Abstract
The enzyme complex KEOPS (Kinase, Endopeptidase and Other Proteins of Small size) installs the universally conserved and essential N6-threonylcarbamoyl adenosine modification (t6A) on ANN-decoding tRNAs in eukaryotes and in archaea. KEOPS consists of Cgi121, Kae1, Pcc1, Gon7 and the atypical kinase/ATPase Bud32. Except Gon7, all KEOPS subunits are needed for tRNA modification, and in humans, mutations in all five genes underlie the lethal genetic disease Galloway Mowat Syndrome (GAMOS). Kae1 catalyzes the modification of tRNA, but the specific contributions of Bud32 and the other subunits are less clear. Here we solved cryogenic electron microscopy structures of KEOPS with and without a tRNA substrate. We uncover distinct flexibility of KEOPS-bound tRNA revealing a conformational change that may enable its modification by Kae1. We further identified a contact between a flipped-out base of the tRNA and an arginine residue in C-terminal tail of Bud32 that correlates with the conformational change in the tRNA. We also uncover contact surfaces within the KEOPS-tRNA holo-enzyme substrate complex that are required for Bud32 ATPase regulation and t6A modification activity. Our findings uncover inner workings of KEOPS including a basis for substrate specificity and why Kae1 depends on all other subunits.
Subject terms: Cryoelectron microscopy, RNA, Kinases, tRNAs, RNA modification
tRNA modifications are vital for their function in protein synthesis, one of the most central processes in all living cells. Here the authors show how KEOPS, a multi-subunit tRNA modifying complex, engages and acts on a substrate tRNA.
Introduction
Post-transcriptional modifications play a major role in controlling tRNA structure and function, and aberrant tRNA modifications underlie a variety of human diseases ranging from developmental disorders to cancer1–4. In particular, modifications in the anticodon loop at positions 34 and 37 are crucial for cellular fitness by modulating translation fidelity3,5. Since each tRNA carries a specific set of modifications amongst over 100 distinct modifications reported to date6, a major question in understanding tRNA biology is uncovering how substrate specificity and catalysis are achieved by their modifying enzymes.
N6-threonylcarbamoyl adenosine (t6A) is a tRNA modification universally present in all domains of life that occurs only at position A37 of tRNAs that read codons that start with an A (ANN-decoding tRNAs, where N is any nucleotide)6. t6A serves to stabilize the structure of the anticodon loop and to increase the binding energy between tRNAs and their cognate codons in mRNA7–11 thereby enhancing translation fidelity and preventing translation initiation at spurious start sites12–14. This modification is essential for cell growth and multicellular organism development12,14–16.
The final step of t6A biosynthesis is catalyzed by the highly conserved Kae1/TsaD/Qri7 enzyme family13,17,18, which is encoded by one of a small group of approximately ~60 essential gene families found in practically all living organisms19 and even in some viral genomes20. As a precursor for t6A modification, Kae1-family enzymes use a threonyl-carbamoyl adenylate (TC-AMP) molecule, which is made by the similarly universally conserved TsaC/Sua5 family enzymes (see Fig. 1A for reaction scheme)21. Curiously, Kae1/TsaD/Qri7 enzymes function in distinct protein complexes22,23. In archaea and in the cytoplasm of eukaryotic cells, Kae1 resides within the KEOPS (Kinase, Endopeptidase and Other Proteins of Small size) complex together with the atypical primordial kinase Bud32 and the non-catalytic subunits Cgi121, Pcc1, and Gon7. All five subunits are needed for efficient tRNA modification in vivo18. In bacteria, TsaD functions with the two auxiliary proteins TsaB and TsaE, the first required for tRNA binding and modification per se and the latter required for multiple rounds of catalysis24–30. In the mitochondria, Qri7 functions without any auxiliary subunits31,32 suggesting that the binding partners of Kae1 and TsaD serve supporting roles such as regulation.
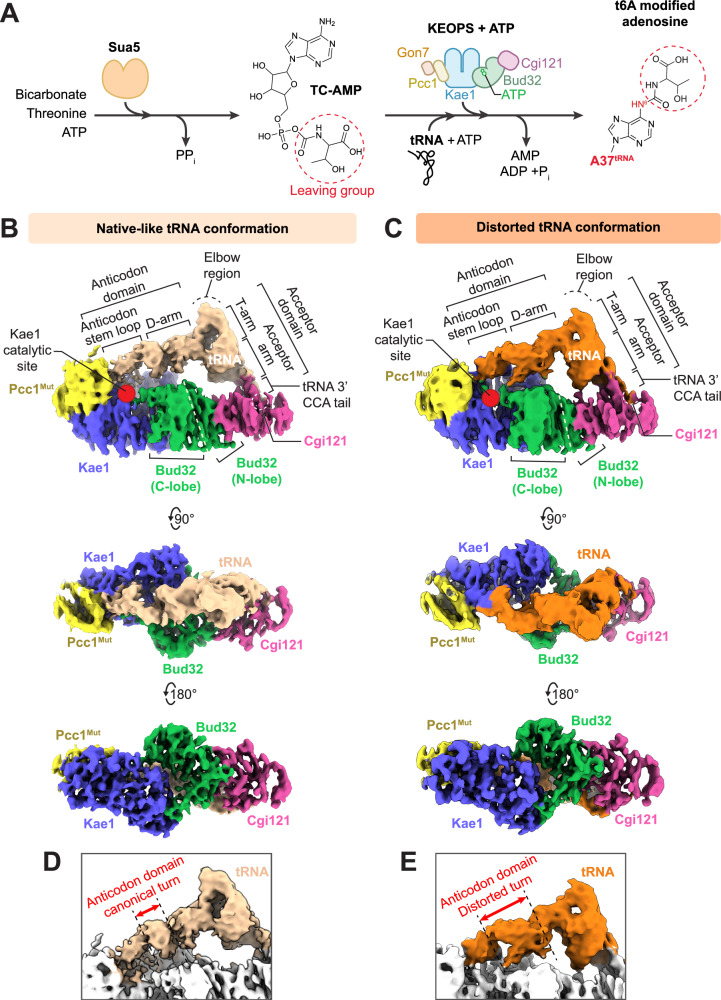
Fig. 1. Substrate tRNA adopts two distinct conformations when bound to KEOPS.
A Schematic of the t6A tRNA modification pathway in archaea and eukaryotes. TsaC/Sua5 enzymes utilize bicarbonate, threonine and ATP to generate threonylcarbamoyl adenylate (TC-AMP). TC-AMP is utilized by the KEOPS subunit Kae1 to t6A modify N6 of A37 with the threonylcarbamoyl group of TC-AMP thereby generating AMP. Kae1 functions with other KEOPS complex members Cgi121, the kinase ATPase Bud32, Pcc1 and Gon7. t6A modification also requires ATP hydrolysis to ADP + Pi by Bud32. B, C Cryo-EM density maps of KEOPS bound to tRNA in a native-like conformation at 3.56 Å resolution (B) and bound to tRNA in a distorted conformation at 3.59 Å resolution (C). D, E Zoom-in side views of the tRNA in its native-like (D) and distorted tRNA conformations (E) highlighting the conformational differences in the anticodon domain when bound to KEOPS.
Genetic studies in humans have identified pathogenic point mutations in the t6A biosynthesis pathway components, including in all 5 KEOPS subunit genes, emphasizing their crucial role in human disease16,33,34. These mutations are associated with renal disfunction, microcephaly, and central nervous system anomalies characteristic of the Galloway-Mowat Syndrome (GAMOS), a severe autosomal recessive disease that leads to early childhood death16,33–35. Further supporting an essential role for KEOPS, knock out of KEOPS genes leads to severe growth and development phenotypes in yeast, fruit flies, zebra fish and mice16,36–41.
The inner workings of KEOPS remain mostly mysterious. Cgi121 recruits tRNA substrates to KEOPS by binding to their 3’ CCA tail42 while the precise role of Bud32, Pcc1 and Gon7 in t6A biosynthesis are mostly not understood. Bud32 is an ancient member of the protein kinase superfamily that is conserved in eukaryotes and archaea43. Unlike most members of the eukaryotic protein kinase superfamily, Bud32 functions as an ATPase rather than a protein kinase and can therefore be classified as a pseudo kinase. Bud32 enzyme activity is strongly potentiated by tRNA binding to KEOPS and its enzyme activity is needed for tRNA modification42,44,45. Interestingly, Bud32 ATPase activation by tRNA depends on the presence of Kae1, hinting at an interplay between the two enzymes’ activities42. Pcc1 is required for tRNA modification in vitro and in cells for unknown reasons18,42 and Gon7, its direct binding partner, is required for cell viability potentially by regulating KEOPS protein levels in vivo33. In the absence of Gon7, Pcc1 proteins form homodimers, which can lead to KEOPS dimerization46,47.
Here we report cryogenic electron microscopy (cryo-EM) structures of an archaeal KEOPS complex without and with tRNA substrate in two distinct conformations. We reveal a set of dynamic interactions between the tRNA and Bud32, Kae1 and Pcc1 that are essential for the activation of Bud32 ATPase activity and for the subsequent modification of tRNA by Kae1. In particular, we show that interactions of Kae1 with the tRNA and with Bud32 are needed to activate Bud32 ATPase activity. We further show that an interaction between Bud32 with the flipped-out base in the substrate tRNA correlates with a conformational rearrangement of the tRNA anticodon domain that can expose A37 for modification by Kae1. We thus provide a first glimpse into the mechanism of action of a t6A modifying enzyme complex and provide a rationale for the dependency of Kae1 on its auxiliary subunits.
Results
Assembling a KEOPS tRNA complex
To understand the inner workings that mediate tRNA modification by KEOPS, we performed single-particle cryo-EM using archaeal KEOPS without Pcc2/Gon7, which is dispensable for tRNA modification in vitro45,46, mixed with substrate tRNALysUUU (denoted tRNA for simplicity; see Supplementary Fig. 1a for tRNA schematic). Preliminary analysis yielded 2D classes representing a mixture of dimeric and monomeric forms of KEOPS with no apparent binding of tRNA (Supplementary Fig. 1b). Wild-type (WT) Pcc1 can itself form a homodimer that dimerizes KEOPS47,48. Therefore, to address monomer-dimer heterogeneity we used a previously described Pcc1 mutant (denoted Pcc1Mut, Supplementary Fig. 1c and methods) that cannot dimerize KEOPS but still supports tRNA modification activity47,48. To promote tRNA binding to KEOPS, we employed a glutamate to arginine substitution at position 152 (E152R) in Bud32 (Supplementary Fig. 1c) that enhances tRNA-binding affinity but impairs tRNA modification function42.
SEC-MALS analysis confirmed that KEOPS containing Pcc1Mut and Bud32E152R was exclusively monomeric, while the WT KEOPS complex displayed a monomer-dimer equilibrium (Supplementary Fig. 1d). KEOPS reconstituted with Pcc1Mut and Bud32E152R displayed ~4-fold enhanced tRNA binding compared to KEOPS WT in a competitive displacement assay (Supplementary Fig. 1e) with only a partial loss of Bud32 ATPase activation in response to tRNA (Supplementary Fig. 1f) and as expected42 a full loss of t6A-modifying activity (Supplementary Fig. 1g). Since a structure of WT KEOPS in complex with tRNA was not attainable, we used KEOPS with Pcc1Mut and Bud32E152R to visualize the tRNA binding mechanism.
Cryo-EM analysis of a KEOPS-tRNA substrate complex reveals two distinct conformations of tRNA
Cryo-EM analysis of KEOPS incorporating Pcc1Mut and Bud32E152R along with tRNA, yielded 2D classes displaying the expected linear architecture of KEOPS49 with tRNA binding parallel to the complex (Supplementary Fig. 2a). Ab initio 3D modeling followed by 3D classification allowed separation of KEOPS without tRNA from KEOPS-tRNA structures. Non-uniform refinement allowed us to generate the structure of the four-subunit archaeal KEOPS complex without tRNA at 2.9 Å resolution (see Supplementary Fig. 2a for workflow, Supplementary Fig. 2b for FSC plot, and Table 1 for statistical validation). This structure corroborated previous models of the linear binding mode of the Cgi121-Bud32-Kae1-Pcc1 subunits47,49,50. 3D variability analysis revealed a high degree of structural flexibility in the tRNA bound to KEOPS (see Supplementary Fig. 2a for workflow and Supplementary Movie 1 highlighting the tRNA flexibility). Structure variation clustering allowed generation of two cryo-EM density maps depicting KEOPS-tRNA complexes at 3.56 Å and 3.59 Å resolution with the tRNA in two distinct conformations (Fig. 1B, C, see Supplementary Fig. 2c, d for FSC plots, Supplementary Fig. 2e for representative cryo-EM density map features, Supplementary Fig. 3 for map anisotropy analysis indicating that the maps were generated by multiple views of the KEOPS-tRNA complex, Table 1 for statistical validations), with overall higher resolution for the KEOPS subunits than for the tRNA (Supplementary Fig. 2f). Due to its apparent flexible mode of binding, density for much of the tRNA in both conformations was limited to ~5–6 Å (Supplementary Fig. 2f), with improved resolution at interfaces with the core protein complex (see below).
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Apo KEOPS | KEOPS + tRNA (native-like conformation) | KEOPS + tRNA (distorted conformation) | |
|---|---|---|---|
| PDB ID | 8UNK | 8UP5 | 9D85 |
| EMDB ID | EMD-43407 | EMD-42443 | EMD-46630 |
| Data Collection and Processing | |||
| Magnification | 105 000 | 105 000 | 105 000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e-/ Å2) | 55.61 | 55.61 | 55.61 |
| Defocus range- tilted (μm) | −3.4 to −6.1 | −3.4 to −6.1 | −3.4 to −6.1 |
| Defocus range- non tilted (μm) | −0.4 to −2.9 | −0.4 to −2.9 | −0.4 to −2.9 |
| Pixel size (Å) | 0.825 | 1.03 | 1.03 |
| Symmetry imposed | C1 | C1 | C1 |
| Final particle images (no.) | 386 585 | 121 003 | 83,710 |
| Map resolution (Å) | 2.91 | 3.56 | 3.59 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Refinement | |||
| Initial model used (PDB codes) | 3ENH/3ENO | 3ENH/3ENO/7KJT | 3ENH/3ENO/7KJT |
| Model resolution (Å) | 3.1 | 3.5 | 3.5 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map sharpening B factor (Å2) | 133.5 | 147.1 | 150.18 |
| Model composition | |||
| Non-hydrogen atoms | 9 865 | 6810 | 7201 |
| Protein residues | 691 | 712 | 739 |
| Nucleotide | 0 | 73 | 65 |
| Ligands | 0 | 0 | 0 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.003 | 0.004 | 0.005 |
| Bond angles (°) | 0.55 | 0.97 | 1.344 |
| Validation | |||
| MolProbity score | 2.19 | 2.3 | 2.35 |
| Clashscore | 22.66 | 24.48 | 27.16 |
| Poor rotamers (%) | 0 | 0.6 | 0.33 |
| Ramachandran plot | |||
| Favored (%) | 95.1 | 93.66 | 93.43 |
| Allowed (%) | 4.9 | 6.34 | 6.43 |
| Disallowed (%) | 0 | 0 | 0.14 |
Atomic models were built into the tRNA containing maps, using known crystal structures of the individual subunits47, the structure of tRNA in isolation42, and the cryo-EM structure of KEOPS in the absence of tRNA (solved here). In both maps, we observed clear L-shaped density with helical ridges and grooves consistent with the backbone structure of the tRNA acceptor domain (composed of the acceptor arm, T-arm, and CCA tail) and anticodon domain (composed of the D-arm, and anticodon stem loop) connected by the elbow region (Fig. 1B C, see also Supplementary Fig. 1a for tRNA schematic). Although we could not unambiguously model the position of all tRNA bases and some protein side chains due to low map resolvability limited by the resolution (see Supplementary Table 1 for the Q scores51 of key residues in this study), these could be inferred in part from the published crystal structures of KEOPS proteins and of tRNALysUUU in isolation.
Refined atomic models of the two KEOPS-tRNA complexes displayed similar tRNA-binding mechanisms (Fig. 1B, C). In both, the 3’ CCA tail (bases 74–76) of tRNA engages a shallow groove on Cgi121, as expected based on the crystal structure of the Cgi121-tRNA complex42. The tRNA acceptor arm and T-arm lie adjacent to the N-lobe of Bud32 and the D-arm lies adjacent to the C-lobe of Bud32. The anticodon stem-loop orients towards the active site of Kae1 and towards Pcc1. Thus, all four KEOPS subunits visualized appear to cradle the tRNA.
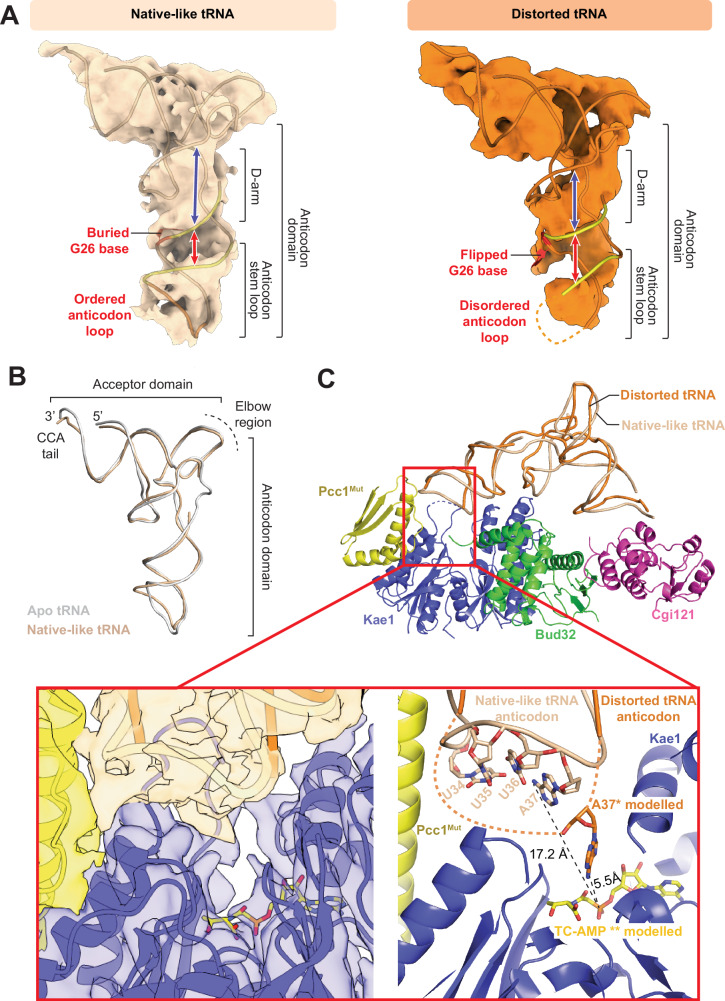
tRNA binding did not cause large-scale structural changes to any of the KEOPS subunits. In contrast, the tRNA exhibited a wide range of structural differences, especially in the anticodon domain (Fig. 1D, E and Supplementary Movie 1). Real space refinement enabled building of models for the two most populated tRNA conformations. In the first conformation, the tRNA exhibits a native-like fold, similar to its crystal structure (PDB 7KJU) (Fig. 2A left side and 2B, RMSD 1.36 Å relative to the native fold). Correspondence between the observed experimental density and the known crystal structure of the tRNA gave higher confidence in the model’s correctness. In the second conformation, the anticodon domain of the tRNA exhibits an unexpected, distorted conformation (Fig. 2A right side, RMSD 4.02 Å relative to the native fold) (Fig. 2A right side, and Fig. 1D, E for a comparison between native like and distorted conformations). Due to limited map resolution and the absence of a pre-existing atomic structure, we could not model the distorted tRNA structure with full confidence and thus interpretations based on the structure were made with caution. Notably, the tRNA in the two conformations makes a series of contacts with Bud32, Kae1 and Pcc1 not visualized previously. The distinguishing features of the two tRNA structures, the interactions of tRNA with KEOPS, and their functional significance are explored below.
Fig. 2. Comparison of native-like and distorted tRNA conformations.
A Cryo-EM density volumes and fitted atomic models in cartoon representation of the tRNA in the native-like conformation (left) and in the distorted conformation (right). Red and blue arrows highlight differences in the D-arm and anticodon stem loop in both structures. Also highlighted are the inferred flipped base of G26 and the disordered anticodon loop of tRNA in the distorted conformation. B Structural similarity of the apo tRNALys crystal structure (PDB ID: 7KJU) with the KEOPS-bound tRNA structure in its native-like conformation. C Disorder of the tRNA anticodon loop can make A37 more accessible to the active site of Kae1. (top) Cartoon representation of KEOPS bound to native-like tRNA (beige) with the distorted tRNA (orange) superimposed. (bottom inset) Detailed view of the active site of Kae1 with cryoEM density shown on left and atomic details shown on right. The dashed line represents the unstructured tRNA anticodon loop region in the distorted conformation. The TC-AMP mimetic BK951 was docked into the active site of Kae1 by superimposing the co-crystal of TC-AMP bound to the Kae1 paralog TsaD (PDB 6Z81, RMSD 0.345 Å). A37 was modeled in the active site of Kae1 by manually adding all missing anticodon nucleotides (U33 to C39, inclusive) using Pymol’s nucleotide builder tool and locally refining the loop using the Coot real space refinement tool, allowing its theoretical positioning within ~5.5 Å of the predicted site of attack in TC-AMP.
Distinguishing features of the two tRNA conformers
Comparison of the two tRNA-bound states of KEOPS revealed major differences in the tRNA anticodon loop and D-arm, oriented towards the Kae1 active site and Bud32 C-lobe respectively. We first focused on structural differences in the anticodon loop region. KEOPS with tRNA in its native-like conformation revealed that although the anticodon loop is directed towards the active site of Kae1 (Fig. 2C) A37 is not suitably positioned for t6A modification (Fig. 2C inset). In the t6A modification reaction, N6 of A37 is modified by TC-AMP positioned in the active site of Kae1 (Fig. 1A)52. Using the published structure of the Kae1 family enzyme TsaD bound to a TC-AMP mimetic molecule (PDB 6Z81)53, we modeled the likely position of TC-AMP in the active site of Kae1 in both KEOPS-tRNA structures. Based on this modeling, in the native-like tRNA conformer, N6 of A37 lies ~17.2 Å away from its site of attack on TC-AMP, thereby ruling out t6A modification (Fig. 2C inset, predicted site of attack in TC-AMP inferred by the catalytic model proposed for the prototypic carbamylating enzyme TobZ54). In the distorted tRNA conformer structure however, 7 nucleotides within the anticodon loop encompassing A37 (U33 to C39, inclusive) are disordered, indicating that this region has higher flexibility in this conformation. Thus, A37 (although not visible due to disorder) in principle would have the ability to more closely approach the active site of Kae1. Although, we cannot discern the precise position of A37, modeling reveals that it could approach within 5.5 angstroms of the inferred position necessary for its modification. Together with additional flexibility of the tRNA structure evident in the 3D variability analysis (Supplementary Movie 1), we reason that the observed disorder of the anticodon loop in the distorted state could possibly position the A37 base into the active site for modification, hinting that this may be an integral step in the tRNA modification reaction.
Next, we focused on the structural differences in the D-arm, a second distinguishing feature between the two tRNA conformers (Fig. 2A). The D-arm in the native-like tRNA conformer exhibited canonical features, while the D-arm in the distorted tRNA conformer exhibited an unusual condensing of its width (Fig. 2A, denoted by blue arrows), a widening of the groove between the D-arm and anticodon stem (denoted by red arrows) and lastly a flipping of a base that we infer is likely to be G26 (Fig. 2A right side).
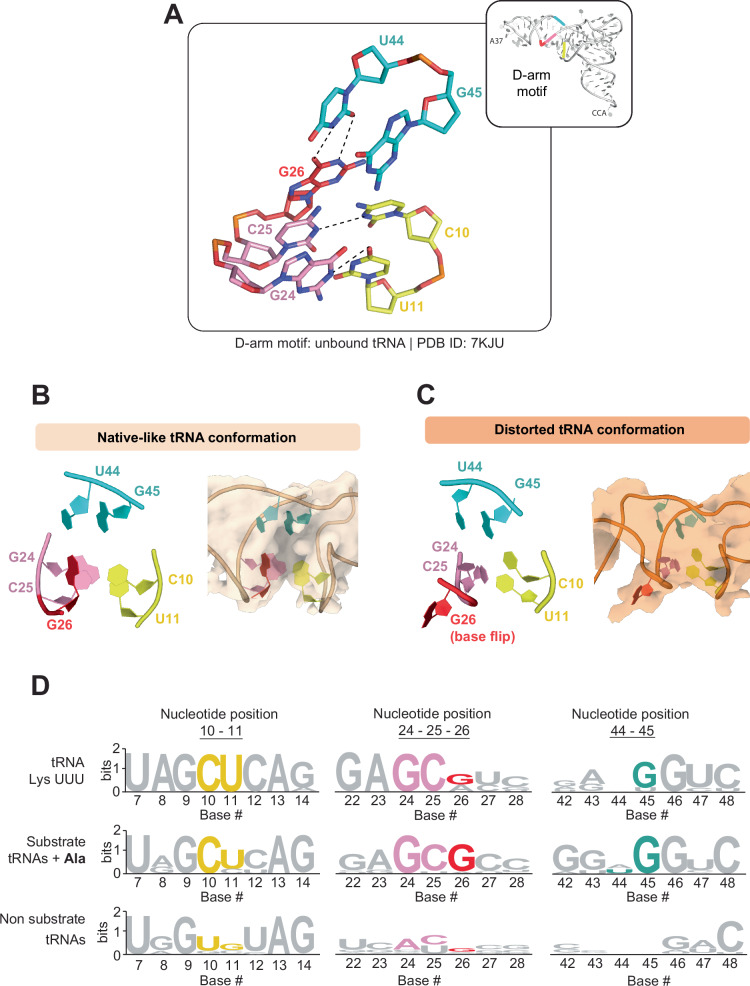
G26 forms a network of interactions in the D-arm that are conserved in KEOPS substrate tRNAs
The conspicuous flipped-out base of G26 raised the question of what intra-molecular interactions it makes in the native-like and distorted conformations. Since the resolution of the tRNA in the cryo-EM structures did not enable unequivocal determination of tRNA atomic positions, we used a crystal structure of the native tRNA as a point of reference (PDB 7KJU). In the native tRNA structure, the G26 base is part of a network of interactions with C10, U11, G24, C25, U44 and G45. G26 base pairs with U44 and stacks with C25 and G24. C25 in turn base pairs with C10, which also interacts with G45, and G24 base pairs with U11 and G45 (Fig. 3A, see Fig. 3B, C for cryo-EM maps of the same region in the native-like and distorted tRNA conformations). The sequence of the D-arm region appears well conserved across KEOPS substrates. Position 26 is conserved as a purine (guanine or adenine) in tRNALysUUU orthologs (Fig. 3D-top and in all t6A substrates in M. jannaschii (Fig. 3D-middle), but not in tRNAs that are not t6A substrates (Fig. 3D-bottom). Five out of six of the nucleotides involved in this interaction network around G26 are conserved in KEOPS substrate tRNAs and not in non-substrates (Fig. 3D) with exception of U44, an observation noted by others previously42,55. For simplicity, we refer to this network of interacting nucleotides as the D-arm motif.
Fig. 3. G26 resides within a network of nucleotides conserved in KEOPS substrate tRNAs.
A Stick representation of the D-arm motif network as observed in the crystal structure of M. jannaschii tRNALys (PDB 7KJU). G26 forms hydrogen bonds with U44, and base stacking interactions with C25 and G24. G24 and C25 base pair with U11 and C10, which stack with G45. (right) Cartoon representation of tRNA structure showing the location of the D-arm nucleotides according to their coloring scheme shown in the left. B, C Atomic models (left) and cryo-EM density volumes (right) of the D-arm motif in tRNAs in their native-like (B) and distorted (C) conformations. D The D-arm-motif is conserved uniquely in KEOPS substrates. Weblogo logo sequences showing the conservation at the indicated positions for tRNALys UUU orthologs (top), the nine substrate ANN-decoding and three tRNAAla sequences (middle), and all 23 other non-substrate tRNA sequences (bottom) from M. jannaschii.
Perturbing G26 interactions within the D-arm alters the ability of KEOPS to t6A modify its tRNA substrate
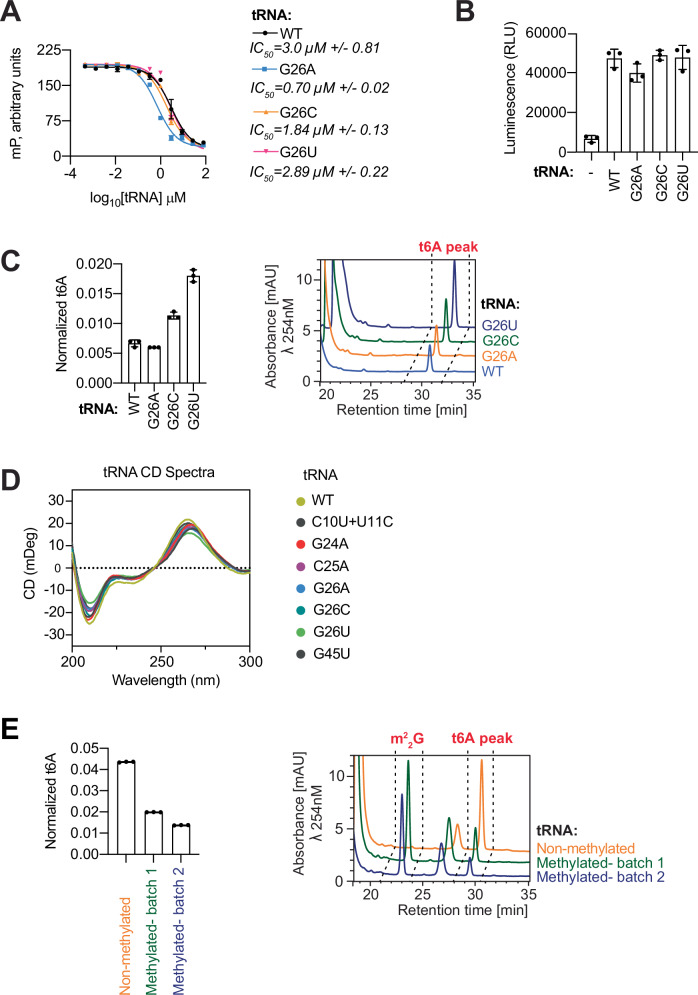
To explore the functional relevance of the flipped-out base at position of G26 and the surrounding D-arm motif more broadly we carried out substitution experiments. We first substituted G26 to A, C and U and tested for effects on tRNA binding, Bud32 activation and KEOPS t6A-modification activity. While all substitutions had no deleterious effect on the ability of KEOPS to bind tRNA (Fig. 4A) or on the ability of tRNA to activate Bud32 (Fig. 4B), some had surprisingly pronounced effects on t6A-modification activity. Substitution of G26 to the smaller C and U, which do not naturally occur in this position in t6A substrates (Fig. 3D), led to ~2- and ~3-fold enhancement of t6A-modification activity respectively (Fig. 4C). Interestingly, substitution of G26 to similarly sized A, which is naturally found in tRNA t6A-accepting substrates at this position (Fig. 3D), had virtually no effect on t6A-modification activity (Fig. 4C). Since U and C are smaller than the naturally occurring G and A at position 26, we reasoned that these substitutions affected the D-arm motif network of interactions, although without perturbing the overall tRNA fold as evidenced by circular dichroism (CD) analysis, since all tRNAs examined exhibited a CD spectra characteristic of a folded tRNA49,56 (Fig. 4D).
Fig. 4. Functional analysis of the role of G26 within the D-arm motif of a KEOPS substrate tRNA.
A Competitive displacement of an Alexa-647 labeled CCA-tail probe (647-CCA) from KEOPS by titration of tRNALys WT or the indicated mutants. Displacement of the 647-CCA probe was monitored by fluorescence polarization (FP). Respective mean IC50 values for the displacement are shown (n = 3 technical replicates, error bars represent ± SD). B ATPase activity analysis of WT KEOPS in the presence and absence of tRNALys WT or the indicated mutants. Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). C t6A modification activity analysis of WT KEOPS with tRNALys WT or the indicated mutants. Representative HPLC profiles of nucleoside composition for each reaction are shown at right. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). D Circular dichroism (CD) spectra of tRNALys WT or the indicated mutants indicates that all tRNAs examined exhibit a similar spectra characteristic of a folded tRNA. E t6A modification activity analysis of WT KEOPS with non-methylated tRNALys WT or m22G26-modified tRNALys WT. Representative HPLC profiles of nucleoside composition for each reaction are shown at right. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). Source data are provided as a Source Data file.
As an alternate to substitutions, we next examined the effect of G26 modification on the ability of KEOPS to t6A modify tRNA. Dimethylation of the G26 base at the N2 position (denoted m22G26) is appreciated to rigidify the tRNA structure at the elbow region between the anticodon and acceptor domains by restricting G26 base pairing capacity57. tRNA bearing quantitative m22G26 modification (Supplementary Fig. 4a, b) displayed ~65% reduction in t6A modification by KEOPS (Fig. 4E). Thus, substitutions that are predicted to destabilize tRNA cause enhanced activity while tRNA stabilization causes reduced activity.
The D-arm motif nucleotides are important for KEOPS activity
Next, we systematically substituted bases at the other positions in the D-arm motif. Unlike the effect of G26 mutation, mutation of G24 to A and C25 to A inhibited tRNA-induced Bud32 ATPase activation (Fig. 5A) and thus not unexpectedly KEOPS t6A modification activity (Fig. 5B). These effects are in line with the effect previously reported on mutation of 10-CU-11 (Fig. 5A, B and ref. 42), the base pairing partners of G24 and C25 (Fig. 3A). Mutation of G45 to U had similar but less pronounced effects. Suggesting that the mutations in the D-arm motif didn’t simply perturb tRNA fold, the tRNA mutants were competent for KEOPS binding (Fig. 5C) and displayed a CD spectrum characteristic of a folded tRNA (Fig. 4D). Thus, the D-arm motif nucleotides are important determinants for both Bud32 activation and t6A modification, yet how these positions contribute to these activities is not obvious.
Fig. 5. Mutational analysis of D-arm motif positions neighboring G26.
A ATPase activity analysis of WT KEOPS in the presence and absence of tRNALys WT or the indicated tRNALys mutants. Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). B t6A modification activity analysis of WT KEOPS with tRNALys WT or the indicated mutants. Representative HPLC profiles of nucleoside composition for each reaction are shown at right. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). C Competitive displacement of an Alexa-647 labeled CCA-tail probe (647-CCA) from KEOPS by titration of tRNALys or the indicated tRNA mutants. Displacement of the 647-CCA probe was monitored by fluorescence polarization (FP). Respective mean IC50 values for the displacement are shown (n = 3 technical replicates, error bars represent ± standard deviation SD). Source data are provided as a Source Data file.
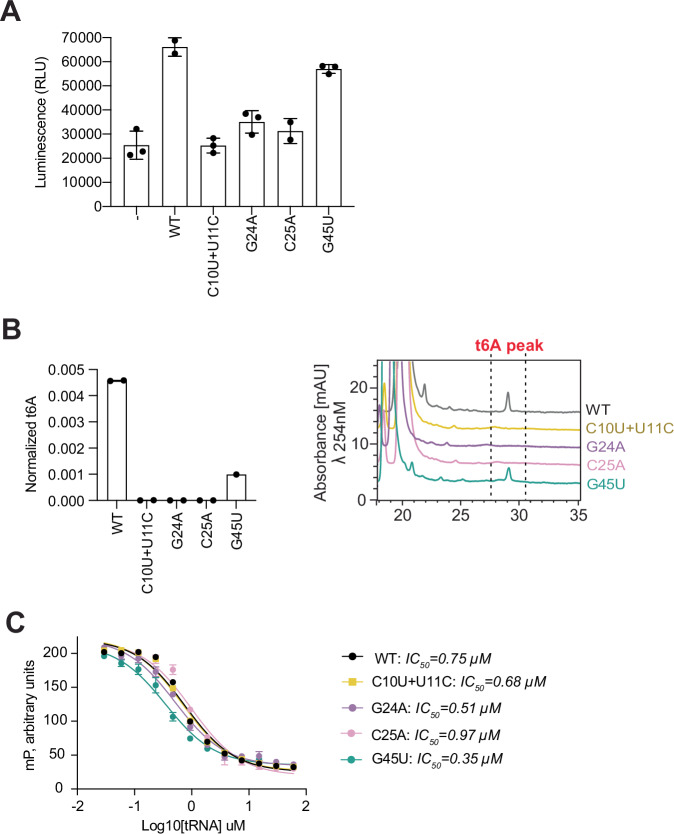
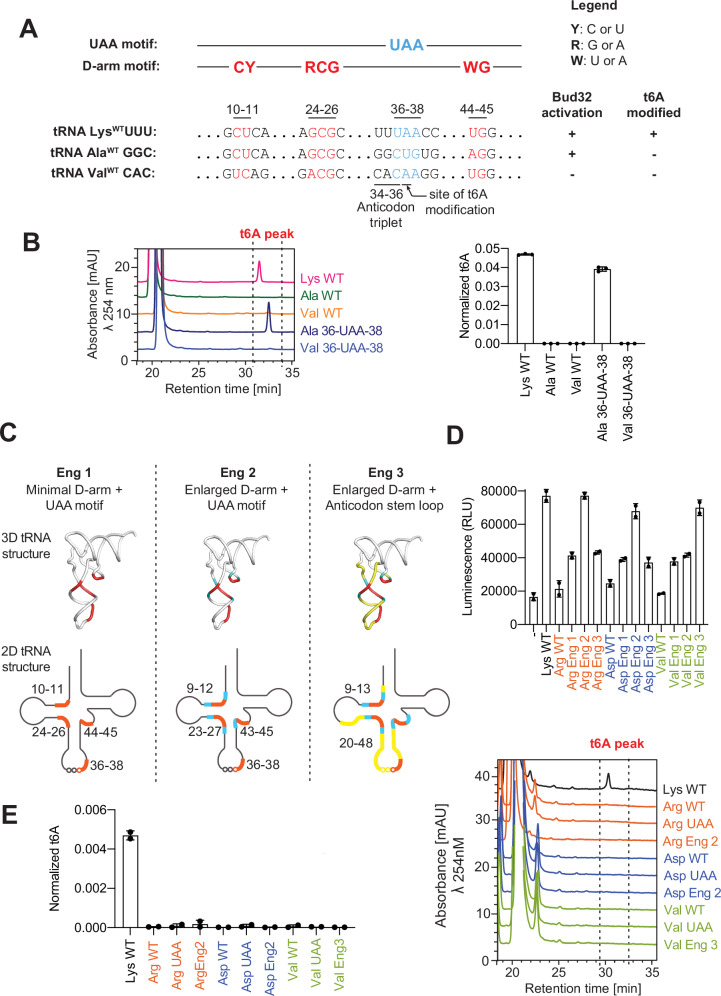
Interestingly, we noticed that the AlaGGC tRNA is the only non-ANN decoding tRNA in M. jannaschii that has an intact D-arm motif but lacks the 36-UAA-38 acceptor motif (Fig. 6A), a motif invariantly present in all t6A-accepting natural substrates in all domains of life that is required for t6A modification6,58,59. Accordingly, tRNA Ala is the only non-substrate in M. jannaschii that can induce Bud32 activation42. However, it can’t be t6A modified (Fig. 6B), presumably because it lacks the UAA acceptor motif. Indeed, addition of a UAA acceptor motif converted tRNA Ala to a KEOPS substrate (Fig. 6B). In contrast, addition of a UAA acceptor motif to ValCAC tRNA, which does not contain a D-arm motif, did not convert it to being a KEOPS substrate. These results suggested that combining a pre-existing D-arm motif with a UAA acceptor motif is sufficient to convert a non-substrate tRNA to a KEOPS substate. To further test this notion, we tried a more drastic approach to substrate engineering. ArgGCG, ValCAC and AspGUC tRNAs lack both the acceptor UAA acceptor motif and a D-Arm motif (see Fig. 6C for schematic representation). To test if we could engineer these into KEOPS substrates, we grafted regions of the anticodon domain of tRNA LysUUU encompassing the D-arm motif and the UAA acceptor motif onto these three non-substrate tRNAs (Fig. 6C). Specifically, each non-substrate tRNA was modified to contain either a minimal D-arm and UAA motif, a slightly enlarged version of these motifs, or the entire anticodon arm of tRNA LysUUU. Remarkably, all engineered tRNAs conferred the ability to activate Bud32 ATPase activity to a higher degree than the parental non-substrate tRNAs (Fig. 6D). Most notably, tRNA Arg Eng#2, tRNA Asp Eng#2 and tRNA Val Eng#3 chimera tRNAs activated Bud32 ATPase activity to levels comparable to the natural substrate tRNA LysUUU. Although the engineered tRNAs activated Bud32, none could be t6A modified by KEOPS (Fig. 6E). Thus, an intact D-arm motif is required and sufficient for the ability of tRNAs to activate Bud32, but combining functional D-arm and UAA motifs is not sufficient to convert a non-substrate tRNA into a t6A-accepting substrate. Whether the lack of t6A modification activity is a result of a missing additional tRNA substrate determinant or a consequence of an imperfect tRNA engineering design, remains to be determined.
Fig. 6. Engineering KEOPS non-substrate tRNAs into substrate tRNAs.
A (top) linear schematic representation of defining elements in KEOPS substrate tRNAs, highlighting the conserved UAA and D-arm motifs. (bottom) Sequence alignments showing the conservation and divergence of the D-arm and UAA motifs in tRNALysUUU, tRNAAlaGGC and tRNAValCAC and their respective abilities to activate Bud32 and to be t6A modified by KEOPS. B t6A modification activity analysis of KEOPS WT on tRNALys, tRNAAla and tRNAVal WT or the indicated mutants. Representative HPLC profiles of nucleoside composition for each reaction are shown at left. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). C Schematic of engineering strategy to convert non-substrate tRNAs Arg, Asp and Val into Bud32 activators and t6A-modification substrates. Engineered (Eng) 1 chimeras contain the D-arm and UAA motifs from tRNALys, shown in red. Eng 2 and Eng 3 chimeras contain extended regions from tRNALys shown in cyan and yellow respectively. D ATPase activity analysis of WT KEOPS in the presence and absence of the indicated WT and engineered tRNAs schematized in (C). Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). E t6A modification activity analysis of KEOPS WT in the presence and absence of the indicated WT and engineered tRNAs schematized in (C). Representative HPLC profiles of nucleoside composition for each reaction are shown at left. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). Source data are provided as a Source Data file.
KEOPS-tRNA structures reveal functionally significant contact surfaces between tRNA and protein subunits
Comparing the two tRNA structures identified important determinants that allow Bud32 activation and tRNA t6A modification. To gain insight into how the tRNA features are sensed by KEOPS we analyzed three previously non-visualized tRNA contact surfaces evident in the two tRNA-KEOPS complex structures (schematized in Supplementary Fig. 5).
The anticodon loop makes different contacts with KEOPS in its distorted and native-like states
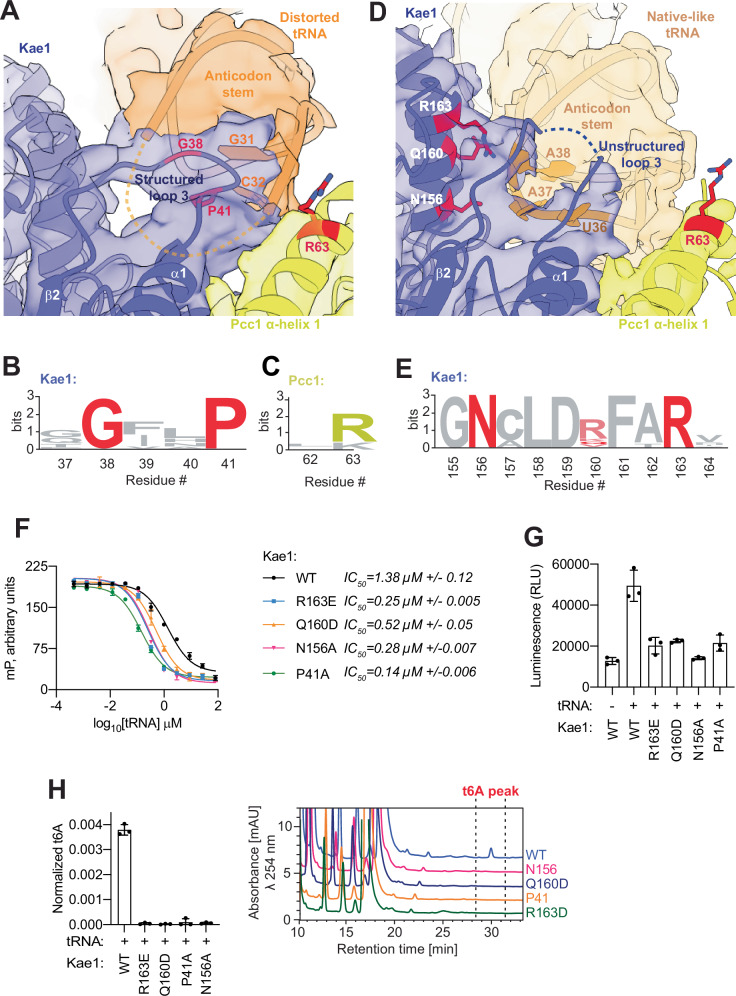
In both tRNA-bound structures, the tRNA anticodon loop is oriented towards the active site of Kae1 and towards Pcc1. In the distorted tRNA conformation, the G31-C32 dinucleotide of tRNA is juxtaposed to loop 3 of Kae1 (residues 23–43, between β-sheet 2 and α-helix 1) and the C’ terminus of Helix 1 of Pcc1 (Fig. 7A). Local resolution did not allow us to determine direct contacts. C32 in the tRNA is a known but not understood determinant for t6A modification by KEOPS55, and Gly38 and Pro41 in Kae1 are invariant (Fig. 7B) suggesting they too are important for function. In Pcc1, Arg63 in this region is highly conserved (Fig. 7C) and its mutation to Asp was shown to completely inhibit KEOPS function in vitro and in yeast42.
Fig. 7. Functional analysis of unique contacts between the anticodon loop of tRNA and Kae1.
A Cryo-EM density in surface representation and fitted atomic model in cartoon representation highlighting contacts between the anticodon stem of the tRNA in its distorted conformation with a structured Kae1 loop 3 (residues 23–43) and Pcc1 α-helix 1 (residues 62–66). B, C Weblogo sequences showing (B) the conservation of Gly38 and Pro41 in Kae1 orthologs and (C) the conservation of Arg63 in Pcc1 orthologs. D Cryo-EM density in surface representation and fitted atomic model in cartoon representation highlighting contacts between the anticodon stem of the tRNA in its native-like conformation and Kae1-specific insert 1 (residues 156–163). E Weblogo logo sequences showing the conservation of the Kae1-specific-insert residues contacting tRNA. F Competitive displacement of an Alexa-647 labeled CCA-tail probe (647-CCA) from KEOPS WT or KEOPS reconstituted with the indicated Kae1 mutants by titration of tRNALys. Displacement of the 647-CCA probe was monitored by fluorescence polarization (FP). Respective mean IC50 values for the displacement are shown (n = 3 technical replicates, error bars represent ± standard deviation SD). G ATPase activity analysis of KEOPS reconstituted with Kae1 WT or the indicated mutants in the presence and absence of tRNALys. Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). H t6A modification activity analysis of KEOPS reconstituted with Kae1 WT or the indicated mutants. tRNALys was used as substrate. Representative HPLC profiles of nucleoside composition for each reaction are shown at left. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). Source data are provided as a Source Data file.
In its native-like conformation, the conserved 36-UAA-38 acceptor motif of tRNA58–60, is positioned to contact a conserved region in Kae1 termed Kae1-specific-insert 1 (Fig. 7D). This insert distinguishes Kae1-family enzymes from other members of the functionally diverse ASKHA-fold family61,62. Although the local resolution does not allow definitive positioning of side chains and bases, Asn156, Gln160 and Arg163 in the Kae1-specific-insert are well placed to interact with A37 (the modification site of the tRNA itself) and A38 (Fig. 7D). Of these residues, Asn156 and Arg163 are invariant (Fig. 7E). We showed previously that a peptide that encompasses Kae1-specific-insert-I exhibits reduced hydrogen-deuterium exchange when KEOPS is incubated with substrate tRNA and that mutation of Gln160 and Arg163 abolishes KEOPS function in vitro and in yeast42.
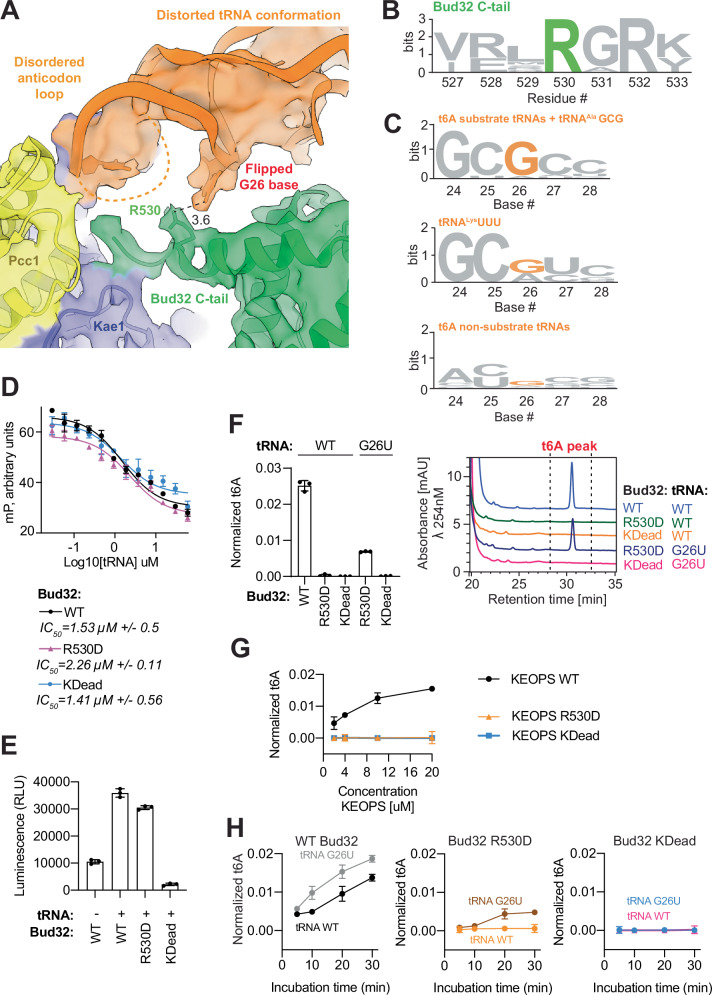
Fig. 8. Functional analysis of unique contacts between the D-arm motif of tRNA and the conserved tail of Bud32.
A Cryo-EM density volume and atomic model of the distorted tRNA highlighting the interaction of the flipped base of G26 with the side chain of Arg530 in the C-terminal tail of Bud32. B Weblogo logo sequences showing conservation of Arg530 in Bud32 orthologs. C Weblogo logo sequences showing conservation of G26 in KEOPS substrate tRNAs and tRNAAlaGCG orthologs (top), in tRNALysUUU orthologs (middle), and in non-KEOPS substate tRNAs (bottom). D Competitive displacement of an Alexa-647 labeled CCA-tail probe (647-CCA) from KEOPS reconstituted with Bud32 WT or the indicated mutants by titration of tRNALys. KDead corresponds to a Bud32 D451A active site mutation that generally disables phospho-transfer activity in the eukaryotic protein kinase superfamily. Displacement of the 647-CCA probe was monitored by fluorescence polarization (FP). Respective mean IC50 values for the displacement are shown (n = 3 technical replicates, error bars represent ± SD). E ATPase activity analysis of KEOPS reconstituted with Bud32 WT or the indicated Bud32 mutants in the presence or absence of tRNALys. Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). F t6A modification activity analysis of KEOPS reconstituted with Bud32 WT or the indicated mutants on tRNALys WT or the G26U mutant. Representative HPLC profiles of nucleoside composition for each reaction are shown at right. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). G t6A modification activity analysis of KEOPS reconstituted with Bud32 WT or the indicated mutants on tRNALys WT as a function of enzyme concentration. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). H t6A modification activity analysis of KEOPS reconstituted with Bud32 WT or the indicated mutants on tRNALys WT or the tRNALys G26U mutant as a function of time. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). Source data are provided as a Source Data file.
Further analysis now shows that the mutation Pro41Ala within Kae1 structured loop 3 (Fig. 7A) and Asn156Ala, Gln160Asp, and Arg163Glu within the Kae1-specific-insert 1 (Fig. 7D) do not outwardly perturb binding of tRNA to KEOPS (Fig. 7F). However, they greatly inhibit the ability of tRNA to activate Bud32 ATPase activity (Fig. 7G), an activity that depends on the presence of the Cgi121-Bud32-Kae1 subunits42, and greatly inhibit the ability of KEOPS to modify tRNA (Fig. 7H). Thus, highly conserved residues in Kae1 that are well positioned to contact the anticodon loop of substrate tRNAs in either the native-like or distorted conformation are needed both for the activation of Bud32 ATPase activity and for the t6A modification of tRNA.
G26 is flipped in the distorted tRNA conformation and interacts with an invariant arginine in the C-tail of Bud32
As noted, in the native-like tRNA conformation, the G26 base is buried in the tRNA core (Fig. 3A, B), whereas in the distorted tRNA conformation the base of G26 appears flipped-out of this core (Figs. 2A and 3C). In its flipped-out state, the G26 base is positioned to interact with Arg530 in the C-terminal tail of Bud32 (Fig. 8A), which is notably an invariant residue (Fig. 8B). The conservation of a purine at position 26 specifically in t6A substrates (Fig. 8C) and its proximity to an invariant residue in the Bud32 tail suggests a functional link. Indeed, we previously showed that truncation of the C-terminal tail of Bud32 encompassing Arg530 inhibited t6A modification activity by KEOPS42 and KEOPS function in yeast47. We now show that the more surgical Arg530Asp mutation in the C-terminal tail of Bud32 phenocopies the full tail deletion in causing loss of t6A activity but without adversely affecting tRNA binding by KEOPS (Fig. 8D) or Bud32 ATPase activity (Fig. 8E). For comparison, KEOPS harboring the Bud32 Arg530Asp mutant is comparably incompetent for t6A modification activity as KEOPS harboring a Bud32 kinase disabled mutant (Asp451 to Ala mutation denoted KDead), even when assessed over a wide range of enzyme concentrations (Fig. 8F, G).
To examine potential functional interplay between G26 of the tRNA and Arg530 of Bud32, we tested the effect of combining the G26 and Arg530 mutations. Interestingly, the G26U activating mutation in tRNA partially overcame the Arg530Asp repressive mutation in Bud32 on t6A modification activity (Fig. 8F, H). In contrast the G26U activating mutation in tRNA displayed no ability to overcome the kinase disabled mutation in Bud32 (Fig. 8F, H). These findings suggest that a guanine at position 26 of tRNA and an arginine at position 530 in the Bud32 tail are functionally linked in a sense that Bud32 kinase/ATPase activity is not.
Bud32 activation mechanism depends on a direct contact with Kae1
Our data and previous work show that binding of substrate tRNAs minimally to the Cgi121-Bud32-Kae1 subcomplex can activate Bud3242. We were somewhat perplexed at understanding how tRNA binding to KEOPS turns on Bud32 ATPase activity, since the contact between tRNA and Bud32 beyond the C-terminal tail is tenuous. This led us to hypothesize that the tRNA might have an indirect effect on Bud32, i.e., that Cgi121 or Kae1 might be more directly involved in the Bud32 activation process.
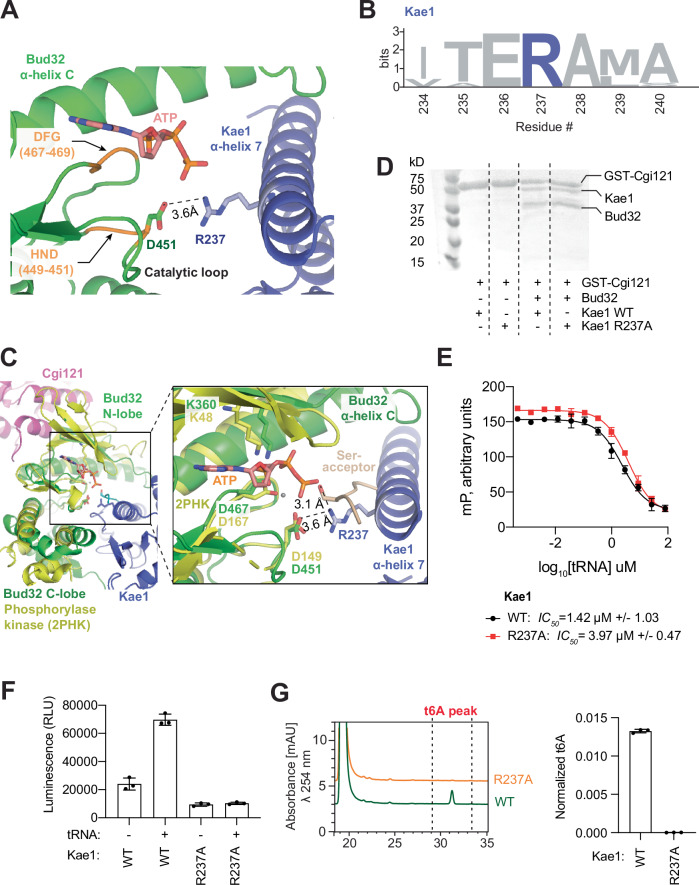
Arg237 of Kae1, which lies close to the active site of Bud32 (Fig. 9A), is notably invariant across all Kae1 orthologs (Fig. 9B). The protein kinase superfamily shares a common catalytic mechanism in which the hydroxyl phospho-acceptor site of a substrate is activated by a conserved catalytic Asp within the HRD motif of the kinase63 (HND in Bud32; see Fig. 9C for structural comparison of Bud32 with the minimalistic prototypical protein kinase phosphorylase kinase). Since Bud32 is an ATPase that transfers phosphate to water rather than to a protein substrate side chain42, the position of the protein substrate hydroxyl would represent the position of the water molecule that serves as an analogous phosphate acceptor. Strikingly, the side chain of Arg237 in Kae1 is adjacent to the anticipated phosphate acceptor site within the active site of Bud32 (Fig. 9C) and thus is ideally placed to influence phosphate transfer from ATP to water. Further highlighting the likely functional importance of Arg237, mutations at this position underlie GAMOS syndrome16. To test if Arg237 plays an essential role in KEOPS function, we examined the effect of its mutation to alanine. While the Arg237Ala mutation in Kae1 had no adverse effect on the binding of Kae1 to Bud32 (Fig. 9D), and only moderately reduced the affinity of tRNA to KEOPS (Fig. 9E), it completely abolished both the activation of Bud32 ATPase by tRNA (Fig. 9F) and the t6A modification activity of KEOPS (Fig. 9G). Interestingly, the Kae1 Arg237Ala mutation also reduced the basal ATPase activity of Bud32 (Fig. 9F), suggesting that its position proximal to the active site of Bud32 also supports the basal ATPase activity of Bud32 even in the absence of tRNA binding. These results support a model whereby binding of tRNA to KEOPS transmits a signal through Kae1 to the active site of Bud32 via Arg237, which leads to an enhancement of its ATPase activity.
Fig. 9. Arg237 of Kae1 is positioned in the active site of Bud32 and is essential for regulating its ATPase activity.
A Cryo-EM density in mesh representation and fitted atomic model in cartoon representation of Kae1 and Bud32 in the structure of KEOPS bound to tRNA in its native-like conformation. Arg237 of Kae1 is ~3.6 Å from the active site residue Asp451 in Bud32. The position of ATP was modeled from the crystal structure of phosphorylase kinase (PDB ID: 2PHK). B Weblogo sequences showing the conservation of Arg237 in Kae1. C Cartoon representation showing the crystal structure of phosphorylase kinase bound to substrate peptide (labeled Ser-acceptor) and the ATP analog AMP-PnP (PDB 2PHK) superimposed on the Bud32 subunit of KEOPS bound to tRNA in a native-like conformation. Box shows detailed view of the active site region of Bud32 and its interaction with Kae1. Arg237 in Kae1 is ~3.1 Å from the phospho-acceptor site of the peptide substrate. D GST pull-down analysis of GST-Cgi121 binding to Kae1 WT and R237A mutant in the presence or absence of Bud32. Samples were analyzed by Coomassie-stained SDS PAGE (n = 2 technical replicates). E Competitive displacement of an Alexa-647 labeled CCA-tail probe (647-CCA) from KEOPS WT or the R237A mutant by titration of tRNALys. Displacement of the 647-CCA probe was monitored by fluorescence polarization (FP). Respective mean IC50 values for the displacement are shown (n = 3 technical replicates, error bars represent ± SD). F ATPase activity analysis of KEOPS reconstituted with Kae1 WT or the Arg237Ala mutant in the presence and absence of tRNALys. Activity was monitored using the ADP Glo assay. Displayed results center represent the mean luminescence for each reaction condition, error bars represent ±SD (n = 3 technical replicates). G t6A modification activity analysis of KEOPS reconstituted with Kae1 WT or the Arg237Ala mutant. tRNALys was used as substrate. Representative HPLC profiles of nucleoside composition for each reaction are shown at left. Quantification shows center of mean t6A content normalized to the content of uridine and error bars represent ±SD (n = 3 technical replicates). Source data are provided as a Source Data file.
Discussion
We have solved structures of KEOPS alone and in complex with a tRNA substrate. We show that Cgi121, Bud32, Kae1 and Pcc1 form an extended surface with high structural complementarity to the L-shaped tRNA substrate, cradling it and guiding A37 to the active site of Kae1. When bound to KEOPS, the tRNA is flexible and likely adopts many conformations, hinting that tRNA deformation may be a requirement for substrate modification. This could explain our inability to obtain higher resolution structures in the absence of further sample manipulation (e.g. crosslinking). Two of the conformations were heavily populated. We termed one of these native-like because it resembles the structure of isolated tRNA and the other distorted since it exhibits unraveling of the anticodon domain. We show that KEOPS substrates display high conservation in the D-arm and anticodon loop suggesting mechanistic importance.
We show that when the tRNA is in its native-like conformation, the anticodon loop is ordered and A37 is not positioned for modification, whereas when it is in the distorted conformation, A37 is released from its ordered state, which potentially make it more accessible to the active site of Kae1. Structures of the KEOPS-tRNA complex visualize specific contact surfaces between the tRNA and the Kae1, Bud32 and Pcc1 subunits. Using mutational analysis, we have probed the functional importance of these contacts discovering varied effects. Some contacts appear vital for tRNA-dependent activation of Bud32 ATPase and tRNA t6A modification, while others affect only t6A modification. This separation of function helps define the key steps in the reaction cycle of tRNA modification activity by KEOPS.
Structural and functional analysis are consistent with four key steps in the catalytic cycle of KEOPS:
Substrate recruitment. The tRNA is recruited by the binding of Cgi121 to its CCA tail. tRNA substrate is then fully engaged by a dynamic set of secondary interactions with Bud32, Kae1 and Pcc1 (depicted in Supplementary Fig. 5).
Activation of Bud32 ATPase activity. Bud32 activation is a requirement for tRNA modification by Kae1. Since Bud32 activation depends on features unique to KEOPS substrates, this dependency might serve to ensure that Kae1 will only be active when a correct substrate is bound. We now show that this key step depends on three factors. Firstly, the interaction between the tRNA anticodon and conserved residues in the Kae1-specific-insert I. These contacts are most evident when the tRNA is in a native-like conformation. Secondly, the positioning of Arg237 of Kae1 in the active site of Bud32, where it facilitates ATP hydrolysis. This could explain why tRNA binding to the Cgi121-Bud32 complex in the absence of Kae1 does not potentiate ATPase activity42. Based on its positioning, we reason that Arg237 could exert its effect by stabilization of the γ–phosphate of ATP or the hydroxy anion (OH−) intermediate generated by deprotonation of water by the catalytic base Asp467 of Bud32. Since Arg237 is visualized in the same position in Kae1-Bud32 structures without tRNA (PDB 3EN9 for example47), we speculate that Bud32 activation may arise from a change in conformational dynamics of its bilobal architecture, as this represents a point of regulation for other protein kinases64. Importantly, mutation of the equivalent Arg residue in human Kae1 (namely Arg247Gln in OSGEP) is causative for GAMOS16. Thus, our findings suggest the basis for this pathogenicity is due to a loss in the ability of OSGEP/Kae1 to activate PRPK/Bud32 in response to tRNA binding. Thirdly, a conserved C10-U11-G24-C25 D-arm motif that is unique to KEOPS substrate tRNAs.
Reordering of the tRNA anticodon loop, liberating A37 to be t6A modified. This step correlates with a base flip at the inferred position of G26. Structural and mutational analysis point to a potential role for Arg530 in the C-terminal tail of Bud32 in interacting with the flipped-out base. We hypothesize that when G26 interacts with Arg530, it is released from its native interactions within the tRNA, which assists to distort the anticodon domain, enabling A37 to access the active site of Kae1. Pcc1 appears to play a role in this step by interacting with the anticodon loop since it is dispensable for the activation of Bud32 but is needed for t6A modification. It is unclear if this step happens in parallel to Bud32 ATPase activation or after it.
The tRNA is modified followed by release from KEOPS, allowing commencement of a new catalytic cycle.
An expanded understanding of the mechanisms that underpin the catalytic cycle KEOPS awaits the determination of additional and higher-resolution structures of the holo-enzyme tRNA substrate complex.
Lastly, one of the most fascinating unanswered questions is why Kae1, Qri7 and TsaD function in such distinct complexes. Most tantalizing is the observation that Qri7 functions without auxiliary subunits. One possible explanation for this is related to the nature of the substrates of each enzyme. Cytoplasmic tRNAs are more heavily modified than mitochondrial tRNAs, and many of these modifications serve to stabilize the tRNA structure65. An important role of the auxiliary subunits of Kae1 may be to make A37 accessible for modification, a function less needed by Qri7 in its mitochondrial environment since it acts upon less heavily modified and thus less structurally rigid substrates. This might explain why high-level overexpression of Qri7 engineered to be retained in the cytoplasm is needed to rescue the Kae1 knockout phenotype in S. cerevisiae31.
Methods
Protein expression and purification
The coding sequences of Methanocaldococcus jannaschii Cgi121 (residues 1–145), Kae1 (residues 1–327 of the naturally occurring Kae1-Bud32 fusion protein mj1130), and Pyrococcus furiosus Pcc1 (residues 1–82) were obtained from the Uniprot website (https://www.uniprot.org/), gene synthesized (Genscript) and cloned into the pGEX2T vector with an ampicillin resistance gene for selection in bacteria. Cgi121, Kae1 and Pcc1 were each individually designed to express as GST fusion proteins with a TEV cleavage site between the GST and the protein of interest coding regions47,66. The Pyrococcus furiosus Pcc1Mut protein expression construct was generated by cloning into the pGEX2T vector two Pcc1 encoding sequences in tandem, connected by 7 repeats of the dipeptide glycine-serine. The second Pcc1 sequence encoded the double mutations Ala75Tyr and Val79Arg, which prevent Kae1 binding47,48. Thus, the Pcc1Mut construct expresses an N-terminal GST (TEV cleavable) fused Pcc1 heterodimer complex, with a Pcc1WT protomer connected to a 7XGS sequence at its C-terminus followed by the Pcc1 Ala75Tyr + Val79Arg protomer. Methanococcus jannaschii Bud32 (residues 328–535 of the naturally occurring Kae1-Bud32 fusion protein mj1130) was cloned into pProEX plasmid with an ampicillin resistance gene for selection in bacteria as an N-terminal hexahistidine (His) tagged protein with a TEV cleavage site between the His-tag and the protein coding region. Schizosaccharomyces pombe Trm1 (residues 1–524) was cloned into the pET28a vector with a kanamycin resistance gene for selection in bacteria as a C-terminal His-tagged protein. All Kae1 and Bud32 mutants described in this study were generated by standard molecular biology approaches and were sequence verified (for a list of primers used, see Supplementary Table 2).
For bacterial expression, Cgi121, Bud32, Kae1 and Pcc1 coding plasmids were each individually transformed to BL21-CodonPlus DE3-RIL bacteria (Agilent Technologies) grown in 2–8 L Terrific Broth media (Sigma). Cells were grown at 37 °C and expression was induced by the addition of 0.3 mM IPTG (UBPBio) to bacterial cultures at OD600 = 0.8–1 at 18 °C overnight. Overnight cultures were spun down (6000 g, 10 min room temperature) and pellets were flash frozen in liquid nitrogen and stored at −80 °C.
GST-fused KEOPS proteins were each individually purified by a similar purification protocol. Cell pellets from overnight cultures were lysed via homogenization in lysis buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5 mM EDTA, 2 mM DTT) supplemented with 0.2 mM PMSF. Lysates were spun down (40,000 g, 30 min at 21 °C) and supernatants were filtered at 45 μm. Cleared supernatants were run over GST-Sepharose resin (GE) at room temperature in a gravity drip column. Bound protein fractions were eluted by TEV-cleavage on resin overnight at room temperature, followed by purification with an S200 sizing column (Cytiva, Superdex 200 Increase 10/300 GL) in sizing buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT).
For Bud32 purification, cells were lysed as described above in lysis buffer (25 mM HEPES pH 7.5, 500 mM NaCl, 25 mM imidazole). Lysates were run over a Ni-NTA column (Cytiva, Hi TrapTM 5 mL chelating HP), bound fraction was eluted with a 0–100 % gradient of lysis buffer with 250 mM imidazole over 20 column volumes. Eluted fractions were pooled and dialyzed to reduce imidazole concentration to 25 mM in parallel to TEV cleavage, followed by subtraction of His-tagged TEV with an Ni-NTA column. The flow-through fraction was further purified by an S200 sizing column in sizing buffer.
Recombinant His-tagged protein Trm1 protein expression was induced in Escherichia coli with 1 mM IPTG at 16 °C for 18 h. Protein was purified over a Ni2+ column (His-TRAP, GE-Amersham), followed by a second round of purification over a heparin column (Hi-TRAP, GE-Amersham). Proteins were concentrated into 1X EMSA buffer (20 mM Tris–HCl pH 7.6, 100 mM KCl, 0.2 mM EDTA pH 8.0, 1 mM DTT) and quantified by relative band intensity on SDS-PAGE67.
T7 in vitro transcription and purification of tRNA
tRNA sequences were obtained from GtRNAdb website (http://gtrnadb.ucsc.edu/)68. tRNA expression constructs were made by gene synthesis (Genscript) by fusing tRNA sequences 3’ to a T7 promoter and 5’ to a hepatitis delta virus ribozyme followed by a BamH1 site and an ampicillin resistance gene for selection in bacteria in the pUC19 plasmid (for tRNA expression construct sequences, see Supplementary Table 3).
T7 run off transcription reactions were done66 using 250 μg of plasmid DNA linearized by BamH1 overnight digestion as a template. In vitro transcription reactions were done in 10 mL of 100 mM Tris-Cl pH 8.0, 4 mM ATP/GTP/CTP/UTP, 10 mM DTT, 1 mM spermidine, 0.1% Triton X-100, 25 mM MgCl2, 0.2 mg/mL T7 RNA polymerase, 10 U/mL thermostable inorganic phosphate (NEB) and 200 U/mL RiboLock (Thermo Scientific) at 37 °C for 4 h. Nucleic acids were purified from the transcription reaction by phenol chloroform extractions followed by ethanol precipitation. Pellets were washed with 80% ethanol and air dried followed by solubilization with 5 mL 8 M urea. RNA was refolded in 45 mL of 10 mM Bis-Tris-Cl pH 6.5, 10 mM MgCl2. tRNA was purified on a 5 mL Q column (GE Healthcare) using DEPC-treated water (buffer A) and 2 M NaCl (buffer B) on an FPLC (GE, Akta) with a purification program starting with 4 column volumes at 25% B followed by a linear gradient over 20 column volumes from 25% to 40% B. Fractions containing tRNA were analyzed via TBE-urea gels, pooled and treated with one volume of isopropanol to precipitate RNA. The precipitated RNA was washed with 80% ethanol and then air dried. The RNA pellet was resuspended in 10 mM Tris-Cl pH 8. Prior to use, the tRNA was refolded by (i) boiling at 95 °C for 2 min, (ii) flash cooling on ice, (iii) warming to 50 °C, (iv) addition of MgCl2 to a final concentration of 2 mM and (v) slow cooling to room temperature.
In vitro t6A assays and HPLC analysis of tRNA modifications
In vitro t6A reactions were performed66 at 55 °C for 20 min unless indicated otherwise in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 2.5 mM DTT, 0.5 mM threonine, 0.5 mM NaHCO3, 2 mM ATP, 0.25 mM MnCl2, 0.25 mM MgCl2, 2.5 mM spermidine, 0.5 μL TIPP. Unless indicated otherwise, reactions contained 2 μM of each Sua5/Cgi121/Kae1/Bud32/Pcc1 proteins together with 80 μM of tRNA substrate in a 20 μL volume. Protein complexes were assembled through mixing individually purified proteins in equimolar concentration based on absorbance readouts at OD 280 nm and theoretical extinction coefficients of each protein.
For HPLC analysis, the tRNA from the t6A reactions were enzymatically digested according to69 by adjusting volume to 50 μL with water, adding 10 μL of Nuclease P1 at 0.2 U/mL (Sigma, N8630) and 5 μL ZnSO4 10 mM over night at 37 °C followed by dephosphorylation of nucleotides by adding 10 μL Tris pH 8.3 0.5 M and 1 μL calf intestinal phosphatase (CIP, NEB) for 2 h at 37 °C. The resulting mononucleosides were analyzed using a Discovery C18 reverse-phase column (15 cm × 4.6 mm × 5 mm, Supelco Analytical) on a Dionex Ultimate 3000 HPLC Unit (Thermo Scientific) with a linear gradient of 98:2 to 87.5:12.5 of 250 mM ammonium acetate pH 6.5 and 40% acetonitrile over 50 min at 1.5 mL per min flow rate. Data graphs were generated by measuring the area underneath the chromatograms for the t6A, m22G and uridine peaks. Normalized t6A values were obtained by dividing the area of t6A by uridine and plotted using GraphPad Prism V8.3 (GraphPad). To identify the relevant peaks, we used standards for t6A (BioLog, C022) and m22G (Cayman chemical, 35347).
Structure based sequence alignments and WebLogo image generation
Protein and tRNA sequences were obtained from the uniport (https://www.uniprot.org/) and GtRNAdb (http://gtrnadb.ucsc.edu/) websites respectively. For sequence alignments showing conservation among KEOPS proteins or tRNA orthologs, the corresponding sequences from M. musculus, C. elegans, D. rerio, H. volcanii, M. Janasschii, S. pombe, A. thaliana, P. furiosus, S. cerevisiae, S. pombe, X. laevis, and H. sapiens orthologs were used. For Qri7 conservation analysis, the corresponding sequences from S. cerevisiae, S. pombe, X. laevis, A. thaliana, C. elegans, D. rerio, and H. sapiens orthologs were used. Protein alignment correctness was verified using available crystal structures or alpha fold models. Logos were generated using the WebLogo software (https://weblogo.berkeley.edu/logo.cgi).
Fluorescence polarization assays
The 647-CCA RNA probe was generated by fusion of an Alexa-647 fluorescent tag to the 5’ of the 5’-CCGCCA-3’ oligonucleotide (IDT). Protein complexes were assembled through mixing individually purified proteins in equimolar concentration based on absorbance readouts at OD 280 nm and theoretical extinction coefficients of each protein. Reactions were performed in 25 μL containing KEOPS proteins (0.3 μM final) and 647-CCA probe (0.3 nM final) and tRNAs at the indicated concentrations in FP buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT) in a 384 well flat bottom black assay plate (Corning, 3573). All data points were done with three technical repeats. FP measurements were done on a BioTek Synergy Neo plate reader (BioTek) with excitation and absorbance at 620/680 nm respectively. Binding graphs and the derived IC50 values were generated using GraphPad Prism V8.3 (GraphPad) using the log(inhibitor) vs response equation (equation 1):
Bottom = maximally inhibited response, Top = maximal response.
ADP GloTM assay
ADP-Glo Kinase assays (Promega) were performed following manufacturer’s guidelines. Reactions were performed in 20 μL containing Bud32 (0.2 μM final) and other KEOPS proteins (0.25 μM final) and tRNA (2 μM final) in reaction buffer (40 mM Tris pH 7.5, 50 mM NaCl, 10 mM MgCl2, 2 mM MnCl2, 0.1 mg/mL BSA, 0.01% Brij 35, and 1 mM DTT). Protein complexes were assembled through mixing individually purified proteins in equimolar concentration based on absorbance readouts at OD 280 nm and theoretical extinction coefficients of each protein. Reactions were initiated by adding ATP (10 μM final) and by incubating at 55 °C for 60 min. Reactions were terminated by transferring 10 μL of the reaction mix to a 384 well white plate (Lumitrac 200, VWR) containing 10 μL of ADP-Glo™ Reagent and incubating at room temperature for 40 min. 20 μL of kinase detection reagent was then added and allowed to incubate at room temperature for 30 min. Luminescence was measured on a BioTek Synergy Neo plate reader (BioTek) using a 1 s integration time. Results were plotted in GraphPad Prism V8.3 (GraphPad).
Size exclusion chromatography - multi angle light scattering analysis (SEC-MALS)
SEC-MALS analysis of protein complexes were conducted using a miniDawn TREOS and Optilab T-rEX detectors (Wyatt Technology) coupled to a 1260 Infinity HPLC system (Agilent Technologies). Protein complexes were assembled through mixing individually purified proteins in equimolar concentration based on absorbance readouts at OD 280 nm and theoretical extinction coefficients of each protein. A WTC-030S5 SEC column (Wyatt Technology) was used for protein separation. Chromatography experiments were conducted in 20 mM HEPES pH 7.5, 100 mM NaCl and 2 mM DTT. All data analysis was conducted using the ASTRA software (Wyatt Technology).
In vitro methylation and primer extension
5 μM tRNA was methylated for 1 h at 32 °C in a 50 μL reaction containing 100 mM Tris HCl pH 7.5, 0.1 mM EDTA pH 8.0, 10 mM MgCl2, 40 mM NH4Cl2, 1 mM DTT, 1.28 mM S-adenosylmethionine (NEB, B9003S), and 5 μg recombinant Trm1. The reaction was stopped by phenol: chloroform: isoamyl alcohol (25:24:1) extraction. Primer extensions were conducted with SuperScript III following standard methods, using the 5’-TGGCGGGCCCGAAGGGATTTG-3’ primer.
Electron microscopy data collection
Individually purified Kae1, Bud32E152R, Pcc1Mut and Cgi121 were mixed in a 1:1:5:5 ratio based on absorbance readouts at OD 280 nm and theoretical extinction coefficients of each protein. Reconstituted KEOPS was obtained by co-purification of the complex on an S200 sizing column. Reconstituted KEOPS (final concentration of 0.5 μM) was mixed with a 100-fold molar excess (final concentration of 50 μM) of tRNALys. 4 μL of sample was applied for 10 s to C-flat carbon grids previously glow-discharged in air (for 10 s at 25 mA). Grids were then blotted using a Vitrobot Mark III (Thermo Fisher Scientific) for 10 s at blot force −3 at 4 °C and 100% humidity before freezing in a liquid ethane–propane mixture.
Cryo-EM non-tilted and tilted images were collected at 300 kV and at 105,000X nominal magnification with an EF-Krios electron microscope equipped with a GatanK3 imaging system (New York University). The calibrated pixel size of 0.4125 Å was used for processing. Movies were collected using Leginon70 at a dose rate of 22.24 e-/Å2/s with a total exposure of 2.50 s, for an accumulated dose of 55.61 e-/Å2. Intermediate frames were recorded every 0.05 s for a total of 50 frames per micrograph. A total of 23,625 images were collected at a nominal defocus range of −0.4 to −2.9 (non-tilted) and −3.4 to −6.1 μm (tilted).
Image processing
Cryo-EM image analysis was performed using cryoSPARC v4.1.2. Movie frames were aligned and patch-based motion corrected for both global motion (stage drift) and local motion (beam-induced anisotropic sample deformation) followed by patch-based contrast transfer function (CTF) estimation. Template selection for particle images was performed and datasets of particle images were extracted in 256 × 256 pixel boxes.
2D classification was then used to remove undesirable particle images. Multiple rounds of ab initio refinement were used to further clean the dataset. Two consensus 3D maps were calculated with non-uniform refinement corresponding to: apo KEOPS at 2.91 Å resolution from 386,585 particle images and KEOPS + tRNA at 3.24 Å resolution from 387,192 particle images.
3D variability analysis in cryoSPARC v4.1.2. was performed on the particle images of the consensus 3D map of KEOPS + tRNA while masking the tRNA with a loose mask, revealing the two distinct conformations of the tRNA (native-like and distorted states). The particle images of both states were then individually subjected to non-uniform refinement using the same mask and reference maps from 3DVA resulting in a final density map of native-like tRNA + KEOPS at 3.56 Å resolution from 121,003 particle images and a final density map of the distorted tRNA + KEOPS conformation at 3.59 Å resolution from 83,710 particle images. To examine map anisotropy and preferential orientation, anisotropy analysis was performed in cryoSPARC v4.1.2.on all three maps (i.e. apo, native-like tRNA, and distorted tRNA conformations) using the orientation diagnostics option. To examine tRNA flexibility, the resulting volume series from the 3D variability analysis were used to create a movie in UCSF Chimera V1.16.
Atomic model building and refinement
All the maps (apo KEOPS, native-like tRNA + KEOPS, and distorted tRNA + KEOPS) were locally sharpened with PHENIX71 before modeling. Previously reported atomic models of Cgi121 (7KJT), Kae1 (3ENO), Bud32 (3ENH), PCC1 (3ENO), and tRNA (7KJT) were docked into the maps using phenix.dock_in_map, adjusted manually to fit the experimental maps in Coot72 and then refined in multiple rounds using, phenix.real_space_refine73. Models were validated with PHENIX and the PDB online validation tool74.
Quantification and statistical analysis
Unless indicated otherwise, all reactions in ADP Glo, t6A assays and FP binding assays were performed with three technical replicates. For deriving Kd and IC50 values, each binding curve was performed three times.
Circular dichroism (CD)
CD spectra were recorded on a JASCO J-1500 circular dichroism spectrophotometer using 1.5 mL quartz cuvettes with a 5 mm path length. tRNA samples were diluted to 10 µM in a buffer containing 10 mM Tris pH 8 and 2 mM MgCl2. The measurements were taken at 25 °C in the 200–300 nm wavelength range with a 1 nm data point interval. CD curves were established as an average of three CD measurements using Spectra Manage and the normalized data was plotted using PRISM.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The authors wish to acknowledge Dr. Samir Benlekbir for help with cryo-EM data collection and for advice regarding specimen preparation. Cryo-EM main data set was collected at NYU Langone Health’s Cryo–Electron Microscopy Laboratory (RRID: SCR_019202) and a preliminary data set was collected at the Toronto High Resolution High Throughput cryo-EM facility, supported by the Canada Foundation for Innovation and Ontario Research Fund. Research was supported by the Canadian Institutes of Health Research (PJT-178026 to F.S.) and the Terry Fox Research Institute (TFRI 1107-04 to F.S.)
Author contributions
S.O., J.B. and F.S. conceived the project, prepared the images and wrote the manuscript, with input from all authors. S.O., J.B., and T.B. prepared all reagents. S.O., J.B., and T.B. performed all in vitro biochemical assays. S.O. and J.P. performed all tRNA methylation experiments. M.B. supervised the methylation experiments. S.O., S.D., and J.Y. prepared and imaged all Cryo-EM samples. S. O., A.K., and M.T.M. analyzed all Cryo-EM data. S.O. solved the structures and performed molecular modeling. F.S. secured funding and supervised the project.
Peer review
Peer review information
Nature Communications thanks James M. Murphy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The biochemical and biophysical data generated in this study are provided in the Source Data file provided with this paper. The Cryo EM density maps and other data used in this study are available in the PDB database under accession codes: 8UNK Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1), EMD-42407 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1); 8UP5 Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in its native-like conformation, EMD-42443 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in its native-like conformation; 9D85 Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in a distorted tRNA conformation, EMD-46630 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in a distorted tRNA conformation. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Samara Mishelle Ona Chuquimarca, Jonah Beenstock.
Contributor Information
Jonah Beenstock, Email: beenstock@lunenfeld.ca.
Frank Sicheri, Email: sicheri@lunenfeld.ca.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54787-w.
References
- 1.Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol.22, 375–392 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Vare, V. Y., Eruysal, E. R., Narendran, A., Sarachan, K. L. & Agris, P. F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules7, 10.3390/biom7010029 (2017). [DOI] [PMC free article] [PubMed]
- 3.Gustilo, E. M., Vendeix, F. A. & Agris, P. F. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol11, 134–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons, S. M., Fay, M. M. & Ivanov, P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett.592, 2828–2844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell161, 1606–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res.46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durant, P. C., Bajji, A. C., Sundaram, M., Kumar, R. K. & Davis, D. R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry44, 8078–8089 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lescrinier, E. et al. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res.34, 2878–2886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy, F. V. T., Ramakrishnan, V., Malkiewicz, A. & Agris, P. F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol.11, 1186–1191 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Stuart, J. W. et al. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry39, 13396–13404 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Seelam Prabhakar, P., Takyi, N. A. & Wetmore, S. D. Posttranscriptional modifications at the 37th position in the anticodon stem-loop of tRNA: structural insights from MD simulations. RNA27, 202–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiaville, P. C. et al. Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Micro Cell3, 29–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daugeron, M. C. et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res.39, 6148–6160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Yacoubi, B. et al. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res.37, 2894–2909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, C. A., Ellis, S. R. & True, H. L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell Biol.30, 354–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun, D. A. et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet.49, 1529–1538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Yacoubi, B. et al. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J.30, 882–893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan, M. et al. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J.30, 873–881 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol1, 127–136 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Jin, M. et al. Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases. Nucleic Acids Res.10.1093/nar/gkad587 (2023). [DOI] [PMC free article] [PubMed]
- 21.Lauhon, C. T. Mechanism of N6-threonylcarbamoyladenonsine (t(6)A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry51, 8950–8963 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Su, C., Jin, M. & Zhang, W. Conservation and diversification of tRNA t6A-modifying enzymes across the three domains of life. Int. J. Mol. Sci.23, 13600 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiaville, P. C., Iwata-Reuyl, D. & de Crecy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol.11, 1529–1539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols, C. E. et al. Structural characterization of Salmonella typhimurium YeaZ, an M22 O-sialoglycoprotein endopeptidase homolog. Proteins64, 111–123 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Nichols, C. E. et al. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ-YjeE complex. Protein Sci.22, 628–640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, W., Collinet, B., Perrochia, L., Durand, D. & van Tilbeurgh, H. The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res.43, 1804–1817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deutsch, C., El Yacoubi, B., de Crecy-Lagard, V. & Iwata-Reuyl, D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem.287, 13666–13673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luthra, A. et al. Conformational communication mediates the reset step in t6A biosynthesis. Nucleic Acids Res.47, 6551–6567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luthra, A. et al. Structure and mechanism of a bacterial t6A biosynthesis system. Nucleic Acids Res.46, 1395–1411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Missoury, S. et al. The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification. Nucleic Acids Res.46, 5850–5860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan, L. C. et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res.41, 6332–6346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, J. B. et al. Molecular basis for t6A modification in human mitochondria. Nucleic Acids Res.48, 3181–3194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arrondel, C. et al. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun.10, 3967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edvardson, S. et al. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet.25, 545–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galloway, W. H. & Mowat, A. P. Congenital microcephaly with hiatus hernia and nephrotic syndrome in two sibs. J. Med Genet5, 319–321 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jobst-Schwan, T. et al. Acute multi-sgRNA knockdown of KEOPS complex genes reproduces the microcephaly phenotype of the stable knockout zebrafish model. PLoS One13, e0191503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, C. J. et al. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA21, 2103–2118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojas-Benitez, D., Eggers, C. & Glavic, A. Modulation of the proteostasis machinery to overcome stress caused by diminished levels of t6A-Modified tRNAs in Drosophila. Biomolecules7, 10.3390/biom7010025 (2017).
- 39.Rojas-Benitez, D., Ibar, C. & Glavic, A. The Drosophila EKC/KEOPS complex: roles in protein synthesis homeostasis and animal growth. Fly. (Austin)7, 168–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rojas-Benitez, D., Thiaville, P. C., de Crecy-Lagard, V. & Glavic, A. The levels of a universally conserved tRNA modification regulate cell growth. J. Biol. Chem.290, 18699–18707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Downey, M. et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell124, 1155–1168 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Beenstock, J. et al. A substrate binding model for the KEOPS tRNA modifying complex. Nat. Commun.11, 6233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannan, N., Taylor, S. S., Zhai, Y., Venter, J. C. & Manning, G. Structural and functional diversity of the microbial kinome. PLoS Biol.5, e17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perrochia, L., Guetta, D., Hecker, A., Forterre, P. & Basta, T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res41, 9484–9499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrochia, L. et al. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res.41, 1953–1964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan, L. C. et al. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res.45, 805–817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao, D. Y. et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell32, 259–275 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan, L. C. et al. Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis. Nucleic Acids Res.44, 6971–6980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, X. et al. Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A. Nucleic Acids Res.52, 4523–4540 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, W. et al. Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res.43, 3358–3372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods17, 328–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beenstock, J. & Sicheri, F. The structural and functional workings of KEOPS. Nucleic Acids Res.49, 10818–10834 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kopina, B. J. et al. Structure of a reaction intermediate mimic in t6A biosynthesis bound in the active site of the TsaBD heterodimer from Escherichia coli. Nucleic Acids Res.49, 2141–2160 (2021). [DOI] [PMC free article] [PubMed]
- 54.Parthier, C. et al. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew. Chem. Int Ed. Engl.51, 4046–4052 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Wang, J. T. et al. Commonality and diversity in tRNA substrate recognition in t6A biogenesis by eukaryotic KEOPSs. Nucleic Acids Res.50, 2223–2239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blum, A. D., Uhlenbeck, O. C. & Tinoco, I. Jr. Circular dichroism study of nine species of transfer ribonucleic acid. Biochemistry11, 3248–3256 (1972). [DOI] [PubMed] [Google Scholar]
- 57.Pallan, P. S., Kreutz, C., Bosio, S., Micura, R. & Egli, M. Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G: A pairs. RNA14, 2125–2135 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, H. et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun.9, 1875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morin, A., Auxilien, S., Senger, B., Tewari, R. & Grosjean, H. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: an in vivo study with Xenopus laevis oocytes. RNA4, 24–37 (1998). [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, Y. et al. A natural non-Watson-Crick base pair in human mitochondrial tRNAThr causes structural and functional susceptibility to local mutations. Nucleic Acids Res46, 4662–4676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hecker, A. et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res35, 6042–6051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hurley, J. H. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev. Biophys. Biomol. Struct.25, 137–162 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Kornev, A. P. & Taylor, S. S. Dynamics-driven allostery in protein kinases. Trends Biochem Sci.40, 628–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiesner, S. et al. A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. Embo J.25, 4686–4696 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lorenz, C., Lunse, C. E. & Morl, M. tRNA Modifications: Impact on structure and thermal adaptation. Biomolecules7, 10.3390/biom7020035 (2017). [DOI] [PMC free article] [PubMed]
- 66.Beenstock, J., Ona, S. M. & Sicheri, F. A suite of in vitro and in vivo assays for monitoring the activity of the pseudokinase Bud32. Methods Enzymol.667, 729–773 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Porat, J. et al. Crosstalk between the tRNA methyltransferase Trm1 and RNA chaperone La influences eukaryotic tRNA maturation. J. Biol. Chem.299, 10.1016/j.jbc.2023.105326 (2023). [DOI] [PMC free article] [PubMed]
- 68.Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gehrke, C. W. & Kuo, K. C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr.471, 3–36 (1989). [DOI] [PubMed] [Google Scholar]
- 70.Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol.151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta. Crystallogr D. Struct. Biol.75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.ww, P. D. B. c. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res.47, D520–D528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The biochemical and biophysical data generated in this study are provided in the Source Data file provided with this paper. The Cryo EM density maps and other data used in this study are available in the PDB database under accession codes: 8UNK Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1), EMD-42407 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1); 8UP5 Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in its native-like conformation, EMD-42443 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in its native-like conformation; 9D85 Structure of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in a distorted tRNA conformation, EMD-46630 Cryo-EM map of the KEOPS complex (Cgi121/Bud32/Kae1/Pcc1) bound to tRNA in a distorted tRNA conformation. Source data are provided with this paper.