Abstract
The class C orphan G-protein-coupled receptor (GPCR) GPR156, which lacks the large extracellular region, plays a pivotal role in auditory function through Gi2/3. Here, we firstly demonstrate that GPR156 with high constitutive activity is essential for maintaining auditory function, and further reveal the structural basis of the sustained role of GPR156. We present the cryo-EM structures of human apo GPR156 and the GPR156–Gi3 complex, unveiling a small extracellular region formed by extracellular loop 2 (ECL2) and the N-terminus. The GPR156 dimer in both apo state and Gi3 protein-coupled state adopt a transmembrane (TM)5/6-TM5/6 interface, indicating the high constitutive activity of GPR156 in the apo state. Furthermore, C-terminus in G-bound subunit of GPR156 plays a dual role in promoting G protein binding within G-bound subunit while preventing the G-free subunit from binding to additional G protein. Together, these results explain how GPR156 constitutive activity is maintained through dimerization and provide a mechanistic insight into the sustained role of GPR156 in maintaining auditory function.
Subject terms: Hair cell, Cryoelectron microscopy, Cell signalling, G protein-coupled receptors
GPR156 has been identified as a regulator in auditory hair cells. Here, authors present the cryo-EM structures of human apo GPR156 and the GPR156–Gi3 complex to reveal the activation mechanism of GPR156 in maintaining auditory function.
Introduction
GPR156 is highly expressed in auditory hair cells (HCs), and inactivation of GPR156 leads to severe hearing impairment1. Variants of GPR156 identified in human pedigrees have been found to be associated with decreased expression levels, causing recessive congenital hearing loss2. Moreover, GPR156 is a well-conserved cell polarity determinant, and the GPR156-Gαi signaling pathway directs the orientation of mechanical sensory HCs in the mouse cochlea, mouse vestibulum, and zebrafish lateral line otolith organ. A recent study suggests the absence of Go expression in auditory HCs and indicates the exclusive involvement of Gi2 and Gi3 in mediating the orientation impact of GPR156 on HCs3. In addition, a study has shown that GPR156 is also widely distributed in the rat central nervous system, but its physiological function in the central nervous system has not been studied further4.
The class C GPCRs encompass metabotropic GABAB (GABAB) receptor, calcium-sensitive receptor (CaSR), metabotropic glutamate (mGlu) receptors, metabotropic glycine (mGly) receptor (GPR158), taste1 receptors, and several orphan receptors5–7, all of which play essential roles in intercellular communication during both physiological and pathological processes8–11. Class C GPCRs differ from other types of GPCRs in that they only function as obligatory homo- or heterodimers rather than as monomers12–15. The structures of class C GPCRs solved to date all share a large and varying Venus Flytrap (VFT) extracellular domain for ligand-binding, binding of which transduces conformational changes from the VFT to the 7-transmembrane domain (TMD)16–18. However, GPR156, as an orphan class C GPCR possesses a unique short N-terminal sequence (45 residues) that is not long enough to form a large extracellular region with ECL2. In addition, GPR156 exhibits high Gi constitutive activity19 and is highly homologous to GABAB, but has been ruled out as a subtype of the latter20, which suggests an unconventional dimeric form and activation mechanism. Nevertheless, little is known about the mechanisms, ligands, and signaling responses associated with GPR156, and its structural organization remains unclear.
In this work, our in vivo experiments show that GPR156 not only plays a role in the establishment of hearing but also has consistent and continuous activity in order to maintain normal hearing function. To shed light on the structural basis of the sustained role of GPR156, we present the cryo-electron microscopy (cryo-EM) structures of apo GPR156 and the GPR156–Gi3 complex at a resolution of 3.09 Å and 2.39 Å, respectively. Our results unveil a dimeric form and activation mechanism of class C GPCRs in which a smaller extracellular region is formed solely by ECL2 and the short N-terminus in the absence of the VFT. Furthermore, the participation of TM6 is key to the activation of class C GPCRs17,21,22. However, in the apo state, a previously unknown homodimer interface locates between TM5 and TM6 of both GPR156 subunits is identified. In contrast to previous reports on class C GPCRs, we show that both subunits of GPR156 exhibit a nearly identical conformation, thus maintaining the active state. Surprisingly, upon G protein coupling, no rearrangement occurs at the dimer interface. Adding to the intrigue, in the GPR156-Gi3 complex, the C-terminus assumes a dual role. It not only participates in G protein binding within the G-bound subunit but also simultaneously occupies the bottom of the TMD in the G-free subunit, potentially in order to impede perturbations from additional G protein binding. These observations highlight the structural and functional diversity of class C GPCRs, with potentially profound implications for advancing our comprehension of the role of GPR156 in maintaining auditory function.
Results
Physiological roles of GPR156-Gi signaling pathway after auditory maturation
Although an important role for GPR156 in hearing development has been reported1, it is still unknown whether GPR156 continues to play a role after auditory maturity is reached in view of the high Gi constitutive activity of GPR15619. The onset of hearing in mice does not occur until postnatal day (P)12–P14, then progresses to full maturity at P2823,24. In order to further explore how GPR156 affects hearing at different ages, we performed in vivo knock-down experiments by using AAV-mediated GPR156-shRNA at three-time points, including the auditory development stage (P2–P3), the mature auditory stage (P30), and the late stage of auditory maturation (P60) (Fig. 1 and Supplementary Fig. 1). Two AAV-mediated GPR156-shRNAs were designed for delivery into the mouse cochlea through the round window membrane (Supplementary Fig. 1b). Of the two, GPR156-shRNA1 was used throughout the study, and it could be successfully delivered to HCs and could knock down GPR156 by about 50% at the transcriptional level (Supplementary Fig. 1b, c).
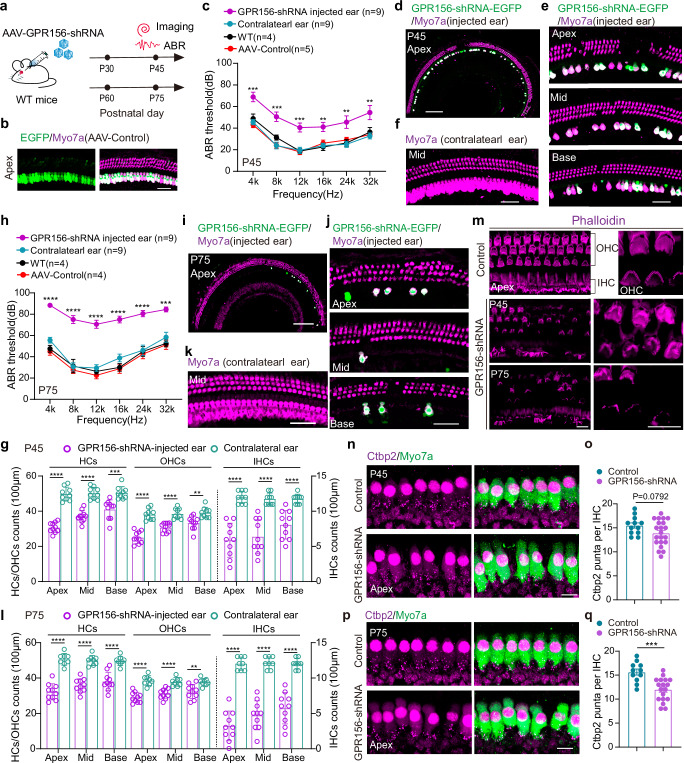
Fig. 1. Knockdown of GPR156 causes hearing loss and hair cell loss in adult mice.
a The experimental design diagram. AAV dose: 6 × 1010 GC /ear. b Representative images of the AAV-control virus infecting inner ear HCs in P30 mice from 6 independent experiments. Scale bar, 40 μm. c The ABR results of the GPR156-shRNA-injected ear (n = 9 mice), the contralateral ear (n = 9 mice), WT mice (n = 4 mice) and AAV-control-injected ear (n = 5 mice). All AAVs were injected into the left ear of P30 mice, and ABR experiments were started at P45. **P < 0.01 and ***P < 0.001 were calculated by a two-tailed unpaired t test. (mean ± SEM (bars), WT vs AAV-control: P = 0.26398(4k), P = 0.07594(8k), P = 0.83978(12k), P = 0.44799(16k), P = 0.50538(24k), P = 0.64441(32k). WT vs contralateral ear: P = 0.50064(4k), P = 0.16799(8k), P = 0.88241(12k), P = 0.72864(16k), P = 0.79457(24k), P = 0.50528(32k). GPR156-shRNA injected ear vs contralateral ear: P = 0.00027(4k), P = 0.00014(8k), P = 0.00064(12k), P = 0.00229(16k), P = 0.00559(24k), P = 0.00794(32k). Source data are provided as a Source Data file.). d, e Low and high magnification representative confocal images of Myo7a signaling in P45 GPR156-shRNA-injected cochleae from 6 independent experiments. Apex, Mid, and Base: apical, middle, and basal turn of the cochlea. Scale bar, 200 μm in (d) and 40 μm in (e). f The representative confocal image of Myo7a signaling in the P45 GPR156-shRNA contralateral cochlea from 6 independent experiments. Scale bar, 40 μm. g The number of HCs, OHCs, and IHCs in the P45 GPR156-shRNA-injected ear and contralateral ear per 100 μm. **P < 0.01, ***P < 0.001, and ****P < 0.0001 were calculated by two-tailed unpaired t test. (mean ± SEM (bars), GPR156-shRNA injected ear(10 regions from 4 mice) vs contralateral ear(9 regions from 4 mice): P = 1.6E-07(Apex-OHC), P = 1.7E-06(Apex-IHC), P = 7.6E-10(Apex-HC), P = 8.6E-06(Mid-OHC), P = 7.4E-06(Mid-IHC), P = 1.8E-08(Mid-HC), P = 0.00181(Base-OHC), P = 7.5E-05 (Base-IHC), P = 0.00034(Base-HC). Source data are provided as a Source Data file.). OHC: the outer hair cell. IHC: the inner hair cell. h The ABR results of the GPR156-shRNA-injected ear (n = 9 mice), the contralateral ear (n = 9 mice), WT mice (n = 4 mice) and AAV-control-injected ear (n = 4 mice). All AAVs were injected into the left ear of P60 mice, and ABR experiments were started at P75. ***P < 0.001 and ****P < 0.0001 were calculated by two-tailed unpaired t test. (mean ± SEM (bars), WT vs AAV-control: P = 0.62022(4k), P = 0.61311(8k), P = 0.38670(12k), P = 0.82908(16k), P = 0.70485(24k), P = 0.82310(32k). WT vs contralateral ear: P = 0.10835(4k), P = 0.92272(8k), P = 0.53047(12k), P = 0.16518(16k), P = 0.89175(24k), P = 0.45458(32k). GPR156-shRNA injected ear vs contralateral ear: P = 6E-09(4k), P = 3E-07(8k), P = 3E-07(12k), P = 1E-06(16k), P = 2E-05(24k), P = 0.0001(32k). Source data are provided as a Source Data file.). i, j Low and high magnification representative confocal images of Myo7a signaling in the P75 GPR156-shRNA-injected cochlea from 6 independent experiments. Scale bar, 200 μm in (i) and 40 μm in (j). k The representative confocal image of Myo7a signaling in the P75 GPR156-shRNA contralateral cochlea from 6 independent experiments. Scale bar, 40 μm. l The number of HCs, OHCs, and IHCs in the P75 GPR156-shRNA-injected ear and contralateral ear per 100 μm. **P < 0.01 and ****P < 0.0001 were calculated by a two-tailed unpaired t test. (mean ± SEM (bars), GPR156-shRNA injected ear(10 regions from 4 mice) vs contralateral ear(8 regions from 4 mice): P = 3E-06(Apex-OHC), P = 7E-09(Apex-IHC), P = 6E-8(Apex-HC), P = 6E-05(Mid-OHC), P = 5E-08(Mid-IHC), P = 2E-07(Mid-HC), P = 0.0037(Base-OHC), P = 1E-06(Base-IHC), P = 3E-05(Base-HC). Source data are provided as a Source Data file.). m The representative image of Phalloidin signaling in the GPR156-shRNA-injected cochlea and control group from 5 independent experiments. Scale bar, 10 μm. n, p The representative images of Ctbp2 staining of GPR156-shRNA-injected cochlea in P45 and P75 mice from 5 independent experiments, respectively. Scale bar, 10 μm. o, q The counts of Ctbp2 from the (n) and (p). ***P < 0.001 was calculated by a two-tailed unpaired t test. (mean ± SEM (bars), GPR156-shRNA injected ear(21 cells from 4 mice) vs control(12cells from 4 mice): P = 0.0792(P45). GPR156-shRNA injected ear(18 cells from 4 mice) vs control(11cells from 4 mice): P = 0.0003(P75). Source data are provided as a Source Data file.).
As expected, the stereocilia of HC in the GPR156 knockdown cochlea showed improper deflection during hearing development, which was consistent with the finding in GPR156 knockout mice1 (Fig. 1m and Supplementary Fig. 1d, e). Of particular interest is the observation that upon maturation of the auditory faculty (P30 and P60), knockdown of GPR156 leads to severe hearing loss (Fig. 1c, h), partial loss of HCs and synapses (Fig. 1d, e, g, i, j, l, n–q). The above in vivo experiments demonstrate that after hearing maturity, appropriate expression of GPR156 and its constitutive activity play an important role in maintaining normal hearing function. Although the specific physiological functions of GPR156-Gαi and its essential role in the auditory establishment have been demonstrated1,3, crucial questions remain regarding the underlying reasons for the high constitutive activity of GPR156 and the structural mechanism by which GPR156 activation triggers downstream signals to maintain the functionality of HCs.
Overall architectures of apo GPR156 and the GPR156–Gi3 complex
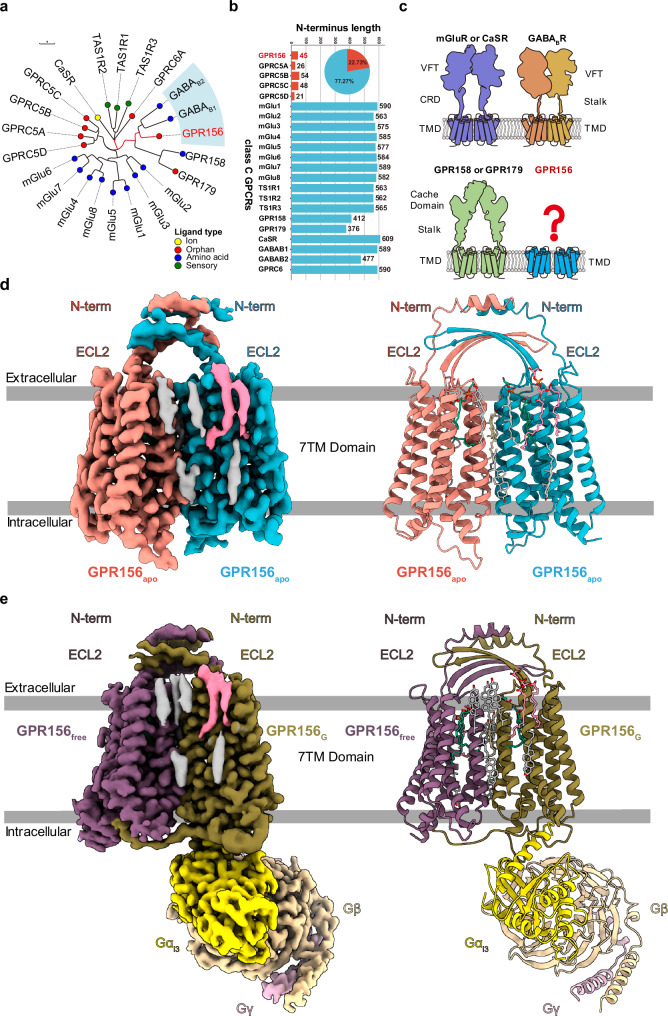
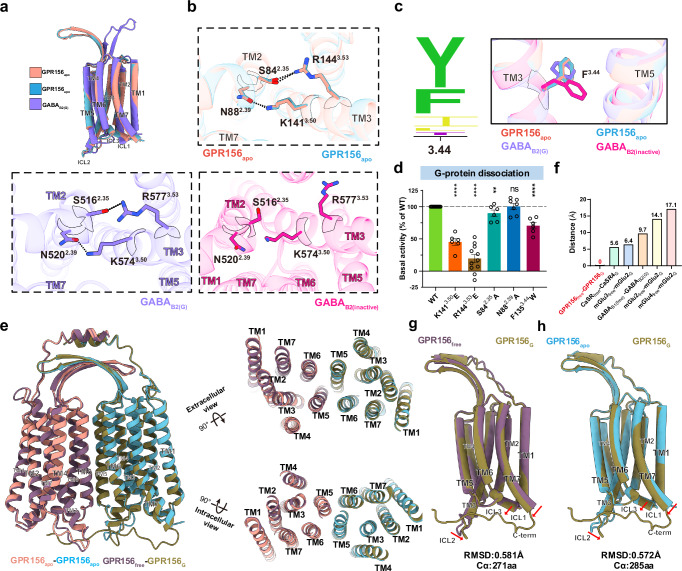
As a class C GPCR, GPR156 exhibits high homology to metabotropic GABA receptors (Fig. 2a). However, GPR156 possesses a distinctively short N-terminal sequence (45 residues), which is insufficient to form a large extracellular region like typical class C GPCRs (Fig. 2b, c). In addition, the characteristic of GPR156’s high constitutive activity further suggests that its structure may adopt an unconventional dimeric form and activation mechanism. To determine the structural basis of the sustained role of GPR156, we expressed and purified human apo GPR156 and GPR156 in complex with the G protein Gi3 (Supplementary Fig. 2 and Supplementary Table 1). The structure of apo GPR156 and the GPR156–Gi3 complex were determined at global resolutions of 3.09 Å and 2.39 Å, respectively (Supplementary Figs. 3 and 4 and Supplementary Table 2).
Fig. 2. Cryo-EM structures of human GPR156 in apo state and in complex with Gi3.
a Phylogenetic tree of the 22 class C GPCR family members with different ligand types. GPR156 is highlighted with a red mark. The scale bar indicates the number of substitutions per site. b Summary of the N-terminal lengths of the 22 class C GPCRs. GPR156 and GPRC5A–D are the only five GPCRs of class C whose N-terminal length is less than 60 residues. VFT: Venus Flytrap, CRD: Cysteine-rich Domain, and TMD: Transmembrane Domain. Source data are provided as a Source Data file. c The structural features of class C GPCRs include mGluR or CaSR (purple), GPR158 or GPR179 (green), GB1 subunit (orange), GB2 subunit (yellow), and GPR156 (blue). d, e Cryo-EM maps (left panel) and models (right panel) of human apo GPR156 (d) and GPR156–Gi3 complex (e).
Significantly different from other class C GPCR dimerization mechanisms, the structure of GPR156 showed a unique homodimer assembly via the ECL2, N-terminus, and TMD (Fig. 2d, e). The conformation of the two subunits was almost the same, with the dimer showing almost mirror symmetry. High-resolution TMDs were observed for each GPR156 protomer (Supplementary Fig. 5). However, N-terminus densities were limited, and side chain densities were not visible due to the high conformational flexibility (Supplementary Figs. 3e, 4e, and 5). Nevertheless, the main organizational features of the N-terminus were identifiable. In addition, the GPR156 dimer as a whole presented a conformation in which the two subunits were close together on the intracellular side (Fig. 2d, e). Moreover, several cholesterol molecules could be seen around the dimer (apo GPR156: 8 cholesterols; the GPR156–Gi3 complex: 10 cholesterols), which is a characteristic consistent with class C GPCRs25. Furthermore, cholesterol was also present within the dimer interface, while lipids packed by hydrophobic residues in the TMD could also be identified (Fig. 2d, e and Supplementary Fig. 5).
The small extracellular region has minimal impact on the constitutive activity of GPR156
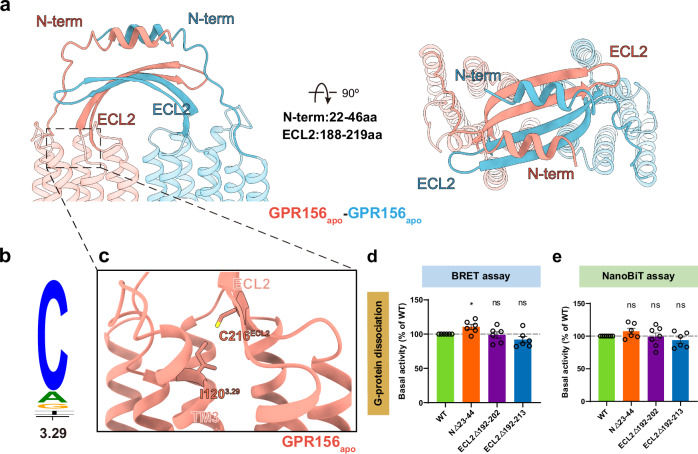
The ECL2 of the currently known class C GPCRs interacts primarily with the elongated stalk–referred to as the cysteine-rich domain (CRD)–in mGlu and CaSR (linking the VFT to the TMD) and forms a disulfide bond with the cysteine of TM317,22,26. However, the N-terminus of most class C GPCRs typically comprises over 300 residues, whereas GPR156 exhibits a significantly shorter N-terminus consisting of only 45 residues (Fig. 2b). Therefore, the N-terminus of both GPR156 protomers without elongated stalks exhibit a unique conformation, forming a double-deck bridge-like structure with the ECL2 (Fig. 3a). In addition, the disulfide bond between ECL2 and TM3 (conserved C3.29; superscript numbers refer to the GPCRdb numbering scheme) was previously considered conserved in class C GPCRs, but no such corresponding disulfide bond exists in GPR156 (I1203.29 in GPR156) (Fig. 3b, c). Thus, the absence of this disulfide bond makes deflection of the ECL2 of GPR156 possible. This is an unprecedented configuration compared to the organization of ECL2 and the N-terminus in other class C GPCRs of various states (active, apo, and inactive states) (Supplementary Fig. 6a–c).
Fig. 3. The unique arrangement of the N-terminus and ECL2.
a Side view (left panel), and a top view (right panel) of the distinct ECL2 and N-terminus conformations. b Sequence alignment logo showing the conservation of C3.29 among family C GPCRs. c Close-up view of the ECL2 and transmembrane region. In GPR156, C3.29 is not conservative and is replaced by I1203.29. d, e Basal activity of WT and three mutant constructs in the N-terminus and ECL2 of GPR156, measured by BRET-based assay (d) (from left to right n = 6, 6, 6, 6; NΔ23-44 vs WT: P = 0.0170; ECL2Δ192-202 vs WT: P = 0.7947; ECL2Δ192-213 vs WT: P = 0.1190) and NanoBiT-based assay (e) (from left to right n = 7, 6, 7, 6; NΔ23-44 vs WT: P = 0.1038; ECL2Δ192-202 vs WT: P = 0.8694; ECL2Δ192-213 vs WT: P = 0.1819). Data are presented as the percentage of WT activity and are shown as the mean ± SEM (bars) from at least six independent experiments performed in technical triplicate with individual data points shown (dots). ns (not significant) = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-tailed unpaired t test compared to WT. Supplementary Fig. 7a, b provides the related surface expression level. Supplementary Tables 3 and 4 provide detailed information related to BRET-based assay (d) and NanoBiT-based assay (e). Source data are provided as a Source Data file.
Given the absence of the VFT region, the elongated stalk, and the disulfide bond between ECL2 and TM3, we hypothesized that GPR156’s ECL2 and N-terminus do not fulfill the conserved roles of all known class C GPCRs in response to ligand activation. To test this, we substituted the ECL2 and N-terminus of GPR156 with a linker composed of glycine and serine, respectively, and examined the effect of these on constitutive activity (Supplementary Fig. 6d). Consistent with our structure, these constructs exhibited slight or no alteration in basal activity when compared to wild type (WT) (Fig. 3d, e, Supplementary Fig. 7a, b and Supplementary Tables 3 and 4), suggesting a minimal impact of GPR156’s ECL2 and N-terminus on its constitutive activity.
A distinct transmembrane homodimer interface in apo GPR156
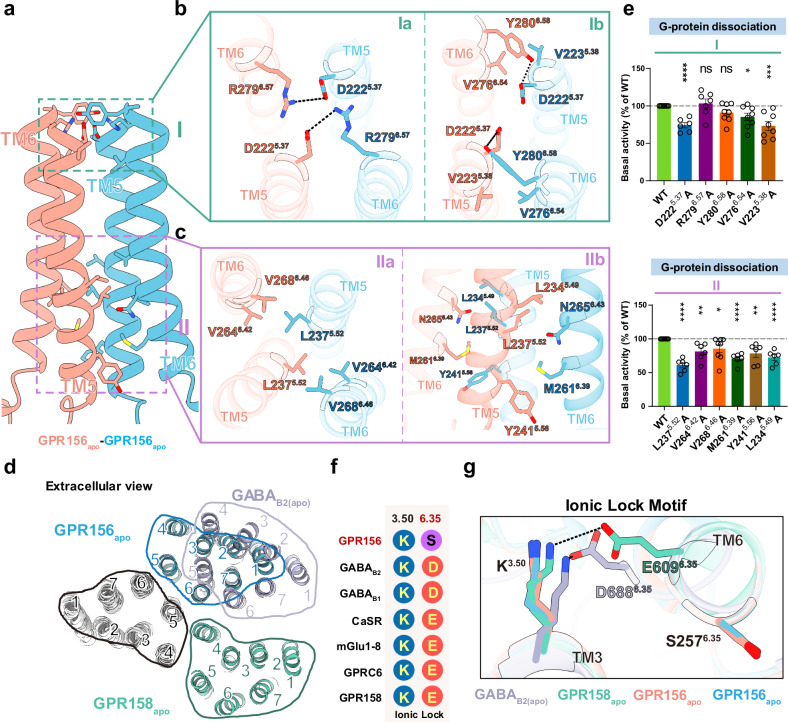
An unknown homodimer interface located between TM5 and TM6 of both protomers was identified in GPR156’s apo conformation (Fig. 4a and Supplementary Fig. 8a). The TM5/6 helices are arranged in a V-shaped orientation at the dimeric interface (Supplementary Fig. 8b). In the homodimer interface, the residues at the top and bottom part of TM5 and TM6 directly contact each other to form two distinct core regions (I-II) (Fig. 4a), whereas the middle region interacts solely through a cholesterol molecule (Supplementary Fig. 8c). Due to the intervention of cholesterol, the area of the interaction interface increased by 165.8 Å2, and cholesterols have also previously been found in the middle of other class C GPCR dimer interfaces13,26,27.
Fig. 4. Transmembrane interface of the apo GPR156 homodimer.
a Detailed interactions of the transmembrane interface in the apo GPR156. The 7TM homodimer interface is formed at two regions (I and II), namely the extracellular and intracellular side. b Detailed interactions on the surface of the region I. The network of electrostatic interactions is shown in Ia, and the affiliated hydrophobic interaction is shown in Ib. c Detailed interactions in the intracellular region II. A hydrophobic contact network is shown in IIa. The van der Waals forces at the homodimer interface are shown in IIb. d Comparison of the dimeric interface of apo GPR156 transmembrane domains with those of apo GABAB receptor (PDB code: 6VJM) and apo GPR158 (PDB code: 7EWL). e The basal activity of WT and versions with mutations in the dimer interface of GPR156, as measured by BRET-based assay (from left to right n = 8, 6, 7, 8, 8, 8, 8, 6, 6, 8, 6, 6, 6; D2225.37A vs WT: P = 2.52817E-06; R2796.57A vs WT: P = 0.5694; Y2806.58A vs WT: P = 0.0560; V2766.54A vs WT: P = 0.0118; V2235.38A vs WT: P = 0.0006; L2375.52A vs WT: P = 5.2657E-08; V2646.42A vs WT: P = 0.0015; V2686.46A vs WT: P = 0.0309; M2616.39A vs WT: P = 3.1286E-07; Y2415.56A vs WT: P = 0.0014; L2345.49A vs WT: P = 6.6258E-06). Data are presented as a percentage of WT activity and are shown as the mean ± SEM (bars) from at least six independent experiments performed in technical triplicate with individual data points shown (dots). ns (not significant) = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-tailed unpaired t test compared to WT. Supplementary Fig. 7c provides the related surface expression level, and Supplementary Table 3 provides detailed information. Source data are provided as a Source Data file. f The key residues in the ionic lock motif (3.50 and 6.35) are aligned among members of class C GPCRs. g Close-up view of the conserved ionic lock motif showing the different conformations between GPR156 and other members of the class C subfamily in the apo state, including GABAB receptor (PDB code: 6VJM) and GPR158 (PDB code: 7EWL).
The core region I, including parts Ia and Ib, encompasses the interface at the extracellular end. Ia represents a network of electrostatic interactions (D2225.37 and R2796.57) that effectively tether the extracellular ends of both of the GPR156 monomers’ TMDs (Fig. 4b). Residue D2225.37 of each subunit also forms a hydrogen bonding interaction with Y2806.58 of the other protomer’s TM6, while V2235.38 of each subunit engages in a hydrophobic interaction with V2766.54 of the other subunit, both of which together establish the affiliated site Ib (Fig. 4b). The core region II covers the intracellular end of the transmembrane homodimer interface (Fig. 4c). A hydrophobic contact network is formed by L2375.52, V2646.42, and V2686.46 of each subunit (IIa in Fig. 4c). In addition, L2375.52, Y2415.56, and L2345.49 on TM5 of one protomer in this region are in contact with M2616.39 and N2656.43 on TM6 of the other protomer, respectively (IIb in Fig. 4c).
While the involvement of TM5 in the class C GPCR apo state homo- or heterodimer interface has been previously identified (apo GABAB receptor28 and apo GPR15826,29) (Fig. 4d), TM6 has previously only been shown to participate in the active class C GPCR dimer interface17,21,22. Here, we also show that TM6 is involved in an apo state dimer interface. The dimerization of GPR156 in the apo state leads to the formation of a TM5/TM6 interface, which may induce steric hindrance between the two subunits’ TM5 helices, thereby facilitating the movement of TM5 and TM3 akin to the positive allosteric modulator (PAM) function observed in the GABAB receptor (Supplementary Fig. 8d). To determine the importance of this specific dimer interface, we mutated the dimer interface and performed BRET-based G-protein dissociation assays (Fig. 4e, Supplementary Fig. 7c and Supplementary Table 3). It is noteworthy that the mutation to alanine in the dimer interface except R2796.57 and Y2806.58 on GPR156 reduced the basal activity of the receptor, which indicated that the TM5/TM6 interface does indeed play a role in the signal transduction of GPR156.
Non-canonical features of the apo GPR156 subunits
Conventionally, the ionic lock motif is conserved in class C GPCRs, but the GPR156 subunit exhibits peculiar differences. The ionic lock is primarily formed by the conserved K3.50 with D6.35 or E6.35, similar to K6653.50–E7706.35 in mGlu530, K5743.50–D6886.35 in GABAB227,31, and K5023.50–E6096.35 in GPR15826,29 (Fig. 4f). In GPR156, S2576.35 replaces the D6.35 or E6.35 that is found in other class C GPCRs, thereby potentially preventing the formation of a stable interaction with K3.50 in the apo or inactive state (Fig. 4f, g).
Given the lack of the ionic lock motif and the high homology between GPR156 and the GABAB receptor, comparing the TMDs of the two subunits in apo GPR156 with the GABAB2 subunit binding the G protein (GABAB2(G)) revealed a high degree of similarity, with the root mean square deviation (RMSD) measuring 1.023 Å and 0.964 Å (Fig. 5a and Supplementary Fig. 8e, f). K1413.50 in both subunits of apo GPR156 forms a hydrogen bond with N882.39, which mirrors the previously observed interaction in the GABAB2(G) subunit (K5743.50–N5202.39)21 (Fig. 5b). In addition, S842.35 and R1443.53 also form a hydrogen bond to further stabilize the conformation, and this previously undiscovered interaction similarly exists in the GABAB2(G) subunit (Fig. 5b). The conformations of the active state feature F3.44 in both apo GPR156 subunits, and these closely resemble the conformation in GABAB2(G), showing marked distinctions from the inactive state of GABAB2 (GABAB2(inactive)) (Fig. 5c). Coinciding with our structure, the mutation of K1413.50, R1443.53, S842.35, and F1353.44 had a substantial impact on the basal activity (Fig. 5d, Supplementary Fig. 7d and Supplementary Table 3). Furthermore, the forementioned findings underscore that both subunits in apo GPR156 exhibit active-state conformations and interactions consistent with the GABAB2(G) subunit, and these are different from the inactive state of GABAB2 (Fig. 5b, c and Supplementary Fig. 8g), implying the ability of both GPR156 subunits to engage with G proteins in the apo state.
Fig. 5. Structural details and Gi-associated transitions of the TMD in GPR156.
a Side views of the superposed structures of the GPR156apo and GABAB2(G) (PDB code: 7EB2) aligned by the TMD of GABAB2(G). b Magnified views of the detailed interactions within the TMD of apo GPR156 (top panel), GABAB2(G) (left panel, PDB code: 7EB2), and GABAB2(inactive) (right panel, PDB code: 6WIV). c Sequence alignment logo showing the conservation of Y3.44 or F3.44 among family C GPCRs (left panel). And magnified views of the critical residue F3.44 in the apo state of GPR156 and different states of GABAB2 (right panel). d The basal activity of WT and mutant versions as measured by BRET-based assay (from left to right n = 12, 6, 11, 6, 7, 6; K1413.50E vs WT: P = 3.9825E-11; R1443.53E vs WT: P = 8.9060E-12; S842.35A vs WT: P = 0.0086; N882.39A vs WT: P = 0.9080; F1353.44W vs WT: P = 1.5807E-07). Data are presented as the percentage of WT activity and are shown as the mean ± SEM (bars) from at least six independent experiments performed in technical triplicate with individual data points shown (dots). ns (not significant) = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-tailed unpaired t-test compared to WT. Supplementary Fig. 7d provides the related surface expression level. Supplementary Table 3 provides detailed information. Source data are provided as a Source Data file. e Side, extracellular, and intracellular views of the superposed structures of apo GPR156 dimer and Gi-bound GRP156 dimer. f Upon G protein coupling, the dimeric TMD of five class C GPCRs (including mGlu3free–mGlu2G (Cα of V699; PDB code: 8JD3), mGlu4free–mGlu2G (Cα of V699; PDB code: 8JD5), mGlu2free–mGlu2G (Cα of V769; PDB code: 7MTS), CaSRfree–CaSRG (Cα of M771; PDB code: 8WPU), and GABAB1(free)–GABAB2(G) (Cα of C614; PDB code: 7EB2)) undergo conformational rearrangements, except for GPR156free–GPR156G. Source data are provided as a Source Data file. g, h Comparison of the GPR156 G-bound subunit and GPR156 G-free subunit (g), and the GPR156 G-bound subunit and apo GPR156 subunit (h). The RMSD levels were calculated.
No rearrangement occurred in the dimer interface after G protein coupling
To assess changes in GPR156 dimerization after G protein coupling, a comparative analysis between apo GPR156 and the active GPR156 dimer revealed striking similarity (RMSD of 0.539 Å for pruned 572aa; RMSD of 0.902 Å for all 596aa) (Fig. 5e). Surprisingly, after G protein coupling, no interface rearrangement occurred in GPR156, a phenomenon unprecedented in class C GPCRs (Fig. 5e, f). When comparing the G-bound subunit with both the G-free and apo subunits (RMSD of 0.581 Å and RMSD of 0.572 Å, respectively), we observed only minor intracellular contraction in intracellular loop 2 (ICL2), ICL1, and TM7 of the G-bound subunit (Fig. 5g, h). Notably, the G-bound subunit exhibited none of the common rotations or translations found in class C GPCRs but showed the presence of the C-terminus (Fig. 5g, h and Supplementary Fig. 5). The subsequent conformation and interactions of the active state GPR156 subunits’ internal TMs mirrored the apo state conformation and interactions of both subunits in the apo GPR156, akin to the GABAB2(G) subunit (Fig. 5h and Supplementary Fig. 8h–j). It can be inferred that a symmetric dimeric form exists in GPR156 (apo state), with both subunits adopting an active-like state. Upon G protein coupling, no substantial rearrangements occur, with only minor intracellular region contraction in the G-bound subunit to accommodate the G protein (Fig. 5g, h). In addition, this work shows a class C GPCR to have a dimeric interface in the active state that exists in a completely symmetric form, as well as the involvement of TM5 in the active state (Supplementary Fig. 9).
Our structure also revealed a kind of endogenous phospholipid within the transmembrane domains of both subunits in apo GPR156 and the GPR156–Gi3 complex structures, also located at the extrahelical site. From the density and pocket size, we inferred that this was a phospholipid with two aliphatic chains, and mass spectrometry was used to further identify this as phosphatidylglycerol (PG) 36:2 (Supplementary Fig. 10a–d and Supplementary Data 1). It is noteworthy that each GPR156 subunit establishes a comprehensive network of contacts with both bound lipids, and this may help maintain the stability of the receptor (Supplementary Fig. 10e, f). In addition, three replicates of the GPR156 dimer system with the internal phospholipids removed were further created, and a 300 ns molecular dynamics (MD) simulation was sampled for each replicate (Supplementary Fig. 10g, h, Supplementary Figs. 11 and 12 and Supplementary Table 5). Based on the calculation results of the cavity volume for all three MD trajectories, a similar collapse of the transmembrane helices was observed, as demonstrated in the GABAB receptor32. The increased RMSD values at the intracellular termini of the TM helix suggest that the depletion of the internal phospholipids hinders G protein binding (Supplementary Figs. 13–16 and Supplementary Table 6). Like the GABAB receptor (C6.50, corresponding to W6.50 of the toggle switch motif)27,31, G6.50 of GPR156 lacks a large side chain (Supplementary Fig. 10i–k), preventing it from responding to the activation of an intramembrane ligand as observed in mGlu233 (Supplementary Fig. 10l).
Distinct conformation of the C-terminus in class C GPCRs
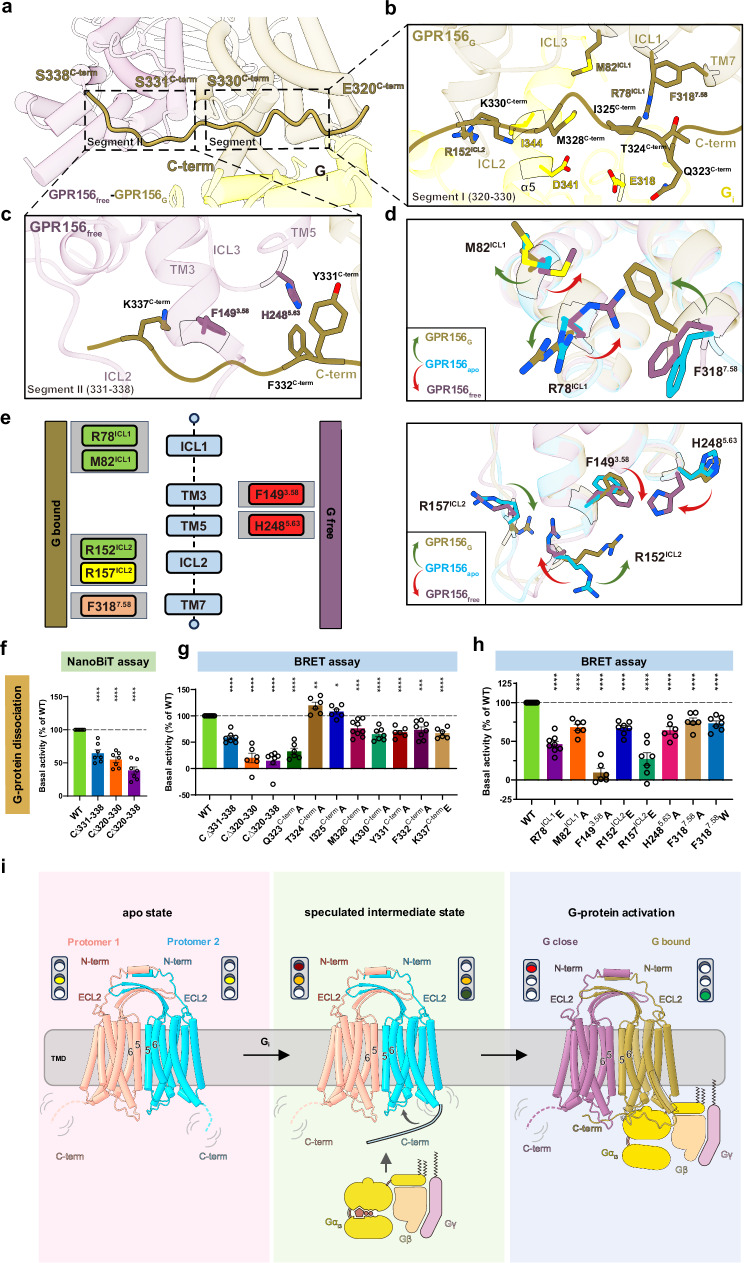
The stark deviation observed from the known G protein coupling mechanisms of class C GPCRs warrants an in-depth investigation into how GPR156 responds to G protein coupling. Hence, the C-terminus observed solely in the G-bound subunit caught our attention. The discernible density of the C-terminus from the G-bound subunit includes residues 320–338 and lies approximately parallel to the intracellular end of the GPR156 dimer, whereas it was absent in the apo state (Figs. 2d, e and 6a and Supplementary Fig. 5). It spans the G protein-binding pocket in the G-bound subunit and extends to the G-free subunit, thus occupying the bottom of the TMD in the G-free subunit (Fig. 6a). This constitutes the initial identification within class C GPCRs of the G-free subunit’s potential G protein pocket being occupied by the C-terminus (Fig. 6a). The G protein pocket of GPR156 aligns with reports on known class C GPCRs21,33,34 by forming a shallow pocket primarily involving ICL1, ICL2, and the C-terminus (Supplementary Fig. 17a–d and Supplementary Table 3). The interaction area between the α5 helix of Gαi3 and GPR156 is 808 Å2 (Supplementary Fig. 17e). Of this, the C-terminus of GPR156 accounts for only 10.6% (86 Å2), which is a notable contrast to the C-terminus of mGlu2 or CaSR (Supplementary Fig. 17f). Furthermore, comparing GPR156’s G-bound subunit with mGlu2’s or CaSR’s G-bound subunit15,33,34 illustrates that, unlike GPR156, their C-terminus extends downward in close proximity to the G protein without extending into the G protein pocket of the G-free subunit (Supplementary Fig. 17g). These findings suggest that the C-terminus of the G-bound subunit of GPR156 may play a previously unknown role.
Fig. 6. GPR156–Gi coupling and the special role of the C-terminus.
a Close-up view of the GPR156-Gi3 complex at the cytoplasmic end, showing a long C-terminal tail (320–338aa). b, c Detailed interactions of the C-terminal tail (320–330aa) of GPR156G with the TMD of GPR156G and Gαi (b), and detailed interactions of the C-terminal tail (331–338aa) of GPR156G with the TMD of GPR156free (c). d Close-up views of the seven key residues in the GPR156G and GPR156free subunits show conformational changes upon G protein coupling. e Schematic representation of the seven key residues’ contacts between the GPR156G and GPR156free subunits and the C-terminal tail of GPR156G and Gαi. Residues from the GPR156G subunit are shown on the left, and those of the GPR156free subunit are shown on the right. The C-terminal tail of GPR156G–GPR156free is in red, the C-terminal tail of GPR156G–GPR156free is in orange, the C-terminal tail of GPR156G–Gαi is in yellow, and both C-terminal tails of GPR156G–GPR156G and GPR156G–Gαi are in green boxes. f The basal activity of WT and the three mutant constructs of the C-terminal tail from GPR156, as measured by NanoBiT-based assay (from left to right n = 7, 7, 7, 7; CΔ331-338 vs WT: P = 2.4064E-05; CΔ320-330 vs WT: P = 2.1818E-07; CΔ320-338 vs WT: P = 9.9574E-08). Data are presented as a percentage of WT activity and are shown as the mean ± SEM (bars) from at least six independent experiments performed in technical triplicate with individual data points shown (dots). *P< 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-tailed unpaired t test compared to WT. Supplementary Fig. 7e provides the related surface expression level, and Supplementary Table 4 provides detailed information. Source data are provided as a Source Data file. g, h The basal activity of WT and mutant versions of GPR156 (including mutant constructs of the C-terminal tail) (g) (from left to right n = 9, 7, 6, 7, 6, 6, 6, 9, 7, 6, 8, 6; C∆331-338 vs WT: P = 7.4009E-09; C∆320-330 vs WT: P = 5.5541E-08; C∆320-338 vs WT: P = 4.6108E-09,; Q323C-termA vs WT: P = 4.2212E-09; T324C-termA vs WT: P = 0.0017; I325C-termA vs WT: P = 0.0470; M328C-termA vs WT: P = 0.0002; K330C-termA vs WT: P = 3.4903E-07; Y331C-termA vs WT: P = 1.8316E-07; F332C-termA vs WT: P = 0.0001; K337C-termE vs WT: P = 9.1879E-08) and mutant constructs of the seven key residues (h) (from left to right n = 9, 8, 6, 6, 7, 7, 6, 6, 7; R78ICL1E vs WT: P = 3.5544E-10; M82ICL1A vs WT: P = 2.1033E-07; F1493.58A vs WT: P = 3.9032E-11; R152ICL2E vs WT: P = 1.4385E-08; R157ICL2E vs WT: P = 7.0588E-08; H2485.63A vs WT: P = 3.5652E-07; F3187.58A vs WT: P = 9.5730E-06; F3187.58W vs WT: P = 3.2740E-07) as measured by BRET-based assay. Data are presented as a percentage of WT activity and are shown as the mean ± SEM (bars) from at least six independent experiments performed in technical triplicate with individual data points shown (dots). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-tailed unpaired t test compared to WT. Supplementary Fig. 7f, g provides the related surface expression level, and Supplementary Table 3 provides detailed information. Source data are provided as a Source Data file. i Schematic diagrams summarizing the conformational changes of the GPR156 homodimers upon G protein coupling.
The dual role of the GPR156 C-terminus in G-protein coupling
To further elucidate the specific interaction and role of the C-terminus of the G-bound subunit, it can be divided into two segments (Fig. 6a). Segment I (320–330aa) is involved in interactions with the G-bound subunit itself and the G protein (Fig. 6b), while segment II (331–338aa) participates in interactions with the G-free subunit (Fig. 6c). We hypothesized that Segment I is involved in G protein binding, somewhat similar to the role seen in mGlu233. Considering the capability of both subunits to bind the G protein, Segment II may prevent the G-free subunit from coupling to the G protein. We found that steric hindrance impeded the possibility of both subunits while simultaneously coupling to the G protein heterotrimer (Supplementary Fig. 18a). It is conceivable that if the potential G protein pocket of the G-free subunit is not occupied by the C-terminus, both subunits would competitively couple with G protein simultaneously, leading to mutual interference. Thus, we posit that Segment II serves the purpose of preventing disturbances from excess G protein binding, thus ensuring the stability of G protein activation. To validate our hypothesis, three constructs (C∆331–338, C∆320–330, and C∆320–338) were constructed (Supplementary Fig. 18b), and the impact of C-terminal deletions on basal activity was assessed (Fig. 6f, g). In comparison to the WT receptor, the basal activity of all three truncation mutants significantly decreased (Fig. 6f, g, Supplementary Fig. 7e, f and Supplementary Tables 3 and 4), indicating that both segments at the C-terminus have an influence on the constitutive activity of GPR156.
In detail, Q323C-term in Segment I forms a potential hydrogen bond with E318G.h4s6.12 of the G protein (Fig. 6b). Additionally, M328C-term interacts with D341G.H5.13 of the G protein and M82ICL1 of the G-bound subunit, while K330C-term engages in interactions with I344G.H5.16 of the G protein and R152ICL2 of the G-bound subunit (Fig. 6b). Furthermore, T324C-term and I325C-term form interactions with F3187.58 and R78ICL1 of the G-bound subunit, respectively (Fig. 6b). In Segment II, F332C-term and K337C-term establish interactions with H2485.63 and F1493.58 of the G-free subunit, respectively, while Y331C-term engages in interactions with H2485.63 of the G-free subunit (Fig. 6c). Compared to the WT receptor, mutations of Y331C-term, F332C-term, and K337C-term in Segment II result in a decrease in basal activity (Fig. 6g, Supplementary Fig. 7f and Supplementary Table 3). Notably, mutation of Q323C-term in Segment I leads to a more than 50% reduction in basal activity (Fig. 6g), underscoring the crucial role of the C-terminus of the G-bound subunit in the G protein pocket of the G-bound subunit in GPR156 coupling to Gi3.
After G protein coupling, the side chains of seven amino acids were found to undergo distinct conformational changes in the G protein pocket of the G-bound subunit and the potential pocket of the G-free subunit (Fig. 6d, e). Mutations of these seven crucial amino acids resulted in varying degrees of reduction in basal activity (Fig. 6h, Supplementary Fig. 7g, and Supplementary Table 3). Notably, F1493.58 underwent conformational change only in the G-free pocket and interacts solely with the K337C-term of the G-bound subunit (Fig. 6c, d). Consistent with this, mutations of F1493.58 led to a reduction in basal activity by over half (Fig. 6h), further highlighting the important role of the C-terminus of the G-bound subunit in occupying the potential G protein pocket of the G-free subunit during GPR156–Gi3 coupling. Combining the aforementioned analysis with functional experiments, we confirm the dual role of the C-terminus of GPR156’s G-bound subunit in the G protein coupling process. On the one hand, the C-terminus promotes G protein coupling in the G protein pocket of the G-bound subunit (320–330aa). On the other hand, the C-terminus occupies the bottom of the TMD of the G-free subunit (331–338aa) so as to impede unwanted perturbations from additional G protein binding.
Discussion
This study presents the cryo-EM structures of apo GPR156 and the GPR156-Gi3 complex. Here, we describe a homotypic dimeric form and provide a dynamic view of G protein coupling that is distinct from all known class C GPCRs, and we metaphorically refer to the dual function of the C-terminus as a traffic light (Fig. 6i). Prior to G protein coupling, GPR156 adopts a symmetrical dimeric form, with both subunits exhibiting nearly identical conformation and possessing the ability to couple with G proteins, analogous to being in a yellow light “ready” state. As G protein coupling initiates, there is no rearrangement at the dimeric interface, and the TMDs of the dimer undergo no significant rotation or translation. We hypothesize that the C-terminus of the GPR156 subunit proximal to the G protein interacts with the G protein (e.g., Q323C-term–E318G.h4s6.12). After G protein coupling, the C-terminus of the G-bound subunit aligns parallel to the intracellular end of the dimer. On the one hand, this facilitates the binding of the G protein of the G-bound subunit, resembling a green light in the open state. On the other hand, it occupies the bottom of the TMD in the G-free subunit, impeding the interference caused by excess G protein binding, akin to a red light in the closed state.
Moreover, the specificity of the small extracellular region of GPR156 is indisputable. The absence of the VFT region, the elongated stalk, and the disulfide bond between ECL2 and TM3 render GPR156 incapable of exerting the conserved functions observed in the activation state of all known class C GPCRs. In addition, there is also a subclass of GPCRs with short N-terminus similar to that of GPR156 in class C GPCRs, including GPRC5A (N-terminus: 26 residues), GPRC5B (N-terminus: 54 residues), GPRC5C (N-terminus: 48 residues), and GPRC5D (N-terminus: 21 residues) (Fig. 2b). A question worth considering is how these class C GPCRs without a large extracellular region form dimers and whether the dimeric form of GPR156 might represent a subclass of class C GPCR dimers, and this may be worthy of further study.
An excellent work has recently been reported on GPR156 in the Go-free and Go-coupled states35, and our physiological and structural analyses presented here have given us further valuable insights into those findings. By comparing the mass spectrometry data with samples of two class A GPCRs known to lack double-chain phospholipids, we have identified the differential enrichment of PG 36:2 in our GPR156 fraction, but not phosphatidylcholine (PC) molecules with corresponding aliphatic chain lengths (Supplementary Fig. 10a, b). Thus, we speculate that the phospholipids bound within the TMD are more likely to be PG rather than PC. Combining the previous studies of phospholipids in GABAB receptors with the highest homology to GPR15627,31,32, our MD simulation analysis (Supplementary Fig. 10g, h), and our structural analysis of the toggle switch motif (Supplementary Fig. 10i–l), we propose that phospholipids within GPR156 act to maintain the integrity and stability of the receptor but might not activate the receptor as recently reported. However, the special dimer interface (TM5/6-TM5/6) and the lack of ionic lock in GPR156 may be the reasons for maintaining the active-like conformation in the apo state.
More importantly, our investigation reveals unique features in GPR156’s interaction with the G protein, highlighting a binding pattern with Gi3 that is distinct from the Go coupling that has recently been observed35. Notably, it has been reported that Go is unable to be detected in auditory HCs, and only Gi2 and Gi3 are involved in the orientation effect of GPR156 on HCs3. Therefore, the dual functional role of the C-terminus in the GPR156–Gi3 complex is more representative of at least the role of GPR156 in auditory function.
Compared with P30, the knockdown of GPR156 at P60 showed more pronounced hearing loss, which may be related to synaptic damage (Fig. 1). Since there is no AAV that can efficiently infect the OHCs of adult mice36–38, and the limitation of age-related hearing loss in C57BL/6 J strain mice39, in order to further investigate the effects of GPR156 on hearing function and its mechanism of action at different time points, subsequent studies using new AAV vectors or transgenic mice are needed.
Taken together, our investigation reveals the indispensable role of GPR156 in ensuring the normal operation of uninterrupted auditory function across developmental and maturation phases. Our observations further provide structural information regarding the atypical activation mechanism of GPR156, which is characterized by rapid and stable constitutive coupling to Gi3 protein, making it well-suited for auditory function.
Methods
Animals
Both sexes, C57BL/6 J Wild-type mice, were used at a 1:1 ratio in all animal experiments. The mice were raised at room temperature of 22 ± 1 °C under a 12 h light-dark cycle with food and water available ad libitum. All experiments were approved by the Institutional Animal Care and Use Committee of Southeast University, China, and all efforts were made to minimize the number of mice used (no.20210606001).
Plasmid design and AAV purification
The GPR156-shRNA sequence was designed with the help of the shRNA-design website (Thermo Fisher Scientific). The GPR156-shRNA was inserted into the AAV vector tagged with the fluorescent protein mNeonGreen. AAV.7m8 was used as the AAV capsid because it can efficiently infect HCs37,40, and this, along with the targeted plasmid and helper plasmid, was delivered to human embryonic kidney (HEK) 293T cells (ATCC, CRL-3216) at molar ratio 1:1:1 by linear polyethylenimine (Yeasen, 40816ES02). AAV purification and titer assays were performed according to the previously published protocol38,41. In brief, The HEK293T cells and medium were collected after plasmid transfection 96 h. Then AAV was obtained by chloroform cleavage and precipitation by PEG8000. The AAV titer was detected by SYBR (Vazyme, Q712). The sense strand sequence of GPR156-shRNA1 and GPR156-shRNA2 were cgg agc atg caa tgt agc ttt and gta ccg ttt cta gtt cac aaa, respectively. And the loop sequence of GPR156-shRNA was tcaagag.
AAV injection in mice
The neonatal mice were anesthetized by cooling on ice. The skin of the neck of the left ear was then clipped, and the fat and muscle were pared away to expose the round window membrane. For the neonatal mice, the AAVs were injected into the cochlea via the round window membrane, and the volume was controlled at 1.5 µL.
The adult mice were anesthetized by tribromoethanol (500 mg/kg). The hair was cleared in the left neck area, and a small cut was made in the neck skin to separate the fat and muscle, thus exposing the posterior semicircular canal into which a small hole was drilled. All of the AAVs were injected into the cochlea via the posterior semicircular canal, and the volume was controlled at 2 µL. After the AAV injection, the wound was sutured and sterilized.
ABR testing
The adult mice were anesthetized by tribromoethanol (500 mg/kg). The closed-field ABR thresholds were measured using tones of different frequencies (4, 8, 12, 16, 24, and 32 kHz) and different sound intensities (15–90 dB) using a TDT system III workstation (Tucker-Davis Technologies, RZ6).
Immunofluorescence staining
Samples were fixed with 4% PFA and decalcified with 0.5 M EDTA. The samples were then blocked with 10% donkey serum for 1–2 h and incubated with primary antibodies overnight at 4 °C. The primary antibodies were Myosin7a (Proteus Bioscience, 25-6790, 1:1000 dilution), CtBP2 (BD Biosciences, 612044, 1:400 dilution). The next day, the secondary antibodies and Phalloidin (Invitrogen, A22287, 1:1000 dilution) were incubated at room temperature for 1 h. The images were obtained on different microscopes (Zeiss LSM 900 and Lecia TCS SP8X STED).
RT-qPCR testing
The mice cochlear samples were collected microscopically in the cold HBSS, and then 1 mL Trizol was added and homogenized. The total RNA was obtained by the chloroform and isopropyl alcohol. Reverse transcription of RNA to cDNA using the kit (Vazyme, R323). GAPDH was used as an internal reference gene, and SYBR (Vazyme, Q712) was utilized to detect GPR156 transcriptome level expression.
Constructs
After codon optimization, the GPR156 gene (UniProt: Q8NFN8), which encodes human full-length (FL) GPR156, was subcloned into a pEG BacMam vector. A hemagglutinin signal peptide (MKTIIALSYIFCLVFA) and 2 × Twin-Strep II tag were inserted at the N-terminus, while a 3 C protease cleavage site, 2 × MBP tag, and 8 × histidine tag were inserted at the C-terminus. GPR156 contains a large extended C-terminal domain (residues 330–814) that is predicted to be unstructured by AlphaFold except for the formation of two loop rings (346–391 and 534–555) (https://www.alphafold.ebi.ac.uk/entry/A0A7K5TS88). Based on this, we tested two constructs of human GPR156 – FL and EM (for cryo-EM, residues 1–346). Finally, GPR156 EM was selected as the WT construct for structural studies due to its better stability without affecting G-protein signaling (Supplementary Fig. 2c). All mutants were generated using site-directed mutagenesis based on the WT (EM (1–346)) construct, and all mutants were confirmed by sequencing.
Expression and purification of apo GPR156
For the expression of apo GPR156, HEK293 GnTI- cells (ATCC, CRL-3022) were grown in suspension culture at 37 °C in 8% CO2 using 293 freestyle media (Sino Biological, M293TII). When the cells had grown to a density of 2.8 × 106 cells/ml, they were infected with recombinant baculoviruses carrying the GPR156 EM (1–346) plasmid and further cultured for 18 h at 37 °C. Then 10 mM sodium butyrate was added, and the cells were further incubated for 72 h at 30 °C. Finally, the cells were collected by centrifugation at 1000 × g, washed in PBS, and stored at − 80 °C until further use.
The cells were homogenized in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.025 mM TCEP, 1/100 DNase, 10% glycerol, and a cocktail of protease inhibitors (Bimake, B14002). The membrane was solubilized for 3 h at 4 °C by adding 0.5% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace, NG310) and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Anatrace, CH210). The cell membrane was then pelleted by ultracentrifugation at 30,000 × g for 45 min. After that, the supernatant was bound to an MBP column for 2 h. The resin was washed with a buffer consisting of 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.01% LMNG, 0.002% CHS, 0.025 mM TCEP, and 10% glycerol. The protein was eluted in the same buffer with 10 mM maltose. The eluted GPR156 was subjected to 3 C protease cleavage for the removal of the 2 × MBP tag. The protein was next loaded onto an anti-Strep affinity resin, and after 1 h binding, the protein was washed with the same wash buffer. The protein was eluted with the same wash buffer with 2.5 mM desthiobiotin. Finally, GPR156 was concentrated in a 100-kDa cutoff filter (Amicon Ultra, UFC910024) and run on a Superose 6 increase 10/300 GL column (GE Healthcare, 29091596) in 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.002% LMNG, and 0.0004% CHS. The peak fractions were collected and concentrated to 9.5 mg/mL for further cryo-EM studies. The peak fractions were also verified by western blot and SDS-PAGE analysis, and the main antibodies used were Anti-Strep-tag II primary antibody (Abcam, ab76950, 1:2000 dilution) and goat anti-rabbit secondary antibody (Abclonal, AS014, 1:5000 dilution).
Expression and purification of the GPR156–Gi3 complex
For expression of the GPR156–Gi3 complex, the recombinant baculoviruses of GPR156, dominant-negative Gαi3 (S47N, G203A, E245A, and A326S), and Gβ1γ2 were prepared using the Bac-to-Bac system (Invitrogen). Spodoptera frugiperda (Sf9) insect cells (Expression Systems, 94-001S) were grown to a density of 1.8 × 106 cells per ml and then infected with GPR156-Gαi3-Gβ1γ2 baculovirus viral stocks at a multiplicity of infection ratio of 6:1:1. After culturing at 27 °C for 48 h, the cells were collected by centrifugation. The purification steps of the GPR156–Gi3 complex were essentially the same as those for apo GPR156, except for incubating the concentrated sample with 1.3 moles of scFv16 (which was expressed and purified as previously described21) at 4 °C for 1 h before running on the Superose6 Increase column.
Cryo-EM sample preparation and data acquisition
For cryo-EM grid preparation, three microliters of the purified apo GPR156 or GPR156–Gi3 complex (apo: 9.5 mg/ml, Gi3-bound: 4.5 mg/ml) were applied onto the glow-discharged carbon grids (Quantifoil, R1.2/1.3, 300 mesh). The grids were blotted for 3.0 s with a blot force of 10 at 4 °C, and 100% humidity and then plunge-frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data collection was performed on a Titan Krios at 300 kV acceleration voltage at the Core Facilities, Liangzhu laboratory, Zhejiang University, equipped with a Cold Field Emission Gun (CFEG) and is equipped with a Falcon 4 direct electron detector. EPU software was used for automated data collection according to standard procedures. Magnification at 130,000 × was used for imaging, yielding a pixel size of 0.93 Å for the images. The defocus range was set from − 1.0 to − 2.5 μm. Each micrograph was dose-fractionated to 40 frames under a dose rate of about 8.7 electrons per pixel per second, with a total exposure time of 6 s, resulting in a total dose of about 52 electrons per Å2. A total of 27,979 and 15,153 movies were collected for apo GPR156 and the GPR156–Gi3 complex, respectively.
Image processing and 3D reconstruction
Cryo-EM image stacks were aligned using RELION 4.042. Contrast transfer function (CTF) parameters were estimated by Gctf v1.1843. The following data processing was performed using RELION 4.0 and CryoSPARC v4.4.044.
For apo GPR156, automated particle selection in RELION produced 17,010,572 particles. The particles were imported to CryoSPARC for several rounds of 2D classification and ab-initio reconstruction to generate the initial reference maps, followed by four rounds of heterogeneous refinement which were classified into four categories. The 675,206 good particles were subjected to further two rounds of classification and were eventually divided into three categories. The best set of particles were reextracted from 27,979 movies in 256 pixels and further carried out non-uniform refinement to generate a map with an indicated global resolution of 3.10 Å in CryoSPARC. To further improve the map quality of apo GPR156, symmetry expansion and local refinement were performed, resulting in a globally refined map with an indicated global resolution of 3.09 Å by 164,823 well-defined particles. The high-quality density map for apo GPR156 were sharpened with deepEMhancer. This map was used for subsequent model building and analysis.
For the GPR156–Gi3 complex, 12,058,641 particles produced from the automated particle picking were subjected to 2D classification, ab-initio reconstruction, and 3D heterogeneous refinement in CryoSPARC, resulting in 1,336,840 well-defined particles. A further two rounds of 3D classification focusing on the alignment of the protein complex generated two high-quality subsets with 1,153,971 particles. The final good particles were subjected to a final round of non-uniform refinement local refinement, generating a map with an indicated global resolution of 2.39 Å at a Fourier shell correlation of 0.143. The final map was sharpened with deepEMhancer and used for subsequent model building and analysis.
Model building and refinement
The structure predicted by AlphaFold2 was used to generate the initial model of GPR15645. The atomic coordinates of the Gi3 protein from the structure of the mGlu4–Gi3 complex (PDB code: 7E9H) were used to generate the initial model of the Gi3 protein46. Models were manually docked into the density maps of apo GPR156 and the GPR156–Gi3 complex using UCSF Chimera v1.17.3. The initial models were then subjected to flexible fitting using Rosetta and were further rebuilt in Coot v0.9.8.1 and real-space refined in Phenix v1.19.2-415847–49. The final refinement statistics were validated using the comprehensive validation (cryo-EM) module in Phenix. The refinement statistics are provided in Supplementary Table 2. Structural figures were created using the UCSF Chimera X package (v1.8)50.
Identification of phospholipid ligands by LC-MS/MS
The sample preparation of the phospholipid specifically bound to GPR156 was performed following previous methods51. The samples of GPR156 were prepared in three independent replicates. In addition, two well-established class A GPCRs (including GPR3452,53 and GPR17453–55), each with two replicates, were employed as control groups where no two-chain phospholipids were present. Briefly, GPR156 and two control proteins (GPR34 and GPR174) were treated with 5 mM TCEP for 30 min at 25 °C and alkylated with 20 mM idoacetamide. The proteins were digested with trypsin (Promega, V5280) (1:50 w/w) at 37 °C overnight. After drying the digests, they were extracted with 400 μl ice-cold methanol:water (9:1) using vortex and sonication. Following centrifugation at 12,000 × g for 15 min at 4 °C, the supernatants were lyophilized, and the lipid extracts were resuspended in methanol:chloroform (9:1) to 2 μM. Then a Dionex U3000 UHPLC (Thermo Fisher Scientific) fitted with Q-Exactive mass spectrometer equipped with a heated electrospray ionization (ESI) source (Thermo Fisher Scientific) was used to analyze the metabolic profiling in both ESI positive and ESI negative ion modes. An ACQUITY UPLC BEH C8 column (1.7 μm, 2.1 × 100 mm) was used in both positive and negative modes. The binary gradient elution system consisted of (A) acetonitrile:water (60:40, v:v, containing 10 mmol/L ammonium formate) and (B) acetonitrile:isopropanol (10:90, v:v, containing 10 mmol/L ammonium formate), and separation was achieved using the following gradient: 0 min, 30% B; 3 min, 30% B; 5 min, 62% B; 15 min, 82% B; 16.5 min, 99% B; 18 min, 99% B; 18.1 min, 30% B; and 22 min, 30% B. The flow rate was 0.26 mL/min, and the column temperature was 55 °C. Positive: Spray voltage (kV): + 3.5; Capillary temperature 300 °C; Aux gas heater 350 °C; Sheath gas flow rate (Arb): 45; Aux gas flow rate (Arb): 10; S-lens RF level: 50; Mass range (m/z): 150–1500, Full ms resolution: 70000; MS/MS resolution: 17500; TopN: 10; NCE/stepped NCE: 25, 35, 45. Negative: Spray voltage (kV): – 3.0; Capillary temperature 300 °C; Aux gas heater temperature 350 °C; Sheath gas flow rate (Arb): 45; Aux gas flow rate (Arb): 10; S-lens RF level: 50; Mass range (m/z): 150–1500, Full ms resolution: 70000; MS/MS resolution: 17500; TopN: 10; NCE/stepped NCE: 25, 35, 45.
The peak extraction and lipid identification were performed using MSDIAL (v.4.9). Candidate phospholipid molecules were annotated by the following parameters: MS/MS assigned = TRUE; Total score ≥ 70 (Total score is based on accurate mass, isotope ratio, retention time, and MS/MS similarities); was detected in all three independent replicates of GPR156; and log2 fold change (logFC) ≥ 2. The identified endogenous phospholipid bound to GPR156 was phosphatidylglycerol (PG) 36:2 (PG (18:1_18:1); International Chemical Identifier (InChI) key: DSNRWDQKZIEDDB-UHFFFAOYSA-N); with MS2 spectra Precursor: Reference Mass = 773.53375: 773.53381.
MD simulations
MD simulations were performed essentially as previously described32,56 with modifications. From the apo structure of the GPR156 dimer complex, the two transmembrane helix core-bound phospholipids were removed, except the PGs (36:2) and the cholesterol molecule in the interface of the GPR156 dimer. The retained GPR156-dimer complex was used as the input conformation for our all-atom simulations by classical molecular dynamics (labeled as with lipid). To assess the impact of the PG molecule on the cavity of the transmembrane helices core, an additional simulation was performed with the two bound PGs (36:2) removed (labeled as no lipid, Supplementary Fig. 11a). For both simulations, the disulfide bridge between C191 and C216 was not consider due to the large distance (Supplementary Fig. 11b). All the GPR156-dimer complex were embedded into a membrane made of 3:1 POPC:POPE and a cholesterol bilayer, which was all built by using the Membrane Builder module on the CHARMM-GUI server57. The resulting system had a total of 240 lipid and 7 cholesterol molecules (added by CHARMM-GUI) in each leaflet and was solvated in a cubic box with TIP3P waters and 0.15 M Na+/Cl– ions. The system sizes of the GPR156 (with lipid) and the GPR156 (no lipid) were 13.4 × 13.4 × 13.1 nm3, which contained a total number of 223,513, and 223,194 atoms. In addition, the corresponding number of water molecules were 49,631 and 49,612, and the number of ions were 135 Na+ and 154 Cl–, and 134 Na+ and 155 Cl–, respectively (Supplementary Table 5). The CHARMM36m forcefield was used to describe the system58, and all MD simulations were performed using GROMACS-2019.459. After 5000 steps of energy minimization performed by the steepest descent algorithm, a 250 ps NVT equilibration simulation was performed at 310 K, with the default restraint potential (kBC/SD/LIP/DIH = 10.0/5.0/2.5/2.5 kcal·mol−1 · Å−2) on the heavy atoms of backbone, sidechain, lipid, and dihedral angle. Subsequently, a cumulatively 1.65 ns NPT equilibration to 1 atm was performed using the Berendsen barostat60, with the restraint potential gradually reduced to zero. Long-range electrostatic interactions were treated with the particle-mesh Ewald method61,62. The short-range electrostatic and van der Waals interactions both used a cutoff of 10 Å. All bonds were constrained by the LINCS algorithm63. During the pre-equilibrium simulation, the vacuum of the cavities of the transmembrane helices core for the GPR156 (no lipid) system was gradually filled with water molecules (Supplementary Fig. 11c). Finally, to escape the uncertainties of the sampling results, three replicates of the production run of 300 ns MD simulation with different initial velocities were performed for both systems: the GPR156 (with lipid) and the GPR156 (no lipid) (Supplementary Fig. 12). The stability of the two systems were evaluated by the RMSD of all the heavy atoms of the GPR156 dimer (Supplementary Fig. 11d). The cavity volume of the transmembrane helices core was calculated with Epock (1.0.5)64 in VMD65. The RMSD of the transmembrane helices (TM3/5/6/7) ends (heavy atoms of 10 residues) in the intracellular side were calculated, and other C alpha atoms of the transmembrane helices were used for structural alignment (Supplementary Figs. 13–16 and Supplementary Table 6). For the RMSD calculation, the first 10 ns was ignored, and only the last 290 ns MD simulation data was used. GROMACS’s rms function was used to calculate RMSD.
ELISA assay
The cell-surface expression of WT GPR156 or its mutants was detected using an Enzyme-linked immunosorbent (ELISA) assay as previously described21. In brief, HEK293T cells were transfected with plasmids of a mixture of WT GPR156 or mutants, Gαi3–LgBiT, Gγ–SmBiT, and Gβ1 using Lipofectamine 3000 (Thermo Fisher Scientific, L3000015) in 500 μl of Opti-MEM (Thermo Fisher Scientific, 31985070). Twenty-four hours post-transfection, cells were distributed into 96-well plates with a white non-transparent bottom and further incubated for 24 h at 37 °C. The HEK293T cells were then fixed with 4% paraformaldehyde and blocked with 1% bovine serum albumin (BSA) for 1 h. Luminescence detection was performed using SuperSignal ELISA Femto Maximum Sensitivity substrate (Thermo Fisher Scientific, 37074) after binding of antibodies coupled to horse-radish peroxidase (Beyotime, A0208, 1:1000 dilution), and luminescence was measured with a Multimode Microplate Reader (Tecan).
Adjusting the transfection levels of both the WT GPR156 and its mutants was essential to ensuring that the mutants exhibited cell surface expression comparable to WT without significant statistical differences. This adjustment facilitated a meaningful comparison in the Gi dissociation assay.
BRET-G-protein dissociation assay
Gi activation was assessed by the Bioluminescence resonance energy transfer (BRET) dissociation assay measuring the proximal interaction between the α and γ subunits of the Gi protein. The G protein BRET probes, including Gαi3-Rluc8, Gβ1, and Gγ2-Venus, were generated according to a previous publication31. HEK293T cells were transfected with WT GPR156 or mutants and Gi protein probes. Twenty-four hours after transfection, the cells were divided into 96-well plates and incubated for an additional 24 h at 37 °C. The BRET signal was quantified following the introduction of the luciferase substrate coelenterazine h (10 μM) employing a Multimode Microplate Reader (Tecan) equipped with BRET filter sets. The BRET signal was determined as the ratio of light emission at 520 nm/460 nm.
NanoBiT-G-protein dissociation assay
Gi activation was also assessed by the NanoBiT-based dissociation assay measuring the proximal interaction between the α and γ subunits of the Gi protein. The transfection system was the same as that in the ELISA assay described above. G protein NanoBiT probes, including Gαi3-LgBiT, Gβ1, and Gγ2-SmBiT, were generated according to a previous publication21. After 1 day of transfection, cells were divided into 96-well plates and incubated for an additional 1 day at 37 °C. The NanoBiT signal was measured using a Multimode Microplate Reader (Tecan).
Statistical analysis
The RT-qPCR data are reported as the mean ± standard deviation (SD), and other data are presented as the mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism 9 software. Adobe Illustrator was used to organize images and graphs. Bar graphs depict the differences of each mutant relative to the WT receptor or contralateral ear/WT mice. In vitro data were from at least six independent experiments, each conducted in triplicate. In vivo experiments related to mice were repeated at least three times. ns (not significant) = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (two-tailed unpaired t test for all data).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The cryo-EM data were collected at the Cryo-Electron Microscopy Facility, Liangzhu laboratory, Zhejiang University. We thank Cheng Ma from the Core Facilities, Zhejiang University School of Medicine for technical support. Cryo‐EM specimens were examined with the help from Shenghai Chang at the Center of Cryo‐Electron Microscopy (CCEM), Zhejiang University. We thank Shanghai OE Biotech for providing mass spectrometry detection. This project was supported by the STI2030-Major Projects (2021ZD0203304); National Key Research and Development Program of China (2023YFC2508402, 2021YFA1101300 (R.C.), 2021YFA1101800 (R.C.), 2020YFA0113600 (J.Y.Q.), and 2020YFA0112503 (R.C.)); the Fundamental Research Funds for the Central Universities (226-2022-00205 (Y.Z.)); the Zhejiang Province Natural Science Fund for Excellent Young Scholars (LR22C050002 (C.M.)); the STI2030-Major Projects (2022ZD0205400 (J.-Y.Q.)); the National Natural Science Foundation of China (82330033 (R.C.), 82030029 (R.C.), 92149304 (R.C.), 82371162 (J.Y.Q.), 32430051 (Y.Z.), 92353303 (Y.Z.), 32141004 (Y.Z.), 32330049 (J.L.), 82320108021 (J.L.), 32300598 (C.S.), 32371249 (C.M.), 32100959 (C.M.), 82322070 (C.M.), U23A200440 (J.Y.Q.), 82401369 (L.Z.), and 82401375 (M.L.)); the Ministry of Science and Technology (2019YFA0508800 (Y.Z.) and 2023YFC2306800 (C.M.)); the Key R&D Projects of Zhejiang Province (2021C03039 (Y.Z.)); the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2020R01006 (Y.Z.)); the “Pioneer” and “Leading Goose” R&D Program of Zhejiang (2024C03147 (Y.Z.)); the China Postdoctoral Science Foundation funded project (2023TQ0056 (M.L.), 2023M730575 (M.L.), GZC20230435 (M.L.), 2024M750455 (L.Z.), and GZB20240145 (L.Z.)); Natural Science Foundation of Jiangsu Province (BK20232007 (R.C.) and BK20241692 (L.Z.)); the Jiangsu Provincial Scientific Research Center of Applied Mathematics under Grant (BK20233002 (R.C.)); Major Project of Guangzhou National Laboratory (GZNL2023A03007 (J.L.)); the Shenzhen Science and Technology Program (JCYJ20210324125608022 (R.C.)); the 2022 Open Project Fund of Guangdong Academy of Medical Sciences to P.N.W. (YKY-KF202201 (R.C.)); Beijing Natural Science Foundation (No. Z200019); the Fundamental Research Funds for the Central Universities (Y.Z.); and Start-up Research Fund of Southeast University (RF1028623028).
Author contributions
R.C. and Y.Z. conceived and supervised the whole project; L.Z. and X.M. designed the AAV strategy; L.Z. performed the animal experiments; X.M. and J.G. designed the constructs and expressed and purified apo GPR156 and the GPR156–Gi3 complex; D.D.S. evaluated the sample by negative-stain EM; L.N.C. prepared the cryo-EM grids; L.N.C., S.K.Z., J.Q., and S.Y.J. collected the cryo-EM data; L.N.C. and C.M. performed cryo-EM map calculation and model building; M.L., X.M., and J.G. generated the mutant constructs; M.L. performed the in vitro cellular functional assays; X.M. and Y.J.D. prepared the samples for the mass spectrometry studies; P.C. and Y.L. performed bioinformatics analysis of the mass spectrometry data; K.X. performed the MD simulations; X.M. and L.N.C. performed the structural analysis; X.M. and L.N.C. prepared the figures; C.S., M.L., L.Z., J.G., Q.S., H.Z., J.Y.Q., L.M., and J.L. participated in data analysis; X.M. and L.N.C. prepared the draft of the manuscript; R.C., Y.Z., C.M., and J.L. wrote the manuscript with inputs from all the authors.
Peer review
Peer review information
Nature Communications thanks Maria Gomez-Casati, Célien Jacquemard, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data generated in this study are included in the manuscript or Supplementary Information. The cryo-EM density maps and the atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) databases under accession codes EMD-39345 and 8YJP for apo-GPR156; and EMD-39356 and 8YK0 for the GPR156–Gi3 complex. The LC-MS/MS data of phospholipid ligands for GPR156 have been deposited NGDC OMIX database (OMIX ID: OMIX007597) [https://ngdc.cncb.ac.cn/omix/release/OMIX007597] and on the Figshare server (10.6084/m9.figshare.25838170.v1). The MD simulation data (cleaned trajectories, start structure, simulation parameters) generated in this study have been deposited in GitHub (https://github.com/Yanzhang-ZJU/GPR156.git) and Zenodo (10.5281/zenodo.13208133). Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiangyu Ma, Li-Nan Chen, Menghui Liao, Liyan Zhang, Kun Xi, Jiamin Guo.
Contributor Information
Jianfeng Liu, Email: jfliu@mail.hust.edu.cn.
Chunyou Mao, Email: maochunyou@zju.edu.cn.
Yan Zhang, Email: zhang_yan@zju.edu.cn.
Renjie Chai, Email: renjiec@seu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54681-5.
References
- 1.Kindt, K. S. et al. EMX2-GPR156-Galphai reverses hair cell orientation in mechanosensory epithelia. Nat. Commun.12, 2861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greene, D. et al. Genetic association analysis of 77,539 genomes reveals rare disease etiologies. Nat. Med.29, 679–688 (2023). [DOI] [PMC free article] [PubMed]
- 3.Jarysta, A. et al. Inhibitory G proteins play multiple roles to polarize sensory hair cell morphogenesis. ELife12, 10.7554/eLife.88186.1 (2023). [DOI] [PMC free article] [PubMed]
- 4.Charles, K. J., Calver, A. R., Jourdain, S. & Pangalos, M. N. Distribution of a GABAB-like receptor protein in the rat central nervous system. Brain Res.989, 135–146 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Chun, L., Zhang, W. H. & Liu, J. F. Structure and ligand recognition of class C GPCRs. Acta Pharmacol. Sin.33, 312–323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laboute, T. et al. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science379, 1352–1358 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patil, D. N. et al. Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179. Sci. Signal16, eadd9539 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettler, B., Kaupmann, K., Mosbacher, J. & Gassmann, M. Molecular structure and physiological functions of GABA(B) receptors. Physiol. Rev.84, 835–867 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Caniceiro, A. B., Bueschbell, B., Schiedel, A. C. & Moreira, I. S. Class A and C GPCR dimers in neurodegenerative diseases. Curr. Neuropharmacol.20, 2081–2141 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, M. et al. G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery. Signal Transduct. Target. Ther.9, 88 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, L. et al. Structure, function and drug discovery of GPCR signaling. Mol. Biomed.4, 46 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kniazeff, J., Prezeau, L., Rondard, P., Pin, J. P. & Goudet, C. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol. Ther.130, 9–25 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, J. et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature594, 589–593 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Wang, X. et al. Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers. Cell Res.33, 762–774 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attwood, T. K. & Findlay, J. B. Fingerprinting G-protein-coupled receptors. Protein Eng.7, 195–203 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Koehl, A. et al. Structural insights into the activation of metabotropic glutamate receptors. Nature566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellaithy, A., Gonzalez-Maeso, J., Logothetis, D. A. & Levitz, J. Structural and biophysical mechanisms of class C G protein-coupled receptor function. Trends Biochem. Sci.45, 1049–1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins, L. R. & Orlandi, C. In vitro profiling of orphan G protein coupled receptor (GPCR) constitutive activity. Br. J. Pharmacol.178, 2963–2975 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Calver, A. R. et al. Molecular cloning and characterisation of a novel GABAB-related G-protein coupled receptor. Brain Res. Mol. Brain Res.110, 305–317 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Shen, C. et al. Structural basis of GABA(B) receptor-G(i) protein coupling. Nature594, 594–598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, Y. et al. Asymmetric activation of the calcium-sensing receptor homodimer. Nature595, 455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberman, L. D. & Liberman, M. C. Postnatal maturation of auditory-nerve heterogeneity, as seen in spatial gradients of synapse morphology in the inner hair cell area. Hear. Res.339, 12–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikaelian, D. O. & Ruben, R. J. Hearing degeneration in shaker-1 mouse. Correlation of physiological observations with behavioral responses and with cochlear anatomy. Arch. Otolaryngol.80, 418–430 (1964). [DOI] [PubMed] [Google Scholar]
- 25.Isu, U. H., Badiee, S. A., Khodadadi, E. & Moradi, M. Cholesterol in Class C GPCRs: Role, Relevance, and Localization. Membranes13, 10.3390/membranes13030301 (2023). [DOI] [PMC free article] [PubMed]
- 26.Patil, D. N. et al. Cryo-EM structure of human GPR158 receptor coupled to the RGS7-Gbeta5 signaling complex. Science375, 86–91 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, J. et al. Structure of human GABA(B) receptor in an inactive state. Nature584, 304–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaye, H. et al. Structural basis of the activation of a metabotropic GABA receptor. Nature584, 298–303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong, E., Kim, Y., Jeong, J. & Cho, Y. Structure of the class C orphan GPCR GPR158 in complex with RGS7-Gbeta5. Nat. Commun.12, 6805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dore, A. S. et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature511, 557–562 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Mao, C. et al. Cryo-EM structures of inactive and active GABA(B) receptor. Cell Res.30, 564–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papasergi-Scott, M. M. et al. Structures of metabotropic GABA(B) receptor. Nature584, 310–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seven, A. B. et al. G-protein activation by a metabotropic glutamate receptor. Nature595, 450–454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He, F. et al. Allosteric modulation and G-protein selectivity of the Ca(2+)-sensing receptor. Nature626, 1141–1148 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin, J. et al. Constitutive activation mechanism of a class C GPCR. Nat. Struct. Mol. Biol. 31, 678–687 (2024). [DOI] [PubMed]
- 36.Gyorgy, B. et al. Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of usher syndrome 3A and transduces hair cells in a non-human primate. Mol. Ther. Methods Clin. Dev.13, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isgrig, K. et al. AAV2.7m8 is a powerful viral vector for inner ear gene therapy. Nat. Commun.10, 427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, F. et al. AAV-ie enables safe and efficient gene transfer to inner ear cells. Nat. Commun.10, 3733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, K. R. et al. Effects of Cdh23 single nucleotide substitutions on age-related hearing loss in C57BL/6 and 129S1/Sv mice and comparisons with congenic strains. Sci. Rep.7, 44450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, J. et al. Refining surgical techniques for efficient posterior semicircular canal gene delivery in the adult mammalian inner ear with minimal hearing loss. Sci. Rep.11, 18856 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, L. et al. AAV-Net1 facilitates the trans-differentiation of supporting cells into hair cells in the murine cochlea. Cell Mol. Life Sci.80, 86 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres, S. H. Processing of structurally heterogeneous Cryo-EM Data in RELION. Methods Enzymol.579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol.193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, S. et al. Structures of G(i)-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature594, 583–588 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Wang, R. Y. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife5, 10.7554/eLife.17219 (2016). [DOI] [PMC free article] [PubMed]
- 48.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu, X. et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature604, 779–785 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia, A. et al. Cryo-EM structures of human GPR34 enable the identification of selective antagonists. Proc. Natl. Acad. Sci. USA120, e2308435120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, G., Li, X., Wang, Y., Zhang, X. & Gong, W. Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174. PLoS Biol.21, e3002387 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang, J. et al. Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation. Nat. Commun.14, 1012 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nie, Y. et al. Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G(s) signaling. Nat. Commun.14, 5901 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, P. et al. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell184, 943–956 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem.29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Huang, J. & MacKerell, A. D. Jr CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem.34, 2135–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular-dynamics with coupling To an external bath. J. Chem. Phys.81, 3684–3690 (1984). [Google Scholar]
- 61.Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys.103, 8577–8593 (1995). [Google Scholar]
- 62.Darden, T., York, D. & Pedersen, L. Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys.98, 10089–10092 (1993). [Google Scholar]
- 63.Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem.18, 1463–1472 (1997). [Google Scholar]
- 64.Laurent, B. et al. Epock: rapid analysis of protein pocket dynamics. Bioinformatics31, 1478–1480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph.14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data generated in this study are included in the manuscript or Supplementary Information. The cryo-EM density maps and the atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) databases under accession codes EMD-39345 and 8YJP for apo-GPR156; and EMD-39356 and 8YK0 for the GPR156–Gi3 complex. The LC-MS/MS data of phospholipid ligands for GPR156 have been deposited NGDC OMIX database (OMIX ID: OMIX007597) [https://ngdc.cncb.ac.cn/omix/release/OMIX007597] and on the Figshare server (10.6084/m9.figshare.25838170.v1). The MD simulation data (cleaned trajectories, start structure, simulation parameters) generated in this study have been deposited in GitHub (https://github.com/Yanzhang-ZJU/GPR156.git) and Zenodo (10.5281/zenodo.13208133). Source data are provided in this paper.