Abstract
The governing laws mandate animal testing guidelines (TG) to assess the developmental and reproductive toxicity (DART) potential of new and current chemical compounds for the categorization, hazard identification, and labeling. In silico modeling has evolved as a promising, economical, and animal-friendly technique for assessing a chemical's potential for DART testing. The complexity of the endpoint has presented a problem for Quantitative Structure-Activity Relationship (QSAR) model developers as various facets of the chemical have to be appropriately analyzed to predict the DART. For the next-generation risk assessment (NGRA) studies, researchers and governing bodies are exploring various new approach methodologies (NAMs) integrated to address complex endpoints like repeated dose toxicity and DART. We have developed four hybrid computational models for DART studies of rodents and rabbits for their adult and fetal life stages separately. The hybrid models were created by integrating QSAR features with similarities-derived features (obtained from read-across hypotheses). This analysis has identified that this integrated method gives a better statistical quality compared to the traditional QSAR models, and the predictivity and transferability of the model are also enhanced in this new approach.
Keywords: Testing guidelines, DART, QSAR, NGRA, NAMs, Read-across
Graphical Abstract
Highlights
-
•
Four Hybrid models are developed for developmental and reproductive toxicity (DART) data.
-
•
The DART data of rodents and rabbits' adult and fetal life stages are used.
-
•
The hybrid models combine physicochemical and similarity-based features.
-
•
The hybrid modeling strategy enhances model quality and prediction.
-
•
The developed hybrid models can utilize a potential substitute approach for evaluating DART toxicity.
1. Introduction
In human toxicology, the reproductive and developmental toxicities (DART) are comparatively poorly characterized endpoints. This is so because a wide range of distinct endpoints are referred to by these common terms. The general phrase "reproductive toxicity," consists of a range of adverse or detrimental effects on adult male and female fertility and sexual functions. In contrast, developmental toxicity includes the impact on offspring, including the impact on or mediated by lactation. As a result, a wide spectrum of endpoints, such as gestational length, sperm quality, neonatal growth, litter size, and functional toxicities, are pertinent [1], [2]. According to the United States Environmental Protection Agency(USEPA) [3] and the European Union's registration, evaluation, authorization, and restriction of chemicals (EU REACH) [4] standards, it is one of the most significant toxicological endpoints; there are strict requirements for DART testing of industrial and consumer chemicals under the EU REACH legislation [5], [6]. Regulatory bodies use the findings of animal studies to establish standards for human exposure [7]. Generally, in vivo animal testing is the standard method for the toxicological evaluation of chemicals. The experimental techniques for assessing a chemical's potential for DART in rats, mice, and rabbits have been described by the USEPA and the Organization for Economic Cooperation and Development (OECD) [3], [8]. DART testing accounted for 90 % of animal use and 70 % of chemical toxicity testing costs connected with finishing the phase one of the REACH regulation. Individual testing methodologies occasionally require up to 3200 animals per chemical [9]. The application of alternative techniques to forecast this kind of toxicity garners much interest [9], [10].
It has become widely acknowledged that New Approach Methodologies (NAMs) are viable substitutes for animal testing in chemical safety assessment [11], [12], [13], [14]. NAMs are in vitro, in chemico, or in silico techniques that enable hazard assessment, enhance knowledge of harmful effects, and either partially or completely replace animal testing with cutting-edge animal-free testing procedures. Several authorities have proposed guidelines, frameworks, and work plans that guarantee trust, consistency, and suitability for generating NAM hazard data for different purposes to promote their development and deployment [15], [16], [17], [18]. There are currently OECD test guidelines (TG) available for a variety of NAMs that can be used to assess local toxicity, such as skin irritation, sensitivity and corrosion, and ocular irritation and corrosion [19], [20], [21], [22], [23], [24]. The need to develop a rational NAM workflow for the toxicological assessment of cosmetic chemicals is crucial as animal testing was banned by the EU in 2013 for these chemicals [25]. The use of NAMs in an integrated manner for the Next Generation Risk Assessment (NGRA) is guided by major principles defined by the International Cooperation on Cosmetics Regulation (ICCR) [26]. Additional guidelines on certain NAMs have also been released. These guidelines can be used in a tiered way to assess the risk of cosmetic constituents in line with the various tiers of the SEURAT-1 (Safety Evaluation Ultimately Replacing Animal Testing) ab initio workflow for systemic repeat-dose toxicity [27], [28]. Case studies are being prepared to demonstrate how these concepts can be used practically in assessing the safety of cosmetic items and to demonstrate how NAMs can provide meaningful information on non-animal safety evaluation [27]; in the future, these frameworks can be utilized for complex endpoints like DART testing. Numerous research studies have assessed the predictive efficacy of a range of various techniques for DART. The creation of NAMs for DART testing has advanced recently due to cooperative initiatives like the ReProTect project [29], [30] and the DART committee of the Health and Environmental Sciences Institute (HESI). An in-silico pre-screening module was also used to minimize the requirement for testing, and toxicokinetic modeling was used to validate the in vitro to in vivo dose comparisons [31], [32]. The researchers are also focusing on developing a transparent and reliable database (DB) for NAM development and constructing a rational framework for DART testing. The in-silico modeling NAMs are considered a promising first step towards addressing the current gaps in DART testing among various alternative non-testing methodologies for the safety evaluation of substances. These computer-assisted techniques include read-across, grouping, structural alerts, and quantitative structure-activity relationship (QSAR) tools. Currently, read-across is one of the most advanced in silico techniques for forecasting systemic toxicity regarding regulatory approval. According to Ball et al., [33] read-across for at least one endpoint is present in over 75 % of REACH dossiers submitted between 2010 and 2013. Other regulatory bodies worldwide, like the International Council of Chemical Associations (ICCA) and USEPA’s high production volume challenge program, also support the read-across procedure. Several case studies on repeated dose toxicity have offered some illustrations of read-across protocols in recent years. Nevertheless, these procedures are limited to compounds with simple structures with readily available analogs. Furthermore, very few reports of read-across studies try to make quantitative predictions like NOAEL values [34], particularly when DART testing fills data gaps.
This work presents a fresh approach to read-across (RA)-based predictions that are reproducible, thorough, systematic, and applicable to a broad variety of chemicals. We apply an integrated approach covering QSAR and RA studies. In this novel method, initially, important features were extracted by the QSAR modeling technique, and these modeled features were again utilized for RA predictions. To address the associated uncertainties, we have computed the RA-derived features based on their similarity measures computed using different methods like the Laplacian kernel, Gaussian kernel, and Euclidean distance). Hence, we have used the RA-derived similarity measures of a defined number of close source chemicals to compute features of a query chemical in the form of error and similarity functions. Based on this, we have developed a final regression model (hybrid model) utilizing both QSAR features and similarity-derived features [35], [36]. Few investigations have been conducted since introducing this hybrid methodology, and those have demonstrated that it performs better for both quantitative and qualitative predictions than the traditional QSAR and read-across methodologies for blood-brain barrier permeability, skin sensitization, acute contact toxicity in bees, mutagenicity, and aquatic toxicity [37], [38], [39], [40], [41]. In the future, this strategy may prove to be a useful method for creating models that accurately predict other chronic toxicity endpoints.
Our current investigation retrieved high-quality prenatal developmental toxicity data from OECD TG 414 studies (specifically lowest observed effect levels (LOELs). We have prepared four datasets based on species and life stages to reduce prediction uncertainty. We have developed four hybrid partial least squares regression (PLS) models containing physicochemical features (from the corresponding QSAR models) and similarity-based features (from RA predictions). Based on the investigation, we have determined that this integrated method has produced superior statistical quality than the conventional QSAR model, and this innovative approach has also boosted the model's predictivity and transferability. Additionally, we have tried to provide a mechanistic interpretation when feasible. This novel approach utilizes the physicochemical as well as the similarity measures that have the potential to precisely categorize chemicals according to their physical, chemical, or structural pattern of the query chemical from the source chemicals that can further be used for regulatory decision-making for complex apical endpoints.
2. Materials and methods
2.1. Collection of database for in vivo prenatal developmental toxicity
To address the demand for curated data and relevant tools to facilitate the development of novel methods for evaluating chemical safety, the US National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) created the Integrated Chemical Environment (ICE) (https://ice.ntp.niehs.nih.gov/). ICE provides unrestricted, free access to a wide range of carefully chosen in vivo, in vitro, and in silico data as well as computational tools to aid in developing and evaluating NAMs [42]. A variety of toxicity endpoint data (e.g., acute oral dermal and inhalational toxicity, cancer DART, endocrine, eye irritation, ADME properties, etc) have been provided by the ICE in the form of different in vitro, in vivo, and curated High Throughput Screening (cHTS) assay data with the description of their testing methods. In the present investigation, we have collected the prenatal developmental toxicity (or DART) data of a diverse set of organic chemicals from the data set page of the ICE (https://ice.ntp.niehs.nih.gov/DATASETDESCRIPTION).
2.2. Preparation of in vivo database for prenatal developmental toxicity
From the initial analysis of the ICE DART database, we have identified that the dataset mainly contains pharmaceutical drugs and excipients, chemicals of agricultural importance, and a few miscellaneous categories of compounds. This database has provided numerous in vitro and in vivo assay results for several endpoints related to DART analysis. Assay results are provided for different life stages (adult, fetal, and juvenile) of rodents (rat and mouse) and rabbits. For both rodents and rabbits, juvenile data has fewer data points that cannot be used for modeling purposes. Therefore, we have removed juvenile data from our study. For 387 molecules,3629 testing results having in vivo ToxRefDB curated LOEL (Lowest Observed Effect Level) data were retrieved from the initial file of 628 molecules having more than 0.1 million test results (in vivo and in vitro). For our study, we have focused on these 3629 LOEL results since other assay methods or endpoints have not provided a definite value for modeling purposes. After thoroughly analyzing the data, we have identified that the LOEL data for adults and fetal life stages of rodents and rabbits provided by ICE strictly follows the OECD regulatory animal test guideline 414. The TG 414 [8] is an in vivo regulatory guideline assay used to evaluate toxic and teratogenic effects that occur when a substance is administered to an animal during gestation. In this guideline, pregnant animals are dosed during the gestation period after implantation to observe the toxic effect on both the mother (adult) and the fetus. For more precise investigation, we have manually divided our data (3629 LOEL data of 387 molecules). In this step, we have segregated the LOEL data based on species where we have divided the data into two parts rodents (rat and mouse) and non-rodent (rabbit). For a more specific study, we have again separated the LOEL data based on the life stages (adult and fetal) of the rodent and rabbit species individually. Finally, four datasets—adult rodent (Dataset 1), fetal rodent (Dataset 2), adult non-rodent/rabbit (Dataset 3), and fetal non-rodent/rabbit (Dataset 4) have been prepared for our investigation. We have manually eliminated the inorganic chemicals from each dataset. A single representative value for each compound was necessary to facilitate the modeling efforts because several chemicals had multiple LOEL values. The representative LOEL values for every molecule were the mean of all observations with response values that deviated less than log scale 1. In summary, we have 236 molecules for Dataset 1, 218 molecules for Dataset 2, 175 molecules for Dataset 3, and 132 molecules for Dataset 4 to facilitate further studies on individual adult and fetal rodent and non-rodent species. We have collected the LOEL data with similar units in mg/kg/day (a small number of studies reporting doses in ppm and mg/m3 implying dietary and inhalation exposure were excluded to ensure the consistency of response data). After this step, the LOEL values for each dataset were converted into a negative logarithmic scale (i.e., -log (LOEL/MW) or pLOEL). The details of the datasets are provided in Supplementary Material 1.
2.3. Representation of structures and descriptors computation
For each of the four DART datasets, the canonical SMILES notions were obtained separately from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), as the ICE database only included the chemical's IUPAC name and CAS number. The retrieved SMILES notions were used for drawing the chemical structure of the molecules by using the Marvin Sketch software (https://chemaxon.com/products/marvin) and transformed into a single.sdf file for each dataset. Before translation into the.sdf file, structures were re-evaluated, and the salt form was removed for required chemicals individually for Dataset 1, Dataset 2, Dataset 3, and Dataset 4. We have calculated 9 classes of 0D-2D molecular descriptors (constitutional indices, ring descriptors, connectivity index, electro-topochemical atom indices, functional group counts, atom-centered fragments, atom-type E-state indices, 2D atom pairs, and molecular properties) and MACCS structural key descriptors by utilizing the “alvaDesc” software [43]. We have computed only these 9 classes of molecular descriptors for the ease of their interpretability and their satisfactory performance in modeling toxicological endpoints based on our experience. Consequently, the initial descriptor pool for each dataset was generated and redundant descriptors were eliminated using the built-in methods of the "alvaDesc" software. The pre-treatment of the computed set of data matrix was carried out for each of the four DART datasets to eliminate the highly inter-correlated descriptors. For that, DataPreTreatmentGUI 1.2 (http://teqip.jdvu.ac.in/QSAR_Tools/DTCLab/), a Java-based tool that is publicly available, was utilized. For data pre-processing, we have selected the variance cut-off of 0.1 and the intercorrelation cut-off of 0.95.
2.4. Random splitting of the training and testing set
One of the most important components of the QSAR modeling technique is the splitting of the data into training and testing sets. For each set of data (Datasets 1,2,3 and, 4), we employed several methods, including random splitting, Euclidean distance-based splitting, Kennard-Stone splitting, response-based Sorted activity-based splitting, and K-medoid clustering algorithm, to derive an optimal splitting of our datasets into training and testing sets. Each of the datasets has been divided into distinct percentages to achieve the best possible division to determine the parameters of machine learning models that best fit the training data for each dataset separately since each data set is different. Varying the splits will help the analysts eventually decide which of the ratios best fits the size of the datasets and produces the highest accuracy. However, we have maintained a standard range of approximately 20–30 percent chemicals in the test set for different datasets. In this rigorous analysis, we identified that random division produced the best statistical outcomes in this study compared to other splitting methods we have performed for each dataset. Therefore, we have taken the randomly divided datasets (training and testing sets) in each case for this study. Finally, for Datasets 1, 2, 3, and 4, we had 194, 168, 133, and 108 compounds in the training sets and 42, 50, 42, and 24 in the testing sets, respectively. A training set was used to develop a model, and a test set was used to validate the model.
2.5. Variable selection and traditional regression-based QSAR model development
Choosing features is a crucial step towards developing a QSAR model. Feature selection allows us to exclude the noisy and unimportant input variables from the original variable spaces and to identify the important feature for the targeted endpoint. For the identification of the best descriptors pool, we have performed feature selection for 0–2D molecular descriptors, MACCS fingerprints, and the combination of (0–2Dmolecular+MACCS) individually. From the initial analysis, we have identified that the 0–2D molecular descriptors perform better compared to the other two descriptor matrix i.e. (0–2D molecular+MACCS) and MACCS fingerprint descriptors. Therefore, we have focused on 0–2D molecular features only for QSAR model generation in this study. Before the model development, we performed the feature selection using different linear regression analysis methods for the DART datasets. Several techniques were used to do the linear regression analysis, including multiple linear regression (MLR) [44], [45] utilizing the stepwise selection or the Best Subset Selection tool (http://teqip.jdvu.ac.in/QSAR_Tools/DTCLab) followed by partial least squares regression (PLS) [46]. We have developed a stepwise regression model of 18, 13, and 14 descriptors from the initial set of pre-treated descriptors for Datasets 1, 2, and 4 respectively using MINITAB” software (version 14) [47]. In the case of Dataset 3, a different strategy of regression analysis has been adopted where we have performed the multilayered stepwise regression for feature selection using “MINITAB” software. The multi-layered selection is carried out in several iterations to extract the important features. To execute this, stepwise regression was performed for the pre-treated set of descriptor matrices (421 descriptors) to develop the stepwise regression model. After this, the selected set of descriptors was removed from the initial set of 421 descriptors, and again, stepwise regression was performed by using a remaining pool of descriptors and so on. In this way, 22 features were selected for further processing. Then, MLR best subset selection was performed using the reduced pool of descriptors (22 descriptors). This model development tool was used to get all the possible subset regression (MLR) models employing software developed in our laboratory and available at http://teqip.jdvu.ac.in/QSAR_Tools/DTCLab. From 22 descriptors (features) obtained by multi-layered stepwise regression, 12 descriptor MLR models of different combinations were generated by using the best subset selection regression analysis from which the best model was selected according to the low mean absolute error (MAE) value [48]. The PLS regression technique (http://www.minitab.com/en-US/default.aspx) was used to develop the final models from the selected descriptors of each dataset. PLS is a generalized version of the multiple linear regression (MLR) method that is applied to obviate multicollinearity and inter-correlation among the descriptors [46]. Therefore, we have reported the PLS model as our final model to remove noise and intercorrelation.
2.6. Quantitative read-across(qRA) predictions and similarity-based descriptor calculation
From a scientific perspective, physicochemical properties alone cannot predict the variety of adverse effects in repeated dose toxicity and DART testing. More scientific evidence is needed for the regulatory acceptance of these complex databases for the most commonly used chemicals to which we are readily exposed. Currently, researchers are looking to identify the correct pattern of similarity of query chemicals. Therefore, the categorization or grouping of chemicals based on the identification of significant similarity patterns of chemicals is now more accepted in terms of regulatory acceptance, and different governing bodies all over the globe are trying to develop new approaches for the categorization of chemicals in a rational way. Driven by this similarity pattern analysis hypothesis and the requirement for an appropriate query chemical classification for complicated endpoints, we have explored a novel methodology that combines the significant physicochemical properties (obtained from QSAR analysis) with the knowledge of the RA-based hypothesis (based on the similarity of chemical structures). Therefore, after the development of the traditional QSAR model for all datasets, the modeled descriptors of each dataset were used for the quantitative read-across (qRA) predictions. The qRA study was performed for query chemicals based on the weighted average prediction of the defined number of closely structure-related chemicals from the training set termed as close source chemicals. The qRA predictions aimed to identify the best similarity algorithm in terms of Q2F1 and mean absolute error (MAE) values for the query set chemicals from a defined number of close source chemicals, which we have further utilized in novel similarity-based descriptor computation. Therefore, qRA predictions were performed using Read-Across-v4.2 (https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home). Similarities were calculated based on three different similarity or distance-based algorithms (Euclidean distance, Gaussian kernel (σ), and Laplacian kernel (γ) approach) [36]. The read-across tool utilizes a set of hyperparameters (σ, γ, and the number of close source compounds) for the optimal read-across-based predictions. Finding the most appropriate similarity-based strategy and optimizing the related hyperparameters using an internal validation set—which is distinct from the test set—is crucial for the ideal computation of similarity functions. Each member of the internal validation set from the training compounds was used as a query compound based on the read-across-based hypotheses. Its prediction was made using its "close" source compounds from the remaining training compounds, and the similarity function that produced the best "predictions" for the internal validation set in terms of MAE or Q2F1 was found [49]. In the current work, we have opted to execute read-across-based predictions with no further optimization using the basic settings to retain homogeneity in all models. Therefore, the software was used in its default setting i.e. σ = 1, γ = 1, and the number of close source compounds (n) = 10). From the analysis, we have identified that for the first three datasets (Datasets 1, 2, and 3), the Laplacian kernel (LK) approach gives the best prediction results based on the Q2F1 MAE values of the validation set chemicals. On the other hand, for Dataset 4, Euclidean distance (ED) showed better results (based on MAE) than the other two similarity calculation methods. The similarity measures that give the optimum RA prediction results were used to compute different similarity-derived features in the form of concordance, error, and similarity measures of chemicals. For the similarity-based descriptor calculation of the training sets, the training set itself served as both source and target data; in contrast, for the similarity-based descriptor computation of test sets, the training and test sets served as source and target data, respectively. Using the same training set as both source and target data increases the likelihood of overfitting when calculating the similarity measures of the training set. The identical training data point was eliminated from the list of nearby training compounds using the "leave-same-out" technique to prevent this. For the computation of similarity-derived features (LK for Dataset 1,2 and 3 and ED for Dataset 4), we have used the freely available java based tool RASAR-Desc-Calc-v3.0.1 (available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home). We obtained 18 RA-derived features that were combined with the modeled QSAR descriptors for each dataset for further analysis [49], [50] (the list and the definitions of the novel similarity-derived features were provided in Table S1 of Supplementary material 2).
2.7. Combining features, selection of features, and final hybrid PLS model development from the “fused” data sets
After computing the novel similarity features based on qRA predictions, they were combined with the modeled physicochemical features to create a comprehensive pool of descriptors for each dataset. Furthermore, it was essential to recognize the core set of attributes needed to create a hybrid model from the entire pool of descriptors. The best subset selection was performed for each dataset using this new descriptor matrix. We have taken the same number of descriptors as in the QSAR model for better comparison. Therefore, different combinations of MLR models were generated for each dataset. We have selected the best MLR model based on MAE (Train) and Q2 (LOO) values for each dataset. Finally, the PLS model (final modeled descriptors with their response values were provided in Supplementary Material 3) was developed for the selected MLR model from each data set (now containing similarity-based descriptors and QSAR descriptors in the model) to compare with the traditional QSAR model fairly. Therefore, in our study, we have developed hybrid models where the non-linear RA predictions (based on similarity measures) were used in the QSAR framework to construct a linear QSAR model for better analysis and prediction of the data. The description of feature selection and model development from the initial descriptor matrix is provided in Table S2 of Supplementary Material 2.
2.8. Development of non-linear models using machine learning (ML)approaches
We have also developed conventional non-linear ML models for all datasets to check the performance of our linear PLS models compared to the ML models for this complex endpoint. Therefore, we have developed Random Forest (RF), AdaBoost, Gradient AdaBoost, Extreme Gradient Boost, and Support Vector Machine (SVM) models for each dataset in the default setting of their respective hyperparameters in Scikit learn package in Python. The conventional non-linear ML models were developed using ML regressor software available at (https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home/machine-learning-model-development-guis). From this analysis, we have identified that the hybrid PLS model which is the blended form of linear (QSAR) and nonlinear (RA hypothesis) analysis performs better than other conventional nonlinearML models. The ML results were provided in Supplementary Material 4. We have also investigated the importance of variables by using random forest (provided in Supplementary Material 4), which also confirms the significance of 0–2D features.
2.9. Statistical model validation
Any predictive technique must be thoroughly evaluated to ensure that the generated model is robust and relevant [51]. It is important to precisely measure the goodness of fit, robustness, and predictability of QSAR models according to the OECD principles [52], [53]. Extensive internal and external validation tests are carried out utilizing the training and testing datasets to evaluate the model's performance. To determine whether or not the developed models meet the acceptability criteria, we computed various internal quality and validation metrics in this study, including determination coefficient (R2), leave-one-out (LOO) cross-validated correlation coefficient (Q2LOO), mean absolute error of training set predictions (MAETrain), etc. External validation metrics, such as external correlation coefficients (Q2F1, Q2F2), and mean absolute error of test set predictions (MAETest), were used to assess the predictability of the resulting models [48], [54].
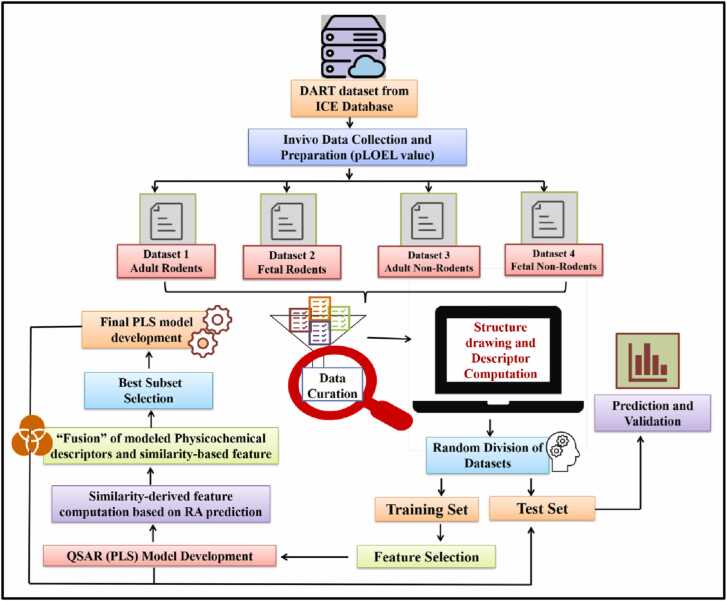
The overall workflow of this study is displayed in Fig. 1.
Fig. 1.
Generalized workflow for the hybridPLS model generation for the DART toxicity.
2.10. Development of PLS Plots using Simca-P software
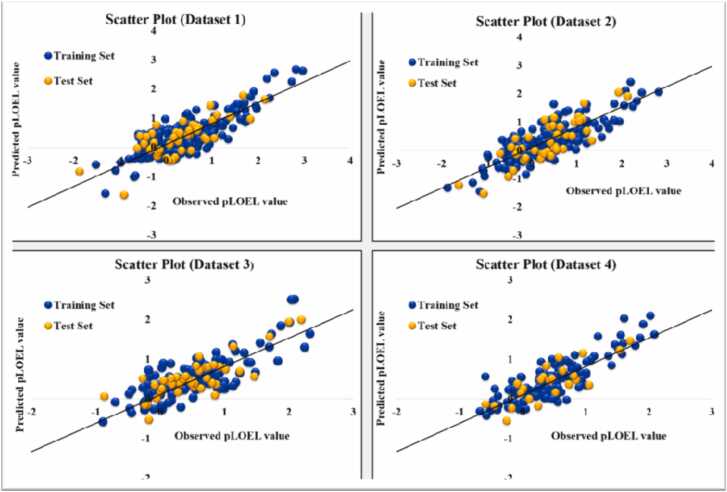
Simca-P v10.0 (https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software/mvda-software/simca) was used to create the characteristic plots associated with the final PLS models [55] The importance of each descriptor concerning the response value, which is reflected in the height of the descriptor bars, is shown by the variable importance plot (VIP) (Shown in Fig. 2).The influence of the descriptors, either positively or negatively, on the response is shown by the color of the bar where the green bar represents a positive and the red bar represents a negative contribution (Shown in Fig. 2). The loading plot (Fig. S1. in Supplementary Material 2), which shows the loading of a specific descriptor in a 2D latent variable space, uses the response and the descriptor's distance from the origin to illustrate how important the descriptor is. The Applicability Domain (AD) Plot DModX distance of the model in X-space) shows the AD status of the compounds considering the X space depicted using bar graphs, where X represents the dimensions (Fig. S2. and Fig. S3. for training and test set, respectively, in Supplementary material 2). An additional graph that shows the AD status of the training set compounds is the score plot (Fig. S4. in Supplementary material 2). Outliers are compounds that are located outside the score plot's ellipse. The DModX and the score plot analysis are different. The former uses all latent variables for outlier identification, while the latter uses only the first two. The Y-randomization graphic provides information on whether the model was developed by chance (Fig. S4. in Supplementary Material 2). To assess the prediction ability of the final hybrid model, a scatter plot of the Predicted pLOEL values v/s the Observed pLOEL values for the training and test set compounds was created (shown in Fig. 3).
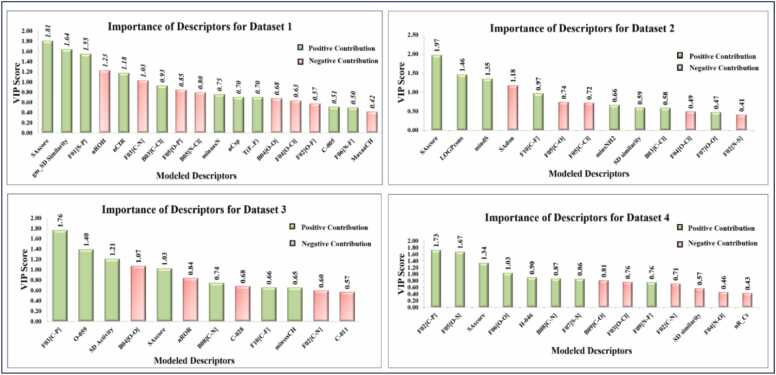
Fig. 2.
Variable importance plots of the final models. Color coding: Green = positive contribution, Red = negative contribution).
Fig. 3.
Scatter plots of observed vs predicted pLOEL values of the final hybrid models for the DART toxicity.
3. Result and discussion
In this investigation, we have developed 4 PLS models using each dataset's pooled set of features (structural and similarity-based features). The equation for the developed traditional QSAR and hybrid models is provided in Table 1. Table 2 provides the various external and internal validation metrics for traditional QSAR, qRA, and hybrid models for individual datasets. The validation results of qRA were given for the best similarity algorithm, which we have used for novel similarity-based feature calculation (discussed in an earlier section). We have reported the hybrid models as our final models for all 4 Datasets because of their better statistical quality than the corresponding QSAR models (as shown in Table 2). All the models' internal and external measures pass the cut-off points (R2 = 0.6, Q2LOO = 0.5, and Q2F1 = 0.5), demonstrating the models' reliability and predictive ability. The cross-validation (Leave-One-Out (LOO) statistics) is used to determine the number of components (LVs) in a PLS model throughout the model's development. The hybrid models may perform somewhat inferior on training data than testing set data due to the combined effect of leave-same-out descriptor computation (during similarity-based descriptors computation discussed in an earlier section) and LOO cross-validation. From the DModX plots (Fig. S2. and Fig. S3. for training and testing sets respectively in Supplementary Material 2), we have identified 9, 7, 2, and 3 molecules as structural outliers for the training sets for Datasets 1, 2, 3, and 4 respectively. For the test sets, on the other hand, the molecules 3, 4, 1, and 0 were outside AD for the respective datasets (i.e. Datasets 1, 2, 3, and 4). From the score plot (Fig. S4. in Supplementary Material 2) analysis, we have found that 5 molecules are present as structural outliers for Datasets 1, 2, and 3 (for training set chemicals) each. However,2 outlier molecules are identified in the case of the Dataset 4. Table 1 describes the developed models for all datasets with their statistical quality for both QSAR and hybrid models. We have also generated various ML models for each dataset using MACCS fingerprints and 2D molecular+MACCS fingerprint descriptors individually before selecting only 0–2D descriptors in this study (results provided in Supplementary Material 4).
Table 1.
The equations of developed QSAR and Hybrid models.
| Datasets | Models | Equation |
|---|---|---|
| Dataset 1 (adult rodents) |
QSAR model (alva descriptors) | |
| Hybrid model (alva+similarity-based descriptors) |
||
| Dataset 2 (Fetal rodents) |
QSAR model (alva descriptors) | |
| Hybrid model (alva+similarity-based descriptors) |
||
| Dataset 3 (adult rabbits) |
QSAR model (alva descriptors) | |
| Hybrid model (alva+similarity-based descriptors) |
||
| Dataset 4 (fetal rabbits) | QSAR model (alva descriptors) | |
| Hybrid model (alva+similarity-based descriptors) |
Table 2.
Various internal and external validation metrics for traditional QSAR, qRA, and hybrid models of all datasets.
|
Data sets |
Models |
Training Set |
Testing Set |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVs | Train | R2 | Q2 | MAE (train) | Test | Q2F1 | Q2F2 | MAE (Test) | ||||
| Adult rodents | Traditional QSAR Model | 7 | 194 | 0.683 | 0.596 | 0.353 | 42 | 0.526 | 0.512 | 0.426 | ||
| RA Model (LK) |
- | - | - | - | - | 42 | 0.481 | 0.465 | 0.461 | |||
| Hybrid Model | 8 | 194 | 0.682 | 0.605 | 0.345 | 42 | 0.572 | 0.559 | 0.393 | |||
| Fetal rodents | Traditional QSAR Model | 5 | 168 | 0.681 | 0.608 | 0.368 | 50 | 0.567 | 0.567 | 0.412 | ||
| RA Model (LK) |
- | - | - | - | - | 50 | 0.475 | 0.475 | 0.439 | |||
| Hybrid Model |
5 | 168 | 0.668 | 0.600 | 0.372 | 50 | 0.594 | 0.594 | 0.398 | |||
| Adult rabbits | Traditional QSAR Model | 8 | 133 | 0.681 | 0.608 | 0.284 | 42 | 0.642 | 0.641 | 0.274 | ||
| RA Model (LK) |
- | - | - | - | - | 42 | 0.582 | 0.580 | 0.307 | |||
| Hybrid Model | 8 | 133 | 0.664 | 0.579 | 0.291 | 42 | 0.653 | 0.652 | 0.271 | |||
| Fetal rabbits | Traditional QSAR Model | 8 | 108 | 0.753 | 0.651 | 0.270 | 24 | 0.633 | 0.620 | 0.287 | ||
| RA Model (ED) |
- | - | - | - | - | 24 | 0.441 | 0.421 | 0.354 | |||
| Hybrid Model | 9 | 108 | 0.756 | 0.659 | 0.266 | 24 | 0.670 | 0.659 | 0.283 | |||
3.1. Insights of modeled descriptors identified in the read-across derived models
Table S3 of Supplementary Material 2 presents the identified descriptors for all datasets along with their class and description. The variable importance plots (VIP) and coefficient plots of the developed pooled descriptor PLS models were obtained using "SIMCA-P" software [55] for additional investigation of the modeled descriptors. The bar height has been used to symbolize the VIP scores of the modeled descriptors, and the color of the bar (green for a positive contribution and red for a negative one) indicates the sort of contribution—positive or negative—to the DART toxicity (depicted in Fig. 2.). The VIP score is a measure of the relative importance of descriptors. In usual practice, descriptors with VIP scores > 1 demonstrate higher importance to the response [56]. Our study has four models with 18, 13, 12, and 14 features. We have focused on the descriptors that contributed the most toward the DART, as indicated by the VIP statistic (VIP score >1), for easy interpretation. Descriptors with a VIP score of less than one are relatively less significant. Our analysis of these less important descriptors has revealed that they either complement or influence the effects of the more significant descriptors in each model. Therefore, for each dataset, we have concentrated on the features that contribute most to DART endpoints (VIP score >1), and we have attempted to present a mechanistic interpretation only for those features in this work.
3.1.1. Features responsible for DART in adult rodents (dataset 1)
There are 18 descriptors in the final equation of the adult rodent model. The VIP statistic (VIP score >1) indicates that six of the descriptors—SAscore, gm*SD Similarity, F01[S-P], nROH, nCIR, F03[C-N]—contributed the most against the DART of adult rodents (shown in Fig. 2.). In our study, we interpreted the significant descriptors with VIP score >1 as discussed earlier in this section. From the VIP plot, SAscore is the most important descriptor against DART for adult rodents. It is a molecular property descriptor that indicates the synthetic accessibility score of the chemical. A molecule’s SAscore is between 1 (easy to synthesize) to 10 (difficult to synthesize) and is calculated from a molecule’s complexity and rarity of natural compounds in its fragment contributions [57]. The reference SAscore values are calculated using a precise method that combines the complexity-based score, which penalizes the presence of ring systems like multiple stereo centers, spiro and fused rings, and macrocycles, with the fragment-based score, which represents the "historical synthetic knowledge". The positive contribution of this descriptor shows that the presence of complex structures and unnatural fragments in a molecule has a toxic effect on adult rodents [58], [59], [60]. For instance, compounds nos.182 and 22 have higher DART toxicity for adult rodents due to their high SAscore or complexity. Conversely, when a molecule's complexity decreases (simpler structure of the molecule), its DART toxicity decreases, as compounds nos.120 and 167 of adult rodents demonstrate.
The second most significant descriptor is gm*SD Similarity, a similarity-derived feature extracted from physicochemical descriptors based on ten near-source compounds. For a given query chemical, the descriptor gm*SD_Similarity, which contributes positively to the toxicity value, is the product of the values of gm and the standard deviation of the similarity (SD_Similarity) values of the close source compounds. The concordance measure, Banerjee-Roy coefficient (gm), determines the reliability of the predictions of a query chemical based on the weight of evidence of the toxicity values/response values, i.e. toxic and non-toxic pattern of close source chemicals. The mathematical expression of gm is given in Eq. 1.
| (1) |
Here, PosFrac (positive fraction) represents the fraction of close source chemicals with a toxicity value more than the training set mean value. By this analysis, we can predict the toxicity of query chemicals based on the toxicity pattern of the close source chemicals. n is an integer whose value is 1 when MaxPos<MaxNeg and 2 when MaxPos ≥ MaxNeg. Here, MaxPos and MaxNeg represent the number of selected close source congeners having a toxicity value greater and less than the training set mean value of toxicity, respectively
Conversely, the descriptor SD_Similarity indicates the standard deviation of the similarity values of the close source chemicals [49]. This feature can be calculated by using the Eq. 2.
| (2) |
Here, = Similarity value of the selected number of close source congener molecule
= average similarity of the selected number of close source congener molecules
= number of selected close source congener molecules
The rationale behind the multiplication of gm and SD_Similarity is to assign negative or positive signs to SD_Similarity values to better categorize toxic and non-toxic chemicals (based on the low and high toxicity value of source molecules from the training set mean response value). A high SD_Similarity value and a positive gm suggest significant dispersion among the close source compounds. It is also likely that as the dispersion among the close source molecules increases, the prediction reliability for the query chemical decreases. So, this feature demonstrates that as the standard deviation of the similarity values of close source chemicals increases, the reliability of predictions decreases for the query chemicals. This is seen in compounds 42 and 46, where the prediction reliability for these query chemicals is reduced due to the higher standard deviation in the similarity values of source chemicals. In contrast, compound nos. 76 and 65 accurately anticipate the query chemical based on a smaller standard deviation of the similarity values for nearby source chemicals.
F01[S-P] is the third most important descriptor for the adult rodent species based on the VIP score. The 2D atom pair descriptor F01[S-P] represents the frequency of phosphorus and sulfur at the topological distance 1. This descriptor has a positive contribution, meaning that the DART toxicity increases with an increase in the frequency of phosphorous and sulfur atoms at topological distance 1 for adult rodents. From the study of organophosphate (OP) compounds (present in our dataset) used as agricultural chemicals, lubricant additives, and softening agents in technology and industry [61], we can see that P S (thion) bonds are present in the majority of OP chemicals. In vivo, they transform into the equivalent oxon derivatives (P O) and become acetylcholinesterase (AChE) enzyme inhibitors. In this manner, the chemical raises the Acetylcholine (Ach) level, which results in convulsions, depression of respiration, circulation muscular paralysis, and death [62], [63]. For example, the frequency of phosphorus and sulfur or the thion group is higher in compounds 26 and 46; as a result, these chemicals are highly toxic for adult rodent species due to AChE inhibition. However, compounds 49 and 50 have no thion group in their structure, showing reduced DART toxicity due to the very low tendency to interact with the binding site of the AChE enzyme.
nROH, the functional group count descriptor, is the next significant feature. This feature indicates the number of hydroxyl groups in a molecule. The DART toxicity of adult rodent species is negatively impacted by this characteristic, indicating that the toxicity of DART decreases in adult rodents as the amount of hydroxyl groups in a molecule increases. An increase in the number of hydroxyl groups in a molecule can make it more polar, thereby facilitating their excretion from the body, making them less harmful to adult rodents [64]. In particular, compounds 61 and 54 show decreased toxicity to DART as the number of hydroxyl groups rises, potentially leading to an increase in the molecule's polarity. Again, compounds 42 and 46 exhibit a high DART value for adult rodents because they lack hydroxyl groups in their structure.
Based on the VIP score, the next most influential descriptor is CIR. This ring descriptor shows the number of circuits (nCIR), a complex descriptor that measures rigidity and is correlated with molecular flexibility. A higher number of circuits indicates a lower degree of flexibility. This descriptor's positive coefficient value implies a rise in the proportion of circuits, which is mainly caused by the presence of more fused aromatic rings [65]. This descriptor positively contributes to the SAscore descriptor, which indicates that a higher level of complexity will result in a higher level of DART toxicity for adult rodents. In this respect, compounds 182 and 3 are more complex and rigid due to an increased fused ring structure, which increases the molecule's toxicity in adult rodents. However, compounds 67 and 36 have a more flexible structure resulting in reduced DART toxicity.
The 2D atom pair descriptor F03[C-N] is another important descriptor based on the VIP score. The frequency of carbon and nitrogen at the topological distance 3 is represented by the symbol F03[C-N]. The polarity of the molecule may also increase with an increase in nitrogen frequency (a strongly electronegative atom). The descriptor's negative contribution suggests that when the frequency of nitrogen increases at the topological distance 3, the molecule's toxicity decreases because of its increased polarity, making it easier for rodent species to eliminate the molecule from their bodies [66]. In this regard, the high frequencies of nitrogen in compounds 94 and 123 make them less detrimental, having lower DART toxicity values. Conversely, compounds 42 and 46 have higher DART toxicity values due to the absence of nitrogen atoms in organic chemicals, which reduces the polarity of the chemical. The other descriptors present in this model are less significant and indirectly influence the effect of the important descriptors. The definitions and classes are provided in Table S3 of Supplementary Material 2.
3.1.2. Features responsible for DART testing in fetal rodents (Dataset 2)
In the case of fetal rodents, we have constructed a 13-descriptor model. The features of model 2 were also interpreted according to the significance of descriptors based on the VIP score of the features (VIP score >1). In this model, four features SAscore, LOGPcons, mindS, and SAdon are the most significant descriptors according to their VIP scores (shown in Fig. 2). Among them, three features, SAscore, LOGPcons, and mindS, contribute positively, whereas SAdon contributes negatively to the DART toxicity of fetal rodents. Similar to the adult rodent endpoint, SAscore [57], [58], [59], [60] is the most important feature of DART toxicity for fetal rodents. The positive contribution of this descriptor suggests that the DART toxicity of a chemical will increase with the increase in the complexity of the molecule, as discussed earlier in Dataset 1. As exemplified in compounds 169 and 7, as the SAscore of the molecule increases, so does their DART toxicity value. On the other hand, in compounds 49 and 113, relatively low SAscore values may also reduce the DART toxicity of these chemicals.
The next influential descriptor is LOGPcons which represents the consensus octanol-water partition coefficient (LogP). This descriptor is a measure of the lipophilicity of a molecule, and lipophilicity is directly related to the toxicity of the chemical as a highly lipophilic molecule is not easily excreted from the body. Lipophilicity was found to be one of the key toxicity factors in several investigations, as it aids in the measurement of the chemicals' tissue distribution. cellular absorption, persistence, and bioavailability—thus their cumulative behavior [67]. A substance with a larger lipophilicity is more quickly absorbed by the body and will thus be more harmful to the organism [68]. In a recent study of the human placenta barrier, it is evident that an increase in the lipophilicity of a molecule may be associated with the tendency to cross the placental barrier showing a detrimental effect on the body of a developing fetus [69]. Therefore, the positive contribution of this descriptor indicates that the DART toxicity of fetal rodent species may rise in parallel with an increase in the lipophilicity of a molecule. As evident from compounds 7 and 135, a rise in LOGPcons value raises the DART toxicity of the concerned chemical. Conversely, a decrease in the LOGPcons value will lessen the DART toxicity, as shown in compounds 59 and 29.
The next important descriptor for fetal rodents is mindS, which indicates the presence of sulfur atoms in the double-bonded form (=S). This descriptor contributes positively which means that an increase in the number of double-bonded sulfurs may increase the DART toxicity of the molecules. This descriptor represents the presence thion group in a molecule (P S) in our database. The thion group is present in the majority of OP chemicals that are in use currently. As discussed earlier in Dataset 1, in vivo, they transform into the equivalent oxon derivatives (P O) and become inhibitors of the AChE enzyme. In this manner, the chemical raises the ACh level, resulting in convulsions, depression of respiration and circulation, muscular paralysis, and death [62], [63]. For example, in compounds 24 and 39, the presence of the thion group may increase the toxicity in fetal rodent species. In contrast, in compounds 29 and 59, the absence of these groups may decrease the DART toxicity due to less susceptibility of the molecules to interact with the binding site of Ach esterase inhibitors.
Another significant descriptor according to the VIP score is SAdon, which is a molecular property descriptor that represents the “surface area of donor atoms from P_VSA-like descriptors”. According to Labute (2000), [70] these molecular descriptors are the percentage of van der Waals surface area (VSA) of the donor atoms. The negative contribution of this descriptor indicates that an increase in the polar surface area of the donor atom may reduce the toxicity value for DART endpoints [71]. In our study, the donor atoms are the hydrogen bond donor atoms. The hydrogen donor in a molecule is the abstractable hydrogen presented adjacent to the highly polar or electronegative atoms (N, O, and F). So, these molecules may tend to form intermolecular hydrogen bonds when present in a close vicinity of water. This might increase the solubility of these chemicals in water thereby increasing the excretion rate of these chemicals. So, we can say that an increase in the polar surface area of the hydrogen bond donor atoms will increase the solubility of the chemical in the water therefore that may be easily excreted from the fetal rodent body. Compounds 59 and 49, for example, have a higher polar surface area for hydrogen bond donor atoms, which may reduce the molecule's toxicity due to stronger intermolecular hydrogen bonding. However, in compounds 39 and 44, the presence of a smaller polar surface area of hydrogen bond donor atoms may increase the toxicity profile of rodent fetuses. The remaining model descriptors are less important and indirectly affect or contribute to the toxicity of rodent fetuses (descriptions of these descriptors are provided in Table S3 of Supplementary Material 2).
3.1.3. Features responsible for DART testing in adult rabbits (Dataset 3)
In the case of Dataset 3, F03[C-P], O-059, SD activity, B04 [O-O], and SAscore are the significant descriptors according to the VIP scores of the modeled descriptors (12 descriptors). The most significant descriptor is F03 [C-P], which indicates the frequency of carbon and phosphorus atoms at the topological distance 3. The positive contribution of this descriptor suggests that an increase in the frequency of carbon and phosphorus atoms at the topological distance three will increase the DART toxicity of adult rabbits. Organophosphorus compounds are characterized by a stable carbon-to-phosphorus (C-P) bond, which usually resists biochemical, thermal, and photochemical decomposition, showing the chemical's persistent nature [72]. The positive contribution of this descriptor represents an increase in the frequency of carbon and phosphorus atoms (as organophosphates), which may enhance the accumulative or persistent nature of the chemicals (since degradation of these chemicals is difficult), thereby increasing the DART toxicity in adult rabbits. For instance, in compound nos. 58 and 60, high carbon and phosphorus frequencies might increase the compounds' DART toxicity (that may be because of the persistent nature of these chemicals). However, in compounds nos. 1, and 142, the absence of this fragment (carbon or phosphorus fragments) may reduce the DART toxicity of adult rabbits.
The second most important descriptor according to the VIP scores is O-059. This is an atom-centered fragment descriptor that indicates the presence of a specific type of atom in a molecule and the connectivity of the atom. This descriptor represents the presence of an aliphatic group on both sides of the oxygen atom i.e. aliphatic ether (representing the local atomic environments) [73]. The molecular descriptor O-059 is related to the phosphorus-group properties by contributing to reducing the electron density of the phosphorus atom, which means that the phosphorus-containing molecules with substructures containing oxygen show a strong electron-withdrawing effect. This makes phosphorus-containing compounds more capable of attacking nucleophiles of biomembranes, hence increasing the toxicity [74]. In compound nos. 104 and 2, as the frequency of this fragment increases, the DART toxicity of the molecules also increases. In contrary in compound nos. 1 and 27, the absence of this fragment reduces the DART toxicity.
The 3rd important descriptor is SD activity [49], a similarity-based feature that represents the standard deviation of the response value of the close source molecules. This descriptor represents the weighted standard deviation of the observed response values of the close source compounds. The mathematical expression for computing this feature is represented in Eq. 3.
| (3) |
| (4) |
Here, n is the number of close source congeners, is the weightage of selected individual close source congeners, is the observed response value of individual close source congeners, is the weighted average prediction value of the query chemical.
In our dataset, this descriptor contributes positively, indicating that as the standard deviation increases in the response value of the close source chemicals, the reliability of prediction is reduced. For example, in compounds 60 and 104, as the standard deviation of the activities of the close source molecules increases, the reliability of predictions for the query chemicals may decrease. On the other hand, in compounds 54 and 110, the close source molecules have low values for the DART toxicity (have a lower standard deviation for the response values); so the reliability of predictions will increase for the query molecule.
Another important descriptor is B04 [O-O], which is a 2D atom pair descriptor that represents the presence or absence of O-O atoms at the topological distance 4. This descriptor contributes negatively indicating that the presence of an electronegative oxygen atom in a molecule will increase the polarity of the molecule and therefore tends to reduce the toxicity profile of the molecules [75]. For example, in compounds 1 and 54, the presence of O-O atoms at the topological distance 4 may increase the polarity thereby decreasing the toxicity profile of the molecule. On the other hand, in molecules 36 and 58, the absence of this fragment shows high DART toxicity in adult rabbits.
The next important descriptor is SAscore [57], [58], [59], [60] which contributes positively. As discussed earlier in the case of Datasets 1 and 2, the positive contribution of this descriptor may increase the DART toxicity for adult rabbits with increased complexity of the molecule as well. This can be seen in compound nos. 117 and 114. In contrast, in compound nos. 132 and 89, comparatively less complexity of the molecule shows less toxicity in adult rabbits. The descriptions of other less modeled descriptors are provided in Table S3 of Supplementary Material 2.
3.1.4. Features responsible for DART testing in fetal rabbits (Dataset 4)
Based on the VIP scores, the most important descriptors for fetal rabbits among the 14 descriptors are F02 [C-P], F05 [O-S], SAscore, and F06 [O-O]. As determined by the VIP score, the most important descriptor is F02 [C-P], which represents the frequency of phosphorus and carbon atoms at the topological distance 2. As discussed earlier for Dataset 3, the positive contribution of this descriptor represents that an increase in the frequency of this feature may reduce the in vivo biodegradability of these chemicals and, therefore, increase the DART toxicity of these chemicals in fetal rabbits, as evident in compounds 26 and 46 [72]. On the other hand, in compounds 55 and 129, an absence of this fragment may reduce the DART toxicity of these chemicals.
The next important descriptor, according to the VIP plot, is the F05 [O-S]. This descriptor represents the frequency of oxygen and sulfur atoms at the topological distance 5. These descriptors contribute positively. In the case of phosphate esters (in the form of thiophosphates), the occurrence of an electronegative sulfur atom at a certain distance from oxygen may increase the toxicity profile [76]. As exemplified in compounds 26 and 46, an increase in this fragment's frequency will increase the chemicals' toxicity. On the other hand, in compounds 129 and 101, the absence of this fragment may reduce systemic toxicity in fetal rabbits.
The 3rd most important descriptor is SAscore [57], [58], [59], [60]. Like the other 3 models, this descriptor contributes positively which shows that as the complexity of the molecule increases, the DART toxicity of fetal rabbits will also increase. As evident from compounds 90 and 26, an increase in the complexity of the structure may increase the DART toxicity. On the other hand, in compounds 5 and 10, the DART toxicity of the compound reduces as the SAscore or the complexity of the molecule decreases.
According to the VIP score, the next important descriptor is the F06 [O-O] descriptor. This descriptor represents the frequency of oxygen atom pair at a topological distance 6. This descriptor also contributes positively, signifying that an increase in the occurrence of this fragment in a molecule will increase the DART toxicity of the molecules [77], [78]. This descriptor complements the effect of F05 [O-S] descriptors where the increase in electronegative atoms (in the presence of organophosphates) in a molecule will enhance the DART toxicity to produce a detrimental effect on the developing rabbits (as a fetus). For example, in compounds 83 and 12, the increase in the electronegative oxygen atom pair will increase the DART toxicity to fetal rabbits. On the other hand, in compounds 129 and 101, the decreased frequency of the fragment will reduce the DART toxicity of the molecules. The effect of this descriptor is also influenced by the presence of other less significant descriptors in the model. Table S3 in Supplementary Material 2 contains the definitions and classes of all the descriptors present in the models since here we have only discussed the most significant features.
3.2. Summary of the combined knowledge from each of the individual hybrid models
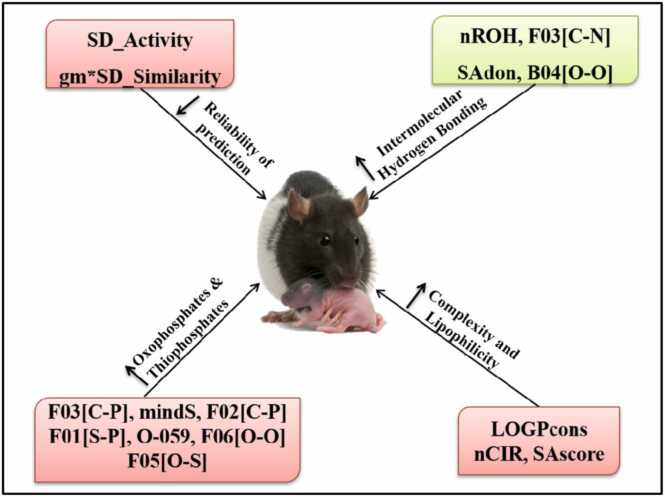
To keep the discussion simple, we have grouped different contributing features of the models into discrete functional nature according to the physicochemical characteristics that increase or decrease the DART toxicity. In general, from our analysis, the chemical features can be briefly categorized into the following classes: 1. Complexity of structures; 2. Presence of Oxophosphates or Thiophosphates; 3. Intermolecular Hydrogen bonding characters (with water molecules); and 4. Lipophilicity is the crucial factor contributing to the DART toxicity of a chemical. Further, the significant similarity measures (descriptors) show the confidence of correctly predicting the query chemicals. Fig. 4 illustrates the contributions of all the influential descriptors for our rodents and rabbit species datasets.
Fig. 4.
The overall contribution of the most influential descriptors for all datasets.
4. Conclusion
There are not enough assay results for DART while employing conventional techniques to close these gaps would require many test animals. Developing in silico NAMs that accurately depict the intricate reproductive cycle at the many crucial junctures is a significant task because the reproductive cycle involves many intricate procedures. We need to understand the similarity pattern of the chemical with its physical or chemical characteristics for a more refined evaluation of the toxicological parameters of a chemical for complex studies (like DART) as suggested by the regulatory bodies. The regulatory bodies are currently focusing on properly categorizing chemicals into their toxicity class, with the prediction of their toxicity based on the pattern analysis of source chemicals. To address this issue, firstly, we have prepared or categorized the LOEL data (extracted from the ICE database) for adult and fetal life stages of rodents and rabbits separately and explored the potential of a novel in silico approach as an alternative method to assess this complicated endpoint rationally. This new approach integrates the two widely accepted in silico techniques (QSAR and RA) used by regulatory authorities to primarily screen chemicals for their toxicological profiles. Therefore, in our study, we have tried to incorporate the information from both QSAR and RA studies to understand the physicochemical and the confidence of prediction for the query chemical (based on the similarity analysis in the form of similarity-based features) for DART testing of rodents and rabbits. The main objective of our study was to check the statistical quality of our developed hybrid models with the traditional QSAR models along with ease of interpretation and transferability for addressing complex endpoints like DART testing by using the knowledge of molecular features and similarity-based features. From the results obtained from this study, we can infer that the hybrid modeling strategy is a viable algorithm for filling the data gap of new chemicals for their DART toxicity assessment in the future. Furthermore, given the different regulatory frameworks and guidelines released in the EU, USA, Canada, Japan, and Australia, and the ongoing and impending efforts to develop alternative testing strategies, the proposed method provides a valuable option for assessing this complex endpoint.
CRediT authorship contribution statement
Kunal Roy: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Sapna Pandey: Writing – original draft, Validation, Investigation, Formal analysis, Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The research was funded by the Indian Council of Medical Research (ICMR), New Delhi (Grant no. BMI/12(73)/ 2022, dated 19.03.23). The authors are grateful for this support.
Handling Editor: Prof. L.H. Lash
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.toxrep.2024.101822.
Appendix A. Supplementary material
Supplementary Material 1 contains the datasets with their response values for adult and fetal rodents and rabbits in Excel format.
Supplementary Material 2 contains Table S1, which provides a detailed list of novel similarity-based descriptors with their definition.; Table S2, which provides the details of model development for each set; and Table S3, which provides the definition and class of modeled QSAR and hybrid features. Figures S1-S5 contain Simca plots for the final hybrid models in the Word file format.
Supplementary Material 3 contains the final descriptor matrix for hybrid models with classified training and testing sets in Excel format.
Supplementary Material 4 contains the conventional nonlinear ML model, ML models for MACCS, and 2D molecular feature + MACCS fingerprint descriptors and the importance of features based on Random Forest.
Data availability
Data will be made available on request.
References
- 1.United Nations. Globally harmonised systems of classification and labelling of chemicals (GHS). Part 3 health hazards; 2007. Available online at: 〈http://www.unece.org/trans/danger/publi/ghs/ghsrev02/English/03epart3.pdf〉〈.(accessed〉 10 May, 2024).
- 2.Hewitt M., Ellison C.M., Enoch S.J., Madden J.C., Cronin M.T.D. Integrating (Q) SAR models, expert systems and read-across approaches for the prediction of developmental toxicity. Reprod. Toxicol. 2010;30(1):147–160. doi: 10.1016/j.reprotox.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Assessment RT. Guidelines for Reproductive Toxicity Risk Assessment.〈https://www.epa.gov/sites/default/files/201411/documents/guidelines_repro_toxicity.pdf〉.(accessed 10 May, 2024).
- 4.Regulation of (EC) No. 1907/2006 of the European Parliament and of the Council, December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No. 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC, Off. J. Eur. Union, L396(2007). 〈https://assets.publishing.service.gov.uk/media/5e1dcebb40f0b610fcf63737/EU_REACH_Regulations_1907_2006__1_.pdf〉. (accessed 10 May, 2024).
- 5.Rovida C., Hartung T. Re-evaluation of animal numbers and costs for in vivo tests to accomplish REACH legislation requirements for chemicals: a report by the transatlantic think tank for toxicology (t (4) Altex. 2009;26(3):187–208. 10.14573/altex.2009.3.187. [PubMed] [Google Scholar]
- 6.Corvi R., Spielmann H., Hartung T. Alternative approaches for carcinogenicity and reproductive toxicity. Hist. Altern. Test. Methods Toxicol. 2019:209–217. doi: 10.1016/B978-0-12-813697-3.00024-X. [DOI] [Google Scholar]
- 7.Marzo M., Roncaglioni A., Kulkarni S., Barton-Maclaren T.S., Benfenati E. In silico model for developmental toxicity: how to use QSAR models and interpret their results. Silico Methods Predict. Drug Toxic. 2016 doi: 10.1007/978-1-4939-3609-0_8. 139-61. [DOI] [PubMed] [Google Scholar]
- 8.Organisation for Economic Co-operation and Development. OECD 414: Prenatal Developmental Toxicity Study. In OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing:Paris, France, 2018, 10.1787/20745788 (accessed 10 May, 2024). [DOI]
- 9.Ciallella H.L., Russo D.P., Sharma S., Li Y., Sloter E., Sweet L., Huang H., Zhu H. Predicting prenatal developmental toxicity based on the combination of chemical structures and biological data. Environ. Sci. Technol. 2022;56(9):5984–5998. doi: 10.1021/acs.est.2c01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basant N., Gupta S., Singh K.P. In silico prediction of the developmental toxicity of diverse organic chemicals in rodents for regulatory purposes. Toxicol. Res. 2016;5(3):773–787. doi: 10.1039/c5tx00493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ECC/HC (2016). Chemicals Management Plan (CMP) Science Committee Objectives Paper Meeting No. 5—Integrating New Approach Methodologies within the CMP: Identifying Priorities for Risk Assessment, Existing Substances Risk Assessment Program. Ottawa, Ontario, Canada: Government of Canada. 〈http://www.ec.gc.ca/ese-ees/default.asp?lang=En&n=172614CE-1〉. (accessed 12 May, 2024).
- 12.ECHA (2016). “New Approach Methodologies in Regulatory Science,” in Proceedings of a Scientific Workshop, Helsinki, Finland, 19–20 April, 2016, 〈https://echa.europa.eu/documents/10162/22816069/scientific_ws_proceedings_en.pdf〉.(accessed 12 May, 2024).
- 13.ECHA (2017). ECHA Strategic Plan 2019–2023. Helsinki, Finland: ECHA. 2017, 〈https://echa.europa.eu/documents/10162/26075800/echa_strategic_plan_2019〉.
- 14.2023_en.pdf/3457ccff-7240-2c1f-3a15-fa6e5e65ac56. (accessed 12 May, 2024).
- 15.EPA (2018). Final Strategic Plan to Promote Development and Implementation of Alternative Test Methods Supporting Toxic Substances Control Act, 83.Washington, DC: Federal Register, 30167–30168. 2018, 〈https://www.epa.gov/sites/default/files/2018-06/documents/epa_alt_strat_plan_6-20-18_clean_final.pdf〉. (accessed 12 May, 2024).
- 16.OECD (2005). Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for hazard Assessment: OECD Series on Testing and Assessment, Number 34 (ENV/JM/MONO(2005)14). 2005, 〈https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV/JM/MONO〉(2018)19&doclanguage=en. (accessed 12 May, 2024).
- 17.OECD (2017). Guidance Document for Describing Non-guideline In Vitro Test Methods, OECD Series on Testing and Assessment, No. 211 (ENV/JM/MONO(2014)35).2017, 〈https://www.oecd-ilibrary.org/docserver/9789264274730en.pdf?expires=1626436326&id=id&accname=guest&checksum=BF5B058705B87255A946A52AC4BE4984〉. (accessed 12 May, 2024).
- 18.OECD (2018). Guidance Document on Good in Vitro Method Practices (GIVIMP). OECD Series on Testing and Assessment, No. 286. 2018, 10.1787/9789264304796-en.(accessed 15 May, 2024). [DOI]
- 19.Parish S.T., Aschner M., Casey W., Corvaro M., Embry M.R., Fitzpatrick S., Kidd D., Kleinstreuer N.C., Lima B.S., Settivari R.S., Wolf D.C. An evaluation framework for new approach methodologies (NAMs) for human health safety assessment. Reg. Toxicol. Pharm. 2020;112 doi: 10.1016/j.yrtph.2020.104592. [DOI] [PubMed] [Google Scholar]
- 20.OECD. (2004) Test No. 430: In Vitro Skin Corrosion: Transcutaneous Electrical Resistance Test (TER).2004, 10.1787/20745788. (accessed 16 May, 2024). [DOI]
- 21.OECD. (2009) Test No. 437: Bovine Corneal Opacity and Permeability Test Method for Identifying Ocular Corrosives and Severe Irritants.2009, 10.1787/9789264076303-en. (accessed 16 May, 2024). [DOI]
- 22.OECD. (2009) Test No. 438: Isolated Chicken Eye Test Method for Identifying Ocular Corrosives and Severe Irritants. 2009, 10.1787/20745788. (accessed 16 May, 2024). [DOI]
- 23.OECD. (2010) Test No. 439: In Vitro Skin Irritation.2010, 〈https://ntp.niehs.nih.gov/sites/default/files/iccvam/suppdocs/feddocs/oecd/oecd-tg439-2013-508.pdf〉. (accessed 16 May, 2024).
- 24.OECD. (2022) Test No. 442C: In Chemico Skin Sensitisation. 2022, 〈https://www.oecd.org/en/publications/test-no-442c-in-chemico-skin sensitisation_9789264229709en.html#:∼:text=This%20Test%20Guideline%20is%20proposed,containing%20either%20lysine%20or%20cysteine〉. (accessed 16 May, 2024).
- 25.OECD. (2022) Test No. 442D: In Vitro Skin Sensitisation. OECD QSAR Toolbox. 2022, 10.1787/9789264229822-en. (accessed 16 May, 2024). [DOI]
- 26.EU. (2009) Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on cosmetic products. Retrieved from 〈http://data〉. europa.eu/eli/reg/2009/1223/oj. Book Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on cosmetic products. 2009, 〈http://data.europa.eu/eli/reg/2009/1223/oj.,City〉. (accessed 16 May, 2024).
- 27.Dent M.P., Vaillancourt E., Thomas R.S., Carmichael P.L., Ouedraogo G., Kojima H., Barroso J., Ansell J., Barton-Maclaren T.S., Bennekou S.H., Boekelheide K. Paving the way for application of next generation risk assessment to safety decision-making for cosmetic ingredients. Reg. Toxicol. Pharm. 2021;125 doi: 10.1016/j.yrtph.2021.105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berggren E., White A., Ouedraogo G., Paini A., Richarz A.N., Bois F.Y., Exner T., Leite S., Grunsven van, Worth L.A., Mahony A. C. Ab initio chemical safety assessment: a workflow based on exposure considerations and non-animal methods. Comput. Toxicol. 2017;4:31–44. doi: 10.1016/j.comtox.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ICCR (2018). Report on Integrated Strategies for Safety Assessment of Cosmetic Ingredients: Part 2.2018, 〈https://www.iccr-cosmetics.org/files/8315/4322/3079/ICCR_Integrated_Strategies_for_Safety_Assessment_of_Cosmetic_Ingredients_Part_2.pdf〉. (accessed 15 May, 2024).
- 30.Hareng L., Pellizzer C., Bremer S., Schwarz M., Hartung T. The integrated project ReProTect: a novel approach in reproductive toxicity hazard assessment. Reprod. Toxicol. 2005;20(3) doi: 10.1016/j.reprotox.2005.04.003. 441-52. [DOI] [PubMed] [Google Scholar]
- 31.Lorenzetti S., Altieri I., Arabi S., Balduzzi D., Bechi N., Cordelli E., Galli C., Ietta F., Modina S.C., Narciso L., Pacchierotti F. Innovative non-animal testing strategies for reproductive toxicology: the contribution of Italian partners within the EU project ReProTect. Ann. I Super. Sanita. 2011;47:429–444. doi: 10.4415/ANN_11_04_16. [DOI] [PubMed] [Google Scholar]
- 32.Piersma A.H., Bosgra S., Van Duursen M.B., Hermsen S.A., Jonker L.R., Kroese E.D., Van der Linden S.C., Man H., Roelofs M.J., Schulpen S.H., Schwarz M. Evaluation of an alternative in vitro test battery for detecting reproductive toxicants. Reprod. Toxicol. 2013;38:53–64. doi: 10.1016/j.reprotox.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 33.van der Burg B., Wedebye E.B., Dietrich D.R., Jaworska J., Mangelsdorf I., Paune E., Schwarz M., Piersma A.H., Kroese E.D. The Chem Screen project to design a pragmatic alternative approach to predict reproductive toxicity of chemicals. Reprod. Toxicol. 2015;55 doi: 10.1016/j.reprotox.2015.01.008. 114-23. [DOI] [PubMed] [Google Scholar]
- 34.Ball N., Cronin M.T., Shen J., Blackburn K., Booth E.D., Bouhifd M., Donley E., Egnash L., Hastings C., Juberg D.R., Kleensang A. T4 report: toward good read-across practice (GRAP) guidance. Altex. 2016;33(2):149. doi: 10.14573/Faltex.1601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hisaki T., Kaneko née, Hirota M.A., Matsuoka M., Kouzuki M. H. Integration of read-across and artificial neural network-based QSAR models for predicting systemic toxicity: a case study for valproic acid. J. Toxicol. Sci. 2020;45(2):95–108. doi: 10.2131/jts.45.95. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee A., Roy K. First report of q-RASAR modeling toward an approach of easy interpretability and efficient transferability. Mol. Divers. 2022;26(5):2847–2862. doi: 10.1007/s11030-022-10478-6. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee M., Banerjee A., De P., Gajewicz-Skretna A., Roy K. A novel quantitative read-across tool designed purposefully to fill the existing gaps in nanosafety data. Environ. Sci. Nano. 2022;9(1):189–203. doi: 10.1039/D1EN00725D. [DOI] [Google Scholar]
- 38.Kumar V., Banerjee A., Roy K. Breaking the barriers: machine-learning-based c-RASAR approach for accurate blood–brain barrier permeability prediction. J. Chem. Inf. Model. 2024 doi: 10.1021/acs.jcim.4c00433. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee A., Roy K. Prediction-inspired intelligent training for the development of classification read-across structure–activity relationship (c-RASAR) models for organic skin sensitizers: assessment of classification error rate from novel similarity coefficients. Chem. Res. Toxicol. 2023;36(9):1518–1531. doi: 10.1021/acs.chemrestox.3c00155. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee M., Banerjee A., Tosi S., Carnesecchi E., Benfenati E., Roy K. Machine learning-based q-RASAR modeling to predict acute contact toxicity of binary organic pesticide mixtures in honey bees. J. Hazard. Mater. 2023;460 doi: 10.1016/j.jhazmat.2023.132358. [DOI] [PubMed] [Google Scholar]
- 41.Pandey S.K., Roy K. Development of a read-across-derived classification model for the predictions of mutagenicity data and its comparison with traditional QSAR models and expert systems. Toxicology. 2023;500 doi: 10.1016/j.tox.2023.153676. [DOI] [PubMed] [Google Scholar]
- 42.Gallagher A., Kar S. Unveiling first report on in silico modeling of aquatic toxicity of organic chemicals to Labeorohita (Rohu) employing QSAR and q-RASAR. Chemosphere. 2024;349 doi: 10.1016/j.chemosphere.2023.140810. [DOI] [PubMed] [Google Scholar]
- 43.Daniel A.B., Choksi N., Abedini J., Bell S., Ceger P., Cook B., Karmaus A.L., Rooney J., To K.T., Allen D., Kleinstreuer N. Data curation to support toxicity assessments using the integrated chemical environment. Front. Toxicol. 2022;4 doi: 10.3389/ftox.2022.987848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauri A. alvaDesc: a tool to calculate and analyze molecular descriptors and fingerprints. Ecotoxicological QSARs. 2020:801–820. doi: 10.1007/978-1-0716-0150-1_32. [DOI] [Google Scholar]
- 45.Ambure P., Aher R.B., Gajewicz A., Puzyn T., Roy K. NanoBRIDGES” software: open access tools to perform QSAR and nano-QSAR modeling. Chemom. Intell. Lab. Syst. 2015;147:1–3. doi: 10.1016/j.chemolab.2015.07.007. [DOI] [Google Scholar]
- 46.Darlington, R.B. Regression and linear models. McGraw-Hill Companies; 1990.
- 47.Wold S., Sjöström M., Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001;58(2):109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 48.Data Analysis, Statistical & Process Improvement Tools | Minitab, 2004.
- 49.Roy K., Das R.N., Ambure P., Aher R.B. Be aware of error measures. Further studies on validation of predictive QSAR models. Chemom. Intell. Lab. Syst. 2016;152:18–33. doi: 10.1016/j.chemolab.2016.01.008. [DOI] [Google Scholar]
- 50.Banerjee A., Roy K. On some novel similarity-based functions used in the ML-based q-RASAR approach for efficient quantitative predictions of selected toxicity endpoints. Chem. Res. Toxicol. 2023;36(3) doi: 10.1021/acs.chemrestox.2c00374. 446-64. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee A., Kar S., Roy K., Patlewicz G., Charest N., Benfenati E., Cronin M.T. Molecular similarity in chemical informatics and predictive toxicity modeling: From quantitative read-across (q-RA) to quantitative read-across structure–activity relationship (q-RASAR) with the application of machine learning. Crit. Rev. Toxicol. 2024:1–26. doi: 10.1080/10408444.2024.2386260. [DOI] [PubMed] [Google Scholar]
- 52.Roy K. On some aspects of validation of predictive quantitative structure–activity relationship models. Expert Opin. Drug Discov. 2007;2(12):1567–1577. doi: 10.1517/17460441.2.12.1567. [DOI] [PubMed] [Google Scholar]
- 53.OECD, 2007. Guidance documents on the validation of (Quantitative) structure-activity relationships [(Q)SAR] models [WWW Document]. OECD Environ. Heal. Saf. Publ. 〈https://www.oecd.org/officialdocuments/〉 publicdisplaydocumentpdf/?doclanguage=en&cote=env/jm/mono(2007)2. (accessed 20 May, 2024).
- 54.OECD, 2004. Validation of (Q)SAR Models - OECD [WWW Document]. 〈https://www.oecd.org/chemicalsafety/risk-assessment/validationofqsarmodels〉. Htm. (accessed 20 May, 2024).
- 55.Roy K., Mitra I., Kar S., Ojha P.K., Das R.N., Kabir H. Comparative studies on some metrics for external validation of QSPR models. J. Chem. Inf. Model. 2012;52(2):396–408. doi: 10.1021/ci200520g. [DOI] [PubMed] [Google Scholar]
- 56.Wu Z., Li D., Meng J., Wang H. Introduction to SIMCA-P and its application.Handbook of partial least squares: concepts. Methods Appl. 2010:757–774. 10.1007/978-3-540-32827-8 33. [Google Scholar]
- 57.Olah M., Bologa C., Oprea T.I. An automated PLS search for biologically relevant QSAR descriptors. J. Comput. Aid. Mol. Des. 2004;18:437–449. doi: 10.1007/s10822-004-4060-8. [DOI] [PubMed] [Google Scholar]
- 58.Ertl P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Chem. Inf. 2009 doi: 10.1186/1758-2946-1-8. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay S., Brylinski M., Bess A., Berglind F., Galliano C., McGrew P.F. Deep drug: applying AI for the advancement of drug discovery. Comsnets. 2022:667–674. doi: 10.1109/COMSNETS53615.2022.9668586. [DOI] [Google Scholar]
- 60.Pu L., Naderi M., Liu T., Wu H.C., Mukhopadhyay S., Brylinski M. e toxpred: a machine learning-based approach to estimate the toxicity of drug candidates. BMC Pharmacol. Toxicol. 2019;20:1–5. doi: 10.1186/s40360-018-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y., Li X., Pu Q., Fu R., Wang Z., Li Y., Li X. Innovative screening for functional improved aromatic amine derivatives: Toxicokinetics, free radical oxidation pathway and carcinogenic adverse outcome pathway. J. Hazard. Mater. 2023;454 doi: 10.1016/j.jhazmat.2023.131541. [DOI] [PubMed] [Google Scholar]
- 62.Maynard R.L., Chilcott R.P. Toxicology of chemical warfare agents. Gen. Appl. Syst. Toxicol. 2009 doi: 10.1002/9780470744307.gat131. [DOI] [Google Scholar]
- 63.Reiner E., Radić Z., Simeon-Rudolf V. Mechanisms of organophosphate toxicity and detoxication with emphasis on studies in Croatia. Arh. Hig. Rada. Toksikol. 2007;58(3):329–338. doi: 10.2478/v10004-007-0026-2. [DOI] [PubMed] [Google Scholar]
- 64.Peter J.V., Sudarsan T.I., Moran J.L. Clinical features of organophosphate poisoning: a review of different classification systems and approaches. Indian J. Crit. Care Med. 2014;18(11):735. doi: 10.4103/0972-5229.144017. https://doi.org/10.4103%2F0972-5229.144017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X., Liu H., Yang Q., Liu J., Chen J., Shi L. Predicting anti-androgenic activity of bisphenols using molecular docking and quantitative structure-activity relationships. Chemosphere. 2016;163:373–381. doi: 10.1016/j.chemosphere.2016.08.062. [DOI] [PubMed] [Google Scholar]
- 66.Cruz-Monteagudo M., Cordeiro N.D., Tejera E., Dominguez Rosa, Borges E. F. Desirability-based multi-objective QSAR in drug discovery. Mini-Rev. Med. Chem. 2012;12(10):920–935. doi: 10.2174/138955712802762329. [DOI] [PubMed] [Google Scholar]
- 67.Mansouri K., Ringsted T., Ballabio D., Todeschini R., Consonni V. Quantitative structure–activity relationship models for ready biodegradability of chemicals. J. Chem. Inf. Model. 2013;53(4):867–878. doi: 10.1021/ci4000213. [DOI] [PubMed] [Google Scholar]
- 68.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: an analysis of ALOGP and CLOGP methods. J. Phys. Chem. A. 1998;102(21):3762–3772. doi: 10.1021/jp980230o. [DOI] [Google Scholar]
- 69.Van Meter R.J., Glinski D.A., Hong T., Cyterski M., Henderson W.M., Purucker S.T. Estimating terrestrial amphibian pesticide body burden through dermal exposure. Environ. Pollut. 2014;193:262–268. doi: 10.1016/j.envpol.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Giaginis C., Tsantili-Kakoulidou A., Theocharis S. Assessing drug transport across the human placental barrier: from in vivo and in vitro measurements to the ex vivo perfusion method and in silico techniques. Curr. Pharm. Biotechnol. 2011;12(5):804–813. doi: 10.2174/138920111795470930. 〈https://www.ingentaconnect.com/content/ben/cpb/2011/00000012/00000005/art00013〉 [DOI] [PubMed] [Google Scholar]
- 71.Labute P. A widely applicable set of descriptors. J. Mol. Graph. Model. 2000;18(4-5):464–477. doi: 10.1016/S1093-3263(00)00068-1. [DOI] [PubMed] [Google Scholar]
- 72.Li T., Huang Y., Wei G., Zhang Y.N., Zhao Y., Crittenden J.C., Li C. Quantitative structure-activity relationship models for predicting singlet oxygen reaction rate constants of dissociating organic compounds. Sci. Total Environ. 2020;735 doi: 10.1016/j.scitotenv.2020.139498. [DOI] [PubMed] [Google Scholar]
- 73.Kafarski, P. Phosphonates: Their natural occurrence and physiological role.Contemporary Topics about Phosphorus in Biology and Materials. Intech Open 2019, 1-9, 〈https://books.google.co.in/books?id=TJYtEAAAQBAJ〉.
- 74.Sayyadi Kord Abadi R., Alizadeh O., Ghasemi G. An Investigation on the QSAR modeling of carfilzomib derivatives using Monte Carlo method and novel modelling-optimization approach. Org. Chemi. Res. 2021;7(1):61–76. doi: 10.22036/org.chem.2022.297270.1261. [DOI] [Google Scholar]
- 75.Daghighi A., Casanola-Martin G.M., Timmerman T., Milenković D., Lučić B., Rasulev B. In silico prediction of the toxicity of nitroaromatic compounds: application of ensemble learning qsar approach. Toxics. 2022;10(12):746. doi: 10.3390/toxics10120746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Worachartcheewan A., Prachayasittikul V., Prachayasittikul S., Tantivit V., Yeeyahya C., Prachayasittikul V. Rational design of novel coumarins: a potential trend for antioxidants in cosmetics. EXCLI J. 2020;19:209. doi: 10.17179/excli2019-1903. https://doi.org/10.17179%2Fexcli2019-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar A., Ojha P.K., Roy K. First report on pesticide sub-chronic and chronic toxicities against dogs using QSAR and chemical read-across. SAR QSAR Environ. Res. 2024;35(3):241–263. doi: 10.1080/1062936X.2024.2320143. [DOI] [PubMed] [Google Scholar]
- 78.Hao Y., Sun G., Fan T., Tang X., Zhang J., Liu Y., Zhang N., Zhao L., Zhong R., Peng Y. In vivo toxicity of nitroaromatic compounds to rats: QSTR modelling and interspecies toxicity relationship with mouse. J. Hazard. Mater. 2020;399 doi: 10.1016/j.jhazmat.2020.122981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1 contains the datasets with their response values for adult and fetal rodents and rabbits in Excel format.
Supplementary Material 2 contains Table S1, which provides a detailed list of novel similarity-based descriptors with their definition.; Table S2, which provides the details of model development for each set; and Table S3, which provides the definition and class of modeled QSAR and hybrid features. Figures S1-S5 contain Simca plots for the final hybrid models in the Word file format.
Supplementary Material 3 contains the final descriptor matrix for hybrid models with classified training and testing sets in Excel format.
Supplementary Material 4 contains the conventional nonlinear ML model, ML models for MACCS, and 2D molecular feature + MACCS fingerprint descriptors and the importance of features based on Random Forest.
Data Availability Statement
Data will be made available on request.