Abstract
Databases of molecules and materials are indispensable for advancing chemical research, especially when enriched with electronic structure information from quantum chemistry methods like density functional theory. In this perspective, we review and analyze the current landscape of materials and molecular databases containing quantum chemical data. Our analysis reveals that the materials community has significantly benefited from data platforms such as the Materials Project, which seamlessly integrate chemical structures, electronic structure data, and open-source software. Conversely, quantum chemical data for molecular systems remains largely fragmented across individual datasets, lacking the comprehensive framework of a unified database. We distilled insights from these existing data resources into seven guiding principles termed QUANTUM, which build upon the foundational FAIR principles of data sharing (Findable, Accessible, Interoperable, and Reusable). These principles are aimed at advancing the development of molecular databases into robust, integrated data platforms. We conclude with an outlook on both short- and long-term objectives, guided by these QUANTUM principles, to foster future advancements in molecular quantum databases and enhance their utility for the research community.
This perspective reviews both materials and molecular data resources and establishes seven guiding principles termed QUANTUM to advance molecular databases toward robust, unified platforms for the research community.
1. Introduction
The dawn of the information age has profoundly transformed how research data is generated, stored, and disseminated. The advent of the World Wide Web in the late 1980s connected scientists like never before, fostering the expansion of chemical repositories such as the Cambridge Structural Database (CSD).1,2 Originally established in 1965 as a compendium of published crystallographic data, the CSD has grown significantly since its inception, now encompassing over 1.25 million curated entries. Similarly, resources like the Crystallographic Open Database (COD)3–5 repository and the PubChem6,7 database have enabled scientists to digitally catalogue and explore millions of unique molecules and materials. The emergence of data-driven platforms, notably the Materials Project,8–11 has marked a significant evolution from traditional data resources to more sophisticated, interconnected platforms.
Quantum chemical (QC) methods, developed in the early 20th century, have empowered researchers to explore and predict the electronic structures of molecules and materials. Foundational approaches such as Hartree–Fock theory and density functional theory (DFT) paved the way for deeper insights into electronic and quantum effects. More advanced methods, including post–Hartree–Fock methods and time-dependent density functional theory (TD-DFT), have enhanced the analysis of electronic excitations and complex spectroscopic properties.12,13 Additionally, computationally less expensive semi-empirical methods like xTB14–16 and PM6/PM7 17–19 have facilitated high-throughput screenings and the manipulation of chemical databases.20,21 The utility of these databases can be greatly enhanced by the integration of QC data, broadening their applicability across various fields.
However, the accuracy of QC data is inherently dependent on the method and system being modelled. For instance, hybrid functionals in DFT, such as ωB97XD,22 which include a percentage of Hartree–Fock exchange, are well-suited for reactivity studies involving systems with some electron correlation. Meanwhile, more accurate methods like coupled-cluster (CC) may be required for highly correlated systems. Additionally, the choice of basis set and the inclusion of relativistic effects are crucial considerations, particularly for systems containing heavy elements.23,24 Thus, benchmarking QC methods against reliable experimental data or higher-level QC calculations is essential for validating predictions. Nevertheless, discrepancies can still arise due to incomplete theoretical models, such as the omission of solvent effects in reaction studies.23 Furthermore, results obtained from QC calculations at different levels of theory are often not directly comparable, which highlights the need for standardized methodologies and cross-validation strategies.
Despite these challenges, recent advances underscore the potential of integrating QC data with large-scale databases. For example, users of the Materials Project have leveraged its QC data to identify efficient electrocatalysts for CO2 reduction through active learning,25 screen solid-state electrolytes for Li-ion batteries,26 and develop interatomic potentials that accurately predict material properties.27 Furthermore, specialized datasets like 2DMatpedia,28,29 a collection of 2D materials, have enabled the development of advanced workflows, such as Gerber et al.'s work on predicting the properties of material interfaces.30 Additionally, chemical data featuring electronic structure information is increasingly employed to train advanced machine learning (ML) algorithms to predict chemically relevant properties, including HOMO–LUMO gaps of molecules and semiconductor bandgaps.31,32
As chemical databases evolve, adherence to data management guidelines like the FAIR principles is becoming increasingly important.33 These principles stipulate that data should be Findable, Accessible, Interoperable, and Reusable. For chemical structure databases, this means indexing entries with unique identifiers and ensuring that data such as molecular mass and formal charge is readily retrievable. Data should also be stored in universally accessible formats such as .xyz or .mol2 for molecular structures, .csv for tabular data, or .gml for graph representations. To promote reuse in subsequent studies, it is essential that data associated with each compound is diverse and abundant, underlying the practical benefits of these principles in modern research.
In this perspective, we review and analyze state-of-the-art QC materials and molecular databases, as well as various related datasets and repositories. These accounts are not intended to provide a holistic evaluation of each database but rather a targeted analysis to learn from their respective merits and limitations. Our review focuses on materials and molecular data resources that are open access, available for download, contain electronic structure information from QC calculations, and exclude macromolecules and reactions. Additionally, while acknowledging the many challenges of implementing and maintaining software and hardware for databases, our work focuses on discussing challenges of molecular and materials databases that are directly relevant to a chemistry audience.
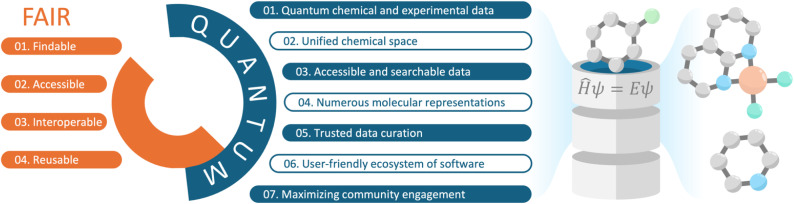
Our analysis reveals that the materials community has benefited immensely from QC databases like the Materials Project, which provides geometric structures, electronic structure data, and associated software under a unified framework. In contrast, while the molecular community relies on several important structural databases and repositories of significant value, these resources would benefit from incorporating QC data and a comprehensive ecosystem of supporting software. Consequently, we propose seven guiding principles for a central molecular QC data platform to support research in the molecular community. These principles build upon the FAIR principles of data management and are collectively referred to as QUANTUM (Fig. 1). Thus, our work discusses key questions for the future development of molecular databases from a chemist's point of view.
Fig. 1. Graphical summary of the proposed QUANTUM principles. The FAIR principles set the standard for scientific data management and sharing (left). We expand upon FAIR to include the QUANTUM principles (centre), which outline seven design guidelines for developing a QC platform for molecular systems (right).
2. Datasets, repositories and databases
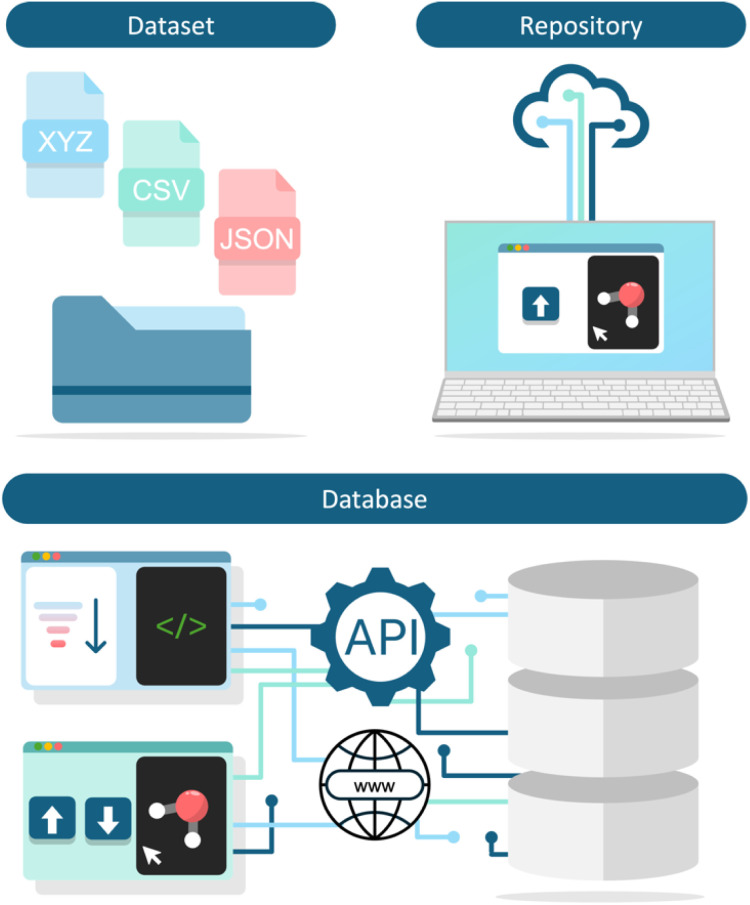
For the purpose of this review, we categorize data resources into three primary groups: datasets, repositories, and databases (Fig. 2). It is important to note that these categories can sometimes overlap.
Fig. 2. Overview of different categories of data resources. Datasets comprise data typically formatted as individual .xyz, .csv or .json files, repositories facilitate the online upload and cataloguing of data, and databases allow users to access entries via online web interfaces and support advanced querying and connectivity via an API.
Datasets are collections of data typically generated and presented by a single set of authors in a publication resulting from a specific research project. Datasets are often formatted as .csv files for tabular data or .json files for more complex data structures, and are commonly uploaded to online portals like Figshare34 or GitHub.35 Due to their specific nature, new datasets emerge frequently, reflecting ongoing advancements in research. In this review, we highlight a selection of notable materials and molecular datasets to illustrate their diversity and utility.
Repositories allow users to upload material and molecular structure information to an online portal, sharing their results with the broader scientific community. Entries in repositories are typically indexed with a unique identifier, which aids in ensuring traceability and reproducibility in scientific research. Each entry in a repository usually represents one user submission, not one molecule. For instance, ioChem-BD is a web-based repository for chemical structures derived from QC calculations and has many entries where the same chemical structure was calculated with different QC methods.36,37 While repositories can offer advanced features similar to those found in databases, the wide variety of user submissions can lead to less consistent entries. Another subtype of repository, referred to as dataset repository, does not contain individual molecules or materials but discrete datasets uploaded by various users, for example the Computational Materials Repository.38,39
Databases generally differ from datasets and repositories by providing enhanced functionalities that facilitate searching, filtering, and querying entries through user-friendly interfaces (e.g. websites), while also being curated and regularly updated. In contrast to repositories, entries in a database usually represent one chemical structure and all data connected to the structure is contained in one entry, such as in the PubChem Compounds database. Databases typically support an application programming interface (API), allowing integration with programming languages such as Python, which fosters a robust ecosystem of software and functionalities for data manipulation and processing. For example, the Materials Project can be easily accessed via the Materials Project API.40 By augmenting data with systems that adhere to the FAIR principles (Findable, Accessible, Interoperable, Reusable), databases significantly increase the impact and utility of their data for the research community. However, developing and maintaining a comprehensive database is often more challenging than creating standalone datasets due to the need for continuous curation and enhancement. In addition to the term database, we will occasionally use the term platform to emphasize a particularly extensive and well-developed database which contains many different functionalities.
3. Materials data resources
In Table 1, we summarize four major general computational materials databases: AFLOW,41–43 OQMD,44–46 the Materials Project, and JARVIS-DFT.47–49 These databases are centralized, housing large amounts of internally curated data computed predominantly using consistent DFT methods to increase comparability between different entries.
Material databases, datasets, repositories, and dataset repositories that contain QC data. The ‘Size’ column indicates the number of entries in each data resource. The ‘Source’ column specifies the origin of the structures.
| Name | Size | Method | Source | Content |
|---|---|---|---|---|
| Material databases | ||||
| AFLOW41–43 | 3.5M | DFT | ICSD, Pauling File, prototypes | Inorganic bulk materials |
| OQMD44–46 | 1.2M | DFT | ICSD & prototypes | Inorganic bulk materials |
| Materials Project8–11 | 1.0M | DFT | ICSD & others | 153k bulk materials (main data), and 222k organic molecules, 4k battery materials, 25k battery electrolytes, 20k MOFs, 560k catalyst surfaces, and 41k synthesis recipes |
| JARVIS-DFT47–49 | 76k | DFT | MP, ICSD, AFLOW, OQMD, COD | 3D, 2D, 1D and 0D materials at varying levels of DFT theory |
| Organic Materials DB50,51 | 41k | DFT | COD | Organic and organometallic materials |
| Material datasets | ||||
| OC20 52 | 1.3M | DFT | MP | Surfaces with N,C,O-containing adsorbates |
| ARC-MOF53 | 280k | DFT | Multiple papers | MOFs |
| InterMatch30 | 199k | DFT | MP | Interfaces of materials |
| Schmidt et al.54 | 175k | DFT | MP & others | Chemically diverse bulks |
| Bare et al.55 | 67k | DFT | ABO3 prototype | ABO3 perovskite bulks |
| OC22 56 | 62k | DFT | MP | Surfaces of oxide materials, coverages, and adsorbates |
| QMOF57 | 20k | DFT | CSD | MOFs |
| ECD-cubic58 | 17k | DFT | MP | Cubic bulks |
| 2DMatpedia28,29 | 6.4k | DFT | MP | 2D materials |
| Emery & Wolverton59 | 5.3k | DFT | ABO3 prototype | ABO3 perovskite bulks |
| C2DB60–62 | 4.0k | DFT | Prototypes | 2D materials |
| C1DB63 | 820 | DFT | ICSD, COD & prototypes | 1D materials |
| Choudhary et al.64 | 430 | DFT | MP | 2D materials |
| CURATED COFs65 | 308 | DFT | Materials Cloud | COFs |
| Material repositories | ||||
| NOMAD66–69 | 12M | DFT & others | Submissions, MP, OQMD, AFLOW, and others | 9M bulk crystals, 75k surfaces; 5k 2D, 33k 1D materials, 2.8M organic and inorganic molecules |
| ioChem-BD36,37 | 356k | DFT | Submissions | 38k materials and 318k molecules, chemically diverse |
| Catalysis-Hub70,71 | 132k | DFT | Submissions | Structures, reaction energies, and barriers for surface reactions, including various tools |
| Material dataset repositories | ||||
| Materials Data Facility72–74 | >650 sets | Mixed | Mixed | Datasets from publications |
| MPContribs75 | 45 sets | Mixed | Mixed | Community contributions to MP |
| Computational Materials Repository38,39 | 31 sets | Mixed | Mixed | Datasets from publications |
| Materials Cloud76,77 | 17 sets | Mixed | Mixed | Datasets from publications |
| MatBench78,79 | 13 sets | Mixed | Mixed | Datasets for benchmarking ML algorithms, hosted by MP |
The AFLOW and OQMD stand out for their significantly large sizes, with 3.5M and 1.2M structures, respectively. Many of these are derived from the ICSD,80,81 a commercial database containing 299k inorganic crystal structures. AFLOW and OQMD further expand their collections by incorporating hypothetical materials, generated by substituting elements in existing structural prototypes, thus extending beyond experimentally confirmed structures.
The JARVIS-DFT database, with 76k structures, distinguishes itself with a diverse range of 3D, 2D, 1D, and 0D materials. This diversity makes it a versatile resource for a broad spectrum of research needs. Moreover, JARVIS-DFT is integrated within the JARVIS infrastructure, which includes a force-field database (JARVIS-FF) and ML tools (JARVIS-ML), offering a suite of resources for computational materials science.
The Materials Project database is particularly notable for its extensive and widely used ecosystem of data, functionalities, and Python tools, all integrated into a unified framework. Launched in 2011 as part of the Materials Genome Initiative,8,10 the Materials Project features a set of 153k bulk materials as its main data resource but has since expanded to include 222k organic molecules, 4k battery materials, 25k battery electrolytes, 20k metal–organic frameworks (MOFs), 560k catalyst surfaces, and 41k synthesis recipes.9 The Materials Project prioritizes consistency between QC calculations, initially employing only two different DFT methods: PBE+U for transition metal oxides and sulfides, and PBE for all other systems.10 The Materials Project also offers numerous utilities to support research, such as tools for generating phase stability diagrams and Pourbaix diagrams. It has released multiple open-source Python packages like Pymatgen,82 Atomate,83 FireWorks,84 and Custodian.82 Additionally, community initiatives such as MPContribs,75 which allows users to contribute their data to existing entries, and MP-Complete,85 which facilitates submission and voting on new structures, have fostered a collaborative research environment.
In addition to these databases, Table 1 displays three repositories of materials QC data: NOMAD,66–69 ioChem-BD and Catalysis-Hub. The ioChem-BD contains 38k submissions of QC calculations for materials and 318k submissions for molecules, some of which correspond to identical chemical structures, while Catalysis-Hub also hosts data on surface reactions and provides tools for analysis. The NOMAD, established in 2015, allows uploads from any user employing supported computational chemistry codes and incorporates substantial data from AFLOW, OQMD, and the Materials Project. Adhering firmly to the FAIR principles, NOMAD ensures all data is universally accessible. At present it features 9M bulk materials, 5k 2D materials, 33k 1D materials, 75k surfaces, and a recent addition of 2.8M organic and inorganic molecules. The extensive coverage of NOMAD spans a large chemical space and includes data calculated with a variety of computational codes and methods. To navigate this vast database, the NOMAD website provides advanced tools to query and filter by chemical space, computational QC code, QC methods, applications, or data origin.
Table 1 also lists various materials datasets that cover specific areas of chemical space not extensively detailed in the major databases, such as surfaces, interfaces, MOFs, covalent organic frameworks (COFs), and 1D or 2D materials. Moreover, dataset repositories such as the Materials Data Facility,72–74 MPContribs,75 Computational Materials Repository,38,39 Materials Cloud,76,77 and MatBench78,79 compile individual materials datasets, facilitating broader access to diverse data.
From Table 1 we can also observe that 7 out of the 14 materials datasets have been generated using and manipulating structures from the Materials Project (MP). The remaining datasets include hypothetical structures or materials from distinct chemical spaces not present in the Materials Project at the time of publication, such as MOFs or COFs. This underscores the significant impact of the Materials Project as a trusted resource, frequently used for downstream research projects. The Materials Project's ecosystem of functionalities and Python packages supports these projects, promoting widespread community engagement.
Overall, the Materials Project exemplifies the concept of a QC platform, a comprehensive database that integrates structures, electronic structure information, software, and community contributions. This concept is central to our perspective, highlighting the substantial benefits the Materials Project provides to the materials community. By promoting a robust ecosystem where data is consistently curated, easily accessible, and actively contributed to by researchers worldwide, the Materials Project not only serves as a vital resource but also accelerates scientific breakthroughs and innovation in materials science.
4. Molecular data resources
The molecular research community benefits from several important databases and repositories that strongly support data sharing and collaboration. These resources provide comprehensive structural data for each entry but typically lack QC information. Table 2 presents a selection of the most prominent molecular structure databases and repositories that do not include QC data. Several of these, like the PubChem database and the CSD repository, are widely used resources in the molecular community, supporting various applications that require molecular structures. However, the absence of electronic structure information limits their broader utility, especially in data-driven applications.
Prominent molecular databases and repositories without QC data. All of them contain 3D structural information.
| Name | Size | Content |
|---|---|---|
| Molecular databases | ||
| HugeMDB86 | 1.7B | Conformers of molecules from PubChem |
| ZINC20 87,88 | 230M | Commercially available compounds |
| ChemSpider89,90 | 129M | Chemically diverse molecules |
| PubChem6,7 | 118M | Chemically diverse molecules |
| ChemDB91,92 | 5.0M | Small commercially available molecules |
| ChEMBL93,94 | 2.0M | Bioactive molecules |
| aDrugBank95,96 | 500k | Pharmaceuticals |
| COCONUT97,98 | 400k | Natural products |
| Molecular repositories | ||
| aCSD1,2 | 1.0M | Small and medium sized organic and inorganic crystallized molecules |
| COD3–5 | 514k | Crystal structures of organic, inorganic, organometallic compounds and minerals, excluding biopolymers |
Not fully open access.
To address this limitation, Nakata and Shimazaki created the PubChemQC dataset by computing QC properties for 94% of all molecules present in the PubChem database as of August 2016.21,99,100 While this effort added significant value, the dataset remains separate from the PubChem database and does not integrate with its search and API functionalities. This separation restricts users, especially in fields like organic photovoltaics, from querying PubChem for molecules with specific HOMO–LUMO gaps.
Table 3 provides an overview of molecular databases, datasets, and repositories that include electronic structure data. While there are multiple comprehensive datasets for monometallic transition metal complexes (TMCs) like the tmQMg133,134 and datasets of extracted ligands,136,143–145 data for other classes of inorganic molecules are less commonly provided. Among datasets containing both organic and inorganic molecules, the PubChemQC dataset covers the largest chemical space by far. Other datasets are either small in scale or contain a large number of data points for a small number of species, such as the DES370K.106 Additionally, these datasets are predominantly focused on organic molecules, with fewer entries for inorganic compounds. Other significant sources of electronic structure data including both organic and inorganic molecules are the two repositories ioChem-BD and NOMAD. While the ioChem-BD features 318k user-submitted QC calculations for chemically diverse molecules, the NOMAD contains the largest number of entries among all molecular data resources, featuring 2.8M organic and inorganic molecules. However, despite these large numbers, the decentralized nature of the ioChem-BD and the NOMAD and the diversity of their entries introduce challenges, such as susceptibility to human errors and inconsistencies, which can complicate downstream research.
Molecular databases, datasets, repositories and dataset repositories that contain QC data. The table is divided into six categories, describing the type of data resource (database, dataset, repository, dataset repository) and the chemical space covered (organic, organic and inorganic, transition metal complexes). An ‘-sp’ in the ‘Method’ column denotes single-point calculations, often preceded by a geometry relaxation using a less computationally intensive method, such as xTB. Computational methods mentioned: semi-empirical (xTB and PM6/PM7), Hartree–Fock, DFT, TD-DFT, Gaussian-4 theory using second-order Møller–Plesset perturbation theory (G4MP2), complete active space self-consistent field (CASSCF), and coupled-cluster (CC).
| Name | Size | Method | Source | Content |
|---|---|---|---|---|
| Organic molecular databases | ||||
| CEPDB101,102 | 2.3M | DFT | Enumerated | Organic compounds for photovoltaics |
| Materials Project8–11 | 1.0M | DFT | ICSD & others | 153k bulk materials (main data), and 222k organic molecules, 4k battery materials, 25k battery electrolytes, 20k MOFs, 560k catalyst surfaces, 41k synthesis recipes |
| OCELOT103,104 | 56k | DFT | CSD, community | Crystalline organic semiconductors |
| Organic + inorganic molecular datasets | ||||
| PubChemQC21,99,100 | 86M | PM6 + DFT-sp | PubChem | Organic and organometallic molecules containing first-row transition metals |
| SPICE105 | 1.1M | DFT | Literature, PubChem, DES370K | Conformations of small molecules, dimers, dipeptide, and solvated amino acids |
| DES370K106 | 370K | DFT + CC-sp | Literature | 370k data points of dimer interactions of 392 mostly organic molecules |
| Alexandria library107 | 2.7k | DFT | PubChem, ChemSpider | Mostly organic molecules |
| CCCBDB108 | 2.2k | DFT | Literature | Gas-phase atoms and small molecules |
| QuestDB109,110 | >500 | CC & others | Literature | Vertical excitation energies for small- and medium-sized molecules |
| Organic molecular datasets | ||||
| GEOM111 | 37M | xTB | AICures, QM9 | 37M conformers of 450k organic molecules |
| Transition1x112 | 10M | DFT-sp | Grambow et al.113 | Molecular configurations along the potential energy surface of 11 961 reactions |
| ANI-1x114 | 5.0M | DFT | GDB11, ChEMBL, generated | Small molecules |
| QM7-X115 | 4.2M | DFT | QM7 | Equilibrium and non-equilibrium structures of small organic molecules |
| QMugs116 | 2.0M | xTB + DFT-sp | ChEMBL | 2M conformers of 665K biologically relevant organic molecules |
| WS22 117 | 1.2M | DFT | Literature | 1.2M data points of equilibrium and non-equilibrium geometries of 10 species |
| VQ24 118 | 836k | DFT & xTB | Generated | Enumerated molecules with up to 5 heavy atoms from C, N, O, F, Si, P, S, Cl, Br |
| Frag20 119 | 566k | DFT | ZINC, PubChem | Small organic molecules from ZINC and PubChem |
| ANI-1ccx114 | 500k | DFT + CC-sp | ANI-1x | Subset of ANI-1x recomputed with CC-sp |
| John et al.120 | 240k | DFT | PubChem | Open- and closed-shell small organic molecules |
| QM-symex121,122 | 173k | DFT & TD-DFT | Generated | Includes point group and excited states of small molecules |
| QM9 123 | 134k | DFT | GDB-17 | Small organic molecules with up to 9 heavy atoms |
| Kim et al.124 | 134k | G4MP2 | QM9 | Refinement of QM9 |
| Narayanan et al.125 | 133k | G4MP2 | QM9 | Refinement of QM9 |
| FORMED126 | 117k | xTB, DFT-sp & TD-DFT | CSD | Organic molecules from the CSD |
| OE62 127 | 62k | DFT | CSD | Organic molecules from the CSD |
| MQMspin128 | 13k | DFT & CASSCF | QM9 | Small organic carbene molecules |
| HOPV15 129 | 6.0k | DFT | Literature | 6k conformers of 353 p-type molecules for organic photovoltaics + exp. data |
| VERDE Materials DB130,131 | 1.8k | DFT | Generated | Light-responsive π-conjugated organic molecules |
| HAB79 132 | 921 | DFT & CASSCF | Literature | Benchmark dataset for DFT |
| Transition metal complex (TMC) datasets | ||||
| tmQM133 | 80k | xTB + DFT-sp | CSD | Monometallic TMCs |
| tmQMg134 | 60k | DFT | tmQM | Subset of tmQM with full DFT and graphs from natural bond orders |
| SC1MC-2022 135 | 7.0k | Hartree–Fock | Generated | TMCs assembled from ligands |
| OHLDB136 | 1.4k | DFT | Enumerated | Homoleptic TMCs |
| divTMC137 | 855 | DFT | CSD | Octahedral TMCs assembled from monodentate ligands |
| 16OSTM10 138 | 160 | DFT | CSD | Open-shell TMCs for conformer benchmark |
| ROST61 139 | 61 | CC | Literature | Open-shell TMCs for DFT functional benchmark |
| MOR41 140 | 41 | CC | Literature | Closed-shell TMCs for DFT functional benchmark |
| Organic + inorganic molecular repositories | ||||
| NOMAD66–69 | 12M | DFT & others | Submissions, MP, OQMD, AFLOW, and others | 9M bulks, 75k surfaces; 5k 2D, 33k 1D materials, 2.8M organic and inorganic molecules |
| ioChem-BD36,37 | 356k | DFT mixed | Submissions | 38k materials and 318k molecules, chemically diverse |
| Organic + inorganic molecular dataset repositories | ||||
| QCarchive141,142 | 47 sets | Mixed | Mixed | Datasets from publications |
For organic molecules, significant efforts have been made to generate extensive datasets with QC information. One of the pioneering examples is the QM9 dataset, which includes DFT properties for all 134k enumerated molecules with up to nine heavy atoms within the chemical space of C, H, O, N, and F.123,146 Other datasets provide electronic structure data for various molecular conformers, non-equilibrium geometries, and open-shell molecules.111,115–117,120
Despite the generation of substantial electronic structure data for predominantly organic molecules, this valuable information largely remains outside the framework of a comprehensive database. The Clean Energy Project Database (CEPDB)101,102 contains 2.3M organic photovoltaic candidates while the Organic Crystals in Electronic and Light-Oriented Technologies (OCELOT)103,104 database contains 56k crystalline organic semiconductors, making both large but specialized databases. Currently, the Materials Project is the only major general database that includes molecules with enriched QC properties.101,102 Initially focused on materials, the Materials Project has since begun expanding to include molecules. It currently contains 222k organic molecules, with plans to include inorganic molecules in the future.11 However, the Materials Project and its ecosystem remain primarily oriented towards materials, affecting its adoption by the molecular research community.
Despite the inclusion of both structural and QC information in the Materials Project database and the NOMAD repository, neither resource is optimized for molecular applications. Widely used molecular repositories such as the CSD and COD still lack electronic structure information. This gap underscores a critical need for a dedicated molecular QC platform, which could significantly enhance research capabilities in fields ranging from pharmaceuticals to organic electronics.
5. Guiding principles for a unified molecular quantum database
Analyzing and comparing the existing materials and molecular databases summarized in Tables 1–3 reveals a significant disparity between the two research communities. The materials community benefits immensely from the Materials Project, a robust QC platform that integrates extensive data, advanced functionalities, and active community engagement. In stark contrast, the molecular community lacks an equivalent comprehensive platform. This gap is further emphasized by the recent expansions of the Materials Project database and the NOMAD repository to incorporate molecular systems, even though both remain primarily focused on materials.
Despite our initial classification of dataset, database, repository, and dataset repository, these distinctions are not always well-defined, especially between a database and a repository. For example, the NOMAD is considered a repository because it collects QC data from many different sources, but it also incorporates data from databases like the Materials Project and features an advanced user interface. While the Materials Project is classified as a database due to its mostly centralized data generation, it also functions as a repository by collecting experimental and computational community data via MPContribs.147 Therefore, a key consideration for developing molecular QC databases is what balance of in-house data generation, curation, and user contribution is novel and needed in the molecular community. In this view, while there are already two major QC repositories for molecular data, the ioChem-BD and the NOMAD, the Materials Project is the only general QC database containing molecular structures. However, these are only a recent addition and are currently limited to organic molecules. Thus, there is a significant opportunity within the molecular community for a QC database encompassing not only organic but also inorganic chemistries.
A general QC molecular database would be well-positioned to evolve into a large platform, similar to the Materials Project, but specifically optimized for molecular structures. This platform could support both experimental and QC user-contributions in the form of analytical spectra such as ultraviolet-visible (UV-Vis) and X-ray diffraction (XRD), as well as QC input and output files. The unification of different chemical systems and the integration of computational and experimental data are central to making data more Findable, Accessible, Interoperable, and Reusable (FAIR). By collecting data in a widely recognized platform, it becomes more visible to researchers across various disciplines and is more likely to be repurposed for different applications. For example, the bulk structures in the Materials Project have been used not only for screening bulk properties, but also as a source for generating surface slabs,52,56 interfaces,30 and 2D materials.28,29,64
The unification of data within a single platform becomes particularly impactful in the context of ML applications, where large and diverse datasets are essential for training robust models. Notably, ML methods such as transfer learning, multi-task learning, and multi-fidelity learning can leverage heterogeneous data to optimize performance predictions for specific targets. For example, Yamada et al. employed transfer and multi-task learning to predict the experimental heat capacity at constant pressure (CP) for 58 polymers. They pre-trained their model on small organic molecules from the QM9 dataset, utilizing QC calculated heat capacities at constant volume (CV) rather than experimental CP values, reducing the mean absolute error (MAE) of predicting the polymeric CP by 35%.148 Similarly, Moore et al. combined QC and experimental data in a transfer learning framework to predict the experimental HOMO–LUMO gap of 26 commercially available polymer donors, achieving a 72% reduction in root mean squared error compared to DFT predictions.149
The potential of ML is further enhanced by multi-fidelity learning, where data of varying reliability, such as calculations performed at multiple levels of theory, is integrated. For instance, Chen et al. used multi-fidelity learning to improve predictions of experimental material band gaps by augmenting experimental datasets with QC data derived from the Materials Project at three different levels of DFT theory reducing the MAE by 22%.150 In each of these studies, a critical yet time-intensive step was the collection and curation of data from multiple sources. A centralized, unified database would have streamlined this process significantly, highlighting the transformative potential of such platforms for accelerating data-driven discoveries.
Despite the potential benefits of unifying data on a single platform, several challenges must be addressed. A significant hurdle is how to incorporate data from different computational and experimental sources in a way that is most useful for users. The Materials Project facilitates this by enabling data annotations via MPContribs,147 while the PubChem handles this issue by identifying new submissions based on their chemical structure and, when possible, linking them to existing entries.151
Another challenge in integrating computational and experimental properties involve semantic issues, where properties with similar names may refer to subtly different concepts. For instance, experimental overpotentials in electrocatalysis are referenced to a specific current density,152 whereas theoretical overpotentials calculated using QC methods are not. These differences need to be clarified for users and can complicate data exchange through standardized, logic-based language (ontologies) such as the PubChemRDF project, which uses ontologies like CHEMINF153 to express the PubChem knowledge in a consistent and machine-understandable format.151
In addition to studying the chemical properties of individual molecules, a major area of interest in chemistry is the interaction between species in chemical reactions, which can be modelled using QC calculations. For instance, the Gibbs energy of H adsorption (ΔGH) is a QC-derived reaction descriptor for the hydrogen evolution reaction that allows the prediction of catalytic performance. However, such values are not intrinsic to a single molecule and often depend on the properties of multiple molecular structures. Similarly, reaction parameters such as temperature, pressure, reactant concentration, and solvent depend on the conditions of the reaction, not just the individual molecules. Consequently, reactions require different organizational structures, such as those provided by the Open Reaction Database154 or the Catalysis-Hub repository for surface reactions.70,71
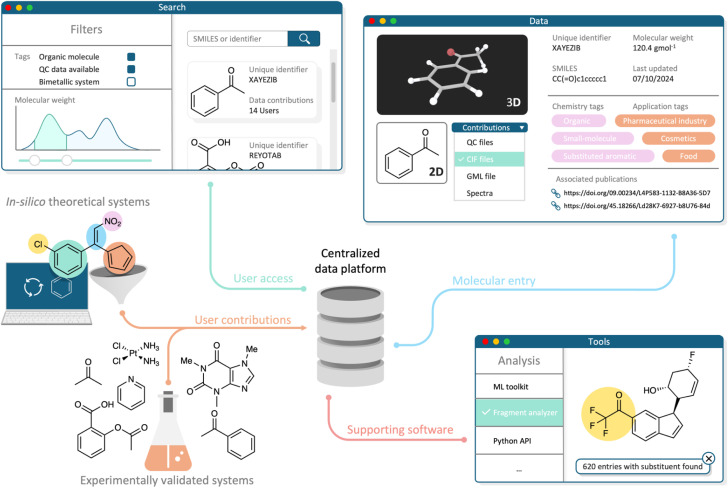
Consequently, our review and evaluation of a diverse range of molecular and material data resources have led us to identify seven key principles crucial for establishing a unified molecular QC database. These principles, which we refer to as the QUANTUM principles, are illustrated in Fig. 3. Designed to build upon the foundational FAIR principles, the QUANTUM principles address the unique needs and challenges in realizing a QC platform for the molecular community. While some of these principles are already partially implemented in existing molecular databases, others highlight critical areas requiring further development and innovation.
Fig. 3. Schematic overview of a molecular QC platform adhering to the QUANTUM principles. The platform allows users to contribute organic and inorganic molecular structures, either experimentally validated or derived from theoretical studies. A web interface enables users to search and query molecules based on diverse properties or tags. Each entry encompasses multiple attributes, including structural details, QC data, and spectral information. The platform also offers integrated software tools for advanced data queries and analyses.
5.1. Quantum chemical and experimental data
The integration of QC and experimental data into a unified molecular database presents both opportunities and challenges. Ideally, a comprehensive database would include a wide range of experimentally measured properties for each molecule, such as nuclear magnetic resonance (NMR), infrared (IR), and UV-Vis spectroscopic data, and XRD analyses, as well as physical properties like melting point, hardness, and even color. However, obtaining such data consistently across a broad chemical space is challenging. For example, difficulties in crystallization can hinder XRD analysis.155 Conversely, QC calculations can be applied to a much broader range of systems, offering valuable insights into the electronic structure of molecules. For instance, Kneiding et al. computed properties such as HOMO–LUMO gaps, polarizability, dipole moments, and Gibbs energies for 60k transition metal complexes using a variety of DFT methods.134 The inclusion of QC data in a database is therefore intended to complement experimental data by filling gaps and providing theoretical insights that can enhance our understanding of molecular properties and reactivity.
However, care must be taken when using and creating QC data to ensure that it is appropriate for the corresponding chemical system and balances both speed and accuracy. It can be more beneficial to focus on fewer, high-quality data points at suitable levels of theory than to amass data with methods that may not be well-suited for the intended purpose. On the other hand, ML techniques can leverage data from computationally inexpensive but less precise QC methods and improve their reliability and speed by incorporating either more accurate QC data or experimental data during training.148–150 These methods, such as multi-fidelity learning, can dramatically enhance the predictive power of models, even when relying on less accurate or incomplete datasets.
5.2. Unified chemical space
A comprehensive molecular database would benefit from covering a wide chemical space, including both organic and inorganic molecules, while recognizing that macromolecules may require special considerations. This enables researchers to explore a diverse array of molecular chemistries, including organometallics, TMCs, main-group organic chemistry, as well as molecules used in medicinal chemistry, catalysis, agrochemicals, and beyond, all while using the same database infrastructure. In addition to benefiting data-driven methods such as ML, unifying chemical systems in a single platform enables the reuse of data across various fields of chemistry. For example, the development of cisplatin illustrates how a compound initially observed for inhibiting the cell division of Escherichia coli in electrochemical experiments eventually became a widely used chemotherapy drug.156,157
Beyond experimentally validated structures, a robust QC platform should also accommodate hypothetical structures generated through various methodologies, such as bottom-up workflows, scaffold diversification inspired by experiments, and generative ML techniques.158 For example, molSimplify159 offers a bottom-up approach by assembling monometallic transition metal complexes from a predefined set of ligands. Similarly, Jin et al. developed a generative ML model that incrementally constructs organic molecules by predicting substructure connections, enabling the exploration of new chemical spaces.160
Evaluating the synthetic feasibility of hypothetical structures is a key challenge, as it involves factors such as byproduct formation, yield, and ease of characterization.158 Computational tools like MegaSyn address this by assessing synthetic viability of organic molecules, using methodologies that evaluate the relative abundance of synthetically accessible molecular fragments within a given compound.161 To the same end, the DART platform allows the generation of bottom-up molecular datasets by assembling novel TMCs from ligands in the CSD with established synthetic precedents, aiming to maximize their synthetic viability.162 These tools help prioritize structures that are more likely to be experimentally realizable, thus streamlining efforts in synthesis and validation.
Nonetheless, hypothetical structures remain valuable even when synthetic feasibility is uncertain. Such systems, especially those with QC data, can serve as training datasets for ML models or as input for high-throughput screenings. By integrating diverse experimental and theoretical molecules from various domains into a unified platform, the QC database can facilitate interdisciplinary innovation, providing access to an expansive and interconnected chemical space.
5.3. Accessible and searchable data
To support public research, the molecular QC platform would benefit from being open access with a modern web interface that facilitates querying and filtering of target molecules. This should include simple descriptors like empirical formula or molecular weight, as well as more complex properties like the HOMO–LUMO gap or sub-structure searches using SMARTS.163 An API should also be available for programmatic batch access to support data-driven applications and extensive computational analyses.
5.4. Numerous molecular representations
To capture the complexity of chemical structures, the database should support multiple molecular representations that complement each other. This includes 3D structures from experimental XRD and optimized 3D structures from QC calculations. Critical structural details of the 2D molecular graph, such as connectivity and bond orders, commonly represented by SMILES164 strings, should also be included.
QC calculations also enable the addition of quantitative information such as atomic charges and spins. If necessary, computationally derived bonds and bond orders can also be assigned using methods such as natural bond orbital analysis,165 as was done for the tmQMg dataset.134 This data can be useful for example in ML applications as molecular features.
To represent molecular structures numerically, various methods are employed, depending on the desired application. For instance, 3D molecular structures can be encoded into fixed-sized vectors using Smooth Overlap of Atomic Positions (SOAP) features,166 while 2D molecular graph representations can be expressed either as a fixed-size vector using autocorrelation167,168 or molecular fingerprints,169 or they can be used to directly train graph neural networks.170 Notably, 2D molecular graphs can incorporate geometric properties such as bond distances and QC-derived properties such as atomic charges. However, these fixed-size vector and graph features are typically not stored in databases due to their computational efficiency and dependence on user-defined hyperparameters. Instead, they are often generated on-the-fly using Python packages such as DScribe,171 RDKit,172 and molSimplify.159 This approach ensures flexibility and adaptability, allowing users to tailor the representations to specific tasks or datasets.
In addition to including 3D coordinates and 2D graph representations of a molecule, it can also be beneficial to include data corresponding to the conformational space of a compound. For instance, Eastman et al.105 emphasized the importance of broad conformational datasets, not limited to only the lowest energy conformers, for training ML potentials. They developed the SPICE dataset, which includes 1.1M conformers and trained a set of ML potentials applicable to a broad region of chemical space.
To effectively collate this data, each entry should also have a unique identifier assigned, as SMILES alone is not always sufficient for defining molecules, especially when capturing different conformations of the same molecule. The database should also enable smart data relations between entries, such as identifying isomers or clustering similar molecules. Additionally, tagging molecules with specific applications (e.g. organic photovoltaics), as is done in the NOMAD and ioChem-BD, and linking them to related publication DOIs, like in the CSD, could significantly boost research efficiency and breakthroughs.
5.5. Trusted data curation
Ensuring that the molecular QC database is a trusted community resource requires regular curation and updates. Integrating community data consistently within the database framework is essential to maintain its reliability. Both the Materials Project and the PubChem provide valuable examples of strongly curated databases managing the inclusion of community data. This can also be supported by automated validation and normalization procedures as described for the PubChem.151
Especially for QC data, inclusion and curation becomes particularly important due to the large range of QC methods and different requirements for different chemical systems. Thus, a QC molecular platform needs to adopt a consistent framework to accept, process, and display data contributions from the community. The implementation and realization must be considered by the developers of the database, considering the target audience, technical details, and available funding, and cannot be imposed, but can develop over time.
5.6. User-friendly ecosystem of software
Offering user-friendly software and functionalities is essential to create an accessible QC platform. For example, the widely used Python package Pymatgen provides API access to the Materials Project and various tools for analyzing and manipulating materials and molecules. A robust ecosystem of web apps and open-source software enhances the database's utility and promotes community contributions to software, reinforcing the database's status within the community.
5.7. Maximizing community engagement
The ultimate value of a QC platform lies in its frequent use by the scientific community. The Materials Project's most relevant accomplishment is not just the diversity of its data but its status as a trusted and widely used resource. This status was achieved by integrating structural and electronic structure data with extensive open-source software, which mobilized the community to further contribute to data and software, forming a positive feedback loop. To cultivate a similar status, a molecular QC platform needs to engage with the community to meet their needs, incentivize contributions to open-source software, and facilitate the incorporation of data from downstream projects by other researchers.
6. Conclusions and outlook
In this perspective, we have reviewed and analyzed the current landscape of materials and molecular databases, datasets, repositories, and dataset repositories, with a particular focus on those incorporating electronic structure properties from QC calculations. Our analysis highlights the considerable benefits that the materials community has gained from robust QC databases like the Materials Project. This platform seamlessly integrates structural data with consistently calculated electronic structure information and supports a vibrant ecosystem of open-source software, driving downstream research and fostering significant community contributions in data and software development. The success of the Materials Project exemplifies the concept and the potential of a well-integrated QC platform.
In contrast, the molecular community, while leveraging several widely used structural databases and repositories, does not benefit from a dedicated platform that includes both electronic structure information and a comprehensive ecosystem of supporting software. To bridge this gap, we propose the seven QUANTUM principles aimed at developing a unified molecular QC platform. These principles draw inspiration from the diverse databases, datasets, repositories, and dataset repositories reviewed herein. Although our focus is on enhancing molecular databases, the QUANTUM principles also offer valuable insights for advancing existing materials databases. They provide a strategic roadmap for researchers in both the molecular and materials communities to collaborate on improving current databases and identifying critical strategies for future developments.
Significant molecular data resources like the PubChem database and the CSD repository already align with several of the QUANTUM principles. However, the most pressing short-term development we have identified is the integration of electronic structure data from QC calculations into these molecular structural databases. The name QUANTUM is therefore not only intended as an acronym but also as a reflection of the urgency of this particular principle. Meanwhile, platforms like the Materials Project and NOMAD, traditionally focused on materials, are beginning to expand their scope to include molecular data, signaling a major shift towards integrating molecular systems into QC platforms.
Looking ahead, we anticipate significant mid-term progress to emerge from the development of associated software that supports and facilitates community contributions of molecular data. In the long term, we envision the establishment of a unified database that fully adheres to all seven QUANTUM principles, serving as the central QC platform for molecular research. This platform would host a vast array of molecular structures, QC calculations, and experimental properties, underpinned by a comprehensive ecosystem of software and functionalities. It would include a subset of highly curated structures with consistent QC calculations while also acting as a repository for users to submit experimental and computational data.
Once established, we foresee that such a QC platform will revolutionize the field of molecular discovery, mirroring the transformative impact that the Materials Project has had on materials research. We therefore urge the research community to unify their efforts and collaborate in establishing a molecular QC platform that will drive future advancements and innovation in chemistry.
Data availability
Data sharing is not applicable to this manuscript as no datasets were generated or analysed in this perspective.
Author contributions
All authors contributed to the initial conceptualization and the outline of the paper. T. S. and C. C. reviewed and analyzed the existing materials and molecular databases and co-wrote the first draft of the manuscript. M. G.-M. supervised the process in all stages and reviewed and edited the first draft. All authors contributed to the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
The authors are very grateful for the financial support provided by the Science Foundation Ireland (SFI-20/FFP-P/8740).
Biographies
Biography
Timo Sommer.

Timo Sommer is a PhD candidate in computational chemistry under the supervision of Prof. Max García-Melchor at Trinity College Dublin, where he develops computational tools and datasets to screen transition metal complexes as catalysts for the oxygen evolution reaction. He earned his master's degree in Theoretical Physics from the Karlsruhe Institute of Technology, where he focused on data-driven methods to predict the critical temperature of superconductors.
Biography
Cian Clarke.

Cian graduated from Trinity College Dublin in 2022 with a BA in Chemistry with Molecular Modelling. Soon after Cian joined the group of Prof. Max García-Melchor in Trinity College Dublin and under his supervision is currently pursuing a PhD in computational chemistry. The focus on Cian's research surrounds the development and in silico screening of novel water oxidation catalysts.
Biography
Max García-Melchor.

Dr Max García-Melchor is an Ikerbasque Research Professor at CIC EnergiGUNE, where he leads the Atomistic & Molecular Modelling for Catalysis group. His research leverages advanced computational methods and artificial intelligence to accelerate the discovery of catalytic systems for sustainable chemical and fuel production. With a PhD in Chemistry from the Universitat Autònoma de Barcelona and over 15 years of experience, he specializes in modelling (electro)catalytic reaction mechanisms and developing rational catalyst design approaches.
References
- Cambridge Structural Database, https://www.ccdc.cam.ac.uk/, accessed 9 May 2024
- Groom C. R. Bruno I. J. Lightfoot M. P. Ward S. C. Acta Crystallogr., Sect. B:Struct. Sci. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystallography Open Database, https://www.crystallography.net/cod/, accessed 9 May 2024
- Gražulis S. Chateigner D. Downs R. T. Yokochi A. F. T. Quirós M. Lutterotti L. Manakova E. Butkus J. Moeck P. Le Bail A. J. Appl. Crystallogr. 2009;42:726–729. doi: 10.1107/S0021889809016690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gražulis S. Daškevič A. Merkys A. Chateigner D. Lutterotti L. Quirós M. Serebryanaya N. R. Moeck P. Downs R. T. Le Bail A. Nucleic Acids Res. 2012;40:D420–D427. doi: 10.1093/nar/gkr900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PubChem, https://pubchem.ncbi.nlm.nih.gov/, accessed 9 May 2024
- Kim S. Chen J. Cheng T. Gindulyte A. He J. He S. Li Q. Shoemaker B. A. Thiessen P. A. Yu B. Zaslavsky L. Zhang J. Bolton E. E. Nucleic Acids Res. 2023;51:D1373–D1380. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. Ong S. P. Hautier G. Chen W. Richards W. D. Dacek S. Cholia S. Gunter D. Skinner D. Ceder G. Persson K. A. APL Mater. 2013;1:011002. [Google Scholar]
- Materials Project, https://next-gen.materialsproject.org/, accessed 8 May 2024
- Jain A., Montoya J., Dwaraknath S., Zimmermann N. E. R., Dagdelen J., Horton M., Huck P., Winston D., Cholia S., Ong S. P. and Persson K., in Handbook of Materials Modeling: Methods: Theory and Modeling, ed. W. Andreoni and S. Yip, Springer International Publishing, Cham, 2020, pp. 1751–1784 [Google Scholar]
- Clark Spotte-Smith E. W. Archer Cohen O. Blau S. M. Munro J. M. Yang R. Guha R. D. Patel H. D. Vijay S. Huck P. Kingsbury R. Horton M. K. Persson K. A. Digital Discovery. 2023;2:1862–1882. [Google Scholar]
- Chrostowska A. and Darrigan C., in Organosilicon Compounds, ed. V. Y. Lee, Academic Press, 2017, pp. 115–166 [Google Scholar]
- Perera A., Park Y. C. and Bartlett R. J., in Comprehensive Computational Chemistry, ed. M. Yáñez and R. J. Boyd, Elsevier, Oxford, 1st edn, 2024, pp. 18–46 [Google Scholar]
- Grimme S. Bannwarth C. Shushkov P. J. Chem. Theory Comput. 2017;13:1989–2009. doi: 10.1021/acs.jctc.7b00118. [DOI] [PubMed] [Google Scholar]
- Bannwarth C. Ehlert S. Grimme S. J. Chem. Theory Comput. 2019;15:1652–1671. doi: 10.1021/acs.jctc.8b01176. [DOI] [PubMed] [Google Scholar]
- Bannwarth C. Caldeweyher E. Ehlert S. Hansen A. Pracht P. Seibert J. Spicher S. Grimme S. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2021;11:e1493. [Google Scholar]
- Stewart J. J. P. J. Mol. Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. J. P. J. Mol. Model. 2013;19:1–32. doi: 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel W. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2014;4:145–157. [Google Scholar]
- Neugebauer H. Bädorf B. Ehlert S. Hansen A. Grimme S. J. Comput. Chem. 2023;44:2120–2129. doi: 10.1002/jcc.27185. [DOI] [PubMed] [Google Scholar]
- Nakata M. Shimazaki T. Hashimoto M. Maeda T. J. Chem. Inf. Model. 2020;60:5891–5899. doi: 10.1021/acs.jcim.0c00740. [DOI] [PubMed] [Google Scholar]
- Chai J.-D. Head-Gordon M. J. Chem. Phys. 2008;128:084106. doi: 10.1063/1.2834918. [DOI] [PubMed] [Google Scholar]
- Bursch M. Mewes J.-M. Hansen A. Grimme S. Angew. Chem., Int. Ed. 2022;61:e202205735. doi: 10.1002/anie.202205735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay P. J. Wadt W. R. J. Chem. Phys. 1985;82:270–283. [Google Scholar]
- Zhong M. Tran K. Min Y. Wang C. Wang Z. Dinh C.-T. De Luna P. Yu Z. Rasouli A. S. Brodersen P. Sun S. Voznyy O. Tan C.-S. Askerka M. Che F. Liu M. Seifitokaldani A. Pang Y. Lo S.-C. Ip A. Ulissi Z. Sargent E. H. Nature. 2020;581:178–183. doi: 10.1038/s41586-020-2242-8. [DOI] [PubMed] [Google Scholar]
- Jun K. Sun Y. Xiao Y. Zeng Y. Kim R. Kim H. Miara L. J. Im D. Wang Y. Ceder G. Nat. Mater. 2022;21:924–931. doi: 10.1038/s41563-022-01222-4. [DOI] [PubMed] [Google Scholar]
- Chen C. Ong S. P. Nat. Comput. Sci. 2022;2:718–728. doi: 10.1038/s43588-022-00349-3. [DOI] [PubMed] [Google Scholar]
- Zhou J. Shen L. Costa M. D. Persson K. A. Ong S. P. Huck P. Lu Y. Ma X. Chen Y. Tang H. Feng Y. P. Sci. Data. 2019;6:86. doi: 10.1038/s41597-019-0097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2D Materials Encyclopedia, http://www.2dmatpedia.org/, accessed 8 May 2024
- Gerber E. Torrisi S. B. Shabani S. Seewald E. Pack J. Hoffman J. E. Dean C. R. Pasupathy A. N. Kim E.-A. Nat. Commun. 2023;14:7921. doi: 10.1038/s41467-023-43496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F. Zhu Z. Lu J. Yan Y. Jiang H. Sun Q. Chem. Phys. Lett. 2023;814:140358. [Google Scholar]
- Dinic F. Neporozhnii I. Voznyy O. Comput. Mater. Sci. 2024;231:112580. [Google Scholar]
- Wilkinson M. D. Dumontier M. Aalbersberg I. J. Appleton G. Axton M. Baak A. Blomberg N. Boiten J.-W. da Silva Santos L. B. Bourne P. E. Bouwman J. Brookes A. J. Clark T. Crosas M. Dillo I. Dumon O. Edmunds S. Evelo C. T. Finkers R. Gonzalez-Beltran A. Gray A. J. G. Groth P. Goble C. Grethe J. S. Heringa J. ’t Hoen P. A. C. Hooft R. Kuhn T. Kok R. Kok J. Lusher S. J. Martone M. E. Mons A. Packer A. L. Persson B. Rocca-Serra P. Roos M. van Schaik R. Sansone S.-A. Schultes E. Sengstag T. Slater T. Strawn G. Swertz M. A. Thompson M. van der Lei J. van Mulligen E. Velterop J. Waagmeester A. Wittenburg P. Wolstencroft K. Zhao J. Mons B. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- figshare, https://figshare.com/, accessed 13 June 2024
- GitHub, https://github.com, accessed 13 June 2024
- ioChem-BD, https://www.iochem-bd.org/, accessed 9 May 2024
- Álvarez-Moreno M. de Graaf C. López N. Maseras F. Poblet J. M. Bo C. J. Chem. Inf. Model. 2015;55:95–103. doi: 10.1021/ci500593j. [DOI] [PubMed] [Google Scholar]
- CMR—Computational Materials Repository, https://cmr.fysik.dtu.dk/, accessed 8 May 2024
- Landis D. D. Hummelshøj J. S. Nestorov S. Greeley J. Dułak M. Bligaard T. Nørskov J. K. Jacobsen K. W. Comput. Sci. Eng. 2012;14:51–57. [Google Scholar]
- The Materials Project API, https://next-gen.materialsproject.org/api, accessed 14 October 2024
- Aflow – Automatic FLOW for Materials Discovery, https://www.aflowlib.org/, accessed 8 May 2024
- Curtarolo S. Setyawan W. Hart G. L. W. Jahnatek M. Chepulskii R. V. Taylor R. H. Wang S. Xue J. Yang K. Levy O. Mehl M. J. Stokes H. T. Demchenko D. O. Morgan D. Comput. Mater. Sci. 2012;58:218–226. [Google Scholar]
- Esters M. Oses C. Divilov S. Eckert H. Friedrich R. Hicks D. Mehl M. J. Rose F. Smolyanyuk A. Calzolari A. Campilongo X. Toher C. Curtarolo S. Comput. Mater. Sci. 2023;216:111808. [Google Scholar]
- OQMD, https://oqmd.org/, accessed 8 May 2024
- Saal J. E. Kirklin S. Aykol M. Meredig B. Wolverton C. JOM. 2013;65:1501–1509. [Google Scholar]
- Shen J. Griesemer S. D. Gopakumar A. Baldassarri B. Saal J. E. Aykol M. Hegde V. I. Wolverton C. JPhys Mater. 2022;5:031001. [Google Scholar]
- NIST-JARVIS, https://jarvis.nist.gov/, accessed 8 May 2024
- Choudhary K. Garrity K. F. Reid A. C. E. DeCost B. Biacchi A. J. Hight Walker A. R. Trautt Z. Hattrick-Simpers J. Kusne A. G. Centrone A. Davydov A. Jiang J. Pachter R. Cheon G. Reed E. Agrawal A. Qian X. Sharma V. Zhuang H. Kalinin S. V. Sumpter B. G. Pilania G. Acar P. Mandal S. Haule K. Vanderbilt D. Rabe K. Tavazza F. npj Comput. Mater. 2020;6:173. doi: 10.1038/s41524-020-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines D. Gurunathan R. Garrity K. F. DeCost B. Biacchi A. J. Tavazza F. Choudhary K. Appl. Phys. Rev. 2023;10:041302. [Google Scholar]
- Organic Materials Database, https://omdb.mathub.io/, accessed 8 May 2024
- Borysov S. S. Geilhufe R. M. Balatsky A. V. PLoS One. 2017;12:e0171501. doi: 10.1371/journal.pone.0171501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanussot L. Das A. Goyal S. Lavril T. Shuaibi M. Riviere M. Tran K. Heras-Domingo J. Ho C. Hu W. Palizhati A. Sriram A. Wood B. Yoon J. Parikh D. Zitnick C. L. Ulissi Z. ACS Catal. 2021;11:6059–6072. [Google Scholar]
- Burner J. Luo J. White A. Mirmiran A. Kwon O. Boyd P. G. Maley S. Gibaldi M. Simrod S. Ogden V. Woo T. K. Chem. Mater. 2023;35:900–916. [Google Scholar]
- Schmidt J. Wang H.-C. Cerqueira T. F. T. Botti S. Marques M. A. L. Sci. Data. 2022;9:64. doi: 10.1038/s41597-022-01177-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare Z. J. L. Morelock R. J. Musgrave C. B. Sci. Data. 2023;10:244. doi: 10.1038/s41597-023-02127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran R. Lan J. Shuaibi M. Wood B. M. Goyal S. Das A. Heras-Domingo J. Kolluru A. Rizvi A. Shoghi N. Sriram A. Therrien F. Abed J. Voznyy O. Sargent E. H. Ulissi Z. Zitnick C. L. ACS Catal. 2023;13:3066–3084. [Google Scholar]
- Rosen A. S. Iyer S. M. Ray D. Yao Z. Aspuru-Guzik A. Gagliardi L. Notestein J. M. Snurr R. Q. Matter. 2021;4:1578–1597. [Google Scholar]
- Wang F. Q. Choudhary K. Liu Y. Hu J. Hu M. Sci. Data. 2022;9:59. doi: 10.1038/s41597-022-01158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery A. A. Wolverton C. Sci. Data. 2017;4:170153. doi: 10.1038/sdata.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C2DB, https://c2db.fysik.dtu.dk/, accessed 9 May 2024
- Haastrup S. Strange M. Pandey M. Deilmann T. Schmidt P. S. Hinsche N. F. Gjerding M. N. Torelli D. Larsen P. M. Riis-Jensen A. C. Gath J. Jacobsen K. W. Mortensen J. J. Olsen T. Thygesen K. S. 2D Mater. 2018;5:042002. [Google Scholar]
- Gjerding M. N. Taghizadeh A. Rasmussen A. Ali S. Bertoldo F. Deilmann T. Knøsgaard N. R. Kruse M. Larsen A. H. Manti S. Pedersen T. G. Petralanda U. Skovhus T. Svendsen M. K. Mortensen J. J. Olsen T. Thygesen K. S. 2D Mater. 2021;8:044002. [Google Scholar]
- Moustafa H. Larsen P. M. Gjerding M. N. Mortensen J. J. Thygesen K. S. Jacobsen K. W. Phys. Rev. Mater. 2022;6:064202. [Google Scholar]
- Choudhary K. Kalish I. Beams R. Tavazza F. Sci. Rep. 2017;7:5179. doi: 10.1038/s41598-017-05402-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongari D. Yakutovich A. V. Talirz L. Smit B. ACS Cent. Sci. 2019;5:1663–1675. doi: 10.1021/acscentsci.9b00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOMAD, https://nomad-lab.eu/nomad-lab/, accessed 8 May 2024
- Draxl C. Scheffler M. MRS Bull. 2018;43:676–682. [Google Scholar]
- Draxl C. Scheffler M. JPhys Mater. 2019;2:036001. [Google Scholar]
- Sbailò L. Fekete Á. Ghiringhelli L. M. Scheffler M. npj Comput. Mater. 2022;8:1–7. [Google Scholar]
- Catalysis-Hub, https://www.catalysis-hub.org/, accessed 8 May 2024
- Winther K. T. Hoffmann M. J. Boes J. R. Mamun O. Bajdich M. Bligaard T. Sci. Data. 2019;6:75. doi: 10.1038/s41597-019-0081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Materials Data Facility (MDF), https://materialsdatafacility.org/, accessed 8 May 2024
- Blaiszik B. Chard K. Pruyne J. Ananthakrishnan R. Tuecke S. Foster I. JOM. 2016;68:2045–2052. [Google Scholar]
- Blaiszik B. Ward L. Schwarting M. Gaff J. Chard R. Pike D. Chard K. Foster I. MRS Commun. 2019;9:1125–1133. [Google Scholar]
- Materials Project, MPContribs Explorer, https://next-gen.materialsproject.org/contribs, accessed 8 May 2024
- The Materials Cloud, https://www.materialscloud.org/home, accessed 8 May 2024
- Talirz L. Kumbhar S. Passaro E. Yakutovich A. V. Granata V. Gargiulo F. Borelli M. Uhrin M. Huber S. P. Zoupanos S. Adorf C. S. Andersen C. W. Schütt O. Pignedoli C. A. Passerone D. VandeVondele J. Schulthess T. C. Smit B. Pizzi G. Marzari N. Sci. Data. 2020;7:299. doi: 10.1038/s41597-020-00637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MatBench, https://matbench.materialsproject.org/, accessed 8 May 2024
- Dunn A. Wang Q. Ganose A. Dopp D. Jain A. npj Comput. Mater. 2020;6:138. [Google Scholar]
- ICSD, https://icsd.products.fiz-karlsruhe.de/, accessed 13 May 2024
- Zagorac D. Müller H. Ruehl S. Zagorac J. Rehme S. J. Appl. Crystallogr. 2019;52:918–925. doi: 10.1107/S160057671900997X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. P. Richards W. D. Jain A. Hautier G. Kocher M. Cholia S. Gunter D. Chevrier V. L. Persson K. A. Ceder G. Comput. Mater. Sci. 2013;68:314–319. [Google Scholar]
- Mathew K. Montoya J. H. Faghaninia A. Dwarakanath S. Aykol M. Tang H. Chu I. Smidt T. Bocklund B. Horton M. Dagdelen J. Wood B. Liu Z.-K. Neaton J. Ong S. P. Persson K. Jain A. Comput. Mater. Sci. 2017;139:140–152. [Google Scholar]
- Jain A. Ong S. P. Chen W. Medasani B. Qu X. Kocher M. Brafman M. Petretto G. Rignanese G.-M. Hautier G. Gunter D. Persson K. A. Concurr. Comput. Pract. Exp. 2015;27:5037–5059. [Google Scholar]
- MP-Complete, https://sciencegateways.org/resources/mp-complete, accessed 6 October 2024
- Huge MDB, https://www.multi-d.com/, accessed 9 May 2024
- ZINC20, https://zinc.docking.org/, accessed 9 May 2024
- Irwin J. J. Tang K. G. Young J. Dandarchuluun C. Wong B. R. Khurelbaatar M. Moroz Y. S. Mayfield J. Sayle R. A. J. Chem. Inf. Model. 2020;60:6065–6073. doi: 10.1021/acs.jcim.0c00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChemSpider, https://www.chemspider.com/, accessed 9 May 2024
- Pence H. E. Williams A. J. Chem. Educ. 2010;87:1123–1124. [Google Scholar]
- ChemDB, https://cdb.ics.uci.edu/, accessed 9 May 2024
- Chen J. H. Linstead E. Swamidass S. J. Wang D. Baldi P. Bioinformatics. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. [DOI] [PubMed] [Google Scholar]
- ChEMBL Database, https://www.ebi.ac.uk/chembl/, accessed 9 May 2024
- Zdrazil B. Felix E. Hunter F. Manners E. J. Blackshaw J. Corbett S. de Veij M. Ioannidis H. Lopez D. M. Mosquera J. F. Magarinos M. P. Bosc N. Arcila R. Kizilören T. Gaulton A. Bento A. P. Adasme M. F. Monecke P. Landrum G. A. Leach A. R. Nucleic Acids Res. 2024;52:D1180–D1192. doi: 10.1093/nar/gkad1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DrugBank, https://go.drugbank.com/, accessed 9 May 2024
- Wishart D. S. Knox C. Guo A. C. Shrivastava S. Hassanali M. Stothard P. Chang Z. Woolsey J. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCONUT: Natural Products Online, https://coconut.naturalproducts.net/, accessed 9 May 2024
- Sorokina M. Merseburger P. Rajan K. Yirik M. A. Steinbeck C. J. Cheminf. 2021;13:2. doi: 10.1186/s13321-020-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M. Maeda T. J. Chem. Inf. Model. 2023;63:5734–5754. doi: 10.1021/acs.jcim.3c00899. [DOI] [PubMed] [Google Scholar]
- Nakata M. Shimazaki T. J. Chem. Inf. Model. 2017;57:1300–1308. doi: 10.1021/acs.jcim.7b00083. [DOI] [PubMed] [Google Scholar]
- CEPDB, https://www.molecularspace.org/, accessed 8 May 2024
- Hachmann J. Olivares-Amaya R. Atahan-Evrenk S. Amador-Bedolla C. Sánchez-Carrera R. S. Gold-Parker A. Vogt L. Brockway A. M. Aspuru-Guzik A. J. Phys. Chem. Lett. 2011;2:2241–2251. [Google Scholar]
- OCELOT – Organic Crystals in Electronic and Light-Oriented Technologies, https://oscar.as.uky.edu/, accessed 2 October 2024
- Ai Q. Bhat V. Ryno S. M. Jarolimek K. Sornberger P. Smith A. Haley M. M. Anthony J. E. Risko C. J. Chem. Phys. 2021;154:174705. doi: 10.1063/5.0048714. [DOI] [PubMed] [Google Scholar]
- Eastman P. Behara P. K. Dotson D. L. Galvelis R. Herr J. E. Horton J. T. Mao Y. Chodera J. D. Pritchard B. P. Wang Y. De Fabritiis G. Markland T. E. Sci. Data. 2023;10:11. doi: 10.1038/s41597-022-01882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donchev A. G. Taube A. G. Decolvenaere E. Hargus C. McGibbon R. T. Law K.-H. Gregersen B. A. Li J.-L. Palmo K. Siva K. Bergdorf M. Klepeis J. L. Shaw D. E. Sci. Data. 2021;8:55. doi: 10.1038/s41597-021-00833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremanpour M. M. van Maaren P. J. van der Spoel D. Sci. Data. 2018;5:180062. doi: 10.1038/sdata.2018.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIST Computational Chemistry Comparison and Benchmark Database, NIST Standard Reference Database Number 101, http://cccbdb.nist.gov/, accessed 8 May 2024
- QUEST: A Database of Highly-Accurate Excitation Energies, https://lcpq.github.io/QUESTDB_website/, accessed 8 May 2024
- Véril M. Scemama A. Caffarel M. Lipparini F. Boggio-Pasqua M. Jacquemin D. Loos P.-F. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2021;11:e1517. [Google Scholar]
- Axelrod S. Gómez-Bombarelli R. Sci. Data. 2022;9:185. doi: 10.1038/s41597-022-01288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner M. Bhowmik A. Vegge T. Busk J. Winther O. Sci. Data. 2022;9:779. doi: 10.1038/s41597-022-01870-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grambow C. A. Pattanaik L. Green W. H. Sci. Data. 2020;7:137. doi: 10.1038/s41597-020-0460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S. Zubatyuk R. Nebgen B. Lubbers N. Barros K. Roitberg A. E. Isayev O. Tretiak S. Sci. Data. 2020;7:134. doi: 10.1038/s41597-020-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoja J. Medrano Sandonas L. Ernst B. G. Vazquez-Mayagoitia A. DiStasio Jr R. A. Tkatchenko A. Sci. Data. 2021;8:43. doi: 10.1038/s41597-021-00812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isert C. Atz K. Jiménez-Luna J. Schneider G. Sci. Data. 2022;9:273. doi: 10.1038/s41597-022-01390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro Jr M. Zhang S. Dral P. O. Barbatti M. Sci. Data. 2023;10:95. doi: 10.1038/s41597-023-01998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan D., Benali A., Kim S. Y. H., von Rudorff G. F. and von Lilienfeld O. A., arXiv, 2024, preprint, arXiv:2405.05961, 10.48550/arXiv.2405.05961 [DOI]
- Lu J. Xia S. Lu J. Zhang Y. J. Chem. Inf. Model. 2021;61:1095–1104. doi: 10.1021/acs.jcim.1c00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John P. C. S. Guan Y. Kim Y. Etz B. D. Kim S. Paton R. S. Sci. Data. 2020;7:244. doi: 10.1038/s41597-020-00588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. Xu Y. Liu R. Zhu X. Sci. Data. 2019;6:213. doi: 10.1038/s41597-019-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. Ye S. Dai T. Zha Z. Gao Y. Zhu X. Sci. Data. 2020;7:400. doi: 10.1038/s41597-020-00746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R. Dral P. O. Rupp M. von Lilienfeld O. A. Sci. Data. 2014;1:140022. doi: 10.1038/sdata.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Park J. Y. Choi S. Sci. Data. 2019;6:109. doi: 10.1038/s41597-019-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan B. Redfern P. C. Assary R. S. Curtiss L. A. Chem. Sci. 2019;10:7449–7455. doi: 10.1039/c9sc02834j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovits J. T. Laplaza R. Vela S. Corminboeuf C. Adv. Mater. 2024;36:2305602. doi: 10.1002/adma.202305602. [DOI] [PubMed] [Google Scholar]
- Stuke A. Kunkel C. Golze D. Todorović M. Margraf J. T. Reuter K. Rinke P. Oberhofer H. Sci. Data. 2020;7:58. doi: 10.1038/s41597-020-0385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwilk M., Tahchieva D. N. and von Lilienfeld O. A., arXiv, 2020, preprint, arXiv:2004.10600, 10.48550/arXiv.2004.10600 [DOI]
- Lopez S. A. Pyzer-Knapp E. O. Simm G. N. Lutzow T. Li K. Seress L. R. Hachmann J. Aspuru-Guzik A. Sci. Data. 2016;3:160086. doi: 10.1038/sdata.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdematerials DB, https://www.verdematerialsdb.com/, accessed 9 May 2024
- Abreha B. G. Agarwal S. Foster I. Blaiszik B. Lopez S. A. J. Phys. Chem. Lett. 2019;10:6835–6841. doi: 10.1021/acs.jpclett.9b02577. [DOI] [PubMed] [Google Scholar]
- Ziogos O. G. Kubas A. Futera Z. Xie W. Elstner M. Blumberger J. J. Chem. Phys. 2021;155:234115. doi: 10.1063/5.0076010. [DOI] [PubMed] [Google Scholar]
- Balcells D. Skjelstad B. B. J. Chem. Inf. Model. 2020;60:6135–6146. doi: 10.1021/acs.jcim.0c01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneiding H. Lukin R. Lang L. Reine S. Pedersen T. B. Bin R. D. Balcells D. Digital Discovery. 2023;2:618–633. [Google Scholar]
- Golub P., Beran P., Antalik A. and Brabec J., arXiv, 2023, preprint, arXiv:2101.06090, 10.48550/arXiv.2101.06090 [DOI]
- Gugler S. Paul Janet J. Kulik H. J. Mol. Syst. Des. Eng. 2020;5:139–152. [Google Scholar]
- Duan C. Ladera A. J. Liu J. C.-L. Taylor M. G. Ariyarathna I. R. Kulik H. J. J. Chem. Theory Comput. 2022;18:4836–4845. doi: 10.1021/acs.jctc.2c00468. [DOI] [PubMed] [Google Scholar]
- Otlyotov A. A. Moshchenkov A. D. Cavallo L. Minenkov Y. Phys. Chem. Chem. Phys. 2022;24:17314–17322. doi: 10.1039/d2cp01659a. [DOI] [PubMed] [Google Scholar]
- Maurer L. R. Bursch M. Grimme S. Hansen A. J. Chem. Theory Comput. 2021;17:6134–6151. doi: 10.1021/acs.jctc.1c00659. [DOI] [PubMed] [Google Scholar]
- Dohm S. Hansen A. Steinmetz M. Grimme S. Checinski M. P. J. Chem. Theory Comput. 2018;14:2596–2608. doi: 10.1021/acs.jctc.7b01183. [DOI] [PubMed] [Google Scholar]
- The MolSSI QCArchive, https://qcarchive.molssi.org/, accessed 2 October 2024
- Smith D. G. A. Altarawy D. Burns L. A. Welborn M. Naden L. N. Ward L. Ellis S. Pritchard B. P. Crawford T. D. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2021;11:e1491. [Google Scholar]
- Gensch T. dos Passos Gomes G. Friederich P. Peters E. Gaudin T. Pollice R. Jorner K. Nigam A. Lindner-D’Addario M. Sigman M. S. Aspuru-Guzik A. J. Am. Chem. Soc. 2022;144:1205–1217. doi: 10.1021/jacs.1c09718. [DOI] [PubMed] [Google Scholar]
- Chen S.-S. Meyer Z. Jensen B. Kraus A. Lambert A. Ess D. H. J. Chem. Inf. Model. 2023;63:7412–7422. doi: 10.1021/acs.jcim.3c01310. [DOI] [PubMed] [Google Scholar]
- Kneiding H. Nova A. Balcells D. Nat. Comput. Sci. 2024;4:263–273. doi: 10.1038/s43588-024-00616-5. [DOI] [PubMed] [Google Scholar]
- Ruddigkeit L. van Deursen R. Blum L. C. Reymond J.-L. J. Chem. Inf. Model. 2012;52:2864–2875. doi: 10.1021/ci300415d. [DOI] [PubMed] [Google Scholar]
- Materials Project, MPContribs Documentation, https://docs.materialsproject.org/services/mpcontribs, accessed 10 October 2024
- Yamada H. Liu C. Wu S. Koyama Y. Ju S. Shiomi J. Morikawa J. Yoshida R. ACS Cent. Sci. 2019;5:1717–1730. doi: 10.1021/acscentsci.9b00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. J. Bardagot O. Banerji N. Adv. Theory Simul. 2022;5:2100511. [Google Scholar]
- Chen C. Zuo Y. Ye W. Li X. Ong S. P. Nat. Comput. Sci. 2021;1:46–53. doi: 10.1038/s43588-020-00002-x. [DOI] [PubMed] [Google Scholar]
- Fu G. Batchelor C. Dumontier M. Hastings J. Willighagen E. Bolton E. J. Cheminf. 2015;7:34. doi: 10.1186/s13321-015-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel A. M. Helm M. L. ACS Catal. 2014;4:630–633. [Google Scholar]
- Hastings J. Chepelev L. Willighagen E. Adams N. Steinbeck C. Dumontier M. PLoS One. 2011;6:e25513. doi: 10.1371/journal.pone.0025513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearnes S. M. Maser M. R. Wleklinski M. Kast A. Doyle A. G. Dreher S. D. Hawkins J. M. Jensen K. F. Coley C. W. J. Am. Chem. Soc. 2021;143:18820–18826. doi: 10.1021/jacs.1c09820. [DOI] [PubMed] [Google Scholar]
- Li H. Li Y. Jiao J. Lin C. Results Chem. 2023;5:100859. [Google Scholar]
- Dasari S. Tchounwou P. B. Eur. J. Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg B. Van Camp L. Krigas T. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- Bilodeau C. Jin W. Jaakkola T. Barzilay R. Jensen K. F. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2022;12:e1608. [Google Scholar]
- Ioannidis E. I. Gani T. Z. H. Kulik H. J. J. Comput. Chem. 2016;37:2106–2117. doi: 10.1002/jcc.24437. [DOI] [PubMed] [Google Scholar]
- Jin W., Barzilay R. and Jaakkola T., in Artificial Intelligence in Drug Discovery, ed. N. Brown, The Royal Society of Chemistry, 2020, pp. 228–249 [Google Scholar]
- Urbina F. Lowden C. T. Culberson J. C. Ekins S. ACS Omega. 2022;7:18699–18713. doi: 10.1021/acsomega.2c01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C., Sommer T., Kleuker F. and García-Melchor M., ChemRxiv, 2024, preprint, 10.26434/chemrxiv-2024-tljj9 [DOI]
- SMARTS – A Language for Describing Molecular Patterns, https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, accessed 21 March 2024
- Weininger D. J. Chem. Inf. Comput. Sci. 1988;28:31–36. doi: 10.1021/ci950169+. [DOI] [PubMed] [Google Scholar]
- Glendening E. D. Landis C. R. Weinhold F. Wiley Interdiscip. Rev.:Comput. Mol. Sci. 2012;2:1–42. [Google Scholar]
- Bartók A. P. Kondor R. Csányi G. Phys. Rev. B:Condens. Matter Mater. Phys. 2013;87:184115. [Google Scholar]
- Janet J. P. Kulik H. J. J. Phys. Chem. A. 2017;121:8939–8954. doi: 10.1021/acs.jpca.7b08750. [DOI] [PubMed] [Google Scholar]
- Morán-González L., Betten J. E., Kneiding H. and Balcells D., ChemRxiv, 2024, preprint, 10.26434/chemrxiv-2023-5wbkr-v2 [DOI] [PMC free article] [PubMed]
- Boldini D. Ballabio D. Consonni V. Todeschini R. Grisoni F. Sieber S. A. J. Cheminf. 2024;16:35. doi: 10.1186/s13321-024-00830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser P. Neubert M. Eberhard A. Torresi L. Zhou C. Shao C. Metni H. van Hoesel C. Schopmans H. Sommer T. Friederich P. Commun. Mater. 2022;3:1–18. doi: 10.1038/s43246-022-00315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen L. Jäger M. O. J. Morooka E. V. Federici Canova F. Ranawat Y. S. Gao D. Z. Rinke P. Foster A. S. Comput. Phys. Commun. 2020;247:106949. [Google Scholar]
- RDKit: Open-source cheminformatics, https://www.rdkit.org/, accessed 14 October 2024
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this manuscript as no datasets were generated or analysed in this perspective.