Abstract
We have determined the nucleotide sequence of a secondary phage lambda attachment site (att) located between the structural genes of the ribitol and D-arabitol catabolic operons of Klebsiella aerogenes. The core region of this secondary attachment site (sequence: GGTTTTTTCGATTAT) shows considerable homology with the 15-base-pair core region common to both the phage att and the primary bacterial att of Escherichia coli K12 (sequence: GCTTTTTTACTAA); however, there is no such clear homology between the sequences flanking the cores of the primary att and this secondary att. Integration of phage lambda into the K. aerogenes secondary att occurred by recombination between the core region of the phage att and an oligo(T.A) stretch located within the K. aerogenes secondary att.
Full text
PDF


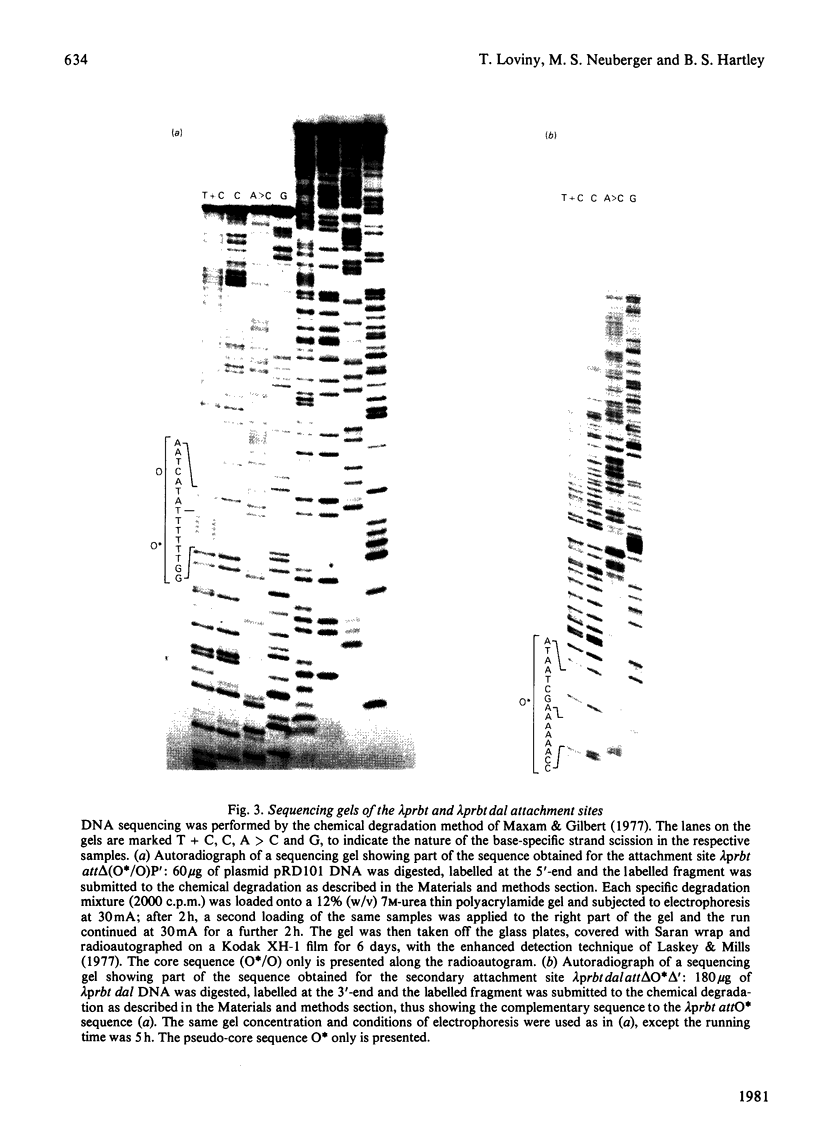



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bidwell K., Landy A. Structural features of lambda site-specific recombination at a secondary att site in galT. Cell. 1979 Feb;16(2):397–406. doi: 10.1016/0092-8674(79)90015-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Christie G. E., Platt T. A secondary attachment site for bacteriophage lambda in trpC of E. coli. Cell. 1979 Feb;16(2):407–413. doi: 10.1016/0092-8674(79)90016-3. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás-Tóth E., Boros I., Venetianer P. Nucleotide sequence of a secondary attachment site for bacteriophage lambda on the Escherichia coli chromosome. Nucleic Acids Res. 1979 Nov 10;7(5):1335–1341. doi: 10.1093/nar/7.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Schreier P. H., Buchel D. E. Nucleotide sequence of the attachment site of coliphage lambda. Nature. 1977 Dec 22;270(5639):757–760. doi: 10.1038/270757a0. [DOI] [PubMed] [Google Scholar]
- Galibert F., Sedat J., Ziff E. Direct determination of DNA nucleotide sequences: structure of a fragment of bacteriophage phiX172 DNA. J Mol Biol. 1974 Aug 15;87(3):377–407. doi: 10.1016/0022-2836(74)90093-x. [DOI] [PubMed] [Google Scholar]
- Guerrini F. On the asymmetry of lambda integration sites. J Mol Biol. 1969 Dec 28;46(3):523–542. doi: 10.1016/0022-2836(69)90194-6. [DOI] [PubMed] [Google Scholar]
- Haggerty D. M., Scheif R. F. Location in bacteriophage lamdba DNA of cleavage sites of the site-specific endonuclease from Bacillus amyloliquefaciens H. J Virol. 1976 May;18(2):659–663. doi: 10.1128/jvi.18.2.659-663.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds L., O'Malley B. W., Nisbet A. D., Fothergill J. E., Givol D., Fields S., Robertson M., Brownlee G. G. Sequence of chicken ovalbumin mRNA. Nature. 1978 Jun 29;273(5665):723–728. doi: 10.1038/273723a0. [DOI] [PubMed] [Google Scholar]
- Neuberger M. S., Hartley B. S. Investigations into the Klebsiella aerogenes pentitol operons using specialised transducing phages lambdaprbt and lambdaprbt dal. J Mol Biol. 1979 Aug 15;132(3):435–470. doi: 10.1016/0022-2836(79)90269-9. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Gething M. J., Hartley B. S. Construction of intergeneric hybrids using bacteriophage P1CM: transfer of the Klebsiella aerogenes ribitol dehydrogenase gene to Escherichia coli. J Bacteriol. 1976 Feb;125(2):728–738. doi: 10.1128/jb.125.2.728-738.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schrenk W. J., Weisberg R. A. A simple method for making new transducing lines of coliphage lambda. Mol Gen Genet. 1975;137(2):101–107. doi: 10.1007/BF00341676. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. III. The components of the secondary attachment sites. J Mol Biol. 1975 Apr 25;93(4):415–429. doi: 10.1016/0022-2836(75)90237-5. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]



