Abstract
In recent decades, 3D bioprinting has garnered significant research attention due to its ability to manipulate biomaterials and cells to create complex structures precisely. However, due to technological and cost constraints, the clinical translation of 3D bioprinted products (BPPs) from bench to bedside has been hindered by challenges in terms of personalization of design and scaling up of production. Recently, the emerging applications of artificial intelligence (AI) technologies have significantly improved the performance of 3D bioprinting. However, the existing literature remains deficient in a methodological exploration of AI technologies' potential to overcome these challenges in advancing 3D bioprinting toward clinical application. This paper aims to present a systematic methodology for AI-driven 3D bioprinting, structured within the theoretical framework of Quality by Design (QbD). This paper commences by introducing the QbD theory into 3D bioprinting, followed by summarizing the technology roadmap of AI integration in 3D bioprinting, including multi-scale and multi-modal sensing, data-driven design, and in-line process control. This paper further describes specific AI applications in 3D bioprinting's key elements, including bioink formulation, model structure, printing process, and function regulation. Finally, the paper discusses current prospects and challenges associated with AI technologies to further advance the clinical translation of 3D bioprinting.
Keywords: 3D bioprinting, Artificial intelligence, Machine learning, Quality by design, Regenerative medicine, Clinical translation
Graphical abstract
Highlights
-
•
AI-driven QbD can enhance 3D bioprinting's quality, rapidity, economy and scalability.
-
•
AI integrated in 3D bioprinting enables multi-scale and multi-modal sensing, data-driven design, and in-line process control.
-
•
Promising development directions of AI technology are proposed to further advance clinical translation of 3D bioprinting.
1. Introduction
3D bioprinting technology can be explored to fabricate well-defined multi-scale structures by precisely manipulating biomaterials and cells within three-dimensional space [[1], [2], [3]]. In the field of regenerative medicine, 3D bioprinted products (BPPs) can be used as patient-specific implants for regenerative repair of damaged organs/tissues or as patient-specific in vitro models for disease modeling and drug screening [4,5]. Despite recent progress in 3D bioprinting technology, clinical cases of BPPs applied in humans remain scarce. We identify several challenges at the R&D and production stages that hinder 3D bioprinting's clinical translation:
-
(ⅰ)
Personalization of design: The BPPs for clinical practice should be patient-specific [6], due to the immunity-, tissue-, structure-, and function-specific nature of repaired parts [[7], [8], [9]]. This necessitates that the design of BPPs replicates the complexity and specificity of natural tissues across multi-materials and multi-scale structures. In this regard, optimized design ensuring effectiveness introduces an extensive range of design parameters, requiring significant trial and error. However, because of the large differences and small batches of BPPs, it is difficult to amortize the R&D costs, resulting in the contradiction of “effectiveness-economy” [9].
-
(ⅱ)
Scaling up of production: Considering regulation, international regulatory frameworks for the commercialization of a medical device or an Advanced Therapeutic Medicinal Product (ATMP) require strict quality control to ensure that BPPs are manufactured in a reproducible and contamination-free manner [10]. However, current BPPs are typically designed and produced by skilled researchers in academic laboratories, which involves a number of complex manual operations. As a result, BPPs are small-scale, poorly repeatable, expensive, and difficult to regulate [6,9].
Therefore, to facilitate the clinical translation of BPPs, it is essential to enhance quality in both the R&D and production stages. The traditional Quality by Testing approach emphasizes post-production testing, which is impractical for the clinical application of BPPs, as changes in the clinical stage are costly and difficult [6]. Furthermore, Quality by Testing typically focuses on optimizing individual variables, making it inadequate for addressing the multi-material and multi-scale design requirements of BPPs. To address the above deficiencies, Quality by Design (QbD) is a promising solution for BPPs requiring effectiveness, economy, and regulatory compliance. This approach has been widely adopted by the U.S. FDA to enhance quality and efficiency, as well as to reduce costs and regulatory burdens, in fields related to 3D bioprinting such as biopharmaceuticals [11]. Compared to Quality by Testing, QbD posits that all problems affecting the quality of the final product are related to its design. Accordingly, in the beginning R&D stage, products should be designed correctly considering quality optimization. In the production stage, QbD uses process control to develop robust and reliable production procedures based on an in-depth understanding of products and processes. Currently, the introduction of QbD into 3D bioprinting is currently being discussed by the academic community, industry, and government [[12], [13], [14]].
In the field of 3D bioprinting, Artificial intelligence (AI), represented by machine learning (ML), has seen widespread application [15]. This revolutionary technology holds great potential in accelerating the deployment of QbD in 3D bioprinting. For example, deep learning can be used to automatically acquire critical quality attributes of BPPs from various sensor data, eliminating the need for extensive manual characterizations and thereby reducing costs. Supervised learning can be used to model the complex mapping relationship between critical material attributes/process parameters and critical quality attributes of BPPs. Given the vast number of design parameters, this approach significantly reduces the need for trial-and-error experiments. Reinforcement learning can be used to construct control strategies of 3D bioprinting, adapting to dynamic working scenarios based on interactions with the environment to meet the needs of scaling-up production. To summarize, AI-driven QbD will accelerate the translation of 3D bioprinting from bench to bedside [4,[16], [17], [18]].
Although recent review papers have elaborated on the utilization of AI in 3D bioprinting [10,[19], [20], [21], [22]], most take a workflow-centric approach, primarily summarizing the specific applications of AI in various steps of 3D bioprinting. In contrast, this paper adopts a clinical product perspective, leveraging the QbD theory from industrial production to propose a systematic framework for applying AI to 3D bioprinting. We begin by analyzing the fundamental methodologies and the technology roadmap of integrating AI with 3D bioprinting within the QbD framework, focusing on multi-scale and multi-modal sensing, data-driven design, and in-line process control. Next, we explore the current research status and application potential of AI across key elements of 3D bioprinting, including bioink formulation, model structure, printing process, and function regulation. Lastly, we propose future directions and challenges for AI in 3D bioprinting. We believe our theoretical framework will further guide the application of AI in broader fields such as tissue engineering, biofabrication, and related domains. We hope this review enables AI scientists to more effectively engage with the 3D bioprinting field, while helping 3D bioprinting researchers deepen their understanding of AI technologies and adopt the latest advancements.
2. AI-driven QbD framework and roadmap for 3D bioprinting
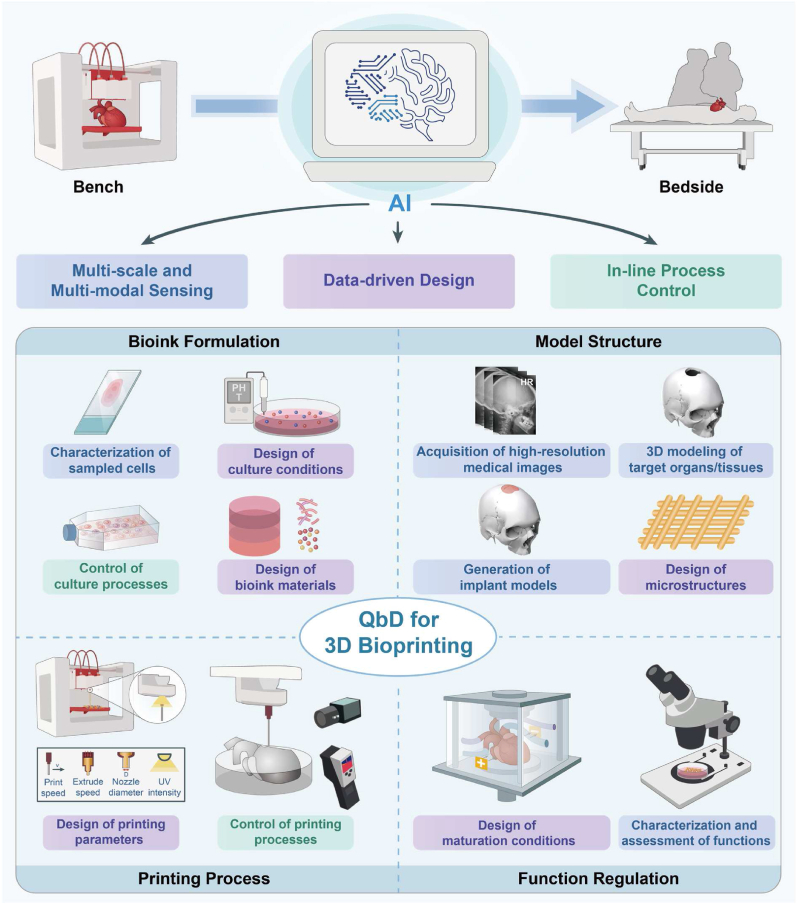
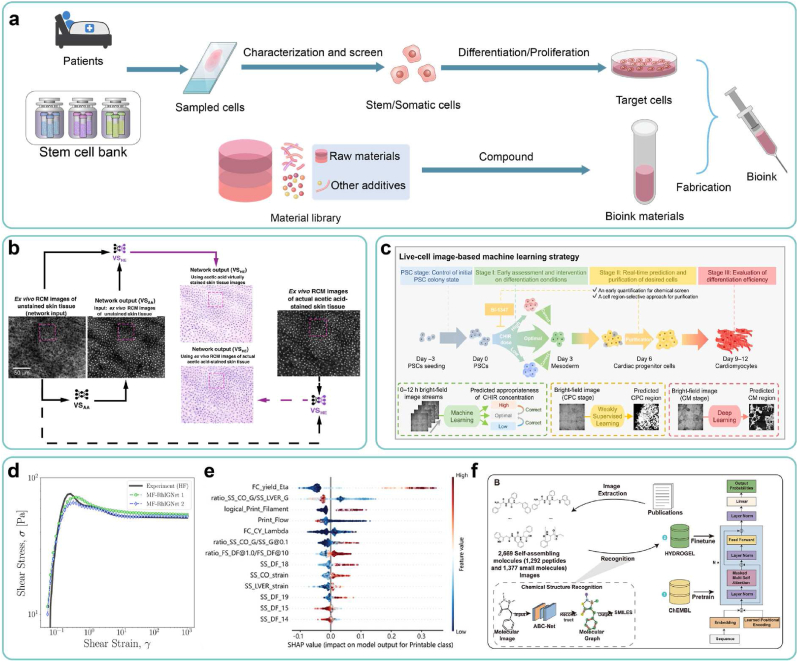
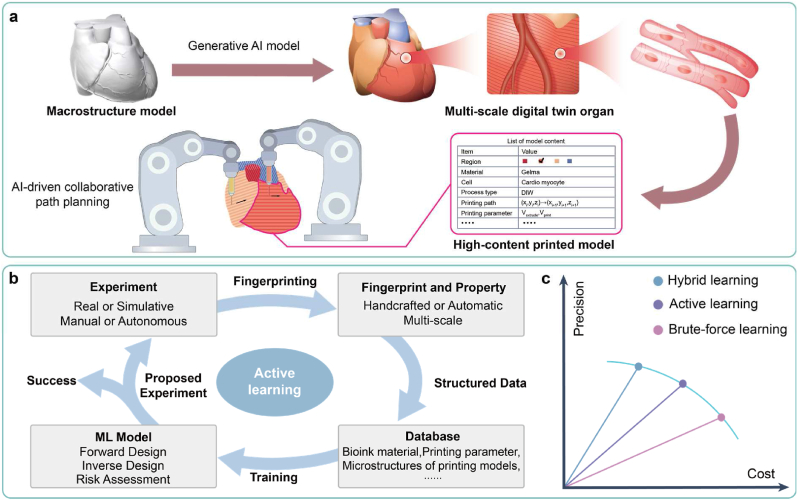
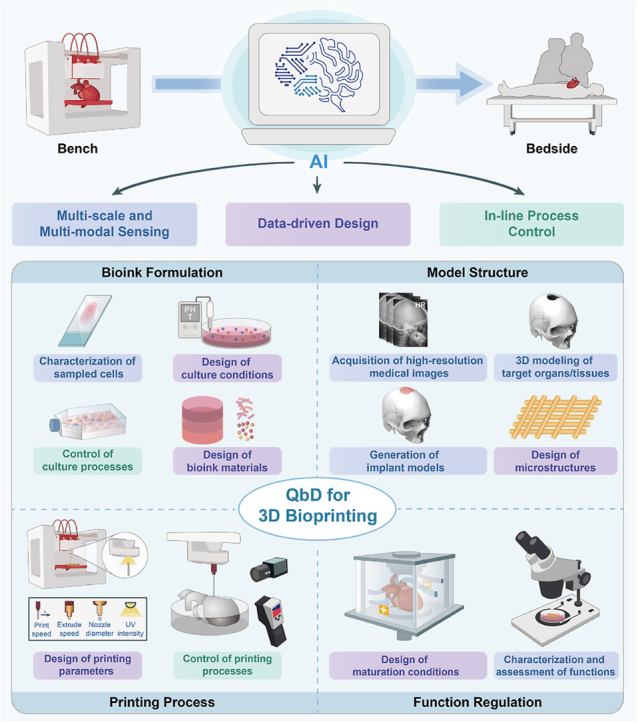
3D bioprinting comprises four key elements: bioink formulation, model structure, printing process, and function regulation. Each element consists of multiple unit operations (UOs) [[23], [24], [25]] within which AI-driven QdD can be integrated, such as design of bioink materials, design of microstructures, control of printing processes, and characterization and assessment of functions (Fig. 1). This chapter aims to offer a comprehensive analysis framework outlining the primary application scenarios of AI technology in 3D bioprinting. Specific applications of AI technology within separate UOs will be detailed in Chapters 3, 4, 5, and 6. Additionally, focusing on the application of AI technologies in 3D bioprinting, this paper has not included a detailed introduction to basic methods and concepts of AI technologies, which can be found in other references [26].
Fig. 1.
Roadmap of AI-driven QbD for 3D bioprinting, containing multi-scale and multi-modal sensing, data-driven design, and in-line process control, which can be used in four key elements, including bioink formation, model structure, printing process and function regulation.
2.1. QbD theory for 3D bioprinting
Given the inherent complexity of original QbD theories, this section simplifies the relevant terms and concepts, with further details of QbD available in other specialized references [[27], [28], [29], [30], [31], [32], [33]]. Following this, integrating 3D bioprinting, we provide a detailed explanation of the simplified QbD terminology:
-
(ⅰ)
Critical quality attributes (CQA): CQA refer to a physical, chemical, or biological property that reflects product quality [33]. In this regard, we propose two approaches: the forming-based and the function-based (Table 1). The forming-based CQA of BPPs are directly evaluated by their printability, which can be predicted by the rheological and gelation properties of bioinks. Printability is a critical category of CQA for mimicking functions of natural organs/tissues, as geometry profoundly determines mechanical and biological properties [[34], [35], [36]]. Further details on printability can be found in other comprehensive reviews [37]. The function-based CQA mainly include transport, mechanical, and biological properties. The transport properties are crucial for the survival and functionalization of BPPs, which ensure the delivery of oxygen, nutrients, biological factors, and drugs, as well as the removal of metabolites [38]. Moreover, it is imperative to form effective vascular networks in large-scale thick tissues [39]. BPPs should possess mechanical responses matching those of natural tissues as well as suitable degradation and swelling properties for in vitro culture and in vivo implantation [40]. The biological properties of BPPs can be assessed at various levels, including tissue, cell, and gene expression. Further details on biological properties can be found in other comprehensive reviews [41].
-
(ⅱ)
Critical material attributes (CMA)/Critical process parameters (CPP): CMA and CPP respectively refer to material attributes and process attributes that have a significant impact on product CQA [33]. For example, in the design of culture conditions, the compositions of the culture medium serve as CMA, and the culture process parameters serve as CPP. In the design of printing parameters, the bioink formulations serve as CMA, and the printing process parameters serve as CPP.
-
(ⅲ)
Design space: In QbD, CQA are determined by CMA and CPP. In this context, the design space describes the distribution of CQA under combinations of CMA/CPP within a certain range. In low-dimensional cases, the design space can be visualized in the form of phase diagrams or process windows, serving as a guide for designing CMA/CPP.
-
(ⅳ)
Control strategy: Control strategy refers to a planned set of controls over CMA/CPP, derived from product and process understanding, ensuring CQA of the production process [33].
-
(ⅴ)
Risk Assessment: Risk assessment refers to a process of quality risk management that can identify the impact of individual CMA/CPP on product CQA and the interactions among CMA/CPP [33]. This process enables a deeper understanding of the underlying process mechanisms.
Table 1.
Examples of CQA in 3D bioprinting.
| Approach | Examples of CQA | |
|---|---|---|
| Forming-based | Printability | Extrudability, filament formation, shape fidelity |
| Rheological properties | Shear-thinning, viscoelasticity, yield stress, constitutive model | |
| Gelation properties | Gelation time, gel fraction | |
| Function-based | Transport properties | Effective mass diffusion rate, vascularization |
| Mechanical properties | Mechanical response: Young's modulus, response curve, constitutive model, strength | |
| Degradation properties: degradation rate | ||
| Swelling properties: swelling rate | ||
| Function-based | Biological properties [41] | Gene expression: cell differentiation and phenotype, cell health, genomic stability |
| Protein expression: matrix production, cell differentiation and phenotype, cell health | ||
| Cell metabolism: nutrient and waste analysis, cell signaling, cellular products, cell health | ||
| Cell properties: viability, morphology, motility, confluence, cell number, cell health | ||
| Tissue properties: morphology, function | ||
2.2. Roadmap of AI-driven QbD for 3D bioprinting
Within the QbD framework, AI technologies enable the faster, more economical, and more scalable design and production of BPPs with higher CQA. This helps address the challenges of personalized design and scaling-up production in 3D bioprinting, accelerating the translation from bench to bedside. Here, we discuss the roadmap of AI technology for 3D bioprinting from three dimensions:
-
(ⅰ)
Multi-scale and multi-modal sensing: The structural and functional features across various scales are extracted by diverse sensors to rapidly and economically acquire CQA, CMA, and CPP.
-
(ⅱ)
Data-driven design: The intricate relationship between CMA/CPP and CQA is modeled through data to precisely determine the optimal design space.
-
(ⅲ)
In-line process control: The control procedures of process quality are then implemented through the established control strategy, which integrates AI technology in the former two dimensions.
In 3D bioprinting, each UO corresponds to the application of AI technology in one of the aforementioned three dimensions (Fig. 1). Thus, it is crucial to clarify the application scope of QbD-related terms. For the latter two dimensions, the corresponding UOs create products (such as cells, printed models, and BPPs), thus defining the application scope of QbD-related terms as the UOs themselves. For the first dimension, the corresponding UOs aim to determine the CQA, CMA, and CPP of products from other UOs, thus defining the application scope of QbD-related terms as their associated products.
2.2.1. Multi-scale and multi-modal sensing
In 3D bioprinting, each UO integrates various sensors to capture multi-modal data, facilitating the acquisition of multi-scale information crucial for personalized design and scaling-up production. The sensing process typically encompasses three sequential stages: (ⅰ) pre-sensing, involving pre-processing of the sensed object, such as tissue section preparation and staining; (ⅱ) sensing, entailing the utilization of various sensors to measure specific attributes of the sensed object and generating corresponding sensor data, and (ⅲ) post-sensing, involving processing and analyzing the collected sensor data to derive quantitative sensing results, including CQA, CMA, and CPP. Traditional sensing methodologies exhibit deficiencies in precision, rapidity, economy, repeatability, safety, and scalability, thereby impeding the clinical translation of BPPs. We attribute these deficiencies primarily to the following three key factors:
-
(ⅰ)
“Scale-depth-precision” contradiction: Imaging, as the primary sensing methodology [42], can sense objects in 3D bioprinting spanning scales from micrometers to centimeters [43]. Typically, larger objects necessitate greater imaging depth but exhibit lower resolution, and vice versa, hindering the extraction of detailed information of large-scale objects and 3D spatial information of small-scale objects (Fig. 2a).
-
(ⅱ)
Insufficient information abundance: Complex objects encompass multiple attributes, yet single sensing modalities excel in detecting specific attributes only, resulting in difficulties in precisely sensing multiple attributes simultaneously and biased sensing results.
-
(ⅲ)
Low automation: Traditional sensing methodologies rely on skilled operators to perform tedious manual operations with professional equipment and reagents in the pre-sensing and post-sensing stages. This reliance results in poor economy and rapidity. Moreover, manual operations entail subjective errors, contamination risks, and scaling-up challenges, further impeding repeatability, safety, and scalability.
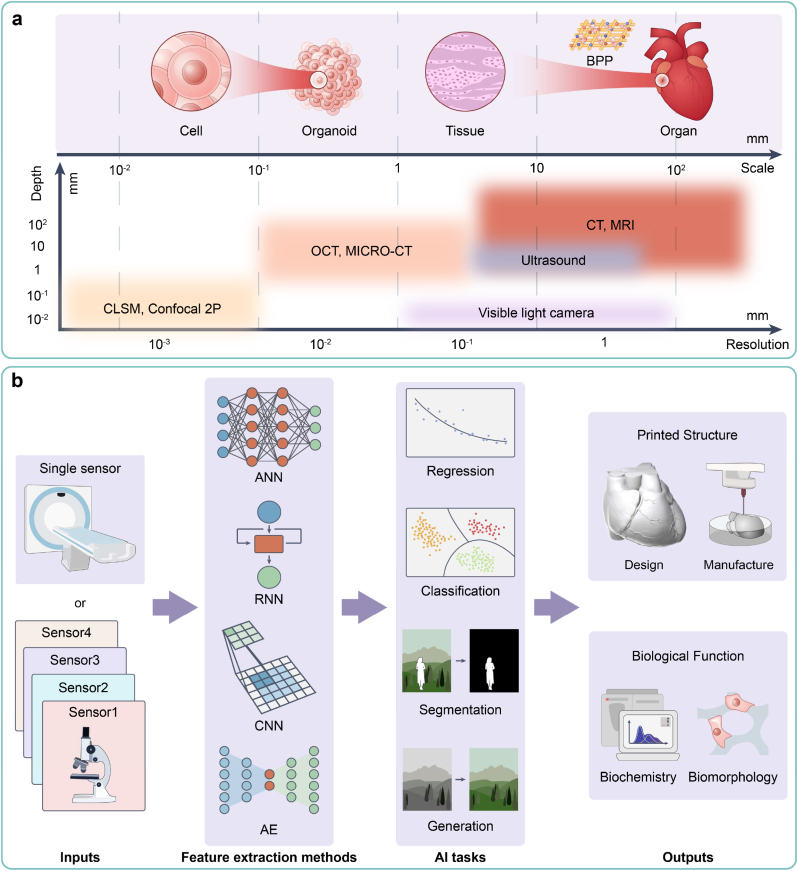
Fig. 2.
Multi-scale and multi-modal sensing. (a) A “scale-depth-precision” contradiction of imaging technologies. (b) A pipeline of AI-driven multi-modal sensing methodologies to obtain comprehensive results.
AI technology, particularly deep learning methodologies, presents viable solutions to the above challenges [41,44]. Based on input data from diverse sensors, specific feature extraction methods such as artificial neural networks (ANNs), convolutional neural networks (CNNs), recurrent neural networks (RNNs), and auto-encoders (AEs) are deployed to execute distinct AI tasks, including regression, classification, segmentation, and generation. Consequently, during the 3D bioprinting process, AI-driven sensing yields comprehensive results encompassing aspects such as the design and manufacturing of printed structures, as well as biochemical and morphological functions (Fig. 2b).
AI technology serves as a remedy for the contradiction of “scale-depth-precision,” enabling precise imaging of objects across various scales. In the realm of large-scale objects, as encountered in medical imaging modalities such as computed tomography (CT) and magnetic resonance imaging (MRI), AI-based super-resolution and denoising can enhance image resolution [45]. Super-resolution enhances overall image clarity, whereas denoising mitigates artifacts induced by patients' motion. Conversely, in the realm of small-scale objects, AI technology facilitates the extraction of 3D spatial information. Notably, studies have demonstrated the efficacy of AI-based automatic segmentation and 3D reconstruction in elucidating the spatial structure of minute tissues from serial sections [46], as well as the spatial distribution of the nucleus from confocal laser scanning microscopy (CLSM) images [47].
AI technology has the capacity to significantly augment information abundance and achieve precise and robust sensing of complex objects. Multi-modal machine learning (MML) stands as a prime example, integrating attribute information gathered from diverse sensors to markedly enhance the sensing precision of complex objects. Numerous studies have explored the application of MML across fields pertinent to 3D bioprinting. Examples include the segmentation of soft tissue sarcomas utilizing four types of medical images including CT, T1 MRI, T2 MRI, and Positron Emission Tomography (PET) [48], virtual staining of tissue sections through non-linear multi-modal imaging (NLM) [49], and monitoring of the printing process leveraging three types of sensor data (layerwise electro-optical, acoustic, and multispectral), alongside off-line process parameters [50].
AI technology can extract complex features and recognize complex patterns to reduce and replace manual operations. On the one hand, AI technology can emulate humans' sensing patterns, replacing humans in repetitive and labor-intensive tasks automatically, such as segmenting medical images [44] and assessing the matching degree of CMA/CPP during the printing process. On the other hand, AI technology can recognize patterns that are challenging for humans to learn from physical phenomena, thereby simplifying the sensing process and manual operations and minimizing reliance on expensive equipment and reagents. For instance, virtual staining technology [[51], [52], [53]] can comprehend the transformation patterns between different stained images, thus eliminating the need for multiple dyeing operations. The “digital rheometer twins” can derive rheological constitutive models from rheological data, reducing the dependence on rheometers [54]. In essence, the replacement and reduction of manual operations by AI technology serve to mitigate subjective errors, contamination risks, and costs while simultaneously enhancing repeatability, safety, and economy. Moreover, automatic sensing processes contribute to enhanced rapidity and scalability.
2.2.2. Data-driven design
The core principle of QbD asserts that CQA is contingent upon CMA/CPP. Consequently, the procedural framework for personalized design within QbD can be succinctly outlined as follows: (ⅰ) modeling of the potential mapping relationship between CMA/CPP and CQA, (ⅱ) determination of the optimal design space of CMA/CPP, taking enhancement of CQA as the primary objective, and (ⅲ) risk assessment to scrutinize the effect of each CMA/CPP on CQA. In the field of 3D bioprinting, personalized design involves various objects, including culture conditions for sampled cells, bioink materials, microstructures of printed models, printing parameters, and maturation conditions of BPPs (Fig. 1). Notably, in tackling modeling problems, the dilemma of “precision-cost” frequently arises [55]. As model precision (or problem complexity) escalates, a corresponding increase in associated costs (such as financial investment, time, and human resources) is observed, while the marginal precision incrementally diminishes (Fig. 3a).
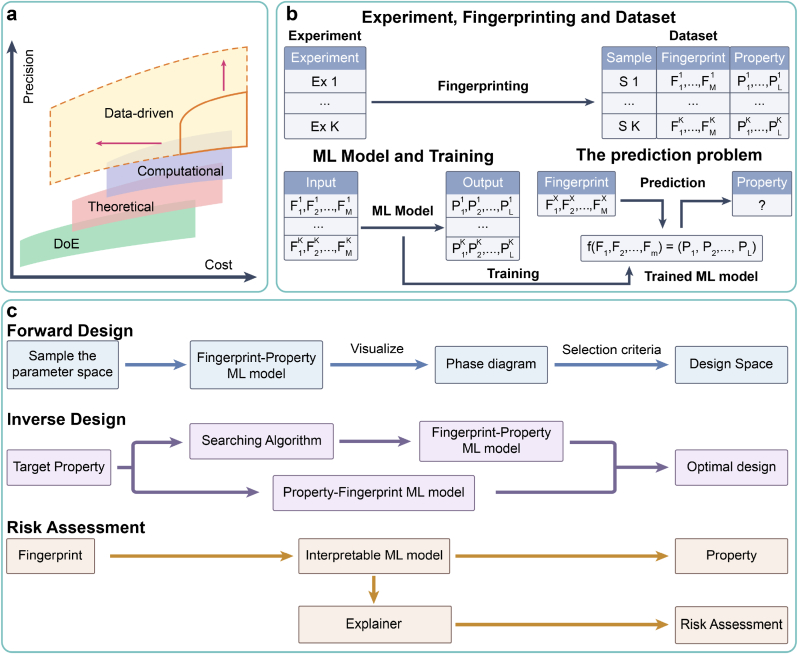
Fig. 3.
Data-driven design. (a) A “precision-cost” landscape of four modeling paradigms. (b) A typical workflow of ML-based data-driven paradigm. (c) Three main ML tasks within data-driven design.
Presently, four modeling paradigms have emerged, including the design-of-experiment (DoE), theoretical, computational, and data-driven paradigms (Fig. 3a) [26,56]. Although widely applicable, the DoE paradigm requires a substantial number of manual experiments to traverse the parameter space [57], resulting in labor-intensive processes. Additionally, the conventional response surface methodology for DoE has a limited ability to model complex relationships. To augment precision and mitigate the necessity for manual experiments, theoretical and computational paradigms have been developed. Both paradigms construct mathematical models based on domain knowledge (such as physics and biology) to expound process mechanisms, offering a “white box” effect [56]. The disparity lies in the approach. The theoretical paradigm entails the manual construction of theoretical formulas, providing prediction with faster speed but lower precision. In contrast, the computational paradigm relies on numerical simulations, such as finite element analysis (FEA) and computational fluid dynamics (CFD). This paradigm necessitates substantial computational resources, providing prediction with higher precision but slower speed. With advancements in computational precision, numerical simulations are progressively superseding manual experiments.
The aforementioned three paradigms have achieved certain advancements in the field of 3D bioprinting [[58], [59], [60], [61], [62]]. However, as 3D bioprinting advances towards clinical translation, especially in constructing substitutes of natural tissues/organs, the demand for higher modeling precision continues to increase. We identify three key dimensions that increasingly highlight the inherent complexity of 3D bioprinting:
-
(ⅰ)
Multi-domain fusion: 3D bioprinting necessitates the integration of knowledge spanning diverse domains, including biology, machinery, materials, and medicine. Constructing mathematical models in the theoretical and computational paradigms has proven challenging due to this multi-disciplinary nature.
-
(ⅱ)
Multi-scale coexistence: Within the realm of 3D bioprinting, factors such as CQA, CMA, and CPP operate across multiple scales. These scales encompass nano scale (such as molecular fragments of bioink materials), micro scale (such as microstructures of printed models), and macro scale (such as mechanical properties of BPPs). The theoretical and computational paradigms encounter difficulties in addressing these multi-scale modeling problems due to a dearth of constitutive models and the burden of excessive computational loads [63].
-
(ⅲ)
Multi-property coupling: For certain design objects within 3D bioprinting, such as bioink materials and microstructures of printed models, conflicting property requirements arise for design parameters. Examples include printability versus biocompatibility necessitating considerations of viscosity [40] and stiffness versus transport properties necessitating considerations of porosity [64]. Furthermore, certain CMA/CPP exhibit coupling, such as extrusion speed versus printing speed. These complexities yield a narrow feasible design space and impose stringent requirements on modeling precision.
Given the complexity of these challenges and the cost constraints, traditional paradigms face bottlenecks and are transitioning toward the data-driven paradigm based on machine learning (ML).
The ML-based data-driven paradigm typically employs supervised learning approaches, which can be succinctly defined as constructing a generalizable mapping model between input fingerprints and output properties (Fig. 3b). It primarily includes three key steps [57]:
-
(ⅰ)
Fingerprinting: The fingerprints (serving as CMA/CPP) and properties (serving as CQA) of samples are digitally represented, and a structured dataset is constructed. Fingerprinting typically requires domain knowledge and can be conducted manually or automatically (see Section 2.2.1 for details of automatic fingerprinting). Depending on the research objectives, fingerprints can be defined at various scales. Generally, smaller-scale fingerprints entail higher costs for the construction of datasets and ML models, but provide deeper insights, and vice versa.
-
(ⅱ)
Training: The mapping model between input fingerprints and output properties is established, predominantly through supervised learning methods such as support vector machines (SVMs), random forests (RFs), k-nearest neighbors (KNNs), and artificial neural networks (ANNs).
-
(ⅲ)
Prediction: Following training, the ML model can output the corresponding predicted property for any input fingerprints.
In the context of personalized design within QbD, we summarize three primary tasks for machine learning (Fig. 3c):
-
(ⅰ)
Forward design: In scenarios where the parameter space is low-dimensional, a forward design approach is effective, where the candidate CMA/CPP serve as inputs and the predicted CQA serve as outputs. Following training, forward design (or fingerprint-property) ML models using fingerprints as inputs and properties as outputs, can predict the property distribution of the design parameter space through traversal, generating visual representations such as process phase diagrams (or windows) [65]. Through visualization, suitable design space meeting property requirements can be determined.
-
(ⅱ)
Inverse design [57]: Conversely, in scenarios where the parameter space is high-dimensional, an inverse design approach is preferable where the expected CQA serve as inputs and the recommended CMA/CPP serve as outputs. To address the challenge of multi-property coupling in 3D bioprinting, Pareto optimal combinations of design parameters can be identified using multi-objective optimization techniques [66]. To this end, two solutions are proposed: ⅰ) forward-design models are initially trained, followed by the utilization of heuristic intelligent algorithms such as genetic algorithms to search for the optimal design parameters; ⅱ) inverse design (or property-fingerprint) ML models using properties as inputs and fingerprints as outputs, are directly designed and trained, such as generative ML models based on AEs [67,68] and generative adversarial networks (GANs) [69].
-
(ⅲ)
Risk assessment: Unlike the “black-box” modeling of traditional machine learning approaches, interpretable machine learning approaches offer a “white-box” effect. Specifically, following training, interpretable ML models use explainers to quantify the impact of each fingerprint on the property, as well as the interactions between fingerprints [70,71]. This interpretability facilitates the risk assessment of CMA/CPP on CQA, enabling a deeper analysis of process mechanisms, such as the printing process and function regulation [72].
2.2.3. In-line process control
To ensure the effectiveness and economy of BPPs for clinical application, two primary considerations govern the production process [6]. The first is quality, requiring the production process to consistently meet regulatory requirements, to ensure safety and effectiveness. The second is scalability, requiring an easily scalable production process to enable large-scale production at an affordable cost.
In 3D bioprinting, continuous production is involved in two key UOs: control of culture processes and control of printing processes. Due to interference factors such as the process drift and model error, the CQA may deviate from expectations in the actual production process, if the optimal CMA/CPP derived from off-line design is continuously adopted. This error probability is particularly heightened for organ-scale BPPs, due to the large number of required cells and the extended printing cycle. Additionally, long-term, low-latency, high-precision monitoring and calibration of the production process pose challenges for human intervention. Manual operations relying on experience, struggle to scale rapidly, posing difficulties and expenses in increasing production capacity.
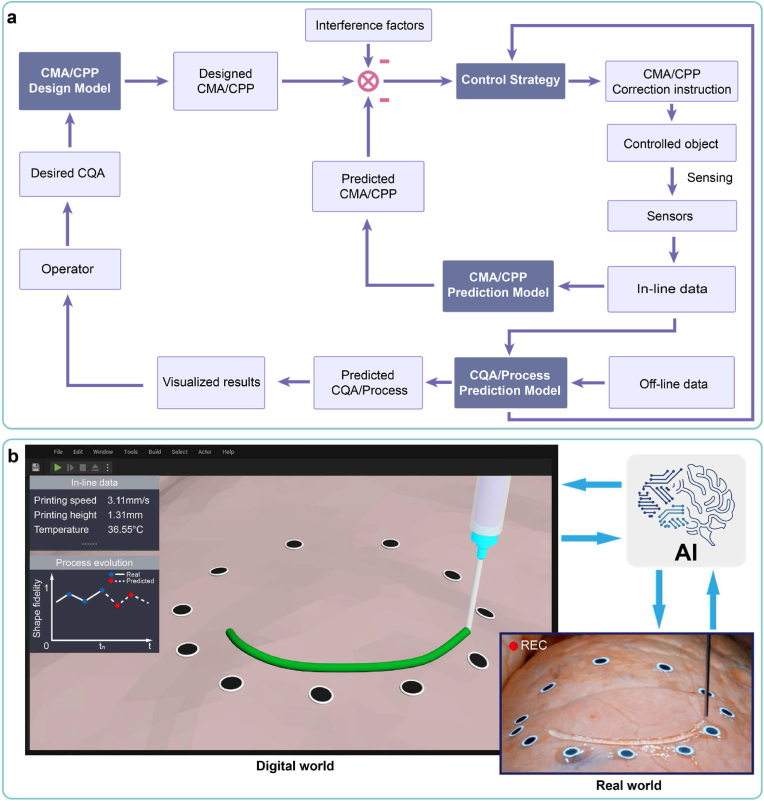
To address the above problems, based on methodologies in Section 2.2.1, 2.2.2, we propose a general AI-based in-line process control pipeline (Fig. 4a). To maintain the CQA at a high level, the CQA, CMA and CPP are monitored in situ by multiple sensors, and the CMA/CPP is corrected in-line according to the reasonable control strategy. We identify four primary categories of AI models involved in the outlined processes:
-
(ⅰ)
CMA/CPP design model: Upon the operator inputting the desired CQA, the AI model outputs the optimal CMA/CPP setting [73].
-
(ⅱ)
CMA/CPP prediction model: Various sensors capture in-line sensor data throughout the production process, based on which the AI model assesses the matching degree of current CMA/CPP (such as whether extrusion speed is too fast or slow) that will be transmitted to the control strategy [74].
-
(ⅲ)
CQA/Process prediction model: Utilizing the in-line data (such as images and numerical data) and off-line data (or CMA/CPP), the AI model predicts CQA or process evolution, which will be transmitted to the control strategy. Following visualizing, the predicted CQA facilitate defect detection and quality monitoring, supporting for the operator’ decision-making. The predicted process evolution offers the operator early warning of errors and a deeper understanding of process mechanisms.
-
(ⅳ)
Control Strategy: The control strategy leverages input information to issue CMA/CPP correction commands, achieving closed-loop correction of the production process. Traditional control strategies rely on rule-based human experience, lacking the ability to learn, which can only address specific and static scenarios [74]. By contrast, the reinforcement learning-based control strategy can learn from interactions with the environment to adapt to complex and dynamic scenarios [75]. The reinforcement learning-based control strategy sets the reward value based on CQA, and takes information such as the matching degree of CMA/CPP and predicted CQA as inputs (or environment state) as well as the correction commands of CMA/CPP as outputs (or action). The reinforcement learning model is trained according to the updated environment state, and will be adopted as the control strategy when the reward value reaches highest. The main algorithms of reinforcement learning include deep q-network (DQN), proximal policy optimization (PPO), and deterministic policy gradient (DPG).
Fig. 4.
In-line process control. (a) An AI-based in-line process control pipeline, containing four categories of AI models. (b) An illustration of in-line digital twin models for 3D bioprinting, linked to the real production process through monitored data and control commands. Copyright 2020, AAAS.
In the field of industrial production, a rapidly emerging research focus is digital twins, which refers to a virtual replica of a physical product or process [6]. This trend has also extended to the field of 3D bioprinting, where digital twin-driven 3D bioprinting is becoming a promising direction [76]. By leveraging the AI models described above, we can establish digital twins for the 3D bioprinting process. Leveraging the aforementioned AI models, the process principles of 3D bioprinting in the digital world of computers can be established, to simulate 3D bioprinting in the real world. Through this approach, we can construct a digital twin of 3D bioprinting, which can operate off-line in the digital world and enable in-line operation through real-time data exchange between the real and digital worlds.
In the design stage, off-line digital twin models enable the rapid execution of numerous virtual experiments in the digital world. Consequently, the design and optimization of CMA/CPP can be accomplished with fewer real experiments, thereby mitigating costs and risks. In the production phase, in-line digital twin models are linked with the real production process through monitored data and control commands, aiming to enhance production efficiency and quality (Fig. 4b). By simulating the process evolution and predicting its outcomes in the digital world, a comprehensive understanding is fostered, facilitating continuous process improvement.
3. AI-driven approaches for bioink formulation
As the primary element of 3D bioprinting, bioinks serve as a crucial foundation for ensuring the immune, tissue, and function specificity of BPPs. Typically, bioinks contain cells and biomaterials. For cells, the preparation process typically involves: first characterizing cells derived from the patient or shared cell bank and screening suitable ones, as described in Section 3.1; then performing differentiation/proliferation to obtain high-quality cells, as described in Sections 3.2, 3.3. For bioink materials, the formulations are designed aimed at specific properties, as described in Section 3.4. AI technology can be applied to each UO in these processes to accelerate the design and production of personalized bioinks for BPPs (Fig. 5a).
Fig. 5.
AI-driven approaches for bioink formulation. (a) A pipeline of personalized design of bioinks. (b) Experimental results of virtual staining for salivary gland tissue based on adversarial learning. Copyright 2021, Nature Publishing Group. (c) A workflow of real time monitoring and regulation of PSCs' differentiation process, using multiple AI algorithms. Copyright 2023, Nature Publishing Group. (d) Prediction results of “digital rheometer twins” on rheology of hydrogels. Copyright 2022, PNAS. (e) Risk assessment and mechanism analysis of bioink materials using interpretable ML models. Copyright 2022, Wiley. (f) A general workflow for the design of self-assembling peptide using HydrogelFinder-GPT. Copyright 2024, Wiley.
3.1. Characterization of sampled cells
Considering the diverse applications of BPPs, selecting appropriate cell sources is a critical factor. Given the inherent immuno-specificity of BPPs, autologous cells derived from the patient are the ideal option. This approach is feasible when fewer cells are required, such as for disease modeling and drug screening. However, for constructing organ-scale in vivo implants, the need for a large number of cells makes this approach less viable. With the continuous advancements in stem cell technology, the issue of immune rejection is being addressed [9]. Several countries and regions have established phenotype-specific stem cell banks [77,78], enabling the rapid provision of large quantities of appropriate cells for patients with varying phenotypes. Allogeneic stem cells obtained through these methods could serve as a new source of cells for constructing organ-scale in vivo implants. Upon selecting the appropriate cell sources, rigorous characterization and screening procedures should be conducted to ensure cell viability and compliance with differentiation and proliferation requirements.
However, conventional destructive characterization methods, such as tissue section preparation and staining, pose numerous challenges in the scaling-up production of cells. Characterized cells often face difficulty undergoing subsequent differentiation, proliferation and other characterizations, which leads to wasting the limited amount of sampled cells and affecting the production efficiency of cells. Furthermore, the extended characterization cycle, spanning from several days to weeks, significantly delays clinical treatment and poses challenges for real-time monitoring of the production process. Additionally, the high cost of specialized equipment and reagents utilized in the characterization process, coupled with labor-intensive and time-consuming manual operations, exacerbates these challenges.
AI-based virtual staining technology offers a solution, enabling non-destructive and rapid characterization of sampled cells [52]. This technology has found application across various organs, such as the liver [[79], [80], [81]], kidney [[82], [83], [84]], stomach [85], and lung [86]. Two types of tasks can be implemented by supervised learning (using paired images for training) or unsupervised learning methods (using unpaired images for training): (ⅰ) generating stained images from the raw images of unstained samples, thus obviating cell-consuming staining procedures [[87], [88], [89]], and (ⅱ) generating diverse and complex staining images from basic staining images, facilitating the characterization of multiple properties through single staining processes [90,91]. For instance, Philip O. Scumpia and Aydogan Ozcan's groups [92] have integrated the aforementioned virtual staining techniques to facilitate the rapid and precise acquisition of multi-modal virtual histology of skin through Al models based on adversarial learning (Fig. 5b). These methods mitigate cell loss, expedite the characterization process, and exhibit significant potential for application in 3D bioprinting.
However, considering the deployment of virtual staining technologies in 3D bioprinting, there are still significant limitations in dataset construction and model evaluation. Regarding datasets, current datasets for virtual staining are primarily focused on pathological/tissue sections, which are insufficient to meet the tailored requirements of 3D bioprinting. For example, the absence of staining data for stem cells and the inability of sectioning methods to provide non-destructive characterization of cell states present significant challenges. Regarding model evaluation, existing evaluation metrics to verify the effectiveness of AI models for virtual staining are mostly based on custom loss functions [92], lacking standardization and generalizability. Considering the safety and regulatory requirements for clinical applications, there is an urgent need to establish a standardized and comprehensive evaluation system to quantitatively assess model performance.
3.2. Design of culture conditions
The inter-patient variation of autologous cells significantly surpasses the batch-to-batch variation observed in mature cell lines for laboratory use. Consequently, ensuring cell quality (serving as CQA) necessitates the personalized design of patient-specific media (serving as CMA) and culture process parameters (serving as CPP) [14,93]. Given the intricate composition of the medium, such as carbon sources, amino acids, vitamins and growth factors, which leads to the expansive parameter space, the DoE paradigm encounters challenges [94], whereas the ML-based data-driven paradigm offers substantial advantages [95]. Currently, studies have utilized ML methods to model mapping relationships between media composition [96]/culture process parameters [97] (such as the temperature and duration) and cell quality (such as viability, cell density, and metabolites), accelerating the design of culture conditions. For instance, Dong-Yup Lee's group has utilized the principal component analysis (PCA) algorithm to screen and optimize the culture medium components for Chinese hamster ovary cells, resulting in a 30–40 % improvement in viable cell density during the early growth phase [98].
However, the aforementioned methods necessitate the construction of independent datasets corresponding to specific patients, which proves impractical for the clinical application of 3D bioprinting. The limited quantity of cells sampled from patients hinders high-throughput dataset generation. Meanwhile, since the time-intensive nature of individual culture experiments (usually taking several days), it's also unfeasible to construct datasets in a low-throughput manner, failing to meet clinical urgency. To address the aforementioned issue, a promising solution is to develop patient-universal ML models untied to specific patients, using electronic health records (EHRs) containing characteristics of the patients themselves as inputs. Trained by a large dataset of patients' EHRs, the ML model can quickly output personalized culture conditions when new patients' EHRs are input, eliminating the need for patient-specific cell experiments. This paradigm has already been widely applied in clinical diagnosis and treatment [99].
3.3. Control of culture processes
Due to factors such as process drift and model error, deviations in cell quality (serving as CQA) may arise during the actual culture process, if the optimal culture conditions (serving as CMA/CPP) derived from off-line design are continuously employed. Hence, implementing in situ monitoring and in-line correction becomes imperative for maintaining cell quality [14,42]. Deep learning-based label-free detection technology, in the form of CQA prediction models, serves to sense cell quality throughout the culture process, both in-line and non-destructively. This technology operates through two avenues: (ⅰ) morphological information, including cell types and cell status, which can be extracted through segmentation of cell images such as cell nucleus, single cells, and cell clusters [[100], [101], [102], [103], [104]]; and (ⅱ) non-morphological information, including the genomic, proteomic, and metabolic [[105], [106], [107], [108], [109], [110]], which can also be gleaned through cell images.
Moreover, the obtained in-line data is input into the CMA/CPP prediction model, offering a real-time assessment of the congruence between ongoing culture conditions and desired outcomes, which will be subsequently fed back to the control strategy for in-line correction. For instance, Yang Zhao's group [111] has employed AI algorithms to intervene in the differentiation process of pluripotent stem cells (PSCs) into cardiomyocytes (CMs). They optimized the initial state of PSC colonies and applied real-time, non-destructive characterization and regional purification of cells during the culture process. As a result, they significantly improved the efficiency of CM induction, increasing the successful differentiation rate from 63 % to 94.7 % (Fig. 5c).
Furthermore, inputting in-line data into a process prediction model can forecast the future status of cell culture, enabling proactive intervention to mitigate risks. Notably, Ming-Dar Tsai's group [112] has employed the RNN algorithm to predict the future status of cell colonies during the reprogramming process of human induced pluripotent stem cells (hiPSCs) based on time-lapse bright-field microscopy images.
3.4. Design of bioink materials
Owing to the tissue- and function-specific nature of the repaired part [113], personalized design for bioink materials is necessary to fulfill specific properties (or CQA) [[114], [115], [116], [117], [118]]. Typically, this entails significant domain expertise and extensive trial and error, which are both time-consuming and expensive. However, ML methods offer avenues for improvement in two key aspects: (ⅰ) reduction of time and cost associated with property characterization for high-throughput screening of bioink materials, and (ⅱ) modeling the intricate mapping relationship between the fingerprints and properties of bioink materials, thereby enabling the property prediction and expediting the design process.
ML-based property characterization of bioink materials: Rheological properties are paramount in characterizing bioink materials, yet traditional characterization methods relying on rheometers suffer from high cost and limited throughput. These challenges can be addressed through ML-based characterization methods. For instance, Min Zhang's group [119] has used characterization data from near-infrared (NIR) spectroscopy and low-field nuclear magnetic resonance (LF-NMR) as inputs of ML models, such as CNN, SVM, long short-term memory (LSTM), and Transformer, to predict the rheological characteristics of hydrogel inks. Blake N. Johnson's group [120] has devised a measurement approach utilizing robots and ML models, enabling high-throughput and cost-effective determination of gelation status. These approaches have offered promising solutions for rapid and high-throughput characterization of rheological properties. Additionally, Safa Jamali's group [54,121] and Gareth H. McKinley's group [122] have employed ML methods, specifically based on physics-informed neural networks (PINNs), to develop precise rheological constitutive models of hydrogels. Termed as “digital rheometer twins”, this approach can accurately predict the complete rheological behavior of hydrogels with minimal experiments (Fig. 5d), offering a potential alternative to physical rheometers and markedly reducing characterization-related time and costs. Furthermore, the rheological constitutive model potentially contributes to a deeper understanding of the rheological mechanism underlying bioink materials.
ML-based design of bioink materials: The properties of bioink materials encompass the form-based and function-based characteristics (Table 1). Bioink materials, represented by hydrogels, typically exhibit multi-scale structures [123]. Consequently, we attribute the construction of ML models to fingerprints at three scales:
-
(ⅰ)
Property-based scale: This approach focuses primarily on composition ratios and rheological properties. Adjusting composition ratios of bioink materials is straightforward for the operator and enables rapid modification of various properties. Studies have utilized ML models to predict a range of properties, including printability [65,124], rheological properties [[124], [125], [126]], gelation properties [[127], [128], [129]], mechanical response [130], degradation properties [131,132], swelling properties [[133], [134], [135], [136]], and cell behavior [126,137,138], with composition ratios of bioink materials serving as inputs. Additionally, as the rheological properties of bioink materials serve as predictors of printability [37], studies employing ML models have explored the relationship between rheological properties and printability [72,125], enhancing the understanding of the printing mechanism. For instance, Jürgen Groll's group [72] has utilized interpretable ML methods to analyze the effect of rheological fingerprints on printability and the interaction between these fingerprints, further elucidating printing behavior. This offers an excellent reference utilizing ML models for risk assessment and mechanism analysis in QbD of 3D bioprinting (Fig. 5e). Besides, the mechanical response has also been used as inputs to predict degradation properties [131] and cell behavior [138]. Furthermore, ML models have been employed to investigate the fabrication process of novel bioinks such as microgel particles [139].
In recent years, the emerging 4D bioprinting has opened an exciting avenue for engineering functional tissues and organs [140]. Through the application of specific external physicochemical stimuli, such as the temperature, pH, ion concentration, electric field, and magnetic field, these 4D BPPs can undergo controlled shape morphing, facilitating the attainment of specific biological functions. This requires bioink materials to exhibit stimuli-responsive swelling properties. In this context, there have been studies using ML models to build the mapping relationship between external physicochemical stimuli (such as the pH, temperature [141], time [142], and external force constraints [143]) and swelling properties of hydrogels (such as the swelling ratio and drug release ratio).
-
(ⅱ)
Structure-based scale: Analogous to the natural extracellular matrix (ECM) [144], bioink materials such as hydrogels possess microscale network structures. ML models have been utilized to predict various properties, including mechanical response [[145], [146], [147]], degradation properties [131], and swelling properties [148], with microstructures of bioink materials serving as inputs. For instance, Linxia Gu's group [146] has integrated finite element analysis with CNN models, employing microstructural images as input to predict the mechanical properties of collagen-based biomaterials.
-
(ⅲ)
Molecule-based scale: Due to the challenges of conventional hydrogel-based bioinks in simultaneously meeting the requirements for both printability and biocompatibility, various reinforcement strategies have emerged to improve bioinks' properties [149]. For example, driven by supramolecular interactions, such as hydrogen bonding, hydrophobic interactions, and electrostatic interactions, peptides can self-assemble to form supramolecular hydrogels [150]. The diversity of molecular structures and the complexity of supramolecular interactions present significant challenges in designing self-assembling peptides. In this context, studies have emerged employing ML models to accelerate the discovery of self-assembling peptides [[150], [151], [152], [153]]. For instance, Junfeng Shi's group [150] has proposed the HydrogelFinder workflow, which utilizes molecular structures as inputs and gelation properties as outputs (Fig. 5f). This workflow has successfully identified nine novel self-assembling peptide hydrogels that had not been previously reported. To summarize, molecular-based modeling has demonstrated significant potential in mechanistically developing bioink materials with novel properties, particularly for supramolecular bioinks.
In contrast to the DoE paradigm, which relies on adjusting composition ratios, the ML-based data-driven paradigm can predict the macroscacle properties of bioink materials using micro-nanoscale fingerprints, such as network structures and molecular structures, achieving multi-scale modeling. Machine learning serves as a powerful tool for comprehending diverse behavioral mechanisms of bioink materials, including rheology, gelation, mechanical response, degradation, swelling, and cell behavior, as well as for designing bioink materials with specific or even potentially groundbreaking properties.
4. AI-driven approaches for model structure
Upon finalizing bioink formulations, another critical element is the design of the printed models' structure. Due to the tissue, structure, and function specificity of BPPs, structures of printed models necessitate personalized design to meet the property requirements. The typical process of designing printed models involves the following steps: first, acquiring medical images of the patient's target organs/tissues using imaging modalities such as CT and MRI, as described in Section 4.1; second, performing 3D modeling based on these medical images to generate the macrostructure model, as described in Section 4.2, 4.3; and finally, designing the internal microstructure, as described in Section 4.4. AI technology can be applied to each UO within these processes to expedite the precise design of printed models with personalized structures (Fig. 6a).
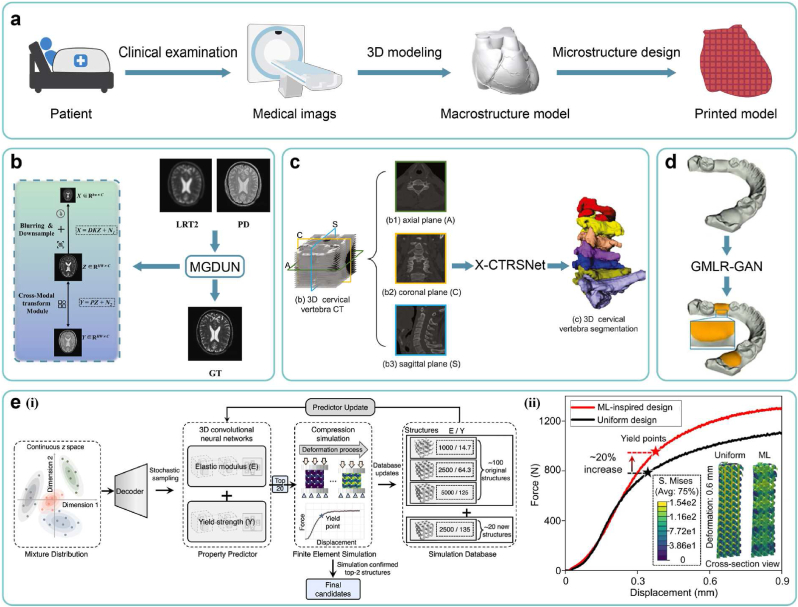
Fig. 6.
AI-driven approaches for model structure. (a) A pipeline of personalized design of printed models. (b) A schematic diagram of super resolution reconstruction based on multi-contrast MRI images. Copyright 2023, Elsevier. (c) A schematic diagram of organ segmentation and 3D reconstruction based on orthogonal CT images. Copyright 2022, Elsevier. (d) Experimental results of tooth gingival margin line reconstruction based on the adversarial learning method. Copyright 2022, Elsevier. (e) (ⅰ) A workflow of active learning loop for high-performance microstructure discovery with 3D-CNN, (ⅱ) Experimental displacement-force curves of the ML-inspired design versus uniform design. Copyright 2023, Nature Publishing Group.
For the precise design of in vivo implants, we summarize multi-scale fingerprints of printed models (serve as CMA/CPP) critical to achieve various properties (serve as CQA) (Table 2). For example, at the macro scale, the external shape of the implant (such as the bone implant) should match the anatomical shape of the defect, which can improve the cosmetic effect and structural support; at the micro scale, the radius and shape of the implant's pores affect the transport properties, mechanical properties, and cell behavior responses; at the nano scale, the nano-topography affects cell behavior responses [13]. AI models aim to sense and optimize these fingerprints to enhance the overall performance of printed models. It is worth mentioning that this chapter focuses on the design of structural fingerprints, where Sections 1, 2, 3, 4 describe the design of the external macrostructure, and Section 4.4 describes the design of the internal microstructure. Additionally, the bioinks used for printed models discussed in this chapter include not only bioinks with cells (such as hydrogels) but also biomaterial inks without cells (such as thermoplastic polymers) [154].
Table 2.
Multi-scale fingerprints of printed models.
| Scale | Examples of fingerprints | Affected properties |
|---|---|---|
| Macro | Macro morphology, physicochemical properties of bioinks, external physicochemical stimuli | Transport, mechanical, biological |
| Micro | Fiber: orientation, size | Transport, mechanical, biological |
| Pores: shape, size, distribution | ||
| Nano | Surface pattern | Biological |
4.1. Acquisition of high-resolution medical images
To acquire the macroscopic morphology of damaged organs/tissues in patients, medical imaging techniques such as CT, MRI, PET, and ultrasound (US) are indispensable. High-resolution medical images serve as the foundation for creating precise printed models. However, the resolution of conventional medical images, typically at the mm level, falls short of that of most 3D bioprinters, typically operating at the um level. Consequently, this limitation hinders 3D bioprinters from fully realizing their manufacturing potential. Moreover, factors such as patients' motion during the imaging process can lead to artifacts in certain areas of images, significantly diminishing the clarity. Despite ongoing advancements in higher-resolution medical imaging equipment [155], widespread clinical application remains challenging due to factors such as bulkiness, cost, and radiation exposure.
Super-resolution technology [156], employing the deep learning methodology, offers remedies to these challenges [45]. By utilizing algorithms such as CNNs [157], convolutional recurrent neural networks (CRNNs) [158], variational networks [159], and attention mechanisms, this technology generates high-resolution images (HRIs) from low-resolution images (LRIs). It has found application across various medical imaging modalities, such as CT [[160], [161], [162]], MRI [[163], [164], [165]], and PET [166] in diverse organs, such as the brain, liver, lung, and abdomen. However, single image super-resolution (SISR) suffers from limited vertical resolution due to the absence of inter-layer information. Additionally, single imaging modalities lack the ability to adequately sense organs with complex tissue distributions, thus compromising resolution. To address these shortcomings, super-resolution technologies utilizing multiple medical images as inputs have emerged. These approaches can enhance the resolution beyond that achievable by SISR alone. Depending on the forms of input images, we categorize these methods into two main categories:
-
(ⅰ)
Based on volumetric images: Leveraging medical volumetric images allows for the consideration of hidden spatial relationships between image layers. Studies have employed AI algorithms such as 3D convolutional neural networks (3D-CNNs) and GANs to perform super-resolution on brain MR images [167] and abdominal CT images [168].
-
(ⅱ)
Based on multi-modal images: In the field of MRI, multi-contrast super-resolution (MCSR) technology amalgamates information from multi-contrast images to produce high-resolution images with improved tissue contrast and reduced noise. This approach has been applied to enhance the resolution of MR images of tissues such as the brain and knee (Fig. 6b) [169].
4.2. 3D modeling of target organs/tissues
Upon acquiring high-resolution medical images, the subsequent task involves identifying and segmenting the region of interest pertaining to the target organs/tissues within the images, followed by constructing a 3D model. AI-based methods for image segmentation and 3D reconstruction offer advantages over manual operations, mitigating subjective errors while providing rapidity and repeatability. Studies have demonstrated the efficacy of employing AI algorithms, such as 3D-CNNs, to segment and reconstruct serial medical images (such as CT and MR images) depicting various organs (such as the abdomen [170,171], liver [172], kidney [173], chest [174], and head [175]) and tissues (such as the vasculature [176], muscles [177], and tumors [[178], [179], [180]]).
However, single-perspective and single-modal imaging methodologies present several limitations. To enhance the precision of segmentation and 3D reconstruction, we summarize two main approaches commonly utilized:
-
(ⅰ)
Based on multi-perspective images: AI algorithms can segment medical images from diverse perspectives, such as orthogonal CT and X-ray images. By voting or weighting the segmentation results, this approach can effectively reveal morphological features of target organs/tissues from multiple perspectives, which has been applied to various targets, including spines (Fig. 6c) [181] and liver tumors [182].
-
(ⅱ)
Based on multi-modal images: Leveraging multi-contrast MR images (such as T1, T1ce, and T2) or cross-sensor images (such as CT, PET, and MRI) and employing MML methods allows for harnessing the strengths of various image types. Studies have successfully achieved segmentation and 3D reconstruction of organs/tissues with the above methods, such as the pancreas [183], breast tumors [184], and brain tumors [185].
Last but not least, existing studies mainly focus on specific performance metrics of AI models (such as ROC), with little attention paid to the regulatory and security aspects of models. In fact, the reproducibility of AI model training results is often poor due to variability stemming from multiple factors, including datasets, optimization processes, hyperparameter choices, model architecture, and hardware configurations. In the context of clinical deployment, the lack of transparency in the training process and the poor reproducibility of results present significant challenges for the regulation of AI models. To address these issues, the first step is to provide a standardized and detailed description of the model design and training process. Then, a robust evaluation system for model performance should be established. Finally, technical measures should be adopted to minimize variability from multiple sources [186].
4.3. Generation of implant models
Upon acquiring the 3D model of the damaged organ/tissue, the design of personalized implants necessitates consideration of the macrostructure to align with the site of damage. In the realm of cranial and tooth restoration, conventional virtual design methods, including mirroring technology, statistical shape models, and deformable templates, are operation-intricated and time-intensive, limited in the application of specific defect types [187]. In light of this, generative AI technology has emerged as a transformative solution for automating the generation of implant macrostructures based on any provided 3D model of damaged organs/tissues. This advancement supplants manual operations, significantly enhancing universality, rapidity, and reproducibility. In cranial restoration, researchers have applied AI algorithms such as GANs [188,189] and AEs [190] to fabricate personalized implants. Similarly, in tooth restoration, investigations have leveraged AI algorithms such as 3D-CNNs [191] and GANs [[192], [193], [194]] to engineer crucial structures such as the occlusal surface and gingival edge of compromised teeth (Fig. 6d).
4.4. Design of microstructures
Following the aforementioned steps, the macro external shape (or macrostructure) of the printed model is obtained, necessitating personalized fine design of its internal microstructure to fulfill property requirements [[195], [196], [197]]. Given the prohibitive cost of clinical trials, the DoE paradigm is not viable. Instead, the computational paradigm can simulate the transport and mechanical properties of the microstructure through numerical simulation methods such as CFD and FEA. However, the high complexity of the microscacle topological structure results in a high-dimensional parameter space, demanding substantial computational resources. By incorporating the ML-based data-driven paradigm, datasets can be constructed from simulation data. The trained ML model can replace numerical calculations, significantly reducing the computational burden.
In the realm of metamaterials, leveraging the two approaches of inverse design outlined in Section 2.2.2, studies have combined machine learning and numerical simulation to inversely design microstructures, further details can be found in other comprehensive reviews [198,199]. Given their multi-scale structures akin to human natural organs/tissues (such as the bone, cartilage, and skin) [[200], [201], [202]], these theories and methods are emerging in the field of 3D bioprinting. Some studies first adopt ML approaches based on ANNs [[203], [204], [205], [206]] or CNNs [68,69,207,208] to establish forward design models, where the design parameters of microstructures (such as geometric parameters of unit cells) serve as inputs, and the resulting mechanical properties (such as the elastic modulus, stiffness, and yield strength) serve as outputs. These forward design models can replace finite element analysis and real experiments, enabling rapid prediction of candidate designs' feasibility. Subsequently, searching strategies (such as particle swarm optimization algorithms [204], genetic algorithms [203], Bayesian optimization [208], and active learning methods [68]) are integrated with the established forward design models to determine the optimal microstructure design with desired mechanical properties. For instance, Peng Wen's group [68] has employed 3D-CNN models as forward design models, with active learning as the search strategy, to design topologies of printed scaffolds' microstructures (Fig. 6e ⅰ). In animal experiments, this approach has ultimately achieved fixed elastic modulus and improved yield strength by 20 % compared to those through uniform design (Fig. 6e ⅱ). Moreover, other studies utilize ML approaches based on AEs [69] and ANNs [205,206] to establish inverse design models, where expected mechanical properties serve as inputs, and the optimal microstructure design serves as outputs.
Notably, the transport and mechanical properties impose conflicting requirements on the microstructure (such as the porosity for stiffness and transport properties), thereby machine learning integrated with multi-objective optimization is necessary for the optimal design of microstructures [204]. In addition, ML models predicting properties rapidly facilitate the discovery of microstructures with rare or extreme properties, which is difficult for traditional paradigms [209,210].
Additionally, the emergence of 4D printing has introduced significant challenges in microstructure design. In 4D bioprinting, the heterogeneous spatial distribution of stimuli-responsive bioinks dictates the unique shape-morphing behaviors of BPPs in response to external stimuli [211]. Given the complexity of these spatiotemporal dynamics, the conventional DoE paradigm faces significant limitations in efficiently and precisely designing the spatial distribution of bioinks to achieve the desired shape-morphing behaviors. Recently, in the field of 4D printing of active composites, studies [[211], [212], [213]] have employed ML methods to design the spatial distribution of ink materials. This offers valuable insights for the application of ML approaches in the 4D bioprinting field.
Last but not least, cell behavior is another critical consideration in microstructure design. Recent studies [214,215] have employed ML approaches to model the mapping relationships between microstructures (such as the fiber diameters/orientations and the pore size distributions) and cell behaviors (such as the cell numbers and cell morphologies). However, the datasets in these studies are derived from post-bioprinting experimental results, which are costly and time-consuming. Concerning this, a promising development direction is multi-scale modeling integrated with spatiotemporal models of cell behavior [216]. Ideally, in the R&D stage, it is necessary to fully predict the properties of the printed model at both the spatial scale (organ, tissue, and cell) and the temporal scale (dynamic evolution over time) for systematic optimization design. However, currently, the design of printed models mainly focuses on the non-biological properties at the organ/tissue scale (such as the transport and mechanical properties), with less attention paid to biological properties at the cell scale. By combining spatiotemporal models of cell behavior and machine learning, it is expected to model the relationship between the microenvironment and the dynamic response of cell behavior. Furthermore, by integrating them into numerical simulations, the multi-scale properties of printed models, including transport properties, mechanical properties and the evolution of cell behavior, can be fully predicted. In this regard, there have been preliminary attempts in relevant studies [217,218].
5. AI-driven approaches for printing process
Upon acquiring the printed model, the printing process element entails the precise manufacture of the designed multi-scale structure while ensuring cell viability. To enhance printing quality, the optimal printing parameters should be designed off-line before the formal printing process, as described in Section 5.1. Subsequently, during the formal printing process, the printing parameters need to be adjusted in real time to maintain control over printing quality, as described in Section 5.2.
5.1. Design of printing parameters
According to the QbD, the bioink parameters (serving as CMA) and printing parameters (serving as CPP) collectively determine the printing quality (serving as CQA), primarily focusing on printability and cell viability. Traditional paradigms have encountered the dilemma of “precision-cost”. In contrast, the ML-based data-driven paradigm can model mapping relationships between CMA/CPP and CQA at a manageable cost, facilitating rapid determination of the optimal design space to enhance printing quality.
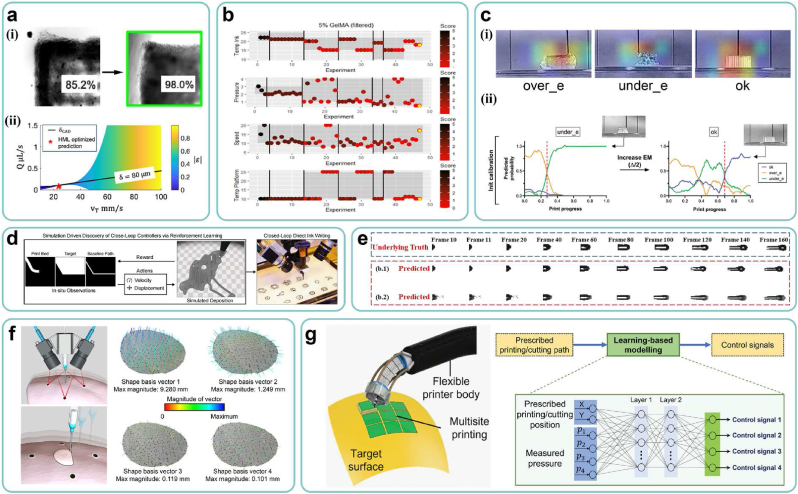
Within the framework of Section 2.2.2, Table 3 provides a summary of examples in which AI technology has been employed for process modeling and parameter optimization across various 3D bioprinting processes. Different types of processes emphasize various aspects of CQA, CMA, and CPP, as documented in prior research [[219], [220], [221], [222], [223]]. For instance, in photocuring printing, digital masks serve as generalized CPP. Shaochen Chen's group [224,225] has employed deep learning methods to optimize the design of digital masks, mitigating the impact of cell scattering on printability. The trained ML models can be used to determine the design space and optimal combinations of printing parameters. For instance, Newell R. Washburn's group [226] has employed the hierarchical machine learning approach to optimize printing parameters, improving printability from 85.2 % to 98 % (Fig. 7a ⅰ). Furthermore, through the trained ML model, phase diagrams of the design space for printing parameters are generated, visually illustrating the distribution of printability (Fig. 7a ⅱ). In addition, Bayesian methods have been utilized as active learning strategies to optimize printing parameters [[227], [228], [229], [230]]. For instance, Gordon Wallace's group [228] has employed Bayesian optimization to guide experimenters in adjusting printing parameters. As the number of experimental iterations increases, printability continuously improves until the optimal combination of printing parameters is found (Fig. 7b).
Table 3.
Examples of AI applications for process modeling and parameter optimization in 3D bioprinting.
| Process category | CMA | CPP | CQA | AI Model | Ref |
|---|---|---|---|---|---|
| EBB | Cink | Q, VT, Dnozzle | Shape fidelity | Hierarchical machine learning (HML) | [226] |
| , | Printing resolution | Rheology-informed hierarchical machine learning (RIHML) | [240] | ||
| GelMA composition | Ink reservoir temperature, pressure, speed, platform temperature | Filament morphology, layer stacking | Bayesian optimization | [228] | |
| – | Air pressure, biomaterial ink temperature, print speed | Print resolution | Bayesian optimization | [227] | |
| FSA concentration | Nozzle size, printing temperature, pneumatic pressure | Printability | Gaussian process regression (GPR) | [241] | |
| – | Rb, Rs, Lu, Ll, Rm (nozzle geometrical parameters) | Maximum shear stress | Gaussian process (GP) | [242] | |
| Material composition | Printing speed, printing pressure, scaffold layer, programmed fiber spacing | Printing quality | RF | [243] | |
| Biomaterial concentration | Nozzle temperature, printing path height | Printability | SVM | [244] | |
| Material concentration, solvent usage | Crosslinking mechanism and duration, printer settings, observation duration | Cell viability, filament diameter, extrusion pressure | Support vector regression (SVR), linear regression (LR), random forest regression (RFR), RF, logistic regression classification, SVM | [245] | |
| Gelatin concentration | Printing speed, flow rate, temperature | Printability, Precision | Fuzzy inference system (FIS) | [246] | |
| Dilution percentage of bioink | Nozzle pressure, printing speed, | Line width | Fuzzy inference system (FIS) | [247] | |
| Viscosity, growth factor concentration | Gauge pressure, build orientation, printing speeds | Print resolution | LSTM | [248] | |
| – | Printing speed, pressure of extrusion, infill percentage | Gel weight, surface area, topographical heterogeneity | SVM, Gaussian model | [249] | |
| Biomaterial type, biomaterial concentration, crosslinker concentration, cell type, cell number | Crosslinking time, printing pressure, movement speed, nozzle size, cartridge temperature, bed temperature | Cell viability | Bayesian optimization, ANN | [229] | |
| Material's weight fraction | Extrusion pressure, print speed, z-height | Filament width | Linear regression | [250] | |
| – | Nozzle temperature, infill density, layer height, printing speed | Tensile strength | Linear regression, RFR, XGB regressor, LGBM regressor, ANN | [251] | |
| – | Air pressure, biomaterial ink temperature, print speed | The width of printed filament | Bayesian optimization | [227] | |
| Material's weight fraction | Extrusion pressure, print speed, nozzle diameter, z-height | Filament width | Linear regression | [250] | |
| – | Layer height, nozzle travel speed, and dispensing pressure | Time, porosity, and geometry precisions | Multi-objective Bayesian Optimization | [252] | |
| – | Printing speed, extrusion pressure | Width average, width variance height average and height variance | SVM | [253] | |
| – | Nozzle tip to collector distance | Jet radius profile | GP | [230] | |
| – | Ratio of the collector speed over the jet speed at the point of interest | Lag distance | |||
| Cell type | Wall shear stress, exposure time | Cell viability | Multi-layer Perceptron (MLP) | [254] | |
| DBB | Viscosity, surface tension | Voltage, diameter of the nozzle | Droplet formation | MLP | [255] |
| Viscosity, surface tension | Voltage, nozzle diameter | Droplet deformation | Fully connected neural network (FCNN) | [256] | |
| Polymer concentration | Voltage, dwell time, rise time | Droplet velocity and volume | Ensemble learning | [257] | |
| – | Standoff height, applied voltage, ink flow rate | Droplet diameter | Regression analysis (RA), backpropagation neural network (BPNN), neural network trained with genetic algorithm (GA-NN) | [258] | |
| The type and concentration of solute and solvent | Inner diameter (Din), outer diameter (Dout), the materials of the nozzle and grounded substrate, volumetric flow rate (Q), the distance (L) between needle and grounded substrate, the environmental gas, the applied voltage (V) between the ground electrode and needle | Spraying patterns | ANN,SVM | [259] | |
| Viscosity (μ), Density (ρ), Conductivity (K), Surface tension (γ), Relative permittivity (κ) | Nozzle internal diameter (Din), nozzle external diameter (Dout), distance between nozzle and grounding electrode (L), applied voltage (V), flow rate (Q) | Droplet diameter | ANN | [260] | |
| Dimensionless number Z | Rise time, drive'voltage, dwell time, fall time | Drop velocity, drop formation | SVM, KNN, RFs, extreme gradient boosting (XGBoost), MLP | [261] | |
| Bioink viscosity, cell concentration | Nozzle size, printing time, printing pressure | Droplet size | DT, RF, PageRank, MLP, LSTM | [262] | |
| LBB | – | Digital mask | Printing fidelity | U-Net-like neural network | [224,225] |
| – | Digital mask | Printing fidelity | 3D U-Net | [263] | |
| – | Digital mask | Printing fidelity | Convolutional Auto-Encoder (CAE) | [264] | |
| – | Digital mask | Printing fidelity | Deep neural networks | [265] | |
| GelMA concentration | UV intensity, UV exposure time, layer thickness | Cell viability | Ensemble learning model | [266] | |
| – | Exposure time, light intensity, print speed, laser current, laser power, infill density | Young's modulus | ANN | [232] | |
| Resin viscosity | Cross section size used for synthetic dataset construction, manufacturing velocity, PDMS thickness, constrained surface type, duration of frame, video projection time, groove width, groove depth, cross section size used for separation force boundary construction | Printing success or failure, optimum printing speed | KNN, SVM, decision tree, logistic regression, quadratic discriminant analysis, GP, naiveBayes, ANN, ensemble learning model, Siamese network |
[267] |
Fig. 7.
AI-driven approaches for printing process. (a) (ⅰ) Experimental results of printability improvement through machine learning optimization, (ⅱ) Phase diagrams of the design space for printing parameters. Adapted with permission from J.M. Bone, C.M. Childs, A. Menon, B. Póczos, A.W. Feinberg, P.R. LeDuc, N.R. Washburn, Hierarchical Machine Learning for High-Fidelity 3D Printed Biopolymers, ACS BIOMATER SCI ENG, 6 (2020) 7021–7031. Copyright 2020, American Chemical Society. (b) Optimization process of printing parameters via Bayesian methods. Copyright 2021, Elsevier. (c) (ⅰ) Prediction results of CNN models on the printing status. (ⅱ) Experimental results on automatic optimization of printing parameters. Copyright 2022, AccScience Publishing. (d) The schematic diagram of a closed-loop control strategy via reinforcement learning. Copyright 2022, ACM. (e) Prediction results on the evolution process of inkjet printing. Copyright 2023, Elsevier. (f) Sensing results of surface deformation using PCA algorithms. Copyright 2020, AAAS. (g) A schematic diagram of the printing head's motion controllers, built by ANN models. Copyright 2023, Wiley.
However, most existing studies are applicable only to single and straightforward working scenarios. As the construction of natural heterogeneous tissues necessitates increasingly complex bioinks and printed models, it is crucial for ML models to address such complexities. We identify two example aspects of these complexities:
-
(ⅰ)
Multiple components: Multi-component bioinks have introduced tremendous diversity to bioink systems. However, existing ML models typically employ composition ratios as input, restricting their applications to specific bioink systems and lacking in universality. Essentially, ML models represent potential mappings between inputs and outputs. Therefore, for bioink systems sharing similar printing mechanisms, such as GELMA-based and HAMA-based used in the direct ink writing process, a universal ML model is theoretically feasible. In this regard, a critical step is the extraction of mechanism-level features that directly influence printing behavior as inputs, such as the rheological curves. Following training, for various bioinks, the single universal ML model can yield satisfactory prediction results with minimal experiments through fine-tuning of transfer learning methods.
-
(ⅱ)
Gradient structures: Naturally, heterogeneous tissues exhibit a zonal gradient structure, requiring dynamic adjustment of printing parameters over a large range in response to gradient changes of structures and properties [231]. Consequently, ML models must possess high prediction accuracy within a broader design space of printing parameters. A relevant study has been conducted in this regard [232].
Although ML methods have made progress in optimizing printing parameters, the process of manually constructing datasets is time-consuming. To expedite the search for optimal printing parameters, an increasingly promising approach involves constructing a fully automatic and universally applicable search procedure, offering significant practical utility. In this approach, the 3D bioprinter autonomously performs local printing tests for any input bioink and printed model, evaluates the printing quality, and adjusts the printing parameters to determine the optimal settings. Notably, the ML model transcends its conventional role of modeling mapping relationships between inputs and outputs; it also assesses printing quality and serves as the adjustment strategy of printing parameters. For instance, Jianhua Zhou's group [233] has automatically traversed three printing parameters in a special-designed 3D bioprinter, employing a CNN-based algorithm to classify printing quality into three categories and derived a process phase diagram to determine the optimal design space of printing parameters. Although this study achieved high-throughput screening of printing parameters, the process was partially automated. Carmelo De Maria's group [234] has employed a CNN-based algorithm to classify printing quality into three categories and iteratively adjusted printing parameters using a dichotomy-like strategy to obtain the optimal settings (Fig. 7c ⅱ). This process was fully automated but did not optimize all printing parameters. Filippos Tourlomousis' group [230] has employed Bayesian optimization and active learning to automatically adjust the ratio of the collector speed over the jet speed in the MEW process, aiming to achieve the minimum Lag distance.
In the broader field of 3D printing, studies [73,75,[235], [236], [237], [238]] have embraced the concept of closed-loop iteration by employing ML methods to automatically search for the optimal printing parameters. Analogously, such methodologies hold promise in the domain of 3D bioprinting. A critical consideration in this context is the selection of printability metrics. During the process of searching for optimal printing parameters prior to formal printing, it is judicious to print simple and standard test models rather than complex and complete input models. Therefore, it becomes imperative to swiftly identify printability metrics [37,239] (such as corners, filament diameters, and layer spacing) that may influence the final printing quality [238] and select test models accordingly, necessitating a comprehensive understanding of the printing process.
5.2. Control of printing processes
Owing to the inevitable occurrence of process drift and model error, deviations in the printing quality from the anticipated standards may arise within the actual printing process, if the optimal printing parameters derived from off-line design is continuously employed [10]. To maintain consistent control over printing quality, the implementation of in situ monitoring and in-line correction becomes imperative.
Within the framework of Section 2.2.3, Table 4 provides a summary of examples, where AI technology integrated into diverse sensors has been employed for in situ monitoring and in-line correction across various 3D bioprinting processes. Among them, the combination of cameras and CNN-based algorithms is the most common technical solution [234,[268], [269], [270]]. Some studies [271,272] have utilized reinforcement learning methods to ascertain adjustment strategies for printing parameters, enabling adaptation to the dynamic environment (Fig. 7d). In addition, other studies [273,274] have leveraged deep learning methods to predict the entire droplet evolution process in inkjet printing, enhancing the understanding of printing behavior and facilitating early defect detection (Fig. 7e).
Table 4.
Examples of AI applications for in situ monitoring and in-line correction in 3D bioprinting.
| Process category | Sensor | Input | Output | AI Model | Type of AI Model | Ref |
|---|---|---|---|---|---|---|
| EBB | Camera | Video frame | Printing status | CNN | CQA prediction model | [234] |
| Camera | Video frame | Extrusion status | LSTM autoencoder | CQA prediction model | [302] | |
| Camera | Image | Printing anomaly | CNN | CQA prediction model | [269] | |
| Camera | Video frame | Velocity of the printing head, offset from the baseline printing path | Reinforcement learning | Control Strategy | [271] | |
| Infrared thermocouples | Three features extracted from the raw sensor signals | Printing status strand width, strand height, strand fusion severity | KNN, SVM, RF, ANN | CQA prediction model | [303] | |
| Material temperature, extrusion pressure, print speed, the location in the strand | Regime classification, width prediction, height prediction | |||||
| Stereo cameras | Surface shape data | Shape basis vectors | PCA | CPP prediction model | [292] | |
| Camera, pressure sensor | Time-varying 2D printing head position (X, Y), SMAMs pressure (p1, p2, p3, p4) | The displacement of syringe plungers (l1, l2, l3, l4) | ANN | Control Strategy | [297] | |
| DBB | Camera | Video frame | Droplet evolution in the printing process | Deep recurrent neural network (DRNN) | Process prediction model | [273] |
| Camera | Video frame | Droplet evolution in the printing process | Network of tensor time series (TTS) | Process prediction model | [274] | |
| Camera | Video frame | Jetting status | MovileNetV2 | CQA prediction model | [270] | |
| Camera | Droplet velocity at two different points | Cell count | LR, SVR, decision tree regressor (DTR), RFR, extra tree regression (ETR) | CQA prediction model | [304] | |
| Camera | Droplet size, aspect ratio, droplet velocity, satellite droplet | Droplet mode | Backpropagation neural network (BPNN) | CQA prediction model | [305] | |
| FPGA module for self-sensing signals acquisition | Two features extracted from the raw sensor signals | Nozzle jetting status | SVM, ANN, Gaussian naïve Bayes model | CQA prediction model | [306] | |
| LBB | Camera | Video frame | Printing status | CNN-LSTM | CQA prediction model | [268] |
| – | Digital Mask | Digital Mask | Reinforcement learning | Control Strategy | [272] |
Currently, various imaging sensors, such as visible light cameras [234,268,270], laser displacement scanners [275], and optical coherence tomography (OCT) devices [[276], [277], [278], [279]], are utilized for in situ monitoring of the printing process. While they share common attributes such as non-destructiveness, low latency, and ease of integration, each possesses distinct advantages and drawbacks. Visible light cameras are prevalent due to their affordability and wide field of view; however, they can only capture 2D external profile information with limited resolution. Laser displacement scanners enable 3D profile imaging but lack the ability to penetrate the surface for internal feature extraction. OCT, which possesses certain penetration capabilities, can detect internal defects such as air bubbles but is characterized by high cost and limited field of view. Combining multiple imaging modalities can enhance the prediction accuracy and robustness of CMA/CPP and CQA/process prediction models. Within the broader realm of 3D printing, there have been studies [50,280] using MML methods to monitor the printing process in situ, combining several types of sensor images, which has important reference significance for 3D bioprinting. Furthermore, while the predominant emphasis remains on printability, it is imperative to underscore the significance of cell viability, especially for the sustained printing of organs/tissues with clinical volumes over extended durations. In this regard, several studies suggested sensing methods for in situ monitoring of cell viability [281,282].
In instances where severe defects are detected during the printing process, discarding the printed part leads to substantial waste, particularly concerning personalized small-batch BPPs. Therefore, compared with detecting the emerged defects, it is more meaningful to provide early warning of possible defects and intervene preemptively to maintain errors within acceptable limits. Analyzing the source of process drift is pivotal for achieving early defect warnings [283], encompassing environment-related (such as temperature fluctuations and mechanical vibrations), material-related (such as rheological nonlinearity of bioinks), system-related (such as motion errors in the driving mechanism) factors [17]. Setting up sensors to detect CMA/CPP related to process drift and collecting in-line data facilitate early defect warnings. ML models such as RNN, LSTM, gated recurrent unit (GRU), and Transformer excel in analyzing sequential information, enabling the prediction of future printing quality by CQA/process prediction models [284]. Moreover, combining off-line and in-line data can enhance prediction accuracy [285]. While these theories and methods are nascent in the realm of 3D bioprinting, relevant studies in 3D printing underscore the importance of similar methods [[284], [285], [286]].
Due to the limitations of traditional in vitro bioprinting [7], the emerging trend of in situ bioprinting technology, which directly prints at the patient's defect site, presents significant potential for clinical application. In contrast to traditional 3D bioprinting operating on static and typically planar workbenches, in situ bioprinting typically operates on dynamic and often curved biological surfaces within the body, such as the breathing lung or beating heart [17]. In this regard, traditional 3D bioprinters face notable challenges in two key aspects:
-
(ⅰ)
Degrees of freedom (DOFs): The 3-DOF XYZ translational motion of traditional 3D bioprinters is difficult to match the 6-DOF surface.
-
(ⅱ)
Path planning strategy: The strategy of planning the printing path in the pre-bioprinting stage struggles to adapt to the surface deformation caused by shrinkage, expansion and bending. This can lead to potential collisions between the printing head and the patient's body, resulting in printing failure or additional damage.
In addressing these challenges, in situ bioprinting technology leveraging robotics and AI offers promising solutions. Surgical robots, renowned for their high DOFs and precise control, have found widespread use in clinical surgery. Integrating printing heads with the ends of surgical robots presents a viable approach for in situ bioprinting [17]. Additionally, considering the high cost of surgical robots, some studies have explored the integration of industrial robots with printing heads [[287], [288], [289], [290], [291]]. Furthermore, employing closed-loop control methods based on predictive AI technology allows for precise real-time planning of the printing path. We present this employment in two aspects:
-
(ⅰ)
Sensing and prediction of dynamic environment: Initially, 3D reconstruction is conducted based on visual features of real-time images to acquire the current printing surface morphology. A prior study utilized the PCA algorithm to extract key morphological features from the lattice information of pigs' lung surfaces marked by stereo cameras, facilitating rapid modeling of surface deformation during in situ bioprinting processes (Fig. 7f) [292]. Subsequently, the prediction of future surface morphology can be achieved based on historical surface morphology. Studies have employed AI algorithms such as LSTM [293,294], attention mechanisms [295], and DenseNet [296] to analyze sequential medical images, enabling tracking and predicting the motion of the 3D surface of target organs/tissues (such as the tumor and abdominal cavity). This approach holds significant potential for predicting complex deformation patterns of organs/tissues during in situ bioprinting processes.
-
(ⅱ)
Optimization of closed-loop control strategy: Upon achieving 3D reconstruction of the printing surface, AI technology facilitates segmentation of the printing head and analysis of its spatial position, aiding in the correction of the printing path. In addition, AI technology can be used to construct control strategies and enable real-time correction of the printing head's position and posture, such as utilizing ANN models to build motion controllers (Fig. 7g) [297]. Reinforcement learning methods have also been employed to automatically plan and adjust the path of surgical robots, enhancing movement precision, efficiency, and adaptability to complex environments [[298], [299], [300], [301]]. These advancements hold significant reference value for in situ bioprinting based on surgical robots.
6. AI-driven approaches for function regulation
Upon completing the printing of high-quality structures, the final element is the function regulation of the printed structures. Primarily, the design of maturation conditions is imperative to functionalize the printed structures, thereby transforming them into BPPs with the requisite biological functions, as described in Section 6.1. Subsequently, for functionalized in vitro models and in vivo implants, their biological functions are characterized and assessed with non-destructive detection methods, facilitating applications such as drug screening, pathological/pharmacological studies, and assessments of clinical functions, as described in Section 6.2.
6.1. Design of maturation conditions
Within bioreactors, specific external physicochemical stimuli (serving as CMA/CPP), including mechanical, electrical, photo, ultrasound, and soluble factors, are employed to modulate the biochemical and mechanical clues (or cell microenvironment) within BPPs [307]. And the cell behavior (serving as CQA) of BPPs is further regulated, such as proliferation, differentiation, and adhesion, to achieve the desired biological functions. Currently, the design of maturation conditions primarily relies on the DoE paradigm, which lacks quantitative theories and models. Recent studies have explored the utilization of the ML-based data-driven paradigm to model the mapping relationships between external physicochemical stimuli (such as biochemical stimulus [308], and drug stimulation [309]) and cell behavior (such as mechanobiological states [308] and drug responses [309]). For example, Yu Yao's group [309] has developed the GlioML workflow, incorporating nine ML models and a weighted ensemble model to predict the treatment response of glioma under different microenvironment characteristics, successfully identifying promising compounds and drugs for glioma treatment (Fig. 8a).
Fig. 8.
AI-driven approaches for function regulation. (a) A flow chart of the GlioML workflow. Copyright 2024, Nature Publishing Group. (b) A workflow of in situ high-throughput characterization of organoids (segmentation, tracking, and classification). Copyright 2023, Nature Publishing Group. (c) A schematic diagram of biodegradable bone implants segmentation based on SRμCT images. Copyright 2021, Nature Publishing Group.
6.2. Characterization and assessment of functions
6.2.1. In vitro models
AI-based sensing technology enables non-destructive characterization of the biological properties (serving as CQA) of in vitro drug/pathological models across multiple scales for drug screening and pathology/pharmacology studies. At the cell scale, studies have performed 3D segmentation and classification of cells/nuclei in images (such as CLSM [47,310] and bright-field microscopy [311]) to evaluate biological properties such as the cell shape and distribution. In addition, some studies have utilized spectral data (such as Raman spectroscopy [312,313], dielectric spectroscopy [314], and dielectric impedance spectroscopy [315]) to assess biological properties including the cell type, shape, distribution, and density. At the organoid scale, studies have conducted 3D segmentation and classification of single organoids in images (such as OCT [316], high-speed live cell interferometry [317], and bright-field microscopy [318,319]) to extract morphological features such as the shape and volume (Fig. 8b).
6.2.2. In vivo implants
Prior to the implantation of the BPP into the patient's body, it is imperative to assess whether the properties of the BPP and the health status of the patient (serving as CMA/CPP) are conducive to surgical implantation (serving as CQA). In the field of organ transplantation, studies have utilized ML models (such as decision trees [320], RFs [321], and ANNs [322,323]) to predict postoperative survival rates based on the healthcare data of donors and recipients. These findings hold significant reference value for the preoperative assessment of BPP implantation suitability.
Following implanting the BPP into the patient's body, monitoring the regeneration status of the damaged organs/tissues (serving as CQA) becomes essential using non-destructive detection methods. Several studies have employed non-destructive medical imaging techniques for continuous monitoring of implant regeneration status. For example, Julian Moosmann's group has employed the U-net algorithm to segment micro-computed tomography (μCT) images, enabling the precise characterization of the morphology of degradable bone implants and facilitating the time-resolved, quantitative assessment of their degradation efficiency (Fig. 8c) [324]. Daniel G. Anderson's group has used a clustering method to track the spatiotemporal morphology of implants encapsulating pancreatic islet cells in MR images, enabling the characterization of quantitative internal oxygen content [325].
In addition to facilitating regeneration of the damaged organs/tissues, implants themselves can function as biosensors for monitoring patients' health status [326]. Smart dressings applied to human wounds can utilize AI technology to sense the health status of the wound (such as the pH, temperature, humidity, and secretion concentration), which can evaluate and monitor wound regeneration status, as demonstrated in several studies [[327], [328], [329]].
7. Future directions
7.1. Construction of natural organs
7.1.1. Non-destructive and rapid construction of digital twin organs
Natural organs exhibit intricate multi-scale heterogeneous structures, while patient-specific digital twin organs aim to capture this multi-scale information to construct transplantable replacements for natural organs [76]. This entails information at organ scale (such as 3D macrostructures), tissue scale (such as microstructures), and cell scale (such as cell types, spatial arrangement, and microenvironment). Currently, non-destructive imaging technologies at the organ scale, such as CT, MRI, and US, are relatively advanced. However, obtaining non-destructive 3D multi-scale models containing information at the tissue and cell scale remains very challenging, for which relevant studies have made preliminary attempts. For instance, the Human BioMolecular Atlas Program (HuBMAP) aims to create a multi-scale spatial atlas of the healthy human body at single-cell resolution [330]. It has already developed spatial atlases of organs/tissues, including the intestine [331], kidney [332], and placenta interface [333]. Techniques such as serial tissue sectioning [46] and optical tissue clearing [334,335] can extend the scale in the depth direction, enabling the acquisition of macro spatial information. These methods hold promise for constructing high-resolution 3D models of large-scale tissues. However, it's worth noting that these approaches rely on destructive sampling of in vitro organs/tissues, rendering them impractical for application in living patients.
Generative AI technology offers a promising solution for the non-destructive and rapid construction of digital twin organs. Multi-scale 3D models derived from in vitro organs/tissues can serve as the dataset to train the generative AI model, enabling it to learn spatial correspondence across different scales. Utilizing the patient's macrostructure model derived from non-destructive imaging as input, the trained AI model automatically populates the information at the tissue and cell scales that conforms to human anatomical principles, generating a digital twin organ enriched with multi-scale information. This approach circumvents the need for destructive sampling of patients' organs/tissues and lays the foundation for designing printed models of natural organ replacements. For instance, there have been relevant studies using AI methods to automatically generate hierarchical 3D vascular networks within organ-scale macrostructure models [[336], [337], [338]].
Another important issue to consider is the scale limitations of biomimicry. The traditional forward-design approach strives to replicate the structure of natural organs as precisely as possible. However, some studies [9] suggest that biomimicry may reach a limit where increased complexity no longer enhances functional outcomes. Additionally, technical and cost constraints render endless biomimicry of natural organs impractical, especially at the micro-nano scale. Considering clinical translation, the inverse-design approach aims to prioritize the regeneration of specific functions and the feasibility of manufacturing rather than emphasizing structural biomimicry. A potential strategy is to strike a balance between these two approaches by employing appropriate AI models at different scales. At the macro scale, the forward-design approach is applied, with AI models focusing on mimicking the macrostructures of natural organs. At the micro-nano scale, the inverse-design approach is used, with AI models prioritizing the enhancement of specific functions. The generated micro-nano structures are artificially designed rather than imitating natural organs, such as triply periodic minimal surfaces (TPMS). In this way, we can construct manufacturable digital twin organs.
7.1.2. Multi-material 3D bioprinting of high-content printed models
Subsequently, the obtained digital twin organs should be converted into printed models that 3D bioprinters can interpret. Given the complexity of natural heterogeneous organs/tissues, multi-material 3D bioprinting, involving various bioinks, cells, and even printing processes, becomes one of the most promising construction techniques [18,339]. Currently, hybrid bioprinters integrating multiple bioinks and processing techniques are emerging [340,341]. In this scenario, the obtained multi-scale information of the digital twin organ needs to be further dissected into a high-content printed model, encompassing details such as the bioink material, cell type, process type, printing path, and printing parameter to guide subsequent multi-material printing (Fig. 9a).
Fig. 9.
Future directions of AI technology in 3D bioprinting. (a) A pipeline of constructing natural organs. (b) A closed-loop active learning pipeline. (c) A “precision-cost” landscape of the brute-force learning, active learning, and hybrid learning.
Meanwhile, the prolonged printing cycle due to the organ scale of printed models poses risks such as cell sedimentation and contamination. To enhance manufacturing efficiency, a promising approach involves utilizing collaborative printing heads that independently deposit various bioinks simultaneously across distinct regions [291,340,342] (Fig. 9a). This collaboration of multiple printing heads introduces significant challenges for path planning, such as avoiding path interference. In the field of collaborative robots, studies have employed AI approaches to optimize path design for multi-robot arm collaboration, such as large language models, reinforcement learning, and computer vision [[343], [344], [345]]. These methods aim to enhance communication between robotic arms, ensuring interference-free operation while identifying more efficient paths, thereby significantly improving manufacturing efficiency.
7.2. Active learning and hybrid learning
Within the realm of 3D bioprinting, the absence of publicly available datasets poses a significant challenge for data-driven design. Consequently, the predominant cost of designing tasks stems from the construction of datasets, typically undertaken by researchers themselves through manual experiments or numerical simulations. In scenarios where datasets can be constructed with high throughput and low cost, conventional brute-force learning approaches (or learning from scratch) can be employed [55]. Initially, the dataset is constructed completely, and the ML model serves solely as a backend tool for automatic data analysis [43], aiding in comprehending underlying mechanisms. However, in scenarios where datasets have to be constructed with low throughput or high cost, the aforementioned methods prove impractical in terms of time or financial burdens.
One promising methodology is active learning [57,[346], [347], [348], [349]]. It can adaptively sample within the high-property region of the parameter space, which is typically the design space-related region of interest, while avoiding inefficient sampling in the low-property region [347]. The closed-loop active learning pipeline is illustrated in Fig. 9b. Initially, a set of preliminary experiments are conducted, and the built dataset is utilized to train the ML model. Subsequently, the trained ML model generates predictions across the parameter space, based on which the next experiment (or sampling point) is selected. The newly conducted experiment is then incorporated into the dataset, fostering a dynamic feedback loop aimed at iteratively enhancing the ML model's precision. Ultimately, once the ML model's precision aligns with the specified requirements, the training process concludes.
The selection of subsequent experiments holds paramount importance. Bayesian methodology, exemplified by Gaussian process (GP) models, can quantify prediction uncertainty. This capability facilitates the delicate balance between exploration and exploitation [57,346]. During the initial stage of active learning, experiments targeting regions of uncertainty are conducted to comprehensively explore the global parameter space. This process aids in discerning the distribution of properties and refining the ML model—a practice termed as exploration. As the ML model's precision improves and uncertainty diminishes, the focus gradually shifts toward conducting experiments aimed at achieving high-property predictions—a practice termed as exploitation. Through this approach, the ML model can efficiently identify high-property regions at a reduced cost and with heightened precision. While the active learning methodology has found widespread applications in the field of materials science [57,[346], [347], [348], [349]], its integration into 3D bioprinting is emerging [68,127,132,152,[227], [228], [229], [230],261].
Compared to brute-force learning, active learning effectively reduces costs through adaptive sampling but remains grounded in data and statistics. Evolving directions of the data-driven paradigm aim to further diminish cost and enhance precision, leading to the development of the hybrid-driven paradigm (Fig. 9c) [55]. The hybrid-driven paradigm amalgamates prior knowledge with experimental data through hybrid learning, utilizing prior knowledge to impose constraints on the functional space of ML models to accelerate the search process, facilitating modeling with small datasets, consequently reducing cost further [55].
Based on the forms of prior knowledge, the hybrid-driven paradigm can be divided into the following three categories:
-
(ⅰ)
Transfer learning: This approach fine-tunes the pre-trained ML models rather than training them from scratch, enabling high precision with minimal experimental data [350,351]. For instance, Jennifer M Bone [352] has employed the transfer learning method based on hierarchical machine learning (HML) to optimize bioinks and support media in embedded bioprinting.
-
(ⅱ)
Multi-fidelity learning: Combining high-cost/high-fidelity data from the DoE paradigm with low-cost/low-fidelity data from the theoretical, computational, and data-driven paradigms for training ML models can effectively address the lack of high-fidelity datasets [57,346]. For instance, Safa Jamali's group [54] has employed the multi-fidelity learning method to train PINNs to construct rheological constitutive models of hydrogels with a limited amount of experimental data.
-
(ⅲ)
Learning integrated with domain knowledge: By incorporating domain knowledge such as expert experience [353,354] and physical laws [355] to constrain the functional space, ML models can achieve higher precision with minimal experimental data. For instance, Salil Desai's group [248] has integrated the physics model into the LSTM algorithm to predict printing resolution.
In summary, the application of the hybrid-drive paradigm in the field of 3D bioprinting is still in its nascent stages but holds significant potential for advancement.
7.3. Integrated automation of entire processes
In data-driven design, the quantity and quality of samples in the dataset are critical for ensuring the precision of the ML model. Conventional manual sampling methods are inefficient and prone to data noise. Currently, across various fields, such as materials science and chemistry, endeavors have been made to integrate AI technology with automation equipment, such as robots, to supplant manual sampling. This integration enables machines to autonomously perform experiments and optimize designs, a concept termed as self-driving laboratory (SDL) [55,[356], [357], [358], [359], [360], [361], [362]]. In UOs of 3D bioprinting, SDL methods hold promise to significantly enhance both the quantity and quality of samples, consequently reducing cost and enhancing precision, as demonstrated in previous studies [230,363].
Looking ahead, the integration of AI with automation is poised to extend beyond discrete UOs to encompass entire processes (Fig. 10). The AI-based systematic design approach will unify the design of various objects, including bioinks, printed models, printing parameters, and maturation conditions. This integrated methodology will effectively consider their interdependent effects on diverse properties. For instance, through AI-driven multi-objective optimization, printed models' microstructures and bioinks' physicochemical properties can be jointly designed to meet the Pareto optimality of BPPs' mechanical properties and cell behavior. Another example can be found in 4D bioprinting, where AI can be utilized to integrate the design of printed models' microstructures, spatial distribution of bioink formulations, and external stimuli, enabling precise spatiotemporal responses of BPPs' mechanical properties [211,212,364,365].
Fig. 10.
A schematic diagram of integrated automation of entire processes.
Furthermore, the establishment of AI-based smart factories will enable efficient management of the material and information flow through advanced technologies such as industrial clouds and digital twins, facilitating full life cycle quality management, which encompasses clinical diagnosis, raw material preparation, model design, 3D printing manufacturing, and efficacy evaluation.
8. Conclusions
The advancement of 3D bioprinting in clinical practice confronts challenges regarding the contraction between effectiveness and economy in personalization of design, compounded by constraints in scaling-up of production due to the numerous manual operations involved. In light of these challenges, AI-driven QbD emerges as a promising solution, enhancing precision, economy, rapidity, repeatability, and scalability. This review seeks to delve into the latest advancements of AI technology applications in 3D bioprinting. Within the QbD framework, the application of AI technology in 3D bioprinting is scrutinized across three principal dimensions: multi-scale and multi-modal sensing, data-driven design, and in-line process control. Subsequently, a detailed overview of the current research status and potential applications of AI technology is provided for key elements of the 3D bioprinting process, spanning bioink formulation, model structure, printing process, and function regulation. Lastly, the development directions of AI technology in 3D bioprinting are discussed from three perspectives: construction of natural organs, active learning and hybrid learning, and integrated automation of entire processes. This comprehensive analysis aims to elucidate the potential of AI-driven approaches in catalyzing a paradigm shift in 3D bioprinting, paving the way for clinical applications.
Despite considerable progress in AI-driven 3D bioprinting, many challenges remain to be addressed. We discuss some typical challenges and their potential solutions as follows:
-
(ⅰ)
Scarce specificity and low quality of characterization datasets: Regarding the characterization issues in 3D bioprinting, existing datasets are rarely tailored to the unique scenarios of this field, impeding the direct implementation of AI models trained on these datasets. For instance, in organ-scale 3D bioprinting, the precise construction of vascular networks is crucial to ensure nutrient transport and cell survival. However, in medical imaging of blood vessels, most studies focus on the retina [366], with limited datasets available for the 3D reconstruction of vascular networks in large-scale organs. Similarly, organ-scale 3D bioprinting requires a large number of cells. Yet, existing datasets for virtual staining are mostly predominantly derived from characterization results of tissue or pathological sections, with a significant lack of datasets available for non-destructive characterization of stem cells during differentiation/proliferation processes. Additionally, current datasets are typically derived from limited experimental results with narrow sources and small scales, resulting in high variability and reducing the generalization capacity of AI models. Furthermore, the performance metrics of existing AI models are often derived from independent test sets, complicating the rigorous comparison of model performance and lowering their reliability for clinical deployment.
To address this issue, ensuring the specificity and quality of datasets is imperative. Given the scale of this task, individual efforts may be insufficient, thereby encouraging collaboration among researchers in the broader 3D bioprinting community. A shared database tailored to 3D bioprinting can be established through the collection of experimental results from various research groups on cloud platforms. In this context, the specialization of the 3D bioprinting field ensures the specificity of datasets, while the diversity of research groups guarantees the multi-source and large-scale nature of datasets. Furthermore, benchmark datasets for 3D bioprinting could be established, akin to the role of ImageNet in the field of computer vision. Last but not least, during the construction of shared databases, it is crucial to address ethical, privacy, and security concerns [367,368], particularly when patients or participants are involved (such as the appropriate use of patient healthcare data). In this regard, relevant legislation should be established to regulate methods of data usage and the processes of dataset creation, accompanied by strict oversight and governance.
-
(ⅱ)
Insufficient universality of data-driven design for clinical deployment: Considering the clinical translation of 3D bioprinting, patient-specific natural organs introduce a wide range of printed structures; the multi-material printing process brings numerous types of bioinks; and various bioprinter brands result in diverse 3D bioprinter configurations. This variability presents a multitude of working scenarios for data-driven design issues. While current ML models and datasets are typically built for specific scenarios, minor variations can render them unusable, requiring costly retraining.
Therefore, establishing universal ML models capable of adapting to various working scenarios will significantly reduce the need to construct datasets tailored to specific working scenarios, thereby resulting in substantial cost savings. Specifically, some design considerations are related to continuous production, such as culture conditions and printing parameters. Across different working scenarios, they follow similar process mechanisms and share relatively fixed categories of CQA, CMA, and CPP, demonstrating a certain level of universality. As an example, consider the design of printing parameters in the direct ink writing process for hydrogel-based bioinks. Across different brands of 3D bioprinters, bioink materials, and printed models, the printing process follows similar physical mechanisms, primarily the rheology of extrusion process. And the concerned CQA (such as shape fidelity of printed structure), CMA (such as rheological properties of bioinks), and CPP (such as the printing speed and extrusion speed) also remain consistent. Consequently, in theory, a universal ML model could be developed to apply across all working scenarios.
Achieving this objective necessitates the accurate identification of CQA, CMA, and CPP, accompanied by their universal and standardized descriptions, which requires a profound understanding of the process mechanism. Subsequently, the acquisition of extensive experimental data under varied working scenarios becomes imperative to construct comprehensive datasets. Ultimately, the successful development of universal ML models hinges upon meticulous architectural design and training methods predicated on the data structure of the inputs/outputs. To summarize, expertise in 3D bioprinting and AI is required to ensure accurate predictions across diverse working scenarios.
-
(ⅲ)
Limited consideration of spatiotemporal dynamics in ML models: With the development of 3D bioprinting technology, the four key elements in this field have increasingly exhibited spatiotemporal dynamics. Firstly, with the advent of dynamic hydrogels, the implanted BPPs undergo multi-scale dynamic interactions with the in vivo environment. At the macro scale, the degradation dynamics of BPPs should align with the host remodeling of the construct; while at the micro scale, the microenvironment provided by hydrogels regulates cell behaviors over time. Similarly, the emerging 4D bioprinting focuses on the stimuli-responsive shape morphing of printed structures over the additional temporal dimension. Additionally, in-situ bioprinting is expected to accommodate the spatiotemporal changes of printing surfaces. In summary, these spatiotemporal dynamics present significant challenges to the design and manufacturing of 3D bioprinting, yet most existing AI models are still confined to static scenarios.
In this context, ML models excelling at handling sequential data, such as RNN, LSTM, GRU, and Transformer, demonstrate strong temporal modeling capabilities, offering substantial potential for effectively managing these spatiotemporal dynamics.
Addressing the above-mentioned challenges will significantly advance the authentic development of human organ substitutes, facilitating the translation of 3D bioprinting from bench to bedside.
CRediT authorship contribution statement
Zhenrui Zhang: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Xianhao Zhou: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Yongcong Fang: Validation, Supervision, Resources, Project administration, Funding acquisition, Formal analysis, Conceptualization, Writing – review & editing. Zhuo Xiong: Validation, Supervision, Resources, Project administration, Funding acquisition, Formal analysis, Conceptualization. Ting Zhang: Validation, Supervision, Resources, Project administration, Funding acquisition, Formal analysis, Conceptualization.
Ethics approval and consent to participate
The current review does not involve any experimental work on human subjects or animals, and thus does not require approval from an ethics committee. As this work solely involves the synthesis and analysis of pre-existing literature, it doesn't necessitate any form of direct involvement or consent from patients or healthy volunteers.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service, and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled, “Artificial intelligence-driven 3D bioprinting for regenerative medicine: From Bench to Bedside".
Acknowledgements
Zhenrui Zhang and Xianhao Zhou contributed equally to this work. We gratefully acknowledge the funding support from National Key Research and Development Program of China (2023YFB4605800) and National Natural Science Foundation of China (Grant No. U21A20394; No.52305314).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Yongcong Fang, Email: fangyc@tsinghua.edu.cn.
Zhuo Xiong, Email: xiongzhuo@tsinghua.edu.cn.
Ting Zhang, Email: t-zhang@tsinghua.edu.cn.
References
- 1.Fang Y., Guo Y., Liu T., Xu R., Mao S., Mo X., Zhang T., Ouyang L., Xiong Z., Sun W. Advances in 3D bioprinting. Addit. Manuf. Front. 2022;1 [Google Scholar]
- 2.Zhou X., Fang Y., Zhang T., Xiong Z. Retrospective: advances and opportunities of 3D bioprinting in China over three decades. Addit. Manuf. Front. 2024 [Google Scholar]
- 3.Wang P., Rui H., Gao C., Wen C., Pan H., Liu W., Ruan C., Lu W.W. Bioprinting living organs: the next milestone in organ transplantation? Innovat. Life. 2023;1 [Google Scholar]
- 4.Prendergast M.E., Burdick J.A. Recent advances in enabling technologies in 3D printing for precision medicine. Adv. Mater. 2020;32:1902516. doi: 10.1002/adma.201902516. [DOI] [PubMed] [Google Scholar]
- 5.Jain P., Kathuria H., Dubey N. Advances in 3D bioprinting of tissues/organs for regenerative medicine and in-vitro models. Biomaterials. 2022;287 doi: 10.1016/j.biomaterials.2022.121639. [DOI] [PubMed] [Google Scholar]
- 6.Claes E., Heck T., Sonnaert M., Donvil F., Schaschkow A., Desmet T., Schrooten J. Tissue Engineering. third ed. Academic Press; 2023. Chapter 20 - product and process design: scalable and sustainable tissue-engineered product manufacturing; pp. 689–716. [Google Scholar]
- 7.Samandari M., Mostafavi A., Quint J., Memić A., Tamayol A. In situ bioprinting: intraoperative implementation of regenerative medicine. Trends Biotechnol. 2022;40:1229–1247. doi: 10.1016/j.tibtech.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bliley J.M., Shiwarski D.J., Feinberg A.W. 3D-bioprinted human tissue and the path toward clinical translation. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abo7047. [DOI] [PubMed] [Google Scholar]
- 9.Murphy S.V., De Coppi P., Atala A. Opportunities and challenges of translational 3D bioprinting. Nat. Biomed. Eng. 2020;4:370–380. doi: 10.1038/s41551-019-0471-7. [DOI] [PubMed] [Google Scholar]
- 10.Bonatti A.F., Vozzi G., De Maria C. Enhancing quality control in bioprinting through machine learning. Biofabrication. 2024;16 doi: 10.1088/1758-5090/ad2189. [DOI] [PubMed] [Google Scholar]
- 11.Mishra V., Thakur S., Patil A., Shukla A. Quality by design (QbD) approaches in current pharmaceutical set-up, EXPERT OPIN DRUG. DEL. 2018;15:737–758. doi: 10.1080/17425247.2018.1504768. [DOI] [PubMed] [Google Scholar]
- 12.Garcia L., Robinson-Zeigler R., Reiterer M.W., Panoskaltsis-Mortari A. Collaborative findings on manufacturing needs for biofabrication of engineered tissues and organs. Regen. Eng. Transl. Med. 2018;4:45–50. [Google Scholar]
- 13.Martinez-Marquez D., Mirnajafizadeh A., Carty C.P., Stewart R.A. Application of quality by design for 3D printed bone prostheses and scaffolds. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adalbert L., Kanti S.P.Y., Jójárt-Laczkovich O., Akel H., Csóka I. Expanding quality by design principles to support 3D printed medical device development following the renewed regulatory framework in europe. Biomedicines. 2022;10:2947. doi: 10.3390/biomedicines10112947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greener J.G., Kandathil S.M., Moffat L., Jones D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022;23:40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen A., Wang W., Mao Z., He Y., Chen S., Liu G., Su J., Feng P., Shi Y., Yan C., Lu J. Multi‐material 3D and 4D bioprinting of heterogeneous constructs for tissue engineering. Adv. Mater. 2024;36:2307686. doi: 10.1002/adma.202307686. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z., Ng D.W.H., Park H.S., McAlpine M.C. 3D-printed multifunctional materials enabled by artificial-intelligence-assisted fabrication technologies. Nat. Rev. Mater. 2021;6:27–47. [Google Scholar]
- 18.Ravanbakhsh H., Karamzadeh V., Bao G., Mongeau L., Juncker D., Zhang Y.S. Emerging technologies in multi‐material bioprinting. Adv. Mater. 2021;33:2104730. doi: 10.1002/adma.202104730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng W.L., Chan A., Ong Y.S., Chua C.K. Deep learning for fabrication and maturation of 3D bioprinted tissues and organs. Virtual Phys. Prototyp. 2020;15:340–358. [Google Scholar]
- 20.Sun J., Yao K., An J., Jing L., Huang K., Huang D. Machine learning and 3D bioprinting. Int. J. Bioprinting. 2023;9:717. doi: 10.18063/ijb.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C., Jiang J. A perspective on using machine learning in 3D bioprinting. Int. J. Bioprinting. 2019;6:253. doi: 10.18063/ijb.v6i1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramesh S., Deep A., Tamayol A., Kamaraj A., Mahajan C., Madihally S. Advancing 3D bioprinting through machine learning and artificial intelligence. Bioprinting. 2024;38 [Google Scholar]
- 23.Datta P., Barui A., Wu Y., Ozbolat V., Moncal K.K., Ozbolat I.T. Essential steps in bioprinting: from pre- to post-bioprinting. Biotechnol. Adv. 2018;36:1481–1504. doi: 10.1016/j.biotechadv.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Mandrycky C., Wang Z., Kim K., Kim D. 3D bioprinting for engineering complex tissues. Biotechnol. Adv. 2016;34:422–434. doi: 10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy S.V., Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 26.Bai L., Wu Y., Li G., Zhang W., Zhang H., Su J. AI-enabled organoids: construction, analysis, and application. Bioact. Mater. 2024;31:525–548. doi: 10.1016/j.bioactmat.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udugama I.A., Badr S., Hirono K., Scholz B.X., Hayashi Y., Kino-oka M., Sugiyama H. The role of process systems engineering in applying quality by design (QbD) in mesenchymal stem cell production. Comput. Chem. Eng. 2023;172 [Google Scholar]
- 28.Lee S., Kim J., Jee J., Jang D., Park Y., Kim J. Quality by Design (QbD) application for the pharmaceutical development process. J. Pharmaceut. Invest. 2022;52:649–682. [Google Scholar]
- 29.Maillot C., Sion C., De Isla N., Toye D., Olmos E. Quality by design to define critical process parameters for mesenchymal stem cell expansion. Biotechnol. Adv. 2021;50 doi: 10.1016/j.biotechadv.2021.107765. [DOI] [PubMed] [Google Scholar]
- 30.Rathore A.S. Roadmap for implementation of quality by design (QbD) for biotechnology products. Trends Biotechnol. 2009;27:546–553. doi: 10.1016/j.tibtech.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Lipsitz Y.Y., Timmins N.E., Zandstra P.W. Quality cell therapy manufacturing by design. Nat. Biotechnol. 2016;34:393–400. doi: 10.1038/nbt.3525. [DOI] [PubMed] [Google Scholar]
- 32.FDA . 2009. Guidance for Industry: Q8(R2) Pharmaceutical Development. USA. [Google Scholar]
- 33.Holm P., Allesø M., Bryde M.C., Holm R. Q8(R2) pharmaceutical development. ICH Quality Guidelines. 2018:535–577. [Google Scholar]
- 34.Armstrong A.A., Norato J., Alleyne A.G., Wagoner Johnson A.J. Direct process feedback in extrusion-based 3D bioprinting. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab4d97. [DOI] [PubMed] [Google Scholar]
- 35.Armstrong A.A., Alleyne A.G., Wagoner Johnson A.J. 1D and 2D error assessment and correction for extrusion-based bioprinting using process sensing and control strategies. Biofabrication. 2020;12 doi: 10.1088/1758-5090/aba8ee. [DOI] [PubMed] [Google Scholar]
- 36.Zandrini T., Florczak S., Levato R., Ovsianikov A. Breaking the resolution limits of 3D bioprinting: future opportunities and present challenges. Trends Biotechnol. 2023;41:604–614. doi: 10.1016/j.tibtech.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Schwab A., Levato R., D Este M., Piluso S., Eglin D., Malda J. Printability and shape fidelity of bioinks in 3D bioprinting. Chem. Rev. 2020;120:11028–11055. doi: 10.1021/acs.chemrev.0c00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prince E., Kumacheva E. Design and applications of man-made biomimetic fibrillar hydrogels. Nat. Rev. Mater. 2019;4:99–115. [Google Scholar]
- 39.Angelopoulos I., Allenby M.C., Lim M., Zamorano M. Engineering inkjet bioprinting processes toward translational therapies. Biotechnol. Bioeng. 2020;117:272–284. doi: 10.1002/bit.27176. [DOI] [PubMed] [Google Scholar]
- 40.Gu Z., Fu J., Lin H., He Y. Development of 3D bioprinting: from printing methods to biomedical applications. Asian J. Pharm. Sci. 2020;15:529–557. doi: 10.1016/j.ajps.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCorry M.C., Reardon K.F., Black M., Williams C., Babakhanova G., Halpern J.M., Sarkar S., Swami N.S., Mirica K.A., Boermeester S., Underhill A. Sensor technologies for quality control in engineered tissue manufacturing. Biofabrication. 2022;15 doi: 10.1088/1758-5090/ac94a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allenby M.C., Woodruff M.A. Image analyses for engineering advanced tissue biomanufacturing processes. Biomaterials. 2022;284 doi: 10.1016/j.biomaterials.2022.121514. [DOI] [PubMed] [Google Scholar]
- 43.Gruneboom A., Kling L., Christiansen S., Mill L., Maier A., Engelke K., Quick H.H., Schett G., Gunzer M. Next-generation imaging of the skeletal system and its blood supply. Nat. Rev. Rheumatol. 2019;15:533–549. doi: 10.1038/s41584-019-0274-y. [DOI] [PubMed] [Google Scholar]
- 44.Zhou S.K., Greenspan H., Davatzikos C., Duncan J.S., Van Ginneken B., Madabhushi A., Prince J.L., Rueckert D., Summers R.M. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE. 2021;109:820–838. doi: 10.1109/JPROC.2021.3054390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umirzakova S., Ahmad S., Khan L.U., Whangbo T. Medical image super-resolution for smart healthcare applications: a comprehensive survey. Inform Fusion. 2024;103 [Google Scholar]
- 46.Kiemen A.L., Braxton A.M., Grahn M.P., Han K.S., Babu J.M., Reichel R., Jiang A.C., Kim B., Hsu J., Amoa F., Reddy S., Hong S.M., Cornish T.C., Thompson E.D., Huang P., Wood L.D., Hruban R.H., Wirtz D., Wu P.H. CODA: quantitative 3D reconstruction of large tissues at cellular resolution. Nat. Methods. 2022;19:1490–1499. doi: 10.1038/s41592-022-01650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao K., Sun J., Huang K., Jing L., Liu H., Huang D., Jude C. Analyzing cell-scaffold interaction through unsupervised 3D nuclei segmentation. Int. J. Bioprinting. 2022;8:495. doi: 10.18063/ijb.v8i1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Z., Li X., Huang H., Guo N., Li Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci. 2019;3:162–169. doi: 10.1109/trpms.2018.2890359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pradhan P., Meyer T., Vieth M., Stallmach A., Waldner M., Schmitt M., Popp J., Bocklitz T. Computational tissue staining of non-linear multimodal imaging using supervised and unsupervised deep learning. Biomed. Opt Express. 2021;12:2280. doi: 10.1364/BOE.415962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrich J., Snow Z., Corbin D., Reutzel E.W. Multi-modal sensor fusion with machine learning for data-driven process monitoring for additive manufacturing. Addit. Manuf. 2021;48 [Google Scholar]
- 51.Shmatko A., Ghaffari Laleh N., Gerstung M., Kather J.N. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nature cancer. 2022;3:1026–1038. doi: 10.1038/s43018-022-00436-4. [DOI] [PubMed] [Google Scholar]
- 52.Bai B., Yang X., Li Y., Zhang Y., Pillar N., Ozcan A. Deep learning-enabled virtual histological staining of biological samples. Light Sci. Appl. 2023;12 doi: 10.1038/s41377-023-01104-7. 57-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geaney A., O Reilly P., Maxwell P., James J.A., McArt D., Salto-Tellez M. Translation of tissue-based artificial intelligence into clinical practice: from discovery to adoption. Oncogene. 2023;42:3545–3555. doi: 10.1038/s41388-023-02857-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahmoudabadbozchelou M., Kamani K.M., Rogers S.A., Jamali S. Digital rheometer twins: Learning the hidden rheology of complex fluids through rheology-informed graph neural networks. Proceedings of the National Academy of Sciences. 2022;119 doi: 10.1073/pnas.2202234119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hippalgaonkar K., Li Q., Wang X., Fisher J.W., Kirkpatrick J., Buonassisi T. Knowledge-integrated machine learning for materials: lessons from gameplaying and robotics. Nat. Rev. Mater. 2023;8:241–260. [Google Scholar]
- 56.Elbadawi M., McCoubrey L.E., Gavins F.K.H., Ong J.J., Goyanes A., Gaisford S., Basit A.W. Harnessing artificial intelligence for the next generation of 3D printed medicines. Adv. Drug Deliv. Rev. 2021;175 doi: 10.1016/j.addr.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 57.Batra R., Song L., Ramprasad R., A.I.U.S. Argonne National Lab. ANL Emerging materials intelligence ecosystems propelled by machine learning. Nat. Rev. Mater. 2021;6:655–678. [Google Scholar]
- 58.Law A.C.C., Wang R., Chung J., Kucukdeger E., Liu Y., Barron T., Johnson B.N., Kong Z. Process parameter optimization for reproducible fabrication of layer porosity quality of 3D-printed tissue scaffold. J. Intell. Manuf. 2023;35:1825–1844. [Google Scholar]
- 59.Shafiee A., Ghadiri E., Ramesh H., Kengla C., Kassis J., Calvert P., Williams D., Khademhosseini A., Narayan R., Forgacs G., Atala A. Physics of bioprinting. Appl. Phys. Rev. 2019;6:021315. [Google Scholar]
- 60.Yang Q., Lv X., Gao B., Ji Y., Xu F. Elsevier; 2021. Chapter Four - Mechanics of hydrogel-based bioprinting: From 3D to 4D, Advances in Applied Mechanics; pp. 285–318. [Google Scholar]
- 61.Soufivand A.A., Abolfathi N., Hashemi S.A., Lee S.J. Prediction of mechanical behavior of 3D bioprinted tissue-engineered scaffolds using finite element method (FEM) analysis. Addit. Manuf. 2020;33 [Google Scholar]
- 62.Mohammadrezaei D., Moghimi N., Vandvajdi S., Powathil G., Hamis S., Kohandel M. Predicting and elucidating the post-printing behavior of 3D printed cancer cells in hydrogel structures by integrating in-vitro and in-silico experiments. Sci. Rep. 2023;13:1211. doi: 10.1038/s41598-023-28286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fish J., Wagner G.J., Keten S. Mesoscopic and multiscale modelling in materials. Nat. Mater. 2021;20:774–786. doi: 10.1038/s41563-020-00913-0. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S., Vijayavenkataraman S., Lu W.F., Fuh J.Y.H. A review on the use of computational methods to characterize, design, and optimize tissue engineering scaffolds, with a potential in 3D printing fabrication. J. Biomed. Mater. Res. B Appl. Biomater. 2019;107:1329–1351. doi: 10.1002/jbm.b.34226. [DOI] [PubMed] [Google Scholar]
- 65.Chen H., Liu Y., Balabani S., Hirayama R., Huang J. Machine learning in predicting printable biomaterial formulations for direct ink writing. Research. 2023;6:0197. doi: 10.34133/research.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emebu S., Ogunleye R.O., Achbergerová E., Vítková L., Ponížil P., Martinez C.M. Review and proposition for model-based multivariable-multiobjective optimisation of extrusion-based bioprinting. Appl. Mater. Today. 2023;34 [Google Scholar]
- 67.Pahlavani H., Tsifoutis-Kazolis K., Saldivar M.C., Mody P., Zhou J., Mirzaali M.J., Zadpoor A.A. Deep learning for size-agnostic inverse design of random-network 3D printed mechanical metamaterials. Adv. Mater. 2023 doi: 10.1002/adma.202303481. [DOI] [PubMed] [Google Scholar]
- 68.Peng B., Wei Y., Qin Y., Dai J., Li Y., Liu A., Tian Y., Han L., Zheng Y., Wen P. Machine learning-enabled constrained multi-objective design of architected materials. Nat. Commun. 2023;14:6630. doi: 10.1038/s41467-023-42415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng Q., Zhao Z., Lei H., Wang P. A deep learning approach for inverse design of gradient mechanical metamaterials. Int. J. Mech. Sci. 2023;240 [Google Scholar]
- 70.Imrie F., Davis R., van der Schaar M. Multiple stakeholders drive diverse interpretability requirements for machine learning in healthcare. Nat. Mach. Intell. 2023;5:824–829. [Google Scholar]
- 71.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019;1:206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nadernezhad A., Groll J. Machine learning reveals a general understanding of printability in formulations based on rheology additives. Adv. Sci. 2022;9 doi: 10.1002/advs.202202638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roach D.J., Rohskopf A., Leguizamon S., Appelhans L., Cook A.W. Invertible neural networks for real-time control of extrusion additive manufacturing. Addit. Manuf. 2023;74 [Google Scholar]
- 74.Brion D.A.J., Pattinson S.W. Generalisable 3D printing error detection and correction via multi-head neural networks. Nat. Commun. 2022;13:4654. doi: 10.1038/s41467-022-31985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung J., Shen B., Law A.C.C., Kong Z.J. Reinforcement learning-based defect mitigation for quality assurance of additive manufacturing. J. Manuf. Syst. 2022;65:822–835. [Google Scholar]
- 76.An J., Chua C.K., Mironov V. Application of machine learning in 3D bioprinting: focus on development of big data and digital twin. Int. J. Bioprinting. 2020;7:342. doi: 10.18063/ijb.v7i1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crook J.M., Hei D., Stacey G. vol. 46. 2010. The international stem cell banking initiative (ISCBI): raising standards to bank on; pp. 169–172. (In Vitro Cellular & Developmental Biology - Animal). [DOI] [PubMed] [Google Scholar]
- 78.O'Shea O., Steeg R., Chapman C., Mackintosh P., Stacey G.N. Development and implementation of large-scale quality control for the European bank for induced Pluripotent Stem Cells. Stem Cell Res. 2020;45 doi: 10.1016/j.scr.2020.101773. [DOI] [PubMed] [Google Scholar]
- 79.Rivenson Y., Liu T., Wei Z., Zhang Y., de Haan K., Ozcan A. PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci. Appl. 2019;8:23. doi: 10.1038/s41377-019-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levy J.J., Azizgolshani N., Andersen M.J., Suriawinata A., Liu X., Lisovsky M., Ren B., Bobak C.A., Christensen B.C., Vaickus L.J. A large-scale internal validation study of unsupervised virtual trichrome staining technologies on nonalcoholic steatohepatitis liver biopsies. Modern Pathol. 2021;34:808–822. doi: 10.1038/s41379-020-00718-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borhani N., Bower A.J., Boppart S.A., Psaltis D. Digital staining through the application of deep neural networks to multi-modal multi-photon microscopy. Biomed. Opt Express. 2019;10:1339. doi: 10.1364/BOE.10.001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., de Haan K., Rivenson Y., Li J., Delis A., Ozcan A. Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue. Light Sci. Appl. 2020;9:78. doi: 10.1038/s41377-020-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Haan K., Zhang Y., Zuckerman J.E., Liu T., Sisk A.E., Diaz M.F.P., Jen K., Nobori A., Liou S., Zhang S., Riahi R., Rivenson Y., Wallace W.D., Ozcan A. Deep learning-based transformation of H&E stained tissues into special stains. Nat. Commun. 2021;12:4884. doi: 10.1038/s41467-021-25221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang X., Bai B., Zhang Y., Li Y., de Haan K., Liu T., Ozcan A. Virtual stain transfer in histology via cascaded deep neural networks. ACS Photonics. 2022;9:3134–3143. [Google Scholar]
- 85.Hong Y., Heo Y.J., Kim B., Lee D., Ahn S., Ha S.Y., Sohn I., Kim K.M. Deep learning-based virtual cytokeratin staining of gastric carcinomas to measure tumor-stroma ratio. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-98857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivenson Y., Wang H., Wei Z., de Haan K., Zhang Y., Wu Y., Gunaydin H., Zuckerman J.E., Chong T., Sisk A.E., Westbrook L.M., Wallace W.D., Ozcan A. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 2019;3:466–477. doi: 10.1038/s41551-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 87.Cao R., Nelson S.D., Davis S., Liang Y., Luo Y., Zhang Y., Crawford B., Wang L.V. Label-free intraoperative histology of bone tissue via deep-learning-assisted ultraviolet photoacoustic microscopy. Nat. Biomed. Eng. 2023;7:124–134. doi: 10.1038/s41551-022-00940-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li X., Zhang G., Qiao H., Bao F., Deng Y., Wu J., He Y., Yun J., Lin X., Xie H., Wang H., Dai Q. Unsupervised content-preserving transformation for optical microscopy. Light Sci. Appl. 2021;10:44. doi: 10.1038/s41377-021-00484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kang L., Li X., Zhang Y., Wong T.T.W. Deep learning enables ultraviolet photoacoustic microscopy based histological imaging with near real-time virtual staining. Photoacoustics. 2022;25 doi: 10.1016/j.pacs.2021.100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie W., Reder N.P., Koyuncu C., Leo P., Hawley S., Huang H., Mao C., Postupna N., Kang S., Serafin R., Gao G., Han Q., Bishop K.W., Barner L.A., Fu P., Wright J.L., Keene C.D., Vaughan J.C., Janowczyk A., Glaser A.K., Madabhushi A., True L.D., Liu J.T.C. Prostate cancer risk stratification via nondestructive 3D pathology with deep learning–assisted gland analysis. Cancer Res. 2022;82:334–345. doi: 10.1158/0008-5472.CAN-21-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu S., Zhang B., Liu Y., Han A., Shi H., Guan T., He Y. Unpaired stain transfer using pathology-consistent constrained generative adversarial networks. IEEE T Med Imaging. 2021;40:1977–1989. doi: 10.1109/TMI.2021.3069874. [DOI] [PubMed] [Google Scholar]
- 92.Li J., Garfinkel J., Zhang X., Wu D., Zhang Y., de Haan K., Wang H., Liu T., Bai B., Rivenson Y., Rubinstein G., Scumpia P.O., Ozcan A. Biopsy-free in vivo virtual histology of skin using deep learning. Light Sci. Appl. 2021;10:233. doi: 10.1038/s41377-021-00674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu X., Zhao N., Liang H., Tan B., Huang F., Hu H., Chen Y., Wang G., Ling Z., Liu C., Miao Y., Wang Y., Zou X. Bone tissue engineering scaffolds with HUVECs/hBMSCs cocultured on 3D-printed composite bioactive ceramic scaffolds promoted osteogenesis/angiogenesis. J Orthop Transl. 2022;37:152–162. doi: 10.1016/j.jot.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cosenza Z., Block D.E., Baar K. Optimization of muscle cell culture media using nonlinear design of experiments. Biotechnol. J. 2021;16 doi: 10.1002/biot.202100228. [DOI] [PubMed] [Google Scholar]
- 95.Hashizume T., Ying B. Challenges in developing cell culture media using machine learning. Biotechnol. Adv. 2024;70 doi: 10.1016/j.biotechadv.2023.108293. [DOI] [PubMed] [Google Scholar]
- 96.Hashizume T., Ozawa Y., Ying B.W. Employing active learning in the optimization of culture medium for mammalian cells. NPJ Syst Biol Appl. 2023;9:20. doi: 10.1038/s41540-023-00284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bashokouh F., Abbasiliasi S., Tan J.S. Optimization of cultivation conditions for monoclonal IgM antibody production by M1A2 hybridoma using artificial neural network. Cytotechnology. 2019;71:849–860. doi: 10.1007/s10616-019-00330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hong J.K., Choi D., Park S., Silberberg Y.R., Shozui F., Nakamura E., Kayahara T., Lee D. Data-driven and model-guided systematic framework for media development in CHO cell culture. Metab. Eng. 2022;73:114–123. doi: 10.1016/j.ymben.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 99.Zhang A., Xing L., Zou J., Wu J.C. Shifting machine learning for healthcare from development to deployment and from models to data. Nat. Biomed. Eng. 2022;6:1330–1345. doi: 10.1038/s41551-022-00898-y. [DOI] [PubMed] [Google Scholar]
- 100.Fu S., Shi W., Luo T., He Y., Zhou L., Yang J., Yang Z., Liu J., Liu X., Guo Z., Yang C., Liu C., Huang Z.L., Ries J., Zhang M., Xi P., Jin D., Li Y. Field-dependent deep learning enables high-throughput whole-cell 3D super-resolution imaging. Nat. Methods. 2023;20:459–468. doi: 10.1038/s41592-023-01775-5. [DOI] [PubMed] [Google Scholar]
- 101.Ounkomol C., Seshamani S., Maleckar M.M., Collman F., Johnson G.R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods. 2018;15:917–920. doi: 10.1038/s41592-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mund A., Coscia F., Kriston A., Hollandi R., Kovacs F., Brunner A.D., Migh E., Schweizer L., Santos A., Bzorek M., Naimy S., Rahbek-Gjerdrum L.M., Dyring-Andersen B., Bulkescher J., Lukas C., Eckert M.A., Lengyel E., Gnann C., Lundberg E., Horvath P., Mann M. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 2022;40:1231–1240. doi: 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wen C., Miura T., Voleti V., Yamaguchi K., Tsutsumi M., Yamamoto K., Otomo K., Fujie Y., Teramoto T., Ishihara T., Aoki K., Nemoto T., Hillman E.M., Kimura K.D. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. Elife. 2021;10 doi: 10.7554/eLife.59187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim M., Namkung Y., Hyun D., Hong S. Prediction of stem cell state using cell image‐based deep learning. Adv. Intell. Syst. 2023;5:2300017. [Google Scholar]
- 105.Eulenberg P., Köhler N., Blasi T., Filby A., Carpenter A.E., Rees P., Theis F.J., Wolf F.A. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 2017;8:463. doi: 10.1038/s41467-017-00623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Blasi T., Hennig H., Summers H.D., Theis F.J., Cerveira J., Patterson J.O., Davies D., Filby A., Carpenter A.E., Rees P. Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat. Commun. 2016;7 doi: 10.1038/ncomms10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin J., Ogawa T., Hojo N., Kryukov K., Shimizu K., Ikawa T., Imanishi T., Okazaki T., Shiroguchi K. Robotic data acquisition with deep learning enables cell image–based prediction of transcriptomic phenotypes. Proceedings of the National Academy of Sciences. 2023;120 doi: 10.1073/pnas.2210283120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lien C., Chen T., Tsai E., Hsiao Y., Lee N., Gao C., Yang Y., Chen S., Yarmishyn A.A., Hwang D., Chou S., Chu W., Chiou S., Chien Y. Recognizing the differentiation degree of human induced pluripotent stem cell-derived retinal pigment epithelium cells using machine learning and deep learning-based approaches. Cells-Basel. 2023;12:211. doi: 10.3390/cells12020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hennig H., Rees P., Blasi T., Kamentsky L., Hung J., Dao D., Carpenter A.E., Filby A. An open-source solution for advanced imaging flow cytometry data analysis using machine learning. Methods. 2017;112:201–210. doi: 10.1016/j.ymeth.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Min R., Wang Z., Zhuang Y., Yi X. Application of semi-supervised convolutional neural network regression model based on data augmentation and process spectral labeling in Raman predictive modeling of cell culture processes. Biochem. Eng. J. 2023;191 [Google Scholar]
- 111.Yang X., Chen D., Sun Q., Wang Y., Xia Y., Yang J., Lin C., Dang X., Cen Z., Liang D., Wei R., Xu Z., Xi G., Xue G., Ye C., Wang L., Zou P., Wang S., Rivera-Fuentes P., Püntener S., Chen Z., Liu Y., Zhang J., Zhao Y. A live-cell image-based machine learning strategy for reducing variability in PSC differentiation systems. Cell Discov. 2023;9 doi: 10.1038/s41421-023-00543-1. 53-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chu S., Sudo K., Yokota H., Abe K., Nakamura Y., Tsai M. Human induced pluripotent stem cell formation and morphology prediction during reprogramming with time-lapse bright-field microscopy images using deep learning methods. Comput. Methods Progr. Biomed. 2023;229 doi: 10.1016/j.cmpb.2022.107264. [DOI] [PubMed] [Google Scholar]
- 113.Wang C., Wang S., Kang D.D., Dong Y. Biomaterials for in situ cell therapy. BmeMat. 2023;1 doi: 10.1002/bmm2.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fang Y., Ji M., Yang Y., Guo Y., Sun R., Zhang T., Sun W., Xiong Z. 3D printing of vascularized hepatic tissues with a high cell density and heterogeneous microenvironment. Biofabrication. 2023;15 doi: 10.1088/1758-5090/ace5e0. [DOI] [PubMed] [Google Scholar]
- 115.Fang Y., Ji M., Wu B., Xu X., Wang G., Zhang Y., Xia Y., Li Z., Zhang T., Sun W., Xiong Z. Engineering highly vascularized bone tissues by 3D bioprinting of granular prevascularized spheroids. Acs Appl Mater Inter. 2023;15:43492–43502. doi: 10.1021/acsami.3c08550. [DOI] [PubMed] [Google Scholar]
- 116.Wang J., Wu Y., Li G., Zhou F., Wu X., Wang M., Liu X., Tang H., Bai L., Geng Z., Song P., Shi Z., Ren X., Su J. Engineering large-scale self-mineralizing bone organoids with bone matrix-inspired hydroxyapatite hybrid bioinks. Adv. Mater. 2024;36 doi: 10.1002/adma.202309875. [DOI] [PubMed] [Google Scholar]
- 117.Han Y., Wu Y., Wang F., Li G., Wang J., Wu X., Deng A., Ren X., Wang X., Gao J., Shi Z., Bai L., Su J. Heterogeneous DNA hydrogel loaded with Apt 02 modified tetrahedral framework nucleic acid accelerated critical-size bone defect repair. Bioact. Mater. 2024;35:1–16. doi: 10.1016/j.bioactmat.2024.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H., Mao B., Zhong J., Li X., Sang H. Localized delivery of metformin via 3D printed GelMA-Nanoclay hydrogel scaffold for enhanced treatment of diabetic bone defects. J Orthop Transl. 2024;47:249–260. doi: 10.1016/j.jot.2024.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tang T., Zhang M., Adhikari B., Li C., Lin J. Indirect prediction of the 3D printability of polysaccharide gels using multiple machine learning (ML) models. Int. J. Biol. Macromol. 2024;280 doi: 10.1016/j.ijbiomac.2024.135769. [DOI] [PubMed] [Google Scholar]
- 120.Zhang J., Liu Y., Chandra Sekhar P D., Singh M., Tong Y., Kucukdeger E., Yoon H.Y., Haring A.P., Roman M., Kong Z.J., Johnson B.N. Rapid, autonomous high-throughput characterization of hydrogel rheological properties via automated sensing and physics-guided machine learning. Appl. Mater. Today. 2023;30 [Google Scholar]
- 121.Mahmoudabadbozchelou M., Kamani K.M., Rogers S.A., Jamali S. Unbiased construction of constitutive relations for soft materials from experiments via rheology-informed neural networks. Proceedings of the National Academy of Sciences. 2024;121 doi: 10.1073/pnas.2313658121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lennon K.R., McKinley G.H., Swan J.W. Scientific machine learning for modeling and simulating complex fluids. Proceedings of the National Academy of Sciences. 2023;120 doi: 10.1073/pnas.2304669120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lei J., Li Z., Xu S., Liu Z. Recent advances of hydrogel network models for studies on mechanical behaviors. Acta Mech Sinica-Prc. 2021;37:367–386. [Google Scholar]
- 124.Qavi I., Halder S., Tan G. Optimization of printability of bioinks with multi-response optimization (MRO) and artificial neural networks (ANN), Progr.Addit. Manuf. 2024:1–26. [Google Scholar]
- 125.Lee J., Oh S.J., An S.H., Kim W.D., Kim S.H. Machine learning-based design strategy for 3D printable bioink: elastic modulus and yield stress determine printability. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab8707. [DOI] [PubMed] [Google Scholar]
- 126.Xu Y., Sarah R., Habib A., Liu Y., Khoda B. Constraint based Bayesian optimization of bioink precursor: a machine learning framework. Biofabrication. 2024;16 doi: 10.1088/1758-5090/ad716e. [DOI] [PubMed] [Google Scholar]
- 127.Martineau R.L., Bayles A.V., Hung C.S., Reyes K.G., Helgeson M.E., Gupta M.K. Engineering gelation kinetics in living silk hydrogels by differential dynamic microscopy microrheology and machine learning. Adv. Biology. 2022;6 doi: 10.1002/adbi.202101070. [DOI] [PubMed] [Google Scholar]
- 128.Karaoglu I.C., Kebabci A.O., Kizilel S. Optimization of gelatin methacryloyl hydrogel properties through an artificial neural network model. Acs Appl Mater Inter. 2023;15:44796–44808. doi: 10.1021/acsami.3c12207. [DOI] [PubMed] [Google Scholar]
- 129.Xi H., Ye X.W., Guo L.G., Xinchao G., Jia M.L., Wai Y.Y. Machine learning-driven prediction of gel fraction in conductive gelatin methacryloyl hydrogels. IJAMD. 2024;1:61–75. [Google Scholar]
- 130.Khalvandi A., Tayebi L., Kamarian S., Saber-Samandari S., Song J. Data-driven supervised machine learning to predict the compressive response of porous PVA/Gelatin hydrogels and in-vitro assessments: employing design of experiments. Int. J. Biol. Macromol. 2023;253 doi: 10.1016/j.ijbiomac.2023.126906. [DOI] [PubMed] [Google Scholar]
- 131.Entekhabi E., Haghbin Nazarpak M., Sedighi M., Kazemzadeh A. Predicting degradation rate of genipin cross-linked gelatin scaffolds with machine learning. Mater. Sci. Eng. C. 2020;107 doi: 10.1016/j.msec.2019.110362. [DOI] [PubMed] [Google Scholar]
- 132.Seifermann M., Reiser P., Friederich P., Levkin P.A. High‐throughput synthesis and machine learning assisted design of photodegradable hydrogels. Small Methods. 2023;7:2300553. doi: 10.1002/smtd.202300553. [DOI] [PubMed] [Google Scholar]
- 133.Islamkulov M., Karakuş S., Özeroğlu C. Design artificial intelligence-based optimization and swelling behavior of novel crosslinked polymeric network hydrogels based on acrylamide-2-hydroxyethyl methacrylate and acrylamide-N-isopropylacrylamide. Colloid Polym. Sci. 2023;301:259–272. [Google Scholar]
- 134.Boztepe C., Daskin M., Erdogan A., Sarici T. Preparation of poly(acrylamide‐co‐2‐acrylamido‐2‐methylpropan sulfonic acid)‐g‐Carboxymethyl cellulose/Titanium dioxide hydrogels and modeling of their swelling capacity and mechanic strength behaviors by response surface method technique. Polym. Eng. Sci. 2021;61:2083–2096. [Google Scholar]
- 135.Soleimani S., Heydari A., Fattahi M. Swelling prediction of calcium alginate/cellulose nanocrystal hydrogels using response surface methodology and artificial neural network. Ind. Crop. Prod. 2023;192 [Google Scholar]
- 136.Sabbagh F., Muhamad I.I., Nazari Z., Mobini P., Taraghdari S.B. From formulation of acrylamide-based hydrogels to their optimization for drug release using response surface methodology. Mater. Sci. Eng. C. 2018;92:20–25. doi: 10.1016/j.msec.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 137.Qiao Q., Zhang X., Yan Z., Hou C., Zhang J., He Y., Zhao N., Yan S., Gong Y., Li Q. The use of machine learning to predict the effects of cryoprotective agents on the GelMA-based bioinks used in extrusion cryobioprinting. Bio-Design Manuf. 2023;6:464–477. [Google Scholar]
- 138.Abalymov A., Van der Meeren L., Skirtach A.G., Parakhonskiy B.V. Identification and analysis of key parameters for the ossification on particle functionalized composites hydrogel materials. Acs Appl Mater Inter. 2020;12:38862–38872. doi: 10.1021/acsami.0c06641. [DOI] [PubMed] [Google Scholar]
- 139.Verheyen C.A., Uzel S.G.M., Kurum A., Roche E.T., Lewis J.A. Integrated data-driven modeling and experimental optimization of granular hydrogel matrices. Matter. 2023;6:1015–1036. [Google Scholar]
- 140.Lai J., Liu Y., Lu G., Yung P., Wang X., Tuan R.S., Li Z.A. 4D bioprinting of programmed dynamic tissues. Bioact. Mater. 2024;37:348–377. doi: 10.1016/j.bioactmat.2024.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boztepe C., Yüceer M., Künkül A., Şölener M., Kabasakal O.S. Prediction of the deswelling behaviors of pH- and temperature-responsive poly(NIPAAm-co-AAc) IPN hydrogel by artificial intelligence techniques. Res Chem Intermediat. 2020;46:409–428. [Google Scholar]
- 142.Boztepe C., Künkül A., Yüceer M. Application of artificial intelligence in modeling of the doxorubicin release behavior of pH and temperature responsive poly(NIPAAm-co-AAc)-PEG IPN hydrogel. J. Drug Deliv. Sci. Technol. 2020;57 [Google Scholar]
- 143.Su H., Yan H., Zhang X., Zhong Z. Multiphysics-informed deep learning for swelling of pH/temperature sensitive cationic hydrogels and its inverse problem. Mech. Mater. 2022;175 [Google Scholar]
- 144.Lou J., Mooney D.J. Chemical strategies to engineer hydrogels for cell culture. Nat. Rev. Chem. 2022;6:726–744. doi: 10.1038/s41570-022-00420-7. [DOI] [PubMed] [Google Scholar]
- 145.Zhu J., Jia Y., Lei J., Liu Z. Deep learning approach to mechanical property prediction of single-network hydrogel. Mathematics. 2021;9:2804. [Google Scholar]
- 146.Shokrollahi Y., Dong P., Gamage P.T., Patrawalla N., Kishore V., Mozafari H., Gu L. Finite element-based machine learning model for predicting the mechanical properties of composite hydrogels. Appl. Sci. 2022;12 [Google Scholar]
- 147.Leng Y., Tac V., Calve S., Tepole A.B. Predicting the mechanical properties of biopolymer gels using neural networks trained on discrete fiber network data. Comput Method Appl M. 2021;387 [Google Scholar]
- 148.Qiu Y., Ye H., Zhang H., Zheng Y. Machine learning-driven optimization design of hydrogel-based negative hydration expansion metamaterials. Comput Aided Design. 2024;166 [Google Scholar]
- 149.Chimene D., Kaunas R., Gaharwar A.K. Hydrogel bioink reinforcement for additive manufacturing: a focused review of emerging strategies. Adv. Mater. 2020;32 doi: 10.1002/adma.201902026. [DOI] [PubMed] [Google Scholar]
- 150.Ren X., Wei J., Luo X., Liu Y., Li K., Zhang Q., Gao X., Yan S., Wu X., Jiang X., Liu M., Cao D., Wei L., Zeng X., Shi J. HydrogelFinder: a foundation model for efficient self-assembling peptide discovery guided by non-peptidal small molecules. Adv. Sci. 2024;11 doi: 10.1002/advs.202400829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li F., Han J., Cao T., Lam W., Fan B., Tang W., Chen S., Fok K.L., Li L. Design of self-assembly dipeptide hydrogels and machine learning via their chemical features. Proceedings of the National Academy of Sciences. 2019;116:11259–11264. doi: 10.1073/pnas.1903376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu T., Wang J., Zhao S., Chen D., Zhang H., Fang Y., Kong N., Zhou Z., Li W., Wang H. Accelerating the prediction and discovery of peptide hydrogels with human-in-the-loop. Nat. Commun. 2023;14:3880. doi: 10.1038/s41467-023-39648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li W., Wen Y., Wang K., Ding Z., Wang L., Chen Q., Xie L., Xu H., Zhao H. Developing a machine learning model for accurate nucleoside hydrogels prediction based on descriptors. Nat. Commun. 2024;15:2603. doi: 10.1038/s41467-024-46866-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mota C., Camarero-Espinosa S., Baker M.B., Wieringa P., Moroni L. Bioprinting: from tissue and organ development to in vitro models. Chem. Rev. 2020;120:10547–10607. doi: 10.1021/acs.chemrev.9b00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nowogrodzki A. The world’s strongest MRI machines are pushing human imaging to new limits. Nature. 2018;563:24–27. doi: 10.1038/d41586-018-07182-7. [DOI] [PubMed] [Google Scholar]
- 156.Dong C., Loy C.C., He K., Tang X. Image super-resolution using deep convolutional networks. IEEE T Pattern Anal. 2016;38:295–307. doi: 10.1109/TPAMI.2015.2439281. [DOI] [PubMed] [Google Scholar]
- 157.Qiu D., Cheng Y., Wang X. Dual U-Net residual networks for cardiac magnetic resonance images super-resolution. Comput. Methods Progr. Biomed. 2022;218 doi: 10.1016/j.cmpb.2022.106707. [DOI] [PubMed] [Google Scholar]
- 158.Qin C., Schlemper J., Caballero J., Price A.N., Hajnal J.V., Rueckert D. Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE T Med Imaging. 2019;38:280–290. doi: 10.1109/TMI.2018.2863670. [DOI] [PubMed] [Google Scholar]
- 159.Chen F., Taviani V., Malkiel I., Cheng J.Y., Tamir J.I., Shaikh J., Chang S.T., Hardy C.J., Pauly J.M., Vasanawala S.S. Variable-density single-shot fast spin-echo MRI with deep learning reconstruction by using variational networks. Radiology. 2018;289:366–373. doi: 10.1148/radiol.2018180445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chi J., Sun Z., Wang H., Lyu P., Yu X., Wu C. CT image super-resolution reconstruction based on global hybrid attention. Comput. Biol. Med. 2022;150 doi: 10.1016/j.compbiomed.2022.106112. [DOI] [PubMed] [Google Scholar]
- 161.Hou H., Jin Q., Zhang G., Li Z. CT image quality enhancement via a dual-channel neural network with jointing denoising and super-resolution. Neurocomputing. 2022;492:343–352. [Google Scholar]
- 162.Chen Y., Zheng Q., Chen J. Double paths network with residual information distillation for improving lung CT image super resolution. Biomed Signal Proces. 2022;73 doi: 10.1016/j.bspc.2021.103412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.M M., G E., Y.C J., S.V S., Z G., X L., M.P J. Deep generative adversarial neural networks for compressive sensing MRI. IEEE T Med Imaging. 2019;38:167–179. doi: 10.1109/TMI.2018.2858752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Berggren K., Ryd D., Heiberg E., Aletras A.H., Hedstrom E. Super-resolution cine image enhancement for fetal cardiac magnetic resonance imaging. J Magn Reson Imaging. 2022;56:223–231. doi: 10.1002/jmri.27956. [DOI] [PubMed] [Google Scholar]
- 165.Chaudhari A.S., Fang Z., Kogan F., Wood J., Stevens K.J., Gibbons E.K., Lee J.H., Gold G.E., Hargreaves B.A. Super‐resolution musculoskeletal MRI using deep learning. Magn. Reson. Med. 2018;80:2139–2154. doi: 10.1002/mrm.27178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu B., Liu J.Z., Cauley S.F., Rosen B.R., Rosen M.S. Image reconstruction by domain-transform manifold learning. Nature. 2018;555:487–492. doi: 10.1038/nature25988. [DOI] [PubMed] [Google Scholar]
- 167.Li Y., Iwamoto Y., Lin L., Xu R., Chen Y. VolumeNet: a lightweight parallel network for super-resolution of MR and CT volumetric data. IEEE T Image Process. 2021;30 doi: 10.1109/TIP.2021.3076285. 1-1. [DOI] [PubMed] [Google Scholar]
- 168.Ge R., Shi F., Chen Y., Tang S., Zhang H., Lou X., Zhao W., Coatrieux G., Gao D., Li S., Mai X. Improving anisotropy resolution of computed tomography and annotation using 3D super-resolution network. Biomed Signal Proces. 2023;82 [Google Scholar]
- 169.Yang G., Zhang L., Liu A., Fu X., Chen X., Wang R. MGDUN: an interpretable network for multi-contrast MRI image super-resolution reconstruction. Comput. Biol. Med. 2023;167 doi: 10.1016/j.compbiomed.2023.107605. 107605-107605. [DOI] [PubMed] [Google Scholar]
- 170.Ma J., Zhang Y., Gu S., An X., Wang Z., Ge C., Wang C., Zhang F., Wang Y., Xu Y., Gou S., Thaler F., Payer C., Štern D., Henderson E.G.A., McSweeney D.M., Green A., Jackson P., McIntosh L., Nguyen Q., Qayyum A., Conze P., Huang Z., Zhou Z., Fan D., Xiong H., Dong G., Zhu Q., He J., Yang X. Fast and Low-GPU-memory abdomen CT organ segmentation: the FLARE challenge. Med. Image Anal. 2022;82 doi: 10.1016/j.media.2022.102616. [DOI] [PubMed] [Google Scholar]
- 171.Roth H.R., Oda H., Zhou X., Shimizu N., Yang Y., Hayashi Y., Oda M., Fujiwara M., Misawa K., Mori K. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imag Grap. 2018;66:90–99. doi: 10.1016/j.compmedimag.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 172.Xie X., Pan X., Shao F., Zhang W., An J. MCI-Net: multi-scale context integrated network for liver CT image segmentation. Comput. Electr. Eng. 2022;101 [Google Scholar]
- 173.Kang L., Zhou Z., Huang J., Han W. Renal tumors segmentation in abdomen CT Images using 3D-CNN and ConvLSTM. Biomed Signal Proces. 2022;72 [Google Scholar]
- 174.Feng X., Qing K., Tustison N.J., Meyer C.H., Chen Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med. Phys. 2019;46:2169–2180. doi: 10.1002/mp.13466. [DOI] [PubMed] [Google Scholar]
- 175.Henderson E.G.A., Vasquez Osorio E.M., van Herk M., Green A.F. Optimising a 3D convolutional neural network for head and neck computed tomography segmentation with limited training data. Phys. Imag.Radiat. Oncol. 2022;22:44–50. doi: 10.1016/j.phro.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang M., Qi S., Wu Y., Sun Y., Chang R., Pang H., Qian W. CE-NC-VesselSegNet: supervised by contrast-enhanced CT images but utilized to segment pulmonary vessels from non-contrast-enhanced CT images. Biomed Signal Proces. 2023;82 [Google Scholar]
- 177.Zhang L., Guo Z., Zhang H., van der Plas E., Koscik T.R., Nopoulos P.C., Sonka M. Assisted annotation in Deep LOGISMOS: simultaneous multi‐compartment 3D MRI segmentation of calf muscles. Med. Phys. 2023;50:4916–4929. doi: 10.1002/mp.16284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kawahara D., Tsuneda M., Ozawa S., Okamoto H., Nakamura M., Nishio T., Saito A., Nagata Y. Stepwise deep neural network (stepwise-net) for head and neck auto-segmentation on CT images. Comput. Biol. Med. 2022;143 doi: 10.1016/j.compbiomed.2022.105295. [DOI] [PubMed] [Google Scholar]
- 179.Zhang B., Qi S., Wu Y., Pan X., Yao Y., Qian W., Guan Y. Multi-scale segmentation squeeze-and-excitation UNet with conditional random field for segmenting lung tumor from CT images. Comput. Methods Progr. Biomed. 2022;222 doi: 10.1016/j.cmpb.2022.106946. [DOI] [PubMed] [Google Scholar]
- 180.Cao Y., Zhou W., Zang M., An D., Feng Y., Yu B. MBANet: a 3D convolutional neural network with multi-branch attention for brain tumor segmentation from MRI images. Biomed Signal Proces. 2023;80 [Google Scholar]
- 181.Ge R., He Y., Xia C., Xu C., Sun W., Yang G., Li J., Wang Z., Yu H., Zhang D., Chen Y., Luo L., Li S., Zhu Y. X-CTRSNet: 3D cervical vertebra CT reconstruction and segmentation directly from 2D X-ray images. Knowl-Based Syst. 2022;236 [Google Scholar]
- 182.Chen Y., Zheng C., Zhou T., Feng L., Liu L., Zeng Q., Wang G. A deep residual attention-based U-Net with a biplane joint method for liver segmentation from CT scans. Comput. Biol. Med. 2023;152 doi: 10.1016/j.compbiomed.2022.106421. [DOI] [PubMed] [Google Scholar]
- 183.Wang F., Cheng C., Cao W., Wu Z., Wang H., Wei W., Yan Z., Liu Z. MFCNet: a multi-modal fusion and calibration networks for 3D pancreas tumor segmentation on PET-CT images. Comput. Biol. Med. 2023;155 doi: 10.1016/j.compbiomed.2023.106657. [DOI] [PubMed] [Google Scholar]
- 184.Rahimpour M., Saint Martin M., Frouin F., Akl P., Orlhac F., Koole M., Malhaire C. Visual ensemble selection of deep convolutional neural networks for 3D segmentation of breast tumors on dynamic contrast enhanced MRI. Eur. Radiol. 2023;33:959–969. doi: 10.1007/s00330-022-09113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Raza R., Ijaz Bajwa U., Mehmood Y., Waqas Anwar M., Hassan Jamal M. dResU-Net: 3D deep residual U-Net based brain tumor segmentation from multimodal MRI. Biomed Signal Proces. 2023;79 [Google Scholar]
- 186.Renard F., Guedria S., Palma N.D., Vuillerme N. Variability and reproducibility in deep learning for medical image segmentation. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Memon A.R., Li J., Egger J., Chen X. A review on patient-specific facial and cranial implant design using Artificial Intelligence (AI) techniques. Expert Rev Med Devic. 2021;18:985–994. doi: 10.1080/17434440.2021.1969914. [DOI] [PubMed] [Google Scholar]
- 188.Xu L., Xiong Y., Guo J., Tang W., Wong K.K.L., Yi Z. An intelligent system for craniomaxillofacial defecting reconstruction. Int. J. Intell. Syst. 2022;37:9461–9479. [Google Scholar]
- 189.Xiong Y., Zeng W., Xu L., Guo J., Liu C., Chen J., Du X., Tang W. Virtual reconstruction of midfacial bone defect based on generative adversarial network. Head Face Med. 2022;18:19. doi: 10.1186/s13005-022-00325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wu C.T., Yang Y.H., Chang Y.Z. Three-dimensional deep learning to automatically generate cranial implant geometry. Sci. Rep. 2022;12:2683. doi: 10.1038/s41598-022-06606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Farook T.H., Ahmed S., Jamayet N.B., Rashid F., Barman A., Sidhu P., Patil P., Lisan A.M., Eusufzai S.Z., Dudley J., Daood U. Computer-aided design and 3-dimensional artificial/convolutional neural network for digital partial dental crown synthesis and validation. Sci. Rep. 2023;13:1561. doi: 10.1038/s41598-023-28442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chau R.C.W., Hsung R.T., McGrath C., Pow E.H.N., Lam W.Y.H. Accuracy of artificial intelligence-designed single-molar dental prostheses: a feasibility study. J. Prosthet. Dent. 2023;131:1111–1117. doi: 10.1016/j.prosdent.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 193.Tian S., Wang M., Ma H., Huang P., Dai N., Sun Y., Meng J. Efficient tooth gingival margin line reconstruction via adversarial learning. Biomed Signal Proces. 2022;78 [Google Scholar]
- 194.Tian S., Wang M., Yuan F., Dai N., Sun Y., Xie W., Qin J. Efficient computer-aided design of dental inlay restoration: a deep adversarial framework. IEEE T Med Imaging. 2021;40:2415–2427. doi: 10.1109/TMI.2021.3077334. [DOI] [PubMed] [Google Scholar]
- 195.Zhang T., Jin L., Fang Y., Lin F., Sun W., Xiong Z. Fabrication of biomimetic scaffolds with oriented porous morphology for cardiac tissue engineering. J Biomater Tiss Eng. 2014;4:1030–1039. [Google Scholar]
- 196.Wang C., Xu Y., Xia J., Zhou Z., Fang Y., Zhang L., Sun W. Multi-scale hierarchical scaffolds with aligned micro-fibers for promoting cell alignment. Biomed. Mater. 2021;16 doi: 10.1088/1748-605X/ac0a90. [DOI] [PubMed] [Google Scholar]
- 197.Wei Y., Wang Z., Lei L., Han J., Zhong S., Yang X., Gou Z., Chen L. Appreciable biosafety, biocompatibility and osteogenic capability of 3D printed nonstoichiometric wollastonite scaffolds favorable for clinical translation. J Orthop Transl. 2024;45:88–99. doi: 10.1016/j.jot.2024.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jiao P., Mueller J., Raney J.R., Zheng X.R., Alavi A.H. Mechanical metamaterials and beyond. Nat. Commun. 2023;14:6004. doi: 10.1038/s41467-023-41679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zheng X., Zhang X., Chen T.T., Watanabe I. Deep learning in mechanical metamaterials: from prediction and generation to inverse design. Adv. Mater. 2023;35:2302530. doi: 10.1002/adma.202302530. [DOI] [PubMed] [Google Scholar]
- 200.Dogan E., Bhusal A., Cecen B., Miri A.K. 3D Printing metamaterials towards tissue engineering. Appl. Mater. Today. 2020;20 doi: 10.1016/j.apmt.2020.100752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang C., Vangelatos Z., Grigoropoulos C.P., Ma Z. Micro-engineered architected metamaterials for cell and tissue engineering. Mater.Today Adv. 2022;13 [Google Scholar]
- 202.Zhang L., Wang B., Song B., Yao Y., Choi S., Yang C., Shi Y. 3D printed biomimetic metamaterials with graded porosity and tapering topology for improved cell seeding and bone regeneration. Bioact. Mater. 2023;25:677–688. doi: 10.1016/j.bioactmat.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu C., Wan B., Xu Y., Al Maruf D.S.A., Cheng K., Lewin W.T., Fang J., Xin H., Crook J.M., Clark J.R., Steven G.P., Li Q. Dynamic optimisation for graded tissue scaffolds using machine learning techniques. Comput Method Appl M. 2024;425 [Google Scholar]
- 204.Asadi-Eydivand M., Solati-Hashjin M., Fathi A., Padashi M., Abu Osman N.A. Optimal design of a 3D-printed scaffold using intelligent evolutionary algorithms. Appl. Soft Comput. 2016;39:36–47. [Google Scholar]
- 205.Kumar S., Tan S., Zheng L., Kochmann D.M. Inverse-designed spinodoid metamaterials. npj Comput. Mater. 2020;6:73. [Google Scholar]
- 206.Bastek J., Kumar S., Telgen B., Glaesener R.N., Kochmann D.M. Inverting the structure–property map of truss metamaterials by deep learning. Proceedings of the National Academy of Sciences. 2022;119 doi: 10.1073/pnas.2111505119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lu Y., Gong T., Yang Z., Zhu H., Liu Y., Wu C. Designing anisotropic porous bone scaffolds using a self-learning convolutional neural network model. Front. Bioeng. Biotechnol. 2022;10:973275. doi: 10.3389/fbioe.2022.973275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wu C., Wan B., Entezari A., Fang J., Xu Y., Li Q. Machine learning-based design for additive manufacturing in biomedical engineering. Int. J. Mech. Sci. 2024;266 [Google Scholar]
- 209.Pahlavani H., Amani M., Saldívar M.C., Zhou J., Mirzaali M.J., Zadpoor A.A. Deep learning for the rare-event rational design of 3D printed multi-material mechanical metamaterials. Comm. Mater. 2022;3:46. [Google Scholar]
- 210.Yu S., Chai H., Xiong Y., Kang M., Geng C., Liu Y., Chen Y., Zhang Y., Zhang Q., Li C., Wei H., Zhao Y., Yu F., Lu A. Studying complex evolution of hyperelastic materials under external field stimuli using artificial neural networks with spatiotemporal features in a small‐scale dataset. Adv. Mater. 2022;34 doi: 10.1002/adma.202200908. [DOI] [PubMed] [Google Scholar]
- 211.Sun X., Yue L., Yu L., Shao H., Peng X., Zhou K., Demoly F., Zhao R., Qi H.J. Machine learning‐evolutionary algorithm enabled design for 4D‐printed active composite structures. Adv. Funct. Mater. 2022;32:2109805. [Google Scholar]
- 212.Sun X., Yu L., Yue L., Zhou K., Demoly F., Zhao R.R., Qi H.J. Machine learning and sequential subdomain optimization for ultrafast inverse design of 4D-printed active composite structures. J Mech Phys Solids. 2024 [Google Scholar]
- 213.Sun X., Yue L., Yu L., Forte C.T., Armstrong C.D., Zhou K., Demoly F., Zhao R.R., Qi H.J. Machine learning-enabled forward prediction and inverse design of 4D-printed active plates. Nat. Commun. 2024;15:5509. doi: 10.1038/s41467-024-49775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sujeeun L.Y., Goonoo N., Ramphul H., Chummun I., Gimié F., Baichoo S., Bhaw-Luximon A. Correlating in vitro performance with physico-chemical characteristics of nanofibrous scaffolds for skin tissue engineering using supervised machine learning algorithms. Roy Soc Open Sci. 2020;7 doi: 10.1098/rsos.201293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tourlomousis F., Jia C., Karydis T., Mershin A., Wang H., Kalyon D.M., Chang R.C. Machine learning metrology of cell confinement in melt electrowritten three-dimensional biomaterial substrates. Microsyst Nanoeng. 2019;5:15. doi: 10.1038/s41378-019-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Devlin B.L., Allenby M.C., Ren J., Pickering E., Klein T.J., Paxton N.C., Woodruff M.A. Materials design innovations in optimizing cellular behavior on melt electrowritten (MEW) scaffolds. Adv Funct Mater, 2024 [Google Scholar]
- 217.G. Drakoulas, T. Gortsas, E. Polyzos, S. Tsinopoulos, L. Pyl, D. Polyzos, An explainable machine learning-based probabilistic framework for the design of scaffolds in bone tissue engineering, Biomech Model Mechan (2024) 1-26. [DOI] [PubMed]
- 218.Wu C., Entezari A., Zheng K., Fang J., Zreiqat H., Steven G.P., Swain M.V., Li Q. A machine learning-based multiscale model to predict bone formation in scaffolds. Nature Comput. Sci. 2021;1:532–541. doi: 10.1038/s43588-021-00115-x. [DOI] [PubMed] [Google Scholar]
- 219.Wang J., Cui Z., Maniruzzaman M. Bioprinting: a focus on improving bioink printability and cell performance based on different process parameters. Int J Pharmaceut. 2023;640 doi: 10.1016/j.ijpharm.2023.123020. [DOI] [PubMed] [Google Scholar]
- 220.Yu K., Zhang X., Sun Y., Gao Q., Fu J., Cai X., He Y. Printability during projection-based 3D bioprinting. Bioact. Mater. 2022;11:254–267. doi: 10.1016/j.bioactmat.2021.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Adhikari J., Roy A., Das A., Ghosh M., Thomas S., Sinha A., Kim J., Saha P. Effects of processing parameters of 3D bioprinting on the cellular activity of bioinks. Macromol. Biosci. 2021;21 doi: 10.1002/mabi.202000179. [DOI] [PubMed] [Google Scholar]
- 222.Tian S., Zhao H., Lewinski N. Key parameters and applications of extrusion-based bioprinting. Bioprinting. 2021;23 [Google Scholar]
- 223.Azizi Machekposhti S., Movahed S., Narayan R.J. Physicochemical parameters that underlie inkjet printing for medical applications. Biophysics Rev. 2020;1 doi: 10.1063/5.0011924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.You S., Guan J., Alido J., Hwang H.H., Yu R., Kwe L., Su H., Chen S. Mitigating scattering effects in light-based three-dimensional printing using machine learning. J. Manuf. Sci. Eng. 2020;142 [Google Scholar]
- 225.Guan J., You S., Xiang Y., Schimelman J., Alido J., Ma X., Tang M., Chen S. Compensating the cell-induced light scattering effect in light-based bioprinting using deep learning. Biofabrication. 2021;14 doi: 10.1088/1758-5090/ac3b92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bone J.M., Childs C.M., Menon A., Póczos B., Feinberg A.W., LeDuc P.R., Washburn N.R. Hierarchical machine learning for high-fidelity 3D printed Biopolymers. ACS Biomater. Sci. Eng. 2020;6:7021–7031. doi: 10.1021/acsbiomaterials.0c00755. [DOI] [PubMed] [Google Scholar]
- 227.Etefagh A.H., Razfar M.R. Bayesian optimization of 3D bioprinted polycaprolactone/magnesium oxide nanocomposite scaffold using a machine learning technique. Proceedings of the Institution of Mechanical Engineers, Part B: Journal of Engineering Manufacture. 2024;238:1448–1462. [Google Scholar]
- 228.Ruberu K., Senadeera M., Rana S., Gupta S., Chung J., Yue Z., Venkatesh S., Wallace G. Coupling machine learning with 3D bioprinting to fast track optimisation of extrusion printing. Appl. Mater. Today. 2021;22 [Google Scholar]
- 229.Mohammadrezaei D., Podina L., Silva J., Kohandel M. Cell viability prediction and optimization in extrusion-based bioprinting via neural network-based Bayesian optimization models. Biofabrication. 2024;16 doi: 10.1088/1758-5090/ad17cf. [DOI] [PubMed] [Google Scholar]
- 230.Oikonomou A., Loutas T., Fan D., Garmulewicz A., Nounesis G., Chaudhuri S., Tourlomousis F. Physics-Informed Bayesian learning of electrohydrodynamic polymer jet printing dynamics. Commun. Eng. 2023;2:20. [Google Scholar]
- 231.Armstrong A.A., Pfeil A., Alleyne A.G., Wagoner Johnson A.J. Process monitoring and control strategies in extrusion-based bioprinting to fabricate spatially graded structures. Bioprinting. 2021;21 [Google Scholar]
- 232.Kiratitanaporn W., Guan J., Berry D.B., Lao A., Chen S. Multimodal three-dimensional printing for micro-modulation of scaffold stiffness through machine learning. Tissue Eng. 2023;30:280–292. doi: 10.1089/ten.TEA.2023.0193. [DOI] [PubMed] [Google Scholar]
- 233.Chen B., Dong J., Ruelas M., Ye X., He J., Yao R., Fu Y., Liu Y., Hu J., Wu T., Zhou C., Li Y., Huang L., Zhang Y.S., Zhou J. Artificial intelligence‐assisted high‐throughput screening of printing conditions of hydrogel architectures for accelerated diabetic wound healing. Adv. Funct. Mater. 2022;32 [Google Scholar]
- 234.Bonatti A.F., Vozzi G., Chua C.K., Maria C. A deep learning quality control loop of the extrusion-based bioprinting process. Int J Bioprint. 2022;8:620. doi: 10.18063/ijb.v8i4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.K C., M L., D E., A P., G A. Improving build quality in laser powder bed fusion using high dynamic range imaging and model-based reinforcement learning. IEEE Access. 2021;9:55214–55231. [Google Scholar]
- 236.Deneault J.R., Chang J., Myung J., Hooper D., Armstrong A., Pitt M., Maruyama B. Toward autonomous additive manufacturing: Bayesian optimization on a 3D printer. MRS Bull. 2021;46:566–575. [Google Scholar]
- 237.Johnson M.V., Garanger K., Hardin J.O., Berrigan J.D., Feron E., Kalidindi S.R. A generalizable artificial intelligence tool for identification and correction of self-supporting structures in additive manufacturing processes. Addit. Manuf. 2021;46 [Google Scholar]
- 238.Druzgalski C.L., Ashby A., Guss G., King W.E., Roehling T.T., Matthews M.J. Process optimization of complex geometries using feed forward control for laser powder bed fusion additive manufacturing. Addit. Manuf. 2020;34 [Google Scholar]
- 239.Gillispie G., Prim P., Copus J., Fisher J., Mikos A.G., Yoo J.J., Atala A., Lee S.J. Assessment methodologies for extrusion-based bioink printability. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab6f0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Oh D., Shirzad M., Chang Kim M., Chung E., Nam S.Y. Rheology-informed hierarchical machine learning model for the prediction of printing resolution in extrusion-based bioprinting. Int. J. Bioprinting. 2023:1280. [Google Scholar]
- 241.Kim N., Lee H., Han G., Kang M., Park S., Kim D.E., Lee M., Kim M.J., Na Y., Oh S., Bang S.J., Jang T.S., Kim H.E., Park J., Shin S.R., Jung H.D. 3D‐Printed functional hydrogel by DNA‐induced biomineralization for accelerated diabetic wound healing. Adv. Sci. 2023;10:2300816. doi: 10.1002/advs.202300816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Reina-Romo E., Mandal S., Amorim P., Bloemen V., Ferraris E., Geris L. Towards the experimentally-informed in silico nozzle design optimization for extrusion-based bioprinting of shear-thinning hydrogels. Front. Bioeng. Biotechnol. 2021;9:701778. doi: 10.3389/fbioe.2021.701778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Conev A., Litsa E.E., Perez M.R., Diba M., Mikos A.G., Kavraki L.E. Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Tissue Eng. 2020;26:1359–1368. doi: 10.1089/ten.tea.2020.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fu Z., Angeline V., Sun W. Evaluation of printing parameters on 3D extrusion printing of pluronic hydrogels and machine learning guided parameter recommendation. Int J Bioprint. 2021;7:434. doi: 10.18063/ijb.v7i4.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tian S., Stevens R., McInnes B., Lewinski N. Machine assisted experimentation of extrusion-based bioprinting systems. Micromachines-Basel. 2021;12:780. doi: 10.3390/mi12070780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sedigh A., Ghelich P., Quint J., Mollocana Lara E.C., Samandari M., Tamayol A., Tomlinson R.E. Approximating scaffold printability utilizing computational methods. Biofabrication. 2023;15 doi: 10.1088/1758-5090/acbbf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sedigh A., DiPiero D., Shine K.M., Tomlinson R.E. Enhancing precision in bioprinting utilizing fuzzy systems. Bioprinting. 2022;25 [Google Scholar]
- 248.Ogunsanya M., Desai S. Physics-based and data-driven modeling for biomanufacturing 4.0. Manuf. Lett. 2023;36:91–95. [Google Scholar]
- 249.Madadian Bozorg N., Leclercq M., Lescot T., Bazin M., Gaudreault N., Dikpati A., Fortin M., Droit A., Bertrand N. Design of experiment and machine learning inform on the 3D printing of hydrogels for biomedical applications. Biomater. Adv. 2023;153 doi: 10.1016/j.bioadv.2023.213533. [DOI] [PubMed] [Google Scholar]
- 250.Limon S.M., Quigley C., Sarah R., Habib A. Advancing scaffold porosity through a machine learning framework in extrusion based 3D bioprinting. Front Mater. 2024;10:1337485. [Google Scholar]
- 251.Ege D., Sertturk S., Acarkan B., Ademoglu A. Machine learning models to predict the relationship between printing parameters and tensile strength of 3D Poly (lactic acid) scaffolds for tissue engineering applications. Biomed. Phys. Eng. Express. 2023;9 doi: 10.1088/2057-1976/acf581. [DOI] [PubMed] [Google Scholar]
- 252.Chen E.S., Ahmadianshalchi A., Sparks S.S., Chen C., Deshwal A., Doppa J.R., Qiu K. Adv. Mater. Technol.; 2024. Machine Learning Enabled Design and Optimization for 3D-Printing of High-Fidelity Presurgical Organ Models. [Google Scholar]
- 253.Arduengo J., Hascoet N., Chinesta F., Hascoet J.Y. Open-loop control system for high precision extrusion-based bioprinting through machine learning modeling. J. Machine Eng. 2024;24:103–117. [Google Scholar]
- 254.Zhang C., Elvitigala K.C.M.L., Mubarok W., Okano Y., Sakai S. Machine learning-based prediction and optimisation framework for as-extruded cell viability in extrusion-based 3D bioprinting. Virtual Phys. Prototyp. 2024;19 [Google Scholar]
- 255.Shi J., Wu B., Song B., Song J., Li S., Trau D., Lu W.F. Learning-based cell injection control for precise drop-on-demand cell printing. Ann. Biomed. Eng. 2018;46:1267–1279. doi: 10.1007/s10439-018-2054-2. [DOI] [PubMed] [Google Scholar]
- 256.Shi J., Song J., Song B., Lu W.F. Multi-objective optimization design through machine learning for drop-on-demand bioprinting. Engineering. 2019;5:586–593. [Google Scholar]
- 257.Wu D., Xu C. Predictive modeling of droplet formation processes in inkjet-based bioprinting. J. Manuf. Sci. Eng. 2018;140:101007. [Google Scholar]
- 258.Ball A.K., Das R., Roy S.S., Kisku D.R., Murmu N.C. Modeling of EHD inkjet printing performance using soft computing-based approaches. Soft Comput. 2020;24:571–589. [Google Scholar]
- 259.Wang J., Dong T., Cheng Y., Yan W. Machine learning assisted spraying pattern recognition for electrohydrodynamic atomization system. Ind. Eng. Chem. Res. 2022;61:8495–8503. [Google Scholar]
- 260.Dong T., Wang J., Wang Y., Tang G., Cheng Y., Yan W. Development of machine learning based droplet diameter prediction model for electrohydrodynamic atomization systems. Chem. Eng. Sci. 2023;268 [Google Scholar]
- 261.Kim S., Cho M., Jung S. The design of an inkjet drive waveform using machine learning. Sci. Rep. 2022;12:4841. doi: 10.1038/s41598-022-08784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shin J., Kang M., Hyun K., Li Z., Kumar H., Kim K., Park S.S., Kim K. Machine learning driven optimization for high precision cellular droplet bioprinting. bioRxiv. 2024 2024.09.04.611131. [Google Scholar]
- 263.S Z., S X., Z M., D X., X G., Y.W F. A learning-based framework for error compensation in 3D printing. IEEE T Cybernetics. 2019;49:4042–4050. doi: 10.1109/TCYB.2019.2898553. [DOI] [PubMed] [Google Scholar]
- 264.Z M., X G., S X., L C., S Z., W H. 2019 IEEE 15th International Conference on Automation Science and Engineering (CASE) 2019. Nonlinear deformation prediction and compensation for 3D printing based on CAE neural networks; pp. 667–672. [Google Scholar]
- 265.Shangting Y., Jiaao G., Shaochen C. Mitigating Scattering Effects in DMD-Based Microscale 3D Printing Using Machine Learning. Proc.SPIE. 2021;11698:1169804. [Google Scholar]
- 266.Xu H., Liu Q., Casillas J., Mcanally M., Mubtasim N., Gollahon L.S., Wu D., Xu C. Prediction of cell viability in dynamic optical projection stereolithography-based bioprinting using machine learning. J. Intell. Manuf. 2022;33:995–1005. [Google Scholar]
- 267.He H., Yang Y., Pan Y. Machine learning for continuous liquid interface production: printing speed modelling. J. Manuf. Syst. 2019;50:236–246. [Google Scholar]
- 268.Men L., Hu N., Deng Y., Zhang W., Yin R., Wen F., Zhao C., Chen Y. Automatic Quality Monitoring of Two-Photon Printed Devices Based on Deep Learning. SPIE. 2023;12709:1094–1100. [Google Scholar]
- 269.Jin Z., Zhang Z., Shao X., Gu G.X. Monitoring anomalies in 3D bioprinting with deep neural networks. ACS Biomater. Sci. Eng. 2021;9:3945–3952. doi: 10.1021/acsbiomaterials.0c01761. [DOI] [PubMed] [Google Scholar]
- 270.Choi E., An K., Kang K. Deep-learning-based microfluidic droplet classification for multijet monitoring. ACS APPL MATER INTER. 2022;14:15576–15586. doi: 10.1021/acsami.1c22048. [DOI] [PubMed] [Google Scholar]
- 271.Piovarči M., Foshey M., Xu J., Erps T., Babaei V., Didyk P., Rusinkiewicz S., Matusik W., Bickel B. Closed-loop control of direct ink writing via reinforcement learning. Acm T Graphic. 2022;41:1–10. [Google Scholar]
- 272.W D., S Z., D X., F Q., W W., D X., X G. 2023 IEEE 19th International Conference on Automation Science and Engineering (CASE) 2023. Deep reinforcement learning for dynamic error compensation in 3D printing; pp. 1–7. [Google Scholar]
- 273.Huang J., Segura L.J., Wang T., Zhao G., Sun H., Zhou C. Unsupervised learning for the droplet evolution prediction and process dynamics understanding in inkjet printing. Addit. Manuf. 2020;35 [Google Scholar]
- 274.Segura L.J., Li Z., Zhou C., Sun H. Droplet evolution prediction in material jetting via tensor time series analysis. Addit. Manuf. 2023;66 [Google Scholar]
- 275.Armstrong A.A., Alleyne A.G., Wagoner J.A. 1D and 2D error assessment and correction for extrusion-based bioprinting using process sensing and control strategies. Biofabrication. 2020;12 doi: 10.1088/1758-5090/aba8ee. [DOI] [PubMed] [Google Scholar]
- 276.Yang S., Chen Q., Wang L., Xu M. In situ defect detection and feedback control with three-dimensional extrusion-based bioprinter-associated optical coherence tomography. Int. J. Bioprinting. 2022;9:624. doi: 10.18063/ijb.v9i1.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Tashman J.W., Shiwarski D.J., Coffin B., Ruesch A., Lanni F., Kainerstorfer J.M., Feinberg A.W. In situ volumetric imaging and analysis of FRESH 3D bioprinted constructs using optical coherence tomography. Biofabrication. 2022;15 doi: 10.1088/1758-5090/ac975e. [DOI] [PubMed] [Google Scholar]
- 278.Yang S., Wang L., Chen Q., Xu M. In situ process monitoring and automated multi-parameter evaluation using optical coherence tomography during extrusion-based bioprinting. Addit. Manuf. 2021;47 [Google Scholar]
- 279.Wang J., Xu C., Yang S., Wang L., Xu M. Continuous and highly accurate multi-material extrusion-based bioprinting with optical coherence tomography imaging. Int. J. Bioprinting. 2023;9:707. doi: 10.18063/ijb.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Snow Z., Scime L., Ziabari A., Fisher B., Paquit V. Scalable in situ non-destructive evaluation of additively manufactured components using process monitoring, sensor fusion, and machine learning. Addit. Manuf. 2023;78 [Google Scholar]
- 281.Poologasundarampillai G., Haweet A., Jayash S.N., Morgan G., Moore J.E., Candeo A. Real-time imaging and analysis of cell-hydrogel interplay within an extrusion-bioprinting capillary. Bioprinting. 2021;23 [Google Scholar]
- 282.Haring A.P., Jiang S., Barron C., Thompson E.G., Sontheimer H., He J., Jia X., Johnson B.N. 3D bioprinting using hollow multifunctional fiber impedimetric sensors. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab94d0. [DOI] [PubMed] [Google Scholar]
- 283.C K., S X., L C., S Z., G H., X G. 2019 IEEE International Conference on Service Operations and Logistics, and Informatics (SOLI) 2019. A kind of accuracy improving method based on error analysis and feedback for DLP 3D printing; pp. 5–9. [Google Scholar]
- 284.Westphal E., Seitz H. Machine learning for the intelligent analysis of 3D printing conditions using environmental sensor data to support quality assurance. Addit. Manuf. 2022;50 [Google Scholar]
- 285.Zhang J., Wang P., Gao R.X. Deep learning-based tensile strength prediction in fused deposition modeling. Comput. Ind. 2019;107:11–21. [Google Scholar]
- 286.Ng W.L., Goh G.L., Goh G.D., Ten J.S.J., Yeong W.Y. Progress and opportunities for machine learning in materials and processes of additive manufacturing. Adv. Mater. 2024 doi: 10.1002/adma.202310006. [DOI] [PubMed] [Google Scholar]
- 287.Fortunato G.M., Rossi G., Bonatti A.F., De Acutis A., Mendoza-Buenrostro C., Vozzi G., De Maria C. Robotic platform and path planning algorithm for in situ bioprinting. Bioprinting. 2021;22 [Google Scholar]
- 288.Fortunato G.M., Batoni E., Bonatti A.F., Vozzi G., De Maria C. Surface reconstruction and tissue recognition for robotic-based in situ bioprinting. Bioprinting. 2022;26 [Google Scholar]
- 289.Zhao W., Chen H., Zhang Y., Zhou D., Liang L., Liu B., Xu T. Adaptive multi-degree-of-freedom in situ bioprinting robot for hair-follicle-inclusive skin repair: a preliminary study conducted in mice. Bioeng. Transl. Med. 2022;7 doi: 10.1002/btm2.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ma K., Zhao T., Yang L., Wang P., Jin J., Teng H., Xia D., Zhu L., Li L., Jiang Q., Wang X. Application of robotic-assisted in situ 3D printing in cartilage regeneration with HAMA hydrogel: an in vivo study. J. Adv. Res. 2020;23:123–132. doi: 10.1016/j.jare.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zhang Z., Wu C., Dai C., Shi Q., Fang G., Xie D., Zhao X., Liu Y., Wang C.C.L., Wang X. A multi-axis robot-based bioprinting system supporting natural cell function preservation and cardiac tissue fabrication. Bioact. Mater. 2022;18:138–150. doi: 10.1016/j.bioactmat.2022.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhu Z., Park H.S., McAlpine M.C. 3D printed deformable sensors. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Weng J., Bhupathiraju S.H.V., Samant T., Dresner A., Wu J., Samant S.S. Convolutional LSTM model for cine image prediction of abdominal motion. Phys. Med. Biol. 2024;69 doi: 10.1088/1361-6560/ad3722. [DOI] [PubMed] [Google Scholar]
- 294.Lombardo E., Rabe M., Xiong Y., Nierer L., Cusumano D., Placidi L., Boldrini L., Corradini S., Niyazi M., Reiner M., Belka C., Kurz C., Riboldi M., Landry G. Evaluation of real-time tumor contour prediction using LSTM networks for MR-guided radiotherapy. Radiother. Oncol. 2023;182 doi: 10.1016/j.radonc.2023.109555. [DOI] [PubMed] [Google Scholar]
- 295.Vázquez Romaguera L., Alley S., Carrier J., Kadoury S. Conditional-based transformer network with learnable queries for 4D deformation forecasting and tracking. IEEE T Med Imaging. 2023;42:1603–1618. doi: 10.1109/TMI.2023.3234046. [DOI] [PubMed] [Google Scholar]
- 296.Bengs M., Sprenger J., Gerlach S., Neidhardt M., Schlaefer A. Real-time motion analysis with 4D deep learning for ultrasound-guided radiotherapy. IEEE T Bio-Med Eng. 2023;70:2690–2699. doi: 10.1109/TBME.2023.3262422. [DOI] [PubMed] [Google Scholar]
- 297.Thai M.T., Phan P.T., Tran H.A., Nguyen C.C., Hoang T.T., Davies J., Rnjak Kovacina J., Phan H.P., Lovell N.H., Do T.N. Advanced soft robotic system for in situ 3D bioprinting and endoscopic surgery. Adv. Sci. 2023;10:2205656. doi: 10.1002/advs.202205656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ou Y., Tavakoli M. Sim-to-Real surgical robot learning and autonomous planning for internal tissue points manipulation using reinforcement learning. IEEE Rob. Autom. Lett. 2023;8:2502–2509. [Google Scholar]
- 299.Shahkoo A.A., Abin A.A. Autonomous tissue manipulation via surgical robot using deep reinforcement learning and evolutionary algorithm. IEEE Trans. Med. Robot. Bionics. 2023;5:30–41. [Google Scholar]
- 300.Iyengar K., Spurgeon S., Stoyanov D. Deep reinforcement learning for concentric tube robot path following. IEEE Trans.Med. Robot. bionics. 2024;6 1-1. [Google Scholar]
- 301.Shahkoo A.A., Abin A.A. Deep reinforcement learning in continuous action space for autonomous robotic surgery. Int. J. Comput. Assist. Radiol. Surg. 2023;18:423–431. doi: 10.1007/s11548-022-02789-8. [DOI] [PubMed] [Google Scholar]
- 302.Bonatti A.F., Vozzi G., Kai Chua C., De Maria C. A deep learning approach for error detection and quantification in extrusion-based bioprinting. Mater. Today Proc. 2022;70:131–135. [Google Scholar]
- 303.Gerdes S., Gaikwad A., Ramesh S., Rivero I.V., Tamayol A., Rao P. Monitoring and control of biological additive manufacturing using machine learning. J. Intell. Manuf. 2023;35:1055–1077. [Google Scholar]
- 304.Huang X., Ng W.L., Yeong W.Y. Predicting the number of printed cells during inkjet-based bioprinting process based on droplet velocity profile using machine learning approaches. J. Intell. Manuf. 2023;35:2349–2364. [Google Scholar]
- 305.Ogunsanya M., Isichei J., Parupelli S.K., Desai S., Cai Y. In-situ droplet monitoring of inkjet 3D printing process using image analysis and machine learning models. Procedia Manuf. 2021;53:427–434. [Google Scholar]
- 306.Phung T.H., Park S.H., Kim I., Lee T., Kwon K. Machine learning approach to monitor inkjet jetting status based on the piezo self-sensing. Sci. Rep. 2023;13:18089. doi: 10.1038/s41598-023-45445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wang G., Yuan Z., Yu L., Yu Y., Zhou P., Chu G., Wang H., Guo Q., Zhu C., Han F., Chen S., Li B. Mechanically conditioned cell sheets cultured on thermo-responsive surfaces promote bone regeneration. Biomater Transl. 2023;4:27–40. doi: 10.12336/biomatertransl.2023.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Bonnevie E.D., Ashinsky B.G., Dekky B., Volk S.W., Smith H.E., Mauck R.L. Cell morphology and mechanosensing can be decoupled in fibrous microenvironments and identified using artificial neural networks. Sci. Rep. 2021;11:5950. doi: 10.1038/s41598-021-85276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tang M., Jiang S., Huang X., Ji C., Gu Y., Qi Y., Xiang Y., Yao E., Zhang N., Berman E., Yu D., Qu Y., Liu L., Berry D., Yao Y. Integration of 3D bioprinting and multi-algorithm machine learning identified glioma susceptibilities and microenvironment characteristics. Cell Discov. 2024;10:39. doi: 10.1038/s41421-024-00650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yao K., Huang K., Sun J., Jing L., Huang D., Jude C. Scaffold-A549: a benchmark 3D fluorescence image dataset for unsupervised nuclei segmentation. Cogn Comput. 2021;13:1603–1608. [Google Scholar]
- 311.Phillip J.M., Han K., Chen W., Wirtz D., Wu P. A robust unsupervised machine-learning method to quantify the morphological heterogeneity of cells and nuclei. Nat. Protoc. 2021;16:754–774. doi: 10.1038/s41596-020-00432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Wang W., Vikesland P.J. SERS-active printable hydrogel for 3D cell culture and imaging. Anal. Chem. 2023;95:18055–18064. doi: 10.1021/acs.analchem.3c02641. [DOI] [PubMed] [Google Scholar]
- 313.Safir F., Vu N., Tadesse L.F., Firouzi K., Banaei N., Jeffrey S.S., Saleh A.A.E., Khuri-Yakub B.P.T., Dionne J.A. Combining acoustic bioprinting with AI-assisted Raman spectroscopy for high-throughput identification of bacteria in blood. Nano Lett. 2023;23:2065–2073. doi: 10.1021/acs.nanolett.2c03015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Shohan S., Hasan M., Starly B., Shirwaiker R. Investigating autoregressive and machine learning-based time series modeling with dielectric spectroscopy for predicting quality of biofabricated constructs. Manuf. Lett. 2022;33:902–908. [Google Scholar]
- 315.Shohan S., Harm J., Hasan M., Starly B., Shirwaiker R. Non-destructive quality monitoring of 3D printed tissue scaffolds via dielectric impedance spectroscopy and supervised machine learning. Procedia Manuf. 2021;53:636–643. [Google Scholar]
- 316.Bao D., Wang L., Zhou X., Yang S., He K., Xu M. Automated detection and growth tracking of 3D bio-printed organoid clusters using optical coherence tomography with deep convolutional neural networks. Front. Bioeng. Biotechnol. 2023;11:1133090. doi: 10.3389/fbioe.2023.1133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Tebon P.J., Wang B., Markowitz A.L., Davarifar A., Tsai B.L., Krawczuk P., Gonzalez A.E., Sartini S., Murray G.F., Nguyen H.T.L., Tavanaie N., Nguyen T.L., Boutros P.C., Teitell M.A., Soragni A. Drug screening at single-organoid resolution via bioprinting and interferometry. Nat. Commun. 2023;14:3168. doi: 10.1038/s41467-023-38832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tröndle K., Miotto G., Rizzo L., Pichler R., Koch F., Koltay P., Zengerle R., Lienkamp S.S., Kartmann S., Zimmermann S. Deep learning-assisted nephrotoxicity testing with bioprinted renal spheroids. Int. J. Bioprinting. 2022;8:528. doi: 10.18063/ijb.v8i2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Benning L., Peintner A., Finkenzeller G., Peintner L. Automated spheroid generation, drug application and efficacy screening using a deep learning classification: a feasibility study. Sci. Rep. 2020;10:11071. doi: 10.1038/s41598-020-67960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Bertsimas D., Kung J., Trichakis N., Wang Y., Hirose R., Vagefi P.A. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am. J. Transplant. 2019;19:1109–1118. doi: 10.1111/ajt.15172. [DOI] [PubMed] [Google Scholar]
- 321.Liu C., Soong R., Lee W., Jiang G., Lin Y. Predicting short-term survival after liver transplantation using machine learning. Sci. Rep. 2020;10:5654. doi: 10.1038/s41598-020-62387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kantidakis G., Putter H., Lancia C., Boer J.D., Braat A.E., Fiocco M. Survival prediction models since liver transplantation - comparisons between Cox models and machine learning techniques. BMC Med. Res. Methodol. 2020;20:277. doi: 10.1186/s12874-020-01153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Briceno J., Cruz-Ramirez M., Prieto M., Navasa M., Ortiz D.U.J., Orti R., Gomez-Bravo M.A., Otero A., Varo E., Tome S., Clemente G., Banares R., Barcena R., Cuervas-Mons V., Solorzano G., Vinaixa C., Rubin A., Colmenero J., Valdivieso A., Ciria R., Hervas-Martinez C., de la Mata M. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: results from a multicenter Spanish study. J. Hepatol. 2014;61:1020–1028. doi: 10.1016/j.jhep.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 324.Baltruschat I.M., Ćwieka H., Krüger D., Zeller-Plumhoff B., Schlünzen F., Willumeit-Römer R., Moosmann J., Heuser P. Scaling the U-net: segmentation of biodegradable bone implants in high-resolution synchrotron radiation microtomograms. Sci. Rep. 2021;11:24237. doi: 10.1038/s41598-021-03542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Spanoudaki V., Doloff J.C., Huang W., Norcross S.R., Farah S., Langer R., Anderson D.G. Simultaneous spatiotemporal tracking and oxygen sensing of transient implants in vivo using hot-spot MRI and machine learning. Proc. Natl. Acad. Sci. USA. 2019;116:4861–4870. doi: 10.1073/pnas.1815909116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Gao Q., Lee J., Kim B.S., Gao G. Three-dimensional printing of smart constructs using stimuli-responsive biomaterials: a future direction of precision medicine. Int. J. Bioprinting. 2022;9:638. doi: 10.18063/ijb.v9i1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Xu Z., Fan J., Tian W., Ji X., Cui Y., Nan Q., Sun F., Zhang J. Cellulose‐based pH‐responsive janus dressing with unidirectional moisture drainage for exudate management and diabetic wounds healing. Adv. Funct. Mater. 2023;34:2307449. [Google Scholar]
- 328.Wang L., Zhou M., Xu T., Zhang X. Multifunctional hydrogel as wound dressing for intelligent wound monitoring. Chem. Eng. J. 2022;433 [Google Scholar]
- 329.Zheng X.T., Yang Z., Sutarlie L., Thangaveloo M., Yu Y., Salleh N.A.B.M., Chin J.S., Xiong Z., Becker D.L., Loh X.J., Tee B.C.K., Su X. Battery-free and AI-enabled multiplexed sensor patches for wound monitoring. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adg6670. eadg6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Jain S., Pei L., Spraggins J.M., Angelo M., Carson J.P., Gehlenborg N., Ginty F., Gonçalves J.P., Hagood J.S., Hickey J.W., Kelleher N.L., Laurent L.C., Lin S., Lin Y., Liu H., Naba A., Nakayasu E.S., Qian W., Radtke A., Robson P., Stockwell B.R., Van de Plas R., Vlachos I.S., Zhou M., Börner K., Snyder M.P., HuBMAP C. Advances and prospects for the human BioMolecular atlas Program (HuBMAP) Nat. Cell Biol. 2023;25:1089–1100. doi: 10.1038/s41556-023-01194-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Hickey J.W., Becker W.R., Nevins S.A., Horning A., Perez A.E., Zhu C., Zhu B., Wei B., Chiu R., Chen D.C., Cotter D.L., Esplin E.D., Weimer A.K., Caraccio C., Venkataraaman V., Schurch C.M., Black S., Brbic M., Cao K., Chen S., Zhang W., Monte E., Zhang N.R., Ma Z., Leskovec J., Zhang Z., Lin S., Longacre T., Plevritis S.K., Lin Y., Nolan G.P., Greenleaf W.J., Snyder M. Organization of the human intestine at single-cell resolution. Nature. 2023;619:572–584. doi: 10.1038/s41586-023-05915-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Lake B.B., Menon R., Winfree S., Hu Q., Melo Ferreira R., Kalhor K., Barwinska D., Otto E.A., Ferkowicz M., Diep D., Plongthongkum N., Knoten A., Urata S., Mariani L.H., Naik A.S., Eddy S., Zhang B., Wu Y., Salamon D., Williams J.C., Wang X., Balderrama K.S., Hoover P.J., Murray E., Marshall J.L., Noel T., Vijayan A., Hartman A., Chen F., Waikar S.S., Rosas S.E., Wilson F.P., Palevsky P.M., Kiryluk K., Sedor J.R., Toto R.D., Parikh C.R., Kim E.H., Satija R., Greka A., Macosko E.Z., Kharchenko P.V., Gaut J.P., Hodgin J.B., Knight R., Lecker S.H., Stillman I., Amodu A.A., Ilori T., Maikhor S., Schmidt I., McMahon G.M., Weins A., Hacohen N., Bush L., Gonzalez-Vicente A., Taliercio J., O Toole J., Poggio E., Cooperman L., Jolly S., Herlitz L., Nguyen J., Palmer E., Sendrey D., Spates-Harden K., Appelbaum P., Barasch J.M., Bomback A.S., D Agati V.D., Mehl K., Canetta P.A., Shang N., Balderes O., Kudose S., Barisoni L., Alexandrov T., Cheng Y., Dunn K.W., Kelly K.J., Sutton T.A., Wen Y., Corona-Villalobos C.P., Menez S., Rosenberg A., Atta M., Johansen C., Sun J., Roy N., Williams M., Azeloglu E.U., He C., Iyengar R., Hansen J., Xiong Y., Rovin B., Parikh S., Madhavan S.M., Anderton C.R., Pasa-Tolic L., Velickovic D., Troyanskaya O., Sealfon R., Tuttle K.R., Laszik Z.G., Nolan G., Sarwal M., Anjani K., Sigdel T., Ascani H., Balis U.G.J., Lienczewski C., Steck B., He Y., Schaub J., Blanc V.M., Murugan R., Randhawa P., Rosengart M., Tublin M., Vita T., Kellum J.A., Hall D.E., Elder M.M., Winters J., Gilliam M., Alpers C.E., Blank K.N., Carson J., De Boer I.H., Dighe A.L., Himmelfarb J., Mooney S.D., Shankland S., Williams K., Park C., Dowd F., McClelland R.L., Daniel S., Hoofnagle A.N., Wilcox A., Bansal S., Sharma K., Venkatachalam M., Zhang G., Pamreddy A., Kakade V.R., Moledina D., Shaw M.M., Ugwuowo U., Arora T., Ardayfio J., Bebiak J., Brown K., Campbell C.E., Saul J., Shpigel A., Stutzke C., Koewler R., Campbell T., Hayashi L., Jefferson N., Pinkeney R., Roberts G.V., Eadon M.T., Dagher P.C., El-Achkar T.M., Zhang K., Kretzler M., Jain S. An atlas of healthy and injured cell states and niches in the human kidney. Nature. 2023;619:585–594. doi: 10.1038/s41586-023-05769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Greenbaum S., Averbukh I., Soon E., Rizzuto G., Baranski A., Greenwald N.F., Kagel A., Bosse M., Jaswa E.G., Khair Z., Kwok S., Warshawsky S., Piyadasa H., Goldston M., Spence A., Miller G., Schwartz M., Graf W., Van Valen D., Winn V.D., Hollmann T., Keren L., van de Rijn M., Angelo M. A spatially resolved timeline of the human maternal-fetal interface. Nature. 2023;619:595–605. doi: 10.1038/s41586-023-06298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Furuta T., Yamauchi K., Okamoto S., Takahashi M., Kakuta S., Ishida Y., Takenaka A., Yoshida A., Uchiyama Y., Koike M., Isa K., Isa T., Hioki H. Multi-scale light microscopy/electron microscopy neuronal imaging from brain to synapse with a tissue clearing method, ScaleSF. iScience. 2022;25 doi: 10.1016/j.isci.2021.103601. 103601-103601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ngo T.B., DeStefano S., Liu J., Su Y., Shroff H., Vishwasrao H.D., Sadtler K. Label‐free cleared tissue microscopy and machine learning for 3D histopathology of biomaterial implants. J. Biomed. Mater. Res. A. 2023;111:840–850. doi: 10.1002/jbm.a.37515. [DOI] [PubMed] [Google Scholar]
- 336.Sedigh A., Tulipan J.E., Rivlin M.R., Tomlinson R.E. Utilizing Q-learning to generate 3D vascular networks for bioprinting bone. bioRxiv. 2020 [Google Scholar]
- 337.Wolterink J.M., Leiner T., Isgum I. Blood Vessel Geometry Synthesis Using Generative Adversarial Networks. arXiv.org. 2018:1804.04381. [Google Scholar]
- 338.Simões J.B., Travasso R., Costa E., Baptista T. In: Generating Vascular Networks: A Reinforcement Learning Approach. Nicosia G., Ojha V., La Malfa E., La Malfa G., Pardalos P., Di Fatta G., Giuffrida G., Umeton R., editors. Springer Nature Switzerland; Cham: 2023. pp. 139–153. [Google Scholar]
- 339.Chen A., Wang W., Mao Z., He Y., Chen S., Liu G., Su J., Feng P., Shi Y., Yan C., Lu J. Multimaterial 3D and 4D bioprinting of heterogenous constructs for tissue engineering. Adv. Mater. 2023;36:2307686. doi: 10.1002/adma.202307686. [DOI] [PubMed] [Google Scholar]
- 340.Zhou H., Liu P., Gao Z., Li Q., Lv W., Yin J., Zhang B., Yang H., Ma L. Simultaneous multimaterial multimethod bioprinting. Bio-Design Manuf. 2022;5:433–436. [Google Scholar]
- 341.Dalton P.D., Woodfield T.B.F., Mironov V., Groll J. Advances in hybrid fabrication toward hierarchical tissue constructs. Adv. Sci. 2020;7 doi: 10.1002/advs.201902953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Noofa S.H., Zainab N.K., Alexander U.V., Charlotte H. A predictive machine learning model to optimize flow rates on an integrated microfluidic pumping system for peptide-based 3D bioprinting. Proc. SPIE. 2023;12374:1237402. [Google Scholar]
- 343.M Z., J S., S S. 2024 IEEE International Conference on Robotics and Automation (ICRA) 2024. RoCo: dialectic multi-robot collaboration with large language models; pp. 286–299. [Google Scholar]
- 344.Li T., Xie F., Zhao Z., Zhao H., Guo X., Feng Q. A multi-arm robot system for efficient apple harvesting: perception, task plan and control. Comput Electron Agr. 2023;211 [Google Scholar]
- 345.Abbas M., Narayan J., Dwivedy S.K. A systematic review on cooperative dual-arm manipulators: modeling, planning, control, and vision strategies. Int. J. Intell. Robot Appl. 2023;7:683–707. [Google Scholar]
- 346.Ramprasad R., Batra R., Pilania G., Mannodi-Kanakkithodi A., Kim C. Machine learning in materials informatics: recent applications and prospects. npj Comput. Mater. 2017;3:54. [Google Scholar]
- 347.Lookman T., Balachandran P.V., Xue D., Yuan R. Active learning in materials science with emphasis on adaptive sampling using uncertainties for targeted design, npj Comput. Mater. 2019;5:21. [Google Scholar]
- 348.Jablonka K.M., Jothiappan G.M., Wang S., Smit B., Yoo B. Bias free multiobjective active learning for materials design and discovery. Nat. Commun. 2021;12:2312. doi: 10.1038/s41467-021-22437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Peng J., Schwalbe-Koda D., Akkiraju K., Xie T., Giordano L., Yu Y., Eom C.J., Lunger J.R., Zheng D.J., Rao R.R., Muy S., Grossman J.C., Reuter K., Gómez-Bombarelli R., Shao-Horn Y. Human- and machine-centred designs of molecules and materials for sustainability and decarbonization. Nat. Rev. Mater. 2022;7:991–1009. [Google Scholar]
- 350.Jha D., Choudhary K., Tavazza F., Liao W., Choudhary A., Campbell C., Agrawal A. Enhancing materials property prediction by leveraging computational and experimental data using deep transfer learning. Nat. Commun. 2019;10:5316. doi: 10.1038/s41467-019-13297-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Gupta V., Choudhary K., Tavazza F., Campbell C., Liao W., Choudhary A., Agrawal A. Cross-property deep transfer learning framework for enhanced predictive analytics on small materials data. Nat. Commun. 2021;12:6595. doi: 10.1038/s41467-021-26921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Bone J.M. Carnegie Mellon University; 2021. Machine Learning Tools For Smarter Automation And Diagnostics In The Development Of Personalized Medicine From Size-Limited Datasets. [Google Scholar]
- 353.Kanarik K.J., Osowiecki W.T., Lu Y., Talukder D., Roschewsky N., Park S.N., Kamon M., Fried D.M., Gottscho R.A. Human–machine collaboration for improving semiconductor process development. Nature. 2023;616:707–711. doi: 10.1038/s41586-023-05773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Biswas A., Liu Y., Creange N., Liu Y., Jesse S., Yang J., Kalinin S.V., Ziatdinov M.A., Vasudevan R.K. A dynamic Bayesian optimized active recommender system for curiosity-driven partially Human-in-the-loop automated experiments. npj Comput. Mater. 2024;10:29. [Google Scholar]
- 355.Karniadakis G.E., Kevrekidis I.G., Lu L., Perdikaris P., Wang S., Yang L. Physics-informed machine learning. Nature Rev.Phys. 2021;3:422–440. [Google Scholar]
- 356.Xie Y., Sattari K., Zhang C., Lin J. Toward autonomous laboratories: convergence of artificial intelligence and experimental automation. Prog. Mater. Sci. 2023;132 [Google Scholar]
- 357.Coley C.W., Thomas D.A., Lummiss J.A.M., Jaworski J.N., Breen C.P., Schultz V., Hart T., Fishman J.S., Rogers L., Gao H., Hicklin R.W., Plehiers P.P., Byington J., Piotti J.S., Green W.H., Hart A.J., Jamison T.F., Jensen K.F. A robotic platform for flow synthesis of organic compounds informed by AI planning. Science. 2019;365 doi: 10.1126/science.aax1566. [DOI] [PubMed] [Google Scholar]
- 358.Steiner S., Wolf J., Glatzel S., Andreou A., Granda J.M., Keenan G., Hinkley T., Aragon-Camarasa G., Kitson P.J., Angelone D., Cronin L. Organic synthesis in a modular robotic system driven by a chemical programming language. Science. 2019;363 doi: 10.1126/science.aav2211. [DOI] [PubMed] [Google Scholar]
- 359.Szymanski N.J., Rendy B., Fei Y., Kumar R.E., He T., Milsted D., McDermott M.J., Gallant M., Cubuk E.D., Merchant A., Kim H., Jain A., Bartel C.J., Persson K., Zeng Y., Ceder G. An autonomous laboratory for the accelerated synthesis of novel materials. Nature. 2023;624:86–91. doi: 10.1038/s41586-023-06734-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Burger B., Maffettone P.M., Gusev V.V., Aitchison C.M., Bai Y., Wang X., Li X., Alston B.M., Li B., Clowes R., Rankin N., Harris B., Sprick R.S., Cooper A.I. A mobile robotic chemist. Nature. 2020;583:237–241. doi: 10.1038/s41586-020-2442-2. [DOI] [PubMed] [Google Scholar]
- 361.Ahmadi M., Ziatdinov M., Zhou Y., Lass E.A., Kalinin S.V. Machine learning for high-throughput experimental exploration of metal halide perovskites. Joule. 2021;5:2797–2822. [Google Scholar]
- 362.MacLeod B.P., Parlane F., Brown A.K., Hein J.E., Berlinguette C.P. Flexible automation accelerates materials discovery. Nat. Mater. 2022;21:722–726. doi: 10.1038/s41563-021-01156-3. [DOI] [PubMed] [Google Scholar]
- 363.Lee N.A., Shen S.C., Buehler M.J. An automated biomateriomics platform for sustainable programmable materials discovery. Matter. 2022;5:3597–3613. doi: 10.1016/j.matt.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Zhang Z., Gu G.X. Finite-element-based deep-learning model for deformation behavior of digital materials. Adv.Theory Simul. 2020;3 [Google Scholar]
- 365.Wang J., Zhang Y., Zheng J., Zhao X., Guo H., Qiu Y., Wang X., Liu L., Yu H. A 4D‐printing inverse design strategy for micromachines with customized shape‐morphing. Small. 2023;19:2302656. doi: 10.1002/smll.202302656. [DOI] [PubMed] [Google Scholar]
- 366.Du C., Zhuang J., Huang X. Deep learning technology in vascular image segmentation and disease diagnosis. J. Intell.Med. 2024 [Google Scholar]
- 367.Trotsyuk A.A., Federico C.A., Cho M.K., Altman R.B., Magnus D. Stronger regulation of AI in biomedicine. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.adi0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Mittal S., Thakral K., Singh R., Vatsa M., Glaser T., Canton Ferrer C., Hassner T. On responsible machine learning datasets emphasizing fairness, privacy and regulatory norms with examples in biometrics and healthcare. Nat. Mach. Intell. 2024;6:936–949. [Google Scholar]