Abstract
The integration of machine learning (ML) classification techniques into migraine research has offered new insights into the pathophysiology and classification of migraine types and subtypes. However, inconsistencies in study design, lack of methodological transparency, and the absence of external validation limit the impact and reproducibility of such studies. This paper presents a framework of six essential recommendations for evaluating ML-based classification in migraine research: (1) group homogenization by clinical phenotype, attack frequency, comorbidity, therapy, and demographics; (2) defining adequate sample size; (3) quality control of collected and preprocessed data; (4) transparent training, testing, and performance evaluation of ML models, including strategies for data splitting, overfitting control, and feature selection; (5) interpretability of results with clinical relevance; and (6) open data and code sharing to facilitate reproducibility. These recommendations aim to balance the trade-off between model generalization and precision while encouraging collaborative standardization across the ML and headache communities. Furthermore, this framework intends to stimulate discussion toward forming a consortium to establish definitive guidelines for ML-based classification research in migraine field.
Keywords: Benchmark, Machine learning classification models, Data quality, Model interpretability, Model reproducibility, Migraine types
Introduction
In recent years, the emerging use of machine learning (ML) classification techniques in headache research has led to promising new understandings about different migraine types and subtypes [1]. ML is a branch of artificial intelligence focused on implementing computational algorithms that enable the recognition of patterns and relationships within data, achieving improved performance through learning and adaptation based on experience [2]. Unlike traditional statistical methods, ML emphasizes pattern recognition and predictive modeling through algorithms uncovering novel insights from intricate datasets [3]. ML comprises a range of task types, including classification, regression, clustering, and others, each fulfilling specific roles and applications in data analysis. Classification is an ML method used to categorize input data into predefined classes, aiming to train a model that effectively distinguishes between these classes based on input features [3]. However, it has been demonstrated that the outcomes of ML-based systems can be subject to systematic errors in their ability to classify subgroups of patients if there are no strategies for mitigating bias in ML research [4].
Moreover, published research papers in the migraine/ML field lack consistency in study design and external validation of presented ML models, which can misrepresent model accuracy and real-world applicability. This is no surprise because there are no recommendations regarding the design of these studies or the criteria for evaluating the quality of research articles dealing with ML classification in the migraine field. Herewith, this work aims to outline recommended criteria for evaluating the quality of those research papers.
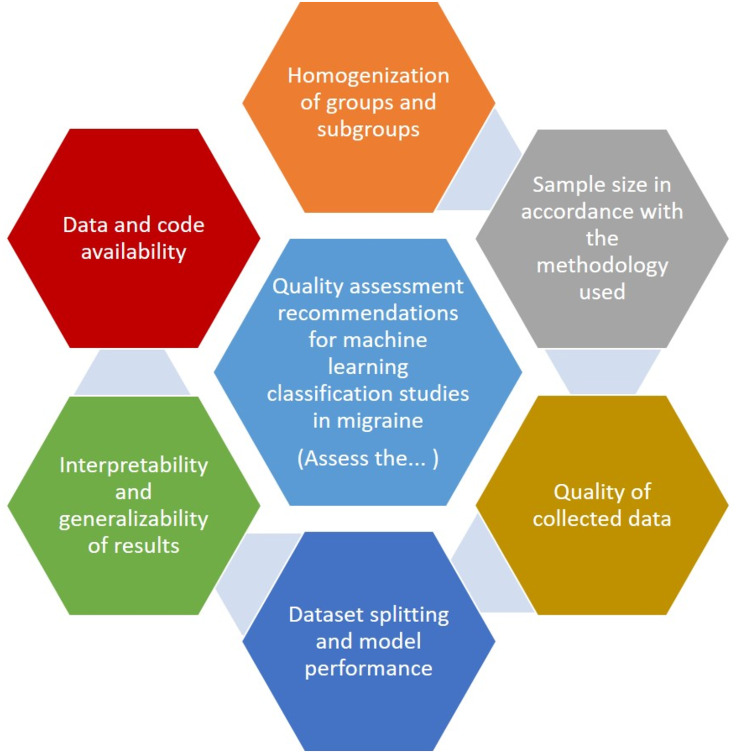
The recommendations are broken down into six categories (Fig. 1) and classified as obligatory (must be included) or preferably (strongly suggested but optional) using a unanimous decision-making approach among all authors.
Fig. 1.
Diagram of quality assessment recommendations for machine learning classification studies in migraine
Homogenized† group and/or subgroups according to
Clinical phenotypes
[5] (migraine without aura; migraine with only visual aura; migraine with somatosensory and dysphasic aura [with or without visual symptoms]; hemiplegic migraine; vestibular migraine, etc.).
Recommendations: Migraine without aura and migraine with aura should be differentiated into separate groups (obligatory); Differentiation into subgroups according to their deep profiled clinical phenotypes like migraine with typical aura with and without somatosensory and dysphasic aura, migraine with and without ictal or interictal allodynia [6], etc. (preferably).
Comment: The headache expert consortium should propose guidelines on what should be considered as a primary group versus a subgroup in ML classification studies within migraine research.
Attack frequency
Recommendations: Episodic and chronic migraine [7] should be differentiated into separate groups (obligatory); Episodic with low frequency (up to 7 days with headache/month), episodic with high frequency (from 8 to 14 days with headache/month), and chronic migraine, distinguishing in with and without medication overuse, should be differentiated into separate subgroups [8] (preferably).
* Patients must be differentiated into active (at least 1 migraine attack in the last year before collecting data for the study) and inactive disease states when included in studies [9].
Comorbidity
Recommendations: Differentiate patients without comorbidity (without neurological, cardiovascular, metabolic, and any other central and systemic conditions, including other types of primary and secondary headache) and patients with comorbidity [10] (obligatory).
Therapy (acute and preventive)
Recommendations: Differentiate patients into groups according to ongoing stable (at least 3 months) preventive treatments (any treatment with potential effect in migraine prophylaxis should be considered, regardless of the indication) [11] or not (obligatory); Differentiate patients into triptans super respondent, triptans respondent, and triptans no respondent (preferably) [12]; Differentiate patients in previous failed preventive treatment (naïve, 1–2 and ≥ 3) (preferably).
Demographic data
Recommendations: Train and validate machine learning models on separate groups according to sex (male and female) and age (< 18, 18 to 65, > 65) (preferably).
Comment: Consortium should propose a range of ages for subgroups.
† Homogenization of migraine patients into groups and subgroups is a challenging process that can lead to a loss of the model’s generalization capability on the one hand and a reduction in the number of patients in the cohort on the other hand. However, not taking all these heterogeneities into account will prevent a better understanding of complex pathophysiological mechanisms in various migraine subtypes and slow down the progress toward precision medicine in migraine treatment. Therefore, most of the recommendations in this category are marked as “preferably” and expert consensus in migraine and ML fields is needed. In our opinion, both ML training model approaches are needed, the ML model trained on large migraine general cohorts consisting of several clinical phenotypes and demographic backgrounds, as well as the ML model trained on highly homogenized migraine cohorts, to advance our knowledge in migraine multifaced pathophysiology and achieve precision medicine at the early stage of treatment [13]. In addition, it would be beneficial to include methods for managing heterogeneous data, such as data integration and batch effect correction techniques (e.g., ENIGMA toolbox that provides standardization and harmonization techniques for multi-site neuroimaging-genetic studies; harmonization via generalized additive models which can model scanner or site effects and adjust data while preserving biological variability; etc.) [14, 15], to enhance model applicability and generalizability.
Number of patients per group/subgroup – sufficient dataset
Recommendations: Sample size should meet the criteria for a confidence level of 95% [16] for the investigated group relative to the training sample (obligatory); Sample size should meet the criteria for a confidence level of 95% for the investigated subgroup relative to the training sample (preferably).
Comment: Consortium should propose sample sizes for groups and subgroups, bearing in mind that calculation should be based on ML classification accuracy, the types of collected data, and balancing sample sizes of groups/subgroups included in the classification task.
Enhancing data quality through data preprocessing techniques
Measurement or labeling of collected data
Recommendations: Measurement or labeling of collected data should be highly reliable and comply with recommended standards for the chosen technique (obligatory).
Comment: The consortium should propose validation strategies relevant to specific techniques, such as neuroimaging and electrophysiological modalities, as well as biomedical data. Considering that it is not one measure but rather several measures, and in most cases, they are not clearly defined, the consortium should recognize and minimize a bias of bad-quality data. Ultimately, a more significant multi-institutional collaborative effort is needed to establish and implement standards for data acquisition methods, ensuring that research results are interoperable and reliable for integration across different practice environments.
Missing data
Recommendations: Collected data should not have missing data, or it should be clearly stated how missing data was handled (such as the exclusion of variables, imputing them using machine learning, or treating missing values as a separate category) (preferably).
Outliers
Recommendations: When handling outliers, the rationale for either including or removing them should be transparently stated to prevent introducing artificial results (obligatory); Outliers may represent natural variations of the collected data and removing them might reduce the model’s ability to generalize. However, if they come from a different distribution, it might be better to treat them separately (preferably).
Comment: Consortium should propose guidelines on what should be defined as outliers in biomedical, neuroimaging, and electrophysiological data collected from migraine patients.
Feature selection
Recommendations: A detailed description of the steps for transforming raw data into features, as well as the techniques performed to reduce the dimensionality of the data, should be provided (obligatory); Approaches to exclude/select features from a dataset should be clearly described to improve transparency. Special attention should be paid to selecting features that could improve the results of the ML because they are a priori highly suggestive of one group/subgroup (i.e., if we build an ML to recognize the patients with migraine from healthy controls with some neurophysiological variables and add among the selected features the number of attacks/months or the Migraine disability assessment test score (MIDAS), the model will overperform not due to its ability to recognize neurophysiological variables per se but because some of the features are a priori highly suggestive of migraine, then resulting in a bias) (obligatory); Furthermore, methods to optimize feature selection (e.g., filter methods, wrapper methods, embedded methods, dimensionality reduction techniques, stability selection, etc.), with a particular focus on reducing information loss and preventing overfitting, should be included in the study design [17, 18] (preferably); Furthermore, the selection of relevant features should be based on plausible biological hypotheses/evidence, experts knowledge and supported by literature (obligatory); When available, guidelines for data preprocessing standardization processes published by specific consortiums for neuroimaging, electrophysiological, and/or biomedical data should be followed [14, 19, 20] (obligatory).
Training and testing data
Data splitting
Recommendations: The dataset should be split into training, validation and testing sets, and the ratio of the division clearly reported (obligatory).
* A common practice is to allocate 70% of the data for training, 15% for validation, and 15% for testing [21]. If alternative ratios are chosen, an explanation should be provided. Proper data splitting is crucial for developing a robust model (training set), fine-tuning model parameters (validation set), and getting an unbiased estimate of model performance (testing set). Furthermore, the use of stratified shuffle split is advised to ensure a proportional representation of categories within each subset of data; afterward, such a dataset could be evaluated using K-fold cross-validation.
Model selection
Recommendations: The criteria used for model selection should be clearly described. This might include the data characteristics, problem domain, and prior performance metrics (preferably).
Model finetuning
Recommendations: Model fine-tuning entails adjusting the model parameters to enhance its performance, often through hyperparameter selection. If hyperparameter tuning is implemented, the methodology should be clearly described (preferably).
* Initial boundaries for hyperparameters should be established through a trial-and-error approach. Subsequently, metaheuristic (e.g., genetic algorithms) or Bayesian methods may be employed to identify optimal hyperparameter values [22]. It should be noted that, according to the no free lunch theorem, a universal ML approach applicable to all datasets does not exist, therefore hyper-parameters’ optimization is necessary and metaheuristics proved to be very efficient in solving this non-deterministic polynomial hard task.
Overfitting
Recommendations: Methods used to control overfitting should be clearly described, such as regularization techniques and model simplification strategies (preferably).
* Overfitting is a common challenge in machine learning where a model learns noise instead of underlying patterns, leading to poor performance on unseen data [23]. To mitigate overfitting, it is recommended to use regularization techniques such as L1 and L2, which add penalties to the model’s complexity, and to implement early stopping, which halts training when validation performance declines. Adopting simpler models, utilizing data augmentation to increase training data diversity, and employing cross-validation methods like K-fold are also beneficial strategies. Continuous monitoring and evaluation of model performance on external datasets further help assess generalizability and reduce overfitting risks. By combining these approaches, more robust and reliable machine learning models can be developed. In case some of the mentioned techniques were used, it is necessary to state and explain the implementation procedure.
Model performance evaluation
Recommendations: Reporting accuracy and confusion matrices for all the datasets (training, validation and test) (obligatory); Reporting the area under the curve (AUC) of the Receiver Operating Characteristic (ROC) curve, F1 score, sensitivity, and specificity (preferably).
* Authors should clearly articulate the accuracy of the model and contextualize this value within the specific clinical or research setting [24]. A discussion should include how accuracy relates to the overall effectiveness of the model and the potential implications of its limitations, especially in scenarios involving imbalanced datasets. The confusion matrix should be leveraged to provide insights into the model’s performance across different classes. Authors are encouraged to analyze the values within the confusion matrix, focusing on true positives, false positives, true negatives, and false negatives. By interpreting these results, authors can identify specific strengths and weaknesses of the model, particularly how it may impact clinical decision-making. In discussing the F1-score, authors should focus on the balance between precision and recall, exploring how the obtained value reflects the model’s reliability. Insights into what the F1-score indicates about the model’s performance should be provided, helping readers understand its relevance in practical applications. Precision and recall should also be highlighted, with authors discussing the values obtained for these metrics and their implications for identifying positive cases. An exploration of the trade-offs between these two metrics can enhance understanding of the model’s performance, particularly in critical scenarios where both false positives and false negatives are significant concerns. The AUC is another vital aspect that should be discussed in terms of its implications for model discrimination. Authors should examine the AUC value and its relevance in assessing how well the model can differentiate between classes, emphasizing its role as a complementary metric to accuracy. Lastly, the discussion of the ROC curve and its AUC should emphasize the insights these tools provide regarding the model’s sensitivity and specificity. Authors should interpret the ROC curve concerning the obtained values, discussing what these results suggest about the model’s practical utility.
External validation
Recommendations: In ML investigations, model validation across new clinical settings is crucial. Many of ML’s biggest issues involve ‘overfitting’, where a model properly explains a training data set but fails to generalize. Showing that a model works in another patient cohort in the same healthcare system is important, but showing it works in a different setting is preferable. Replication is just the start of a protracted validation and dissemination process that depends on decades of diagnostics development experience [25–27] (preferably).
Feature importance
Recommendations: It is recommended that feature importance metrics be clearly presented and interpreted to enhance the transparency and interpretability of the model (preferably).
* Common methods for assessing feature importance include permutation importance, Gini importance, or Mean Decrease Impurity [28]. Understanding feature importance is crucial for identifying key predictors, guiding feature selection, and uncovering potential biomarkers or insights that can inform clinical decision-making. Additionally, presenting visualizations of feature importance, such as bar plots, can facilitate better understanding and aid in the interpretation of model predictions. Finally, ML models need to produce transparent explanations to effectively manage the benefits of ML methodology and allow the discovery of biomarkers and new predictors.
Interpretability of results
Interpretation
Recommendations: To enhance clinical applicability, the results should be interpreted in terms of their clinical relevance. Additionally, the most significant features should be analysed in relation to their corresponding underlying pathological changes and compared with relevant literature. Finally, the impact of these features on the discovery of new biomarkers should be discussed and address how findings could improve clinical practice (obligatory).
* Regardless of the ML methods employed during analysis, results should be interpreted clinically and in the context of the evaluation metrics defined in the study design. Moreover, high-impact features (e.g., key defining clinical features) should be presented in a summary along with a narrative rationale for focusing on these variables [29].
Future perspective
Recommendations: When study results are interpreted, future perspective should be offered. This could include plans for external validation using datasets from different centers or proposing new methodologies to enhance the explanation of results and validate feature importance. These approaches illuminate the next step toward a better understanding of migraine pathophysiology and may aid in discovering biomarkers for specific migraine subtypes (preferably).
Limitations
Recommendations: Limitations of the study should be recognized and discussed, such as reporting any unexplained model behaviors (preferably).
Data and code availability
Data Availability
Recommendations: The dataset should be made available upon request using data use agreements under clearly defined privacy rules (respecting general data protection regulation (GDPR)) approved by ethical oversight bodies [30] (obligatory); Anonymized data should be shared on the platform respecting FAIR principles (a set of guidelines for data management designed to make scientific data more Findable, Accessible, Interoperable, and Reusable) and protocols to secure patient information while allowing researchers to access valuable datasets [31, 32] (preferably).
Code Availability
Recommendations: Code should be shared on the platform or as a supplementary document (obligatory).
The above-proposed recommendations are only our reflection on this highly actual and important topic, born from our personal experience and profound study of existing literature. We hope this letter will echo in the headache and ML community and stimulate further discussions, which will lead to the formation of a consortium and definitive recommendations.
Acknowledgements
IP and AS are supported by the Ministry of Science, Technological Development and Innovation, Republic of Serbia (contract number for IP: 451-03-66/2024-03/200146; contract number for AS: 451-03-66/2024-03/200103).
Author contributions
I.P., A.S., K.M., N.B., G.S., D.S., G.C. - Conceptualization and writing.
Funding
This article received no external funding.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
IP serves as Junior and Guest Editor of The Journal of Headache and Pain and Head of Imaging Section of SN Comprehensive Clinical Medicine journal. GS serves as editorial board member of the Resident & Fellow Section of Neurology. GC serves as Associate Editor for The Journal of Headache and Pain, Cephalalgia, Cephalalgia Reports, BMC Neurology (Pain section), Frontiers in Neurology (Neurotechnology section), and Frontiers in Human Neuroscience (Brain Imaging and Stimulation section).
Footnotes
The original version of this article was revised: “Author name correction”.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/19/2024
A Correction to this paper has been published: 10.1186/s10194-024-01940-x
References
- 1.Ihara K, Dumkrieger G, Zhang P et al (2024) Application of Artificial Intelligence in the Headache Field. Curr Pain Headache Rep 28:1049–1057. 10.1007/s11916-024-01297-5 [DOI] [PubMed] [Google Scholar]
- 2.Jordan MI, Mitchell TM (2015) Machine learning: Trends, perspectives, and prospects. Science 349:255–260. 10.1126/science.aaa8415 [DOI] [PubMed] [Google Scholar]
- 3.Jovel J, Greiner R (2021) An introduction to Machine Learning Approaches for Biomedical Research. Front Med (Lausanne) 8:771607. 10.3389/fmed.2021.771607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vokinger KN, Feuerriegel S, Kesselheim AS (2021) Mitigating bias in machine learning for medicine. Commun Med (Lond) 1:25. 10.1038/s43856-021-00028-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Headache Classification Committee of the International Headache Society (IHS) (2018) The International Classification of Headache Disorders, 3rd edition. Cephalalgia 38:1–211. 10.1177/0333102417738202 [DOI] [PubMed]
- 6.Louter MA, Bosker JE, van Oosterhout WPJ et al (2013) Cutaneous allodynia as a predictor of migraine chronification. Brain 136:3489–3496. 10.1093/brain/awt251 [DOI] [PubMed] [Google Scholar]
- 7.Ford JH, Jackson J, Milligan G et al (2017) A real-world analysis of migraine: a cross-sectional study of Disease Burden and treatment patterns. Headache: J Head Face Pain 57:1532–1544. 10.1111/head.13202 [DOI] [PubMed] [Google Scholar]
- 8.Jedynak J, Eross E, Gendolla A et al (2021) Shift from high-frequency to low-frequency episodic migraine in patients treated with Galcanezumab: results from two global randomized clinical trials. J Headache Pain 22:48. 10.1186/s10194-021-01222-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccininni M, Brinks R, Rohmann JL, Kurth T (2023) Estimation of migraine prevalence considering active and inactive states across different age groups. J Headache Pain 24:1–10. 10.1186/S10194-023-01624-Y/FIGURES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altamura C, Coppola G, Vernieri F (2024) The evolving concept of multimorbidity and migraine. Handb Clin Neurol 199:535–566. 10.1016/B978-0-12-823357-3.00014-8 [DOI] [PubMed] [Google Scholar]
- 11.Puledda F, Sacco S, Diener H-C et al (2024) International Headache Society global practice recommendations for the acute pharmacological treatment of migraine. Cephalalgia 44:3331024241252666. 10.1177/03331024241252666 [DOI] [PubMed] [Google Scholar]
- 12.Sacco S, Lampl C, Amin FM et al (2022) European Headache Federation (EHF) consensus on the definition of effective treatment of a migraine attack and of triptan failure. J Headache Pain 23:133. 10.1186/s10194-022-01502-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrušić I, Ha W-S, Labastida-Ramirez A et al (2024) Influence of next-generation artificial intelligence on headache research, diagnosis and treatment: the junior editorial board members’ vision – part 1. J Headache Pain 25:151. 10.1186/s10194-024-01847-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larivière S, Paquola C, Park B-Y et al (2021) The ENIGMA Toolbox: multiscale neural contextualization of multisite neuroimaging datasets. Nat Methods 18:698–700. 10.1038/s41592-021-01186-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaramillo-Jimenez A, Tovar-Rios DA, Mantilla-Ramos Y-J et al (2024) ComBat models for harmonization of resting-state EEG features in multisite studies. Clin Neurophysiol 167:241–253. 10.1016/j.clinph.2024.09.019 [DOI] [PubMed] [Google Scholar]
- 16.Serdar CC, Cihan M, Yücel D, Serdar MA (2021) Sample size, power and effect size revisited: simplified and practical approaches in pre-clinical, clinical and laboratory studies. Biochem Med (Zagreb) 31:010502. 10.11613/BM.2021.010502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur A, Guleria K, Kumar Trivedi N (2021) Feature Selection in Machine Learning: Methods and Comparison. In: 2021 International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE). pp 789–795
- 18.Jović A, Brkić K, Bogunović N (2015) A review of feature selection methods with applications. In: 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO). pp 1200–1205
- 19.Rodrigues J, Weiß M, Hewig J, Allen JJB (2021) EPOS: EEG Processing Open-Source scripts. Front Neurosci 15. 10.3389/fnins.2021.660449 [DOI] [PMC free article] [PubMed]
- 20.Chahid I, Elmiad AK, Badaoui M (2023) Data Preprocessing For Machine Learning Applications in Healthcare: A Review. In: 2023 14th International Conference on Intelligent Systems: Theories and Applications (SITA). pp 1–6
- 21.Shujaaddeen AA, Mutaher Ba -Alwi F, Zahary AT, Sultan Alhegami A (2024) A Model for Measuring the Effect of Splitting Data Method on the Efficiency of Machine Learning Models: A Comparative Study. In: 2024 4th International Conference on Emerging Smart Technologies and Applications (eSmarTA). pp 1–13
- 22.Koul N, Manvi SS (2021) Framework for classification of cancer gene expression data using bayesian hyper-parameter optimization. Med Biol Eng Comput 59:2353–2371. 10.1007/s11517-021-02442-7 [DOI] [PubMed] [Google Scholar]
- 23.López OAM, López AM, Crossa J (2022) Verfitting, Model tuning, and evaluation of Prediction Performance. In: López OAM, López AM, Crossa J (eds) Multivariate Statistical Machine Learning methods for genomic prediction, 1st edn. Springer Cham, pp 109–139 [PubMed]
- 24.Hicks SA, Strümke I, Thambawita V et al (2022) On evaluation metrics for medical applications of artificial intelligence. Sci Rep 12:5979. 10.1038/s41598-022-09954-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabitza F, Campagner A, Soares F et al (2021) The importance of being external. Methodological insights for the external validation of machine learning models in medicine. Comput Methods Programs Biomed 208:106288. 10.1016/j.cmpb.2021.106288 [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Chen P-HC, Krause J, Peng L (2019) How to read Articles that Use Machine Learning: users’ guides to the Medical Literature. JAMA 322:1806–1816. 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 27.Doshi-Velez F, Perlis RH (2019) Evaluating machine learning articles. JAMA 322:1777–1779. 10.1001/jama.2019.17304 [DOI] [PubMed] [Google Scholar]
- 28.Saarela M, Jauhiainen S (2021) Comparison of feature importance measures as explanations for classification models. SN Appl Sci 3:272. 10.1007/s42452-021-04148-9 [Google Scholar]
- 29.Stevens LM, Mortazavi BJ, Deo RC et al (2020) Recommendations for reporting machine learning analyses in Clinical Research. Circ Cardiovasc Qual Outcomes 13:e006556. 10.1161/CIRCOUTCOMES.120.006556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.General Data Protection Regulation (GDPR) – Legal Text. In: General Data Protection Regulation (GDPR). https://gdpr-info.eu/. Accessed 4 Nov 2024
- 31.Wilkinson MD, Dumontier M, Aalbersberg IJ et al (2016) The FAIR Guiding principles for scientific data management and stewardship. Sci Data 3:160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinaci AA, Núñez-Benjumea FJ, Gencturk M et al (2020) The FAIRification Workflow for Health Research. Methods Inf Med 59:e21–e32. 10.1055/s-0040-1713684. From Raw Data to FAIR Data: [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Data Availability Statement
Data Availability
Recommendations: The dataset should be made available upon request using data use agreements under clearly defined privacy rules (respecting general data protection regulation (GDPR)) approved by ethical oversight bodies [30] (obligatory); Anonymized data should be shared on the platform respecting FAIR principles (a set of guidelines for data management designed to make scientific data more Findable, Accessible, Interoperable, and Reusable) and protocols to secure patient information while allowing researchers to access valuable datasets [31, 32] (preferably).
Code Availability
Recommendations: Code should be shared on the platform or as a supplementary document (obligatory).
The above-proposed recommendations are only our reflection on this highly actual and important topic, born from our personal experience and profound study of existing literature. We hope this letter will echo in the headache and ML community and stimulate further discussions, which will lead to the formation of a consortium and definitive recommendations.
Recommendations: The dataset should be made available upon request using data use agreements under clearly defined privacy rules (respecting general data protection regulation (GDPR)) approved by ethical oversight bodies [30] (obligatory); Anonymized data should be shared on the platform respecting FAIR principles (a set of guidelines for data management designed to make scientific data more Findable, Accessible, Interoperable, and Reusable) and protocols to secure patient information while allowing researchers to access valuable datasets [31, 32] (preferably).
No datasets were generated or analysed during the current study.