Abstract
The actions of endogenous opioids and nociceptin/orphanin FQ are mediated by four homologous G protein-coupled receptors that constitute the opioid receptor family. However, little is known about opioid systems in cyclostomes (living jawless fish) and how opioid systems might have evolved from invertebrates. Here, we leveraged de novo transcriptome and low-coverage whole-genome assembly in the Pacific hagfish (Eptatretus stoutii) to identify and characterize the first full-length coding sequence for a functional opioid receptor in a cyclostome. Additionally, we define two novel endogenous opioid precursors in this species that predict several novel opioid peptides. Bioinformatic analysis shows no closely related opioid receptor genes in invertebrates with regard either to the genomic organization or to conserved opioid receptor-specific sequences that are common in all vertebrates. Furthermore, no proteins analogous to vertebrate opioid precursors could be identified by genomic searches despite previous claims of protein or RNA-derived sequences in several invertebrate species. The presence of an expressed orthologous receptor and opioid precursors in the Pacific hagfish confirms that a functional opioid system was likely present in the common ancestor of all extant vertebrates some 550 million years ago, earlier than all previous authenticated accounts. We discuss the premise that the cyclostome and vertebrate opioid systems evolved from invertebrate systems concerned with antimicrobial defense and speculate that the high concentrations of opioid precursors in tissues such as the testes, gut, and activated immune cells are key remnants of this evolutionary role.
Keywords: enkephalin, evolution, MAP Kinase, opiate, opioid peptide
1 |. INTRODUCTION
The opioid system plays a central role in controlling the transmission of pain and emotional responses, including stress, affect, and complex social behaviors such as bonding in mammals (Lutz & Kieffer, 2013a, 2013b). Opioids are also involved in a myriad of other physiological, defensive, and behavioral processes, including autonomic regulation, control of breathing, immune function, gut transit, and itch (Bodnar, 2018). The actions of endogenous opioids and the closely related peptide nociceptin/orphanin FQ are mediated by four homologous G protein-coupled receptors (GPCRs) denoted mu (MOP), delta (DOP), kappa (KOP), and opioid receptor-like/nociceptin (NOP). There is an extensive literature concerning the evolution of opioid systems in vertebrates, which has been confirmed at multiple levels including at the transcriptome and genomic level (Khalap et al., 2005). There is likewise an extensive literature of opioid systems in different invertebrate species, with claims of opioids systems even at the level of the bacteria (Cadet & Stefano, 1999; Harrison et al., 1994; Salzet & Stefano, 1997; Stefano et al., 2003; Stefano & Salzet, 1999; Stefano, Salzet-Raveillon, & Salzet, 1998). However, despite the multiple reports of opioid-like transcripts and proteins, it is difficult to accept that authentic opioid systems are indeed present in invertebrates without definitive genomic identification of opioid peptide sequences, precursors, and receptors in these species.
A combined approach using phylogenetics and synteny suggests that the four genes encoding the opioid receptors arose out of two whole-genome duplication events that preceded the radiation of jawed vertebrates (Dreborg, Sundstrom, Larsson, & Larhammar, 2008; Larhammar, Dreborg, Larsson, & Sundstrom, 2009; Sundstrom, Dreborg, & Larhammar, 2010). However, the precise timing of these duplication events remains unresolved; it is not clear whether the second round (2R) occurred before or after the divergence of cyclostomes and gnathostomes (Janvier, 2010; Smith & Keinath, 2015; Takahashi et al., 2006). A comprehensive characterization of the opioid system within this key evolutionary taxon could therefore provide valuable insight into the origin and diversification of opioid function in higher vertebrates.
There is evidence to suggest a functional opioid system exists in cyclostomes. Opioid precursors have been cloned in the lamprey (Petromyzon marinus; Takahashi et al., 2006). The recently completed genome sequence (Smith et al., 2013) of this species suggests several different loci that may encode for opioid receptor-like sequences. However, these sequences, derived from projection of orthologous protein-coding genes, are incomplete. In hagfish, we previously identified three short, distinct partial opioid receptor-like sequences (within exon 2) from genomic DNA using degenerate primers (Li, Keith, & Evans, 1996b), and a single partial sequence was subsequently isolated from brain cDNA (Li, Keith, & Evans, 1996a). As yet, however, no opioid precursors in hagfish have been identified.
In this study, we report the identification of the first full-length coding sequence of a MOP-like opioid receptor from the brain of a cyclostome, the Pacific hagfish (Eptatretus stoutii). Genomic analysis shows considerable homology to the genomic organization of the mammalian opioid receptors and conservation of key functional elements. Additionally, we identify two novel endogenous opioid precursors in this species that potentially encode several novel opioid peptides flanked by canonical prohormone cleavage sites (paired basic residues, particularly lysine–arginine or single arginine residues) and that activate the hagfish MOP-like opioid receptor during heterologous expression in mammalian cells. The repertoire of genes and functional confirmation described here firmly establishes that all elements of a true multifaceted opioid system are present in cyclostomes and were likely in the common ancestor of cyclostomes and gnathostomes. This extends the origins of the opioid system to at least 550 million years ago, earlier than all previous authenticated reports.
2 |. MATERIALS AND METHODS
2.1 |. Animals
Pacific hagfish E. stoutii (30–60 cm long) were acquired from Marinus Scientific (Long Beach, CA) and maintained at 14–17°C in artificial sea water.
2.2 |. Library construction and sequencing
Hagfish were anesthetized with iced water before tissue dissection. For RNAseq libraries, total RNA was extracted from homogenized brain tissue samples taken from a single adult hagfish using the RNeasy kit (Qiagen). Paired-end Illumina RNA libraries were generated from each tissue sample from 1 ug of total RNA with the TruSeq RNA Sample Preparation v2 kit (Illumina) using random hexamer primers followed by poly-A selection. For the genomic library, total genomic DNA was isolated from a liver tissue using the DNeasy kit (Qiagen). One ug of total genomic DNA was used to construct a TruSeq Genomic Library (Illumina). Quality of all libraries was assessed using a Bioanalyzer 2200 TapeStation (Agilent). Each RNAseq library was sequenced in single HiSeq 2500 lane. The genomic library was sequenced in four lanes. All sequencing was performed using 100 bp paired-end reads.
2.3 |. Data processing and sequence assembly
Using a custom Perl script, the FAST-Q files generated from Illumina sequencing were first filtered for quality and trimmed of adapter sequence. The reads were trimmed of leading or trailing N bases and clipped of adapter sequences. Low quality base-calls with a Sanger-scale Phred quality score Q < 15 were removed from the 3′ ends using Trimmomatic. Reads with length less than 36 were removed, and unpaired reads were separated into a single-ended library prior to assembly using Trinity (Alrubaian et al., 2006), a program developed for efficient de novo transcriptome assembly using a de Bruijn graph algorithm assembler. SOAPdenovo (Bojnik et al., 2009) was used for the genomic sample, with the parameter pair_num_cutoff set to 5 to adjust for low sequencing depth.
2.4. |. Isolation of opioid receptor and peptide transcripts
Total RNA was extracted from brain tissue as described previously and Superscript III (Invitrogen) was used to construct a cDNA library using random hexamer primers. Transcripts were amplified using Phusion polymerase (New England Biolabs) with the following primer pairs: esMOP_fwd 5′-TGTGTTCCGATCGAACCTT-3′ and esMOP_rev 5′-TGCGACATTTCTTTTCACCA-3′, and esPENK1_fwd 5′-TCGCCTTCACTTTTGTGCCCTTG-3′ and esPENK1_rev 5′-TATGAGACCGAGATTCTGTTAAC-3′, encoding the full-length cds of esMOP and esPENK1, respectively. Amplicon identity was verified by Sanger sequencing (Retrogen).
2.5 |. Gene identification from assembled transcripts
The transcript assembly for the hagfish brain was used to generate a sequence database using BLAST v. 2.2.29. This nucleotide database was queried using a tBLASTn search with peptide sequences corresponding to the mouse MOP, DOP, and KOP sequences (Swiss-Prot accessions P35372, P41143, and P41145, respectively), as well as the reported receptor fragment identified by our group (Li et al., 1996a). The “best-hit” nucleotide sequences were then translated to their corresponding peptide sequences using the ExPASy translate tool (http://web.expasy.org/) to determine the putative open reading frames. The sequence of the most complete transcript was verified by manual inspection. To determine the genomic structure of the transcripts, a separate database was compiled with all scaffolds and contigs generated by the SOAP de novo assembler. We then performed a BLAST search using the assembled transcript that encoded for esMOP, esPENK1, and esPENK2. Putative intron/exon boundaries were determined by the overlap of the transcriptome and genomic alignments, and manual verification for the presence of canonical splice donor/acceptor sites.
2.6 |. Homology modeling
The sequence for esMOP was submitted to the SWISS-MODEL workspace server (Keith et al., 1996) for homology modeling and subjected to Gapped Blast using BLOSUM 62 matrix with an E-value cutoff of 0.000001 to identify the closest homologous structure. The closest template was 4DKL, corresponding to murine OPRM-1 bound to β-FNA. A homology model was then generated using a MEMOIR (Ebejer, Hill, Kelm, Shi, & Deane, 2013), a pipeline tailored specifically for the molecular modeling of transmembrane proteins. The resultant structure was visualized using pyMOL.
2.7 |. Multiple sequence alignment and phylogenetic analysis
Multiple sequence analysis was performed using CLUSTAL-W for esMOP against all six peptide sequences available for the MOP present in HomoloGene (http://www.ncbi.nlm.nih.gov/homologene) corresponding to full-length cDNA clones isolated from human, cow, mouse, chicken, frog, and zebra fish. Transmembrane domains were predicted using the TSPRED server. The analysis involved all 12 peptide sequences available for the MOP, DOP, KOP, and ORL sequences present in HomoloGene. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 (Danielson & Dores, 1999) using the Maximum likelihood (ML) method. Five hundred Bootstrap iterations were performed to assess significance.
2.8 |. cDNA library construction, subcloning, and cell-line generation
The PCR product was cloned using the Zero-blunt TOPO PCR cloning kit (ThermoFisher Scientific) and inserted into the pCMV-tag2A vector (Stratagene) to fuse an N-terminal Flag epitope tag to the ORF of clone of esMOP. The fusion protein esMOP-FLAG was then shuttled into vector pRRL-sin-cPPT-CMV-MCS-IRES-mCherry, which encodes a HIV-derived lentiviral vector with a cytomegalovirus (CMV) promoter to drive expression. We verified the insertion using Sanger sequencing (Retrogen, Inc.) and coined the resultant vector pRRL-esMOP. pRRL-esMOP was packaged viral particles (UCLA Vector Core). Untitered lysates were used to treat human embryonic kidney cells expressing the large-T antigen (HEK293T, ATCC) in serum-free media. Note, HEK293T cells do not express opioid receptors (Keith et al., 1996). The infected cells were sorted by fluorescence-activated cell sorting (FACS) gated on mCherry expression with the BD FACSAria platform (UCLA Flow Cytometry core) to generate a heterogenous line of cells that stably express flag-tagged hagfish MOP, which we called 293T-FLAG-esMOP.
2.9 |. Phospho-MAPK assay
293T-FLAG-esMOP cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. Cells were cultured in six-well plate for 24 hr prior to treatment, washed and then incubated in serum-free medium overnight before ligand stimulation as indicated in the figure legends. Cell extracts were prepared on ice by lysing the cells in the lysis buffer (10 mM HEPES, 1mM EDTA, 250 mM sucrose, 10 mM Tris-HCL, 1 mM Na3PO4, 1 μg leupeptin, 1 mM PMSF) with protease inhibitor. Protein concentration was assessed using Bio-Rad Protein Assay. For Western Blot, equal amounts of protein (20 ug) were loaded onto pre-cast 10% polyacrylamide gels and transferred to nitrocellulose membranes. For dot blot, equal amounts of protein (as above) were loaded onto a nitrocellulose membrane within a manifold dot-blot system. The membrane was left to dry. For both dot blot and Western Blot procedures, membranes were incubated with phospho-MAPK rabbit antibody (Cell Signaling, Beverly, MA) and visualized with a goat anti-rabbit antibody conjugated to a horseradish peroxidase (HRP; Jackson Immunoresearch, West Grove, PA). Some blots were re-stained with anti-beta-actin (1:5,000; Sigma-Aldrich) to validate equivalent loading and protein transfer. One point in the 1 μM morphine treatment was removed due to it being more than 2 SD away from the mean. Data for the MAPK signal were analyzed by one-way ANOVA with Dunnett’s post hoc analysis to identify specific differences from the 0.01 μM dose. Data are presented as means ± SD.
2.10 |. MOP internalization assay
293T-FLAG-esMOP was maintained in DMEM supplemented with 10% fetal bovine serum and plated on a 24-well plate lined with glass coverslips. Immediately prior to the addition of a ligand, cells were switched to a serum-free DMEM to avoid premature degradation of Met-enkephalin. In a subset of wells, cells were treated with 1 μM naloxone for 15 min prior to the addition of agonist. Following this, cells were treated with either: sterile saline (control), 1 μM Met-enkephalin, 1 μM morphine, or 1 μM etorphine, and incubated at 37°C for 1 hr. Cells were then fixed for 15 min with 4% paraformaldehyde in 0.1 M phosphate-buffered saline followed by washing thoroughly in 0.1 M phosphate-buffered saline. In order to facilitate visualization of FLAG-tagged esMOP, cells were labeled with an anti-FLAG antibody (Sigma; 1:500) followed by a goat anti-mouse IgG secondary antibody conjugated to an Alexa 488 fluorophore (Life Technologies; 1:1,000). Coverslips were mounted on glass slides with a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vectashield). Slides were imaged with a multiphoton confocal microscope with an oil-immersion 40x objective (Leica TCS SP2).
2.11 |. Synthetic peptides
The following peptides were purchased from either Phoenix Pharmaceuticals, Inc. or Antagene, Inc.: YGGFMRS (Y7S), YGGFMRRFFGVAV (Y13V), YGGFMRRVGGPL (Y12L), YGRFMSN (Y7N), and YGGFM (Met-enkephalin).
3 |. RESULTS
3.1 |. Identification of a hagfish MOP
We hypothesized that genes encoding for an opioid receptor would likely be expressed in the brain of the hagfish. We therefore generated two RNAseq libraries derived from both hindbrain and forebrain tissue taken from the same adult animal. As no reference genome exists for this species, this sequence data were combined and used for de novo transcriptome assembly. This consolidated set of assembled transcripts was subsequently used to generate a searchable BLAST database.
Using a partial sequence we identified previously (Li et al., 1996a, 1996b), we performed a tBLASTn-based search against our transcriptome database and found the sequence fragment to be completely contained within an 815 bp transcript (Figure 1). The coding sequence of this assembled transcript was incomplete as it did not contain a stop codon. To augment our database of assembled transcripts, we created an additional BLAST database comprised of contigs generated from de novo assembly of a low-coverage genomic library derived from hagfish liver tissue (Table 1).
FIGURE 1.

Genomic structure of the esMOP gene. The nucleotide sequence shown is comprised of the three genomic contigs (alternately colored) that aligned to the cDNA clone of esMOP, with the deduced amino acid translation shown above. Regions that overlap with esMOP are in uppercase, with mismatches shown as red letters and alterative cDNA nucleotides underneath in black. Gray boxes denote overlap with the transcript identified from de novo assembly, The region that overlaps the peptide sequence published by Li et al. (1996a) is boxed in green. Translation start/stop sites are bolded. Pink boxes denote canonical GU/AG splice donor/acceptor motifs. Intronic regions excluded for clarity are showed as “||”
TABLE 1.
Statistics for de novo assembly
| Assembly | Hagfish brain | Hagfish gDNA |
|---|---|---|
| Tissue(s) | Whole brain, post brain | Liver |
| Library type | Poly-A RNA | Genomic DNA |
| Software | Trinity 2.0.3 | SOAPdenovo 2.04 |
| Raw reads | 203,168,548 | 530,037,086 |
| Raw GB | 20.32 | 53.00 |
| Trimmed reads | 182,200,622 | 489,943,570 |
| Trimmed GB | 17.40 | 45.97 |
| N50 | 2,189 | 1,677 |
|
| ||
Aligning our transcript against the assembled contigs allowed us to infer the remaining coding region by analysis of the adjacent genomic sequence. We then designed PCR primers and isolated a single amplification product from a hagfish brain cDNA library. Sequencing of the cloned product confirmed a single insert with a longest open reading frame (ORF) of 1,044 bp, whose deduced amino acid translation most closely resembles (in the mouse genome) the MOP with a homology of 63.92% (compared to 57.95% and 53.98% for murine DOP and KOP, respectively). We labeled this transcript as esMOP (GenBank: KY860575).
Aligning the cDNA sequence of esMOP to our genomic hagfish assembly database, we achieved complete and contiguous coverage across three different contigs, indicating that the cDNA sequence of esMOP consists of three exons. Inspection of the genomic sequence flanking the aligned regions also showed that full-length cDNA sequence is mediated by canonical GT/AG splice donor/acceptor sites (Figure 1). The first coding exon spans amino acids (aa) 1–62, containing the N-terminal extracellular region as well as the first transmembrane domain. The second exon encodes the majority of the first intracellular loop and transmembrane domains 2–4, and the third exon contains the rest of the protein (Figures 1 and 2). This three-exon genetic structure of esMOP is universally conserved among all members of the opioid receptor family in every vertebrate species thus far analyzed (Herrero-Turrion & Rodriguez, 2008).
FIGURE 2.
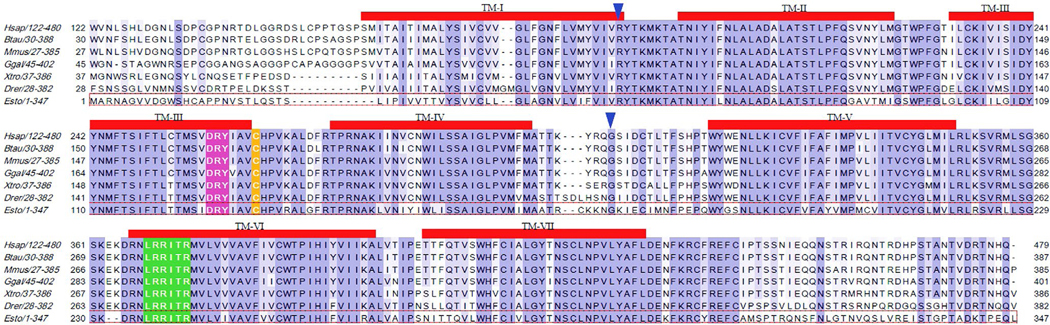
Comparison of MOP sequences from representative vertebrate species. A sequence alignment of the translated coding sequence from human (Homo sapiens), cow (Bos taurus), mouse (Mus musculus), chicken (Gallus gallus), western clawed frog (Xenopus (Silurana) tropicalis), zebra fish (Danio rerio), and Pacific hagfish (Eptatretus stoutii) is depicted above. Red bars denote transmembrane domains predicted by TsPred. However, in should be recognized that residues transitioning transmembrane domains to extracellular or intracellular domains are speculative and likely to be regulated by both posttranslational modifications and state of activation of the receptor. Blue triangles denote intron–exon boundaries. Conserved regions highlighted are the “DRY” motif (pink), “xRRxxRx” (green), and palmitoylation site (orange). Gaps are shown as “−.” The alignment is anchored to the hagfish sequence (red-dotted box)
3.2 |. Structural features of the hagfish MOP
The translated amino acid sequence of the cDNA clone of hagfish receptor has several conserved structural features consistent among all vertebrate opioid receptors (Figure 2). As with other members of the seven transmembrane domain GPCR family, esMOP has a highly conserved DRY (Asp-Arg-Tyr) sequence as well as the xRRxxR motif, which is critical for MOP receptor activation of Gi proteins (Wang, 1999), both motifs are highlighted in Figure 2. It was recently demonstrated that the palmitoylation site in MOP is next to the DRY domain adjacent to TM-III (Zheng et al., 2012), not in the C-terminus as in most other GPCRs, which corresponds to position C130 in esMOP. Also, there is a single consensus N-linked glycosylation site within the N-terminal extracellular region (Figure 2).
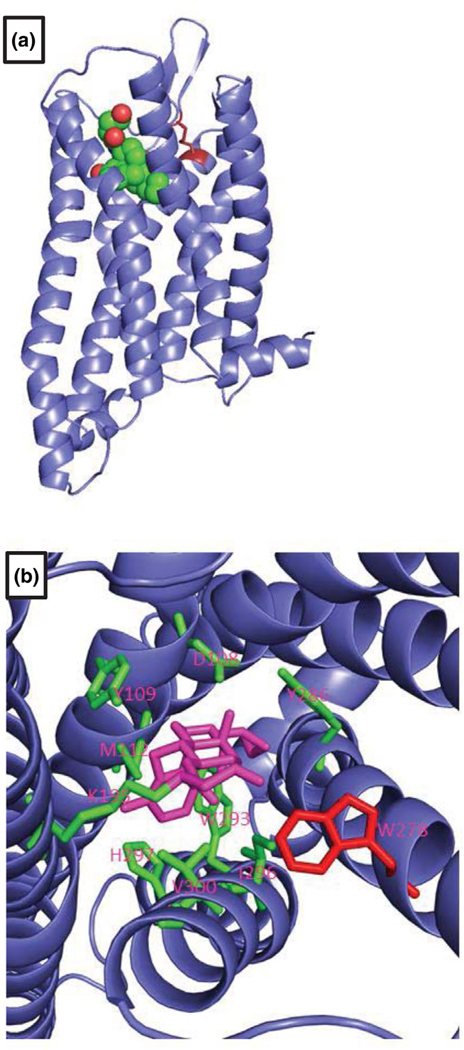
A homology model of esMOP exhibits typical GPCR structure, with seven transmembrane domains arranged in a counterclockwise fashion forming a ligand binding pocket (Figure 3a). Our 3D model shows the conservation of a key disulfide bond that is critical for proper ligand recognition (Shi & Javitch, 2004), formed between residues Cys-101 in TM-III and Cys-192 in EL-II of esMOP (Figure 3a). Additionally, all of the nine residues within the ligand binding pocket identified by x-ray crystallography that form polar interactions with the irreversible MOP inhibitor, βFNA, are conserved and correctly oriented in esMOP (Figure 3b). Solving of the structure for the murine MOP determined that a single tryptophan residue W318 in TM-7 prohibited the binding of the DOP-specific agonist, naltridole (Manglik et al., 2012). This also has a corresponding residue aa W278 in the esMOP sequence (Figure 3b) suggesting a degree of conserved receptor selectivity may exist through the entire vertebrate subphylum.
FIGURE 3.
Homology model of hagfish MOP. The predicted structure of esMOP the mouse MOP bound to a morphinan agonist as a template (PDB: 4DKL) is shown. (a) esMOP exhibits typical seven-transmembrane structure and a conserved disulfide bond between TM-III and EL-II shown in red. (b) View of the ligand binding pocket from extracellular side shows conserved residues involved in polar interactions with the morphinan agonist, beta-funaltrexamine. W318, which prevents the binding of the DOP-specific agonist, Naltrindole, is shown in red
3.3 |. Phylogenetic analysis of the cyclostome opioid receptors
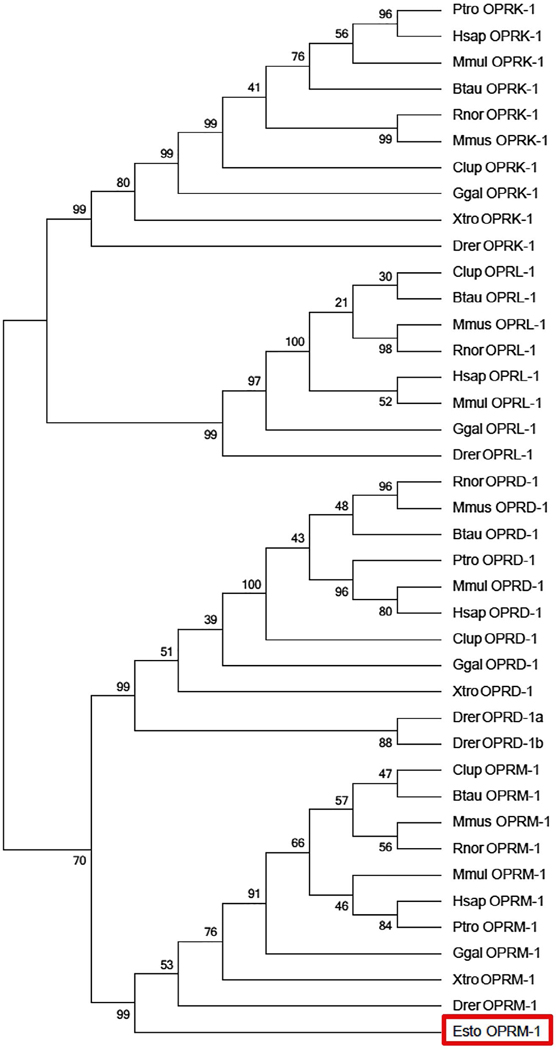
To examine evolutionary relationship of the esMOP among other opioid receptor types, we conducted phylogenetic analysis from a diverse set of vertebrate species corresponding to full-length coding sequences available for each of the four opioid receptors and generated a consensus tree, rooted to rhodopsin, using the ML method with 500 bootstrap replicates. The overall tree topology is consistent with previous studies of opioid receptor evolution among higher vertebrates, demonstrating that the MOP and DOP first diverged from the KOP and NOP types (Pasternak & Pan, 2013; Stevens, 2009). The hagfish opioid receptor clusters confidently with MOP sequences from other sampled species, with a bootstrap support of 99%, similar to the value obtained for clades consisted of the other opioid receptor types (Figure 4).
FIGURE 4.
Phylogenetic relationship between the hagfish MOP and other Opioid Receptor Types. The tree topology generated by maximum likelihood using aligned amino acid positions and excluding all gaps is shown, condensed to the 50% bootstrap level after 500 iterations, rooted using the sequence for rhodopsin (RHO). esMOP (red box) forms a clear clade with MOP from other species
The lamprey genome contains three loci annotated by projection of protein-coding genes that potentially encode for opioid receptors. We also examined esMOP in relation to the opioid receptors present in the lamprey genome by constructing a second tree including the three incomplete opioid receptor sequences in lamprey genome, as annotated by Ensembl v78. Although bootstrap support for this topology is modest at 59%, all partial lamprey opioid receptors cluster within the MOP clade (Figure 5).
FIGURE 5.
Phylogenetic close relationship between the hagfish MOP and lamprey opioid receptor-like sequences. The tree topography generated by maximum likelihood using aligned amino acid positions and excluding all gaps is shown, condensed to the 50% bootstrap level after 500 iterations, rooted using the sequence for rhodopsin (RHO). esMOP and putative lamprey opioid receptor-like sequences from Ensembl release 65 (red box) form a clear clade with MOP from all other considered species. Species are represented as the first letter of the genus followed by the first three letters of the species
Further informatic analysis showed no close genomic homology in the invertebrate genomes of the protochordates; the sea squirt (Ciona intestinalis) or lancelet (Branchiostoma floridae), with the most homologous gene being a single exon gene belonging to the GPCR family, and with closest homology to the somatostatin receptors. GPCRs in several invertebrates that are annotated as “opioid receptors” showed less than 25% homology with esMOP or other vertebrate opioid receptors.
3.4 |. Identification of novel hagfish opioid precursors
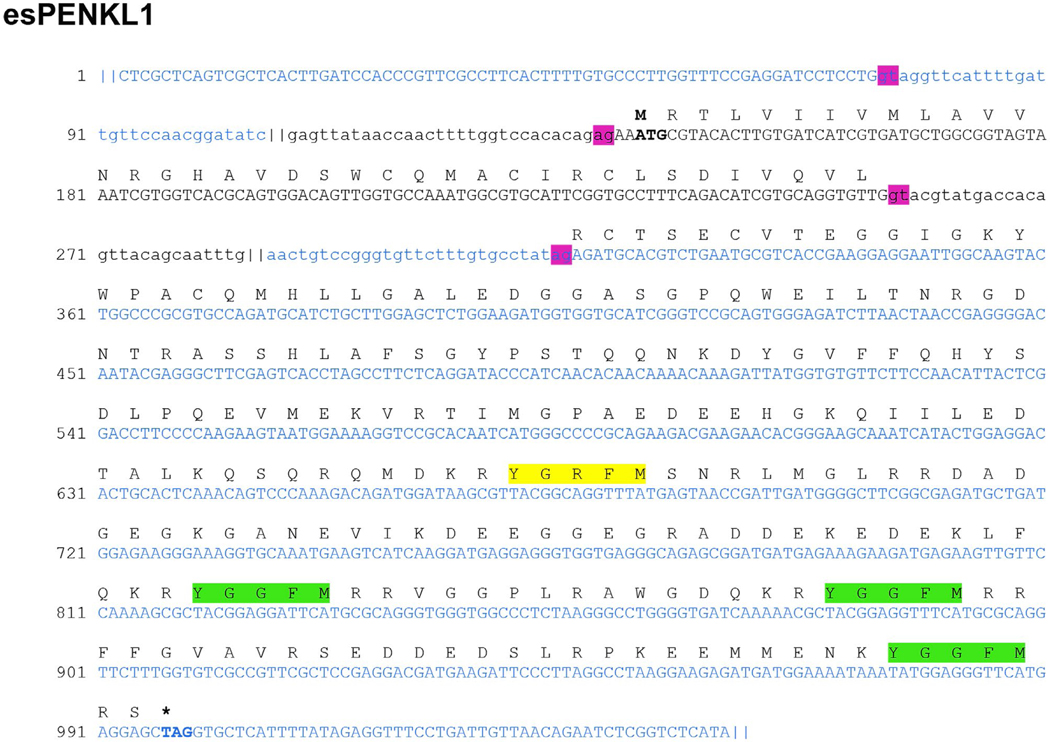
We further searched our assembly database for evidence of endogenous opioid precursors expressed in the brain of the hagfish. This yielded two candidates, each of which contained multiple instances of the canonical opioid Tyr-Gly-Gly-Phe (Met, Ile) sequence. The first of these, which we label E. stoutii proenkephalin-like 1 (esPENKL1), is a 934bp transcript fragment whose translated sequence revealed a complete 266 aa ORF (GenBank: KY860576). Alignment of esPENKL1 back to our hagfish genomic database showed the transcript is encoded by three separate exons, with splice sites located inside of the 5′ UTR and separating the coding region (Figure 6), analogous to mammalian PENK. The putative opioid precursor contains a predicted hydrophobic signal peptide of 18 residues in length followed immediately by a stretch of amino acids containing six cysteine residues computationally predicted to form disulfide bonds between them (Figure 7). This cysteine-rich portion is found in other hormone precursor proteins and notably present in all other currently known endogenous opioid peptide precursors (Mollereau et al., 1996; Sossin, Fisher, & Scheller, 1989). In proenkephalin, this species-conserved cysteine-rich region (syn-enkephalin) has been shown to be co-stored and co-released with enkephalins; however, its function as a peptide in its own right or in sorting or processing of proenkephalin is less clear (Albert & Liston, 1993).
FIGURE 6.
Genomic structure of the esPENKL1 gene. The nucleotide sequence shown is comprised of the three genomic contigs (alternately colored) that aligned to the assembled transcript of esPENKL1, with the deduced amino acid translation shown above. Regions that aligned with the esPENKL1 transcript are in uppercase; intronic regions are shown in lowercase. Pink boxes denote canonical GU/AG splice donor/acceptor motifs. Green boxes denote Met-enkephalin motifs preceded by canonical basic cleavage sites. Yellow boxes denote additional closely homologous sequences to enkephalin also preceded by KR residues. Nucleotide regions excluded for readability are shown as “||”
FIGURE 7.
Deduced amino acid sequence of hagfish PENKL1. The translated sequence of hagfish PENKL1 ORF is shown. The functional motifs are highlighted: signal peptide (purple), disulfide-bonded cysteine (blue), degenerate enkephalin (pink), dibasic cleavage site (blue), met-enkephalin (maroon). Boxed regions denote potential products that were synthesized and tested for activity, labeled in blue letters
The hagfish PENKL1 protein contains two YGGFM (Met-enkephalin) motifs nested within canonical dibasic cleavage sites at aa positions 207–211 and 227–231 (Figure 7). It has an additional Met-enkephalin motif at the C-terminus for which the proceeding dibasic cleavage site is absent, and no Leu-enkephalin YGGFL (Leu-enkephalin) which is present in mammalian proenkephalin. There are two additional degenerate motifs that differ from a known enkephalin: YGVFF at aa 105–109 and YGRFM at aa 157–161 (Figure 7). As determined by BLAST, PENKL1 has the highest aa homology (30% identity, 43% positive) to the full-length proenkephalin of the Oriental fire-bellied toad, Bombina orientalis (GenBank: AAU95755.1). Upon exclusion of the invariant Met-enkephalin motifs, however, the percent of identity is below 20%, indicating low conservation of the intervening sequence between Met-enkephalin motifs.
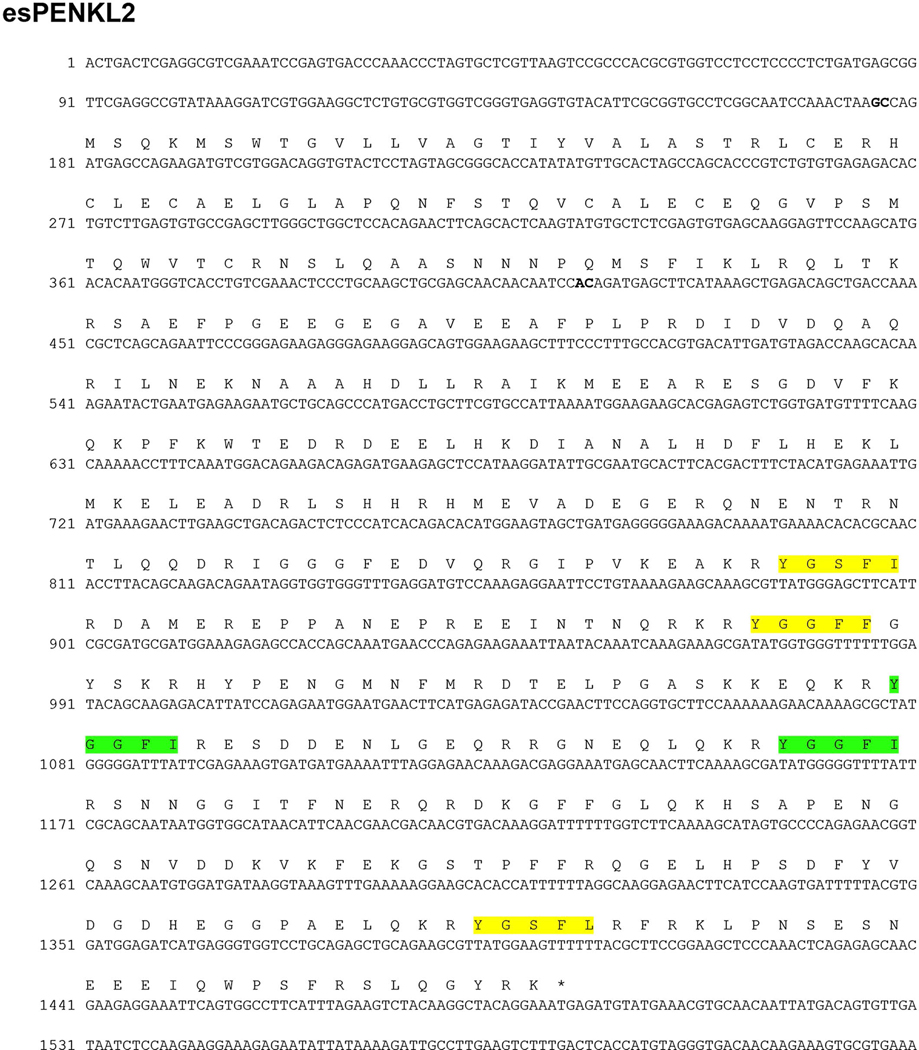
We uncovered a second genomic sequence that encodes a putative opioid precursor with a complete ORF of 437 aa (Figure 8). This sequence (esPENKL2) contains a predicted signal peptide followed by a cysteine-rich region. Interestingly and homologous with the esPENKL1 construct, esPENKL2 has an intron–exon boundary linking the cysteine-rich N-terminal domain with the enkephalin containing exon. esPENKL2 contains two Ile-enkephalin (YGGFI) opioid peptides, and both of these have only a single basic residue at the C-terminus to the enkephalin motif. Ile-enkephalin has been previously described in frog, eel, and tilapia (Alrubaian et al., 2006; Mollereau et al., 1996).
FIGURE 8.
Structure of the esPENKL2 transcript and intron–exon boundaries. The esPENKL2 sequence was identified from the whole transcriptome assembly library and completely aligned within two genomic contigs in three distinct regions, with putative intron–exon boundaries indicated in bold. A complete ORF of 437 amino acids is shown above and includes several YGGFI (Ile-enkephalin) sequences highlighted in green, which are preceded by paired basic (KR) residues. Motifs that are closely homologous sequences to enkephalin (YGGF-M/L/I) and also preceded by KR residues are highlighted in yellow
3.5 |. Hagfish MOP is activated by opioid peptides
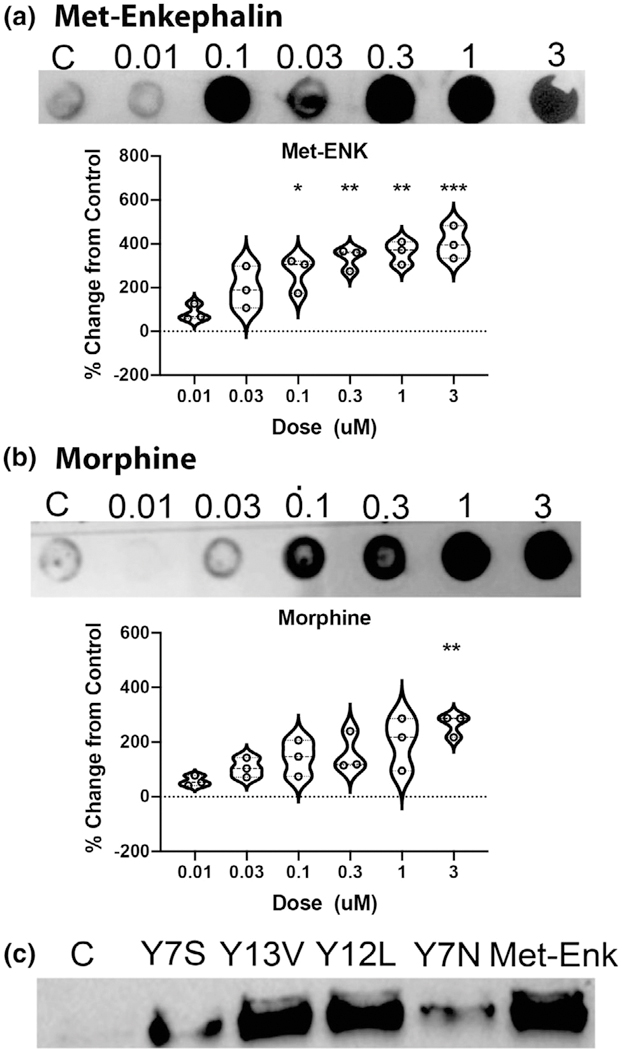
Endogenous opioid receptor ligands are produced from their precursors by enzymatic actions of endopeptidases (the prohormone convertases and furins cleaving at the C-terminus of basic residues) and carboxypeptidase E removing basic residues from the C-terminus. The exact profile of cleavage sites is quite complex and has been shown to vary in a tissue and region-specific manner, allowing for fine-tuned control of ligand-dependent activation of opioid receptors. This inherent variability in processing is evidenced by the fact that many C-terminal extended forms of Met-enkephalin derived from proenkephalin are present in the murine brain that bind DOP, MOP, and KOP with high affinity. The sequence of esPENKL1 suggests the possible production of several novel endogenous opioid peptides, some of which were synthesized namely Y7S: YGGFMRS, Y13V: YGGFMRRFFGVAV, and Y12L: YGGFMRRVGGPL (Figures 7 and 9c). We also generated a potential cleavage product, which based on opioid peptide structure–activity relationships, we predicted would not be a functional opioid, YGRFMSN (Y7N, Figure 9c).
FIGURE 9.
esMOP MAPK signaling following agonist binding. (a) esMOP expressing HEK293T cells were serum starved overnight, and then treated with 0–3 μM Met-enkephalin or morphine for 5 min. Cell extracts were assayed by dot blot for phospho-MAPK, and intensity measured and expressed as percent change from control. Data represent means ± standard deviation, n = 3. (b) esMOP expressing HEK293T cells were cultured as above and treated with putative hagfish opioid peptides from esPENKL1: Y7S: YGGFMRS, Y13V: YGGFMRRFFGVAV, Y12L: YGGFMRRVGGPL, Y7N: YGRFMSN (1 μM) or Met-Enkephalin (Met-Enk; 1 μM) for 5 min. Phospho-MAPK was assessed with Western Blot
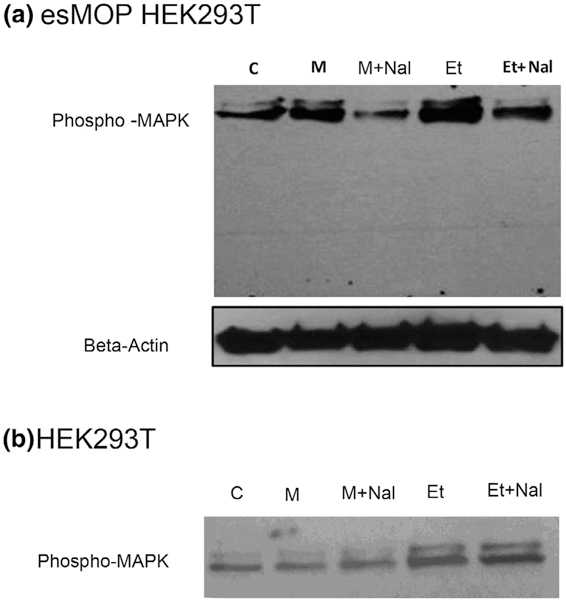
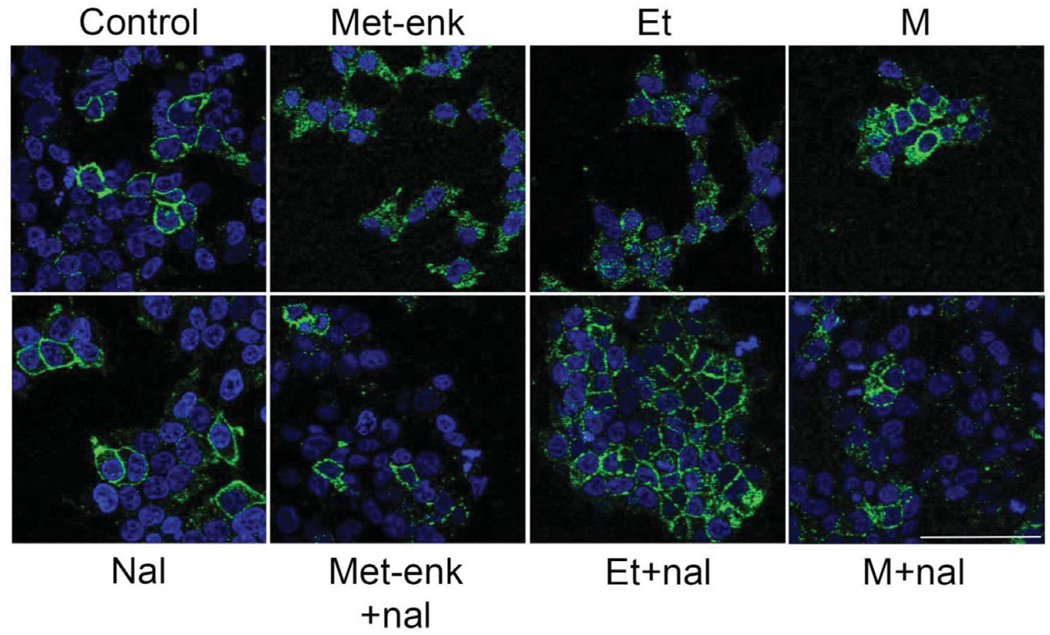
We employed a mammalian cell model to test if these peptides might function as novel opioids. HEK293T cells, which express no endogenous opioid receptors, were infected with a lentiviral vector esMOP. We treated these cells with morphine or etorphine, both in the presence or absence of the general opioid antagonist naloxone (Figures 9 and 10). We also performed additional treatments of the synthetic putative endogenous hagfish opioids and the putative non-opioid Y7N peptide. Signaling of esMOP was assayed by immunoblotting with a phospho-MAPK antibody. This assay we used to assess opioid receptor signaling has been previously described (Li et al., 2003). Consistent with the presence of a functional opioid receptor, phosphorylated levels of MAPK increased significantly when treated with morphine compared to controls. This increase was substantially higher in the presence of Met-enkephalin (Figure 9a and 9b). Of the predicted products derived from esPENKL1, phosphorylated MAPK was observed in cells treated with the putative hagfish opioid peptides Y13V, Y12L similar to that with Met-Enk, and to a lesser extent, Y7S (Figure 9c). As expected, there was no increase in activity in cells treated with the predicted non-opioid peptide Y7N (Figure 9 c). Functionality of esMOP was further confirmed by agonist-induced trafficking of HA-tagged esMOP expressed in HEK293T cells (Figure 11). Under control conditions, esMOP appeared associated with the extracellular membrane. Treatment with MOP agonists Met-enkephalin or the high efficacy opioid agonist etorphine produced robust internalization of the HA-tagged esMOP. Internalization could be blocked by pretreatment with the nonspecific opioid agonist, naloxone. Morphine, a non-internalizing MOP agonist, did not induce esMOP internalization, analogous to mammalian MOP treated with morphine (Keith et al., 1996).
FIGURE 10.
esMOP activation following agonist binding. (a) esMOP expressing HEK293T cells were serum starved overnight and then treated with morphine (M, 1 μM) or etorphine (Et, 1 μM) with or without naloxone (Nal, 10 μM). Cell extracts were assayed by Western Blot for phospho-MAPK. (b) Control HEK293T cells not expressing any opioid receptors were treated, as above
FIGURE 11.
MOP internalization after agonist treatment. HEK 293T cells expressing esMOP were plated in serum-free DMEM and treated with saline (control), Met-enkephalin (Met-enk;1 μM), etorphine (E; 1 μM), or morphine (M; 1 μM) for 1 hr (top row). A separate group was treated with naloxone (Nal; 1 μM) 15 min prior to agonist treatment (bottom row). Met-enkephalin and etorphine treatment caused robust MOP internalization (MOP-Flag = green, DAPI = blue) that was blocked by naloxone pretreatment. Morphine, a noninternalizing opioid agonist, failed to stimulate MOP internalization in these cells. Scale bar = 75 μm
4 |. DISCUSSION
This is the first report to describe the complete coding sequence and functional activity for an opioid receptor (esMOP) expressed in the brain of a cyclostome, the Pacific hagfish, one of only two species in the earliest branch of extant vertebrate lineages. In addition, we also present the isolation of two endogenous opioid-like precursors, esPENKL1 and esPENKL2. These precursors encode gene-duplicated copies of Met or Ile-enkephalin-like sequences preceded by basic residues that represent canonical prohormone-convertase cleavage sites. We demonstrate that an intact, opioid system is not only present in the genome of a cyclostome, but also expressed. Furthermore, in vitro esMOP functions in a similar fashion to mammalian opioid receptors, showing signaling via MAP-kinases and agonist-induced internalization. In this, we also illustrate the utility in leveraging de novo transcriptome assembly coupled with low-coverage genomic contig assembly for directed gene discovery in species that lack an established reference genome.
Prior analyses of opioid receptor-like sequences have been unable to conclusively determine which of the four known opioid receptor types are present in hagfish (Li et al., 1996a, 1996b), due to the paucity of sequencing data available, none of which represented full-length coding sequence. Based on sequence similarity, structural comparison, and phylogenetic analysis, we conclude that the expressed hagfish opioid receptor identified in this paper most closely resembles MOP. Other partial opioid receptor homologous sequences were observed in our analysis of the hagfish genome or transcript libraries and the sequences confirmed for multiple opioid-like receptor exon 2-derived sequences that were described in our earlier study (Li et al., 1996a). However, full-length sequences could not be assembled from our libraries and we cannot at this point confirm these are full-length expressed transcripts in the hagfish transcriptome.
Opioid receptor type was historically determined based on ligand receptor selectivity derived from pharmacological studies performed predominantly in mammals. Opioid receptor type selectivity is gained during evolution of vertebrates (Stevens, 2004). Phylogenetic analysis incorporating opioid receptor gene annotations derived from the lamprey genome assembly placed all cyclostome opioid receptors together in the MOP clade. This suggests a lack of receptor diversity, rather than specificity, could also contribute to the decrease in type-specificity observed in lower vertebrates.
Overall, esMOP is highly conserved among all vertebrates in which the receptor has been characterized, especially within transmembrane domains. Although the overall homology of the opioid precursors we identified in the hagfish is not high compared to with mammalian opioid precursors, there is conservation of a hydrophobic N-terminal signal peptide followed by a cysteine-rich region then an intron/exon boundary. There are just three copies of Met-enkephalin present in esPENKL1 compared with seven Met/Leu-enkephalin sequences in the mammalian forms, and only two of these are flanked by dibasic cleavage sites. However, there are multiple “close to enkephalin” sequences within the identified precursor which is suggestive of a gene duplication within enkephalin-like domains. Indeed, it has been suggested that the addition of precursor products arose through the mechanism of tandem duplication locally with the gene (Danielson & Dores, 1999; Khalap et al., 2005). Core features of the esPENK are retained in all vertebrates including hagfish, reinforcing that PENK emerged prior to divergence of the cyclostomes from the vertebrate lineage.
Interestingly, none of the opioid precursors we identified in cyclostomes thus far encoded Leu-enkephalin. However, immunoreactive forms of both Met- and Leu-enkephalins have been found in the brain extracts of both lamprey and hagfish, at ratios of 3:1 and 2:1, respectively (Dores & Gorbman, 1990). It is unclear if the antisera used for Leu-enkephalin in this study would distinguish leucine from isoleucine-enkephalin as we would predict would be a cleavage product of esPENKL2. A separation of the genes encoding of Met-enkephalin and Leu-enkephalin into two genes has already been observed in several non-mammalian vertebrate species (Dores et al., 2004).
The question remains as to the evolutionary etiology of the mammalian opioid system. Several invertebrate genomic sequences have previously been assigned as “opioid receptors,” for example in the sea slug (Aplysia californica), the round worm (Brugia malayi), and the oyster (Crassostrea gigas), (Liu et al., 2015). However, homology of these invertebrate annotated opioid receptors is extremely low with mammalian opioid receptors, with less than 25% homology and identity at the amino acid level. Furthermore, the pharmacology, if it has been performed, is not convincing of their designation as “opioid receptors.” PCR has revealed several opioid receptor-like sequences in the leech and mussel, that are strikingly similar to their human and vertebrate counterparts (Stefano et al., 1998). A transcript with greater than 95% identity to the human MOP transcript was identified from blue mussel (Mytilus edulis) transcripts (Cadet & Stefano, 1999). Unexplained is why the putative mussel mu transcript has a markedly higher homology with human than to lower vertebrate species (including the hagfish we report in this paper). At this juncture genomic analysis is required to validate these transcripts.
Although genomic analysis thus far reveals no endogenous opioid precursors in invertebrates that are closely related to any vertebrate opioid peptide precursor, PCR and Edmund sequencing of purified proteins has revealed several putative opioids and precursors in leech and mussel that are highly homologous with their vertebrate counterparts (Salzet et al., 1997; Salzet & Stefano, 1997). These sequences are in core areas 100% identical to their vertebrate homologues. However, these highly conserved core sequences between human and leech are not observed in any sequenced protostome genomes including aplysia, fly, C. elegans, or bee, nor are they present in the genomes of the pre-vertebrate lineage genomes of Ciona (sea squirts) and Amphioxus. These sequences remain speculative and waiting for genomic confirmation. Many invertebrate claims of opioid systems have relied on bioassays or antibody recognition (Carpenter et al., 1995; Ewadinger, Ridgway, Syed, Lukowiak, & Bulloch, 1996; Gutierrez & Asai, 1991; Kemenes, Rozsa, Stefano, & Carpenter, 1992; Leon-Olea et al., 2013; Liu et al., 2015; Nieto-Fernandez et al., 2009; Ollivaux, Dircksen, Toullec, & Soyez, 2002). However, given unusual pharmacology in the invertebrate bioassays (some opioid-like effects require very high ligand concentrations and some are non-naloxone reversible or naloxone has agonist effects) and the inherent problems with immune detection (Baker, 2015), genome sequence authentication is necessary to rigorously confirm signal identity.
Recent articles identify an opioid-like system in C. elegans involved in aversion and feeding behaviors (Cheong, Artyukhin, You, & Avery, 2015; Harris et al., 2010). In the feeding assays there is evidence for activation of a GPCR, NPR-17 by morphine (0.5 millimolar) and blockade by naloxone (10 millimolar). The data are compelling of the NLP-17 receptor interaction with some kappa and mu opioid alkaloids especially given that naloxone blocks GPYGYG-amide activity and NPL-17 is required for animal responses to opiate drugs. NLP-24 has repetitive YGG motifs and peptide products from this precursor activate NPR-17, a GPCR homologous to an annotated opioid receptor. However, peptides activating the NPR-17 receptor include peptides that do not possess characteristics to activate vertebrate opioid receptors, and NPR-17 is activated by peptides that will not activate opioid receptors, questioning NPR-17 as truly an opioid receptor or perhaps an invertebrate precursor of an opioid receptor. Furthermore, NRP-17 shows minimal homology with vertebrate opioid receptors and GenBank BLASTp searches identify other GPCRs with higher homology to NPR-17 than opioid receptors. Finally, in contrast to opioid receptors, NRP-17 has a single exon-coding region.
Opioid-like and opioid receptor-active sequences are found in invertebrate toxins such as the scorpion (Zhang et al., 2012) and immune components of several invertebrates including the C. elegans immune peptides NP24–34 (Husson, Lindemans, Janssen, & Schoofs, 2009). These precursors contain YGG repeats. Importantly, these peptides are not flanked by basic residues, which act as the canonical cleavage sites for neuropeptide precursors in both vertebrates and invertebrates and release the free N-terminal tyrosine residue that is essential for opioid peptide activity. It is likely that these precursors have an antimicrobial role in innate immune functions, as elegant studies in C. elegans have demonstrated (McVeigh et al., 2008). Noteworthy, is that several peptides derived from proenkephalin, namely Proenkephalin 1–73 constituting the cysteine-rich N-terminal fragment (synkephalin) and proenkephalin 209–237 (enkelytin) are both potent antimicrobial peptides (Goumon et al., 1998; Metz-Boutigue, Goumon, Lugardon, Strub, & Aunis, 1998). Together these data point to a potential origin of opioid precursors from proteins generating antimicrobial peptides.
Despite the multiple reports indicating otherwise, it is difficult to accept that authentic opioid precursors and opioid receptors are found in invertebrates. Without genomic identification of opioid peptide precursors which contain opioid sequences flanked by protease processing signals, as well as characterized receptors for the endogenous opioids, key authentication for invertebrate opioid systems are missing. Careful analysis of the pre-vertebrate lineages comprising the genomes of the sea squirt and the lancelet reveals no opioid-like precursors (defined as sequences with Tyr-Gly-Gly-Phe-Met-(Leu/Ile) preceded by canonical basic or paired basic residues to generate the N-terminal Tyr residue with the free amine that is required for binding opioid receptors). Furthermore, the sea hare (A. californica) and the lancelet (B. floridae) genomes reveal no opioid receptor sequences (defined as receptors with closer homology to opioid than somatostatin receptors and with conserved primary structural features found in all vertebrate opioid receptors including conserved intron/exon boundaries). The question of how the opioid system arose is still unresolved, but our analysis points to the period of vertebrate expansion greater than 550 million years ago. The creation of the opioid receptors from other GPCRs is not difficult to envisage but the hagfish receptor reported here reveals the earliest opioid receptor sequence with high homology, receptor features, and genomic intron–exon structure that is analogous to the mammalian opioid receptors. The evolution of opioid peptides is more difficult to explain since no like-peptide sequences are present in pre-vertebrate genomes. Horizontal gene transfer is one speculative possibility, whereby a gene from a species secreting enkephalin-like antimicrobial peptides was transferred into a pre-vertebrate genome.
5 |. CONCLUSION
Here, we have conclusively demonstrated that a functional opioid system is indeed present in all extant vertebrate lineages. Further elucidation of all the components of the opioid system in both hagfish and lamprey would be invaluable to the understanding of how this system evolved and ultimately diversified into the wide-ranging of functions that are observed in mammals.
Supplementary Material
Significance.
We report the identification and characterization of the first full-length coding sequence for a functional opioid receptor in a cyclostome, the Pacific Hagfish (Eptatretus stoutii). We identify two novel opioid precursors in this species that predict novel opioid peptides. Informatic analysis of invertebrate genomes did not reveal any genes or predicted transcripts as clear evolutionary antecedents of opioid receptors or opioid peptide precursors. Our study establishes the presence of a functional opioid system as a vertebrate-specific trait which likely emerged at least 550 million years ago in a common ancestor of cyclostomes and gnathostomes, earlier than all validated accounts.
ACKNOWLEDGMENTS
The Shirley Hatos Foundation supports CJE, AG, CMC, and AMWT. NIH Grant numbers K99DA40016 (AMWT), R01DA041781 (CMC), 1UG3TR003148–01 (CMC) and 2P50 DA005010 (CMC, CJE), and the Department of Defense Grant number W81XWH-15–1-0435 (CMC). We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691) and Departmental funds from the UCLA Semel Institute for Neuroscience and Human Behavior.
Funding information
National Institutes of Health; Jane and Terry Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles; Shirley and Stefan Hatos Foundation
Footnotes
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
CONFLICT OF INTEREST
The authors declare they have no conflict of interest and nothing to declare.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
Transparent Science Questionnaire for Authors
Transparent Peer Review Report
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Albert PR, & Liston D (1993). Deletions of the synenkephalin domain which do not alter cell-specific proteolytic processing or secretory targeting of human proenkephalin. Journal of Neurochemistry, 60, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Alrubaian J, Lecaude S, Barba J, Szynskie L, Jacobs N, Bauer D,… Dores RM (2006). Trends in the evolution of the prodynorphin gene in teleosts: Cloning of eel and tilapia prodynorphin cDNAs. Peptides, 27, 797–804. [DOI] [PubMed] [Google Scholar]
- Baker M (2015). Reproducibility crisis: Blame it on the antibodies. Nature, 521, 274–276. [DOI] [PubMed] [Google Scholar]
- Bodnar RJ (2018). Endogenous opiates and behavior: 2016. Peptides, 101, 167–212. [DOI] [PubMed] [Google Scholar]
- Bojnik E, Magyar A, Toth G, Bajusz S, Borsodi A, & Benyhe S (2009). Binding studies of novel, non-mammalian enkephalins, structures predicted from frog and lungfish brain cDNA sequences. Neuroscience, 158, 867–874. [DOI] [PubMed] [Google Scholar]
- Cadet P, & Stefano GB (1999). Mytilus edulis pedal ganglia express mu opiate receptor transcripts exhibiting high sequence identity with human neuronal mu1. Brain Research. Molecular Brain Research, 74, 242–246. [DOI] [PubMed] [Google Scholar]
- Carpenter DO, Kemenes G, Elekes K, Leung M, Stefano G, Rozsa KS, & Salánki J (1995). Opioid peptides in the nervous system of Aplysia: A combined biochemical, immunocytochemical, and electrophysiological study. Cellular and Molecular Neurobiology, 15, 239–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MC, Artyukhin AB, You Y-J, & Avery L (2015). An opioid-like system regulating feeding behavior in C. elegans. Elife, 4, e06683. 10.7554/eLife.06683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson PB, & Dores RM (1999). Molecular evolution of the opioid/orphanin gene family. General and Comparative Endocrinology, 113, 169–186. [DOI] [PubMed] [Google Scholar]
- Dores RM, & Gorbman A (1990). Detection of Met-enkephalin and Leu-enkephalin in the brain of the hagfish, Eptatretus stoutii, and the lamprey, Petromyzon marinus. General and Comparative Endocrinology, 77, 489–499. [DOI] [PubMed] [Google Scholar]
- Dores RM, Sollars C, Lecaude S, Lee J, Danielson P, Alrubaian J, … Vaudry H (2004). Cloning of prodynorphin cDNAs from the brain of Australian and African lungfish: Implications for the evolution of the prodynorphin gene. Neuroendocrinology, 79, 185–196. [DOI] [PubMed] [Google Scholar]
- Dreborg S, Sundstrom G, Larsson TA, & Larhammar D (2008). Evolution of vertebrate opioid receptors. Proceedings of the National Academy of Sciences of the United States of America, 105, 15487–15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebejer J-P, Hill JR, Kelm S, Shi J, & Deane CM (2013). Memoir: Template-based structure prediction for membrane proteins. Nucleic Acids Research, 41, W379–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewadinger NM, Ridgway RL, Syed NI, Lukowiak K, & Bulloch AG (1996). Identification and localization of a [Met5]-enkephalin-like peptide in the mollusk, Lymnaea stagnalis. Brain Research, 737, 1–15. [DOI] [PubMed] [Google Scholar]
- Goumon Y, Lugardon K, Kieffer B, Lefevre JF, Dorsselaer AV, Aunis D, & Metz-Boutigue MH (1998). Characterization of antibacterial COOH-terminal proenkephalin-A-derived peptides (PEAP) in infectious fluids. Importance of enkelytin, the antibacterial PEAP209–237 secreted by stimulated chromaffin cells. Journal of Biological Chemistry, 273, 29847–29856. [DOI] [PubMed] [Google Scholar]
- Gutierrez R, & Asai M (1991). IR-Met and IR-leu-enkephalin content in the perioesophageal ganglia of Helix aspersa seasonal variations. Comparative Biochemistry and Physiology C, 100, 609–613. [DOI] [PubMed] [Google Scholar]
- Harris G, Mills H, Wragg R, Hapiak V, Castelletto M, Korchnak A, & Komuniecki RW (2010). The monoaminergic modulation of sensory-mediated aversive responses in Caenorhabditis elegans requires glutamatergic/peptidergic cotransmission. Journal of Neuroscience, 30, 7889–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison LM, Kastin AJ, Weber JT, Banks WA, Hurley DL, & Zadina JE (1994). The opiate system in invertebrates. Peptides, 15, 1309–1329. [DOI] [PubMed] [Google Scholar]
- Herrero-Turrion MJ, & Rodriguez RE (2008). Bioinformatic analysis of the origin, sequence and diversification of mu opioid receptors in vertebrates. Molecular Phylogenetics and Evolution, 49, 877–892. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Lindemans M, Janssen T, & Schoofs L (2009). Comparison of Caenorhabditis elegans NLP peptides with arthropod neuropeptides. Trends in Parasitology, 25, 171–181. [DOI] [PubMed] [Google Scholar]
- Janvier P (2010). microRNAs revive old views about jawless vertebrate divergence and evolution. Proceedings of the National Academy of Sciences of the United States of America, 107, 19137–19138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, … von Zastrow M (1996). Morphine activates opioid receptors without causing their rapid internalization. Journal of Biological Chemistry, 271, 19021–19024. [DOI] [PubMed] [Google Scholar]
- Kemenes G, Rozsa KS, Stefano G, & Carpenter DO (1992). Distinct receptors for Leu- and Met-enkephalin on the metacerebral giant cell of Aplysia. Cellular and Molecular Neurobiology, 12, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalap A, Bagrosky B, Lecaude S, Youson J, Danielson P, & Dores RM (2005). Trends in the evolution of the proenkephalin and prodynorphin genes in gnathostomes. Annals of the New York Academy of Sciences, 1040, 22–37. [DOI] [PubMed] [Google Scholar]
- Larhammar D, Dreborg S, Larsson TA, & Sundstrom G (2009). Early duplications of opioid receptor and Peptide genes in vertebrate evolution. Annals of the New York Academy of Sciences, 1163, 451–453. [DOI] [PubMed] [Google Scholar]
- Leon-Olea M, Miller-Perez C, Sanchez-Islas E, Mendoza-Sotelo J, Garduno-Gutierrez R, de Gortari P, & Amaya MI (2013). The nociceptin/orphanin FQ-like opioid peptide in nervous periesophageal ganglia of land snail Helix aspersa. Brain Research, 1505, 22–46. [DOI] [PubMed] [Google Scholar]
- Li X, Keith DEJ, & Evans CJ (1996a). Mu opioid receptor-like sequences are present throughout vertebrate evolution. Journal of Molecular Evolution, 43, 179–184. 10.1007/PL00006076 [DOI] [PubMed] [Google Scholar]
- Li X, Keith DEJ, & Evans CJ (1996b). Multiple opioid receptor-like genes are identified in diverse vertebrate phyla. FEBS Letters, 397, 25–29. 10.1016/S0014-5793(96)01126-X [DOI] [PubMed] [Google Scholar]
- Li Y, Eitan S, Wu J, Evans CJ, Kieffer B, Sun X, & Polakiewicz RD (2003). Morphine induces desensitization of insulin receptor signaling. Molecular and Cellular Biology, 23, 6255–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou Z, Wang L, Jiang S, Wang W, Zhang R, & Song L (2015). The immunomodulation mediated by a delta-opioid receptor for [Met(5)]-enkephalin in oyster Crassostrea gigas. Developmental and Comparative Immunology, 49, 217–224. [DOI] [PubMed] [Google Scholar]
- Lutz P-E, & Kieffer BL (2013a). Opioid receptors: Distinct roles in mood disorders. Trends in Neurosciences, 36, 195–206. 10.1016/j.tins.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz P-E, & Kieffer BL (2013b). The multiple facets of opioid receptor function: Implications for addiction. Current Opinion in Neurobiology, 23, 473–479. 10.1016/j.conb.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, … Granier S (2012). Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature, 485, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVeigh P, Alexander-Bowman S, Veal E, Mousley A, Marks NJ, & Maule AG (2008). Neuropeptide-like protein diversity in phylum Nematoda. International Journal for Parasitology, 38, 1493–1503. [DOI] [PubMed] [Google Scholar]
- Metz-Boutigue MH, Goumon Y, Lugardon K, Strub JM, & Aunis D (1998). Antibacterial peptides are present in chromaffin cell secretory granules. Cellular and Molecular Neurobiology, 18, 249–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau C, Simons MJ, Soularue P, Liners F, Vassart G, Meunier JC, & Parmentier M (1996). Structure, tissue distribution, and chromosomal localization of the prepronociceptin gene. Proceedings of the National Academy of Sciences of the United States of America, 93, 8666–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Fernandez F, Andrieux S, Idrees S, Bagnall C, Pryor SC, & Sood R (2009). The effect of opioids and their antagonists on the nocifensive response of Caenorhabditis elegans to noxious thermal stimuli. Invertebrate Neuroscience, 9, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollivaux C, Dircksen H, Toullec J-Y, & Soyez D (2002). Enkephalinergic control of the secretory activity of neurons producing stereoisomers of crustacean hyperglycemic hormone in the eyestalk of the crayfish Orconectes limosus. Journal of Comparative Neurology, 444, 1–9. [DOI] [PubMed] [Google Scholar]
- Pasternak GW, & Pan Y-X (2013). Mu opioids and their receptors: Evolution of a concept. Pharmacological Reviews, 65, 1257–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzet M, Salzet-Raveillon B, Cocquerelle C, Verger-Bocquet M, Pryor SC, Rialas CM, … Stefano GB (1997). Leech immunocytes contain proopiomelanocortin: Nitric oxide mediates hemolymph proopiomelanocortin processing. Journal of Immunology, 159, 5400–5411. [PubMed] [Google Scholar]
- Salzet M, & Stefano GB (1997). Invertebrate proenkephalin: Delta opioid binding sites in leech ganglia and immunocytes. Brain Research, 768, 224–232. [DOI] [PubMed] [Google Scholar]
- Shi L, & Javitch JA (2004). The second extracellular loop of the dopamine D2 receptor lines the binding-site crevice. Proceedings of the National Academy of Sciences of the United States of America, 101, 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, & Keinath MC (2015). The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Research, 25, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, … Li W (2013). Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics, 45, 415–421, 421e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, Fisher JM, & Scheller RH (1989). Cellular and molecular biology of neuropeptide processing and packaging. Neuron, 2, 1407–1417. [DOI] [PubMed] [Google Scholar]
- Stefano GB, Cadet P, Rialas CM, Mantione K, Casares F, Goumon Y, & Zhu W (2003). Invertebrate opiate immune and neural signaling. Advances in Experimental Medicine and Biology, 521, 126–147. [PubMed] [Google Scholar]
- Stefano GB, & Salzet M (1999). Invertebrate opioid precursors: Evolutionary conservation and the significance of enzymatic processing. International Review of Cytology, 187, 261–286. [DOI] [PubMed] [Google Scholar]
- Stefano GB, Salzet-Raveillon B, & Salzet M (1998). Mytilus edulis hemolymph contain prodynorphin. Immunology Letters, 63, 33–39. [DOI] [PubMed] [Google Scholar]
- Stevens CW (2004). Opioid research in amphibians: An alternative pain model yielding insights on the evolution of opioid receptors. Brain Research. Brain Research Reviews, 46, 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CW (2009). The evolution of vertebrate opioid receptors. Frontiers in Bioscience, 14, 1247–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom G, Dreborg S, & Larhammar D (2010). Concomitant duplications of opioid peptide and receptor genes before the origin of jawed vertebrates. PLoS One, 5, e10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Nakata O, Moriyama S, Nozaki M, Joss JMP, Sower SA, & Kawauchi H (2006). Occurrence of two functionally distinct proopiomelanocortin genes in all modern lampreys. General and Comparative Endocrinology, 148, 72–78. [DOI] [PubMed] [Google Scholar]
- Wang HL (1999). A conserved arginine in the distal third intracellular loop of the mu-opioid receptor is required for G protein activation. Journal of Neurochemistry, 72, 1307–1314. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu J, Wang Z, Zhang X, Liang X, & Civelli O (2012). BmK-YA, an enkephalin-like peptide in scorpion venom. PLoS One, 7, e40417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Pearsall EA, Hurst DP, Zhang Y, Chu J, Zhou Y, … Law PY (2012). Palmitoylation and membrane cholesterol stabilize mu-opioid receptor homodimerization and G protein coupling. BMC Cell Biology, 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.