Abstract
Objectives: Ovarian cancer is a gynecologic tumor with the highest mortality rate worldwide. Nonetheless, chemoresistance remains a significant obstacle in treating ovarian cancer. PARP inhibitors (PARPis) are effective drugs approved for maintenance therapy in ovarian cancer. However, the development of natural or acquired resistance to PARPis poses a major challenge for ovarian cancer treatment. Methods: Public database analysis of cGAS expression in relation to PARPi resistance. cCK-8 assay was used to determine cell survival. qPCR assay with Western Blot was implemented to determine gene expression and protein activation status. Results: Analysis of public databases revealed significantly higher cGAS expression in Olaparib-resistant cells and in recurrent ovarian tumors. Furthermore, high cGAS expression significantly promoted Olaparib tolerance in ovarian cancer cells. Our findings demonstrate that Olaparib treatment induces activation of the TBK1-IRF3 signaling axis downstream of cGAS, leading to the production of type I interferon. This, in turn, activates NF-κB and IL-6-STAT3 signaling, contributing to inflammation and PARPi resistance. Consequently, targeting cGAS effectively counteracts Olaparib resistance and enhances its efficacy in suppressing cancer cell growth, ultimately leading to cell death. Conclusions: Our study highlights the crucial function of cGAS signaling in mediating PARPi resistance in ovarian cancer cells. These findings provide valuable novel therapeutic strategies targeting cGAS to improve the efficacy of PARPi-based treatments for ovarian cancer.
Keywords: Ovarian cancer, PARP inhibitor, cGAS, inflammation
Introduction
Ovarian cancer is the third most common type of gynecological tumor, having the highest mortality rate worldwide, with high-grade serous ovarian cancer (HGSOC) being the most prevalent subtype [1]. Regrettably, effective management of HGSOC remains a significant challenge. Platinum-based chemotherapy constitutes the initial treatment modality for advanced ovarian cancer [2]. However, the efficacy of this treatment remains limited, characterized by modest response rates. In the past, radiotherapy and chemotherapy aimed to eliminate cancer cells by targeting specific gene expression profiles. However, this approach can lead to collateral damage of healthy cells. Nearly, approximately 50% of HGSOC patients, particularly those harboring BRCA1/2 mutations, exhibit a deficiency in homologous recombination (HR). PARP inhibitors (PARPis), such as Olaparib, have demonstrated promising therapeutic effects and have received regulatory approved for the maintenance treatment of patients with ovarian cancer [3]. Poly(ADP-ribose) polymerases (PARPs) play an essential role in DNA damage repair, specifically in detecting and responding to single-strand breaks (SSBs). They produce substantial quantities of poly(ADP-ribose) polymers (PARs) to recruit other DNA damage repair factors. Notably, the synthetic lethal mechanism observed between key HR genes, namely BRCA1/2 and PARP1, has been identified as the most promising therapeutic strategy to date [4,5]. PARPi bind to the catalytic domain of PARP, inhibit its enzymatic activity, thereby blocking single-strand DNA repair. Unrepaired SSBs are converted to double-strand breaks (DSBs) at replication forks in cells with HR deficiencies such as those lacking BRCA1. Consequently, BRCA2-deficient tumor cells can only undergo double-strand repair via non-homologous end joining (NHEJ), ultimately leading to cell death. This phenomenon is known as the synthetic lethal process. The presence of inherent HR deficiency allows the addition of PARPis to inhibit single-strand repair. The absence of either of these two factors alone does not result in cell death. Consequently, the use of PARPis has been established as an antitumor strategy termed synthetic lethality, particularly in BRCA-mutant tumor cells [6,7]. Notably, PARP inhibitors have demonstrated therapeutic benefits for all ovarian cancer patients, regardless of their BRCA mutation or HR status [8,9].
Despite demonstrating strong clinical efficacy, a significant proportion of ovarian cancer patients treated with PARPis experience cancer recurrence due to primary or acquired drug resistance. To date, several mechanisms have been discovered that lead to resistance against PARP inhibitors. The restoration of HR function has been identified as a prominent mechanism underlying PARPi resistance. Secondary mutations or epigenetic alterations in BRCA1/2, such as reversal of BRCA1 promoter methylation and re-expression of functional HR proteins, can contribute to the development of PARPi resistance [10]. In addition to mutations in BRCA1/2, HR-associated secondary mutations in RAD51C and PALB2 have been documented [11]. Beyond the reversal of mutations in key genes, HR restoration can also be achieved through compensatory mutations. For instance, cancer cells can restore HR repair capacity by downregulating 53BP1, thus bypassing the lethal consequences of DNA damage accumulation [12,13]. Moreover, increased drug efflux and disruptions in the cell cycle pathways represent significant factors contributing to PARPi resistance. It is noteworthy that over 50% of HGSOC patients with intact HR activity in tumor cells can still clinical benefit from PARPi therapy [14]. This observation suggests the existence of additional pathways, beyond DNA damage repair, that contribute to resistance against PARPi treatment.
Prolonged PARPi administration induces inherent DNA damage within tumors, leading to an accumulation of DNA errors and the release of DNA into the cytoplasm. Cyclic GMP-AMP synthase (cGAS) is an intracellular DNA sensor [15]. Initially, cGAS was found to be located in the cytoplasm, where it directly binds to cytoplasmic double-stranded DNA (dsDNA). DNA binding promotes cGAS liquid-phase dissociation and aggregation, activating cGAS to produce cGAMP [16]. Subsequently, cGAMP is detected by a positioning adapter protein on the endoplasmic reticulum (ER), called stimulator of interferon genes (STING) [17]. Upon recognizing cGAMP, STING translocates to the Golgi apparatus and recruits TANK-binding kinase 1 (TBK1) and the transcription factor IRF3 to promote their phosphorylation and activation [18]. Furthermore, the STING binding to IκB kinase can stimulate the transcriptional activity of NF-κB and proinflammatory signals, promoting the production of cytokines, including IFN-α and IFN-β [19]. cGAS can also be located in the nucleus, where its activity is inhibited by chromatin anchoring and cell cycle-dependent phosphorylation, potentially promoting DNA damage [20,21]. The cGAS-STING pathway plays a crucial role in the innate immune response and cancer immunity; however, its role in ovarian cancer and PARPi resistance remains unexplored.
In this study, we elucidated the key role of cGAS in mediating Olaparib resistance in ovarian cancer cells. Our findings demonstrate a significant increase in both cGAS expression and inflammatory response gene expression in Olaparib-resistant ovarian cancer cells. Elevated cGAS further promotes tumor cell growth by stimulating interferon expression and activating STAT1 signaling, thus contributing to Olaparib resistance. Our findings suggest that combined inhibition of the cGAS-STING signaling pathway and Olaparib treatment may overcome PARPi resistance and enhance antitumor efficacy.
Materials and methods
Cell culture
Human OVCAR-8 cells and SK-OV-3 were purchased from the Wuhan Pricella Biotechnology, Wuhan, China. OVCAR-8 cells were cultured in RPMI-1640 (Biological Industries) supplemented with 10% FBS (Biological Industries), 100 U/ml penicillin, and 100 μg/ml streptomycin. SK-OV-3 cells were cultured in McCoy’s 5A (Biological Industries) supplemented with 10% FBS (Biological Industries), 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured at 37°C in a humid atmosphere (5% CO2-95% air).
Immunoblot analysis
The cells were rinsed with chilled PBS and cell lysates were produced using SDS lysis buffer (Beyotime) that included a mixture of protease and phosphatase inhibitors. The quantification of proteins was performed using the BCA protein assay kit from Beyotime. All samples underwent normalization of total protein concentrations. Subsequently, the proteins were subjected to a temperature of 95°C for a duration of 5 minutes in the presence of the 5× SDS loading buffer (Beyotime). Gel electrophoresis using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was employed to segregate protein samples. Subsequently, the proteins were deposited onto a nitrocellulose (NC) membrane (PALL) and then blocked using QuickBlock Western Blocking Buffer (Beyotime). Subsequently, the membranes were subjected to an overnight incubation at 4°C with primary antibodies. The membranes were rinsed three times with TBST and then exposed to a peroxidase-labeled secondary antibody for 1 hour at room temperature. The Pico PLUS Chemiluminescent Substrate reagent (Thermo Scientific) was employed to visualize protein bands following the second round of wash. Antibodies for GAPDH (60004-1-Ig) and TBK1 (28397-1-AP) were purchased from Proteintech. Antibodies for p-TBK1 (#54382), IRF3 (#4302), p-IRF3 (#29047), p-P65 NF-κB (#3033), p-STAT3 (#9145) were purchased from Cell Signaling Technology. Peroxidase-labeled Anti-Mouse IgG (H+L) (No. 5220-0338) and peroxidase-labeled Anti-Rabbit IgG (H+L) (No. 5220-0335) were purchased from Seracare.
Cell viability assay
The cells were distributed onto a 96-well plate with a concentration of 5000 cells per well. The cells were subjected to drug treatment for a duration of 24 hours, followed by treatment with 10 μl of Cell Counting Kit-8 solution (Beyotime). Following a 1-hour incubation period at a temperature of 37°C, the absorbance was quantified at a wavelength of 450 nm using a microplate reader (Biotek). Olaparib was administered for drug therapy at dosages of 1.25 μM, 2.5 μM, 5 μM, and 10 μM. Olaparib (S1060), and RU.521 (S6841) were purchased from Selleck.
Cytokine detection
The supernatants of cultured cells treated with Olaparib were collected. The IFN-β concentrations in the supernatants were analysed using a AuthentiKine Human IFN-beta ELISA Kit (Proteintech, KE00195) according to the manufacturer’s instructions. The IL-6 concentrations in the supernatants were analysed using an AuthentiKine Human IL-6 ELISA Kit (Proteintech, KE00139). Signals were obtained using a microplate reader.
Plasmidand transfection and quantitative PCR (qPCR) assays
The human genes MB21D1 and TMEM173 were inserted into the pcDNA3.1 expression vector and introduced into cells by transfection using jetPrime Transfection Reagent (Polyplus). The synthesis of cDNA for quantitative PCR (qPCR) experiments was performed using the ReverTra Ace qPCR RT Kit (TOYOBO). The qPCR analysis was conducted using SYBR Green and the ABI Q5 System (Thermo Fisher). The mRNA levels of desired genes were standardized by comparing them to the amount of β-actin (ACTB). The following are the primers utilized for amplifying the target genes: ACTB-F: 5’-CATGTACGTTGCTATCCAGGC-3’, ACTB-R: 5’-CTCCTTAATGTCACGCACGAT-3’; IFNB1-F: 5’-ATGACCAACAAGTGTCTCCTCC-3’, IFNB1-R: 5’-GGAATCCAAGCAAGTTGTAGCTC-3’; CXCL10-F: 5’-GTGGCATTCAAGGAGTACCTC-3’, CXCL10-R: 5’-TGATGGCCTTCGATTCTGGATT-3’; IL6-F: 5’-GGATTCAATGAGGAGACTTGC-3’, IL6-R: 5’-GTTGGGTCAGGGGTGGTTAT-3’.
Bioinformation analysis
The data for the TCGA cohort of Ovarian cancer were obtained by downloading them from the website https://xenabrowser.net/. The RNA-sequencing (RNA-seq) data for Olaparib-resistant and parental cells were acquired from GSE153867 and GSE117765. Gene expression across samples was assessed using transcripts per million (TPM).
Statistical analysis
The data were reported as means ± standard error of the mean (SEM) and were analyzed using GraphPad Prism software version 8.0 (GraphPad). The Student’s t-test was employed to compare the two groups, while a one-way ANOVA with Dunnett’s multiple comparisons test was utilized to compare several groups. Each experiment was conducted a minimum of three times, and statistical significance was indicated by P < 0.05 (*P < 0.05), P < 0.01 (**P < 0.01), while non-significance was designated by P > 0.05 (ns) in the figures.
Results
Upregulated cGAS contributes to Olaparib resistance
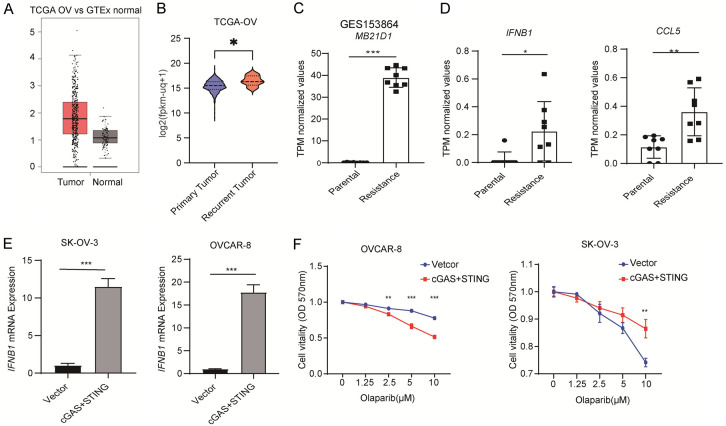
At first, we observed the presence of cGAS in ovarian cancer and observed that the level of cGAS expression in the tumor tissues of ovarian cancer patients was considerably higher compared to normal ovarian tissues (Figure 1A). Moreover, cGAS expression was significantly higher in recurrent ovarian tumors compared to primary tumors, suggesting a potential role for cGAS in mediating drug resistance in recurrent ovarian cancer (Figure 1B). Furthermore, cGAS was significantly elevated in PARPi drug-resistant cells compared to parental cells (Figure 1C). As cGAS plays a key role in the production of type I interferons, we observed significantly higher expression of IFNB1 and the expression of the interferon-stimulated gene CCL5 in PARPi-resistant cells compared to parental cells (Figure 1D). To further investigate the role of cGAS in PARPi resistance, both cGAS and its downstream effector protein STING were overexpressed in SK-OV-3 and OVCAR-8 cells. This overexpression resulted in a significant upregulation of type I interferons in ovarian cancer cells (Figure 1E). Furthermore, high expression of cGAS and STING promoted Olaparib resistance in ovarian cancer cells (Figure 1F). These findings suggest that cGAS and its downstream signaling pathways may contribute to ovarian cancer tolerance to PARPi.
Figure 1.
High expression of cGAS promotes Olaparib resistance in Ovarian cancer cells. A, B. RNA-seq violin plot exhibiting the expression of cGAS in primary and recurrent tumors from ovarian cancer patients. The RNA-seq data was collected from Santa Cruz Xena platform using the TCGA datasets. C. Transcripts per million (TPM) values of MB21D1 genes in parental and Olaparib-resistant cells were shown. D. Transcripts per million (TPM) values of IFNB1 and CCL5 genes in parental and Olaparib-resistant cells were shown. E. qRT-PCR analysis of IFNB1 expression in SK-OV-3 (left) and OVCAR-8 (right) cells transfected with cGAS and STING plasmid (n=3). F. CCK-8 assay is used to detect the sensitivity of SK-OV-3 (left) and OVCAR-8 (right) cells overexpressing cGAS and STING. C-E. Data are the mean ± SD; Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001 and nsP > 0.05. F. Data are the mean ± SD; one-way ANOVA with Dunnett’s multiple comparisons test; **P < 0.002, ***P < 0.001.
Olaparib treatment promotes TBK1-IRF3 signaling activation in ovarian cancer cells
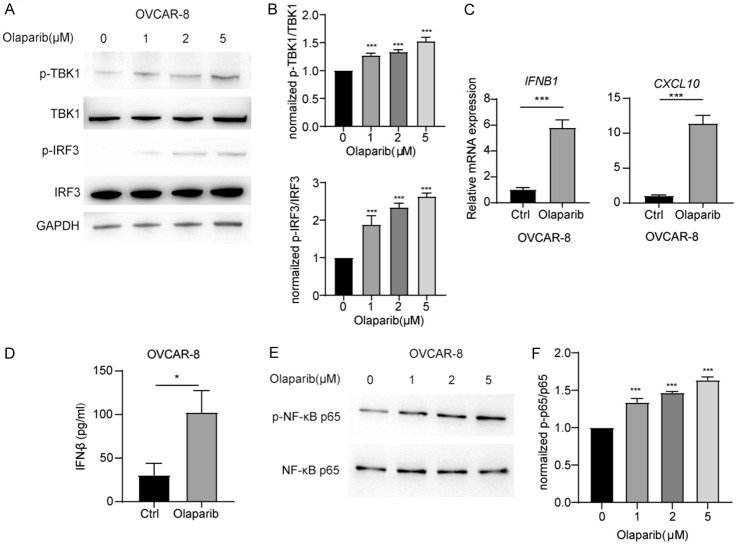
cGAS-STING signaling plays a crucial role in triggering innate immunity by activating the TBK1-IRF3 signaling axis, which promotes the expression of inflammatory cytokines. Elevated levels of these cytokines are a significant factor contributing to drug resistance in ovarian cancer. Previous findings demonstrated a significant increase in cGAS in PARPi-resistant cells. To investigate whether PARPi activates innate immune signals and contributes to PARPi tolerance through feedback in ovarian cancer cells, we conducted further studies. Treatment of OVCAR-8 cells with Olaparib resulted in a dose-dependent increase in the levels of phosphorylated TBK1 and phosphorylated IRF3 (Figure 2A and 2B). These findings suggest that Olaparib induces the activation of IRF3 signaling. As IRF3 is a key transcription factor that promotes the production of type I interferons, Olaparib treatment of OVCAR-8 cells led to a significant increase in IFNB1 expression and substantial rise in the secretion of IFN-β (Figure 2C and 2D). Furthermore, the interferon-stimulated gene CXCL10 was also upregulated (Figure 2C). These findings demonstrate that Olaparib activates the TBK1-IRF3 signaling pathway, leading to increased type I interferon production in ovarian cancer cells.
Figure 2.
Olaparib treatment promotes TBK1-IRF3 signaling activation in ovarian cancer cells. A. Immunoblot analysis of total and p-TBK1, total and p-IRF3 in OVCAR-8 cells, followed by treatment Olaparib. B. Densitometric analysis of p-TBK1 and p-IRF3 protein expression levels was quantitated by “ImageJ” software. C. qRT-PCR analysis of IFNB1 and CXCL10 mRNA in OVCAR-8 cells treated with Olaparib. D. ELISA quantification of IFN-β secretion in OVCAR-8 cells treated with Olaparib. E. Immunoblot analysis of total and p-NF-κB-p65 in OVCAR-8 cells, followed by treatment Olaparib. F. Densitometric analysis of p-NF-κB-p65 protein expression levels was quantitated by “ImageJ” software. Data are the mean ± SD. C and D. Data are the mean ± SD; Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001 and nsP > 0.05. B and F. One-way ANOVA with Dunnett’s multiple comparisons test; *Data are the mean ± SD; P < 0.033, ***P < 0.001 and nsP > 0.033.
Olaparib triggers the IL-6-STAT3 pathway
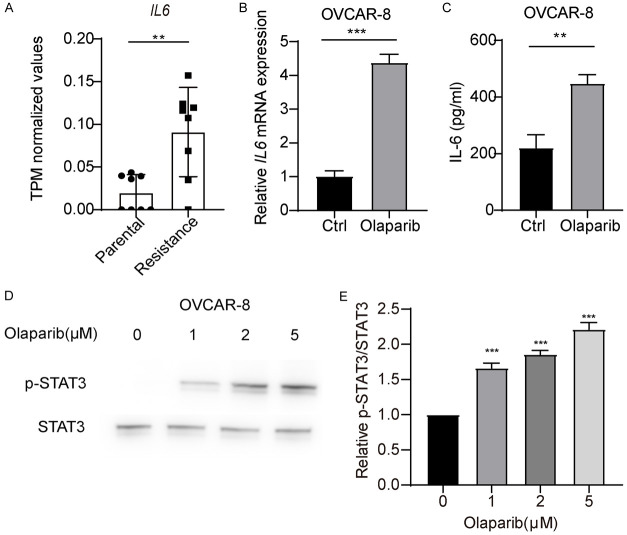
Inflammation is a key mechanism of ovarian cancer cell resistance to PRAPi, and NF-κB, a core transcription factor involved in inflammation, is a critical downstream signaling molecule of type I interferons. Therefore, we further studied whether Olaparib promotes activation of NF-κB in ovarian cancer cells. We found that treatment of OVCAR-8 cells with Olaparib resulted in a significant increase the phosphorylation level of NF-κB p65 (Figure 2E and 2F). These findings suggest that Olaparib promotes the activation of NF-κB. IL-6, a key inflammatory cytokine downstream of NF-κB, can promote tumor proliferation and drug resistance. RNA-seq data revealed significantly higher levels of IL6 expression in PARPi drug-resistant cells compared to parental cells (Figure 3A). We further detected whether Olaparib could promote IL6 expression. The results showed that treatment of OVCAR-8 cells with Olaparib resulted in a significant increase in IL-6 transcription and protein secretion (Figure 3B and 3C). Furthermore, the phosphorylation of downstream signaling molecule STAT3, which is activated by IL-6, was significantly increased (Figure 3D and 3E). These data demonstrated that Olaparib promotes IL-6 expression, providing a feedback mechanism in ovarian cancer cells.
Figure 3.
Olaparib promotes activation of IL-6-STAT3 pathway in ovarian cancer cells. A. Transcripts per million (TPM) values of IL6 gene in parental and Olaparib-resistant cells. B. qRT-PCR analysis of IL6 mRNA in OVCAR-8 cells treated with Olaparib. C. ELISA quantification of IL-6 secretion in OVCAR-8 cells treated with Olaparib. D. Immunoblot analysis of total and p-STAT3 OVCAR-8 cells, followed by treatment with Olaparib. E. Densitometric analysis of p-STAT3 protein expression levels was quantitated by “ImageJ” software. Data are the mean ± SD. A-C. Student’s t-test, **P < 0.01 and ***P < 0.001. E. One-way ANOVA with Dunnett’s multiple comparisons test; **P < 0.002.
cGAS inhibition synergizes with Olaparib to suppress tumor cell proliferation
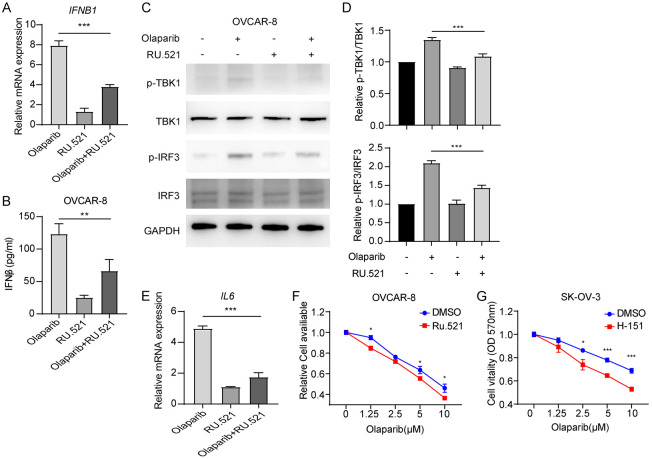
Our previous findings demonstrated that Olaparib activates cGAS-STING signaling, leading to promote the expression of type I interferon signaling and IL-6, which subsequently mediate feedback resistance to Olaparib in ovarian cancer cells. To investigate whether inhibiting cGAS could enhance the sensitivity of ovarian cancer cells to Olaparib, we conducted further studies. We first treated OVCAR-8 cells with the cGAS inhibitor Ru.521 and Olaparib. Ru.521 significantly inhibited Olaparib-induced IFNB1 expression and IFN-β secretion (Figure 4A and 4B). Ru.521 rescued Olaparib-induced phosphorylation of TBK1 and IRF3 in OVCAR-8 cells (Figure 4C and 4D). Furthermore, Ru.521 inhibited the Olaparib-induced increases in IL6 expression (Figure 4E). Our findings demonstrate that Ru.521 enhanced the sensitivity of the ovarian cancer cell lines OVCAR-8 and SK-OV-3 to Olaparib (Figure 4F and 4G). These findings suggest that Olaparib promotes the expression of inflammatory factors in ovarian cancer cells, and that Olaparib tolerance is dependent on cGAS.
Figure 4.
Combination of PARPi with cGAS inhibition exhibits synergetic effects to overcome PARPi resistance. A. qRT-PCR analysis of IFNB1 and CXCL10 mRNA in OVCAR-8 cells treated with Olaparib or/and cGAS inhibitor RU.521. B. ELISA quantification of IFN-β secretion in OVCAR-8 cells treated with Olaparib or/and RU.521. C. Immunoblot analysis of total and p-TBK1, total and p-IRF3 in OVCAR-8 cells, followed by treatment Olaparib or/and RU.521. D. Densitometric analysis of p-TBK1 and p-IRF3 protein expression levels was quantitated by “ImageJ” software. E. qRT-PCR analysis of IL6 mRNA in OVCAR-8 cells treated with Olaparib or/and cGAS inhibitor RU.521. F. CCK-8 detection of Olaparib sensitivity in OVCAR-8 or SK-OV-3 cells treated with DMSO or cGAS inhibitor RU.521. A, B, D-G. Data are the mean ± SD; one-way ANOVA with Dunnett’s multiple comparisons test; *P < 0.033, **P < 0.002 and ***P < 0.001.
Discussion
Olaparib, the first FDA-approved PARP inhibitor, is widely used to treat ovarian cancer. However, the development of drug resistance and subsequent relapse are common occurrences after PARPi treatment, including with Olaparib. Understanding the mechanisms underlying Olaparib resistance in ovarian cancer cells is crucial. In this study, we elucidate the key role of cGAS in mediating resistance to Olaparib. Our findings demonstrate that Olaparib treatment induces upregulation of cGAS expression in ovarian cancer cells. Upregulated cGAS activates STAT1 signaling through the induction of type I interferons and promotes the downstream expression of IL-6 to contributing to cell growth, and migration, thereby leading to cancer cell resistance to Olaparib. Therefore, inhibiting of cGAS signaling could synergize with Olaparib to suppress tumor growth, offering potential for the developing of novel anticancer therapies.
Prior research has primarily focused on investigating alternative strategies of DNA damage repair when studying the mechanisms of PARPi resistance. However, PARPis have also shown effectiveness in people who have an intact ability to repair DNA damage [22]. Furthermore, prolonged PARP inhibition in HR-proficient tumor cells can induce DNA damage, leading to the activation of the type I interferon response and inflammation associated DNA recognition. Therefore, beyond the natural repair of DNA damage, alternative mechanisms of resistance to PARP inhibitors may exist, potentially linked to the generation of new DNA damage and subsequent inflammation [23]. Our findings highlight a significant increase in the inflammatory response in Olaparib-resistant cells following prolonged treatment with PARPi. This increase can be attributed to substantial upregulation of the innate immune adaptor molecule cGAS in Olaparib-resistant cells. Furthermore, Olaparib further stimulated the expression of cGAS. During tumor treatment, the accumulation of DNA in the plasma of cells can trigger the activation of cGAS, resulting in an increase in inflammatory levels [24].
The activation of cGAS-STING in cancer cells induced by targeted or conventional chemotherapeutic drugs, enables these cells to counter drug challenge through adaptive and autonomous responses. Drug pressure directly or indirectly damages DNA, leading to its leakage into the cytoplasm. DNA accumulated in the cytoplasm is sensed and triggers the cGAS-STING signaling pathway. Cancer cells utilize this pathway to resist drug stress and acquire resistance to these drugs through the downstream TBK1-IRF3/NF-κB signaling pathway [25]. Our analysis revealed significantly increased cGAS expression in tumor tissues from patients with ovarian cancer, correlating with tumor recurrence. Furthermore, our findings show that Olaparib activates the downstream signaling molecules TBK1 and IRF3, leading to the production of type I interferons. Type I interferons are potent inflammatory cytokines with a wide range of effects. NF-κB signaling is a crucial downstream signaling pathway. Our findings demonstrate that Olaparib enhances the production of IL-6, leading to the activation of STAT3. The cGAS-type I IFN-IL-6-STAT3 signaling plays a crucial role in mediating Olaparib resistance. Inhibiting this pathway may offer advantages for enhancing the anticancer activity of Olaparib. In our study, the cGAS inhibitor Ru.521 demonstrated a stronger antitumor effect in combination with Olaparib.
In conclusion, our study highlights the critical role of cGAS and type I interferon signaling in mediating Olaparib-resistant ovarian cancer cells through the activation of inflammatory signaling pathways. Our findings emphasize the importance of targeting cGAS to overcome Olaparib resistance. This finding holds significance for better understanding cGAS-STING function and cancer treatment. The clinical development of cGAS and STING inhibitors suggests that combining these agents with PARP inhibitors could potentially enhance the efficacy of ovarian cancer treatment.
Limitations
This study has a few limitations. We explored the role of the cGAS pathway in mediating PARPi drug resistance. cGAS is a critical intrinsic immune signaling molecule, and its activation typically promotes anti-tumor immunity. Consequently, cGAS activation may exhibit a dual role, promoting immune cell infiltration while simultaneously contributing to drug resistance through chronic inflammation. A significant limitation of this study lies in its focus on intrinsic immune-inflammatory factors generated by cGAS activation in promoting ovarian cancer cell survival. The study did not investigate the potential contribution of acquired immunity to PARPi resistance. Further research is warranted to elucidate the role of cGAS in PARPi treatment and its interplay with the immune microenvironment. Furthermore, this study primarily investigated the role of the cGAS pathway in mediating PARPi drug resistance at the cellular level. It lacked in vivo experimental validation. Future research should prioritize validating these findings in vivo and in clinical settings.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. doi: 10.3322/caac.21820. [DOI] [PubMed] [Google Scholar]
- 2.Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371:m3773. doi: 10.1136/bmj.m3773. [DOI] [PubMed] [Google Scholar]
- 3.Hirschl N, Leveque W, Granitto J, Sammarco V, Fontillas M, Penson RT. PARP inhibitors: strategic use and optimal management in ovarian cancer. Cancers (Basel) 2024;16:932. doi: 10.3390/cancers16050932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 5.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 6.Ramakrishnan N, Weaver TM, Aubuchon LN, Woldegerima A, Just T, Song K, Vindigni A, Freudenthal BD, Verma P. Nucleolytic processing of abasic sites underlies PARP inhibitor hypersensitivity in ALC1-deficient BRCA mutant cancer cells. Nat Commun. 2024;15:6343. doi: 10.1038/s41467-024-50673-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi T, Feng Y, Sundaram R, Tie Y, Zheng H, Qian Y, You D, Yi T, Wang P, Zhao X. Antitumor efficacy of PARP inhibitors in homologous recombination deficient carcinomas. Int J Cancer. 2019;145:1209–1220. doi: 10.1002/ijc.32143. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Martin A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, McCormick C, Lorusso D, Hoskins P, Freyer G, Baumann K, Jardon K, Redondo A, Moore RG, Vulsteke C, O’Cearbhaill RE, Lund B, Backes F, Barretina-Ginesta P, Haggerty AF, Rubio-Perez MJ, Shahin MS, Mangili G, Bradley WH, Bruchim I, Sun K, Malinowska IA, Li Y, Gupta D, Monk BJ PRIMA/ENGOT-OV26/GOG-3012 Investigators. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 9.Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, Fujiwara K, Vergote I, Colombo N, Maenpaa J, Selle F, Sehouli J, Lorusso D, Guerra Alia EM, Reinthaller A, Nagao S, Lefeuvre-Plesse C, Canzler U, Scambia G, Lortholary A, Marme F, Combe P, de Gregorio N, Rodrigues M, Buderath P, Dubot C, Burges A, You B, Pujade-Lauraine E, Harter P PAOLA-1 Investigators. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 10.Ter Brugge P, Kristel P, van der Burg E, Boon U, de Maaker M, Lips E, Mulder L, de Ruiter J, Moutinho C, Gevensleben H, Marangoni E, Majewski I, Jozwiak K, Kloosterman W, van Roosmalen M, Duran K, Hogervorst F, Turner N, Esteller M, Cuppen E, Wesseling J, Jonkers J. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djw148. [DOI] [PubMed] [Google Scholar]
- 11.Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI, Kuiper MJ, Ho GY, Barker H, Jasin M, Prakash R, Kass EM, Sullivan MR, Brunette GJ, Bernstein KA, Coleman RL, Floquet A, Friedlander M, Kichenadasse G, O’Malley DM, Oza A, Sun J, Robillard L, Maloney L, Bowtell D, Giordano H, Wakefield MJ, Kaufmann SH, Simmons AD, Harding TC, Raponi M, McNeish IA, Swisher EM, Lin KK, Scott CL AOCS Study Group. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7:984–998. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker JR, Cuella-Martin R, Barazas M, Liu R, Oliveira C, Oliver AW, Bilham K, Holt AB, Blackford AN, Heierhorst J, Jonkers J, Rottenberg S, Chapman JR. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat Commun. 2018;9:5406. doi: 10.1038/s41467-018-07855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, Zhou Y, Pan P, Yu H, Wang Z, Li LL, Wang B, Yan Y, Pan Y, Ye Q, Liu T, Feng X, Xu S, Wang K, Wang X, Jian Y, Ma B, Fan Y, Gao Y, Huang H, Li L. TRABID overexpression enables synthetic lethality to PARP inhibitor via prolonging 53BP1 retention at double-strand breaks. Nat Commun. 2023;14:1810. doi: 10.1038/s41467-023-37499-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Duval AJ, Adli M, Matei D. Biology-driven therapy advances in high-grade serous ovarian cancer. J Clin Invest. 2024;134:e174013. doi: 10.1172/JCI174013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaauboer SM, Gabrielle VD, Jin L. MPYS/STING-mediated TNF-alpha, not type I IFN, is essential for the mucosal adjuvant activity of (3’-5’)-cyclic-di-guanosine-monophosphate in vivo. J Immunol. 2014;192:492–502. doi: 10.4049/jimmunol.1301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R, Jungblut P, Pei G, Dorhoi A, Yan Q, Zhang F, Zheng R, Liu S, Liang H, Liu Z, Yang H, Chen J, Wang P, Tang T, Peng W, Hu Z, Xu Z, Huang X, Wang J, Li H, Zhou Y, Liu F, Yan D, Kaufmann SHE, Chen C, Mao Z, Ge B. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 21.Fang L, Hao Y, Yu H, Gu X, Peng Q, Zhuo H, Li Y, Liu Z, Wang J, Chen Y, Zhang J, Tian H, Gao Y, Gao R, Teng H, Shan Z, Zhu J, Li Z, Liu Y, Zhang Y, Yu F, Lin Z, Hao Y, Ge X, Yuan J, Hu HG, Ma Y, Qin HL, Wang P. Methionine restriction promotes cGAS activation and chromatin untethering through demethylation to enhance antitumor immunity. Cancer Cell. 2023;41:1118–1133. e1112. doi: 10.1016/j.ccell.2023.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Richardson DL, Quintanilha JCF, Danziger N, Li G, Sokol E, Schrock AB, Ebot E, Bhardwaj N, Norris T, Afghahi A, Frachioni A, Washington C, Dockery L, Elvin J, Graf RP, Moore KN. Effectiveness of PARP inhibitor maintenance therapy in ovarian cancer by BRCA1/2 and a scar-based HRD signature in real-world practice. Clin Cancer Res. 2024;30:4644–4653. doi: 10.1158/1078-0432.CCR-24-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi L, Huan L, Zhang C, Wang H, Lu J. Adenosine receptor A2b confers ovarian cancer survival and PARP inhibitor resistance through IL-6-STAT3 signalling. J Cell Mol Med. 2023;27:2150–2164. doi: 10.1111/jcmm.17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Zhang S, Han J, Nie X, Qi Y, Han Y, Chen X, He C. Activation of STING pathway contributed to cisplatin-induced cardiac dysfunction via promoting the activation of TNF-alpha-AP-1 signal pathway. Front Pharmacol. 2021;12:711238. doi: 10.3389/fphar.2021.711238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv QM, Lei HM, Wang SY, Zhang KR, Tang YB, Shen Y, Lu LM, Chen HZ, Zhu L. Cancer cell-autonomous cGAS-STING response confers drug resistance. Cell Chem Biol. 2023;30:591–605. e594. doi: 10.1016/j.chembiol.2023.05.005. [DOI] [PubMed] [Google Scholar]