Abstract
Background
Immune checkpoint inhibitors (ICIs) have greatly improved cancer treatment, but the role of genes related to T cell-mediated tumor killing (TTK) sensitivity in ovarian cancer (OV) is unclear.
Methods
This study analyzed 1367 OV patients and 11 independent cohorts. The unsupervised clustering was conducted to identify three tumor subtypes based on genes that regulate tumor cell sensitivity to TTK (GSTTKs). The biological characteristics, genetic variations, immunological landscape, and therapeutic evaluation for each subtype were further explored.
Results
Patients were divided into three reproducible subtypes based on 61 GSTKKs associated with prognosis. C1 was likely to be an invasive subtype with the worst prognosis and highly unstable genome. C2 might be an immune-active subtype with the best prognosis, high immune infiltration and preferable response to immunotherapy. C3 might be a metabolic subtype, resistant to immunotherapy, but sensitive to drug therapy. Following an extensive exploration into a variety of distinct molecular features between the three subtypes, the results suggested that C2 patients were considered to derive significant efficacy from immunotherapy. For C1 and C3 patients, chemotherapy might be an ideal treatment strategy.
Conclusions
In this study, three GSTKKs-based subtypes were identified by assessing TTK patterns in OV. These new insights further improved our understanding of GSTTKs and might refine clinical treatment strategies for OV patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01635-4.
Keywords: Ovary cancer, T cell-mediated tumor killing, Immune microenvironment, Somatic mutation analysis, Metabolism analysis, Cancer treatment
Introduction
Ovarian cancer, which primarily affects the female reproductive system, is predominantly of the epithelial cell type, accounting for approximately 90% of cases. Epithelial ovarian cancer comprises various histologic subtypes, each with distinct molecular changes, clinical behaviors, and treatment outcomes. The remaining 10% of ovarian cancers are non-epithelial, including mainly germ cell tumors, sex cord-stromal tumors, and rare entities such as small cell carcinomas. Germ cell tumors, in particular, differ from epithelial ovarian cancers with their earlier age of incidence, faster growth rate, unilateral localization in 95% of cases, and generally favorable prognosis [1]. According to the statistics in the United States, there were 19,880 new cases of OV in 2022, accounting for approximately 2.1% of all new cancers in women. Meanwhile, there were approximately 12,810 deaths, accounting for 5% [2]. Owing to the lack of specific early symptoms and effective means of early detection, most OV patients were diagnosed at an advanced stage [3]. Currently, CA125 and HE4 are the only approved biomarkers for use in epithelial ovarian cancer, but they are inadequate for early detection. To address the limitations of single serum biomarkers, multivariate index (MVI) assays have been developed, especially for the pre-surgical evaluation of adnexal masses. The Risk of Malignancy Algorithm (ROMA), which integrates menopausal status with CA125 and HE4 concentrations, has been utilized to diagnose women with pelvic masses. Additionally, miRNAs show great potential in predicting epithelial ovarian cancer. However, further work is needed to standardize the processing of samples and refine detection platforms before they could be reliably used as clinical biomarkers [4]. Patients have a dismal prognosis as invasive high-grade serous carcinoma predominates, with a 47% five-year survival rate [5]. Patients have a dismal prognosis as invasive high-grade serous carcinoma predominates, with a 47% five-year survival rate. This poor prognosis has driven investigations into economic and cost-effective strategies for early detection and prevention of ovarian cancer over the past decade. Notably, the cost of treatment per patient remains among the highest across all cancer types, with initial costs in the first year averaging around USD 80,000 and final-year costs approaching USD 100,000 [6]. In the past, the primary classification of ovarian cancer was based on morphology, categorizing the disease into two types: Type I epithelial ovarian cancers, which are relatively indolent, genetically stable tumors that typically arise from precursor lesions such as endometriosis or borderline tumors with low malignant potential, and Type II epithelial ovarian cancers, which are biologically aggressive from the outset and prone to metastasis even from small-volume primary lesions. High-grade serous ovarian cancer, the most common subtype, falls under the Type II category and is characterized by frequent p53 and BRCA mutations [7]. Therefore, a more accurate approach is needed to achieve the goal of precision medicine.
In recent decades, more and more treatment options have been applied to treat cancer, such as surgical resection, chemoradiotherapy, and targeted therapy. Currently, the clinical treatment of OV is mainly cytoreductive surgery combined with cisplatin and paclitaxel chemotherapy [8]. Notably, BRCA1/2 germline mutations are the strongest known genetic risk factors for epithelial ovarian cancer, found in 6–15% of diagnosed women. Importantly, BRCA1/2 mutation carriers with epithelial ovarian cancer tend to respond better to platinum-based chemotherapies than non-carriers, leading to improved survival outcomes, despite the typically late-stage and higher-grade diagnosis [9]. Some targeted drugs have also been applied to OV patients. However, the mortality rate of advanced patients is as high as 70%, as a result of treatment resistance and the strong probability of recurrence [10]. Substantial evidence suggests that the immune system plays a critical role in cancer development [11]. OV has also been found to be an immunogenic neoplasm [12]. It was reported that Avastin (bevacizumab), a recombinant antibody targeting VEGF/VEGFR signal transduction, has been used to treat some patients with advanced OV [13]. In cancer immunotherapy, tumor immune checkpoint inhibitors are the latest cutting-edge treatment, and their clinical research is the most mature and sufficient. Their application is also the most extensive [14]. However, due to the high heterogeneity of OV, the therapeutic effect of immunotherapy remains largely unknown.
T cell-mediated tumor killing plays an essential role in cancer destruction as the basis for immune checkpoint inhibitor therapy, and therapies that enhance anti-tumor T cell responses have achieved encouraging results in clinical practice [15]. In the meanwhile, researchers are gradually delving into the processes behind GSTKKs' anti-tumor immunological capabilities. Pan et al. showed that human tumors with inactivating mutations in PBRM1, ARID2, and BRD7 might be more sensitive to PD-1 blockade and other forms of immunotherapy with cytotoxic T cells as the primary effector mechanism [16]. Similarly, KLRB1 gene inactivation or antibody-mediated CD161 blocking agents can enhance both the anticancer function in vivo and the TTK of glioma cells in vitro [17, 18]. Subsequent studies have validated this idea [19]. Ru et al. identified genes, such as PTPN2 and CD47 that may lead to drug resistance or increase the tumor cells' susceptibility to TTK by integrating high-throughput screening technology [20]. Nevertheless, so far, no studies have systematically analyzed T cell-mediated tumor killing in OV, including more in-depth molecular characterization and more detailed molecular classification, which may facilitate the purpose of diagnosis and treatment.
For this reason, our study identified three OV subtypes by consensus clustering with non-negative matrix factorization (NMF) based on a set of identified GSTKKs and verified the stability of typing in three independent cohorts with the nearest template prediction (NTP) algorithm. At the same time, distinctions between the three molecular subtypes were also revealed in biological functional characteristics, genetic variations, and immunological microenvironments. For each type of patient, we further evaluated the effect of immunotherapy and developed potential therapeutic agents to achieve precise treatment and improve prognosis. Consequently, the findings of our study might provide a more practical approach to the clinical treatment for OV patients.
Materials and methods
Data source
The TCGA-OV (n = 354) RNA-seq data was acquired from the UCSC Xena platform. Other microarray datasets, including GSE32062 (n = 270), GSE53963 (n = 174), and GSE140082 (n = 380) were obtained from the Gene Expression Omnibus database (GEO). Collecting genes that have a favorable response to TTK from the TISIDB database, which were further used to establish a gene set defined as GSTTKs. The raw count expression was processed and converted to transcripts per kilobase million (TPM) for subsequent analysis.
The mutation data was downloaded from the GDC website. Copy number variation (CNV) data of GISTIC_2.0 level was collected from FireBrowse. A total of seven cohorts matching the expression and therapeutic information were also retrieved for clinical treatment assessment, encompassing GSE100797 (n = 21), GSE126004 (n = 16), GSE115821 (n = 37), GSE131987 (n = 39), GSE23554 (n = 28), GSE9455 (n = 20), and GSE51373 (n = 28). The basic features of all patients were illustrated in Supplementary Table S1-2.
Identification and validation of GSTKKs-based subtypes
The univariate Cox regression analysis was conducted to detect GSTTKs genes, which were significantly related to the overall survival (OS) of OV patients. Then, whether there was a correlation between these GSTKKs and immune cells was further analyzed. Meanwhile, GSTKKs score were also calculated and employed to explore the relationship with immune scores. Subsequently, according to the expression matrix of GSTTKs genes described above, an unsupervised NMF clustering method was performed to identify various subtypes [21, 22].
Since the NTP algorithm is flexible in evaluating each patient's confidence level of category prediction, it was implemented to evaluate the reliability and stability of subtypes based on the signature gene list [23]. Differentially expressed genes (DEGs) among GSTKKs-based subtypes in OV were first screened by limma package. Subsequently, the top 500 genes with the most considerable log2FoldChange (log2FC) value in each subtype were filtrated as the characteristic genes and combined into a gene signature for NTP validation. According to previous studies [23], samples with FDR < 0.2 were retained.
Associations of GSTKKs-based subtypes with clinical and molecular features
Kaplan–Meier (K-M) analysis was carried out to examine the difference in survival among the three GSTKKs-based subtypes in all cohorts. The multivariate cox regression analysis was employed to identify the characteristics of independent prognostic significance in the cohorts. In addition, clinical grade and stage were vital indicators for evaluating prognosis in the clinic practice. The proportion of different clinical grades and stages in GSTKKs-based subtypes were separately explored.
The gene set enrichment analysis (GSEA) was conducted to explore the biological features in the three subtypes based on the gene signature mentioned above. Biological function was significant when FDR < 0.05. Based on 50 Hallmark gene sets, the Gene Set Variation Analysis (GSVA) was carried out to evaluate the biological pathways in subtypes to further support the results of GSEA.
Genomic alterations analysis of GSTKKs-based subtypes
In MAF files, the top 20 genes with the highest mutation frequency were selected for subsequent analysis. Using Fisher's exact test, we compared the frequency of mutated genes and identified significantly different genes. Meanwhile, levels of tumor mutational burden (TMB), homologous recombination defects (HRD), and tumor neo-antigens were compared among subtypes.
Copy number variations (CNVs) were analyzed by counting the total number of genes with copy number alterations (gains or losses) at the focal and arm levels. After that, the percentage of mutation rate was quantized, encompassing fraction of genome alteration (FGA), fraction of genomic gained (FGG), and fraction of genome lost (FGL). After that, the overall proportion of genome changes in the three subtypes was quantified, and the differences between different subtypes were compared using clinical information.
Immune landscapes of GSTKKs-based subtypes
The TIMER, CIBERSORT_ABS, EPIC, ESTIMATE, MCPcounter, quanTIseq, CIBERSORT, and Xcell algorithms were utilized to evaluate the immune cell components in distinct subtypes [24]. In addition, the ssGSEA was also implemented to analyze immune infiltration. Calculating the coordination degree of specific genes in a single sample can decode the differences in immune characteristics among various subtypes. The immune score, stromal score, and tumor purity were compared through the ESTIMATE algorithm among three subtypes [25].
A total of 51 immunomodulators and three types of immune checkpoints were recruited [26]. Further research into expression profiling of the aforementioned genes, as well as leukocyte fractions in the three subtypes, will allow demarcation of the immunological state of the three subtypes, paving the way for better treatment choices. Meanwhile, in order to evaluate the tumor inflammation Signature (TIS) score of each type, GSVA was conducted based on the TIS gene set.
To further validate the different subtypes in response to immunotherapy, the ssGSEA algorithm was conducted to calculate enrichment scores for each subtype's seven anti-tumor immunity cycle steps. Knowledge-based functional gene expression signatures (Fges) were selected to characterize the TME of the three subtypes. After that to assess the sensitivity of OV patients to immune checkpoint blockers (ICBs), unsupervised subclass mapping (SubMap) was applied to evaluate the similarity of expression configuration files [27]. Adjusted P < 0.05 indicated a significant difference in response to ICBs. Additionally, some pharmacotherapy cohorts, including platinum and paclitaxel, were similarly included in the study to implement customized treatment plans for various individuals.
Pharmacotherapy prediction for GSTKKs-based subtypes
The CMAP database was conducted to explore subtypes of potentially sensitive drugs and further revealed patterns (MOA) of pathways or related molecular pathways targeted by drugs exerting their effects. Connectivity scores were calculated from the amount of gene expression by querying differentially expressed genes for each subtype in the CMAP database. Drugs with a score > 95 were considered potential therapeutic agents, and the mechanism of action shared by each potential therapeutic agent was also represented. The pRRophetic package was based on the amount of gene expression and drug sensitivity to establish a ridge regression model, which could achieve the goal of predicting clinical chemotherapy response [28]. Therefore, it was conducted to predict the potential efficacy of drugs. The therapeutic sensitivity of potential drugs or targeted molecular drugs, as judged by the half-maximal inhibitory concentration (ICI) of patients with OV.
Statistic analysis
All statistical analysis is conducted in R statistical environment (R version 4.1.3). The correlation coefficients of the two variables were calculated by Spearman correlation analysis. The normality of the data was tested by Kolmogorov–Smirnov test. The quantitative data of the variance between multiple groups in accordance with normal distribution and homogeneity were tested by one-way ANOVA; data not meeting normal distribution and or heterogeneity of variance were analyzed by Kruskal–Wallis test. Chi-square test was conducted to compare qualitative data among multiple groups. The significance of the difference was determined through Log-rank test, and the prognosis analysis was compared in a curve generated by Kaplan–Meier method. The risk ratio of each subtype was calculated by using univariate Cox regression analysis, and the independent prognostic factors were determined by multivariate Cox regression analysis. The Benjamin Hochberg (BH) multiple test method was utilized to adjust the P value. Statistically, bilateral P < 0.05 was considered significant.
Results
Identification of subtypes based on GSTTKs gene
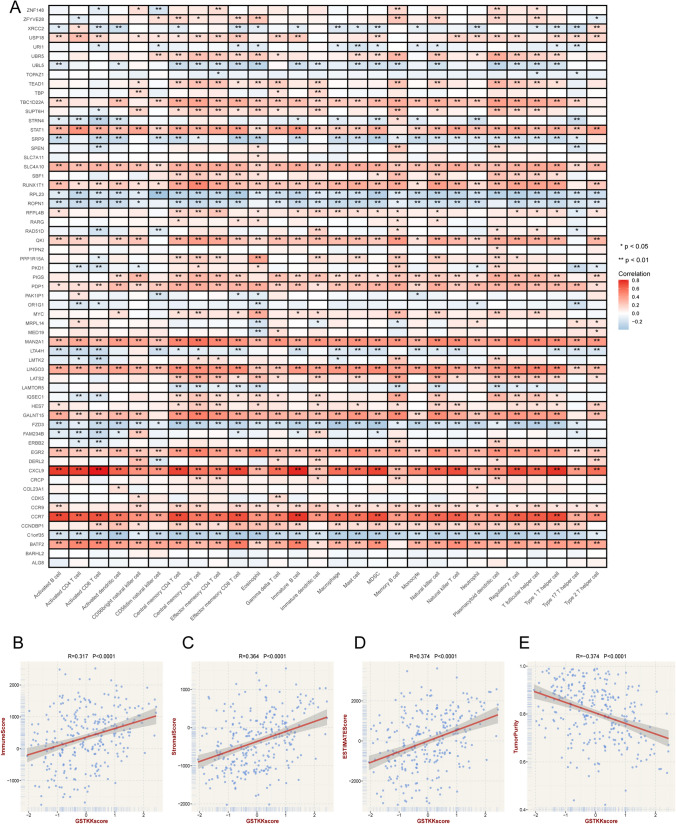
The flow chart of this study was shown in Fig. 1. A total of 1310 GSTTKs were collected and enrolled in our study (Supplementary Table S3). The Univariate Cox regression analysis suggested 61 GSTTKs seemed to have substantial prognostic value (Supplementary Table S4). The tight association between 61 GSTTKs and immune cells was demonstrated as shown in Fig. 2A. Furthermore, GSTKKs score presented substantial positive correlations with immune score, stromal score, and estimation score, as well as a significant negative connection with tumor purity (Fig. 2B–E). This suggested that GSTTKs may play a crucial role in tumor microenvironment and tumor immunity.
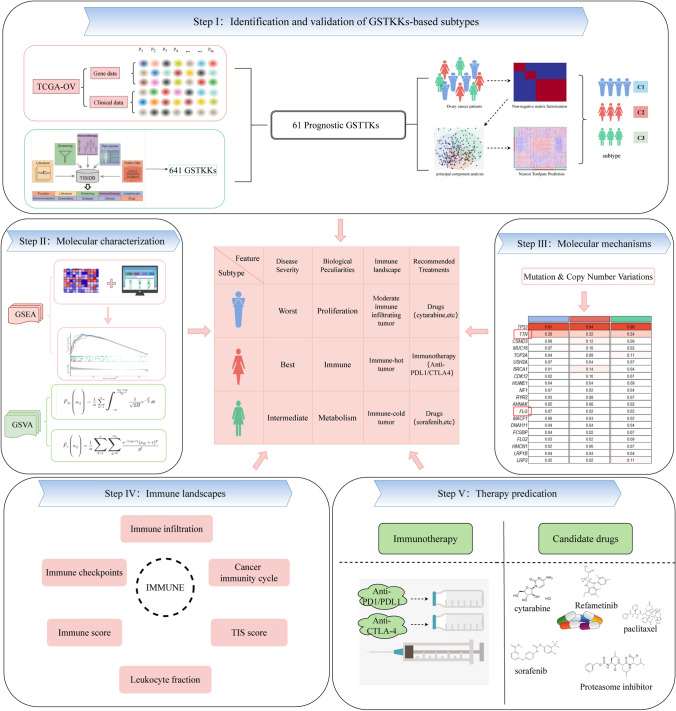
Fig. 1.
The flow chart of our research
Fig. 2.
The relationship between prognostic GSTKKs and immune landscape. A The correlation between 61 prognostic GSTKKs and immune cells, Red indicated positive correlations, and blue indicated negative correlations. Asterisks denoted p-value (**p < 0.01, *p < 0.05). Blank cells represented no statistical significance of the correlation. (B–E). Correlation of GSTKKs score with immune score (B), stromal score (C), estimate score (D), and tumor purity (E) generated by estimate algorithm
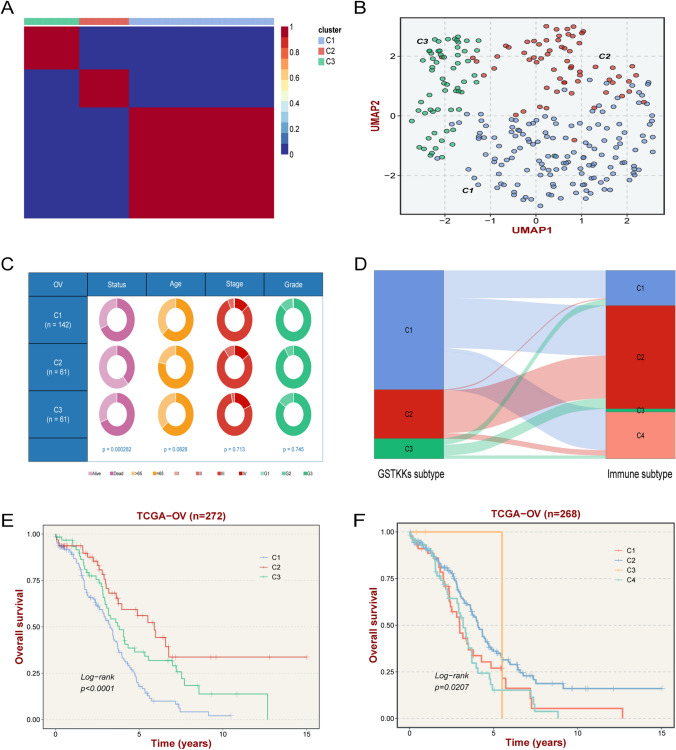
Three molecular subtypes constructed based on the expression amounts of the 61 GSTKKs by the NMF algorithm were exhibited in Fig. 3A. The UMAP and PCA analysis also confirmed that the three types were robust (Fig. 3B, Supplementary Fig. S1A). Previous studies have reported that a larger value of the contour coefficient indicates a higher matching relationship between the target and the cluster in which he or she is located, illustrating when it is close to 0 that he should be on the boundary points of the two clusters. Thus, patients with a contour coefficient > 0 were included in the study to achieve more precise patient stratification [29]. By comparison, there was a strong correlation between GSTKKs-based subtypes and published classical molecular subtypes, especially C2 was similar to Immunoreactive (Fig. 3D).
Fig. 3.
Identification of GSTKKs-based subtypes. A The consensus map after NMF clustering revealed three clusters with no overlap between clusters. B The UMAP algorithm displayed the two-dimensional principal component diagram of three subtypes, with each point representing a single sample. C The pie chart showed variations of clinical characteristics between the three subtypes by Fisher’s exact test. D The links between GSTKKs-based subtypes and immune subtypes (TCGA 2014). E Kaplan–Meier curve of OS according to GSTKKs-based subtypes in TCGA-OV cohort. F Kaplan–Meier curve of OS according to immune subtypes in TCGA-OV cohort
Consent with the previous result, the K-M analysis displayed significant prognoses differences among the three types of samples (Fig. 3E). The survival curve of classical molecular subtypes also displayed similar results (Fig. 3F). Among them, C1 got the worst prognosis, C2 possessed the most excellent prognosis, and C3 seemed to have an intermediate prognosis.
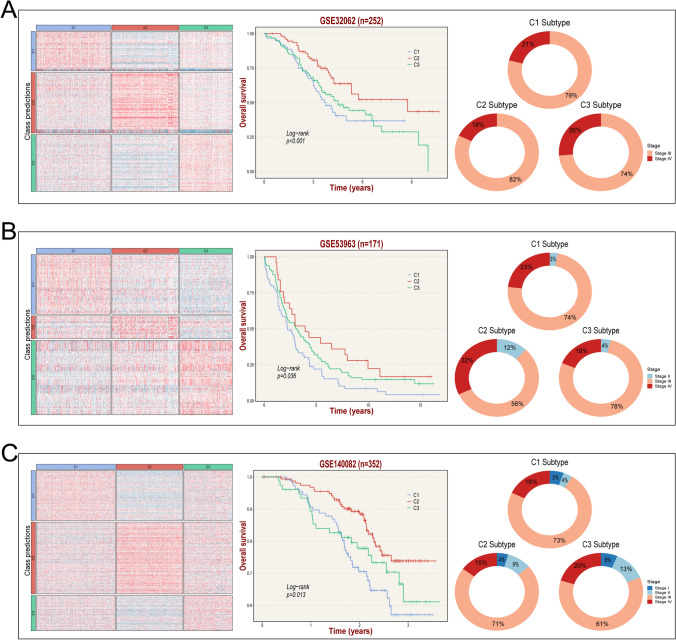
To further validate the accuracy and stability of NMF results, NTP approach was performed on three independent cohorts based on the gene signature (Supplementary Table S5), including GSE32062, GSE53963, and GSE140082 (Fig. 4A–C). Similarly, K-M analysis revealed that C1 had the worst prognosis and C2 had the best prognosis in each cohort. These results indicated that the subtypes based on GSTKKs were repeatable and practical.
Fig. 4.
Validation of GSTKKs-based subtypes in three independent cohorts by the nearest template prediction (NTP) approach. A-C From left to right: verification of the three GSTKKs-based subtypes in GSE32062, GSE53963, and GSE140082 cohorts with NTP analysis, Kaplan–Meier curves of OS according to the GSTKKs-based subtypes in GSE32062, GSE53963, and GSE140082 datasets, and proportion of different stages in the subtypes
Analysis of clinical characteristics related to subtypes prognosis
The relevance between clinical features of the TCGA cohort and the GSTKKs-based subtypes was manifested in Fig. 3C. The independent prognostic elements of TCGA-OV and three GEO cohorts were displayed in Supplementary Fig. S1C-E, G. In the TCGA cohort, C1 subtype was an independent prognostic factor; In the GSE140082 cohort, age, stage, and C2 subtype were independent prognostic factors; There were none both in GSE53963 and GSE32062 cohorts. The proportion of each stage and grade in the three subtypes were also performed (Fig. 4, Supplementary Fig. S1F, H). Consistent with previous studies, advanced patients were the majority in each subtype. To sum up, patients with favorable prognoses were mainly distributed in the C2 subtype, while poor prognoses were mainly distributed in the C1 subtype.
Biological characteristics of GSTKKs-based subtypes
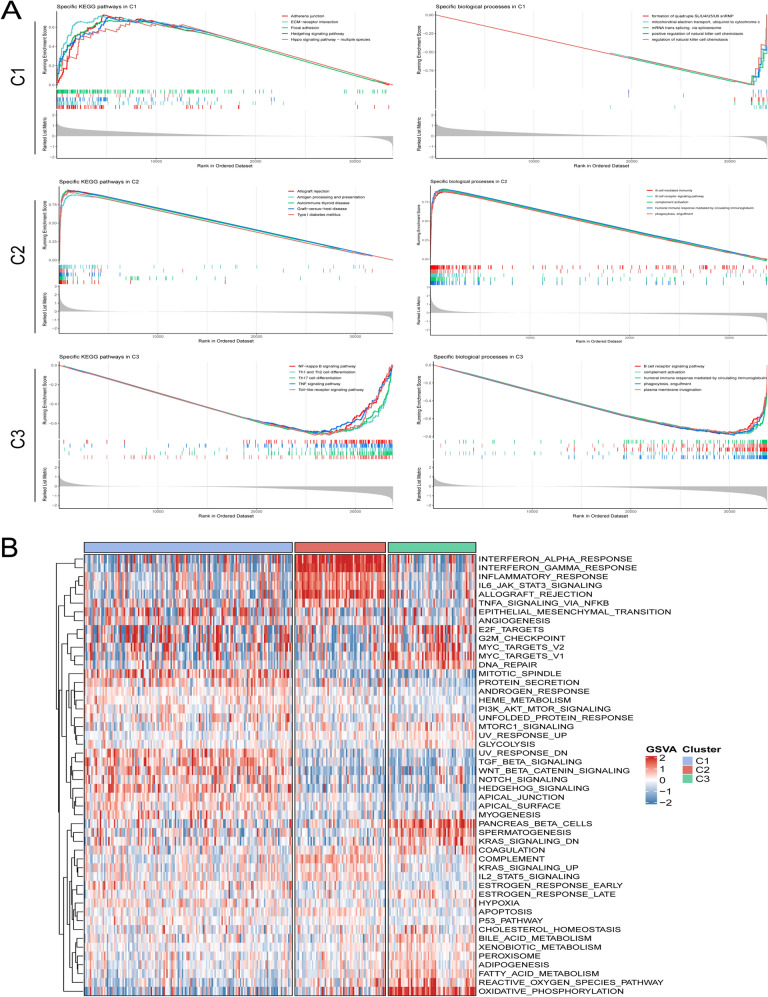
GSEA enrichment analysis was utilized to characterize metabolic pathways and specific biological processes among the three subtypes (Fig. 5A). Proliferation-associated pathways were enriched in C1, such as Hedgehog signaling pathway. Immune-related pathways were enriched in C2, such as autoimmune thyroid disease. Metabolism pathways were enriched in C3, such as Th1 and Th2 cell differentiation Thus, molecular features of C1 were defined as proliferation, C2 as immune, and C3 as immune and metabolism. Further gene set variation analysis (GSVA) also validated the above molecular features (Fig. 5B), C1 was significantly associated with proliferative activity, C2 was mainly related to immune pathways, and C3 was significantly enriched in metabolic processes.
Fig. 5.
Biological characteristics of the three subtypes. A From top to bottom, GSEA enrichment analysis respectively revealed activated marker pathways of C1, C2, and C3. The FDR of the biological function was < 0.05. B Heatmap based on the score of each subtype in 50 Hallmark gene sets. The higher the score, the higher the pathway activity
Genomic alterations analysis of GSTKKs-based subtypes
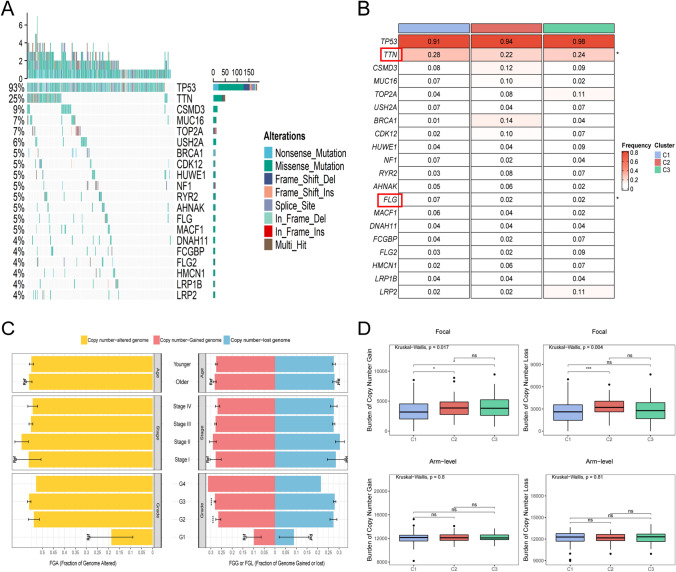
As illustrated in Fig. 6A, the landscapes of gene mutations and copy number alterations in each subtype were explored. Top 20 frequently mutated genes were exhibited in Fig. 6B. Among these genes, TTN and FLG showed differences in the three subtypes. Their mutation frequency in C1 was highest, suggesting that TTN and FLG might play a critical role in tumor development. Notably, while some mutations were not statistically different, such as LRP2 and BRCA1, which separately had the highest mutation rate in C3 and C2, these genes still deserve attention.
Fig. 6.
The mutational landscape of three subtypes. A Mutation landscape of top 20 frequently mutated genes (FMGs) in the three clusters. B The mutation frequency of top 20 FMGs among three subtypes. P values are shown as *P < 0.05. C The differences of fraction of genome altered (FGA), fraction of genome gained (FGG), and fraction of genome lost (FGL) in clinical characteristics. D The burden of copy number gain or loss in arm and focal levels. P values are shown as *P < 0.05; **P < 0.01; ***P < 0.001
The association between copy number variants and the clinical characteristics of patients was further identified (Fig. 6C). Fraction of genome altered (FGA) and fraction of genome lost (FGL) did not differ in clinical characteristics. Grade2 and Grade3 were associated with fraction of genome gained (FGG) only when graded diagnoses were made (P < 0.001). FGA was a measure of genomic instability and represented more specific mechanisms of chromosome alterations. After that, the genomic variants of the three isoforms were further analyzed (Fig. 6D). C1 had fewer deletions and amplifications at the focal level, whereas at the chromosome arm level, there was no significant difference among subtypes. Further evaluation of the mutational landscape found that there was no difference among the three subtypes of HRD, new antigen and TMB (Supplementary Fig. S2B).
Altogether, in the analysis of genomic variants, differences brought about by mutations and copy number alterations might have contributed to the diversified outcomes of the three heterogeneous molecular subtypes.
Immunocyte infiltration landscape of GSTKKs-based subtypes
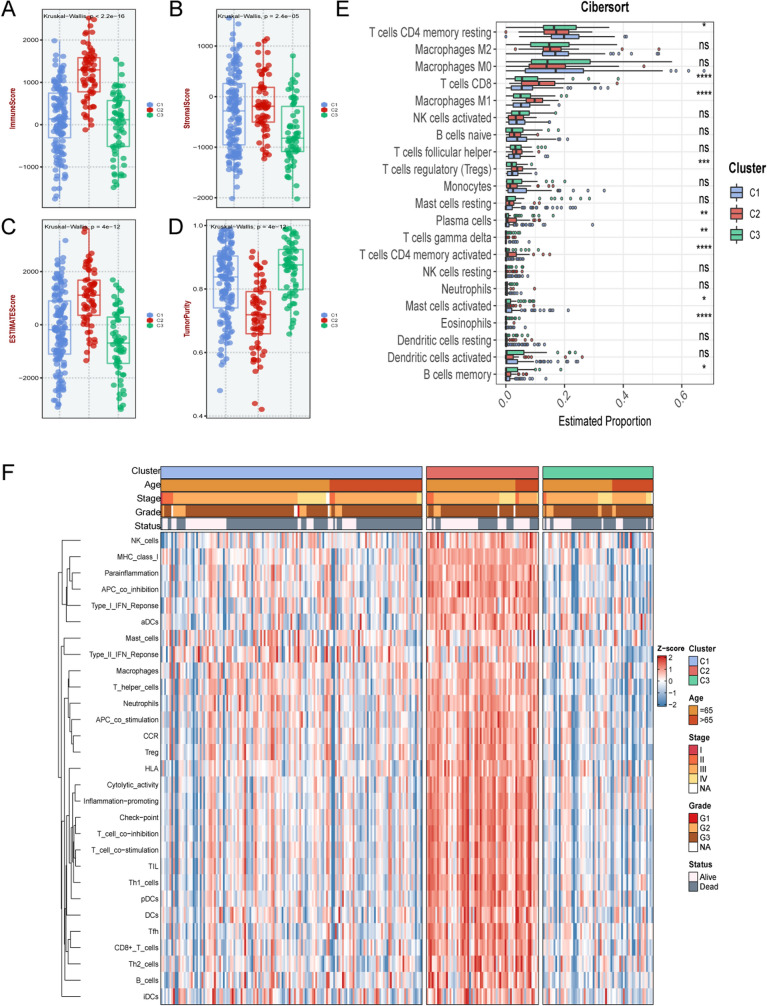
Because the three subtypes were enriched on significantly different biologically relevant pathways, further exploration of the immune landscape of each isoform was of great value in determining the best treatment. As illustrated in Fig. 7A-D, the immune score, stromal score and estimated score were the highest in C2, while the tumor purity was the lowest. This might proclaim that C2 was more prone to immunotherapy. The immune infiltrate status of the three subtypes was assessed using eight algorithms (Supplementary Fig. S2A). Respectively, C1, C2, and C3 were seemed as the moderate immune infiltrating tumor, immune-hot tumor, and immune-cold tumor.
Fig. 7.
Analysis of the proportion of immune cells in three subtypes. A–D The immune score (A), stromal score (B), estimate score (C), and tumor purity (D) in three subtypes. E Estimated proportion of immune cells among the three subtypes. P values are shown as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns was the abbreviation of no significance. F The heatmap illustrated the correlation between three subtypes and clinical characteristics, and the infiltration abundance of 28 immune cell subsets evaluated by ssGSEA algorithm
Assessing the proportion of immune cell components in each subtype using the CIBERSORT algorithm illustrated that all three subtypes were certified with a higher proportion of macrophages and T cells (Fig. 7E). Meanwhile, the proportion of immune cells was the highest in C2, such as M1 macrophages and activated CD8 memory T cells. This may be related to better immunotherapeutic efficacy. Figure 7F revealed the correlation between subtypes and immune cells, C2 displayed a significant positive correlation in most immune cells, suggesting that C2 might be associated with immune activation, harboring a more vital anti-tumor killing ability. C3 was deemed immune evasion because the infiltration was lowest in almost all immune cells and was significantly negatively correlated with most immune cells. C1 was in an intermediate state, which only significantly negatively correlated with activated B cells and natural killer T cells.
Immune checkpoints analysis of GSTKKs-based subtypes
In co-inhibited and co-stimulated ICPs, C2 was expressed at the highest levels in most cases (Supplementary Fig. S3A, B). Due to C2 had the highest expression level of HLA molecules, it was presumed that its patients had better antigen delivery ability (Supplementary Fig. S3C). In line with these results, shown in a heatmap of 27 ICP molecules expression (Supplementary Fig. S3D), C2 was expressed at the highest levels, followed by C1 and C3. Also, in both leukocyte fraction and TIS score, the C2 subtype was significantly different from the other two groups (P < 0.001), which means C2 is more responsive to immunotherapy (Supplementary Fig. S3E, F).
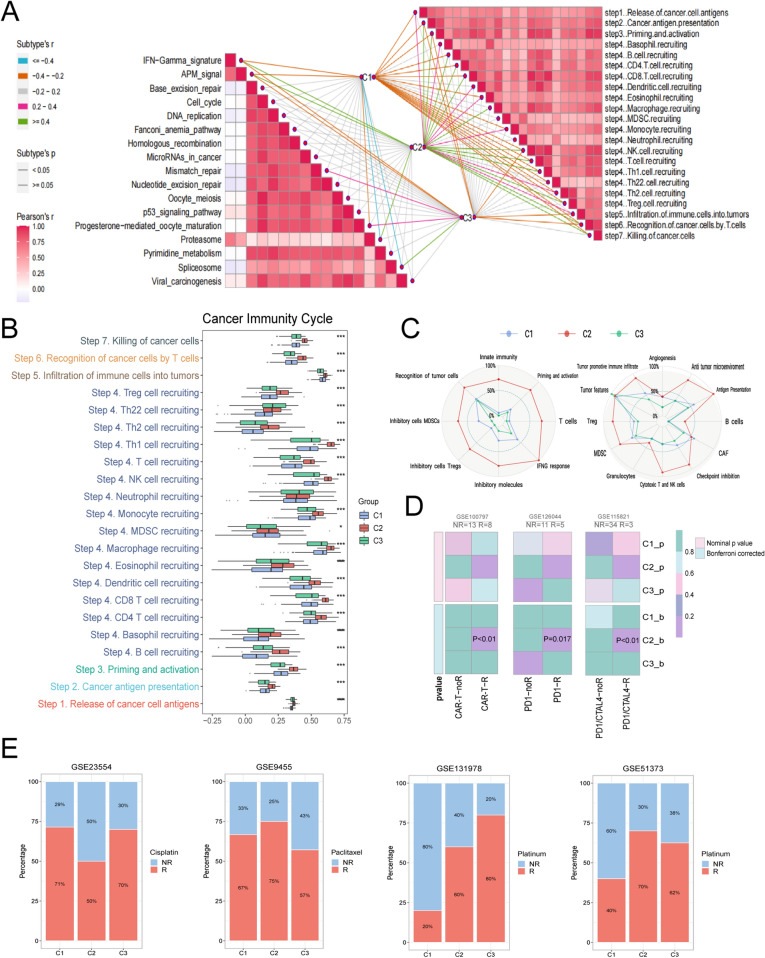
The use of immune checkpoint inhibitors (ICIs) has revolutionized the treatment paradigm for various cancers. Thus, enrichment scores for the seven anti-tumor immune cycle steps were calculated using the ssGSEA algorithm, with all steps showing higher scores (P < 0.001) in C2 but lower levels in C1 and C3 (Fig. 8A, B). The TME was most abundant in C2 according to the results of Fges enrichment, indicating that C2 would respond positively to immunotherapy. Notably, CAFs (cancer associated fibroblasts) was considerably enriched in C1 (Fig. 8C), which can secrete a range of growth factors, cytokines, extracellular matrix, etc. These factors are crucial for promoting tumorigenesis, proliferation, tumor angiogenesis, invasion, and metastasis. This might also imply a poorer prognosis in C1. Additionally, the results of Submap demonstrated that C2 subtype might achieve excellent clinical effectiveness from an immunotherapy response (Fig. 8D). Overall, the evidence presented above all indicated that C2 was an immunological subtype, and immunotherapy for C2 patients might be a more potent weapon.
Fig. 8.
Characteristics of immune circulation GSTKKs-based subtypes and clinical treatment cohorts. A The butterfly diagram illustrated the distribution of metabolic pathway and cancer immune cycle among the three subtypes. B Enrichment scores for the seven anti-tumor immune cycle steps were calculated with ssGSEA algorithm. P values are shown as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. C The radar map displayed the proportion of the immune-related characteristics and immune molecules in the three subtypes. D Submap analysis of the three subtypes, including GSE100797 cohort with detailed CAR-T therapy information, GSE126044 cohort with detailed anti-PD1 therapy information and GSE115821 with detailed anti-PD1 and anti-CTLA4 therapy information. E Analysis of the response of three subtypes to clinical drugs in the treatment cohorts, including cisplatin, paclitaxel, and platinum
Pharmacotherapy prediction for GSTKKs-based subtypes
The outcomes of the clinical medication treatment cohort were displayed in Fig. 8E. Compared with C2, C1 was more sensitive to cisplatin, and C3 patients might benefit from platinum treatment. According to the CMap analysis results, 29 drugs harbored individualized therapeutic potential for subtypes (Supplementary Fig. S2D), and the molecular pathways and genes they target were exhibited (Supplementary Fig. S2E). In addition, based on the ridge regression model of pRRophetic package, candidate drugs for different subtypes were developed by calculating half-maximal inhibitory concentration (IC50) and quantifying drug sensitivity data (Supplementary Fig. S2C, Supplementary Fig. S4A-C). Altogether, all these candidates may bring better efficacy to specific OV patients.
Discussion
Ovarian cancer remains one of the most prevalent malignancies among women, yet effective screening and management strategies are still lacking. Despite extensive research, current screening methods, such as CA125 levels and transvaginal ultrasound, have not significantly reduced mortality rates. This highlights the urgent need for novel management strategies and the integration of emerging biomarkers, such as miRNAs, into clinical practice to improve early detection and patient outcomes [30]. In recent years, immunotherapy with immune checkpoint blockade has achieved encouraging results in cancer treatment [31]. However, OV is regarded as a "cold tumor" due to the absence of cytotoxic T lymphocytes and immunosuppression [32]. Previous studies have shown that the objective remission rate (ORR) of immune checkpoint inhibitors in patients with relapsed or drug-resistant OV is between 10 and 15% [31]. Meanwhile, the latest NCCN guidelines only recommend Pembrolizumab as a grade II treatment for platinum-sensitive/platinum-resistant relapsed patients with MSI-H/dMMR. Previous research has demonstrated that gene expression profiling and microarray analysis are valuable tools for classifying and predicting the prognosis of a variety of malignancies, particularly colorectal cancer [33, 34]. Therefore, a novel classifier of immunological correlates in OV is urgently required to enable tailored therapy and enhance patient clinical outcomes. As the basis for immune system function, cytotoxic T cells play a significant role in killing tumors procession. Studying T cell-associated genes may be an effective weapon in attacking cancer. As previously described, the worst prognosis was C1, which was characterized by proliferation and differentiation. Immunity, heightened sensitivity to immunotherapy, and the best prognosis were all traits of C2. Additionally, C3 was described as metabolic and chemotherapy-sensitive.
The pathway enrichment analysis reflects the heterogeneity of the three subtypes regarding biological function. C1 patients are mainly associated with several proliferation and differentiation signaling pathways: dysregulation and abnormal deposition or loss of ECM components have been implicated in OV progression [35]. Meanwhile, recent studies suggest that the ECM-receptor interaction pathway is strongly associated with survival and prognosis in patients with high-grade plasmacytoid OV [36]. C2 patients are mainly enriched in several autoimmune disease pathways. The immune landscape of C2 also demonstrates that C2 patients are rich in immune cells, suggesting that immunotherapy is a very effective tool. A large number of metabolic pathways are significantly enriched in C3 patients, suggesting that this subtype may be more sensitive to metabolic therapy, which is effective in cancer chemotherapy. Among others, high OXPHOS was found to increase the responsiveness of high-grade plasma ovarian cancer to conventional chemotherapy [37].
In recent years, numerous mutations have been identified, and several molecular inhibitors have been developed for treating ovarian cancer. Many of these inhibitors, targeting pathways such as PI3K, PARP, and immune checkpoints, are currently being evaluated in clinical trials, showing promise for improving patient outcomes and offering new hope in the fight against this challenging disease [38]. By analyzing somatic mutation profiles, two genes with significantly different mutation frequencies were identified in the three subtypes: TTN and FLG. They were both mutated most frequently in C1. TTN is the most prominent polypeptide encoded by the human genome 18, which is expressed in many functional cell types in tumorigenesis and is highly associated with cancer development [39]. FLG mutations are associated with ichthyosis vulgaris and atopic eczema. Previous studies identified it as a biomarker associated with poor prognosis in glioma [25]. Furthermore, although not statistically different, the highest mutation rate in C2 was drawn to BRCA1, a tumor suppressor gene that acts in different pathways to suppress tumorigenesis [40]. Somatic BRCA1 mutations have been identified as an essential feature of high-grade serous ovarian cancer [41]. Meanwhile, mutations in TP53 and BRCA1 have been reported to enhance the sensitivity of cancer cells to cytotoxic or targeted therapies, but the molecular mechanisms are not yet precise [42]. Therefore, further analysis to determine the functional role of these mutations in OV is of great significance in identifying new prognostic biomarkers and developing new therapeutic targets.
Immune score was a powerful predictor of survival in OV patients. High immune scores are associated with a higher frequency of BRCA1/2 mutations in OV [41]. As mentioned earlier, C2 had the highest immune score. Studies have confirmed that higher immune scores correspond to a better prognosis, consistent with previous findings. Tumor-infiltrating lymphocytes (TILs) have improved survival and delayed disease recurrence in OV patients [43]. Further analysis of immune cell infiltration similarly supported this result. Compared with C1 and C3, CD8 + T cells were significantly increased in the C2 subtype. Killer T cells to the rescue in OV, higher CD8 + T cell infiltration can significantly improve the clinical outcome of patients [44].
The discovery and clinical implementation of ICIs have revolutionized cancer treatment. However, most patients receiving these therapies, even when combined, do not achieve clinical benefit [45]. Recent studies have explored combining PARP inhibitors with immunotherapies, such as anti-CTLA-4 and PD-1/PD-L1, based on the hypothesis that BRCA1/2-mutated and homologous recombination (HR)-deficient tumors exhibit a higher neoantigen load, which can enhance the anti-tumor immune response. Additionally, BRCA deficiency may activate a STING-dependent innate immune response by inducing type I interferon and pro-inflammatory cytokine production. Moreover, clinical models have shown that PARP inhibition can inactivate GSK3 and upregulate PD-L1 in a dose-dependent manner, leading to T-cell activation suppression and enhanced cancer cell apoptosis [46]. To seek a better therapeutic regimen, immune checkpoints, costimulatory and coinhibitory T cell receptors were further estimated in each subtype. A wide variety of immune checkpoints are abundantly expressed in C2, implying more immunotherapeutic targets and tremendous therapeutic potential [47]. For example, CTLA4 binds to ligands, producing inhibitory signals in T cells. BTLA within the TNF superfamily regulates all stages of T-cell activation, and blocking them enhances immune responses [48]. In addition, high expression levels of HLA molecules and leukocyte proportions in C2 may further enhance the response to immunotherapy [49, 50]. Therefore, in addition to the prognostic value of these checkpoints, further understanding their underlying biological mechanisms and functions may lay the foundation for new cancer immunotherapies.
All research focuses on making cancer treatment more individualized. As mentioned above, the heterogeneity of the three subtypes is impressive. ICBs have been shown to have promising therapeutic efficacy in the C2 subtype, but the status of chemotherapy should not be underestimated due to their low objective remission rate. Consequently, identifying potential therapeutic agents for patients with various subtypes is a complex issue. Representative therapeutic agents and potential mechanisms of action for various subtypes were identified from CMap and pRRophetic analyses. TW.37, QS11, Cytarabine and Doxorubicin of C1; AZD6482, Z.LLNle.CHO, CGP.082996 and Pazopanib of C2; NVP.BEZ235, RDEA119, and Bortezomib of C3. Among these drugs, Doxorubicin intercalates DNA and inhibits nucleic acid synthesis, and although cardiotoxic [51], the combined use of QS11 counteracts this side effect [52]. Pazopanib is a small molecule inhibitor of multiple protein tyrosine kinases with potential anti-tumor activity, and it is an effective drug approved by the US FDA and EU for the treatment of advanced OV [53]; RDEA119 is a solid and selective inhibitor of MEK [54]. To conclude, these potential drugs constitute a reference for the precise treatment of ovarian cancer patients. Additionally, it is important to note that the PI3K pathway is frequently upregulated in epithelial ovarian cancer, playing a significant role in chemoresistance and maintaining genomic stability. Inhibiting PI3K may induce genomic instability and mitotic catastrophe by reducing the activity of the spindle assembly checkpoint protein Aurora kinase B, leading to an increased occurrence of lagging chromosomes during prometaphase[55]
However, it must be recognized that our study still contains some limitations. First, this study was retrospective and additional prospective studies are necessary for a comprehensive analysis. Secondly, intra-tumoral heterogeneity was not included in this study. Further investigation at the single-cell level is critical. Finally, further studies are needed to investigate the impact of combination regimens of ICIs on the patient's prognosis in order to provide more individualized immunotherapy.
Conclusions
Our study explored three stable GSTKKs-based subtypes with various clinical prognoses, functional phenotypes, genome alterations and immune landscape. Furthermore, we propose possible individualized treatment strategies for the various subtypes. In conclusion, this study could be helpful for predicting the prognosis of OV patients and uncovering new therapeutic approaches.
Supplementary Information
Acknowledgements
Not Applicable.
Author contributions
Cungeng Fan, Ruiying Liao, Long Liu, and Jinhai Deng contributed study design and paper revisiting. Cungeng Fan, Ruiying Liao, and Jinhai Deng contributed project oversight. Beining Yin, Xinglin Wen, and Li Lin contributed data analysis, visualization, and paper writing. Beining Yin, Xinglin Wen, Li Lin, Long Liu, Siyuan Weng, Hui Xu, and Yuyuan Zhang contributed paper revisiting. All authors approved this manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
All data in our study are available upon request. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. The TCGA-OV (n = 354) RNA-seq data was acquired from the UCSC Xena platform (https://xenabrowser.net/datapages/). Other microarray datasets, including GSE32062 (n = 270), GSE53963 (n = 174), and GSE140082 (n = 380) were obtained from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/). Collecting genes that have a favorable response to TTK from the TISIDB database (http://cis.hku.hk/TISIDB/), which were further used to establish a gene set defined as GSTTKs. The raw count expression was processed and converted to transcripts per kilobase million (TPM) for subsequent analysis. The mutation data was downloaded from the GDC website (https://portal.gdc.cancer.gov/). Copy number variation (CNV) data of GISTIC_2.0 level was collected from FireBrowse (http://firebrowse.org/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinglin Wen, Beining Yin and Li Lin have contributed equally to this work and share first authorship.
Contributor Information
Ruiying Liao, Email: 351921628@qq.com.
Cungeng Fan, Email: 13970771415@163.com.
References
- 1.Saani I, Raj N, Sood R, Ansari S, Mandviwala HA, Sanchez E, et al. Clinical challenges in the management of malignant ovarian germ cell tumours. Int J Environ Res Public Health. 2023. 10.3390/ijerph20126089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A, et al. Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19(2):191–226. 10.6004/jnccn.2021.0007. [DOI] [PubMed] [Google Scholar]
- 4.Ghose A, McCann L, Makker S, Mukherjee U, Gullapalli SVN, Erekkath J, et al. Diagnostic biomarkers in ovarian cancer: advances beyond CA125 and HE4. Ther Adv Med Oncol. 2024;16:17588359241233224. 10.1177/17588359241233225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millstein J, Budden T, Goode EL, Anglesio MS, Talhouk A, Intermaggio MP, et al. Prognostic gene expression signature for high-grade serous ovarian cancer. Ann Oncol. 2020;31(9):1240–50. 10.1016/j.annonc.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghose A, Bolina A, Mahajan I, Raza SA, Clarke M, Pal A, et al. Hereditary ovarian cancer: towards a cost-effective prevention strategy. Int J Environ Res Public Health. 2022. 10.3390/ijerph191912057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlidis N, Rassy E, Vermorken JB, Assi T, Kattan J, Boussios S, et al. The outcome of patients with serous papillary peritoneal cancer, fallopian tube cancer, and epithelial ovarian cancer by treatment eras: 27 years data from the SEER registry. Cancer Epidemiol. 2021;75: 102045. 10.1016/j.canep.2021.102045. [DOI] [PubMed] [Google Scholar]
- 8.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280–304. 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 9.Shah S, Cheung A, Kutka M, Sheriff M, Boussios S. Epithelial ovarian cancer: providing evidence of predisposition genes. Int J Environ Res Public Health. 2022. 10.3390/ijerph19138113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371: m3773. 10.1136/bmj.m3773. [DOI] [PubMed] [Google Scholar]
- 11.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–45. 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baci D, Bosi A, Gallazzi M, Rizzi M, Noonan DM, Poggi A, et al. The ovarian cancer tumor immune microenvironment (TIME) as target for therapy: a focus on innate immunity cells as therapeutic effectors. Int J Mol Sci. 2020. 10.3390/ijms21093125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, et al. Olaparib plus Bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381(25):2416–28. 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 14.Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398(10304):1002–14. 10.1016/S0140-6736(21)01206-X. [DOI] [PubMed] [Google Scholar]
- 15.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol. 2019;234(6):8509–21. 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 16.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–5. 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed]
- 17.Drapkin BJ, Farago AF. Unexpected synergy reveals new therapeutic strategy in SCLC. Trends Pharmacol Sci. 2019;40(5):295–7. 10.1016/j.tips.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Mathewson ND, Ashenberg O, Tirosh I, Gritsch S, Perez EM, Marx S, et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell. 2021;184(5):1281-98 e26. 10.1016/j.cell.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Du J, Liu C, Zeng H, Chen Y, Liu L, et al. A pan-cancer analysis of CD161, a potential new immune checkpoint. Front Immunol. 2021;12: 688215. 10.3389/fimmu.2021.688215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2. 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 21.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinf. 2010;11:367. 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Weng S, Dang Q, Xu H, Ren Y, Guo C, et al. Gene interaction perturbation network deciphers a high-resolution taxonomy in colorectal cancer. Elife. 2022. 10.7554/eLife.81114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Xu H, Weng S, Ren Y, Han X. Stemness refines the classification of colorectal cancer with stratified prognosis, multi-omics landscape, potential mechanisms, and treatment options. Front Immunol. 2022;13: 828330. 10.3389/fimmu.2022.828330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi Q, Pu Y, Chao F, Bian P, Lv L. ACAP1 deficiency predicts inferior immunotherapy response in solid tumors. Cancers. 2022. 10.3390/cancers14235951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Lu T, Wang L, Liu L, Li L, Han X. Comprehensive molecular analyses of a novel mutational signature classification system with regard to prognosis, genomic alterations, and immune landscape in glioma. Front Mol Biosci. 2021;8: 682084. 10.3389/fmolb.2021.682084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The Immune Landscape of Cancer. Immunity. 2018;48(4):812-30 e14. 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2(11): e1195. 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9(9): e107468. 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Liu Z, Gao J, Liu X, Weng S, Guo C, et al. CD8+ T cell trajectory subtypes decode tumor heterogeneity and provide treatment recommendations for hepatocellular carcinoma. Front Immunol. 2022;13: 964190. 10.3389/fimmu.2022.964190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrari F, Giannini A. Approaches to prevention of gynecological malignancies. BMC Womens Health. 2024;24(1):254. 10.1186/s12905-024-03100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49. 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Zhao T, Chen Q, Li Y, Xiao Z, Xiang Y, et al. Nanomedicine strategies for heating “Cold” ovarian cancer (OC): next evolution in immunotherapy of OC. Adv Sci (Weinh). 2022;9(28): e2202797. 10.1002/advs.202202797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrera M, Berral-Gonzalez A, Lopez-Cade I, Galindo-Pumarino C, Bueno-Fortes S, Martin-Merino M, et al. Cancer-associated fibroblast-derived gene signatures determine prognosis in colon cancer patients. Mol Cancer. 2021;20(1):73. 10.1186/s12943-021-01367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35(2):161–8. 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdul Pari AA, Singhal M, Augustin HG. Emerging paradigms in metastasis research. J Exp Med. 2021. 10.1084/jem.20190218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luengo A, Gui DY, Vander Heiden MG. Targeting metabolism for cancer therapy. Cell Chem Biol. 2017;24(9):1161–80. 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentric G, Kieffer Y, Mieulet V, Goundiam O, Bonneau C, Nemati F, et al. PML-regulated mitochondrial metabolism enhances chemosensitivity in human ovarian cancers. Cell Metab. 2019;29(1):156–7310. 10.1016/j.cmet.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrone C, Angioli R, Luvero D, Giannini A, Di Donato V, Cuccu I, et al. Targeting BRAF pathway in low-grade serous ovarian cancer. J Gynecol Oncol. 2024;35(4): e104. 10.3802/jgo.2024.35.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Duanmu J, Fu X, Li T, Jiang Q. Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment. J Transl Med. 2020;18(1):324. 10.1186/s12967-020-02491-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Luo M, Yang H, Zhu S, Cheng X, Qing C. Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in Chinese primary epithelial ovarian cancer patients. J Ovarian Res. 2019;12(1):19. 10.1186/s13048-019-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao D, Liu J, Chen M, Li J, Wang L, Li X, et al. Immunogenomic analyses of advanced serous ovarian cancer reveal immune score is a strong prognostic factor and an indicator of chemosensitivity. Clin Cancer Res. 2018;24(15):3560–71. 10.1158/1078-0432.CCR-17-3862. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–13. 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 43.Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. 2017;28(8):viii1–7. 10.1093/annonc/mdx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muthuswamy R, McGray AR, Battaglia S, He W, Miliotto A, Eppolito C, et al. CXCR6 by increasing retention of memory CD8(+) T cells in the ovarian tumor microenvironment promotes immunosurveillance and control of ovarian cancer. J Immunother Cancer. 2021. 10.1136/jitc-2021-003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B, Zhao H, Zhao J. Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther Adv Med Oncol. 2020;12:1758835920937612. 10.1177/1758835920937612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boussios S, Karathanasi A, Cooke D, Neille C, Sadauskaite A, Moschetta M, et al. PARP inhibitors in ovarian cancer: the route to “Ithaca.” Diagnostics. 2019. 10.3390/diagnostics9020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Zhang J, Tu H, Liang D, Chang DW, Ye Y, et al. Soluble immune checkpoint-related proteins as predictors of tumor recurrence, survival, and T cell phenotypes in clear cell renal cell carcinoma patients. J Immunother Cancer. 2019;7(1):334. 10.1186/s40425-019-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotsias F, Cebrian I, Alloatti A. Antigen processing and presentation. Int Rev Cell Mol Biol. 2019;348:69–121. 10.1016/bs.ircmb.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 50.Crayne CB, Albeituni S, Nichols KE, Cron RQ. The immunology of macrophage activation syndrome. Front Immunol. 2019;10:119. 10.3389/fimmu.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173(3):649-64 e20. 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Z, Wang W, Zhao L, Wang X, Gimple RC, Xu L, et al. Plasma cells shape the mesenchymal identity of ovarian cancers through transfer of exosome-derived microRNAs. Sci Adv. 2021. 10.1126/sciadv.abb0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peter S, Alven S, Maseko RB, Aderibigbe BA. Doxorubicin-based hybrid compounds as potential anticancer agents: a review. Molecules. 2022. 10.3390/molecules27144478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim HY, Merle P, Weiss KH, Yau T, Ross P, Mazzaferro V, et al. Phase II studies with refametinib or refametinib plus sorafenib in patients with RAS-mutated hepatocellular carcinoma. Clin Cancer Res. 2018;24(19):4650–61. 10.1158/1078-0432.CCR-17-3588. [DOI] [PubMed] [Google Scholar]
- 55.Aliyuda F, Moschetta M, Ghose A, Sofia Rallis K, Sheriff M, Sanchez E, et al. Advances in ovarian cancer treatment beyond PARP inhibitors. Curr Cancer Drug Targets. 2023;23(6):433–46. 10.2174/1568009623666230209121732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in our study are available upon request. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. The TCGA-OV (n = 354) RNA-seq data was acquired from the UCSC Xena platform (https://xenabrowser.net/datapages/). Other microarray datasets, including GSE32062 (n = 270), GSE53963 (n = 174), and GSE140082 (n = 380) were obtained from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/). Collecting genes that have a favorable response to TTK from the TISIDB database (http://cis.hku.hk/TISIDB/), which were further used to establish a gene set defined as GSTTKs. The raw count expression was processed and converted to transcripts per kilobase million (TPM) for subsequent analysis. The mutation data was downloaded from the GDC website (https://portal.gdc.cancer.gov/). Copy number variation (CNV) data of GISTIC_2.0 level was collected from FireBrowse (http://firebrowse.org/).