Abstract
Background
Efforts toward tuberculosis management and control are challenged by the emergence of Mycobacterium tuberculosis (MTB) resistance to existing anti-TB drugs. This study aimed to explore the potential of machine learning algorithms in predicting drug resistance of four anti-TB drugs (rifampicin, isoniazid, streptomycin, and ethambutol) in MTB using whole-genome sequence and clinical data from Uganda. We also assessed the model’s generalizability on another dataset from South Africa.
Results
We trained ten machine learning algorithms on a dataset comprising of 182 MTB isolates with clinical data variables (age, sex, HIV status) and SNP mutations across the entire genome as predictor variables and phenotypic drug-susceptibility data for the four drugs as the outcome variable. Model performance varied across the four anti-TB drugs after a five-fold cross validation. The best model was selected considering the highest Mathews Correlation Coefficient (MCC) and Area Under the Receiver Operating Characteristic Curve (AUC) score as key metrics. The Logistic regression excelled in predicting rifampicin resistance (MCC: 0.83 (95% confidence intervals (CI) 0.73–0.86) and AUC: 0.96 (95% CI 0.95–0.98) and streptomycin (MCC: 0.44 (95% CI 0.27–0.58) and AUC: 0.80 (95% CI 0.74–0.82), Extreme Gradient Boosting (XGBoost) for ethambutol (MCC: 0.65 (95% CI 0.54–0.74) and AUC: 0.90 (95% CI 0.83–0.96) and Gradient Boosting (GBC) for isoniazid (MCC: 0.69 (95% CI 0.61–0.78) and AUC: 0.91 (95% CI 0.88–0.96). The best performing model per drug was only trained on the SNP dataset after excluding the clinical data variables because intergrating them with SNP mutations showed a marginal improvement in the model’s performance. Despite the high MCC (0.18 to 0.72) and AUC (0.66 to 0.95) scores for all the best models with the Uganda test dataset, LR model for rifampicin and streptomycin didn’t generalize with the South Africa dataset compared to the GBC and XGBoost models. Compared to TB profiler, LR for RIF was very sensitive and the GBC for INH and XGBoost for EMB were very specific on the Uganda dataset. TB profiler outperformed all the best models on the South Africa dataset. We identified key mutations associated with drug resistance for these antibiotics. HIV status was also identified among the top significant features in predicting drug resistance.
Conclusion
Leveraging machine learning applications in predicting antimicrobial resistance represents a promising avenue in addressing the global health challenge posed by antimicrobial resistance. This work demonstrates that integration of diverse data types such as genomic and clinical data could improve resistance predictions while using machine learning algorithms, support robust surveillance systems and also inform targeted interventions to curb the rising threat of antimicrobial resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-10282-7.
Keywords: Machine learning, Antimicrobial resistance, Whole-genome sequence, Mutations, Mycobacterium tuberculosis, Clinical, Drug resistance, Genes
Introduction
The growing challenge of antimicrobial resistance (AMR) is a global public health emergency posing a great threat to modern medicine [1, 2]. AMR is associated with a negative effect on the economies of communities and countries most especially low-income and middle-income countries (LMICs) having a higher burden of infectious diseases [3]. Previous estimates in 2022, estimated 4.95 million deaths to be associated with AMR, including 1.27 million deaths attributed to bacterial AMR in 2019 [4]. While some researchers critique these forecasts [5], the World Health Organization (WHO) and numerous organizations recognize AMR as a pressing issue requiring a coordinated global response [4]. The WHO has established the Global Antimicrobial Resistance Surveillance System in 2015, marking the first global collaborative effort for AMR surveillance [6].
Mycobacterium tuberculosis (MTB) is the causative agent of tuberculosis (TB), a leading infectious disease with an estimated 1.6 million deaths annual deaths globally [7]. MTB is part of the Mycobacterium tuberculosis Complex which encompasses several lineages (L) causing TB in both humans and animals. While some of the MTBC human associated lineages are geographically widespread, others are more restricted and they are often referred to as M. tuberculosis sensu stricto (L1 - L4 and L7)), Mycobacterium africanum (L5, and L6), and a recently discovered L 8 [8, 9]. Standard TB treatment involves a 6-month regimen of four first-line drugs: isoniazid (INH), rifampicin (RIF), ethambutol (EMB) and pyrazinamide (PZA) [10]. However, the increasing prevalence of first-line drug resistance necessitates the use of second-line drugs for longer durations (at least 9 to 20 months) to treat multidrug-resistant TB (MDR-TB) [10].
The emergence and spread of drug-resistant TB (DR-TB) poses a major challenge to global TB control efforts [11]. In 2022, the WHO estimated that 410,000 people developed MDR-TB/Rifampicin-Resistant TB (RR-TB) and these accounted for 3.9 of the 10.6 million estimated incident TB cases for that year [11]. Effective management of DR-TB management requires multi-pronged approach encompassing rapid accurate detection, treatment, prevention, surveillance, and continuous program evaluation [12].
Current diagnostic methods for TB and DR-TB strains in Uganda include the GeneXpert MTB/RIF assay and conventional culture-based tests like Phenotypic Drug Susceptibility Testing [12–14]. However, these methods have limitations including focusing only on the principal mutations associated with rifampicin-resistance and have extended turnaround times [10, 12]. Recently, the WHO TB Supranational Reference Laboratory in Uganda has started performing Next-generation sequencing (NGS) for DR-TB especially for national drug resistance survey samples and potential extensively drug-resistant TB cases [15].
To address these limitations and enable faster identification and prediction of antibiotic resistance, researchers have explored various approaches. Conventional association rule methods based on whole genome sequencing (WGS) data have been used to identify variants associated with AMR but are also limited in detecting resistance by unknown mechanisms as they depend on pre-existing databases [10, 16]. In recent years, machine learning (ML) has emerged as a powerful tool for predicting resistance to different antibiotics using WGS data. A study in 3 African countries applied ML for E. coli resistance prediction suggesting broader applicability for DR-TB diagnosis in resource limited settings [17]. This aligns with existing research demonstrating the potential of ML for DR-TB prediction.
For instance, Green et al. developed a comprehensive framework utilizing a neural network to predict resistance patterns in Mycobacterium tuberculosis from genomic data [18]. Similarly, a study in the United Kingdom developed and compared traditional ML methods including support vector machine (SVM), logistic regression (LR) and random forests (RF) for predicting resistance to eight anti-TB drugs in a cohort of 1839 MTB isolates [19]. Using a cohort of 13,402 MTB isolates collected from 16 countries across 6 continents, several ML classifiers and linear dimension reduction techniques were developed to predict DR-TB for 11 drugs [20]. Also another study by Zhang et al. employed a deep convolutional neural network model for resistance diagnosis and a SVM model to identify resistance genes and mutations utilizing clinically collected MTB genomic data of MTB for PZA resistance [21]. Kuang et al. developed 24 binary classifiers for MTB drug resistance status across eight anti-MTB drugs using LR, RF and 1D CNN with a training dataset of 10,575 MTB isolates [10].
Despite the growing application of ML in TB research, most of this has been applied in the high-income countries with limited research done in the Low- and Middle-Income Countries (LMICs). Furthermore, prior methods often lack integration of clinical data, which is critical in LMIC contexts for capturing the full spectrum of factors influencing resistance. This study aims to evaluate the performance of ML algorithms in predicting drug resistance of MTB isolates using both genomic and clinical data from Uganda.
Materials and methods
Study design
This was a cross-sectional study utilizing data collected in the past years (2013 to 2023) to explore associations between predictors and outcomes from drug resistance using machine learning algorithms.
Sample size
In this study, we used two datasets referred to as the Uganda (UG) data and the South Africa (SA) (Table 1) and (Fig. 1).
Table 1.
Overview of the antimicrobial resistance phenotype data for the four drugs: the number of isolates that are resistant or susceptible
| Drug | RIF | INH | EMB | STM | ||||
|---|---|---|---|---|---|---|---|---|
| Source | UG | SA | UG | SA | UG | SA | UG | SA |
| Resistant | 131 | 210 | 93 | 173 | 68 | 128 | 80 | 141 |
| Susceptible | 75 | 26 | 113 | 63 | 138 | 108 | 126 | 95 |
| Total | 206 | 236 | 206 | 236 | 206 | 236 | 206 | 236 |
Abbreviations* UG – Uganda; SA – South Africa
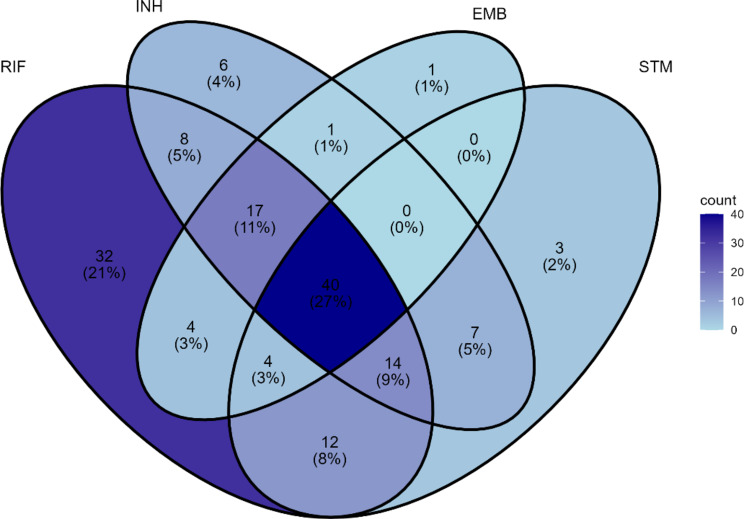
Fig. 1.
Venn diagram quantifying the number of instances of co-occurrence of resistance between the four drugs in the UG dataset
Data description
In this study we used WGS data and corresponding clinical data consisting of age, sex, and HIV status. We then focused on four anti-TB drugs; rifampicin (RIF), isoniazid (INH), ethambutol (EMB) and Streptomycin (STM). Three of these drugs (RIF, INH, EMB) are known as first-line drugs for TB treatment and STM belongs to the aminoglycoside class of drugs. These drugs were used because data on resistance was available in the two datasets and also because increasing prevalence to resistance to these drugs.
Uganda dataset
The UG dataset comprised of WGS of 226 MTB isolates that was corresponded to phenotypic information for the anti-TB drugs and associated clinical data (age, sex and HIV status) (Table 2). This data was made publicly available from the sequence read archive database [22] by various studies (Table 3).
Table 2.
Overview of the patient characteristics from the Uganda dataset
| Characteristic | Frequency(N) |
|---|---|
| Age Range (years) | |
| 13–29 | 74 |
| 30–39 | 51 |
| 40+ | 44 |
| SEX | |
| Male | 96 (45.07%) |
| Female | 74 (34.74%) |
| Missing | 43 (20.19%) |
| HIV Status | |
| Positive | 75 (35.21%) |
| Negative | 94 (44.13%) |
| Missing | 44 (20.66%) |
Table 3.
Description of the studies from which UG data was obtained
| Study | Bio project | Sample size | Country |
|---|---|---|---|
| Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing [23] | PRJEB2424 | 51 | Uganda |
| High Genotypic Discordance of Concurrent Mycobacterium tuberculosis Isolates from Sputum and Blood of HIV-Infected Individuals [24] | PRJEB10577 | 26 | Uganda |
| Whole genome sequencing to complement drug resistance surveys in Uganda [25] | PRJEB10533 | 90 | Uganda |
| Whole Genome Sequencing-based Characterization of Mycobacterium tuberculosis Isolated from HIV-Seropositive Ugandans with Tuberculosis and CD4 + T-Cell Counts of 0-1150 Cells/µL | PRJNA481638 | 59 | Uganda |
| First report of Whole-genome analysis of an extensively drug-resistant Mycobacterium tuberculosis clinical isolate with Bedaquiline, Linezolid and Clofazimine resistance from Uganda [15] | PRJNA749651 | 2 | Uganda |
South Africa dataset
The SA dataset consisted of WGS data obtained from patients in KwaZulu-Natal, South Africa who were recruited with drug-resistant TB as they had previously experienced the worst outcomes and most problems with acquired resistance, as well the drug susceptible cohort [26]. Out of the 399 MTB isolates, only 236 were included in the study as they had complete phenotypic information from the four drugs. All WGS data are available under the NCBI SRA Bio Project PRJNA559528.
Genome processing
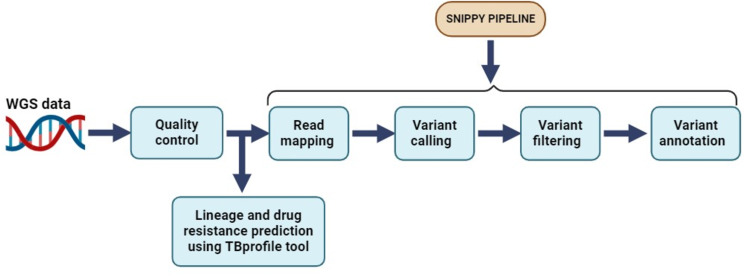
The quality of the raw WGS reads was assessed using FastQC v0.11.3 [27] and trimmomatic v0.39 for adapter clipping, quality trimming and minimum length exclusion (LEADING:3, TRAILING:3 SLINDINGWINDOW:4:20 MINLEN:25) [28]. Read taxonomy investigation using kraken v.2.1.3 [29] and bracken v.2.9 [30] was performed to assess for read contamination and a total of 23 reads were excluded with less 95% proportion of MTB. The trimmed reads were then mapped to a reference genome (H37Rv; NC_000962.3) using Snippy v4.6.0 which is a variant calling and core genome alignment pipeline [31]. This pipeline begins by mapping the reads to the reference genome using BWA MEM v0.7.17 [32] generating a sequence alignment file (SAM). The SAM file is then processed by samtools v.1.18 [33] generating a binary alignment file (BAM) from which variants are called using Freebayes v.1.3.6 [34] with default values for key parameters (--mincov –minfrac –minqual). These variants were subsequently annotated using SnpEff v.5.0 [35]. Additionally, we used the default parameters of TB Profiler too v4.3.0 for lineage and drug resistance prediction using the trimmed reads [36]. The entire bioinformatics analysis workflow (Fig. 2) was executed on the Open Science Grid High Throughput Computing infrastructure [37, 38].
Fig. 2.
Illustration of the bioinformatics analysis workflow
Phylogenetics analysis
Using the Snippy-core, we generated a core SNP alignment file that we used to obtain the phylogenetic tree. Prior to obtaining the core SNP alignment file we masked repetitive regions (PE/PPE/PGRS genes) of the genome using the bed file provided by Snippy. We also filtered for recombination regions using gubbins [39] and obtained the core snp alignment file containing the polymorphic sites using snp-sites v.2.5.1 [40] tools. Using FastTree v.2.1.11 [41], we generated the maximum likelihood tree with a generalized time-reversible model. The phylogenetic tree obtained was visualized using iTOL v6 (Interactive Tree Of Life) (https://itol.embl.de/).
SNPs pre-processing and encoding
We utilized the bcftools [42] to extract the reference alleles, alternate alleles and their chromosome positions from the individual filtered VCF files obtained from the snippy pipeline and merged all the isolates based on position of the reference alleles [17]. The final output was a SNP matrix which consisted of samples represented in the rows and the variant alleles as columns.
One hot encoding was employed to encode the SNP matrix, where each sequence variation was represented as a binary vector. The presence of a specific allele at a given chromosome position was denoted by a ‘1’, while the absence was denoted by a ‘0’. This approach allowed us to effectively capture variants across the entire genome for each of the MTB isolates included in the study and also transformed the categorical data indicated by A, C, G, T into a numerical format which is suitable for the ML analyses. We also denoted the missing values represented as N in the SNP matrix with a ‘0’. To assess the generalizability of the ML models on the unseen data, we obtained a homogeneous set of variants between the training (UG dataset) and the validation dataset (SA dataset). Using the similar technique, the antibiotic phenotypes for key drugs were represented a binary vector where the ‘S’ for susceptible was denoted by a ‘0’ and ‘R’ for resistant by a ‘1’.
Machine learning for antimicrobial resistance prediction
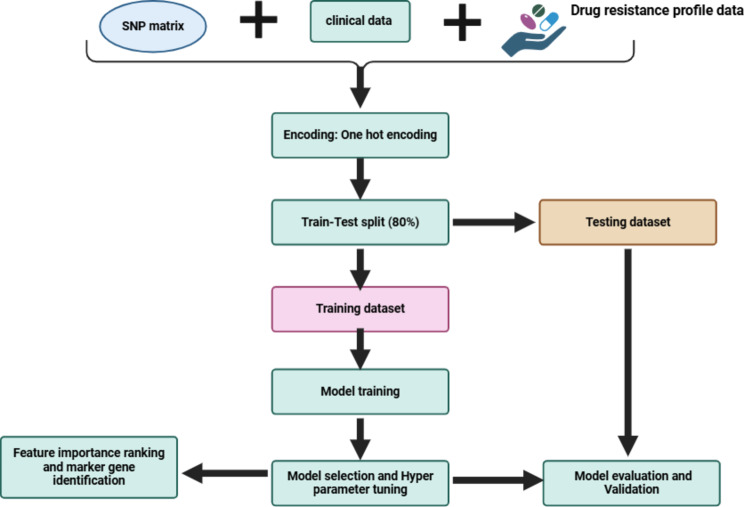
We trained ten machine learning algorithms implemented in the Scikit-learn version 1.3.2 [43]on a combined dataset consisting of 4994 variants across the entire MTB genome and clinical data variables (age, sex and HIV status) (Fig. 3). However, since we had missing values within our dataset, we excluded 21 MTB isolates of which 19 had no antimicrobial resistance phenotype data for the four drugs (RIF, INH, EMB and STM) and 2 had empty VCF files after filtering for low quality variants. In addition, we imputed the missing data for the clinical data variables using the simple imputer module with either mean strategy for numerical data or most frequent strategy for categorical data in order to obtain a complete dataset. For each drug, the dataset consisting of 182 MTB isolates was split into training set (145 MTB isolates) that comprised 80% of the resistant and susceptible isolates respectively and the remaining 20% to the testing set (37 MTB isolates).
Fig. 3.
Illustration of machine learning workflow
All the ML models included had unique capabilities and were implemented using the default parameters, focusing on each antibiotic at a time to ensure accurate predictions. Logistic Regression (LR) is a simple, fast and easy model to implement for binary classification tasks. Decision Trees (DT), Extra Trees Classifier (ETC) and Random Forest are tree-based models that construct tree-like structures to make predictions. However, ETC and RF aggregate multiple decision trees, reducing overfitting which is one of the problems arising from using DT. Support Vector Machines (SVM) can perform well with small datasets, handle high dimensional data and have a good generalization performance on unseen data. Boosting classifiers such as Adaptive Boosting (AdaBoost) and Gradient Boosting (GBC) improve the predictive performance by training a sequence of weak models, each compensating the weaknesses of its predecessors in order to make a strong ML model. CatBoost are employed for their state -of-art performance in handling categorical data variables and Extreme Gradient Boosting (XGBoost) is a powerful gradient-boosting algorithm known for its versatility and effectiveness in handling large datasets and complex feature interactions. Multilayer Perceptron (MLP), a neural network captures complex non-linear relationships in the data.
All models were trained on both originally imbalanced and balanced datasets. First, we trained the ML models on SNP data only and combined dataset to assess if integration of the clinical data improves the model performance. For balancing, we applied the up-sampling strategy using the Synthetic Minority Over-sampling Technique (SMOTE) technique on the training dataset. This balancing technique is crucial most especially when dealing with data in real-world scenarios as it prevents models from becoming biased towards the majority class, thereby enhancing performance.
Model selection and evaluation on the Uganda test dataset
The performance of the ML models was evaluated using various key metrics such as accuracy, precision, recall (sensitivity), specificity, F1 score, MCC, receiver operating characteristics curve (ROC) and the area under the curve (AUC) after a five-fold cross-validation was performed on the training set. Each metric was calculated along with its 95% confidence intervals (CI) using the bootstrap method. The best model was selected considering the highest MCC and AUC score as key metrics, the parameters of the model were then optimized through cross-validation on the training set while using the gridsearchCV technique This is because the MCC metric considers all the elements of a confusion matrix for instance the True Negative (TN), True Positive (TP), False Positive (FP) and False Negative (FN). Additionally, the AUC score shows the ability of the ML models to distinguish between the classes and it’s plotted as the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings.
The LR model achieved the optimal parameters as a regularization strength (C) of 10 and maximum number of iterations (max_iter:300) for the RIF drug and C of 5 and maximum number of iterations (max_iter:500) for the STM drug. For EMB, XGBoost had optimal parameters as learning rate: 0.3, maximum depth(max_depth):3, number of estimators (n_estimators):100 and subsample:1.0. Finally, for INH, GBC model achieved the optimal parameters as learning rate: 0.5, maximum depth(max_depth):9, number of estimators (n_estimators):100 and subsample:0.7. The performance of these best models for each drug were then evaluated on the test Uganda dataset while using the optimal parameters obtained.
Model evaluation on the external dataset (South Africa)
To assess the generalizability of the best ML models, we assessed the performance of the best ML models in predicting DR on the SA dataset (validation dataset). This dataset consisted of 236 samples with a severe class imbalance issue that varied across the antibiotics.
Feature importance ranking and marker gene identification
To identify the top ten most important features for the best model, we used the feature importance attribute for the tree-based models like XGBoost, GBC which quantifies the contribution of each feature to the model’s prediction. For LR, we calculated the absolute values of the coefficients to identify feature importance scores. We annotated the identified the SNPs and extracted the corresponding gene annotations from annotated VCF files in order to obtain the functional consequence of these genes and to further investigate their potential contribution to drug resistance mechanisms in MTB.
Results
Genetic diversity of the UG sequence data: MTB lineages, sublineage distribution and drug resistance types
The MTB UG isolates were classified into five Lineages(L) where L4 was the most predominant with n = 149/203 followed by L3 (46/203), L2(6/203), L3&L4(1/203) and finally L1(1/203) (Table 4) The most predominant sublineage observed was L4.6.1.1(T2-Uganda or Uganda II) with n = 36/203 (17.73%) belonging to the Uganda genotype (L4.6.1 or T2-Uganda; T2) compared to the other sub lineages Among the 203 MTB isolates, 37.93% were susceptible to all anti TB drugs, 2.96% were purely resistant to Rifampicin (RR-TB), 0.49% were Pre-XDR-TB, 32.51% were MDR-TB, 0.99% were XDR-TB, 22.66% were mono resistant to Isoniazid (HR-TB) and 2.46% were classified as Others (Table 5).
Table 4.
MTB lineage and sublineage distribution
| Lineage (L) | N (203) | Sublineage | N (203) |
|---|---|---|---|
| L4 | 149 | L4.6.1.1/Uganda II | 36 |
| L4.6.1.2/Uganda I | 27 | ||
| L4.4.1.1 | 17 | ||
| L4.3.4.2.1/LAM | 15 | ||
| L4.1.1.3/X | 12 | ||
| L4.3.4.2 | 12 | ||
| L4.8 | 8 | ||
| L4.6.1 | 5 | ||
| L4.3.3/LAM | 4 | ||
| L4.2.2.2 | 3 | ||
| L4.1.2.1/Haarlem | 2 | ||
| L4.9 | 2 | ||
| L4.1.1.1/X | 1 | ||
| L4.6.1.1; L4.3.4.2 | 1 | ||
| L4.6.1.2; L4.2.2.2 | 1 | ||
| L4.6.1.2; L4.3.4.2.1 | 1 | ||
| L4.6.1.2; L4.6.1.1 | 1 | ||
| L4.7 | 1 | ||
| L3 | 46 | L3 | 27 |
| L3.1.2.1 | 12 | ||
| L3.1.1 | 7 | ||
| L2 | 6 | L2.2.1 | 6 |
| L3; L4 | 1 | L4.6.1.2; L3 | 1 |
| L1 | 1 | L1.1.2 | 1 |
Table 5.
Distribution of the drug-resistant TB profiles
| DR-TB type | N = 203(%) |
|---|---|
| HR-TB | 46(22.66) |
| MDR-TB | 66(32.51) |
| Other | 5(2.46) |
| Pre-XDR-TB | 1(0.49) |
| RR-TB | 6(2.96) |
| Sensitive | 77(37.93) |
| XDR-TB | 2(0.99) |
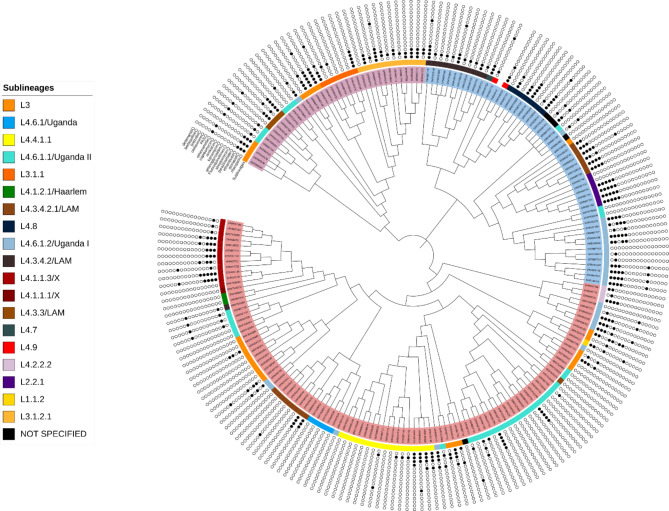
A maximum likelihood phylogenetic tree of 203 MTB isolates constructed using all genome-wide SNPs revealed the expected clustering by sub lineage (Fig. 4).
Fig. 4.
A maximum likelihood phylogenetic tree of 203 MTB UG isolates showing three major clades alongside the sublineage and individual drug resistance profile for the anti TB drugs obtained from TB profiler with the white colour indicating absence and the black colour indicating presence of resistance mutation for the drug
Comparison of the performance of the ML models in predicting AMR on the SNP data only and a combined dataset (SNP data and clinical data)
The mean scores for each of the metrics (accuracy, recall, precision, MCC, F1_score, ROC_AUC), after a five-fold cross validation on the training set consisting of SNP data only and combined dataset respectively (Tables 6 and 7). In this study we focused on the MCC score and AUC score as our metrics of performance. Generally, there were marginal changes in ML model performance (increase or decrease by 0.01or 0.02) across all metrics which was consistent across all the drugs. For drugs INH and RIF, there a notable increase MCC score (INH:0.17 and RIF:0.26) and AUC score (INH:0.60 and RIF:0.66) after Intergrating the clinical and SNP data compared to the other drugs (EMB and STM).
Table 6.
Performance of the best three ML models on only SNP data
| Drug | Model | Recall (Sensitivity) (95% CI) |
Specificity (95% CI) |
ROC_AUC (95% CI) |
MCC (95% CI) |
|---|---|---|---|---|---|
| RIF | LR | 0.88 (0.80–0.93) | 0.84 (0.69–0.95) | 0.92 (0.89–0.96) | 0.71 (0.64–0.78) |
| CatBoost | 0.91 (0.87–0.94) | 0.73 (0.52–0.86) | 0.90 (0.88–0.92) | 0.65 (0.52–0.74) | |
| GBC | 0.88 (0.83–0.93) | 0.74 (0.61–0.88) | 0.89 (0.87–0.91) | 0.63 (0.55–0.71) | |
| INH | GBC | 0.74 (0.68–0.82) | 0.89 (0.79–0.99) | 0.89 (0.85–0.93) | 0.65 (0.52–0.78) |
| XGBoost | 0.77 (0.68–0.86) | 0.79 (0.67–0.92) | 0.86 (0.81–0.90) | 0.58 (0.42–0.69) | |
| CatBoost | 0.66 (0.58–0.75) | 0.85 (0.75–0.96) | 0.85 (0.80–0.89) | 0.54 (0.42–0.68) | |
| EMB | CatBoost | 0.59 (0.38–0.74) | 0.91 (0.84–0.97) | 0.84 (0.74–0.93) | 0.53 (0.28–0.72) |
| AdaBoost | 0.63 (0.42–0.78) | 0.87 (0.83–0.92) | 0.83 (0.77–0.89) | 0.52 (0.35–0.63) | |
| GBC | 0.58 (0.41–0.72) | 0.87 (0.82–0.92) | 0.83 (0.75–0.91) | 0.47 (0.31–0.64) | |
| STM | GBC | 0.47 (0.40–0.57) | 0.80 (0.74–0.85) | 0.70 (0.64–0.76) | 0.29 (0.26–0.31) |
| XGBoost | 0.50 (0.33–0.66) | 0.75 (0.66–0.83) | 0.70 (0.63–0.78) | 0.25 (0.08–0.45) | |
| CatBoost | 0.34 (0.26–0.42) | 0.81 (0.74–0.85) | 0.65 (0.55–0.73) | 0.17 (0.06–0.27) |
Table 7.
Performance of the best three ML models on a combined dataset (SNP data and clinical data)
| Drug | Model | Recall (Sensitivity) (95% CI) |
Specificity (95% CI) |
ROC_AUC (95% CI) |
MCC (95% CI) |
|---|---|---|---|---|---|
| RIF | XGBoost | 0.92 (0.87–0.96) | 0.77 (0.67–0.89) | 0.93 (0.90–0.97) | 0.71 (0.64–0.80) |
| CatBoost | 0.91 (0.87–0.94) | 0.72 (0.62–0.82) | 0.93 (0.90–0.95) | 0.66 (0.60–0.71) | |
| LR | 0.88 (0.81–0.95) | 0.84 (0.70–0.95) | 0.93 (0.89–0.97) | 0.73 (0.65–0.81) | |
| INH | GBC | 0.73 (0.65–0.83) | 0.91 (0.83–0.99) | 0.89 (0.85–0.93) | 0.66 (0.57–0.76) |
| XGBoost | 0.77 (0.70–0.84) | 0.82 (0.73–0.91) | 0.87 (0.83–0.92) | 0.60 (0.44–0.70) | |
| CatBoost | 0.68 (0.57–0.79) | 0.88 (0.79–0.96) | 0.85 (0.80–0.89) | 0.57 (0.45–0.72) | |
| EMB | CatBoost | 0.58 (0.42–0.74) | 0.90 (0.83–0.97) | 0.84 (0.73–0.94) | 0.51 (0.25–0.70) |
| XGBoost | 0.64 (0.51–0.73) | 0.87 (0.80–0.94) | 0.82 (0.75–0.89) | 0.54 (0.36–0.68) | |
| GBC | 0.60 (0.47–0.72) | 0.84 (0.79–0.89) | 0.82 (0.74–0.90) | 0.46 (0.33–0.60) | |
| STM | XGBoost | 0.54 (0.45–0.64) | 0.75 (0.67–0.81) | 0.71 (0.69–0.73) | 0.30 (0.23–0.36) |
| GBC | 0.47 (0.35–0.58) | 0.84 (0.77–0.90) | 0.70 (0.67–0.74) | 0.36 (0.23–0.49) | |
| DT | 0.60 (0.53–0.70) | 0.75 (0.68–0.82) | 0.68 (0.61–0.75) | 0.33 (0.24–0.41) |
Evaluation of the ML models on the up sampled UG dataset
The models showed a substantial increase in all the metrics after a five-fold cross validation on the up sampled dataset for all the drugs (Table 8). For RIF, the best model was LR, it had the highest MCC (0.83) with AUC score as (0.96), recall (0.90), precision (0.93), specificity (0.93) and f1score (0.91). For INH, GBC achieved the highest MCC (0.69) with AUC score as (0.91), recall (0.76), precision (0.90), specificity (0.91) and f1 score (0.82). For EMB, XGBoost achieved the highest MCC (0.65), with AUC score as (0.90), recall (0.79), precision (0.85), specificity (0.85) and f1 score (0.81). For STM, LR and Adaboost shared the same MCC score (0.44) which was the highest. However, LR was selected as the best model as it had a higher AUC score (0.80) compared to that of Adaboost (0.77). LR also had recall (0.70), precision (0.73), specificity (0.73) and f1 score (0.71) (Table 8).
Table 8.
Evaluation of the ML models on the up sampled UG dataset
| Drug | Model | Recall (sensitivity) (95% CI) |
Specificity (95% CI) |
ROC_AUC (95% CI) |
MCC (95% CI) |
|---|---|---|---|---|---|
| RIF | XGBoost | 0.89 (0.83–0.94) | 0.91 (0.86–0.95) | 0.97 (0.95–0.98) | 0.79 (0.72–0.87) |
| LR | 0.90 (0.86–0.92) | 0.93 (0.86–0.96) | 0.96 (0.95–0.98) | 0.83 (0.73–0.86) | |
| GBC | 0.83 (0.77–0.93) | 0.90 (0.85–0.95) | 0.96 (0.93–0.98) | 0.73 (0.64–0.85) | |
| INH | GBC | 0.76 (0.67–0.87) | 0.91 (0.81–0.97) | 0.91 (0.88–0.96) | 0.69 (0.61–0.78) |
| CatBoost | 0.74 (0.62–0.85) | 0.85 (0.72–0.96) | 0.89 (0.84–0.95) | 0.61 (0.49–0.76) | |
| XGBoost | 0.76 (0.69–0.84) | 0.81 (0.63–0.92) | 0.88 (0.83–0.94) | 0.59 (0.46–0.72) | |
| EMB | XGBoost | 0.79 (0.74–0.83) | 0.85 (0.76–0.95) | 0.90 (0.83–0.96) | 0.65 (0.54–0.74) |
| GBC | 0.82 (0.77–0.86) | 0.77 (0.69–0.85) | 0.88 (0.83–0.92) | 0.59 (0.50–0.67) | |
| CatBoost | 0.83 (0.71–0.87) | 0.80 (0.73–0.91) | 0.87 (0.83–0.91) | 0.63 (0.46–0.74) | |
| STM | XGBoost | 0.71 (0.64–0.78) | 0.68 (0.57–0.78) | 0.82 (0.77–0.85) | 0.40 (0.30–0.48) |
| GBC | 0.77 (0.71–0.76) | 0.64 (0.61–0.68) | 0.80 (0.77–0.84) | 0.42 (0.36–0.41) | |
| LR | 0.70 (0.59–0.79) | 0.73 (0.65–0.83) | 0.80 (0.74–0.82) | 0.44 (0.27–0.58) |
Performance of the best performing ML algorithms for predicting DR on Uganda data
The best performing models for each drug were trained on up sampled SNP dataset only because adding the clinical data features didn’t significantly improve the model performance. Generally, all the models demonstrated good predictive performance with MCC scores ranging from 0.18 to 0.72 and AUC scores from 0.66 to 0.95 across the four drugs (Table 9). Performance metrics with confidence intervals for each ML model on this dataset are shown in Table 9.
Table 9.
Performance of the ML algorithms in predicting DR on the UG dataset (test dataset)
| Drug | Best ML model | Recall (Sensitivity) (95% CI) | Specificity (95%CI) | ROC AUC (95% CI) | MCC (95% CI) |
|---|---|---|---|---|---|
| RIF | LR | 0.75(0.57–0.91) | 1.00(1.00–1.00) | 0.95(0.87–0.99) | 0.72(0.54–0.90) |
| INH | GBC | 0.76(0.54–0.94) | 0.75(0.55–0.94) | 0.82(0.67–0.94) | 0.51(0.23–0.78) |
| EMB | XGBoost | 0.77(0.53- 1.00) | 0.92(0.79- 1.00) | 0.89(0.72- 1.00) | 0.69(0.43–0.90) |
| STM | LR | 0.41(0.15–0.67) | 0.77(0.58–0.94) | 0.66(0.48–0.84) | 0.18(-0.14- 0.48) |
Performance of the best performing ML algorithms for predicting DR on South Africa data
The LR model didn’t generalize well on the SA dataset achieving an MCC of 0.02 (95% CI: -0.25 -0.01) to -012 (95% CI: -0.25 -0.01) and ROC-AUC of 0.63 (95% CI: 0.52–0.73) to 0.46 (95% CI: 0.39–0.53) when compared to GBC for INH and XGBoost for EMB (Table 10).
Table 10.
Performance of the ML algorithms in predicting DR on the SA dataset (validation dataset)
| Drug | Best ML model | Recall (Sensitivity) (95% CI) | Specificity (95%CI) | ROC AUC (95% CI) | MCC (95% CI) |
|---|---|---|---|---|---|
| RIF | LR | 0.94(0.90–0.97) | 0.08(0.00- 0.20) | 0.63(0.52–0.73) | 0.02(-0.09- 0.17) |
| INH | GBC | 0.68(0.60–0.74) | 0.81(0.71–0.91) | 0.73(0.65–0.80) | 0.43(0.32–0.54) |
| EMB | XGBoost | 0.51(0.42–0.60) | 0.83(0.76–0.90) | 0.76(0.69–0.81) | 0.35(0.23–0.47) |
| STM | LR | 0.43(0.35–0.51) | 0.45(0.35–0.55) | 0.46(0.39–0.53) | -0.12(-0.25-0.01) |
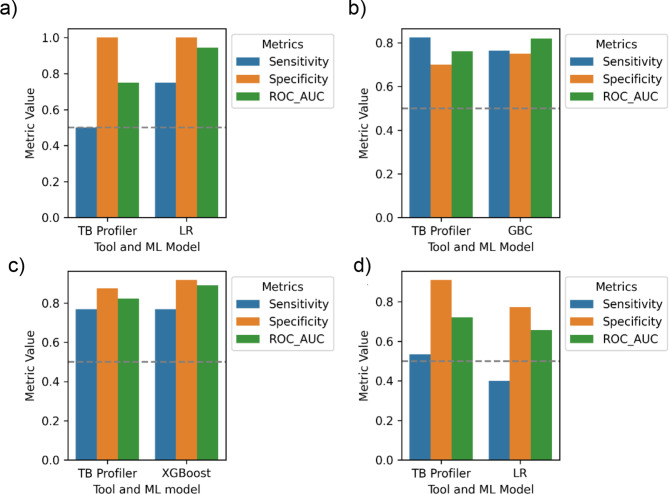
Benchmarking the best ML models against TB profiler on the UG dataset
We compared the predictive performance of the best ML models for four drugs (RIF, INH, EMB and STM) using key metrics sensitivity, specificity and ROC-AUC against WHO catalogue based tool TB profiler. Generally, all the best models had a higher ROC-AUC score compared to TB profiler (Fig. 5) with exception of LR model for STM. The mean LR sensitivity (0.75) for RIF is significantly higher than that of TB profiler (0.5). XGBoost for EMB showed a similar performance as TB profiler in terms of mean sensitivity (0.77) but achieved a higher mean specificity of 0.92 compared to TB profiler (0.88). Despite the fact that, TB profiler achieved a higher mean sensitivity (0.82) than GBC (0.76) for INH, it’s mean specificity (0.7) was slower than that of GBC (0.75).
Fig. 5.
The comparison of the performance of the best ML models and TB profiler in predicting DR on the UG dataset across different drugs. (a) RIF, (b) INH, (c) EMB, (d) STM
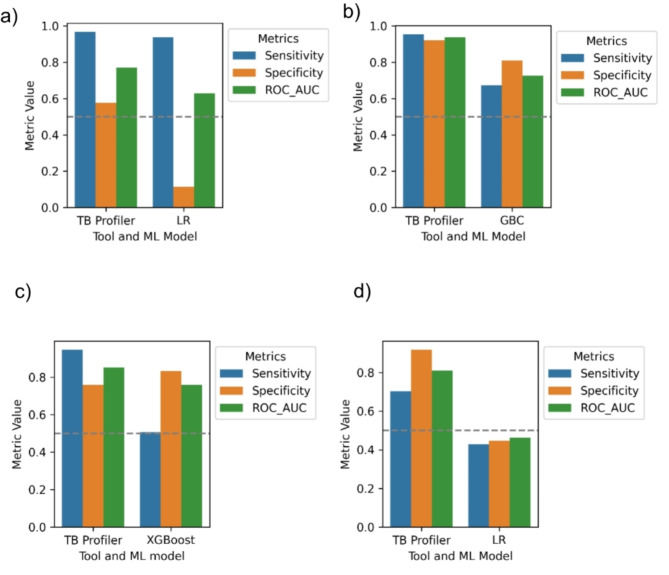
Benchmarking the best ML models against TB profiler on the SA dataset
Generally, the TB profiler tool achieved a higher sensitivity, specificity and ROC-AUC score when compared to the LR model for RIF and STM, GBC for INH and XGBoost for EMB (Fig. 6).
Fig. 6.
The comparison of the performance of the best ML models and TB profiler in predicting DR on the SA dataset across different drugs. (a) RIF, (b) INH, (c) EMB, (d) STM
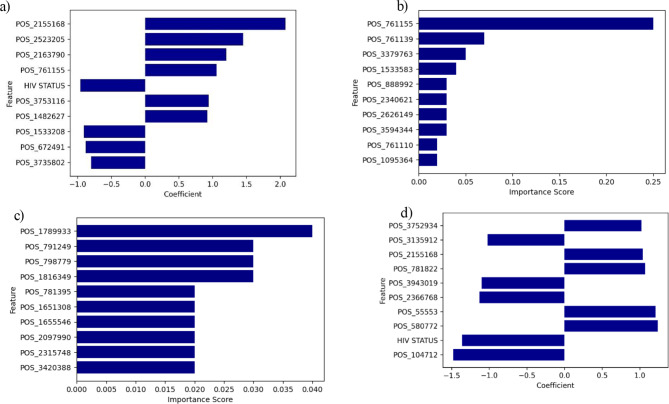
Identification of the top significant features for the best model
Figure 7 shows the top ten important features selected for each drug by considering the top performing models. In addition to the SNP positions, HIV status was identified among the top 10 important features for RIF and STM with best models as LR however it had a negative coefficient score value. Some features were shared across some drugs such as SNP position 761,155 (gene annotations) (RIF, EMB and INH) and 2,523,205 (gene annotations) (EMB and RIF) (Fig. 7).
Fig. 7.
Feature importance scores for the best performing models across different drugs. (a) RIF, (b) INH, (c) EMB, (d) STM
Marker genes and mutations associated with drug resistance
The ten most important SNPs for each antibiotic as shown in (Fig. 7) where annotated and analyzed. The corresponding genes and mutations identified are shown in the (Table 11). Some of the mutations we identified are in well-known genes conferring antibiotic resistance, such as rpoB which confers RIF resistance, katG which is related to INH resistance and rpsL gene which is associated with STM resistance.
Table 11.
SNP mutations and corresponding marker genes associated with drug resistance
| Drug | SNP Position | SNP Annotation | Mutation | Gene name | |
|---|---|---|---|---|---|
| INH, RIF | 761,155 | missense | p.Ser450Trp | rpoB | |
| INH | 761,139 | missense | p.His445Asp | rpoB | |
| INH | 3,379,763 | intergenic_region | n.3379763G > A | Rv3020c-Rv3022A | |
| INH | 1,533,583 | synonymous | p.Tyr17Tyr | PPE19 | |
| INH | 888,992 | intergenic_region | n.888,992 A > C |
Rv0794c- Rv0797 |
|
| INH | 2,340,621 | missense | p.Pro638Arg | Rv2082 | |
| INH | 2,626,149 | synonymous | p.Gly8Gly | esxO | |
| INH | 3,594,344 | intergenic_region | n.3594344G > A | Rv3217c-Rv3218 | |
| INH | 761,110 | missense | p.Asp435Val | rpoB | |
| INH | 1,095,364 | missense | p.Asn363Thr | PE_PGRS18 | |
| RIF | 2,163,790 | synonymous | p.Pro1174Pro | PPE34 | |
| RIF | 2,523,205 | intergenic_region | n.2523205_2523206insCGC | Rv2248-Rv2249c | |
| RIF | 2,155,168 | missense | p.Ser315Thr | katG | |
| RIF | 1,533,208 | synonymous | p.Gly142Gly | PPE19 | |
| RIF | 672,491 | synonymous | p.Gly1142Gly | PE_PGRS7 | |
| RIF | 3,735,802 | synonymous | p.Thr378Thr | PPE54 | |
| EMB | 1,789,933 | intergenic | n.1789933G > A | Rv1588c-Rv1589 | |
| EMB | 791,249 | missense | p.Ala140Thr | Rv0691c | |
| EMB | 798,779 | intergenic | n.798779T > C | Rv0697-Rv0698 | |
| EMB | 1,816,349 | missense | p.Ala54Val | pykA | |
| EMB | 1,651,308 | missense | p.Ala54Val | pykA | |
| EMB | 1,655,546 | intergenic | n.1655546T > G | Rv1467c-Rv1468c | |
| EMB | 2,097,990 | synonymous | p.Ala10Ala | ureC | |
| EMB | 2,315,748 | missense | p.Asp192Ala | Rv2059 | |
| EMB | 3,420,388 | synonymous | p.Phe299Phe | cyp136 | |
| STM | 3,752,934 | missense | p.Val84Glu | PPE55 | |
| STM | 3,135,912 | missense | p.Thr141Arg | Rv2828c | |
| STM | 781,822 | missense | p.Lys88Arg | rpsL | |
| STM | 3,943,019 | intergenic | n.3,943,019 C > G | Rv3511-Rv3512 | |
| STM | 2,366,768 | intergenic | n.2,366,768 A > G | Rv2104a-Rv2107 | |
| STM | 55,553 | missense | p.Pro631Ser | Rv0050 | |
| STM | 580,772 | intergenic |
n.580772_ 580,797 delTGGGGGCACCAC CCGCTTGCGGGGGA insAG |
Rv0490-Rv0491 | |
| STM | 104,712 | intergenic | n.104,712 C > T | Rv0094c-Rv0095c | |
Discussion
The advent of WGS has increased the availability genomic data, necessitating the application of advanced analytical approaches, such as ML, to address complex challenges in disease diagnosis and prognosis, including TB management [44]. The increasing availability of bacterial genome sequences offers an unprecedented opportunity to detect antimicrobial AMR in silico, by identifying resistance-conferring patterns [45]. In our study, we identified lineage L4 and sub lineage as L4.6.1.1 (T2-Uganda and Uganda II) from the MTB UG isolates consistent with previous studies conducted in the region [8, 46, 47].
We evaluated the predictive performance of ten different ML models (RF, LR, SVM, MLP, CatBoost, GBC, AdaBoost, XGBoost, DT, and ETC) to infer resistance to four first-line antibiotics in MTB using both WGS and clinical data. Comparisons of model performance between SNP-only datasets and combined SNP-clinical datasets revealed only marginal differences (± 0.01 or 0.02) in performance across the drugs. This suggests that while some clinical features such as age or sex might not provide clear insights into drug resistance mechanisms, incorporating diverse data sources remains beneficial as it could capture interactions between features that may not be apparent from a single data type.
Performance varied across the four drugs, with LR outperforming other models for RIF and STM, GBC excelling for INH and XGBoost for EMB. Our findings of the LR and GBC among the best performing models in predicting drug resistance are comparable to those from published studies [20, 44]. These results likely reflect the unique resistance-conferring mutations associated with each drug and the varying capabilities of the different ML models.
The best performing models particularly LR for RIF and STM showed strong predictive performance on the Ugandan dataset but didn’t generalize well on South African dataset compared to the boosting classifier models, GBC for INH and XGBoost for EMB. This observation aligns with previous findings from Nsubuga et al. where boosting classifiers exhibited strong generalizability on an African dataset for AMR prediction in E. coli [17]. This result could be due to the different proportions of resistant or susceptible isolates in the Uganda test dataset and South Africa dataset and lineage-specific genomic variations associated with DR.
Benchmarking the best ML models for each drug against TB profiler on the UG test dataset and SA dataset (validation set) showed varied results in terms of sensitivity, specificity and ROC-AUC scores. Generally, the best ML models achieved a higher ROC-AUC score compared to TB profiler on the UG dataset with exception of the LR model on STM indicating that ML methods have the ability to discriminate between the resistant and susceptible strains of MTB. The LR model for RIF achieved a higher sensitivity than TB profiler indicating that it successfully identified snp mutations associated with rifampicin resistance with a low false negative rate. In contrast whereas the XGBoost for EMB is as sensitive as TB profiler it achieved a higher specificity as the GBC model for INH. In our study we also observed the trade-off between sensitivity and specificity in the performance of the predictive models. For instance, although GBC model achieved a higher specificity compared to the TB profiler, its sensitivity was low. The high specificity exhibited by the boosting algorithms (GBC and XGBoost) makes them to correctly identify susceptible cases thereby minimising the false positive cases. This could be explained by their serial learning process which enables them to capture more complex patterns, enhancing their robustness across diverse datasets [48].
Comparatively, TB profiler outperformed all the best models in predicting drug resistance on the SA dataset as it achieved a higher mean sensitivity. This observation could be attributed to the fact it uses tbdb mutation library which is a combination of the original list of mutations associated with resistance from literature and WHO catalogue of mutations [36].
Our models identified HIV status as a key feature influencing resistance to RIF and STM albeit with a negative coefficient. These findings corroborate previous studies that suggest while the HIV epidemic amplifies TB outbreaks by increasing the pool of susceptible hosts, but that HIV co-infection does not directly drive for the emergence of resistant MTB strains [49].
In addition to well-known resistance genes such as rpoB, katG and rpsL (associated with RIF, INH and STM resistance), our study detected novel SNPS within Proline-Glutamate (PE) and Proline-Glutamate-Polymorphic Guanine-Cytosine Rich Sequence (PE-PGRS) and proline-glutamate/ proline-proline-glutamate (PE/PPE) genes, including PE_PGRS18 associated with INH resistance, PE_PGRS7, PPE34 and PPE54 associated with RIF resistance, PPE 19 associated with RIF resistance, and PPE55 for STM resistance. This family of proteins is implicated in host-pathogen interactions, virulence, and the development of drug resistance [50–53]. Our findings of mutations in PPE19 gene associated with INH resistance and PPE 54 gene associated with RIF resistance were consistent with those from other studies [52, 54, 55].
Additionally, mutations in the pykA gene (Rv1617) were identified to be associated with EMB resistance. Our findings are in contrast with though from recent studies highlighting a potential association between mutations in the pykA gene and resistance to cycloserine [56]. We observed mutations in the ureC (Rv1850) gene, cyp136 (Rv3059) associated with EMB resistance, and also mutations in genes (Rv2059, Rv2082, Rv2828A, and Rv2828c), all of which are conserved hypothetical proteins to be associated with INH and EMB resistance. In addition, while we also observed mutations in genes in intergenic regions to be associated with resistance to INH, EMB and STM, there precise role and mechanisms associated with resistance in MTB is not defined. However our findings were consistent with those from previous studies that observed genes in intergenic regions associated with drug resistance, such as the oxyR-ahpC intergenic region for INH resistance [57], and embC-embA Intergenic Region for EMB resistance [58].
Further research is needed to establish if the novel markers identified as associated with resistance in this study are causally involved in mediating resistance. The substantial drop in performance on the South African dataset highlights the challenges of generalizing models trained on one dataset to others with distinct characteristics. Future studies should investigate the role of lineage-specific genomic variations and their impact on model performance, which could provide deeper insights into the interplay between lineage diversity and resistance mechanisms. This will be crucial in establishing their robustness and reliability as predictive indicators of drug resistance in Mycobacterium tuberculosis. Moreover, our approach can also be applied to other biomedical areas, e.g. drug repurposing, drug response prediction, for cancer resistance prediction etc. More importantly, ML approaches have a great promise in systems medicine, to improve the diagnosis, targeted therapy and disease prevention.
Our study is not without limitations. First, we used SNP data derived from a single reference genome (H37Rv reference genome), which may exclude important genomic regions related to resistance. Incorporating a pseudo-pan-genome or selecting more suitable reference genomes representing different resistance phenotypes could improve feature selection and ML performance. Additionally, the limited availability of patient clinical data constrained the models’ ability to capture complex biological interactions that may underlie resistance mechanisms. Expanding the dataset to include additional drugs and clinical factors could enhance the predictive power of these models. Multicollinearity was not extensively assessed, however, boosting classifiers like Gradient Boosting and XGBoost are robust to multicollinearity, effectively minimizing the impact of correlated features [59]. Future studies with larger datasets and more numerical variables should consider explicit collinearity assessments.
Conclusion
In summary, our study identified potential markers associated with resistance in MTB and demonstrates the potential of ML algorithms to predict resistance using diverse data types. While our models focused on WGS data and clinical data, future efforts could benefit from integrating multiple omics layers, such as transcriptomics, proteomics, and metabolomics, alongside comprehensive clinical information. Achieving a balance between dataset size, class distribution, and model complexity will be critical to avoiding overfitting and ensuring robust model performance.
The observed variation in model performance across the Ugandan and South African datasets underscores the importance of developing generalizable ML models that can be deployed across diverse populations. By building more scalable and adaptable models, we can enhance their utility in global TB control efforts and inform targeted treatment strategies in the fight against AMR.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The author SRB was funded by the Makerere University Data Science Research Training Program to Strengthen Evidence Based Health Innovation, Intervention and Policy (MakDARTA) under the Fogarty International Center of the National Institutes of Health (NIH) under Award Number U2RTW012116. as a Masters scholar. The authors would also like to acknowledge the Open Science Grid (OSG) consortium which provided computational resources to carry out this study. The OSG is supported by the National Science Foundation award number 2030508 and 1836650. The author RG is funded by a grant from the Wellcome Trust through the Centers for Antimicrobial Optimisation Network (CAMO-Net) (Award No. 226692/Z/22/Z). RG is also supported by the She Data Science (SHEDS) program: Empowering Uganda’s Women in Health Data Science: Identifying Barriers, Bridging Knowledge and Innovation for Tangible Impact; through a collaborative agreement with the University of California San Francisco (UCSF) (Agreement No. UFRA-460).
Abbreviations
- AMR
Antimicrobial resistance
- AUC
Area Under the Curve
- DR
Drug resistance
- DT
Decision trees
- EMB
Ethambutol
- GBC
Gradient Boosting
- INH
Isoniazid
- LR
Logistic regression
- MCC
Mathews Correlation Coefficient
- ML
Machine learning
- MLP
Multi-layer Perceptron classifier
- MTB
Mycobacterium tuberculosis
- RF
Random Forest
- RIF
Rifampicin
- ROC
Receiver Operating Curve
- SNP
Single Nucleotide Polymorphism
- STM
Streptomycin
- SVM
Support Vector Machine
- TB
Tuberculosis
- WGS
Whole-genome sequence
- XGBoost
Extreme Gradient boosting classifier
Author contributions
SRB: Data analysis, data interpretation, writing-Original draft; DPK: Conception, writing-Final draft; GM: Conception, writing-Final draft; MN: Data interpretation, writing-Final draft, RG: Data interpretation, writing-Final draft and CB: writing-Final draft. All authors read and approved the final manuscript.
Funding
Not applicable.
Data availability
The datasets analysed during the current study are available in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under the following Bio project IDs; PRJEB10533, PRJNA481638, PRJEB10577, PRJEB2424, PRJNA559528 and PRJNA74965.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim JI, Maguire F, Tsang KK, Gouliouris T, Peacock SJ, McAllister TA et al. Machine learning for Antimicrobial Resistance Prediction: current practice, limitations, and clinical perspective. Clin Microbiol Rev. 2015;35:e00179–21. [DOI] [PMC free article] [PubMed]
- 2.Kariuki S. Global burden of antimicrobial resistance and forecasts to 2050. Lancet. 2024;0. [DOI] [PubMed]
- 3.Kariuki S, Kering K, Wairimu C, Onsare R, Mbae C. Antimicrobial Resistance Rates and Surveillance in Sub-saharan Africa: where are we now? Infect Drug Resist. 2022;15:3589–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet Lond Engl. 2022;399:629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kraker MEA, Stewardson AJ, Harbarth S. Will 10 million people die a year due to Antimicrobial Resistance by 2050? PLoS Med. 2016;13:e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tornimbene B, Eremin S, Escher M, Griskeviciene J, Manglani S, Pessoa-Silva CL. WHO Global Antimicrobial Resistance Surveillance System early implementation 2016–17. Lancet Infect Dis. 2018;18:241–2. [DOI] [PubMed] [Google Scholar]
- 7.Bagcchi S. WHO’s Global Tuberculosis Report 2022. Lancet Microbe. 2023;4:e20. [DOI] [PubMed] [Google Scholar]
- 8.Ngabonziza JCS, Loiseau C, Marceau M, Jouet A, Menardo F, Tzfadia O, et al. A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region. Nat Commun. 2020;11:2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscolla M, Gagneux S, Menardo F, Loiseau C, Ruiz-Rodriguez P, Borrell S, et al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb Genomics. 2021;7:000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuang X, Wang F, Hernandez KM, Zhang Z, Grossman RL. Accurate and rapid prediction of tuberculosis drug resistance from genome sequence data using traditional machine learning algorithms and CNN. Sci Rep. 2022;12:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farhat M, Cox H, Ghanem M, Denkinger CM, Rodrigues C, El Abd MS et al. Drug-resistant tuberculosis: a persistent global health concern. Nat Rev Microbiol. 2024;:1–19. [DOI] [PubMed]
- 12.Horne DJ, Pinto LM, Arentz M, Lin S-YG, Desmond E, Flores LL, et al. Diagnostic accuracy and reproducibility of WHO-Endorsed phenotypic drug susceptibility testing methods for first-line and second-line antituberculosis drugs. J Clin Microbiol. 2020;51:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabugo J, Namutebi J, Mujuni D, Nsawotebba A, Kasule GW, Musisi K, et al. Implementation of GeneXpert MTB/Rif proficiency testing program: a case of the Uganda national tuberculosis reference laboratory/supranational reference laboratory. PLoS ONE. 2021;16:e0251691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ssengooba W, de Dieu Iragena J, Komakech K, Okello I, Nalunjogi J, Katagira W, et al. Discordance of the repeat GeneXpert MTB/RIF Test for Rifampicin Resistance Detection among patients initiating MDR-TB treatment in Uganda. Open Forum Infect Dis. 2021;8:ofab173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabahita JM, Kabugo J, Kakooza F, Adam I, Guido O, Byabajungu H, et al. First report of whole-genome analysis of an extensively drug-resistant Mycobacterium tuberculosis clinical isolate with bedaquiline, linezolid and clofazimine resistance from Uganda. Antimicrob Resist Infect Control. 2022;11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20:356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nsubuga M, Galiwango R, Jjingo D, Mboowa G. Generalizability of machine learning in predicting antimicrobial resistance in E. Coli: a multi-country case study in Africa. BMC Genomics. 2024;25:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green AG, Yoon CH, Chen ML, Ektefaie Y, Fina M, Freschi L, et al. A convolutional neural network highlights mutations relevant to antimicrobial resistance in Mycobacterium tuberculosis. Nat Commun. 2022;13:3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Niehaus KE, Walker TM, Iqbal Z, Walker AS, Wilson DJ, et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics. 2018;34:1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouchaki S, Yang Y, Walker TM, Sarah Walker A, Wilson DJ, Peto TEA, et al. Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinformatics. 2019;35:2276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang A, Teng L, Alterovitz G. An explainable machine learning platform for pyrazinamide resistance prediction and genetic feature identification of Mycobacterium tuberculosis. J Am Med Inf Assoc. 2021;28:533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leinonen R, Sugawara H, Shumway M, on behalf of the International Nucleotide Sequence Database Collaboration. The sequence read Archive. Nucleic Acids Res. 2011;39 suppl1:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark TG, Mallard K, Coll F, Preston M, Assefa S, Harris D, et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS ONE. 2013;8:e83012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ssengooba W, Cobelens FG, Nakiyingi L, Mboowa G, Armstrong DT, Manabe YC, et al. High genotypic discordance of Concurrent Mycobacterium tuberculosis isolates from Sputum and blood of HIV-Infected individuals. PLoS ONE. 2015;10:e0132581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ssengooba W, Meehan CJ, Lukoye D, Kasule GW, Musisi K, Joloba ML, et al. Whole genome sequencing to complement tuberculosis drug resistance surveys in Uganda. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2016;40:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimmo C, Brien K, Millard J, Grant AD, Padayatchi N, Pym AS, et al. Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine. 2020;55:102747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews S. s-andrews/FastQC. 2024.
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. [Google Scholar]
- 31.Seemann T. tseemann/snippy. 2024.
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012.
- 35.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verboven L, Phelan J, Heupink TH, Van Rie A. TBProfiler for automated calling of the association with drug resistance of variants in Mycobacterium tuberculosis. PLoS ONE. 2022;17:e0279644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pordes, TOSGEB on behalf of the OC, Petravick D, Kramer B, Olson D, Livny M, Roy A, et al. The open science grid. J Phys Conf Ser. 2007;78:012057. [Google Scholar]
- 38.The open science grid - IOPscience. 10.1088/1742-6596/78/1/012057. Accessed 24 Sep 2024.
- 39.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics. 2016;2:e000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a Distance Matrix. Mol Biol Evol. 2009;26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O et al. Scikit-learn: Machine Learning in Python. 2018.
- 44.Deelder W, Christakoudi S, Phelan J, Benavente ED, Campino S, McNerney R et al. Machine learning predicts accurately Mycobacterium tuberculosis Drug Resistance from whole genome sequencing data. Front Genet. 2019;10. [DOI] [PMC free article] [PubMed]
- 45.Aytan-Aktug D. Machine learning of antimicrobial resistance. 2021.
- 46.Micheni LN, Kassaza K, Kinyi H, Ntulume I, Bazira J. Diversity of Mycobacterium tuberculosis Complex Lineages Associated with Pulmonary Tuberculosis in Southwestern, Uganda. Tuberc Res Treat. 2021;2021:5588339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wampande EM, Mupere E, Debanne SM, Asiimwe BB, Nsereko M, Mayanja H, et al. Long-term dominance of Mycobacterium tuberculosisUganda family in peri-urban Kampala-Uganda is not associated with cavitary disease. BMC Infect Dis. 2013;13:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanha J, Abdi Y, Samadi N, Razzaghi N, Asadpour M. Boosting methods for multi-class imbalanced data classification: an experimental review. J Big Data. 2020;7:70. [Google Scholar]
- 49.Eldholm V, Rieux A, Monteserin J, Lopez JM, Palmero D, Lopez B, et al. Impact of HIV co-infection on the evolution and transmission of multidrug-resistant tuberculosis. eLife. 2016;5:e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Guo X, Xiang D, Pitt ME, Bainomugisa A, Coin LJM. Computational analysis and prediction of PE_PGRS proteins using machine learning. Comput Struct Biotechnol J. 2022;20:662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian J, Chen R, Wang H, Zhang X. Role of the PE/PPE family in host–Pathogen interactions and prospects for Anti-tuberculosis Vaccine and Diagnostic Tool Design. Front Cell Infect Microbiol. 2020;10:594288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehtram A, Shariq M, Ali S, Quadir N, Sheikh JA, Ahmad F, et al. Teleological cooption of Mycobacterium tuberculosis PE/PPE proteins as porins: role in molecular immigration and emigration. Int J Med Microbiol IJMM. 2021;311:151495. [DOI] [PubMed] [Google Scholar]
- 53.Kanji A, Hasan Z, Ali A, McNerney R, Mallard K, Coll F, et al. Characterization of genomic variations in SNPs of PE_PGRS genes reveals deletions and insertions in extensively drug resistant (XDR) M. Tuberculosis strains from Pakistan. Int J Mycobacteriology. 2015;4:73–9. [DOI] [PubMed] [Google Scholar]
- 54.Cui Z-J, Yang Q-Y, Zhang H-Y, Zhu Q, Zhang Q-Y. Bioinformatics Identification of Drug Resistance-Associated gene pairs in Mycobacterium tuberculosis. Int J Mol Sci. 2016;17:1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hang NTL, Hijikata M, Maeda S, Thuong PH, Ohashi J, Van Huan H, et al. Whole genome sequencing, analyses of drug resistance-conferring mutations, and correlation with transmission of Mycobacterium tuberculosis carrying katG-S315T in Hanoi, Vietnam. Sci Rep. 2019;9:15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trisakul K, Nonghanphithak D, Chaiyachat P, Kaewprasert O, Sakmongkoljit K, Reechaipichitkul W, et al. High clustering rate and genotypic drug-susceptibility screening for the newly recommended anti-tuberculosis drugs among global extensively drug-resistant Mycobacterium tuberculosis isolates. Emerg Microbes Infect. 2022;11:1857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bakhtiyariniya P, Khosravi AD, Hashemzadeh M, Savari M. Detection and characterization of mutations in genes related to isoniazid resistance in Mycobacterium tuberculosis clinical isolates from Iran. Mol Biol Rep. 2022;49:6135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z, et al. Mutations in the embc-emba Intergenic Region Contribute to Mycobacterium tuberculosis Resistance to Ethambutol. Antimicrob Agents Chemother. 2014;58:6837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fdez-Díaz L, Quevedo JR, Montañés E. Regularized boosting with an increasing coefficient magnitude stop criterion as meta-learner in hyperparameter optimization stacking ensemble. Neurocomputing. 2023;551:126516. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study are available in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under the following Bio project IDs; PRJEB10533, PRJNA481638, PRJEB10577, PRJEB2424, PRJNA559528 and PRJNA74965.