Abstract
Background: The emergence of immune resistance and a lack of effective therapeutic targets have become significant challenges in immunotherapy, highlighting the urgent need for new molecular markers and treatment targets. Moreover, the significance and mechanisms of PGRN (Progranulin) in gastric cancer remain ambiguous. Objective: To identify differentially expressed proteins in gastric cancer and elucidate the function and mechanism of PGRN. Methods: The data-independent acquisition proteomics was used to identify the differentially expressed proteins in gastric adenocarcinoma and the corresponding paraneoplastic tissues, providing a comprehensive dataset of gastric cancer-related proteins. The function and mechanism of PGRN in gastric cancer were further explored using a series of experiments, including RT-qPCR (Real Time-Quantitative Polymerase Chain Reaction), cell transfection, cell viability assays, cell scratch, immunohistochemistry and Transwell assays, Western blot, and a mouse tumor-bearing model. These investigations were combined with bioinformatics analyses to examine the relationship between PGRN expression and clinical-pathological characteristics, confirming its high expression of PGRN in gastric cancer tissues. Results: We identified a large number of differentially expressed proteins between gastric cancer and adjacent tissues and conducted an initial functional analysis. Further studies on PGRN showed that it was associated with gastric cancer prognosis and lymph node metastasis. The inhibition of PGRN expression led to reduced cell viability, migration, and invasion, with corresponding changes in related genes and proteins. In a mouse tumor-bearing model, the tumor growth of the subcutaneously transplanted tumors in nude mice was reduced after the inhibition of PGRN expression. An in-depth functional analysis of PGRN was performed using bioinformatics to predict protein interactions, miRNA regulation, and relationships with multiple immune cell types. Enrichment analysis indicated that PGRN is involved in multiple signaling pathways, with the MAPK (Mitogen-Activated Protein Kinase) pathway selected for validation. In AGS and HGC27 cells, PGRN inhibition led to increased expression of phosphorylated p38 (p-p38) in the MAPK pathway, suggesting that PGRN may promote gastric cancer development by regulating p-p38. Conclusions: This study identified significant differences in protein expression between gastric adenocarcinoma and adjacent tissues, with PGRN emerging as a key protein influencing gastric cancer proliferation, migration, and invasion. These findings suggest that PGRN could serve as a potential therapeutic target for gastric cancer.
Keywords: Gastric cancer, quantitative proteomics, bioinformatics, functional analysis, PGRN
Introduction
Gastric cancer is a prevalent malignancy, ranking fifth in incidence and fourth in mortality worldwide [1]. In China, it ranks third in both incidence and mortality [2]. Despite some improvements, the 5-year overall age-standardized relative survival rate for gastric cancer remains low at 42.9%, with the long-term survival rate is as low as 10% [3]. With advancements in immunotherapy, the diagnosis and treatment of gastric cancer have moved toward precision medicine. However, there remain challenges, such as limited treatment strategies, unclear mechanisms of action, and the emergence of immune drug resistance. Therefore, identifying novel therapeutic targets and molecular markers is urgently needed to expand treatment strategies, investigate mechanisms, and improve patient prognosis.
As proteomics and bioinformatics continue to advance, their roles in studying cancer development and regulation has been increasingly recognized. Emerging techniques, such as data-independent acquisition (DIA) quantitative proteomics are increasingly being used to screen and identify differentially expressed proteins between tumor and adjacent tissues and cells. Several studies have identified meaningful differentially expressed molecules in various cancers, such as prostate, liver, colorectal, and pancreatic cancers [4-7]. Some of these molecules are closely associated with tumor invasion and metastasis, making them promising therapeutic targets. Therefore, proteomics holds great potential for discovering new biomarkers and therapeutic targets in cancer research.
Although some progress has been made in the application of proteomics to gastric cancer research [8-10], key molecules that can serve as clinical tumor markers in novel therapeutic strategies remain limited. Additionally, the functions and mechanisms of action of these molecules are still unclear. We screened the top 100 proteins with up-regulated expression in cancer tissues. Through bioinformatics analysis and literature review, we identified PGRN (Progranulin) as a molecule with limited studies in gastric cancer but significant potential research value. As a result, PGRN was selected for further investigation.
Progranulin, a protein encoded by the GRN (granulin) gene, serves multiple functions as a neurotrophic factor, tumor growth factor, and anti-inflammatory cytokine. It is implicated in the development of various cancers, including cervical, rectal, bladder and bile duct cancers, through various mechanisms of action [11-15]. For example, the PGRN/EphA2 (Ephrin type-A receptor 2) axis has been identified as a novel oncogenic pathway in bladder cancer, offering potential for the discovery of novel therapeutic targets [11]. In pancreatic ductal adenocarcinoma, PGRN mediates the immune escape of tumors by regulating MHCI (Major histocompatibility complex class I) expression [12]; while in colorectal cancer, PGRN promotes tumor proliferation and angiogenesis through the TNFR2/Akt (Tumor necrosis factor receptor-2/Protein kinase B) and ERK (Extracellular regulated protein kinases) signaling pathways [14]. Previous studies have confirmed the correlation between PGRN and Helicobacter pylori [16,17], suggesting that PGRN may be a potential marker of gastric cancer [18]. However, its precise mechanism in gastric cancer remains unclear.
In this study, we employed quantitative proteomics techniques to screen for differentially expressed proteins, identifying PGRN as a key differentially expressed protein in gastric cancer. We performed an in-depth analysis and validation of the function and significance of PGRN using bioinformatics studies in conjugation with in vivo and ex vivo experiments. These studies hold great promise for identifying potential biomarkers and therapeutic targets and elucidating associated mechanisms in gastric cancer.
Materials and methods
Collection of clinical tissue samples
Five patients with gastric cancer who underwent radical surgery at the Department of Surgery, Fourth Hospital of Hebei Medical University between September 2021 and September 2022 were selected for this study. Inclusion criteria: adenocarcinoma confirmed by postoperative pathology, no previous treatment before surgery, and no other concomitant cancer. From each patient, tumor tissues (specimen C) and corresponding paraneoplastic tissues (specimen N) of the same size were collectd after isolation. These tissues were placed in labeled cryotubes and initially stored in a liquid nitrogen tank before being transferred to a -80°C freezer. The tumor tissue samples were verified to contain more than 90% tumor cells before initiating further experiments. This study was approved by the Institutional Review Board of the Fourth Hospital of Hebei Medical University (approval no.: 2019ME0039), and all patients provided written informed consent.
Protein extraction
The normal and cancer tissues were placed into a tissue extractor (Jingxin, China) and mixed with 2 mL of PBS (0.01 M) containing a protease inhibitor cocktail (Roche, Germany) for rapid impact grinding. After the tissues were fully crushed, the mixture was centrifuged in a 15 mL centrifuge tube at 12,000×g and 4°C for 10 min. The resulting supernatant was transferred to a new 15 mL centrifuge tube, and an equal volume of pre-cooled Tris-saturated phenol was added. The mixture was shaken vigorously and centrifuged at 4°C for 10 min, separating the solution into layers. The upper layer was discarded, leaving the proten layer. An equal volume of pre-cooled Tris-HCl (pH = 8.0) was added to the remaining mixture, shaken and mixed thoroughly, and then centrifuged (12,000×g, 4°C) for 10 min. The upper Tris-HCl layer was discarded, and the operation was repeated again. Five times the volume of ammonium acetate (0.1 M) methanol solution was then added to the mixture, shaken, and mixed well, and left to precipitate at a low temperature for 12 h. Following precipitation, the sample was centrifuged at 12,000×g at 4°C for 10 min, and the supernatant was discarded. One milliliter of pre-cooled methanol was added to the protein at the bottom of the tube, shaken and mixed thoroughly, and then centrifuged at 12,000×g at 4°C for 10 min. After discarding the liquid, the protein pellet was lyophilized and stored at -80°C until further use.
Trypsin digestion
The protein samples awaiting treatment were quantitatively analyzed using a BCA (Bicinchoninic acid) protein analysis kit (Jining, China), and the total amount of protein in each tube was adjusted to 100 μg. Dithiothreitol was added to each sampel to a final concentration of 0.01 M, followed by incubation at 37°C for 1 h. IAA (Iodoacetamide) was added to each tube to a final concentration of 0.02 M, and the samples were incubated in the dark at room temperature for 30 min. Next, sequence-grade trypsin (Promega, USA) was added (1:50 w/w) to each tube for enzymatic digestion at 37°C for 12 h, with an additional trypsin supplement at the 6-hour mark. The enzymatically-digested peptides were preliminarily purified using a C18 solid phase extraction column (Waters, USA). The peptide concentration was determined using the BCA protein analysis kit (Jining, China), and the enzymatic efficiency was monitored using a mass spectrometer (Q Exactive HF, Thermo Fisher Scientific, USA).
Quantitative proteomic analysis
We utilized the DIA method for quantitative proteomic analysis of normal and tumor tissues. The iRT reagent (Biognosys, Switzerland) was added to each peptide sample, which was then re-suspended in 10 μL LC/MS (Liquid Chromatography Mass Spectrometry) water containing 0.1% FA. Quantitative proteomic analysis was performed using an LC/MS consisting of an ultimate 3000 RS LC nano and Q Exactive HF mass spectrometer (Thermo Fisher, USA). The peptides were separated on a C18 reverse-phase (RP) column (1.8 μm particle size, 75 μm × 250 mm, Waters, USA). The linear Acetonitrile gradient was as follows: elution with 8-15% Acetonitrile (containing 0.1% formic acid) for 50 min, followed by elution with 15-40% Acetonitrile for 70 min; the flow rate was set to 0.3 μL/min. The samples were electrosprayed into the Q Exactive HF mass spectrometer (voltage set at 2 kV and heat capillary temperature set at 290°C). Other parameters were set as follows: (1) Full MS range: 350-1200 m/z, resolution: 60,000, and AGC (Automatic Gain Control) set to 3 × 106; the MS/MS resolution was 30,000, and AGC was set to 1 × 106; (2) IT (injection time) was set to 50 ms; (3) 27% HCD (Higher-energy collisional dissociation) collision energy; (4) The DIA method was set as follows: the primary spectrum scanning was 350-1250 m/z, followed by 20 DIA secondary fragmentations, and each window was set at 10 m/z; (5) The raw data were analyzed using Spectronaut software version 15.0 (Biognosys, Switzerland) with default settings. The database of human proteins derived from Swissport (downloaded on January 2, 2020, containing 50,442 sequences) was utilized for protein identification. The selected search parameters were as follows: (1) Trypsin with two missed cleavages; (2) Variable modification was set as N-terminal acetylation and methionine oxidation; (3) Fixed modifier was set for reduction alkylation of cysteine.
Bioinformatics analysis
We performed bioinformatics analysis using various online tools. Venn diagrams were generated using the Bioinformatics & Evolutionary Genomics software (http://bioinformatics.psb.ugent.be/webtools/Venn/). Additionally, we used the Panther online software (http://pantherdb.org/) for GO (Gene Ontology) annotation analysis and omicsolution online software (https://www.omicsolution.org/wkomics/main/) for volcano drawing. Furthermore, the online software KOBAS (KEGG Orthology Based Annotation System) (http://kobas.cbi.pku.edu.cn/genelist/) was utilized for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis. PGRN, identified from the protein screen, was then analyzed using bioinformatics. GEPIA2 (Gene Expression Profiling Interactive Analysis 2) (http://gepia2.cancer-pku.cn) was applied to compare differential expression between TCGA (The Cancer Genome Atlas) cancer data and GTEx (Genotype Tissue Expression) normal data. The STRING database (https://www.string-db.org/) was utilized for PPI (Protein-Protein Interaction Networks) analysis. GEPIA2 was used for survival analysis, and Metascape (https://metascape.org) was used for the enrichment analysis of the PPI network genes. Finally, TIMER (Tumor Immune Estimation Resource) (http://timer.cistrome.org/) was used for the analysis of immune infiltrating cells and their associated gene expression.
Culture and passage of cell lines
Human gastric cancer cell lines HGC27, AGS, MKN74, and MKN45 and the gastric epithelial cell line GES-1 were obtained from the NICR (National Infrastructure of Cell Line Resource) platform of the National BMCR (Biomedical Cell Resource). These cell lines were cultured and passaged in our laboratory. All cell lines were maintained in RPMI-1640 medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37°C and 5% CO2 in a sterile incubator. The cells were monitored daily for growth and contamination; the culture media was changed once every two days. The cells were passaged by digestion with 0.25% trypsin, and only cells in the logarithmic growth phase were used for experiments. The RPMI-1640 culture medium, trypsin, fetal bovine serum (Gibco, USA), PBS (phosphate buffer saline), and penicillin/streptomycin (Sigma, USA) were used in the cell culture procedures.
Cell transfection and treatment
Single-cell suspensions were prepared from each group at a concentration of 1 × 106 cells/mL and inoculated in 6-well plates, followed by culture until the cells were completely confluent. siRNA (Small interfering RNA) (negative control siRNA and PGRN siRNA) (RiboPharm, Guangzhou, China) was introduced into the cells using LipofectamineTM 2000 (11668019, Thermo Fisher Scientific) according to the manufacturer’s instructions. PGRN-shRNAs and PGRN-shRNAs were purchased from GenePharma Company (Shanghai, China) for knockdown of PGRN. Then, plasmids psPAX2, pMD2G, and pcDNA3.1/PGRN were cotransfected into 293T cells to generate lentivirus. The lentivirus was harvested after 48 h of transfection. The virus was then co-cultured with target cells, and 48 hours after infection, puromycin was added to select successfully transfected cells. Cell death was monitored, and Western blotting was used to test the knockdown efficiency of different sequences. Cells with effective knockdown were selected for subsequent in vivo experiments. PGRN sequence: PGRN KO1: CCAGTGCCCTGATAGTCAGTTCGAA, PGRN KO2: TGACACGCAGAAGGGTACCTGTGAA, PGRN KO3: TGATCCAGAGTAAGTGCCTCTCCAA. SB203580 (S8307, Sigma) was diluted to 10 μg/mL, and cells in the logarithmic growth phase were seeded into 96-well plates at 5 × 103 per well. 10 μL of diluted SB203580 was added to each well in the treatment group while the control group received the same volume of DMSO. Each group was set up in triplicate wells. After continuous culture for 24 hours, the cells were used for subsequent experiments.
Immunohistochemistry
The tumor and paraneoplastic tissues were fixed, dehydrated, and embedded in paraffin. The tissues were then sectioned, deparaffinized in xylene, and rehydrated using an ethanol gradient. Heat-induced epitope retrieval was performed using a microwave oven, followed by treatment with hydrogen peroxide for 20 min to block endogenous peroxidase activity. Non-specific antigens were blocked by incubating the sections with serum. The sections were then incubated with a primary antibody (anti-PGRN, AF2420, R&D Systems) in a humidified chamber overnight at 4°C, followed by incubation with a secondary antibody (ab6741, Abcam) for 30 min. Finally, the sections were stained using a DAB kit, counterstained with hematoxylin, dehydrated, and mounted with neutral gum, as previously described [19]. The intensity score of cell staining at high magnification × percentage of stained cells was used as the immunohistochemical scoring criterion. A total score ≤ 3 was defined as a low expression, and a total score ≥ 4 was defined as a high expression.
Western blotting
The total protein from each group of cells was collected and extracted following standard protocols. The total protein concentration was determined using the BCA protein assay kit (23225, Thermo Fisher Scientific). The proteins were separated using 10% SDS-PAGE gel electrophoresis and transferred onto a PVDF membrane (IPVH00010, Millipore). All antibodies were diluted to 1:2000 in antibody dilution buffer before use. The membrane was blocked for 30 min with TBS containing 5% skim milk powder. The corresponding primary antibody was added, and the membrane was incubated at 4°C overnight. Next day, the membrane was washed three times with TBS-T and then incubated with the secondary antibody at room temperature for 1 h. After washing the membrane three times with TBS-T, the membrane was imaged using an Odyssey two-color infrared fluorescence imaging system. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, the relative expression levels of the target genes were determined by measuring the grayscale values of the protein bands. The primary antibodies used were anti-PGRN (AF2420, AF2557, R&D Systems), and all other primary and secondary antibodies were obtained from Abcam.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues or cells using TRIzol (15596018, Thermo Fisher Scientific). RNA purity and concentration were determined using a spectrophotometer. The cDNA was synthesized using the RevertAid First Strand cDNA synthesis kit containing DNase I (K16215, Thermo Fisher Scientific) according to the manufacturer’s instructions. Corresponding primers (synthesized by Sangon Biotech, Shanghai, China) and SYBRTM Green PCR Premix (A25742, Thermo Fisher Scientific) were added to the single-strand cDNA, and qPCR amplification was performed using a StepOne real-time fluorescence quantitative PCR system (4376357, Thermo Fisher Scientific) following the manufacturer’s instructions. The relative expression levels of the detected genes were calculated using the 2-∆∆Ct method, with GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) as the internal reference gene.
Cell Counting Kit-8 viability assay
The cells were prepared as single-cell suspensions and inoculated into a 96-well plate at a density of approximately 5 × 103 cells per well. The plate was then placed in a 37°C, 5% CO2 incubator for 48 h. After cell adhesion, 10 μL of CCK-8 (Cell Counting Kit-8) reagent (K1018, APExBIO) was added to each well, and the plates were incubated for an additional 4 h. The OD (Optical density) at 450 nm of each well was measured using a microplate reader, and the cell growth curves were plotted.
Cell scratch assay
Straight lines were drawn 1 cm apart at the bottom of a 6-well plate using a marker and a straightedge. Then, each group of cells was prepared as a single-cell suspension and inoculated into the 6-well plate. The cells were cultured until they formed a confluent monolayer, and a sterile 1-mL pipette tip was used to scratch the cell monolayer along the marked lines. The suspended cells were washed away with PBS, followed by replacing the culture medium with a serum-free medium. The healing of the scratch was observed after 0 and 24 h using a microscope, and the degree of healing was recorded by imaging.
Transwell chamber invasion assay
A sterile 24-well plate was used to hold a Transwell chamber, and 60 μL of Matrigel was diluted to 1 mg/mL using serum-free medium (356234, BD, USA) and added to the upper chamber. The Matrigel was placed in a 37°C incubator for solidification. Next, 100 μL of the single-cell suspension prepared from each group of cells was inoculated into each upper chamber, and 600 μL of culture medium containing 10% fetal bovine serum was added to the lower chamber. The cells were then cultured for an additional 48 h. The cells that did not cross the membrane were removed, and the invading cells were fixed and stained with 0.1% crystal violetTop of Form.
Establishment of a tumor-bearing model for gastric cancer
Male BALB/c nude mice, approximately 4-5 weeks of age, were randomly divided into two groups. Single-cell suspensions (5 × 106 cells/100 μL) were prepared from control and transfected cell lines. Subsequently, 200 μL of each suspension was subcutaneously injected into the right axilla of the mice in each group. After the formation of subcutaneous tumors, the long diameter (a) and short diameter (b) of the tumors were measured every three days using a vernier caliper, and the volume of the transplanted tumor was calculated using the formula (V (mm3) = 0.5 × a × b2). On day 24, mice were euthanized by cervical dislocation with anesthesia (all mice weighed less than 200 g), and tumor tissues were taken en bloc and weighed. The expression of relevant proteins was determined using western blot analysis. This experiment was approved by the Institutional Animal Care and Use Committee of the Fourth Hospital of Hebei Medical University (approval no.: 2022256).
Statistical analysis
IBM SPSS Statistics 25.0 was utilized for statistical analysis, GraphPad Prism 8 was used for data visualization, and Adobe Photoshop was used for image quantification. For data conforming to normal distribution, t-tests were used to compare the means of two samples, with standard t-test used for homoscedastic data and approximate t-test used for heteroscedastic data. The Wilcoxon test was used to compare the means of two samples that did not conform to the normal distribution. ANOVA (Analysis of variance) was employed for comparisons among multiple groups with homoscedastic data conforming to the normal distribution, and the LSD (least significant difference) method was used for pairwise comparisons between multiple groups. The Kruskal-Wallis H test was used for heteroscedastic data and data that did not conform to the normal distribution. The chi-square test was used to analyze the relationship between PGRN expression levels and clinical and pathological characteristics. The Kaplan-Meier method was used to plot survival curves, and the log-rank method was used for the comparison of survival data. A p-value < 0.05 was considered statistically significant. All experiments were repeated at least three times.
Data availability
The original mass spectrometry proteomics data has been uploaded and deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD035058.
Results
Identification and quantification of proteins and preliminary analysis
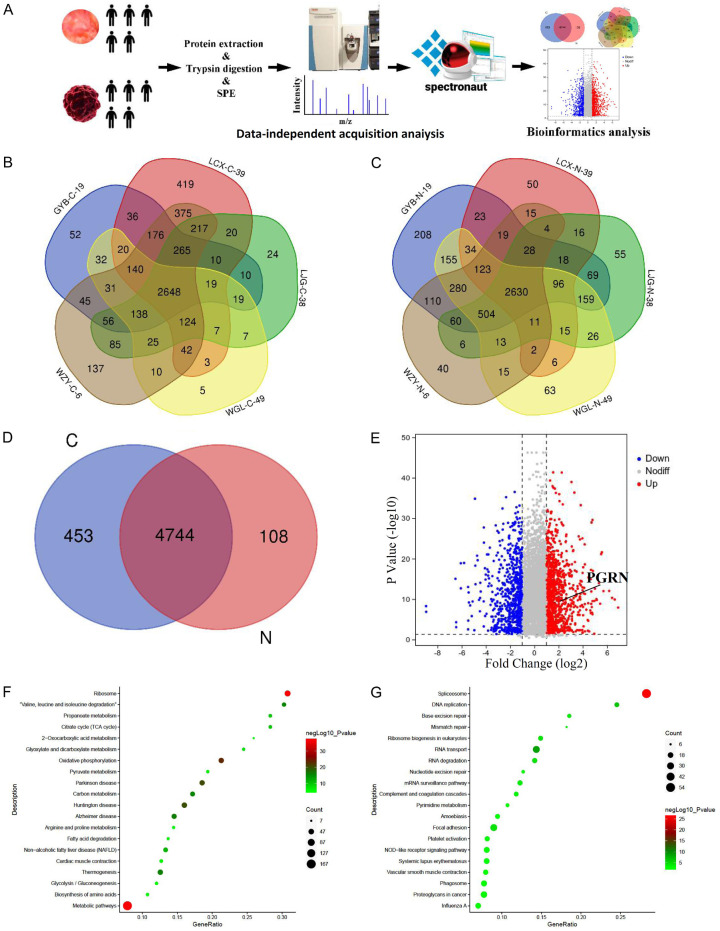
DIA quantitative proteomics was utilized to quantitatively analyze five normal tissue samples and five tumor tissue samples. The experimental methods and procedures are illustrated in Figure 1A. The quantitative results of five gastric cancer tissues showed that 3,697, 4,521, 3,674, 3,270, and 4,514 proteins were identified in GYB-C-19, LCX-C-39, LJG-C-38, WGL-C-49, and WZY-C-6, respectively. The intersection proteins across these five groups of data totaled 2,648 (Figure 1B). Additionally, 4,517, 3,090, 3,711, 4,133, and 3,816 proteins were identified in the corresponding normal tissues (GB-N-19, LCX-N-39, LJG-N-38, WGL-N-49, and WZY-N-6), with 2,630 intersection proteins across the five groups (Figure 1C). A comparison of unique and overlapping proteins between the tumor (C) and normal (N) groups was conducted (Figure 1D). Among the intersection proteins, those with expression levels that changed by ≥ 2 folds or ≤ 0.5 folds were defined as differentially expressed proteins. The differential proteins were visualized using a volcano plot (Figure 1E).
Figure 1.
Identification and quantification of proteins and preliminary analysis. A. Quantitative proteomic analysis of five paraneoplastic and five tumor tissue samples using the DIA (Data-independent acquisition) quantitative proteomics method. The figure illustrates the experimental workflow. B. In the five tumor tissue samples, 3,697, 4,521, 3,674, 3,270, and 4,514 proteins were identified, with 2,648 proteins at the intersection of these five datasets. C. In the five paraneoplastic tissue samples, 4,517, 3,090, 3,711, 4,133, and 3,816 proteins were identified, with 2,630 proteins at the intersection of these five datasets. D. 5,198 proteins were identified in tumor tissues, and 4,853 proteins were identified in paraneoplastic tissues, with 4,744 proteins intersecting between the two groups. E. Differentially expressed proteins were defined as proteins with a fold change of expression ≥ 2 or ≤ 0.5. The volcano plot includes 693 upregulated proteins and 1,006 downregulated proteins that were identified. F. The metabolic pathway was found to have the smallest p-value among the upregulated proteins, and other proteins involved in various signaling pathways are also depicted. G. The spliceosome pathway was found to have the smallest p-value among the downregulated proteins, and other proteins involved in various signaling pathways are also shown.
KEGG annotation analysis on the differentially expressed proteins revealed that the upregulated proteins were enriched in 272 pathways (including 54 pathways with a p-value of < 0.05), and the downregulated proteins were enriched in 285 pathways (including 38 pathways with a p-value of < 0.05). Detailed KEGG analysis of upregulated and downregulated proteins is shown in Figure 1F and 1G. Additionally, GO annotation analysis categorized the differentially expressed proteins into three major categories: molecular function, biological process and cell component. The results of up-regulated and down-regulated proteins are shown in Figure S1A and S1B.
Analysis of the relationship between PGRN expression and prognostic, clinical, and pathological features
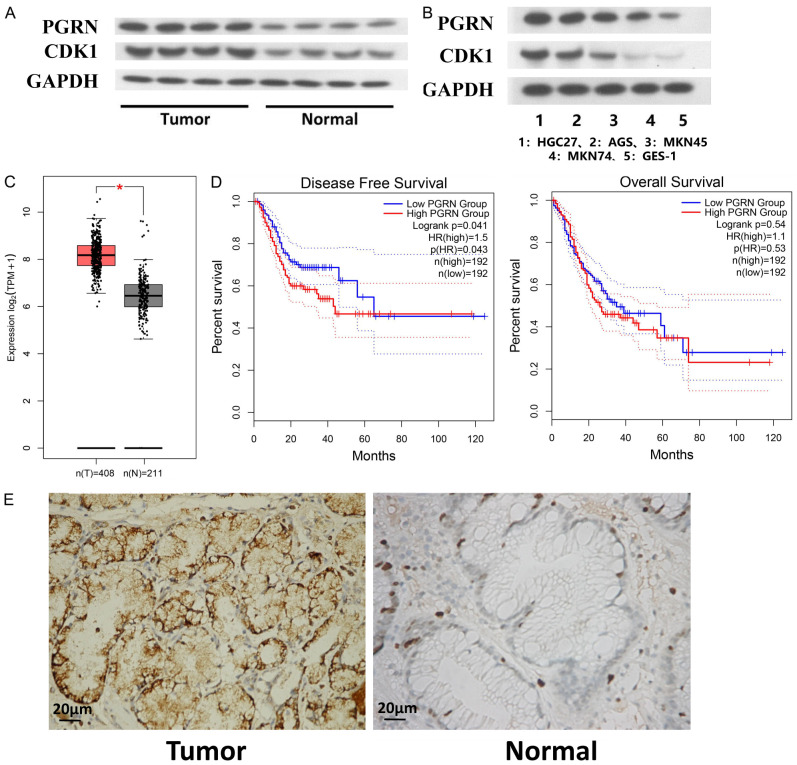
We initially measured the expression levels of PGRN and CDK1 (Cyclin Dependent Kinase 1) in 30 pairs of samples using WB. The results showed that the expression levels of these molecules were higher in gastric cancer tissues than in paraneoplastic tissues (Figure 2A). Moreover, the expression levels of PGRN and CDK1 were lower in GES-1 cells than those in gastric cancer cell lines. Among the cell lines, PGRN expression was highest in HGC27, followed by AGS, MKN45, MKN74, and GES-1 (Figure 2B).
Figure 2.
Relationship between PGRN (Progranulin) expression and prognostic, clinical, and pathological features. A. Western blotting results indicated that the expression levels of PGRN and CDK1 (Cyclin Dependent Kinase 1) were higher in gastric cancer tissues than in paraneoplastic tissues (n = 30). B. PGRN and CDK1 expression was lower in GES-1 cells than in any of the gastric cancer cell lines, with the cell lines in decreasing order of PGRN expression being HGC27, AGS, MKN45, MKN74, and GES-1 (n = 6). C. Analysis of differentially expressed proteins indicated that PGRN was highly expressed in gastric cancer tissues (P < 0.05). D. Survival analysis showed that PGRN expression correlated with the disease-free survival of patients (P < 0.05). E. Immunohistochemistry showed that PGRN expression was significantly higher in gastric cancer tissues than in paraneoplastic tissues (400×) (n = 30).
Bioinformatics analysis revealed that PGRN was highly expressed in gastric cancer tissues, with a log2 > 1.7216 and P < 0.05 (Figure 2C). Moreover, the survival analysis indicated that elevated PGRN expression was correlated with poorer disease-free survival of patients (Figure 2D). We employed immunohistochemistry to investigate the relationship between PGRN expression (cytoplasmic staining) and clinical and pathological features of different types of gastric cancer (Figure 2E). Furthermore, PGRN expression (cytoplasmic staining) was associated with clinical and pathological features, particularly lymph node metastasis (Table 1).
Table 1.
Relationship between Progranulin expression and clinicopathological features in gastric cancer tissues (n = 30)
| Characteristics | Number (n) | Expression of Progranulin | Χ2 | P | |
|---|---|---|---|---|---|
|
| |||||
| Low (5) | High (25) | ||||
| Gender | |||||
| Male | 21 | 3 | 18 | 0.286 | 0.593 |
| Female | 9 | 2 | 7 | ||
| Age | |||||
| ≥ 60 | 18 | 4 | 14 | 1.001 | 0.317 |
| < 60 | 12 | 1 | 11 | ||
| Tumor location | |||||
| Upper | 17 | 2 | 15 | 0.679 | 0.410 |
| Lower | 13 | 3 | 10 | ||
| Tumor diameter | |||||
| ≥ 5 cm | 11 | 1 | 10 | 0.718 | 0.397 |
| < 5 cm | 19 | 4 | 15 | ||
| Tumor invasion depth | |||||
| In the serous | 14 | 2 | 12 | 0.107 | 0.743 |
| Serous outside | 16 | 3 | 13 | ||
| Tumor differentiation degree | |||||
| Well and mid differentiated | 18 | 4 | 14 | 1.001 | 0.317 |
| Poorly differentiated | 12 | 1 | 11 | ||
| Lymph node metastasis | |||||
| Positive | 22 | 1 | 21 | 8.727 | 0.003* |
| Negative | 8 | 4 | 4 | ||
| Vascular tumors thrombus | |||||
| Positive | 21 | 2 | 19 | 2.571 | 0.109 |
| Negative | 9 | 3 | 6 | ||
| Distant metastasis | |||||
| With | 4 | 1 | 3 | 0.231 | 0.631 |
| Without | 26 | 4 | 22 | ||
P < 0.05.
Selection of gastric cancer cell lines with high PGRN expression for inhibition studies using siRNA
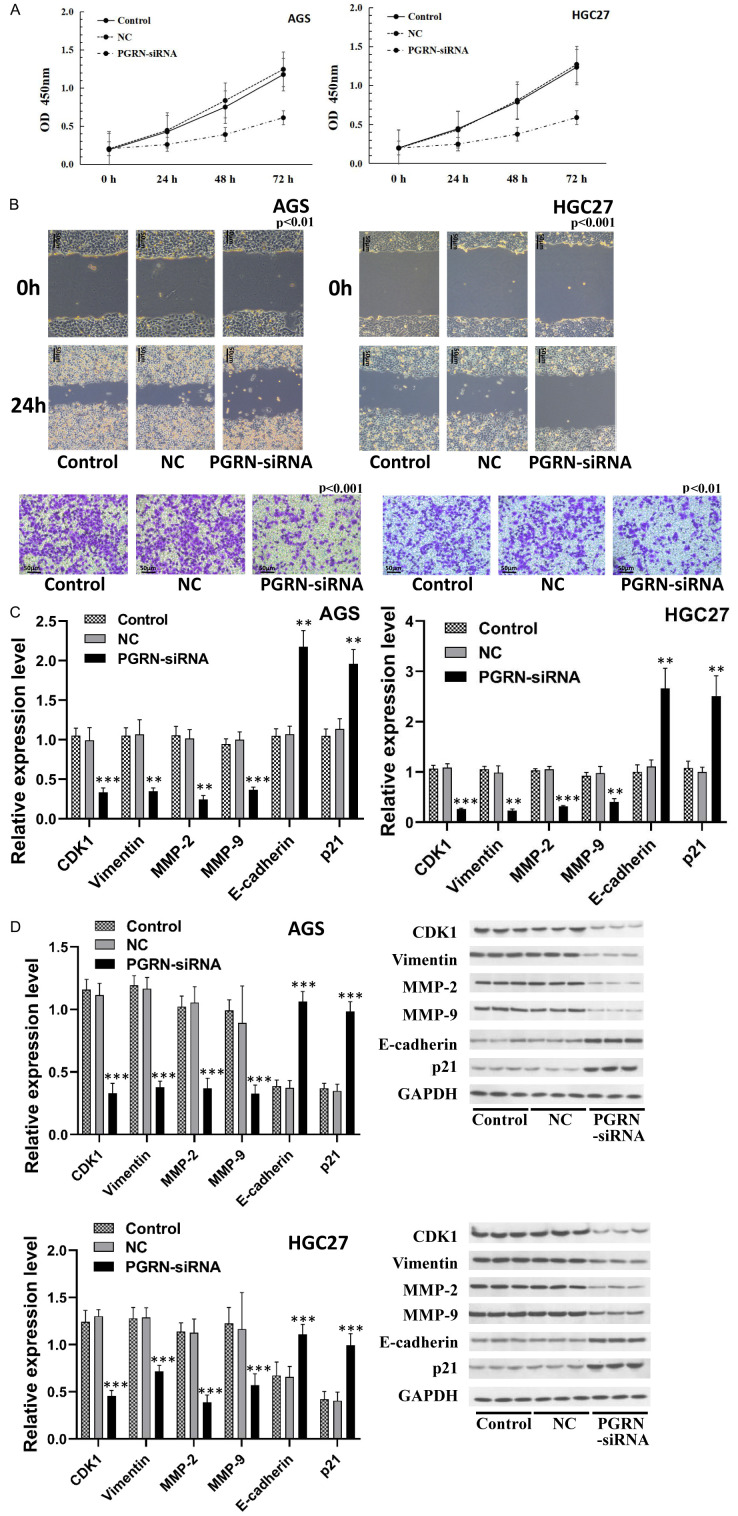
The gastric cancer cell lines AGS and HGC27, with a high PGRN expression, were selected for further studies. The CCK-8 assay showed a significant decrease in cell viability following PGRN inhibition in AGS and HGC27 cells (Figure 3A). The cell scratch and Transwell invasion assays demonstrated a significant reduction in the invasion and migration abilities of AGS and HGC27 cells after PGRN inhibition (P < 0.01) (Figure 3B). These results suggest that PGRN can promote the viability, invasion and migration of gastric cancer cells.
Figure 3.
Relationship between PGRN expression and cell viability, migration, and invasion. (A) CCK-8 results showed that cell viability was significantly reduced after inhibition of PGRN expression in AGS and HGC27 cells (P < 0.05). (B) Scratch and Transwell invasion assays showed that the invasion and migration of cells were significantly reduced after inhibition of PGRN expression in AGS and HGC27 cells (200×) (P < 0.01). (C) RT-qPCR and (D) western blotting showed that genes and proteins associated with proliferation, invasion, and migration were significantly altered in AGS and HGC27 cells after PGRN inhibition (P < 0.01). All experiments were repeated three times.
Furthermore, inhibiting PGRN expression in AGS and HGC27 cells led to significant alterations in the expression of genes and proteins associated with proliferation, invasion, and migration. The expression of CDK1, MMP (Matrix metallopeptidase)-2, MMP-9, and vimentin was significantly decreased, whereas the expression of p21 and E-cadherin was significantly increased (all P < 0.01) (Figure 3C, 3D). These findings demonstrate that PGRN is closely related to cell proliferation, invasion and migration.
Bioinformatics analysis for functional enrichment of PGRN and experimental validation
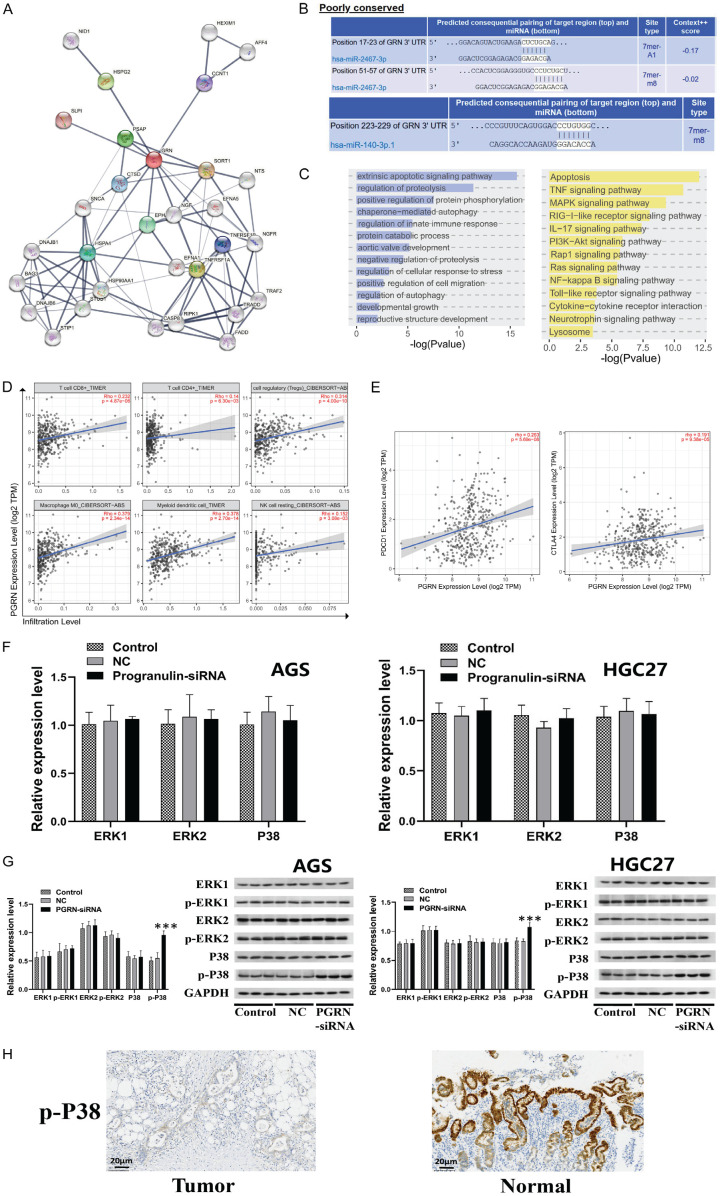
We conducted a PPI network analysis of PGRN using the three-step neighbor method, which revealed interactions with CTSD, PSAP, HSPG2, CCNT1, SORT1, NGF, EPH, and HSPA4 (Figure 4A). Additionally, we predicted miRNA regulation of PGRN and identified binding sites for hsa-miR-2467-3p and hsa-miR-140-3p.1 (Figure 4B). Enrichment analysis of the genes in the PPI network showed their involvement in the intrinsic apoptotic signaling pathway, regulation of proteolysis, and positive regulation of protein phosphorylation in the GO analysis. Furthermore, there was a correlation with several signaling pathways, such as apoptosis, TNF, MAPK, and RIG-I-like, in the KEGG enrichment analysis (Figure 4C). We also analyzed the correlation between PGRN expression and immune cell abundance, showing a positive correlation between PGRN expression and the abundance of CD8+ T cells, Treg cells, M0 macrophages, and myeloid dendritic cells (Figure 4D). Additionally, a positive correlation between PGRN expression and PD-1 immune checkpoint gene expression was observed (Figure 4E). The KEGG enrichment analysis revealed that the MAPK signaling pathway was among the functionally enriched signaling pathways of PGRN. Upon inhibiting PGRN expression in AGS and HGC27 cells, there was no significant change in the expression of ERK1/2, phosphorylated ERK1/2 (p-ERK1/2), or P38-MAPK in the MAPK signaling pathway. However, the expression of phosphorylated P38 (p-P38) increased significantly (Figure 4F, 4G). Immunohistochemistry analysis showed that p-P38 expression was significantly lower in gastric cancer tissues than in paraneoplastic tissues (200×) (Figure 4H).
Figure 4.
Functional analysis and mechanism of action of PGRN. A. PPI network of PGRN (three-step neighbor method). B. Prediction of miRNA regulation of PGRN. C. Enrichment analysis of genes in the PPI network by GO and KEGG analyses. D. Analysis of correlation between PGRN expression and immune cell abundance. E. Analysis of the correlation between PGRN expression and immune checkpoint gene expression. F, G. RT-qPCR and WB showed that the expression of ERK1/2, p-ERK1/2, or P38-MAPK in the MAPK signaling pathway was not significantly changed after inhibiting the expression of PGRN in AGS and HGC27 cells; however, the expression of p-P38 was significantly increased (P < 0.001). H. Immunohistochemistry detection of p-P38 expression in tissues showed that p-P38 expression was significantly lower in gastric cancer tissues than in paraneoplastic tissues (200×). All experiments were repeated three times.
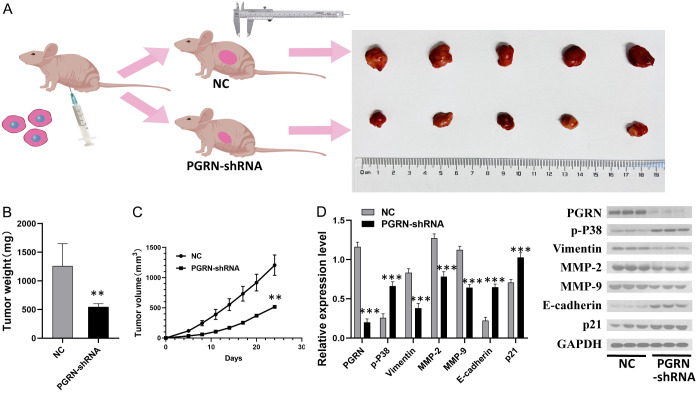
Inhibition of PGRN expression and its effects on subcutaneously transplanted tumor growth in nude mice
The average weight of the transplanted tumors in the PGRN-shRNA transfected group was significantly lower than that of the tumors in the empty vector-transfected group (P < 0.05). The tumor growth curve showed a marked delay (Figure 5B, 5C), and the eventual subcutaneous tumors were noticeably smaller (Figure 5A). The determination of related protein expression in both groups using WB showed that MMP-2, MMP-9, and vimentin expression was decreased, whereas P21 and E-cadherin expression was increased in the PGRN-shRNA transfected group compared with those in the empty vector-transfected group (all P < 0.001) (Figure 5D). These results suggest that inhibiting PGRN expression in gastric cancer cells can hinder growth of xenografts.
Figure 5.
Effect of PGRN expression on subcutaneously transplanted tumor growth in nude mice. A. The transplanted tumors in the PGRN-shRNA transfected group was smaller than those in the empty vector-transfected group. B, C. The average weight of transplanted tumors in the PGRN-shRNA transfected group was significantly lower than that in the empty vector-transfected group, and the growth curve showed a delay (P < 0.01). D. Western blotting analysis of related proteins in both groups showed that the protein expression of MMP-2, MMP-9, and vimentin decreased, whereas P21 and E-cadherin expression increased in the PGRN-shRNA transfected group compared with those in the empty vector-transfected group (all P < 0.001) (n = 3). These results confirm that inhibiting PGRN expression can suppress tumor formation in gastric cancer cells in vivo.
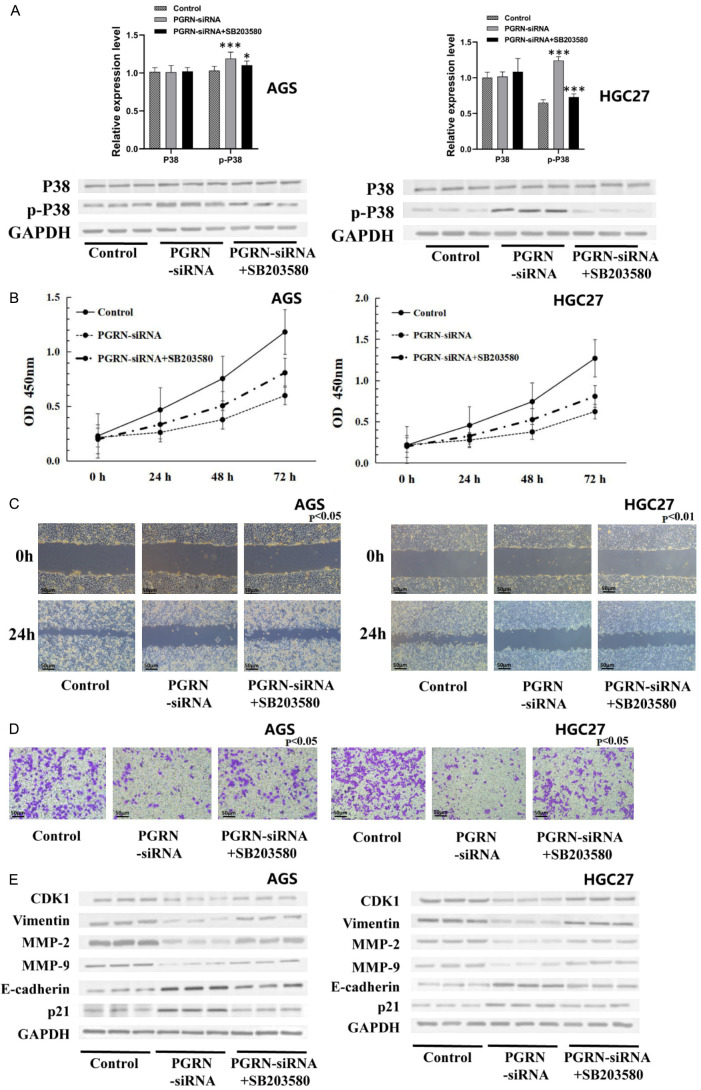
SB203580 alleviated the effects of inhibiting PGRN expression on cell activity, migration, invasion, and related gene and protein expression by inhibiting p-P38 in MAPK
SB203580, a selective inhibitor of MAPK pathway, was used to investigate the role of p-P38 proten in PGRN regulation. The expression of p-P38 in the PGRN-siRNA group was significantly higher than that in the control group, and the expression of p-P38 in the SB203580 combined with PGRN-siRNA group was lower than that in the PGRN-siRNA group (Figure 6A).
Figure 6.
SB203580 alleviated the effects of inhibiting PGRN expression on cell activity, migration, invasion, and related gene and protein expression by inhibiting p-P38 in MAPK pathway. A. The expression of p-P38 in the PGRN-siRNA group was higher than that in the control group, and the expression of p-P38 in the SB203580 combined with PGRN-siRNA group was lower than that in the PGRN-siRNA group. B-D. Compared with the PGRN-siRNA group, the SB203580 combined with PGRN-siRNA group showed increased cell activity, migration, and invasion abilities (200×). E. Compared with the PGRN-siRNA group, the expression of genes and proteins related to proliferation, invasion, and migration was altered after treatment with SB203580 combined with PGRN-siRNA in AGS and HGC27 cells (P < 0.05). All experiments were repeated three times.
The CCK-8 assay, the scratch test and the transwell chamber invasion assay showed that the cell viabilty, migration and invasion in the SB203580 combined with PGRN-siRNA group was notably higher than those in the PGRN-siRNA group but lower than those in the control group (all P < 0.05) (Figure 6B-D). The proteins related to cell proliferation, invasion and migration changed correspondingly (Figure 6E). These results indicate that the effects of PGRN on the proliferation, invasion and migration of gastric cancer cells are closely related to the MAPK pathway.
Discussion
Gastric cancer is one of the most prevalent gastrointestinal malignancies worldwide, with a generally poor prognosis. The molecular mechanisms udnerlying gastric cancer progression remain unclear. Recent progress in mechanistic studies has shown great potential for translation into clinical diagnosis and treatment. DIA proteomics is a newly developed global mass spectrometry (MS)-based proteomics strategy that offers broad protein coverage, high reproducibility, and high sensitivity, allowing for better protein quantification [20]. DIA and HPLC-MS were used for quantitative proteomic analysis of gastric adenocarcinoma tissues and corresponding accessory tumor tissues, forming a rich protein expression dataset for gastric cancer studies. Following preliminary functional and pathway analyses, Progranulin (PGRN) was identified as a key protein for further investigation.
PGRN is a multifunctional protein implicated in the development and progression of various tumors through multiple mechanisms of action [11-15,21]. It has been associated with the prognosis of multiple tumors [22-24] and is considered a potential biomarker and therapeutic target [25,26]. Previous studies have confirmed the correlation between PGRN and Helicobacter pylori infection [17,27], suggesting that PGRN may be a potential marker of gastric cancer [18]. However, the mechanism of PGRN in gastric cancer is still unclear. Thus, we conducted an in-depth study on the function and significance of PGRN in gastric cancer, which has been rarely reported in previous studies.
We initially conducted database analysis and showed that PGRN was highly expressed in gastric cancer tissues, a finding further corroborated by experimental results. Survival analysis demonstrated that PGRN expression levels were correlated with the disease-free survival rate of patients. Additionally, we determined the relationship between PGRN expression (cytoplasmic staining) and clinical and pathological features of gastric cancer using immunohistochemistry, showing that higher PGRN expression is significantly associated with lymph node metastasis. These results suggest that PGRN could serve as a biomarker for gastric cancer, which is consistent with findings from breast cancer studies. In breast cancer, subgroups with high PGRN expression and sortilin were significantly associated with breast cancer-specific mortality, tumor size, grade, and lymph node positivity, suggesting their use as prognostic biomarkers [28]. In our study, inhibition of PGRN expression resulted in significantly decreased cell viability, invasion and migration, along with significant alterations in the expression of associated genes and proteins. This is consistent with previous studies on bladder [11] and thyroid cancer [29]. Therefore, PGRN is closely associated with the proliferation and invasive metastasis of gastric cancer.
Apart from determining the specific mechanism of PGRN’s role in gastric cancer, we further conducted data mining using bioinformatics. PGRN interacts with various proteins, among which NGF, SORT1, and EPH are associated with the development of gastric cancer [30-32]. Next, PGRN was predicted to have a regulatory relationship with hsa-miR-2467-3p and hsa-miR-140-3p. Notably, Hsa-miR-140-3p is known to regulate the progression of gastric cancer [33]. Enrichment analysis of the genes in the PPI network further revealed associations with multiple signaling pathways, such as MAPK. These data mining results offer valuable insights to guide future studies.
Further analysis revealed a positive correlation between PGRN expression and the infiltration of several key immune cell types, as well as the expression of PD-1 and CTLA4. These findings align with previous studies, showing that PGRN promotes tumor immune evasion by upregulating PD-L1 expression in TAM (Tumor-associated macrophages) and faciliating CD8+ T cell rejection [34]. This result suggests that the development and progression of gastric cancer may also be associated with PGRN-mediated immune pathways. Functionally, PGRN was found to be enriched in the MAPK signaling pathway. We verified that inhibiting PGRN expression in gastric cancer cells did not significantly alter the expression of ERK1/2, phosphorylated ERK1/2 (p-ERK1/2), or P38-MAPK but increased p-P38 (phosphorylated P38) expression in the MAPK signaling pathway. Our experiments demonstrate that PGRN regulates the expression of p-P38 in the MAPK pathway, influencing the activity, invasion, and migration of gastric cancer cells. Furthermore, in a tumor bearing model of gastric cancer cells, we found that the tumor growth was significantly slowed after PGRN expression was inhibited, and the expression of invasion and migration related proteins changed correspondingly. Although additional in-depth studies and examination of PGRN levels in peripheral blood are necessary, the results of this study indicate that PGRN holds promise as a novel therapeutic target for the treatment of gastric cancer.
Conclusions
This study identified significant differences in protein expression between gastric adenocarcinoma tissues and adjacent paraneoplastic tissues. Among the differentially expressed proteins, PGRN emerged as a major differentially expressed protein involved in the proliferation, migration, and invasion of gastric cancer, indicating that it could serve as a potential novel therapeutic target for gastric cancer.
Acknowledgements
We would like to express our gratitude to the Institutional Review Board of the Fourth Hospital of Hebei Medical University for their assistance.
Written informed consent was obtained from all patients.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2016. J Natl Cancer Cent. 2022;2:1–9. doi: 10.1016/j.jncc.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Feng A, Zheng S, Chen C, Lyu J. Recent estimates and predictions of 5-year survival in patients with gastric cancer: a model-based period analysis. Cancer Control. 2022;29:10732748221099227. doi: 10.1177/10732748221099227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, Cai S, Cui P, Song G, Rao D, Yi X, Wu Y, Song N, Liu F, Zou Y, Zhang S, Zhang X, Wang X, Qiu S, Zhou J, Wang S, Zhang X, Shi Y, Figeys D, Ding L, Wang P, Zhang B, Rodriguez H, Gao Q, Gao D, Zhou H, Fan J. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40:70–87. e15. doi: 10.1016/j.ccell.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Puttamallesh VN, Deb B, Gondkar K, Jain A, Nair B, Pandey A, Chatterjee A, Gowda H, Kumar P. Quantitative proteomics of urinary bladder cancer cell lines identify UAP1 as a potential therapeutic target. Genes (Basel) 2020;11:763. doi: 10.3390/genes11070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Mouradov D, Wang X, Jorissen RN, Chambers MC, Zimmerman LJ, Vasaikar S, Love CG, Li S, Lowes K, Leuchowius KJ, Jousset H, Weinstock J, Yau C, Mariadason J, Shi Z, Ban Y, Chen X, Coffey RJC, Slebos RJC, Burgess AW, Liebler DC, Zhang B, Sieber OM. Colorectal cancer cell line proteomes are representative of primary tumors and predict drug sensitivity. Gastroenterology. 2017;153:1082–1095. doi: 10.1053/j.gastro.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, Andersson R, Hu D, Bauden M, Kristl T, Sasor A, Pawlowski K, Pla I, Hilmersson KS, Zhou M, Lu F, Marko-Varga G, Ansari D. Quantitative proteomics identifies brain acid soluble protein 1 (BASP1) as a prognostic biomarker candidate in pancreatic cancer tissue. EBioMedicine. 2019;43:282–294. doi: 10.1016/j.ebiom.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Repetto O, De Re V, Giuffrida P, Lenti MV, Magris R, Venerito M, Steffan A, Di Sabatino A, Cannizzaro R. Proteomics signature of autoimmune atrophic gastritis: towards a link with gastric cancer. Gastric Cancer. 2021;24:666–679. doi: 10.1007/s10120-020-01148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Q, Polom K, Williams C, de Oliveira FMS, Guergova-Kuras M, Lisacek F, Karlsson NG, Roviello F, Kamali-Moghaddam M. A targeted proteomics approach reveals a serum protein signature as diagnostic biomarker for resectable gastric cancer. EBioMedicine. 2019;44:322–333. doi: 10.1016/j.ebiom.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y, Wang J, Sun J, Chen X, Shi J, Wu Z, Yu D, Zhang F, Wang Z. Screening of potential biomarkers for gastric cancer with diagnostic value using label-free global proteome analysis. Genomics Proteomics Bioinformatics. 2020;18:679–695. doi: 10.1016/j.gpb.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buraschi S, Neill T, Xu SQ, Palladino C, Belfiore A, Iozzo RV, Morrione A. Progranulin/EphA2 axis: a novel oncogenic mechanism in bladder cancer. Matrix Biol. 2020;93:10–24. doi: 10.1016/j.matbio.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung PF, Yang J, Fang R, Borgers A, Krengel K, Stoffel A, Althoff K, Yip CW, Siu EHL, Ng LWC, Lang KS, Cham LB, Engel DR, Soun C, Cima I, Scheffler B, Striefler JK, Sinn M, Bahra M, Pelzer U, Oettle H, Markus P, Smeets EMM, Aarntzen EHJG, Savvatakis K, Liffers ST, Lueong SS, Neander C, Bazarna A, Zhang X, Paschen A, Crawford HC, Chan AWH, Cheung ST, Siveke JT. Progranulin mediates immune evasion of pancreatic ductal adenocarcinoma through regulation of MHCI expression. Nat Commun. 2022;13:156. doi: 10.1038/s41467-021-27088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frampton G, Invernizzi P, Bernuzzi F, Pae HY, Quinn M, Horvat D, Galindo C, Huang L, McMillin M, Cooper B, Rimassa L, DeMorrow S. Interleukin-6-driven progranulin expression increases cholangiocarcinoma growth by an Akt-dependent mechanism. Gut. 2012;61:268–77. doi: 10.1136/gutjnl-2011-300643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang D, Wang LL, Dong TT, Shen YH, Guo XS, Liu CY, Liu J, Zhang P, Li J, Sun YP. Progranulin promotes colorectal cancer proliferation and angiogenesis through TNFR2/Akt and ERK signaling pathways. Am J Cancer Res. 2015;5:3085–3097. [PMC free article] [PubMed] [Google Scholar]
- 15.Yue S, Ye X, Zhou T, Gan D, Qian H, Fang W, Yao M, Zhang D, Shi H, Chen T. PGRN-/- TAMs-derived exosomes inhibit breast cancer cell invasion and migration and its mechanism exploration. Life Sci. 2021;264:118687. doi: 10.1016/j.lfs.2020.118687. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Sun Y, Liu S, Yu H, Li W, Zeng J, Chen C, Jia J. Upregulation of progranulin by Helicobacter pylori in human gastric epithelial cells via p38MAPK and MEK1/2 signaling pathway: role in epithelial cell proliferation and migration. FEMS Immunol Med Microbiol. 2011;63:82–92. doi: 10.1111/j.1574-695X.2011.00833.x. [DOI] [PubMed] [Google Scholar]
- 17.Ren Z, Li J, Du X, Shi W, Guan F, Wang X, Wang L, Wang H. Helicobacter pylori-induced progranulin promotes the progression of the gastric epithelial cell cycle by regulating CDK4. J Microbiol Biotechnol. 2022;32:844–854. doi: 10.4014/jmb.2203.03053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Li R, Wang H, Wang J, Han L, Pan L, Li X, Kong Q, Wang G, Su X. Clinical implications of progranulin in gastric cancer and its regulation via a positive feedback loop involving AKT and ERK signaling pathways. Mol Med Rep. 2017;16:9685–9691. doi: 10.3892/mmr.2017.7796. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Yuan H, Zhao L, Guo S, Hu S, Tian M, Nie Y, Yu J, Zhou C, Niu J, Wang G, Song Y. Targeting the miR-34a/LRPPRC/MDR1 axis collapse the chemoresistance in P53 inactive colorectal cancer. Cell Death Differ. 2022;29:2177–2189. doi: 10.1038/s41418-022-01007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barkovits K, Pacharra S, Pfeiffer K, Steinbach S, Eisenacher M, Marcus K, Uszkoreit J. Reproducibility, specificity and accuracy of relative quantification using spectral library-based data-independent acquisition. Mol Cell Proteomics. 2020;19:181–197. doi: 10.1074/mcp.RA119.001714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Li E, Luo L, Zhao S, Liu L, Wang J, Kang R, Luo J. A PSCA/PGRN-NF-kappaB-integrin-alpha4 axis promotes prostate cancer cell adhesion to bone marrow endothelium and enhances metastatic potential. Mol Cancer Res. 2020;18:501–513. doi: 10.1158/1541-7786.MCR-19-0278. [DOI] [PubMed] [Google Scholar]
- 22.Carlson AM, Maurer MJ, Goergen KM, Kalli KR, Erskine CL, Behrens MD, Knutson KL, Block MS. Utility of progranulin and serum leukocyte protease inhibitor as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:1730–1735. doi: 10.1158/1055-9965.EPI-12-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do IG, Jung KU, Koo DH, Lee YG, Oh S, Kim K, Kim DH, Sohn JH, Son BH, Lee SR, Shin JH, Kim HO, Kim H, Chun HK, Serrero G, Yoo CH. Clinicopathological characteristics and outcomes of gastrointestinal stromal tumors with high progranulin expression. PLoS One. 2021;16:e0245153. doi: 10.1371/journal.pone.0245153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greither T, Steiner T, Bache M, Serrero G, Otto S, Taubert H, Eckert AW, Kappler M. GP88/PGRN serum levels are associated with prognosis for oral squamous cell carcinoma patients. Biology (Basel) 2021;10:400. doi: 10.3390/biology10050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guha R, Yue B, Dong J, Banerjee A, Serrero G. Anti-progranulin/GP88 antibody AG01 inhibits triple negative breast cancer cell proliferation and migration. Breast Cancer Res Treat. 2021;186:637–653. doi: 10.1007/s10549-021-06120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Juarez CE, Arechavaleta-Velasco F, Zeferino-Toquero M, Alvarez-Arellano L, Estrada-Moscoso I, Diaz-Cueto L. Inhibition of PI3K/AKT/mTOR and MAPK signaling pathways decreases progranulin expression in ovarian clear cell carcinoma (OCCC) cell line: a potential biomarker for therapy response to signaling pathway inhibitors. Med Oncol. 2019;37:4. doi: 10.1007/s12032-019-1326-5. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Xiang M, Zhou J, Ren Z, Shi W, Du X, Fu X, Li P, Wang H. Progranulin inhibits autophagy to facilitate intracellular colonization of Helicobacter pylori through the PGRN/mTOR/DCN axis in gastric epithelial cells. Front Cell Infect Microbiol. 2024;14:1425367. doi: 10.3389/fcimb.2024.1425367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger K, Rhost S, Rafnsdottir S, Hughes E, Magnusson Y, Ekholm M, Stal O, Ryden L, Landberg G. Tumor co-expression of progranulin and sortilin as a prognostic biomarker in breast cancer. BMC Cancer. 2021;21:185. doi: 10.1186/s12885-021-07854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong Y, Tan H, Wang L, Liu Z. Progranulin promoted the proliferation, metastasis, and suppressed apoptosis via JAK2-STAT3/4 signaling pathway in papillary thyroid carcinoma. Cancer Cell Int. 2023;23:191. doi: 10.1186/s12935-023-03033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou N, Yang D, Yu S, Wu B, Gao Y, Li Y. SNRPA enhances tumour cell growth in gastric cancer through modulating NGF expression. Cell Prolif. 2018;51:e12484. doi: 10.1111/cpr.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PC, Chen ST, Kuo TC, Lin TC, Lin MC, Huang J, Hung JS, Hsu CL, Juan HF, Lee PH, Huang MC. C1GALT1 is associated with poor survival and promotes soluble Ephrin A1-mediated cell migration through activation of EPHA2 in gastric cancer. Oncogene. 2020;39:2724–2740. doi: 10.1038/s41388-020-1178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z, Guo W, Wang H, Dong Z, Lin M, Zhou X, Zheng Y. Circular RNA hsa_circ_0110389 promotes gastric cancer progression through upregulating SORT1 via sponging miR-127-5p and miR-136-5p. Cell Death Dis. 2021;12:639. doi: 10.1038/s41419-021-03903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Cai S, Gu T, Song F, Xue Y, Sun D. MiR-140-3p impedes gastric cancer progression and metastasis by regulating BCL2/BECN1-mediated autophagy. Onco Targets Ther. 2021;14:2879–2892. doi: 10.2147/OTT.S299234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang W, Zhou T, Shi H, Yao M, Zhang D, Qian H, Zeng Q, Wang Y, Jin F, Chai C, Chen T. Progranulin induces immune escape in breast cancer via up-regulating PD-L1 expression on tumor-associated macrophages (TAMs) and promoting CD8+ T cell exclusion. J Exp Clin Cancer Res. 2021;40:4. doi: 10.1186/s13046-020-01786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectrometry proteomics data has been uploaded and deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD035058.