Abstract
Carbon nanomaterials (CNMs) are a heterogeneous class of advanced materials. Their widespread use is associated with human safety concerns, which can be addressed by safe-by design strategies. This implies a deep knowledge of how physico-chemical properties drive biological effects. The ability of CNMs to interact with cytochrome c (cyt c), a heme-protein playing a key role in the respiratory chain, in apoptosis and in cellular redox homeostasis, has been reported in some studies. However, the consequences of this interaction on the cyt c functions are controversial. Here the mechanism of interaction of carbon nanoparticles (CNPs), chosen as model of redox-active CNMs, with cyt c has been studied with the aim to shed light into these discrepancies. The effect of CNPs on the redox state of cyt c was monitored by UV–vis spectroscopy and 1D 1H NMR, while the effect on the primary, secondary, and tertiary cyt c structure was investigated by FIA/LC-MS and Circular Dichroism (CD). Finally, the peroxidase activity of cyt c and the involvement of superoxide radicals was evaluated by EPR spectroscopy. We demonstrate the existence of two mechanisms, one leading to the suppression of the cyt c peroxidase activity following the NADH-independent reduction of the heme-iron, and the other resulting in the irreversible protein unfolding. Overall, the results suggest that these two processes might be independently modulated by redox and surface properties respectively.
Keywords: Carbon nanomaterials, Cytochrome c, Peroxidase activity, Heme iron, Electron transfer, Conformational changes
Graphical abstract
1. Introduction
Carbon nanomaterials (CNMs) are a wide family of advanced materials that find application in several industrial fields. Graphene, graphene oxide (GO), single walled-carbon nanotubes (SWCNT) are the most studied, but other emerging materials like carbon dots [1] and hydrothermal carbon (HTC) nanoparticles attract increasing interest in different fields [2]. As consequence, accidental exposure to CNM is likely, either in the environment or in occupational settings [3].
CNMs are also extensively studied in medicine not only as versatile carriers of drugs but also as antimicrobial or antioxidant agents, and in photodynamic therapy [4]. Nevertheless, few CNMs have reached the clinical trials, and none is currently in the market [5]. Like for other nanomaterials (NMs), significant challenges in terms of biocompatibility, stability and toxicity hamper the translation to clinics. Moreover, due to the poor understanding of the interaction of NMs with living systems at a molecular level, Safe by Design (SbD) approaches cannot be applied, and therefore expensive and time-consuming case-by-case approaches are necessary [6]. This is particularly relevant for CNMs, for which several different adverse pathways have been described [7].
Properties like shape, size, presence of impurities, structural defects, surface modifications and surface reactivity are known to affect the biological activity of CNMs [7]. Among them, redox activity is particularly relevant under a toxicological and medical point of view [8].
CNMs have been shown to be able to interact with the cellular redox system in different ways [8]. For example, fullerenes, carbon dots, graphene carbon dots are able to release Reactive Oxygen Species (ROS) when activated by light [9]. For this reason, they have been proposed as active agents in photodynamic therapy of cancer and infectious diseases. In other cases, oxidant activity is observed without light activation. For example, the ability of GO to release ROS or to reduce the antioxidant defences by oxidizing glutathione has been reported [8]. The latter reaction has been demonstrated also for carbon black, multiwalled carbon nanotubes (MWCNT) [10] and carbon nanoparticles. On the other hand, fullerenes and other CNMs act as antioxidant agents due to their ability to scavenge ROS [11,12]. Furthermore, some CNM have been reported to be antioxidant agents through a peroxidase-like activity [8].
An alternative pathway with potential to induce adverse effects to cells is the interaction of NMs with redox active enzymes involved in the maintenance of the cellular redox equilibrium like cytochrome c (cyt c) [13]. Cyt c is a small structural flexible globular protein composed by a single polypeptide chain of about 100 amino acid residues and heme-iron (III) as prosthetic group. It is a multifunctional protein involved in cellular redox homeostasis maintenance, in the respiratory chain, and in the intrinsic apoptotic pathway [14].
There is evidence that some CNMs can interact with cyt c altering its biological functions. An early study by Ma and co-workers [15] reported SWCNT to be able to induce an increase of cyt c redox potential thus facilitating the reduction to Fe(II) cyt c. This interaction was claimed to induce mitochondrial dysfunction that turned in metabolic hypoxia and cell death [15]. Other studies reported the ability of CNMs to increase the peroxidase activity of cyt c [16,17]. In other cases, CNMs were shown to interact with this enzyme without any alteration of its activity [16,17]. These contrasting data suggest that the interaction might be modulated by the physico-chemical properties of CNMs. Unfortunately, the poor knowledge on the mechanism of interaction prevents the definition of clear correlations between each property and the effect.
With the aim to contribute to fill this gap, hydrothermal carbon nanoparticles (CNPs) chosen as model of redox-active CNMs were synthetized and tested for their ability to interact with cyt c. The mechanism of the interaction was studied by UV–vis spectrophotometry, Circular Dichroism (CD), 1D 1H NMR and Electron Paramagnetic Resonance (EPR) spectroscopy and Mass Spectrometry (MS).
2. Materials and methods
2.1. Materials and reagents
Carbon black (CB1) was obtained by oxidation with H2SO4/H2O2 of activated charcoal (Sigma-Aldrich, Germany). Carbon black (CB2) was a kind gift of Dr. Michela Alfé (Istituto di Ricerche sulla Combustione – CNR, Naples, Italy). D-(+)-glucose, Phosphate Buffer Saline (PBS), bovine cytochrome c Fe(III) from bovine heart, sodium polyacrylate, xanthine and xanthine oxidase were purchased from Merck (Darmstadt, Germany). DEPMPO were purchased by ENZO Lifesciences (20016 Pero (MI) Italy). In all experiments, fresh ultrapure water was used (Milli Q Plus system -Millipore, Bedford, MA, USA).
2.2. Synthesis of carbon nanoparticles
Synthesis of carbon nanoparticles was achieved by using hydrothermal carbonization method previously described [18]. Briefly, 5 g of glucose were dissolved in 50 mL of Milli-Q water and solubilized using a magnetic stirrer, sodium polyacrylate (15 mg) was then added in order to prevent the typical cross-linking of the nanoparticles during the synthesis. The solution was transferred in a Teflon-lined stainless-steel autoclave (100 ml, Büchi AG), and placed in a preheated oven at 190 °C for 8 h. After the synthesis, the nanoparticles suspension was concentrated, purified with Milli-Q water by ultrafiltration using Hydrosart® Vivaflow 50R cassettes (Sartorius, 30 kDa cut-off). The concentration of carbon nanoparticles (34.8 mg/mL) was determined gravimetrically. The stock suspension was centrifuged at 10000g for 15 min, the solvent removed and replaced with 0.01 M PBS, the procedure was repeated three times. A final CNP working suspension at 10 mg/mL was prepared in PBS.
2.3. Characterization of carbon nanoparticles
The mean hydrodynamic diameter and size distribution analysis were performed by the ZetaView® PMX-120 (Particle Metrix GmbH, Germany) nanoparticle tracking analyser (NTA), equipped with a light source set to a wavelength of 488 nm. After the instrumental parameters optimization, the sensitivity, the shutter, and the frame rate were set at 70, 100 and 30, respectively; 3 × 33 videos of 1 s for each sample were recorded. The dilution of the CNP suspension in double-filtered Milli-Q water was set at 1:40000. The results were reported as mean size ± standard deviation of three measurements and as standard deviation of the population (SDP), an indicator of the size polydispersity.
Electron microscopy measurements were performed using a Field Emission Scanning Electron Microscopy (FESEM) TESCAN S9000G equipped with a Schotty-type field emission gun (FED) operating at 15 keV. Size distribution analysis was performed using ImageJ software. The number of measured particles in the images were 310.
ζ-potential of CNP suspensions was measured using the electrophoretic light scattering (ELS) instrument Zetasizer (Nano ZS Malvern Instruments, Worcestershire, UK) diluting the sample 1:200 in Milli-Q water (pH 5.05, conductivity 0.08 mS/cm) or PBS (pH 7.4, conductivity 17.00 mS/cm). The results were expressed as mean values of three independent measurements ± standard deviation. The parameters used for the carbon materials were as follows: refractive index = 2.417; and absorption = 1.00.
2.4. Preparation of Fe(II) cyt c
Fe(II) cyt c was prepared by incubating for 30 min Fe(III) cyt c with ascorbic acid in a ratio 1:10 (0.120 mM and 1.2 mM, respectively in PBS 10 mM). For CD experiments, the reduction was performed with a solution of reduced glutathione (0.6 mM) in water to avoid interference.
2.5. Spectroscopic evaluation of the cyt c reduction by the interaction with CNPs
2 mL of the 10 mg/mL CNP suspension was then transferred into a glass vial sealed with a rubber stopper. An appropriate amount of Fe(III) cyt c was weighed in a glass vial and then subsequently solubilized in 2 ml of 0.1 M PBS to obtain a concentration of 24 or 240 μM solution. The solution was then mixed with the cyt c solution (final concentrations: Fe(III) cyt c 12 or 120 μM; CNPs 0.5 mg/mL or 5 mg/mL) and then magnetically stirred for 10 min. The suspensions were then filtered through a cellulose acetate filter (0.45 μm, Millipore, Bedford, MA, USA) and the filtrate was analysed by UV–vis spectroscopy (Uvikon, Kontron Instruments, Inc., Everett, MA). Solutions of Fe(III) cyt c and Fe(II) cyt c magnetically stirred at room temperature for 10 min were used as controls. To monitor the effect of oxygen the reaction was also conducted by incubating the suspension for 3 h in open air, or by fluxing oxygen or nitrogen.
The reaction was also performed on the solution obtained by filtrating the CNP suspension (prior the incubation with the Fe(III) cyt c) through a cellulose acetate filter (0.45 μm, Millipore, Bedford, MA, USA) and then filtrate again with 3 kDa (Amicon® Ultra-4 - Merck). In this case, the UV–vis spectra were recorded at different time points. Results were validated by almost three independent experiments.
2.6. Persistence of the reductive activity
2 mL of the 10 mg/mL CNPs suspension was then transferred into a glass vial sealed with a rubber stopper. An appropriate amount of Fe(III) cyt c was weighed in a glass vial and then subsequently solubilized in 2 mL of 0.01 M PBS to obtain a concentration of 240 μM solution. The solution was then mixed with the cyt c solution (final concentrations: Fe(III) cyt c 120 μM; CNPs 5 mg/mL) sample was magnetically stirred for 10 min and then centrifuged at 1000 g for 40 min, the supernatant removed, and pallet re-suspended in 0.01 M PBS. In order to remove all cyt c, the procedure was repeated 3 times. An aliquot of 1 mL was taken from the batch and mixed with 1 mL of fresh Fe(III) cyt c (240 μM) and incubated for 10 min. The same procedure was applied for four times. 990 μL of the last solution was then added to 10 μL of a (120 mM) solution of L-ascorbic acid and the UV–vis spectra recorded. All measurements has been repeated almost three times.
2.7. 1D 1H NMR analysis
All NMR experiments were conducted at 298 K on a JEOL JNM-ECZR NMR spectrometer equipped with a ROYAL probe. 1D 1H spectra were acquired with acquisition time of 30 ms, spectral width of 180 ppm using 4096 complex points. The number of scans was 4096 with a recycle delay of 2 s. All NMR experimental data were processed with JEOL Delta software 6.1.0. Samples were prepared in phosphate buffer 10 mM, 137 mM NaCl, 10 % D2O at pH 7.4. The samples analysed are cyt c oxidized and reduced (by ascorbic acid), and CNP nanoparticles at 0.12 mM concentration. A mixture of cyt c and CNP nanoparticles in 5 mg/mL:120 μM ratio was also analysed by NMR spectroscopy.
2.8. LC-MS analysis
cyt c samples were analysed by flow injection analysis or LC-MS after protein digestion with trypsin in a weighted (w/w) ratio of 1/20 at 37 °C for 20 h in 50 mM ammonium bicarbonate solution (pH 8). Digestion was stopped with 1 % aqueous formic acid and an aliquot was transferred in a plastic vial for LC-MS. The analysis was achieved using a nanoHPLC-HRMS instrument. FIA was performed using a syringe pump (5 μl/min flow). The nanoHPLC system used was a Thermo Dionex Ultimate 3000 chromatograph coupled with an Orbitrap Fusion MS analyzer (Thermo Scientific, Milan, Italy). The chromatographic separation was achieved with a PepMap™ RSLC C18 column (2 μm, 100 Å, 75 μm × 50 cm; Thermo Scientific, Milan, Italy) headed by a pre-concentration column (C18 PepMap trap cartridge 100 Å, 5 μm, 0.3 mm × 5 mm; Thermo Scientific, Milan, Italy). Eluents were 0.1 % formic acid in water (A) and 0.1 % formic acid in acetonitrile/water 8/2 (B) for the separation column, and 0.05 % trifluoracetic acid (TFA) in water/acetonitrile 98/2 (C) for the pre-concentration one. Gradient run (0.3 μL/min) started with 5 % of B for 3 min up to 50 % in 30 min raising 90 % in 1 min and held for 5 min. Column equilibration time at 5 % B was 21 min. C eluent flow was set at 5 μL/min.
A nanoESI ion source was used with the following parameters: spray positive voltage 2000 V and ion transfer tube temperature 275 °C. A full scan experiment was performed in a mass range of m/z 200–2000 with a resolution of 120k in the positive ion mode. A subsequent (each 3 s) higher-energy collisional dissociation (HCD) MS2 experiment was executed with an AGC target threshold of 40k. A quadrupole was used as an ion filter with an isolation window of m/z 1.6 and a HCD activation mode at 30 % collision energy was used with an OT resolution of 50k. Mass accuracy of recorded ions (vs. calculated) was ±2 milli mass units.
Obtained spectra were analysed with Protein Prospector 6.5.0 software (https://prospector.ucsf.edu/prospector/mshome.htm UCSF, San Francisco, CA, USA) in order to identify cyt c methionine-containing peptides (M80IFAGIK and GITWGEETLM65EYLENPK). SwissProt 2021.06.18 database, Bos Taurus taxonomy and ESI-EThcD-high-res instrument parameters were used with Met-loss (protein N-term M) and oxidation (M) as variable modification.
2.9. Determination of the amount of cyt c adsorbed to CNPs
1 mL of a 0.5 mg/mL CNPs suspension was mixed with 1 mL of cyt c solution 12 μM in 0.01 M PBS. The mixture was then incubated for 10 min and centrifuged at 15000g for 20 min. The supernatant was filtered using a cellulose acetate filter (0.45 μm, Millipore, Bedford, MA, USA). Subsequently, a 500 μL aliquot of filtered solution was mixed with 500 μL of Coomassie Brilliant Blue G-250. The mixture was incubated for 5 min at room temperature. The concentration of cyt c was evaluated by measuring the absorbance at 595 nm using a calibration curve. The amount of cyt c adsorbed was calculated as a difference from the starting concentration. All measurements were validated by almost three independent experiments.
2.10. Detection of superoxide radicals
47 μL of a suspension of CNPs (0.5 mg/ml) in PBS 10 mM was added to 23 μL of a 1 mM solution of 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline-N-oxide (DEPMPO) and the suspension magnetically stirred at RT for 10 min. The EPR spectra was recorded on the suspension (Miniscope MS100, Magnettech, Berlin, Germany). As positive control, superoxide radicals were generated in a solution containing xanthine 2.2 mM, xanthine oxidase 0.02 U/ml, NaOH 2.2 mM and DEPMPO 50 mM.
2.11. Circular dichroism
30 μL of Fe(III) cyt c 0.6 mM were added in an Eppendorf to 1.470 mL of a suspension of CNPs (0,5 mg/ml) in MilliQ H2O. The suspension was magnetically stirred at room temperature for 10 min, filtered (cellulose acetate, pores size 0.2 μm) and the filtrate transferred in SUPRASIL quartz cell with a pathlength of 1 mm. 0.012 mM solutions of Fe(III) cyt c and Fe(II) cyt c magnetically stirred at room temperature for 10 min were used as controls. The spectra were recorded on a Jasco J-815 Spectrometer in the range from 190 to 460 nm with a scanning speed of 100 nm/min, a data pitch of 0.1 nm, a bandwidth of 1.0 nm. 20 spectra were accumulated per sample at 20 °C.
Deconvolution of the spectra has been performed by using by Dichroweb online analysis software. The algorithm used was CDSS3R, reference set 4, and the calculated wavelength range was 190–240 nm. All measurements were validated by almost three independent experiments.
2.12. Crystal structure of cyt c
The crystal structure of cytochrome c from bovine heart (PDB ID: 2B4Z) was achieved from the RCSB protein database (Mirkin 2015, 2008) and the image generated using Chimera X software.
2.13. Peroxidase activity
250 μL of 0,4 mM Fe (III) cyt c in water, 250 μL H2O2 4 mM in water, 250 μL DMPO 0,176 M in water, 250 μL of solution 0,5 - 0,025 mg/ml CNPs in 10 mM PBS. The EPR spectra was recorded on the suspension (Miniscope MS100, Magnettech, Berlin, Germany) after 10 min of incubation. As positive control, PBS was used instead of CNPs.
2.14. Simulation of the EPR spectra
The simulation of the EPR experimental signals were performed by using the software Winsim 2002. The hyperfine splitting constants obtained by optimization of the simulation have been compared with those reported in the US National Institute of Environmental Health Sciences (NIESH) database.
2.15. Formation of the biomolecular corona
1.5 mL of CNPs (0.5 mg/mL) in water was centrifuged (11000g) and the supernatant removed. The pellet was suspended in 5 mg/mL bovine serum albumin or foetal bovine serum (10 % or 80 %), and incubated for 1 h at 37 °C. The suspensions were centrifuged and suspended in 10 mM PBS for three times to remove the weakly bound proteins.
3. Results and discussion
3.1. Synthesis and characterization of carbon nanoparticles
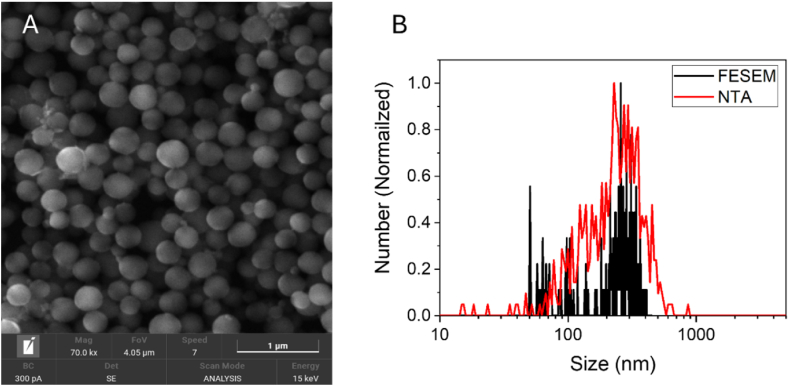
Carbon nanoparticles (CNPs) have been synthetized by following a protocol previously reported [18] by hydrothermal decomposition of glucose. This synthesis leads to particles composed by elemental carbon, mainly amorphous with some crystalline domains [18]. The particles size has been evaluated by NTA and FESEM (Fig. 1).
Fig. 1.
Size of CNPs. A) Representative FESEM image; B) FESEM (red) and NTA (black) size distribution. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
NTA revealed the presence of a population of particles having a mean size around 240 nm, with a moderate degree of polydispersity, as inferred by the low value of SDP (Table 1). The size distribution evaluated by FESEM was close to that obtained by NTA, suggesting that the particles do not agglomerate forming a stable colloidal suspension in water. This is due to the highly negative ζ-potential in water (Table 1), a consequence of the presence of dissociated acidic functional groups at the surface as previously reported [19]. The acidic character of CNPs was further confirmed by the pH of the suspension (Table 1). CNPs exhibited a negative ζ-potential also when suspended in PBS at pH7.4 (Table 1).
Table 1.
Size distribution of CNPs.
| Mean size (nm) NTA∗ | SDP (nm) NTA∗ | Mean size (nm), FESEM | SDP (nm) FESEM | ζ potential (mV) ELS∗ |
|---|---|---|---|---|
| 243.5 ± 4.2 | 114.1 ± 4.5 | 241.8 | 86.1 | −52.2 ± 0.6 (H2O, pH 5) −28.1 ± 1.9 (PBS, pH 7.4) |
SDP: standard deviation of the population; ∗data are reported as mean of three measurements ± standard deviation.
3.2. NADH-independent cyt c reductase activity of carbon nanoparticles
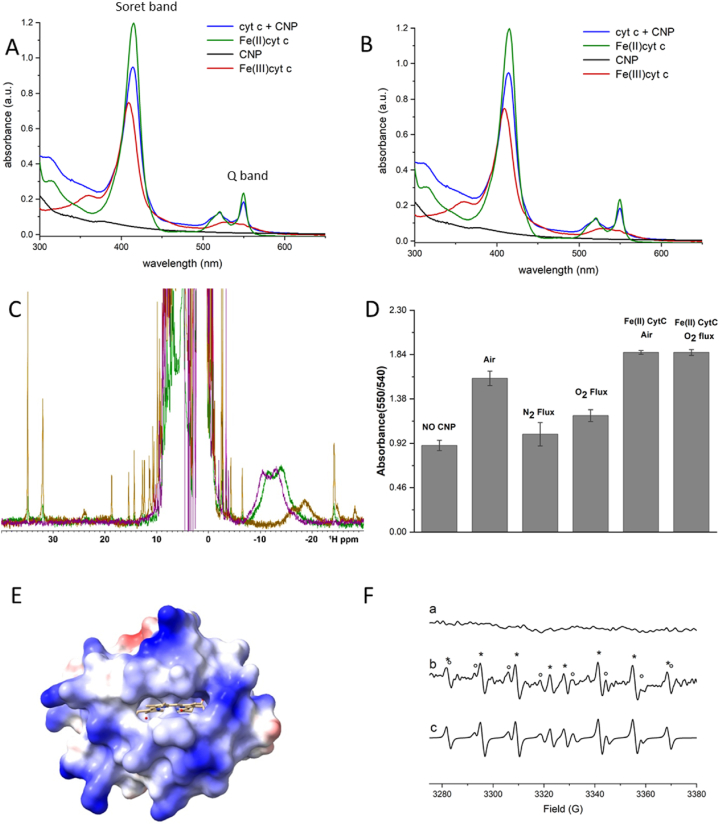
The nanoparticles were incubated with a buffered suspension of Fe(III) cyt c for 10 min, removed by filtration and the supernatant analysed by UV–vis spectrophotometry. In Fig. 2A the UV/vis spectra obtained is compared with that of the Fe(II) cyt c and Fe(III) cyt c. The appearance of the Q band at 550 nm reveals the ability of the particles to reduce the heme iron in cyt c. The reduction is confirmed by the red-shift of the Soret band from 409 to 415 nm [20]. Note that cyt c was not completely reduced. This was confirmed also for time of incubation up to 22 h and for higher concentration of CNPs. (data not reported). To investigate if the reaction was due to the particles or to molecular contaminants, the reaction was repeated on the filtrate obtained by removing CNPs with a 3 kDa filter (Fig. 2B). In this case no reduction was observed. Two samples of carbon black having crystallinity higher than CNPs [18] were also tested for their reducing activity (SI, Fig. S1). These materials were found to be inert.
Fig. 2.
A) UV–Vis spectra obtained following incubation of Fe(III) cyt c (12 μM) and CNPs (0.5 mg/mL) in PBS (10 mM, pH 7.4) for 10 min compared with the spectra recorded on 12 μM Fe(III) cyt c, 12 μM Fe(II) cyt c and 0.5 mg/mL CNPs; All solutions were stirred for 10 min in air; B) UV–Vis spectra at different time points obtained following incubation of Fe(III) cyt c (120 μM) with the solution obtained by filtrating the suspension of CNPs (5 mg/mL) through a 3 kDa dialysis membrane; C) Overlap of the 1D 1H NMR spectra of the paramagnetic species of cyt c: green spectrum corresponds to cyt c (120 μM) in presence of CNPs (5 mg/mL), violet spectrum corresponds to Fe(II) cyt c and brown spectrum corresponds to oxidized Fe(III) cyt c. D) Amount of Fe(II) cyt c formed expressed as absorbance ratio (550 nm/540 nm) following incubation of Fe(III) cyt c (12 μM) with CNPs (0.5 mg/mL) in PBS (10 mM, pH 7.4) in air, under nitrogen atmosphere and by fluxing oxygen; The amount of Fe(II) cyt c formed with ascorbic acid in air and under oxygen flux are reported for comparison (A.A.: ascorbic acid); bars represent the mean value ± standard deviation. E) Crystal structure of cyt c. Colour indicate the coulombic electrostatic potential, red negative, white to blue positive; F) EPR spectra recorded on a) a suspension of CNPs (0.34 mg/mL) and DEPMPO (1 mM) in PBS 10 mM, pH 7.4 after 10 min of incubation; b) a solution containing xanthine 2.2 mM, xanthine oxidase 0.02 U/mL, NaOH 2.2 mM and DEPMPO 50 mM c) Simulation of the EPR spectra in b). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The ability of CNPs to reduce cyt c was further confirmed by NMR (Fig. 2C). The NMR proton spectrum of Fe(III) cyt c (brown spectrum) shows several resonances, among them the most shifted are 8- and 3-methyl heme signals at 33 and 35 ppm and protons of the heme-iron bound Met-80 around −24 and −29 ppm [21,22]. Two broad signals at −18.6 and −16.4 ppm are also visible. This unknown signal is certainly due to protons of the heme ring or residues in proximity of the iron, as their chemical shifts is modulated by the paramagnetic centre. In Fe(II) cyt c (violet spectrum) the shifted peaks disappeared since cyt c is in the low-spin form [23]. In the presence of CNPs (green spectrum) the intensity of the heme signals were reduced but not disappeared, confirming a not complete reduction. The amount of cyt c reduced in presence of CNP was estimated above 90 %, in agreement with the UV–Vis data. Note that shift of the two unknown broad signals at −18.6 and −16.4 ppm was observed in both Fe(II) cyt c and CNP-treated cyt c.
To get insights into the mechanism of reduction, the reaction was performed in air, or by fluxing nitrogen or oxygen (Fig. 2D). In anaerobic condition, no reduction was observed, suggesting a role of oxygen in the reaction. This also suggest a mechanism of reduction different from that of ascorbic acid, since oxygen had no effect in this case. Unexpectedly, when oxygen was fluxed, the amount of cyt c reduced was also lower than in air. This in consistent with a double effect of oxygen, which on one hand participates to the CNP-induced reduction cyt c, on the other oxidize it back to Fe(III) cyt c. This also explain why cyt c is not completely reduced by CNPs in air. To confirm this further, the UV–vis spectra was recorded on cyt c after removal of CNPs (SI, Fig. S2). A slow re-oxidation was observed.
The need of oxygen for the reaction suggests the possible involvement of superoxide radicals. These radicals are in fact known to be able to reduce cyt c [24]. Moreover, they should have a facilitated access to the small heme pocket (Fig. 2E). To explore this hypothesis, the possible generation of superoxide radicals by CNPs was evaluated by EPR spectroscopy (Fig. 2F) by using the spin trap DEPMPO. This spin trap generates with the conjugated acid of superoxide (the hydroperoxyl radical) the specie DEPMPO/OOH. This specie can be detected as eight-peaks EPR signal, as shown in the positive control (Fig. 2F, spectra b, hyperfine splitting constants in Tables S1 and SI) obtained by generating superoxide by the xanthine/xanthine oxidase system. An additional low intense signal correspondent to the specie DEPMPO/OH, due to a partial decomposition of the DEPMPO/OOH is also observed. In the presence of CNPs, the signal was not observed (spectra a), suggesting that no free superoxide radicals were involved in the reaction.
3.3. Conformational changes of cyt c
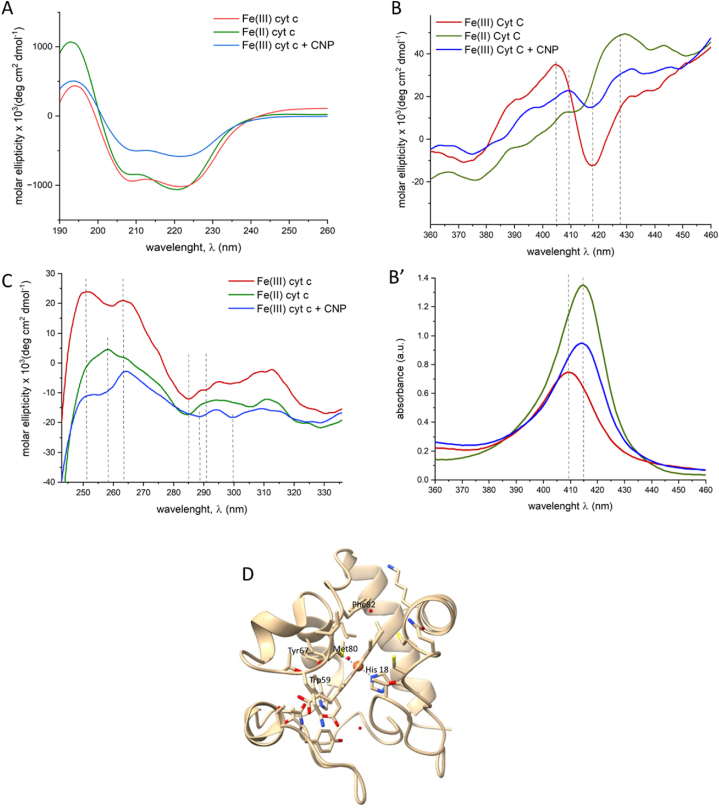
The occurrence of conformational changes induced by CNPs was evaluated by circular dichroism (CD). The CD spectra recorded in the far-UV region (190–260 nm, Fig. 3A) gives information of the secondary structure of proteins. The spectra of Fe(III) cyt c shows two negative peaks at 208 and 222 nm and a positive one at 190 nm. These features arise from the electronic transitions of the peptide bond in the α-helix secondary structures. In the spectra of Fe(II) cyt c a decrease of the positive peak and an increase of the intensity of the component at 208 nm was observed, suggesting that the reduction of the heme-iron induces small modifications of the secondary structure, confirming what previously reported [25]. Following incubation with CNPs, a clear decrease in negative molar ellipticity was observed in the CD spectra recorded on the supernatant. The deconvolution of the CD spectra (SI, Fig. S3) confirmed a significant loss of helical content. These modifications appeared irreversible since the protein did not return to the native conformation with time (Fig. S4). Note that the CD spectra were recorded on the free protein, not bound to CNPs. On the other hand, only a minor fraction (5 %) of the total cyt c was found associated to CNPs (SI, Fig. S5).
Fig. 3.
Effect of CNPs on the conformation of cyt c. A) CD spectrum in the far-UV region; B) CD spectrum in the visible region; the electronic spectra is reported in B′ for comparison; C) CD spectrum in the near-UV region; D) Crystal structure of cyt c, relevant amino acid residues are labelled.
The CD spectra of the visible region, correspondent to the Soret band at 360–460 nm (Fig. 3B) gives information on the structure of the heme group. In fact, it arises from interactions of the heme electronic transitions with those of nearby aromatic amino acid residues [26]. The spectra of Fe(III) cyt c is characterized by a positive peak at 405 nm and a negative peak at 418 nm due to Cotton effect [26]. In the spectra of Fe(II) cyt c a predominant positive peak around 427 nm is present, in agreement with previous studies [27,28]. The spectra of cyt c after contact with CNPs appeared different from that of Fe(II) cyt c: in this case a dramatic decrease of the intensity of the peaks was detected, suggesting an alteration of the protein close to the heme group. In particular, the decrease of the intensity of the negative band at 418 nm suggests a variation on the distance or orientation of the Phe82 residue positioned on the Met80 side of the heme [29] (Fig. 3D).
The CD spectra of the near-UV region (240–320 nm, Fig. 3C) gives information on the tertiary structure close to aromatic amino acids. Bovine cyt c has, like the human one, four Phe, four Tyr, and one Trp [30]. Tyr-67, Trp-59 and Phe-82 are among the conserved residues since they support essential structural and functional roles [31]. In particular, Tyr-67 is known to tune the electron transfer by stabilizing the Met80-iron bond through hydrogen bonding [32] (Fig. 3D). The spectra of Fe(III) cyt c exhibits two minima at 285 and 291 nm that can be assigned to the tertiary structure close to Trp-59 [32], while the large positive component centred at 263 nm can be attributed to the porphyrin and Phe. The band centred at 251 nm reflects a contribution from Tyr side chains [20]. The Fe (II) cyt c spectrum is different at the lower wavelength by that of Fe (III) cyt c, with a positive component at 258 nm and a negative one at 284 nm. The spectrum recorded following incubation of Fe (III) cyt c with CNPs appeared also in this case different from both Fe (III) cyt c and Fe (II) cyt c spectra. In fact, the band at 251 nm largely decreased, suggesting modifications close to the Tyr residues. Moreover, two new negative bands at 289 and 300 nm suggested the presence of modifications of the conformation close to Trp59 [20].
Conformational changes close to the heme pocket are known to induce the weakening of the Met80-iron bond. However, this is not the case here, as inferred by the appearance in the electronic absorption spectrum of the cyt c incubated with CNPs and re-oxidized (SI, Fig. S6) of the band at 695 nm due to charge-transfer characteristic of Met80-iron bond in native Fe(III) cyt c [33].
Conformational changes might occur following adsorption of proteins onto material surfaces, but also following modifications of the amino acid residues involved in structural stabilization. To explore the latter hypothesis, FIA-MS and LC-MS analyses were performed on the cyt c treated with CNP in comparison with the pristine one. The deconvolution of MS spectrum of cyt c (base peak 765.3998 m/z, z = 16) revealed the presence of a major peak corresponding to a mass of 12230 in both pristine and CNP treated cyt c (SI Figure S7). Minor peaks were found corresponding to mass 12327, likely due to the phosphorylation of a Tyr residue, a common post-translational modification in cyt c [31], and 12246, likely due to small amount of Met oxidation. No significant modification of the spectrum was observed after treatment with CNPs.
Since CNP was reported to be able to oxidize glutathione and to release singlet oxygen [18], the analysis deepened the possible oxidation of the sulphur containing amino acids, and in particular on the methionine residues. The protein was digested with trypsin, and two peptides (MIFAGIK and GITWGEETLMEYLENPKK) containing respectively the two methionine residues Met 80 and Met 65 were analysed. No significant oxidation of both Met 80 and Met 65 occurring after incubation with CNPs was detected. In fact, unchanged MIFAGIK only was detected both in pristine and in treated cyt c (SI Figure S8). As regards GITWGEETLMEYLENPKK, both the unmodified (SI Figure S9) and the oxidized-Met 65 (SI Figure S10) peptides were detected in the two cytochrome-digested samples. No significant quantitative increase of GITWGEETLM(Ox)EYLENPKK was observed.
3.4. Peroxidase activity
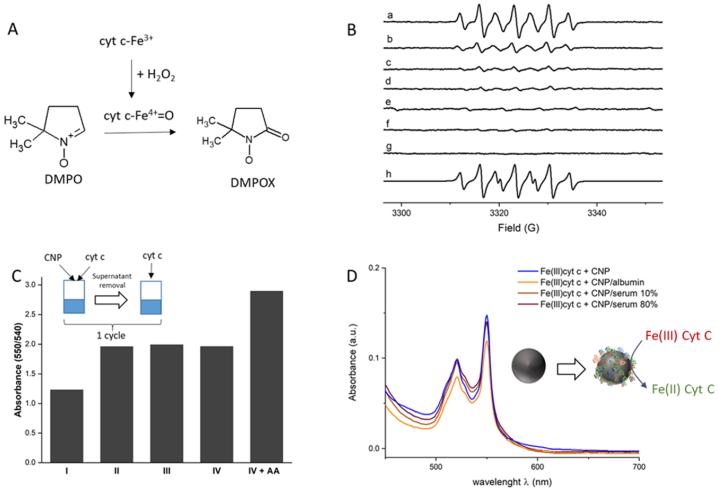
The occurrence of conformational changes might implicate the possible modification of the cyt c enzymatic activity [33,34]. We therefore investigate the possible variation of the peroxidase activity of cyt c by EPR spectroscopy in the presence of hydrogen peroxide by using DMPO as substrate. In fact, DMPO was reported to be oxidized by cyt c in the presence of H2O2 to give as final product the specie 5,5-dimethyl-1-pyrrolidone- 2-oxyl (DMPOX) as described in the scheme in Fig. 4A [35].
Fig. 4.
A) Scheme of the generation of DMPOX catalysed by cyt c; B) EPR spectra recorded on a) a solution of Fe(III) cyt c (50 μM) in the presence of 0.5 mM H2O2 and 22 mM DMPO; b–g: EPR spectra recorded on a solution of Fe(III) cyt c (50 μM) H2O2 (0.5 mM) and DMPO (22 mM) and increasing concentration of CNPs (3.1–62 μg/mL). h) Simulation of the EPR spectra in a). C) Amount of Fe(II) cyt c formed expressed as absorbance ratio (550 nm/540vnm) following 4 cycles of incubation of Fe(III) cyt c (120 μM) and CNPs (5 mg/mL). D) UV–Vis spectra obtained following incubation of Fe(III) cyt c (120 μM) and CNPs (5 mg/mL) pre-treated with serum albumin or foetal bovine serum at two different concentrations (10 and 80 %) in PBS (10 mM, pH 7.4) for 10 min.
When cyt c was incubated with H2O2 and DMPO, an intense signal was observed (Fig. 4B, spectrum a). The simulation (Fig. 4B, spectrum h, Tables S1 and SI) allowed to assign this signal to that of DMPOX. After incubation with increasing concentration of CNPs a decrease of the signal was observed indicating a suppression of the peroxidase activity (Fig. 4B, spectra b-g).
3.5. Persistence of the reductive activity
To monitor if the reductive activity was persistent, Fe(III) cyt c was incubated with CNPs, removed, and the CNPs put in contact again with fresh Fe(III) cyt c. The UV–vis spectra were recorded on the supernatant after each cycle. The results are reported in Fig. 4C. CNPs remained active in the reaction following four incubations with Fe(III) cyt c suggesting a catalytic activity. When ascorbic acid was added in the last supernatant, the intensity of the peak at 550 nm increased, indicating that CNPs was not able to reduce all Fe(III) cyt c.
3.6. Effect of the biomolecular corona
To investigate if the CNPs were still active in the presence of a biomolecular corona, CNPs were pre-treated with bovine serum albumin or foetal bovine serum at two different concentrations (10 and 80 %) and incubated with Fe(III) cyt c (Fig. 4D). The pre-treatment with proteins only slightly decreases the ability of CNPs to reduce Fe(III) cyt c, as inferred by the presence of the band at 550 nm.
4. Discussion
Nanomaterials (NMs) are known to interact with biological systems in many ways. Among them, the alteration of the endogenous systems regulating the intracellular redox equilibrium is considered one of the processes with the highest potential to induce adverse outcomes [36,37]. One of the possible pathways is the perturbation of enzymes involved in redox homeostasis like cyt c. The alteration of this multifunctional protein is of high toxicological relevance since it could have multiple consequences like mitochondrial damage, and modification of the intrinsic apoptosis pathway [33]. The ability of carbon nanomaterials (CNMs) to interfere with the cyt c functions has been reported in some studies [[15], [16], [17]]. However, the mechanistic understanding of the process, and of the related physico-chemical determinants, are limited, preventing the control of derived adverse effects by safe-by design approaches [38]. Here we report that CNMs can affect the cyt c functions by direct, NADH-independent reduction of heme-iron, inhibition of the peroxidase activity and unfolding, and demonstrate that these processes occur through independent mechanisms.
As model CNM, carbon nanoparticles (CNPs) synthesized by hydrothermal carbonization of glucose have been selected. This synthetic method allows to produce spherical monodisperse particles made of elemental carbon, having a prevalent disordered structure with some graphitic domains [18]. These particles are particularly interesting since exhibit multiple redox properties that can be found individually in other kinds of CNMs. CNPs are in fact able to scavenge free radicals, oxidize thiols [18], and reduce iron and copper ions [39]. Moreover, when activated with NIR light, CNPs act as photosensitizers generating singlet oxygen. [18], a property that makes them interesting for medical applications.
4.1. CNPs exhibit a NADH-independent cyt c reductase activity
UV–vis spectroscopy and NMR data showed that CNPs are able to reduce cyt c. Reduction of cyt c was previously observed with carboxylated SWCNT [15] and carbon nanoparticles produced by electrochemistry [40]. However, while in the first case an increase of cyt c redox potential was observed, a slight decrease was observed in the latter case [15,40]. Oppositely, no reduction was found with surface modified MWCNT and GO [16] or with SWCNT [17]. In two studies, CNMs were reported to be able to reduce cyt c only in the presence of NADH or other endogenous reducing agents [41,42] thus acting as electron transfer agents. In the present case, no reducing agent was necessary for the reaction demonstrating that CNMs can reduce cyt c in cells also through an alternative mechanism in which they act themselves as reductants.
Albeit oxygen was found to be necessary for the reaction, EPR spectroscopy data demonstrate here that free superoxide radicals, which are known to reduce cyt c by reacting with the heme moiety [24], are not involved. Studies reporting the generation of superoxide free radicals by CNMs not activated by light are conflicting [8], and the generation was clearly demonstrated only in the presence of NADH or other reducing agents [41,42]. Alternatively, superoxide radicals bound to the NPs surface might be involved, albeit, to the best of our knowledge, such species have been reported to exist only at graphene edges at very low temperature [43].
CNPs have a highly disordered structure. Therefore, the identification of the nature of the reactive moieties able to react with oxygen to generate superoxide is not straightforward. One possible kind of reactive sites are unpaired electrons. They have been reported to be redox active, e.g. by inducing oxidative damage to lipids or by reducing iron ions through a direct electron transfer mechanism [44]. The observed inertness of the carbon black samples, which are expected to have a lower abundance of unpaired electrons having a more ordered structure, is consistent with a role of these reactive species. Crystallinity modulates not only the abundance, but also the properties of these species: less ordered structure exhibit mainly isolated paramagnetic species, while increasing the graphitic structure radicals stabilized by conjugation with π bonds become more abundant [45]. Differences in type and density of carbon centred radical species might account for the discrepancies found for other CNMs, like SWCNTs [[15], [16], [17]]. Note that the modulation of the CNMs reactivity by structural defects is in line with what found by Bengalli et al. which observed a higher ability of defective CNMs to reduce cyt c [42].
Albeit we were unable to identify the surface reactive species, overall, the cyt c reduction appears to occur by the electron transfer directly from the surface to the heme iron. This limits the reaction to CNMs able to approach the cyt c close to the heme. In the present case, the negative surface of CNPs should facilitate the orientation of the more positive side of cyt c (Fig. 2D) positioned close to the heme pocket toward the surface.
4.2. CNPs induces cyt c unfolding
Unfolding is a common process occurring in proteins interacting with materials and generally follows strong adsorption of the molecule at the surface [37]. Unfolding has been reported for cyt c following incubation with surface modified MWCNT and GO [16] or carbon dots [45], while no alteration was reported for cyt c electrostatically conjugated to SWCNT [17] and for a type of hydrophilic carbon nanoparticles [46]. Properties like surface curvature, and roughness or hydrophobicity might, in principle, justify the differences encountered in the different studies. For example, in the study by Shang and co-workers, conformational changes were reported only with 35 nm amorphous silica nanoparticles, while no effect was observed on 15 nm silica nanoparticles having a higher surface curvature [26]. In the present case, the low surface curvature of the CNPs surface makes cyt c unfolding likely. Moreover, since cyt c exhibits a net positive charge at neutral pH (pI 10.2–10.5) [30], the adsorption following electrostatic interaction with the negative CNP surface is highly favoured. However, cyt c-nanoparticles association appeared to be unexpectedly reversible, since alteration of both secondary and tertiary cyt c structure was observed on the free protein. We can exclude that the observed structural modifications arise from cyt c reduction. In fact, the CD spectrum observed for CNPs-reduced cyt c is different by that of Fe(II) cyt c. Moreover, while cyt c reduced by CNPs return to the oxidized form with time, the conformational modification appeared irreversible.
Oxidation of the sulphur containing amino acids, which can result in conformational modifications, was not detected. Despite a possible modification on amino acid residues other than Met cannot rule out, our experimental evidences suggest a process following reversible physisorption. Unfolding is generally believed to occur following strong adsorption of the proteins at the surface [47], while desorbed proteins commonly return to the native conformation. However, some examples of conformational memory of desorbed proteins has been reported. For example, Sanchez-Guzman and co-workers [48][50] demonstrated partial unfolding on oxyhemoglobin interacting with silica nanoparticles, while Sabatino and co-workers reported irreversible conformational modifications of albumin desorbed by Chrysotile asbestos [49].
4.3. CNPs induces suppression of cyt c peroxidase activity
The enzymatic activity of cyt c is modulated by conformational changes. A key step in the intrinsic apoptotic pathway is the association of cyt c to the membrane component cardiolipin (CL), which induces a transition to an alternative conformation and a decrease of the redox potential due to the Fe(III)-Met80 axial bond weakening [50], and an increase of the peroxidase activity [33]. Alkaline pH [33] or negatively charged nanoparticles like liposomes [51] induces a similar effect. Amorphous silica nanoparticles [26] and carbon dots [45] were also reported to induce conformational changes resulting in an increase of peroxidase activity.
In the present case, CD spectroscopy supports the occurrence of α-helix loss and modifications close to aromatic amino acids of the cyt c. Nevertheless, CNPs unexpectedly induced a suppression of the peroxidase activity. This data might be consistent with a conformation alternative to that observed with other NMs, or more likely with the reduction of the heme, and it is partially in agreement with what reported by Ma and co-workers for SWCNT [15].
4.4. Biological relevance of CNMs-mediated reduction of cyt c
The capability of CNMs to interfere with the cyt c functions might have different biological significance: the alteration of mitochondrial cyt c could lead to mitochondrial respiratory chain disruption, while the interaction with cyt c released in cytoplasm in activated cells might affect the apoptosis. In fact, cyt c is capable of mediating apoptosis in the oxidized form only [[52], [53], [54]]. Furthermore, the interaction with both mitochondrial and cytosolic cyt c might affect the redox homeostasis in cells [33]. However, to be relevant, the reaction must occur in the presence of biomolecular coronas and the effect must be persistent. Here we demonstrated that the incubation with serum proteins to generate a hard protein corona [55] did not reduce the surface reactivity of CNPs, probably because part of the surface of the particles was still exposed to the solvent. This agrees with a previous study, in which CNPs were reported to be still redox active in the presence of a foetal bovine serum-derived biomolecular corona [56]. The reactivity was also found to be persistent, suggesting a potential of CNPs to induce cyt c alteration in cells. However, this is not the case: in fact, CNPs do not interfere with the redox homeostasis of cells [15] likely because they do not came in contact with mitochondrial or cytosolic cyt c; in fact, they were reported to accumulate in endo/phagosomes in RAW 264.7 macrophages and human A549 lung adenocarcinoma cell [18,19]. In fact, CNPs do not induce oxidative stress or cell death in macrophages. On the other hand, the interaction with cyt c may occur with other CNMs like fibre-shaped SWCNTs, which are released into cytosol and penetrate into mitochondria [15].
5. Conclusion
The data reported in the present study demonstrate the potential of CNMs to affect the cyt c functions by direct, NADH-independent reduction of heme-iron, inhibition of the peroxidase activity and unfolding. These effects do not imply the presence of cyt c in the hard corona, since they were detected on the free protein. Finally, we demonstrate that these processes occur through distinct mechanisms, which might be independently modulated by tuning the electronic or the surface properties of the particles.
CRediT authorship contribution statement
Ivana Fenoglio: Writing – review & editing, Supervision, Resources, Conceptualization. Shagufta Gul: Writing – original draft, Investigation. Francesco Barbero: Supervision, Investigation, Data curation. Enrica Mecarelli: Investigation. Claudio Medana: Supervision, Funding acquisition, Data curation. Angelo Gallo: Investigation, Data curation. Carlotta Polizzi: Investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. Shagufta Gul is recipients of PhD grant from Ministero dell’Università' e della Ricerca (MUR), Italy.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40587.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Tu L., Li Q., Qiu S., Li M., Shin J., Wu P., Singh N., Li J., Ding Q., Hu C., Xiong X., Sun Y., Kim J. Recent developments in carbon dots: a biomedical application perspective. J. Mater. Chem. 2023;11:3038–3053. doi: 10.1039/D2TB02794A. [DOI] [PubMed] [Google Scholar]
- 2.Titirici M.M., Antonietti M. Chemistry and materials options of sustainable carbon materials made by hydrothermal carbonization. Chem. Soc. Rev. 2010;39:103–116. doi: 10.1039/b819318p. [DOI] [PubMed] [Google Scholar]
- 3.Bergamaschi E., Garzaro G., Wilson Jones G., Buglisi M., Caniglia M., Godono A., Bosio D., Fenoglio I., Guseva Canu I. Occupational exposure to carbon nanotubes and carbon nanofibres: more than a cobweb. Nanomaterials. 2021;11:745. doi: 10.3390/nano11030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sell M., Lopes A.R., Escudeiro M., Esteves B., Monteiro A.R., Trindade T., Cruz-Lopes L. vol. 13. 2023. p. 2887. (Application of Nanoparticles in Cancer Treatment: A Concise Review Nanomaterials). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbero F., Gul S., Perrone G., Fenoglio I. Photoresponsive inorganic nanomaterials in oncology technol. Cancer. Res. Treat. 2023;22 doi: 10.1177/15330338231192850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaldoss M.J.N., Yang J.-L.P., Koshy, Unnikrishnan A., Sorrell C.C. Inorganic nanoparticle-based advanced cancer therapies: promising combination strategies Drug Discovery. Today Off. 2022;27 doi: 10.1016/j.drudis.2022.103386. [DOI] [PubMed] [Google Scholar]
- 7.Yuan X., Zhang X., Sun L., Wei Y., Wei X. Cellular toxicity and immunological effects of carbon-based nanomaterials part. Fibre Toxicol. 2019;16:1–27. doi: 10.1186/s12989-019-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims C.M., Hanna S.K., Heller D.A., Horoszko C.P., Johnson M.E., Bustos A.R.M., Reipa V., Riley K.R., Nelson B.C. Redox-active nanomaterials for nanomedicine applications. Nanoscale. 2017;9:15226–15251. doi: 10.1039/c7nr05429g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan M., Zhao S., Liu W., Lee C.S., Zhang W., Wang P. Photosensitizers for photodynamic therapy. Adv. Healthc. Mater. 2019;8 doi: 10.1002/adhm.201900132. [DOI] [PubMed] [Google Scholar]
- 10.Liu X., Sen S., Liu J., Kulaots I., Geohegan D., Kane A., Puretzky A.A., Rouleau C.M., More K.L., Palmore G.T.R. vol. 7. 2011. pp. 2775–2785. (Antioxidant Deactivation on Graphenic Nanocarbon Surfaces Small). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenoglio M. Tomatis, Lison D., Muller J., Fonseca A., Nagy J.B., Fubini B. Pro- and anti-oxidant properties of near-infrared (NIR) light responsive carbon nanoparticles. Free Radical Biol. Med. 2006;40:1227–1233. doi: 10.1016/j.freeradbiomed.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira C.A., Ni D., Rosenkrans Z.T., Cai W. Scavenging of reactive oxygen and nitrogen species with nanomaterials. Nano Res. 2018;11:4955–4984. doi: 10.1007/s12274-018-2092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Ji Z., Xia T., Meng H., Low-Kam C., Liu R., Pokhrel S., Lin S., Wang X., Liao Y.-P., Wang Li L., R. Rallo M., Damoiseaux R., Telesca D., Mädler L., Cohen Y.J., Zink J., Nel A. Use of metal oxide nanoparticle band gap to develop a predictive paradigm for oxidative stress and acute pulmonary inflammation. ACS Nano. 2012;6:4349–4368. doi: 10.1021/nn3010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santucci R., Sinibaldi F., Cozza P., Polticelli F., Fiorucci L. Cytochrome c: an extreme multifunctional protein with a key role in cell fate. Int. J. Biol. Macromol. 2019;136:1237–1246. doi: 10.1016/j.ijbiomac.2019.06.180. [DOI] [PubMed] [Google Scholar]
- 15.Ma X., Zhang L.-H., Wang L.-R., Xue X., Sun J.-H., Wu Y., Zou G., Wu X., Wang P.C., Wamer W.G., Yin W., Zheng K., Liang X.-J. Single-walled carbon nanotubes alter cytochrome c electron transfer and modulate mitochondrial function. ACS Nano. 2012;6:10486–10496. doi: 10.1021/nn302457v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patila M., Pavlidis I.V., Diamanti E.K., Katapodis P., Gournis D., Stamatis H. Enhancement of cytochrome c catalytic behaviour by affecting the heme environment using functionalized carbon-based nanomaterials. Process Biochem. 2013;48:1010–1017. doi: 10.1016/j.procbio.2013.04.021. [DOI] [Google Scholar]
- 17.Shukla A.K., Abidi S.M., Sharma C., Saini T.C., Acharya A. Single-walled carbon nanotube conjugated cytochrome c as exogenous nano catalytic medicine to combat intracellular oxidative stress Free. Radical Biol. Med. 2022;193:238–252. doi: 10.1016/j.freeradbiomed.2022.10.276. [DOI] [PubMed] [Google Scholar]
- 18.Kokalari I., Gassino R., Giovannozzi A.M., Croin L., Gazzano E., Bergamaschi E., Rossi A.M., Perrone G., Riganti C., Ponti J. Pro- and anti-oxidant properties of near-infrared (NIR) light responsive carbon nanoparticles. Free Radical Biol. Med. 2019;134:165–176. doi: 10.1016/j.freeradbiomed.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Kokalari I., Keshavan S., Rahman M., Gazzano E., Barzan G., Mandrile L., Giovannozzi A., Ponti J., Antonello G., Monopoli M., Bergamaschi E., Riganti C., Bengt F., Fenoglio I. vol. 16. 2021. pp. 689–707. (Efficacy, Biocompatibility and Degradability of Carbon Nanoparticles for Photothermal Therapy of Lung CancerNanomedicine). [DOI] [PubMed] [Google Scholar]
- 20.Fedurco M., Augustynski J., Indiani C., Smulevich G., Antalík M., Bánó M., Sedlák E., Glascock M.C., Dawson J.H. The heme iron coordination of unfolded ferric and ferrous cytochrome c in neutral and acidic urea solutions. Spectroscopic and electrochemical studies Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2004;1703:31–41. doi: 10.1016/j.bbapap.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Chevance S., Le Rumeur E., De Certaines J., Simonneaux G., Bondon A. 1H NMR structural characterization of the cytochrome c modifications in a micellar. Environment Biochemistry. 2003;42:15342–15351. doi: 10.1021/bi035044. [DOI] [PubMed] [Google Scholar]
- 22.Battistuzzi G., Borsari M., Rossi G., Sola M. Effects of solvent on the redox properties of cytochrome c: cyclic voltammetry and 1H NMR experiments in mixed water-dimethylsulfoxide solutions. Inorg. Chim. Acta. 1998;272:168–175. doi: 10.1016/S0020-1693(97)05937-9. [DOI] [Google Scholar]
- 23.Oellerich S., Wackerbarth H., Hildebrandt P. Spectroscopic characterization of nonnative conformational states of cytochrome c. J. Phys. Chem. B. 2002;106:6566–6580. doi: 10.1021/jp013841g. [DOI] [Google Scholar]
- 24.Tarpey M.M., Fridovich I. Methods of detection of vascular reactive species. Circ. Res. 2001;89:224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- 25.Calvert J.F., Hill J.L., Dong A. Arch. Redox-dependent conformational changes are common structural features of cytochrome c from various species. Biochem. Biophys. 1997;346:287–293. doi: 10.1006/abbi.1997.0324. [DOI] [PubMed] [Google Scholar]
- 26.Shang W., Nuffer J.H., Muñiz‐Papandrea V.A., Colón W., Siegel R.W., Dordick J.S. Cytochrome c on silica nanoparticles: influence of nanoparticle size on protein structure. Stability, and Activity Small. 2009;5:470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- 27.Mugnol K.C., Ando R.A., Nagayasu R.Y., Faljoni-Alario A., Brochsztain S., Santos P.S., Nascimento O.R., Nantes I.L. Spectroscopic, structural, and functional characterization of the alternative low-spin state of horse heart cytochrome c. Biophys. J. 2008;94:4066–4077. doi: 10.1529/biophysj.107.116483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serpas L., Milorey B., Pandiscia L.A., Addison A.W., Schweitzer-Stenner R.J. Autoxidation of reduced horse heart cytochrome c catalyzed by cardiolipin-containing membranes. Phys. Chem. B. 2016;120:12219–12231. doi: 10.1021/acs.jpcb.6b05620. [DOI] [PubMed] [Google Scholar]
- 29.Pielak G.J., Oikawa K., Mauk A.G., Smith M., Cyril M.K. Elimination of the negative Soret Cotton effect of cytochrome c by replacement of the invariant phenylalanine using site-directed mutagenesis. J. Am. Chem. Soc. 1986;108:2724–2727. [Google Scholar]
- 30.Nakashima T., Higa H., Matsubara H., Benson A.M., Yasunobu K.T.J. The amino acid sequence of bovine heart cytochrome c. Biol. Chem. 1966;241:1166–1177. doi: 10.1016/S0021-9258(18)96817-1. [DOI] [PubMed] [Google Scholar]
- 31.Hannibal L., Tomasina F., Capdevila D.A., Demicheli V., Tórtora V., Alvarez-Paggi D.N., Jemmerson R., Murgida D.H., Radi R. Alternative conformations of cytochrome c: structure, function, and. Detection Biochemistry. 2016;55:407–428. doi: 10.1021/acs.biochem.5b01385. [DOI] [PubMed] [Google Scholar]
- 32.Xu Q., Keiderling T.A. top-flow kinetics studies of the interaction of surfactant, sodium dodecyl sulfate, with acid-denatured cytochrome c Proteins: structure. Function, and Bioinformatics. 2006;63:571–580. doi: 10.1002/prot.20926. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Paggi D.N., Hannibal L., Castro M.A., Oviedo-Rouco S., Demicheli V., Tórtora V., Tomasina F., Radi R., Murgida D.H. Multifunctional cytochrome c: learning new tricks from an old dog. Chem. Rev. 2017;117:13382–13460. doi: 10.1021/acs.chemrev.7b00257. [DOI] [PubMed] [Google Scholar]
- 34.Parakra R.D., Kleffmann T., Jameson G.N., Ledgerwood E.C. The proportion of Met80-sulfoxide dictates peroxidase activity of human cytochrome c. Dalton Trans. 2018;47:9128–9135. doi: 10.1039/c8dt02185f. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence A., Jones C.M., Wardman P., Burkitt M.J. Evidence for the role of a peroxidase compound i-type intermediate in the oxidation of glutathione, NADH, ascorbate, and dichlorofluorescin by cytochrome c/H2O2 implications for oxidative stress during apoptosis. J. Biol. Chem. 2003;278:29410–29419. doi: 10.1074/jbc.M300054200. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Cai R., Chen C. The nano–bio interactions of nanomedicines: understanding the biochemical driving forces and redox reactions. Acc. Chem. Res. 2019;52:1507–1518. doi: 10.1021/acs.accounts.9b00126. [DOI] [PubMed] [Google Scholar]
- 37.Fenoglio I., Fubini B., Ghibaudi E.M., Turci F. Multiple aspects of the interaction of biomacromolecules with inorganic surfaces. Adv. Drug Deliv. Rev. 2011;63:1186–1209. doi: 10.1016/j.addr.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Fadeel B., Keller A.A. Nanosafety: a perspective on nano-bio interactions. Small. 2024 doi: 10.1002/smll.202310540. [DOI] [PubMed] [Google Scholar]
- 39.Tacu I., Kokalari I., Abollino O., Albrecht C., Malandrino M., Ferretti A.M., Schins R.P., Fenoglio I. Mechanistic insights into the role of iron, copper, and carbonaceous component on the oxidative potential of ultrafine particulate matter. Chem. Res. Toxicol. 2021;34:767–779. doi: 10.1021/acs.chemrestox.0c00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L., Hao J., Xu L., Meng X., Li X., Nie C., Xie F., Liu K., Peng X., Xie J., Liang T., Guo Z. Spectroscopic approach for the interaction of carbon nanoparticles with cytochrome c and BY-2 cells: protein structure and mitochondrial function. Int. J. Biol. Macromol. 2019;138:29–36. doi: 10.1016/j.ijbiomac.2019.07.076. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Chen J., Dong Q., Sun X., Liu Q., Li D., Wang E., Wang J. N, P, or S-doped carbon nanotubes as dual mimics of NADH oxidase and cytochrome c reductase. Nano Res. 2023;16:6615–6621. doi: 10.1007/s12274-023-5393-8. [DOI] [Google Scholar]
- 42.Bengalli R.D., Zerbi G., Lucotti A., Catelani T., Mantecca P. Carbon nanotubes: structural defects as stressors inducing lung cell toxicity. Chem. Biol. Interact. 2023;382 doi: 10.1016/j.cbi.2023.110613. [DOI] [PubMed] [Google Scholar]
- 43.Osipov V.Y., Boukhvalov D.W., Takai K. Structure and magnetic properties of superoxide radical anion complexes with low binding energy at the graphene edges russ. J. Coord. Chem. 2020;46:738–745. doi: 10.1134/S107032842011007X. [DOI] [Google Scholar]
- 44.Qin Y., Zhang L., An T. Hydrothermal carbon-mediated fenton-like reaction mechanism in the degradation of alachlor: direct electron transfer from hydrothermal carbon to Fe(III) ACS Appl. Mater. Interfaces. 2017;9:17115–17124. doi: 10.1021/acsami.7b03310. [DOI] [PubMed] [Google Scholar]
- 45.Essner J.B., McCay R.N., Smith Ii C.J., Cobb S.M., Laber C.H., Baker G.A. A switchable peroxidase mimic derived from the reversible co-assembly of cytochrome c and carbon dots. J. Mater. Chem. B. 2016;4:2163–2170. doi: 10.1039/c6tb00052e. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Hao J., Xu L., Meng X., Li X., Nie C., Xie F., Liu K., Peng X., Xie J., Liang T., Guo Z. Spectroscopic approach for the interaction of carbon nanoparticles with cytochrome c and BY-2 cells: protein structure and mitochondrial function. Int. J. Biol. Macromol. 2019;138:29–36. doi: 10.1016/j.ijbiomac.2019.07.076. [DOI] [PubMed] [Google Scholar]
- 47.Casals E., Vitali M., Puntes V. The nanoparticle-Protein Corona untold history (1907–2007) Nano Today. 2024;58 doi: 10.1016/j.nantod.2024.102435. [DOI] [Google Scholar]
- 48.Sanchez-Guzman D., Giraudon-Colas G., Marichal L., Boulard Y., Wien F., Degrouard J., Baeza-Squiban A., Pin S., Renault J.P., Devineau S. In situ analysis of weakly bound proteins reveals molecular basis of soft corona formation. ACS Nano. 2020;14:9073–9088. doi: 10.1021/acsnano.0c04165. [DOI] [PubMed] [Google Scholar]
- 49.Sabatino P., Casella L., Granata A., Iafisco M., Lesci I.G., Monzani E., Roveri N. Synthetic chrysotile nanocrystals as a reference standard to investigate surface-induced serum albumin structural modifications. J. Colloid Interf. Sci. 2007;314:389–397. doi: 10.1016/j.jcis.2007.05.081. [DOI] [PubMed] [Google Scholar]
- 50.Zaidi S., Hassan M.I., Islam A., Ahmad F. The role of key residues in structure, function, and stability of cytochrome-c Cell. Mol Life Sci. 2014;71:229–255. doi: 10.1007/s00018-013-1341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandiscia L.A., Schweitzer-Stenner R. Coexistence of native-like and non-native partially unfolded ferricytochrome c on the surface of cardiolipin-containing liposomes. J. Phys. Chem. B. 2015;119:1334–1349. doi: 10.1021/jp5104752. [DOI] [PubMed] [Google Scholar]
- 52.Suto D., Sato K., Ohba Y., Yoshimura T., Fujii J. Suppression of the pro-apoptotic function of cytochrome c by singlet oxygen via a haem redox state-independent mechanism. Biochem. J. 2005;392:399–406. doi: 10.1042/BJ20050580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hancock J.T., Desikan R., Neill S.J. Does the redox status of cytochrome C act as a fail-safe mechanism in the regulation of programmed cell death? Biochem. Soc. Trans. 2001;31:697–703. doi: 10.1016/S0891-5849(01)00646-3. [DOI] [PubMed] [Google Scholar]
- 54.Vaughn A.E., Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat. Cell Biol. 2008;10:1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soddu L., Trinh D.N., Dunne E., Kenny D., Bernardini G., Kokalari I., Marucco A., Monopoli M.P., Fenoglio I. Identification of physicochemical properties that modulate nanoparticle aggregation in blood Beilstein. J. Nanotechnol. 2020;11:550–567. doi: 10.3762/bjnano.11.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonello G., Marucco A., Gazzano E., Kainourgios P., Ravagli C., Gonzalez-Paredes A., Sprio S., Padín-González E., Soliman M.G., Beal D., Barbero F., Gasco P., Baldi G., Carriere M., Monopoli M.P., Charitidis C.A., Bergamaschi E., Fenoglio I., Riganti C. vol. 19. 2022. p. 49. (Changes of Physico-Chemical Properties of Nano-Biomaterials by Digestion Fluids Affect the Physiological Properties of Epithelial Intestinal Cells and Barrier Models Part Fibre Toxicol). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.