Abstract
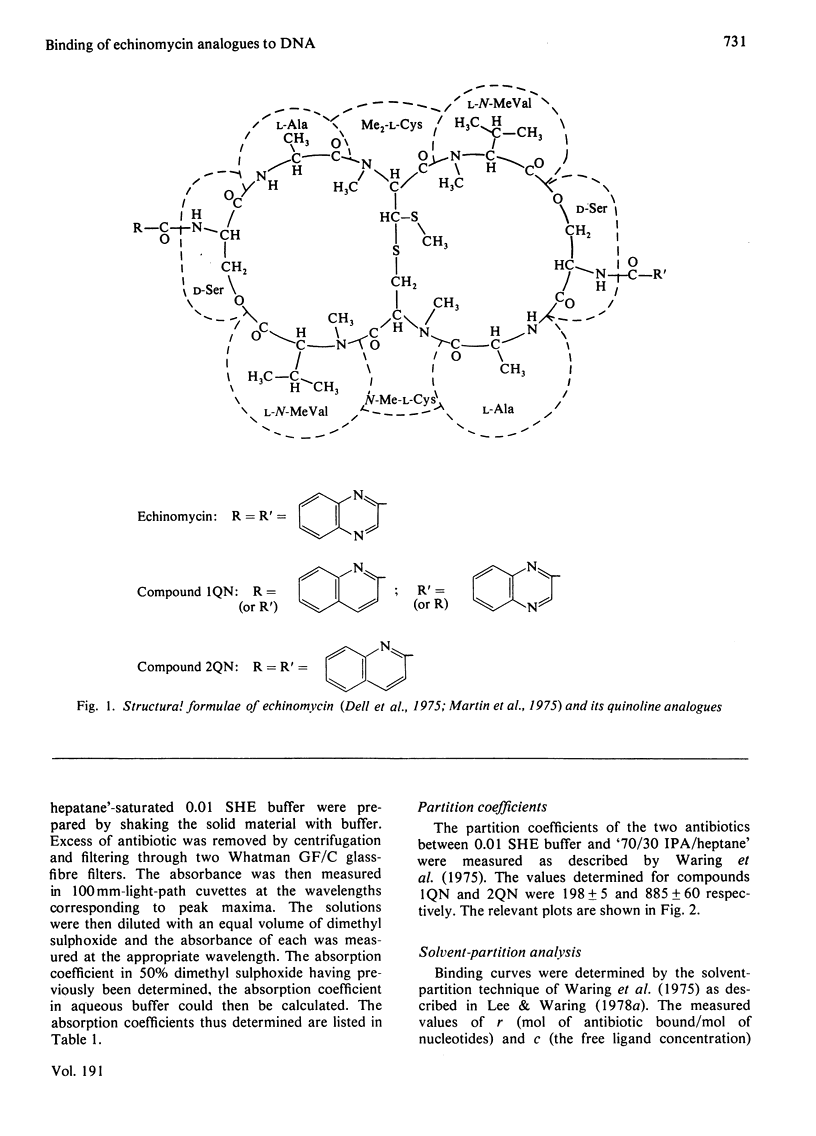
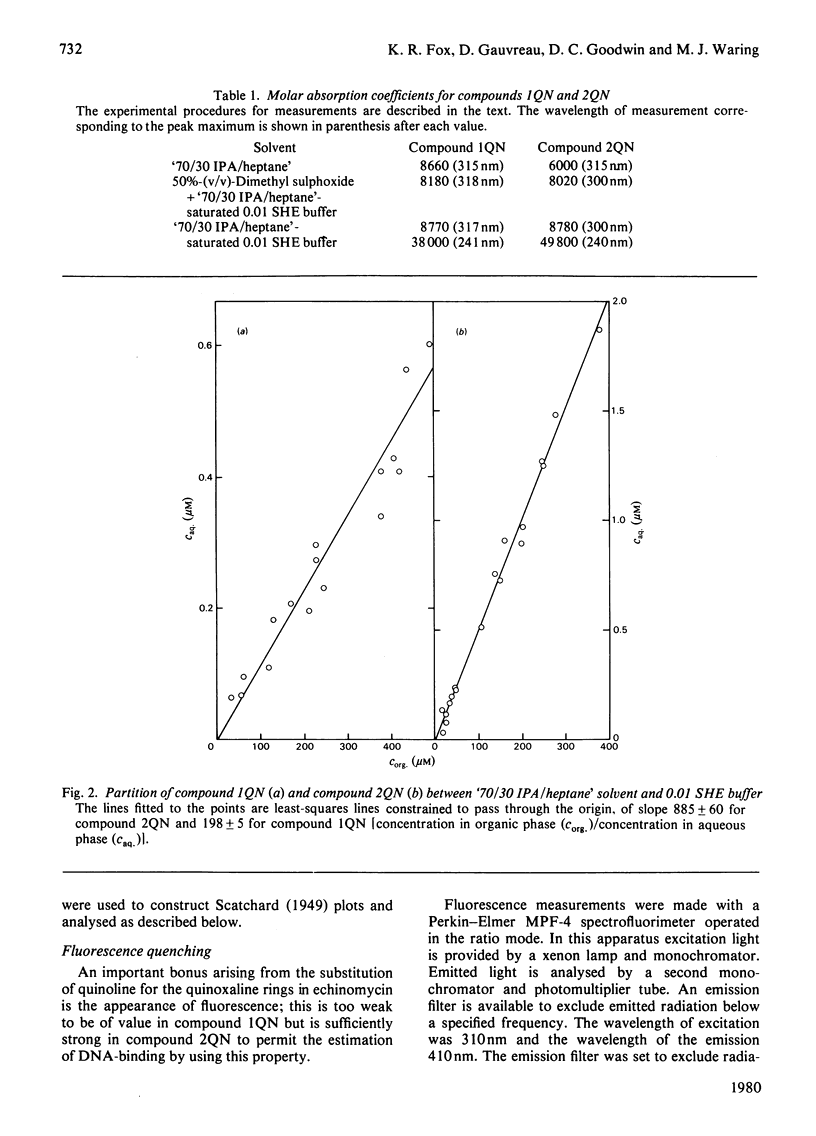
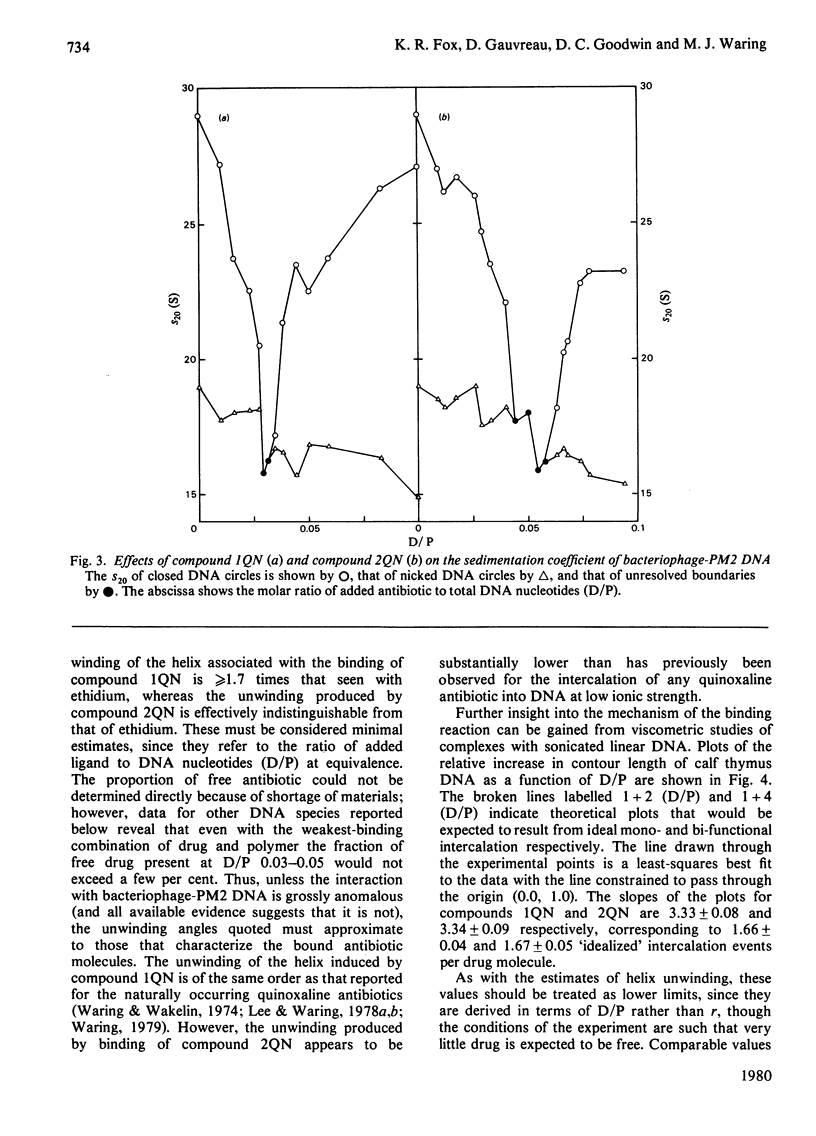
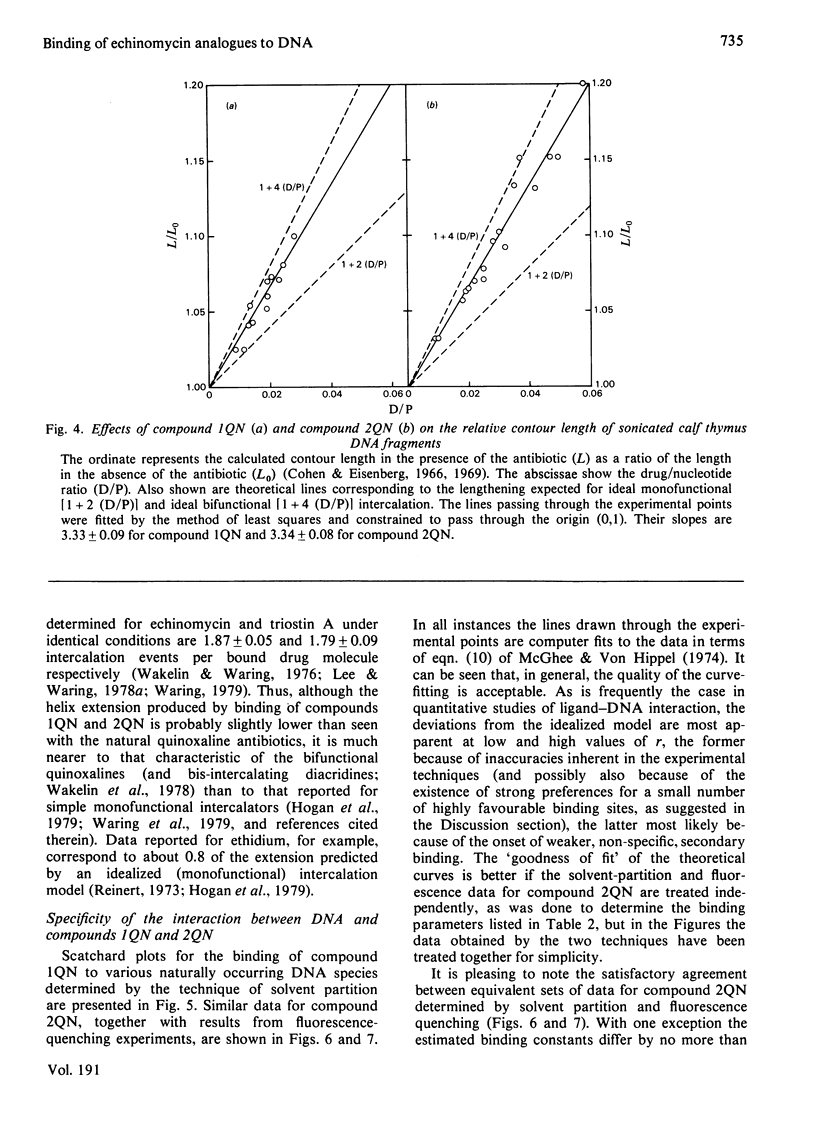
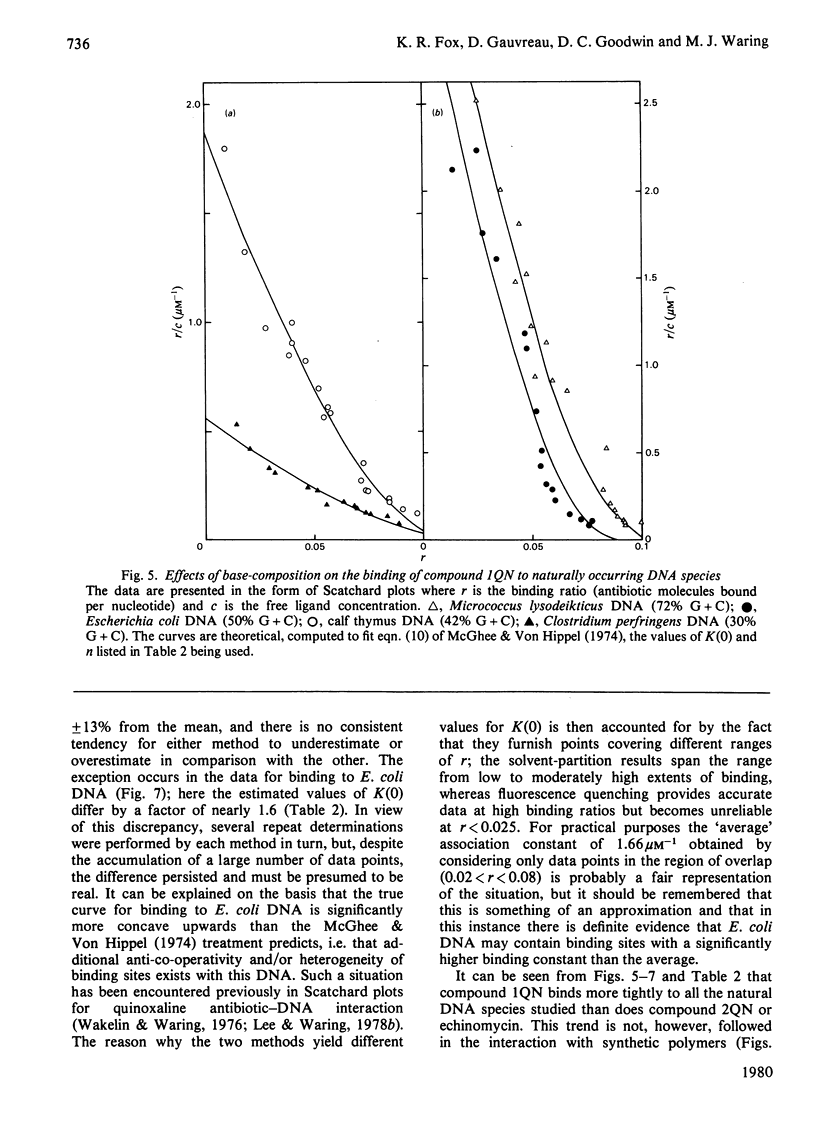
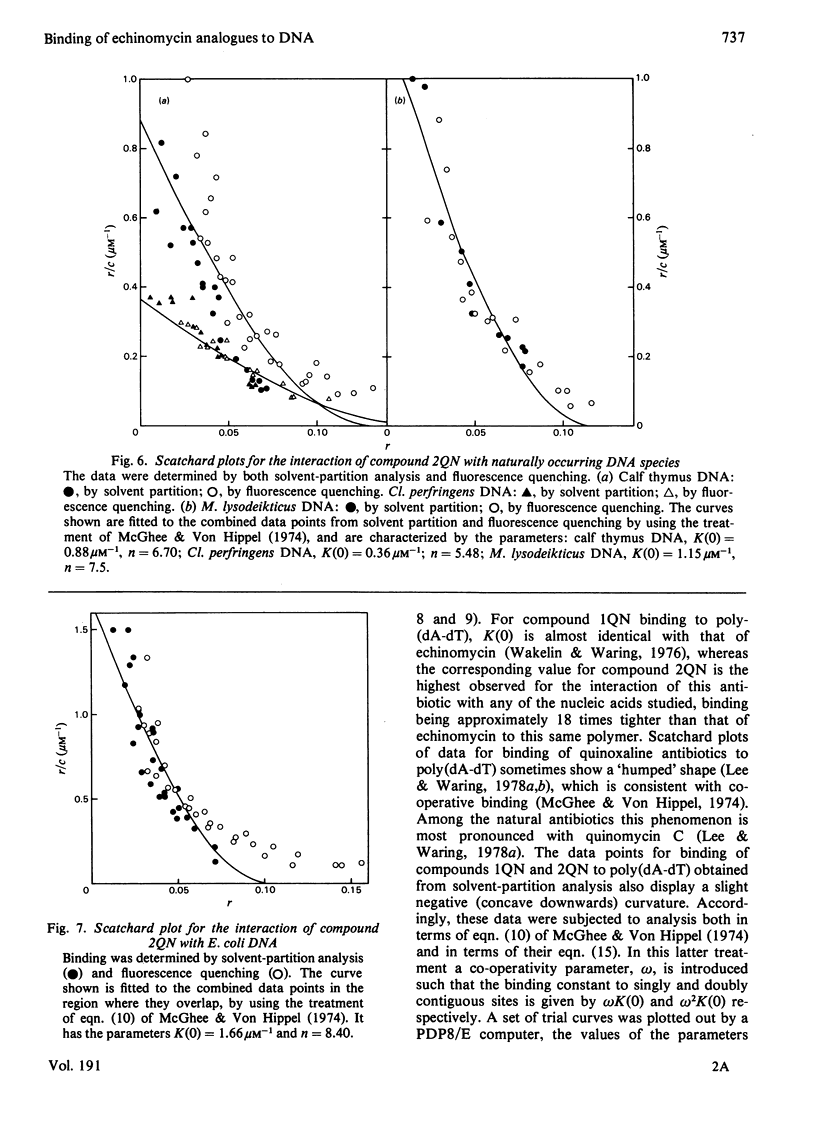
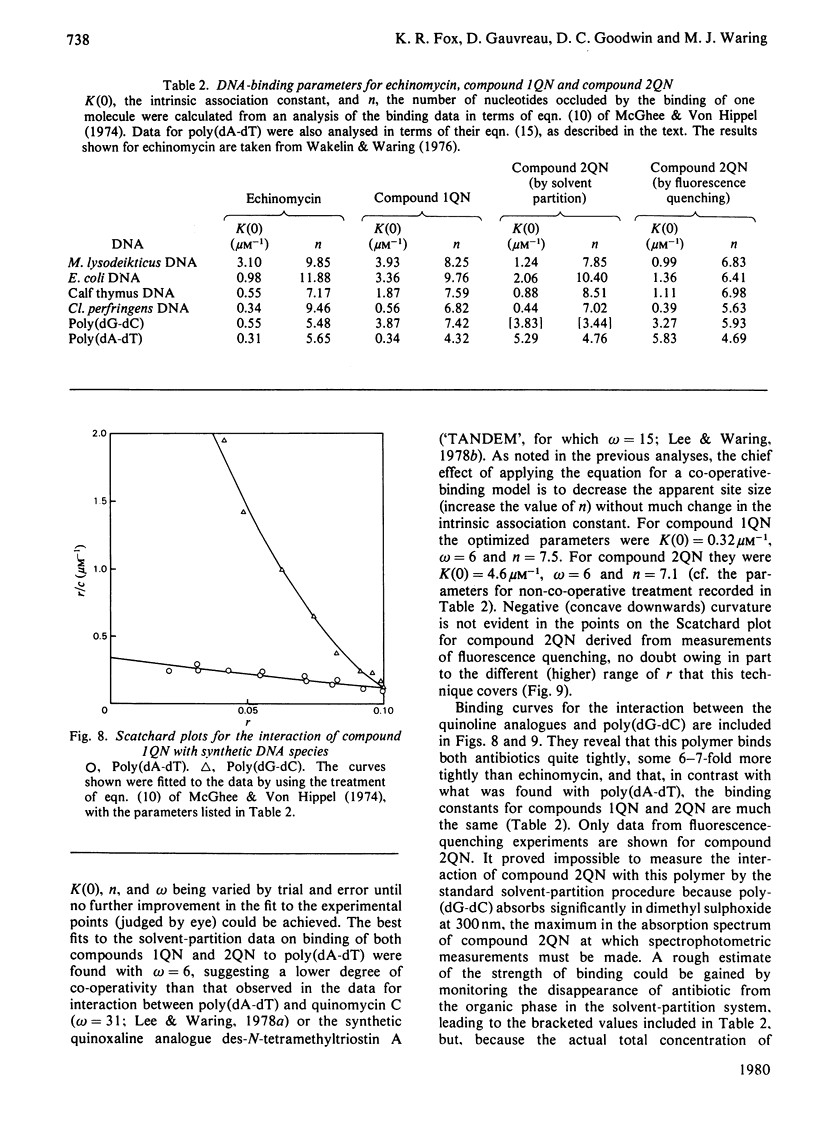
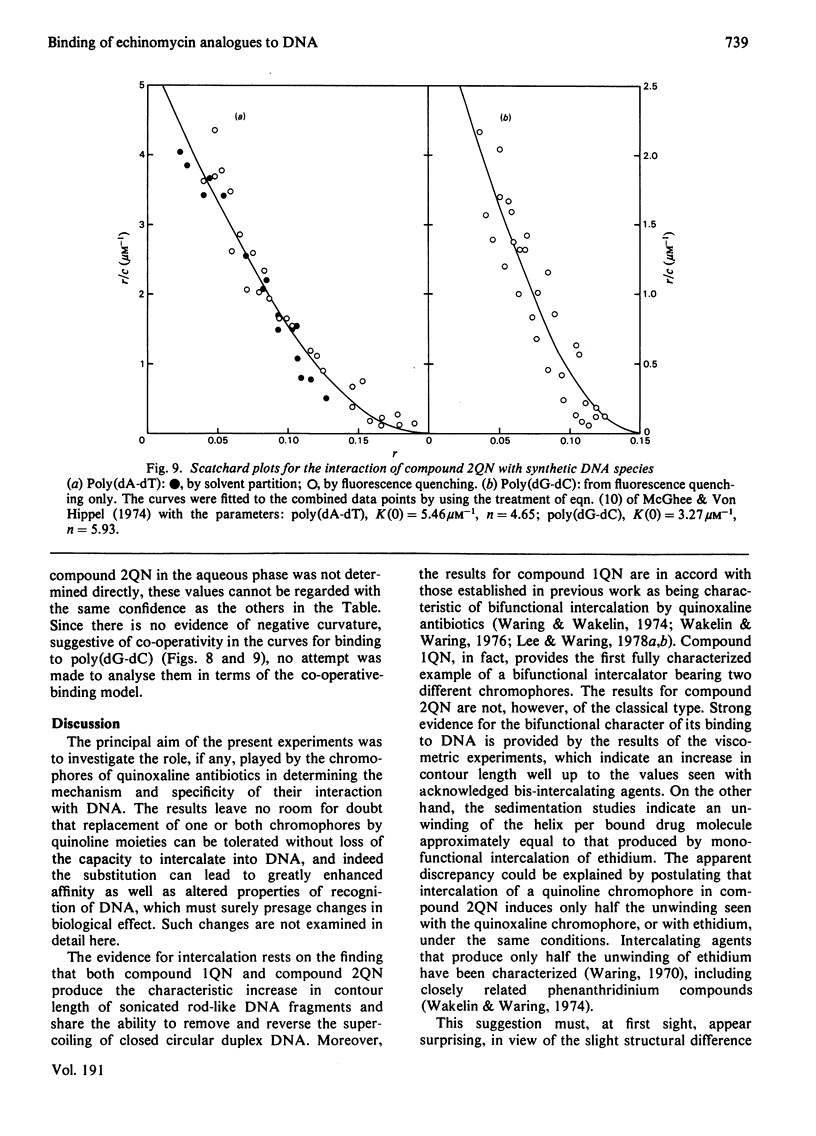
Two novel antibiotics were isolated, designated compounds 1QN and 2QN respectively, having quinoline rings in place of one or both of the quinoxaline chromophores of echinomycin. Each removes and reverses the supercoiling of closed circular duplex DNA from bacteriophage PM2 in the fashion characteristic of intercalating drugs. For compound 1QN, the unwinding angle at I0.01 is almost twice that of ethidium, whereas for compound 2QN the value is indistinguishable from that of ethidium. Binding of both analogues produced changes in the viscosity of sonicated rod-like DNA fragments corresponding to double the helix extension found with ethidium, a feature characteristic of bifunctional intercalation by quinoxaline antibiotics. These results suggest that both compounds 1QN and 2QN behave as bifunctional intercalators but that compound 2QN produces only half the helix unwinding seen with compound 1QN and the natural quinoxalines. Binding curves for the interaction of both analogues with a variety of synthetic and naturally occurring nucleic acids were determined by solvent-partition analysis. Values for compound 2QN were also obtained by a fluorimetric method and found to agree well with the solvent-partition measurements. Compound 1QN bound most tightly to Micrococcus lysodeikticus DNA and, like echinomycin, exhibited a broad preference for (G + C)-rich DNA species. For compound 2QN no marked (G + C) preference was indicated, and the tightest binding among the natural DNA species studied was found with DNA from Escherichia coli. The two analogues also displayed different patterns of specificity in their interaction with synthetic nucleic acids. Compound 2QN bound to poly(dA-dT) slightly more tightly than to poly-(dG-dC), whereas compound 1QN displayed a large (approx. 11-fold) preference in the opposite sense. There was evidence of co-operativity in the binding to poly(dA-dT). It may be concluded that the chromophore moieties play an active role in determining the capacity of quinomycin antibiotics to recognize and bind selectively to specific sequences in DNA.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Canellakis E. S., Shaw Y. H., Hanners W. E., Schwartz R. A. Diacridines: bifunctional intercalators. I. Chemistry, physical chemistry and growth inhibitory properties. Biochim Biophys Acta. 1976 Feb 5;418(3):277–289. doi: 10.1016/0005-2787(76)90290-2. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Conformation studies on the sodium and cesium salts of calf thymus deoxyribonucleic acid (DNA). Biopolymers. 1966 Apr-May;4(4):429–440. doi: 10.1002/bip.1966.360040404. [DOI] [PubMed] [Google Scholar]
- Dell A., Williams D. H., Morris H. R., Smith G. A., Feeney J., Roberts G. C. Structure revision of the antibiotic echinomycin. J Am Chem Soc. 1975 Apr 30;97(9):2497–2502. doi: 10.1021/ja00842a029. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism studies of the structure of the DNA complex with intercalated drugs. Biochemistry. 1979 Jan 23;18(2):280–288. doi: 10.1021/bi00569a007. [DOI] [PubMed] [Google Scholar]
- Le Pecq J. B., Le Bret M., Barbet J., Roques B. DNA polyintercalating drugs: DNA binding of diacridine derivatives. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2915–2919. doi: 10.1073/pnas.72.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Waring M. J. Bifunctional intercalation and sequence specificity in the binding of quinomycin and triostin antibiotics to deoxyribonucleic acid. Biochem J. 1978 Jul 1;173(1):115–128. doi: 10.1042/bj1730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Waring M. J. Interaction between synthetic analogues of quinoxaline antibiotics and nucleic acids. Changes in mechanism and specificity related to structural alterations. Biochem J. 1978 Jul 1;173(1):129–144. doi: 10.1042/bj1730129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. G., Mizsak S. A., Biles C., Stewart J. C., Bacynsky L., Meulman P. A. Structure of quinomycin antibiotics. J Antibiot (Tokyo) 1975 Apr;28(4):332–336. doi: 10.7164/antibiotics.28.332. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Müller W., Bünemann H., Dattagupta N. Interactions of heteroaromatic compounds with nucleic acids. 2. Influence of substituents on the base and sequence specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):279–291. doi: 10.1111/j.1432-1033.1975.tb04138.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Interactions of heteroaromatic compounds with nucleic acids. 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Gautier F. Interactions of heteroaromatic compounds with nucleic acids. A - T-specific non-intercalating DNA ligands. Eur J Biochem. 1975 Jun;54(2):385–394. doi: 10.1111/j.1432-1033.1975.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Pyeritz R. E., Schlegel R. A., Thomas C. A., Jr Hydrodynamic shear breakage of DNA may produce single-chained terminals. Biochim Biophys Acta. 1972 Jul 31;272(4):504–509. doi: 10.1016/0005-2787(72)90505-9. [DOI] [PubMed] [Google Scholar]
- Reinert K. E. DNA stiffening and elongation caused by the binding of ethidium bromide. Biochim Biophys Acta. 1973 Aug 24;319(2):135–139. doi: 10.1016/0005-2787(73)90004-x. [DOI] [PubMed] [Google Scholar]
- TUBBS R. K., DITMARS W. E., Jr, VANWINKLE Q. HETEROGENEITY OF THE INTERACTION OF DNA WITH ACRIFLAVINE. J Mol Biol. 1964 Aug;9:545–557. doi: 10.1016/s0022-2836(64)80226-6. [DOI] [PubMed] [Google Scholar]
- Wakelin L. P., Romanos M., Chen T. K., Glaubiger D., Canellakis E. S., Waring M. J. Structural limitations on the bifunctional intercalation of diacridines into DNA. Biochemistry. 1978 Nov 14;17(23):5057–5063. doi: 10.1021/bi00616a031. [DOI] [PubMed] [Google Scholar]
- Wakelin L. P., Waring M. J. The unwinding of circular deoxyribonucleic acid by phenanthridinium drugs: structure-activity relations for the intercalation reaction. Mol Pharmacol. 1974 May;10(3):544–561. [PubMed] [Google Scholar]
- Wakelin S. P., Waring M. J. The binding of echinomycin to deoxyribonucleic acid. Biochem J. 1976 Sep 1;157(3):721–740. doi: 10.1042/bj1570721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J., González A., Jiménez A., Vázquez D. Intercalative binding to DNA of antitumour drugs derived from 3-nitro-1,8-naphthalic acid. Nucleic Acids Res. 1979 Sep 11;7(1):217–230. doi: 10.1093/nar/7.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P., Lee J. S. A solvent-partition method for measuring the binding of drugs to DNA. Application to the quinoxaline antibiotics echinomycin and triostin A. Biochim Biophys Acta. 1975 Oct 1;407(2):200–212. doi: 10.1016/0005-2787(75)90285-3. [DOI] [PubMed] [Google Scholar]
- Waring M., Makoff A. Breakdown of pulse-labeled ribonucleic acid and polysomes in Bacillus megaterium: actions of streptolydigin, echinomycin, and triostins. Mol Pharmacol. 1974 Mar;10(2):214–224. [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Kimura Y., Katagiri K. Novel quinomycins: biosynthetic replacement of the chromophores. J Antibiot (Tokyo) 1968 Jul;21(7):465–467. doi: 10.7164/antibiotics.21.465. [DOI] [PubMed] [Google Scholar]


