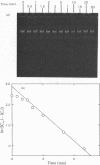
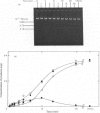
Abstract
The kinetics of the reactions of the EcoRI restriction endonuclease at individual recognition sites on the DNA from bacteriophage lambda were found to differ markedly from site to site. Under certain conditions of pH and ionic strength, the rates for the cleavage of the DNA were the same at each recognition site. But under altered experimental conditions, different reaction rates were observed at each recognition site. These results are consistent with a mechanism in which the kinetic stability of the complex between the enzyme and the recognition site on the DNA differs among the sites, due to the effect of interactions between the enzyme and DNA sequences surrounding each recognition site upon the transition state of the reaction. Reactions at individual sites on a DNA molecule containing more than one recognition site were found to be independent of each other, thus excluding the possibility of a processive mechanism for the EcoRI enzyme. The consequences of these observations are discussed with regard to both DNA-protein interactions and to the application of restriction enzymes in the study of the structure of DNA molecules.
Full text
PDF

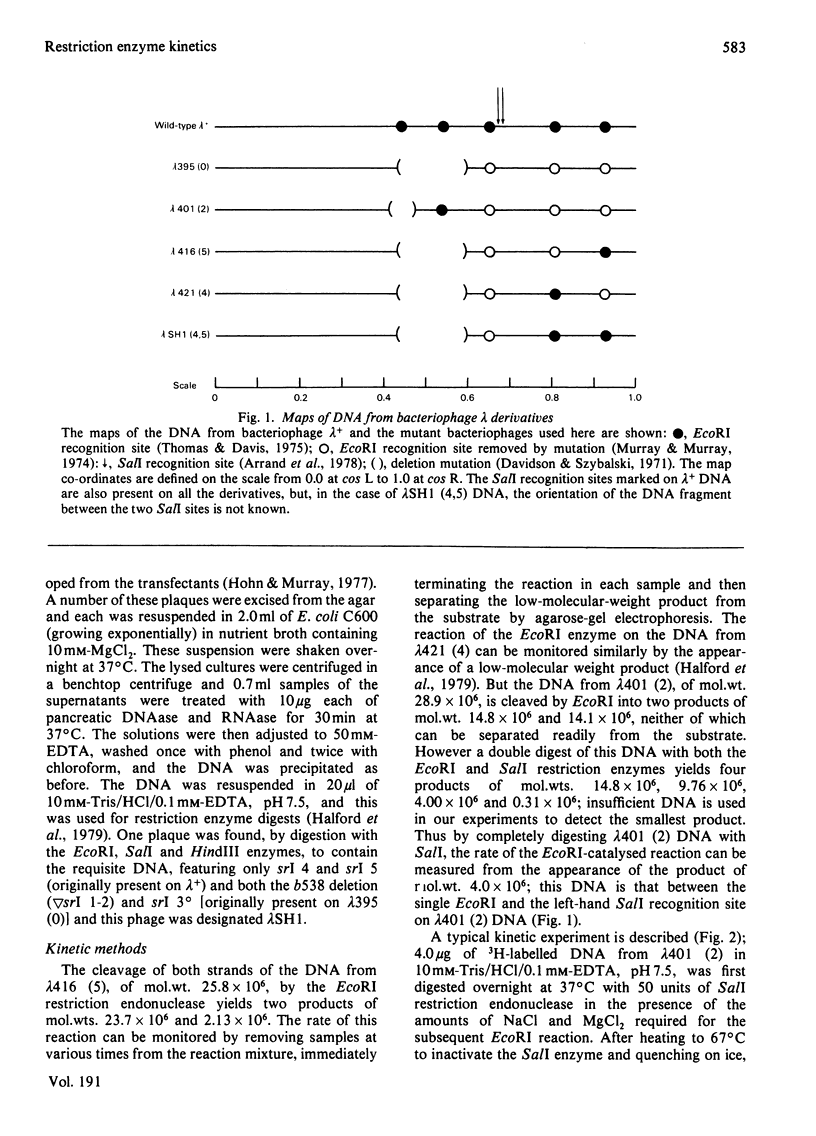
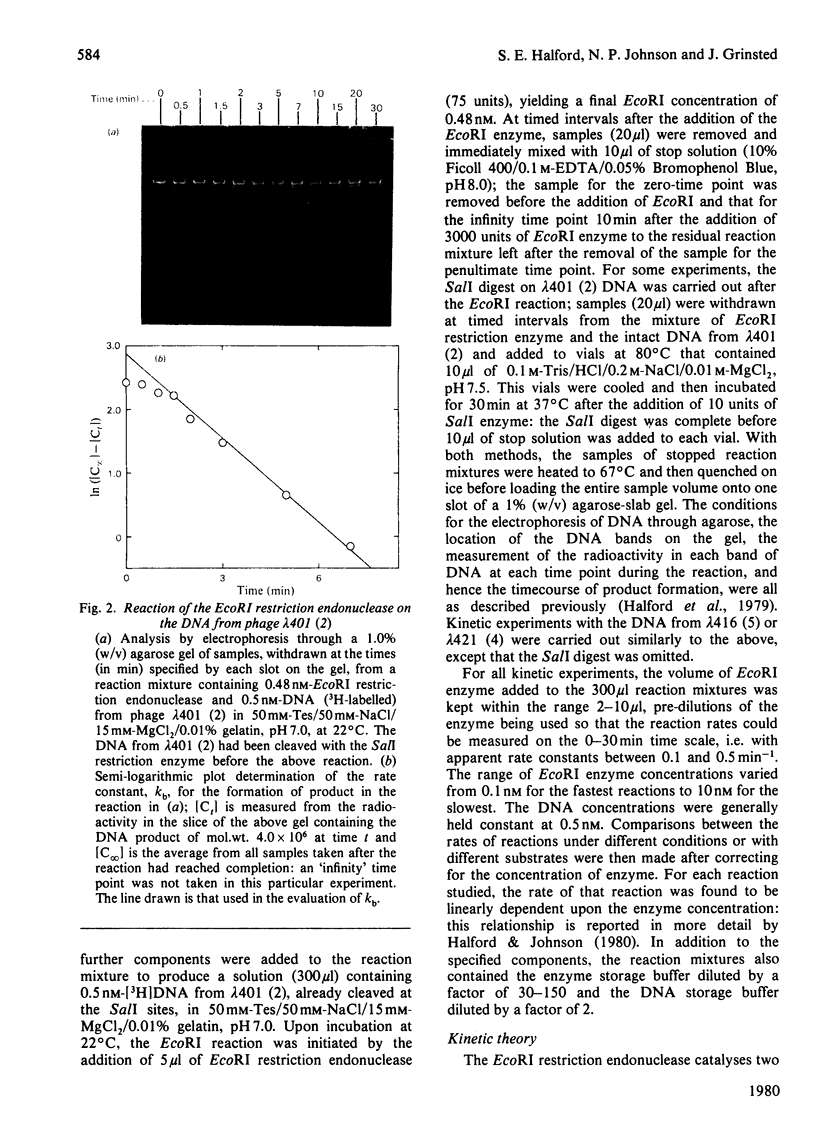





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrand J. R., Myers P. A., Roberts R. J. A new restriction endonuclease from Streptomyces albus G. J Mol Biol. 1978 Jan 5;118(1):127–135. doi: 10.1016/0022-2836(78)90249-8. [DOI] [PubMed] [Google Scholar]
- Forsblom S., Rigler R., Ehrenberg M., Philipson L. Kinetic studies on the cleavage of adenovirus DNA by restriction endonuclease Eco RI. Nucleic Acids Res. 1976 Dec;3(12):3255–3269. doi: 10.1093/nar/3.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfin D. E., Boyer H. W., Goodman H. M. Sequences spanning the EcoRI substrate site. Nucleic Acids Res. 1975 Oct;2(10):1851–1865. doi: 10.1093/nar/2.10.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P., Grinsted J. The reactions of the EcoRi and other restriction endonucleases. Biochem J. 1979 May 1;179(2):353–365. doi: 10.1042/bj1790353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Equilibrium binding studies. Biochem J. 1980 Nov 1;191(2):593–604. doi: 10.1042/bj1910593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn A. G., Hindley J. Small-scale techniques for the analysis of recombinant plasmids. J Biochem Biophys Methods. 1979 Oct;1(5):299–308. doi: 10.1016/0165-022x(79)90004-6. [DOI] [PubMed] [Google Scholar]
- Hohn B., Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M. Recognition mechanisms of DNA-specific enzymes. Annu Rev Biochem. 1976;45:889–920. doi: 10.1146/annurev.bi.45.070176.004325. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. The intrinsic pKa-values of functional groups in enzymes: improper deductions from the pH-dependence of steady-state parameters. CRC Crit Rev Biochem. 1976 Nov;4(2):165–173. doi: 10.3109/10409237609105457. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Murray N. E., Murray K. Manipulation of restriction targets in phage lambda to form receptor chromosomes for DNA fragments. Nature. 1974 Oct 11;251(5475):476–481. doi: 10.1038/251476a0. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter P. H., Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor--operator association and membrane bound enzymes. Biophys Chem. 1974 Oct;2(3):255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Dickerson R. E., Greene P. J., Boyer H. W. Preliminary x-ray diffraction analysis of crystalline EcoRI endonuclease. J Mol Biol. 1978 Jun 25;122(2):241–245. doi: 10.1016/0022-2836(78)90039-6. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Williams J. The formation of iron-binding fragments of hen ovotransferrin by limited proteolysis. Biochem J. 1974 Sep;141(3):745–752. doi: 10.1042/bj1410745. [DOI] [PMC free article] [PubMed] [Google Scholar]