Abstract
Long non-coding RNAs (lncRNAs) have recently emerged as critical regulators of oncogenic or tumor-suppressive pathways in human cancers. LINC01133 is a lncRNA that has exhibited dichotomous roles in various malignancies but to the best of our knowledge, the role of LINC01133 in laryngeal squamous cell carcinoma (LSCC) has not been previously investigated. This study aimed to investigate the expression, clinical significance, and potential functions of the LINC01133 in LSCC. Integrative bioinformatics analysis of sequencing data obtained from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets revealed LINC01133 as a differentially expressed lncRNA in head and neck/laryngeal cancers. Experimental validation via quantitative real-time PCR in 41 pairs of stage III and IV LSCC tissues and normal tissues adjacent to the tumor (NAT) demonstrated significant downregulation of LINC01133 in tumors (p<0.0001). Decreased LINC01133 expression associated with advanced tumor stage (p=0.0206) and lymph node metastasis (p=0.0203). The receiver operating characteristic analysis indicated potential diagnostic utility (AUC=0.7115, p=0.001). Bioinformatic predictions and literature mining suggested two potential competing endogenous RNA (ceRNA) mechanisms whereby LINC01133 may act as a tumor suppressor by sponging miR-205-5p to derepress the leucine-rich repeat kinase 2 (LRRK2) and androgen receptor, leading to dysregulation of cancer-related signaling cascades. This study provides initial evidence that loss of lncRNA LINC01133 expression may promote LSCC tumorigenesis, possibly by dysregulating microRNA interactions. Further verification of its regulatory mechanisms and diagnostic value is warranted.
Key Words: Laryngeal Squamous cell carcinoma, Long non-coding RNA, LINC01133LRRK2, Competing endogenous RNA, Androgen Receptor
INTRODUCTION
Laryngeal squamous cell carcinoma (LSCC) represents the second most prevalent form of head and neck squamous cell carcinoma (HNSCC), accounting for approximately 95% of laryngeal malignancies. Despite advancements in surgery, chemotherapy, and radiotherapy, the clinical management of LSCC has not led to satisfactory patient outcomes, with over 180,000 new cases and nearly 100,000 deaths worldwide in 2020 [1, 2]. Therefore, it is crucial to understand the underlying mechanisms involved in the development and progression of LSCC, which can lead to identifying novel diagnostic, therapeutic, and prognostic biomarkers.
In recent years, long non-coding RNAs (lncRNAs) have emerged as one of the major regulators of cellular homeostasis and contributors to tumor biology across human cancers. LncRNAs are defined as non-protein coding transcripts greater than 200 nucleotides in length that regulate gene expression at various levels [3-6]. Numerous studies have revealed that lncRNAs play vital roles in cancer tumorigenicity and progression [7]. lncRNAs are aberrantly expressed in a variety of cancers and participate in many biological functions, such as cell proliferation, apoptosis, epithelial-to-mesenchymal transition (EMT), invasion, migration, angiogenesis, and differentiation [8, 9]. Despite various investigations on the function of lncRNAs in laryngeal cancer, the expression landscape and functional roles of many lncRNAs in the pathogenesis of LSCC remain largely unexplored. In the present study, focusing on the expression profiles of lncRNAs in The Cancer Genome Atlas (TCGA) repository and the Gene Expression Omnibus (GEO) database we performed an integrative data analysis to obtain differentially expressed (DE) lncRNAs, mRNAs, and related miRNAs. According to the results, LINC01133 lncRNA was selected for the next steps of the study.
Long intergenic non-coding RNA 01133 (LINC01133), located on chromosome 1q23.2, has gained considerable attention due to its dysregulated expression and involvement in numerous cancer types. LINC01133 has context-dependent activities in cancer, with evidence for both tumor- suppressive and oncogenic properties [10]. It is demonstrated that LINC01133 acts as a tumor suppressor in gastric cancer [11], nasopharyngeal cancer [12], oral [13] esophageal squamous cell carcinoma (ESCC) [14], and colorectal cancer [15]. However, studies have also shown potential oncogenic functions for LINC01133 whereby it promotes proliferation, invasion, and metastasis of hepatocellular carcinoma [16], renal cell carcinoma [17], cervical cancer [18], lung cancer [19], and pancreatic cancer [20] by modulating various pathways. The role of LINC01133 in ovarian [21, 22] and breast [23, 24] cancers remains contentious, as some studies suggest tumor-suppressive effects while others indicate possible pro-oncogenic activities.
LINC01133 expression has been used as a biomarker to predict prognosis across various cancer types. However, as LINC01133 can have opposing roles in different tissues, the impact of its dysregulation on clinical outcomes is tissue-dependent [25]. For example, in gastric cancer [11], ESCC [14], and colorectal cancer [15], low LINC01133 levels correlate with lymph node metastasis (LNM) and advanced TNM stage. In contrast, increased expression of LINC01133 is associated with shorter overall survival and poorer prognosis in pancreatic [20, 26, 27] and lung cancers [19, 28]. The disparate relationships between LINC01133 levels and clinical outcomes highlight its complex, context-specific functions in human malignancies.
Exploring the molecular functions and clinical impact of LINC01133 may uncover new biomarkers and therapeutic approaches for laryngeal carcinoma. However, to the best of our knowledge, the expression of LINC01133 and its possible roles in LSCC remain unclear. In this study, we assessed LINC01133 expression in LSCC using quantitative real-time polymerase chain reaction (qRT-PCR) and examined its correlations with clinicopathological characteris-tics. Finally, using bioinformatics databases and systems biology approaches, we analyzed the potential roles of LINC01133 as a competing endogenous RNA (ceRNA).
MATERIALS AND METHODS
Patients and tissue samples: In this investigation, 41 pairs of LSCC tissue specimens at pathological stages III and IV, along with corresponding normal tissues adjacent to the tumor (NAT), were procured from the tumor bank of the Otorhinolaryngology Research Center at AmirAlam Hospital in Tehran, Iran. The study received approval from the ethical committee of the AmirAlam Hospital Complex (Ethical code: IR.TUMS.AMIRALAM.REC.1401.034). Additionally, written informed consent was obtained from all patients before their tissue samples being deposited in the tumor bank. Patients who had undergone any preoperative chemotherapy, radiotherapy, or immunotherapy prior to recruitment, as well as those with a history of previous or secondary malignancy, were excluded from the study. Samples were also evaluated by expert pathologists to determine the stages, based on clinicopathological characteristics.
Transcriptome expression profiling in LSCC and HNSC through GEO and TCGA data analysis
Data source and analysis: The TCGA-HNSC project transcriptomic data (https://portal. gdc.cancer.gov) was downloaded using the GDCRNATools package (R software). Data consisted of the Gene expression profile data (564 samples, 44 normal samples, and 520 primary tumors), miRNA expression profile data (567 samples, 43 normal and 524 primary tumors), and clinical data of patients. In addition, from the GEO database (https://www.ncbi. nlm.nih.gov/geo/), (GPL20301 platform), GSE130605 dataset which included sequencing data from the whole transcriptome of 50 LSCC and 50 paired NAT were analyzed using GEO2R.
Identification of DE mRNAs, DE lncRNAs and DEmiRs: The TCGA-HNSC data was investigated to determine differentially expressed genes (mRNAs and lncRNAs) and miRNAs between tumor samples and normal head and neck tissue samples using R software, the limma package. Differentially expressed genes and miRNAs with log2 fold change |log2FC| ≥ 1 and adj.p.value <0.05 were considered for further analysis. The GSE130605 dataset was also analyzed to identify DE genes (mRNAs, lncRNAs, and miRNAs) between LSCC and paired NAT using GEO2R which utilizes the DESeq2. DE genes with the significance level cut-off < 0.05 and log2 fold change threshold ≥|1| were selected. To reduce false positive results, the results of the GSE130605 dataset analysis and TCGA-HNSC project were merged and intersection values were extracted using the Venn diagram tool (http://bioinformatics.psb.ugent. be/webtools/Venn/) and the final DEmRNAs, DElncRNAs, and DEmiRNAs were obtained.
Construction of ceRNA regulatory network hypothesis: There were 16 DElncRNA obtained from previous steps and all of them upregulated in both HNSCC (TCGA-HNSC) and laryngeal data (GSE130605) except LINC01133. This lncRNA showed a contradictory expression in bioinformatics data and previously published studies. Therefore, LINC01133 was selected to construct the lncRNA/miRNA/mRNA network.
Expression Study and survival analysis: Total RNA was extracted from 50 mg of fresh frozen tissue stored in liquid nitrogen using the Kiazol Reagent (KIAZIST Life Sciences, Iran) following the manufacturer’s protocol. The quantity and purity of the extracted total RNA were evaluated using a Nanodrop 2000C spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed by visualizing the 28S:18S rRNA ratio via agarose gel electrophoresis. cDNA synthesizing from the isolated total RNA was performed using the ExcelRTTM Reverse Transcription Kit II (SMOBIO Technology, Inc., Taiwan). The relative expression levels of lncRNA were assessed in both cancerous and non-cancerous tissues via qRT-PCR using the RealQ Plus 2x Master Mix Green (Amplicon, Denmark) on a Roche LightCycler® 96 System. The following cycling conditions were considered: initial denaturation at 95°C for 15 min, followed by 45 cycles of 95°C for 20 sec and 60°C for 50 sec. Primer specificity and reaction efficiency were evaluated via melt curve analysis (60-99°C) and agarose gel electrophoresis of the PCR products. The RPL30 gene was utilized as the reference for normalization of gene expression, as it has been identified as a suitable and stable housekeeping gene for quantitative gene expression analyses in HNSCC [29]. The primers used for LINC01133 and RPL30 were as follows: LINC01133-F: 5′-GGGGAGAGTAGGTGAAAAGATGA-3′, R: 5′-GCTGGACTTT GGAGAACTTTGC-3′; RPL30-F: 5′-TGGCTATCATTGATCCAGGTGAC-3′, R: 5′- GCAGG TTTAAGGTTTGCAGGTG-3′. The data for each sample was collected in duplicate. The 2−ΔΔCT method was used to calculate the relative expression levels. Also, the correlation between LINC01133 expression and patient survival was determine using GEPIA2.
Constructing protein-protein interaction (PPI) network of the downregulated DEmRNAs: The search tool for retrieval of interacting genes (STRING) (https://string-db.org) database was used to investigate the physical and functional interactions between downregula- ted mRNAs (proteins). The active interaction sources considered were according to text- mining, experiments, databases, coexpression, neighborhood, gene fusion, and cooccurrence and the minimum required interaction score was 0.400 for PPI network construction. The visualization of the obtained PPI network was done using Cytoscape software version 3.10. and the following criteria were considered for the selection of hub genes in the PPI network. 12 hub-genes genes with i) the highest degree (11-24) and ii) the highest corresponding BetweennessCentrality (0.04-0.1) were selected as downregulated hub-genes for further analysis.
Functional and pathway enrichment analysis of hub-genes: Enrichr (http://amp.pharm. mssm.edu/Enrichr/) were used to investigate functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of 12 hubs downregulated common genes. P-value and adjusted p-value of <0.05 for KEGG pathway terms, were considered for selecting the significant terms respectively.
ceRNA network construction: The potential interactions between LINC01133 and DEmiRNAs were predicted using RNA22 tool (https://cm.jefferson.edu/rna22/) and the interactions between DEmiRNAs and potential genes were also identified by miRtarbase (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php). Subsequently, the DEmRNAs which are interacted with common DEmiRNAs in association with LINC01133 were selected. Cytoscape software (version 3.10.1). was used to construct the potential regulatory networks involved LINC01133/miRNA/mRNA axis.
Statistical Analysis: In the current study, GraphPad Prism 9.1.0 software (GraphPad Software, Inc., San Diego, CA) was used for all statistical analyses. The data were expressed as mean ± standard deviation. Normality was assessed using the Kolmogorov-Smirnov test and Shapiro-Wilk test. The paired samples t-test was employed to compare the relative expression of lncRNA between LSCC tissues and matched NAT. The association between lncRNA expression level and clinicopathological characteristics was analyzed using the independent samples t-test. The receiver operating characteristic (ROC) curve was plotted to assess the diagnostic performance of studied lncRNA as a potential biomarker. P-values below 0.05 were considered statistically significant.
RESULTS
The TCGA-HNSC project transcriptomic data and whole transcriptome sequencing of the GSE130605 dataset were analyzed. The transcriptomic data between tumoral tissues and normal or adjacent tissues revealed 4137 DE mRNAs, 153 DElncRNAs, and 37 DEmiRNAs in the GSE130605 dataset and 2016 DE mRNAs, 127 DElncRNAs and 169 DEmiRNAs in TCGA-HNSC project. After merging the sequencing data, the deferentially expressed results were as follows: 1185 DE mRNAs (547 downregulated), 16 DElncRNAs (15 upregulated and one controversial LINC01133), and 4 DEmiRNAs (3 upregulated and one downregulated) (Fig.1 and Tables S1-S3).
Figure 1.
Venn diagrams of intersected deferentially expressed sequencing data. a. 1185 DE mRNAs (547 downregulated), b. 16 DElncRNAs (15 upregulated and one controversial LINC01133), and c. 4 DEmiRNAs. See also tables S1-S3.
Considering the dysregulation pattern of DElncRNAs after analysis of two transcriptomic data, it is revealed that among 16 DElncRNAs only LINC01133 had a controversial expression pattern between two transcriptomic data. The results of several previous studies also indicated a double-edged sword-like properties for LINC01133 which can act as a tumor suppressor or an oncogene in a context-dependence manner. Fifteen other DElncRNAs demonstrated an upregulated expression pattern. So, in the current study, LINC01133/miRNA/mRNA regulatory network is considered for further analysis.
LINC01133 expression levels were assessed in 41 LSCC tissues with stages III and IV and compared with their corresponding NAT. The results indicated a significant downregulation of LINC01133 expression in tumoral tissues compared to NAT (Mean of differences = -1.538 ± 1.896, p<0.0001) (Fig. 2A). Furthermore, the expression level of LINC01133 was remarkably lower in 73% (30/41) of tumor samples compared to that in NAT (Fig. 2B). The correlation between LINC0113 expression and patient survival in TCGA- HNSC was determine using GEPIA2 and is shown in Figure S2.
Figure 2.
LINC01133 expression in LSCC. A. Lower expression of LINC01133 (****p<0.0001) in LSCC tissues compared with adjacent non-cancerous tissues. B. The corresponding relationship of LINC01133 expression between each pair of LSCC tissues and adjacent non-tumor tissues (****p<0.0001). C. LINC01133 expression was obviously lower in stage IV than in stage III (*p=0.020). D. Expression of LINC01133 was significantly lower in patients with lymph node metastasis (LNM) (*p=0.020).
The clinicopathological characteristics of the included patients are summarized in Table 1. These features include age, tumor size, tumor stage, histological grade, presence of LNM and lymphovascular/perineural invasion, and smoking/drinking status. As shown in Table 1, there is a significant association between decreased expression of LINC01133 and stage IV LSCC (p=0.0206) (Fig. 2C) as well as the presence of lymph node metastasis (p=0.0203) (Fig. 2D). No significant associations were observed between LINC01133 expression and other clinicopatho-logical factors, including age, tumor size, histological grade, and the presence of lymphovascu-lar and perineural invasion (p>0.05).
Table 1.
Correlation of LINC01133 expression with clinicopathological features in LSCC patients
| Characteristics | No. of cases (%) | Difference between means (A-B) ± SEM | 95% confidence interval | p -value |
|---|---|---|---|---|
|
Gender
Male Female |
41 (100%) 0 (0%) |
- | - | - |
|
Age (years)
< 57 ≥ 57 |
21 (51.22%) 20 (48.78%) |
0.7708 ± 0.6037 | -0.4502 to 1.992 | 0.209 |
|
Tumor diameter (cm)
< 4 ≥ 4 |
21 (51.22%) 20 (48.78%) |
-0.2869 ± 0.6144 | -1.530 to 0.9559 | 0.643 |
|
Differentiation
Well + Moderate Poor |
28 (68.29%) 13 (31.71%) |
-0.01158 ± 0.7441 | -1.517 to 1.493 | 0.987 |
|
Lymphovascular invasion
Yes No |
13 (31.71%) 28 (68.29%) |
0.09102 ± 0.6617 | -1.247 to 1.429 | 0.891 |
|
Perineural invasion
Yes No |
17 (41.46%) 24 (58.54%) |
-0.1156 ± 0.6249 | -1.379 to 1.148 | 0.854 |
|
Lymph node metastasis
Yes No |
20 (48.78%) 21 (51.22%) |
-1.390 ± 0.5745 | -2.552 to -0.2276 | 0.020 |
|
Clinical stage
III IV |
22 (53.66%) 19 (46.34%) |
1.390 ± 0.5761 | 0.2251 to 2.556 | 0.020 |
|
Smoking
Yes No |
37 (90.24%) 4 (9.76%) |
-0.05865 ± 1.038 | -2.158 to 2.041 | 0.955 |
|
Alcohol consumption
Yes No |
3 (7.32%) 38 (92.68%) |
0.1057 ± 1.183 | -2.286 to 2.498 | 0.929 |
To assess the potential diagnostic and prognostic value of the expression status of the studied lncRNA for distinguishing LSCC tissues from NAT, the ROC curve analysis was conducted (Fig. 3). The ROC curve analysis revealed that the LINC01133 expression level could be considered a promising discriminating marker for LSCC patients, with a sensitivity of 51.22% and specificity of 82.93% (AUC=0.7115, 95% CI=0.6004 to 0.8226, p= 0.0010).
Figure 3.
The receiver operating characteristic (ROC) curve of LINC01133 expression for discrimination of LSCC from adjacent tissues. AUC indicates area under the ROC curve.
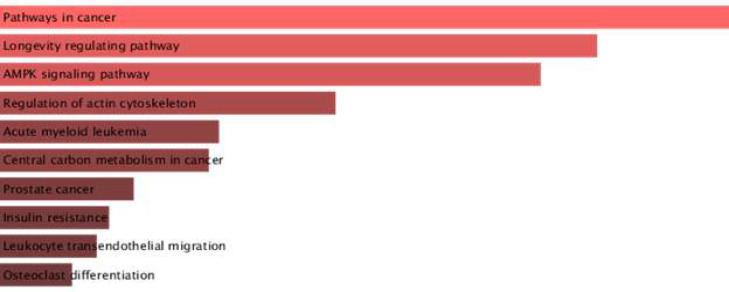
After the expression study, it was revealed that LINC01133 was downregulated in our samples. We hypothesize that there are some crucial downregulated hub genes that are regulated by some miRNAs and LINC01133 acts as a ceRNA related to these miRNAs. Accordingly, we considered the LINC01133/miRNA/mRNA regulatory network in which LINC01133 is downregulated and there are upregulated and downregulated miRNAs and mRNAs, respectively. The PPI network for downregulated common mRNAs was constructed using STRING. The resulting network included 546 nodes and 1192 edges (Fig. S1). In the next step, 12 Hub genes were identified according to the degree and Betweenness Centrality score using Cytoscape version 3.10.1 (Table 2). Enrichment analysis for functions and pathways of the hub genes revealed that they are active in pathways such as pathways in cancer, longevity regulating pathway, AMPK signaling pathway, prostate cancer, and regulation of actin cytoskeleton (Fig. 4).
Table 2.
12 Hub genes in the PPI network
| Hub genes | Degree | BetweennessCentrality | |
|---|---|---|---|
| 1 | PPARG coactivator 1 alpha (PPARGC1A) | 19 | 0.068002851 |
| 2 | clusterin(CLU) | 14 | 0.055599654 |
| 3 | leucine rich repeat kinase 2(LRRK2) | 11 | 0.046535115 |
| 4 | elastin (ELN) | 15 | 0.05588649 |
| 5 | forkhead box A1(FOXA1) | 13 | 0.054779315 |
| 6 | androgen receptor (AR) | 19 | 0.091613674 |
| 7 | KIT proto-oncogene, receptor tyrosine kinase (KIT) | 21 | 0.084867494 |
| 8 | mucin 1, cell surface associated (MUC1) | 21 | 0.062050042 |
| 9 | myosin heavy chain 11(MYH11) | 14 | 0.061951599 |
| 10 | phosphoinositide-3-kinase regulatory subunit 1(PIK3R1) | 18 | 0.079871036 |
| 11 | peroxisome proliferator activated receptor gamma (PPARG) | 24 | 0.107825272 |
| 12 | C-X-C motif chemokine ligand 12(CXCL12) | 24 | 0.076872587 |
Figure 4.
Enrichment analysis according to Enrichr database and according to the p-value for 12 hub-genes.
LINC01133- mediated ceRNA network was constructed based on the experimental results which approved the downregulation of LINC01133 in LSCC tissues in comparison to NAT. According to the RNA22 results, among 3 up-regulated common DEmiRNAs achieved in the bioinformatics phase, only the interaction of hsa-miR-205-5p, hsa-miR-205-3p, and hsa-miR-4652-5p with LINC01133 was approved. On the other hand, using miRtarbase it is revealed that among 12 hub-genes extracted from common downregulated DEmRNAs, AR and LRRK2 genes have interaction sites only with hsa-miR-205-5p. Accordingly, these two final networks can be proposed for LINC01133 activity in the context of LSCC development: LINC01133/hsa-miR-205-5p/LRRK2 and LINC01133 /hsa-miR-205-5p/AR (Fig. 5).
Figure 5.
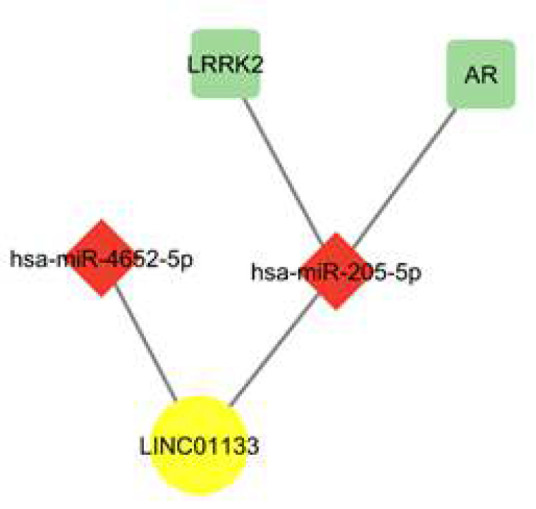
Two potential networks for LINC01133 activity
Discussion
LSCC is one of the most prevalent forms of HNSCC [1]. LncRNAs are crucial regulators of gene expression in human malignancies through diverse mechanisms [4]. In particular, they can act as ceRNAs that sequester microRNAs and indirectly regulate target gene expression levels [30]. LncRNAs have been implicated as promising diagnostic and prognostic biomarkers across different types of cancers, including laryngeal cancer [4]. Many lncRNAs, such as FOXD2-AS1 [31], SNHG1 [32], and HOXA11-AS [33], have been shown to mediate therapy resistance, proliferation, and invasion in LSCC. Regarding LINC01133, contrasting effects in promoting or suppressing tumors across various cancer types have been reported [10, 34]. The impact of LINC01133 is believed to be influenced by its distinct interacting partners and signaling consequences in different tissues [10, 34]. Despite extensive research on dysregulated lncRNAs and their clinical utility as biomarkers, the expression and functional significance of LINC01133 in LSCC pathogenesis remain unexplored to the best of our knowledge.
In the current investigation, bioinformatics analysis revealed a contradictory expression pattern of LINC01133 between sequencing data obtained from TCGA-HNSC and GEO-LSCC datasets. LINC01133 expression was downregulated in the TCGA-HNSC dataset but upregulated in the GEO-LSCC dataset. This discrepancy might arise from potential biases in patient cohorts between TCGA-HNSC and GSE130605 datasets, including factors such as the tumor's origin, race/ethnicity, and other demographic or clinical variables. Additionally, technical issues, including platform variations, RNA contamination, or other experimental factors, could contribute to the observed inconsistencies. Furthermore, previous studies on other head and neck cancers, such as oral squamous cell carcinoma (OSCC) [13] and nasopharyngeal cancer [12], have reported downregulation of LINC01133 in tumor tissues compared to normal adjacent tissues, aligning with our bioinformatic finding from the TCGA-HNSC dataset. To elucidate the expression profile of LINC01133 and predict its possible role in laryngeal carcinoma, we performed expression analysis of LINC01133 in LSCC and adjacent tissues through qRT-PCR.
The expression analysis in 41 pairs of stage III-IV LSCC tissues and matched NAT showed significant downregulation of LINC01133 in tumor tissues versus NAT (p<0.0001). In the present study, significant associations were observed between low expression of LINC01133 and aggressive features of LSCC, including advanced TNM stage (p=0.0206) and LNM (p=0.0203), indicating its tumor suppressive function in LSCC. This aligns with some prior studies in other malignancies where LINC01133 inhibited proliferation, invasion, and treatment resistance [11-15]. The ROC curve analysis indicated that LINC01133 can be considered as a promising discriminatory biomarker in LSCC. If these findings are validated in clinical settings, assessing the expression level of this gene could assist in prognosis prediction and guide therapeutic decision-making.
In the subsequent step, considering the downregulation of LINC001133 in LSCC tissues at the experimental level, further bioinformatics analysis was performed to investigate the hypothetical molecular mechanism of LINC01133’s function as a ceRNA. Two hypothetical pathways were proposed, including the LINC01133/hsa-miR-205-5p/LRRK2 and the LINC01133 /hsa-miR-205-5p/AR pathways.
MiR-205-5p displays heterogeneous dysregulation across various cancer subtypes, acting either as an oncogene or tumor suppressor through various downstream mechanisms [35]. This miRNA, known as a marker of epithelial phenotype, has been observed to decrease during the process of EMT, a crucial stage in facilitating tumor invasion and metastasis. Mechanistically, it can target EMT transcriptional regulators like ZEB1/ZEB2 to inhibit EMT [35, 36]. However, in endometrial carcinoma [37, 38], ovarian cancer [39, 40], and nasopharyngeal carcinoma [41, 42], miR-205-5p is overexpressed and promotes malignant phenotypes. In endometrial cancer, it targets tumor suppressors PTEN and ESRRG to inhibit apoptosis and drive proliferation [37, 38]. In nasopharyngeal carcinoma, the overexpression of miR-205 leads to PTEN down-regulation and the activation of the AKT pathway. This process enhances tumor progression and contributes to resistance against radiation therapy [41, 42]. Interestingly, in the lung [43] and esophageal [44] cancers, the high miR-205-5p expression indicates squamous cell carcinoma whereas low expression marks adenocarcinoma, highlighting utility as a diagnostic marker. There is contradictory evidence for miR-205 expression in LSCC. Tian et al. demonstrated that miR-205 could suppress proliferation and promote apoptosis in LSCC [45]. Conversely, other studies have reported elevated levels of miR-205 in HNSCC [46] and in LSCC, where it promotes cell proliferation and invasion by downregulating CDK2AP1 expression [47] or through AKT-mediated EMT [48].
Consistent with our bioinformatic findings, some other investigations have revealed that miR-205 can be a negative regulator for the expression of Leucine-rich repeat kinase 2 (LRRK2) [49-51]. LRRK2 is a large, multi-domain protein that exhibits both GTPase and kinase enzymatic activities. As a result, this protein engages in multifunctional activities through interactions with various proteins and the display of catalytic functions as a GTPase and kinase. LRRK2 is found to act in several key cellular activities, primary stages of autophagy, endocytosis, and the functions related to mitochondria and cytoskeleton [52, 53]. Substantial studies revealed the association of LRRK2 genetic variations with Parkinson’s disease as well as immune-mediated conditions like inflammatory bowel disease, leprosy, and tuberculosis [52, 53]. In recent years, several studies have identified LRRK2 mutations or dysregulation of its expression in various types of human malignancies including thyroid cancer [54], intrahepatic cholangiocarcinoma [55], lung cancer [56, 57], and renal carcinoma [58]. Intriguingly, in a cancer-type dependent manner, LRRK2 appears to affect tumor development -both promoting and suppressing them- by affecting crucial cellular pathways [53]. Proposed mechanisms for a potential LRRK2 tumor suppressor function include the phosphorylation of p53 [59] and activation of ERK and JNK-dependent autophagy [60]. In contrast, LRRK2 may also have an oncogenic activity via MET signaling induction [58]. Notably, in ovarian cancer, elevated miR-205 levels are associated with a decrease in its lncRNA sponge LINC01133. This imbalance leads to reduced expression of the miR-205 target LRRK2 and enhances the proliferative, migration, and invasion of ovarian cancer cells. Thus, the LINC01133/miR-205/LRRK2 axis represents a novel pathway driving ovarian cancer pathogenesis [21]. According to our bioinformatic analysis, dysregulation of this pathway could also contribute to LSCC oncogenesis. However, further validation analyses examining the expression and functional impacts of the LINC01133/miR-205/LRRK2 network specifically in LSCC models are needed. Elucidating whether this signaling cascade is truly aberrant and a driver in LSCC pathogenesis could unveil new prognostic or therapeutic opportunities.
As previously described, our bioinformatic analysis predicted another potential ceRNA network involving LINC01133, hsa-miR-205-5p, and the androgen receptor (AR). In this hypothesized pathway, LINC01133 acts as a ceRNA that sequesters hsa-miR-205-5p, effectively derepressing AR, a validated target of hsa-miR-205-5p. Several studies have demonstrated that miR-205 directly targets and downregulates AR expression across different cell types [61, 62]. AR is a sex steroid hormone receptor that is activated by ligands such as dihydrotestosterone (DHT) and plays a key role in multiple physiological processes, such as reproductive system development. In its inactive state, AR is stabilized in the cytoplasm through binding to chaperone proteins. Upon ligand binding, AR dissociates from chaperones, after conformational changes, it moves to the nucleus and binds DNA at androgen responsive elements (AREs) to modulate target gene expression. In addition to this genomic signaling, AR can activate non-genomic cascades like the PI3K/AKT pathway through cytoplasmic interactions [63]. Given its influence over various oncogenic mechanisms including the PI3K/AKT, EGFR, Src, and WNT pathways, AR dysregulation plays crucial roles in promoting the proliferation and progression of certain cancers such as prostate, breast, liver, and ovarian malignancies [63, 64]. While the oncogenic functions of AR signaling have been well documented, numerous evidence indicates AR can also exhibit tumor-suppressive roles in certain cancer contexts. For example, studies in prostate cancer [65-70], renal cell carcinoma [71], liver cancer [72], and breast cancer [73-75] have shown that AR activation can suppress cell proliferation, migration, and metastasis or induce apoptosis and differentiation programs counteracting malignant phenotypes. The exact mechanisms determining AR's dichotomous context-dependent roles are not fully understood but may relate to differences in interacting cofactors, relative expression of AR variants, epigenetic and post-translational control of AR activity, crosstalk with other cellular signaling networks, and influence of the tumor microenvironment. Further investigation is warranted to elucidate the specific factors that dictate when AR signaling promotes or impedes tumorigenesis across distinct cancer types.
AR signaling has been implicated as potentially playing a role in HNSCC; however, its mechanism of action remains poorly understood [76, 77]. While some studies have reported upregulation of the AR in HNSCC [78, 79], other analyses [80, 81], as well as our bioinformatic findings, indicate that downregulation of AR can also occur in HNSCC compared to normal tissue. Additionally, it has been reported that AR expression is significantly lower in poorly differentiated laryngeal SCC and cases with lymphatic invasion, indicating low AR may promote more aggressive carcinoma progression [82]. Overall, the available evidence highlights the complex, context-specific nature of AR signaling in HNSCC/LSCC pathogenesis.
In summary, the lncRNA LINC01133 appears to act as a tumor suppressor in LSCC. Our experimental analysis revealed that LINC01133 is downregulated in LSCC tissues compared to NAT. Decreased expression associated with the presence of LNM and advanced TNM stages supporting its inhibitory impact on aggressive cancer phenotypes. Bioinformatics prediction and literature evidence suggest the tumor suppressive functions of LINC01133 may occur through sponging miR-205-5p to derepress the miR-205-5p targets LRRK2 and AR. Additional studies validating and exploring these ceRNA networks would offer further insight into LSCC pathogenesis. Given its dysregulation in LSCC and correlation with unfavorable prognostic features, LINC01133 merits investigation as a novel biomarker or even a potential therapeutic target in laryngeal carcinoma. Overall, this study unveils the clinical relevance and hypothetical molecular mechanisms of LINC01133 in LSCC that warrant future research.
Acknowledgment:
We would like to express our gratitude to the AmirAlam Hospital Otorhinolaryngology Research Center for providing the patient samples that enabled this study. This work was supported by a research grant from the Faculty of Medicine, Tehran University of Medical Sciences (TUMS), Tehran, Iran. We sincerely appreciate this funding opportunity. Additionally, we would like to extend our appreciation to all the individuals who participated in our research.
Conflict of Interest:
The authors declare no conflict of interest.
Authors’ Contribution:
MR: Conceptualization, methodology, investigation, formal analysis, visualization, writing–original draft; ZJ: Conceptualization, methodology, investigation, formal analysis, visualization, writing–original draft; SS: resources; FH: Resources; ML: Resources; EGh: Investigation; MA: Investigation; MM: Investigation; LS: Formal analysis; ASh: Supervision, project administration, funding acquisition.
Supplementary materials
References
- 1.Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67:31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Richard Boland C. Non-coding RNA: It's not junk. Dig Dis Sci. 2017;62:1107–1109. doi: 10.1007/s10620-017-4506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, Liu H, Fan T. Long non-coding rNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol. 2020;10:598817. doi: 10.3389/fonc.2020.598817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. mechanisms and functions of long non-coding rNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding rNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of LncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 8.Bartonicek N, Maag JLV, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal S. Long noncoding rNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghafouri-Fard S, Khoshbakht T, Mahmud Hussen B, Taheri M, Mokhtari M. A review on the role of LINC01133 in cancers. Cancer Cell Int. 2022;22:270. doi: 10.1186/s12935-022-02690-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Du M, Wang T, Chen W, Wu J, Li Q, Tian X, Qian L, Wang Y, Peng F, Fei Q, Chen J, He X, Yin L. Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am J Cancer Res. 2019;9:779–790. [PMC free article] [PubMed] [Google Scholar]
- 13.Kong J, Sun W, Zhu W, Liu C, Zhang H, Wang H. Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J Surg Oncol. 2018;118:1326–1334. doi: 10.1002/jso.25278. [DOI] [PubMed] [Google Scholar]
- 14.Yang XZ, He QJ, Cheng TT, Chi J, Lei ZY, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. Predictive value of lINC01133 for unfavorable prognosis was impacted by alcohol in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2018;48:251–262. doi: 10.1159/000491724. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JH, Li AY, Wei N. Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:2103–2107. [PubMed] [Google Scholar]
- 16.Yin D, Hu ZQ, Luo CB, Wang XY, Xin HY, Sun RQ, Wang PC, Li J, Fan J, Zhou ZJ, Zhou J, Zhou SL. LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin Transl Med. 2021;11:e409. doi: 10.1002/ctm2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai X, Wu Y, Wang Z, Zhao D, Li H, Chong T, Zhao J. Long noncoding rNA LINC01133 promotes the malignant behaviors of renal cell carcinoma by regulating the miR-30b-5p/Rab3D axis. Cell Transplant. 2020;29:963689720964413. doi: 10.1177/0963689720964413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Zhang Y, Sun X. LINC01133 promotes the progression of cervical cancer via regulating miR-30a-5p/FOXD1. Asia Pac J Clin Oncol. 2021;17:253–263. doi: 10.1111/ajco.13451. [DOI] [PubMed] [Google Scholar]
- 19.Zang C, Nie FQ, Wang Q, Sun M, Li W, He J, Zhang M, Lu KH. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016;7:11696–11707. doi: 10.18632/oncotarget.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CS, Chu J, Zhu XX, Li JH, Huang XT, Cai JP, Zhao W, Yin XY. The C/EBPβ-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett. 2018;421:63–72. doi: 10.1016/j.canlet.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Shen C, Wang C. Long noncoding RNA LINC01133 confers tumor-suppressive functions in ovarian cancer by regulating Leucine-Rich repeat kinase 2 as an miR-205 sponge. Am J Pathol. 2019;189:2323–2339. doi: 10.1016/j.ajpath.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Xi X. LINC01133 contribute to epithelial ovarian cancer metastasis by regulating miR-495-3p/TPD52 axis. Biochem Biophys Res Commun. 2020;553:1088–1094. doi: 10.1016/j.bbrc.2020.09.074. [DOI] [PubMed] [Google Scholar]
- 23.Tu Z, Schmollerl J, Cuiffo BG, Karnoub AE. Microenvironmental regulation of long noncoding RNA LINC01133 promotes cancer stem Cell-Like phenotypic traits in triple-negative breast cancers. Stem Cells. 2019;37:1281–1292. doi: 10.1002/stem.3055. [DOI] [PubMed] [Google Scholar]
- 24.Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med. 2019;23:7554–7565. doi: 10.1111/jcmm.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong C, Zhang Q, Zhang M, Qi Y, Duan S. LINC00662: A new oncogenic LncRNA with great potential. J Cell Physiol. 2022;237:1105–1118. doi: 10.1002/jcp.30599. [DOI] [PubMed] [Google Scholar]
- 26.Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis. 2018;39:1016–1025. doi: 10.1093/carcin/bgy069. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Tang T, Yang X, Qin P, Wang P, Zhang H, Bai M, Wu R, Li F. Tumor-derived exosomal long noncoding RNA LINC01133, regulated by Periostin, contributes to pancreatic ductal adenocarcinoma epithelial-mesenchymal transition through the Wnt/β-catenin pathway by silencing AXIN2. Oncogene. 2021;40:3164–3179. doi: 10.1038/s41388-021-01762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36:7465–7471. doi: 10.1007/s13277-015-3460-9. [DOI] [PubMed] [Google Scholar]
- 29.Palve V, Pareek M, Krishnan NM, Siddappa G, Suresh A, Kuriakose MA, Panda B. A minimal set of internal control genes for gene expression studies in head and neck squamous cell carcinoma. PeerJ. 2018;6:e5207. doi: 10.7717/peerj.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu ZS, Wang WH, Dong XN, Tian LML. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol. 2020;26:4240–4260. doi: 10.3748/wjg.v26.i29.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Chen S, Zhan J, Li X, Liu W, Sheng X, Lu Z, Zhong R, Chen L, Luo X, Hu Y, Ouyang Y, Liu T, Zhang Q, Zhang S. Long noncoding RNA FOXD2-AS1 enhances chemotherapeutic resistance of laryngeal squamous cell carcinoma via STAT3 activation. Cell Death Dis. 2020;11:41. doi: 10.1038/s41419-020-2232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin SX, Jiang H, Xiang GZ, Zhang WR, Weng YH, Qiu FD, Wu J, Wang HG. Up-regulation of long non-coding RNA SNHG1 contributes to proliferation and metastasis in laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:1333–1341. doi: 10.26355/eurrev_201803_14475. [DOI] [PubMed] [Google Scholar]
- 33.Qu L, Jin M, Yang L, Sun C, Wang P, Li Y, Tian L, Liu M, Sun Y. Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res. 2018;10:573–580. [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S, Zhang Q, Li J, Raziq K, Kang X, Liang S, Sun C, Liang X, Zhao D, Fu S, Cai M. New sights into long Non-Coding rNA LINC01133 in cancer. Front Oncol. 2022;12:908162. doi: 10.3389/fonc.2022.908162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari E, Gandellini P. Unveiling the ups and downs of miR-205 in physiology and cancer: transcriptional and post-transcriptional mechanisms. Cell Death Dis. 2020;11:980. doi: 10.1038/s41419-020-03192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 37.Su N, Qiu H, Chen Y, Yang T, Yan Q, Wan X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol Rep. 2013;29:2297–2302. doi: 10.3892/or.2013.2400. [DOI] [PubMed] [Google Scholar]
- 38.Zhang G, Hou X, Li Y, Zhao M. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer ishikawa cells. BMC Cancer. 2014;14:440. doi: 10.1186/1471-2407-14-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iorio MV, Visone R, Di Leva G, Donati V, Pertocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 40.He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J, Wu X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9:8206–8220. doi: 10.7150/thno.37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu C, Liang Z, Huang J, Zhao R, Su C, Wang S, Wang X, Zhang R, Lee MH, Yang H. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Y, Wu S, Zhao R, Deng Q. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J Int Med Res. 2016;44:231–240. doi: 10.1177/0300060515576556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charkiewicz R, Pilz L, Sulewska A, Kozlowski M, Niklinska W, Moniuszko M, Reszec J, Manegold C, Niklinski J. Validation for histology-driven diagnosis in non-small cell lung cancer using hsa-miR-205 and hsa-miR-21 expression by two different normalization strategies. Int J Cancer. 2016;138:689–697. doi: 10.1002/ijc.29816. [DOI] [PubMed] [Google Scholar]
- 44.Hezova R, Kovarikova A, Srovnal J, Zemanova M, Harustiak T, Ehrmann J, Hajduch M, Sachlova M, Svoboda M, Slaby O. MiR-205 functions as a tumor suppressor in adenocarcinoma and an oncogene in squamous cell carcinoma of esophagus. Tumour Biol. 2016;37:8007–8018. doi: 10.1007/s13277-015-4656-8. [DOI] [PubMed] [Google Scholar]
- 45.Tian L, Zhang J, Ge J, Xiao H, Lu J, Fu S, Liu M, Sun Y. MicroRNA-205 suppresses proliferation and promotes apoptosis in laryngeal squamous cell carcinoma. Med Oncol. 2014;31:785. doi: 10.1007/s12032-013-0785-3. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Z, Liu C, Liu K, Lv M, Li B, Lan Z, Chen W, Kang M. Expression and Possible Molecular Mechanisms of microRNA-205-5p in Patients With Head and Neck Squamous Cell Carcinoma. Technol Cancer Res Treat. 2020;19:1533033820980110. doi: 10.1177/1533033820980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong G, Xiong X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol Res. 2015;48:60. doi: 10.1186/s40659-015-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Lv K, Chen W, Zhao J, Luo J, Wu J, Li Z, Qin H, Wong TS, Yang W, Fu QL, Lei W. miR-375 and miR-205 Regulate the Invasion and Migration of Laryngeal Squamous Cell Carcinoma Synergistically via AKT-Mediated EMT. Biomed Res Int. 2016;2016 doi: 10.1155/2016/9652789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Li J, Tao L, Lv L, Sun J, Zhang T, Wang H, Wang J. MiR-205 regulates LRRK2 expression in dopamine neurons in parkinson's disease through methylation modification. Iran J Public Health. 2022;51:1637–1647. doi: 10.18502/ijph.v51i7.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Q, Huang X, Li R. lncRNA MALAT1/miR-205-5p axis regulates MPP(+)-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am J Transl Res. 2018;10:563–572. [PMC free article] [PubMed] [Google Scholar]
- 51.Cho HJ, Li G, Jin SM, Parisiadou L, Xie C, Yu J, Sun L, Ma B, Ding J, Vacraenenbroeck R, Lobbestael E, Baekelandt V, Taymans JM, He P, Troncoso JC, Shen Y, Cai H. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan J, Zhao W, Yu W, Cheng H, Zhu B. LRRK2 correlates with macrophage infiltration in pan-cancer. Genomics. 2022;114:316–327. doi: 10.1016/j.ygeno.2021.11.037. [DOI] [PubMed] [Google Scholar]
- 53.Lopez G, Lazzeri G, Rappa A, Isimbaldi G, Cribiu FM, Guerini-Rocco E, Ferrero S, Vaira V, Di Fonzo A. Comprehensive genomic analysis reveals the prognostic role of LRRK2 copy-number variations in human malignancies. Genes (Basel) 2020;11:846. doi: 10.3390/genes11080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang ZC, Chen XJ, Zhou Q, Gong XH, Chen X, Wu WJ. Downregulated LRRK2 gene expression inhibits proliferation and migration while promoting the apoptosis of thyroid cancer cells by inhibiting activation of the JNK signaling pathway. Int J Oncol. 2019;55:21–34. doi: 10.3892/ijo.2019.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu S, Chen J, Zhou Q, Yan M, He J, Han X, Qiu Y. LRRK2 is associated with recurrence-free survival in intrahepatic cholangiocarcinoma and downregulation of LRRK2 suppresses tumor progress in vitro. Dig Dis Sci. 2020;65:500–508. doi: 10.1007/s10620-019-05806-0. [DOI] [PubMed] [Google Scholar]
- 56.Lebovitz C, Wretham N, Osooly M, Milne K, Dash T, Thornton S, Tessier-Cloutier B, Sathiyaseelan P, Bortnik S, Go NE, Halvorsen E, Cederberg RA, Chow N, Santos ND, Bennewith KL, Nelson BH, Bally MB, Lam WL Gorski SM. Loss of parkinson's susceptibility gene LRRK2 promotes carcinogen-induced lung tumorigenesis. Sci Rep. 2021;11 doi: 10.1038/s41598-021-81639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng F, Zhang L, Ren Y, Ma Q. The genomic alterations of lung adenocarcinoma and lung squamous cell carcinoma can explain the differences of their overall survival rates. J Cell Physiol. 2019;234:10918–10925. doi: 10.1002/jcp.27917. [DOI] [PubMed] [Google Scholar]
- 58.Looyenga BD, Furge KA, Dykema KJ, Koeman J, Swiatek PJ, Giordano TJ, West AB, Resau JH, The BT, Mackeigan JP. Chromosomal amplification of leucine-rich repeat kinase-2 (LRRK2) is required for oncogenic MET signaling in papillary renal and thyroid carcinomas. Proc Natl Acad Sci U S A. 2011;108:1439–1444. doi: 10.1073/pnas.1012500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho DH, Kim H, Kim J, Sim H, Ahn H, Kim J, Seo H, Chung KC, Park BJ, Son I, Seol W. Leucine-Rich repeat kinase 2 (LRRK2) phosphorylates p53 and induces p21(WAF1/CIP1) expression. Mol Brain. 2015;8 doi: 10.1186/s13041-015-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leong YQ, Koh RY, Chye SM, Ng KY. Unravelling the genetic links between Parkinson's disease and lung cancer. Biol Chem. 2023;404:551–567. doi: 10.1515/hsz-2022-0228. [DOI] [PubMed] [Google Scholar]
- 61.Wen Y, Liu G, Zhang Y, Li H. MicroRNA-205 is associated with diabetes mellitus-induced erectile dysfunction via down-regulating the androgen receptor. J Cell Mol Med. 2019;23:3257–3270. doi: 10.1111/jcmm.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagman Z, Haflidadottir BS, Ceder JA, Larne O, Bjartell A, Lilja H, Edsjo A, Ceder Y. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br J Cancer. 2013;108:1668–1676. doi: 10.1038/bjc.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You CP, Tsoi H, Man EPS, Leung MH, Khoo US. Modulating the activity of androgen receptor for treating Breast Cancer. Int J Mol Sci. 2022;23:15342. doi: 10.3390/ijms232315342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antonarakis ES. AR Signaling in human malignancies: prostate cancer and beyond. Cancers (Basel) 2018;10:22. doi: 10.3390/cancers10010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu SQ, Han BM, Shao Y, Wu JT, Zhao FJ, Liu HT, Sun XW, Tang YQ, Xie SJ. Androgen receptor functioned as a suppressor in the prostate cancer cell line PC3 in vitro and in vivo. Chin Med J (Engl) 2009;122:2779–2783. [PubMed] [Google Scholar]
- 66.Han W, Liu M, Han D, Toure AA, Li M, Besschetnova A, Wang Z, Patalano S, Macoska JA, Lam HM, Corey E, He HH, Gao S, Balk SP, Cai C. Exploiting the tumor-suppressive activity of the androgen receptor by CDK4/6 inhibition in castration-resistant prostate cancer. Mol Ther. 2022;30:1628–1644. doi: 10.1016/j.ymthe.2022.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niu Y, Altuwaijri S, Lai KP, Wu CT, Ricke WA, Messing EM, Yao J, Yeh S, Chang C. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc Natl Acad Sci U S A. 2008;105:12182–12187. doi: 10.1073/pnas.0804700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao S, Gao Y, He HH, Han D, Han W, Avery A, Macoska JA, Liu X, Chen S, Ma F, Chen S, Balk SP, Cai C. Androgen receptor tumor suppressor function is mediated by recruitment of retinoblastoma protein. Cell Rep. 2016;17:966–976. doi: 10.1016/j.celrep.2016.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grosse A, Bartsch S, Baniahmad A. Androgen receptor-mediated gene repression. Mol Cell Endocrinol. 2012;352(1-2):46–56. doi: 10.1016/j.mce.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 70.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Chen S, Nelson PS, Liu XS, Brown M, Balk SP. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang Q, Sun Y, Ma X, Gao Y, Li X, Niu Y, Zhang X, Chang C. Androgen receptor increases hematogenous metastasis yet decreases lymphatic metastasis of renal cell carcinoma. Nat Commun. 2017;8:918. doi: 10.1038/s41467-017-00701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang N, Dou X, You X, Li Y, Li X, Liu G. Androgen receptors Act as a tumor suppressor gene to suppress hepatocellular carcinoma cells progression via miR-122-5p/RABL6 signaling. Front Oncol. 2021;11:756779. doi: 10.3389/fonc.2021.756779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hickey TE, Selth LA, Chia KM, Laven-Law G, Milioli HH, Roden D, Jindal S, Hui M, Finlay-Schultz J, Ebrahimie E, Birrell SN, Stelloo S, Iggo R, Alexandrou S, Caldon CE, Abdel-Fatah TM, Ellis IO, Zwart W, Palmieri C, Sartorius CA, Swarbrick A, Lim E, Carroll JS, Tilley WD. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat Med. 2021;27:310–320. doi: 10.1038/s41591-020-01168-7. [DOI] [PubMed] [Google Scholar]
- 74.Wei L, Gao H, Yu J, Zhang H, Nguyen TTL, Gu Y, Passow MR, Carter JM, Qin B, Boughey JC, Goetz MP, Weinshiboum RM, Ingle JN, Wang L. Pharmacological Targeting of androgen receptor elicits context-specific effects in estrogen receptor-positive ereast cancer. Cancer Res. 2023;83:456–470. doi: 10.1158/0008-5472.CAN-22-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, He X, Yu Q, Eng C. Androgen receptor-induced tumor suppressor, KLLN, inhibits breast cancer growth and transcriptionally activates p53/p73-mediated apoptosis in breast carcinomas. Hum Mol Genet. 2013;22:2263–2272. doi: 10.1093/hmg/ddt077. [DOI] [PubMed] [Google Scholar]
- 76.Qin C, Lu Y, Zhang H, Zhang Z, Xu W, Wen S, Gao W, Wu Y. Biological roles and clinical significance of estrogen and androgen receptors in head and neck cancers. J Cancer. 2022;13:2189–2199. doi: 10.7150/jca.66707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Čonkaš J, Sabol M, Ozretić P. 'Toxic Masculinity': what is Known about the role of androgen receptors in head and neck squamous cell carcinoma. Int J Mol Sci. 2023;24:3766. doi: 10.3390/ijms24043766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batelja-Vuletic L, Tomasovic-Lancaric C, Ceppi M, Bruzzone M, Fucic A, Krstanac K, Vucicevic VB. Comparison of androgen receptor, VEGF, HIF-1, Ki67 and MMP9 expression between non-metastatic and metastatic stages in stromal and tumor cells of oral squamous cell carcinoma. Life (Basel) 2021;11:336. doi: 10.3390/life11040336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomasovic-Loncaric C, Fucic A, Andabak A, Andabak M, Ceppi M, Bruzzone M, Vrdoljak D, Vucicevic-Boras V. Androgen receptor as a biomarker of oral squamous cell carcinoma progression risk. Anticancer Res. 2019;39:4285–4289. doi: 10.21873/anticanres.13593. [DOI] [PubMed] [Google Scholar]
- 80.Colella G, Izzo G, Carinci F, Campisi G, Muzio LL, D'Amato S, Mazzotta M, Cannavale R, Ferrara D, Minucci S. Expression of sexual hormones receptors in oral squamous cell carcinoma. Int J Immunopathol Pharmacol. 2011;24(2 Suppl):129–132. doi: 10.1177/03946320110240S222. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto H, Uryu H, Segawa Y, Tsuneyoshi M. Aggressive invasive micropapillary salivary duct carcinoma of the parotid gland. Pathol Int. 2008;58:322–326. doi: 10.1111/j.1440-1827.2008.02231.x. [DOI] [PubMed] [Google Scholar]
- 82.Atef A, El-Rashidy MA, Elzayat S, Kabel AM. The prognostic value of sex hormone receptors expression in laryngeal carcinoma. Tissue Cell. 2019;57:84–89. doi: 10.1016/j.tice.2019.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.