Abstract
Congenital heart disease (CHD) affects approximately 1% of live births. Although genetic and environmental etiologic contributors have been identified, the majority of CHD lacks a definitive cause, suggesting the role of gene-environment interactions (GxEs) in disease pathogenesis. Maternal diabetes mellitus (matDM) is among the most prevalent environmental risk factors for CHD. However, there is a substantial knowledge gap in understanding how matDM acts upon susceptible genetic backgrounds to increase disease expressivity. Previously, we reported a GxE between Notch1 haploinsufficiency and matDM leading to increased CHD penetrance. Here, we demonstrate a cell lineage–specific effect of Notch1 haploinsufficiency in matDM-exposed embryos, implicating endothelial/endocardial tissues in the developing heart. We report impaired atrioventricular cushion morphogenesis in matDM-exposed Notch1+/– animals and show a synergistic effect of NOTCH1 haploinsufficiency and oxidative stress in dysregulation of gene regulatory networks critical for endocardial cushion morphogenesis in vitro. Mitigation of matDM-associated oxidative stress via superoxide dismutase 1 overexpression did not rescue CHD in Notch1-haploinsufficient mice compared to wild-type littermates. Our results show the combinatorial interaction of matDM-associated oxidative stress and a genetic predisposition, Notch1 haploinsufficiency, on cardiac development, supporting a GxE model for CHD etiology and suggesting that antioxidant strategies alone may be ineffective in genetically susceptible individuals.
Keywords: Cardiology, Development
Keywords: Embryonic development, Endothelial cells, Genetic diseases
Gene-environment etiology for congenital heart disease is studied. Maternal diabetes and Notch1 haploinsufficiency hinder endocardial cushion development but antioxidants show a differing ability to rescue disease.
Introduction
Congenital heart disease (CHD) affects approximately 1% of live births and is the leading cause of birth defect–related infant mortality (1). Although high-throughput genome-sequencing technologies have made remarkable advances in uncovering genetic etiologies for CHD, the underlying cause for more than half of CHD cases is still unknown (2–5). A subset of these cases have long been proposed to be the combinatorial effect of genetic predisposition and environmental influences, resulting in complex inheritance patterns and variable CHD expressivity (6, 7). Among environmental contributors, maternal pregestational diabetes mellitus is a highly prevalent and well-established risk factor for CHD, increasing the risk of having an infant with CHD by 3- to 5-fold (8–13). The influence of maternal diabetes mellitus (matDM) on cardiac development has been studied in animal models, shedding light on key cellular and molecular pathways particularly vulnerable to this environmental milieu (14). Recently, matDM has been shown to have cell lineage–specific effects during cardiac morphogenesis, specifically affecting the second heart field and neural crest lineages to disrupt cardiomyocyte differentiation and anterior-posterior specification (15, 16). The incidence of CHD in a pregnancy complicated by pregestational diabetes mellitus has remained elevated even with substantial advancements in prenatal care, suggesting that matDM may cause CHD by acting on susceptible genetic backgrounds, i.e., gene-environment interaction (GxE) etiology. However, the cellular and molecular basis for GxE in CHD as postulated in the multifactorial hypothesis for CHD remain limited (6, 17, 18).
Cardiac development is a dynamic process with tight spatiotemporal regulation of several multipotent cardiac cell lineages (19, 20). Endothelial-mesenchymal transition (EndMT) is a crucial process that occurs early in heart development wherein endocardial cells lining the atrioventricular canal (AVC) and developing outflow tract (OFT) detach from the monolayer and migrate into the preformed cardiac jelly to form endocardial cushions (21–23). Subsequent remodeling of the endocardial cushions at the AVC leads to the formation of the atrioventricular valve leaflets, atrioventricular septum, and membranous portion of the ventricular septum (24, 25). At the OFT, endocardial cushions receive additional contributions from the migrating cardiac neural crest cells to form the semilunar valves and aorticopulmonary septum (26). Although matDM is associated with a spectrum of CHD phenotypes, the most commonly observed include septal and conotruncal (involving OFT and great vessels) heart defects, suggesting EndMT and endocardial cushion morphogenesis are particularly susceptible to disruption by the abnormal diabetic environment (10, 11). This is supported by several published studies in animal models showing dysregulation of key EndMT signaling pathways in embryonic hearts exposed to matDM, including BMP, TGF-β, and Notch signaling (27–31). Additionally, oxidative stress is a hallmark of diabetic embryopathy, and impaired redox signaling has been implicated in dysregulation of several cardiac developmental pathways, suggesting that matDM-associated oxidative stress may be culpable for the elevated risk of CHD (31, 32).
Previously, we found matDM interacts with Notch1 haploinsufficiency in mice to increase the incidence of membranous ventricular septal defects (VSDs), supporting a GxE between Notch1 and matDM (27). Pathogenic NOTCH1 variants are associated with a spectrum of CHD in humans, and Notch signaling is known to interact with other signaling pathways such as BMP and TGF-β to facilitate EndMT in the developing heart (33–38). To better characterize the Notch1-matDM GxE and place it in the context of cardiac development, we hypothesized that matDM and Notch1 haploinsufficiency functionally converge to disrupt endocardial cushion morphogenesis and EndMT to increase the risk of CHD.
In this study, we demonstrate that matDM and Notch1 haploinsufficiency interact within the developing endothelial/endocardial and endocardially derived mesenchyme to increase the incidence of membranous VSD. We report abnormal atrioventricular (AV) cushion morphogenesis in matDM-exposed Notch1-haploinsufficient (Notch1+/–) embryonic hearts compared with nondiabetic controls. In a human induced pluripotent stem cell–based (iPSC-based) in vitro model, we show that NOTCH1 haploinsufficiency sensitizes endothelial cells to the effects of oxidative stress and disrupts a network of genes and biological processes underlying EndMT and endocardial cushion morphogenesis. Consistent with this, we find matDM-exposed Notch1+/– embryos are insensitive to antioxidant-based therapeutic strategy, despite rescue of VSD observed in matDM-exposed WT littermates. Overall, the results from this study elucidate mechanisms by which matDM interacts with a genetic susceptibility, i.e., Notch1 haploinsufficiency, to bring about cell lineage–specific effects and increase penetrance of CHD. This work serves as experimental proof supporting a multifactorial etiology for CHD, underscoring the need to identify novel genetic modifiers that act in conjunction with environmental teratogens to increase the incidence of CHD, and shows the variability in phenotypic rescue with antioxidant therapies in a genetically susceptible background.
Results
GxE between endothelial Notch1 haploinsufficiency and matDM causes CHDs.
Previously, we identified a novel GxE between Notch1 haploinsufficiency and matDM in mice, wherein Notch1+/– embryos exposed to matDM had significantly higher incidence of VSD compared with WT littermates at embryonic day (E) 13.5 (27). We first sought to determine if this GxE is evident at a later developmental stage in mice. For this, we generated streptozotocin-induced (STZ-induced) diabetic WT females for timed mating with nondiabetic Notch1+/– males. Analysis of matDM-exposed embryos at E14.5 (n = 3 litters) verified a previously reported GxE resulting in a significantly higher incidence of membranous VSD in Notch1+/– (10/14, 71%) embryos compared with WT littermates (1/10, 10%, Fisher’s exact test P value = 0.005) (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.183516DS1).
The expression pattern of activated NOTCH1 in the developing heart has been well characterized, being highly expressed within endocardial and endocardially derived mesenchymal cells, which form the endocardial cushions at the AVC and OFT (39, 40). Subsequently, we sought to determine if endothelium-specific Notch1 haploinsufficiency is sufficient to sensitize the effects of matDM on cardiac development. For this experiment, STZ-induced diabetic and nondiabetic Notch1fl/fl females were bred with nondiabetic Tie2-Cre+ males to generate control and matDM-exposed Notch1fl/wt Tie2-Cre+ embryos, which are conditionally heterozygous for Notch1 in all endothelial cells, including endocardial cells and their derived tissues in the heart (Figure 1A). While no VSD was found in nondiabetic Notch1fl/wt Tie2-Cre+ or Notch1fl/wt littermates, we found a significantly higher incidence of membranous VSD in E14.5 matDM-exposed Notch1fl/wt Tie2-Cre+ embryos (7/17, 41%) compared with non-Cre+ littermates (3/31, 9.7%, Fisher’s exact test P value = 0.022) (Figure 1, B and C). To account for variability across multiple matDM-exposed litters (n = 6), we used a general linear mixed model and included litter as the random effect, and the probability of VSD for Notch1fl/wt Tie2-Cre+ was found to be higher compared with Notch1fl/wt (P value = 0.06, see Supporting Data Values) (41). While this is slightly higher than the traditional 0.05 cut point, this still small P value provides evidence that the data are not compatible with the null hypothesis (42). We also noted that the incidence of VSD in matDM-exposed Notch1fl/wt Tie2-Cre+ (41%) was found to be lower than the incidence observed in matDM-exposed Notch1+/– (71%). We attribute this difference to potential roles of Notch1 haploinsufficiency in non-endothelium-derived cells of the heart in the GxE with matDM, or this variability may be due to incomplete deletion of Notch1 in all endothelial cells. Overall, these results suggest that the observed GxE between Notch1 haploinsufficiency and matDM can act within the developing endothelium, including the endocardium- and endocardium-derived cells to impair cardiac morphogenesis, particularly ventricular septation.
Figure 1. Endothelial/endocardial haploinsufficiency of Notch1 is sufficient for GxE with matDM.
(A) Breeding scheme to generate diabetic and nondiabetic E14.5 Notch1fl/wt and Notch1fl/wt Tie2-Cre+ mice for histological analysis. (B) Table showing incidence of VSD in E14.5 embryos. (C) Representative images of matDM-exposed and nondiabetic Notch1fl/wt and Notch1fl/wt Tie2-Cre+ embryonic hearts. Asterisk denotes VSD. P value obtained from Fisher’s exact test; scale bar = 200 μm. ND, nondiabetic; DM, diabetic; RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; IVS, interventricular septum; VSD, ventricular septal defect.
Notch1 haploinsufficiency exacerbates the effects of matDM on AV cushion development.
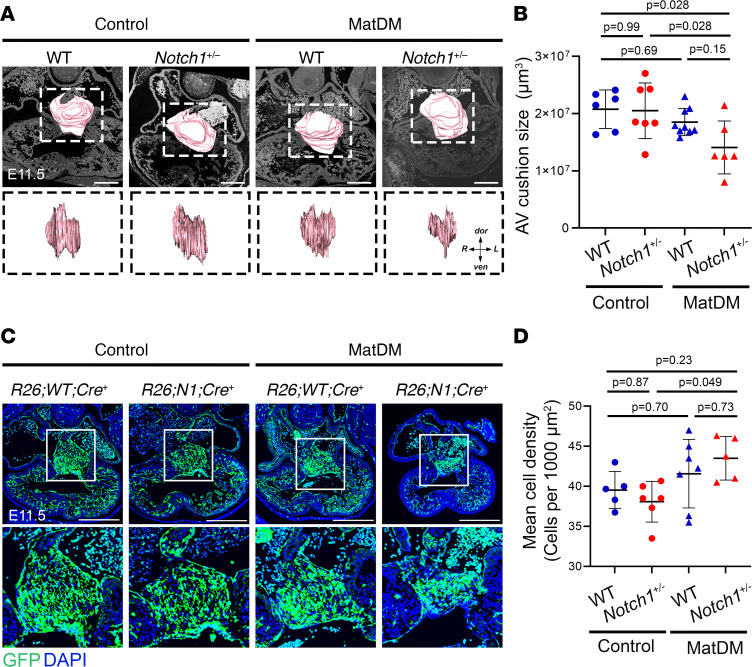
During cardiac development, endocardial cells undergo EndMT to form endocardial cushions at the AVC and OFT. Subsequent remodeling of the proximal OFT and AV cushions gives rise to the AV septal complex, which includes the mitral and tricuspid valve leaflets and the membranous ventricular septum (24, 25, 43). Here, we sought to determine if interaction between Notch1 haploinsufficiency and matDM impairs endocardial cushion morphogenesis at the OFT and AVC. As Notch signaling is known to regulate EndMT during endocardial cushion development, we utilized the Rosa26mT/mG locus to trace endothelial cells that have undergone EndMT in endocardial cushions of matDM-exposed Notch1+/– embryos compared with nondiabetic controls. We analyzed histological sections of OFT and AV cushions at E11.5; volumetric analysis using 3D reconstruction of serial histological sections revealed matDM-exposed Notch1+/– hearts had significantly smaller AV cushion size compared with nondiabetic Notch1+/– and nondiabetic WT hearts (Figure 2, A and B). On the other hand, matDM-exposed WT hearts did not show a statistically significant difference in AV cushion size compared to nondiabetic controls. We found no significant difference in OFT cushion size across the 4 groups, suggesting that this GxE affects AV cushion morphogenesis to contribute to membranous VSD (Supplemental Figure 2, A and B). Upon examination of GFP+ EndMT-derived cells within the AV cushions, GFP+ cells appeared more compacted within the AV cushion of matDM-exposed Notch1+/– hearts, which was likely due to the smaller AV cushion size (Figure 2C). This was quantified by counting the number of GFP+ cell nuclei within the AV cushion, and a statistically significant increase in cell density was noted between matDM-exposed Notch1+/– embryos compared with nondiabetic Notch1+/– control (Figure 2D). Again, this effect was not observed in the matDM-exposed WT littermates, and we noted a high degree of variability in this group, possibly due to the low penetrance of disease in matDM-exposed WT embryos. These results show that the overall size of the AV cushion is significantly smaller in the matDM-exposed Notch1+/– compared with nondiabetic controls, causing EndMT-derived cells to become more densely packed within the AV cushion.
Figure 2. matDM impairs AV cushion size in Notch1-haploinsufficient embryonic hearts.
(A) Three-dimensional (3D) projection of AV cushions in E11.5 control and matDM-exposed WT and Notch1+/– embryonic hearts. Top panel shows overlay of 3D projection on corresponding 2D transverse section of the heart; scale bar = 100 μm. Bottom panel shows AV cushion 3D projections rotated 90° to show AV cushion span right to left across AVC. (B) Quantification of AV cushion volume from 3D projections. (C) Control and matDM-exposed WT and Notch1+/– embryonic hearts showing Tie2-Cre–driven RosamT;mG-based GFP expression. GFP+ cells in the heart represent all endocardially and endocardially derived cells. Top panel shows GFP expression throughout the embryonic heart; scale bar = 200 μm. Bottom panel shows corresponding 240 × 240 µm zoomed-in images of AV cushions for each genotype. (D) Quantification of GFP+ cell nuclei within AV cushion based on DAPI staining. Each point represents a single animal of corresponding genotype/condition. P values obtained from t tests for the estimated marginal means for all pairwise comparisons using Tukey’s correction to adjust for multiple testing. AV, atrioventricular; dor, dorsal; R, right; L, left; ven, ventral.
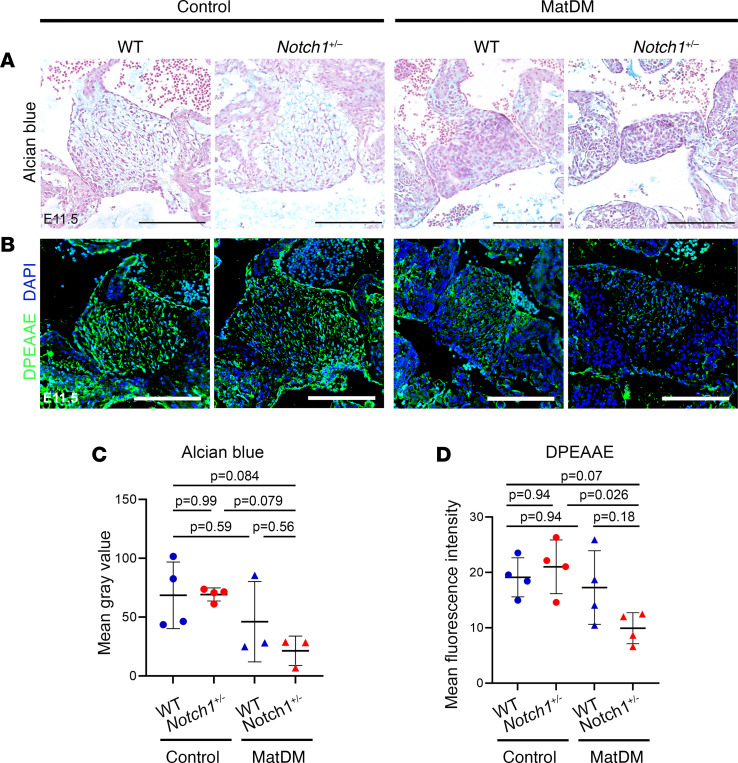
The deposition and distribution of extracellular matrix (ECM) components, including sulfated proteoglycans such as hyaluronic acid and versican, are crucial for endocardial cushion formation and remodeling into the AV valvuloseptal complex, a process highly regulated by both endocardial and myocardial signaling adjacent to the AV cushion (44–46). Alcian blue staining of WT and Notch1+/– embryos of diabetic and nondiabetic dams at E11.5 showed reduced proteoglycan deposition in matDM-exposed Notch1+/– AV cushion compared with nondiabetic control while matDM-exposed WT embryos did not show a difference compared to nondiabetic controls (Figure 3, A and C). During AV cushion development, full-length versican undergoes proteolytic cleavage by the action of matrix metalloproteinases MMP2 and ADAMTS1 (47). Similarly, expression of cleaved versican, labeled by an antibody against neo-epitope DPEAAE, was significantly decreased in the matDM-exposed Notch1+/– AV cushion compared with nondiabetic Notch1+/– control (Figure 3, B and D). Similar to the AV cushion size and cellular density data, we found a high degree of variability in the matDM-exposed WT littermates, and as a result no statistical significance was reached between matDM-exposed WT and nondiabetic controls in either the Alcian blue staining or DPEAAE expression. Overall, these results indicate that GxE between Notch1 haploinsufficiency and matDM impairs AV cushion morphogenesis potentially via disruption of ECM organization.
Figure 3. matDM impairs proteoglycan distribution within AV cushion of Notch1-haploinsufficient embryonic hearts.
(A) Alcian blue staining of control and matDM-exposed E11.5 WT and Notch1+/– AV cushion. (B) Immunostaining of cleaved versican (DPEAAE) in control and matDM-exposed WT and Notch1+/– AV cushion. (C) Quantification of Alcian blue staining from A. (D) Quantification of DPEAAE staining from B. Each point represents a single animal of corresponding genotype/condition. P values obtained from t tests for the estimated marginal means for all pairwise comparisons using Tukey’s correction to adjust for multiple testing. Scale bar = 100 μm.
NOTCH1 haploinsufficiency acts synergistically with oxidative stress to dysregulate genes involved in EndMT and endocardial cushion morphogenesis.
Previously published research from us and others have proposed matDM-associated oxidative stress as the key driver of cardiac maldevelopment, leading to increased incidence of CHD. To determine if there is elevated oxidative stress in matDM-exposed Notch1+/– embryos compared with nondiabetic controls, we probed E11.5 matDM-exposed and nondiabetic WT and Notch1+/– embryonic hearts with an antibody against 4-hydroxynoneal (4-HNE), a stable by-product of lipid peroxidation and a robust biomarker for cellular oxidative stress (48). We detected increased 4-HNE labeling in matDM-exposed Notch1+/– and WT littermates compared with nondiabetic controls while no difference was detected between Notch1+/– and WT matDM-exposed hearts, including in the AV cushions, suggesting there are comparable levels of oxidative stress in matDM-exposed WT and Notch1+/– littermates at this time point (Supplemental Figure 3, A and C). Increased oxidative stress can induce cellular apoptosis in diabetic embryos, particularly during the neurulation stage at E8.75 (49, 50). To probe for increased apoptosis in matDM-exposed WT and Notch1+/– embryonic hearts, we performed TUNEL assay on E11.5 matDM-exposed and nondiabetic Notch1+/– and WT embryonic hearts. However, no significant differences were observed across groups, suggesting there is no increase in apoptosis in matDM-exposed cardiac tissues in either Notch1+/– or WT embryos at this time point (Supplemental Figure 3, B and D).
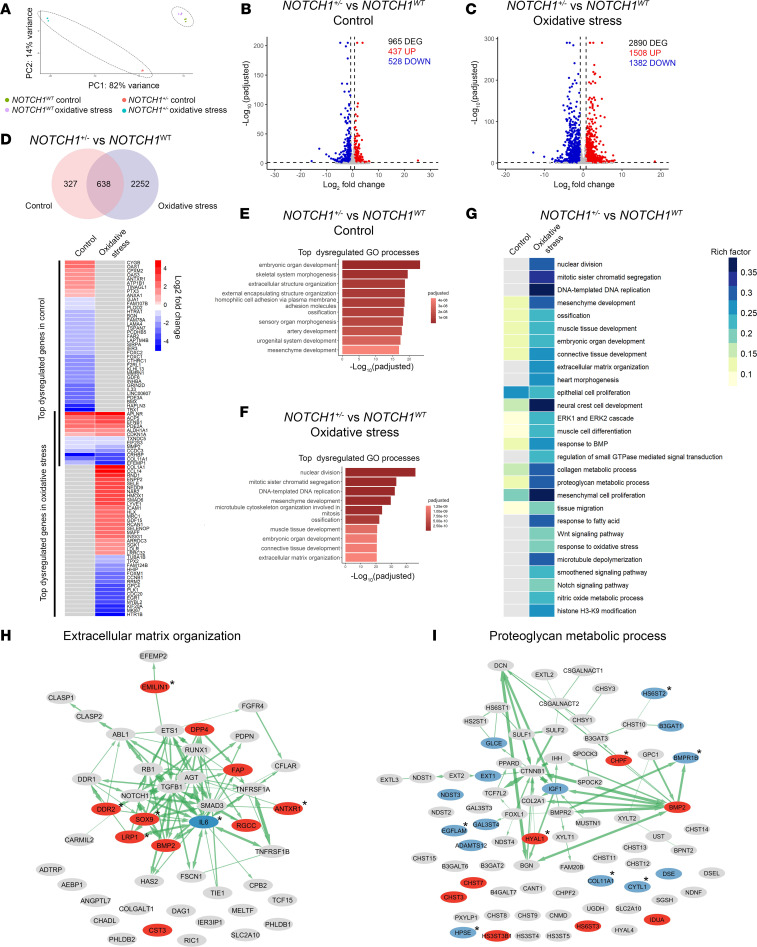
To determine the molecular mechanisms underlying GxE between endothelial Notch1 and matDM-associated oxidative stress, we utilized NOTCH1WT and isogenic NOTCH1+/– iPSC lines, generated as previously described (51, 52). We differentiated NOTCH1WT and NOTCH1+/– iPSCs to induced endothelial cells (iECs) using a previously published differentiation protocol (53). Following differentiation, subsets of NOTCH1WT and NOTCH1+/– iECs were either exposed to oxidative stress (50 μM H2O2) or left untreated as control, to mimic matDM-associated oxidative stress in vitro. After 4 days, total RNA was isolated from each iEC subset and bulk RNA sequencing performed for differential gene expression analysis. Principal component analysis of the sequenced samples revealed distinct clustering of biological replicates (n = 3 per genotype/condition), with the largest variance observed between NOTCH1+/– and NOTCH1WT in oxidative stress (Figure 4A). As expected, we observed high variance between NOTCH1+/– and NOTCH1WT control samples, suggesting there are intrinsic transcriptomic differences between NOTCH1+/– and NOTCH1WT iECs at baseline untreated condition. Differential gene expression analysis revealed there were 965 genes significantly differentially expressed (FDR-adjusted P value < 0.05, absolute log2 fold-change > 0.75) between NOTCH1+/– and NOTCH1WT iECs in control condition (437 upregulated and 528 downregulated) (Figure 4B and Supplemental Table 1). Next, we turned our attention to how these genotypes differed from each other during exposure to oxidative stress. Differential gene expression analysis revealed there were 2,890 genes significantly differentially expressed (FDR-adjusted P value < 0.05, absolute log2 fold-change > 0.75) between NOTCH1+/– and NOTCH1WT iECs under oxidative stress (1,508 upregulated and 1,382 downregulated) (Figure 4C and Supplemental Table 2). We verified approximately 40% downregulation of Notch1 mRNA in NOTCH1+/– iECs under both conditions (log2 fold-change = –0.64, FDR-adjusted P value < 0.05 in control; log2 fold-change = –0.61, FDR-adjusted P value < 0.05 in oxidative stress); however, it was not assigned as a DEG because of our stringent log2 fold-change cutoff of 0.75. Comparison of DEGs from control and oxidative stress revealed 638 genes were dysregulated in NOTCH1+/– iECs in both control and oxidative stress while 2,252 genes were dysregulated in NOTCH1+/– iECs only under oxidative stress (Figure 4D). Overrepresentation analysis of DEGs in NOTCH1+/– iECs under each condition was performed. Top significant biological processes in NOTCH1+/– iECs in control condition included embryonic organ development, skeletal system morphogenesis, and extracellular structure organization (Figure 4E and Supplemental Table 3). In contrast, top dysregulated biological processes in NOTCH1+/– in oxidative stress included terms related to cell division (MKI67, MYBL2, CDC20, PLK1, CCNB1), mesenchyme development (APLNR, EFNB1), and extracellular matrix organization (COL1A1, COL11A1, MMP2) (Figure 4F and Supplemental Table 4). While several biological pathways were commonly dysregulated in NOTCH1+/– iECs in both control and oxidative stress, including GO terms mesenchyme development, connective tissue development, collagen metabolic processes, and proteoglycan metabolic processes, the number of DEGs in each process was higher in NOTCH1+/– iECs in oxidative stress compared with control condition (Figure 4G). Additionally, many biological processes were found to be dysregulated in NOTCH1+/– iECs in oxidative stress, including the terms nuclear division, mitotic sister chromatid segregation, extracellular matrix organization, heart morphogenesis, Notch signaling, and nitric oxide signaling pathway among others (Figure 4G). Considering that extracellular matrix organization and proteoglycan metabolic processes are highly relevant processes in the context of endocardial cushion morphogenesis, we investigated the effect of oxidative stress in NOTCH1+/– iECs on genes within these terms, reporting several interconnected genes to be dysregulated across both processes (Figure 4, H and I). Among DEGs affecting ECM organization, the majority were upregulated, and many of these genes are known to be negative regulators of ECM organization (DPP4, FAP, ANTXR1, CST3, EMILIN1), while the majority of genes in proteoglycan metabolic processes were downregulated (BMPR1B, IGF1, COL11A1). Taken together, these results suggest Notch1 haploinsufficiency acts synergistically with oxidative stress to dysregulate gene regulatory networks crucial for endocardial cushion morphogenesis.
Figure 4. NOTCH1 haploinsufficiency and oxidative stress act synergistically to dysregulate processes involved in endocardial cushion morphogenesis.
(A) Principal component analysis shows the grouping of biological replicates and high variance between genotype and response to oxidative stress treatment. (B) Volcano plot showing upregulated and downregulated DEGs in NOTCH1+/– versus NOTCH1WT iECs in control condition. (C) Volcano plot showing upregulated and downregulated DEGs in NOTCH1+/– and NOTCH1WT iECs in oxidative stress. (D) Venn diagram showing number of common and unique DEGs in NOTCH1+/– iECs versus NOTCH1WT in control and oxidative stress condition followed by heatmap showing top 50 significant DEGs in NOTCH1+/– versus NOTCH1WT in control and oxidative stress condition. (E) Top significantly enriched GO BP terms in NOTCH1+/– iECs versus NOTCH1WT in control. (F) Top significantly enriched GO BP terms in NOTCH1+/– iECs versus NOTCH1WT in oxidative stress. (G) Heatmap of dysregulated pathways in NOTCH1+/– versus NOTCH1WT in control and oxidative stress. (H) DEGs in NOTCH1+/– iECs versus NOTCH1WT in oxidative stress that affect GO term extracellular matrix organization. Asterisk indicates genes that were also dysregulated in NOTCH1+/– iECs versus NOTCH1WT in control. (I) DEGs in NOTCH1+/– iECs versus NOTCH1WT in oxidative stress that affect GO term proteoglycan metabolic processes. Asterisk indicates genes that are also significantly differentially expressed in NOTCH1+/– iECs versus NOTCH1WT in control. DEGs, differentially expressed genes; FC, fold-change; GO, gene ontology; BP, biological processes; Padj, FDR-adjusted P value.
Overexpression of antioxidant gene, SOD1, does not reduce incidence of matDM-associated CHD in the setting of Notch1 haploinsufficiency.
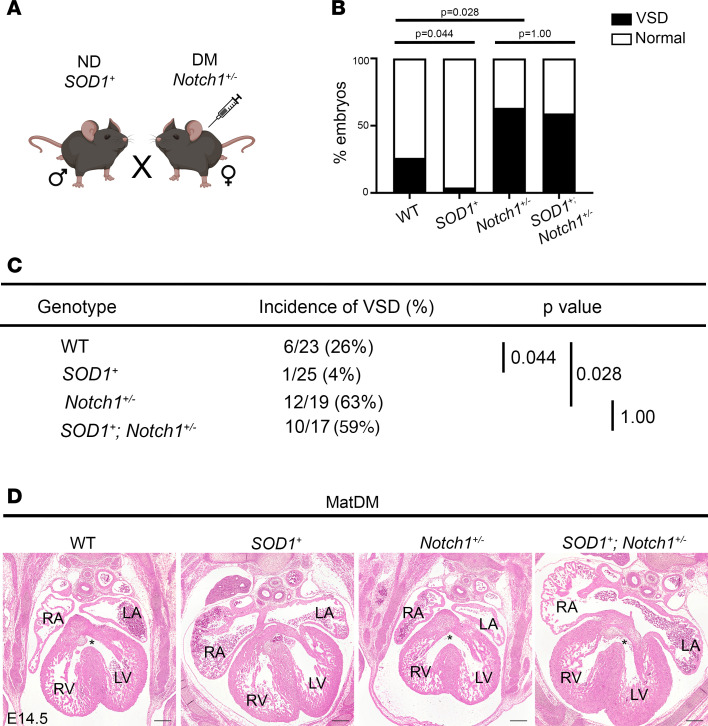
Oxidative stress is characterized by an imbalance of reactive oxygen species (ROS) production and antioxidant defense response. Superoxide dismutases (SODs) are crucial for neutralizing superoxide radicals, converting them to H2O2, which can be degraded to H2O and O2 by endogenous catalase and glutathione peroxidase. To determine if a genetic antioxidant strategy via overexpression of SOD1 is effective in rescuing CHD in a Notch1+/– background, we bred SOD1-overexpressing transgenic male mice (SOD1+) to diabetic Notch1+/– females to generate WT, Notch1+/–, SOD1+, and SOD1+ Notch1+/– compound-mutation embryos (n = 11 litters) (Figure 5A). We performed histological analysis of E14.5 embryonic hearts and found a significantly decreased incidence of VSD in SOD1+ (1/25) compared with WT (6/23, Fisher’s exact test P value = 0.044) (Figure 5, B–D). This is consistent with prior publications showing overexpression of SOD1 in embryonic hearts can reduce the incidence of matDM-associated CHD in a WT setting (31, 32). However, no significant reduction in the incidence of VSD was noted when comparing Notch1+/– (12/19) and SOD1+ Notch1+/– embryos (10/17, Fisher’s exact test P value = 0.99). To account for litter variability, we used binomial regression with litter as a random effect to further analyze these results, and while statistical significance was not achieved, a similar trend was noted (see Supporting Data Values). We compared the level of oxidative stress via 4-HNE labeling between the above genotypes and found decreased immunostaining of 4-HNE in SOD1+ compared with WT while no difference was observed between the Notch1+/– and SOD1+ Notch1+/– (Supplemental Figure 4, A and B), suggesting SOD1 overexpression cannot reduce oxidative stress in a Notch1+/– background. The failure of SOD1 overexpression to reduce the incidence of VSD resulting from the Notch1-matDM interaction is consistent with our in vitro results showing an exacerbated effect on cardiac developmental process in NOTCH1+/– iECs exposed to H2O2. Overall, our results solidify the role of Notch1 haploinsufficiency as a genetic modifier in matDM-associated CHD, highlighting the presence of GxEs and their mechanisms in CHD pathogenesis.
Figure 5. SOD1 overexpression does not rescue matDM-associated VSD in the setting of Notch1 haploinsufficiency.
(A) Breeding scheme to generate E14.5 DM WT, SOD1+, Notch1+/–, and Notch1+/– SOD1+ embryos. (B and C) Graph and table showing incidence of VSD in E14.5 embryos by genotype. (D) Representative images of matDM-exposed WT, SOD1+, Notch1+/–, and Notch1+/– SOD1+ embryonic hearts. Asterisk denotes VSD. P values obtained from Fisher’s exact test; scale bar = 200 μm. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; IVS, interventricular septum; VSD, ventricular septal defect; DM, diabetic.
Discussion
matDM is an established risk factor for CHD; however, its effect on genetic susceptibilities has not been well described. In this study, we show matDM interacts with Notch1 haploinsufficiency in vivo to increase the disease penetrance of a membranous VSD phenotype. We find that this GxE acts within the developing endothelial/endocardial and endocardially derived cells and that Notch1 haploinsufficiency sensitizes the effects of matDM on EndMT and AV cushion morphogenesis, affecting deposition of ECM components including versican. Using a human iPSC-based model, we demonstrate that oxidative stress–exposed NOTCH1+/– endothelial cells have exacerbated effect on cardiac developmental processes compared with oxidative stress–exposed NOTCH1WT iECs. We show effects of oxidative stress and NOTCH1 haploinsufficiency converge to dysregulate networks underlying endocardial cushion morphogenesis. Finally, we find that targeting matDM-associated oxidative stress via overexpression of SOD1 is unable to rescue VSD in a Notch1-haploinsufficient background as compared with successful rescue in WT littermates. Taken together, these results reveal that susceptible genetic variation acts in combination with effects of matDM to disrupt cardiac development and increase the incidence of CHD.
Complex inheritance patterns and incomplete penetrance of CHD are often explained by genetic heterogeneity among individuals, with strong speculation as to the requirement of specific GxE or gene-gene interaction for disease manifestation (6, 54). In humans, pathogenic NOTCH1 variants are associated with a spectrum of CHD with variable expressivity (33–35). The Notch signaling pathway is highly conserved, and Notch1 heterozygosity in mice has been reported to interact with both environmental factors (e.g., gestational hypoxia, maternal diabetes) as well as genetic factors (e.g., deletion of Nos3) to increase the incidence of heart defects (27, 55, 56). Hence, NOTCH1 variation may play a crucial role in multifactorial cases of CHD, and results from this study corroborate the role of NOTCH1 as both a genetic modifier and driver in CHD pathogenesis.
During cardiac development, activated NOTCH1 (N1CD) is expressed in the developing endocardium lining the AVC and OFT; in synchrony with myocardial BMP signaling, N1CD coordinates initiation of EndMT and mesenchyme development at these regions to form the endocardial cushions (37, 57). EndMT-derived mesenchymal cells communicate with adjacent endocardial and myocardial layers to dynamically regulate organization and stratification of the ECM to form the valvuloseptal complex (24, 25). Based on our in vivo results, matDM-exposed Notch1+/– endocardial cushions do not show any overt signs of initial EndMT failure as lineage tracing does not show a reduction in number of EndMT-derived cells in either matDM-exposed WT or Notch1+/– littermates compared with nondiabetic controls. Instead, our results suggest that disruption in AV cushion formation results in smaller overall size of the AV cushion in matDM-exposed Notch1+/– with altered ECM deposition, specifically deposition of cleaved versican. This is consistent with recent reports that have revealed a requirement of N1CD to induce secretion of ECM remodeling and structural proteins within the developing AV cushion (58). Versican is essential for ventricular septum formation, and mice harboring mutant Versican exhibit smaller AV cushions and VSD (59). In comparison, matDM-exposed WT littermates displayed great variability in their AV cushion size, cell density, and proteoglycan deposition, with only a subset trending similar to matDM-exposed Notch1+/– animals, consistent with the low penetrance of disease observed in these animals. In addition, our in vitro findings revealed dysregulation of BMP2 and MMP2 expression in NOTCH1+/– iECs in oxidative stress. While Notch signaling is known to modulate expression of MMP2, required for proteolytic cleavage of versican, matDM has also been independently linked to decreased MMP2 expression in the AV cushions (28, 60). Endocardial BMP2 has also been shown to be essential for later AV cushion morphogenesis, regulating deposition of ECM components such as Col9a1, Has2, Postn, and Vcan (61). Our results suggest Notch1 haploinsufficiency can sensitize the genetic network underlying ECM organization during AV cushion development with exposure to matDM-associated oxidative stress.
Although diabetes mellitus is characterized by chronic hyperglycemia, diabetic embryopathy is likely driven by a combination of metabolic imbalances occurring within the intrauterine diabetic milieu. Diabetes mellitus is associated with disruption of several metabolic pathways, including glycolysis, sorbitol production, ketone formation, and increased synthesis of advanced glycation end-products, all of which result in increased ROS production and cellular oxidative stress (62–64). As expected, matDM-exposed Notch1+/– embryos exhibit oxidative stress comparable to matDM-exposed WT littermates, as shown by 4-HNE labeling. Antioxidant therapy has been tested by several research groups, including us, in an attempt to decrease the incidence of matDM-associated CHD, but lack of consensus on route of administration and/or strategy (genetic versus pharmacologic) has led to inconsistent results (27, 32, 65). In this study, we find SOD1 overexpression can only decrease the incidence of VSD in a WT background and cannot rescue VSD in a Notch1-haploinsufficient setting. Based on 4-HNE labeling across matDM-exposed littermates, this seems to be due to inability of SOD1 to effectively reduce ROS levels below the required threshold for rescue in matDM-exposed Notch1+/– mice. Based on our in vitro studies, processes such as cell division and apoptosis were among the top significantly affected pathways in NOTCH1-haploinsufficient iECs exposed in H2O2 along with ECM organization. Proliferation and apoptosis of mesenchymal cells within the endocardial cushions is carefully coordinated with the start of ECM stratification (66). Previous studies have shown increased apoptosis and decreased proliferation in embryonic hearts exposed to maternal diabetes, particularly at later stages of endocardial cushion development. However, we did not find increased TUNEL+ apoptotic cells in matDM-exposed WT and Notch1+/– compared to nondiabetic controls, possibly due to small sample size and timing during embryonic development. Our in vitro results show decreased expression of BMP4 specifically in NOTCH1+/– iECs exposed to H2O2, and BMP4 has been reported to mediate regulation of endocardial cushion apoptosis (30, 67). As such, we speculate that a combination of dysregulated ROS/Notch signaling contributes to the magnified effect on cell division and cell death in NOTCH1 haploinsufficiency and may be acting synergistically to cause VSD in vivo, thus making it unable to be rescued by SOD1 overexpression alone.
Results from the in vitro experiments demonstrated relevant transcriptomic changes related to endocardial cushion morphogenesis in NOTCH1+/– compared with NOTCH1WT iECs in the control condition, although they were not as affected as found in oxidative stress conditions. This supports the notion that a genetic hit in NOTCH1 can “prime” the cell to a secondary environmental hit. Although Notch1+/– mice are not reported to have a congenital cardiac phenotype, they are known to have impaired lipid metabolism and develop aortic aneurysms with age (68, 69). The lack of CHD in nondiabetic Notch1+/– mice may be explained by compensatory mechanisms occurring in vivo that cannot be recapitulated in our in vitro endothelial cell-autonomous system.
In summary, our work uncovers a composite effect of genetic variation and exposure to an environmental teratogen on cardiac morphogenesis. We report that these effects can disrupt specific cardiac developmental networks in a cell lineage–specific manner. Furthermore, we have shown that broad therapeutic strategies, such as the use of antioxidants, may not be effective in genetically sensitized populations. Future studies should aim to uncover other potential GxEs that may be contributing to matDM-associated CHD. Studies of this nature will pave the way for personalized therapy to improve outcomes from diabetic pregnancies in genetically susceptible individuals and reduce the risk of matDM-associated CHD.
This study has limitations. In this study, we defined diabetic mice as those having fasting blood glucose greater than 200 mg/dL. However, we were not able to monitor or control blood glucose levels throughout pregnancy and attribute a degree of variability in the matDM-exposed samples due to differences in maternal glucose levels. We have performed binomial regression analysis with litter as a random factor to increase confidence in our results. In this study, we used C57BL/6J mice for all in vivo studies. However, strain-related differences may lead to variable phenotypic results. In the context of GxE, it is important to determine whether Notch1 haploinsufficiency in a different strain/genetic background can also interact with matDM to increase CHD incidence. Consequently, we focused on the effect of GxE in AV cushion development as our phenotype was limited to membranous VSDs, and we found no significant difference in OFT cushions. Future studies with other animal strains may identify additional cardiac phenotypes that may require investigation into other cardiac developmental processes. Additionally, we focused on endothelial/endocardial and endocardially derived cell–specific GxE as Notch1 expression is enriched in this cell lineage. However, NOTCH1 can also play a role during cardiomyocyte differentiation and specification, and investigation of the nonendothelial roles of Notch1 in the context of matDM is beyond the scope of this paper and will be reserved for future investigations. For our in vitro studies, all experiments were conducted using iPSC lines derived from a single healthy donor. Whether similar results will be observed with multiple donor cell lines is yet to be determined.
Methods
Sex as a biological variable.
Our study did not consider sex to be a biological variable as our studies involved evaluation of embryonic hearts.
Experimental mouse models.
WT C57BL/6 (Jackson Laboratory [Jax], 000664), Rosa26mTmG (Jax, 007676), Tie2-Cre+ (Jax, 004128), and SOD1-tg (Jax, 002297) animals were purchased. Notch1+/– and Notch1fl/fl mice were generated and genotyped as described previously (70, 71). Rosa+/+ Notch1+/– compound mutants were generated by intercrossing Rosa26mTmG with Notch1+/– mice. Six- to 8-week-old female WT, Notch1fl/fl, Rosa+/+ Notch1+/–, and Notch1+/– mice were used to induce diabetes mellitus by intraperitoneal injection of STZ (NC0146241, Thermo Fisher Scientific) dissolved in 0.01 mol/L citrate buffer, pH 4.5, at 75 mg/kg body weight for 3 consecutive days. Two weeks after STZ injection, mice were fasted for 8 hours during the light cycle, and glucose levels were measured using the AlphaTrak 2 veterinary blood glucometer calibrated specifically for rodents from tail vein blood (Abbott Laboratories). Mice with fasting blood glucose greater than 200 mg/dL (11 mmol/L) were defined as diabetic and used for timed breeding and embryo collections.
Mouse embryo collection.
Timed mating was performed using the following pairs: nondiabetic Notch1+/– males and diabetic WT females, nondiabetic Tie2-Cre+ males with diabetic and nondiabetic Notch1fl/fl females or diabetic and nondiabetic Rosa+/+ Notch1+/– females, and nondiabetic SOD1-tg males with diabetic Notch1+/– females. Mice were maintained on a 12-hour-light/12-hour dark cycle, with noon of the day of vaginal plug observation defined as E0.5. Pregnant dams were euthanized using isoflurane, and embryos were collected at E11.5 and E14.5.
Embryo processing and histological analysis.
Harvested embryos were fixed in 4% paraformaldehyde at 4°C overnight followed by alcohol gradient dehydration and paraffin embedding. Serial tissue sections (6 μm) of the embryonic heart were collected for histological analysis. H&E staining (Vector Laboratories) was used to visualize gross morphology and Alcian blue staining (Vector Laboratories) to visualize proteoglycan deposition. Images were taken using Keyence BZ-X810 Fluorescence Microscope.
Amira3D reconstruction of developing endocardial cushions and cell counts.
Serial sections of embryonic hearts from diabetic and nondiabetic group were imaged at 10× original magnification using Olympus BX51, and images were loaded onto Amira3D v2023.2 for alignment and segmentation of AV endocardial cushions. Volume measurement was recorded using the MaterialStatistics module. To count EndMT-derived cells within the AV cushion, diabetic and nondiabetic E11.5 RosamTmG; WT; Tie2-Cre+ and RosamTmG; Notch1+/–; Tie2-Cre+ (Figure 2C) sections were used. Following immunostaining against GFP, the number of cells in the AV cushion was quantified by counting nuclei stained with DAPI of GFP+ cells within the defined area of the AV cushion. At least 3 readings were taken from each image, and 3 serial sections were used for each biological replicate to obtain mean cell count/area within the AV cushion.
Immunofluorescence and immunohistochemistry.
Histological sections of embryonic hearts were deparaffinized using xylene and gradient ethanol washes, followed by antigen retrieval using heat-mediated, citrate-based Antigen Unmasking solution (H-3300, Vector Laboratories). For immunofluorescence, tissue sections were blocked with 2% normal horse serum in 1× phosphate-buffered saline with 0.1% Tween 20 (PBST) for 1 hour and then incubated with rabbit anti-GFP (Abcam ab290, 1:200) and rabbit anti-DPEAAE (Invitrogen PA1-1748A, 1:100) overnight at 4°C. Following 1× PBST wash, sections were incubated with donkey anti-rabbit secondary antibody conjugated to Alexa Fluor 488 for 1 hour at room temperature in the dark (Invitrogen A-21206). Sections were washed 3 times with 1× PBST and counterstained with VECTASHIELD Vibrance Mounting Medium with DAPI (Vector Laboratories). Fluorescence images were visualized using the Keyence BZX-810 Fluorescence Microscope. Relative fluorescence intensity was measured using ImageJ software (NIH). For immunohistochemistry, unmasked tissue sections were incubated in 3% H2O2 followed by blocking in 1% bovine serum albumin in 1× PBST for 1 hour and then incubated with mouse anti–4-HNE (R&D Systems, Bio-Techne; MAB3249, 1:100) overnight at 4°C. Following 1× PBST wash, sections were incubated with SignalStain Boost IHC detection reagent (HRP anti-mouse; Cell Signaling Technology 8125) for 1 hour at room temperature. SignalStain DAB substrate kit (Cell Signaling Technology 8059) was used to develop the stain. Images were taken using the Keyence BZX-810 Microscope in bright-field. Mean gray value of each image was measured using ImageJ software.
Cellular apoptosis.
TUNEL assay was performed to detect cellular apoptosis using TUNEL Assay Kit (Cell Signaling Technology 64936S) according to manufacturer’s instructions. For nuclear staining, slides were mounted using VECTASHIELD Vibrance Mounting Medium with DAPI. Fluorescence images were taken using Keyence BZ-X810 Fluorescence Microscope, and percentage of TUNEL+ cells within the embryonic heart was counted using ImageJ software.
Culture of iPSCs, directed differentiation into endothelial lineage, and oxidative stress treatments.
NOTCH1WT and NOTCH1+/– iPSCs were generated as previously described (51, 52). For differentiation into endothelial cells, NOTCH1WT and NOTCH1+/– iPSCs were cultured in complete E8 media until approximately 90% confluent and then changed to differentiation media following a published protocol (53). For oxidative stress studies, H2O2 was added to EGM2 media at a final concentration of 50 μM, and cells were exposed for 4 days, with media refreshment every 24 hours.
RNA-Seq analysis.
Following oxidative stress treatment for 4 days, total RNA was collected from NOTCH1WT and NOTCH1+/– iECs using Total RNA Purification Kit (Norgen Biotek) according to manufacturer’s instructions. RNA was prepared for sequencing following provider’s instructions (Novogene Corporation) and sequenced on a NovaSeq 6000 (Illumina). Primary processing and expression quantification of the reads were performed by DataPrudence (Carboctet). FASTQ files were trimmed to remove Illumina adaptor sequences and low-quality reads using TrimGalore! in “paired” mode (72). Paired, trimmed reads were then aligned against Homo sapiens assembly version GRCh38.p14, GENCODE 44 (http://Jul2023.archive.ensembl.org/Homo_sapiens/Info/Annotation), using the STAR alignment tool (v2.7.10b) with “--quantMode TranscriptomeSAM” option (73). The transcriptome-aligned BAM was used to collect gene-level and transcriptome-level count estimates using RSEM (v1.3.1) (74). The gene count estimates were used to identify DEGs using DESeq2 (v1.34.0) (75). Overrepresentation analysis was performed using R package genekitr (https://CRAN.R-project.org/package=genekitr) and cross-validated with clusterProfiler (76). GO terms were simplified using GO semantic similarity analysis using built-in GOsim() function in genekitr (77). Volcano plots were created using ggplot2 (78). Gene regulatory networks were visualized and annotated using Cytoscape (79).
Statistics.
Categorical data are presented with count and percentage while continuous data are presented as mean and SD. Our experiments included embryos from multiple diabetic dams, which introduces the possibility that correlation of outcomes from embryos from the same litter could impact results. For binomial outcomes (incidence of VSD in Figures 1 and 5), Fisher’s exact test was used, and binomial regression models with robust standard errors accounting for litter were conducted as sensitivity analyses (Supporting Data Values). For continuous outcomes, linear regression with random effects for litter were conducted, and 2-tailed t tests for the estimated marginal means for all pairwise comparisons were carried out using Tukey’s correction to adjust for multiple testing. A P value less than 0.05 was considered statistically significant.
Study approval.
All animal use was approved and monitored on protocol AR09-00056 by the Institutional Animal Care and Use Committee at the Research Institute at Nationwide Children’s Hospital and conducted in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Data availability.
Values for all data points shown in graphs are reported in the Supporting Data Values file. The RNA-Seq data produced in this study were deposited to the NCBI’s Gene Expression Omnibus database (accession no. GSE279323).
Author contributions
VG and TZC conceived the study and experimental design with input from DL and MTZ. TZC and SCG performed and analyzed the experiments. The murine studies were performed by TZC, SCG, HBG, and EMC. MTZ generated and provided iPSC lines. Bioinformatics analyses were performed by TZC, ASR, and YB. Statistical analysis was performed by TZC, SC, and VG. TZC and VG wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
The authors acknowledge funding support from NIH National Heart, Lung, and Blood Institute (R01HL144009) to VG, (R01 HL155282) to MTZ, and (T32HL166149) to ASR. We thank members of the Biomorphology Core at Nationwide Children’s Hospital for histology support as well as S. Adamczak, C. Boettcher, and the Animal Resource Core at Nationwide Children’s Hospital for animal support. We would also like to acknowledge T. Vetter in the Microscopy Core at Nationwide Children’s Hospital for imaging support. We thank S. Manivannan, Novogene Corporation, and Carboctet LLC for RNA-Seq and bioinformatics analysis support. The authors would like to acknowledge D. Lewandowski for manuscript editing assistance. We would like to acknowledge all members in the laboratories of VG, DL, MTZ, and J. Jang (Nationwide Children’s Hospital) for technical support.
Version 1. 10/22/2024
In-Press Preview
Version 2. 12/06/2024
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2024, Choudhury et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2024;9(23):e183516.https://doi.org/10.1172/jci.insight.183516.
Contributor Information
Talita Z. Choudhury, Email: talita.choudhury2@nationwidechildrens.org.
Sarah C. Greskovich, Email: sarah.greskovich1@nationwidechildrens.org.
Holly B. Girard, Email: hollygirard2015@gmail.com.
Anupama S. Rao, Email: anupama.rao@nationwidechildrens.org.
Yogesh Budhathoki, Email: yogesh.budhathoki@nationwidechildrens.org.
Emily M. Cameron, Email: emily.cameron@nationwidechildrens.org.
Sara Conroy, Email: sara.conroy@nationwidechildrens.org.
Deqiang Li, Email: deqiang.li@nationwidechildrens.org.
Ming-Tao Zhao, Email: Mingtao.Zhao@NationwideChildrens.org.
Vidu Garg, Email: vidu.garg@nationwidechildrens.org.
References
- 1.Tsao CW, et al. Heart disease and stroke statistics-2022 update: a report from the American Heart Association. Circulation. 2022;145(8):e153–e639. doi: 10.1161/CIR.0000000000001052. [DOI] [PubMed] [Google Scholar]
- 2.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sifrim A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48(9):1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morton SU, et al. Genomic frontiers in congenital heart disease. Nat Rev Cardiol. 2022;19(1):26–42. doi: 10.1038/s41569-021-00587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasuhara J, Garg V. Genetics of congenital heart disease: a narrative review of recent advances and clinical implications. Transl Pediatr. 2021;10(9):2366–2386. doi: 10.21037/tp-21-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nora JJ. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases. The genetic-environmental interaction. Circulation. 1968;38(3):604–617. doi: 10.1161/01.CIR.38.3.604. [DOI] [PubMed] [Google Scholar]
- 7.Vecoli C, et al. Congenital heart disease: the crossroads of genetics, epigenetics and environment. Curr Genomics. 2014;15(5):390–399. doi: 10.2174/1389202915666140716175634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simeone RM, et al. Proportion of selected congenital heart defects attributable to recognized risk factors. Ann Epidemiol. 2016;26(12):838–845. doi: 10.1016/j.annepidem.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalisch-Smith JI, et al. Environmental risk factors for congenital heart disease. Cold Spring Harb Perspect Biol. 2020;12(3):a037234. doi: 10.1101/cshperspect.a037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loffredo CA, et al. Maternal diabetes: an independent risk factor for major cardiovascular malformations with increased mortality of affected infants. Teratology. 2001;64(2):98–106. doi: 10.1002/tera.1051. [DOI] [PubMed] [Google Scholar]
- 11.Helle EI, et al. First trimester plasma glucose values in women without diabetes are associated with risk for congenital heart disease in offspring. J Pediatr. 2018;195:275–278. doi: 10.1016/j.jpeds.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoang TT, et al. Original findings and updated meta-analysis for the association between maternal diabetes and risk for congenital heart disease phenotypes. Am J Epidemiol. 2017;186(1):118–128. doi: 10.1093/aje/kwx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Øyen N, et al. Prepregnancy diabetes and offspring risk of congenital heart disease: a nationwide cohort study. Circulation. 2016;133(23):2243–2253. doi: 10.1161/CIRCULATIONAHA.115.017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhury TZ, et al. Impact of maternal hyperglycemia on cardiac development: insights from animal models. Genesis. 2021;59(11):e23449. doi: 10.1002/dvg.23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino T, et al. Single cell multimodal analyses reveal epigenomic and transcriptomic basis for birth defects in maternal diabetes. Nat Cardiovasc Res. 2023;2(12):1190–1203. doi: 10.1038/s44161-023-00367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manivannan S, et al. Single-cell transcriptomic profiling unveils dysregulation of cardiac progenitor cells and cardiomyocytes in a mouse model of maternal hyperglycemia. Commun Biol. 2022;5(1):820. doi: 10.1038/s42003-022-03779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland TW, et al. Congenital heart disease in infants of diabetic mothers. J Pediatr. 1973;83(5):815–820. doi: 10.1016/S0022-3476(73)80374-9. [DOI] [PubMed] [Google Scholar]
- 18.Correa A. Pregestational diabetes mellitus and congenital heart defects. Am Heart Assoc. 2016;133(23):2219–2221. doi: 10.1161/CIRCULATIONAHA.116.022960. [DOI] [PubMed] [Google Scholar]
- 19.Christoffels V, Jensen B. Cardiac morphogenesis: specification of the four-chambered heart. Cold Spring Harb Perspect Biol. 2020;12(10):a037143. doi: 10.1101/cshperspect.a037143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018;15(11):705–724. doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- 21.Kinsella MG, Fitzharris TP. Origin of cushion tissue in the developing chick heart: cinematographic recordings of in situ formation. Science. 1980;207(4437):1359–1360. doi: 10.1126/science.7355294. [DOI] [PubMed] [Google Scholar]
- 22.Mjaatvedt CH, Markwald RR. Induction of an epithelial-mesenchymal transition by an in vivo adheron-like complex. Dev Biol. 1989;136(1):118–128. doi: 10.1016/0012-1606(89)90135-8. [DOI] [PubMed] [Google Scholar]
- 23.Markwald RR, et al. Structural development of endocardial cushions. Am J Anat. 1977;148(1):85–119. doi: 10.1002/aja.1001480108. [DOI] [PubMed] [Google Scholar]
- 24.Wessels A, et al. The development of the atrioventricular junction in the human heart. Circ Res. 1996;78(1):110–117. doi: 10.1161/01.RES.78.1.110. [DOI] [PubMed] [Google Scholar]
- 25.Webb S, et al. Formation of the atrioventricular septal structures in the normal mouse. Circ Res. 1998;82(6):645–656. doi: 10.1161/01.RES.82.6.645. [DOI] [PubMed] [Google Scholar]
- 26.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77(2):211–215. doi: 10.1161/01.RES.77.2.211. [DOI] [PubMed] [Google Scholar]
- 27.Basu M, et al. Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease. JCI Insight. 2017;2(20):e95085. doi: 10.1172/jci.insight.95085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enciso JM, et al. Elevated glucose inhibits VEGF-A-mediated endocardial cushion formation: modulation by PECAM-1 and MMP-2. J Cell Biol. 2003;160(4):605–615. doi: 10.1083/jcb.200209014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S, et al. Restoring BMP4 expression in vascular endothelial progenitors ameliorates maternal diabetes-induced apoptosis and neural tube defects. Cell Death Dis. 2020;11(10):859. doi: 10.1038/s41419-020-03078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar SD, et al. Maternal diabetes induces congenital heart defects in mice by altering the expression of genes involved in cardiovascular development. Cardiovasc Diabetol. 2007;6:34. doi: 10.1186/1475-2840-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, et al. Oxidative stress is responsible for maternal diabetes-impaired transforming growth factor beta signaling in the developing mouse heart. Am J Obstet Gynecol. 2015;212(5):650.e1–650.e11. doi: 10.1016/j.ajog.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, et al. Superoxide dismutase 1 in vivo ameliorates maternal diabetes mellitus-induced apoptosis and heart defects through restoration of impaired Wnt signaling. Circ Cardiovasc Genet. 2015;8(5):665–676. doi: 10.1161/CIRCGENETICS.115.001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 34.McBride KL, et al. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17(18):2886–2893. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerstjens-Frederikse et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med. 2016;18(9):914–923. doi: 10.1038/gim.2015.193. [DOI] [PubMed] [Google Scholar]
- 36.Meester JA, et al. Overlapping but distinct roles for NOTCH receptors in human cardiovascular disease. Clin Genet. 2019;95(1):85–94. doi: 10.1111/cge.13382. [DOI] [PubMed] [Google Scholar]
- 37.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Monte G, et al. Monitoring Notch1 activity in development: evidence for a feedback regulatory loop. Dev Dyn. 2007;236(9):2594–2614. doi: 10.1002/dvdy.21246. [DOI] [PubMed] [Google Scholar]
- 40.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golub MS, Sobin CA. Statistical modeling with litter as a random effect in mixed models to manage “intralitter likeness”. Neurotoxicol Teratol. 2020;77:106841. doi: 10.1016/j.ntt.2019.106841. [DOI] [PubMed] [Google Scholar]
- 42.Greenland S, et al. Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol. 2016;31(4):337–350. doi: 10.1007/s10654-016-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gittenberger-de Groot AC, et al. Morphogenesis and molecular considerations on congenital cardiac septal defects. Ann Med. 2014;46(8):640–652. doi: 10.3109/07853890.2014.959557. [DOI] [PubMed] [Google Scholar]
- 44.Camenisch TD, et al. Regulation of cardiac cushion development by hyaluronan. Exp Clin Cardiol. 2001;6(1):4–10. [PMC free article] [PubMed] [Google Scholar]
- 45.Mjaatvedt C, et al. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev Biol. 1998;202(1):56–66. doi: 10.1006/dbio.1998.9001. [DOI] [PubMed] [Google Scholar]
- 46.Deepe R, et al. The mesenchymal cap of the atrial septum and atrial and atrioventricular septation. J Cardiovasc Dev Dis. 2020;7(4):50. doi: 10.3390/jcdd7040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kern CB, et al. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev Dyn. 2006;235(8):2238–2247. doi: 10.1002/dvdy.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jinsmaa Y, et al. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol. 2009;22(5):835–841. doi: 10.1021/tx800405v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong D, et al. Maternal diabetes triggers DNA damage and DNA damage response in neurulation stage embryos through oxidative stress. Biochem Biophys Res Commun. 2015;467(2):407–412. doi: 10.1016/j.bbrc.2015.09.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, et al. Oxidative stress-induced JNK1/2 activation triggers proapoptotic signaling and apoptosis that leads to diabetic embryopathy. Diabetes. 2012;61(8):2084–2092. doi: 10.2337/db11-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye S, et al. Impaired human cardiac cell development due to NOTCH1 deficiency. Circ Res. 2023;132(2):187–204. doi: 10.1161/CIRCRESAHA.122.321398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aljuhani M, et al. Generation and characterization of a human induced pluripotent stem cell line heterozygous for a NOTCH1 mutation (NCHi014-A) Stem Cell Res. 2024;74:103281. doi: 10.1016/j.scr.2023.103281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joachim A, et al. Generation of cardiomyocytes and endothelial cells from human iPSCs by chemical modulation of Wnt signaling. Methods Mol Biol. 2022;2549:335–344. doi: 10.1007/7651_2021_427. [DOI] [PubMed] [Google Scholar]
- 54.Zhu H, et al. Importance of gene-environment interactions in the etiology of selected birth defects. Clin Genet. 2009;75(5):409–423. doi: 10.1111/j.1399-0004.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- 55.Bosse K, et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J Mol Cell Cardiol. 2013;60:27–35. doi: 10.1016/j.yjmcc.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapman G, et al. Functional genomics and gene-environment interaction highlight the complexity of congenital heart disease caused by Notch pathway variants. Hum Mol Genet. 2020;29(4):566–579. doi: 10.1093/hmg/ddz270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luxán G, et al. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118(1):e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 58.Torregrosa-Carrion R, et al. NOTCH activation promotes valve formation by regulating the endocardial secretome. Mol Cell Proteomics. 2019;18(9):1782–1795. doi: 10.1074/mcp.RA119.001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatano S, et al. Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology. 2012;22(9):1268–1277. doi: 10.1093/glycob/cws095. [DOI] [PubMed] [Google Scholar]
- 60.Fischer A, et al. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 2007;100(6):856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- 61.Saxon JG, et al. BMP2 expression in the endocardial lineage is required for AV endocardial cushion maturation and remodeling. Dev Biol. 2017;430(1):113–128. doi: 10.1016/j.ydbio.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Ramirez CA, et al. Atlas of fetal metabolism during mid-to-late gestation and diabetic pregnancy. Cell. 2024;187(1):204–215. doi: 10.1016/j.cell.2023.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos-Riera KP, et al. Oxidative stress: a common imbalance in diabetes and epilepsy. Metab Brain Dis. 2023;38(3):767–782. doi: 10.1007/s11011-022-01154-7. [DOI] [PubMed] [Google Scholar]
- 64.Yan Lj. Redox imbalance stress in diabetes mellitus: role of the polyol pathway. Animal Model Exp Med. 2018;1(1):7–13. doi: 10.1002/ame2.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moazzen H, et al. N-Acetylcysteine prevents congenital heart defects induced by pregestational diabetes. Cardiovasc Diabetol. 2014;13:46. doi: 10.1186/1475-2840-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rabkin E, et al. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104(21):2525–2532. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Z, Rivkees SA. Programmed cell death in the developing heart: regulation by BMP4 and FGF2. Dev Dyn. 2000;217(4):388–400. doi: 10.1002/(SICI)1097-0177(200004)217:4<388::AID-DVDY6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi K, et al. Notch1 haploinsufficiency in mice accelerates adipogenesis. Sci Rep. 2021;11(1):16761. doi: 10.1038/s41598-021-96017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koenig SN, et al. Notch1 haploinsufficiency causes ascending aortic aneurysms in mice. JCI Insight. 2017;2(21):e91353. doi: 10.1172/jci.insight.91353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conlon RA, et al. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 71.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/S1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 72. Krueger F, et al. FelixKrueger/TrimGalore: v0. 6.10-add default decompression path (0.6. 10). https://github.com/FelixKrueger/TrimGalore/releases/tag/0.6.10 Accessed October 23, 2024.
- 73.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu T, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J Artif Intell Res. 1999;11:95–130. doi: 10.1613/jair.514. [DOI] [Google Scholar]
- 78.Wilkinson L. ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics. 2011;67(2):678–679. doi: 10.1111/j.1541-0420.2011.01616.x. [DOI] [Google Scholar]
- 79.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Values for all data points shown in graphs are reported in the Supporting Data Values file. The RNA-Seq data produced in this study were deposited to the NCBI’s Gene Expression Omnibus database (accession no. GSE279323).