Abstract
Legumes establish endosymbioses with arbuscular mycorrhizal (AM) fungi or rhizobia bacteria to improve mineral nutrition. Symbionts are hosted in privileged habitats, root cortex (for AM fungi) or nodules (for rhizobia) for efficient nutrient exchange. To reach these habitats, plants form cytoplasmic cell bridges, key to predicting and guiding fungal hyphae or rhizobia-filled infection thread (IT) root entry. However, the underlying mechanisms are poorly studied. Here we show that unique ultrastructural changes and calcium (Ca2+) spiking signatures, closely associated with Medicago truncatula Annexin 1 (MtAnn1) accumulation, accompany rhizobia-related bridge formation. Loss of MtAnn1 function in M. truncatula affects Ca2+ spike amplitude, cytoplasmic configuration and rhizobia infection efficiency, consistent with a role of MtAnn1 in regulating infection priming. MtAnn1, which evolved in species establishing intracellular symbioses, is also AM-symbiosis-induced and required for proper arbuscule formation. Together, we propose that MtAnn1 is part of an ancient Ca2+-regulatory module for transcellular endosymbiotic infection.
Subject terms: Plant symbiosis, Rhizobial symbiosis, Calcium signalling
This study describes ultrastructural changes and cell-specific calcium responses that accompany cytoplasmic bridge formation in legume root cells committed to symbiotic rhizobial infection, with fine-tuning by a calcium-binding annexin.
Introduction
Plants benefit from associations with microbes in the soil which improve the acquisition of essential nutrients for growth. These associations include the ancestral symbiosis with arbuscular mycorrhizal (AM) fungi providing phosphorous, or the more recently evolved interactions with endosymbiotic bacteria for nitrogen acquisition1–3. Microbial endosymbionts are hosted in privileged intracellular niches, where optimal conditions are met for efficient nutrient exchange (e.g. the root cortex hosting arbuscules formed by AM fungi, or root nodules, hosting nitrogen-fixing bacteria). Extensive studies on model legumes such as Medicago truncatula, combined with evolution-based studies across a broad species range revealed that nodule-forming nitrogen-fixing endosymbiosis (RNS) evolved ~100 million years ago in species from related angiosperm plant lineages, by co-opting signalling components from the ancient AM symbiosis and recruiting new key genes through neo-functionalization and/or rewiring of expression1,3,4. These endosymbioses share a common symbiotic pathway that uses calcium (Ca2+) as a key secondary messenger to trigger downstream signalling5,6. Upon perception of mycorrhizal Myc and rhizobial Nod factor signals, Ca2+ oscillations (spiking) are triggered in and around the root hair nucleus through the concerted action of nuclear envelope channel/pump complexes5–7. While Ca2+ spiking is critical in the nucleus (after its decoding by Ca2+/calmodulin-dependent protein kinases CCaMK or Does not Make Infection, DMI3)8,9 to regulate host transcriptional reprogramming10–12, the functional relevance of sustained Ca2+ spiking in the cytoplasmic compartment has not yet been established. The rhizobia- and AM-induced MtAnn113 and MtAnn214 genes, which encode cytosolic Ca2+-binding annexins that can bind to membranes to modulate a variety of Ca2+-regulated processes (e.g. ion conductance, exocytosis/endocytosis)15,16, are potential players in this process.
Following signal exchange to reach compatibility, infection of root tissues by AM fungi or rhizobia bacteria occurs in most legumes transcellularly, through the de novo construction of apoplastic compartments, delimited by host cell wall/ membrane interfaces, which physically separate microsymbionts from the host cytoplasm17,18. The hyphae of AM fungi progress through epidermal and cortical root tissues until the fungus reaches the inner cortex to form highly branched arbuscules inside plant cells. Arbuscules comprise a massive network of membrane tubules at the symbiotic host-fungus interface19,20, which likely ensures the release of nutrients, particularly phosphate, to the plant. In RNS, rhizobia enter the roots of most legume species via apoplastic tubular structures called infection threads (ITs)18,21,22 that grow from root hairs to the developing nodule underneath23 under the influence of key plant hormones, intercellular signalling and long-distance regulatory pathways24–26.
ITs are formed in root hairs after rhizobia are entrapped and enclosed in a globular infection chamber27. This plant-driven process requires targeted cell wall remodelling at the IT interface27,28 for polar growth, under the control of the infectosome protein complex29–31. Transcellular IT infection also imposes tight regulation of the plant’s cell cycle status through the recruitment of mitotic regulators in root hairs and acquisition of specific pre-mitotic states before and during IT passage in the cortex32–34. Finally, the route of IT entry is also fine-tuned by the host, which creates a broad cytoplasmic column filled with endoplasmic reticulum (ER) and surrounded by microtubule arrays to guide IT progression35,36. In root hairs, this bridge connects the tip of the IT to the nucleus to direct IT elongation. In the adjacent cortex, differentiated root cells with a large central vacuole enter an activated state (hereafter termed pre-infection priming), with characteristic transvacuolar cytoplasmic strands emanating from the nucleus before a transvacuolar cytoplasmic bridge, the pre-infection thread (PIT37), forms in anticipation of future IT passage. PIT bridges have been reported to occur in nodules of several legume and actinorhizal species37–39. Moreover, PIT bridges closely resemble Pre-Penetration Apparatus (PPA)17,40 cytoplasmic columns that precede AM fungal hyphae transcellular passage. The fact that PIT/PPA-like remodelling is not seen in pathogen interactions41, suggests their specific and probably ancient recruitment for endosymbiosis. Parallels have been drawn between PIT/PPA and premitotic phragmosome transvacuolar bridges formed in cells preparing for mitosis, as these processes share similarities in tightly controlling plant cell division status, membrane trafficking and microtubule dynamics32,34,39,42.
The development of fluorescent protein fusions labelling the ER network has made it possible to visualise the dynamics of ER-enriched bridge remodelling in vivo during PPA/PIT formation35,40,43. Complementary in vivo studies using Cameleon Ca2+ sensors revealed that primed cortical cells exhibit a low-frequency Ca2+ spiking signature that switches to high frequency at the onset of infection by AM fungi or rhizobia43. CCaMK/DMI3 is critical for PPA formation in Medicago41. Moreover, a gain-of-function variant in Lotus can trigger cytoplasmic remodelling in the cortex in the absence of symbionts44. Thus, CCaMK/DMI3-mediated signalling can bypass early symbiotic activation to trigger the priming response in the cortex. These studies suggest that cytoplasmic bridge reprogramming involves fine-tuning of Ca2+ spiking signatures, but the underlying genetic pathways or molecular players have not been identified.
In this study, using resolutive microscopy methods in different M. truncatula genetic backgrounds, we provide in-depth insights on ultrastructural rearrangements and in vivo Ca2+ signalling dynamics in plant cells preparing for rhizobia infection. We show that an intimate link between specific Ca2+ spiking signatures and the accumulation of annexin MtAnn1 characterises this priming response. MtAnn1 is a clear indicator of cytoplasmic reprograming for infection, and its expression in this context is induced by the master symbiotic transcription factor Nodule Inception (NIN)1. Through phylogenetics and mutant phenotyping, we provide evidence that MtAnn1, recruited during evolution in plants establishing root endosymbioses, likely plays an ancestral role for successful rhizobia and AM fungi infection. We propose that MtAnn1 was recruited to shape Ca2+-regulated transvacuolar cytoplasmic rearrangements for efficient endosymbiotic infection.
Results
Cell-specific ultrastructural changes in plant cells prior to IT entry
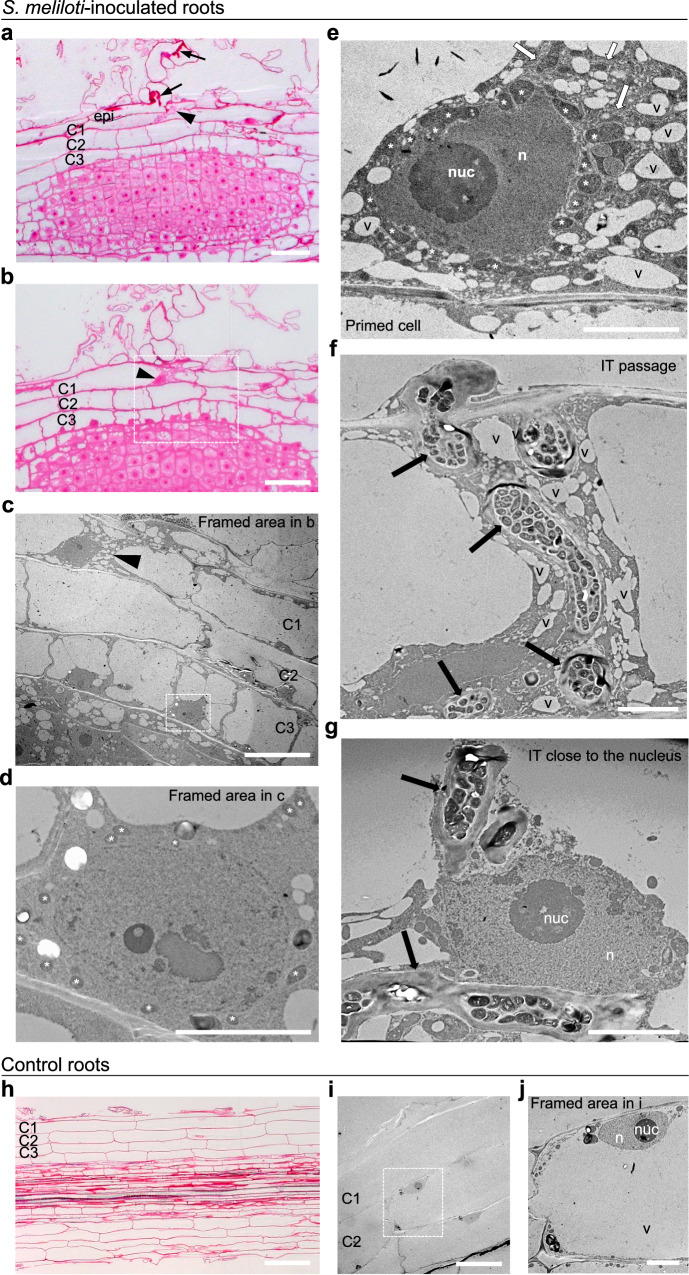
PIT bridge formation is a widespread process in RNS nodules37,38, but detailed studies of this process are lacking. Previous work in M. sativa and M. truncatula skl mutant roots showed that microtubule rearrangements accompany PIT formation39, however overall cellular features underlying this process remain unknown. Here, we set up an adapted methodology to gain access to ultrastructural changes underlying rhizobia pre-infection priming in M. truncatula roots. Segments of early infected roots, harvested 5–6 dpi with rhizobia, were prepared for light (Fig. 1a, b and Supplementary Fig. 1a) and transmission electron microscopy (TEM) (Fig. 1c–g) using an Epon-embedding procedure. These analyses revealed specific cytoplasmic rearrangements in primed outer cortical cells (C1) next to an infected root hair site (Fig. 1a–c) compared to mock control (Fig. 1h–j) root samples. TEM analyses showed that the thick cytoplasmic bridge was highly enriched in ER (white arrows, Fig. 1e) and large quantities of small vacuole-like structures with sizes ranging from 0.5 to 5 µm (v in Fig. 1e). These activated cells also featured significant and unusually high mitochondrial clustering (asterisks, Fig. 1e) around the nucleus. This mitochondria-nuclei association was consistently observed in independent pre-infection (PI) primed cells compared to actively dividing cells in the nodule primordia (NP) (Fig. 1d, e and Supplementary Fig. 1b–k). At a later stage, when the IT had progressed across this small vacuole- and ER-rich cytoplasmic bridge (Fig. 1f), the nucleus was often found in close proximity to the IT (Fig. 1g). Thus, the plant nucleus not only guides the progression of ITs but also establishes close physical interactions with them. Overall, drastic ultrastructural changes take place specifically in outer cortical cells (C1 or C2) primed for rhizobia infection, namely a unique transcellular cytoplasmic organisation, enriched with ER, small vacuole-like structures and a significant increase in mitochondria-nucleus association.
Fig. 1. Cell-specific structural changes in M. truncatula cells primed for infection.
a, b Representative images of longitudinal sections of M. truncatula A17 roots 5 days after inoculation with S. meliloti. These are consecutive 1 µm sections stained with Basic Fuchsin to reveal cell outlines and contents. Arrows indicate ITs in root hair epidermal cells and arrowheads indicate a C1 cortical cell exhibiting pre-infection priming. After analysis of 200–250 sections (1 µm) per S. meliloti-inoculated root segment, 7 individual pre-infection priming events were captured, 5 of which were further analysed by TEM (data are from 3 independent experiments). The primed cell (arrowhead) and framed area in (b) are shown in 80 nm TEM sections in (c–e). Note that the primed C1 outer cortical cell (arrowhead in a–c and enlarged image in e) exhibits a unique organisation compared with neighbouring cells. Primed cells have a cytoplasmic bridge enriched in endoplasmic reticulum (white arrows in e), and numerous vesicles or small vacuole-like structures (v). They also have a large nucleus (n) with mitochondria (asterisks) clustering around (quantified in Supplementary Fig. 1). f, g At a later stage, when the IT (arrows) has progressed across the cytoplasmic bridge towards inner tissues, the nucleus of the crossed cell appears closely associated to the IT (n = 4, 2 independent experiments) (g). h–j Representative images of M. truncatula A17 root sections after mock (water) treatment for 5 days (2 independent experiments). h A 1 µm longitudinal section of a mock control root stained with Basic Fuchsin. i, j TEM of 80 nm sections of a mock control root. The framed area in (i) is shown in (j). Note that non-symbiotically activated cells have a large vacuole and nuclei near the cell periphery. Abbreviations: cortical cells (C1, C2, C3), nucleolus (nuc). Scale bars: a, b, h = 50 µm, c, i = 20 µm, d–g, j = 5 µm. See also Supplementary Fig. 1. Source data are provided as a Source Data file.
MtAnn1 and Ca2+ spiking dynamics mark symbiotically engaged and infected plant cells
Ca2+ spiking responses are transiently regulated in cortical cells preparing for infection43, but how these responses relate to cytoplasmic remodelling is unknown. To study this, we used an in vivo approach based on co-imaging a fluorescent Ca2+ sensor and a cytoplasmic marker to simultaneously monitor Ca2+ spiking and cytoplasmic remodelling in individual cells. We used M. truncatula A17 (wild-type) and the supernodulating mutant sunn, which shows wild-type infection but at higher levels, facilitating live imaging studies of early rhizobia infection events27,29,45. These in vivo analyses cannot provide as much data from the wild-type background in which infection thread events are more rare than in sunn. However, results were confirmed in the two genetic backgrounds (A17, sunn) by independent experiments. Furthermore, as sites of interest were individually imaged multiple times in living roots, responses were dynamically monitored and confirmed overtime by follow-up observations.
We used the red fluorescent nuclear NR-GECO1 sensor for Ca2+ imaging because of its better compatibility with the green fluorescence cytoplasmic marker and higher sensitivity to detect symbiotic Ca2+ spiking, as compared to a Förster resonance energy transfer (FRET) cameleon sensor46. To validate this sensor in our system, we first monitored Ca2+ spiking in S. meliloti-responsive root hairs of the nodulation susceptible zone, just below the infection zone, in both A17 and sunn, 1–4 dpi with rhizobia. Using NR-GECO1, we confirmed that S. meliloti triggers periodic, high-frequency Ca2+ spiking in these root hairs with profiles indistinguishable from those induced by purified Nod factors, as previously reported46,47 (Supplementary Fig. 2 and Movies 1, 2). Subsequent analysis of rhizobia-infected sites in sunn at 2–7 dpi, revealed specific low frequency Ca2+ spiking profiles in the outer cortex underlying rhizobia infection sites (Supplementary Fig. 3). Cortical cells that spiked included those that later became infected (arrowhead, Supplementary Fig. 3). Furthermore, only cortical cells in close contact with the infected epidermal cell spiked, while other distant cortical cells did not. This suggests that potential short distance signalling from the infected cell is required for pre-infection priming of cortical cells. Our results are consistent with previous data using a FRET-cameleon Ca2+ probe43, and confirm that low-frequency Ca2+ spiking is associated with rhizobial pre-infection priming of cortical cells.
MtAnn1, encoding a cytosolic Ca2+ and membrane-binding annexin protein, is induced by rhizobia both in root hairs and in outer cortical cells preparing for infection13,48, where Ca2+ spiking responses are coincidently transiently regulated (Supplementary Fig. 3)43. Thus, the use of a cytosolic MtAnn1-GFP fusion expressed under its native rhizobia-induced promoter seemed a suitable tool to monitor cytoplasmic remodelling during rhizobia infection. Comparative localisation analyses were performed in M. truncatula A17 and/or sunn genotypes using a GFP-ER marker, which labels the cytoplasmic ER endomembrane network and the MtAnn1-GFP fusion. This revealed similar labelling of root hair cytoplasmic bridges by both markers (Supplementary Fig. 4). Co-localisation studies confirmed that MtAnn1-GFP and a red mcherry-ER fusion co-labelled cytoplasmic bridges in both root hair (Supplementary Fig. 5) and cortical cells (Supplementary Fig. 6). MtAnn1-GFP fusion expressed under native MtAnn1 promoter specifically labelled cytoplasmic bridges of pre-infection-primed (C1a in Supplementary Fig. 6) and infected OC cells (C1b in Supplementary Fig. 6), whereas no labelling was observed in surrounding OC cells expressing cytoplasmic red mCherry-ER marker (driven by p35S). Combined, these data support the conclusion that the MtAnn1-GFP fusion specifically marks cytoplasmic bridges, where it partially co-localises with the ER network.
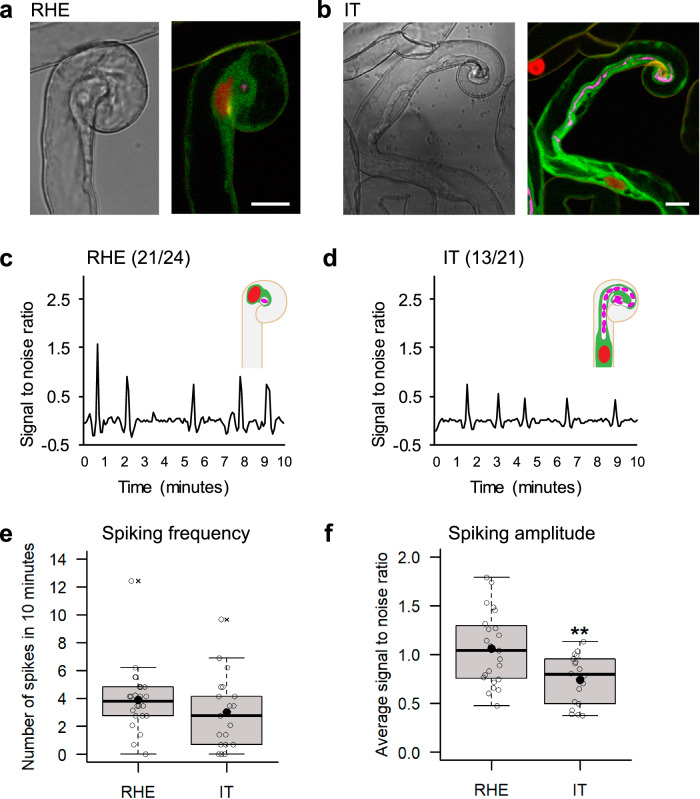
Co-expression of red NR-GECO1 Ca2+ sensor and green MtAnn1-GFP fusion was then used to live track the spatio-temporal dynamics of Ca2+ spiking and MtAnn1 accumulation simultaneously in M. truncatula A17 (Fig. 2). The MtAnn1/NR-GECO1 double-labelling greatly facilitated detection of root hairs in early stages of bacterial entrapment (RHE) and IT polar growth (IT) (Fig. 2a, b). RHE- or IT- root hairs consistently showed both MtAnn1-GFP fusion signal and high frequency Ca2+ spiking profiles (Fig. 2a–e). However, the amplitude of Ca2+ spikes dropped significantly from RHE to IT stage root hairs (Fig. 2c, d, f). Conversely, relative levels of MtAnn1-GFP fluorescence tended to increase in IT versus RHE root hairs (Fig. 2a, b and Supplementary Fig. 7), consistent with root hair transcriptome data48. Overall, our data suggest that decreased Ca2+ spiking amplitude and increased MtAnn1 levels are associated with polar IT growth.
Fig. 2. Ca2+ spiking amplitude drops along root hair IT development.
a, b Representative bright-field and corresponding confocal fluorescence images of a a root hair with entrapped CFP expressing (magenta)-S. meliloti (RHE) and b a root hair with a growing infection thread (IT) in sunn. Nuclei expressing the NR-GECO1 Ca2+ sensor appear in red, and the MtAnn1-GFP fluorescence (green) labels the cytoplasmic zone around the IT and the cytoplasmic bridge connecting and surrounding the nucleus. Representative Ca2+ spiking traces of RHE (c) and growing IT stages (d) in sunn. The relative concentration of Ca2+ ions in the nucleus is reflected by the intensity of NR-GECO1 fluorescence, expressed as signal-to-noise ratio (SNR, cf. ‘Methods’ section). The number of root hairs with nuclear spiking/total number of root hairs are indicated between parentheses. Nuclei are counted as spiking when showing more than 2 peaks in 10 min. Quantification of nuclear Ca2+ spiking: spiking frequency (e), expressed as number of spikes in 10 minutes per nucleus, and spiking amplitude (f), expressed as average SNR of spikes per nucleus. Box plots represent the distribution of individual values (indicated by open circles) from root hairs with entrapped rhizobia RHE, (n = 24 in e and n = 23 in f) or with an IT (n = 21 in e and n = 18 in f) in sunn, 2–4 dpi with S. meliloti from 3 independent experiments. First and third quartile (horizontal box edges), minimum and maximum (outer whiskers), median (centreline), mean (solid black circle) and outliers (crosses) are indicated. Differences were not significant in spiking frequency in IT vs. RHE in (e) (p = 0.1531, two-tailed Mann-Whitney test). Asterisks indicate statistically significant differences in spiking amplitude in RHE vs. IT in (f) (p = 0.0037, two-tailed t-test). Scale bars: a, b = 10 µm. See also Supplementary Fig. 2 and Movies 1, 2 for Ca2+ spiking responses in A17 and/or sunn root hairs, and Supplementary Figs. 4–7 for MtAnn1-GFP localisation and fluorescence quantification in root hair cytoplasmic bridges. Source data, including split channels for merged fluorescence, are provided as a Source Data file.
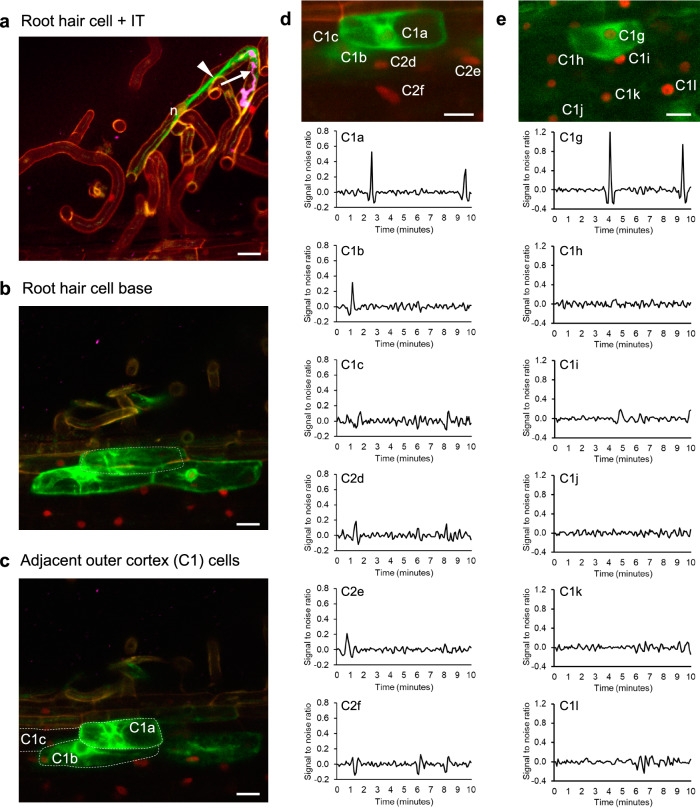
MtAnn1-GFP also labelled cytoplasmic bridges of cortical cells primed for infection in both wild-type A17 and sunn (Fig. 3a–c, Supplementary Figs. 6 and 8). MtAnn1-GFP-labelled cells were those in close contact with the infected root hair, which consistently showed low frequency Ca2+ spiking (Fig. 3d, e). Furthermore, this low-frequency Ca2+ spiking profile switches to a high-frequency pattern when the cortical cell is infected (Supplementary Fig. 8), consistent with previously published data43. Strikingly, nearby cortical cells not labelled with MtAnn1 did not show any spike in both A17 and sunn (Fig. 3b–e and Supplementary Fig. 8). Collectively, our data establish that IT development and pre-infection priming involve stage- and cell-type-specific Ca2+ spiking signatures closely linked to MtAnn1-GFP bridge accumulation.
Fig. 3. Strong MtAnn1-GFP fusion fluorescence and low frequency Ca2+ spikes are hallmarks of pre-infection priming in the cortex.
a–e Representative images of rhizobia infection sites in roots co-expressing MtAnn1-GFP (green) and NR-GECO1 (red) in M. truncatula A17. a–c Confocal images illustrate an infected root hair (CFP-labelled rhizobia in the IT, arrow, in magenta) and the nucleus (n) in front, guiding the growth of the IT in a cytoplasmic bridge (arrowhead) (a), the base of the infected root hair cell (b, grey dotted line) and the neighbour C1a-c outer cortical cells (c, dotted lines). C1a-C1c are also shown in (d). d, e Representative Ca2+ spiking traces, expressed as signal-to-noise ratio (SNR), from outer cortical cells (C1a-c, C2d-f, C1g-l, top panels) adjacent to two independent infected root hair sites. Cortical cells C1a-c, C2d-f are adjacent to the infection site shown in (a, b) (C1a-c are also shown in c), while C1g-l cells are adjacent to another root hair infection site (not shown). Traces of cortical cells that are in direct contact (C1a, C1b, C1g) or not (C1b-c, C2d-f in d and C1h-l in e) with the infected root hair site are shown. Images were obtained from A17 roots 4 dpi (a–d) or 3 dpi (e) with CFP-labelled S. meliloti. Data were obtained from 2 independent experiments after the analysis of Ca2+ traces in n = 12 individual cells. Note that only cortical cells co-expressing MtAnn1-GFP (C1a-b in d, C1g in e) show detectable low frequency Ca2+ spiking. Images in a-c are maximal z-projections of sub-stacks and images in d-e are maximal projections of whole time series. Abbreviations: first and second outer cortex layers (C1, C2), infection thread (IT). Scale bars: a–e = 20 µm. See also Supplementary Figs. 3, 6 and 8, 9 for Ca2+ spiking responses and MtAnn1-GFP dynamics in primed cells in the cortex in sunn and in infection-defective ern1 and dmi3 mutants. Source data, including split channels for merged fluorescence, are provided as a Source Data file.
Rhizobia root hair infection is required for pre-infection priming of adjacent cortical cells
MtAnn1-GFP labelling and Ca2+ spiking mark primed outer cortical cells (Fig. 3 and Supplementary Fig. 8), but it is unclear if IT formation is required for this process. Co-expression of NR-GECO1 and MtAnn1-GFP in infection-defective ern1 (ERF Required for Nodulation 1)49–51, revealed that Ca2+ spiking and cytoplasmic-bridge formation are still seen in root hairs with arrested chambers (Supplementary Fig. 9a–c). MtAnn1-GFP labelling and Ca2+ spiking were also seen in the outer cortex adjacent to root hairs with arrested-infections in ern1 (Supplementary Fig. 9d–f), but the pattern appeared to be deregulated (more cells labelled with MtAnn1-GFP or Ca2+ spiking sometimes in MtAnn1 unlabelled cells, never seen in the wild-type control A17 or sunn, Fig. 3 and Supplementary Fig. 8). This suggests that ERN1 is not required to promote pre-infection priming, but in its absence the priming response is deregulated.
CCaMK/DMI3 is critical for PPA formation in Medicago41, however, it is unknown if it is also required for PIT formation. Since dmi38 is impaired in root hair infection, it is not possible to access PIT formation in this genetic background. We thus took advantage of stable transgenic lines complemented with DMI3 only in epidermis52, to investigate if Ca2+ spiking and MtAnn1-GFP could still label cortical cells in this line compared to wild-type A17 control (Fig. 3). Complemented dmi3 pEXT:DMI3 lines showed Ca2+ spiking, rescued IT development and MtAnn1 labelling in root hairs (Supplementary Fig. 9g and Movie 3). However, no Ca2+ spiking or MtAnn1-GFP labelling was seen in the cortex (Supplementary Fig. 9h), compared to the control A17 or sunn (Fig. 3 and Supplementary Fig. 8). No cytoplasmic remodelling in the cortex of dmi3 pEXT:DMI3 could be detected by further analysis of a constitutively expressed GFP (Supplementary Fig. 9i, j). Thus, MtAnn1-labelled cytoplasmic bridges were clearly visible only in infected root hairs of dmi3 pEXT:DMI3 and not in adjacent cortical cells (no MtAnn1 labelling or GFP-labelled cytoplasm remodelling, Supplementary Fig. 9). This suggests that PIT infection priming uses a DMI3-dependent pathway that must be active in the cortex.
Taken together, these analyses indicate that root hair infection chamber formation is sufficient in ern1 to trigger MtAnn1-GFP-labelled bridge in the root hair itself and adjacent cortex. The absence of cortical priming in dmi3 is consistent with the conclusion that, like in PPA, DMI3 is required for PIT reprogramming.
NIN controls pre-infection priming and the associated expression of MtAnn1
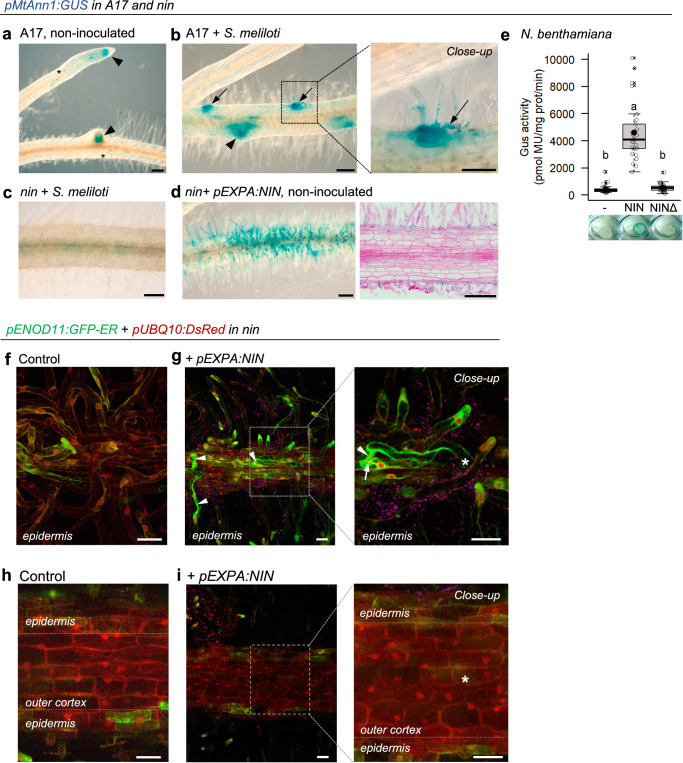
The master symbiotic transcription factor Nodule INception (NIN) is genetically downstream of DMI3 and its mutation abolishes rhizobia-induced MtAnn1 expression48,53. To test if NIN is required for MtAnn1 expression, we compared the spatio-temporal expression profile of a pMtAnn1:GUS fusion in M. truncatula A17 wild-type and nin roots. In control conditions, the pMtAnn1:GUS fusion was active in root tips, lateral roots and some endodermal cells (Fig. 4a). Upon S. meliloti inoculation, strong promoter activity was detected at rhizobial infection sites, both in root hairs and outer cortex (Fig. 4b), and later in other nodule primordium cortical cells13 but not in nin (Fig. 4c). This confirms the NIN-dependent control of MtAnn1 expression. Conversely, epidermal expression of NIN (pEXPA:NIN 54) resulted in pMtAnn1:GUS fusion activity in the absence of rhizobia in nin (Fig. 4d). This indicates that NIN can bypass early symbiotic signalling to promote pMtAnn1:GUS activity. Transactivation assays in Nicotiana benthamiana (Fig. 4e and Supplementary Fig. 10) showed that NIN can transcriptionally activate pMtAnn1:GUS. This activation was abolished when a NIN mutated version was used (Fig. 4e and Supplementary Fig. 10). These data show that the expression of MtAnn1 in rhizobia infection sites is under NIN dependence and suggests that NIN is a possible direct MtAnn1 regulator.
Fig. 4. NIN is required for pre-infection priming and MtAnn1 expression.
pMtAnn1:GUS activity in roots of M. truncatula A17 (a, b) or nin (c, d) inoculated or not with S. meliloti at 4 dpi. GUS activity (blue) is visualised in endodermis (asterisks), root tips or lateral roots (arrowheads) and rhizobial infection sites (arrows). LacZ-expressing rhizobia are in magenta (Close-up in b). c Infection-induced pMtAnn1:GUS activity is abolished in nin. d NIN under pEXPA promoter induces pMtAnn1:GUS activity in root epidermis without rhizobia, also shown in Basic Fuchsin counterstained 10 µm sections. Data are from two independent experiments (A17, n = 41; nin, n = 31; nin + pEXPA:NIN, n = 37). e Transactivation assay of pMtAnn1:GUS with NIN or NINΔ (DNA-binding domain deletion version) in N. benthamiana leaves. Box plots show distribution of values (open circles) of individual plants (n = 25 per sample) from 4 independent experiments. First and third quartile (horizontal box edges), minimum and maximum (outer whiskers), median (centerline), mean (solid black circle) and outliers (crosses) are indicated. Different letters above boxes indicate statistically significant differences (p < 2e-16, by One-way ANOVA α = 5% followed by Tukey honest significant difference tests). Representative leaf disc images are shown. Expression of pENOD11:GFP-ER and pUBQ10:DsRed fusions in nin control (+pEXPA:GUS) (f, h) and complemented (+pEXPA:NIN) roots (g, i). Red DsRed fluorescence labels cytosol and nucleoplasm. Green GFP-ER fluorescence labels ER network and growing root hair tips (f, g), and rhizobia-induced cytoplasmic columns in complemented roots (arrowhead in g). Arrow indicates enclosed rhizobia (RHE). Cortical cells of control or epidermally-complemented nin do not show GFP-ER or DsRed-labelled cytoplasmic rearrangements (h, i), whereas these are clearly visible with DsRed in sunn (see Supplementary Fig. 11). Data are from 2 independent experiments (n = 7 for nin + pEXPA:GUS and n = 19 for nin + pEXPA:NIN). Asterisks mark xy positions of infected root hair base. Scale bars: a–d = 100 µm, f–i = 40 µm. See also Supplementary Figs. 10, 11. Source data, including split channels for merged fluorescence, are provided as a Source Data file.
nin fails to form infection chambers and ITs but can still trap rhizobia in a curled root hair27. The question remains if a cytoplasmic bridge can still form in nin. To assess this, we examined cytoplasmic bridge formation in nin transformed with pEXPA:NIN or a pEXPA:GUS control (Fig. 4f–i). Since MtAnn1 is transcriptionally dependent on NIN (Fig. 4a–e), we could not use the pMtAnn1:MtAnn1-GFP fusion for these studies, but instead used a pENOD11:GFP-ER fusion co-expressed with pEXPA:NIN or pEXPA:GUS (control) in nin. Green GFP-ER fluorescence marked the perinuclear ER network and the tip of growing root hairs in control and NIN-complemented samples (Fig. 4f, g). However, thick cytoplasmic bridges marked by GFP-ER were only detected in NIN-complemented root hairs (Fig. 4g). No GFP-ER signal was observed in the root cortex of either control or NIN-complemented roots (Fig. 4h, i). Moreover, the constitutive DsRed marker was also unable to detect cytoplasmic remodelling in nin (Fig. 4h, i), whereas this marker allowed visualisation of remodelling in control sunn roots (as illustrated in Supplementary Fig. 11). This suggests that a functionally active NIN in the cortex is required to trigger cytoplasmic remodelling.
Together, these data infer that NIN acts downstream of DMI3 to control MtAnn1 gene transcription (possibly directly) and pre-infection priming.
MtAnn1 mutation affects efficiency of rhizobial infection and nodule differentiation
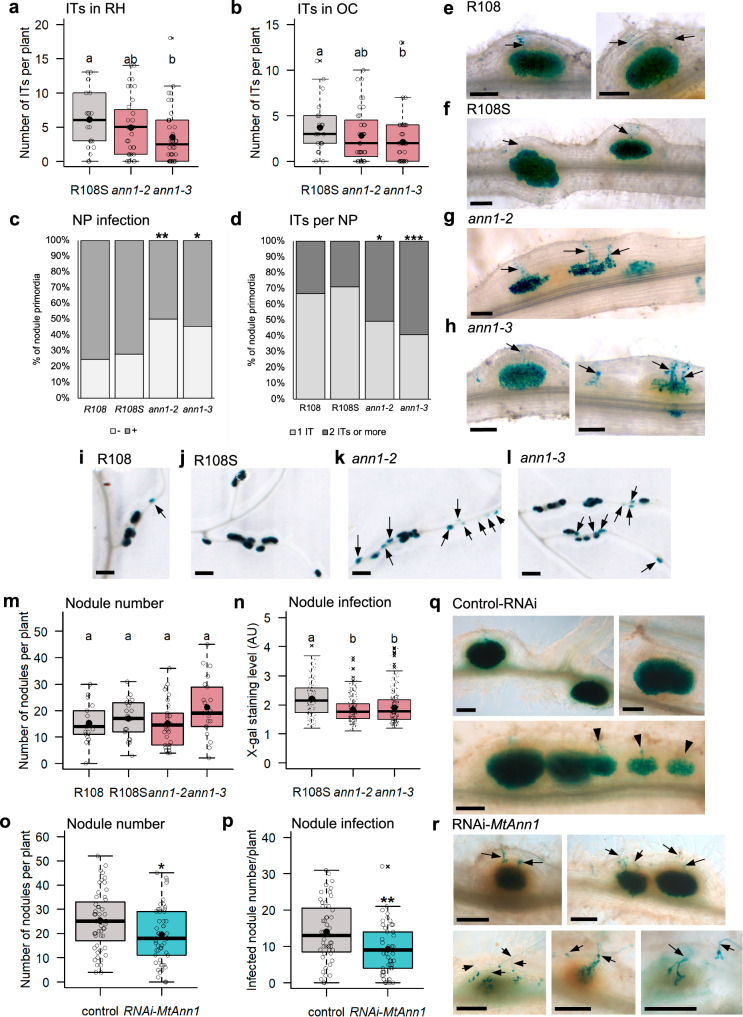
Since MtAnn1 expression is closely linked to early stages of rhizobial entry, we investigated the impact of its mutation on rhizobial infection by analysing M. truncatula Tnt1 R108 insertion mutant lines (Supplementary Fig. 12). Mutant lines showed reduced (ann1-1) or nearly abolished (ann1-2 and ann1-3) MtAnn1 but not MtAnn2 expression compared to R108 and R108S, a wild-type sibling from the same mutant population (Supplementary Fig. 12a–c). Rhizobia infection was assessed in ann1-2 and ann1-3, where MtAnn1 expression was most reduced, by monitoring bacterial β-galactosidase reporter activity in root and nodule tissues. ITs formed in root hairs and progressed into the cortex in mutant roots, but with lower efficiency (Fig. 5a, b). This did not affect nodule numbers, but their infection levels and relative VPY expression (Fig. 5c–n and Supplementary Fig. 12d, e). At 4 dpi, there was a consistent decrease in nodule primordia (NP) colonisation in mutants (50–55% of infected NP (+) in mutants, compared to 70–75% in wild-type lines) (Fig. 5c). These NPs were often infected by multiple ITs compared with controls (Fig. 5d–h), suggesting an inefficient process. At 10 dpi, mutant roots showed a mixture of well and poorly colonised nodules, not seen in wild-type R108/R108S (Fig. 5i–l). At this stage, nodule numbers were unaffected in mutant roots, but infection levels were significantly reduced (Fig. 5m, n). RNAi knockdown of MtAnn1 in M. truncatula A17 resulted in a significant reduction in both NP/nodule number and infection at 5 dpi (Fig. 5o, p), consistent with the mutant infection phenotype. The further reduction in nodule number, not clearly seen in ann1-3 mutant, may be due to the double MtAnn1 and MtAnn2 knockdown in RNAi roots (Supplementary Fig. 12f, g). Infected nodules/NP in RNAi roots show the same multiple ITs above as in ann1 mutants (Fig. 5q, r), as well as abnormal NPs with poor or no colonisation, with several associated root hair ITs (arrow, in Fig. 5r), not seen in control NPs (Fig. 5q, arrowhead). Overall, impaired MtAnn1 function in mutant and RNAi contexts affected rhizobial root and nodule infection, in R108 and A17 M. truncatula backgrounds.
Fig. 5. Root infection is negatively impacted in ann1 mutant and RNAi transgenic roots.
Infection of M. truncatula R108 mutant (a–n) and A17 RNAi roots (o–r) for MtAnn1 with lacZ-expressing (blue) S. meliloti. At 4 dpi, quantification of the number of ITs in root hairs (a) and reaching outer cortex (b), proportion of non-infected (-) vs. infected (+) nodule primordia (NP) (c) and NPs with 1 or more ITs (d), as depicted in (e–h) (R108 n = 17, R108S n = 21, ann1-2 n = 36 and ann1-3 n = 38). At 10 dpi (i–l), quantification of nodule number/plant (R108 n = 18, R108S n = 21, ann1-2 n = 30, ann1-3 n = 21) (m) and infection level (X-gal staining intensity) (n) (R108S n = 196, ann1-2 n = 295, ann1-3 n = 310). Arrows indicate ITs (e–h) and poorly-infected nodules (i–l). Quantification of number of nodules/NP (o) and infection level (p) in pMtAnn1:GUS control (n = 51) and pMtAnn1:RNAi-MtAnn1 (n = 50) roots at 5 dpi (q, r). Control (arrowheads) and RNAi ITs (arrows) are indicated. Box plots (a, b, m–p) show the distribution of values (dots or circles) from 2 (a–d, m, n) or 3 (o, p) independent experiments. First and third quartiles (horizontal box edges), minimum and maximum (outer whiskers), median (centerline), mean (solid black circle) and outliers (crosses) are shown. Classes with the same letter (a, b, m, n) are not significantly different (p = 0.0388 in a, p = 0.0449 in b, p = 9,17e-13 in n, Kruskal-Wallis α = 5%; p = 0,076 in m, one-way ANOVA). Asterisks (c, d, o, p) indicate statistical difference relative to R108S (in c, p = 0.8408 for R108, p = 0,0027 for ann1-2, p = 0,0198 for ann1-3; in d, p = 0.7947 for R108, p = 0.0226 for ann1-2, p = 0.001 for ann1-3, two-tailed Fisher’s exact tests) or control (p = 0.0176, two-tailed Student t-test in o; p = 0.0023, two-tailed Mann-Whitney test in p). Scale bars: e–h, i–l = 100 µm, q, r = 1 mm. See Supplementary Figs. 12, 13. Source data are provided as a Source Data file.
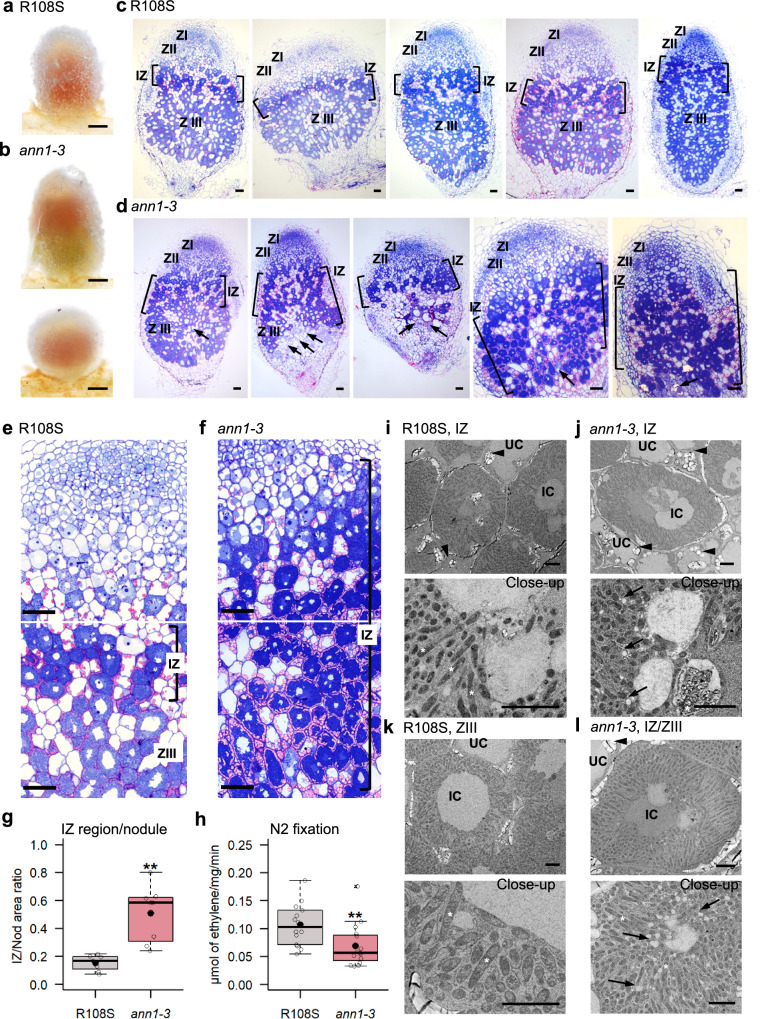
While ann1 mutants showed no major changes in root architecture or nodule number, striking variations in shape (less elongated overall) and colour (paler pink) were seen at 14–16 and 21 dpi (Fig. 6a, b and Supplementary Fig. 13), reminiscent of potential differentiation and/or nitrogen fixation defects55. To better understand why ann1 nodules were smaller and less elongated, we analysed thin and ultrathin sections of wild-type and mutant nodules. As the two mutant alleles led to similar nodulation defects and the wild-type sibling (R108S) behaved reliably as R108 (see Fig. 5 and Supplementary Figs. 12, 13), we chose to perform sensitive thin/ultrathin sections on one representative of each genotype (R108S and ann1-3) (Fig. 6c–f). Meristematic (ZI), infection (ZII), amyloplast-rich interzone (IZ) and nitrogen fixation (ZIII) zones were clearly distinguished in R108S nodules (Fig. 6c, e and Supplementary Fig. 14). However, ann1-3 nodules showed variable zonation patterns, either wild-type-like or with significantly abnormally large IZ regions, sometimes combined with senescent cells (arrows) (Fig. 6d, f and Supplementary Fig. 14). This was accompanied by a significant difference in the IZ area and nitrogen-fixing capacity of mutant nodules compared to wild-type R108S nodules (Fig. 6g, h), which ultimately led to reduced plant growth (Supplementary Fig. 13). Although ann1-3 nodules formed bacteroids, they did not appear to complete their differentiation in Zone III-like bacteroids and instead remained interzone-type bacteroids56, imbedded in a white vesicle-enriched region (black arrows) (Fig. 6j, I).
Fig. 6. MtAnn1 mutants show impaired nodule differentiation and function.
Representative images (a, b) and 1 µm longitudinal sections stained with Toluidine Blue and Basic Fuchsin (c–f) of wild-type R108S and ann1-3 mutant nodules 14–16 dpi. Meristematic (ZI), infection (ZII), interzone (IZ) and nitrogen fixing (ZIII) zones are labelled. The amyloplast-rich IZ is indicated between brackets (c–f). Arrows (d) indicate senescing cells. g Nodule IZ area was measured in 1 µm nodule section images (see Supplementary Fig. 14) of 14–16 dpi R108S (n = 7) and ann1-3 (n = 8) nodules from 2 independent experiments. h Nitrogen fixation capacity was measured in R108S (n = 14 roots with 365 nodules) and ann1-3 (n = 15 roots with 417 nodules) 21 dpi nodulated root segments from 3 independent experiments. Box plots in g, h represent the distribution of individual values (open circles). First and third quartiles (horizontal box edges), minimum and maximum (outer whiskers), median (centerline), mean (solid black circle) and outliers (crosses) are shown. Asterisks in g, h indicate significant differences in ann1-3 mutant vs R108S control (p = 0.0012, two-tailed Welsh t-test in g, p = 0.0049, two-tailed Student t-test in h). i–l TEM ultrastructural analyses of IZ or ZIII zones in R108S and ann1-3 14–16 dpi nodules (n = 5 for R108S and n = 4 for ann1-3 from 2 independent experiments) and close-up views of IZ and ZIII or IZ/ZIII of R108S or ann1-3 nodules with uninvaded cells (UC) and bacteroid-filled invaded cells (IC). Black arrowheads indicate amyloplasts located near intercellular spaces of IC and within UC cells in the IZ of R108S and ann1-3, and in the IZ/ZIII of ann1-3. Differentiated bacteroids (white asterisks) are seen in both R108S and ann1-3, but in ann1-3 nodules they resemble interzone-type bacteroids55 imbedded in white vesicle-enriched regions (black arrows). Scale bars: a, b = 1 mm, c–f = 50 µm, i–l = 5 µm. See also Supplementary Figs. 12-14 for complementary molecular and phenotypic description of MtAnn1 mutants. Source data are provided as a Source Data file.
MtAnn1-dependent modulation of Ca2+ spiking and ultrastructural organisation of the cytoplasmic bridge
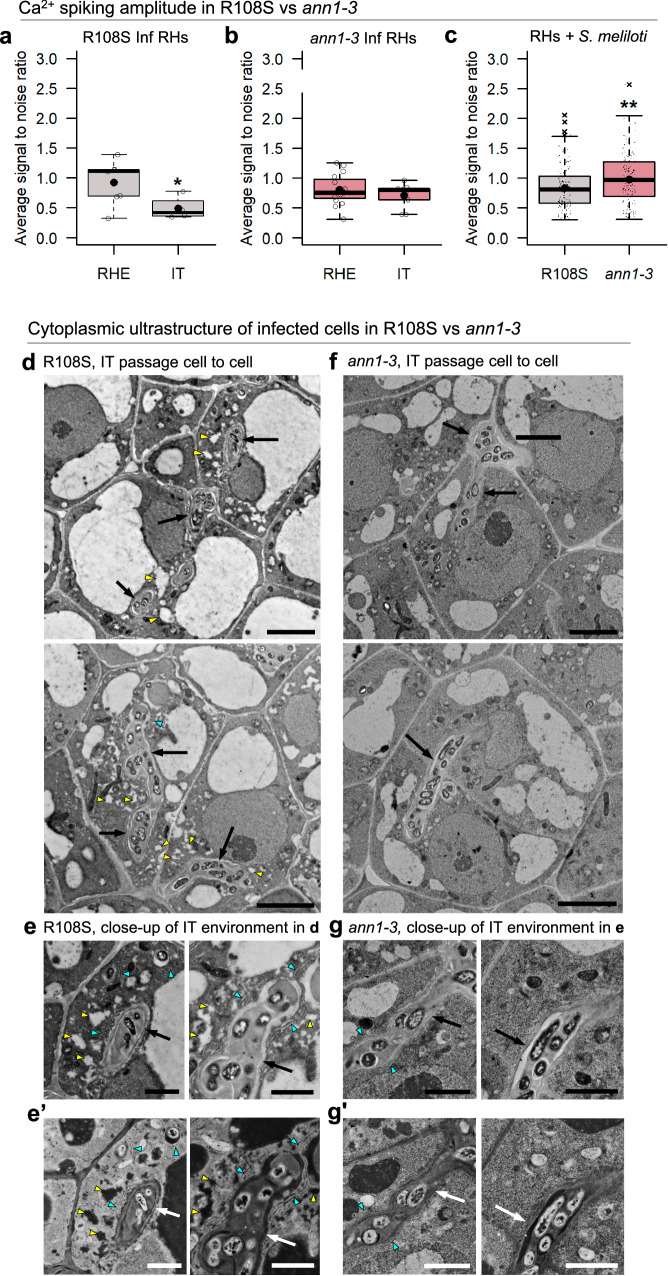
As Ca2+ spiking and MtAnn1 dynamics were closely linked in the peri-nuclear regions of primed cells (Figs. 2 and 3), we assessed if MtAnn1 mutation impacts symbiotic Ca2+ spiking. Although more challenging than previous live analysis in the hyper infectious sunn context (Fig. 2), we succeeded in monitoring Ca2+ spiking responses at infection sites in both wild-type R108S and ann1-3 transgenic roots expressing the NR-GECO1 Ca2+ sensor. Similarly, as seen in sunn (Fig. 2), a drop in Ca2+ spiking amplitude was observed in R108S in IT-containing RHs compared with RHE root hairs (Fig. 7a). Conversely, this drop was not clearly observed in ann1-3 (Fig. 7b). Likewise, a global deregulated Ca2+ spiking amplitude pattern was observed in many S. meliloti-responsive root hairs of ann1-3 compared with wild-type R108S, while Ca2+ spiking frequency was not significantly changed in any root hair categories (Fig. 7c and Supplementary Fig. 15). Together, these data indicate a global deregulation of Ca2+ spiking amplitude in ann1-3, suggestive of the need of MtAnn1 for modulating symbiotic Ca2+ spiking responses.
Fig. 7. Modified Ca2+ spiking and IT cytoplasmic environment in the absence of MtAnn1.
a–c Transgenic roots expressing the NR-GECO1 Ca2+ sensor were generated to monitor nuclear Ca2+ oscillations in ann1-3 mutant or wild-type (R108S) plants. Variations in Ca2+ ion concentration, reflected by changes in the relative intensity of NR-GECO1 fluorescence, are expressed as signal-to-noise ratio (SNR, cf. ‘Methods’ section). Box plots represent average amplitude of spikes (spike SNR), calculated separately for each nucleus of root hairs with entrapped rhizobia (RHE) or with a growing IT (IT) in (a) R108S (RHE, n = 7; IT, n = 4) or in (b) ann1-3 (RHE, n = 15; IT, n = 9) or in S. meliloti-responsive root hairs in the close competence zone (c) (R108S, n = 160 and ann1-3, n = 156) at 1–4 dpi. Box plots show the distribution of values (open circles or black dots) obtained from three independent experiments. First and third quartiles (horizontal box edges), minimum and maximum (outer whiskers), median (centerline), mean (solid black circle) and outliers (crosses) are shown. Asterisks indicate statistical difference in RHE vs. IT in R108S RHs in a (p = 0.0285, one-tailed Student t-test) and in RHs of R108S vs. ann1-3 in (c) (p = 0.0018, two-tailed Mann-Whitney test). Differences were not significant in RHE vs. IT in ann1-3 RHs in (b) (p = 0.1735, one-tailed Student t-test). (d-g) TEM analysis of infected cells from apical zone II of R108S (d, e, e’) or ann1-3 (f, g, g’) were performed in 80 nm sections of 14–16 dpi nodules (n = 5 R108S nodules and n = 4 ann1-3 nodules derived from 2 independent experiments). ITs (arrows) in wild-type R108S are embedded in vesicles (yellow arrowheads) and ER-rich (blue arrowheads) cytoplasmic bridges, which are less visible in ann1-3. e’, g’ are complementary inverted LUT images of (e-g) that were generated by ImageJ. Scale bars: d, f = 5 µm, e, e’, g, g’ = 2 µm. See also Supplementary Fig. 15. Source data are provided as a Source Data file.
Ca2+ spiking and MtAnn1 dynamics accompany cytoplasmic bridge formation (Figs. 2 and 3), so we wondered how the deregulated Ca2+ spiking response in ann1-3 (Fig. 7b) could impact cytoplasmic bridge formation. MtAnn1 is expressed during root infection (Fig. 4), but also in the nodule pre-infection and infection zones57. Thus, the defective nodule differentiation phenotype of ann-1-3 could be related to a possible deregulated infection in the nodule. We thus analysed the cytoplasmic composition of infected cells in R108S and ann1-3 apical nodule zones by TEM (Fig. 7d–g), which can reveal ultrastructural organelle features and distribution that confocal imaging cannot. In wild-type R108S, ITs progressed through a small vacuole-like enriched cytoplasmic bridge containing a dense ER network (Fig. 7d, e, e’), as shown similarly in A17 (Fig. 1). In ann-1-3, the small vacuole-like and ER-enriched cytoplasmic environment around the IT was strongly attenuated (Fig. 7f, g, g’). Thus, mutation in MtAnn1 resulted in a deregulated Ca2+ spiking response and a change in the ER and vesicle composition of the cytoplasmic bridge.
MtAnn1 is required for efficient mycorrhizal root colonisation
Recently developed genomic and transcriptomic databases from around 300 plant species establishing different types of endosymbioses enabled to address key endosymbiosis-related ancestral gene functions58. We have thus exploited these extensive resources to trace the evolutionary history of MtAnn1. Previous studies of the monophyletic plant annexin group, mainly composed of angiosperm sequences, suggested their emergence through duplications from a common ancestor prior to the monocot/eudicot split59. Consistent with these data, we found that the MtAnn1 annexin cluster likely arose from sequential pre-Angiosperms, Eudicots and Papilionoideae-specific duplications (Supplementary Fig. 16 and Supplementary Data 1). MtAnn1 belongs to one of the Papilionoideae duplicated clades, which mainly includes species that establish intracellular endosymbioses, except for Caryophyllales and Cephalotus (status uncertain). Ann1 orthologs were also detected in Papilionoideae species that lost RNS but maintained AMS. This indicates that the function of Ann1 is not restricted to RNS but could expand to AMS. Strikingly, Ann1 from different species show consistent transcriptional activation during both root nodule and AM symbioses4, suggesting an ancestral role in root endosymbiotic infection.
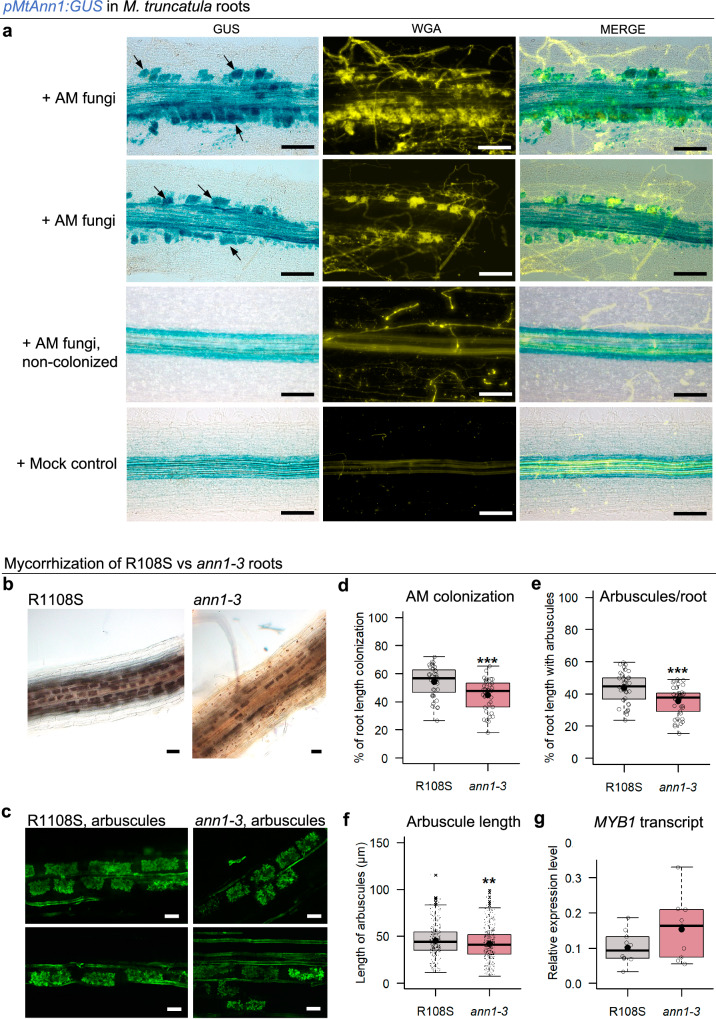
To determine if MtAnn1 indeed plays a role during AMS, we first investigated if MtAnn1 promoter activity was spatio-temporally regulated in M. truncatula roots colonised by the AM fungus Rhizophagus irregularis. Strong activation of the pMtAnn1:GUS fusion was only observed in root segments colonised by R. irregularis at 4 wpi, especially in cortical cells containing arbuscules (Fig. 8a). R. irregularis colonisation rates were then monitored in ann1-3 compared to R108S wild-type roots at 6 wpi. While R108S roots had dense colonisation by R. irregularis, with cortex cells fully filled with arbuscules, mutant roots had a lower proportion of colonised root sectors and arbuscule density (Fig. 8b–f). In addition, arbuscules formed in the mutant showed a slight but significant reduction in length and this was associated with a trend increase in the expression of the arbuscule senescence-related marker MYB160 (Fig. 8c, f–g). These data highlight the importance of MtAnn1 for the efficient establishment of the ancient AM symbiosis.
Fig. 8. MtAnn1 is required for efficient mycorrhization of M. truncatula roots.
a pMtAnn1:GUS activity in M. truncatula A17 roots 4 wpi with R. irregularis (+AM fungi, n = 10) or mock control (n = 8) was analysed in 2 independent experiments. Representative images of root sectors inoculated with R. irregularis (+AM fungi) with or without fungal colonisation are shown. pMtAnn1:GUS (GUS) blue staining in root cortex (arrows) correlates with areas colonised by R. irregularis, visualised yellow by WGA-Alexa Fluor 488 fluorescence staining (WGA) and merged images (MERGE). b–g Phenotypic analysis of R108S or ann1-3 M. truncatula roots 6 wpi with R. irregularis. Representative images of ink-stained AM-colonised roots (b) or WGA-stained arbuscules (c) are shown. d, e Intraradical colonisation by R. irregularis was determined in R108S wild-type (n = 36) and ann1-3 (n = 38) using the gridline intersect method. Box plots show the relative surface of the root system with vesicles, arbuscules and/or intraradical hyphae (d) and the relative area corresponding specifically to arbuscules (e). f Arbuscule lengths were compared in R108S (n = 371, from 8 plants) and ann1-3 (n = 440, from 7 plants) roots at 6 wpi. g Q-RT-PCR expression analysis of MYB1 in total RNA samples from independent R108S (n = 10) or ann1-3 (n = 9) plants inoculated with R. irregularis (6 wpi). Box plots (d–g) show single values distributions (open circles or black dots) from 2 independent experiments. First and third quartiles (horizontal box edges), minimum and maximum (outer whiskers), median (centerline), mean (solid black circle) and outliers (crosses) are shown. Asterisks indicate statistical difference between R108S and ann1-3 in (d–f) (p = 0.0004 in d, p = 0.0002 in e, two-tailed Student t-tests; p = 0.0046 in f, two-tailed Mann-Whitney test). Although there is a trend towards higher MYB expression in ann1-3 vs. R108S (g), the difference is not significant (p = 0.1228, two-tailed Student t-test). Scale bars: a = 100 µm, b = 50 µm, c = 20 µm. See also Supplementary Fig. 16. Source data are provided as a Source Data file.
Discussion
Rhizobia root infection occurs transcellularly in most legumes via apoplastic IT structures that grow polarly to reach the developing nodule. Plant cells engaged in this process form cytoplasmic bridges that precede and accompany the route for ITs to progress. This process, analogous to PPA bridges formed during AM symbiosis, is likely governed by common ancestral cellular mechanisms, though knowledge remains limited. Here we report cell-specific Ca2+ responses and cytoplasmic ultrastructural rearrangements that prime M. truncatula plant cells for rhizobia infection. We show that this priming state, under DMI3-NIN genetic control, is marked by an intimate cellular link between Ca2+ spiking and annexin MtAnn1 accumulation. Moreover, MtAnn1 is needed for modulation of Ca2+ spiking responses and creation of an infection-permissive, dense, ER- and small vacuole-rich cytoplasmic environment. Ultimately, MtAnn1 is required for successful root nodule differentiation and efficient AM root colonisation, implying an ancient recruitment of MtAnn1 in endosymbiosis. Our work supports the need to fine-tune cell-specific Ca2+-spiking responses by annexin Ca2+ binding proteins for the transcellular passage of endosymbionts.
Organelle dynamics shaping rhizobia pre-infection priming
Ultrastructure dissection of plant cells preparing for transcellular rhizobia infection revealed marked enrichment and distribution of organelles during this process (Fig. 1). This priming state is seen only in a few outer cortical cells in close contact with the infected root hair cell (Figs. 1, 3, Supplementary Figs. 6 and 8), highlighting the likely need for signals from the infected cell to promote it. A marked clustering of mitochondria around the nucleus is observed in primed cells (Fig. 1 and Supplementary Fig. 1). This may reflect the energy-demanding nature of this process. However, dividing nodule primordia cells with presumably high metabolic activity show no such enrichment. Alternatively, perinuclear mitochondria may mark a specific regulated cell cycle status, likely to occur in these cells32,34,61 or contribute to modulating cytosolic Ca2+ oscillations, as shown in other systems62.
Consistent with previous findings35, TEM analysis revealed that a dense ER-rich network occupy the transcellular cytoplasmic bridge in primed or rhizobia infected M. truncatula cells (Figs. 1 and 7). The bridge is also rich in small vacuole-like structures63, but we cannot exclude that multivesicular bodies (MVBs), seen in the PPA bridge, are part of this organelle population. Future use of molecular markers63 may help discriminate between them. Dynamic changes in plant vacuole size have been reported to occur in different cell types, and in early meristematic cells this is thought to increase cytosolic area for subsequent daughter cell formation events63,64. An analogous need for increased cytosolic area is possibly required for IT passage. The concomitant reduction of small vacuoles and ER (involved in vacuole biogenesis65) in MtAnn1 mutants (Fig. 7) raises the question of their functional interconnection in this process.
Role of MtAnn1 in modulating cell- and stage-specific symbiotic Ca2+ responses
High frequency Ca2+ spiking, triggered by Nod factors in root hairs is key for early rhizobia recognition5,6,46. Here, we showed that a second Ca2+ spiking phase occurs in root hairs undergoing rhizobia infection and involves a decrease in Ca2+ peak amplitude as ITs extend into root hairs (Fig. 2). This drop was also reported in root outer cortical cells crossed by an IT43, consistent with this switch being key for IT progression. Ca2+ amplitude regulation fine-tunes different biological processes ranging from gene expression, stomatal closure, gravitropic responses to synaptic plasticity in animals66–68. The deregulated Ca2+ amplitude profile and defective infection caused by MtAnn1 mutation (Figs. 5 and 7) points to a functional link between MtAnn1 and regulation of Ca2+ spiking amplitude during rhizobia infection.
We determined that Ca2+ spiking and strong MtAnn1-GFP labelling are hallmarks of rhizobia infection and pre-infection priming (Fig. 3 and Supplementary Fig. 8) and rely on the cell-autonomous activity of DMI3 and NIN (Fig. 4 and Supplementary Fig. 9). Thus, spatially distinct Ca2+ spiking responses are first triggered in non-infected root hairs independently of DMI3-NIN69,70, before DMI3-NIN-dependent Ca2+ spiking is induced in cells engaged for infection. Using the NR-GECO1 sensor, we observe a unique low frequency Ca2+ spiking only in primed cells of the cortex in both A17 and sunn backgrounds (Fig. 3 and Supplementary Fig. 8), in agreement with previous findings using a FRET-cameleon sensor43. Although root hairs show similar remodelling and MtAnn1 labelling, they never show such low frequency Ca2+ spiking, a signature likely reflecting a commitment state to infection.
In Medicago, symbiotic Ca2+ spiking is generated in the nucleoplasm and perinuclear cytoplasm by the concerted action of nuclear envelope ion channels (DMI1, CNGC15), a Ca2+ pump (MCA8) and under negative feedback regulation by a nuclear calmodulin (CaM2)6,71. MtAnn1 is a cytosolic phospholipid- and Ca2+-interacting protein of the annexin family13, viewed as dynamic regulators that associate with membranes when intracellular Ca2+ levels change15,16. Annexin family members are involved in the regulation of ion transport, modulation of cytoplasmic Ca2+ signals and ROS signalling in diverse abiotic or biotic responses72–76. Although predominantly cytoplasmic, annexins can also topologically associate with the ER77,78 and modulate the activity of Ca2+-release channels79. As MtAnn1 partially localises to the ER network in cytoplasmic bridges (Supplementary Figs. 5, 6), it may exert such a regulatory function. The radical change in the cytosolic distribution of the ER and small vacuoles by the MtAnn1 mutation (Fig. 7) also establishes a functional link between MtAnn1 and the creation of this specific cytoplasmic environment. Ca2+ orchestrates membrane trafficking and fusion, while vacuoles, emitters and receivers of Ca2+ signals, can be directly affected by altered cytosolic Ca2+ levels80,81. Annexins are Ca2+ sensing proteins involved in endomembrane trafficking in plants82, either in conjunction with the membrane fusion machinery or by regulating exocytosis. Thus, MtAnn1 may directly or indirectly impact Ca2+-regulated vesicle trafficking and/or vacuole morphology. However, the biological relevance of such a dense cytoplasmic strand for IT passage remains to be determined.
Evolutionary recruitment of MtAnn1 for root endosymbioses
In Mtann1 mutants, root infection by rhizobia is reduced (Fig. 5) and this culminates in mature nodules with reduced size, altered shape, abnormal zonation and reduced nitrogen fixation ability. These phenotypes are reminiscent of the reduced infection levels and nodule size observed after PvAnn1 RNAi knock-down in common bean83. Such phenotypes probably stem from the early deregulated Ca2+ spiking, impaired cytoplasmic configuration and consequently defective infection caused by the MtAnn1 mutation. Furthermore, as MtAnn1 is expressed in nodule pre-infection and infection zones57, the defective nodule differentiation phenotype is likely a consequence of maintained deregulated infection in the nodule. Finally, the close paralogue MtAnn214 may act redundantly with MtAnn1 in early root infection (Fig. 5) where they are both co-expressed48, but it is unlikely to compensate for MtAnn1 loss of function in nodules as it has a distinct spatial expression profile.
We showed that the subclade containing MtAnn1 emerged mainly in plant species establishing intracellular symbioses. Induced expression of MtAnn1 in rhizobia and AM symbioses and its requirement for proper formation of nitrogen-fixing nodules and cortical arbuscules (Figs. 5 and 8) suggest ancestral and shared roles for MtAnn1 in endosymbiotic infection. We propose that MtAnn1 fine-tunes oscillatory Ca2+ responses to create an optimal dense cytoplasmic environment for rhizobia IT and AM fungi hyphae to progress. As MtAnn1 is transcriptionally regulated by NIN, even in the absence of symbiotic signalling (Fig. 4), we hypothesise that MtAnn1 is a direct target of NIN and that its evolutionary recruitment for RNS involved the acquisition of putative NIN cis-regulatory motifs, which are indeed present in the MtAnn1 promoter. Since NIN is a major regulator in RNS84, the AM-associated expression of MtAnn1 is possibly under the control of another, so far unidentified regulator.
In conclusion, we showed that cell specific Ca2+ spiking signals shape rhizobia pre-infection priming to prepare the optimal dense cytoplasmic environment for IT passage in Medicago, and MtAnn1 emerges as a modulator of this process. MtAnn1, necessary for efficient rhizobia and AM colonisation, may have been recruited during evolution to integrate an ancestral Ca2+-regulatory module for guiding endosymbiotic infection. MtAnn1, with a conserved K/H/RGD protein-interacting motif15, may work in partnership with other players and discovering them could help identify other critical components in this process.
Methods
Plant materials and microbial strains
M. truncatula Jemalong A17 and R108 wild-type ecotypes, a regenerated transgenic line (referred to as A2) expressing the p35S:GFP-ER construct35 as well as sunn (sunn-2 allele)85, dmi3 (dmi3-1 allele)8, nin (nin-1 allele)70 and ern1 (bit1 allele)49,51 mutants were used in this work. The dmi3 line carrying the pEXT:DMI3 construct52, hereafter called dmi3 + pEXT:DMI3, was kindly provided by C. Remblière (LIPME, Toulouse). The three R108 Tnt1 mutants carrying insertions in MtAnn1 were obtained from Oklahoma State University and are designated ann1-1 (NF0830), ann1-2 (NF17737) and ann1-3 (NF4963). These lines were self-crossed and genotyped by PCR and sequencing at each generation using primers −344 pMtAnn1-Fw, Mtr14183-Rev, −914 pMtAnn1-Fw, LTR6- and LTR4- (Supplementary Table 1). Homozygous mutants and sibling wild-type lines (referred to as R108S) were selected for seed multiplication and phenotyping. R108S is a wild-type sibling derived from line NF4963. Seeds were scarified and surface-sterilised prior to germination on inverted soft agar plates45 and used for A. rhizogenes transformation or nodulation experiments. N. benthamiana seeds were germinated in potting soil in glass-covered trays before transfer to pots (7 cm × 7 cm × 6.5 cm) at 21 °C in a growth chamber under a 16 h photoperiod and 70 μE/m/s light intensity for 3 weeks until bacterial infiltration. The strains A. rhizogenes ARquA1, A. tumefaciens GV3103 & GV3101 and S. meliloti 2011 (Sm2011-lacZ), constitutively expressing a hemA-lacZ fusion, have been previously described45. The S. meliloti 2011 strain Sm2011-cCFP, constitutively expressing a cyan fluorescent protein was kindly provided by P. Smit, Wageningen (The Netherlands). R. irregularis spores (DAOM197198) were purchased from Agronutrition (Labège, France).
DNA constructs
pMtAnn1:MtAnn1-GFP13 in pLP100, pENOD11:GFP-ER35 and p35S:GFP-ER86 in pBIN121 and p2x35S:mCherry-ER in pBIN2087 were used to study live pre-infection priming responses in M. truncatula. p2x35S:NR-GECO1 in pCAMBIA-CR1ΔDsRed46 was used to monitor Ca2+ spiking in M. truncatula. pMtAnn1:GUS in pLP10013, pEXPA:NIN in pK7WG2-R, p35S:3xHA-NIN and p35S:3xHA-NINΔ in PAMPAT-GTW54, were used in transcription activation studies in M. truncatula or N. benthamiana. p35S:MtAnn1-GFP and p35S:GFP in pBIN12113, were used in mutant complementation studies. The pMtAnn1:RNAi-MtAnn1 and pMtAnn1:GUS (control) constructs were generated here by Golden Gate cloning into the pCAMBIA-CR1 vector46. Briefly, DNA fragments corresponding to 2231 bp of the genomic MtAnn1 sequence upstream of the ATG, 942 bp of the MtAnn1 coding sequence in sense and antisense orientations, and the GUS coding sequence, were generated by PCR amplification using the primer pairs pMtAnn1-GG-A-Fw/pMtAnn1-GG-B-Rev, MtAnn1-ATG-GG-B-Fw/MtAnn1-STOP-GG-C-Rev, MtAnn1 antisens-STOP-GG-X-Fw/MtAnn1 antisens-ATG-GG-D-Rev and GUS-GG-B-Fw/GUS-GG-D-Rev, respectively (Supplementary Table 1). DNA fragments were then cloned into a pBluescript SK vector and validated by DNA sequencing prior to Golden Gate assembly. Golden Gate reactions with plasmids comprising (i) the MtAnn1 promoter, MtAnn1 coding sequences (sense and antisense) and a 1300 bp intron spacer24 or (ii) the MtAnn1 promoter and the GUS coding sequence, were used to generate RNAi MtAnn1 (pMtAnn1:MtAnn1sens-intron-MtAnn1antisens) or GUS control (pMtAnn1:GUS) constructs in the binary pCAMBIA-CR1 vector. All binary vectors used for A. rhizogenes transformation of M. truncatula (pLP100, pCAMBIA-CR1, pK7WG2-R, pBIN121) include a kanamycin resistance cassette in the T-DNA region. pK7WG2-R and pCAMBIA-CR1 vectors also include a DsRed cassette.
Generation of M. truncatula transgenic material
A. rhizogenes-mediated transformation of M. truncatula A17 (wild-type, sunn, ern1 and dmi3) or R108 (R108S wild-type sibling and ann1-3) was performed as described45. Briefly, germinated seedlings had their root tips removed with a scalpel and were placed on Fahraeus medium plates (12 cm × 12 cm) supplemented with 0.5 mM NH4NO3 and 20 mg/L kanamycin, before inoculation with a drop (~3 µL) of A. rhizogenes aqueous suspension (OD600nm 0.5). For co-transformation experiments with strains pMtAnn1:MtAnn1-GFP + p2x35S:NR-GECO1, p2x35S:NR-GECO1 + pENOD11:GFP-ER, pEXPA:NIN + pENOD11:GFP-ER or pEXPA:NIN + pMtAnn1:GUS (in pLP100), equal volumes of bacterial suspensions were mixed prior to seedling inoculation. The lower ¾ part of the plates were sealed with parafilm and placed vertically in plastic boxes under controlled 16 h light/ 8 h dark photoperiod conditions, first at 20 °C for 1 week, then at 25 °C for two to three weeks. Composite M. truncatula plants with kanamycin resistant roots were selected. In some experiments, kanamycin-resistant composite roots were re-selected on the basis of constitutive fluorescence of the DsRed marker (for pEXPA:NIN in pK7WG2-R and pMtAnn1:RNAi-MtAnn1 and pMtAnn1:GUS in pCAMBIA-CR1 constructs), fusion fluorescence in lateral roots (for pMtAnn1:MtAnn1-GFP and pENOD11:GFP-ER constructs) or in the nucleus (for p2x35S:NR-GECO1). Selected composite plants were then transferred to appropriate nitrogen- and antibiotic-free plates or pots for subsequent inoculation with rhizobia.
Plant growth and microbial inoculation procedures
For in vivo imaging experiments, kanamycin-resistant and fluorescent-positive composite plants of A17 (wild-type, sunn, ern1 and dmi3) or R108 (R108S and ann1-3) carrying pMtAnn1:MtAnn1-GFP + p2x35S:NR-GECO1, pEXPA:NIN + pENOD11:GFP-ER or p2x35S:NR-GECO1 + pENOD11:GFP-ER constructs were transferred to nitrogen-free Fahraeus plates supplemented with Amoxycillin sodium/Clavulanate potassium 5:1 (200 mg/L) for 3–7 days, then transferred to nitrogen-free 0.5% [w/v] phytagel Fahraeus plates supplemented with 50 nM 2-amino ethoxyvinyl glycine (AVG) for 3 days, as described previously27. Whole root systems were then inoculated with 0.5 to 1 mL of the Sm2011-cCFP suspension (OD600nm 0.001), applied between the medium and the LUMOX film (Sarstedt, UK). The inoculated root systems were kept protected from light, by wrapping ¾ of the plates with dark plastic bags in a culture room at 20 °C or 25 °C with a 16-h photoperiod and a light intensity of 70 mE/s/m2, until microscopy observations.
For high-resolution microscopy analyses of pre-infection priming in A17 and nodule differentiation in R108S/ann-3, germinated seedlings of M. truncatula A17 and R108 (R108S and ann1-3) were grown for 3 days on nitrogen-free paper/Fahraeus Kalys HP696 agar plates45 before roots were either spot or flood-inoculated with water (control) or a Sm2011-lacZ (OD600nm 0.01) suspension, as described24. For expression analyses of pMtAnn1:GUS (in pLP100) in A17 and nin (co-expressing or not pEXPA:NIN), kanamycin-resistant and/or DsRed fluorescence-positive composite plants were transferred 3 weeks after A. rhizogenes transformation to the same nitrogen-free paper/Fahraeus Kalys HP696 agar plates and grown for 3 days prior to flood inoculation (for 1 h) of the entire root system with Sm2011-lacZ (OD600nm 0.01). Plants grown in vitro on plates were all cultivated at 25 °C under a 16-h photoperiod and a light intensity of 70 mE/s/m2 with their root systems protected from light by wrapping ¾ of the plates with dark plastic bags.
For phenotyping experiments, germinated seedlings of M. truncatula R108 lines (R108, R108S and ann1-1 to ann1-3 mutants) and selected composite plants (2–3 weeks after transformation) of A17-transformed with RNAi-MtAnn1/control constructs or R108 (R108S or ann1-3) transformed with p35S:GFP/p35S:MtAnn1-GFP constructs, were transferred to 8 × 8 × 7 cm pots (3 plants/pot for germinated seedlings and 2 plants/pot for composite plants) filled with inert attapulgite substrate (Oil Dri US Special; http://www.oildri.com/), supplemented with 10 mL nitrogen-free Fahraeus medium. For longer growth periods (>3 wpi), plants were grown in 9 × 9 × 8 cm pots (3 plants/pot) supplemented with 14 mL medium. Pots were placed in small greenhouses at 25 °C, with a 16 hours photoperiod and a light intensity of 100 mE/s/m2 and inoculated with a suspension of Sm2011-lacZ (OD600nm 0.1), after 3 days of nitrogen starvation.
To assess mycorrhizal root colonisation of R108S and ann1-3 mutant lines, germinated seedlings were transferred to 8 × 8 × 7 cm pots (1 plant per pot), filled with a 1:1 mix of Zeolite substrate fractions 1.0–2.5 mm and 0.5–1.0 mm (Symbiom LTD, Lanskroun, Czech Republic), and inoculated with R. irregularis spores (strain DAOM197198, 150 spores per pot). Pots were placed in 60 × 40 × 12 cm trays in a 16-h photoperiod chamber (light intensity: 300 μmol/s/m2) at a day-time temperature of 22 °C and a night-time temperature of 20 °C, and 70% humidity. Plants were watered weekly with a modified low-phosphate and low-nitrogen Long Ashton solution (7.5 μM Na2HPO4, 750 μM KNO3, 400 μM Ca(NO3)2, 200 mg/L MES buffer, pH 6.5).
β-glucuronidase (GUS) and β-galactosidase enzymatic assays
Root segments from M. truncatula composite plants expressing pMtAnn1:GUS (in pLP100) or pEXPA:NIN + pMtAnn1:GUS constructs were collected from control (non-inoculated) or rhizobia-inoculated roots and incubated in 0.5% paraformaldehyde/0.1 M potassium phosphate buffer pH 7.0, for 1 h, prior to histochemical (blue) staining for GUS activity for 2–5 h at 37 °C using 1 mM of the substrate X-Gluc (5-bromo-4-chloro-3-indoxyl-b-D-GlcA, cyclohexylammonium salt, B7300; Biosynth, Staad, Switzerland) as described51. Histochemical GUS staining of mycorrhizal roots expressing pMtAnn1:GUS or N. benthamiana leaf discs was carried out in the same X-Gluc substrate but supplemented with 0.1% Triton X-100, first under vacuum for 20–25 min at room temperature before incubation at 37 °C for 1-3 hours. Enzymatic GUS fluorimetric assays of N. benthamiana leaf discs were done according to ref. 45. Briefly, tissues were ground using a MM400 grinder (Retsch) and homogenised in GUS extraction buffer before 1 µg of total protein extracts were used for enzymatic reactions at 37 °C using 1 mM of the 4-MUG substrate (4-Methylumbelliferyl-β-D-glucuronide hydrate, Biosynth M-5700). GUS activity was measured using a FLUOstar Omega 96 microplate reader (BMG LABTECH, France) through quantification of the fluorescence of 4-MU (4-Methylumbelliferone, Sigma) reaction product. To reveal the constitutive β-galactosidase activity of the S. meliloti strain Sm2011-lacZ in ITs, root samples (sometimes pre-stained for GUS activity) were rinsed and fixed for 1 h in 1.25% glutaraldehyde/Z buffer (10 mM KCl, 1 mM MgCl2 and 0.1 M phosphate buffer, pH 7.0) as described45. After rinsing in Z-buffer, root samples were incubated overnight in the dark at 28 °C in Z-buffer containing 2 mM Magenta-Gal (5-bromo-6-chloro-3-indoxyl-b-D-galactopyranoside; B7200; Biosynth) or X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside, W5376C; Thermo Fisher Scientific, Guilford, CT). GUS and/or LacZ-stained tissues were cleared for 30 seconds with 12% sodium hypochlorite solution before microscopy observations.
Tissue harvesting and microscopy methods
Roots of M. truncatula composite plants expressing different fluorescent fusions and grown under LUMOX film were observed using a Leica TCS SP8 AOBS laser scanning confocal microscope equipped with a 40x long distance water immersion objective (HCX APO L U-V-I 40x/0.80 WATER). Confocal images were recorded using Leica LAS-X software, before and after rhizobia inoculation (from 1 to 7 dpi). For GFP, CFP and DsRed or mCherry fluorescent proteins, 458 nm and 488 nm Argon laser lines and a 561 nm diode were used for excitation, respectively, with emission windows set at 465–495 nm, 500–525 nm and 570–600 nm (DsRed) or 600–630 nm (mCherry) (hybrid detector), respectively. To avoid potential interference, sequential mode was used to acquire CFP and red fluorescent protein fluorescence separately from GFP fluorescence and bright-field images. For confocal imaging of Ca2+ responses, the 561 nm diode was used to excite the mApple red fluorescent protein from the NR-GECO1 sensor, and the emitted fluorescence was recovered using a hybrid detector in the 600–643 nm emission window. Time series were acquired at 5 s intervals for 10–15 min, with pinhole diameter set to 3 Airy units, as described46.
Roots or nodulated roots of M. truncatula A17 and R108 (R108S and ann1-3) or composite plants were selected based on their fluorescence and/or analysed before or after GUS and/or β-galactosidase staining using stereomicroscopy (Leica S6E) or light microscope (AxioPlan II Imaging; Carl Zeiss, Oberkochen, Germany). For pMtAnn1:GUS (in pLP100) expression analysis, samples were harvested before and after (4 dpi) with rhizobia. For comparative phenotyping of wild-type and MtAnn1 mutant or RNAi roots, β-galactosidase stained nodulated root samples were harvested 4–5 or 8–10 dpi with rhizobia. Roots expressing pEXPA:NIN + pMtAnn1:GUS were dehydrated in ethanol series and embedded in glycol methacrylate (Technovit 7100; Haereus-Kulzer) according to the manufacturer’s instructions. Sections of 10 µm were counterstained with aqueous Basic Fuchsin solution (0.007%) for observations. To quantify nodule size and rhizobia infection levels, β-galactosidase-stained nodulated root systems (10 dpi) from R108S and ann1-3 plants were scanned (Objectscan 1600, Microtek) and the acquired images (TIFF) were used for image quantification (see details in the following part of the methods).
For high-resolution microscopy analyses of pre-infection priming responses in A17, control roots and rhizobia-infected root regions or nodule primordia were carefully isolated from spot-inoculated or flood-inoculated root systems 5–6 dpi, after histochemical revelation of rhizobial β-galactosidase activity. Harvested root sections or nodule primordia were fixed in 2% glutaraldehyde, diluted in 0.2 M cacodylate buffer pH=7.3 for a few hours at room temperature or 1–3 days at 4 °C, then rinsed with buffer, before progressive dehydration in an ethanol series and final inclusion in LR White resin following manufacturer’s instructions (EMS). For comparative ultrastructural analysis of isolated R108S and ann1-3 nodules, nodule samples were carefully isolated from rhizobia-inoculated root systems 14–16 dpi and fixed under vacuum in 2 successive batches of 2.5% glutaraldehyde diluted in 0.2 M cacodylate buffer pH=7.3, the first batch containing saponin or triton (0.1% final) for 3–5 days at 4 °C. After rinsing in buffer, post-fixation in 2% osmium tetroxyde (diluted in 0.2 M sodium cacodylate), rinsing again in buffer, progressive dehydration in an ethanol series and incubation in propylene oxide (2 × 1 h), samples were finally embedded in Epon 812 resin (EMS) following the manufacturer’s instructions. For all samples, semi-thin (1 µm) and ultra-thin (80 nm) sections were generated using an Ultracut E ultramicrotome (Reichert Jung). Semi-thin sections of root/nodule primordia (5–6 dpi) were stained with Basic Fuchsin (0.07% in water) to help visualise ITs, while mature nodule sections (14–16 dpi) were stained in an aqueous solution with methylene blue (0.2%), Toluidine Blue (1%), borax (1%) and Basic Fuchsin (0.07%) before observation by bright field microscopy (Axioplan II Imaging; Carl Zeiss). Ultrathin sections (80 nm) were contrasted with Uranyless and lead citrate (Delta microscopy) and observed using a Hitachi 7700 electron microscope (Hitachi High-Tech).
For histochemical GUS staining of mycorrhizal roots, selected composite plants transformed with pMtAnn1:GUS were transferred to 7 × 7 × 8 cm pots (5 plants/pot) filled with washed quartz sand (grain size 0.7–1.2 mm) pre-watered with modified half-strength Hoagland solution containing 20 µM phosphate88. Each plant was inoculated with 500 spores of R. irregularis DAOM197198 (C-grade, Agronutrition, Toulouse, France). Plants were grown in a Polyklima cabinet at 22 °C constant temperature, 60% air humidity and 16-h-light/8-h-dark cycles, at 200 µE/s/m2 light intensity and a combination of warm and cold LED ‘True DayLight’ at a 40:25 ratio. Pots were watered twice per week with 30–40 ml of autoclaved de-ionised tap water and fertilised once per week with 30–40 ml of modified half-strength Hoagland solution containing 20 µM phosphate. To visualise the fungal structures, the GUS staining solution was exchanged for 10% KOH and the roots incubated for 15 minutes at 95 °C. After KOH removal, the roots were washed 3 times with de-ionised water. After addition of 0.1 M HCl, the roots were further incubated for 2 h in the dark at room temperature. HCl was removed and the roots were washed 3 times with de-ionised water and once with PBS. This was followed by an overnight incubation at room temperature in the dark with 200 ng/µL WGA Alexa Fluor 488 (Invitrogen W11261) in PBS. Roots were imaged with a LEICA DM6B epifluorescence microscope. To compare the extent of mycorrhizal colonisation of roots between R108S and ann1-3 lines, whole root systems were collected 6 weeks post-inoculation with R. irregularis and cleared by boiling (95 °C) in 10% KOH for 5 min, rinsed with water, then stained by boiling in an acidic ink solution (5% acetic acid, 5% Sheaffer black ink #94321, in water) to reveal fungal structures. Overall intraradical colonisation rates and arbuscular density were estimated in ink-stained roots under a stereomicroscope (Leica S6E) using the gridline intersect method.
Microscopy image analyses
Analysis of confocal time-series was performed using the Fiji software89. Intensity data were calculated from selected regions of interest (ROIs), corresponding to single nuclei, imaged in independent roots, in 2–3 independent experiments. Maximal z-projections of stacks and merged confocal images for illustration purpose were prepared using Leica confocal software or Fiji. To quantify relative amplitudes of Ca2+ spiking, the mean fluorescence of each ROI was measured as a function of time and used to determine the Signal-to-Noise Ratio (SNR). The SNR measures the difference between the fluorescence at each time-point (Ft) and the fluorescence baseline, calculated as the average of 4 fluorescence values taken 10 and 15 s before and after t (Ft-15; Ft-10; Ft+10; Ft+15). A threshold of 0.3 was applied to the SNR to define individual peaks, and the amplitude of Ca2+ spiking was calculated for each nucleus as the mean of peak SNR values. Spiking frequency, defined as the number of peaks observed in 10 minutes, was also calculated. In each experiment where Ca2+ spiking was monitored, nuclei displaying no spiking were also included, resulting in the sample size (n) being higher in the frequency graph than in the associated amplitude graph.
For quantification of MtAnn1-GFP fluorescence levels, root hairs were imaged at either the RHE or IT stages with identical acquisition settings in 3 independent experiments. In each acquired confocal z-stack, 4–6 successive confocal sections encompassing the cytoplasmic aggregation around the site of rhizobia entrapment (RHE) or the cytoplasmic bridge characteristic of the IT stage were selected. In ImageJ (https://imagej.net/ij/), the threshold function was used to generate a mask based on a maximal z-projection of the selected sub-stack, from which the contours of the cytoplasmic zone (ROI) were defined. The mean grey value was then measured in the defined ROI on an average z-projection of the same sub-stack.
Scanned images of β-galactosidase-stained root systems from R108S and ann1 mutants (10 dpi) were imported to Ilastik software90 to enable machine-learning recognition of blue β-galactosidase-stained nodules (pixel-based classification method). Objects identified as nodules were exported in a new single segmentation file (.TIFF format) for analysis in ImageJ to measure specific nodule features (size and infection level, based on the blue intensity of rhizobial β-galactosidase activity).
Transient expression assays in N. benthamiana leaves
An A. tumefaciens GV3101 strain carrying the pMtAnn1:GUS fusion construct (in pLP100) was used for infiltration studies in N. benthamiana alone or with A. tumefaciens GV3103 strains carrying p35S:3HA-NIN or p35S:3HA-NINΔ constructs54, designed to constitutively express NIN or NINΔ under the 35S promoter in transactivation experiments. Briefly, A. tumefaciens strains grown overnight at 28 °C in LB medium with appropriate antibiotics were harvested and resuspended in Agromix (10 mM MgCl2, 10 mM MES/KOH pH 5.6 and Acetosyringone in DMSO Sigma-Aldrich 150 µM) and kept in the dark for at least 2 h, at room temperature. Equal volumes of A. tumefaciens cultures (OD600nm = 0.25) were used to co-infiltrate leaves of 3-week-old N. benthamiana plants. Infiltrated plants were kept at 21 °C in a growth chamber (16-h photoperiod and a light intensity of 70 mE/s/m2) for 36 h prior to collecting discs from infiltrated leaves, and subsequent histochemical GUS assays or storage at −70 °C prior to protein extraction for Western-blot or fluorimetric GUS assays.
Western blot analyses
N. benthamiana leaf discs, previously stored at −70 °C, were ground using a MM400 (Retsch) crusher and resuspended in Laemmli 2X sample buffer for SDS-PAGE (Bio-Rad). Samples were then placed at 95 °C for 3 min and centrifuged at 16,000 × g for 1 min. For each sample, 10 µL of the supernatant was loaded in a polyacrylamide 4–15% Mini-PROTEAN precast gel (Bio-Rad) along with 5 µL of pre-stained protein ladder marker (ThermoScientific, Lithuania). The Mini-Protean tank Electrophoresis System (Bio-Rad, USA) was used for gel migration in 1X Tris/Glycine/SDS Buffer (Bio-Rad). Protein transfer to nitrocellulose membranes (Bio-Rad) was performed using a Trans-Blot Turbo semi-dry transfer system (Bio-Rad). Membranes were stained with Ponceau Red before incubation for 1 h in blocking 1 × TBS solution (Tris base, NaCl and H2O), 0.1% Tween, 5% milk at room temperature, with mild agitation, before rinsing 3× in TBS-Tween 0.1% for 10 min and incubated overnight in the same solution at 4 °C with a 6000X dilution of anti-HA-peroxidase antibodies (Sigma-Aldrich) in 0.5% milk in TBS-Tween 0.1% to reveal HA-tagged NIN proteins following chemiluminescence revelation using the Clarity Western ECL Substrate (Bio-Rad) and the ChemiDoc Touch imaging system (Bio-Rad).
Acetylene reduction assay
To assess nitrogenase activity, grouped nodules were isolated from root systems of plants grown in pots 3 weeks after inoculation with Sm2011-lacZ and tested for acetylene reduction. Nodulated root fragments were incubated at 25 °C in sealed 60 mL vials containing 0.5 mL of nitrogen-free Fahraeus liquid medium in the presence of 10% (v/v) acetylene for 3 h. 400 µL of gas was then collected from each vial and ethylene production was quantified by gas chromatography (model no. GC 7280A; Agilent Technologies, Lexington, MA).
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from M. truncatula control and rhizobia-inoculated roots (4–5 dpi) using the Macherey-Nagel total RNA isolation kit according to the manufacturer’s instructions. DNA-free RNA samples were quantified, and RNA integrity was verified by Agilent RNA Nano Chip (Agilent Technologies). First-strand complementary DNA synthesis was performed using 1 µg of total RNA using an anchored oligo (dT) and Transcriptor Reverse Transcriptase (Roche) following the manufacturers’ protocol. Quantitative RT-PCR was performed in 384-well plates, with Roche LightCycler 480 or Bio-Rad CFX OPUS 384 Real-Time PCR systems and using the SYBR Green MasterMixes from Roche or Takyon (Eurogentec), according to manufacturer’s instructions. Reactions and cycling conditions were performed as described before24 using Primer pairs listed in Supplementary Table 1. The specificity of the PCR amplification was verified by analysing the PCR dissociation curve and sequencing of the PCR product. Transcript levels were normalised to the endogenous Ubiquitin reference.
Phylogenetic analysis
Protein sequence of MtAnn1 (MtrunA17_Chr8g0352611) was used as query to search against a database containing 227 plant genomes covering the main lineages of green plants and five SAR genomes as outgroups using the BLASTp v2.14.0+ with an e-value threshold of 1e-1058. Homologous proteins were then aligned using Muscle v5.1 with the ‘super5’ option91 and trimmed to remove positions with more than 60% of gaps using trimAl v1.4.rev2292. The trimmed alignment served as matrix for phylogenetic reconstruction using FastTree v2.1.11-OpenMP93. To gain insights about the evolution of Ann1 in relation with the nitrogen-fixing symbiosis, the flowering clade containing orthologs of MtAnn1 has been extracted and proteins re-aligned using Muscle with default parameters before trimming as described above. Few spurious sequences with obvious mispositioning and abnormal short sequences were removed. Then, maximum likelihood tree has been reconstructed using IQ-TREE v2.1.294 after testing the best evolution model using ModelFinder95 as implemented in IQ-TREE2 and according to the Bayesian Information Criteria. Branch supports have been tested with 10,000 replicates of both SH-aLRT96 and UltraFast Bootstraps with UFBoot297. Finally, trees were visualised and annotated in the iTOL platform v6.8.198.
Graph generation and statistical analyses
Graph preparation and statistical analyses were performed using R, except for graphs in Fig. 5c, d, which were generated using Microsoft Excel and for which contingency analyses were performed using GraphPad Prism 10. For all other analyses, data were presented as box plots, normal distribution of the data was evaluated using the Shapiro-Wilk test and homogeneity of variance was assessed using Fisher or Bartlett tests. When applicable, a transformation was performed to normalise data distribution (Log10 or BoxCox). Parametric statistical tests (t-test, ANOVA) were used to analyse data with a normal distribution, while non-parametric equivalents (Mann-Whitney, Kruskal-Wallis) were used for data with a non-normal distribution. Number of individually analysed samples (n), replicates and p significance levels are indicated in Figure legends. In detail, data was analysed as follows: In Fig. 2e values do not follow a normal distribution and were thus analysed using a two-tailed Mann-Whitney test (W = 315, p = 0.1531). Values in Fig. 2f follow a normal distribution and display homogeneous variance, so a two-tailed Student t-test was performed (t = −3.0927, df = 39, p = 0.0037). Log10-transformed values of Fig. 4e follow a normal distribution and variance homogeneity and were thus analysed using one-way ANOVA followed by Tukey honest significant difference (HSD) tests (F = 149.5, df = 2, p < 2e-16). Values in Figs. 5a, b do not follow a normal distribution, hence Kruskal-Wallis tests were carried out (K = 6.499, p = 0.0388 for 5a and K = 6.204, p = 0.0449 for 5b). Figure 5c, d were built from contingency tables reporting the number of nodule primordia belonging to different categories: not infected (−) or partially to fully infected (+) in c, or with a single IT extending to the cortex (1 IT) or multiple ITs (≥2 ITs) in d. Relative proportions of these categories were compared one-on-one between R108S and each other genotypes (R108, ann1-2 and ann1-3), by running separate Fisher’s exact tests. Results are for Fig. 5c, R108S vs R108 p = 0,8408, R108S vs ann1-2 p = 0.0027, R108S vs ann1-3 p = 0.0198, and for Fig. 5d, R108S vs R108 p = 0.7947, R108S vs ann1-2 p = 0.0226, R108S vs ann1-3 p = 0.001. Values in Fig. 5m follow a normal distribution and variance homogeneity and were thus analysed using one-way ANOVA followed by Tukey HSD tests (F = 2.372, df = 3, p = 0.076), and values in Fig. 5n do not follow a normal distribution and were thus analysed by a Kruskal-Wallis test (K = 55.44 and p = 9,17e-13). In Fig. 5o, values follow a normal distribution and variance homogeneity, hence a two-tailed Student t-test was carried out (t = 2.4153, df = 99, p = 0.0176), while values in Fig. 5p do not follow a normal distribution, thus two-tailed Mann-Whitney tests were applied (W = 1724, p = 0.0023). Values in Fig. 6g follow a normal distribution but not homogeneity of variances, thus a Welsh two-sample t-test was carried out (t = 4.8433, df = 8.2905, p = 0.0012). Log10-transformed values in Fig. 6h show a normal distribution and variance homogeneity and were thus analysed using a two-tailed Student t-test (t = −3.066, df = 27, p = 0.0049). Values in Fig. 7a, b, which show a normal distribution and variance homogeneity were analysed using one-tailed Student t-tests, due to the low number of values (7a: t = 2.1821, df = 9, p = 0.0285; 7b: t = 0.9612, df = 22, p = 0.1735). In Fig. 7c, values do not follow a normal distribution, thus a two-tailed Mann-Whitney test was performed (W = 9944, p = 0.0018). Values in 8d and BoxCox-transformed values in 8e (λ = 1.353535) show a normal distribution and homogeneity of variance and were thus analysed using two-tailed Student t-tests (8d: t = 3.6816, df = 72, p = 0.00045; 8e: t = 3.8831, df = 72, p = 0.0002). Distribution of values is not normal in Fig. 8f, so data was analysed via a two-tailed Mann-Whitney test (W = 91032, p = 0.0046), values of Fig. 8g follow a normal distribution so a two-tailed Student t-test was applied (t = −1.6238, df = 17, p = 0.1228). In Supplementary Fig. 1b, values do not follow a normal distribution, thus a Mann-Whitney test was carried out (W = 35, p = 0.005349), while in Supplementary Fig. 1c values show a normal distribution and variance homogeneity, so a two-tailed Student t-test was performed (t = 3.9724, df = 10, p = 0.0026). In Supplementary Fig. 2e–h, values show a non-normal distribution and were thus analysed using two-tailed Mann-Whitney tests (Supplementary Fig. 2e: W = 853, p = 1.811e-07; 2f: W = 205, p = 2.893e-05, Supplementary Fig. 2g: W = 348, p = 3.377e-09, 2h: W = 237, p = 1.926e-09). In Supplementary Fig. 7c, values follow a normal distribution and variance homogeneity, hence a two-tailed Student t-test was performed (t = 1.9307, df = 6, p = 0.1017). Supplementary Fig. 12b, c, Log10-transformed data follow a normal distribution and variance homogeneity, so one-way ANOVAs followed by Tukey HSD tests were carried out (respectively F = 321.1, df = 9, p < 2e-16 for Supplementary Fig. 12b and F = 1.701, df = 9, p = 0.106 for Supplementary Fig. 12c). In Supplementary Fig. 12d, values do not have a normal distribution, so a Kruskal-Wallis test was used (K = 43.0318, p = 4.5265e-10). In Supplementary Fig. 12e, data follows a normal distribution and variance homogeneity, so a one-way ANOVA followed by Tukey HSD tests was carried out (F = 4.593, df = 2, p = 0.0143). Log10-transformed data in Supplementary Fig. 12f, and non-transformed data in Supplementary Fig. 12g follow a normal distribution and variance homogeneity, hence two-tailed Student t-tests were employed (respectively t = 4.4053, df = 22, p = 0.0002 for Supplementary Fig. 12f, and t = 2.0775, df = 22, p = 0.0496 for Supplementary Fig. 12g). In Supplementary Fig. 13c, e, data follow a normal distribution (after a Log10-transformation for Supplementary Fig. 13c) and variance homogeneity, hence one-way ANOVAs followed by Tukey HSD tests were used for comparisons (respectively F = 2.223, df = 4, p = 0.0765 for Supplementary Fig. 13c and F = 4.398, df = 4, p = 0.0034 for Supplementary Fig. 13e). Values in Supplementary Fig. 13d do not follow a normal distribution, so statistical analysis was done using a Kruskal-Wallis test (K = 9.15, p = 0.0575). In Supplementary Fig. 13f, values follow a normal distribution and variance homogeneity, hence a two-tailed Student t-test was performed (t = −3.51, df = 89, p = 0.0007). Values in Supplementary Fig. 15c do follow a normal distribution and variance homogeneity, and were analysed using a two-tailed Student t-test (t = −1.0271, df = 10, p = 0.3286). In Supplementary Fig. 15d, e, values do not follow a normal distribution and were thus analysed using two-tailed Mann-Whitney tests (W = 86, p = 0.57 for Supplementary Fig. 15d, and W = 18215, p = 0.3726 for Supplementary Fig. 15e).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The M. truncatula R108 mutant plants utilised in this research project, which are jointly owned by the Centre National De La Recherche Scientifique, were obtained from Noble Research Institute, LLC and were created through research funded, in part, by grants from the National Science Foundation, NSF #DBI-0703285 and NSF #IOS-1127155. We thank the Toulouse CMEAB platform service for assistance with electron microscopy, S. Bensmihen for providing the pEXT:DMI3 construct52 and Celine Remblière for providing seeds of the M. truncatula dmi3 line carrying the pEXT:DMI3 construct. This work was supported by French National Research grants TULIP ANR-10-LABX-41, COME-IN ANR-14-CE35-0007-01 to F.d.C.N. and LIVE-SWITCH ANR-19-CE20-0026-01 to F.d.C.N. and J.F. and by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant No. 759731) to C.G. A.G. was funded by an ANR postdoctoral grant (ANR-19-CE20-0026-01), A.K., A.D. and L.P. were funded by PhD grants from the French Ministry of National Education and Research. We acknowledge the TRI-FRAIB imaging facility, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04).
Author contributions
Coordinated the work: F.d.C.N.; designed experiments: F.d.C.N., A.G., J.F., A.K., K.H., M.C.A., L.F., N.F.D.F., C.G.; performed experiments: A.G., J.F., A.K., K.H., M.C.A., L.F., A.D., N.F.D.F.; Machine learning methods development: A.G., A.L.R.; phylogenetic analysis: J.K., P.M.D.; analysed data: A.G., J.F., A.K., K.H., M.C.A., L.F., A.D., L.P., J.K., P.M.D., C.G., N.F.D.F., F.d.C.N.; wrote the manuscript: F.d.C.N., A.G. and J.F.
Peer review
Peer review information
Nature Communications thanks Manish Tiwari and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data supporting the results of this study are available within the article and its Supplementary Information Files. Materials generated in this study, including the ImageJ macro used for quantification of nodule size and X-gal staining, are available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ambre Guillory, Joëlle Fournier, Audrey Kelner.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55067-3.
References
- 1.Griesmann, M. et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science361, eaat1743 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Lanfranco, L., Fiorilli, V. & Gutjahr, C. Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytol.220, 1031–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Huisman, R. & Geurts, R. A roadmap toward engineered nitrogen-fixing nodule symbiosis. Plant Commun.1, 100019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libourel, C. et al. Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programmes. Nat. Plants9, 1067–1080 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genre, A. & Russo, G. Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci.7, 96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charpentier, M. Calcium signals in the plant nucleus: origin and function. J. Exp. Bot.10.1093/jxb/ery160 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Krönauer, C. & Radutoiu, S. Understanding Nod factor signalling paves the way for targeted engineering in legumes and non-legumes. Curr. Opin. Plant Biol.62, 102026 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Lévy, J. et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science303, 1361–1364 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Tirichine, L. et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature441, 1153–1156 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Singh, S., Katzer, K., Lambert, J., Cerri, M. & Parniske, M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe15, 139–152 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Cerri, M. R. et al. The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol.215, 323–337 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Pimprikar, P. & Gutjahr, C. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol.59, 678–695 (2018). [DOI] [PubMed] [Google Scholar]
- 13.De Carvalho-Niebel, F., Timmers, A. C., Chabaud, M., Defaux-Petras, A. & Barker, D. G. The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J.32, 343–352 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Manthey, K. et al. Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol. Plant Microbe Interact.17, 1063–1077 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Davies, J. M. Annexin-mediated calcium signalling in plants. Plants3, 128–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerke, V. et al. Annexins-a family of proteins with distinctive tastes for cell signaling and membrane dynamics. Nat. Commun.15, 1574 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutjahr, C. & Parniske, M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol.29, 593–617 (2013). [DOI] [PubMed] [Google Scholar]
- 18.de Carvalho-Niebel, F., Fournier, J., Becker, A. & Marín Arancibia, M. Cellular insights into legume root infection by rhizobia. Curr. Opin. Plant Biol.81, 102597 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Ivanov, S., Austin, J., Berg, R. H. & Harrison, M. J. Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat. Plants5, 194–203 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Roth, R. et al. Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat. Plants5, 204–211 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Gao, J. P., Liang, W., Liu, C. W., Xie, F. & Murray, J. D. Unraveling the rhizobial infection thread. J. Exp. Bot.10.1093/jxb/erae017 (2024). [DOI] [PubMed]
- 22.Zhang, G. & Ott, T. Cellular morphodynamics and signaling around the transcellular passage cleft during rhizobial infections of legume roots. Curr. Opin. Cell Biol.91, 102436 (2024). [DOI] [PubMed] [Google Scholar]
- 23.Xiao, T. T. et al. Fate map of Medicago truncatula root nodules. Development141, 3517–3528 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Gaudioso-Pedraza, R. et al. Callose-regulated symplastic communication coordinates symbiotic root nodule development. Curr. Biol.28, 3562–3577.e3566 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Lin, J., Frank, M. & Reid, D. No home without hormones: how plant hormones control legume nodule organogenesis. Plant Commun.1, 100104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaulagain, D. & Frugoli, J. The regulation of nodule number in legumes is a balance of three signal transduction pathways. Int. J. Mol. Sci.22, 1117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fournier, J. et al. Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol.167, 1233–1242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su, C. et al. Transcellular progression of infection threads in Medicago truncatula roots is associated with locally confined cell wall modifications. Curr. Biol.33, 533–542.e535 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, C. W. et al. A protein complex required for polar growth of rhizobial infection threads. Nat. Commun.10, 2848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lace, B. et al. RPG acts as a central determinant for infectosome formation and cellular polarization during intracellular rhizobial infections. Elife12, e80741 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, X. et al. RPG interacts with E3-ligase CERBERUS to mediate rhizobial infection in Lotus japonicus. PLoS Genet.19, e1010621 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, W. C. et al. Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell6, 1415–1426 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao, J. P. et al. Intracellular infection by symbiotic bacteria requires the mitotic kinase AURORA1. Proc. Natl. Acad. Sci. USA119, e2202606119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batzenschlager, M. et al. Competence for transcellular infection in the root cortex involves a post-replicative, cell-cycle exit decision in Medicago truncatula. Elife12, RP88588 (2023).
- 35.Fournier, J. et al. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol.148, 1985–1995 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrine-Walker, F. M., Lartaud, M., Kouchi, H. & Ridge, R. W. Microtubule array formation during root hair infection thread initiation and elongation in the Mesorhizobium-Lotus symbiosis. Protoplasma251, 1099–1111 (2014). [DOI] [PubMed] [Google Scholar]
- 37.van Brussel, A. A. et al. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science257, 70–72 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Berg, R. H. Cytoplasmic bridge formation in the nodule apex of actinorhizal root nodules. Can. J. Bot.77, 1351–1357 (1999). [Google Scholar]
- 39.Timmers, A. C., Auriac, M. C. & Truchet, G. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development126, 3617–3628 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Genre, A., Chabaud, M., Faccio, A., Barker, D. G. & Bonfante, P. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell20, 1407–1420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genre, A., Ortu, G., Bertoldo, C., Martino, E. & Bonfante, P. Biotic and abiotic stimulation of root epidermal cells reveals common and specific responses to arbuscular mycorrhizal fungi. Plant Physiol.149, 1424–1434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo, G. & Genre, A. Divide and be conquered-cell cycle reactivation in arbuscular mycorrhizal symbiosis. Front. Plant Sci.12, 753265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sieberer, B. J., Chabaud, M., Fournier, J., Timmers, A. C. & Barker, D. G. A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J.69, 822–830 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Takeda, N., Maekawa, T. & Hayashi, M. Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell24, 810–822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerri, M. R. et al. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol.160, 2155–2172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelner, A., Leitão, N., Chabaud, M., Charpentier, M. & de Carvalho-Niebel, F. Dual color sensors for simultaneous analysis of calcium signal dynamics in the nuclear and cytoplasmic compartments of plant cells. Front Plant Sci.9, 245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wais, R. J., Keating, D. H. & Long, S. R. Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol.129, 211–224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breakspear, A. et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell26, 4680–4701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Middleton, P. H. et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell19, 1221–1234 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andriankaja, A. et al. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell19, 2866–2885 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerri, M. R. et al. The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol.171, 1037–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rival, P. et al. Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development139, 3383–3391 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Schiessl, K. et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol.29, 3657–3668.e3655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vernie, T. et al. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula Root. Plant Cell27, 3410–3424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, H. et al. A genetic screen for plant mutants with altered nodulation phenotypes in response to rhizobial glycan mutants. New Phytol.220, 526–538 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Vasse, J., de Billy, F., Camut, S. & Truchet, G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol.172, 4295–4306 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Carvalho-Niebel, F., Lescure, N., Cullimore, J. V. & Gamas, P. The Medicago truncatulaMtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol. Plant Microbe Interact.11, 504–513 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Radhakrishnan, G. V. et al. An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants6, 280–289 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Clark, G. B., Morgan, R. O., Fernandez, M. P. & Roux, S. J. Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol.196, 695–712 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Floss, D. S. et al. A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol.27, 1206–1212 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Seguí-Simarro, J. M., Coronado, M. J. & Staehelin, L. A. The mitochondrial cycle of Arabidopsis shoot apical meristem and leaf primordium meristematic cells is defined by a perinuclear tentaculate/cage-like mitochondrion. Plant Physiol.148, 1380–1393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa, A., Navazio, L. & Szabo, I. The contribution of organelles to plant intracellular calcium signalling. J. Exp. Bot.10.1093/jxb/ery185 (2018). [DOI] [PubMed]
- 63.Cui, Y. et al. A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat. Plants5, 95–105 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Cui, Y., Zhao, Q., Hu, S. & Jiang, L. Vacuole biogenesis in plants: how many vacuoles, how many models? Trends Plant Sci.25, 538–548 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Viotti, C. ER and vacuoles: never been closer. Front. Plant Sci.5, 20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whalley, H. J. & Knight, M. R. Calcium signatures are decoded by plants to give specific gene responses. New Phytol.197, 690–693 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Evans, R. C. & Blackwell, K. T. Calcium: amplitude, duration, or location? Biol. Bull.228, 75–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, T., Niu, J. & Jiang, Z. Sensing mechanisms: calcium signaling mediated abiotic stress in plants. Front. Plant Sci.13, 925863 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wais, R. J. et al. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA97, 13407–13412 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marsh, J. F. et al. Medicago truncatulaNIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol.144, 324–335 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Del Cerro, P. et al. Engineered CaM2 modulates nuclear calcium oscillation and enhances legume root nodule symbiosis. Proc. Natl. Acad. Sci. USA119, e2200099119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laohavisit, A. et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca²+- and K+-permeable conductance in root cells. Plant Cell24, 1522–1533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kodavali, P. K. et al. Structural and functional characterization of annexin 1 from Medicago truncatula. Plant Physiol. Biochem.73, 56–62 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Richards, S. L. et al. Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. Plant J.77, 136–145 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Liao, C., Zheng, Y. & Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol.216, 163–177 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Malabarba, J. et al. ANNEXIN1 mediates calcium-dependent systemic defense in Arabidopsis plants upon herbivory and wounding. New Phytol.231, 243–254 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Tichá, M. et al. Advanced microscopy reveals complex developmental and subcellular localization patterns of ANNEXIN 1 in Arabidopsis. Front. Plant Sci.11, 1153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, Z. X. et al. ANNEXIN 8 negatively regulates RPW8.1-mediated cell death and disease resistance in Arabidopsis. J. Integr. Plant Biol.63, 378–392 (2021). [DOI] [PubMed] [Google Scholar]
- 79.Vais, H. et al. ER-luminal [Ca2+] regulation of InsP3 receptor gating mediated by an ER-luminal peripheral Ca2+-binding protein. Elife9, 10.7554/eLife.53531 (2020). [DOI] [PMC free article] [PubMed]
- 80.Peiter, E. The plant vacuole: emitter and receiver of calcium signals. Cell Calcium50, 120–128 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Himschoot, E., Pleskot, R., Van Damme, D. & Vanneste, S. The ins and outs of Ca. Curr. Opin. Plant Biol.40, 131–137 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Konopka-Postupolska, D. & Clark, G. Annexins as overlooked regulators of membrane trafficking in plant cells. Int. J. Mol. Sci.18, 10.3390/ijms18040863 (2017). [DOI] [PMC free article] [PubMed]
- 83.Carrasco-Castilla, J. et al. Down-regulation of a Phaseolus vulgaris annexin impairs rhizobial infection and nodulation. Environ. Exp. Bot.153, 108–119 (2018). [Google Scholar]
- 84.Liu, J. & Bisseling, T. Evolution of NIN and NIN-like genes in relation to nodule symbiosis. Genes11, 10.3390/genes11070777 (2020). [DOI] [PMC free article] [PubMed]
- 85.Schnabel, E., Journet, E. P., de Carvalho-Niebel, F., Duc, G. & Frugoli, J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol.58, 809–822 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Haseloff, J., Siemering, K. R., Prasher, D. C. & Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA94, 2122–2127 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelson, B. K., Cai, X. & Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J.51, 1126–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Zeng, T. et al. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J.94, 411–425 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Edgar, R. C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun.13, 6968 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol.59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 97.Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol.35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all data supporting the results of this study are available within the article and its Supplementary Information Files. Materials generated in this study, including the ImageJ macro used for quantification of nodule size and X-gal staining, are available from the corresponding author upon request. Source data are provided with this paper.