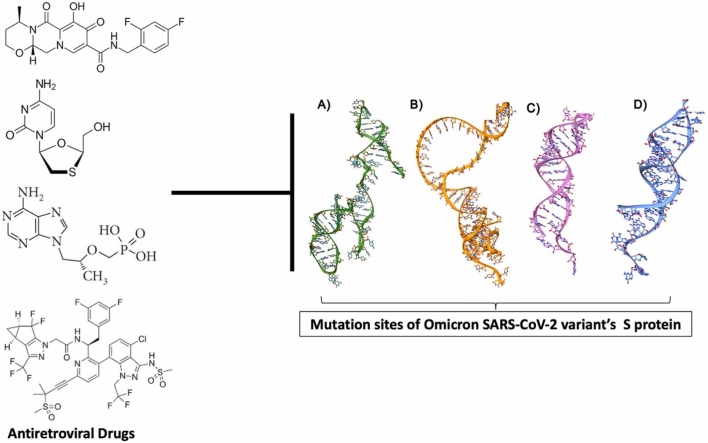
Abstract
The multiple mutation of the spike (S) protein of the Omicron SARS-CoV-2 variant is a major concern, as it has been implicated in the severity of COVID-19 and its complications. These mutations have been attributed to COVID-19-infected immune-compromised individuals, with HIV patients being suspected to top the list. The present study investigated the mutation of the S protein of the omicron variant in comparison to the Delta and Wuhan variants. It also investigated the molecular interactions of antiretroviral drugs (ARVd) vis-à-vis dolutegravir, lamivudine, tenofovir-disoproxilfumarate and lenacapavir with the initiation and termination codons of the mRNAs of the mutated proteins of the omicron variant using computational tools. The complete genome sequences of the respective S proteins for omicron (OM066778.1), Delta (OK091006.1) and Wuhan (NC 045512.2) SARS-CoV-2 variants were retrieved from the National Center for Biotechnology Information (NCBI) database. Evolutionary analysis revealed high trends of mutations in the S protein of the omicron SARS-CoV-2 variant compared to the delta and Wuhan variants coupled with 68 % homology. The sequences of the translation initiation sites (TISs), translation termination sites (TTSs), high mutation region-1 (HMR1) and high region mutation-2 (HMR2) mRNAs were retrieved from the full genome of the omicron variant S protein. Molecular docking analysis revealed strong molecular interactions of ARVd with TISs, TTSs, HMR1 and HMR2 of the S protein mRNA. These results indicate mutations in the S protein of the Omicron SARS-CoV-2 variant compared to the Delta and Wuhan variants. These mutation points may present new therapeutic targets for COVID-19.
Keywords: Antiretroviral, COVID-19, Omicron, And SARS-CoV-2
Graphical Abstract
Highlights
-
•
There were trends of mutations in the S protein of the omicron SARS-CoV-2 variant.
-
•
The studied ARVds exhibited adequate solubility, distribution and partition coefficient.
-
•
ARVds molecularly interacted with target sites of mutations.
1. Introduction
The emergence of the coronavirus disease 2019 (COVID-19) pandemic in 2019, with its origin from Wuhan, China has caused the world a major blow in the health sector, as it contributed to the devastating global mortality. The disease is a highly transmissible infection which is caused by the novel β-coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). It was first observed as a cluster of pneumonia in patients in Wuhan, China in December 2019 and subsequently in the world [7]. Over time, there were reports on the mutations of the Wuhan virus leading to the emergence of new variants of SARS-CoV-2 [14], [9]. These mutations and new variants coupled with their potential resistance to COVID-19 vaccines have further raised concerns about the treatment and management of the disease. Among the emerging variants are the delta (B.1.617.2), alpha (B.1.1.7), gamma (P.1), beta (B.1.351) and omicron (B.1.1.529) variants [1], [9]. This is apparently not unconnected with the current increase of COVID-19 cases and hospitalizations even among the fully vaccinated and boosted individuals.
Interestingly, the omicron variant ranks among the most mutated SARS-CoV-2 viruses, with the multiple mutations of its spike (S) protein being a major concern. The S protein enables the re-entry of SARS-CoV-2 into host cells, and it is a major target for neutralization by vaccines [11], [26]. Thus, changes in the genomic sequence of the S protein of the omicron variant arising from these mutations are a cause of concern as they allow the virus to evade antibodies, including vaccine-induced antibodies as well as increase transmissibility [11]. Since its discovery in South Africa in November 2021, the omicron variant has spread across the globe and has been ranked among the dominant variants [12], [19]. The rapid spread of the variant has however been attributed to the mobility of infected individuals across borders [30]. Certain COVID-19 treatments, like antiviral drugs, may focus on particular parts of the spike protein initially translated from the S gene. However, the mutations found in the S protein of the omicron variant might affect how well these drugs work. If the mutations appear in essential drug-binding regions, it could diminish the drugs' ability to effectively neutralize the virus. This emphasizes the importance of exploring alternative drugs that specifically target these mutated regions, aligning with the focus of our ongoing research [17].
Mutations in the omicron variant have been attributed to COVID-19-infected immune-compromised individuals, with HIV patients being suspected to top the list [23], [6]. South Africa accounts for the highest number of people living with HIV (PLHIV) in the world, with a prevalence rate of 19 % in people aged 15–49 [23]. Inconsistency in the usage of antiretroviral drugs (ARVds) leading to a suppressed immune system and viral resistance in PLHIV may be a contributing factor [21]. PLHIV have been documented for their high susceptibility to COVID-19 and its complications, as well as high rates of hospitalization and mortality [28]. Several studies have suggested the repurposing of ARDs for the treatment and management of COVID-19, especially in PLHIV [2], [20], [29].
Although the efficacy of these drugs in the management of the disease is controversial [18], [20], we hypothesize that selectively targeting the mutation points of the S protein of the omicron variant using ARVd may be a strategic therapeutic mechanism in the treatment and management of the disease. Thus, the present study was carried out to investigate the mutation trends of the S protein of the omicron variant in comparison to the Delta and Wuhan variants. The study also investigated the molecular interactions of selected ARVd vis-à-vis dolutegravir, lamivudine, tenofovir-disoproxilfumarate and lenacapavir with the initiation and termination codons of the mRNAs of the mutated proteins of the omicron variant using computational tools. Our findings highlighted the mutations in the S protein of the Omicron SARS-CoV-2 variant compared to that of the Delta and Wuhan variants; the pharmacokinetics and molecular interactions of the said drugs as well as their potential S protein translational suppression of mRNA to protein within and beyond the regulatory sites.
2. Materials and methods
2.1. Antiretroviral drugs
The ARVd investigated in the present study were dolutegravir, lamivudine, tenofovir-disoproxilfumarate and lenacapavir. These are current approved mainstream antiretroviral therapeutics.
2.2. Viral genome sequences
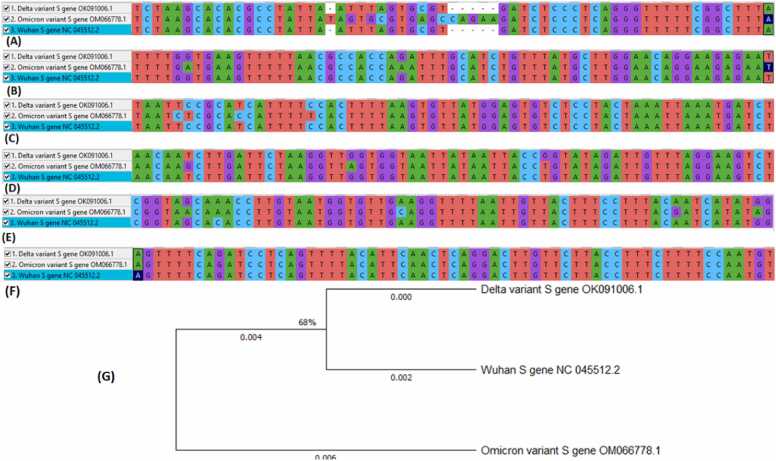
The complete genome sequences of the respective S proteins for omicron (OM066778.1), Delta (OK091006.1) and Wuhan (NC 045512.2) variants of SARS-CoV-2 were retrieved from the National Center for Biotechnology Information (NCBI) database and thereafter subjected to alignment using CLUSTALW X Software, version 10.1.8. as shown in Fig. S1 [14]. The aligned sequences were then used to plot a phylogenetic tree.
2.3. Evolutionary relationships of taxa
The evolutionary history was inferred using the neighbor-joining method [24]. The optimal tree is shown (next to the branches). The evolutionary distances were computed using the Maximum Composite Likelihood method [27] and are in the units of the number of base substitutions per site. The proportion of sites where at least 1 unambiguous base is present in at least 1 sequence for each descendent clade is shown next to each internal node in the tree. This analysis involved 3 nucleotide sequences. The codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair using the pairwise deletion option. There was a total of 3828 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [14].
2.4. Pharmacokinetic and drug-likeness studies
The pharmacokinetic and drug-likeness properties of the ARVd and standard compound were predicted using the ADMETlab 2.0 platform, which is a completely redesigned version of the widely used AMDETlab web server for the prediction of the drug-likeness, pharmacokinetics and toxicity properties of lead compounds (https://admetmesh.scbdd.com/) according to a previous description [33]. The conical SMILES of the ARVd were obtained from PubChem® (https://pubchem.ncbi.nlm.nih.gov/), while that of harpagide5-O-β-D-glucopyranoside was obtained from a previous study [8]. The prediction was conducted based on robust quantitative structure-property relationship (QSPR) modeling of a high-quality experimental ADMET dataset from numerous open-access bioactivity databases, such as ChEMBL, PubChem, OCHEM, peer-reviewed literature, and freely accessible software toxicity estimation software tools (TEST) developed by the U.S. Environmental Protection Agency by the multitask graph attention (MGA) framework. The server uses a total of 53 prediction models which include 40 classification and 13 regression models. Datasets from each endpoint were split into training, validation and test sets, and stratified sampling was used to maintain a balance between positive and negative instances as in the case of classification models. The larger part of the datasets was used as the training set, and the validation and test sets were used to optimize the hyperparameters and test the predictive capacity of each model, respectively.
2.5. Molecular modeling studies on the TIS and TTS of the omicron S protein
Molecular modelling studies were conducted to gain deeper insights into the interacting pattern of the various ARVd on SARS-CoV-2 omicron variant S mRNA sites. The sequences of the translation initiation sites (TISs), translation termination sites (TTSs), high mutation region-1 (HMR1) and high region mutation-2 (HMR2) were retrieved from the full genome of the omicron variant S protein from the experiments presented in this study, as shown in Table 1.
Table 1.
Selected sequence of initiation, termination, and high mutation region sites of target protein mRNAs from SARS-CoV-2 whole genome.
| Sites | Sequences |
|---|---|
| TIS | 5'-AUGUUACUUGGUUCCAUGUUAUCUCUGGGACCAAUGGUACUAAG−3′ |
| TTS | 5'-ACUGUGAUGUUGUAAUAGGAAUUGUCAACAACACAGUUUAUGA−3′ |
| High mutation region 1 | 5'- AACAAACCUUGUAAUGGUGUUGCAGGUUUUAAUUGUUACUUUCCUUUACGAUCAUAUAGUUUCCGACCCACUUAUGGUGUUGGUCA−3′ |
| High mutation region 2 | 5'- AGAUGUGGUCAACCAUAAUGCACAAGCUUUAAACACGCUUGUUAAACAACUUAGCUCCAAAUUUGGUGCAAUUUCAAGUGUUUUAAAUGAUAUCUU-−3′ |
The sequences of TIS, TTS, HMR1 and HMR2 mRNAs were initially converted to 2D configurations using the RNA Fold webserver with the default input parameters [http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi] according to a previous protocol [8]. The retrieved 2D mRNA structures were then converted to single-stranded 3D mRNA structures employing the RNA Composer webserver [3]. The structures of dolutegravir, lamivudine, tenofovir, disoproxilfumarate, and lenacapavir were obtained from PubChem® [13]. The novel compound harpagide-5-O-β-D-glucopyranoside was built using a 2D-sketcher tool in Maestro software, part of the Schrodinger suite. Molecular docking studies were performed using Atuodock4 (AD4) along with AutoDockTools GUI [22]. The 3D mRNA structures were then prepared and converted from PDB to PDBQT formats using scripts such as prepare_ligand4.py and prepare_receptor4.py, part of AutoDockTools. The grid box was parameterized in such a way that the whole 3D mRNA structure could be accommodated as its binding site is unknown. For TIS, a set of grids with 110 Å × 64 Å × 80 Å along with its grid centers (23.675, −9.743, −10.522) at x, y, z coordinates with a spacing constraint of 0.667 Å. For TTS, a different set of grids with 92 Å × 98 Å × 110 Å with grid centers (-6.26, −10.933, −1.754) on their x, y, z coordinates and spacing of 0.667 Å. Whereas RM1, a set of grids 52 Å × 114 Å × 72 Å with grid centers (-0.382, 0.473, 12.039) with spacing constraint of 1.000 Å and RM2 with grids 82 Å × 114 Å × 90 Å and grid centers (-10.35, −0.935, 17.42) along with its spacing constraint of 1.000 Å using an AutoGrid4 element. For each ligand against all other mRNAs, a set of 100 independent dockings were run. Each independent docking consists of 20 million energy evaluations using the Lamarckian Genetic Algorithm Local Search (GALS) method for the purpose of the conformational search. The docking poses of all the ligands were clustered based on the root mean square deviation (rmsd) of 2 Å. Additionally, visual inspections and the scoring of the binding poses was calculated based on the binding free energy (ΔGAD4).
3. Results and discussion
3.1. Phylogenetic and evolutionary relationships of Omicron, Delta, and Wuhan variants
The analysis of the evolutionary relationship of complete nucleotide sequences of the S gene for the Omicron (OM066778.1), Delta (OK091006.1) and Wuhan (NC_045512.2) variants of SARS-CoV-2 indicates trends of mutations in the S gene of the SARS-CoV-2 omicron variant in comparison to the Delta and Wuhan variants. The phylogenetic tree clustered the SARS-CoV-2 omicron variant distantly from other variants (SARS-CoV-2 Delta and SARS-CoV-2 Wuhan) with merely 68 % homology. In addition, sequence multiple alignments of the S protein showed that the SARS-CoV-2 omicron variant has multiple mutations compared to other variants. Basically, twentyfour (24) transitions/transversions point mutations and deletion types of mutations were observed at different regions (A, B, C, D, and E), as shown in Fig. 1. Fascinatingly and relative to Delta and Wuhan variants, two higher regions (1326–1411 and 2780–28870) of mutation were equally observed and subsequently selected for molecular modelling studies. However, a conserved region was also observed in the S protein at the F region (Fig. 1F).
Fig. 1.
The regions of transitions/transversions point mutations and deletion types (A) 1, (B) 2, (C) 3, (D) 4, (E) 5 and (F) conserved S protein regions of SARS-CoV-2 omicron variant compared to delta and Wuhan variants; (G) Phylogenetic tree depicting the evolutionary relationships between the S protein of the studied SARS-CoV-2 variants.
As SARS-CoV-2 undergoes at least two genomic changes in a month, new SARS-CoV-2 variants are continuously and rapidly emerging [31], [32], [5]. Compared to SARS-CoV-2 Delta and Wuhan variants, the Omicron variant of the SARS-CoV-2 virus has the greatest S gene mutations (Fig. 1). This is consistent with a prior work [15], which found that the Omicron’s S gene had more mutations in the receptor binding domain than the Delta variant. Equally concurrent findings also validate the fact that the omicron variant strains encode large numbers of changes in the spike protein relative to initial SARS-CoV-2 isolates [4]. As a result, the research suggested that the Omicron variant may be involved in immune resistance to antibody-mediated protection. However, due to multiple mutations in the S gene region, the omicron variant may be employed as a potential therapeutic candidate for the development of antiretroviral drugs. Evidently, S protein plays a vital role in coronavirus entry into the host cell by binding to the ACE2 host receptor in the cell membrane and gaining entry through the endocytosis pathway [10]. Thus, the S protein and its RBD constitute a fascinating and captivating target for antiviral research for viral-based treatment possibilities. The SARS-CoV-2 virus relies on the S protein to re-enter host cells, making it a crucial target for neutralization by vaccines [11], [26]. However, our research has taken a different approach by investigating the potential of certain drugs to not only target the HMR1 and 2 regions within the coding sequences but also suppress the translation of the S gene into the S protein (both TIS and TTS). Based on our findings, we speculate that these drugs, if further explored, could offer benefits through translational suppression of the S gene to S protein in individuals infected with the omicron variant.
3.2. Pharmacokinetic and drug-likeness studies
Several molecular descriptors have been reported with utmost relevance in the prediction of druggable character through numerical values comparison and physical/chemical behavior of a molecule. These descriptors have been proven useful in performing similarity searches in molecular libraries or datasets based on physical and chemical properties and molecular structures.
The physicochemical properties of the ARVd indicated that most of the drugs exhibit adequate solubility based on solubility, distribution and partition coefficient (Fig. S1). Lower solubility was observed in lenacapavir, a lower partition and distribution coefficient in lamivudine and harpagide-5-O-β-D-glucopyranoside, and a moderately lower partition and distribution coefficient in tenofovir and disoproxilfumarate. Lenacapavir showed a higher violation of all physicochemical properties with the exception of a number of hydrogen acceptors and donors, formal charges and number of atoms in the largest ring, while no violation was observed in dolugravir and lamivudine with the exception of the partition and distribution coefficient mentioned earlier. The absorption of the compounds by human intestinal and human epithelial colorectal adenocarcinoma cells and Madin-Darby canine kidney human cell-lineabsorption models indicated very good absorption and bioavailability with respect to all the compounds apart from harpagide 5-O-β-D-glucopyranoside in the human intestine and epithelial colorectal adenocarcinoma cells, respectively (Table 2). The prediction also showed that all the compounds can be efficiently distributed in the body, with a lower unbound fraction and high plasma binding observed in dolutegravir and lamivudine. Lamivudine was predicted to be a potential substrate of CYP1A2, dolutegravir as a CYP2C9 substrate and lenacapavir as a substrate of CYP1A2, CYP2C9, CYP3A4 and an inhibitor of CYP2C19, CYP2C9, CYP2D6 and CYP3A4. The compounds further indicated a low and moderate clearance rate, as well as a very short half-life.
Table 2.
Predicted pharmacokinetic properties of studied antiretroviral drugs and harpagide 5-O-β-D-glucopyranoside.
| Category | Property (unit) | Inference / Reference Range | |||||
|---|---|---|---|---|---|---|---|
| DTG | LVD | TDF | LCP | HGG | |||
| Absorption | Caco−2 permeability (cm/s) | −4.692 | −5.52 | −5.538 | −5.792 | −6.334 | Optimal: higher than −5.15 or −4.70 |
| MDCK permeability | 1.3e−05 | 1.35e−4 | 1.76e−4 | 2.2e−5 | 4.33e−4 | low permeability: < 2 × 10–6 cm/s medium permeability: 2–20 × 10–6 cm/s high passive permeability: > 20 × 10–6 cm/s |
|
| Pgp-inhibitor | 0.008 | 0.002 | 0.0 | 1.0 | 0.0 | +ve value inhibitor and –ve non inhibitor | |
| Pgp-substrate | 0.053 | 0.003 | 0.146 | 0.001 | 0.971 | +ve value substrate and –ve non substrate | |
| F20 % | 0.097 | 0.769 | 0.116 | 0.004 | 0.995 | +ve value bioavailability < 20 % and –ve value ≥ 20 % | |
| F30 % | 0.084 | 0.24 | 0.891 | 0.002 | 1.0 | +ve value bioavailability < 30 % and –ve value ≥ 30 % | |
| HIA (Human Intestinal Absorption) (%) | 0.008 | 0.064 | 0.002 | 0.082 | 0.549 | +ve value HIA > 30 % and –ve value HIA < 30 % | |
| Distribution | PPB (Plasma protein binding) (%) | 93.66 % | 14.08 | 6.066 % | 101.5 % | 12.23 | Optimal: < 90 %. |
| Fu | 2.840 % | 80.84 | 78.70 % | 0.807 % | 56.21 | Low: <5 %; Middle: 5–20 %; High: > 20 % | |
| VD | 1.089 | 0.823 | 0.918 | 0.643 | 0.216 | Optimal: 0.04–20 L/kg | |
| BBB (Blood brain barrier) (%) | 0.293 | 0.636 | 0.171 | 0.156 | 0.35 | +ve value BBB positive and –ve value BBB negative. | |
| Metabolism | CYP1A2-inhibitor | 0.034 | 0.028 | 0.033 | 0.063 | 0.0 | >0.5: An inhibitor |
| CYP1A2-substrate | 0.135 | 0.778 | 0.129 | 0.556 | 0.034 | >0.5: Substrate | |
| CYP2C19-inhibitor | 0.245 | 0.069 | 0.096 | 0.597 | 0.002 | >0.5: An inhibitor | |
| CYP2C19-substrate | 0.168 | 0.077 | 0.053 | 0.18 | 0.087 | >0.5: Substrate | |
| CYP2C9-inhibitor | 0.452 | 0.006 | 0.048 | 0.961 | 0.0 | >0.5: An inhibitor | |
| CYP2C9-substrate | 0.89 | 0.156 | 0.045 | 0.932 | 0.063 | >0.5: Substrate | |
| CYP2D6-inhibitor | 0.027 | 0.011 | 0.062 | 0.624 | 0.0 | >0.5: An inhibitor | |
| CYP2D6-substrate | 0.197 | 0.25 | 0.036 | 0.137 | 0.072 | >0.5: Substrate | |
| CYP3A4-inhibitor | 0.079 | 0.006 | 0.316 | 0.982 | 0.011 | >0.5: An inhibitor | |
| CYP3A4-substrate | 0.119 | 0.095 | 0.289 | 0.934 | 0.002 | >0.5: Substrate | |
| Excretion | Clearance (mL/min/kg) | 2.721 | 4.71 | 5.251 | 5.458 | 0.951 | High: >15 mL/min/kg; moderate: 5–15 mL/min/kg; low: <5 mL/min/kg |
| T1/2 (Half life) (H) | 0.109 | 0.858 | 0.897 | 0.001 | 0.449 | long half-life: >3 h; short half-life: <3 h | |
| Toxicity | hERG (hERG blockers) | 0.031 | 0.007 | 0.005 | 0.262 | 0.049 | >0.5: Active <0.5: Non-active |
| H-HT (Human Hepatotoxicity) | 0.98 | 0.959 | 0.991 | 0.999 | 0.158 | >0.5: HHT positive <0.5: HHT negative | |
| DILI | 0.918 | 0.991 | 0.979 | 0.989 | 0.076 | >0.5 high risk, <0.5 no risk | |
| FDAMDD | 0.888 | 0.014 | 0.337 | 0.973 | 0.039 | +ve value FDAMDD (+), -ve value FDAMDD (-) | |
| Carcinogenicity | 0.154 | 0.84 | 0.256 | 0.435 | 0.751 | >0.5 carcinogens; <0.5 non-carcinogens | |
| Respiratory toxicity | 0.151 | 0.018 | 0.947 | 0.639 | 0.792 | >0.5: toxicants; <0.5: nontoxicants | |
| Acute toxicity rule | 0 alerts | 0 alerts | 0 alerts | 0 alerts | 0 alerts | 20 substructures | |
| Genotoxic carcinogenicity rule | 0 alerts | 0 alerts | 1alerts (A) | 0 alerts | 2 alerts (F$G) | 117 substructures | |
| Non-genotoxic carcinogenicity rule | 0 alerts | 0 alerts | 0 alerts | 1 alerts (C) | 0 alerts | 23 substructures | |
| SureChEMBL Rule | 0 alerts | 0 alerts | 1alerts (B) | 1 alerts (D) | 0 alerts | 164 substructures | |
| AMES (Ames mutagenicity) | 0.046 | 0.038 | 0.891 | 0.424 | 0.093 | >0.5: Positive <0.5: Negative |
|
DTG = Dolutegravir, LVD = Lamivudine, TDF = Tenofovir-disoproxil fumarate, LCP = Lenacapavir, HGG = Harpagide 5-O-β-D-glucopyranoside, DILI = Drug Induced Liver Injury, FDAMDD = FDA Maximum Recommended Daily Dose, hERG = Ppg = P-glycoprotein, MDCK = Madin-Darby canine kidney human cell-line, VD = Distribution volume, Fu = Fraction unbound, CL = Clearance, H-HT = Human Hepatotoxicity.
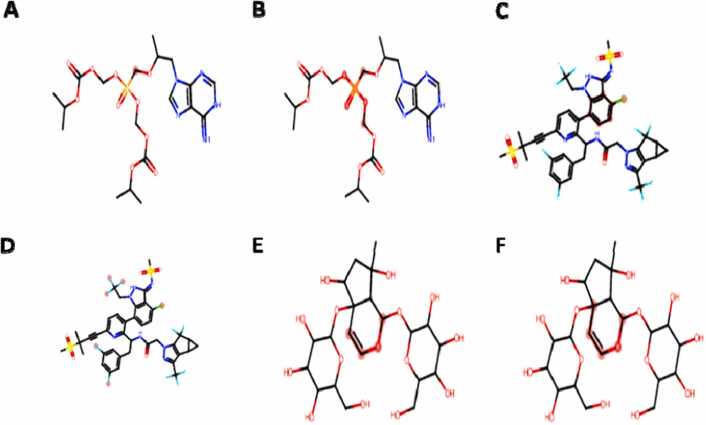
Hepatotoxicity was predicted in all the compounds with the exception of harpagide 5-O-β-D-glucopyranoside, carcinogenicity in lamivudine and harpagide 5-O-β-D-glucopyranoside, respiratory toxicity in lenacapavir, harpagide 5-O-β-D-glucopyranoside and tenofovir-disoproxilfumarate. One genotoxic alert in tenofovir-disoproxilfumarate, harpagide 5-O-β-D-glucopyranoside and a nongenotoxic status in lenacapavir were observed (Fig. 2).
Fig. 2.
Geometric clash of some undesirable substructures based on ADMET prediction.
Clash atoms indicated in oily red dots, A = Tenofovir-disoproxil fumarate genotoxic carcinogencity, B = Tenofovir-disoproxil fumarate medicinal chemistry status, C = Lenacapavir non-genotoxic carcinogenicity, D = Lenacapavir medicinal chemistry status, E&F = Harpagide 5-O-β-D-glucopyranoside genotoxic carcinogenicity.
Ames mutagenicity prediction indicated that only tenofovir-disoproxilfumarate had mutagenic potential. Drug likeness prediction shows that only dolutegravir and lamivudine have an attractive quantitative estimate of drug likeness value, while all the compounds exhibited a low synthetic accessibility score (Table 3). The synthetic accessibility score is formulated to evaluate the drug-like characteristics of compounds for virtual screening purposes, and it is calculated as a sum of fragment scores and complexity penalty [25].
Table 3.
Predicted medicinal chemistry and drug-likeness properties of studied antiretroviral drugs and harpagide 5-O-β-D-glucopyranoside.
| Properties | Dolutegravir | Lamivudine | Tenofovir-disoproxil fumarate | Lenacapavir | Harpagide 5-O-β-D-glucopyranoside | Inference / Reference Range |
|---|---|---|---|---|---|---|
| Predicted Scores/Comment | ||||||
| QED | 0.782 | 0.621 | 0.224 | 0.112 | 0.155 | > 0.67 (Attractive) 0.49–0.67 (unattractive) < 0.34 (too complex) |
| SAscore | 3.622 | 4.401 | 4.198 | 5.609 | 5.454 |
≥6 difficult, <6, easy |
| Fsp3 | 0.35 | 0.5 | 0.632 | 0.385 | 0.905 | ≥0.42 suitable |
| MCE−18 | 84.259 | 38.5 | 36 | 185.741 | 86.8 | ≥45 suitable |
| NPscore | −0.925 | 1.245 | −0.195 | −0.773 | 2.103 | −5–5 |
| Lipinkis Rule | Accepted | Accepted | Rejected | Rejected | Rejected | MW ≤500; logP ≤5; Hacc ≤10; Hdon ≤5 |
| Pfizer Rule | Accepted | Accepted | Accepted | Accepted | Accepted | logP > 3; TPSA < 75 |
| GSK Rule | Rejected | Accepted | Rejected | Rejected | Rejected | MW ≥400; logP ≥4 |
| Golden triangle | Accepted | Accepted | Rejected | Rejected | Rejected | 200 ≤MW ≤50; −2 ≤logD ≤5 |
| PAINS | 0 alerts | 0 alerts | 0 alerts | 0 alerts | 0 alerts | Pan Assay Interference Compounds |
| ALARM NMR rule | 0 alerts | 2 alerts (A&B) | 0 alerts | 2 alerts (D&E) | 0 alerts | Thiol reactive compounds. |
| BMS Rule | 0 alerts | 0 alerts | 1 alerts (C) | 0 alerts | 1 alerts (F) | Undesirable and reactive compounds |
| Chelator Rule | 0 alerts | 0 alerts | 0 alerts | 0 alerts | 0 alerts | Chelating compounds |
QED = Quantitative Estimate of Druglikeness, Fsp3 = number of sp3 hybridized carbons / total carbon count, PAINS = Pan Assay Interference Compounds SAscore = Synthetic accessibility score, MCE = medicinal chemistry evolution, NPscore = Natural product-likeness score.
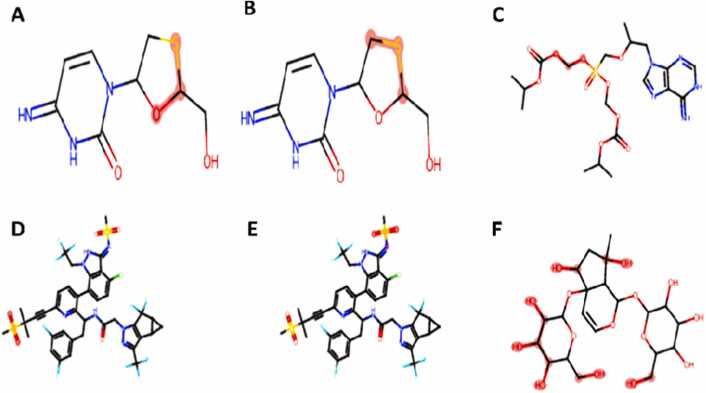
All the compounds were accepted by Pfizer's rule of druggable properties; only dolutegravir and lamivudine were accepted by Lipinkis rule, and golden triangle and lamivudine were accepted by GSK rule. Two reactive thiol alerts were recorded in lamivudine and lenacapavir, one undesirable reactive conformer in tenofovir-disoproxilfumarate and harpagide 5-O-β-D-glucopyranoside, respectively (Fig. 3).
Fig. 3.
Geometric clash on some functional groups based on Medicinal Chemistry and Drug Likeness properties. Clash atoms indicated in oily red dots, A&B = Lamivudine reactive thiol, C = Tenofovir-disoproxil fumarate undesirable reactive conformer, D&E = Lenacapavir reactive thiol, F = Harpagide 5-O-β-D-glucopyranoside undesirable reactive conformer.
Biotransformation of drugs in the body involves both activation to a pharmacologically active form or deactivation to the inactive form. One of the most influential enzymes in the metabolism of foreign substances is cytochrome P450, and stimulation of these enzymes is less likely to increase drug interactions through the formation of an active metabolite, which could lead to more efficiency (Kamel and Harriman, 2013). The compounds examined in this study have indicated a differential mode of interaction with various CYP450 enzymes. Smaller-sized compounds can be easily removed from the body through urine; therefore, molecular weight, including other factors such as solubility, lipophilicity and protein binding capacity, determines the drug removal mechanism [16]. Effective drugs must maintain a steady-state concentration within a therapeutic concentration, which is determined by several factors, such as clearance, volume of distribution, half-life, and bioavailability of a drug. The compounds in this study were predicted to have a lower clearance rate and short half-life. A drug can reach a steady-state concentration after several doses since many drugs are eliminated in an exponential decay with first-order kinetics. Most of the molecules examined in this study indicated potential toxicity to hepatocytes, except for harpagide 5-O-β-D-glucopyranoside and some traces of genotoxic and nongenotoxic carcinogenicity, respiratory toxicity, and mutagenicity. Functional groups in a drug molecule assume several conformational orientations that can interact best with target proteins. Out of 112 substructures, one conformer of tenofovir-disoproxilfumarate and two harpagide 5-O-β-D-glucopyranoside indicated genotoxic carcinogenicity potentials, and one conformer of lenacapavir out of 23 and 164 substructures indicated nongenotoxic carcinogenicity potential and an unfriendly drug likeness status. This may be attribute for the observed flexible scaffolds and indicates a potential to interact with other off-target receptors. Thus, cause side effects or toxicity.
3.3. Molecular modeling studies
To obtain in-depth insights into the various interaction patterns and the binding modes of the small molecules (ARVds) considered in this study against the translation initiation site (TIS), translation termination site (TTS), HMR1 and HMR2 mRNAs of the omicron variant S protein. In molecular modelling studies, to further perform molecular dockings, the 3D structures of all the mRNAs such as TIS TTS, HMR1 and HMR2 were modelled using RNA Fold and RNA Composer as shown in Fig. 4A – 4D. The structures were further visualized using the molecular visualization software, UCSF Chimera. The binding energies of the molecular docking results are shown in Table 4.
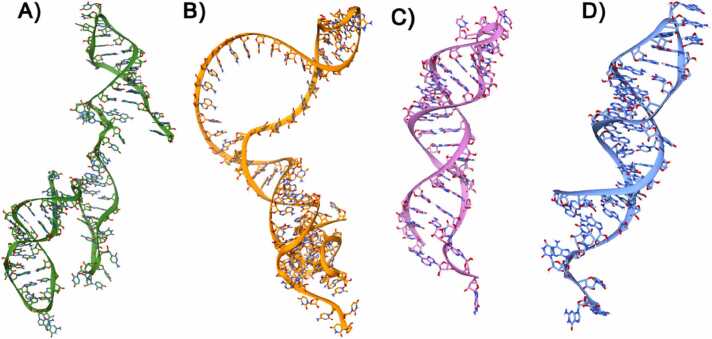
Fig. 4.
A)Modeled 3D-structure of HMR-1 depicted in Ribbons and Sticks and colored in green, B)Modeled 3D-structure of HMR-2 depicted in Ribbons and Sticks and colored in orange, C)Modeled 3D-structure of TIS depicted in Ribbons and Sticks and colored in pink, D)Modeled 3D-structure of TTS depicted in Ribbons and Sticks and colored in blue.
Table 4.
Binding energies of the molecular interactions of studied AVRd and harpagide 5-O-β-D-glucopyranoside with HMR-1, HMR-2, TIS and TTS mRNAs of the S protein of the omicron variant of SARS-CoV-2.
| Compound/AVRd |
HMR-1 |
HMR-2 |
TIS |
TTS |
|---|---|---|---|---|
| Binding Energy (kcal/mol) | ||||
| Dolutegravir | −5.62 | −5.30 | −6.24 | −6.24 |
| Harpagide 5-O-β-D-glucopyranoside | −0.45 | −0.34 | −1.68 | −2.89 |
| Lamivudine | −3.87 | −3.28 | −5.18 | −4.46 |
| Lenacapavir | −2.91 | −2.86 | −4.63 | −5.13 |
| Tenofovir-disoproxil fumarate | −1.55 | −0.83 | −2.92 | −2.19 |
TIS: translation initiation site; TTS: translation termination site; HMR1: high mutation region-1; and HMR2: high region mutation-2 mRNAs.
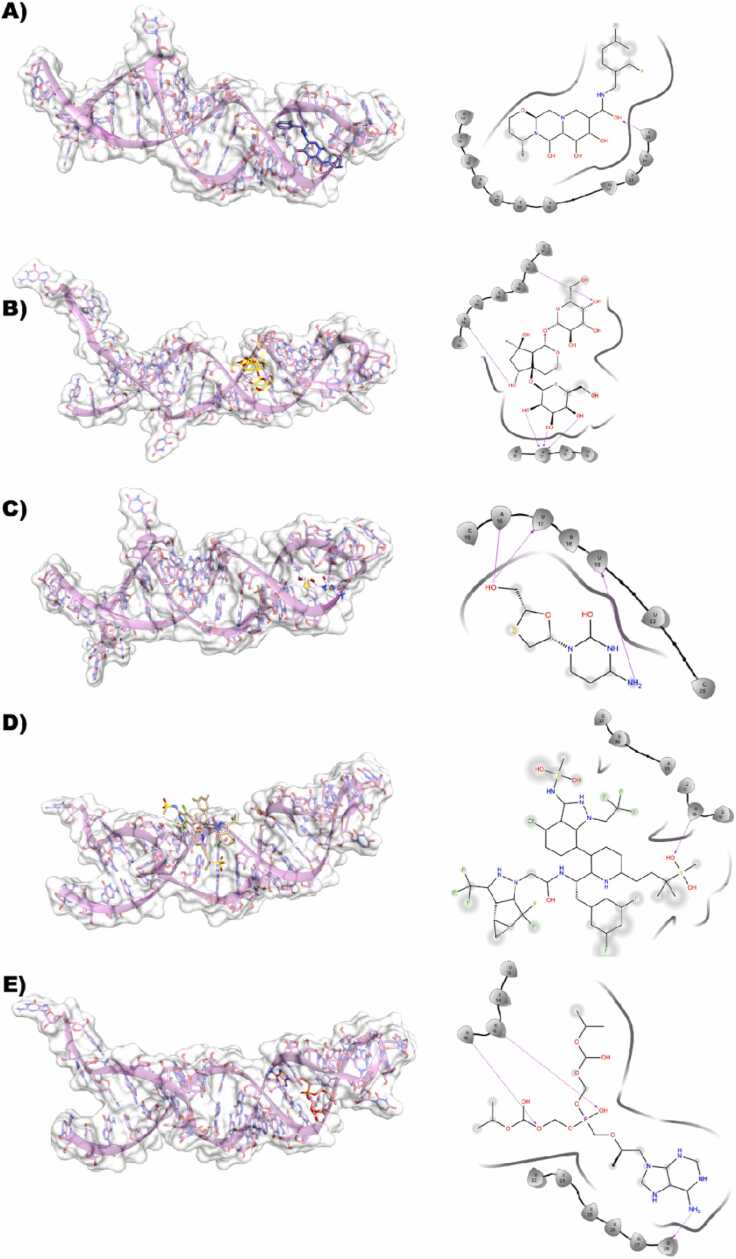
3.3.1. HMR1-dolutegravir
This particular binding site where Dolutegravir is bound seems to be almost equally distributed with A, C, G, and U nucleotides. In the lowest energy docking pose of Dolutegravir against HRM-1, it is evident that most of the ligand is solvent-exposed, whereas the parts of the ligands such as the multiple OH from 9,12-diox attached to the diazatricyclo ring structure accept an H-bond with the side chain NH of G76 and the side chain NH of G77 (Fig. 4A).
3.3.2. HMR1- harpagide 5-O-β-D-glucopyranoside
This binding site is heavily dominated by the C and U nucleotides, except for one nucleotide while the others are C and U. The compound, harpagide 5-O-β-D-glucopyranoside primarily consists of only hydroxyl groups attached to the ring structure. Multiple H-bonds are formed between C39, which is donating an H-bond to the OH of the R-hydroxyl group attached to the 7-methyl group. The methyl OH attached to the cycle pentane group is seen to be donating an H-bond to the backbone oxygen of U42, and the OH of 3,4,5 triol is also donating an H-bond to the backbone oxygen of U45 (Fig. 4B)
3.3.3. HMR1- lamivudine
This binding site is dominated by the G nucleotide; lamivudine is bound tightly to HMR1 by forming various H-bonds such as the 4-amino group donating an H-bond to the backbone O of G79, and the methyl OH attached to the oxothiolane group is involved in multiple H-bonds donating an H-bond to the backbone O of G77 and accepting an H-bond from the side chain NH of G76 (Fig. 4C)
3.3.4. HMR1- lenacapavir
This binding site is lined with nucleotides such as U36, U37, A38, C39, U47, A48 and C49. The lenacapavir-bound state is mostly solvent-exposed, and it is hardly seen to be in the vicinity of having a feasible H-bond interaction with any of the nucleotides present in the binding site of HMR1 (Fig. 4D)
3.3.5. HMR1- tenofovir
The binding site of HMR1 with tenofovir is primarily lined by the A, U and C nucleotides. Most of the part in tenofovir is seen to be solvent-exposed, but it is still involved in several H-bond interactions with HMR1 such as the OH attached to the oxo-dimethyl group accepting a H-bond from the NH side chain of C49 and the adjacent O atom accepting a H-bond from the side chain NH of A48. Additionally, the NH attached to the purine is involved in double H-bonds by donating H-atoms to the side chain O of U46 and the side chain O of A31 (Fig. 4E)
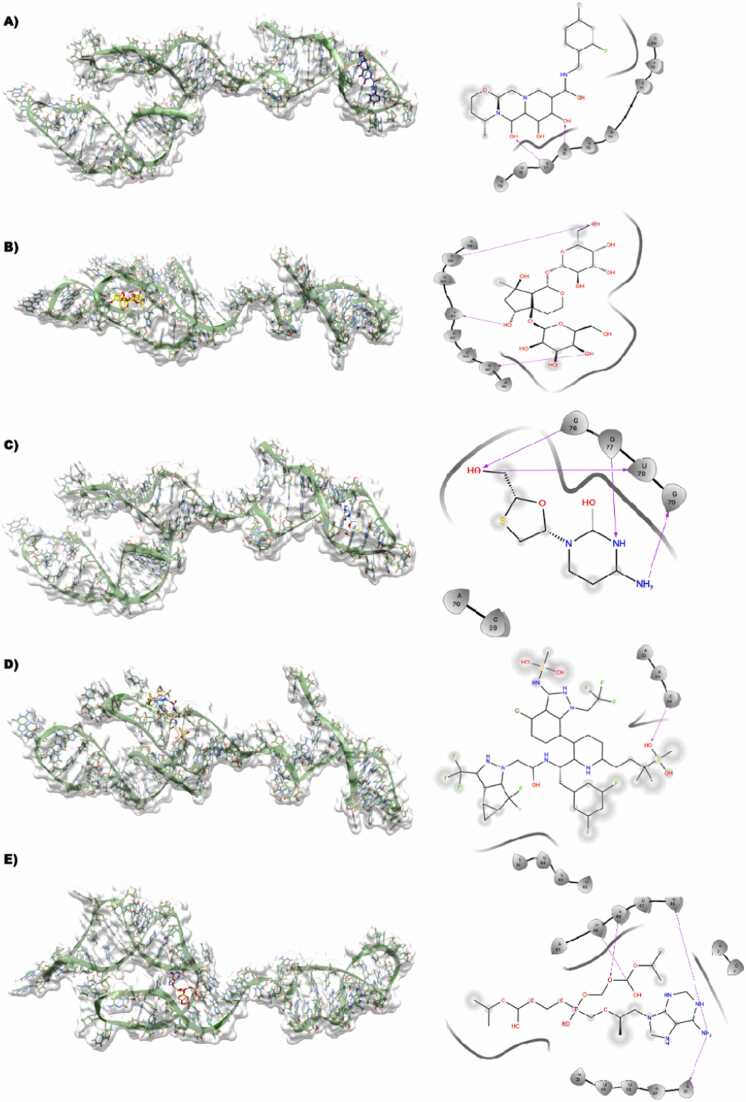
3.3.6. HMR2-dolutegravir
In the binding site of HMR1 with bound Dolutegravir equally distributed with A and U. In the lowest energy docking pose of Dolutegravir against HRM-1, it is clear that the 2,4-difuorophenyl methyl part of the ligand is solvent-exposed, whereas the parts of the ligands are well surrounded by the various nucleotides of HMR2. The carboxamide NH donates an H-bond with the side chain OH of U84 (Fig. 5A)
Fig. 5.
A,B,C,D,E Dolutegravir (dark blue), Harpagide 5-O-β-D-glucopyranoside (yellow), Lamivudine (white), Lenacapavir (tan), Tenofovir-disoproxil fumarate (Salmon) docked against modeled HMR1-mRNA(green) is represented along with its interactions shown in 2D-ligplot.
3.3.7. HMR2- harpagide 5-O-β-D-glucopyranoside
The compound harpagide 5-O-β-D-glucopyranoside with only hydroxy groups could be seen sharing multiple H-bonds formed between C39 that is donating an H-bond to the OH of the R-hydroxyl group attached to the 7-methyl, the methyl OH attached to the cycle pentane group is seen to be donating an H-bond to the backbone oxygen of U42 and the Oh of 3,4,5 triol is also donating an H-bond to the backbone oxygen of U45 (Fig. 5B)
3.3.8. HMR2- lamivudine
In comparison to HMR1, this binding site is equally distributed by the A, U, and G nucleotides. Lamivudine is bound tightly to HMR1 by forming an H-bond, such as the 4-amino group donating an H-bond to the backbone O of U64 (Fig. 5C)
3.3.9. HMR2- lenacapavir
This binding site is aligned with nucleotides such as A, U and C, with A and U being the most abundant. The lenacapavir-bound state is mostly solvent-exposed and accepts a feasible H-bond interaction between the O group present in the sulfonyl group and the side chain NH of C34, while most of the region apart from the core region of the molecule is solvent-exposed (Fig. 5D)
3.3.10. HMR2- tenofovir
The binding site of HMR2 with tenofovir is primarily lined by the A, U and C nucleotides. Most of the part in tenofovir is seen to be solvent-exposed but is also involved in several H-bond interactions with HMR2, such as the OH attached to the oxo-dimethyl group accepting an H-bond from the NH side chain of A22, and the O atom in the phosphonyl acid group also accepting an H-bond from the side chain NH of the same A24 nucleotide (Fig. 5E)
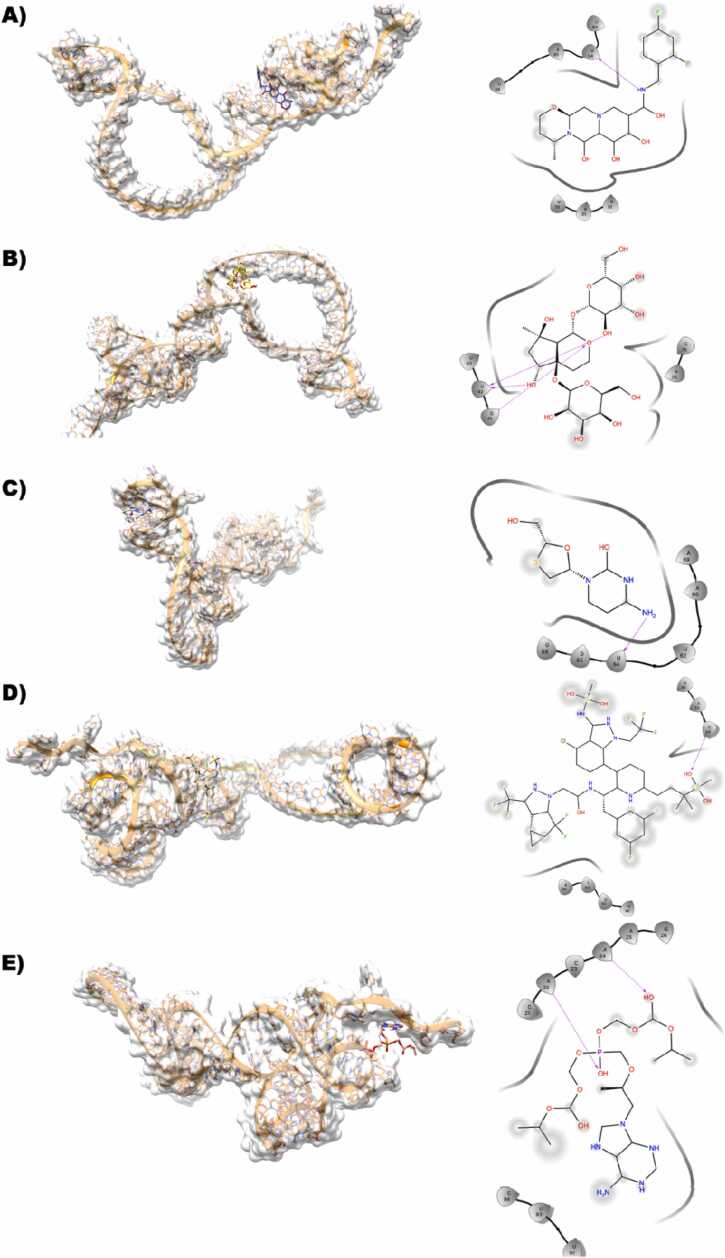
3.3.11. TIS-dolutegravir
This binding site of the TIS and dolutegravir complex seems to be almost equally distributed with C and U. It is evident that the 2,4-difluorophenyl ring is solvent-exposed, whereas the parts of the ligand are well-defined in the binding site. OH from 9,12-diox attached to the diazatricyclo ring structure accepts an H-bond with the side chain NH of C25 (Fig. 6A).
Fig. 6.
A,B,C,D,E Dolutegravir (dark blue), Harpagide 5-O-β-D-glucopyranoside (yellow), Lamivudine (white), Lenacapavir (tan), Tenofovir-disoproxil fumarate (Salmon) docked against modeled HMR2-mRNA(orange) is represented along with its interactions shown in 2D-ligplot.
3.3.12. TIS-harpagide 5-O-β-D-glucopyranoside
In the binding site of TIS, nucleotide G11 plays a very important role. However, harpagide 5-O-β-D-glucopyranoside primarily consists of only hydroxy groups attached to the ring structure. There are multiple H-bonds formed between G11 that accept triple H-bonds from the various OH groups of the ligand, and there are also some H-bonds that are shared between the ligand and the U26 and A30 side chains and main chains (Fig. 6B).
3.3.13. TIS- lenacapavir
This binding site is dominated by the G nucleotide, and lamivudine is bound tightly to the TIS by forming an H-bond, such as the O present in the sulfonyl group, which accepts an H-bond from the side chain NH of the G10 nucleotide. As the ligand is quite large in size, the ligand is seen to be quite solvent-exposed. (Fig. 6C).
3.3.14. TIS- lamivudine
This binding site has all the nucleotides such as A, U, G, and C. The lamivudine is bound tightly to the TIS by forming various H-bonds, such as the 4-amino group donating an H-bond to the sidechain O of U19, and the hydroxy methyl group is involved in multiple H-bonds by donating an H-bond to the U17 and accepting an H-bond from the side chain A16 (Fig. 6D).
3.3.15. TIS- tenofovir
The binding site of TIS with tenofovir is primarily lined by the A, U, G and C nucleotides. In this binding, it is seen that only a partial part in tenofovir is seen to be solvent-exposed but still it is involved in several H-bond interactions with TIS such as the phosphonyl O is accepting an H-bond from the NH side chain of C15 and the consecutive nucleotide A16 is also donating an H-bond. Additionally, the NH attached to the purine is involved in double H-bonds by donating H-atoms to the backbone O of G28 (Fig. 6E).
3.3.16. TTS-dolutegravir
This binding site complex seems to be almost equally distributed with A, U, G, and C. It is evident that the compound is heavily solvent-exposed and is not in a very close vicinity to be involved in any H-bond interaction with any of the nucleotides in TTS (Fig. 7A).
Fig. 7.
A,B,C,D,E Dolutegravir (dark blue), Harpagide 5-O-β-D-glucopyranoside (yellow), Lamivudine (white), Lenacapavir (tan), Tenofovir-disoproxil fumarate (Salmon) docked against modeled TIS-mRNA(pink) is represented along with its interactions shown in 2D-ligplot.
3.3.17. TTS-harpagide 5-O-β-D-glucopyranoside
In the binding site of TTS, nucleotide G9 plays a very important role. However, the compound harpagide 5-O-β-D-glucopyranoside primarily consists of only electronegative groups attached to the ring structure. Multiple H-bonds are formed between G9 and A30 and are involved in the H-bond formation (Fig. 7B).
3.3.18. TTS-lamivudine
This binding site has all the nucleotides such as A, U, G and C. G12 is an important nucleotide, as it is involved in the multiple H-bond sharing between the OH of the oxathiolane group and the OH attached to the pyrimidine ring. The hydroxy methyl group attached to the oxathiolane group is also involved in the formation of multiple H-bonds with U13 and G24. The amino group attached to the pyrimidine ring is also involved in H-bond formation with the A28 nucleotide (Fig. 7C).
3.3.19. TTS-lenacapavir
This binding site is dominated by the G nucleotide, and lamivudine is bound tightly to the TTS by forming a H-bond. For example, the O present in the sulfonyl group accepts an H-bond from the sidechain NH of the G9 nucleotide. As the ligand is quite large in size, the ligand is quite solvent-exposed (Fig. 7D).
3.3.20. TTS-tenofovir
The binding site of TTS with tenofovir is primarily lined by the A, U, G and C nucleotides. In this binding, it is seen that only a partial part in tenofovir is seen to be solvent-exposed but still it is involved in several H-bond interactions with TTS such as the phosphonyl O is accepting an H-bond from the NH sidechain of G12 nucleotide. Additionally, the NH attached to the purine is involved in double H-bonds by donating H-atoms to the backbone O of A27 (Fig. 7E).
4. Conclusion
In this study, we have computationally investigated the interaction of the small molecules (ARVds) against the translation initiation site (TIS), translation termination site (TTS), HMR1 and HMR2 mRNAs of the omicron variant S protein. The results obtained in this study, indicate mutations in the S protein of the Omicron SARS-CoV-2 variant compared to that of the Delta and Wuhan variants. These mutation points may present a therapeutic target for COVID-19 and its complications, with ARVd as potential drugs. This is theoretically evident by the potent molecular interactions of the studied ARVd with the initiation and termination codons of the mRNAs of the mutated proteins. However, further wet studies are recommended to confirm these in silico results.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Rahul Ravichandran: Writing – original draft, Visualization, Software, Methodology, Investigation. Musa M. Abarshi: Writing – original draft, Visualization, Software, Methodology, Investigation. Sanusi B. Katsayal: Writing – original draft, Methodology, Investigation. Murtala B. Abubakar: Writing – original draft, Software, Methodology, Investigation. Priyanka Banerjee: Writing – review & editing, Supervision, Software, Resources, Project administration, Methodology, Formal analysis. Ochuko L. Erukainure: Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Conceptualization. Aliyu Muhammad: Writing – original draft, Validation, Supervision, Software, Methodology, Investigation, Data curation, Conceptualization. Ya'qub U. Abiodun: Writing – review & editing, Software, Methodology, Investigation. Olubunmi Atolani: Writing – review & editing, Validation, Project administration, Methodology. Robert Preissner: Writing – review & editing, Validation, Supervision, Software, Resources, Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Handling Editor: Prof. L.H. Lash
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.toxrep.2024.101825.
Contributor Information
Ochuko L. Erukainure, Email: loreks@yahoo.co.uk.
Aliyu Muhammad, Email: amachida31@gmail.com.
Appendix A. Supplementary material
Supplementary material
Data availability
The authors are unable or have chosen not to specify which data has been used.
References
- 1.Alkhatib M., Svicher V., Salpini R., et al. SARS-CoV-2 variants and their relevant mutational profiles: update summer 2021. Microbiol. Spectr. 2021;9(3) doi: 10.1128/Spectrum.01096-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauso L.V., Imbesi C., Irene G., Calì G., Bitto A. New approaches and repurposed antiviral drugs for the treatment of the SARS-CoV-2 infection. Pharmaceuticals. 2021;14(6):503. doi: 10.3390/ph14060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesiada M., Purzycka K.J., Szachniuk M., Blazewicz J., Adamiak R.W. Automated RNA 3D structure prediction with RNAcomposer. Methods Mol. Biol. 2016;1490:199–215. doi: 10.1007/978-1-4939-6433-8_13. [DOI] [PubMed] [Google Scholar]
- 4.Case J.B., Mackin S., Errico J.M., et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 omicron lineage strains. Nat. Commun. 2022;13(1):1–11. doi: 10.1038/s41467-022-31615-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6(2) doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyer O. Covid-19: South Africa’s surge in cases deepens alarm over omicron variant. Br. Med. J. 2021;375:n3013. doi: 10.1136/bmj.n3013. [DOI] [PubMed] [Google Scholar]
- 7.Erukainure O.L., Atolani O., Muhammad A., et al. Targeting the initiation and termination codons of SARS-CoV-2 spike protein as possible therapy against COVID-19: the role of novel harpagide 5-O-β-D-glucopyranoside from Clerodendrum volubile P Beauv.(Labiatae) J. Biomol. Struct. Dyn. 2022;40(6):2475–2488. doi: 10.1080/07391102.2020.1840439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erukainure O.L., Atolani O., Muhammad A., et al. Translational suppression of SARS-COV-2 ORF8 protein mRNA as a viable therapeutic target against COVID-19: computational studies on potential roles of isolated compounds from Clerodendrum volubile leaves. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ettaboina S.K., Nakkala K., Laddha K. A mini review on SARS-COVID-19-2 omicron variant (B. 1.1. 529) SciMedicine J. 2021;3(4):399–406. [Google Scholar]
- 10.Gil C., Ginex T., Maestro I., et al. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63(21):12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann M., Krüger N., Schulz S., et al. The omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185(3):447–456. doi: 10.1016/j.cell.2021.12.032. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson A.G. COVID-19 incidence and death rates among unvaccinated and fully vaccinated adults with and without booster doses during periods of delta and omicron variant emergence—25 US Jurisdictions, April 4–December 25, 2021. Morb. Mortal. Wkly. Rep. 2022;71(12):466–473. doi: 10.15585/mmwr.mm7104e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S., Thiessen P.A., Bolton E.E., et al. Shoemaker (2016) BAJNar. PubChem Subst. Compd. Databases. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94(4):1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 16.Lamberti G., Cascone S., Marra F., Titomanlio G., d’Amore M., Barba A.A. Gastrointestinal behavior and ADME phenomena: II. In silico simulation. J. Drug Deliv. Sci. Technol. 2016;35:165–171. [Google Scholar]
- 17.Liu B., Zhou H., Tan L., Siu K.T.H., Guan X.-Y. Exploring treatment options in cancer: tumor treatment strategies. Signal Transduct. Target. Ther. 2024;9:175. doi: 10.1038/s41392-024-01856-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liuzzo G., Patrono C. Re-purposed antiviral drugs without a purpose in COVID-19: a valuable lesson for clinicians. Eur. Heart J. 2021 doi: 10.1093/eurheartj/ehab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallapaty S. The hunt for the origins of omicron. Nature. 2022;602:26–28. doi: 10.1038/d41586-022-00215-2. [DOI] [PubMed] [Google Scholar]
- 20.Martinez M.A. Efficacy of repurposed antiviral drugs: lessons from COVID-19. Drug Discov. Today. 2022;27(7):1954–1960. doi: 10.1016/j.drudis.2022.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirzaei H., McFarland W., Karamouzian M., Sharifi H. COVID-19 among people living with HIV: a systematic review. AIDS Behav. 2021;25(1):85–92. doi: 10.1007/s10461-020-02983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris G., Huey R., Lindstrom W., Sanner M., Belew R. DS goodsell i AJ olson. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Msomi N., Lessells R., Mlisana K., de Oliveira T. Nature Publishing Group; 2021. Africa: tackle HIV and COVID-19 together600; pp. 33–36. [DOI] [PubMed] [Google Scholar]
- 24.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Skoraczyński G., Kitlas M., Miasojedow B., Gambin A. Critical assessment of synthetic accessibility scores in computer-assisted synthesis planning. J.Cheminformatics. 2023;15(1):6. doi: 10.1186/s13321-023-00678-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sternberg A., Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. N. Engl. J. Med. 2021;384(6):497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO . World Health Organisation. WHO; 2021. WHO Global Clinical Platform for COVID-19. [Google Scholar]
- 30.Wong S.-C., Au A.K.-W., Chen H., et al. Transmission of omicron (B. 1.1. 529)-SARS-CoV-2 variant of concern in a designated quarantine hotel for travelers: a challenge of elimination strategy of COVID-19. Lancet Reg. Health–West. Pac. 2022;18 doi: 10.1016/j.lanwpc.2021.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo H.G., Shah M. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. Front. Immunol. 2021:6031. doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worobey M., Pekar J., Larsen B.B., et al. The emergence of sars-cov-2 in europe and north america. Science. 2020;370(6516):564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong G., Wu Z., Yi J., et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The authors are unable or have chosen not to specify which data has been used.