Abstract
Background
The tumor immune microenvironment, particularly tumor-infiltrating lymphocytes (TILs), plays a critical role in disease progression and treatment response in triple-negative breast cancers (TNBCs). This study was aimed to characterize the composition of TILs and investigate their clinicopathological and prognostic significance with a special focus on the spatial distribution of TILs in TNBCs.
Methods
We analyzed TNBC samples through PanCancer Immune Profiling using NanoString nCounter assays to identify immune-related genes that are expressed differentially in relation to TIL levels and evaluated protein expression of selected markers through immunohistochemical staining on tissue microarrays. For a comprehensive assessment of the expression of cytotoxic T lymphocyte (CTL) and natural killer (NK) cell markers, a CTL-NK score was devised based on CD8+, CD56+, CD57+, GNLY+, and GZMB+ TIL levels.
Results
Gene expression analysis revealed significant upregulation of CTL and NK cell-associated genes including GNLY, KLRC2, and GZMB in TIL-high TNBCs. Immunohistochemical validation confirmed that TNBCs with higher TILs had a greater amount of CD56+, CD57+, GNLY+, and GZMB+ TILs not only in absolute number but also in proportion relative to CD4+ or CD8+ TILs. High TIL and its subset (CD4+, CD8+, CD56+, CD57+, GNLY+, and GZMB+ TIL) infiltration correlated with favorable clinicopathological features of tumor. In survival analysis, high CTL-NK score was found to be an independent prognostic factor for better disease-free survival (DFS) of the patients. Furthermore, uniformly high TIL infiltration was linked to better DFS, whereas cases with heterogeneous TIL infiltration showed no difference in survival compared to those with uniformly low TIL infiltration.
Conclusion
Our study showed that CTL and NK cell-associated gene expression and protein levels differ significantly according to TIL levels and that CTL-NK score and distribution of TILs within tumors have a prognostic value. These findings emphasize the importance of CTLs and NK cells as well as the spatial uniformity of TIL infiltration in clinical outcome of TNBC patients, providing valuable insights for refining prognostic assessments and guiding immunotherapeutic strategies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13058-024-01932-4.
Keywords: Tumor infiltrating lymphocyte, Triple-negative breast cancer, Cytotoxic T lymphocyte, NK cell, Spatial distribution
Background
Breast cancer is a heterogeneous group of diseases that exhibit various clinical characteristics, course of disease, and response to treatment; it is broadly categorized into luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched, and basal-like (or triple-negative) subtypes [1, 2]. Triple-negative breast cancer (TNBC), defined by negativity for estrogen receptor, progesterone receptor, and HER2, is a heterogeneous disease with different molecular characteristics and clinical outcome [3, 4]. Compared to the other subtypes of breast cancer, TNBC exhibits a worse prognosis [2] and has had limited therapeutic options [5].
However, recent advances in immunotherapy for the treatment of TNBC have shown the importance of tumor immune microenvironment (TIME). The TIME, which plays a crucial role in the progression of tumors as well as response to therapies [6], can vary significantly across different breast cancer subtypes [7] and even within TNBCs [8]. It encompasses tumor-infiltrating lymphocytes (TILs) as well as their subtype and activation state, programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1)-positive immune cells, macrophages, and myeloid-derived suppressor cells. It also includes immune-related soluble factors such as cytokines and chemokines [6]. Since TNBC has relatively more TILs than the other subtypes [9], it is a primary focus of research on TIME in breast cancer. The role of TILs in prognosis and treatment response varies by subtype [10]. In TNBC, higher TIL infiltration is associated with a better prognosis and a better response to neoadjuvant chemotherapy [10, 11].
TILs identified via hematoxylin and eosin (H&E) staining permit neither a comprehensive assessment of their overall directional impact on tumor suppression/promotion nor an evaluation of their activation status. Therefore, ongoing research continues to focus on studying subtypes of TILs. However, studies related to TIL subsets other than CD4+ [12], CD8+ [12–14], and FOXP3+ T cells [15, 16] are limited; thus, there are needs for a more comprehensive evaluation of the TIME. Detailed profiling of immune-related genes and proteins in TNBC tissues can provide insights into the mechanisms underlying immune responses and tumor progression.
Moreover, the way TIME affects cancer often varies by region [17]. In breast cancer, TIL infiltration is assessed in a representative tumor block according to the guidelines of the International Working Group for TILs (www.tilsinbreastcancer.org). However, the distribution of immune cell infiltration can be uneven within a tumor. A study on TNBC reported that there were transcriptomic differences in immune system pathways between regions with low versus. high TILs via whole RNA sequencing [18]. In addition, another study has revealed that intratumoral spatial heterogeneity of TILs is an important prognostic factor in microsatellite instability-high (MSI-H) colorectal cancer [19]. In breast cancer, however, there is a lack of research on prognostic differences based on spatial heterogeneity of TIL infiltration. Understanding this heterogeneity could potentially refine prognostic stratification and therapeutic decisions.
In this study, we aimed to conduct a comprehensive analysis of TIME in TNBC by comparing high versus low TIL-infiltrated tumors. We investigated the composition of TILs in TNBC and correlated it with clinicopathological features as well as clinical outcomes. In addition, we explored spatial heterogeneity of TIL and its subset infiltration within tumors and their impact on prognosis of TNBC patients.
Methods
Tissue samples
All tissue samples used in this study were from patients with primary TNBC who underwent surgical resection at Seoul National University Bundang Hospital. Two sets of tumor samples were used as a discovery set and a validation set, respectively. The first discovery set was composed of 36 cases of TNBC and was used for PanCancer Immune Profiling. To address the issue of RNA quality, the samples were restricted to those which were acquired between 2019 and 2022. They were intentionally matched with regards to recurrence and level of TIL infiltration of a similar number. The baseline clinicopathological characteristics of the 36 cases are presented in Additional file 1: Table S1.
The second validation set was composed of 172 cases of primary TNBC without recurrence at the time of diagnosis that were consecutively resected from 2003 to 2012, and immunohistochemical analyses were performed using tissue microarrays. Of the 172 cases, 17 had received neoadjuvant chemotherapy; these cases were excluded from the analysis because the infiltration of TIL and its subsets was significantly lower compared to those who did not receive neoadjuvant chemotherapy, indicating alteration of the TIME due to treatment. Thus, the final analysis was conducted on 155 cases. The clinicopathological characteristics of the 155 TNBC patients are summarized in Additional file 2: Table S2.
NanoString nCounter assay
Representative formalin-fixed paraffin-embedded tissue blocks of each tumor were sectioned at a thickness of 8 μm. The sections containing the tumors were marked and dissected, and RNA was extracted using RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, Grand Island, NY). The isolated RNA was used for the NanoString nCounter multiplex assay (NanoString Technologies, Seattle, WA). PanCancer Immune Profiling Panel for human (NanoString Technologies) was used for this study which targets and quantitates 770 mRNAs, 730 of which are immune-related genes and 40 of which are housekeeping genes that allow sample-to-sample normalization. After hybridization of RNA with the target-specific capture and reporter probes, excess probes were eliminated and the probe/target complexes were then aligned on the nCounter Cartridge (NanoString Technologies). The cartridges were placed into the nCounter Digital Analyzer (NanoString Technologies) for image acquisition and data capture. The automated fluorescence microscope of the analyzer scanned the sample, and the labeled barcodes were directly counted.
Evaluation of clinicopathological characteristics
Clinicopathological characteristics of the tumors were retrieved from the medical records and from reviewing the H&E or immunohistochemical slides. The following information was recorded: age at diagnosis, sex, histologic type (by WHO classification), TNM stage (by 8th American Joint Committee on Cancer staging system), presence of lymphovascular invasion (LVI), histologic grade including nuclear pleomorphism (by Nottingham histologic grading system), presence of ductal carcinoma in situ (DCIS) component, tumor border (pushing vs. infiltrative), multicentricity, level of TIL infiltration (< 10%, ≥ 10% and < 50%, or ≥ 50%), Ki-67 index, p53 overexpression, and basal phenotype. Basal phenotype was defined as positive membranous staining for either cytokeratin 5/6 or epidermal growth factor receptor. In addition, using the clinical follow-up data in the patients’ medical records, we collected the following information pertinent to survival as well: type or site of recurrence, date of diagnosis for recurrent cancers, date of last contact or death, and type of adjuvant therapy.
Tissue microarray construction
The tumor section slides were reviewed, and one to two representative slides of each tumor were selected. From these slides, three areas with varying levels of TIL infiltration were chosen. Paired three 2 mm-diameter tissue cores from these areas were collected to create a tissue microarray (TMA) using a trephine apparatus (SuperBioChips Laboratories, Seoul, South Korea).
Immunohistochemical staining and scoring
Immunohistochemical staining for CD4 (clone SP35; ready to use; Ventana Medical Systems, Tucson, AZ), CD8 (clone C8/144B; ready to use; Dako, Carpinteria, CA), CD56 (clone MRQ-42; ready to use; Ventana Medical Systems), CD57 (clone NK1; ready to use; Abcam, Cambridge, United Kingdom), granulysin (GNLY; polyclonal; 1:200; Abcam), and granzyme B (GZMB; Clone EPR22645-206; 1:500; Abcam) was performed using 4 μm-thick sections of TMA blocks. It was carried out on BenchMark XT autostainer (Ventana Medical Systems) using ultraView Universal DAB Detection Kit (Ventana Medical Systems).
Immunohistochemically-stained TMA slides were scanned using Aperio GT 450 DX (Leica, Wetzlar, Germany), and an open-source digital image analyzer QuPath (version 0.4.3) was used to count TILs expressing each marker. A high-power field (HPF) with the highest number of infiltration was selected in each TMA core, and the numbers of the TIL subsets were counted in each HPF. As a result, counts from the hotspots of three paired cores per case were obtained, and an average value was calculated for each case. The median value of these averages of all 155 cases were used as cut-offs when categorizing the cases into low-expression (≤ median) and high-expression (> median) groups.
Using the average numbers of TIL subset infiltration in each case, the ratios of TIL subsets over CD4+ or CD8+ TILs were calculated as follows: CD4+/CD8+, CD56+/CD8+, CD57+/CD8+, GNLY+/CD8+, GZMB+/CD8+, CD56+/CD4+, CD57+/CD4+, GNLY+/CD4+, and GZMB+/CD4+ TIL ratio.
For a comprehensive assessment of the expression levels of markers associated with cytotoxic T lymphocyte (CTL) or natural killer (NK) cells, a CTL-NK score was devised. For each five markers (CD8, CD56, CD57, GNLY, and GZMB), a score of 0 was assigned for cases classified into the low-expression group and a score of 1 for those categorized into the high-expression group. These scores were then summed up to derive the final CTL-NK score for each case. Consequently, the score ranged from 0 to 5.
Evaluation of heterogeneity of TIL infiltration
The overall TIL infiltration was scored using the guidelines of the International Working Group for TILs in breast cancer. Heterogeneity of TIL infiltration was determined by examining the degrees of TIL infiltration in each of the three TMA cores. When it was either low (< 10%) or high (≥ 10%) in all three cores, it was considered to be homogeneously low or high; otherwise, it was described as heterogeneous.
Similarly, evaluation of heterogeneity in TIL subset infiltration was based on whether the extent of infiltration in each of the three hotspots was consistently low (below median) or high (above median). Any instances where there was a mixture of low and high infiltration levels were classified as heterogeneous. When one of the three TMA cores was fragmented or lost during the immunohistochemical staining process and made evaluation of one core impossible, cases were deemed heterogeneous only when TIL subset infiltration showed values below and above the median in two cores, respectively. Cases whose infiltration levels consistently fell below or exceeded the median in both remaining cores were considered inconclusive.
Statistical analysis
In PanCancer immune profiling using Nanostring nCounter assay, collected data were checked for quality control, normalized, and analyzed by nSolver analysis software (version 4.0) and nCounter Advanced Analysis version 2.0 (NanoString Technologies). R software was used for comparison of the mRNA expression between two groups. Differential gene expression between groups was described with a log2 fold change, and adjusted p value using the Benzamini-Yekutieli procedure was presented.
Other statistical analyses were performed using IBM SPSS Statistics for Windows, version 27.0 (IBM Corporation, Armonk, New York). To examine the correlation between TIL, TIL subset, and TIL subset ratio, Spearman’s rank correlation test was employed, and the Rho correlation coefficient was presented. Mann-Whitney U test was used to compare the differences in TIL subset infiltration and TIL subset ratio between low and high TIL groups and to assess the differences in TIL subset infiltration and TIL subset ratio between groups divided by clinicopathological parameters. Comparison of clinicopathological features according to TIL infiltration (low vs. high-TIL) and according to CTL-NK score (< 2 vs. ≥2) was performed by Chi-square test or Fisher’s exact test. Survival curves were plotted by the Kaplan-Meier method, and their significance of differences was assessed using the log-rank test. Corrections for multiple testing were made by Bonferroni method, and adjusted p values were calculated. To identify independent factors associated with survival, multivariate analysis using Cox proportional hazard model was conducted with backward stepwise selection model. Hazard ratio (HR) and 95% confidence interval (CI) were calculated for each variable. All p values were two-sided, and p values less than 0.05 were considered statistically significant.
Results
Comprehensive immune profiling using NanoString nCounter assay
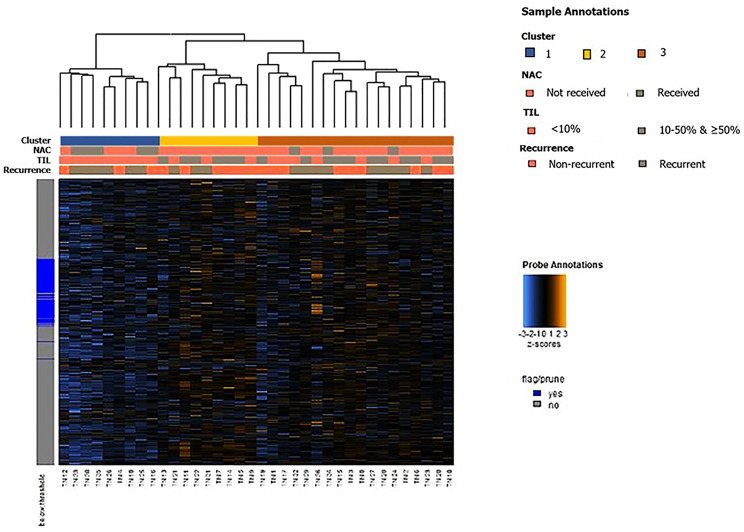
As a whole, unsupervised clustering analysis revealed that the samples could be broadly categorized into three clusters based on the overall expression patterns of immune-related genes (Fig. 1): one cluster (cluster 1) characterized by a decrease in the expression of immune-related genes, another cluster (cluster 2) exhibiting an increase in the expression of immune-related genes, and the other cluster (cluster 3) displaying an intermediate level of immune-related gene expression.
Fig. 1.
Heatmap of immune-related genes generated by unsupervised clustering. Samples were grouped into three based on their immune-related gene expression; one cluster had lower expression (cluster 1), another one had higher expression (cluster 2), and the last one showed intermediate expression (cluster 3). The clustering of each group was not determined by recurrence status. However, a potential relationship between immune-related gene expression patterns, TIL levels, and prior neoadjuvant chemotherapy (NAC) was observed. Cluster 1 consistently exhibited tumor-infiltrating lymphocyte infiltration of less than 10%. Additionally, none of the cases in Cluster 2 had undergone NAC. Color keys for sample annotation are displayed on the right
Although the samples were not clustered based on recurrence status, an association between the clusters and the level of TIL infiltration was observed. Cluster 1, which was characterized by a decrease in immune-related gene expression, uniformly exhibited TIL infiltration level of less than 10%. In addition, none of Cluster 2 had undergone neoadjuvant chemotherapy. These findings suggest a potential relationship between the immune-related gene expression patterns, TIL levels, and a history of neoadjuvant chemotherapy within our cohort.
Comparing groups with low (< 10%) and high (≥ 10%) levels of TILs, the top 20 genes exhibiting differential expression are presented in Table 1. Notably, genes showing substantial differences in expression (log2 fold change ≥ 1.5) and statistically-significant adjusted p values (< 0.05) included GNLY, KLRC2, and GZMB. This finding suggests that there may be differences in cytotoxicity and the distribution of NK cells between the groups.
Table 1.
List of top 20 genes with a significant fold change in high-TIL group compared to low-TIL group
| Gene | Log2 fold change | p value | Adjusted p value |
|---|---|---|---|
| GNLY-mRNA | 1.78 | 1.60E-06 | 0.0063 |
| KLRC2-mRNA | 2.22 | 9.93E-06 | 0.0170 |
| CD83-mRNA | 0.80 | 1.29E-05 | 0.0170 |
| TGFB2-mRNA | -1.46 | 2.25E-05 | 0.0222 |
| CCL5-mRNA | 1.42 | 3.69E-05 | 0.0291 |
| CCR6-mRNA | 1.38 | 5.02E-05 | 0.0318 |
| SELL-mRNA | 1.36 | 5.64E-05 | 0.0318 |
| IL2RB-mRNA | 1.47 | 6.89E-05 | 0.0338 |
| ISG20-mRNA | 0.99 | 7.72E-05 | 0.0338 |
| GZMB-mRNA | 1.82 | 9.45E-05 | 0.0372 |
| TAP1-mRNA | 1.32 | 0.000183 | 0.0608 |
| PRF1-mRNA | 1.39 | 0.000185 | 0.0608 |
| MICB-mRNA | 1.03 | 0.000204 | 0.0619 |
| CCR7-mRNA | 1.61 | 0.000242 | 0.0668 |
| PSMB9-mRNA | 1.19 | 0.000254 | 0.0668 |
| SOCS1-mRNA | 1.15 | 0.000305 | 0.0704 |
| KLRK1-mRNA | 1.33 | 0.000308 | 0.0704 |
| NOD2-mRNA | 0.90 | 0.000343 | 0.0704 |
| CD247-mRNA | 1.23 | 0.000355 | 0.0704 |
| GZMM-mRNA | 1.27 | 0.000387 | 0.0704 |
Log2 fold changes were calculated using tumors with low levels of TIL infiltration as the baseline
p values were adjusted by Benzamini-Yekutieli procedure
TIL, tumor-infiltrating lymphocyte
Evaluation of differentially expressed genes and related TIL subsets
Based on the findings from comprehensive immune profiling of TNBC, we selected immune-related markers GNLY and GZMB for further analysis. Given the association of these markers with CTL and NK cells, CD8+, CD56+ and CD57+ TILs were also evaluated in the validation set. Additionally, CD4+ TILs were evaluated to obtain baseline data.
As a whole, TIL infiltration ranged from 1 to 90% with median of 30%. While 48 cases (31.0%) showed less than 10% of TIL infiltration, 107 cases (69.0%) showed greater than or equal to 10% of TIL infiltration. Among the TIL subsets, CD4+ and CD8+ TILs comprised the majority of the TILs. The median of the average number of infiltrated cells per HPF was 223.7 (range, 0.0-1238.3) for CD4+ TIL and 215.0 (range, 0.0-1022.3) for CD8+ TIL. On the other hand, CD56+, CD57+, GNLY+, and GZMB+ TILs accounted for a smaller portion with the median values of 1.7 (range, 0.0-22.3), 6.7 (0.0-116.7), 13.0 (0.0-313.0), and 7.3 (0.0-244.0), respectively.
When analysis was done based on CD4+ or CD8+ TILs, the ratios of TIL subsets, expressed as median (inter-quartile range) were as follows: CD4+/CD8+, 0.93 (0.62–1.42); CD56+/CD8+, 0.01 (0.00-0.02); CD57+/CD8+, 0.03 (0.01–0.06); GNLY+/CD8+, 0.06 (0.02–0.15); GZMB+/CD8+, 0.03 (0.01–0.14); CD56+/CD4+, 0.01 (0.00-0.02); CD57+/CD4+, 0.03 (0.01–0.06); GNLY+/CD4+, 0.06 (0.03–0.14); GZMB+/CD4+ TIL ratio, 0.04 (0.01–0.15).
Correlation between TIL and its subsets infiltration or CTL-NK score
Next, we examined the correlation between TIL and TIL subset infiltration. Infiltration of TIL and its subsets including CD4+, CD8+, CD56+, CD57+, GNLY+, and GZMB+ TIL showed positive correlations with Rho correlation coefficient ranging from 0.534 to 0.806 (p < 0.001; Additional file 3: Table S3). When the infiltration levels of TIL subsets between low-TIL (TIL < 10%) and high-TIL (TIL ≥ 10%) groups were compared, a significant association was also observed for all TIL subsets (p < 0.001; Table 2; Fig. 2).
Table 2.
Comparison of TIL subset and TIL ratio between low-TIL and high-TIL groups
| TIL subset or ratio | Low-TIL group (n = 48) | High-TIL group (n = 107) | p value |
|---|---|---|---|
| CD4+ TIL | 63.7 (31.9-121.2) | 288.3 (200.0-427.3) | < 0.001 |
| CD8+ TIL | 83.3 (43.7-142.3) | 279.7 (161.3-386.3) | < 0.001 |
| CD56+ TIL | 0.0 (0.0-0.7) | 3.3 (0.7–7.7) | < 0.001 |
| CD57+ TIL | 0.7 (0.0-3.2) | 10.7 (5.0-18.7) | < 0.001 |
| GNLY+ TIL | 1.3 (0.1–6.3) | 22.7 (11.3–46.0) | < 0.001 |
| GZMB+ TIL | 1.15 (0.0-5.2) | 15.0 (2.3–39.3) | < 0.001 |
| CD4+/CD8+ TIL | 0.64 (0.39–1.18) | 0.99 (0.73–1.53) | 0.002 |
| CD56+/CD8+ TIL | 0.00 (0.00-0.01) | 0.01 (0.00-0.02) | < 0.001 |
| CD57+/CD8+ TIL | 0.01 (0.00-0.03) | 0.04 (0.02–0.06) | < 0.001 |
| GNLY+/CD8+ TIL | 0.02 (0.00-0.06) | 0.09 (0.04–0.19) | < 0.001 |
| GZMB+/CD8+ TIL | 0.01 (0.00-0.06) | 0.06 (0.01–0.16) | 0.001 |
| CD56+/CD4+ TIL | 0.00 (0.00-0.01) | 0.01 (0.01–0.02) | < 0.001 |
| CD57+/CD4+ TIL | 0.01 (0.00-0.04) | 0.04 (0.02–0.07) | < 0.001 |
| GNLY+/CD4+ TIL | 0.03 (0.01–0.07) | 0.08 (0.04–0.18) | < 0.001 |
| GZMB+/CD4+ TIL | 0.02 (0.00-0.08) | 0.04 (0.01–0.16) | 0.029 |
Data are presented as median (inter-quartile range), and p values were calculated by Mann-Whitney U test
TIL, tumor-infiltrating lymphocyte; GNLY, granulysin; GZMB, granzyme B
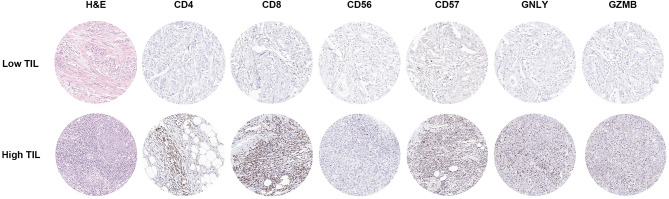
Fig. 2.
Representative images of H&E and immunohistochemical staining from one case showing low TIL and the other case showing high TIL. The case with low TIL infiltration also shows a low level of CD4+, CD8+, CD56+, CD57+, GNLY+, and GZMB+ cell infiltration. On the other hand, the case with high TIL infiltration shows abundant CD4+, CD8+, CD56+, CD57+, GNLY+, and GZMB+ cell infiltration. Each circle represents one high power field
We also examined the correlation between TIL infiltration and TIL subset ratios. All ratios except for CD4+/CD8+ TIL ratio showed moderate correlations with TIL infiltration (Rho correlation coefficients of 0.351–0.528; p < 0.001; Additional file 4: Table S4). When TIL subset ratios were compared between low-TIL and high-TIL groups, there was a statistically significant difference between the two groups in all TIL subset ratios (Table 2).
As for the CTL-NK score, 60 patients (38.7%) had a CTL-NK score of less than 2, while 95 patients (61.3%) had a score of 2 or higher. The group with a CTL-NK score of less than 2 exhibited a TIL median of 5.0% (inter-quartile range, 5.0-18.8%), whereas those with a CTL-NK score of 2 or higher showed a substantially higher TIL median of 45.0% (inter-quartile range, 20.0–70.0%) (p < 0.001).
Clinicopathological features associated with infiltration of TILs and its subsets
We compared clinicopathological characteristics of low- and high-TIL groups (Additional file 5: Table S5). Notably, the high-TIL group (TIL ≥ 10%) exhibited significant associations with lower T stage (p = 0.030), lower rates of LVI (p = 0.006), higher nuclear pleomorphism score (p = 0.036), absence of DCIS component (p = 0.006), and pushing tumor border (p = 0.022) compared to the low-TIL group (TIL < 10%). A trend towards higher Ki-67 index (≥ 50%) was observed in the high-TIL group (p = 0.051). There were no significant associations between TIL levels and other factors such as age, histologic type, N stage, American Joint Committee on Cancer stage, histologic grade, tumor multicentricity, p53 overexpression, or basal phenotype.
When we examined the relationship between clinicopathological characteristics and TIL subsets (CD8+, CD56+, CD57+, GNLY+, GZMB+ TIL), we observed similar findings (Table 3). High infiltration of CD8+, GNLY+, and GZMB+ TIL subsets showed a significant association with lower T stage (all p < 0.05), and high infiltration of CD56+, CD57+, GNLY+, and GZMB+ TIL subset was associated with an absence of LVI (all p < 0.05). Especially, high Ki-67 index was associated with higher levels of CD8+, CD56+, GNLY+, and GZMB+ TIL infiltration (all p < 0.05).
Table 3.
Relationship between clinicopathological characteristics and CD8+, CD56+, CD57+, GNLY+, and GZMB+ TIL subsets
| Clinicopathological characteristics | CD8+ TIL | CD56+ TIL | CD57+ TIL | GNLY+ TIL | GZMB+ TIL | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of TILs | p value | No. of TILs | p value | No. of TILs | p value | No. of TILs | p value | No. of TILs | p value | |
| Age (year) | 0.990 | 0.027 | 0.306 | 0.052 | 0.473 | |||||
| < 50 | 198.2 (113.7-332.3) | 2.3 (0.1–6.9) | 7.7 (1.4–15.7) | 17.8 (6.1–36.5) | 7.7 (1.0–31.0) | |||||
| ≥ 50 | 220.3 (80.3-377.3) | 0.7 (0.0-4.3) | 5.7 (0.7–13.7) | 11.3 (2.0–30.0) | 5.3 (1.3–22.3) | |||||
| Histologic type | 0.422 | 0.755 | 0.779 | 0.938 | 0.979 | |||||
| IC-NST | 200.0 (89.1-362.2) | 1.7 (0.0–6.0) | 6.7 (1.0-15.7) | 13.4 (3.3–34.1) | 7.5 (1.0-25.5) | |||||
| Othersa | 255.3 (144.2-331.9) | 1.7 (0.2–6.4) | 5.0 (0.7–13.4) | 10.7 (1.0-48.4) | 7.3 (1.5–31.4) | |||||
| T stage | 0.044 | 0.066 | 0.098 | 0.005 | 0.024 | |||||
| T1-2 | 217.3 (112.4-362.5) | 2.0 (0.0–6.0) | 6.7 (1.1–15.7) | 14.7 (4.3–35.7) | 7.7 (1.1–26.0) | |||||
| T3-4 | 70.0 (5.7-256.7) | 0.0 (0.0-0.3) | 0.7 (0.0-8.7) | 1.3 (0.0–7.0) | 0.3 (0.0-9.7) | |||||
| N stage | 0.540 | 0.343 | 0.374 | 0.083 | 0.125 | |||||
| N0 | 221.7 (87.2-373.3) | 2.0 (0.0–6.0) | 6.9 (1.1–15.8) | 13.9 (4.7–39.9) | 8.5 (1.1–26.0) | |||||
| N1-3 | 188.3 (118.3-294.7) | 1.0 (0.0–5.0) | 5.7 (1.0-12.3) | 12.0 (2.0-19.3) | 2.3 (0.3–20.0) | |||||
| AJCC stage | 0.410 | 0.087 | 0.161 | 0.147 | 0.030 | |||||
| I-II | 219.8 (102.3-365.2) | 2.0 (0.0–6.0) | 6.9 (1.0-15.9) | 14.2 (3.6–36.5) | 8.2 (1.3–26.0) | |||||
| III | 153.3 (115.7-294.7) | 0.3 (0.0–4.0) | 3.3 (0.5-8.0) | 10.7 (2.0-16.7) | 1.0 (0.0-7.3) | |||||
| LVI | 0.169 | 0.001 | 0.002 | 0.001 | 0.021 | |||||
| Absent | 232.3 (117.3–374.0) | 2.7 (0.0-7.3) | 8.3 (1.3–18.3) | 20.3 (4.7–46.0) | 9.7 (1.3–31.7) | |||||
| Present | 170.3 (86.4-315.8) | 0.6 (0.0-3.3) | 5.0 (0.4–8.5) | 11.0 (2.0-19.2) | 2.3 (0.4–20.0) | |||||
| Histologic grade | 0.335 | 0.066 | 0.080 | 0.009 | 0.041 | |||||
| II | 144.7 (67.5-288.5) | 0.0 (0.0-3.7) | 1.3 (0.2–10.7) | 1.3 (0.3–17.2) | 1.3 (0.0-4.5) | |||||
| III | 217.3 (109.3-362.2) | 2.0 (0.0–6.0) | 6.9 (1.2–15.8) | 14.7 (4.7–36.7) | 8.2 (1.0–26.0) | |||||
| Nuclear pleomorphism | 0.104 | 0.516 | 0.505 | 0.032 | 0.382 | |||||
| 2 | 127.3 (46.0-292.3) | 0.0 (0.0-7.3) | 7.3 (0.3–11.7) | 0.7 (0.3–22.7) | 1.7 (0.0-39.3) | |||||
| 3 | 219.8 (110.6–357.0) | 1.7 (0.0–6.0) | 6.5 (1.1–15.6) | 14.0 (4.7–35.7) | 7.7 (1.0-24.7) | |||||
| DCIS component | 0.157 | 0.006 | 0.066 | 0.002 | 0.020 | |||||
| Absent | 254.7 (113.8-369.1) | 3.0 (0.5–9.2) | 8.7 (1.6–18.7) | 24.5 (9.6–56.7) | 14.5 (2.7–35.3) | |||||
| Present | 179.3 (103.7-331.3) | 1.0 (0.0–5.0) | 6.0 (0.7–12.7) | 11.3 (2.4–27.7) | 3.7 (0.7–22.3) | |||||
| Tumor border | 0.229 | 0.038 | 0.011 | < 0.001 | 0.002 | |||||
| Pushing | 234.0 (120.7–366.0) | 2.0 (0.0-7.7) | 8.0 (2.0–17.0) | 21.3 (6.7–46.3) | 14.3 (1.0–35.0) | |||||
| Infiltrative | 185.5 (87.4-319.7) | 1.4 (0.0–4.0) | 3.7 (0.7–10.3) | 6.9 (1.5–17.8) | 2.2 (1.0-15.4) | |||||
| Multicentricity | 0.524 | 0.951 | 0.645 | 0.994 | 0.911 | |||||
| Absent | 217.3 (86.3–347.0) | 1.9 (0.0–6.0) | 6.7 (1.0-15.7) | 13.0 (2.5–36.0) | 7.3 (1.0-23.4) | |||||
| Present | 190.3 (140.3-345.2) | 1.0 (0.2–4.2) | 6.3 (0.9–13.0) | 14.3 (5.8–31.5) | 8.7 (1.0-32.5) | |||||
| Ki-67 index | 0.028 | 0.011 | 0.111 | 0.003 | 0.015 | |||||
| < 50% | 154.6 (77.8–302.0) | 0.9 (0.0-4.1) | 5.9 (0.7–13.2) | 11.0 (1.3–22.7) | 2.7 (0.7–20.1) | |||||
| ≥ 50% | 250.3 (122.9-377.8) | 3.0 (0.3-7.0) | 8.0 (2.0-16.5) | 21.3 (6.5–42.7) | 14.7 (1.3–36.7) | |||||
| p53 overexpression | 0.568 | 0.815 | 0.629 | 0.339 | 0.831 | |||||
| Absent | 192.3 (84.5-362.4) | 1.7 (0.0–6.0) | 6.7 (0.9–16.5) | 11.7 (1.3–32.8) | 8.7 (0.7–22.4) | |||||
| Present | 221.7 (109.3-334.5) | 1.7 (0.0-5.4) | 6.5 (1.2–15.0) | 14.5 (5.2–36.0) | 6.8 (1.2–26.6) | |||||
| Basal phenotype | 0.385 | 0.515 | 0.397 | 0.844 | 0.552 | |||||
| Absent | 155.5 (86.3-292.3) | 2.0 (0.0–7.0) | 7.0 (0.3–13.0) | 16.7 (0.7–34.3) | 7.3 (0.5–22.7) | |||||
| Present | 219.8 (107.6–357.0) | 1.7 (0.0-5.3) | 6.5 (1.1–15.6) | 13.0 (4.3–35.3) | 7.5 (1.0-27.7) | |||||
Data are presented as median (inter-quartile range), and p values were calculated by Mann-Whitney U test
aInclude metaplastic carcinoma and carcinoma with apocrine differentiation
TIL, tumor-infiltrating lymphocyte; GNLY, granulysin; GZMB, granzyme B; IC-NST, invasive carcinoma of no special type; AJCC, American Joint Committee on Cancer; LVI, lymphovascular invasion; DCIS, ductal carcinoma in situ
Analysis of the relationship between the clinicopathological features and the TIL subset ratios (CD4+/CD8+, CD56+/CD8+, CD57+/CD8+, GNLY+/CD8+, GZMB+/CD8+, CD56+/CD4+, CD57+/CD4+, GNLY+/CD4+ and GZMB+/CD4+ TIL ratio) also yielded similar results (Additional file 6 and 7: Tables S6 and S7).
Impact of TIL, TIL subsets, and CTL-NK score on patient survival
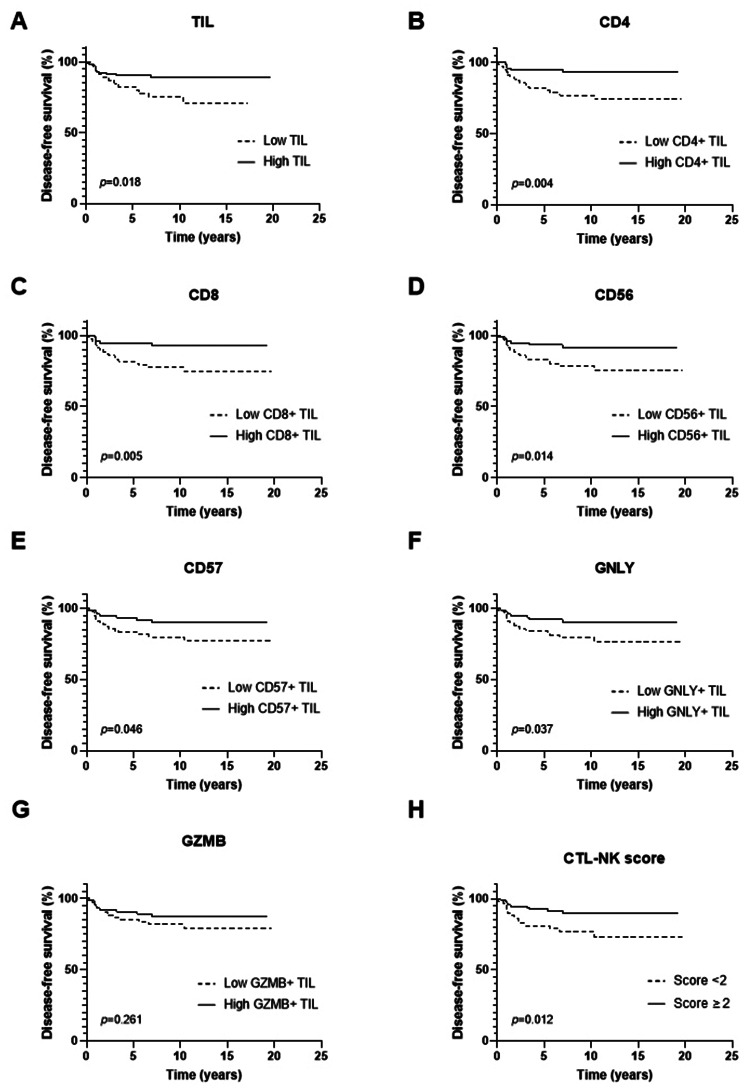
Survival analysis was conducted in the validation set composed of 155 patients. The median follow-up period was 10.04 years (range, 0.17–19.68). During this period, there were 23 recurrences and 16 deaths. The Kaplan-Meier survival curves for disease-free survival (DFS) are shown in Fig. 3. Higher infiltration of TILs and TIL subset (CD4+, CD8+, CD56+, CD57+, and GNLY+ TIL) along with CTL-NK score of 2 or higher were associated with better DFS of patients (all p < 0.05 by log-rank test) while GZMB+ TIL did not show a significant association (p = 0.261).
Fig. 3.
Kaplan-Meier survival curves for disease-free survival according to TIL, TIL subsets, and CTL-NK score. (A-F) High infiltration of TIL and CD4+, CD8+, CD56+, CD57+ and GNLY+ TIL subset was associated with increased disease-free survival (p = 0.018, 0.004, 0.005, 0.014, 0.046, and 0.037, respectively). (G) GZMB+ TIL subset infiltration was not associated with disease-free survival (p = 0.261). (H) CTL-NK score of 2 or higher was associated with increased disease-free survival (p = 0.012)
Next, we conducted survival analyses by combining TIL levels and CTL-NK scores in order to investigate whether DFS differed depending on CTL-NK score (< 2 vs. ≥2) even within the patients with similar TIL levels. The patients were divided into four groups: TIL-low and CTL-NK score-low, TIL-low and CTL-NK score-high, TIL-high and CTL-NK score-low, and TIL-high and CTL-NK score-high. For these analyses, TIL levels were divided using cutoffs of 10% and 30% (median of TIL levels). Although statistically significant differences were not found among the four groups (p = 0.054 for TIL 10% cutoff; p = 0.089 for TIL 30% cutoff; Additional file 8: Figure S1), there were some trends of survival differences between two groups. TIL-high and CTL-NK score-high group tended to have better DFS than TIL-high and CTL-NK score-low group (p = 0.106 for TIL 10% cutoff, p = 0.102 for TIL 30% cutoff).
In addition, the impacts of clinicopathological features, TIL, TIL subsets, and CTL-NK score on the DFS and overall survival (OS) were collectively analyzed using univariate Cox proportional hazard model (Table 4). The clinicopathological variables associated with DFS included T stage (p = 0.049) and LVI (p = 0.003). TIL, its subsets (except for CD57+ and GZMB+ TIL), and CTL-NK score showed a significant association with DFS (all p < 0.05). However, none of TIL, TIL subsets, or CTL-NK score showed a significant association with OS of patients.
Table 4.
Univariate analyses of disease-free survival and overall survival
| Variables | Category | Disease-free survival | Overall survival | ||
|---|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | ||
| Age | < 50 yrs vs. ≥50 yrs | 0.775 (0.335–1.792) | 0.552 | 0.964 (0.359–2.588) | 0.941 |
| Histologic type | IC-NST vs. othersa | 1.609 (0.478–5.416) | 0.443 | 0.697 (0.092–5.281) | 0.727 |
| T stage | T1-2 vs. T3-4 | 3.386 (1.003–11.436) | 0.049 | 1.513 (0.199–11.500) | 0.689 |
| N stage | N0 vs. N1-3 | 1.935 (0.837–4.473) | 0.123 | 1.759 (0.639–4.843) | 0.274 |
| LVI | Absent vs. Present | 3.643 (1.544–8.595) | 0.003 | 3.054 (1.109–8.407) | 0.031 |
| Histologic grade | II vs. III | 1.015 (0.238–4.329) | 0.984 | 1.494 (0.197–11.316) | 0.697 |
| DCIS | Absent vs. Present | 3.005 (0.892–10.120) | 0.076 | 1.395 (0.449–4.338) | 0.565 |
| Tumor border | Pushing vs. Infiltrative | 0.885 (0.375–2.089) | 0.781 | 1.740 (0.652–4.645) | 0.269 |
| Multicentricity | Absent vs. Present | 1.482 (0.550–3.993) | 0.437 | 1.718 (0.554–5.328) | 0.349 |
| TIL | < 10% vs. ≥10% | 0.387 (0.171–0.878) | 0.023 | 0.693 (0.252–1.911) | 0.479 |
| CD4+ TIL | Low vs. High | 0.258 (0.096–0.696) | 0.007 | 0.439 (0.152–1.263) | 0.127 |
| CD8+ TIL | Low vs. High | 0.266 (0.099–0.716) | 0.009 | 0.587 (0.213–1.617) | 0.303 |
| CD56+ TIL | Low vs. High | 0.328 (0.129–0.833) | 0.019 | 0.578 (0.210–1.592) | 0.289 |
| CD57+ TIL | Low vs. High | 0.416 (0.171–1.012) | 0.053 | 0.593 (0.215–1.631) | 0.311 |
| GNLY+ TIL | Low vs. High | 0.401 (0.165–0.976) | 0.044 | 0.563 (0.204–1.551) | 0.266 |
| GZMB+ TIL | Low vs. High | 0.621 (0.269–1.436) | 0.266 | 0.985 (0.370–2.625) | 0.976 |
| CTL-NK score | 0–1 vs. 2–5 | 0.357 (0.154–0.825) | 0.016 | 0.451 (0.168–1.212) | 0.114 |
p values were calculated by Cox proportional hazards model
aInclude metaplastic carcinoma and carcinoma with apocrine differentiation
HR, hazard ratio; CI, confidence interval; Yrs, years; IC-NST, invasive carcinoma of no special type; LVI, lymphovascular invasion; DCIS, ductal carcinoma in situ; TIL, tumor-infiltrating lymphocyte; DCIS, ductal carcinoma in situ; GNLY, granulysin; GZMB, granzyme B; CTL, cytotoxic T lymphocyte; NK, natural killer
As factors such as TIL, its subsets, and CTL-NK score showed moderate to strong correlations with each other (ρ = 0.534–0.853, p < 0.001), multivariate analyses were conducted using different models (Table 5). Across various models, LVI was consistently found to be an independent prognostic factor for worse DFS. Additionally, specific subsets of TILs, notably CD4+ (HR, 0.307; 95% CI, 0.112–0.841; p = 0.022) and CD8+ TILs (HR, 0.312; 95% CI, 0.115–0.844; p = 0.022), and the CTL-NK score (HR, 0.420; 95% CI, 0.181–0.978; p = 0.044) were revealed as independent prognostic factors of better DFS.
Table 5.
Multivariate analyses of disease-free survival
| Model | Variables | Category | Multivariate analysis | |
|---|---|---|---|---|
| HR (95% CI) | p value | |||
| A | T stage | T1-2 vs. T3-4 | 2.008 (0.573–7.036) | 0.276 |
| N stage | N0 vs. N1-3 | 1.129 (0.449–2.842) | 0.796 | |
| LVI | Absent vs. Present | 2.784 (1.050–7.385) | 0.040 | |
| TIL | < 10% vs. ≥10% | 0.546 (1.050–7.385) | 0.163 | |
| B | T stage | T1-2 vs. T3-4 | 1.677 (0.480–5.859) | 0.418 |
| N stage | N0 vs. N1-3 | 1.007 (0.395–2.563) | 0.989 | |
| LVI | Absent vs. Present | 3.055 (1.160–8.041) | 0.024 | |
| CD4+ TIL | Low vs. High | 0.307 (0.112–0.841) | 0.022 | |
| C | T stage | T1-2 vs. T3-4 | 2.170 (0.624–7.549) | 0.223 |
| N stage | N0 vs. N1-3 | 1.143 (0.456–2.869) | 0.776 | |
| LVI | Absent vs. Present | 2.822 (1.086–7.331) | 0.033 | |
| CD8+ TIL | Low vs. High | 0.312 (0.115–0.844) | 0.022 | |
| D | T stage | T1-2 vs. T3-4 | 2.012 (0.570–7.094) | 0.277 |
| N stage | N0 vs. N1-3 | 1.140 (0.441–2.952) | 0.787 | |
| LVI | Absent vs. Present | 2.676 (0.985–7.275) | 0.054 | |
| CD56+ TIL | Low vs. High | 0.441 (0.169–1.150) | 0.094 | |
| E | T stage | T1-2 vs. T3-4 | 2.486 (0.718–8.602) | 0.150 |
| N stage | N0 vs. N1-3 | 1.210 (0.483–3.029) | 0.684 | |
| LVI | Absent vs. Present | 2.853 (1.098–7.413) | 0.031 | |
| CD57+ TIL | Low vs. High | 0.462 (0.188–1.132) | 0.091 | |
| F | T stage | T1-2 vs. T3-4 | 1.752 (0.489–6.275) | 0.389 |
| N stage | N0 vs. N1-3 | 1.211 (0.486–3.021) | 0.681 | |
| LVI | Absent vs. Present | 2.838 (1.085–7.423) | 0.034 | |
| GNLY+ TIL | Low vs. High | 0.525 (0.207–1.332) | 0.175 | |
| G | T stage | T1-2 vs. T3-4 | 2.239 (0.651–7.709) | 0.201 |
| N stage | N0 vs. N1-3 | 1.173 (0.46–2.957) | 0.735 | |
| LVI | Absent vs. Present | 2.829 (1.081-7.400) | 0.034 | |
| CTL-NK score | 0–1 vs. 2–5 | 0.420 (0.181–0.978) | 0.044 | |
p values were calculated by Cox proportional hazards model using the backward stepwise selection method
HR, hazard ratio; CI, confidence interval; TIL, tumor-infiltrating lymphocyte; GNLY, granulysin; GZMB, granzyme B
Heterogeneity of TIL and its subset infiltration and their impact on patient survival
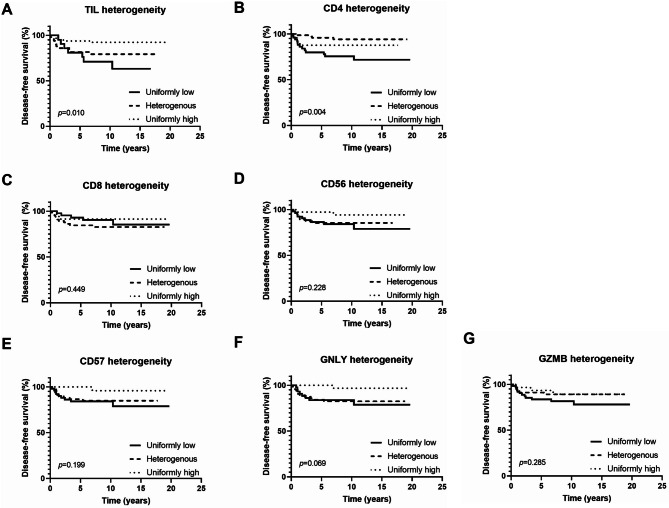
Heterogeneity in TIL levels was observed in 32.3% of cases while heterogeneity in the infiltration of CD4+, CD8+, C56+, CD57+, GNLY+, and GZMB+ TIL was observed in 47.7%, 43.2%, 36.8%, 44.5%, 41.9%, and 36.8% of cases, respectively (Additional file 9: Table S8). Figure 4 shows DFS of different groups based on heterogeneity in TILs and their subset infiltration levels. When cases were divided into three groups based on the heterogeneity of TIL infiltration, a significant difference in DFS was observed among the groups (p = 0.010). Groups with heterogeneous TIL levels seemed to exhibit survival intermediate between uniformly high and low groups. However, post-hoc analysis revealed no significant difference between the group showing uniformly low infiltration and the group showing heterogeneous infiltration (adjusted p = 1.125). Statistically significant differences were observed between the group with uniformly high infiltration and the group with uniformly low infiltration as well as between the group showing heterogeneous infiltration and the group showing uniformly high infiltration (adjusted p = 0.009 for both).
Fig. 4.
Kaplan-Meier survival curves for disease-free survival according to heterogeneity in TIL infiltration and its subset infiltration. (A) A significant difference in disease-free survival was noted between the group with uniformly high infiltration and the group with uniformly low infiltration as well as between the group with heterogeneous infiltration and the group exhibiting uniformly high infiltration (adjusted p = 0.009 for both). (B) Regarding CD4+ TIL subset infiltration, better prognosis was observed in groups with heterogeneous levels compared to those with consistently low levels (adjusted p = 0.003). (C) When disease-free survival was compared among the groups with different levels of heterogeneity of CD8+ TIL subset infiltration, no significant differences were observed between the groups. (D-F) Although not statistically significant, similar trends for poor survival were noted in both uniformly low and heterogeneous infiltration groups of CD56+, CD57+, and GNLY+ TIL subset. (G) When the disease-free survival was assessed among the groups divided based on the heterogeneity of GZMB+ TIL subset infiltration, no significant differences were observed among them
A significant difference in DFS was also observed between the groups based on heterogeneity in CD4+ TIL infiltration (p = 0.004). When a post-hoc analysis was conducted, a difference in survival was observed only between tumors with uniformly low infiltration and those with heterogeneous infiltration (adjusted p = 0.003). Although not statistically significant, for CD56+, CD57+, and GNLY+ TILs, tumors with uniformly low levels of infiltration and those with heterogeneous levels of infiltration tended to exhibit a similar degree of poor prognosis.
Discussion
This study provides an in-depth characterization of TILs and their clinicopathological and prognostic significance in TNBC with a particular focus on CTLs and NK cells as well as their spatial distributions. We employed the NanoString nCounter PanCancer Immune Profiling Panel to assess the expression of immune-related genes in primary TNBC samples. Comparing TNBC with low TIL levels to those with high TIL levels revealed significantly different expression of the genes GNLY, KLRC2, and GZMB, which encode proteins GNLY, killer cell lectin-like receptor subfamily C member 2 (KLRC2), and GZMB, respectively. GNLY is a pore-forming protein found in the cytotoxic granules of human killer cells such as CTLs and NK cells. Human GNLY is initially present as a 15 kDa protein which is later cleaved to generate a 9 kDa molecule [20], which enables cytotoxic immune cells to directly induce cell death including tumor cells [21]. On the other hand, the 15 kDa form promotes chemo-attraction of additional immune cell populations and maturation of antigen-presenting cells [22]. GZMB is the most prevalent serine protease found in the secretory granules of CTLs and NK cells. GZMB-induced cell death is a key mechanism by which CTLs and NK cells eliminate harmful cells including allogeneic, virus-infected, and tumor cells [23]. In addition, GZMB directly modulates inflammation by enhancing the pro-inflammatory activity of cytokines such as interleukin-1 (IL-1) and interleukin-18 (IL-18) [24, 25]. NK cells have receptors for major histocompatibility complex (MHC) class I molecules that regulate cell-mediated cytotoxicity and cytokine production [26]. One such receptor, encoded by the KLRC2 gene, is KLRC2, also known as NK cell Group 2 isoform C (NKG2C). NKG2 receptors are heterodimers expressed on NK cells and a small subset of T cells with their function dependent on the isoform. Specifically, NKG2C expression leads to cell activation and enhances cytotoxic function [27]. Therefore, from the different expression of GNLY, KLRC2, and GZMB genes in TNBC with low TIL levels and those with high TIL levels, it could be inferred that NK cells and CTLs are more numerous or more active in TNBCs with high TILs.
Next, we validated the expression of selected immune-related markers through immunohistochemical staining in a larger cohort of TNBC cases. Since KLRC2 expression in tissue has not been confirmed for immunohistochemistry, immunohistochemical staining was performed only for GNLY and GZMB. As mentioned above, it was notable that all three of these genes encoded proteins that are expressed in CTLs and NK cells. Therefore, in order to determine whether a high expression of these genes is associated with abundance of cytotoxic TIL subsets in TNBC with high TIL levels, we performed immunohistochemistry for CD8, CD56, and CD57, the latter two of which were selected as markers to identify NK cells. By definition, NK cells are identified by the presence of CD56 and/or CD16 markers and the lack of the CD3 marker found on T cells [28]. CD57 is recognized as a marker of terminal differentiation with CD57+ NK cells exhibiting reduced proliferation yet heightened cytotoxicity against tumor cells compared to CD57− NK cells [29]. Lastly, as CD4+ T cells are known to constitute a substantial portion of TILs in invasive breast carcinoma after CD8+ T cells [30], we performed immunohistochemistry for CD4 and CD8 as a baseline comparison. During TMA validation, CD4+, CD8+, CD56+, CD57+, GZMB+, and GNLY+ TILs exhibited a significant positive correlation with overall TIL levels. In addition, when we calculated the TIL subset ratios of CD56+, CD57+, GZMB+, and GNLY+ TILs using CD4+ and CD8+ TILs as denominators, a correlation with overall TIL levels was also observed. These findings suggest that as TILs increase, the numbers of NK cells and cytotoxic molecules increase more significantly compared to CD4+ and CD8+ T cells. These findings collectively suggest that CTLs and NK cells may play a crucial role in TNBC with high TIL levels. NK cells are innate immune effectors exhibiting natural cytotoxicity against primary tumor cells and also play a role in immune-surveillance of metastasis [31, 32]. In addition to their cytotoxic functions, NK cells secrete numerous cytokines, primarily interferon-γ (IFN-γ), which modulate adaptive immune responses [33]. Using the validation set, we demonstrated that TNBC with high TIL levels also has a high number of CTLs and NK cells. Considering that they have antitumor functions, it may be inferred that TNBC with high TIL levels have an association with prolonged survival or favorable prognostic features as discussed below.
High-TIL TNBC was associated with several favorable clinicopathological features including lower T stage and absence of LVI. Similarly, TIL subsets including CD8+, CD56+, CD57+, GNLY+, and GZMB+ TIL showed a significant association with lower T stage or absence of LVI. These correlations suggest a possibility that a robust immune response, as evidenced by high TIL levels or high CTL and NK cell infiltration, may inhibit tumor progression. This finding is in line with a study by Burnstein et al. [3] who showed that the basal-like immune-suppressed subtype of TNBC, which shows downregulation of B cell, T cell, and NK cell immune-regulating and cytokine pathways, had the least favorable prognosis while the basal-like immune-activated subtype, which exhibits upregulation of immune-related genes, had the best prognosis. However, it is also possible that tumor progression leads to a decrease in TIL levels. A recent study on TNBC showed that during metastasis, there is a general decrease in immune-related gene expression and a reduction in stromal TILs [34]. Another study using flow cytometry revealed that NK cell activity in breast cancer decreases with advancing cancer stage [35]. Thus, it seems that the TIME of TNBC shifts towards an immune-suppressed state as the disease progresses.
In order to evaluate the degree of infiltration of CTL and NK cells comprehensively, we devised a CTL-NK score based on the expression levels of CD8+, CD56+, CD57+, GNLY+, and GZMB+ TILs. The fact that this score was shown to be a significant independent prognostic factor for improved DFS in the current study indicates the prognostic value of combined CTL and NK cell infiltration. CD8+ T cells have been widely reported to be associated with a favorable prognosis in TNBC [10, 11], and NK cells are reported to be linked to a favorable prognosis in many solid tumors [36] including colorectal cancer [37], gastric cancer [38], lung cancer [39], renal cell carcinoma [40], and melanoma [41]. One meta-analysis found that high levels of NK cell markers CD56 and CD57 in solid tumor tissues could predict favorable prognosis and serve as an independent prognostic marker for favorable OS [42]. In breast cancer, one study found that high NK cell infiltration was significantly associated with TILs and that high CD56 expression (≥ 5 cells/10 HPFs) correlated with improved OS and DFS [43]. GNLY and GZMB have been reported to be associated with a better prognosis in solid tumors as well. One study showed that upregulated mRNA expression of intratumoral GNLY and GZMB was associated with a favorable outcome in patients with colorectal cancer [44]. Another study on primary untreated breast cancer patients revealed that high levels of GNLY mRNA predict a low risk of local recurrence but a high risk of distant metastasis [45]. Increased numbers of GZMB+ cells in the TIME have been linked to a favorable prognosis in some tumors including cervical intraepithelial neoplasia [46], oral squamous cell carcinoma [47], and melanoma [48]. Our findings that are consistent with the results of previous studies emphasize the need for an integrated assessment of multiple immune cell types in TNBC beyond the traditional focus on overall TILs.
The spatial heterogeneity of TIL infiltration within tumors was also found to be a crucial factor associated with patient prognosis. Uniformly high TIL infiltration was associated with better DFS while heterogeneous TIL infiltration did not confer the same survival benefit. This is a significant finding since it suggests that in addition to the quantity of TILs, the distribution of TILs within the TIME impacts clinical outcomes. This is in line with a previous study on MSI-H colon cancer where TIL-low/heterogeneity-high, TIL-low/heterogeneity-low, and TIL-high subgroups were significantly associated with poor, intermediate, and good DFS [19].
There are some limitations in this study. First, the relatively small sample size, particularly in the discovery set, may limit generalization of the results. Further research is needed to confirm the findings of our study in a large cohort. Secondly, the discovery set included some post-neoadjuvant chemotherapy samples which showed significantly lower TIL and its subset infiltration compared to the non-treated samples, whereas the final validation set was only composed of non-treated samples. This may lead to a bias in the results. Thirdly, although we selected representative tumor areas to construct TMAs, they may not reflect the TIME of the entire tumor due to a sampling issue. Furthermore, we selected a hotspot in a TMA core to evaluate TIL subset infiltration. Scanning of the whole slide of the tumor with subsequent digital analyses of spatial heterogeneity of TIL and its subset infiltration may be a next step to extend this study.
Conclusion
In summary, this study showed that CTL and NK cell-associated gene expression and protein levels differ significantly according to TIL levels in TNBCs, with higher CTLs and NK cells in the high-TIL group. We developed a CTL-NK score which reflects overall CTL and NK cell infiltration that also has a prognostic value in TNBCs. More importantly, our study showed that spatial distribution of TILs within a tumor has a prognostic implication. While tumors with homogeneously high TIL infiltration showed favorable clinical outcome, those with heterogeneous TIL infiltration was associated with poor clinical outcomes similar to those with homogeneously low TIL infiltration. Collectively, this study demonstrates the importance of NK cells as well as CTLs and the spatial distribution of TILs in prognosticating TNBC patients, providing valuable insights into understanding the TIME as well as potential therapeutic strategies in TNBCs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Additional file 1: Table S1. Clinicopathological characteristics of 36 TNBC patients in the test set. Additional file 2: Table S2. Clinicopathological characteristics of 155 TNBC patients in the validation set. Additional file 3: Table S3. Correlation between tumor infiltrating lymphocyte and its subsets. Additional file 4: Table S4. Correlation between tumor infiltrating lymphocyte and its subset ratios. Additional file 5: Table S5. Relationships between TIL levels and clinicopathological features of tumors. Additional file 6: Table S6. Relationship between clinicopathological characteristics and TIL subset ratios using CD8+ TIL. Additional file 7: Table S7. Relationship between clinicopathological characteristics and TIL subset ratios using CD4 + TIL. Additional file 9: Table S8. Distribution of tumor infiltrating lymphocyte and its subsets.
Supplementary Material 2: Additional file 8: Figure S1. Kaplan-Meier survival curves for disease-free survival according to combined analyses of TIL levels (A, < 10% or ≥ 10%; B, < 30% or ≥ 30%) and CTL-NK scores (< 2 or ≥ 2).
Acknowledgements
Not applicable.
Abbreviations
- CTL
Cytotoxic T lymphocyte
- DCIS
Ductal carcinoma in situ
- DFS
Disease-free survival
- GNLY
Granulysin
- GZMB
Granzyme B
- H&E
Hematoxylin and eosin
- HPF
High-power field
- HER2
Human epidermal growth factor receptor 2
- KLRC2
Killer cell lectin-like subfamily C member 2
- LVI
Lymphovascular invasion
- MSI-H
Microsatellite instability-high
- NK cell
Natural killer cell
- NKG2C
NK cell Group 2 isoform C
- TIL
Tumor-infiltrating lymphocyte
- TIME
Tumor immune microenvironment
- TMA
Tissue microarray
- TNBC
Triple-negative breast cancer
- OS
Overall survival
Author contributions
EH participated in the interpretation and analysis of data and drafted the manuscript. HYC carried out the experiments. HJK and YRC participated in the acquisition and interpretation of pathologic data. HCS, EKK, KJS, SHK and JHK participated in the acquisition of clinical data. SYP conceived of the study, and participated in its design and was responsible for preparation of the manuscript. All authors reviewed and approved the final manuscript.
Funding
This study was supported by a grant from the National Research Foundation of Korea (NRF)’s Basic Science Research Program to Park SY by the Ministry of Science, ICT and Future Planning (Grant No. 2022R1F1A1065468) and a grant from Seoul National University Bundang Hospital to Park SY (Grant No. 02-2021-0029).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was approved by the institutional review board (IRB No B-2210-786-303), and the requirement of informed consent was waived.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leon-Ferre RA, Goetz MP. Advances in systemic therapies for triple negative breast cancer. BMJ. 2023;381:e071674. [DOI] [PubMed] [Google Scholar]
- 6.Li JJ, Tsang JY, Tse GM. Tumor microenvironment in breast Cancer-updates on therapeutic implications and pathologic Assessment. Cancers (Basel). 2021; 13(16). [DOI] [PMC free article] [PubMed]
- 7.Li J, Wu JS, Han JW. Analysis of Tumor Microenvironment heterogeneity among breast Cancer subtypes to identify subtype-specific signatures. Genes-Basel. 2023; 14(1). [DOI] [PMC free article] [PubMed]
- 8.Bareche Y, Buisseret L, Gruosso T, Girard E, Venet D, Dupont F, Desmedt C, Larsimont D, Park M, Rothe F, et al. Unraveling triple-negative breast Cancer Tumor Microenvironment Heterogeneity: towards an Optimized Treatment Approach. J Natl Cancer Inst. 2020;112(7):708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J. Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med. 2016;14(1):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, Budczies J, Huober J, Klauschen F, Furlanetto J, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19(1):40–50. [DOI] [PubMed] [Google Scholar]
- 11.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25(8):1544–50. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto H, Thike AA, Li H, Yeong J, Koo SL, Dent RA, Tan PH, Iqbal J. Increased CD4 and CD8-positive T cell infiltrate signifies good prognosis in a subset of triple-negative breast cancer. Breast Cancer Res Treat. 2016;156(2):237–47. [DOI] [PubMed] [Google Scholar]
- 13.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR. Tumor-infiltrating CD8 + lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29(15):1949–55. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8 + lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14(2):R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, Banham AH. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24(34):5373–80. [DOI] [PubMed] [Google Scholar]
- 16.West NR, Kost SE, Martin SD, Milne K, Deleeuw RJ, Nelson BH, Watson PH. Tumour-infiltrating FOXP3(+) lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br J Cancer. 2013;108(1):155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–54. [DOI] [PubMed] [Google Scholar]
- 18.Quintana A, Arenas EJ, Bernado C, Navarro JF, Gonzalez J, Esteve-Codina A, Moline T, Marti M, Curigliano G, Schmid P, et al. Evaluation of triple negative breast cancer with heterogeneous immune infiltration. Front Immunol. 2023;14:1149747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung M, Lee JA, Yoo SY, Bae JM, Kang GH, Kim JH. Intratumoral spatial heterogeneity of tumor-infiltrating lymphocytes is a significant factor for precisely stratifying prognostic immune subgroups of microsatellite instability-high colorectal carcinomas. Mod Pathol. 2022;35(12):2011–22. [DOI] [PubMed] [Google Scholar]
- 20.Krensky AM, Clayberger C. Biology and clinical relevance of granulysin. Tissue Antigens. 2009;73(3):193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang LP, Lyu SC, Clayberger C, Krensky AM. Granulysin-mediated tumor rejection in transgenic mice. J Immunol. 2007;178(1):77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayberger C, Finn MW, Wang T, Saini R, Wilson C, Barr VA, Sabatino M, Castiello L, Stroncek D, Krensky AM. 15 kDa granulysin causes differentiation of monocytes to dendritic cells but lacks cytotoxic activity. J Immunol. 2012;188(12):6119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousalova I, Krepela E. Granzyme B-induced apoptosis in cancer cells and its regulation (review). Int J Oncol. 2010;37(6):1361–78. [DOI] [PubMed] [Google Scholar]
- 24.Omoto Y, Yamanaka K, Tokime K, Kitano S, Kakeda M, Akeda T, Kurokawa I, Gabazza EC, Tsutsui H, Katayama N, et al. Granzyme B is a novel interleukin-18 converting enzyme. J Dermatol Sci. 2010;59(2):129–35. [DOI] [PubMed] [Google Scholar]
- 25.Afonina IS, Tynan GA, Logue SE, Cullen SP, Bots M, Luthi AU, Reeves EP, McElvaney NG, Medema JP, Lavelle EC, et al. Granzyme B-dependent proteolysis acts as a switch to enhance the proinflammatory activity of IL-1alpha. Mol Cell. 2011;44(2):265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanier LL. Follow the leader: NK cell receptors for classical and nonclassical MHC class I. Cell. 1998;92(6):705–7. [DOI] [PubMed] [Google Scholar]
- 27.Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35(3):263–78. [DOI] [PubMed] [Google Scholar]
- 28.Robertson MJ, Ritz J. Biology and clinical relevance of human natural killer cells. Blood. 1990;76(12):2421–38. [PubMed] [Google Scholar]
- 29.Nielsen CM, White MJ, Goodier MR, Riley EM. Functional significance of CD57 expression on Human NK cells and relevance to Disease. Front Immunol. 2013;4:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong PP, Mohammad R, Ibrahim N, Ithnin H, Abdullah M, Davis WC, Seow HF. Phenotyping of lymphocytes expressing regulatory and effector markers in infiltrating ductal carcinoma of the breast. Immunol Lett. 2006;102(2):229–36. [DOI] [PubMed] [Google Scholar]
- 31.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of Metastasis by NK Cells. Cancer Cell. 2017;32(2):135–54. [DOI] [PubMed] [Google Scholar]
- 33.Razeghian E, Kameh MC, Shafiee S, Khalafi F, Jafari F, Asghari M, Kazemi K, Ilkhani S, Shariatzadeh S, Haj-Mirzaian A. The role of the natural killer (NK) cell modulation in breast cancer incidence and progress. Mol Biol Rep. 2022;49(11):10935–48. [DOI] [PubMed] [Google Scholar]
- 34.Hutchinson KE, Yost SE, Chang CW, Johnson RM, Carr AR, McAdam PR, Halligan DL, Chang CC, Schmolze D, Liang J, et al. Comprehensive profiling of poor-risk paired primary and recurrent triple-negative breast cancers reveals Immune phenotype shifts. Clin Cancer Res. 2020;26(3):657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piroozmand A, Hassan ZM. Evaluation of natural killer cell activity in pre and post treated breast cancer patients. J Cancer Res Ther. 2010;6(4):478–81. [DOI] [PubMed] [Google Scholar]
- 36.Nersesian S, Schwartz SL, Grantham SR, MacLean LK, Lee SN, Pugh-Toole M, Boudreau JE. NK cell infiltration is associated with improved overall survival in solid cancers: a systematic review and meta-analysis. Transl Oncol. 2021; 14(1). [DOI] [PMC free article] [PubMed]
- 37.Coca S, Perez-Piqueras J, Martinez D, Colmenarejo A, Saez MA, Vallejo C, Martos JA, Moreno M. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer. 1997;79(12):2320–8. [DOI] [PubMed] [Google Scholar]
- 38.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, Aridome K, Hokita S, Aikou T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000;88(3):577–83. [PubMed] [Google Scholar]
- 39.Villegas FR, Coca S, Villarrubia VG, Jimenez R, Chillon MJ, Jareno J, Zuil M, Callol L. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer. 2002;35(1):23–8. [DOI] [PubMed] [Google Scholar]
- 40.Geissler K, Fornara P, Lautenschlager C, Holzhausen HJ, Seliger B, Riemann D. Immune signature of tumor infiltrating immune cells in renal cancer. Oncoimmunology. 2015;4(1):e985082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S, Behren A, Huntington ND, Davis MJ. A gene signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma patients. Cancer Immunol Res. 2019;7(7):1162–74. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Liu W, Hu B, Wang P, Lv X, Chen S, Shao Z. Prognostic significance of Tumor-Infiltrating Natural Killer cells in solid tumors: a systematic review and Meta-analysis. Front Immunol. 2020;11:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouzidi L, Triki H, Charfi S, Kridis WB, Derbel M, Ayadi L, Sellami-Boudawara T, Cherif B. Prognostic value of natural killer cells besides tumor-infiltrating lymphocytes in breast Cancer tissues. Clin Breast Cancer. 2021;21(6):e738–47. [DOI] [PubMed] [Google Scholar]
- 44.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353(25):2654–66. [DOI] [PubMed] [Google Scholar]
- 45.Milovanovic J, Todorovic-Rakovic N, Vujasinovic T, Greenman J, Mandusic V, Radulovic M. Can granulysin provide prognostic value in primary breast cancer? Pathol Res Pract. 2022; 237. [DOI] [PubMed]
- 46.Kondo MC, Ribalta JCL, da Silva IDG, Alves MTS, Focchi GRD, Martins NV, Focchi J. Granzyme B as a prognostic marker of cervical intraepithelial neoplasia. Eur J Gynaecol Oncol. 2005;26(1):87–9. [PubMed] [Google Scholar]
- 47.Costa NL, Alencar Rde C, Valadares MC, Silva TA, Mendonca EF, Batista AC. The clinicopathological significance of the expression of Granzyme B in oral squamous cell carcinoma. Oral Oncol. 2010;46(3):185–9. [DOI] [PubMed] [Google Scholar]
- 48.van Houdt IS, Sluijter BJR, Moesbergen LM, Vos WM, de Gruijl TD, Molenkamp BG, van den Eertwegh AJM, Hooijberg E, van Leeuwen PAM, Meijer CJLM, et al. Favorable outcome in clinically stage II melanoma patients is associated with the presence of activated tumor infiltrating T-lymphocytes and preserved MHC class I antigen expression. Int J Cancer. 2008;123(3):609–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Additional file 1: Table S1. Clinicopathological characteristics of 36 TNBC patients in the test set. Additional file 2: Table S2. Clinicopathological characteristics of 155 TNBC patients in the validation set. Additional file 3: Table S3. Correlation between tumor infiltrating lymphocyte and its subsets. Additional file 4: Table S4. Correlation between tumor infiltrating lymphocyte and its subset ratios. Additional file 5: Table S5. Relationships between TIL levels and clinicopathological features of tumors. Additional file 6: Table S6. Relationship between clinicopathological characteristics and TIL subset ratios using CD8+ TIL. Additional file 7: Table S7. Relationship between clinicopathological characteristics and TIL subset ratios using CD4 + TIL. Additional file 9: Table S8. Distribution of tumor infiltrating lymphocyte and its subsets.
Supplementary Material 2: Additional file 8: Figure S1. Kaplan-Meier survival curves for disease-free survival according to combined analyses of TIL levels (A, < 10% or ≥ 10%; B, < 30% or ≥ 30%) and CTL-NK scores (< 2 or ≥ 2).
Data Availability Statement
No datasets were generated or analysed during the current study.