of Burkina Faso. Revue de Médecine Vétérinaire, 2001; 152(2): 165–170. [Google Scholar]
- 45.Purja R, Maharjan M. Gastrointestinal parasites in goat (Capra hircus) of Puranchaur VDC, Pokhara. Int. J. Res. Studies Zool. 2017; 3(4): 39–45. 10.20431/2454-941X.0304005. [DOI] [Google Scholar]
- 46.Mullen GR, OConnor BM. Mites (Acari). In: Mullen GR, Durden LA, editors. Medical and veterinary entomology. 3rd ed. New York: Academic Press; 2019. pp. 533–602. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GenBank repository, with Accession numbers PQ009197-203 for Moniezia expansa https://www.ncbi.nlm.nih.gov/nuccore/ PQ009197 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009198 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009199 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009200 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009201 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009202 https://www.ncbi.nlm.nih.gov/nuccore/ PQ009203.