Abstract
Background
The emergence of induced pluripotent stem cells (iPSCs) offers a promising approach for replacing damaged neurons and glial cells, particularly in spinal cord injuries (SCI). Despite its merits, iPSC differentiation into spinal cord progenitor cells (SCPCs) is variable, necessitating reliable assessment of differentiation and validation of cell quality and safety. Phenotyping is often performed via label-based methods including immunofluorescent staining or flow cytometry analysis. These approaches are often expensive, laborious, time-consuming, destructive, and severely limits their use in large scale cell therapy manufacturing settings. On the other hand, cellular biophysical properties have demonstrated a strong correlation to cell state, quality and functionality and can be measured with ingenious label-free technologies in a rapid and non-destructive manner.
Method
In this study, we report the use of Magnetic Resonance Relaxometry (MRR), a rapid and label-free method that indicates iron levels based on its readout (T2). Briefly, we differentiated human iPSCs into SCPCs and compared key iPSC and SCPC cellular markers to their intracellular iron content (Fe3+) at different stages of the differentiation process.
Results
With MRR, we found that intracellular iron of iPSCs and SCPCs were distinctively different allowing us to accurately reflect varying levels of residual undifferentiated iPSCs (i.e., OCT4+ cells) in any given population of SCPCs. MRR was also able to predict Day 10 SCPC OCT4 levels from Day 1 undifferentiated iPSC T2 values and identified poorly differentiated SCPCs with lower T2, indicative of lower neural progenitor (SOX1) and stem cell (Nestin) marker expression levels. Lastly, MRR was able to provide predictive indications for the extent of differentiation to Day 28 spinal cord motor neurons (ISL-1/SMI-32) based on the T2 values of Day 10 SCPCs.
Conclusion
MRR measurements of iPSCs and SCPCs has clearly indicated its capabilities to identify and quantify key phenotypes of iPSCs and SCPCs for end-point validation of safety and quality parameters. Thus, our technology provides a rapid label-free method to determine critical quality attributes in iPSC-derived progenies and is ideally suited as a quality control tool in cell therapy manufacturing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-04070-y.
Keywords: Cell manufacturing, Cell therapy, Spinal cord injury, Induced pluripotent stem cells, Neural progenitors, Magnetic resonance relaxometry, Label-free technology
Introduction
Spinal cord injury (SCI) is an extremely debilitating medical condition and is severely lacking in effective clinical treatments [1]. In particular, the current therapies of SCI mainly focus on surgeries for spinal realignment and subsequent rehabilitation, which are unable to reverse the damage to injured spinal cords. Consequently, there is a severe need for more efficacious treatments [2]. The use of iPSCs is a highly promising alternative since iPSCs can differentiate into neural stem/progenitor cells (NSC/NPC), which may facilitate effective spinal cord regeneration as these cells can differentiate in neurons and glial cells [3]. The NSC/NPCs are usually implanted directly into the injury site of the spinal cord or as part of a biomaterial scaffold [4]. In this regard, several research groups have successfully demonstrated the transplantation of iPSC-derived NSC/NPCs into rodent and monkey models of SCI, resulting in the restoration of their motor function [3, 5, 6].
Although the differentiation process of iPSCs into NSC/NPCs is relatively efficient (typically > 80% differentiation efficiency) [7–9], there are major safety concerns over the presence of residual undifferentiated iPSCs, which presents a risk of tumorigenicity post-transplantation [10, 11]. Specifically, the variations in differentiation efficiency and tumor-like overgrowth are still recurring problems in iPSC-derived NSC/NPCs that were transplanted into injured spinal cords [11]. Only a small number of residual undifferentiated iPSC-like cells are required to pose a significant safety risk due to their potential neoplasticity in the implanted tissue [12]. Hence, the detection of these residual undifferentiated iPSC-like cells is of paramount importance to the safety of iPSC-related cell therapies.
Current assays that are used to detect the undifferentiated iPSCs are done by quantifying phenotypes of iPSCs and NSC/NPCs via label-based fluorescent markers. The labeled cells are then analyzed via a flow cytometer or visualized via a fluorescent microscope [13]. Unfortunately, these assays often perturb or destroy the cells due to terminal fixation, which renders the cells unusable for subsequent applications. Furthermore, given that these assays are expensive, laborious, and time-consuming, they are ultimately inadequate for quality control of iPSCs and iPSC-derived progenies in bioproduction processes. On the other hand, label-free technologies can characterize cellular biophysical attributes, which correlate well to cell quality and safety. These assays allow rapid and non-destructive assessment of cell batch quality and are ideally suited for large-scale cell manufacturing. Cellular biophysical attributes, including cell size [14, 15], cell deformability [16], auto-fluorescing endogenous metabolites [17], impedance properties [18, 19], and intracellular iron levels [20], have been investigated for their abilities to obtain critical and reliable information of cells. In particular, the levels of intracellular iron in biological samples have indicated strong correlations with cell quality and patient health [21, 22]. Furthermore, several studies pertaining to the role of iron in iPSCs have strongly suggested that iron homeostasis is critical to the maintenance of iPSC pluripotency and downstream functionalities [23, 24]. However, there is no assay that can rapidly detect intracellular iron levels reliably in the field of iron biology.
Previously, we demonstrated that micro magnetic resonance relaxometry (MRR) can rapidly measure intracellular iron levels in a label-free manner, and its readouts have been used to detect malaria infection [21], measure oxidated stress [22] in the blood, and very recently to detect senescence in mesenchymal stem cells (MSCs) [20]. The spin–spin relaxation time (T2) measurement is sensitive to the paramagnetic content of the sample, and in the context of cell biology, it is correlated with the intracellular content of iron (Fe3+) [25]. Here, we hypothesize that the intracellular iron content of iPSCs and spinal cord progenitor cells (SCPCs) can be distinctively different, which allows our MRR system to provide indications on the level of residual undifferentiated iPSCs after differentiation to SCPCs with T2 measurements.
In this study, we report a novel use of MRR to rapidly analyze differentiation outcomes of iPSCs to SCPCs, a subtype of NSC/NPCs with a spinal cord identity, and sought to demonstrate that T2 measurements are a good surrogate for label-based quantification of iPSC and SCPC phenotypes. Moreover, several different manufacturing scenarios for generating these SCPCs were implemented in this study to mimic batch-to-batch variations that may arise in typical cell therapy manufacturing pipelines. Lastly, we demonstrated that MRR can provide predictive indications at different stages of the differentiation process of iPSC to SCPCs. We were able to predictively indicate Day 10 SCPC OCT4 levels from the MRR readouts of undifferentiated iPSCs and the level of Day 28 motor neuron marker expression from the MRR readouts of Day 10 SCPCs. Altogether, our results demonstrate that MRR heralds a considerable potential for determining critical quality attributes (CQAs) in iPSC-derived progenies and is ideally suited as a quality control tool in cell therapy manufacturing.
Materials and methods
iPSC culture
Two human iPSC lines were routinely cultured and maintained on Matrigel (83.3 µg/mL, Corning, USA) coated tissue culture plastic dishes in StemMACS™ iPS-Brew XF (Miltenyi Biotech, Germany). (1) Healthy umbilical cord-lining epithelial cell (CLEC23)-derived iPSCs were kindly provided by Dr Kah-Leong Lim and CellResearch Corporation Pte Ltd [26]. CLEC-derived iPSCs were hypothesized to exhibit immune-privileged properties similar to that of CLECs [27, 28]. (2) BJ-iPSCs were derived from BJ-fibroblasts with the use of modified mRNA [29]. Specifically, the BJ-iPSC line was meant to represent fibroblast-derived iPSCs that were previously used to generate spinal motor neurons [30, 31]. Tissue culture plates were coated with 3 mL of Matrigel for a minimum of 15 min at 37 °C before passaging. Briefly, the cells had a daily change of medium and were passaged via ReleSR™ (STEMCELL Technologies, Canada) upon reaching 70–80% confluency.
SCPC differentiation and culture
SCPCs are NPC/NSCs with a spinal cord identity. Specifically, SCPCs were shown to interconnect with host neural circuits and achieve motor function recovery in SCI mice models as compared to NPC/NSCs with other regional identities [32]. CLEC23 iPSC and BJ-iPSC cell lines were used to differentiate into SCPCs. The method for generating SCPCs is derived from [33]. Briefly, SCPCs were generated from iPSCs via the use of three small molecules that mimic the native developmental pathway of the spinal cord [32, 34]. Quantification of cellular phenotypes upon differentiation was obtained via FACS or immunocytochemistry staining. Hence, the differentiation efficiency of SCPCs on Day 10 of differentiation was quantified via the expression of pluripotent marker (OCT4) and neural progenitor marker (SOX1).
The differentiation process began by lifting iPSCs off tissue culture dishes at 70–80% confluency with Accutase (Nacalai Tesque Inc., Japan) and plated onto Matrigel-coated (1 mL of 83.3 µg/mL for a minimum of 15 min at 37 °C) tissue culture plastic 6-well plates at 800,000 cells/well with neural induction media (NIM) supplemented with ROCK inhibitor Y-27632 (ROCKi, 5 µM, Miltenyi Biotech). NIM consisted of DMEM/F12 (50%, Thermo Fisher Scientific, Waltham, MA, USA), neural medium (50%, Miltenyi Biotech), NeuroBrew-21 (1x, Miltenyi Biotech), N2 (1x, Miltenyi Biotech), Non-Essential Amino Acid (1x, Thermo Fisher Scientific), Glutamax (0.5x, Thermo Fisher Scientific), LDN-193189 (0.5 µM, Miltenyi Biotech) and CHIR-99021 (CHIR, 4.25 µM, Miltenyi Biotech). On Day 3 of differentiation, retinoic acid (RA, 1 µM, Sigma, USA) was added to the NIM to induce caudalisation. On Day 4, Accutase was used to dissociate the semi-differentiating cells and the cells were subsequently transferred onto a Matrigel-coated tissue culture plastic dish at 2.5 × 106 cells/dish with NIM + RA and ROCKi. On Day 5, NIM + RA and ROCKi were aspirated and replenished with NIM + RA. From Day 6 to Day 9, the cells received a daily media change of NIM + RA. On Day 10, Accutase was used to dissociate and lift the differentiated cells and the cells were subsequently characterized with various assays.
Motor neuron differentiation and culture
A subset of Day 10 SCPCs that were harvested were subjected to further differentiation into motor neurons. The method for generating motor neurons was adapted from [33]. Briefly, motor neurons are generated from iPSC-derived SCPCs by firstly adding a sonic hedgehog agonist to ventralise the SCPCs and secondly by adding growth factors to support the differentiating motor neurons to maturation. Quantification of motor neurons upon differentiation was obtained via immunocytochemistry staining. The motor neurons on Day 28 of differentiation was quantified via the expression of motor neuron marker (ISL-1) and neurofilament protein marker (SMI-32).
The differentiation process began by seeding the Day 10 SCPCs onto Matrigel-coated (1 mL of 83.3 µg/mL for a minimum of 15 min at 37 °C) tissue culture plastic 6-well plates at 2 × 106 cells/well with neural differentiation media (NDM) supplemented with ROCK inhibitor Y-27632 (ROCKi, 5 µM, Miltenyi Biotech). NDM consisted of DMEM/F12 (50%), neural medium (50%), NeuroBrew-21 (1x), N2 (1x), Non-Essential Amino Acid (1x), Glutamax (0.5x), purmophamine (1 µM, Miltenyi Biotech) and retinoic acid (1 µM). From Day 11 to Day 16, the cells received a daily media change of NDM. On Day 17 of differentiation, Accutase was used to dissociate and lift the differentiated cells.
The differentiated cells were reseeded onto Matrigel-coated (1 mL of 83.3 µg/mL for a minimum of 15 min at 37 oC) tissue culture plastic 6-well plates at 1.5 × 106 cells/well with neural maturation media (NMM) supplemented with ROCKi (5 µM). NMM consisted of DMEM/F12 (50%), neural medium (50%), NeuroBrew-21 (1x), N2 (1x), Non-Essential Amino Acid (1x), Glutamax (0.5x), brain-derived neurotrophic factor (BDNF, 10 ng/mL, Miltenyi Biotech), glial-derived neurotrophic factor (GDNF, 10 ng/mL, Miltenyi Biotech), ascorbic acid (AA, 200 µM, Sigma), and sodium pyruvate (1x, Thermo Fisher Scientific). On Day 18 to Day 21, the cells received a daily media change of NMM. On Day 22, Accutase was used to dissociate the differentiated cells as the culture is reaching full confluency. The differentiated cells were reseeded onto Matrigel-coated (1 mL of 83.3 µg/mL for a minimum of 15 min at 37 oC) tissue culture plastic 6-well plates at 1.5 × 106 cells/well with NMM supplemented with ROCKi (5 µM). On Day 23 to Day 27, the cells received a daily media change of NMM. On Day 28, Accutase was used to dissociate and lift the differentiated motor neurons and the cells were seeded in a 96-well plate for immunohistochemistry staining.
Fe3+ staining and quantification of iPSCs and SCPCs
Quantification of intracellular iron (Fe3+) in iPSCs and SCPCs was performed via a reversible fluorescent Fe3+ sensor (RPE) [35]. For Fe3+ staining, a stock solution of RPE (1 mM in acetonitrile) was diluted to a concentration of 20 μM in PBS. 3 × 105 suspended cells were harvested and centrifuged to remove the supernatant. The cells were then incubated with PBS containing RPE (20 μM) at 37 °C for 20 min. After incubation, the cells were washed twice and suspended in PBS to measure their fluorescent intensity via flow cytometry.
MRR measurement
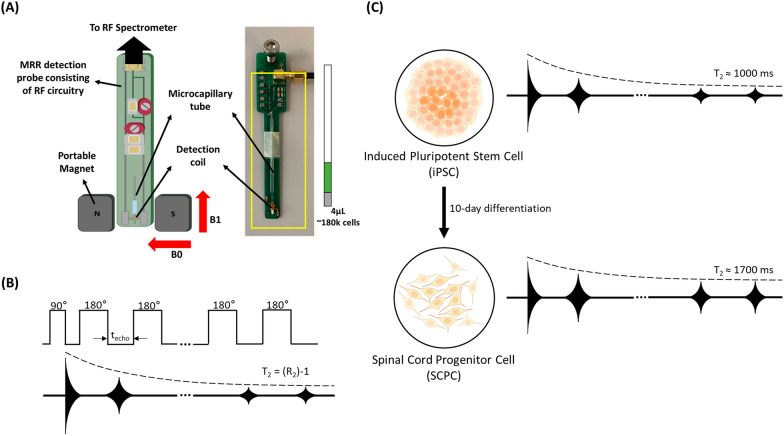
The apparatus was established in our previous works [20–22]. Briefly, the MRR system consists of a portable, permanent magnet (Metrolab Instruments, Switzerland) with B0 = 0.5 T and a bench-top type nuclear magnetic resonance (NMR) console (Kea Magritek, New Zealand). MRR measurements of 1H were performed with a resonance frequency of 21.015 MHz inside the magnet. MRR samples are loaded into a microcapillary tube (Fisherbrand, USA) fitted into a 900-μm inner diameter detection micro-coil within the single resonance proton MRR probe. The electronic components and the coil are all parts of a printed circuit board in the MRR probe (Fig. 1A). Every MRR experiment was performed at a temperature of 26.3 °C within the magnet, as maintained by a temperature controller (RS component, UK).
Fig. 1.
Using MRR system to measure changes in T2 when iPSCs differentiate into SCPCs. A MRR system consists of a 0.5 T permanent and portable magnet that has a radiofrequency (RF) detection probe hooked up to an RF spectrometer. iPSC and SCPC samples of 4 µL consisting of about 180,000 cells are loaded into a microcapillary tube. The microcapillary tube is sealed with critoseal and fitted into the RF probe with the sample directly within the RF detection coil. The actual RF probe is shown on the right of the schematic for illustration purposes. B Illustration of CPMG (Carr-Purcell-Meiboom-Gill) pulse sequence for obtaining T2 measurements. C Illustration of spin–spin relaxation time of iPSCs changing as they differentiate into SCPCs. iPSC and SCPC cell figures are adapted from the icon library of “Cell Types” by BioRender.com
For MRR experiments regarding operator and batch-to-batch variations, the concentration of iPSCs and SCPCs used were fixed at 120,000 cells in a 4 μL volume. For all other MRR experiments, the concentration of iPSCs and SCPCs used were fixed at 180,000 cells in a 4 μL volume. iPSC or SCPC samples were spun down at 300 g for 5 min and 1000 rpm at 3 min, respectively, with the supernatant being aspirated subsequently. PBS was then added, and the tube was spun down at 1500 rpm for 3 min. The supernatant was aspirated, and the cell pellet was transferred to an Eppendorf tube. The number of cells was counted (INCYTO, South Korea), and a sample of 30,000 or 45,000 cells/µL in 50 µL of cell solution was prepared. 4 µL of cell solution was pipetted into a micro-capillary tube, sealed with critoseal (Leica Microsystems) and mounted into the MRR system to obtain a readout. The measurement of proton transverse relaxation times (T2) was performed by a standard Carr-Purcell-Meiboom-Gill (CPMG) pulse program (Fig. 1B). For all experiments, an inter-echo time of 500 μs with 4000 echoes was applied in the CPMG train of pulses. To allow sufficient time for all spins to return to thermal equilibrium, a recycle delay of 3 s was applied. Signal averaging was performed with a total of 24 scans for all MRR experiments.
Immunocytochemistry staining
SCPCs and motor neurons were seeded onto Matrigel-coated (100 μL of 83.3 µg/mL for a minimum of 15 min at 37 oC) 96-well plates at 80,000 cells/well. SCPCs were stained with SOX1, HOXB4 and OCT4 antibodies whilst motor neurons were stained with ISL-1 and SMI-32 antibodies. Briefly, SCPCs and motor neurons were fixed with 4% Paraformaldehyde (PFA, Biotium, USA) for 15 min. The fixative was removed and washed twice with PBS. The fixed wells were then permeabilized with 0.1% Triton-X in PBS for 15 min. After permeabilization, blocking buffer (2% Bovine Serum Albumin (BSA), 5% Fetal Bovine Serum) was added and incubated at room temperature for 1 h. Thereafter, blocking buffer was aspirated and primary antibodies were added to the samples and subjected to 4 °C overnight incubation. The primary antibodies that were used includes SOX1 (1:250, Cell Signalling Technology, #4194), HOXB4 (1:250, Abcam, ab133521), OCT4 (1:500, Santa Cruz Biotechnology, SC-5279), ISL-1 (1:1500, Abcam, ab109517) and SMI-32 (1:1000, Biolegend, 801701) and were all diluted in blocking buffer at their specific ratios. PBS was used to wash the wells twice after the removal of primary antibodies. Secondary antibodies with the corresponding host species and DAPI were then added to the wells and incubated at room temperature without light exposure for 1 h. The secondary antibodies and DAPI used were: donkey AlexaFluor488-conjugated anti-rabbit IgGs (1:500, Thermo Fisher Scientific, A21206), donkey AlexaFluor555-conjugated anti-Mouse IgGs (1:500, Thermo Fisher Scientific, A31570) and DAPI (1:1000, Thermo Fisher Scientific). PBS was used to wash the wells twice after removing primary antibodies. A Leica DMi8 Microscope was used to image the stained SCPCs and the staining images were quantified using CellProfiler. An Opera Phenix Plus High-Content Screening System was used to image the stained motor neurons and the staining images were quantified with the Columbus Image Data Storage and Analysis System.
Flow cytometry analysis
SCPCs were collected into 15 mL centrifuge tubes and adjusted to a density of 5 × 106 cells/mL. The cells were then fixed in 4% PFA for 15 min at room temperature. Once the cells were fixed, they were centrifuged at 3000 rpm for 5 min and washed once with PBS. Thereafter, the cells were stained by incubation at room temperature for a minimum of 2 h with primary antibodies reconstituted in permeabilization/blocking buffer (0.5% saponin, 1% BSA). The primary antibodies used were: SOX1 (1:250, Cell Signalling Technology, #4194), Nestin (1:200, Abcam, ab22035), HOXB4 (1:250, Abcam, ab133521) and OCT4 (1:500, Santa Cruz Biotechnology, SC-5279). After staining, the cells were centrifuged at 3000 rpm for 5 min and washed twice with PBS. They were then stained by secondary antibodies reconstituted in permeabilization/blocking buffer in the absence of light and at room temperature for 45 min. The secondary antibodies used were: donkey AlexaFluor488-conjugated anti-rabbit IgGs (1:500, Thermo Fisher Scientific, A21206) and donkey AlexaFluor555-conjugated anti-Mouse IgGs (1:500, Thermo Fisher Scientific, A31570). Lastly, the cells were centrifuged at 3000 rpm for 5 min, washed once with PBS and resuspended in 300 µL PBS. Cytoflex flow cytometer (Beckman Coulter, USA) was used to analyze the cells.
iPSC spiking and colony culture assay
SCPCs that were harvested were spiked with iPSCs at different percentages, including 1%, 5% and 10%. Thereafter, the SCPCs were counted, and a fixed number of cells was removed to account for the addition of iPSCs. The cells were seeded in a 6-well plate at an initial density of 1 × 107 cells/well. They were cultured in 2 mL of hiPSC medium for six days with a daily change of the medium. Cells were fixed and stained with DAPI and OCT4 antibodies before imaging with a Leica DMi8 microscope. Colony sizes were analyzed with an ImageJ particle size analyzer.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9. All data were presented as the mean ± standard deviation (SD). p value < 0.05 was considered statistically significant, *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001. Statistical significance for Fig. 2A, B and D was determined by a Mann–Whitney u-test. Statistical significance for Fig. 3C was determined by a Kruskal–Wallis test followed by a Dunn post-hoc test. Statistical significance for Fig. 4A, B, 5A, B, 6C, D, and 7C was determined by a Pearson’s correlation test.
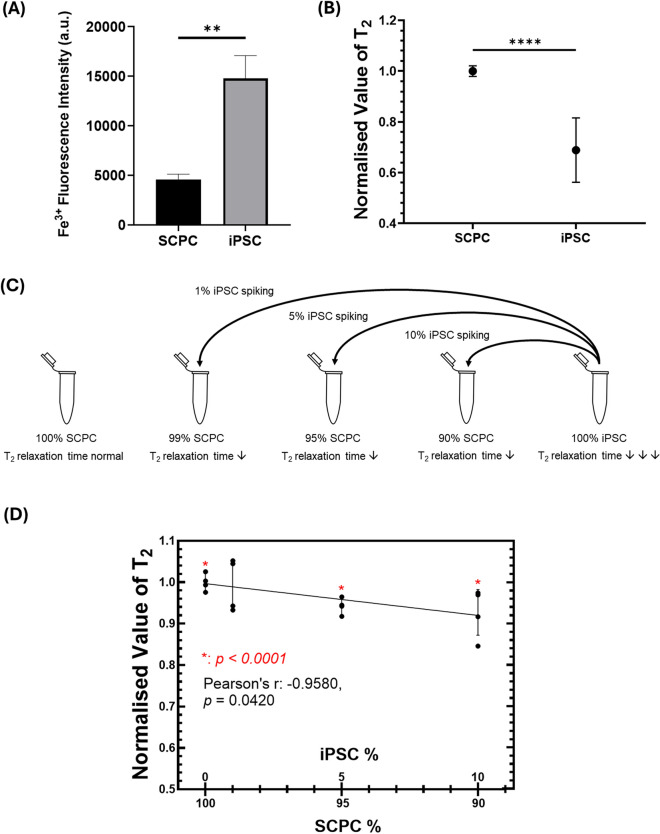
Fig. 2.
Difference in iPSCs and SCPCs intracellular iron levels allows MRR to pick up varying levels of iPSCs in SCPCs via T2 measurements A Fe3+ fluorescent intensity of iPSCs and SCPCs analyzed via FACS showed a significant difference in Fe3 between the 2 cell types. B T2 values of SCPCs were significantly higher than iPSCs. C Schematic of artificial spiking of iPSCs in SCPCs at 1%, 5% and 10% concentrations to alter SCPC T2 measurements. D Decrement of T2 values with increasing spiked iPSC concentration at 1%, 5% and 10% (n = 4). Statistical significance was determined by a Mann–Whitney u-test
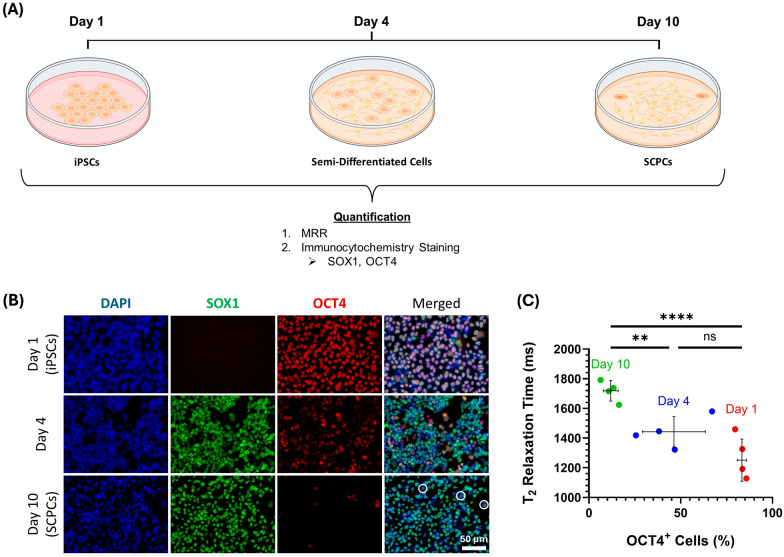
Fig. 3.
T2 value increased as iPSCs differentiated into SCPCs with correspondence to phenotypic changes (SOX1 and OCT4). A Multi-timepoint measurement of T2 values to compare with immunofluorescent staining as iPSCs differentiate to SCPCs across 3 different time points (Day 1, Day 4 and Day 10). B Immunofluorescent staining to illustrate changes in SOX1 and OCT4 phenotypes during the differentiation of iPSCs into SCPCs. DAPI (blue, nuclear), SOX1 (green, neural progenitor marker) and OCT4 (red, pluripotent marker) (Scale bar: 50 µm). B Increase in T2 with decreasing level of OCT4+ cells across a 10-day differentiation period. At least 10 ROIs (more than 60,000 cells) were quantified for each experimental repeat (n = 4). Statistical significance was determined by a Kruskal–Wallis test followed by a Dunn post-hoc test
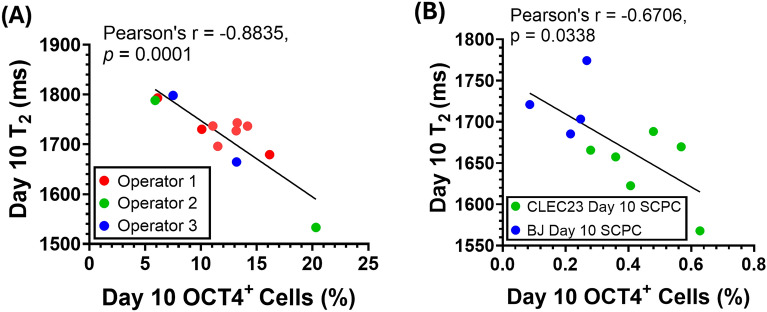
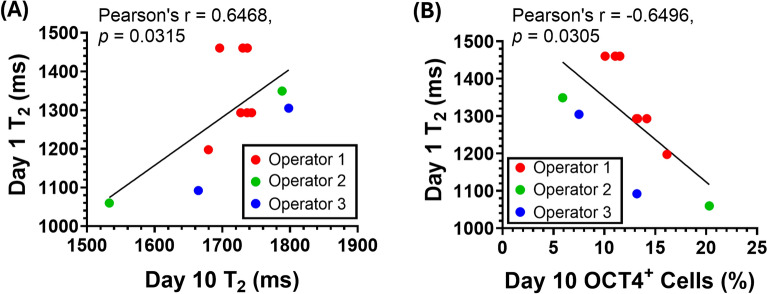
Fig. 4.
T2 decreased with increasing OCT4 levels in day 10 SCPCs corresponding with manufacturing considerations, including batch-to-batch, operator and cell-line variations. A Correlation of day 10 SCPC T2 values with the percentage of day 10 OCT4+ cells quantified by immunofluorescent staining. 3 different operators generated SCPCs, and each data point refers to a different batch of SCPCs that was generated. At least 10 ROIs (more than 100,000 cells) were quantified for each experimental repeat (n = 12). B Day 10 BJ-SCPCs had lower OCT4 levels and higher T2 values as compared to day 10 CLEC23 SCPCs. At least 25 ROIs (more than 60,000 cells) were quantified for each experimental repeat (n = 9)
Fig. 5.
T2 values in Day 1 iPSCs could predictively indicate Day 10 T2 and OCT4 levels. A Differentiating Day 1 iPSC with lower T2 values resulted in Day 10 SCPCs with lower T2. B Differentiating Day 1 iPSC with lower T2 values resulted in Day 10 SCPCs with higher levels of OCT4+ cells as quantified by immunofluorescent staining. At least 10 ROIs (more than 60,000 cells) were quantified for each experimental repeat (n = 11)
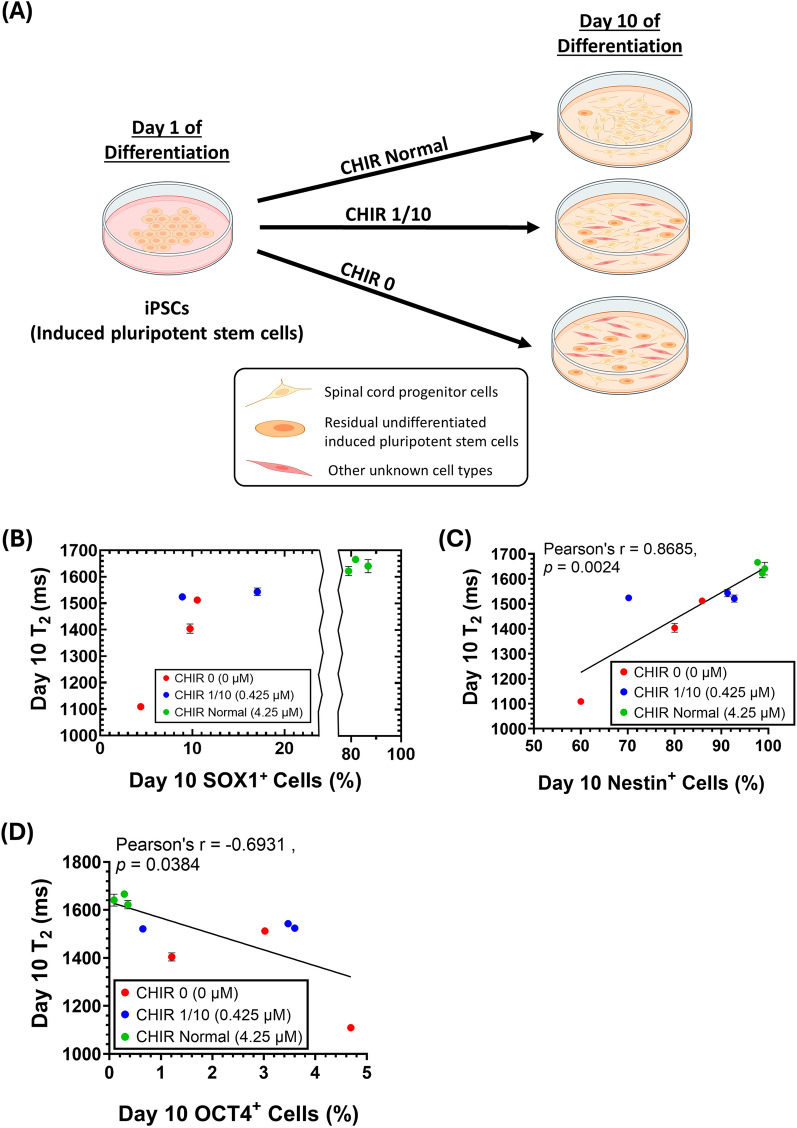
Fig. 6.
T2 value decreased in day 10 SCPCs with poor differentiation outcomes. A Reduction of CHIR resulted in a negative effect on SCPC differentiation to simulate poor manufacturing outcomes. Cell figures were adapted from the icon library of “Cell Types” by BioRender.com. B Day 10 T2 decreased with decreasing day 10 SCPC SOX1+ levels (n = 3). C Day 10 T2 decreased with decreasing day 10 SCPC Nestin+ levels (n = 3). D Day 10 T2 decreased with increasing day 10 SCPC OCT4+ levels (n = 3)
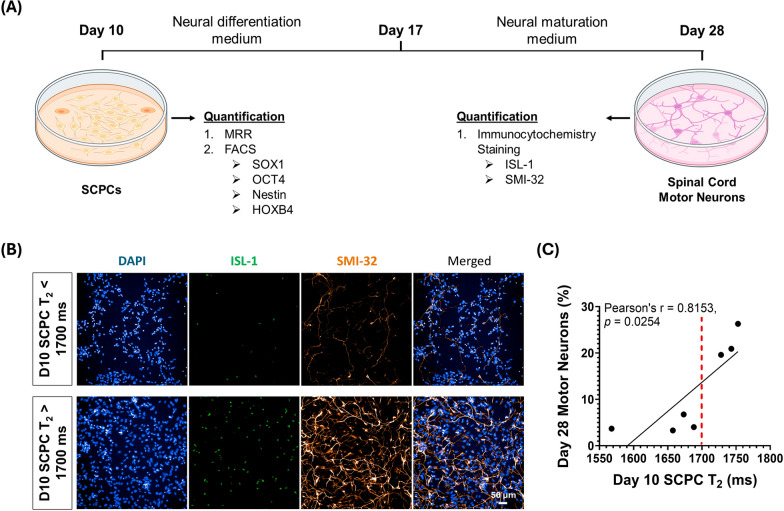
Fig. 7.
Day 10 SCPCs with T2 higher than 1700 ms has better extent of differentiation to Day 28 spinal cord motor neurons. A Day 10 SCPCs were differentiated into Day 28 spinal cord motor neurons and compared both T2 and Day 28 motor neuron marker expression. Cell figures were adapted from the icon library of “Cell Types” by BioRender.com. B Immunofluorescent staining of ISL-1 and SMI-32 markers for identification and quantification of motor neurons. A motor neuron is stained positive for both ISL-1 and SMI-32 (Scale bar: 50 µm). C Increasing level of expression in Day 28 motor neurons found in batches of day 10 SCPCs that had T2 values higher than 1700 ms as quantified by immunofluorescent staining. Red line separates the high and low motor neuron expressing groups. At least 25 ROIs (more than 80,000 cells) were quantified for each experimental repeat (n = 7)
Results
MRR can detect varying iPSC levels in SCPCs
To utilize intracellular iron as a biomarker to determine the levels of residual iPSCs in SCPCs, the levels of intracellular iron content within these cell types were analyzed. Specifically, a Fe3+ stain was utilized to quantify the intracellular iron levels of iPSCs and SCPCs via FACS. The results clearly indicated that iPSCs had a higher amount of Fe3+ than SCPCs (Fig. 2A).
To demonstrate that MRR can serve as a quality control method, the MRR readings from iPSCs and SCPCs were also evaluated. Correspondingly, the initial T2 measurements indicated that the values of iPSCs were 20–40% lower than SCPCs (p < 0.0001) (Fig. 2B), which corroborated with the results in Fig. 2A. Next, to demonstrate that T2 measurements are a good surrogate for label-free based quantification of residual iPSCs in SCPCs, an artificial spiking study was performed (Fig. 2C), by spiking the SCPCs with different known amounts of iPSCs.
The spiking of iPSCs resulted in a significant decrease in SCPC T2 (p < 0.0001) as the spiking ratio increased to 5% and beyond (Fig. 2D). Furthermore, T2 values correlated with increasing spiked iPSCs percentages (Pearson’s r = 0.958, p < 0.05). The results indicated that T2 measurements can reflect the varying levels of iPSCs in a SCPC culture.
T2 value increased as iPSCs differentiated into SCPCs
Since iPSCs and SCPCs have different MRR readings, we sought to better understand how T2 value changed as iPSCs differentiated into SCPCs (Fig. 3A). Changes in OCT4 and SOX1 expression were observed as iPSCs differentiated into SCPCs across three different time points, including Day 1 (undifferentiated iPSCs), Day 4 (mid-point differentiated iPSCs), and Day 10 (SCPCs). Figure 3B illustrates the immunostaining results of OCT4 and SOX1 on Day 1, Day 4, and Day 10. As iPSCs began to differentiate into SCPCs, high expression of SOX1 was observed on Day 4, while the expression of OCT4 decreased. On the final day of differentiation, SOX1 was still highly expressed with a relatively low expression of OCT4. This small cell population that expressed OCT4 was typically noted to be residual undifferentiated iPSCs (marked by white circles in the Day 10 merged image, Fig. 3B).
Figure 3C illustrates the relationship between the percentage of OCT4+ cells within a cell population and the corresponding T2 values. As iPSCs differentiated into SCPCs, a significant decrease (p < 0.0001) in OCT4+ levels was observed (Day 1 vs. Day 10, p < 0.0001, Fig. 3C). Correspondingly, a significant increase (p < 0.0001) in T2 was seen across the 10-day differentiation period (Fig. 3C).
MRR can detect residual OCT4+ iPSCs that arose from batch-to-batch; operator; and cell-line variations
To show that MRR can robustly detect varying levels of residual OCT4+ iPSCs in Day 10 SCPCs (the product), several manufacturing variations were evaluated, including batch-to-batch, operator and cell-line variations. Figure 4A illustrates the correlation between the T2 values and OCT4 expression levels that were assessed in Day 10 SCPCs. Specifically, a linear negative correlation was observed, where T2 values decreased with an increased number of OCT4+ cells in Day 10 SCPCs, regardless of operator variations (Fig. 4A).
The robustness of MRR in responding to cell line variations was also evaluated by characterizing the T2 values of another iPSC line (BJ-iPSC) and its differentiation outcomes. In Fig. 4A, OCT4+ cell percentages ranged from 5 to 20%, aligning well with MRR's effective detection range of 1000–2000 ms. However, the BJ-iPSC line exhibited significantly fewer OCT4+ cells after differentiation to SCPCs, causing MRR readings to fall outside the detection range. In Fig. 4B, by increasing the cell concentration to 180,000 cells per measurement, MRR was able to detect OCT4 levels below 1% with corresponding T2 values that were within the MRR detection range. Importantly, even though Day 10 BJ-SCPCs showed lower OCT4 levels than CLEC23-SCPCs, a negative linear correlation between OCT4 levels and T2 values was still observed (Fig. 4B).
Day 1 T2 values predicted OCT4 expression levels of Day 10 SCPCs
To extend the use of MRR as a tool to predict the final cellular differentiation outcomes based on measurements at an early time point, the T2 values of iPSCs after one day of SCPC differentiation were measured and compared against Day 10 SCPC differentiation extent.
Specifically, Day 1 undifferentiated iPSC T2 values were correlated to the corresponding Day 10 SCPC T2 values. The correlation was found to be significant and clearly indicated that Day 1 undifferentiated iPSCs with lower T2 values resulted in Day 10 SCPCs with lower T2 values (Fig. 5A). This predictive analysis was further extended to the OCT4 levels of Day 10 SCPCs. Hence, the results clearly indicated that Day 1 undifferentiated iPSCs with lower T2 values resulted in Day 10 SCPCs with higher OCT4+ cells (Fig. 5B). This suggests that MRR can provide a predictive indication of Day 10 outcomes via its Day 1 T2 values.
Low Day 10 SCPC T2 values indicates poor SCPC differentiation
In addition to detecting residual iPSCs, it is also crucial to ensure that iPSCs differentiate specifically into SOX1+ SCPCs and not other cell types. To evaluate the efficacy of MRR in identifying suboptimal differentiation outcomes, the extent of SCPC differentiation was modulated by reducing CHIR-99021 concentration in the neural differentiation media. CHIR-99021 is a potent GSK3 inhibitor that is responsible for the level of activation of the Wnt signaling pathway, which controls the differentiation efficiency of iPSCs to various progenitor cell types [8, 17].
The experimental groups included the absence of CHIR-99021 (CHIR 0, 0 µM), 1/10 the amount of CHIR-99021 that is used in a typical SCPC differentiation process (CHIR 1/10, 0.425 µM) and the control group (CHIR Normal, 4.25 µM) (Fig. 6A). Correspondingly, a decrease in CHIR concentration affected the levels of SOX1+, Nestin+ (neural stem cell marker) and OCT4+ cells in Day 10 SCPCs (Supplementary Figure S5A, S5B & S5C).
When the SCPCs were evaluated with MRR, T2 values were significantly lower in CHIR 0 (p < 0.01) and CHIR 1/10 (p < 0.0001) as compared to CHIR Normal (Supplementary Figure S5D). Importantly, the decrease in T2 values corresponded to the phenotypic changes (decrease in SOX1 and Nestin expressions and increase in OCT4 expression) in both CHIR 0 and CHIR 1/10 as compared to CHIR Normal (Fig. 6B–D). Notably, CHIR 0 led to a significant reduction of SOX1+ cells (< 10%), but the residual OCT4+ cells were only 5%. This suggests that MRR is also able to identify the poor differentiation outcomes in which the cells differentiate into non-SOX1+ SCPCs.
Day 10 T2 values of SCPCs predicts level of ISL-1/SMI-32 expression in Day 28 motor neurons
In addition to detecting poorly differentiated SCPCs, it is also crucial to ensure that the SCPCs can differentiate into spinal cord motor neurons which are essential to successful spinal cord repair and regeneration. To extend the use of MRR in providing an indication on SCPC quality, different batches of Day 10 SCPCs had their T2 values measured and compared against the level of motor neuron marker expression in Day 28 spinal cord motor neurons (Fig. 7A).
Interestingly, immunostaining images showed that Day 10 SCPCs with T2 values above 1700 ms had higher expression of motor neuron markers, ISL-1/SMI-32, as compared to those with T2 values below 1700 ms (Fig. 7B). We further confirmed this difference by identifying that the T2 values and motor neuron marker expression between the high and low motor neuron groups were significantly different (Supplementary Figure S6A and S6B). A red dashed line was drawn on Fig. 7C to indicate a point where both the T2 values and motor neuron marker expression significantly differs.
Moreover, there was also a significant correlation between the number of Day 28 motor neurons and their Day 10 SCPC T2 values clearly indicating that Day 10 SCPCs with higher T2 values resulted in Day 28 cells having a higher expression of motor neuron markers (Fig. 7C). This suggests that MRR can provide a predictive indication of Day 28 motor neuron marker expression outcomes via its Day 10 SCPC T2 values.
Discussion
In the context of cell manufacturing for clinical applications, the purity of cell therapy products is an important release criterion that is used to determine if the product is well-manufactured and ready for use [36]. For iPSC-based regenerative cell therapy products, technologies must be robust and consistent in detecting residual iPSCs or cells with the potential for tumorigenicity (safety), as well as quantifying the purity of the target cells as compared to other unwanted differentiated cells (quality). This is especially important as recent work has shown that iPSC-derived progenies can result in the development of tumors post-transplantation, which presents a significant roadblock to iPSC-based regenerative cell therapy applications [37, 38]. More importantly, these tumors were found to express pluripotency markers, including OCT4 and SOX2 [11, 39, 40]. Hence, the detection of OCT4+ cells in SCPCs is vital for ensuring and validating the safety of the SCPCs for use in SCI cell therapy applications. In this study, we hypothesized that cellular properties, such as intracellular iron content of iPSCs and SCPCs, can be distinctively different, which allows our MRR system to provide indications on the level of residual undifferentiated iPSCs after differentiation to SCPCs with T2 measurements.
Using our MRR system, we confirmed our hypothesis by observing a noticeable difference in T2 values of iPSCs and SCPCs (Fig. 2B). Spiking of iPSCs in SCPCs demonstrated that various percentages of iPSCs can be detected using the MRR system (Fig. 2C). We also demonstrated the safety risks in SCPCs that were associated with lower T2 values (due to an increasing presence of residual undifferentiated iPSCs) with a colony culture assay (Supplementary Figure S2). These results confirmed that MRR was able to evaluate safety levels via the T2 values of SCPCs.
To better test MRR in more practical scenarios, we further applied it to detect variations in residual OCT4+ iPSCs that arose from batch-to-batch, operator and cell line differences (Fig. 4). The correlations indicate that MRR is a robust, consistent, and reproducible endpoint analytical technique that can determine the level of residual undifferentiated iPSCs in Day 10 SCPCs. Moreover, we demonstrated that T2 values of a batch of undifferentiated iPSCs can provide predictive indications of its Day 10 SCPC OCT4 levels (Fig. 5). This further indicates that T2 measurements of iPSCs, as early as day 1 in the differentiation process, may be used to predict the differentiation efficiency outcome of iPSCs to SCPCs. Such a predictive correlation is highly beneficial in determining if a batch of iPSCs would result in a high-quality batch of SCPCs. Such an early-stage quality assessment tool would be highly desirable in iPSC manufacturing workflows, which may suffer from significant line-to-line and batch-to-batch quality variations.
Even though the lack of undifferentiated iPSCs within SCPC populations, as measured by MRR, could serve as a safety CQA, it does not automatically translate into detecting batches of SCPCs with high levels of purity. To test if MRR can detect the presence of non-SOX1+ cells (i.e., differentiated cells that are non-SCPCs), we extended the use of MRR to detect poor manufacturing outcomes by negatively disrupting iPSC differentiation towards SCPCs. Our findings indicate that poorly differentiated SCPCs had a significant decrement of T2. Our previous observations in Fig. 4A, B suggest that the increased presence of OCT4+ cells possibly contributed to the decrease in T2 of poorly differentiated SCPCs. However, SOX1+ cells in poorly differentiated SCPCs were reduced to less than 10%, whilst the residual OCT4+ cells only increased to 5%. Hence, this huge decrement of T2 (~ 400 ms) should have been largely contributed by the presence of non-SOX1+ cells. These findings indicate that MRR was also able to identify the poor differentiation outcomes in which the cells differentiate into non-SOX1+ SCPCs, and its T2 values can indicate if the batch was of poor quality.
As the MRR system measures the T2 of live cells, it provides a unique window to the iron biology of cells. T2 measurements of iPSCs and SCPCs in this study were well correlated to markers that require terminal fixation for analysis (e.g., OCT4, SOX1, Nestin). Moreover, this method requires a relatively small number of cells (< 2 × 105) and does not require any chemical or biological processing, allowing the same cells to be subjected to subsequent biological or functional measurements that can be compared or correlated to their T2 values. This was demonstrated in previous works where MRR was used to indicate MSC quality by quantifying their senescence levels, which had a direct effect on its chondrogenic differentiation efficiency [20].
In this study, we used MRR to assess SCPC quality by determining if a batch could efficiently differentiate into spinal cord motor neurons, a key cell type required for effective spinal cord regeneration and functional recovery [32, 41, 42]. Notably, we demonstrated that the use of MRR as a quality metric was more definitive than the use of gene expression markers (requires destructive measurements) as an indicator of cell fate commitment. We noted that there was no correlation between any of the cellular markers that were quantified for Day 10 SCPCs with the level of Day 28 motor neuron marker expression (Supplementary Figure S6C–S6F). On the other hand, MRR was able to provide predictive indications of its Day 28 motor neuron marker expression via its Day 10 SCPC T2 values (Fig. 7C). Hence, predictive correlations as such will be highly beneficial in determining if any given batch of SCPCs will have a high differentiation efficiency towards Day 28 motor neurons.
Iron homeostasis has been shown to play a role in regulating pluripotency in iPSCs [23, 24] previously, and this could potentially provide a connection between changes in iron levels and the differentiation efficiency of iPSCs to SCPCs. Although the exact mechanistic relationship between intracellular iron level and cellular phenotype (such as stem cells, progenitors, and fully differentiated cells) is poorly understood, several recent studies have suggested the importance of intracellular iron content on cell fate commitment. For example, the differentiation of iPSCs into neural and cardiac progenitors has been enhanced with iron-treated media or the addition of iron-binding proteins [43, 44]. The degradation of ferritin was also shown to significantly affect the differentiation efficiency of MSCs to neural cells [45]. Since ferritin stores up to 4500 atoms of Fe3+, such observation suggests the involvement of iron in MSC neural lineage commitment. Hematopoietic stem cells (HSCs) also utilise readily accessible iron in the cytoplasm as a key cellular rheostat enabling them to regulate their cell fate whilst in quiescent and active states [46]. Hence, this suggests that the use of MRR to provide indications of subsequent cell fate commitment via iron-related measurements heralds significant potential.
Cellular iron content has been shown to have numerous potential implications in many physiological and pathophysiological processes such as aging [46–48], immune cell activation [49, 50], cellular senescence [51, 52], cancer [53–55], and inflammation [56–58]. Yet, there is no systematic understanding regarding the exact roles of Fe2+/Fe3+ in these critical biological contexts. This knowledge gap is further exacerbated by difficulties in obtaining accurate measurements of Fe2+ and Fe3+. The conventional assays and methods for detecting Fe2+ [59–61], Fe3+ [62, 63] and even total iron concentration [64–66], are largely inaccurate due to the unstable nature of iron ions, inaccurate correlations and the limitation of current technologies. Using MRR, we confirmed that T2 measurements of iPSCs, Day 4 semi-differentiated cells and SCPCs corroborate with their intracellular Fe3+ levels via a Fe3+ stain analyzed with FACS (Supplementary Figure S7). These findings clearly indicated that T2 measurements from MRR are a good surrogate for the current methods of measuring intracellular Fe3+ content.
Even though MRR can provide cell quality and safety information whilst being label-free and non-destructive, it is not without its limitations. Although we report that T2 measurements can be an indicator of cell quality, cell differentiation changes were evaluated only by phenotypic marker expressions. Hence, future works should focus on applying MRR to provide predictive indications for actual in vivo tissue repair and regeneration from the use of iPSC-based cell therapy products. In addition to assessing cell batches, there is potential for MRR to be adapted into a safety monitoring method for in vivo applications. Magnetic resonance imaging, which relies on MRR as one of its core mechanisms, has been extensively used to diagnose tissue and cellular changes, especially for the detection of neoplastic lesions [67–69]. This suggests that our MRR platform could be engineered to monitor tumorigenicity in an in vivo model by scanning tissues from the surface. Alternatively, standard MRI scanners with high spatial resolution (~ 100 µm) could be used, although this approach would be relatively more costly.
It is also essential to note that variations in differentiation efficiency likely stem from the initial quality of iPSCs. Specifically, iron overload or underload conditions in iPSCs can severely impact the maintenance of their pluripotency [23, 24]. Therefore, future studies may explore the use of MRR to elucidate the roles of iron homeostasis in iPSCs for quality control purposes. Also, given that mechanical stresses, such as external physical vibration of cell cultures, have significant downstream impacts on the maintenance of iPSCs [70], future works may also explore the applicability of MRR in monitoring the corresponding iPSC quality variations. The MRR technology may significantly impact the manufacturing processes of iPSC-based cell products and will be of considerable benefit to the cell therapy manufacturing industry.
Conclusion
Using intracellular Fe3+ content to quantify iPSC and SCPC phenotypes, we demonstrated the capabilities of our label-free MRR technology to perform rapid and non-destructive endpoint assessment of the safety and quality of SCPCs. T2 measurements were shown to correlate well with conventional biochemical assays of iPSC and SCPC phenotyping. Furthermore, T2 measurements were also able to provide predictive indications of its day 10 SCPC OCT4 levels based on day 1 iPSC T2 values and the differentiation extent towards day 28 motor neurons based on day 10 SCPC T2 values. This indicates that T2 measurements via the MRR system is ideally suited to monitor and assess the safety and quality of iPSC-derived SCPCs that are produced for spinal cord injury cell therapy applications.
Supplementary Information
Acknowledgements
We would like to express our sincere gratitude to Professor Lim Kah Leong and Dr Chai Chou, SingHealth, Singapore and Associate Professor Phan Toan Thang, CellResearch Corporation, Singapore for providing us with their proprietary CLiPS cells. The authors declare that they have not used Artificial Intelligence in this study.
Author contributions
J.T., S.Y.C., J.H.: conceptualized and designed the overall experimental plan. J.T.: carried out all the MRR experiments and data analysis with support from J.C., D.N.R., W.H.C., T.D.N. Financial support, administrative support, data analysis and interpretations were provided by S.Y.N., S.Y.C., J.H. The manuscript was written by J.T. All authors participated in editing and revision of the manuscript.
Funding
This research was supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme (IntraCREATE grant award number: NRF2019-THE002-0001) and Singapore MIT Alliance for Research and Technology (SMART): Critical Analytics for Manufacturing Personalised-Medicine (CAMP) Inter-Disciplinary Research Group. J.T. would like to acknowledge NTU for providing the Nanyang Research Scholarship that made it possible to carry out the research works.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study utilized deidentified iPSC lines and does not involve animals, clinical experiments, or primary patient-derived human tissues. Derivation of the CLEC23 iPSC line had received prior IRB approval (document no.: IRB-2020-05-026, date of approval: 02/06/2020, name of institution: NTU). BJ-iPSCs were derived from the BJ (ATCC CRL-2522) cell line obtained from ATCC (https://www.atcc.org/products/crl-2522) and does not require approvals for derivation as it does not involve the use of patient-derived material. NTU-IRB waived the need for ethical approvals regarding this study in accordance with relevant guidelines and regulations following the Singapore’s Human Tissue Framework under the Human Biomedical Research Act. In accordance to Singapore’s Human Tissue Framework, any form of human biological material which has been substantially manipulated and de-identified are no longer considered human material, and, therefore, ethical approvals or consent to participate does not apply.
Consent for publication
Not applicable.
Competing interests
J.H., S.Y.C., D.N.R, J.T. filed an intellectual property with a patent that is co-owned by SMART Singapore-MIT Alliance for Research and Technology and NTU. The other authors declared no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jongyoon Han, Email: jyhan@mit.edu.
Sing Yian Chew, Email: sychew@ntu.edu.sg.
References
- 1.Yousefifard M, Rahimi-Movaghar V, Nasirinezhad F, Baikpour M, Safari S, Saadat S, et al. Neural stem/progenitor cell transplantation for spinal cord injury treatment. A systematic review and meta-analysis. Neuroscience. 2016;13(322):377–97. [DOI] [PubMed] [Google Scholar]
- 2.Nagoshi N, Tsuji O, Nakamura M, Okano H. Cell therapy for spinal cord injury using induced pluripotent stem cells. Regen Ther. 2019;13(11):75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura M, Okano H. Cell transplantation therapies for spinal cord injury focusing on induced pluripotent stem cells. Cell Res. 2013;23(1):70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin JE, Jung K, Kim M, Hwang K, Lee H, Kim IS, et al. Brain and spinal cord injury repair by implantation of human neural progenitor cells seeded onto polymer scaffolds. Exp Mol Med. 2018;50(4):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nori S, Okada Y, Yasuda A, Tsuji O, Takahashi Y, Kobayashi Y, et al. Grafted human-induced pluripotent stem-cell–derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci. 2011;108(40):16825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuda A, Tsuji O, Shibata S, Nori S, Takano M, Kobayashi Y, et al. Significance of remyelination by neural stem/progenitor cells transplanted into the injured spinal cord. Stem Cells. 2011;29(12):1983–94. [DOI] [PubMed] [Google Scholar]
- 7.Qu Q, Li D, Louis KR, Li X, Yang H, Sun Q, et al. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat Commun. 2014;5(1):3449. [DOI] [PubMed] [Google Scholar]
- 8.Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 2015;6(1):6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balafkan N, Mostafavi S, Schubert M, Siller R, Liang KX, Sullivan G, et al. A method for differentiating human induced pluripotent stem cells toward functional cardiomyocytes in 96-well microplates. Sci Rep. 2020;10(1):18498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11(4):268–77. [DOI] [PubMed] [Google Scholar]
- 11.Nori S, Okada Y, Nishimura S, Sasaki T, Itakura G, Kobayashi Y, et al. Long-term safety issues of iPSC-based cell therapy in a spinal cord injury model: oncogenic transformation with epithelial-mesenchymal transition. Stem Cell Rep. 2015;4(3):360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlos J, L. A. Safety assessment of reprogrammed cells prior to clinical applications: potential approaches to eliminate teratoma formation. In: Bhartiya D, editor. Pluripotent stem cells [Internet]. InTech; 2013 [cited 2022 Jul 19]. Available from: http://www.intechopen.com/books/pluripotent-stem-cells/safety-assessment-of-reprogrammed-cells-prior-to-clinical-applications-potential-approaches-to-elimi.
- 13.Kuroda T, Yasuda S, Kusakawa S, Hirata N, Kanda Y, Suzuki K, et al. Highly sensitive in vitro methods for detection of residual undifferentiated cells in retinal pigment epithelial cells derived from human iPS cells. PLoS ONE. 2012;7(5): e37342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin L, Wu Y, Yang Z, Denslin V, Ren X, Tee CA, et al. Characterization and application of size-sorted zonal chondrocytes for articular cartilage regeneration. Biomaterials. 2018;165:66–78. [DOI] [PubMed] [Google Scholar]
- 15.Dai T, Hon W. Label-free and high-throughput removal of residual undifferentiated cells from iPSC-derived spinal cord progenitor cells. Stem Cells Transl Med. 2024;13:387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeming KK, Vernekar R, Chua MT, Quek KY, Sutton G, Krüger T, et al. Label-free biophysical markers from whole blood microfluidic immune profiling reveal severe immune response signatures. Small. 2021;17(12):2006123. [DOI] [PubMed] [Google Scholar]
- 17.Qian T, Heaster TM, Houghtaling AR, Sun K, Samimi K, Skala MC. Label-free imaging for quality control of cardiomyocyte differentiation. Nat Commun. 2021;12(1):4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petchakup C, Yang H, Gong L, He L, Tay HM, Dalan R, et al. Microfluidic impedance-deformability cytometry for label-free single neutrophil mechanophenotyping. Small. 2022;18(18):2104822. [DOI] [PubMed] [Google Scholar]
- 19.He L, Tan J, Ng SY, Li KHH, Han J, Chew SY, et al. Label-free impedance analysis of induced pluripotent stem cell-derived spinal cord progenitor cells for rapid safety and efficacy profiling. Adv Mater Technol. 2024;5:2400589. [Google Scholar]
- 20.Thamarath SS, Tee CA, Neo SH, Yang D, Othman R, Boyer LA, et al. Rapid and live-cell detection of senescence in mesenchymal stem cells by micro magnetic resonance relaxometry. Stem Cells Transl Med. 2023;9:szad014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng WK, Kong TF, Ng CS, Chen L, Huang Y, Bhagat AAS, et al. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat Med. 2014;20(9):1069–73. [DOI] [PubMed] [Google Scholar]
- 22.Peng WK, Chen L, Boehm BO, Han J, Loh TP. Molecular phenotyping of oxidative stress in diabetes mellitus with point-of-care NMR system. Npj Aging Mech Dis. 2020;6(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z, Yu Y, Xu J, Bao Z, Xu Z, Hu J, et al. Iron homeostasis determines fate of human pluripotent stem cells via glycerophospholipids-epigenetic circuit. Stem Cells. 2019;37(4):489–503. [DOI] [PubMed] [Google Scholar]
- 24.Han Z, Xu Z, Chen L, Ye D, Yu Y, Zhang Y, et al. Iron overload inhibits self-renewal of human pluripotent stem cells via DNA damage and generation of reactive oxygen species. FEBS Open Bio. 2020;10(5):726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petronek MS, St-Aubin JJ, Lee CY, Spitz DR, Gillan EG, Allen BG, et al. Quantum chemical insight into the effects of the local electron environment on T2*-based MRI. Sci Rep. 2021;11(1):20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Gan SU, Lin G, Lim YT, Masilamani J, Mustafa FB, et al. Characterization of human umbilical cord lining-derived epithelial cells and transplantation potential. Cell Transplant. 2011;20(11–12):1827–41. [DOI] [PubMed] [Google Scholar]
- 27.Saleh R, Reza HM. Short review on human umbilical cord lining epithelial cells and their potential clinical applications. Stem Cell Res Ther. 2017;8(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim RHG, Liew JXK, Wee A, Masilamani J, Chang SKY, Phan TT. Safety evaluation of human cord-lining epithelial stem cells transplantation for liver regeneration in a porcine model. Cell Transplant. 2020;29:963689719896559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winanto N, Khong ZJ, Soh BS, Fan Y, Ng SY. Organoid cultures of MELAS neural cells reveal hyperactive Notch signaling that impacts neurodevelopment. Cell Death Dis. 2020;11(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng SY, Soh BS, Rodriguez-Muela N, Hendrickson DG, Price F, Rinn JL, et al. Genome-wide RNA-seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell. 2015;17(5):569–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hor JH, Santosa MM, Lim VJW, Ho BX, Taylor A, Khong ZJ, et al. ALS motor neurons exhibit hallmark metabolic defects that are rescued by SIRT3 activation. Cell Death Differ. 2021;28(4):1379–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajikawa K, Imaizumi K, Shinozaki M, Shibata S, Shindo T, Kitagawa T, et al. Cell therapy for spinal cord injury by using human iPSC-derived region-specific neural progenitor cells. Mol Brain. 2020;13(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tay SH, Winanto, Khong ZJ, Koh YH, Ng SY. Generation of cortical, dopaminergic, motor, and sensory neurons from human pluripotent stem cells. In: Methods in molecular biology. New York: Springer; 2021. Available from: 10.1007/7651_2021_399. [DOI] [PubMed]
- 34.Kumamaru H, Kadoya K, Adler AF, Takashima Y, Graham L, Coppola G, et al. Generation and post-injury integration of human spinal cord neural stem cells. Nat Methods. 2018;15(9):723–31. [DOI] [PubMed] [Google Scholar]
- 35.Wei Y, Aydin Z, Zhang Y, Liu Z, Guo M. A turn-on fluorescent sensor for imaging labile Fe3+ in live neuronal cells at subcellular resolution. ChemBioChem. 2012;13(11):1569–73. [DOI] [PubMed] [Google Scholar]
- 36.Stroncek DF, Jin P, Ren J, Feng J, Castiello L, Civini S, et al. Quality assessment of cellular therapies: the emerging role of molecular assays. Korean J Hematol. 2010;45(1):14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19(8):998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lezmi E, Jung J, Benvenisty N. High prevalence of acquired cancer-related mutations in 146 human pluripotent stem cell lines and their differentiated derivatives. Nat Biotechnol. 2024;9:1–5. [DOI] [PubMed] [Google Scholar]
- 39.Han L, He H, Yang Y, Meng Q, Ye F, Chen G, et al. Distinctive clinical and pathologic features of immature teratomas arising from induced pluripotent stem cell-derived beta cell injection in a diabetes patient. Stem Cells Dev. 2022;31(5–6):97–101. [DOI] [PubMed] [Google Scholar]
- 40.Sugai K, Sumida M, Shofuda T, Yamaguchi R, Tamura T, Kohzuki T, et al. First-in-human clinical trial of transplantation of iPSC-derived NS/PCs in subacute complete spinal cord injury: study protocol. Regen Ther. 2021;1(18):321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki H, Imajo Y, Funaba M, Nishida N, Sakamoto T, Sakai T. Current concepts of neural stem/progenitor cell therapy for chronic spinal cord injury. Front Cell Neurosci. 2022;3(15): 794692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosseini SM, Borys B, Karimi-Abdolrezaee S. Neural stem cell therapies for spinal cord injury repair: an update on recent preclinical and clinical advances. Brain. 2024;147(3):766–93. [DOI] [PubMed] [Google Scholar]
- 43.Lu D, Chen EYT, Lee P, Wang YC, Ching W, Markey C, et al. Accelerated neuronal differentiation toward motor neuron lineage from human embryonic stem cell line (H9). Tissue Eng Part C Methods. 2015;21(3):242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, Qiu H, Dong X, Wang C, Na J, Zhou J, et al. Transferrin improved the generation of cardiomyocyte from human pluripotent stem cells for myocardial infarction repair. J Mol Histol. 2021;52(1):87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HN, Ko KN, Kim HJ, Rosebud Aikins A, Kim CW. Ferritin is associated with neural differentiation of bone marrow-derived mesenchymal stem cells under extremely low-frequency electromagnetic field. Cell Mol Biol Noisy Gd Fr. 2015;61(7):55–9. [PubMed] [Google Scholar]
- 46.Kao YR, Chen J, Kumari R, Ng A, Zintiridou A, Tatiparthy M, et al. An iron rheostat controls hematopoietic stem cell fate. Cell Stem Cell. 2024;31(3):378-397.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen WJ, Kung GP, Gnana-Prakasam JP. Role of iron in aging related diseases. Antioxidants. 2022;11(5):865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato T, Shapiro JS, Chang HC, Miller RA, Ardehali H. Aging is associated with increased brain iron through cortex-derived hepcidin expression. Elife. 2022;11(11):e73456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuvibidila S, Warrier RP, Surendra BB. An overview of the role of iron in T cell activation. J Trace Elem Exp Med. 2003;16(4):219–25. [Google Scholar]
- 50.Ni S, Yuan Y, Kuang Y, Li X. Iron metabolism and immune regulation. Front Immunol. 2022. 10.3389/fimmu.2022.816282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Killilea DW, Wong SL, Cahaya HS, Atamna H, Ames BN. Iron accumulation during cellular senescence. Ann N Y Acad Sci. 2004;1019(1):365–7. [DOI] [PubMed] [Google Scholar]
- 52.Cozzi A, Orellana DI, Santambrogio P, Rubio A, Cancellieri C, Giannelli S, et al. Stem cell modeling of neuroferritinopathy reveals iron as a determinant of senescence and ferroptosis during neuronal aging. Stem Cell Rep. 2019;13(5):832–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manz DH, Blanchette NL, Paul BT, Torti FM, Torti SV. Iron and cancer: recent insights. Ann N Y Acad Sci. 2016;1368(1):149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Q, Li L, Hou S, Yuan Z, Li C, Zhang W, et al. The role of iron in cancer progression. Front Oncol. 2021. 10.3389/fonc.2021.778492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basak T, Kanwar RK. Iron imbalance in cancer: intersection of deficiency and overload. Cancer Med. 2022;11(20):3837–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessling-Resnick M. Iron homeostasis and the inflammatory response. Annu Rev Nutr. 2010;21(30):105–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kell DB, Pretorius E. Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics. 2014;6(4):748–73. [DOI] [PubMed] [Google Scholar]
- 58.Marques O, Weiss G, Muckenthaler MU. The role of iron in chronic inflammatory diseases: from mechanisms to treatment options in anemia of inflammation. Blood. 2022;140(19):2011–23. [DOI] [PubMed] [Google Scholar]
- 59.Hirayama T, Nagasawa H. Chemical tools for detecting Fe ions. J Clin Biochem Nutr. 2017;60(1):39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenopoulou M, Kurz T, Doulias PT, Galaris D, Brunk UT. Does the calcein-AM method assay the total cellular ‘labile iron pool’ or only a fraction of it? Biochem J. 2007;403(Pt 2):261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abbasi U, Abbina S, Gill A, Bhagat V, Kizhakkedathu JN. A facile colorimetric method for the quantification of labile iron pool and total iron in cells and tissue specimens. Sci Rep. 2021;16(11):6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohrer JS, Joo MS, Dartyge E, Sayers DE, Fontaine A, Theil EC. Stabilization of iron in a ferrous form by ferritin. A study using dispersive and conventional x-ray absorption spectroscopy. J Biol Chem. 1987;262(28):13385–7. [PubMed] [Google Scholar]
- 63.Knovich MA, Storey JA, Coffman LG, Torti SV. Ferritin for the clinician. Blood Rev. 2009;23(3):95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fiorito V, Geninatti Crich S, Silengo L, Altruda F, Aime S, Tolosano E. Assessment of iron absorption in mice by ICP-MS measurements of (57)Fe levels. Eur J Nutr. 2012;51(7):783–9. [DOI] [PubMed] [Google Scholar]
- 65.Song C, Wang J, Mo C, Mu S, Jiang X, Li X, et al. Use of ferritin expression, regulated by neural cell-specific promoters in human adipose tissue-derived mesenchymal stem cells, to monitor differentiation with magnetic resonance imaging in vitro. PLoS ONE. 2015;10(7): e0132480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arı B, Can SZ, Bakırdere S. Traceable and accurate quantification of iron in seawater using isotope dilution calibration strategies by triple quadrupole ICP-MS/MS: characterization measurements of iron in a candidate seawater CRM. Talanta. 2020;1(209): 120503. [DOI] [PubMed] [Google Scholar]
- 67.Essig M, Anzalone N, Combs SE, Dörfler A, Lee SK, Picozzi P, et al. MR imaging of neoplastic central nervous system lesions: review and recommendations for current practice. AJNR Am J Neuroradiol. 2012;33(5):803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qu J, Pan B, Su T, Chen Y, Zhang T, Chen X, et al. T1 and T2 mapping for identifying malignant lymph nodes in head and neck squamous cell carcinoma. Cancer Imaging. 2023;23(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martucci M, Russo R, Schimperna F, D’Apolito G, Panfili M, Grimaldi A, et al. Magnetic resonance imaging of primary adult brain tumors: state of the art and future perspectives. Biomedicines. 2023;11(2):364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanie K, Sakai T, Imai Y, Yoshida K, Sugimoto A, Makino H, et al. Effect of mechanical vibration stress in cell culture on human induced pluripotent stem cells. Regen Ther. 2019;15(12):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.