Abstract
Introduction/Aims:
Needle impedance-electromyography (iEMG) assesses the active and passive electrical properties of muscles concurrently by using a novel needle with six electrodes, two for EMG and four for electrical impedance myography (EIM). Here, we assessed an approach for combining multifrequency EMG and EIM data via machine learning (ML) to discriminate D2-mdx muscular dystrophy and wild-type (WT) mouse skeletal muscle.
Methods:
iEMG data were obtained from quadriceps of D2-mdx mice, a muscular dystrophy model, and WT animals. EIM data were collected with the animals under deep anesthesia and EMG data collected under light anesthesia, allowing for limited spontaneous movement. Fourier transformation was performed on the EMG data to provide power spectra that were sampled across the frequency range using three different approaches. Random forest-based, nested ML was applied to the EIM and EMG data sets separately and then together to assess healthy versus disease category classification using a nested cross-validation procedure.
Results:
Data from 20 D2-mdx and 20 WT limbs were analyzed. EIM data fared better than EMG data in differentiating healthy from disease mice with 93.1% versus 75.6% accuracy, respectively. Combining EIM and EMG data sets yielded similar performance as EIM data alone with 92.2% accuracy.
Discussion:
We have demonstrated an ML-based approach for combining EIM and EMG data obtained with an iEMG needle. While EIM-EMG in combination fared no better than EIM alone with this data set, the approach used here demonstrates a novel method of combining the two techniques to characterize the full electrical properties of skeletal muscle.
Keywords: disease classification; electrical impedance myography; electromyography; machine learning, muscle; muscular dystrophy
1. | INTRODUCTION
Distinguishing subtle myopathic abnormalities from healthy muscle has remained one of the more challenging problems of standard qualitative needle electromyography (EMG), with one recent study showing only a 70% accuracy.1 One reason for this is that mild myopathic states may cause only subtle alterations to motor unit potentials (MUPs), making them only of slightly shorter duration and lower amplitude with minimal polyphasia. Similarly, subtle alterations in recruitment patterns can be especially difficult to identify as the ratio of MUPs to firing rate remains preserved in myopathy.2 The most useful electrodiagnostic measure is what has been termed “early recruitment”—namely an increase in the number of MUPs recruited for generated force. More challenging still, chronic myopathies may be associated with changes more typically considered neurogenic in nature, such as some enlarged MUPs with decreased MUP recruitment.3 The presence of fibrillation potentials, positive sharp waves, myotonic potentials, or complex repetitive discharges may, of course, assist with disease diagnosis, but also are not specific for myopathy, as they can occur in neurogenic disorders as well.
To improve upon this, a large number of quantitative approaches to EMG analysis have been taken, including manual and automated MUP analysis,3,4 macro-EMG analysis,5–7 and power spectrum analysis, among others.3,8,9 The last has typically used tools to quantify the electromyographic signal such as through Fourier transformation8 on the acquired EMG signal or using principal component analysis.10,11 Myopathic disorders typically demonstrate fewer lower frequency components, as the MUPs are of shorter duration and polyphasic. More recently, machine learning (ML) approaches have also been applied to the assessment of the EMG signal and studies have suggested that ML can improve our ability to identify subtle myopathic alterations over standard approaches.12–14
Electrical impedance myography (EIM) is a newer quantitative approach to neuromuscular disease assessment.15,16 EIM is a bioimpedance-based technique in which a weak, high-frequency electrical current is applied to a discrete area of muscle and the consequent voltages are measured. Alterations in the composition and structure of the tissue change the resistive and reactive components of the tissue, impacting the measured voltages. As there are no devices approved by the US Food and Drug Administration for performing EIM, its development and testing remain confined to the research realm currently. Whereas much of the earlier focus and application of EIM has been in tracking disease progression or response to therapy,17 its potential as an improved diagnostic technique also remains of interest. Indeed, like EMG, EIM offers a rich array of measures including multiple primary parameters and a complex frequency dependence. And like EMG, ML has been employed in EIM as another approach to data interpretation, improving its capability in disease discrimination in mouse models beyond that obtainable with just single frequency measurements.18 Recently, EIM technology has been combined into a concentric needle EMG to create a single “impedance-EMG” or “iEMG” device to allow concurrent collection of both signals and improve diagnostic outcomes.19,20
In this study, we sought to assess the potential of ML-based iEMG for discriminating healthy muscle from dystrophic muscle by studying a mouse model of Duchenne muscular dystrophy, the D2-mdx mouse. Our goal was to present an approach for doing so to establish proof-of-principle for use in future more challenging classification problems. We hypothesized that iEMG would provide more accurate classification than either technique alone as it would provide a holistic electrophysiologic representation of the muscle, with EMG assessing the active properties and EIM the passive. The intent of this work was to set the stage for future investigations of this combination technology in both animal and clinical work.
2. | METHODS
2.1. | Animal procedures
Experimental procedures were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center. A total of 12 male wild-type (WT) animals were available for study (11 DBA/2J, strain #000671 and 1 B6SJLF1/J, Strain #100012) aged 16–35 wk. Thirteen D2-mdx mice (D2.B10-Dmdmdx/J; strain #013141) aged to 35 wk, an age at which major dystrophic features are present,21 were also available for study. All animals were obtained from Jackson Laboratories (Bar Harbor, ME), and fed standard chow ad libitum up until the time of study. During data collection, the animals were anesthetized with isoflurane (0.5%–3%) delivered by a nosecone. Blinding of the individual collecting the data (S.B.R.) to animal category (healthy vs. diseased) was not possible given the considerably smaller size of the D2-mdx mice.
With the animal under deep anesthesia, the fur overlying the quadriceps was removed bilaterally with a standard rechargeable clipper. We studied quadriceps as it shows considerable pathology in this disease.22 To facilitate the iEMG needle insertion, a small hole was then placed in the skin using a 21-gauge angiocath needle just proximal to the left knee. A prototype iEMG needle20 (Haystack Diagnostics, Inc, Boston, MA, USA) was then passed through the hole in the skin and into the center of quadriceps with an effort to keep the needle parallel to the long axis of the muscle so that all six electrodes remained within the muscle group itself. Impedance measurements were then made with the mView impedance system (Myolex, Inc, Boston, MA, USA) interfaced to an iEMG needle connector. The impedance device was configured to measure resistance R and reactance X values at 41 logarithmically spaced frequencies from 1 kHz to 10 MHz.
Once the impedance data were collected, the mView device was disconnected and the Natus Ultrapro S100 EMG system with Synergy Software (Natus. Middleton, WI, USA) was connected to the EMG leads on the connector and an adhesive ground electrode was placed on the tail. Filter settings were set at standard levels 10 Hz and 10 kHz (high-pass and low-pass, respectively) at a sampling frequency of 50 kHz. The anesthesia was then lightened to the point that voluntary movements of the animal could be observed. After a few minutes, stereotypical flexion/extension of the entire hindlimbs occurred. Approximately 10–20 s of EMG activity was collected.
Impedance data were directly exported from the mView system for analysis. Similarly, the EMG data from the evaluation of the voluntary activity were exported as a text file. These were then reconstructed into a single trace. In order to increase sample size, we considered the measurements of the limbs of same animals as independent of each other.
2.2. | Data analysis
EIM data were obtained at 41 frequencies of applied current and included the 3 standard measures: resistance, reactance, and phase values at each of the measured frequencies; all of these values were included in our analysis. In terms of analyzing the EMG signal, we performed a standard Fourier transformation using R 4.1.023 including frequencies from 1 Hz to 25 kHz. To reduce computational time while promoting robustness in the model, and to balance the number of samples measured between EIM and EMG used in our ML model, we downsampled the EMG power spectrum at selected frequencies aiming for approximately the same number of frequencies as EIM. We explored various approaches to downsampling the EMG data including principal component analysis and the least absolute shrinkage and selection operator (LASSO). However, these approaches showed worse discrimination than the following somewhat simpler sampling approaches, including:
Equal spacing. We chose frequencies that were evenly spaced beginning at approximately 1 Hz through 25 kHz. Using these equally distant sets of points, we included 44 frequencies in our final ML model from this procedure.
Fibonacci’s sequence. Fibonacci’s sequence is a naturally progressing series of values and has been used by others in analyzing EMG data.24 We chose to use this series to include a more extensive set of lower frequency values, which are likely to be reduced in power in a myopathic model. From this procedure, the final model contained 48 frequencies.
Geometric sequence. A geometric sequence is a series of sequences that comes from a power of a given number. We chose two as the base number with linear power that increases by one. Through this procedure, we included 44 frequencies in our final model.
ML-based diagnostic outcomes were conducted using three frequency data sets: (1) EIM data alone, (2) EMG data alone, and (3) both EIM and EMG data sets together. We chose these three separate strategies with the final goal of our analysis to evaluate the comparative performance of each of these three models.
To conduct our statistical analysis, we used R 4.1.0 where we applied the “caret” package to build our learning algorithm and “eegkit” package for the Fourier transformation. Our data set was then tested and evaluated with numerous sets of supervised algorithms, including support vector machines, K-nearest neighbor, an individual decision trees. Based on their comparative performance, we found that the random forest algorithm appeared to offer the best performance for our data sets. The random forest algorithm is based on a mixture of decision trees.25 A decision tree is one approach that helps model the data visually using “if and else” conditions. Such visualization helps in identifying explicit patterns that can be used to classifya given group. Random forest constitutes a collection of a multitude of decision trees that are created to predict probable class outputs. The class output that is chosen in most of these decision trees will be the outcome result of the entire random forest.
We used a nested cross-validation procedure to conduct our analysis, as previously described.26,27 Here we split the data into two parts where 80% of it was assigned to the training data and 20% to the test set. Then we used a nested 10-fold cross-validation method for classifying the condition of each mouse with random forest. The nested procedure works in two loops: (1) an inner loop that was allocated to determine the individual training data estimates and their performance within the training data and (2) an outer loop assigned for checking the performance of these estimates on the test set. The final evaluation of the model was based on combining the classifier’s performance in each test set from all outer loops.
The learning algorithm’s performance was based on the average receiver operator characteristic (ROC) curves generated by these outer loops. The AUC values in these curves gave the probability of the learning model’s ability to classify the condition (i.e., healthy or diseased) of each mouse correctly. To compare the ROC curves from different data sets, we used Delong’s ROC comparison that has been extended for unpaired ROC curves.28,29
3. | RESULTS
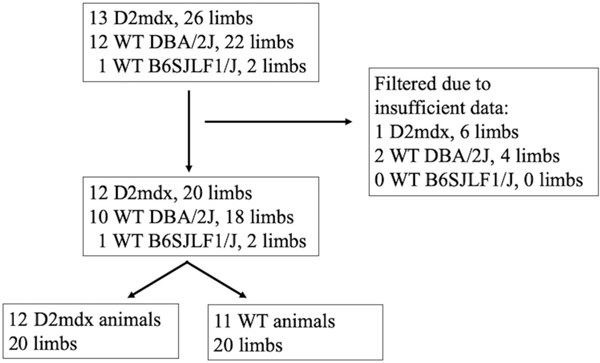
From the entire group of animals described in the methods section, we were able to achieve successful data collection on a set of 20 D2-mdx limbs and 20 WT limbs. The specific breakdown of how this was accomplished, and which animals were used in the analysis below, is diagrammed in Figure 1. We note that whereas obtaining the EIM data from the animals at rest was relatively straightforward, obtaining sufficient active EMG data was usually the most challenging aspect of the data collection and simply could not be achieved successfully in a number of animals on both quadriceps.
FIGURE 1.
Flow chart showing animal and limb selection used in the analysis from the original set of animals.
1. EIM and Fourier transformed EMG data.
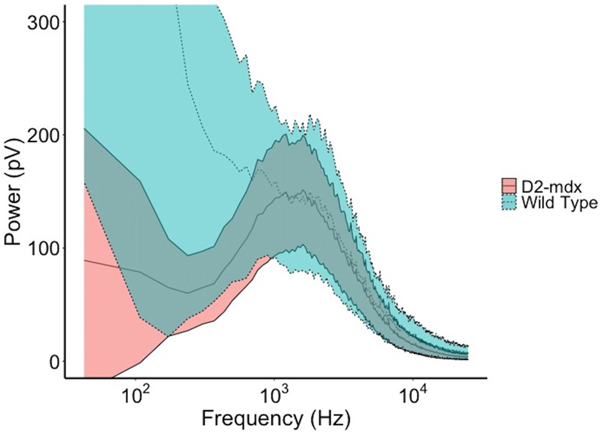
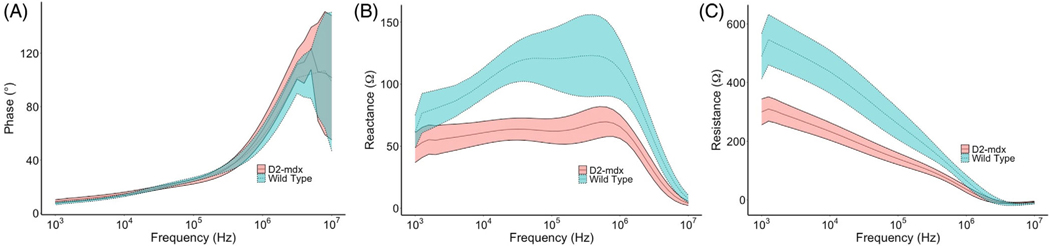
Figure 2 shows the averaged EMG power spectra from the Fourier transformed EMG signals from all WT and diseased animals. In addition, the power spectra were smoothed using an average sequence of 500 frequencies. Figure 3 shows the raw multifrequency EIM data (i.e., phase, resistance, and reactance) averaged across all WT versus diseased animals. As expected, the EMG power spectra show greater power at higher frequency in the animals with muscular dystrophy and fewer low frequency components, consistent with short-duration, polyphasic motor units, characteristic of myopathy. Similarly, the EIM data reveal, as expected, generally higher reactance and resistance values in the healthy animals with an overall greater frequency dependence. Although it is difficult to compare these data sets directly, it is worth noting here the separation between the EIM data for the healthy versus the diseased mice, as compared to the considerable overlap between healthy and diseased EMG spectra. This provides some indication of the anticipated performance of our ML-based analyses, shown next.
FIGURE 2.
Comparison of EMG power spectrum averaged across all WT and D2-mdx mice. Every segment of 500 frequency sequence were averaged for smoothing. Note shift to right (higher frequencies) apparent in the D2-mdx mice, consistent with myopathic motor units. The inner dashed/solid lines represent the mean activity with the outer limits representing the 95% confidence intervals of the data across the 20 limbs studied.
FIGURE 3.
Comparison of EIM reactance spectra between the two WT and D2-mdx mice. Data show mean across the animals and standard error. Note the good separation in resistance and reactance values between the groups.
2. ROC analysis for EIM and equally spaced EMG frequencies.
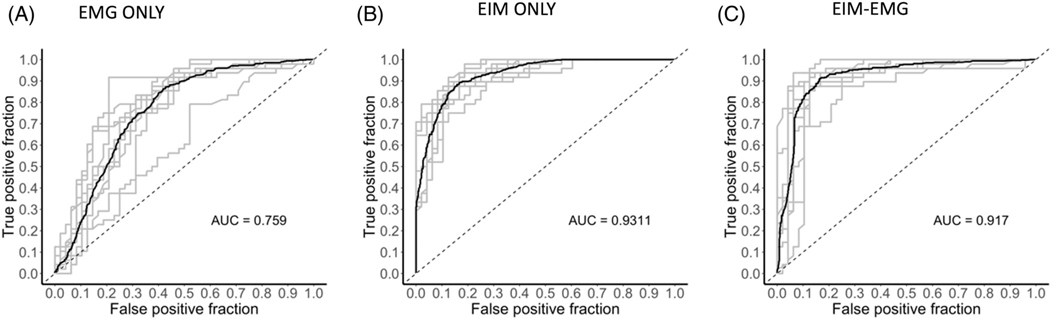
Figure 4 provides the ROC-AUC values for sampled power of EMG frequencies (44 frequencies) that are equally spaced and the multifrequency EIM (using values from all 41 frequencies). The EMG frequencies were 589 Hz apart, evenly spaced beginning at a frequency of 1 Hz and ending at a frequency of 24,754 Hz. This number was chosen through a series of tests with various gap sizes that yielded the data sets’ highest AUC value for EMG alone. The results show that EIM-only data had the greater power of discrimination with EMG performing considerably less well. The combined EIM-EMG data set, rather than providing discrimination even more robustly than either one alone, performed slightly worse than the EIM data set alone. This is confirmed by ROC test comparisons, which demonstrated that EMG performed worse than EIM and the EIM-EMG. EIM alone was also better than combined EIM-EMG.
FIGURE 4.
EMG ROC curves for our test using ML for (A) EMG data alone (equally spaced frequency selection). (B) EIM data alone. (C) EIM and EMG data together. EMG data alone were significantly different than both EIM only data (p < .001) and EIM-EMG data (p < .001). EIM only data were slightly significantly different than EIM-EMG data (p = .047).
3. ROC-AUC curves for EIM and Fibonacci sequenced EMG frequencies.
Figure S1 gives the ROC-AUC values for EMG frequencies that are spaced by Fibonacci’s sequence with identical assessment of the multifrequency EIM data as in Figure 4. The results are similar to that presented in Figure 4 with EIM-only data and EIM-EMG data outperforming EMG-only data in this analysis. The combined EIM-EMG analysis had a non-significant difference compared to EIM data alone.
4. ROC analysis for EIM and geometrically sequenced EMG frequencies.
Figure S2 shows the analogous figure for geometric sequence sampling. The results here are quite like that obtained using the Fibonacci series, with EIM alone outperforming EMG alone with the combined data set analysis performing similarly to EIM alone.
4. | DISCUSSION
Here we sought to integrate EIM and EMG data collected with needle iEMG to provide a “complete” picture of muscle’s passive (EIM) and active (EMG) electrical properties in the context of a common electrodiagnostic classification problem: differentiating healthy from myopathic muscle. Whereas we had hypothesized that the combined data set would perform better than either the EIM or the EMG data alone, the ultimate combined classifier performed similarly to that of EIM alone. This occurred despite our utilizing several different approaches for sampling the EMG power spectra. Regardless of this somewhat unexpected result, we believe further investigation into the concept of EIM-EMG data integration using ML would be worthwhile pursuing, especially in human disease, as discussed below. We briefly review several of the issues that may have influenced our results here and that may not hold true when this approach is applied to human data.
Studying mice limited our ability to obtain active, voluntary EMG data since all EMG signals had to be acquired in brief time periods while the animals were under light anesthesia. This resulted in having considerably less data available for analysis than might be anticipated in human research. The animals demonstrated stereotypical movements in this state, allowing us to successfully perform interference pattern analysis, a standard approach which has been described in the literature since the 1980s.9 Performing single MUP analysis would also have been valuable, but virtually impossible given our inability to control the level of voluntary contraction.
In terms of transforming the EMG data for analyses, we also experimented using several approaches, including principal component analysis, but the results were ultimately weaker in terms of EMG classification data than using the Fourier transform approach described here. Accordingly, we have excluded the details of these preliminary investigations here. Regardless of the approach chosen, in reviewing Figure 2, there is a clear loss of the lowest frequencies in the D2-mdx mice as compared to the WT animals, consistent with expectations. In short, while study of a rodent model with this new needle is a necessary first step to its wider application, we anticipate that human application of this technique will be more straightforward simply because we will be able to obtain much better-quality active EMG data for analysis. It is also possible that the mouse MUPs, which are smaller than those of a human muscle,30 may be difficult for our equipment to differentiate as it was designed from human diagnostics.
Another important point is technical. While this iEMG needle has E1 at the tip and E2 is the barrel around it, it is not a typical concentric needle in which the entire barrel serves as a reference electrode. As we described previously,20 in this device, only the distal most rim of the needle along the tip serves as the E2 electrode here. This much smaller recording area near the concentric inner electrode may impact the EMG data itself. Thus, it would have been ideal to also perform a second study comparing the needle EIM data alone with EMG data obtained with a standard concentric needle in these animals. The study we completed here is decidedly unique to this needle design and should be interpreted as such.
We had more constrained sampling choices with the EIM data set than the EMG since measurements occur in the frequency domain and not in time-domain as is the case with EMG. Nevertheless, the findings are consistent with what we have described previously for ML using EIM data in distinguishing healthy and myopathic muscle ex vivo, with AUCs of over 0.90,18 supporting outstanding disease discrimination.
How is it possible that the combined impedance-EMG data set does not surpass the EIM data alone? The expectation is that any predictive model that is developed will be only as good as the predictors included in it. The final AUCs we report here are based on the average of the repeated test sets only, implying that the training sets, when using both EIM and EMG together, may be overfitting the training data. As the AUCs that are provided represent an average of the test data and not the original model, if there is more noise in the original training set as might be the case with the EMG signal, the combined data set could have a slightly worse performance than a model using the EIM data alone. Alternatively, our EMG frequency selection in the model may be falsely underestimating the diagnostic power of EMG. While this is certainly possible, as we have noted, we are including only the most successful outcomes here. Moreover, simply visually inspecting and comparing Figures 2 and 3 reveals the substantial overlap between the healthy and myopathic EMG spectra (Figure 2) versus the clear separation for resistance and reactance data in the EIM spectra (Figure 3). This supports that our findings of poorer performance of the EMG data alone versus the EIM data alone are not simply due to undersampling of the EMG power spectrum. Finally, our AUCs for EMG data alone are fairly similar to those in a recently published human study looking at the accuracy of qualitative needle EMG in the diagnosis of myopathy.1
As with all ML approaches, there is an uncomfortable “black box” aspect to the modeling adopted here. ML does not offer any insights into why EIM fared so much better than EMG. However, one possibility is that EIM is simply far more sensitive to the major histological and compositional abnormalities that occur in severely dystrophic muscle,31,32 which are considerable in this disease model, as compared to the relatively modest motor unit and recruitment abnormalities observed on EMG. Performing a similar study in ALS animals, for example, may have produced a far different outcomes, with EMG potentially outperforming EIM, given the stark EMG abnormalities observed in any advanced neurogenic disorder.
While the focus here was classification, iEMG could also be used as a biomarker to assess disease progression over time or therapy effect. Surface EIM techniques have already shown value in this,33,34 but an iEMG approach could also be advantageous. For example, it could be useful in obese patients or where a specific deep muscle needs to be evaluated. The addition of a quantified EMG signal over EIM alone could offer potential improved sensitivity to disease alteration over EIM alone.
There are several additional limitations to this work beyond the simple choice of using a mouse. First, the total number of animals, both D2-mdx and WT, was relatively low. Although we took approaches for mitigating this by using a nested ML model, there is always the risk of overfitting when the data set is very limited. We assessed reducing the data set by reducing the number of values by 3/4 and observed the anticipated reduction in the AUC, especially for the EMG signal (e.g., Fibonacci sequence AUC decreasing from 0.73 to 0.70). However, given the preliminary nature of these analyses, we put this forward as a proof-of-principle analysis and are willing to accept the potential of overfitting. Second, given the relatively small number of animals, we considered each hind limb as a separate and independent data set; ideally it would have been better to have data from more individual animals; this may have resulted in stronger models with better overall classification. Third, spontaneous activity was not evaluated. If this assessment were also included, then the presence of fibrillation potentials and positive sharp waves likely would have improved our ability to detect differences between the sets of animals on EMG. Fourth, the iEMG needle remains in a prototype state, and thus our ability to obtain good data was limited and the size of the muscle relative to the size of the needle is quite small meaning that we could not easily reposition the needle to view MUPs most advantageously. Fifth, the EMG data obtained were limited as it was obtained while awakening from anesthesia. Presumably more extended EMG signals, as would be possible in humans, would add EMG discriminatory capability. Finally, we do note that all the D2-mdx mice were approximately 35 wk of age whereas some of the WT were younger. We do not believe that this introduced any significant confounding since they are all adult animals, and age-related muscle changes in mice do not become apparent until 1.5–2 y.
We briefly reflect on potential challenges and applications moving forward. First, mouse studies represent a model that is not entirely reflective of human disease, with these D2-mdx animals often showing findings not typical of human DMD, including minimal fat deposition and heterotopic calcification.22 Moreover, the muscle pathology is severe, being typical of dystrophic disease; a milder, non-dystrophic myopathy may have yielded different results with EIM perhaps not showing such dramatic differences compared to WT animals. Third, it remains uncertain how well this approach compares to standard qualitative EMG. Fourth, at this point, it is not clear how this technology would be employed in a clinical setting. Would an individual undergo standard qualitative EMG and then EIM assessments only if the EMG were unrevealing? And then would ML be used immediately in some fashion, or would it be possible to still use qualitative EMG along with the quantified analyses of EIM? At this point, all possibilities remain valid, and we must stay agnostic as to the best future approach. Finally, we have not assessed neurogenic disorders here. Clearly, that would be another important direction to explore.
This work represents only an initial foray into a new field of muscle electrophysiology, where we seek to quantitatively evaluate the passive and active properties of the tissue simultaneously and then meld them into improved diagnostic assays. Only by dedicated study in a range of neuromuscular conditions in both animal models and diseased and healthy humans can the potential of this new technology in clinical diagnosis and disease characterization be fully realized.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health Grants R41NS112029–01A1 and R01NS055099.
Funding information
National Institutes of Health, Grant/Award Numbers: R41NS112029–01A1, R01NS055099
Abbreviations:
- AUC
area under the curve
- EIM
electrical impedance myography
- EMG
electromyography
- iEMG
impedance-electromyography
- ML
machine learning
- MUP
motor unit potential
- ROC
receiver operating characteristic
- WT
wild-type
Footnotes
ETHICS STATEMENT
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
CONFLICT OF INTEREST STATEMENT
Dr. Rutkove has equity in and serves a consultant and scientific advisor to Myolex, Inc and Haystack Diagnostics, companies that design impedance devices for clinical and research use; he is also a member of Myolex’s Board of Directors. The companies also have an option to license patented impedance technology of which Dr. Rutkove is named as an inventor.
Dr. Sanchez holds equity in Haystack Diagnostics, a company that develops clinical needle impedance technology for neuromuscular evaluation. The company has an option to license patented needle impedance technology where the author is named an inventor. He also holds equity and serves as Scientific Advisory Board Member of Ioniq, a company that develops clinical impedance technology for early cancer detection. Dr. Sanchez serves as Scientific Advisory Board Member of B-Secur, a company that develops wearable ECG and impedance technology. He consults for Myolex, Inc., a company that develops surface impedance technology. The company has an option to license patented surface EIM technology where the author is named an inventor. Dr. Sanchez also serves as a consultant to Impedimed, a company that develops clinical impedance technology for early detection of secondary lymphedema, and Texas Instruments, Happy Health, and Maxim Integrated, companies that develop impedance related technology for consumer use.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Rutkove Laboratory Webiste at https://www.rutkovelab.org/data-repository/.
REFERENCES
- 1.Constantinides VC, Papahatzaki MM, Papadimas GK, et al. Diagnostic accuracy of muscle biopsy and electromyography in 123 patients with neuromuscular disorders. In Vivo. 2018;32:1647–1652. doi: 10.21873/invivo.11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimura J. Electrodiagnosis in Diseases of Nerve and Muscle: Principles and Practice. Third ed. Oxford University Press; 2001. [Google Scholar]
- 3.Fuglsang-Frederiksen A. The role of different EMG methods in evaluating myopathy. Clin Neurophysiol. 2006;117:1173–1189. doi: 10.1016/j.clinph.2005.12.018 [DOI] [PubMed] [Google Scholar]
- 4.Tankisi H, Burke D, Cui L, et al. Standards of instrumentation of EMG. Clin Neurophysiol. 2020;131:243–258. doi: 10.1016/j.clinph.2019.07.025 [DOI] [PubMed] [Google Scholar]
- 5.Finsterer J, Fuglsang-Frederiksen A. Concentric-needle versus macro EMG: II. Detection of neuromuscular disorders. Clin Neurophysiol. 2001;112:853–860. doi: 10.1016/S1388-2457(01)00499-0 [DOI] [PubMed] [Google Scholar]
- 6.Cengiz B, Ozdag F, Ulas UH, Odabasi Z, Vural O. Discriminant analysis of various concentric needle EMG and macro-EMG parameters in detecting myopathic abnormality. Clin Neurophysiol. 2002;113:1423–1428. doi: 10.1016/S1388-2457(02)00170-0 [DOI] [PubMed] [Google Scholar]
- 7.Barkhaus PE, Nandedkar SD, Sanders DB. Quantitative EMG in inflammatory myopathy. Muscle Nerve. 1990;13:247–253. doi: 10.1002/mus.880130312 [DOI] [PubMed] [Google Scholar]
- 8.Fuglsang-Frederiksen A, Rnager J. EMG power spectrum, turns-amplitude analysis and motor unit potential duration in neuromuscular disorders. J Neurol Sci. 1990;97:81–91. doi: 10.1016/0022-510X(90)90100-2 [DOI] [PubMed] [Google Scholar]
- 9.Rnager J, Christensen H, Fuglsang-Frederiksen A. Power spectrum analysis of the EMG pattern in normal and diseased muscles. J Neurol Sci. 1989;94:283–294. doi: 10.1016/0022-510X(89)90237-2 [DOI] [PubMed] [Google Scholar]
- 10.Güler NF, Koçer S . Classification of EMG signals using PCA and FFT. J Med Syst. 2005;29:241–250. doi: 10.1007/s10916-005-5184-7 [DOI] [PubMed] [Google Scholar]
- 11.Nandedkar SD, Sanders DB. Principal component analysis of the features of concentric needle EMG motor unit action potentials. Muscle Nerve. 1989;12:288–293. doi: 10.1002/mus.880120406 [DOI] [PubMed] [Google Scholar]
- 12.Nodera H, Osaki Y, Yamazaki H, Mori A, Izumi Y, Kaji R. Classification of needle-EMG resting potentials by machine learning. Muscle Nerve. 2019;59:224–228. doi: 10.1002/mus.26363 [DOI] [PubMed] [Google Scholar]
- 13.Yousefi J, Hamilton-Wright A. Characterizing EMG data using machine-learning tools. Comput Biol Med. 2014;51:1–13. doi: 10.1016/j.compbiomed.2014.04.018 [DOI] [PubMed] [Google Scholar]
- 14.Karlik B. Machine learning algorithms for characterization of EMG signals. Int J Inf Electron Eng. 2014;4:189–194. doi: 10.7763/ijiee.2014.v4.433 [DOI] [Google Scholar]
- 15.Sanchez B, Rutkove SB. Electrical impedance myography and its applications in neuromuscular disorders. Neurotherapeutics. 2017;14:107–118. doi: 10.1007/s13311-016-0491-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutkove SB, Sanchez B. Electrical impedance methods in neuromuscular assessment: an overview. Cold Spring Harb Perspect Med. 2019; 9. doi: 10.1101/cshperspect.a034405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shefner JM, Rutkove SB, Caress JB, et al. Assessing ALS progression with a dedicated electrical impedance myography system. Amyotroph Lateral Scler. 2018;19:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandeya SR, Nagy JA, Riveros D, et al. Using machine learning algorithms to enhance the diagnostic performance of electrical impedance myography. Muscle Nerve. 2022;66:354–361. doi: 10.1002/mus.27664 [DOI] [PubMed] [Google Scholar]
- 19.Kwon H, Rutkove SB, Sanchez B. Recording characteristics of electrical impedance myography needle electrodes. Physiol Meas. 2017;38:1748–1765. doi: 10.1088/1361-6579/aa80ac [DOI] [PubMed] [Google Scholar]
- 20.Rutkove SB, Le M, Ruehr SA, Nagy JA, Semple C, Sanchez B. Design and pilot testing of a 26-gauge impedance-electromyography needle in wild-type and ALS mice. Muscle Nerve. 2022;65:702–708. doi: 10.1002/mus.27551 [DOI] [PubMed] [Google Scholar]
- 21.Coley WD, Bogdanik L, Vila MC, et al. Effect of genetic background on the dystrophic phenotype in mdx mice. Hum Mol Genet. 2016;25:130–145. doi: 10.1093/hmg/ddv460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Putten M, Putker K, Overzier M, et al. Natural disease history of the D2-mdx mouse model for Duchenne muscular dystrophy. FASEB J. 2019;33:8110–8124. doi: 10.1096/fj.201802488R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna Austria. [Google Scholar]
- 24.Simeoni RJ, Mills PM. Biocoherence analysis of quadriceps electromyogram during isometric knee extension. Australas Phys Eng Sci Med. 2003;26:12–17. doi: 10.1007/BF03178691 [DOI] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 26.Pandeya SR, Nagy JA, Riveros D, et al. Predicting myofiber cross-sectional area and triglyceride content with electrical impedance myography: a study in db/db mice. Muscle Nerve. 2021;63:127–140. doi: 10.1002/mus.27095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross-validation pitfalls when selecting and assessing regression and classification models. J Chem. 2014;6. doi: 10.1186/1758-2946-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Xu W. Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process Lett. 2014;21:1389–1393. doi: 10.1109/LSP.2014.2337313 [DOI] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a non-parametric approach. Biometrics. 1988;44:837. doi: 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 30.Manuel M, Chardon M, Tysseling V, Heckman CJ. Scaling of motor output, from mouse to humans. Phys Ther. 2019;34:5–13. doi: 10.1152/physiol.00021.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chrzanowski SM, Nagy JA, Pandeya S, Rutkove SB. Electrical impedance Myography correlates with functional measures of disease progression in D2-mdx mice and boys with Duchenne muscular dystrophy. J Neuromuscul Dis. 2023;10:81–90. do 10.3233/JND-210787 [DOI] [PubMed] [Google Scholar]
- 32.Pandeya SR, Nagy JA, Riveros D, et al. Estimating myofiber cross-sectional area and connective tissue deposition with electrical impedance myography: a study in D2-mdx mice. Muscle Nerve. 2021;63:941–950. doi: 10.1002/mus.27240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shefner JM, Rutkove SB, Caress JB, et al. Reducing sample size requirements for future ALS clinical trials with a dedicated electrical impedance myography system. Amyotroph Lateral Scler. 2018;19:555–561. doi: 10.1080/21678421.2018.1510008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkove SB, Kapur K, Zaidman CM, et al. Electrical impedance myography for assessment of Duchenne muscular dystrophy. Ann Neurol. 2017;81:622–632. doi: 10.1002/ana.24874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Rutkove Laboratory Webiste at https://www.rutkovelab.org/data-repository/.