Abstract
Microsatellite instability-high (MSI-H) is a critical biomarker for immunotherapy, yet primary resistance remains a significant challenge. Current MSI-H detection methods evaluate the proportion of MSI-H loci, termed molecular MSI-H score, which can be affected by intratumoral heterogeneity (ITH). To address this limitation, we propose evaluating MSI-H at the cellular level to improve the prediction of immunotherapy outcomes. Using bulk tissue (TCGA-CRC) and cell line (CCLE-CRC) datasets, we identified genes highly expressed in MSI-H and MSS samples. These signatures were applied to a single-cell RNA sequencing (scCRC) dataset for enrichment analysis, enabling classification of tumor cells into MSI-H, MSS, and microsatellite dual (MSD) clusters using a Gaussian finite mixture model. Validation showed that MSI-H and MSS enrichment scores were higher in mismatch repair-deficient (MMRd) and mismatch repair-proficient (MMRp) patients, respectively. Functional enrichment analysis revealed that MSI-H cells were associated with pathways such as carboxylic acid catabolism, inflammatory responses, and IL-6/JAK2/STAT3 signaling. We developed a cellular MSI-H signature using genes specifically expressed in the MSI-H cell cluster and transformed the scCRC dataset into a cell-type-specific pseudobulk expression matrix. Using this matrix as a reference, we performed reference-based deconvolution on TCGA-CRC data. We defined the deconvolution score of MSI-H cell as cellular MSI-H score. This score strongly correlated with the molecular MSI-H score (R = 0.55, P < 0.001) and showed modest correlations with macrophage (MoMac, R = 0.14) and CD8+ T-cell (R = 0.11). To investigate its potential for clinical application, we applied the cellular MSI-H signature to the BJ-cohort, comprising 97 immunotherapy-treated gastrointestinal patients sequenced with a 395-gene panel. The cellular MSI-H score was significantly higher in responders (P = 0.002), positively correlated with tumor reduction percentage (R = 0.29, P = 0.006), and associated with improved progression-free survival (PFS) (HR: 0.00, 95% CI: 0.00-0.31, P = 0.021). In summary, the cellular MSI-H score reflects the MSI-H cell level within a tumor and demonstrates superior accuracy compared to molecular MSI-H status in predicting immunotherapy response and PFS. This underscores its potential as a more robust biomarker for guiding immunotherapy decisions.
Keywords: MSI, single-cell transcriptome, deconvolution, immunotherapy, prognosis
Introduction
Gastrointestinal cancer is a prevalent malignancy worldwide [1]. One well-described genetic subset of gastrointestinal cancers, ranging from 10-15%, is characterized by mismatch repair-deficient (MMRd)/microsatellite instability-high (MSI-H) status. Immune-checkpoint inhibitors (ICIs) have demonstrated notable clinical efficacy as first-line treatment for metastatic MMRd/MSI-H gastrointestinal cancer [2]. Despite these encouraging results, about half of the patients do not respond to ICIs, showing primary resistance to immunotherapy. Recent evidence suggests that misdiagnosis of MMRd/MSI-H status may account for a substantial portion of these primary resistant cases [3,4]. Besides the misinterpreted diagnostic results, the intra-tumoral heterogeneity (ITH) is the major reason for the misdiagnosis of MMRd/MSI-H status [5,6], and the most immediate indicator of ITH lies in the proportional composition of various cell types within the tumor microenvironment.
MSI detection currently relies on assessing the ratio of unstable microsatellite loci to the total tested microsatellite loci in the genome, yielding a molecular MSI-H score. Tissues are then qualitatively classified as MSI-H or MSS based on predefined molecular MSI-H score thresholds. Approximately 20 million microsatellite (MS) loci exist in the entire genome [7], with only about 1 in 40 loci being detectable through whole exome sequencing (WES). Various panel-based algorithms have been developed, but most only encompass a fraction of MS loci. For instance, the FoundationOne 395-gene panel includes around 1800 MS loci [8], the Guardant360 74-gene panel incorporates 99 MS loci [9], and the Burning Rock 36-gene panel covers 18 MS loci [10]. These methods initially assess the instability of each MS locus based-on length distribution, then calculate the proportion of MSI-H loci in the genome. Generally, when this ratio exceeds 40%, the sample is considered MSI-H tissue [10,11]. The resulting MSI score obtained through this approach reflects the molecular-level proportion of MSI-H loci, indirectly indicating the presence of MSI-H cells in the tumor microenvironment. However, the accuracy of this method is influenced by ITH [5] as indicated by the relatively high primary resistance rates. With the maturation of single-cell technologies, it is now possible to analyze the tumor microenvironment at the cellular level. This suggests that by identifying the MSI status of each cell, we can decipher the proportion of MSI-H cells in the tumor microenvironment, overcoming ITH, providing a basis for evaluating the effectiveness of immunotherapy.
Materials and methods
Bulk tissue, cell-line, and single-cell RNA sequencing data acquisition and preprocess
We retrieved public transcriptome datasets from three dimensions: bulk tissues from The Cancer Genome Atlas (TCGA-CRC) and Gene Expression Omnibus (GEO, GSE39582), cell lines from the Cancer Cell Line Encyclopedia (CCLE-CRC), and single-cell data from GEO (scCRC). The TCGA-CRC dataset (N = 360) was obtained from UCSC-Xena (http://xena.ucsc.edu/), with molecular MSI-H scores provided by MANTIS [11]. Among them, 64 cases exhibited MSI-H, while 296 cases were MSS. The CRC cell line data (N = 57) was downloaded from CCLE (https://depmap.org/portal) with version 2020Q4, comprising 38 with MSS and 19 with MSI-H. Single-cell transcriptome data (scCRC) from 28 MMRp and 34 MMRd primary treatment-naïve CRC patients were acquired from GEO (GSE178341), excluding one MMRp patient and one MMRd patient lacking tumor tissues. This comprised 105,122 epithelial cells and many other cells. The GSE39582 dataset, comprised of 74 MMRd and 441 MMRp patients, were download and utilized as an independent validation dataset.
Patients, treatment, and sample sequencing in the immunotherapy cohort
We utilized an immunotherapy cohort, the BJ-cohort, to investigate the role of cellular MSI-H score in immunotherapy. A total of 96 patients with metastatic gastrointestinal cancers, who failed standard therapy and received anti-PD-1/PD-L1 therapy or a combination with a CTLA-4 inhibitor between January 2016 and January 2018, were enrolled. MMR/MSI status was determined by IHC or PCR. IHC was performed on archival formalin-fixed, paraffin-embedded (FFPE) tissue sections using monoclonal antibodies targeting MLH1, MSH2, MSH6, and PMS2. Tumors with a loss of expression in any of these four mismatch repair (MMR) proteins were classified as MMRd, while those retaining expression were categorized as MMRp. In some cases, PCR-based molecular testing was conducted to assess MSI at five specific loci: BAT-25, BAT-26, D2S123, D5S346, and D17S250. Tumors demonstrating instability at two or more markers were defined as MSI-H, whereas those with instability at fewer than two markers were considered MS. Gene expression profiling was conducted through a panel sequencing platform assessing the expression of 395 human genes (395-panel).
Establishment of a single-cell level MSI classification system
Establishment of MSI-H signature and MSS signature
We conducted an analysis of differentially expressed genes (DEGs) between MSI-H and MSS tumor samples in the TCGA-CRC dataset. Genes meeting the criteria of an adjusted p-value < 0.05 and log2(fold change) > 2 were identified as candidate DEGs. Subsequently, we examined the expression differences of these candidate DEGs in MSI-H and MSS cell lines from the CCLE-CRC dataset, selecting genes with significant differences to form the potential set of DEGs. Finally, genes exhibiting non-zero expression values in at least 1% of the tumor cells in scCRC dataset were retained as the final set of DEGs. Based on whether these genes were highly expressed in MSI-H or MSS, we defined the molecular MSI-H and MSS signatures.
Scoring the enrichment of MSI-H and MSS signatures in each tumor cell
Utilizing the acquired molecular MSI-H and MSS signatures, we computed the MSI and MSS scores for each tumor cell through AUCell, which employs the “Area Under the Curve (AUC)” to assess whether a critical subset of the input gene set is enriched within the expressed genes for each cell. Analyzing the distribution of AUC scores across all cells enables exploration of the relative expression of the signatures.
Determining the MSI status of each tumor cell
Based on the MSI-H and MSS enrichment scores of each single cell, we performed cell clustering with mclust [7,12], which uses the Gaussian finite mixture model (GMM) to fit data. GMM assumes a multivariate Gaussian distribution for each component, i.e. fx(x;θk)∽N(μk;Σk). Thus, clusters are ellipsoidal, centered at the mean vector μk and with other geometric features determined by the covariance matrix Σk. We employed all 14 models applied in this software and utilized the Bayesian Information Criterion (BIC) to determine the optimal number of mixing components and the covariance parameterization.
Functional enrichment analysis
We conducted functional enrichment analysis at two levels. Firstly, the DEGs-based. We computed DEGs and performed enrichment analysis using reference gene sets from Gene Ontology biological process (GO-BP). The p-values were calculated through over-representation analysis. Secondly, Gene Set Enrichment Analysis (GSEA), leveraging gene expression ranks of all genes rather than focusing solely on DEGs using clusterprofiler [13].
Trajectory analysis
Trajectory analysis in the scCRC dataset was carried out using Monocle3. The union of the top 3000 highly variable genes and all genes from the MSI-H and MSS signatures were employed for principal component analysis (PCA). The top 30 dimensions were selected and graph-based clustering was performed, with the appropriate number of clusters determined by adjusting the resolution parameter. Evolution trajectory was inferred by estimating the sequence of gene expression change. To order the cells in pseudotime, the beginning of the biological process was identified by specifying the root of the trajectory programmatically.
Reference-based and signature-based deconvolution
Deconvolution, employed to estimate cell type fractions in bulk RNA-seq data, utilized two common approaches in this study: reference-based and signature-based. The reference matrix was constructed by aggregating gene counts for each cell type in the scCRC dataset, and reference-based deconvolution was performed in TCGA-CRC using seven methods [14]: ordinary least squares (ols), non-negative least squares (nnls), quadratic programming with non-negativity and sum-to-one constraint (qprogwc), quadratic programming without constraints (qprog), re-weighted least squares (rls), linear mixing model (dtangle), and support vector regression (svr). Otherwise, signature-based deconvolution was conducted in the BJ-cohort using the intersection of the 395-panel and MSI-H DEGs derived in scCRC.
Cell-cell communication analysis
We employed package CellChat to analyze cell-cell communication. We modeled the probability of communication between cells using the law of mass action by integrating gene expression with prior knowledge of the interactions between signaling ligands, receptors, and their cofactors. Subsequently, we inferred biologically significant cell-cell communications by assigning each interaction a probability value and conducting a permutation test.
Statistical analysis and data visualization
All data analysis and visualization were carried out using R version 4.2.0. Continuous data were presented as median (range), while categorical data were presented as number (%). The Wilcoxon Rank Sum Test was the preferred method for hypothesis testing, with the t-test used only when the data did not reject a normal distribution. All reported p-values were two-tailed, and a p-value of < 0.05 was considered statistically significant unless otherwise specified. Multiple comparisons correction adjusting the false discovery rate was considered when necessary. R packages ggvenn, pheatmap, clusterProfiler, and ggplot2 were utilized for general visualization.
Results
Determining MSI status of each tumor cells
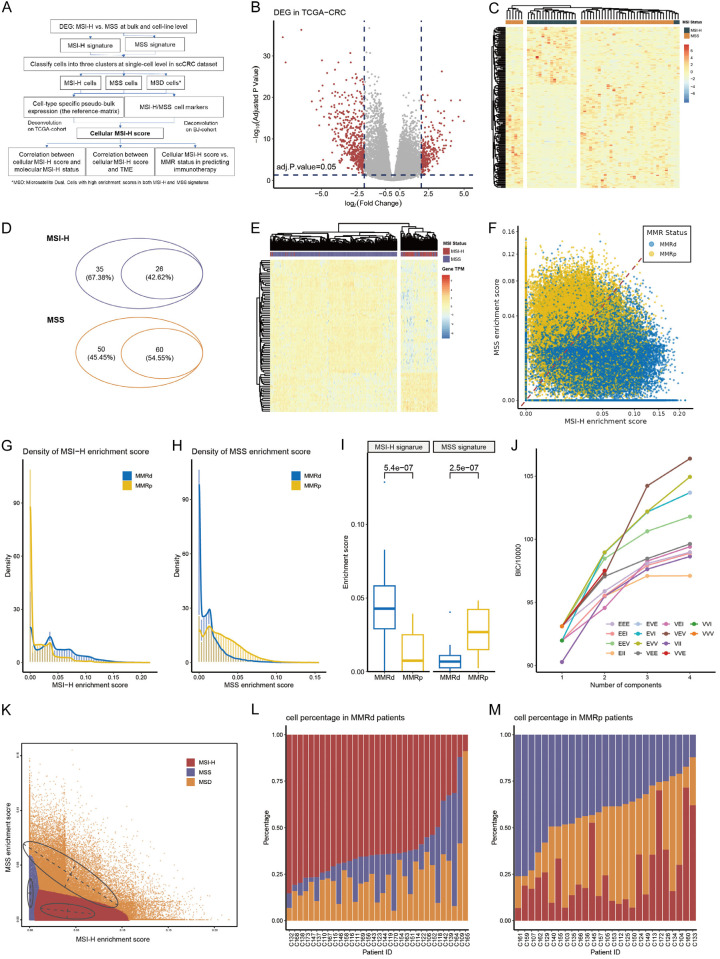
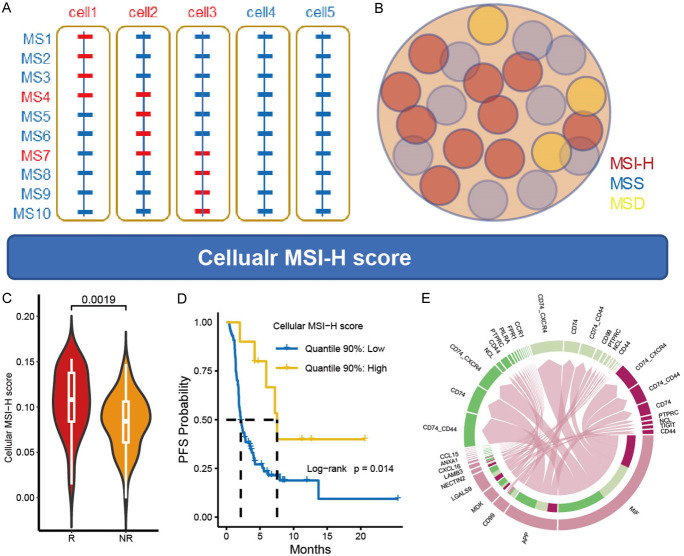
The flow chart of this study is presented in Figure 1A. Through analysis of gene expression data from 296 MSS patients and 64 MSI-H patients in the TCGA-CRC cohort (with baseline characteristics in Supplementary Table 1), a total of 794 DEGs were identified. This set comprised 520 upregulated genes in MSS patients and 274 upregulated genes in MSI-H patients (Figure 1B; Supplementary Table 2). Subsequently, the expression differences of these DEGs were validated in MSS and MSI-H CRC cell lines (with baseline characteristics shown in Supplementary Table 3), resulting in the exclusion of 624 insignificant genes, leaving 171 significant ones (Figure 1C; Supplementary Table 4). Further filtering was performed using single-cell data (with baseline characteristics in Supplementary Table 5), requiring each gene to be expressed in at least 1% of tumor cells. Ultimately, a total of 26 and 60 genes were obtained and utilized for constructing MSI-H and MSS status-related signatures, respectively (Figure 1D; Supplementary Table 6). These signatures were further validated employing unsupervised clustering. They could effectively distinguish between MSI-H and MSS samples in TCGA-CRC (Figure 1E), and between MMRd and MMRp samples in GSE178341, an independent CRC dataset (Supplementary Figure 1A; Supplementary Table 7). Also, in GSE178341 dataset, the enrichment scores of MSI-H signature were significantly higher in MMRd samples than that in MMRp samples (Supplementary Figure 1B).
Figure 1.
Constructing MSI-H and MSS signatures and determining MSI status of each tumor cells. (A) Diagram of the study design. (B) DEGs between MSI-H and MSS tumors in TCGA-CRC. The red dots represent candidate DEGs. (C) DEGs filtered by CCLE-CRC and their performance in discriminating MSI-H and MSS cell lines. (D) Filtering DEGs inherited from CCLE-CRC in scCRC dataset. The smaller sets represent genes with expression values above 0 in at least 1% of tumor cells and were used as MSI-H signature and MSS signature. (E) Clustering TCGA samples using the final MSI-H signature and MSS signature. (F) The MSI-H enrichment score and MSS enrichment score in the scCRC dataset colored by patient origin. (G, H) Density distribution of MSI-H (G) and MSS (H) enrichment score stratified by patient origin. (I) Difference of MSI-H and MSS enrichment score between MMRd and MMRp patients. (J) Change of Bayesian Information Criterion (BIC) with respect to the number of mixing components in each Gaussian finite mixture model. (K) The three clusters determined by the VVE model. (L, M) Percentage of MSI-H, MSS, and MSD cells in MMRd (L) and of MMRp (M) patients.
Using these two signatures, we calculated MSI-H and MSS enrichment scores for all 105,122 tumor cells from the single-cell RNA-seq data of 27 MMRp and 33 MMRd CRC individuals. Generally, most tumor cells from MMRd individuals exhibited a higher MSI-H enrichment score compared to those from MMRp individuals, while most tumor cells from MMRp individuals showed a higher MSS enrichment score than those from MMRd individuals (Figure 1F). The probability density distribution of MSI-H enrichment scores in cells originating from MMRd patients presented as a bimodal distribution, with peaks near 0 and 0.04, while cells originating from MMRp patients displayed a prominent peak near 0 (Figure 1G). On the other hand, the probability density distribution of MSS enrichment scores in cells originating from MMRp patients showed a right-skewed normal distribution with a peak around 0.025, whereas cells originating from MMRd patients had a small peak near 0.02 (Figure 1H). By taking median value of the enrichment score in each patient, we validated that MSI-H enrichment scores were significantly higher in MMRd group, while the MSS enrichment scores were significantly higher in MMRp group (Figure 1I).
Utilizing the MSI-H and MSS enrichment scores, we classified tumor cells into clusters, selecting the optimal number of clusters based on the Bayesian Information Criterion (BIC) using a Gaussian finite mixture model (GMM). As depicted in Figure 1J, with the increase in the number of clusters, BIC initially rose rapidly, then tended to stabilize. When K equaled 3, the BIC corresponding to several models, such as VVE, approached the high point. Therefore, we chose the optimal model VEV (variable volume, equal shape, and variable covariance) when K = 3 and identified three tumor cell clusters. Based on the MSI-H and MSS enrichment scores, we annotated the three clusters as MSI-H, MSS, and MSD (microsatellite dual positive, Figure 1K).
After determining the MSI status of each tumor cell, we further examined the distribution of cell states in patients with MMRp and MMRd origins. As expected, MMRd patients predominantly exhibited MSI-H cell proportions (median percentage: 65.17%, Figure 1L), while MMRp patients primarily showed MSS proportions (median percentage: 44.69%, Figure 1M). However, we also noted that in 3 out of 33 MMRd patients (10%), MSS cells were predominant (C164, C109, C139), suggesting potential primary resistance. Conversely, in 8 out of 27 MMRp patients (30%), MSI-H cells were the most abundant (C160, C172, C133, C145, C126, C124, C113, and C104), hinting at potential benefit from immunotherapy.
Intercellular communication between MSI-H cells and immune cells
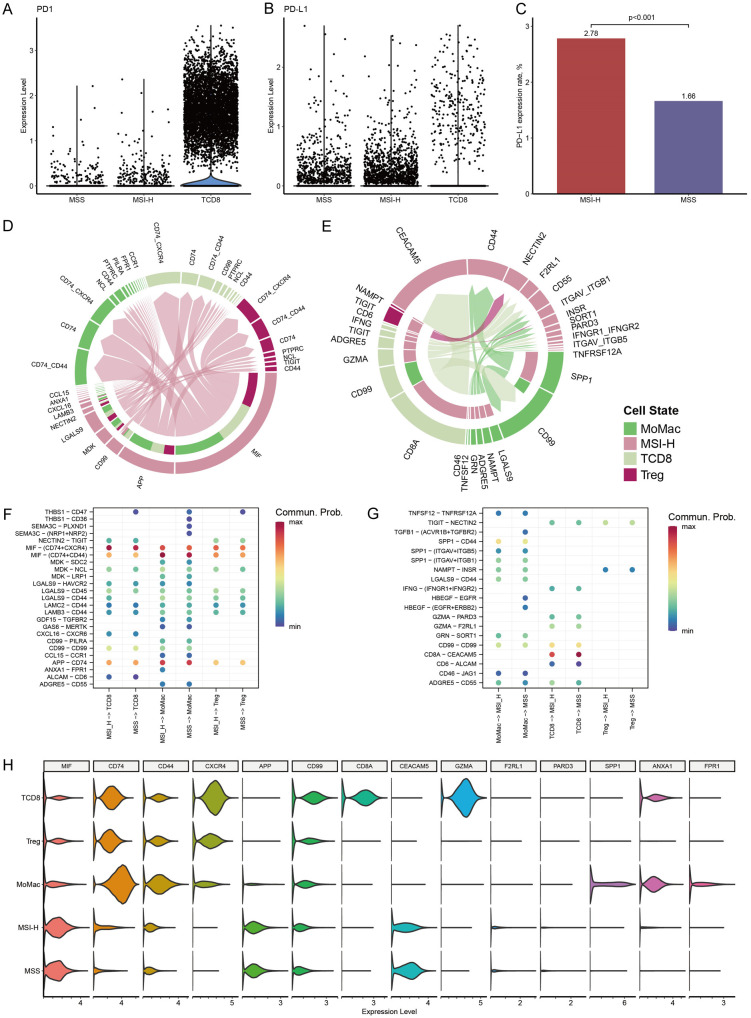
We explored the expression of PD1 in CD8+ T cells and PD-L1 in MSI-H and MSS cells within the scCRC dataset. As anticipated, PD1 expression was predominantly detected on CD8+ T cells rather than tumor cells (Figure 2A). Although PD-L1 expression was observed in only a minority of tumor cells (Figure 2B), we observed that approximately 2.78% of cells in the MSI-H cluster expressed PD-L1, a significantly higher proportion compared to the expression of PD-L1 in MSS cells (2.78% vs. 1.66%, P < 0.001, Figure 2C).
Figure 2.
Ligand-receptor interactions between MSI-H cells and immune cells in scCRC dataset. (A, B) PD1 and PD-L1 expression in MSI-H, MSS, and CD8+ T cells. (C) Percentage of PD-L1 expression cells in MSI-H cluster and MSS cluster. (D, E) Ligand-receptor interaction from MSI-H cells to immune cells (D), and from immune cells to MSI-H cells (E). Cell types are denoted by colors, arrows signify communication direction, and arrow thickness symbolizes communication probabilities. (F, G) Communication probability of ligand-receptor pairs between MSI-H and immune cells, and between MSS and immune cells. Labels on x-axis labels indicate the communication direction. (H) Density of gene expression mediating the crosstalk.
We then researched the interplay between MSI-H cells and CD8+ T, MoMac, and regulatory T cells (Treg) by examining the co-expression of ligand-receptor (L/R) pairs (Supplementary Table 8). The L/R pairs involving MIF-(CD74+CD44)/(CD74+CXCR4) and APP-CD74 were identified as the most prominent interactions facilitating signal transduction from MSI-H cells to all three types of immune cells (Figure 2D). It is widely acknowledged that extracellular MIF interacts with CD74, forming a hetero-complex with CD44, CXCR2, CXCR4, and/or CXCR7, thereby initiating downstream MAPK and/or PI3K pathway effectors [15]. However, the outcome of this interaction, whether it leads to pro-inflammatory, anti-inflammatory, or immune evasion responses, depends on the prevailing conditions. The amyloid precursor protein (APP) has been demonstrated to be upregulated in various cancer types [16], yet the role of the APP-CD74 interaction in the tumor microenvironment has been scarcely reported. Additionally, other ligands originating from MSI-H cells included CD99, MDK, LGALS9, et al (Figure 2D).
Regarding the signals received by MSI-H cells, the signaling pathways initiated by CD8A and GZMA from CD8+ T cells were identified as the most robust communications (Figure 2E). GZMA is a well-known granzyme associated with inducing cell death [17]. Its binding to CEACAM5 on CD8A has been reported to be involved in the activation of CD8-associated LcK (lymphocyte-specific protein tyrosine kinase) [18-20]. Among the communication pairs between MoMac and MSI-H cells, SPP1-CD44 was the predominant interaction, followed by LGALS9-CD44, NAMPT-INSR, and ADGRE5-CD55 (Figure 2E). CD44-mediated signals, particularly SPP1-CD44 interactions, are generally considered to be immunosuppressive in the tumor microenvironment.
To elucidate the distinction between MSI-H and MSS cells in the crosstalk network, we compared their interactions with the aforementioned immune cells. Overall, MSI-H and MSS cells exhibited similar crosstalk networks in both signal sending (Figure 2F) and receiving (Figure 2G). Upon closer examination, the ANXA1-FPR1 signal pair, known for its anti-inflammatory properties [21], was notably expressed between MSI-H cells and MoMac. Additionally, the SPP1-CD44 signal transmission from MoMac to tumor cells was evidently higher in MSI-H cells compared to MSS cells (communication probability: 0.14 vs. 0.11). The expression levels of critical molecules involved in these communication pathways within the scCRC dataset were illustrated in Figure 2H.
Signaling pathway activity and evolutionary trajectory of the MSI-H cells
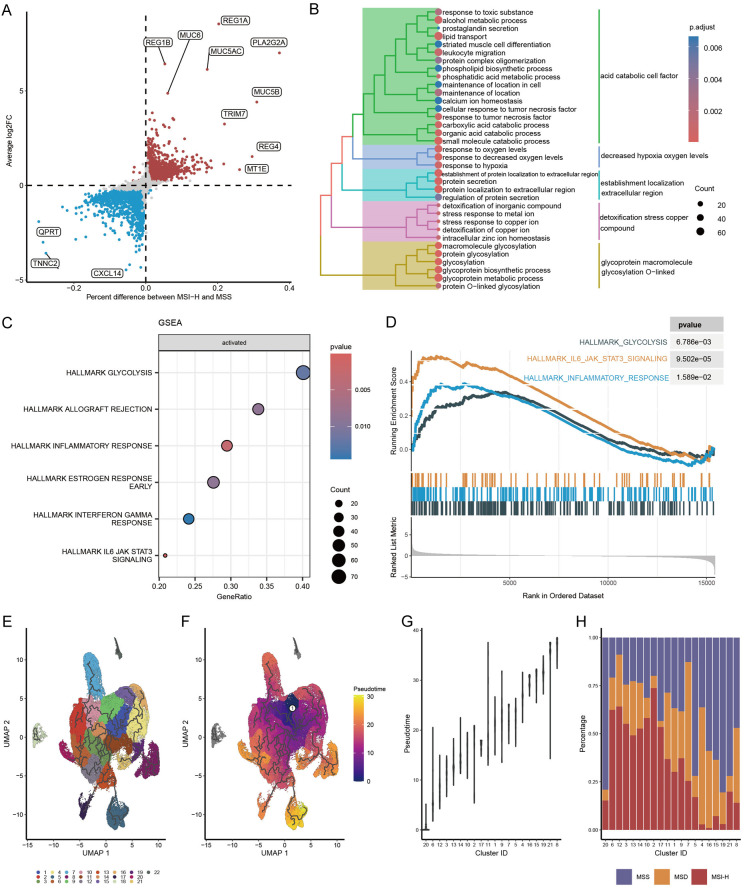
With the establishment of this relatively homogeneous MSI-H cell cluster, we were able to investigate the MSI-H cell properties unaffected by other cellular components such as stromal cells and immune cells. DEG analysis was conducted between MSI-H and MSS clusters. We imposed stringent criteria for significant differential gene expression, requiring genes to meet three criteria simultaneously: a proportion difference in expression of the gene between the two groups greater than 1%, statistically significant differences in expression levels between the two groups, and a fold change in expression levels between the two groups greater than 0.25. Consequently, 1996 genes were found to be significantly upregulated in the MSI-H cluster, while 2464 genes were significantly upregulated in the MSS cluster (Figure 3A; Supplementary Table 9). The top five upregulated protein-coding genes in the MSI-H cluster were REG1A, PLA2G2A, REG1B, MUC5AC, and MUC6, whereas in the MSS cluster, they were CXCL14, SFTPB, NOTUM, SFTA3, and WIF1.
Figure 3.
DEGs, signaling pathway activity, and evolutionary trajectory of MSI-H cells. A. DEGs between MSI-H and MSS cells. B. Enriched GO items of DEGs in MSI-H cells. C. Enriched hallmark pathways in MSI-H cells using GSEA. D. Running score and preranked list of three hallmark pathways visualized by GSEA plot. E. Graph-based clustering of scCRC tumor cells. F. Trajectory and pseudotime analysis of scCRC tumor cells. The black lines show the structure of the trajectory graph. White circle denotes the initial time point. G. Ordering clusters by evolutionary pseudotime. H. Percentage of MSI-H, MSS, and MSD cells in clusters ordered by pseudotime.
The DEGs upregulated in the MSI-H cluster were notably enriched in Gene Ontology Biological Process (GO-BP) terms such as carboxylic acid catabolic process, response to decreased oxygen levels, stress response to copper ion, and protein glycosylation (Figure 3B). Conversely, in the MSI-H bulk tissue, the enriched pathways associated with DEGs primarily included positive regulation of macrophage-derived foam cell differentiation, microtubule-based movement, and hydrogen peroxide catabolic process (Supplementary Figure 2A), indicating potential influences of immune cell infiltration on the enriched pathways within the MSI-H bulk tissue. Moreover, the upregulated DEGs in the MSS cluster were significantly enriched in the WNT signaling pathway and carboxylic acid transport pathways. Meanwhile, the pathways enriched in the MSS bulk tissue primarily included carboxylic acid transport and intermediate filament-based processes (Supplementary Figure 2B, 2C), suggesting a higher similarity between the MSS bulk tissue and the MSS cluster.
We conducted additional analysis to assess the signaling pathway activity of the MSI-H cluster utilizing Gene Set Enrichment Analysis (GSEA). Specifically focusing on the HALLMARK gene set, our findings revealed significant enrichment of pathways including glycolysis, inflammatory response, allograft rejection, and the IL-6/JAK2/STAT3 signaling pathway within the MSI-H cluster (Figure 3C, 3D). Notably, aberrant activation of the IL-6/JAK2/STAT3 pathway has been previously reported to promote PD-L1 expression and consequently contribute to resistance against immune-mediated killing [16,22].
To elucidate the temporal dynamics of MSS, MSI-H, and MSD cell populations during tumor progression, trajectory analysis was conducted. A total of 22 clusters were identified for tumor cells (Figure 3E). The trajectory was inferred based on the sequential patterns of gene expression changes at the cluster level, and pseudotime was assigned to position cells along the trajectory (Figure 3F). Subsequently, the 22 clusters were sorted according to their median pseudotime values (Figure 3G) to represent tumor differentiation at the cluster level. Following the calculation of MSI-H, MSS, and MSD cell proportions within each cluster, it was observed that, concomitant with tumor progression, the percentage of MSI-H cells decreased, while the percentage of MSS cells increased, with the proportion of MSD cells remaining relatively unchanged (Figure 3H). This observation suggests that tumor cells, under the selective pressure of high mutational adaptation, eventually evolve into a stable state, thereby abandoning the highly mutated state that is susceptible to immune recognition.
Correlation between cellular MSI-H score, molecular MSI score, and immune cell infiltration in TCGA-CRC
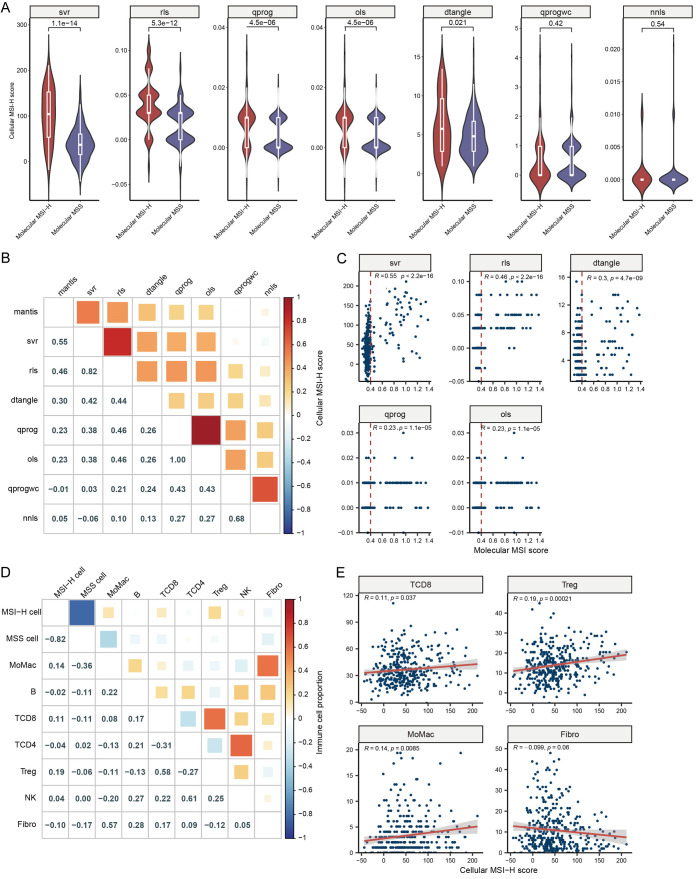
After establishing the MSI status of each cell, we generated cell-type specific gene expression data (Supplementary Table 10), commonly referred to as pseudobulk expression, utilizing the scCRC data. Using this as reference gene set, we performed deconvolution of the TCGA-CRC dataset using seven different methods to determine the score of each cell type within the bulk tissue (Supplementary Table 11). We defined the deconvolution score that quantifies the proportion of MSI-H cells within the tumor as cellular MSI-H score. To validate the applicability of the methods employed, we examined the correlation between cellular MSI-H cell score and the molecular MSI-H status and score calculated by the MANTIS algorithm. Five out of the seven methods yielded similar results, demonstrating that cellular MSI-H score was significantly higher in molecular MSI-H tissues compared to MSS tissues (Figure 4A). Notably, the SVR method resulted in cellular MSI-H score following a bimodal distribution in both molecular MSI-H and MSS tissues, with the smallest p-value between the two groups, suggesting that the SVR method may be more suitable for this dataset. Furthermore, the molecular MSI score showed significant correlations with the cellular MSI-H score derived from all five methods, with particularly strong correlation reaching 0.55 with the SVR method (Figure 4B, 4C). Additionally, cellular MSI-H score determined by the five deconvolution methods also demonstrated high correlations among themselves. It is noteworthy that some MSS tissues identified by the molecular MSI score exhibited higher cellular MSI-H score, while some MSI-H tissues had lower cellular MSI-H score (Figure 4C).
Figure 4.
Deconvolution of TCGA-CRC to derive cellular MSI-H score. (A) Assessment of the correlation between cellular MSI-H score, determined by seven algorithms, and molecular MSI status using the MANTIS algorithm. (B, C) Evaluation of correlation scores (B) and significance (C) between molecular MSI score and cellular MSI-H score. (D, E) Examination of correlation scores (D) and significance (E) between cellular MSI-H score and levels of immune cell infiltration. ols, ordinary least squares; nnls, non-negative least squares; qprogwc, quadratic programming with non-negativity and sum-to-one constraint; qprog, quadratic programming without constraints; rls, re-weighted least squares; dtangle, linear mixing model; svr, support vector regression.
Since MSI-H cells generate a higher number of neoantigens, they are expected to more effectively activate the immune system. We conducted an analysis to examine the correlation between cellular MSI-H score and immune cell infiltration. We found a weak but significant positive correlation between cellular MSI-H score and MoMac (R = 0.14, P = 0.008), CD8+ T cells (R = 0.11, P = 0.037), and regulatory T cells (Treg, R = 0.19, P < 0.001) (Figure 4D, 4E). Conversely, the cellular MSS score displayed negative correlations with MoMac (R = -0.36), CD8+ T cells (R = -0.11), and regulatory T cells (R = -0.06). Additionally, cellular MSI-H score exhibited a negative correlation with cellular MSS score (R = -0.82) and a trend towards a negative correlation with fibroblasts (R = -0.10, P = 0.06). Furthermore, we observed correlations among various immune cell types, such as CD8+ T cells and Treg (R = 0.58), CD4+ T cells and NK cells (R = 0.61), which highlight the complexity of the immune microenvironment.
Predictive value of cellular MSI-H score on immunotherapy
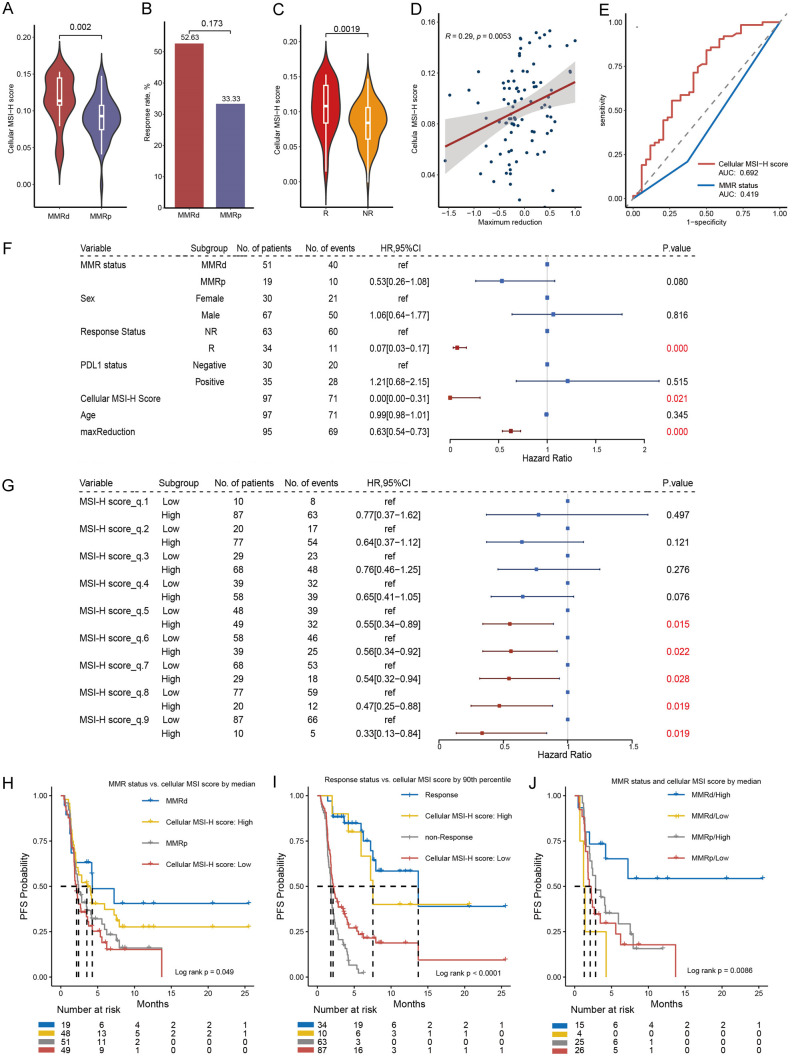
Our BJ-cohort comprises 96 cases of digestive tract tumor patients who underwent immunotherapy. RNA-sequencing using a 395-panel were performed. We intersected the up-regulated DEGs in MSI-H cells with this 395-panel, resulting in a set of 39 genes, which we refer to as the cellular MSI signature (Supplementary Table 12). We conducted deconvolution of the BJ-cohort using this signature. Of note, the deconvolution score calculated for both the TCGA and BJ cohorts reflects the proportion of MSI-H cells within the tumor microenvironment and is referred to as the cellular MSI-H score. The primary distinction between the two lies in the foundational data used for deconvolution: in the TCGA cohort, a cell-type expression matrix is utilized, while in the BJ cohort, the cellular MSI-H signature serves as the reference. We observed that cellular MSI-H score was significantly higher in the MMRd compared to the MMRp group (P = 0.002, Figure 5A), providing evidence of the applicability of our method. The MMRd group exhibited a slightly higher response rate compared to MMRp group (P = 0.173, Figure 5B), which is a reflection of intratumoral heterogeneity. However, the cellular MSI-H score exhibited a significant correlation with response status, being significantly higher in the response group compared to the non-response group (P = 0.002, Figure 5C). Additionally, a significant correlation was found between the cellular MSI-H score and the maximum reduction percentage (R = 0.29, P = 0.005, Figure 5D). When using the cellular MSI-H score to predict patient response status, we achieved an AUC of 0.69 (Figure 5E), while the AUC was 0.419 using MMR status.
Figure 5.
Correlation between cellular MSI-H score and immunotherapy response and PFS in the BJ-cohort. A. Comparison of cellular MSI-H score between MMRd and MMRp samples. B. Response rate comparison between molecular MMRd and MMRp patients. C. Density distribution of cellular MSI-H score between response and non-response patients. D. Correlation between cellular MSI-H score and tumor maximum regression. E. Predicting response status using cellular MSI-H score and MMR status by ROC curve. F. Univariate-cox survival analysis against PFS. G. Categorizing the BJ-cohort into high and low cellular MSI-H score groups using each decile of cellular MSI-H score as a threshold and compare the PFS between groups. H. Kaplan-meier (KM) analysis between groups defined by MMR status, and by median value of cellular MSI-H score. I. KM analysis between groups defined by response status, and by upper decile value of cellular MSI-H score. J. KM curve by combining MMR status and cellular MSI-H score discretized by median.
Since the cellular MSI-H score is significantly correlated with response, it is expected to predict PFS. As anticipated, as a continuous variable, the cellular MSI-H score showed a significant association with PFS (HR: 0.00, 95% CI 0.00-0.31, P = 0.021, Figure 5F). Comparatively, only response status and maximum reduction percentage had smaller p-values than the cellular MSI-H score, and both are variables after treatment. Furthermore, when we dichotomized the cellular MSI-H score into binary variables based on each decile, we observed a continuous decrease in HR for the MSI-H high-score group as the threshold of the cellular MSI-H score increased (Figure 5G). This indicates the robustness of the cellular MSI-H score as a predictive biomarker, as it performs well across relatively broad threshold limits. Using the correlation between MMR status and PFS as a reference (log rank P = 0.074, Figure 5H), when we set the cellular MSI score threshold at the 50th percentile, the p-value for the difference in PFS between the two groups was 0.038 (Figure 5H). However, it’s important to note that approximately 40% of patients still experienced rapid progression. When setting the cellular MSI-H score threshold at the 90th percentile, the p-value for the difference in PFS between the two groups decreased to 0.014, and, the MSI-H high-score group displayed similar survival probability compared to the response group (P = 0.509, Figure 5I). Furthermore, by combining the two biomarkers - MMR status and cellular MSI-H score (using the median value as the cutoff) - we were able to refine PFS) prediction (Figure 5J). The four groups, ranked by median survival time from highest to lowest, were as follows: MMRd/Hig, MMRp/High, MMRp/Low, and MMRd/Low. The PFS difference between the MMRd/High and MMRp/Low groups (log-rank P = 0.011) suggests that the cellular MSI-H score can be used to further differentiate between sensitive and primary resistant patients within the MMRd cohort. This refinement highlights the potential of integrating MMR status and cellular MSI-H score for more accurate prediction of therapeutic response in MSI-H tumors.
Discussion
In this study, we established molecular MSI-H and MSS signatures separately by integrating bulk tissue and cell line RNA expression datasets. By evaluating the enrichment score of these two signatures in single-cell dataset, we determined the MSI status of each individual tumor cell as MSI-H, MSS, or MSD. Then we analyzed cell-type specific expression matrix including MSI-H, MSS, and various immune cells. Using this matrix, we deconvoluted TCGA-CRC, obtained various cell-type proportion in each sample. Meanwhile, we analyzed genes highly expressed in MSI-H cells, which were used to deconvolute the BJ-cohort. The deconvolution score, in both TCGA-CRC and BJ-cohort, was called cellular MSI-H score. We explored the correlation between cellular MSI-H score and MoMac, CD8+ T, and Treg, demonstrated the robust predictive role of cellular MSI-H score on immunotherapy response and PFS.
Theoretically, the high mutational burden of MSI-H cells provides opportunities for rapid adaptation to the environment but also results in the generation of numerous new antigens [27,28], therefore, more susceptible to immune killing. For balance, MSI-H cells are expected to exhibit distinct biological characteristics. The top-5 up-regulated genes in MSI-H clusters included REG1A, REG1B, MUC5AC, MUC6, and PLA2G2A. The major pathways that these DEGs enriched to were carboxylic acid catabolic process, protein glycosylation, response to decreased oxygen levels, and stress response to copper ion. The relevance of the former three to tumorigenesis has been extensively reported [29-31]. Previous studies have indicated that exposure to heavy metals can disrupt DNA repair mechanisms, indirectly contributing to the development of MSI-H tumors [32-34]. Moreover, GSEA identified the up-regulation of the IL-6/JAK2/STAT3 signaling pathway, which has been reported to promote PD-L1 expression in various cancer types [22], suggesting that MSI-H cells may evade immune elimination by activating this pathway, which was corroborated by the higher expression of PD-L1 observed in MSI-H cells. Trajectory analysis of MSS, MSI-H, and MSD cell populations during tumor progression also highlighted the balance between mutational burden and immune escape. The reduction in MSI-H cells may reflect tumor adaptation to evade immune recognition under mutational stress.
By comparing the interactions between MSI-H and various immune cells, we identified two potential immune evasion pathways in MSI-H cell. First, upregulating co-inhibitory molecule PD-L1 to suppress CD8+ T cells; second, interaction with immunosuppressive macrophages to adapt to the environment. The role of PD-L1 in immune evasion is well-established, while the modulation of macrophages is still under extensive investigation. Our findings mainly involve two signal pairs, SPP1-CD44 and ANXA1-FPR1. SPP1 secreted by tumor-associated macrophages may regulate the mesenchymal phenotype of glioma by interacting with CD44 on tumor cells [35], activate downstream target genes in epithelial cells to promote dynamic changes in intratumor heterogeneity [36]. ANXA1 has been reported to induce the production of M2-like macrophages via its surface receptor FPR1 [21], thereby establishing a Treg cell-driven immunosuppressive tumor microenvironment.
Currently, MMRd/MSI-H is the only well-recognized biomarker that can guide the immunotherapy of gastrointestinal cancer. Nevertheless, its prediction efficiency is relatively limited mainly attributed to the ITH [3,4]. Burgeoning single-cell RNA-seq has been widely applied to explore tumor heterogeneity [23-25], for example, the identification of a specific subtype of MSS, the iCMS3_MSS, which is more similar to MSI-H cancers [26]. Unlike iCMS3_MSS and many other methods that rely solely on statistical approaches to predict immune therapy outcomes [37-39], we propose a cellular-level MSI-H identification method that is based on the cellular composition of the tumor microenvironment. We explored the correlation between cellular MSI-H score, molecular MSI status, and immune cell infiltration in TCGA, demonstrated the association between cellular MSI-H score and immunotherapy response and PFS in the BJ cohort. Additionally, by progressively increasing the threshold, we validated the robustness of the cellular MSI-H score as a PFS predictor (Figure 6).
Figure 6.
A. Molecular MSI scores are influenced by cell distribution of MSI-H loci. For example, according to the length distribution of sequencing reads covering the MS sites, we consider MS4 and MS7 as MSI-H loci, i.e., the molecular level MSI score is 20% (2/10), therefore the tissue is determined as MSS tissue. However, at cellular level, suppose a cell with greater than 40% MSI-H loci could be regarded as MSI-H cell, we find 60% (3/5) cells are MSI-H cells. Theoretically, we cannot overlook the possibility that tissues dominated by MSI-H cells may be considered as MSS tissues at the molecular level. B. Intra tumor heterogeneity (ITH) caused by cellular composition, MSI-H, MSS, and MSD. C, D. The predictive role of cellular MSI-H score on immunotherapy response and PFS. E. Signals mediating the crosstalk from MSI-H cells to immune cells.
Our study has some limitations. First, the subjectivity in the trajectory analysis. Since we inferred starting point based on gene expression patterns and cell state distribution, the trajectory interpretation would benefit from further validation. Second, we identified a subset of cells, termed MSD cells, which exhibited characteristics of both MSI-H and MSS phenotypes, we speculated that these cells maybe transitional states between MSS and MSI-H. However, further experimental investigations are required.
In summary, we identified an improved immunotherapy predictive biomarker, the cellular MSI-H score. We created it from single-cell data, applied it to bulk tissue data, and demonstrated its outstanding correlation with immunotherapy response and PFS. We believe it could influence clinical practice in the future.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82172973), Key Research and Development Project of Shaanxi Province (2022ZDLSF03-04), Key Clinical HighTech Project of Xijing Hospital (No. XJZT24LY32), the Beijing Natural Science Foundation (Z230008), Beijing Xisike Clinical Oncology Research Foundation (Y-2022HER2AZQN-0198) and Beijing Nova Program (20240484524).
Informed consent was obtained from all subjects involved in the study.
Disclosure of conflict of interest
None.
Supplementary Figures 1 and 2
Supplementary Tables 1-12
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA Jr KEYNOTE-177 Investigators. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 3.Cohen R, Hain E, Buhard O, Guilloux A, Bardier A, Kaci R, Bertheau P, Renaud F, Bibeau F, Fléjou JF, André T, Svrcek M, Duval A. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019;5:551–555. doi: 10.1001/jamaoncol.2018.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Wang X, Xu Y, Li J, Zhang X, Peng Z, Hu Y, Zhao X, Dong K, Zhang B, Gao C, Zhao X, Chen H, Cai J, Bai Y, Sun Y, Shen L. Mutations of PI3K-AKT-mTOR pathway as predictors for immune cell infiltration and immunotherapy efficacy in dMMR/MSI-H gastric adenocarcinoma. BMC Med. 2022;20:133. doi: 10.1186/s12916-022-02327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cusnir M, Cavalcante L. Inter-tumor heterogeneity. Hum Vaccin Immunother. 2012;8:1143–1145. doi: 10.4161/hv.21203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitale I, Sistigu A, Manic G, Rudqvist NP, Trajanoski Z, Galluzzi L. Mutational and antigenic landscape in tumor progression and cancer immunotherapy. Trends Cell Biol. 2019;29:396–416. doi: 10.1016/j.tcb.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 8.Trabucco SE, Gowen K, Maund SL, Sanford E, Fabrizio DA, Hall MJ, Yakirevich E, Gregg JP, Stephens PJ, Frampton GM, Hegde PS, Miller VA, Ross JS, Hartmaier RJ, Huang SA, Sun JX. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn. 2019;21:1053–1066. doi: 10.1016/j.jmoldx.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, Catenacci DVT, Fakih M, Barbacioru C, Zhao J, Sikora M, Fairclough SR, Lee H, Kim KM, Kim ST, Kim J, Gavino D, Benavides M, Peled N, Nguyen T, Cusnir M, Eskander RN, Azzi G, Yoshino T, Banks KC, Raymond VM, Lanman RB, Chudova DI, Talasaz A, Kopetz S, Lee J, Odegaard JI. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res. 2019;25:7035–7045. doi: 10.1158/1078-0432.CCR-19-1324. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Huang Y, Fang X, Liu C, Deng W, Zhong C, Xu J, Xu D, Yuan Y. A novel and reliable method to detect microsatellite instability in colorectal cancer by next-generation sequencing. J Mol Diagn. 2018;20:225–231. doi: 10.1016/j.jmoldx.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, Roychowdhury S. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017;8:7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfister S, Kuettel V, Ferrero E. granulator: rapid benchmarking of methods for *in silico* deconvolution of bulk RNA-seq data. 10.18129/B9.bioc.granulator 2023. [Google Scholar]
- 15.Noe JT, Mitchell RA. MIF-dependent control of tumor immunity. Front Immunol. 2020;11:609948. doi: 10.3389/fimmu.2020.609948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi M, Niu M, Xu L, Luo S, Wu K. Regulation of PD-L1 expression in the tumor microenvironment. J Hematol Oncol. 2021;14:10. doi: 10.1186/s13045-020-01027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachmann MF, Gallimore A, Linkert S, Cerundolo V, Lanzavecchia A, Kopf M, Viola A. Developmental regulation of Lck targeting to the CD8 coreceptor controls signaling in naive and memory T cells. J Exp Med. 1999;189:1521–1530. doi: 10.1084/jem.189.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie HY, Mong MS, Itano A, Crooks ME, Littman DR, Burakoff SJ, Robey E. The cytoplasmic domain of CD8 beta regulates Lck kinase activation and CD8 T cell development. J Immunol. 1998;161:183–191. [PubMed] [Google Scholar]
- 20.Roda G, Jianyu X, Park MS, DeMarte L, Hovhannisyan Z, Couri R, Stanners CP, Yeretssian G, Mayer L. Characterizing CEACAM5 interaction with CD8alpha and CD1d in intestinal homeostasis. Mucosal Immunol. 2014;7:615–624. doi: 10.1038/mi.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Jiang H, Yang N, Shen S, Huang D, Jia L, Ling J, Xu L, Li M, Yu K, Ren X, Cui Y, Lan X, Lin S, Lin X. Glioma-derived ANXA1 suppresses the immune response to TLR3 ligands by promoting an anti-inflammatory tumor microenvironment. Cell Mol Immunol. 2024;21:47–59. doi: 10.1038/s41423-023-01110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Lang X, Li X. The role of IL-6/JAK2/STAT3 signaling pathway in cancers. Front Oncol. 2022;12:1023177. doi: 10.3389/fonc.2022.1023177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren X, Kang B, Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol. 2018;19:211. doi: 10.1186/s13059-018-1593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, Deschler DG, Varvares MA, Mylvaganam R, Rozenblatt-Rosen O, Rocco JW, Faquin WC, Lin DT, Regev A, Bernstein BE. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624. e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, Eng CLP, Macalinao DC, Kahraman M, Srinivasan H, Lakshmanan V, Verbandt S, Tsantoulis P, Gunn N, Venkatesh PN, Poh ZW, Nahar R, Oh HLJ, Loo JM, Chia S, Cheow LF, Cheruba E, Wong MT, Kua L, Chua C, Nguyen A, Golovan J, Gan A, Lim WJ, Guo YA, Yap CK, Tay B, Hong Y, Chong DQ, Chok AY, Park WY, Han S, Chang MH, Seow-En I, Fu C, Mathew R, Toh EL, Hong LZ, Skanderup AJ, DasGupta R, Ong CJ, Lim KH, Tan EKW, Koo SL, Leow WQ, Tejpar S, Prabhakar S, Tan IB. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet. 2022;54:963–975. doi: 10.1038/s41588-022-01100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shlyakhtina Y, Moran KL, Portal MM. Genetic and non-genetic mechanisms underlying cancer evolution. Cancers (Basel) 2021;13:1380. doi: 10.3390/cancers13061380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie N, Shen G, Gao W, Huang Z, Huang C, Fu L. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther. 2023;8:9. doi: 10.1038/s41392-022-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian X, Qian Y, Tan B, Li K, Hong X, Wong CC, Fu L, Zhang J, Li N, Wu JL. In-depth mapping carboxylic acid metabolome reveals the potential biomarkers in colorectal cancer through characteristic fragment ions and metabolic flux. Anal Chim Acta. 2020;1128:62–71. doi: 10.1016/j.aca.2020.06.064. [DOI] [PubMed] [Google Scholar]
- 30.Ho WL, Hsu WM, Huang MC, Kadomatsu K, Nakagawara A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J Hematol Oncol. 2016;9:100. doi: 10.1186/s13045-016-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Han F, Du Y, Shi H, Zhou W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct Target Ther. 2023;8:70. doi: 10.1038/s41392-023-01332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartwig A, Asmuss M, Blessing H, Hoffmann S, Jahnke G, Khandelwal S, Pelzer A, Bürkle A. Interference by toxic metal ions with zinc-dependent proteins involved in maintaining genomic stability. Food Chem Toxicol. 2002;40:1179–1184. doi: 10.1016/s0278-6915(02)00043-1. [DOI] [PubMed] [Google Scholar]
- 33.Hartwig A, Schwerdtle T. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett. 2002;127:47–54. doi: 10.1016/s0378-4274(01)00482-9. [DOI] [PubMed] [Google Scholar]
- 34.Yildiz A, Kaya Y, Tanriverdi O. Effect of the interaction between selenium and zinc on DNA repair in association with cancer prevention. J Cancer Prev. 2019;24:146–154. doi: 10.15430/JCP.2019.24.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He C, Sheng L, Pan D, Jiang S, Ding L, Ma X, Liu Y, Jia D. Single-cell transcriptomic analysis revealed a critical role of SPP1/CD44-mediated crosstalk between macrophages and cancer cells in Glioma. Front Cell Dev Biol. 2021;9:779319. doi: 10.3389/fcell.2021.779319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie W, Cheng J, Hong Z, Cai W, Zhuo H, Hou J, Lin L, Wei X, Wang K, Chen X, Song Y, Wang Z, Cai J. Multi-transcriptomic analysis reveals the heterogeneity and tumor-promoting role of SPP1/CD44-mediated intratumoral crosstalk in gastric cancer. Cancers (Basel) 2022;15:164. doi: 10.3390/cancers15010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z, Chen H, Jiao X, Zhou W, Han W, Li S, Liu C, Gong J, Li J, Zhang X, Wang X, Peng Z, Qi C, Wang Z, Li Y, Li J, Li Y, Brock M, Zhang H, Shen L. Prediction of immune checkpoint inhibition with immune oncology-related gene expression in gastrointestinal cancer using a machine learning classifier. J Immunother Cancer. 2020;8:e000631. doi: 10.1136/jitc-2020-000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao X, Wei X, Li S, Liu C, Chen H, Gong J, Li J, Zhang X, Wang X, Peng Z, Qi C, Wang Z, Wang Y, Wang Y, Zhuo N, Zhang H, Lu Z, Shen L. A genomic mutation signature predicts the clinical outcomes of immunotherapy and characterizes immunophenotypes in gastrointestinal cancer. NPJ Precis Oncol. 2021;5:36. doi: 10.1038/s41698-021-00172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatalica Z, Vranic S, Xiu J, Swensen J, Reddy S. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer. 2016;15:405–412. doi: 10.1007/s10689-016-9884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.