Significance
Variants in ion channel genes controlling neuronal excitability can cause severe neuropsychiatric disorders including developmental and/or epileptic encephalopathies (DEEs) characterized by developmental delay or regression with or without epilepsy. In this manuscript, we reveal that DEE-causing missense variants affecting the activation gate of Kv7.2 and Kv7.3 voltage-gated potassium channel subunits encoded, respectively, by KCNQ2 and KCNQ3 genes, strongly enhance potassium channel function by selectively stabilizing the channel open configuration of the pore; by means of molecular modeling and simulations, we also reveal a plausible atomistic explanation for the observed results. The molecular mechanism revealed may provide additional classification criteria for patients with KCNQ2- and KCNQ3-related neurodevelopmental disorders which may guide prognostic and therapeutic decisions.
Keywords: potassium channels, developmental and epileptic encephalopathies, Genotype–phenotype correlations, channel gating, Molecular dynamics
Abstract
Pathogenic variants in KCNQ2 encoding Kv7.2 voltage-gated potassium channel subunits cause developmental encephalopathies (KCNQ2-encephalopathies), both with and without epilepsy. We herein describe the clinical, in vitro, and in silico features of two encephalopathy-causing variants (A317T, L318V) in Kv7.2 affecting two consecutive residues in the S6 activation gate that undergoes large structural rearrangements during pore opening; the disease-causing A356T variant in KCNQ3, paralogous to the A317T variant in KCNQ2, was also investigated. Currents through KCNQ2 mutant channels displayed increased density, hyperpolarizing shifts in activation gating, faster activation and slower deactivation kinetics, and resistance to changes in the cellular concentrations of phosphatidylinositol 4,5-bisphosphate (PIP2), a critical regulator of Kv7 channel function; all these features are consistent with a strong gain-of-function effect. An increase in the probability of single-channel opening, with no change in membrane abundance or single-channel conductance, was responsible for the observed gain-of-function effects. All-atom molecular dynamics simulations revealed that the mutations widened the inner pore gate and stabilized a constitutively open channel configuration in the closed state, with minimal effects on the open conformation. Thus, mutation-induced stabilization of the inner pore gate open configuration is a molecular pathogenetic mechanism for KCNQ2-related encephalopathies.
Variants in the KCNQ2 or KCNQ3 genes, encoding Kv7.2 and Kv7.3 voltage-gated potassium (K+) channel subunits, respectively, can cause a spectrum of mostly neonatal-onset phenotypes ranging from self-limited familial neonatal epilepsy (SLFNE) to developmental and epileptic encephalopathies (DEEs) (1–5). DEEs are the most severe group of epilepsies characterized by early-onset drug-resistant seizures, epileptiform activity on EEG, developmental delay or regression, and an increased risk of early death (6). DEEs often have a genetic etiology identified in 50% of cases, with KCNQ2 and KCNQ3 pathogenic variants being responsible for the largest fraction of neonatal-onset DEEs (7).
Kv7.2 and Kv7.3 subunits are mainly expressed in neurons, where they can assemble as homo- or heterotetrameric channels underlying the M-current (IKM), a noninactivating K+ current with slow activation and deactivation kinetics, that regulates the resting membrane potential and suppresses repetitive firing (8, 9). Kv7 subunits contain six transmembrane (TM) segments (S1−S6), with S1−S4 forming the voltage-sensing domain (VSD), and S5 and S6 together with the S5−S6 intervening linker forming the pore domain (PD) (9, 10). The latter can be divided into three structurally and functionally distinct regions: two constricted areas located toward the extracellular or the intracellular sides, represented by the selectivity filter (SF) and the activation gate (AG), respectively, and a central cavity (CC) in between (11). The cytosolic C-terminal region contains four α-helices (HA, HB, HC, and HD), where critical sites for assembly, interaction with regulatory molecules, subcellular localization, and binding of accessory proteins such as calmodulin (CaM) (9) have been identified. In addition to membrane depolarization, Kv7 channel opening requires endogenous ligands such as the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2), which binds to distinct channel regions in the VSD and the PD (12) and induces large and complex structural rearrangements (13, 14).
Over the years, functional in vitro studies in heterologous expression systems revealed that the majority of pathogenic variants in KCNQ2 or KCNQ3 cause loss-of-function (LoF) effects by reducing the maximal current density, decreasing subunit expression and/or trafficking to the membrane, shifting the activation gating in the depolarizing direction, and/or altering the response to regulatory proteins such as kinases (2). A decreased IKM density has also been observed in neurons differentiated from human-induced pluripotent stem cells harboring distinct LoF variants associated with KCNQ2-DEE (15). A correlation between the extent of LoF in vitro and the clinical phenotype has been reported, with SLFNE-causing variants being associated with a milder degree of current impairment (haploinsufficiency), whereas most DEE-causing pathogenic variants exert more dramatic, dominant-negative functional consequences (16, 17). This genotype/phenotype correlation was also observed in neuronal populations from murine models of SFLNE, which exhibit a relatively small decrease in IKM density (~25%), compared to those from a mouse model carrying a Kv7.2 recurrent pathogenic variant causing DEE, in which an ~50% decrease was found (18).
In addition to LoF, few heterozygous missense variants enhancing channel function (gain-of-function, GoF) have been described in both KCNQ2 and KCNQ3 (3, 4). For KCNQ2, the phenotypes associated with GoF variants differ from the classical KCNQ2-DEE phenotype, ranging from severe ID without neonatal seizures but with characteristic nonepileptic myoclonus and poor prognosis, to moderate/severe ID and infantile epilepsy (19–23). Similarly, pathogenic GoF variants in KCNQ3 are associated with global developmental delay evolving to ID, sleep-activated near-continuous multifocal spikes, and autistic features without neonatal seizures (24). The mechanisms by which KCNQ2 GoF variants cause such diverse clinical phenotypes are complex and not completely understood (2, 25). Differential effects on the excitability of distinct neuronal populations of cortical neurons have been recently described in transgenic mice harboring a Kv7.2 GoF variant, in which layer 2/3 pyramidal neurons become hyperexcitable and CA1 pyramidal neurons are hypoexcitable (26).
All pathogenic GoF variants, except one (G239S) (20), are missense and are located within the VSD of either Kv7.2 (19, 27, 28) or Kv7.3 (24) subunits. These GoF variants cause a hyperpolarizing shift in voltage-dependent activation gating by stabilizing the activated configuration of S4 and impeding VSD repositioning upon hyperpolarization in the physiological voltage range (29, 30).
A correlation between genotype, clinical phenotype, and in vitro functional consequences has been revealed not only for KCNQ2- and KCNQ3-related disorders but also for several other channelopathies causing neurodevelopmental disorders (31, 32); therefore, identifying the specific molecular mechanism(s) by which each variant affects channel function may provide critical insights into phenotypic heterogeneity and prognosis and may direct precision medicine treatments.
Here, by utilizing a translational multidisciplinary approach linking clinical phenotyping to mutagenesis, electrophysiology, biochemistry, and all-atom Molecular Dynamics (MD), we investigated the functional and structural consequences of three missense variants affecting residues in the pore AG of the Kv7.2 and Kv7.3 subunits that cause severe developmental encephalopathy. These include 1) the KCNQ2 A317T variant identified in a patient with ID and autism spectrum disorders (ASD) but without seizures; 2) the KCNQ2 L318V variant found in a DEE-affected patient with infantile epileptic spasms without preceding neonatal seizures (33); and 3) the KCNQ3 A356T variant, paralogous to the A317T variant in KCNQ2, identified in a large cohort of patients with ID (34). Taken together, our results revealed that these variants exert strong GoF effects by stabilizing the open-state configuration of the PD, thus revealing an additional molecular pathogenetic mechanism for KCNQ2- and KCNQ3-related neurodevelopmental disorders.
Results
Clinical and Genetic Features of the 3 Patients Included in the Present Study.
Patient 1 is a 5-y-old boy with severe ASD and severe ID. He had global developmental delay and was a placid infant. He sat at 8 mo, crawled at 14 mo, and walked at 2 y 11 mo after a long period of knee walking. He vocalized complex babble at 8 mo, but this phenomenon regressed and occurred intermittently. By 5 y, he was nonverbal and had no signs. He could follow a single command and was extremely hyperactive with impulsivity and a fascination with water. He never had an unprovoked seizure; an awake and sleep EEG at 10 mo of age was normal. Trio exome sequencing identified a de novo heterozygous KCNQ2 pathogenic variant (c.949G>A; p.A317T). In silico predictions (SIFT, Polyphen-2, and MutationTaster) were consistent with a deleterious effect. This variant was not detected in control populations in gnomAD (https://gnomad.broadinstitute.org/).
Patient 2 was identified in a cohort study of individuals with DEEs with spike-wave activation in sleep (DEE-SWAS) (33, 35). This patient (case 6) was a 4 y and 4 mo old boy who presented with infantile epileptic spasm syndrome and hypsarrhythmia at 5 mo of age. At 1.8 y, Rolandic epileptiform discharges were observed, which evolved to SWAS at 2.3 y. There was a global developmental delay from birth, with regression after seizure onset, resulting in language delay and impairment of fine motor skills. A clinical gene panel identified a de novo KCNQ2 variant (c.952C>G; p.L318V). In silico predictions (SIFT, Polyphen-2, MutationTaster) were consistent with a deleterious effect, and the variant was classified as pathogenic according to the ACMG/AMP criteria (36).
Patient 3 is a female identified in a cohort of 4,293 individuals with severe undiagnosed developmental disorders, recruited as part of the Deciphering Developmental Disorders study via the genetics services of the UK National Health Service and the Republic of Ireland (patient ID DDD4K.02558). She carried the c.1066C>T; p.A356T variant in KCNQ3. Human Phenotype Ontology terms reported for this patient were as follows: abnormal repetitive mannerisms, bimanual synkinesia, delayed speech and language development, microcephaly, moderate global developmental delay, and poor motor coordination. No epilepsy at any age was reported, and no additional clinical detail is available for this patient (34). Table 1 summarizes the main clinical findings from the 3 patients whose KCNQ2 and KCNQ3 pathogenic variants have been herein investigated.
Table 1.
Main clinical features of patients
| Patient | 1 | 2 | 3 |
|---|---|---|---|
| Age at last follow up | 5y | 4y 4mo | unknown |
| Gene; variant | KCNQ2; c.949G > A, p.A317T | KCNQ2; c.952C > G, p.L318V | KCNQ3; c.1066C > T, p.A356T |
| Early development | Delayed motor milestones | Global DD | Delayed speech development |
| Regression | Yes. Complex babble at 8 mo, nonverbal by 5y | Yes, after seizure onset, resulting in language delay and impairment of fine motor skills | Not reported |
| Neurobehavioral problems | Severe ASD, extremely hyperactive, and impulsive | Not reported | Abnormal repetitive mannerisms, bimanual synkinesia |
| Current level of functioning | Severe ID | Not reported | Moderate global DD |
| Seizures | No | Epileptic spasms (5mo), Controlled with ACTH at 10mo | Not reported |
| EEG | Awake and sleep EEG at 10mo normal | HS between 5 to 10mo; Rolandic discharges at 1y10mo; SWAS at 2y3mo | Not reported |
| Other | Corpus callosum agenesis on MRI | Poor motor coordination | |
| Reference | This study | 33 (case 6) | 34 (patient DDD4K. 02558) |
Abbreviations: ASD: autism spectrum disorder; ACTH: adrenocorticotrophic hormone; DD: Developmental delay; HS: hypsarrhythmia; ID: intellectual disability; mo: months; SWAS: Spike and wave activation in sleep; y: years.
Homomeric or Heteromeric Kv7.2 Channels Carrying Pathogenic Variants in the Pore AG Display a Strong GoF In Vitro Phenotype.
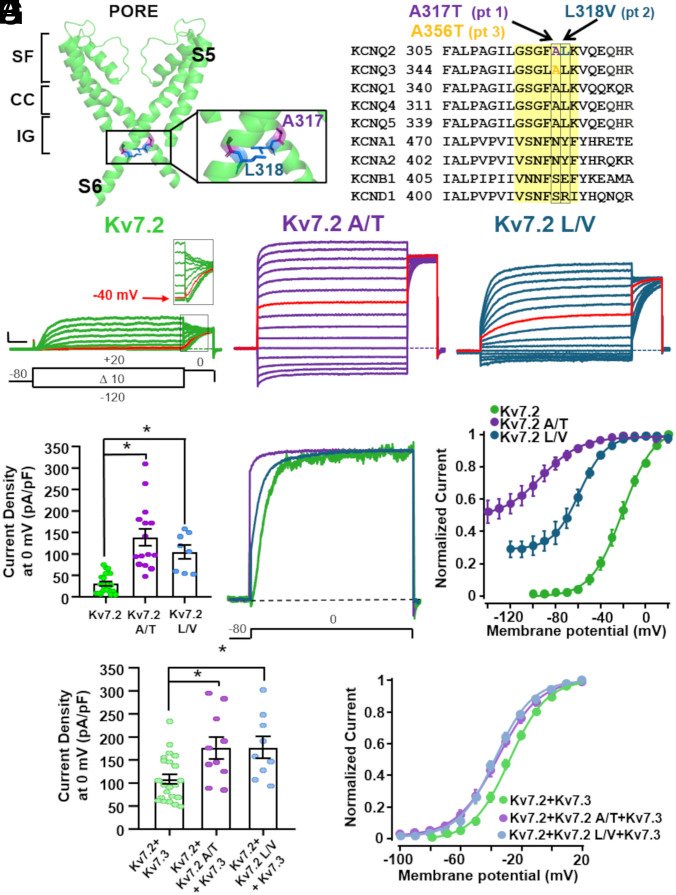
The A317T and L318V variants found in patients 1 and 2, respectively, affect two consecutive residues located at the distal end of the S6 segment of the Kv7.2 subunit, falling within the pore AG, a highly conserved region that undergoes large structural rearrangements during pore opening (Fig. 1 A and B) (13, 14, 37, 38). To evaluate the functional consequences of these two variants, Chinese Hamster Ovary (CHO) cells were transfected with cDNA plasmids encoding the Kv7.2, Kv7.2 A317T, or Kv7.2 L318V subunits, and the resulting currents were recorded with the whole-cell configuration of the patch-clamp technique. As shown in Fig. 1C, when depolarized from −140/−120 mV to 20 mV in 10-mV increments, CHO cells expressing Kv7.2 channels generated voltage-dependent K+-selective currents with an activation threshold of approximately −50 mV and a rather slow time-course of activation and deactivation (Table 2). The half-activation potential (V½) and the slope of the I/V relationship (k) calculated from equation 1 (Methods) were −20.13 ± 1.09 mV and 12.03 ± 0.49 mV/e − fold, respectively (Table 2). At a holding voltage of −80 mV, the Kv7.2 channel was closed, and the ratio between the current measured at the beginning of the depolarization step and that at the end of the 0-mV depolarization was close to 0 (Table 2).
Fig. 1.
Biophysical properties of K+ currents from Kv7.2 channels harboring variants in the pore AG. (A) Kv7.2 structure; only the PD from two opposite subunits is shown for clarity. The enlargement highlights the A317 (purple) and the L318 (blue) residues. The SF, CC, and AG regions are indicated. (B) Sequence alignment of the indicated Kv subunits in the S6 region; the investigated variants are highlighted. The region in yellow indicates the pore AG. (C) Macroscopic whole-cell currents from Kv7.2 (green), Kv7.2 A317T (purple), and Kv7.2 L318V (blue) homomeric channels in response to the indicated voltage protocol. In red are the traces recorded at −40 mV, corresponding to the threshold voltage in Kv7.2 currents (see Inset). Current scale: 200 pA; time scale: 200 ms. (D) Quantification of the current densities recorded from cells transfected with the indicated cDNA constructs. * = P < 0.05. (E) Superimposed normalized current traces at 0 mV from the Kv7.2, Kv7.2 A317T, and Kv7.2 L318V homomeric channels. Note the instantaneous current component in the Kv7.2 A317T (purple) and Kv7.2 L318V channels at the beginning of the pulse. (F) Conductance/voltage curves for the indicated channels; continuous lines represent Boltzmann fits of the experimental data to equation 1 in the Methods section. (G) Quantification of current densities recorded from cells transfected with the indicated cDNA constructs. * = P < 0.05. (H) Conductance/voltage curves for the indicated channels; continuous lines represent Boltzmann fits of the experimental data to equation 1 in the Methods section.
Table 2.
Biophysical properties of Kv7.2 and Kv7.3 mutant channels
| n | pA/pF at 0 mV | V½ (mV) | k | Iinst/Iss | Activation τ @0 mV (ms) | Deactivation τ @−120 mV (ms) | |
|---|---|---|---|---|---|---|---|
| Nontransfected | 5 | 0.50 ± 0.2 | - | - | - | - | |
| Kv7.2 | 25 | 31.4 ± 4.6 | −20.1 ± 1.1 | 12 ± 0.5 | 0.03 ± 0.01 | 177.7 ± 10.2 | 43.9 ± 4.0 |
| Kv7.2 A317T | 15 | 138.7 ± 20.3* | −92.9 ± 3.6* | 13.6 ± 1.9 | 0.81 ± 0.04* | 131.3 ± 12.7* | 100.8 ± 10.7* |
| Kv7.2 L318V | 8 | 105.3 ± 16.6* | −58.3 ± 1.3* | 11.9 ± 11.1 | 0.33 ± 0.03* | 104.1 ± 9.8* | 142.1 ± 15.7* |
| Kv7.2 + Kv7.3 | 20 | 109.1 ± 11.9 | −27.4 ± 1.2 | 11.4 ± 0.6 | 0.01 ± 0.003 | 225.1 ± 17.6 | 22.3 ± 1.9 |
| Kv7.2 + Kv7.2 A317T+Kv7.3 | 12 | 178.0 ± 25.0† | −36.7 ± 2.1† | 13.1 ± 2.1 | 0.12 ± 0.03† | 147.1 ± 9.6† | 35.5 ± 2.2† |
| Kv7.2 + Kv7.2 L318V+Kv7.3 | 9 | 178.1 ± 26.3† | −37.5 ± 1.4† | 12 ± 0.8 | 0.04 ± 0.01 | 77.0 ± 8.7† | 29.2 ± 4.8 |
| Kv7.3 | 8 | 25.1 ± 5.9 | −31.1 ± 2.0 | 9.5 ± 1.2 | - | 93.0 ± 6.4 | - |
| Kv7.3 A356T | 8 | 0.4 ± 0.1‡ | - | - | - | - | - |
| Kv7.3 + pcDNA3.1 | 6 | 24.7 ± 7.8 | −34.4 ± 2.3 | 6.7 ± 0.7 | - | - | |
| Kv7.3 + Kv7.3 A356T | 8 | 7.9 ± 2.7‡ | −34.6 ± 1.2 | 6.5 ± 0.7 | - | 111.5 ± 18.8 | - |
| Kv7.2 + Kv7.3 A356T | 9 | 371.9 ± 64.9† | −41.6 ± 3.7† | 12 ± 0.7 | 0.08 ± 0.03† | 109.7 ± 9.0† | 29.5 ± 2.7 |
| Kv7.2 + Kv7.3 + Kv7.3 A356T | 9 | 261.6 ± 50.3† | −44.1 ± 1.2† | 11.8 ± 1.2 | 0.05 ± 0.01† | 86.2 ± 8.4† | 24.6 ± 6.3 |
* P < 0.05 versus Kv7.2.
† P < 0.05 versus Kv7.2 + Kv7.3.
‡ P < 0.05 versus Kv7.3.
Compared to those of Kv7.2, currents from both Kv7.2 A317T- and L318V-transfected cells showed a significant 3-4-fold increase in the maximal current density (Fig. 1 C and D; Table 2). In both mutants, the activation process revealed two distinct kinetic components: an instantaneous voltage-independent component followed by a slower voltage- and time-dependent component. At 0 mV, the instantaneous voltage-independent fraction accounted for 81 ± 4% and 33 ± 3% of the total current in the Kv7.2 A317T and Kv7.2 L318V channels, respectively (Fig. 1E; Table 2). The V½ of the voltage-dependent component in Kv7.2 A317T and Kv7.2 L318V channels was hyperpolarized by 70 mV and 40 mV, respectively (Fig. 1F). In addition, currents from both mutant channels exhibited faster activation and slower deactivation kinetics (Table 2). Despite these dramatic changes in gating, both mutant channels retained their selectivity for K+ ions; indeed, the reversal potential (Erev) of the currents from Kv7.2, Kv7.2 A317T, and Kv7.2 L318V channels was close to that of a K+-selective pore (SI Appendix, Table S1). Moreover, currents carried by mutant channels retained their sensitivity to the open-pore blocker tetraethylammonium (TEA) (39) (SI Appendix, Table S1), indicating that they were flowing through the central pore. Experiments under bi-ionic conditions with extracellular 15 mM K+, Rb+, or Cs+ ions revealed no difference in relative permeability between the Kv7.2 and Kv7.2 A317T mutant channels (SI Appendix, Fig. S1).
In adult neurons, the M-current is mainly formed by heteromeric Kv7.2/Kv7.3 channels (2). To replicate in vitro the genetic status of patients 1 and 2, who were heterozygous for the KCNQ2 pathogenic allele, functional studies were also carried out upon transfection of KCNQ2 + KCNQ2 A317T + KCNQ3 and KCNQ2 + KCNQ2 L318V + KCNQ3 cDNAs at a 0.5:0.5:1 cDNA ratio. Under these conditions, when compared to those of WT Kv7.2 + Kv7.3 heteromeric channels, the functional effects of heteromeric channels incorporating mutant subunits were similar to but quantitatively smaller than those observed for homomeric channels, with a 50% increase in the maximal current density (Fig. 1G), a ~10 mV hyperpolarizing shift in activation gating (Fig. 1H), and faster activation kinetics, and slower deactivation kinetics (reaching statistical significance only for KCNQ2+KCNQ2 A317T+KCNQ3 channels; Table 2). When compared to WT channels, no change in Erev or TEA sensitivity was observed in heteromeric channels incorporating mutant subunits (SI Appendix, Table S1). Overall, these results suggest that, when compared to Kv7.2, channels harboring the A317T or L318V variant exhibit enhanced maximal current density, hyperpolarized activation gating, and a significant fraction of current at hyperpolarizing potentials; all these effects are consistent with an in vitro GoF phenotype.
Notably, the A356 residue in Kv7.3 is paralogous to A317 in Kv7.2 (Fig. 1B), and the KCNQ3 A356T variant, corresponding to A317T in KCNQ2, has been found in a DEE patient (34). In contrast to the strong in vitro GoF phenotype of homomeric Kv7.2 A317T channels, Kv7.3 A356T subunits did not form functional channels and exhibited reduced currents when coexpressed with WT Kv7.3 (1:1 cDNA ratio) (SI Appendix, Fig. S2A and Table 2). However, a marked increase in current density and an ~15 mV hyperpolarizing shift in activation gating were observed in heteromeric currents recorded from cells that were cotransfected with KCNQ2 + KCNQ3 A356T (1:1 cDNA ratio) or KCNQ2 + KCNQ3 + KCNQ3 A356T (1:0.5:0.5 cDNA ratio) compared to KCNQ2 + KCNQ3-transfected cells (1:1 cDNA ratio) (SI Appendix, Fig. S2 B and C and Table 2). Thus, Kv7.3 A356T subunits, similar to Kv7.2 subunits carrying the paralogous A317T variant, promoted strong GoF when expressed together with Kv7.2 and/or Kv7.3 subunits.
Kv7.2 Pore AG Variants Increase the Probability of Opening without Affecting Single Channel Currents or Plasma Membrane Subunit Expression.
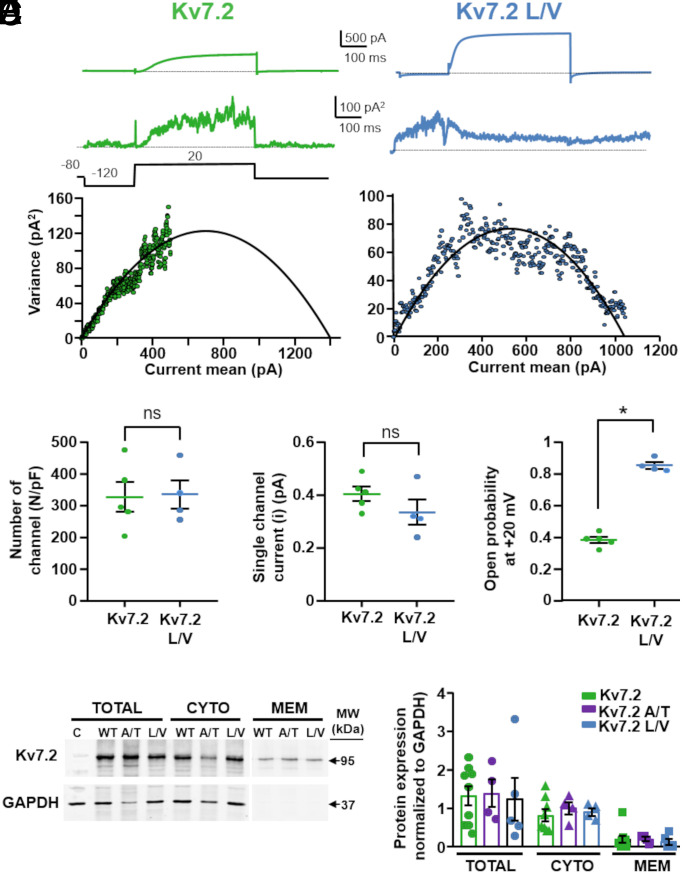
Both the Kv7.2 A317T and Kv7.2 L318V variants caused a large increase in macroscopic current density, which depends on the number of functional channels (N), the single-channel current (i), and the channel opening probability (Po). To investigate variant-induced changes in each of these parameters, nonstationary noise analysis was performed. Mean currents and their respective variances were measured for Kv7.2-, Kv7.2 A317T-, and Kv7.2 L318V-expressing cells. When the variance was plotted against the average current and the data were fitted with the parabolic equation 2, as described in the Methods (Fig. 2A), no differences in the number of functional channels or in the single-channel current (Fig. 2 B and C) between the Kv7.2 and Kv7.2 L318V channels were observed. In contrast, compared to Kv7.2, the maximal Po of Kv7.2 L318V channels increased 2.5/3-fold (Fig. 2D). These data suggest that the Kv7.2 L318V variant caused a strong GoF in vitro phenotype mainly through an increase in the probability of channel opening, without affecting the number of functional channels or their unitary conductance. While the data from the Kv7.2 and Kv7.2 L318V channels allowed a detailed exploration of the entire open probability space, a similar approach in Kv7.2 A317T channels was limited by the small time-dependent component observed for the currents carried by these channels, leading to a less rigorous definition of the single-channel estimates (40) (SI Appendix, Fig. S3A). Nevertheless, unrestrained fitting of the available data revealed that, compared to Kv7.2, Kv7.2 A317T, similar to L318V, also displayed an increased probability of opening with no changes in conductance or number (SI Appendix, Fig. S3 B–D).
Fig. 2.
Nonstationary noise analysis of K+ currents from Kv7.2 channels carrying variants in the pore AG. (A) Representative average response to 100 pulses at +20 mV (Top traces), variance (Middle traces), and variance versus current mean plot (Bottom traces) for Kv7.2 and Kv7.2 L318V, as indicated. The continuous lines in the variance/mean plots are the parabolic fits of the experimental data to equation 2 in the Methods section. (B–D) Quantification of the number of channels divided by the capacitance (b), the single-channel current (c), and the opening probability at 20 mV (d) for the indicated channels. * = P < 0.05. (E) Representative western blot image of total, cytosolic, or plasma membrane protein fractions from CHO cells transfected with pcDNA3.1 (empty vector, C), Kv7.2 (WT), Kv7.2 A317T (A/T), or Kv7.2 L318V (L/V) subunits. On the right, the positions of the estimated molecular masses of the Kv7.2 (95 kDa) and GAPDH (37 kDa) bands are shown. (F) Densitometric quantification of the 95 kDa band intensity in the indicated experimental groups. The data are expressed as the means ± SEMs.
Consistent with the noise analysis results, western blotting experiments revealed no quantitative difference in the expression of the Kv7.2, Kv7.2 A317T, or Kv7.2 L318V subunits in total, plasma membrane, or cytosolic fractions isolated from transfected CHO cells (Fig. 2 E and F). In addition, compared to that of Kv7.3, the plasma membrane expression of Kv7.3 A356T mutant subunits was slightly greater (SI Appendix, Fig. S2D), suggesting that the lack of functional current in homomeric Kv7.3 A356T channels is not attributable to mutation-induced impairment of membrane targeting or expression of the mutant subunits.
Currents from Kv7.2 Pore AG Variants Are Resistant to Changes in Cellular PIP2 Concentrations.
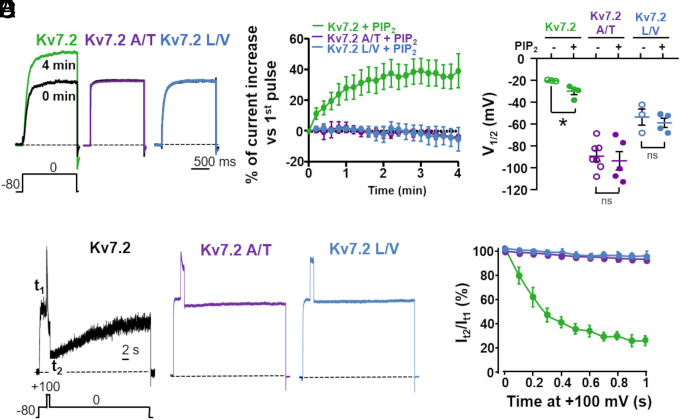
Channel opening of all Kv7 channels relies on the plasma membrane abundance PIP2; PIP2 is required for coupling VSD movement to PD opening of Kv7 channels during electromechanical coupling, and depleting PIP2 results in a closed pore with normal VSD activation (41). Several PIP2-binding regions, including the bottom part of the S6 segment, have been described in Kv7 channels (12, 38, 42). In Kv7.2 channels, increased PIP2 availability causes GoF consequences similar to those described above for the DEE variants affecting the AG, including an increase in maximal conductance and a hyperpolarizing shift in activation gating (43, 44). To investigate the sensitivity of the Kv7.2 A317T and Kv7.2 L318V currents to changes in cellular PIP2 concentrations, PIP2 levels were increased by adding the water-soluble PIP2 analog dic8-PIP2 to the intracellular solution (43), or decreased by coexpressing a voltage-sensitive phosphatase from Danio rerio (DrVSP), which transiently reduces intracellular PIP2 levels when activated by depolarizations (45).
Increasing cellular PIP2 levels by Dic8-PIP2 led to a progressive, time-dependent increase in Kv7.2 currents, together with a negative shift in the half-activation potential. By contrast, pipette inclusion of Dic8-PIP2 did not affect the current density in Kv7.2 A317T- or Kv7.2 L318V-transfected cells, and no shift in the current half-activation potential occurred in these two mutants (Fig. 3 A–C). Moreover, in CHO cells expressing Dr-VSP, depolarization to 100 mV time-dependently reduced Kv7.2 currents; in contrast, currents carried by the Kv7.2 A317T and Kv7.2 L318V channels were completely insensitive to DrVSP-mediated inhibition at all investigated time points (Fig. 3 D and E).
Fig. 3.
Effect of PIP2 levels manipulations on Kv7.2, Kv7.2 A317T, and Kv7.2 L318V currents. (A) Macroscopic currents recorded in the presence of 100 µM Dic8-PIP2 in an intracellular pipette solution at 0 min (immediately after patch rupture, black traces) and after 4 min of whole-cell intracellular dialysis (colored traces). (B) Normalized whole-cell currents for the three channels are indicated as a function of time; 100 µM Dic8-PIP2 was added to the pipette solution. (C) V½ values comparison between cells recorded in the absence (−) or in the presence (+) of 100 µM Dic8-PIP2 in the intracellular solution after 4 min of whole-cell intracellular dialysis. * = P < 0.05. (D) Currents recorded in response to the indicated voltage protocol in cells expressing DrVSP and Kv7.2, Kv7.2 A317T, or Kv7.2 L318V. Time scale, 2 s. (E) Time-dependent current changes in cells coexpressing the indicated channels and DrVSP. The data are expressed as the ratio between the current values recorded at 0 mV immediately after (t2) and before (t1) the Dr-VSP–activating +100 mV depolarizing step as a function of time.
Molecular Modeling and MD Simulations of the Kv7.2 AG Variants.
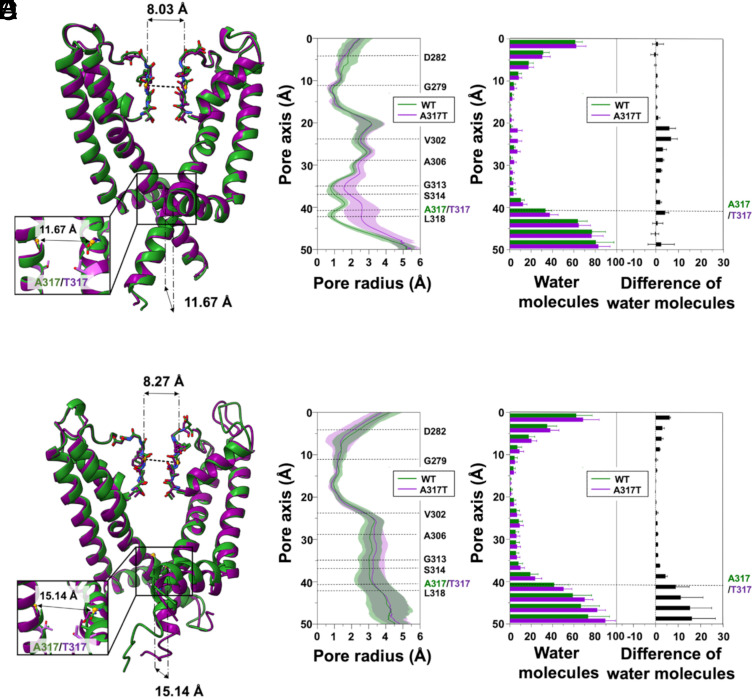
To gain insight into the molecular mechanism underlying the GoF effect of the described Kv7.2 pathogenic variants, all-atom MD simulations were performed. In addition to A317T and L318V, we also modeled the Kv7.2 G313S variant, which is paralogous to a pathogenic Kv7.5 variant that also causes strong GoF effects (46). Given the relevance of allosteric coupling between the AG and the SF regions in K+ channels and the occurrence of DEE-causing mutations at the filter residue D282 (34, 47, 48), we modeled the WT sequence with either a charged or protonated neutral aspartic acid at this site. WT and mutant protein pores were simulated in two distinct conformations, each with a different degree of AG opening, quantified by cross-distances (CDs) between pairs of Cα or side-chain atoms from diagonally opposed subunits. Five distances were used in total, one characterizing the SF via the Cα atoms of G279 (d1) and four characterizing the AG, using the Cα atoms of G313 (d2) and A317 (d3), the Oγ atoms of S314 (d4), and the Cδ atoms of L318 (d5). The latter two residues form the main AG constrictions in closed Kv7 structures (37, 49).
The first conformation simulated was derived from the cryoelectron microscopy (cryo-EM) structure of human Kv7.2 (hKv7.2, PDB ID: 7CR0) (37) and showed a conductive filter (d1 = 8.03 Å) and a constricted gate, with d2 = 11.67 Å, d3 = 13.69 Å, d4 = 4.00 Å and d5= 7.14 Å (SI Appendix, Fig. S4). Since no experimental structure of Kv7.2 with an open AG was available at the beginning of this project, the open Kv7.2 conformation was homology modeled using open human Kv7.1 (hKv7.1, PDB ID: 6V01) as a template (38), resulting in a conducting filter (d1=7.97 Å) and a wide AG (d2 = 13.55 Å, d3 = 19.62 Å, d4 = 12.51 Å, and d5 = 17.30 Å). This modeled structure is nearly identical to that of four recently reported cryo-EM hKv7.2 structures with open AG conformations (PDB ID: 8J01, 8J02, 8W4U, and 8IJK), each obtained in the presence of both small-molecule activators and PIP2 (14, 49). In fact, the comparison resulted in RMSDs between 0.331 Å and 0.889 Å, calculated for all 115 Cα atoms of the simulated chains, and very similar values of CDs (SI Appendix, Fig. S5).
The channels were embedded in explicit POPC membrane bilayers, solvated with water and ions, and simulated in five independent replicates for both closed and open AG conformations, obtaining a cumulative time length of 24.5 μs. All protein structures were stable during the MD simulations and did not substantially deviate from the starting conformations, as evidenced by backbone RMSDs averaged over the replicas that are stationary around values between 2.5 and 3 Å (see SI Appendix, Figs. S6 and S7 for closed systems and SI Appendix, Figs. S8 and S9 for open systems).
To investigate the impact of the mutations on the protein structure, we first examined the size of the channel pore along the simulated trajectories. For the WT channel, the open and closed pore radius profiles, averaged over the replicas, are superimposed on those of the corresponding starting structures, confirming that the pore geometry is maintained throughout the simulations, even with neutral D282 (SI Appendix, Fig. S10). When we compared the pore profiles of A317T, L318V, and G313S to those of the WT Kv7.2, the results for starting closed and open AG structures differed. In particular, the A317T mutation induced a widening of the closed AG region, from above the level of G313 down to the cytoplasm, which was highly pronounced in correspondence of the mutated residue (Fig. 4 A and B). Interestingly, we observed significant loosening of the two distinctive closed-gate constrictions at residues S314 and L318. This localized broadening of the pore makes it more accessible to water, as shown by the average number of water molecules computed along the channel axis (Fig. 4C). The difference is most noticeable in the CC region, particularly around V302, where the WT system is essentially dry, while A317T accommodates approximately eight water molecules. In contrast, the WT and A317T open structure trajectories showed no difference in pore profile (Fig. 4 D and E) or extent of hydration of the CC (Fig. 4F). In line with the enhanced hydrophilicity of the substitution, more water molecules entered the cytoplasmic side of the mutated pore at A317T and below, but the difference remained limited to that region.
Fig. 4.
MD simulations of the A317T Kv7.2 variant. (A) Superposition of WT (green) and A317T (purple) representative Kv7.2 closed structures after equilibration and before MD production simulations. (B) Channel radius profiles along the pore axis of the two proteins, averaged over all the simulated replicas. Shaded regions indicate standard deviations (SD). (C) Distribution of water molecules along the channel axis. Averages and SD were calculated for all replicates. (D–F) Same as (A–C) but for the open structures.
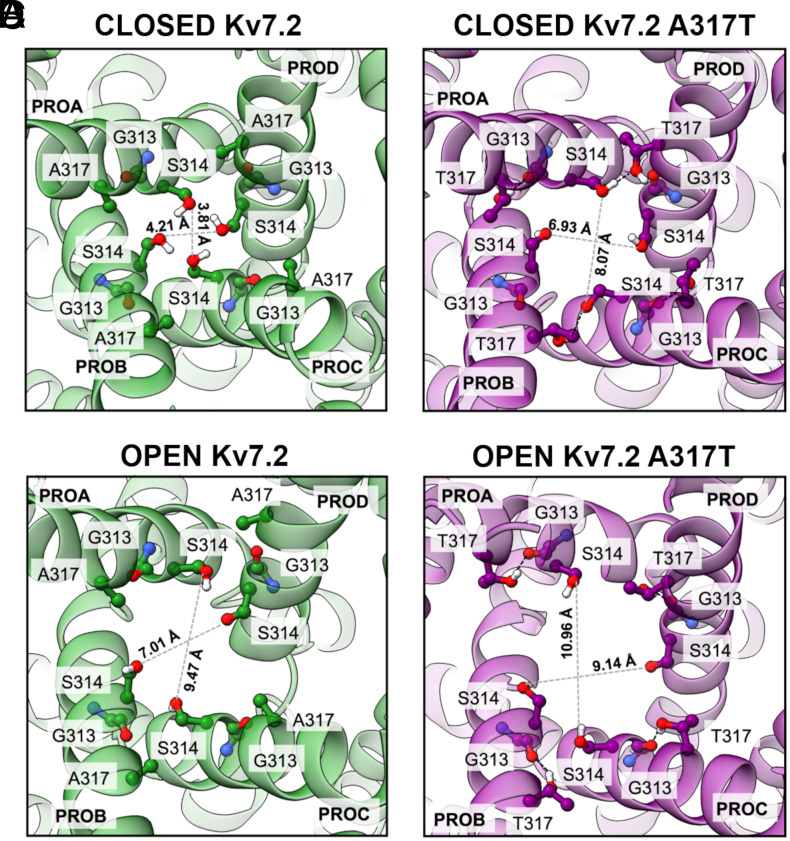
Given the rearrangement of the closed A317T AG into a wider conformation, we searched for interactions involving the mutated amino acid that could be responsible for its stabilization. We found that the inserted threonine sidechains can form hydrogen bonds (HBs) with S314 from adjacent subunits (average persistence times of 54.8%, 68.0%, 64.3%, and 45.1% for the four pairs of chains; SI Appendix, Fig. S11), which causes the serine sidechains to point away from the pore lumen, removing the stable constriction they form in the WT closed conformation (Fig. 5 A and B). Notably, this S314 orientation is similar to that observed in a recently determined open Kv7.2 structure bound to the Ebio1 activator (49) (SI Appendix, Fig. S12). In summary, the substituted T317 is directly involved in interchain bonds that cause partial widening of the closed AG, generating a conformation that allows more water entry and may be more prone to open completely. Conversely, in the open conformations, the chains are more distant from each other, the T317-S314 intersubunit interaction is absent, and the mutated threonine mostly connects to G313 from the same subunit (Fig. 5 C and D and SI Appendix, Fig. S11). Notably, no current could be recorded from cells expressing Kv7.2 channels in which the S314 residue (volume of 88.6 Å3) was substituted with a bulkier (227.8 Å3) tryptophan residue, as well as by an alanine residue which has a similar volume (89 Å3), but a much larger hydropathy index (1.8 for alanine vs −0.8 and −0.9 for serine and tryptophan, respectively), both alone (Kv7.2 S314W and Kv7.2 S314A channels) and in combination with the A317T mutation (Kv7.2 S314W/A317T and Kv7.2 S314A/A317T channels) (SI Appendix, Fig. S13). These results may support the conclusion achieved in our MD simulations that the strong GoF in vitro phenotype prompted by the A317T variant is due to a specific intersubunit interaction between the side chain of the serine at position 314 and the -OH group of T317.
Fig. 5.
Close-up cytosolic views of AG conformations from WT and A317T Kv7.2 channel simulations. (A) Closed WT. (B) Closed A317T variant. (C) Open WT. (D) Open A317T variant. Cross distances are indicated as gray dashed lines. Where present, black dashed lines indicate HBs.
To verify that the structural effects observed in the A317T channel simulations do not depend on the membrane model, we performed additional calculations for the mutated closed pore embedded in bilayers of different lipid compositions (pure POPE, POPG, DMPC, and a mixture of POPC-CHOL-PIP2). In all the cases, we observed a broadened AG and an enhanced hydration of the CC, comparable to those found with the pure POPC bilayer (SI Appendix, Figs. S14 and S15).
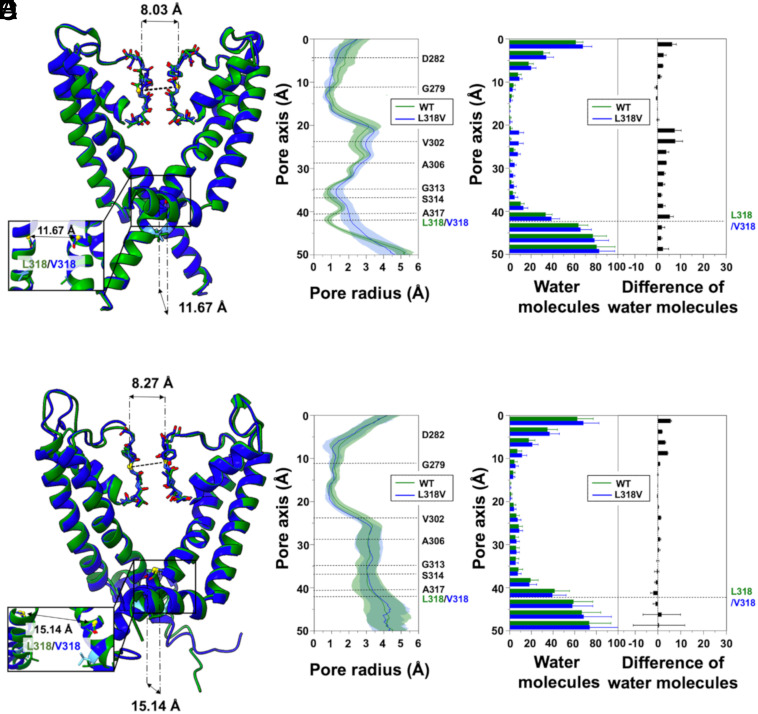
For the L318V variant, we also found broadening of the AG region in the closed conformation, although to a lesser extent than in A317T (Fig. 6 A and B). In L318V, the widening is mostly pronounced at the mutated residue, corresponding to the lower constriction site. Like in A317T, widening of the closed AG results in enhanced hydration of the CC (Fig. 6C). Moreover, the WT and L318V open structure trajectories showed no difference in pore profile (Fig. 6 D and E) or hydration of the CC (Fig. 6F).
Fig. 6.
MD simulations of the L318V Kv7.2 variant. (A) Superposition of WT (green) and L318V (blue) representative Kv7.2 closed structures after equilibration and before MD production simulations. (B) Channel radius profiles along the pore axis of the two proteins averaged over all the simulated replicas. Shaded regions indicate SD. (C) Distribution of water molecules along the channel axis. Averages and SD were calculated for all replicates. (D–F) Same as (A–C) but for the open structures.
To further investigate the structural consequences associated with GoF Kv7.2 variants, we also simulated the G313S variant; this variant, never found in a clinical case, is paralogous to the DEE-causing G347S variant in Kv7.5 (46) and affects a residue immediately before the S314 which interacts with T317 in our MD simulations (Fig. 5). Similar to the A317T and the L318V variants, the Kv7.2 G313S variant also induces GoF effects by increasing the probability of single-channel opening (46). The MD results obtained (SI Appendix, Fig. S16 A–F) revealed that, similarly to the A317T and L318V variants, the G313S mutation also induced a widening of the closed AG (at the level of G313 and A317) and, in this case, also of the CC (around V302), which was associated with increased hydration of this region. The broadened AG conformation is stabilized by a highly persistent hydrogen bond formed in all four S6 by the introduced S313 with A309, which causes a tilting and a rotation of the helices bringing S314 side-chains out of the pore lumen (SI Appendix, Fig. S16 G–I). Conversely, in the open conformation, no major difference was detected in pore size or CC hydration. A comparison between four representative Kv7.2 snapshots of the WT, G313S, A317T, and L318V closed AG extracted from the respective simulations is reported in SI Appendix, Fig. S17, highlighting the enhanced pore opening and cavity hydration of the variants.
To further characterize the WT and mutant channel structures, we examined the temporal evolution of the CDs at the level of the SF (d1) and the AG (d2, d3, d4, and d5). The latter measure the separation between backbone atoms (d2 and d3), or side-chain atoms of specific closed-state constrictions of the WT (d4 and d5). Results for d1 reveal that the mutations did not affect the SF conformation, as all the starting closed AG structures showed d1 values around 8 Å, which are generally considered indicative of a conductive conformation (SI Appendix, Fig. S18). Smaller fluctuations and a preponderance of less constricted (i.e., conducting) filter structures are observed in the mutants (A317T and L318 in particular). For the AG distances, closed A317T and G313S display higher and more fluctuating d2 and d3 values than the WT (SI Appendix, Figs. S19 and S20), while L318V had a less clear signal. Conversely, d4 and d5 measurements clearly indicate a widening of the closed AG constrictions in all three variants, consistent with those of pore profiles (SI Appendix, Figs. S21 and S22, respectively). Hence, while in A317T and G313S the AG widening originates from both side-chain reorientation and separation of the helices, in L318V it is due to side-chains displacements. Finally, in the open AG simulations, no significant differences were detected between the WT and mutants in terms of CDs (SI Appendix, Figs. S23–S25).
Discussion
In the present study, we describe pathogenic variants affecting residues in the pore AG of Kv7.2 leading to GoF features with a marked hyperpolarizing shift of voltage-dependent gating, a significant increase in current density, and faster activation and slower deactivation kinetics. Complementary MD simulation analysis revealed that the substituted residues widen the pore AG by promoting a complex network of intra- and intersubunit interactions, thus providing a plausible atomistic explanation for the observed results.
Phenotypic Spectrum and Genotype–Phenotype Correlations in KCNQ2/3-Related Diseases.
Pathogenic KCNQ2 variants have been identified in patients with a broad spectrum of predominantly neonatal-onset epileptic phenotypes, including SLFNE at the mild end and KCNQ2 early-infantile DEE at the severe end (2, 3, 5). When expressed in vitro, the largest majority of pathogenic variants in KCNQ2 cause LoF effects (2, 16, 17); however, few KCNQ2 GoF variants have also been described, mostly in patients with rarer phenotypes of developmental disorders with moderate to severe ID in the absence of neonatal seizures (3). Indeed, the phenotypic characteristics of the three patients described herein are consistent with this stratification, with GoF effects associated with ASD, global developmental delay, and no seizures in Patient 1 and delayed development with infantile epileptic spasms syndrome followed by DEE-SWAS in Patient 2 (33); neither of these patients had neonatal-onset seizures. Similarly to KCNQ2, LoF pathogenic variants in the closely related KCNQ3 gene cause phenotypes ranging from SLFNE to more severe neurodevelopmental disorders with or without seizures (4, 50, 51), whereas KCNQ3, de novo missense pathogenic variants causing strong GoF effects (28) have also been described in children with global developmental delay (likely similar to that justifying inclusion of Patient 3 in the NDD cohort) (34), ASD, and frequent SWAS (24).
Changes in VSD stability or VSD–PD Coupling Underlie GoF Effects by Known Variants Causing DEEs in KCNQ2 and KCNQ3.
Recent structural evidence for Kv7.1 (38, 52–55), Kv7.2 (14, 37), and Kv7.4 (13, 56) revealed important clues regarding their voltage-dependent gating mechanism, as well as their regulation by CaM and PIP2, providing a framework for the interpretation of the molecular mechanism by which disease-causing variants affect channel function. Although some details are still unknown and specific structural features differentiate Kv7s from other voltage-gated K+ channels, the overall mechanism for voltage-dependent gating can be summarized as follows (10). Membrane depolarization induces a vertical shift in the S4 segment of the VSD, with the first two positively charged arginine residues (R1, R2) passing through the gating charge transfer center formed by a phenylalanine residue in S2 and two negatively charged residues in S2 and S3. This upward dislocation of S4 induces a lateral rotation and an upward motion of the S4–S5 linker which bends the S6 C-terminal half of an adjacent subunit and leads to the opening of the pore. In the closed pore state of Kv7.2 (PDB ID: 7CR0), the diagonal atom-to-atom distance is ~4 Å at the constriction-lining residue S314. In contrast, in the open state, the constriction-lining residue is G310, which has a diagonal atom-to-atom distance of 8 Å. Pore opening is also accompanied, and possibly facilitated, by the fact that, upon depolarization, the HA and HB helices in the proximal C-terminus, where CaM binds in the closed channel configuration, undergo an almost 180° rotation, causing the HA and S6 helices to join in a continuous helix and CaM to be released from its attachment site in the S2–S3 linker (10, 14).
With the exception of G239S in S5 (20), all the described KCNQ2 GoF variants affect the VSD of Kv7.2. For example, recurrent variants at R2 in S4 of Kv7.2 increase the maximal current density, causing a marked hyperpolarizing shift in the voltage dependence of activation and accelerating activation kinetics, suggesting an increased sensitivity of the opening process to voltage (22). Given that R2 forms an intricate network of electrostatic interactions with neighboring negatively charged residues in the resting configuration of the VSD, while no interaction involving R2 occurs when the VSD occupies the active configuration, these GoF effects occur as a consequence of a mutation-induced destabilization of the resting VSD configuration, leading to constitutive channel activation and pore opening (28). A similar molecular mechanism likely explains the GoF effects in vitro caused by variants affecting other VSD residues in both KCNQ2 (19, 21) and KCNQ3 (23, 28). Although no direct structural confirmation for such a mechanism is currently available, voltage-clamp fluorometry studies of both Kv7.2 (29) and Kv7.3 (30) confirm that variants affecting charged residues in the proximal part of S4, including R1 and R2, alter channel function by directly impacting S4 activation, whereas those in the distal part of S4 do not interfere with VSD movement during activation gating but rather with VSD–PD coupling.
Constitutive Pore Opening by KCNQ2/3 Pathogenic Variants in the AG.
Both A317 and L318 residues are located in a highly flexible region of S6 where the pore opens as a consequence of the mechanical forces imposed by VSD repositioning during activation. When expressed as homomers or heteromers, channels carrying the A317T or the L318V variants displayed a GoF in vitro phenotype, with a dramatic increase in current density and a marked facilitation of the voltage-dependent opening process. Nonstationary noise analysis revealed that these changes, highly consistent with the in vitro GoF phenotype, were due to an increase in the probability of channel opening, with no changes in the number of functional channels or in the amplitude of single-channel currents. MD simulations indicate that, with respect to the WT closed conformation, the mutations induce a broadened pore and an increase in CC hydration. Both of these features are consistent with the observed GoF effects, since a broadened gate could facilitate the transition of the mutated channel toward the open state, and the concomitant increase in hydration of the cavity may facilitate channel gating (11, 57). In the A317T variant, the formation of HBs between the -OH group of T317 and the side chain of S314 from an adjacent subunit removes the AG constrictions, inducing a constitutive enlargement of its diameter. Such stabilization of the pore in an open-like conformation, which occurs independently of voltage-dependent VSD repositioning or VSD–PD coupling, is also observed in simulations of L318V and G313S. Notably, Kv7.3 subunits carrying the A356T variant, corresponding to A317T in Kv7.2 and also causing neurodevelopmental delay (34), induced strong GoF effects, although only when expressed with Kv7.2 or Kv7.2 and Kv7.3 subunits. A similar lack of functional current in the homomeric state but strong GoF when coexpressed with Kv7.2 subunits, has been previously reported for another KCNQ3 variant affecting the turret domain in the S5–S6 linker (Kv7.3 A315V) (58).
Kv7.2 Channels Carrying GoF Variants in the AG Are Insensitive to Changes in Cellular PIP2 Levels.
The endogenous lipid PIP2 is essential for the activation of KCNQ channels (12, 59). Although PIP2-bound structures of the Kv7.1 (52), Kv7.2, (14) and Kv7.4 (47) channels have been recently described, the mechanism of PIP2 activation is still unclear (10). PIP2 seems to facilitate voltage-dependent pore opening by binding to the S2–S3 linker, where it disrupts the (likely inhibitory) interaction between the VSD and CaM (site I in Kv7.1 and Kv7.4) and/or stabilizes the channel in the open state by direct interactions with positively charged residues in the distal S6 or in the proximal C-terminus (site II in Kv7.2 and Kv7.4) (10).
Currents from Kv7.2 A317T and L318V channels are fully insensitive to changes in cellular PIP2 levels, as mutant channels are resistant to inhibition or potentiation when PIP2 levels are decreased or increased, respectively, with the experimental approaches herein performed; whether the A317T and L318V variants in Kv7.2 trigger a complete loss of PIP2-dependent regulation or a strong enhancement of PIP2 affinity remains to be investigated. Nonetheless, the L351K substitution in Kv7.1, which affects the paralogous residue to L318 in Kv7.2, also destabilizes the closed pore state and renders the channels insensitive to PIP2 depletion (12). The fact that pore opening in the Kv7.2 A317T and L318V channels is insensitive to changes in PIP2 availability allows us to hypothesize that these variants promote structural changes leading to a pore configuration that recapitulate the open pore structure promoted by PIP2 in Kv7.2 channels (14). This hypothesis finds support on the identical permeability ratios for K+/Rb+ and K+/Cs+ between the Kv7.2 and Kv7.2 A317T channels, as well as by our MD simulations showing no marked difference in the average CDs among the open pores of the Kv7.2 and Kv7.2 A317T and L318V channels.
Conclusions
In vitro functional features of disease-causing ion channel variants are often associated with specific clinical traits, such as disease severity, comorbidities, and therapeutic response. Thus, defining the molecular pathomechanism(s) responsible for variant-induced functional changes may unveil prognostic clues and allow patient stratification for personalized treatment approaches. We herein provide evidence that pathogenic variants in the pore AG of the Kv7.2 (and Kv7.3) channels cause strong GoF effects through a VSD-independent stabilization of the open pore configuration, thereby expanding genotype-’phenotype correlations and providing additional hypotheses for Kv7 channel gating and regulation deserving structural testing. The present results have also relevant pharmacological implications; in fact, both the Kv7.1 activator ML277 (60, 61), as well as the potent and selective Kv7.2 activator Ebio1 (49), trigger structural changes in the inner pore gate similar to those herein-described. Thus, drug-induced manipulation of the pore AG network may be exploited for the treatment of diseases linked to Kv7 channel dysfunction, including severe neurodevelopmental disorders.
Methods
Patient Recruitment and Genetic Analyses.
Patient 1 was recruited at the Epilepsy Research Centre, Department of Medicine, University of Melbourne, Australia. Detailed phenotyping and trio exome sequencing were performed. The study was approved by the Human Research Ethics Committee, Austin Health. Written informed consent for study participation was obtained from the patient’s parents. Patient 2 (33) patient 3 (34) have been reported previously in large cohort studies of neurodevelopmental disorders and DEEs (Table 1).
Mutagenesis and Heterologous Expression of KCNQ2 and KCNQ3 cDNAs.
Mutations were engineered in human KCNQ2 and KCNQ3 cloned into pcDNA3 by site-directed mutagenesis. Channel subunits were expressed in CHO cells by transient transfection. CHO cells were transfected using Lipofectamine 2000 (16).
Whole-Cell Electrophysiology and Nonstationary Noise Analysis.
Currents from CHO cells were recorded at room temperature (20–22 °C) 1 to 3 d after transfection. The extracellular solution contained (in mM) 138 NaCl, 5.4 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES, pH 7.4, with NaOH. The intracellular solution contained (in mM) 140 KCl, 2 MgCl2, 10 EGTA, 10 HEPES, and 5 Mg-ATP at pH 7.3 to 7.4 with KOH. To evaluate PIP2 sensitivity, the water-soluble, synthetic PIP2 analog dic8-PIP2 (100 μM) was added to the intracellular solution. pCLAMP software (version 10.0.2) was used for data acquisition and analysis. Linear cell capacitance (C) and series-resistance (RS) calculations, as well as cell capacitance compensation, were performed as described previously (62). Current densities (expressed in pA/pF) were calculated as peak K+ currents divided by C. Data were acquired at 0.5 to 2 kHz and filtered at 1 to 5 kHz with the 4-pole lowpass Bessel filter of the amplifier. No corrections were made for the liquid junction potentials. To generate conductance-’voltage (G/V) curves, the cells were held at −80 mV and then depolarized for 1.5 s from −80 mV to 20/80 mV in 10 mV increments, followed by an isopotential pulse at 0 mV for a duration of 300 ms. The current values recorded at the beginning of the 0 mV pulse were measured, normalized, and expressed as a function of the preceding voltages. The data were then fit to a Boltzmann distribution of the following equation 1:
where V is the test potential, V½ is the half-activation potential, and k is the slope factor (16).
Nonstationary noise analysis data were acquired and analyzed as previously described (46). Currents were lowpass filtered at 10 kHz and sampled at 100 kHz. Variance was obtained by averaging the squared difference of consecutive records after appropriate scaling. The baseline variance was subtracted, and the variance-mean plot was fitted by equation 2:
where σ2 is the variance, <I> is the mean current, i is the single-channel current, and N is the number of channels. During fitting, i and N were not constrained, and points were not weighted by their SE. The maximal open probability was calculated as Pmax= <I> max/(Ni). The GePulse and Ana programs (http://users.ge.ibf.cnr.it/pusch/programs-mik.htm) were used for data acquisition and analysis, respectively.
Permeability to monovalent cations was measured as previously described (63). Briefly, an extracellular solution containing (in mM) 127.4 NaCl, 15 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES (pH 7.4 with NaOH) was used to measure K+ permeability (PK). KCl was replaced by 15 mM RbCl or CsCl to estimate the permeabilities of Rb+ and Cs+. Currents were activated by 500-ms pulses at 20 mV and deactivated by 1,500 ms steps from −120 mV to −30 mV to estimate their reversal potential under each bi-ionic condition. The permeability of the outer monovalent cations relative to K+ (PX/PK) was determined using the Goldman–Hodgkin–Katz equation: PX/PK = ([K]o/[X]o)exp – (FΔErev/RT), where ΔErev is the shift in reversal potential when K+ is replaced by the test cation X. The constants F, R, and T in the equation also adhere to their conventional thermodynamic meanings.
Cell-Surface Biotinylation and Western Blotting.
The plasma membrane expression of wild-type and mutant Kv7.2 or Kv7.3 subunits in CHO cells was investigated by surface biotinylation of membrane proteins in transfected cells one day after transfection as described previously (28). Briefly, following incubation with Sulfo-NHS-LC-Biotin (0.5 mg/mL; Thermo Fisher, Monza, Italy), a cell membrane impermeable reagent, for 20 min at 4 °C, the cells were lysed, and the protein lysates were incubated with streptavidin beads (Thermo Fisher, Monza, Italy) to isolate the biotinylated proteins. Channel subunits in streptavidin precipitates and total lysates were analyzed via western blotting on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels and transferred onto a polyvinylidene fluoride membrane (Merck, Milan, Italy).
Membranes were incubated overnight at 4 °C with rabbit polyclonal anti-Kv7.2 (1:1,000 dilution; GeneTex, Irvine, USA) or anti-Kv7.3 (1:1,000 dilution; Alomone Lab., Jerusalem, Israel); secondary antibodies conjugated to the fluorescent dyes StarBright Blue 520 or StarBright Blue 700 (Bio-Rad, Milan, Italy) were incubated for 1 h at RT. An anti-GAPDH antibody (dilution 1:2,000: Thermo Fisher, Monza, Italy) was used to check for equal protein loading and to assess the purity of the plasma-membrane preparation. Blot images, as well as fluorescence signals, were acquired with a ChemiDoc™ Touch Imaging System (Bio-Rad, Milan, Italy). The images were analyzed by using Image Lab Software (version 6.1; Bio-Rad, Milan, Italy). The intensities of the Kv7.2 and Kv7.3 bands were normalized to that of GAPDH. The plasma-surface Kv7 protein concentration was normalized to the total Kv7 protein concentration.
Statistics.
The data are expressed as the means ± SEMs for the number of independent samples (n). A normal distribution was assessed using the D’Agostino–Pearson normality test. To compare two normally distributed sample groups, the unpaired two-tailed Student’s t test was used. To compare more than two normally distributed sample groups, one-way ANOVA followed by post hoc Tukey’s multiple comparisons test was used. P < 0.05 was considered to indicate statistical significance. Statistical analysis was carried out using Prism v9 (GraphPad Software).
Modeling and MD Simulations of Kv7.2 Channels with a Starting Closed AG.
To model the Kv7.2 pore with a closed AG, we used the cryo-EM structure of the human channel (hKv7.2, PDB ID: 7CR0, apo-state) (37). This configuration has a conductive SF, as confirmed by the CDs between G279 Cα atoms of pairs of diagonally opposed monomers across the pore (distance d1 = 8.03 Å). Moreover, the AG is characterized by short distances between the Cα atoms facing the G313 (distance d2 = 11.67 Å) and A317 (distance d3 = 13.69 Å) residues.
Two other relevant closed-AG constriction points are identified by the distances between S314 Oγ atoms (d4 = 4.00 Å) and L318 Cδ atoms (d5 =7.14 Å). All five distances are illustrated in SI Appendix, Fig. S4. The segment of each subunit defining the TM PD (residues G215 to E330) was retained for MD simulations, while the VSDs were removed to reduce the size of the system.
The CHARMM-GUI Membrane Builder server (64) was used to prepare all the necessary files for the simulations. The five Kv7.2 structures (WT Kv7.2 with either charged or neutral D282; variants A317T, L318V, and G313S) were inserted into a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane and solvated with explicit water molecules. The total charge was neutralized with a 150 mM KCl solution, obtaining approximately 155,000 atoms in total. We used the NAMD software (65) and the CHARMM36 (66–68) force field for proteins and lipids and used an integration time step ∆t = 2 fs. Before production, as previously described (69, 70), all the systems were relaxed with ∼30 ns of equilibration, extending the CHARMM-GUI equilibration protocol and allowing proper hydration of solvent-exposed regions of the pore cavity. Then, five statistically independent replicates of each system were simulated for 500 ns each in the NPT ensemble at 1 atm and 310 K.
To investigate the effect of the membrane lipid composition, we performed additional MD simulations of the Kv7.2 A317T channel embedded in membranes of different lipid species (SI Appendix, Table S2), such as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and a mixture of POPC-cholesterol and PIP2. To speed up the calculations, the hydrogen mass repartitioning (HMR) method with an integration time step of 4 fs was used in these runs (71–73). Before production, each system was relaxed beyond the default CHARMM-GUI equilibration procedure with an additional ∼70 ns in the NPT ensemble. All simulations were performed in the absence of a TM potential, i.e. at 0 mV. Additional details about the MD setup and accompanying results can be found in SI Appendix.
Modeling and MD Simulations of Kv7.2 Channels with a Starting Open AG.
When we started this project, no experimental structure of Kv7.2 with an open IG was available. We thus relied on homology modeling using open human Kv7.1 (hKv7.1, PDB ID: 6V01) as a template (38). Our model comprises only the pore TM domain (residues G215 to E330) and was constructed in two steps following previously described procedures (69, 70, 74). First, we modeled each of the four TM segments separately using the structure prediction method included in the SWISS-MODEL server (75). Then, the Kv7.2 PD was assembled by structurally aligning the four subunits to the template in a clockwise order viewed from the extracellular side using UCSF Chimera (76). After the standard CHARMM-GUI relaxation, an additional 30 ns equilibration was performed with positional restraints on the protein and on the membrane atoms. The equilibrated channel has a conductive SF (d1 = 7.97 Å) and an open AG (d2 = 13.55 Å; d3 = 19.62 Å). At this point, to avoid sudden occlusion of the AG, a specific additional equilibration procedure was designed: three independent replicates of the system were further simulated for 500 ns, each by applying a set of harmonic restraints on the atomic distances between pairs of Cα atoms of residues G313 and A317 from diagonally opposed subunits and then for 100 ns completely unrestrained. At the end of these simulations, the gate was stably open, and the final conformations were used as starting structures for five 500 ns-long MD simulations of each of the channels investigated.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Alessia Vignolo, Mattia Pini, and Sergio Decherchi for their kind assistance at the Italian Institute of Technology (IIT) computing center and Michael Pusch at the Biophysics Institute at the National Research Council in Genova (Italy) for sharing noise analysis software. We are grateful to Diego Moruzzo for useful help and technical assistance. Computing time allocations were granted by the CINECA supercomputing center under the ISCRA initiative. We also gratefully acknowledge the High Performance Computing infrastructure and the Support Team at Fondazione IIT. This work was supported by the Italian Ministry for University and Research with PRIN2020 (project 2020XBFEMS to LM), PRIN2022 (project 2022M3KJ4N to MT), and PRIN20 22PNRR (project P2022FJXY5 to MVS and FM; project P2022ZANRF to MT); the European Union - Next Generation EU, Mission 4, Component 2, CUP E63C2 2002170007 (Project “A multiscale integrated approach to the study of the nervous system in health and disease”, MNESYS) (to MT); the Italian Ministry of Health with Ricerca Finalizzata (RF) Projects RF-2019-12370491 to MT and PNRR-MR1-2022-12376528 to MVS, FB, and MT; the FWO (grants 1861419 N and G041821N to SW); and the European Joint Programme on Rare Disease JTC 2020 (TreatKCNQ, to SW and MT). The DDD study includes independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003]. This study used DECIPHER (http://www.deciphergenomics.org), which is funded by Wellcome [grant number WT223718/Z/21/Z]. https://www.ddduk.org/ for full acknowledgment.
Author contributions
S.W., I.E.S., F.B., L.M., F.M., and M.T. designed research; M.N., G.A., A.B., A.R., M.V.S., I.S., V.B., T.-G.A.C., I.E.S., L.M., and F.M. performed research; M.N., G.A., A.B., A.R., M.V.S., I.S., V.B., S.W., T.-G.A.C., I.E.S., F.B., L.M., F.M., and M.T. analyzed data; and M.N., G.A., F.B., L.M., F.M., and M.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission J.C. is a guest editor invited by the Editorial Board.
Contributor Information
Luca Maragliano, Email: l.maragliano@staff.univpm.it.
Maurizio Taglialatela, Email: mtaglial@unina.it.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Weckhuysen S., et al. , KCNQ2 encephalopathy: Emerging phenotype of a neonatal epileptic encephalopathy. Ann. Neurol. 71, 15–25 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Nappi P., et al. , Epileptic channelopathies caused by neuronal Kv7 (KCNQ) channel dysfunction. Pflugers Arch. 472, 881–898 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Miceli F., Soldovieri M. V., Weckhuysen S., Cooper E., Taglialatela M., “KCNQ2-Related Disorders” in GeneReviews®, Adam M. P., et al. , Eds. (University of Washington, Seattle, 1993). [PubMed] [Google Scholar]
- 4.Miceli F., Soldovieri M. V., Weckhuysen S., Cooper E. C., Taglialatela M., “KCNQ3-Related Disorders” in GeneReviews®, Adam M. P., et al. , Eds. (University of Washington, Seattle, 1993). [PubMed] [Google Scholar]
- 5.Zuberi S. M., et al. , ILAE classification and definition of epilepsy syndromes with onset in neonates and infants: Position statement by the ILAE task force on nosology and definitions. Epilepsia 63, 1349–1397 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Scheffer I. E., et al. , ILAE classification of the epilepsies: Position paper of the ILAE commission for classification and terminology. Epilepsia 58, 512–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Symonds J. D., et al. , Incidence and phenotypes of childhood-onset genetic epilepsies: A prospective population-based national cohort. Brain 142, 2303–2318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrese V., Stott J. B., Greenwood I. A., KCNQ-Encoded potassium channels as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 58, 625–648 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Soldovieri M. V., Miceli F., Taglialatela M., Driving with no brakes: Molecular pathophysiology of Kv7 potassium channels. Physiology 26, 365–376 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Huang Y., Ma D., Yang Z., Zhao Y., Guo J., Voltage-gated potassium channels KCNQs: Structures, mechanisms, and modulations. Biochem. Biophys. Res. Commun. 689, 149218 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Gu R.-X., de DeGroot B. L., Central cavity dehydration as a gating mechanism of potassium channels. Nat. Commun. 14, 2178 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaydman M. A., Cui J., PIP2 regulation of KCNQ channels: Biophysical and molecular mechanisms for lipid modulation of voltage-dependent gating. Front. Physiol. 5, 195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y., et al. , Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel. Neuron 110, 237–247.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Ma D., et al. , Ligand activation mechanisms of human KCNQ2 channel. Nat. Commun. 14, 6632 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosa F., et al. Electrophysiological signatures of a developmental delay in a stem cell model of KCNQ2 developmental and epileptic encephalopathy. bioRxiv [Preprint] (2024). https://www.biorxiv.org/content/10.1101/2024.03.13.584717v1 (Accessed 3 April 2024).
- 16.Miceli F., et al. , Genotype-phenotype correlations in neonatal epilepsies caused by mutations in the voltage sensor of K(v)7.2 potassium channel subunits. Proc. Natl. Acad. Sci. U.S.A. 110, 4386–4391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orhan G., et al. , Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Ann. Neurol. 75, 382–394 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Brun L., Viemari J.-C., Villard L., Mouse models of Kcnq2 dysfunction. Epilepsia 63, 2813–2826 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millichap J. J., et al. , Infantile spasms and encephalopathy without preceding neonatal seizures caused by KCNQ2 R198Q, a gain-of-function variant. Epilepsia 58, e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayat A., et al. Phenotypic and functional assessment of two novel KCNQ2 gain-of-function variants Y141N and G239S and effects of amitriptyline treatment. Neurotherapeutics 21, e00296 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miceli F., et al. , KCNQ2 R144 variants cause neurodevelopmental disability with language impairment and autistic features without neonatal seizures through a gain-of-function mechanism. eBioMedicine 81, 104130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulkey S. B., et al. , Neonatal nonepileptic myoclonus is a prominent clinical feature of KCNQ2 gain-of-function variants R201C and R201H. Epilepsia 58, 436–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mary L., et al. , Pathogenic variants in KCNQ2 cause intellectual deficiency without epilepsy: Broadening the phenotypic spectrum of a potassium channelopathy. Am. J. Med. Genet. Part A 185, 1803–1815 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Sands T. T., et al. , Autism and developmental disability caused by KCNQ3 gain-of-function variants. Ann. Neurol. 86, 181–192 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Niday Z., Tzingounis A. V., Potassium channel gain of function in epilepsy: An unresolved paradox. Neuroscientist 24, 368–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varghese N., et al. , KCNQ2/3 Gain-of-function variants and cell excitability: Differential effects in CA1 versus L2/3 pyramidal neurons. J. Neurosci. 43, 6479–6494 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devaux J., et al. , A Kv7.2 mutation associated with early onset epileptic encephalopathy with suppression-burst enhances Kv7/M channel activity. Epilepsia 57, e87 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Miceli F., et al. , Early-onset epileptic encephalopathy caused by gain-of-function mutations in the voltage sensor of Kv7.2 and Kv7.3 potassium channel subunits. J. Neurosci. 35, 3782–3793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edmond M. A., Hinojo-Perez A., Wu X., Perez Rodriguez M. E., Barro-Soria R., Distinctive mechanisms of epilepsy-causing mutants discovered by measuring S4 movement in KCNQ2 channels. eLife 11, e77030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barro-Soria R., Epilepsy-associated mutations in the voltage sensor of KCNQ3 affect voltage dependence of channel opening. J. Gen. Physiol. 151, 247–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masnada S., et al. , Clinical spectrum and genotype–phenotype associations of KCNA2-related encephalopathies. Brain 140, 2337–2354 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Brunklaus A., et al. , Gene variant effects across sodium channelopathies predict function and guide precision therapy. Brain 145, 4275–4286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong P., Xue J., Jiao X., Zhang Y., Yang Z., Genetic etiologies in developmental and/or epileptic encephalopathy with electrical status epilepticus during sleep: Cohort study. Front. Genet. 12, 607965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deciphering Developmental Disorders Study, Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Specchio N., et al. , International league against epilepsy classification and definition of epilepsy syndromes with onset in childhood: Position paper by the ILAE Task Force on nosology and definitions. Epilepsia 63, 1398–1442 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Richards S., et al. , Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Gen. Med. 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., et al. , Molecular basis for ligand activation of the human KCNQ2 channel. Cell Res 31, 52–61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J., MacKinnon R., Structural basis of human KCNQ1 modulation and gating. Cell 180, 340–347.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadley J. K., et al. , Differential tetraethylammonium sensitivity of KCNQ1–4 potassium channels. Br. J. Pharmacol. 129, 413–415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez O., Gonzalez C., Latorre R., Counting channels: A tutorial guide on ion channel fluctuation analysis. Adv. Physiol. Ed. 26, 327–341 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Kim R. Y., Pless S. A., Kurata H. T., PIP2 mediates functional coupling and pharmacology of neuronal KCNQ channels. Proc. Natl. Acad. Sci. U.S.A. 114, E9702–E9711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pant S., et al. , PIP2-dependent coupling of voltage sensor and pore domains in Kv7.2 channel. Commun. Biol. 4, 1189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falkenburger B. H., Jensen J. B., Hille B., Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135, 99–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soldovieri M. V., et al. , Early-onset epileptic encephalopathy caused by a reduced sensitivity of Kv7.2 potassium channels to phosphatidylinositol 4,5-bisphosphate. Sci. Rep. 6, 38167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hossain M. I., et al. , Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J. Biol. Chem. 283, 18248–18259 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Nappi M., et al. , Gain of function due to increased opening probability by two KCNQ5 pore variants causing developmental and epileptic encephalopathy. Proc. Natl. Acad. Sci. U.S.A. 119, e2116887119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millichap J. J., et al. , KCNQ2 encephalopathy: Features, mutational hot spots, and ezogabine treatment of 11 patients. Neurol. Genet. 2, e96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson H. E., et al. , Genetics and genotype-phenotype correlations in early onset epileptic encephalopathy with burst suppression. Ann. Neurol. 81, 419–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S., et al. , A small-molecule activation mechanism that directly opens the KCNQ2 channel. Nat. Chem. Biol. 20, 847–856, (2024), 10.1038/s41589-023-01515-y. [DOI] [PubMed] [Google Scholar]
- 50.Miceli F., et al. , A novel KCNQ3 mutation in familial epilepsy with focal seizures and intellectual disability. Epilepsia 56, e15 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Ambrosino P., et al. , Kv7.3 Compound heterozygous variants in early onset encephalopathy reveal additive contribution of C-terminal residues to PIP2-dependent K+ channel gating. Mol. Neurobiol. 55, 7009–7024 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Sun J., MacKinnon R., Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandala V. S., MacKinnon R., The membrane electric field regulates the PIP2-binding site to gate the KCNQ1 channel. Proc. Natl. Acad. Sci. U.S.A. 120, e2301985120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willegems K., et al. , Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277. Nat. Commun. 13, 3760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma D., et al. , Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers. Proc. Natl. Acad. Sci. U.S.A. 119, e2207067119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li T., et al. , Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell 81, 25–37.e4 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Trick J. L., et al. , Functional annotation of ion channel structures by molecular simulation. Structure 24, 2207–2216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaika O., Hernandez C. C., Bal M., Tolstykh G. P., Shapiro M. S., Determinants within the turret and pore-loop domains of KCNQ3 K+ channels governing functional activity Biophys. J. 95, 5121–5137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H., et al. , PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron 37, 963–975 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Mattmann M. E., et al. , Identification of (R)-N-(4-(4-methoxyphenyl)thiazol-2-yl)-1-tosylpiperidine-2-carboxamide, ML277, as a novel, potent and selective K(v)7.1 (KCNQ1) potassium channel activator. Bioorg. Med. Chem. Lett. 22, 5936–5941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennan S., et al. , Slowly activating voltage-gated potassium current potentiation by ML277 is a novel cardioprotective intervention. Proc. Natl. Acad. Sci. U.S.A. Nexus 2, pgad156 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soldovieri M. V., et al. , Atypical gating of M-type potassium channels conferred by mutations in uncharged residues in the S4 region of KCNQ2 causing benign familial neonatal convulsions. J. Neurosci. 27, 4919–4928 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prole D. L., Marrion N. V., Ionic permeation and conduction properties of neuronal KCNQ2/KCNQ3 potassium channels. Biophys. J. 86, 1454–1469 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jo S., Kim T., Im W., Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLOS One 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J., MacKerell A. D., CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alberini G., Benfenati F., Maragliano L., Structural mechanism of ω-currents in a mutated Kv7.2 voltage sensor domain from molecular dynamics simulations. J. Chem. Inf. Model. 61, 1354–1367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alberini G., et al. , Molecular dynamics simulations of ion permeation in human voltage-gated sodium channels. J. Chem. Theory Comput. 19, 2953–2972 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feenstra K. A., Hess B., Berendsen H. J. C., Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20, 786–798 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Hopkins C. W., Le Grand S., Walker R. C., Roitberg A. E., Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Balusek C., et al. , Accelerating membrane simulations with hydrogen mass repartitioning. J. Chem. Theory Comput. 15, 4673–4686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y., et al. , Structural modelling and mutant cycle analysis predict pharmacoresponsiveness of a Nav1.7 mutant channel. Nat. Commun. 3, 1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen E. F., et al. , UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.