Abstract
Gut microbiome dysbiosis is associated with gestational diabetes mellitus (GDM), and its modulation represents a promising approach for enhancing glycemic control. In this study, we aimed to discover specific alterations in the gut microbiome through lifestyle management. We performed metagenome sequencing on fecal samples and measured short-chain fatty acid (SCFA) in plasma samples from 27 well-controlled GDM pregnancies before and after glycemic control. At the same time, 38 normal glucose tolerance (NGT) samples served as controls. Additionally, we employed two-sample Mendelian Randomization (MR) to validate our findings against Genome-Wide Association Study (GWAS) database. Our dynamic analysis revealed Bifidobacterium genus increased in GDM patients after intervention. The MR analysis confirmed that the family of Bifidobacteriaceae (OR 0.929, 95% CI, 0.886–0.975; P = 0.003) was the only negatively associated family with GDM. Further analysis indicated the increased abundance of Bifidobacterium species were negatively correlated with glycemic traits (Spearman rho mean − 0.32 ± 0.34) but positively correlated with plasma SCFA levels (Spearman rho mean 0.24 ± 0.19). Functional analysis revealed that the quorum-sensing pathway had the strongest effect on the ability of Bifidobacterium to promote glucose homeostasis (Spearman rho = -0.34), suggesting its role in regulating intestinal microbiota. Finally, the multivariable MR analysis demonstrated that two pathways, COLANSYN PWY and PWY 7323, responsible for cell surface compound synthesis in gram-negative bacteria, mediated 14.83% (P = 0.017) and 16.64% (P = 0.049) of the protective effects of Bifidobacteriaceae against GDM, respectively. In summary, Bifidobacterium is an effective gut microbiota regulator for GDM-related glucose homeostasis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03680-z.
Keywords: Bifidobacterium, Gestational diabetes mellitus, Glycemic control, Mendelian randomization, Gut microbiome regulator
Introduction
Gestational diabetes mellitus (GDM) is a hyperglycemic condition with onset during pregnancy [1] and its increasing prevalence has raised public concerns [2, 3]. The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study revealed that gestational hyperglycemia is positively correlated with various negative outcomes including large-for-gestational-age infants, cesarean section, and neonatal hypoglycemia [4]. Importantly, women with GDM have an increased risk of developing type 2 diabetes and cardiovascular diseases later in life [5, 6] and their offspring also experience increased long-term risks of metabolic disorders, obesity and cardiovascular complications [7, 8]. Therefore, glycemic control in individuals with GDM is imperative for improving maternal and neonatal pregnancy outcomes.
Currently, the first-line interventions are medical nutrition therapy (MNT) and physical exercise [2, 9]. If conservative management fails, medical therapy is needed, leading to extra cost, insulin injection pain, the risk of hypoglycemia and poor compliance [10, 11]. Moreover, evidence of associations between gut microbiome dysbiosis and the prediction [12], development [13], progression [14, 15], and prognosis [16] of GDM has accumulated. Therefore, auxiliary approaches such as gut microbiome modulation may enhance the effects of nutrition and physical exercise and improve glycemic control. For instance, a previous randomized controlled trial has demonstrated the dietary supplementation of the potential health-related bacteria Lactobacillus rhamnosus HN001 (6 × 109 colony-forming units, n = 184) for 14–16 weeks reduced GDM prevalence in 26–28 weeks’ gestation (relative rates 0.59, 95% CI 0.32–1.08, P = 0.08), compared to a placebo consuming maize-derived maltodextrin (n = 189) [17]. Moreover, studies have focused on dynamic changes in the gut microbiome during pregnancy and its associations with GDM-related glucose metabolism [18]. For example, a recent study revealed that Bacteroides plebeius contributed to SCFA elevation from first to second pregnant trimester through the ANAGLYCOLYSIS-PWY pathway and increased GDM predictive performance [18]. Inspired by the above studies, dynamic research on the gut microbiome before and after successful glycemic control of GDM could help elucidate the interplay between the gut microbiome and host glucose homeostasis and provide a potential targeted intervention reference.
Besides, the causal relationship between the gut microbiota and GDM remains unclear. Tow-sample Mendelian randomization (MR) uses genetic variations as instrumental variables (IVs) to evaluate causal associations between exposures and outcomes [19]. Therefore, MR could facilitate the investigation of the causal relationship between the gut microbiota and GDM, providing genetic evidence for gut microbiome therapy.
In this study, we enrolled NGT and GDM pregnancies at the second (T2, before GDM diagnosis) and third (T3, after glycemic control) trimesters and collected both fecal and plasma samples. The glycemic traits of our GDM cohort were controlled effectively, belonging to the GDM A1 group (A1: well-controlled GDM by lifestyle management; A2: GDM requiring medication) [2]. Through integrative analysis of gut metagenome sequencing and metabolic data, we explored the dynamic changes in the gut microbiota in GDM patients and the underlying mechanisms contributing to successful glycemic control. Further, we performed MR and mediation analyses using summary statistics from genome-wide association studies (GWASs) of the gut microbiota, gut bacterial pathways and GDM to validate the discovered associations.
Materials and methods
Study design and sample collection
We conducted a nested case‒control study at Peking University First Hospital between October 2017 and July 2019. Pregnant women were enrolled in the first trimester and followed up throughout the entire pregnancy. Eligible participants had singleton fetuses and were able to provide informed consent. The major exclusion criteria included a history of type 1 or 2 diabetes, gastrointestinal diseases, preeclampsia, hypertension disorders, smoking or alcohol consumption habits, and long-term medicine or prebiotic use. Together, 38 NGT and 30 GDM pregnancies were enrolled in the second trimester. Our goal was to focus on GDM A1 patients whose glucose levels were well controlled without the use of exogenous medication; therefore, we excluded 3 GDM patients who were receiving medication. Ultimately, our study design included 38 NGT and 27 GDM A1 patients. Consequently, we collected 130 fecal and plasma samples from these 65 patients during the T2 and T3 stages.
The samples were collected as previously described according to standard operating procedures [20]. Briefly, the middle of the fecal core (2–3 g) was self-collected at home and placed into a labeled sterile fecal sampling tube (Sarstedt, Germany, 80.734.311) containing RNA storage reagent (Tiangen, China, DP409-02). The fecal samples were transported to the hospital within 3 h. Maternal fasting blood was drawn in prechilled EDTA tubes by well-trained staff and centrifuged (2000 rpm, 20 min) to prepare the plasma. The feces and plasma were aliquoted and stored at -80 °C until laboratory analysis. This project was approved by the Ethics Committee of Peking University First Hospital (V2.0/201504.20), and informed consent was obtained from all participants.
Diagnosis and subtypes of gestational diabetes mellitus
According to the International Association of Diabetes and Pregnancy Study Groups (IADPSG) criteria, the 75 g oral glucose tolerance test (OGTT) was performed between 24 and 28 weeks of gestation. GDM was diagnosed if any single threshold value was met or exceeded (fasting-5.1 mmol/L, 1 h-10.0 mmol/L, 2 h-8.5 mmol/L). All the pregnancies diagnosed in our hospital were directed toward lifestyle treatment and self-glucose monitoring. With intervention, GDM pregnancies could be further categorized into two subtypes: A1, which is well-controlled through lifestyle management, and A2, which requires medication.
Anthropometrics and biochemical evaluation
Clinical information such as demographic factors (maternal age; prepregnancy weight and height) and biochemical examination results (white blood cell count; neutrophil percentage, OGTT) were collected from medical records. The fasting glucose levels, hemoglobinA1c (HbA1c), insulin levels and total triglyceride (TG) levels in the second (T2) or third trimester (T3) were obtained from medical records or remeasured in our hospital. Maternal prepregnancy BMI (pre-BMI) was calculated as weight divided by height squared (kg/m2). The area under the curve (AUC) for glucose was calculated following the trapezoidal rule.
Measurement of SCFAs by LC‒MS/MS
SCFA contents were measured in the plasma. Briefly, 100 µl of plasma was thawed at 4 °C and mixed with 400 µL of cold methanol/acetonitrile (1:1, v/v) to remove the protein. The mixture was centrifuged for 20 min (14000 × g, 4 °C). The supernatants were dried in a vacuum centrifuge, resuspended in 100 µL of acetonitrile/water (1:1, v/v) and adequately vortexed. After centrifugation for another 15 min (14000 × g, 4 °C), the supernatants were collected for LC‒MS/MS analysis. Analyses were performed using a UHPLC (1290 Infinity LC, Agilent Technologies, Palo Alto, CA, USA) coupled to a QTRAP (AB Sciex 5500) using an ACQUITY UPLC BEH Amide column (2.1*100 mm, 1.7 μm, Waters MS Technologies, Manchester, UK). MS/MS analysis (MRM) was performed in ESI negative mode. Data acquisition and processing were accomplished using Multiquant software (AB SCIEX, Boston, MA, USA).
DNA extraction and metagenomic sequencing
Total bacterial DNA was extracted from the fecal samples using a QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s standards. The genomic DNA was randomly sonicated into fragments of appropriately 350 bp for Illumina sequencing on a NovaSeq 6000 platform with a 150 bp paired-end sequencing strategy. The libraries were analyzed for size distribution using an Agilent 2100 Bioanalyzer and quantified with real-time PCR.
Metagenomic sequencing data analysis
The quality control process of whole-genome shotgun sequencing data was performed by KneadData (https://bitbucket.org/biobakery/kneaddata). After quality control, the taxonomic and functional profiles were determined by MetaPhlAn 3 and HUMAnN 3 [21], respectively. The functions were then annotated to pathways according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
We identified the microbiome biomarkers by LEfSe (version 1.1.01) with Linear discriminant analysis (LDA) scores > 2.0 and linear mixed models. For linear mixed models, the influences of pre-BMI and age were adjusted with MaAsLin2 (version 1.15.1) R package [22].
To identify the hub species associated with GDM-related gut microbiome dysbiosis, weighted gene coexpression network analysis (WGCNA) was performed using the R package WGCNA (version 1.72-5) [23]. The co-occurrence network correlated with Bifidobacterium was visualized by Cytoscape 3.9.1 (http://cytoscape.org/).
SNP calling and filtering
Two tools, GATK (version 4.4.0.0) [24] and VarScan2 (version 2.3.9) [25], were applied to identify SNPs in the metagenome sequencing data. After data quality control process by KneadData and based on the reference genome of Faecalibacterium prausnitzii. (reference strain KLE1255, GenBank accession no. GCA_000166035.1), we conducted alignment using BWA-MEM [26] and filtered duplicates with Picard (http://broadinstitute.github.io/picard/).
For GATK, we used the HaplotypeCaller module (parameters: --do-not-run-physical-phasing --max-alternate-alleles 2 --sample-ploidy 1) and generated the GVCF files. For Varscan2, SAMtools (version 1.17) [27] was used to generate “mpileup” files from the SAM-formatted alignment files. Then, mpileup files were employed as input files [27] for VarScan2 to further call SNPs (parameters: pileup2snp min-coverage 10, p value 0.05, min-avg-qual15). The major SNPs (≥ 0.5 mutated alleles) detected by both tools were selected.
Phylogenetic tree construction
First, genome regions with > 20% samples not having valid coverage (≥ 10× depth) were discarded. The nucleotides at SNP sites from the samples were subsequently extracted. Phylogenetic trees were constructed based on whole-genome level aligned SNPs using randomized axelerated maximum likelihood (RAxML) v8.2.9 (100 bootstrap replicates), with a GTR model of nucleotide substitution, γ-distributed rates among sites, and Felsenstein correction for ascertainment bias [27, 28]. Trees were drawn with the ggtree (version 3.6.2) R package [28].
GWAS data source
The summary level of GDM GWAS data was obtained from the 10th release of the FinnGen project (European ancestry, released on December 18, 2023, https://r10.finngen.fi/). The 10th FinnGen GDM study summarized 21,306,157 SNPs based on 14,718 GDM individuals and 215,592 GDM individuals [29, 30]. The detailed project information was described in a previous study [29]. Summary data on the gut microbiota and bacterial pathways were obtained from a previous study comprising 7,738 individuals from the Dutch Microbiome Project cohort [31].
Instrumental variables
For the exposure data, single nucleotide polymorphisms (SNPs) that met the locus-wide significance level (P < 5 × 10− 5) were selected as instrumental variables (IVs). Next, we kept only independent significantly associated IVs without linkage disequilibrium (r2 < 0.001 and a clump distance > 10,000 kb window).
MR analysis
For two-sample univariable Mendelian randomization (UVMR) analysis to explore the causal effect between the gut microbiota or bacterial pathway and GDM, we adopted inverse-variance weighted (IVW) and MR‒Egger analysis modes and utilized the conventional IVW method to determine the genetic casual effect (P < 0.05). The results are reported as odds ratios (ORs) with 95% confidence intervals (CIs). For the heterogeneity test, the Cochran Q statistic was employed for the IVW and MR‒Egger modes (P < 0.05 indicated the presence of heterogeneity). For the pleiotropy test, the MR‒Egger regression method reported an intercept to represent an average pleiotropic effect (a nonzero intercept and P < 0.05 indicated potential pleiotropy). For the sensitivity test, leave-one-out analysis was performed by removing one IV sequentially from the instrumental variables.
For bacterial pathways that are causally associated with GDM, we conducted UVMR between the significant gut microbiota and the above pathways and determined the final significant pathway mediators for the microbiota‒GDM relationship. Finally, we performed Multivariable Mendelian Randomization (MVMR) to explore the causal effect of the gut microbiota after adjusting for each pathway and the mediating effect of each pathway.
Data organization, analysis and visualization were conducted using R Software (version 4.2.2). The MR analysis was performed with the TwoSampleMR (version 0.6.3) and MVMR (version 0.4) R packages. Plots were generated using the ggplot2 (version 3.5.1) and forestploter (version 1.1.2) R packages.
Statistical analysis
We used species-level Bray‒Curtis distances to calculate β diversity between samples and visualized with principal coordinate analysis (PCoA). Then, the microbial composition difference between sample groups was calculated by Analysis of Similarities (ANOSIM) algorithm with a permutation of 999 times via vegan (version 2.6–6.1) R package.
The results are expressed as means ± standard deviations (SDs) for continuous variables. Differences in normally distributed clinical and baseline data were calculated with Student’s t test. Non-parametric tests, including the Mann–Whitney U test for comparisons between two groups and the Kruskal-Wallis test followed by Dunn’s post-hoc test for pairwise comparisons within multiple groups, were performed to assess the differences in the relative abundances of the main bacteria (phyla, genera, and species). Spearman’s correlation analysis was carried out between microbiome constitution and clinical parameters. R v4.2.2 (R Foundation for Statistical Computing) was used for statistical analysis. A P value < 0.05 was considered statistically significant.
Results
Basic characteristics of the study participants
At the 75 g OGTT test time, we prospectively enrolled 30 GDM patients and 38 NGT and the clinical measurements and fecal and plasma samples were collected and appropriately stored. During follow-up, all GDM patients underwent MNT and physical exercise management in our “one-day” GDM clinics. Among the 30 recruited GDM patients, 3 were excluded because their glycemic control was ineffective because they were receiving medication. During the third trimester, the fecal and plasma samples of 27 GDM A1 and 38 NGT patients were collected again with standard protocols, as well as clinical measurements (Fig. 1A). Our fecal and plasma samples were then used for metagenome sequencing and plasma SCFA detection. SCFAs are the metabolites of dietary fiber that promote host glucose homeostasis [32] and are utilized here as metabolic indicators for glycemic control.
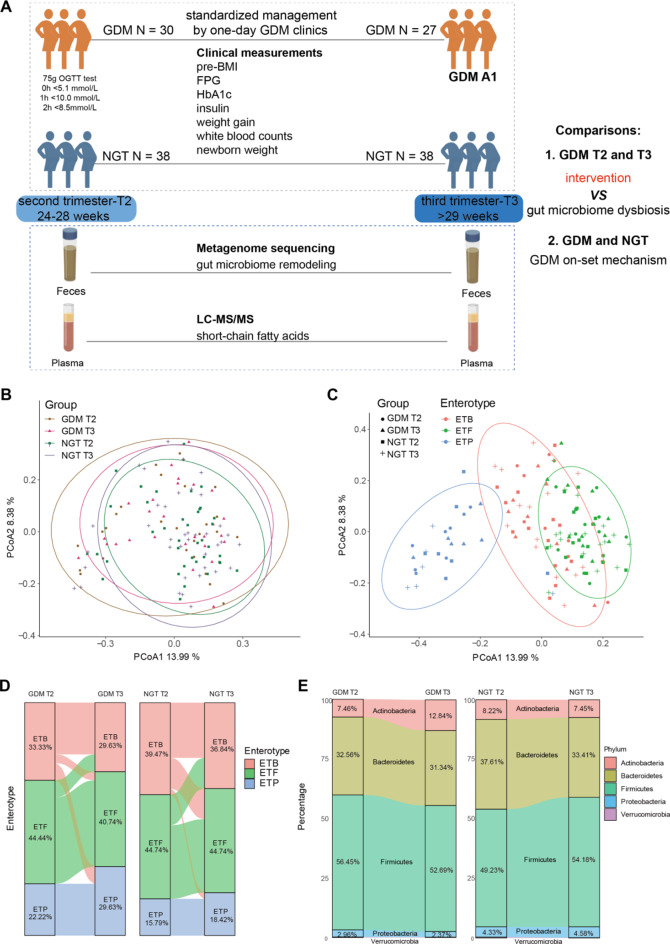
Fig. 1.
Study design and general gut microbiome communities. (A) Flow chart of the prospective study design. We divided patients according to the 75 g OGTT test. The GDM pregnancies achieved effective glycemic control in our one-day GDM clinics under standardized MNT and exercise management, belonging to the GDM A1 category. Together, 65 patients with 130 samples were included in this study, and glycemic traits as well as other clinical covariates were measured. Fecal and plasma samples from each trimester were collected for metagenome sequencing and plasma SCFA detection, respectively. (B) Principal coordinate visualization of the four groups of samples using Bray–Curtis distance at the species level. (C) Principal coordinate visualization of all samples annotated by both enterotype and group. (D) Sankey plot showing individual dynamic enterotype changes from T2 to T3 in GDM (left) and NGT (right) patients. (E) Sankey plot showing dynamic composition changes in the 4 main phyla from T2 to T3 in GDM (left) and NGT (right) patients
In general, the GDM group presented greater maternal ages and earlier delivery gestational weeks than the NGT group (Table 1). For the 75 g OGTT test, the GDM patients had significantly higher 0 h, 1 h, and 2 h blood glucose levels than the NGT patients (all P < 0.0001). In addition, elevated insulin levels, white blood cell counts and neutrophil percentages in the blood were observed in the GDM group relative to the NGT group, indicating that GDM patients have systematic inflammatory disorders. Besides, the information of SCFAs of our 65 samples are listed in Table S1. Specifically, the butyric acid of GDM (0.0046 ± 0.0012) was significantly lower than that of NGT (0.0052 ± 0.0012) at T3 (P = 0.011), consistent with our previous study [33].
Table 1.
Clinical information of the study participants
| Variables | GDM (N = 27) | Control (N = 38) | P value |
|---|---|---|---|
| Age (year) | 33.37 ± 3.42 | 31.32 ± 3.05 | 0.015 |
| BMI (kg/m2) | |||
| Prepregnancy (pre-BMI) | 22.32 ± 2.47 | 20.98 ± 2.36 | < 0.001 |
| OGTT | 25.14 ± 2.55 | 23.56 ± 2.87 | 0.022 |
| Delivery | 26.79 ± 2.55 | 26.82 ± 3.53 | 0.970 |
| OGTT test | |||
| 0 h (mmol/L) | 5.17 ± 0.52 | 4.61 ± 0.28 | < 0.001 |
| 1 h (mmol/L) | 10.30 ± 1.23 | 7.60 ± 1.27 | < 0.001 |
| 2 h (mmol/L) | 8.71 ± 1.2 | 6.09 ± 1.05 | < 0.001 |
| AUC | 17.2 ± 1.61 | 12.95 ± 1.63 | < 0.001 |
| Weight gain (kg) | |||
| before OGTT | 7.64 ± 4.38 | 6.14 ± 3.2 | 0.130 |
| after OGTT | 4.19 ± 3.54 | 7.11 ± 3.48 | 0.002 |
| Insulin (uIU/ml) | |||
| T2 | 13.4 ± 11.76 | 23.98 ± 19.86 | 0.015 |
| T3 | 32.59 ± 49.95 | 15.00 ± 11.59 | 0.110 |
| TG (mmol/L) | |||
| T2 | 2.26 ± 0.58 | 2.11 ± 0.67 | 0.350 |
| T3 | 2.78 ± 1.02 | 2.74 ± 0.78 | 0.870 |
| WBC (109/L) | |||
| T2 | 10.61 ± 2.24 | 9.04 ± 1.71 | 0.004 |
| T3 | 10.03 ± 1.92 | 8.19 ± 1.49 | < 0.001 |
| NE (109/L) | |||
| T2 | 8.00 ± 1.88 | 6.57 ± 1.36 | 0.002 |
| T3 | 7.62 ± 1.71 | 5.88 ± 1.26 | < 0.001 |
| HbA1c (%) | |||
| T2 | 5.30 ± 0.32 | ||
| T3 | 5.41 ± 0.28 | ||
| Newborn weight (g) | 3304.81 ± 390.01 | 3306.97 ± 306.68 | 0.980 |
Notably, our GDM A1 group achieved satisfactory glycemic control. First, although the BMIs before pregnancy and during the OGTT of GDM patients were greater than those of the NGT, the BMIs at delivery were matched between the two groups (Table 1). Second, the GDM group also presented significantly lower gestational weight gains than the NGT group after GDM diagnosis. Third, the newborn weights of the GDM group were almost the same as those of the NGT group. Therefore, our study represents the significant on-set hyperglycemic but well-controlled GDM population, which are the pursued goals in the management of GDM.
Overall gut microbiome structure
In total, 718 well-annotated bacterial species were detected by Metaphlan3 in all 130 samples. Previous studies have reported inconsistent results concerning the differences in overall microbial structure between GDM patients and healthy controls [13, 16, 34]. In our study, based on principal coordinate analysis of Bray‒Curtis distances among 718 species, we detected no obvious differences in β diversity between GDM and NGT patients at either T2 or T3 (Fig. 1B, ANOSIM test). However, we reannotated our samples according to enterotype at the genus level and clearly identified three enterotype clusters [35] (Fig. 1C). Further analysis revealed that the major enterotype was ETF (Firmicutes), followed by ETB (Bacteroides) and ETP (Prevotella) in both the GDM and NGT groups (Fig. 1D). The ETP enterotype has been reported to be associated with type 2 diabetes. Notably, there were more ETP individuals in the GDM group (T2 = 22.2%, T3 = 26.3%) than in the NGT group (T2 = 15.8%, T3 = 18.4%), indicating that the overall microbiome profiles reflected the disease state.
We next focused on the main gut microbial compositions and differentially abundant microbiota from T2 to T3 in each group. The five primary phyla were Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia (Fig. 1E). Among these, Bacteroidetes were reduced from T2 to T3 in both the GDM and NGT groups, while Firmicutes exhibited opposite trends between the GDM (decreased from T2 to T3) and NGT (increased from T2 to T3) groups. Notably, Actinobacteria significantly increased after glycemic control in the GDM group (Figure S1, 32.56% vs. 12.84%, P < 0.01), which is worthy of further detailed investigation.
Dynamic intestinal microbiota shifts revealed gut microbiome dysbiosis in GDM
To determine the influence of glycemic control on the gut microbiome we further conducted a dynamic analysis of the GDM-related gut microbiome from T2–T3. We defined β diversity distance as the Euclidean distance between T2 and T3 paired samples to represent direct gut microbiome shifts (Fig. 2A). The correlations of β diversity distances with dynamic alterations in clinical measurements were subsequently analyzed.
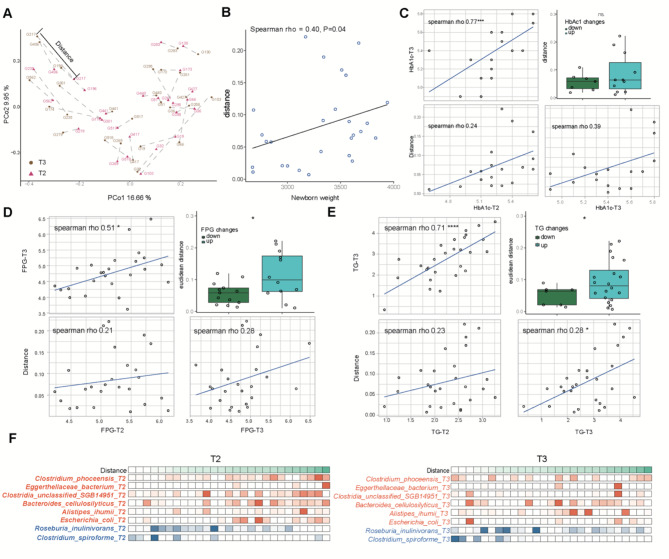
Fig. 2.
β diversity distances were positively related to the progression of GDM-related glucose disorders. (A) Principal coordinate visualization of 27 GDM paired samples using the Bray–Curtis distance at the species level, and β diversity distances are marked by lines between two paired samples. (B) The association between β diversity distance and newborn weight. (C) The association between β diversity distances and HbA1c. The four panels clockwise from the upper left show the associations between T3 and T2 HbA1c, β-diversity distances and T2 HbA1c, β-diversity distances and T3 HbA1c and the comparison of β-diversity distances between the HbA1c down and up groups. (D) The associations between β diversity distances and FPG. The four panels clockwise from the upper left show the associations between T3 and T2 FPG, β-diversity distances and T2 FPG, β-diversity distances and T3 FPG and the comparison of β-diversity distances between the lower and upper FPG groups. (E) The associations between β diversity distances and TG. The four panels clockwise from the upper left show associations with T3 and T2 TG, β diversity distances and T2 TG, β diversity distances and T3 TG, and comparisons of β diversity distances between the TG lower and upper groups. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (F) Abundance distributions of specific species ordered by the β-diversity Euclidean distance. An increase in orange species and a decrease in blue species may increase the β-diversity Euclidean distance
We found that the β diversity distances were positively associated with newborn weight (Fig. 2B, Spearman rho = 0.4, P = 0.04). For glycemic traits, the HbA1c (Fig. 2C) and FPG (Fig. 2D) levels at T2 and T3 were both positively correlated with β diversity distances. When the GDM subgroup was further divided into downregulated and upregulated subgroups according to the changes in HbA1c and FPG, greater β diversity distances were observed in the elevated HbA1c and FPG subgroups. The relationships between β diversity distances and the lipid metabolic indicator TG were the same as those between HbA1c and FPG (Fig. 2E). Therefore, the smaller β diversity distances here represented more effective glycemic control. Moreover, corresponding correlations were not observed in the NGT group (Figure S2), indicating the close relationship between the gut microbiome and GDM pathogenesis.
Furthermore, as the gut microbiome dysbiosis that occurs at T2 affects the effects of the intervention, we discovered associations between T2 microbial species and β diversity distances. By WGCNA, we detected significantly negatively correlated “gray” and positively correlated “blue” modules (Figure S3A). In the gray module, the hub microbial species were Clostridium spiroforme and Roseburia inulinivorans, representing potentially beneficial species (Figure S3B). In the blue module, the hub microbial species included Clostridium phoceensis, Clostridia unclassified SGB14951, Bacteroides cellulosilyticus, Alistipes ihumii, Escherichia coli and Eggerthellaceae bacteria, representing potentially harmful species (Figure S3B). When tracking the distributions of the abundances of the above hub species, we discovered that the proportions of less beneficial (blue) and more harmful (orange) species at either T2 or T3 contributed to increases in β diversity distances (Fig. 2F), reflecting the importance of microbial community balance.
Overall, we found that dynamic shifts in the intestinal microbiota from T2 to T3 were positively associated with GDM metabolism progression. Moreover, the above findings suggest that the causal role of glycemic control in the gut microbiome may be attributed to changes in specific genera, species or even strains.
The abundance of Bifidobacterium species specifically increased after intervention
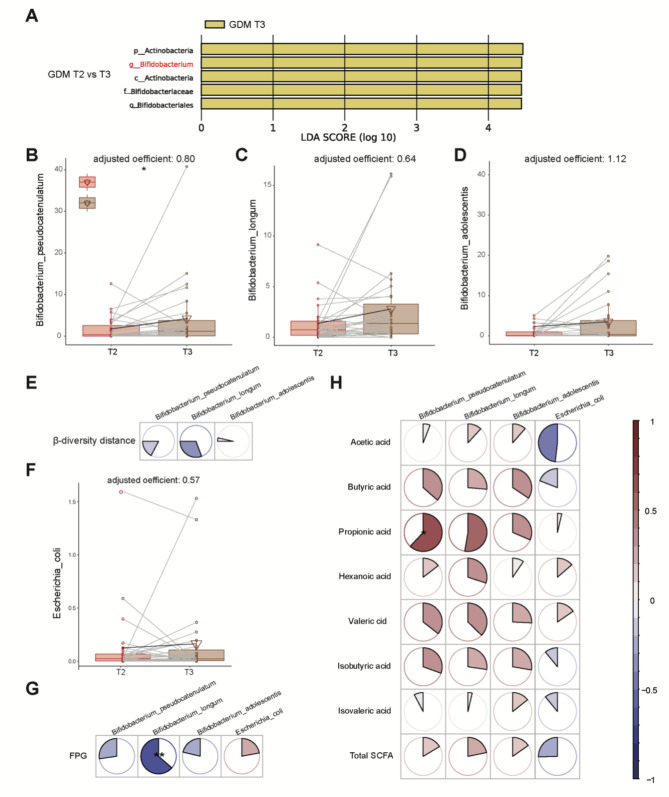
We next focused on specific altered microbial species associated with our standardized glycemic control in GDM patients. As shown in Fig. 1E, the Actinobacteria phylum was significantly increased after GDM intervention, and further LEfSe analysis revealed that Bifidobacterium was the main enriched genus (Fig. 3A). As controlling for confounding variables could help identify the associated microbial species related to the disease itself, we utilized a MaAsLin2 linear mixed model with age and pre-BMI as random effects and further identified elevated Bifidobacterium pseudocatenulatum, Bifidobacterium adolescentis and Bifidobacterium longum in GDM-T3 pregnancies (Fig. 3B‒D, coefficient > 0.5). LEfSe further revealed that Bifidobacterium adolescentis was also enriched in the GDM group compared with the NGT group at T3 (Figure S4), indicating that alterations in Bifidobacterium species may be specific to our glycemic intervention.
Fig. 3.
Bifidobacterium was the enriched beneficial genus from GDM T2 to T3. (A) LEfSe analysis revealed that the microbes differed significantly between the GDM-T2 and GDM-T3 groups. The findings with respect to phylum, family, and genus are shown in the plot (LDA score > 2, p < 0.05). (B-D) Massalin-2 was used to evaluate three main Bifidobacterium species including Bifidobacterium pseudocatenulatum (B), Bifidobacterium longum (C) and Bifidobacterium adolescentis (D). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (E) Correlations of dynamic changes in Bifidobacterium species with β diversity distances. (F) Changes in the abundance of Escherichia coli from T2–T3. (G) The dynamic correlations of Bifidobacterium species and E. coli with glycemic traits. (H) The dynamic correlations of Bifidobacterium species and E. coli with plasma SCFA levels. Spearman correlation analysis, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001
As expected, dynamic changes in Bifidobacterium species were negatively correlated with β diversity distance (Fig. 3E). To further distinguish the pathogenic or probiotic functions of our three Bifidobacterium species, we investigated the correlations of the abundances of B. pseudocatenulatum, B. longum and B. adolescentis with plasma SCFAs and crucial glycemic traits. Moreover, the harmful LPS producer Escherichia coli was used as a reference (Fig. 3F). Unlike Escherichia coli, the dynamic changes of Bifidobacterium species were positively correlated with SCFAs (Spearman rho mean 0.24 ± 0.19), but negatively correlated with FPG (Spearman rho mean − 0.32 ± 0.34) (Fig. 3G-H). The above results confirmed that the increases in B. pseudocatenulatum, B. longum and B. adolescentis were health-promoting changes caused by our standardized management.
Advantageous functions of Bifidobacterium species
To better understand how Bifidobacterium species affect glucose homeostasis, we utilized the KEGG database to explore the differentially abundant pathways. By fitting a linear model, we identified 98 significantly enriched KEGG orthologs (KOs) in T3, referring to 74 pathways (Fig. 4A, coefficient > 0 and P < 0.05, still robust after adjustment for age and pre-BMI). Among these, most were related to amino acid biosynthesis or metabolism (Fig. 4A, in purple) or carbon or glucose utilization (Fig. 4A, in red), indicating the important influence of the intestinal microbiota on energy metabolism.
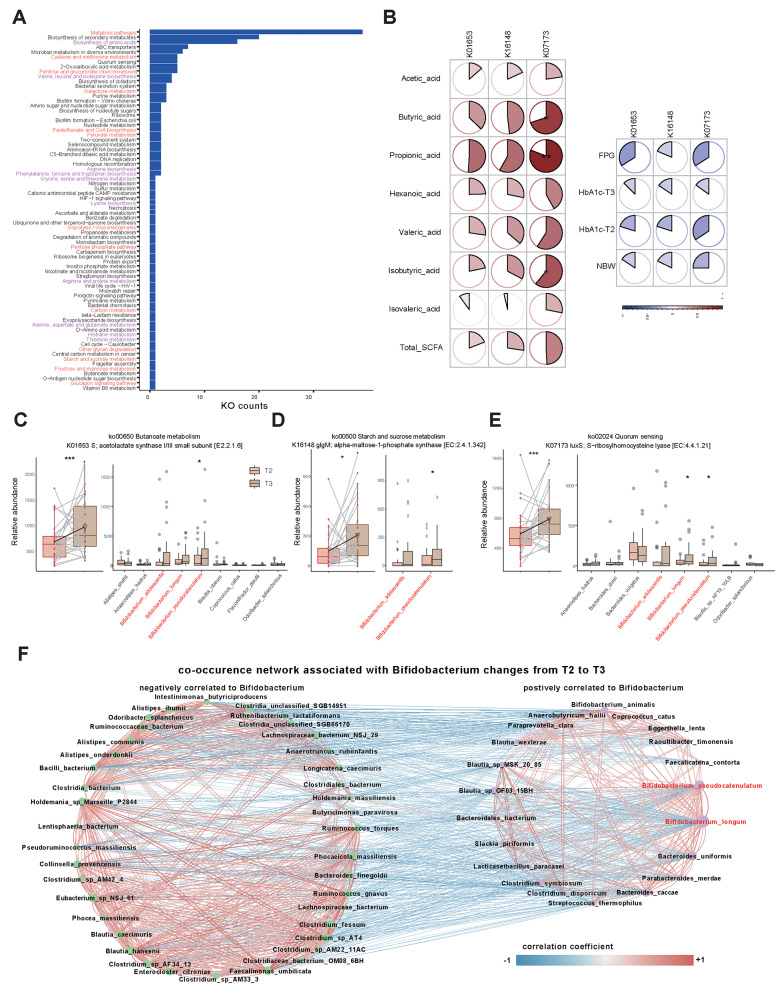
Fig. 4.
Advantageous functions and co-occurrence network associated with Bifidobacterium species. (A) The categories of KEGG-mapped pathways associated with significantly increased KOs from T2 to T3. (B) Dynamic correlations of three enzymes with SCFAs and glycemic traits. (C) Boxplot of the relative abundance of the K16148 enzyme (belonging to starch and sucrose metabolism) in GDMs T2 and T3 and the relative abundances of the main contributing species. (D) Boxplot of K01653 enzyme abundance (belonging to butanoate metabolism) in GDMs T2 and T3 and the relative abundances of the main contributing species. (E) Boxplot of K07173 enzyme abundance (belonging to the quorum sensing pathway) in GDMs T2 and T3 and the relative abundances of the main contributing species. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (F) The co-occurrence network associated with dynamic changes in B. pseudocatenulatum and B. longum from T2 to T3. The left part shows the negatively associated species (marked with blue lines, Spearman correlation test, P < 0.05, rho < -0.3), and the right part shows the positively associated changed species (marked with red lines, Spearman correlation test, P < 0.05, rho > 0.3)
We then selected the pathways enriched with three Bifidobacterium species. In total, 71, 44 and 1 pathways were elevated in B. pseudocatenulatum, B. longum and B. adolescentis, respectively. Next, we correlated the alterations in Bifidobacterium-related KOs with those in SCFAs and glycemic traits. We discovered that three key enzymes involved in butanoate metabolism (K01653, ALS: acetolactate synthase I/III small subunit), starch and sucrose metabolism (K16148, glgM: alpha-maltose-1-phosphate synthase) and quorum-sensing (K07173, luxS: S-ribosylhomocysteine lyase) pathways were positively correlated with SCFA tendencies (Spearman rho mean 0.35 ± 0.22), yet negatively correlated with glycemic trait tendencies (Spearman rho mean − 0.22 ± 0.07) (Fig. 4B). Further analysis demonstrated that the increases in the three enzymes were caused mainly by Bifidobacterium species (Fig. 4C-E). These findings revealed that the increase in Bifidobacterium abundance after intervention may regulate glucose homeostasis through the production of SCFAs, the synthesis of glycogen and the balance of the microbial community. Notably, K07173 in the Quorum-sensing pathway exhibited the strongest correlation with glycemic alterations (Spearman rho = -0.34), suggesting that Bifidobacterium is a potential microbiome regulator of SCFA-producing microbiota and is worthy of further investigation.
Co-occurrence network associated with Bifidobacterium species
Since B. pseudocatenulatum and B. longum significantly contributed to the elevation of K07173 (luxS, crucial enzyme for the quorum-sensing pathway), we next explored the co-occurrence of gut microbes associated with dynamic changes in the above two species (Fig. 4F, Spearman’s rho > 0.3). We noticed a positive correlation with SCFA-producing species such as Blautia wexlerae [36], Bacteroides uniformis [37], Bacteroides caccae [38], Anaerobutyricum hallii [39] and Coprococcus catus [40]. Conversely, reported GDM pathogenic hallmarks, such as Ruminococcus torques, Ruminococcus gnavus and Ruminococcaceae bacterium [41, 42], were negatively correlated. The results suggest that Bifidobacterium species may enhance the growth of beneficial SCFA-producing bacteria, inhibit pathogens via the luxS/AI-2 system, and help alleviate hyperglycemia.
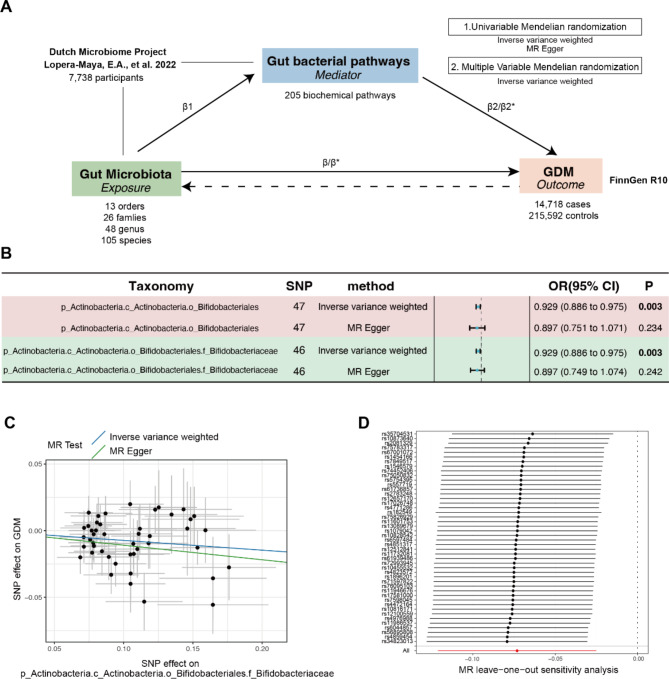
Genetic causality and correlation between Bifidobacteriaceae and GDM
We further conducted two-sample MR analysis to validate the relationship between Bifidobacterium and GDM and potential associated pathways (Fig. 5A). With respect to the causal effects of the gut microbiota on GDM, o_Bifidobacteriales and f_Bifidobacteriaceae (OR 0.929, 95% CI, 0.886–0.975; P = 0.003) were the only negatively associated orders and families, respectively (Figure S5-6 and Fig. 5B). In addition, no heterogeneity or pleiotropy was observed (Table S2). The scatter plot and results of the leave‒one-out analyses are shown in Fig. 5C and D. Furthermore, reverse MR analysis revealed no causal effect of GDM on o_Bifidobacteriales or f_Bifidobacteriaceae (Table S3). Therefore, Bifidobacteriaceae is a genetically casual protective factor for GDM.
Fig. 5.
The MR study design and UVMR revealed causal effects between Bifidobacteriaceae and GDM. (A) The design of the mediation Mendelian randomization (MR) analyses. First, two-sample bidirectional MR was performed to investigate the causal relationships between the gut microbiota (exposure) and GDM (outcome). Second, 205 pathways (mediators) were utilized for subsequent mediation analyses. Specifically, we investigated the causal effect of pathways on GDM. For significant pathways, we further studied the causal effects of the gut microbiota on pathways. Finally, UVMR was used to validate the mediation relationship. β1 is the causal effect of the gut microbiota on the bacterial pathway. β represents the total effect of the gut microbiota on GDM. β* and β2* represent the adjusted direct effects of the gut microbiota and bacterial pathway on GDM. (B) The causal effect of Bifidobacteriaceae on GDM. (C) Scatter plot of the causal effect between f_Bifidobacteriaceae and GDM. (D) Leave-one-out plot of the causal effect between f_Bifidobacteriaceae and GDM
Furthermore, we evaluated the causal effects of gut bacterial pathways on GDM and identified 11 significantly associated pathways (Table S4). Among these, f_Bifidobacteriaceae was causally associated with six bacterial pathways (Table 2). Finally, we performed MVMR to validate the mediating effects of the above six bacterial pathways. We found that the roles of COLANSYN PWY-colanic acid building block biosynthesis, the PWY01415-superpathway of heme biosynthesis from uroporphyrinogen III and the PWY 7323-superpathway of GDP mannose-derived O antigen building blocks remained significant after adjusting for f_Bifidobacteriacea (Table 3). Overall, the mediated proportions of COLANSYN PWY, PWY01415 and PWY 7323 on the f_Bifidobacteriacea-GDM relationship were 14.83% (P = 0.017), 8.501% (P = 0.029) and 16.64% (P = 0.049), respectively. The effects of LACTOSECAT PWY-lactose and galactose degradation I, the PWY 5173-superpathway of acetyl-CoA biosynthesis and PWY 7446-sulfoglycolysis were insignificant after adjusting for f_Bifidobacteriacea.
Table 2.
Mendelian randomization analyses of the causal effects between the f_Bifidobacteriaceae-bacterial pathway and the bacterial pathway-GDM
| Pathway | Method | f_Bifidobacteriaceae (Exposure) | GDM (Outcome) | ||||
|---|---|---|---|---|---|---|---|
| SNP | β ± SE | P | SNP | β ± SE | P | ||
|
COLANSYN PWY colanic acid building blocks biosynthesis |
Inverse variance weighted | 39 | -0.146 ± 0.037 | < 0.001 | 36 | 0.069 ± 0.027 | 0.016 |
| MR Egger | 0.035 ± 0.165 | 0.832 | -0.134 ± 0.114 | 0.246 | |||
|
LACTOSECAT PWY lactose and galactose degradation I |
Inverse variance weighted | 52 | 0.606 ± 0.031 | < 0.001 | 35 | -0.059 ± 0.029 | 0.045 |
| MR Egger | 0.593 ± 0.125 | < 0.001 | -0.004 ± 0.110 | 0.973 | |||
|
PWY0141 superpathway of heme biosynthesis from uroporphyrinogen III |
Inverse variance weighted | 52 | -0.152 ± 0.050 | 0.002 | 37 | 0.043 ± 0.017 | 0.014 |
| MR Egger | -0.172 ± 0.199 | 0.391 | 0.138 ± 0.071 | 0.059 | |||
|
PWY 5173 superpathway of acetyl-CoA biosynthesis |
Inverse variance weighted | 52 | -0.106 ± 0.042 | 0.011 | 37 | 0.045 ± 0.023 | 0.046 |
| MR Egger | 0.058 ± 0.165 | 0.728 | -0.049 ± 0.086 | 0.571 | |||
|
PWY 7323 superpathway of GDP mannose derived O antigen building blocks biosynthesis |
Inverse variance weighted | 52 | -0.201 ± 0.031 | < 0.001 | 34 | 0.073 ± 0.032 | 0.021 |
| MR Egger | -0.029 ± 0.123 | 0.811 | 0.041 ± 0.116 | 0.728 | |||
|
PWY 7446 sulfoglycolysis |
Inverse variance weighted | 52 | -0.172 ± 0.065 | 0.008 | 28 | 0.034 ± 0.016 | 0.031 |
| MR Egger | -0.119 ± 0.262 | 0.651 | 0.146 ± 0.055 | 0.013 | |||
Beta (β), standard error (SE), and P values were obtained from the univariate Mendelian randomization analysis
Table 3.
Multivariate mendelian randomization analyses of the causal effects between f_Bifidobacteriaceae, bacterial pathways and GDM
| Mediator-pathway | Direct effect (β* ± SE) | Direct effect (β2* ± SE) | Indirect effect (β1 × β2* ± SE) | P | Mediated proportion (β1 × β2*/β) |
|---|---|---|---|---|---|
|
COLANSYN PWY colanic acid building blocks biosynthesis |
-0.037 ± 0.031 | 0.074 ± 0.031 | -0.011 ± 0.005 | 0.017 | 14.83 |
|
LACTOSECAT PWY lactose and galactose degradation I |
-0.043 ± 0.043 | -0.033 ± 0.046 | -0.019 ± 0.003 | 0.479 | 27.08 |
|
superpathway of heme biosynthesis from uroporphyrinogen III |
-0.057 ± 0.026 | 0.041 ± 0.019 | -0.006 ± 0.003 | 0.029 | 8.501 |
|
PWY 5173 superpathway of acetyl-CoA biosynthesis |
-0.074 ± 0.025 | 0.024 ± 0.021 | -0.003 ± 0.002 | 0.258 | 3.505 |
|
PWY 7323 superpathway of GDP mannose derived O antigen building blocks biosynthesis |
-0.056 ± 0.026 | 0.061 ± 0.031 | -0.012 ± 0.006 | 0.049 | 16.64 |
|
PWY 7446 sulfoglycolysis |
-0.071 ± 0.026 | 0.029 ± 0.015 | -0.005 ± 0.003 | 0.051 | 6.995 |
β* and β2* represent the adjusted direct effects of f_Bifidobacteriaceae and each bacterial pathway on GDM
β1 is the causal effect of f_Bifidobacteriaceae on each bacterial pathway
The indirect effect (β1 × β2*) is the effect of exposure on GDM via the corresponding mediator
β is the total effect of f_Bifidobacteriaceae on GDM
The mediated proportion is calculated as the “indirect effect/total effect”
The differential gut microbiome signatures between two groups at T2
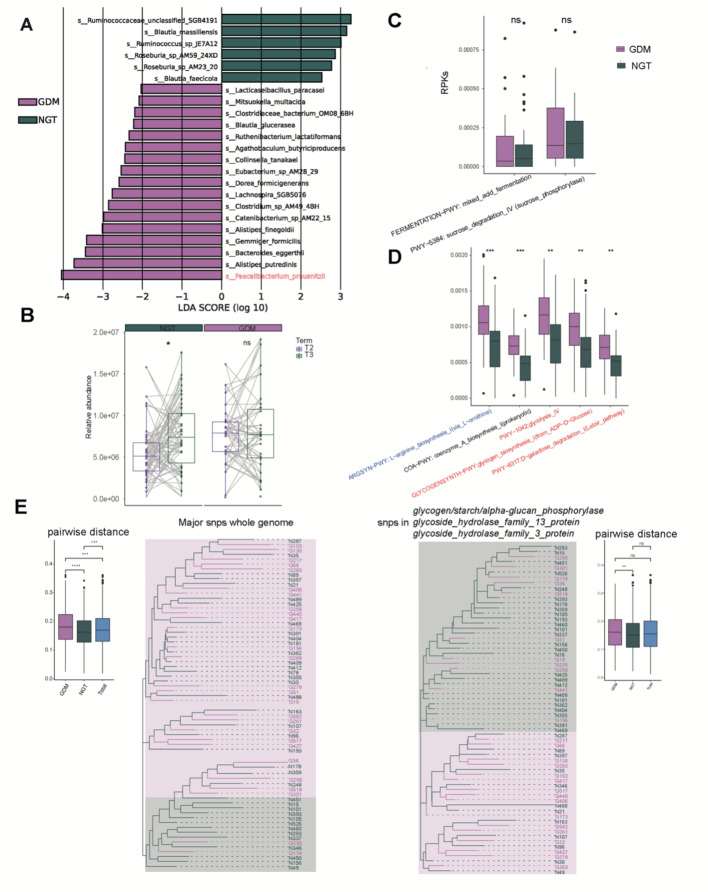
Finally, we focused on the mechanism by which the gut microbiome affects GDM onset by comparing the differences between GDM and NGT at T2. First, the Firmicutes/Bacteroidetes ratios were greater in the GDM-T2 group than in the NGT-T2 group (Figure S7), consistent with a previous study [43]. Further LEfSe analysis revealed that the abundance of Ruminococcaceae_unclassified_SGB4191 and Faecalibacterium prausnitzii was the most significantly enriched species in NGT and GDM T2 group, respectively (Fig. 6A-B).
Fig. 6.
The differential gut microbiome signatures between two groups at T2. (A) LEfSe revealed that the microbes significantly differed from those in the GDM-T2 to T3 samples. The findings with respect to species are shown in the plot (LDA score > 2, p < 0.05). (B) Abundances of F. prausnitzii in the GDM and NGT samples at both T2 and T3. (C) Abundances of SCFA-related metabolic pathways contributed by F. prausnitzii in the GDM and NGT samples at T2. (D) The significantly differential pathways affected by F. prausnitzii in the GDM and NGT samples at T2. (E) The phylogenetic trees constructed from F. prausnitzii’s major SNPs and three genes with significantly biased SNPs between groups
The increase of F. prausnitzii was also observed in the GDM cohort in our previous study and was positively correlated with inflammatory factors [20]. Interestingly, two SCFA-related pathways contributed by F. prausnitzii. were reduced in the GDM group while pathways related to glucose and amino metabolism were upregulated (Fig. 6C-D). Further, we analyzed all the differentially eggNOG enzymes in the GDM-T2 group and revealed that the carbon-nitrogen hydrolase COG0010 was significantly enriched, with F. prausnitzii being the dominant contributor, which positively correlated with the OGTT AUC and WBC (Figure S8). According to a previous study, the same species but with single-nucleotide polymorphism (SNP) level differences may have distinct biological functions [44]. We constructed a phylogenetic tree using major SNPs of F. prausnitzii in 65 T2 samples. The phylogenetic tree revealed a biased F. prausnitzii. distribution between the GDM and NGT groups (Fig. 6E). The intratree distance of the GDM (0.182) was greater than that of the NGT (0.164) and the total sample pool (0.172). We subsequently selected the genes of F. prausnitzii with the most biased SNP distributions (hypergeometric test, q < 0.05) and identified 3 genes responsible for the hydrolysis of glycosidic bonds in complex sugars. Phylogenetic trees constructed based on the 3 genes revealed that GDM and NGT also belonged to two distinct clusters (average pairwise distance of GDM vs. NGT: 0.164 vs. 0.158).
Discussion
To date, studies have focused on how the gut microbiome affects GDM development [12, 13, 18, 45]. However, research on the impact of GDM management on the intestinal microbiota is still lacking. In this study, we aimed to investigate the dynamic changes in the gut microbiome associated with intervention in the GDM A1 group. We first investigated β diversity distances to explore the associations between shifts in metabolic traits and the intestinal microbiota, which exhibited positive correlations. These findings support those of a previous study in which gut microbiome dysbiosis occurring at the time of GDM diagnosis remained throughout pregnancy [18]. Accordingly, lifestyle education and physical exercise work on delicate changes in the gut microbiota.
We discovered that three Bifidobacterium species, B. longum, B. pseudocatenulatum and B. adolescentis, specifically increased after our glucose intervention. Enzyme analysis revealed that three Bifidobacterium species were the main contributors to K01653 (ALS: acetolactate synthase I/III small subunit), K16148 (glgM: alpha-maltose-1-phosphate synthase) and K07173 (luxS: S-ribosylhomocysteine lyase), which are key enzymes in butanoate (butyric acid) metabolism, starch and sucrose metabolism and quorum-sensing pathways, respectively. Among the three pathways, quorum-sensing pathway (luxS/AI-2 system) had the strongest effects on both plasma SCFA elevation and glycemic trait reduction. A further co-occurrence network also confirmed the interactions of B. longum and B. pseudocatenulatum with SCFA producers and pathogens. Therefore, the gut microbiome regulatory capacity of Bifidobacterium is more important for glycemic control.
Furthermore, we validated the genetic causal effect of Bifidobacterium on GDM via MR methodology. The mediation analysis revealed that COLANSYN PWY-colanic acid building block biosynthesis, the PWY01415-superpathway of heme biosynthesis from uroporphyrinogen III and the PWY 7323-superpathway of GDP mannose-derived O antigen building blocks remained significant after adjustment for f_Bifidobacteriaceae. Notably, COLANSYN PWY and PWY 7323 are two pathways responsible for the synthesis of cell surface compounds in gram-negative bacteria such as Escherichia coli. The above mediating effect again emphasized the importance of Bifidobacterium as a gut microbiome regulator.
Several studies have shown an increased abundance of Bifidobacterium in normal pregnancies [18, 34, 46]. However, its function in regulating gestational glucose has not been well described. Recent studies have shown that B. adolescentis and B. longum are able to relieve T2DM by restoring the homeostasis of the gut microbiota, increasing the abundance of SCFA-producing microbiota, and alleviating inflammation [47, 48]. Overall, we speculated that the potential mechanism by which Bifidobacterium promotes glucose homeostasis is as follows: (1) increasing potential health-related bacteria and inhibiting pathogens through the quorum-sensing system; (2) producing SCFAs and further alleviating hyperglycemia through reducing inflammation; and (3) improving glycogenesis. Moreover, Bifidobacterium has been demonstrated to be the dominant microbiota in healthy infants, and its reduction is related to an increased prevalence of obesity, diabetes, and metabolic disorders later in life [49–51]. Therefore, a special bond related to Bifidobacterium may exist between the glycemic regulation of mothers and newborns, and further investigations into the mechanism of Bifidobacterium intervention in hyperglycemic pregnant mouse models and their offspring are warranted.
Our second goal was to explore the effects of the gut microbiome on GDM pathology and identify enriched Faecalibacterium prausnitzii in GDM patients at the onset stage. F. prausnitzii is the most popular butyric acid producer, and several studies have reported depletion in GDM or metabolism disorder patients [13, 43]; however, controversial phenomena exist: for example, a recent study showed that F. prausnitzii was enriched in obese pregnant women [52]. Our previous research also revealed increased Faecalibacterium in the gut of germ-free mice after transplantation with GDM donor feces [20]. With metagenome sequencing, we could better understand the deeper and complicated reasons. We detected a greater abundance of amino hydrolases and SNP differences in the glycosyl hydrolases of F. prausnitzii in the GDM group than in the control group. The above results highlight the complex nature of bacteria, and strain-level and even SNP-level research is worthwhile before their clinical application [44, 53].
Our study has several limitations. First, the sample size was limited, and further large-cohort validation is needed. Second, although our GDM patients were educated and managed on a standardized basis, we did not directly quantify the management measurements, especially for the nutrient intake indicators. Third, our patients were mainly from North China, which limits the interpretability of our findings. Fourth, our MR analysis was based on European populations, and further investigations are still needed in Chinese and East Asian population.
In conclusion, we first depicted the dynamic microbiome changes in well-controlled GDM patients before and after intervention. We found that the Bifidobacterium species were specific health-related bacteria for standardized glycemic management, highlighting the regulatory capacity of the gut microbiome. Further MR analysis validated that Bifidobacteriacea could serve as a potentially protective target for GDM. Our findings enhance the understanding of the interplay between the gut microbiome and host glucose metabolism, offering potential health-related bacteria for targeted gut microbiome interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Z.C. and S.W. designed the study. Z.C. performed the analysis and drafted the manuscript. S.W. and J.N. participated in patient recruitment and sample collection. H.Y. and J.M. conceived of the study, participated in its design and coordination and helped draft the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by National High Level Hospital Clinical Research (22cz020401-4811009) Funding, the National Natural Science Foundation of China (81830044) and the National Key Research and Development Program of China (No. 2021YFC2700700) Funding awarded to H.Y.
Data availability
The datasets generated during the current study are available in the NCBI SRA repository under the PRJNA1142935/SRP545576 project.
Declarations
Ethics approval and consent to participate
This project was approved by the Ethics Committee of Peking University First Hospital (V2.0/201504.20), and informed consent was obtained from all participants.
Consent for publication
Not applicable.
Clinical trial number
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zifeng Cui and Shuxian Wang contributed equally to this work.
Contributor Information
Jingmei Ma, Email: jingmeima@bjmu.edu.cn.
Huixia Yang, Email: yhxktz2018@163.com.
References
- 1.Juan JYH, Su R, Kapur A. Diagnosis of gestational diabetes Mellitus in China: Perspective, Progress and prospects. Maternal-Fetal Med 2019. 2019;1(1):31–7. [Google Scholar]
- 2.Wang CJJ, Yang H. A summary of Chinese guidelines on diagnosis and management of hyper-glycemia in pregnancy (2022). Maternal-Fetal Med 2023. 2023;5(1):4–8. [Google Scholar]
- 3.Zhu WW, Yang HX, Wang C, Su RN, Feng H, Kapur A. High prevalence of gestational diabetes Mellitus in Beijing: effect of maternal birth weight and other Risk factors. Chin Med J (Engl). 2017;130(9):1019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Group HSCR, Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, et al. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358(19):1991–2002. [DOI] [PubMed] [Google Scholar]
- 5.Vounzoulaki E, Khunti K, Abner SC, Tan BK, Davies MJ, Gillies CL. Progression to type 2 diabetes in women with a known history of gestational diabetes: systematic review and meta-analysis. BMJ. 2020;369:m1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, Wang Y, Xiao S, Qiu L, Yu Y, Zhang Z. Association of gestational diabetes mellitus with overall and type specific cardiovascular and cerebrovascular diseases: systematic review and meta-analysis. BMJ. 2022;378:e070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe WL Jr., Scholtens DM, Kuang A, Linder B, Lawrence JM, Lebenthal Y, et al. Hyperglycemia and adverse pregnancy outcome follow-up study (HAPO FUS): maternal gestational diabetes Mellitus and Childhood glucose metabolism. Diabetes Care. 2019;42(3):372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tam WH, Ma RCW, Ozaki R, Li AM, Chan MHM, Yuen LY, et al. In Utero exposure to maternal hyperglycemia increases Childhood Cardiometabolic risk in offspring. Diabetes Care. 2017;40(5):679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ElSayed NA, Aleppo G, Aroda VR, Bannuru RR, Brown FM, Bruemmer D, et al. Management of diabetes in pregnancy: standards of Care in Diabetes-2023. Diabetes Care. 2023;46(Suppl 1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juan J, Yang H. Prevalence, Prevention, and Lifestyle intervention of gestational diabetes Mellitus in China. Int J Environ Res Public Health 2020, 17(24). [DOI] [PMC free article] [PubMed]
- 11.D’Anna R, Corrado F, Loddo S, Gullo G, Giunta L, Di Benedetto A. Myoinositol plus alpha-lactalbumin supplementation, insulin resistance and birth outcomes in women with gestational diabetes mellitus: a randomized, controlled study. Sci Rep. 2021;11(1):8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng W, Xu Q, Huang W, Yan Q, Chen Y, Zhang L et al. Gestational diabetes Mellitus is Associated with reduced dynamics of Gut Microbiota during the first half of pregnancy. mSystems 2020, 5(2). [DOI] [PMC free article] [PubMed]
- 13.Ye D, Huang J, Wu J, Xie K, Gao X, Yan K, et al. Integrative metagenomic and metabolomic analyses reveal gut microbiota-derived multiple hits connected to development of gestational diabetes mellitus in humans. Gut Microbes. 2023;15(1):2154552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, Yin P, Chu S, Gao W, Cui S, Guo S et al., Correlation Analysis between GDM and Gut Microbial Composition in Late Pregnancy. J Diabetes Res. 2021, 2021: 8892849. [DOI] [PMC free article] [PubMed]
- 15.Ye G, Zhang L, Wang M, Chen Y, Gu S, Wang K et al., The Gut Microbiota in Women Suffering from Gestational Diabetes Mellitus with the Failure of Glycemic Control by Lifestyle Modification. J Diabetes Res. 2019, 2019: 6081248. [DOI] [PMC free article] [PubMed]
- 16.Crusell MKW, Hansen TH, Nielsen T, Allin KH, Ruhlemann MC, Damm P, et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. 2018;6(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickens KL, Barthow CA, Murphy R, Abels PR, Maude RM, Stone PR, et al. Early pregnancy probiotic supplementation with Lactobacillus rhamnosus HN001 may reduce the prevalence of gestational diabetes mellitus: a randomised controlled trial. Br J Nutr. 2017;117(6):804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z, Pan XF, Li X, Jiang L, Hu P, Wang Y, et al. The Gut Microbiome Dynamically Associates with Host Glucose Metabolism throughout pregnancy: longitudinal findings from a Matched Case-Control Study of Gestational Diabetes Mellitus. Adv Sci (Weinh). 2023;10(10):e2205289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Qin S, Feng Y, Song Y, Lv N, Liu F, et al. Perturbations of gut microbiota in gestational diabetes mellitus patients induce hyperglycemia in germ-free mice. J Dev Orig Health Dis. 2020;11(6):580–8. [DOI] [PubMed] [Google Scholar]
- 21.Beghini F, McIver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S et al., Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021, 10. [DOI] [PMC free article] [PubMed]
- 22.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. 2021;17(11):e1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinf 2013, 43(1110): 11 10 11–11 10 33. [DOI] [PMC free article] [PubMed]
- 25.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50(6):913–25. [DOI] [PubMed] [Google Scholar]
- 27.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10(2). [DOI] [PMC free article] [PubMed]
- 28.Xu S, Dai Z, Guo P, Fu X, Liu S, Zhou L, et al. ggtreeExtra: Compact visualization of richly annotated phylogenetic data. Mol Biol Evol. 2021;38(9):4039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keikkala E, Mustaniemi S, Koivunen S, Kinnunen J, Viljakainen M, Mannisto T, et al. Cohort Profile: the Finnish gestational diabetes (FinnGeDi) study. Int J Epidemiol. 2020;49(3):762–g763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurki MI, Karjalainen J, Palta P, Sipila TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopera-Maya EA, Kurilshikov A, van der Graaf A, Hu S, Andreu-Sanchez S, Chen L, et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat Genet. 2022;54(2):143–51. [DOI] [PubMed] [Google Scholar]
- 32.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Liu Y, Qin S, Yang H. Composition of maternal circulating short-chain fatty acids in gestational diabetes Mellitus and their associations with placental metabolism. Nutrients 2022, 14(18). [DOI] [PMC free article] [PubMed]
- 34.Chen T, Zhang Y, Zhang Y, Shan C, Zhang Y, Fang K, et al. Relationships between gut microbiota, plasma glucose and gestational diabetes mellitus. J Diabetes Investig. 2021;12(4):641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosomi K, Saito M, Park J, Murakami H, Shibata N, Ando M, et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat Commun. 2022;13(1):4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Almela I, Romani-Perez M, Bullich-Vilarrubias C, Benitez-Paez A, Gomez Del Pulgar EM, Frances R, et al. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes. 2021;13(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shon HJ, Kim YM, Kim KS, Choi JO, Cho SH, An S, et al. Protective role of colitis in inflammatory arthritis via propionate-producing Bacteroides in the gut. Front Immunol. 2023;14:1064900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70(10):5810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhtar M, Chen Y, Ma Z, Zhang X, Shi D, Khan JA, et al. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim Nutr. 2022;8:350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mokkala K, Roytio H, Munukka E, Pietila S, Ekblad U, Ronnemaa T, et al. Gut microbiota richness and composition and dietary intake of overweight pregnant women are related to serum zonulin concentration, a marker for intestinal permeability. J Nutr. 2016;146(9):1694–700. [DOI] [PubMed] [Google Scholar]
- 42.Cortez RV, Taddei CR, Sparvoli LG, Angelo AGS, Padilha M, Mattar R, et al. Microbiome and its relation to gestational diabetes. Endocrine. 2019;64(2):254–64. [DOI] [PubMed] [Google Scholar]
- 43.Cunningham AL, Stephens JW, Harris DA. Gut microbiota influence in type 2 diabetes mellitus (T2DM). Gut Pathog. 2021;13(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Li Z, Hu S, Zhang J, Wu J, Shao N, et al. Gut metagenomes of type 2 diabetic patients have characteristic single-nucleotide polymorphism distribution in Bacteroides coprocola. Microbiome. 2017;5(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, Bible PW, Long S, Ming WK, Ding W, Long Y, et al. Metagenomic analysis reveals gestational diabetes mellitus-related microbial regulators of glucose tolerance. Acta Diabetol. 2020;57(5):569–81. [DOI] [PubMed] [Google Scholar]
- 46.Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, He JR, et al. Connections between the human gut microbiome and gestational diabetes mellitus. Gigascience. 2017;6(8):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian X, Si Q, Lin G, Zhu M, Lu J, Zhang H et al., Bifidobacterium adolescentis Is Effective in Relieving Type 2 Diabetes and May Be Related to Its Dominant Core Genome and Gut Microbiota Modulation Capacity. Nutrients. 2022, 14(12). [DOI] [PMC free article] [PubMed]
- 48.Hao J, Zhang Y, Wu T, Liu R, Sui W, Zhu J, et al. The antidiabetic effects of Bifidobacterium longum subsp. Longum BL21 through regulating gut microbiota structure in type 2 diabetic mice. Food Funct. 2022;13(19):9947–58. [DOI] [PubMed] [Google Scholar]
- 49.Hiraku A, Nakata S, Murata M, Xu C, Mutoh N, Arai S et al. Early probiotic supplementation of healthy term infants with Bifidobacterium longum subsp. infantis M-63 is safe and leads to the development of Bifidobacterium-predominant gut microbiota: a Double-Blind, placebo-controlled trial. Nutrients 2023, 15(6). [DOI] [PMC free article] [PubMed]
- 50.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J et al. The First Microbial colonizers of the human gut: composition, activities, and Health implications of the infant gut microbiota. Microbiol Mol Biol Rev 2017, 81(4). [DOI] [PMC free article] [PubMed]
- 51.Korpela K, Zijlmans MA, Kuitunen M, Kukkonen K, Savilahti E, Salonen A, et al. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome. 2017;5(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dreisbach C, Alhusen J, Prescott S, Dudley D, Trinchieri G, Siega-Riz AM. Metagenomic characterization of the maternal prenatal gastrointestinal microbiome by pregravid BMI. Obes (Silver Spring). 2023;31(2):412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blatchford P, Stoklosinski H, Eady S, Wallace A, Butts C, Gearry R, et al. Consumption of kiwifruit capsules increases Faecalibacterium prausnitzii abundance in functionally constipated individuals: a randomised controlled human trial. J Nutr Sci. 2017;6:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI SRA repository under the PRJNA1142935/SRP545576 project.