Abstract
The clonal evolution model of cancer was developed in the 1950–1970s, and became central to cancer biology in the 21st century, largely through studies of cancer genetics. Although it has proved its worth, its structure has been challenged by observations of phenotypic plasticity, non-genetic forms of inheritance, non-genetic determinants of clone fitness, and non-tree-like transmission of genes. There is even confusion about the definition of a clone, which we aim to resolve. The performance and value of the clonal evolution model depends on the empirical extent to which evolutionary processes are involved in cancer, and on its theoretical ability to account for those evolutionary processes. Here, we identify limits in the theoretical performance of the clonal evolution model and provide solutions to overcome those limits. While we do not claim that clonal evolution can explain everything about cancer, we show how many of the complexities that have been identified in the dynamics of cancer can be integrated into the model to improve our current understanding of cancer.
ToC blurb
Clonal evolution is now a central theoretical framework in cancer research. In this Perspective, Laplane and Maley identify challenges to that theory. There are some non-evolutionary phenomena in cancer that cannot be captured by the theory. Other challenges such as non-genetic hereditary, phenotypic plasticity, reticulate evolution and clone diversity, can be included in an expanded evolutionary theory of cancer.
Introduction
Cancers are populations of cells that are heterogeneous across space and through time. This diversity is currently a major clinical issue, limiting the efficiency of most cancer treatments as there is often a subset of cells that is resistant to whatever treatment is being used.1,2 The diversity also limits the accuracy of prognosis and our ability to predict how it will respond to an intervention because a biopsy may not be representative of the entire neoplasm and there is a strong stochastic component to how it changes through time. A better understanding of the mechanisms involved in this diversification are thus urgently needed to better manage cancer. An explanatory mechanism of cell diversification that has gained traction in the 21st century is clonal evolution: cancer cells diversify through the accumulation of genetic and epigenetic alterations, which can change the cellś relative fitness and consequently lead to clonal expansion or contraction by natural selection. The principles of evolution and the tools of population genetics can be and have been successfully applied to cancer cells.3,4 However, the evolutionary view of cancer progression has been challenged5 and it was shown that phenotypic heterogeneity within cancers can be largely independent of the genetics of clones.6 Given that evolution by natural selection acts on the phenotype and relies on its heritability, these data question the relevance of the clonal evolution model. It is high time that we evaluate the strengths, weaknesses and opportunities for improving the clonal evolution model.
To achieve this, we here first explore what is known about the evolution of cancer cells through a historical and contemporary review of the literature. We then analyze the theoretical structure of the clonal evolution model by highlighting its underlying assumptions. By doing so, we intend to locate the theoretical and conceptual tensions and difficulties inherent to the current model. We conclude with suggestions and perspectives that can help address some of the issues and enhance the explanatory relevance of clonal evolution in cancer.
What is the clonal evolution model?
Historical perspective
The clonal evolution model is an abstract model depicting the evolution of cancer cells in a patient. It is often attributed to Peter Nowell, but Nowell himself always made it clear that he was summarizing ideas that were developed by the community.7 The observation of intratumor heterogeneity, and the idea that cancer cells evolve was already present before 1976, and its success occurred later.
In the 1950s, the notion of evolution was often used in oncology to describe cancer progression (see for example refs8,9), though not always implying a Darwinian process.10 During this time, studies on the cancer cell karyotype supported the hypothesis first introduced by Theodor Boveri that tumors could emerge from chromosomal defects caused by abnormal mitoses, which predicted the clonal origin of cancer.11 Karyotyping also led to the observation of a variation in the number of chromosomes contained by cancer cells, and, as a single cell whole genome assay, played a crucial role in revealing the evolution of cancer cells. Karyotypic observations directly raised the question of whether tumors are composed of multiple strains of cells, each having a fixed number of chromosomes, or whether the karyotype of tumor cells could change (see for example refs12,13) and if so, whether such changes were stochastic or heritable and selected (see for example refs14,15). T.S. Hauschka directly refers to “mutation-selection sequence analogous to phylogeny” and argues that “specific tumor karyotypes have competitive survival value.”15 Many techniques were used to resolve the question of the clonal origin of cancer, including the discovery of the Philadelphia chromosome,16 random somatic inactivation of one of the two alleles of glucose-6-phosphate dehydrogenase enzyme in women17,18 or immunoglobulin heavy chain rearrangements.19,20
All of those experiments converged toward the hypothesis of a clonal origin of cancer, with intratumoral heterogeneity originating from subsequent variations that may undergo selection. In most of these papers, evolutionary notions are kept in the background,10 but some directly discussed evolutionary principles applied to cancer. The English cytogeneticist C.E. Ford dedicated a paper to selective pressure in healthy, irradiated, or cancerous somatic cells defending the hypothesis that “unbalanced karyotypic changes have an effect on the probability of survival and proliferation” and thus that “the karyotypic structure of a cell population would then be the resultant of the operation of selective forces on the variability arising within it.”21 J. Lejeune argued that the hypothesis of selection was necessary to the coherence of the clonal evolution model and explicitly mentioned the use of karyotypes to reconstruct the “natural history of the clone”, comparing such reconstruction to “the approach of paleontologists reconstructing, from form to form, the history of the phylum”22 (p. 76, translated from French).
After Nowell’s elegant summary of the evolutionary model of cancer, we got… crickets. The evolutionary biology of cancer lay dormant for decades with only a few scientists23–28 building on Nowell’s framework. It was not until the 21st century that the field gained momentum. It is probably not a coincidence that interest in cancer evolution accelerated when high throughput sequencing started generating extensive amounts of genetic data from cancers. Observations of the clinical importance of intratumor heterogeneity29–34 and clonal expansions35–38 soon followed, along with the confirmation that therapy often selects for pre-existing clones with mutations that render them resistant to therapy.1,39–44 Although clinical impact remains limited, further studies demonstrated that an evolutionary approach to cancer therapy can lead to dramatic improvements in time to progression and overall survival.45–51 Amidst all this, reviews of how evolutionary biology and ecology could be productively applied to cancer29,52–55 may have helped to lay a foundation for future progress.
How do cancer cells evolve?
Any population of entities with a diversity of heritable properties that can result in differential fitness between entities can evolve by natural selection.56 Somatic cells are such entities and can be subject to evolution by natural selection because mutations cause heritable diversity among cells and (at least some) can alter cell fitness. The open question is whether and how much they evolve by natural selection, a question that has been debated by scientists and philosophers.57–60
It is important to recognize that cancer cells can evolve through a plurality of mechanisms. While they can evolve by natural selection, some tumors show little evidence of natural selection and appear to be mainly evolving by neutral evolution.61–64 A mathematical neutral evolution model is consistent with genetic data in approximately a third of solid tumors4, of multiple myeloma65 and chronic myelomonocytic leukemia.66 Another pan-analysis also highlighted that negative selection, which is predominant in the germline, is nearly absent in cancer and somatic evolution.64 Although clonal evolution was initially considered as a continuous gradual process, it has been shown that bursts of changes can also happen.67–70 Clonal evolution can occur through stasis, gradualism, or punctuation, with different molecular clocks ticking at different speeds,71,72 and can proceed by linear evolution or branched evolution.31,73–77 There is a diversity of processes that underlie the clonal evolution of cancer cells.
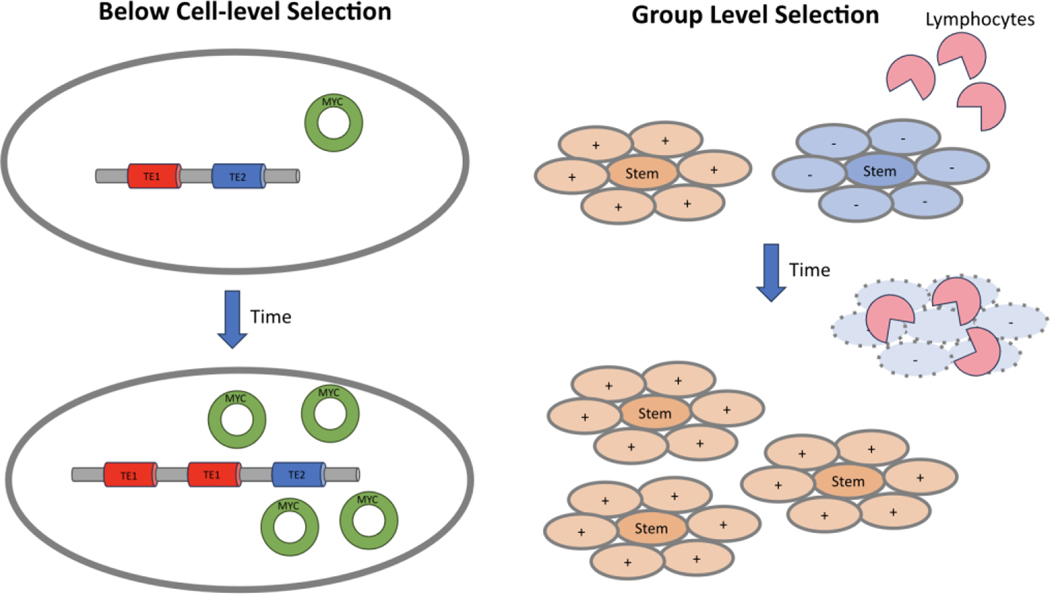
In addition, although the clonal evolution model traditionally focuses on the evolution of cancer (stem) cells as the unit of selection,78,79 selection may occur at higher levels (Figure 1a), on groups of cells rather than individual cells. For example, there is a debate on whether there is evolution by natural selection between metastases, each metastasis counting as a reproductive entity, generating further secondary metastases.59,60,80 Higher level selection could also select among colonies of cancer stem cells and their non-stem cell progeny81 or among epithelial proliferative units like colonic crypts, which may divide and die.52,82 Selection in cancer may also occur at levels below the cell, through the evolution of transposable elements,83 or extrachromosomal DNA84 and perhaps micronuclei,85 if they can replicate independently of cell replication (Figure 1b). The fact that selection may act at additional levels, above and below the cell, does not violate the clonal evolution model. It adds complexity to it, in the same way that multilevel selection,86,87 above and below the level of the organism, does not violate the theory of evolution, but rather adds complexity to it.
Figure 1|. Cancer evolution by natural selection can operate at multiple levels of biological organization at once.
a) There may be natural selection on groups of cells, here shown as stem cells and their non-stem cell progeny. In this example, the blue non-stem cells have a property (marked by a ´+´) that protects their stem cells from destruction by lymphocytes, and allows the colonies of blue cells to replicate. Yellow progeny lack (-) these properties, leading to the destruction of the yellow population by the lymphocytes. b) Below the level of the cell, extrachromosomal DNA (ecDNA) may increase the copy number of cancer-related genes (for example MYC) independent of the chromosomes. Similarly, transposable elements (TE1 and TE2) in the genome may replicate within the genome.
What is the value of the clonal evolution model?
The clonal evolution model integrates current knowledge about cancer cell evolution. It is a highly theoretical framework. It aims not just at describing but also at reconstructing past evolution, predicting potential future evolution (for example resistance to treatment), explaining phenomena occurring in patients, and providing the foundation for novel therapeutic interventions. We refer to these properties as the theoretical value of the clonal evolution model.
A primary theoretical value of the model is to explain how and why cancers change over time and in response to therapy.53,88 Phylogenetic reconstruction provides a description of the natural history of a cancer. For example, clonal mutations (also known as ´truncaĺ mutations) tend to be different from subclonal mutations (also known as mutations that only appear on a ‘branch´). Phylogenetic reconstructions can highlight changes in exposures and mutational processes over time, exemplified by the loss of the aflatoxin B1-related mutational signature in African migrants with hepatocellular carcinoma in the years after they arrived in France.89 It also showed that contrary to expectation, metastases can occur early in carcinogenesis.75,90–92 Lessons learned from these phylogenetic reconstructions also facilitate drug targeting (for example targeting a clonal mutation rather than a subclonal one), and help predict the risk of therapeutic resistance through selection of resistant mutations, such as the T790M mutation of epidermal growth factor receptor (EGFR) that causes resistance to first-generation EGFR inhibitors in lung cancer.93 Many mutations have been associated with resistance to treatment in ways that now allow clinicians to better decide which treatment to apply. The clonal evolution model has also been used in risk stratification and prognosis, using measures of the clonal evolution, such as intratumor heterogeneity, to predict which precancers are likely to progress to invasive disease, and which cancers are likely to be lethal.1,94–96
The clonal evolution model predicts that different environments will change the ability of mutated clones to expand. In the hematopoietic system for example, mutated clones emerge with aging, a process referred to as clonal hematopoiesis. Different environmental changes may lead to clonal hematopoiesis, such as aging, chemotherapy, infections, smoking. But each tend to select different types of mutations.97 Clonal hematopoiesis also comes with various mutations, and various dynamics through age.98,99 This also helps to explain the late occurrence of cancer, as the decrease of healthy cell fitness with aging provides weaker competitors for the selection of mutated clones.100–103 Note that the promotion theory is sometime thought as in opposition to the somatic mutation theory, but these two theories are compatible in the context of clonal evolution, as fitness depends on both intrinsic cell properties and extrinsic properties.104,105
More fundamentally, the clonal evolution model generated a profound conceptual switch that views cancer as a dynamic process. As such, it contributed to discrediting the therapeutic strategy of searching for a magic bullet and required changes in research and treatment practices. The model predicts that most advanced cancers are likely to be able to escape any treatment. This has led to various propositions for how to develop therapies based on evolutionary principles, such as drug holidays, changing drugs through time, and adaptive therapies that manage the clonal dynamics in order to prevent therapeutically resistant clones from expanding out of control.46–49
Years of observations and experiments testing aspects of the clonal evolution model have supported its theoretical value. Yet, it does not explain everything about the phenotype of cancer. Putting cancer cells in a different microenvironment gets them to behave differently. An extreme illustration of this are the examples where placing cancer cells in a healthy environment can ńormalizé their behavior.106–111 Phenotypic intratumoral heterogeneity may largely be independent of the genotype6 and phenotypic plasticity is increasingly recognized as an important contributor to intra-tumoral heterogeneity, and treatment escape.112–115
Model assumptions and limitations
The performance of the clonal evolution model depends on two different aspects, one factual, the extent to which evolutionary processes contribute to cancer, and one theoretical, the extent to which the current clonal evolution model captures these processes. The model’s premise is that evolutionary processes do play a role in cancer. This is supported by decades of evidence. However, the importance of the model is an open question. The more evolutionary processes influence cancer, the more relevant the clonal evolution model becomes. The theoretical performance of the clonal evolution model is a different issue. The current model may only partially capture the actual involvement of evolutionary processes in cancer. It is important to separate the challenges related to the factual involvement of evolutionary processes from those related to the model’s theoretical performance. The latter can be addressed by extending the current model.
We see two important but questionable underlying assumptions in the current clonal evolution model: (1) the phenotype of cancer cells, in particular their fitness, is assumed to largely rely on their genotype (studies of clonal evolution have been historically dominated by genetic approaches); (2) the ancestral relationships between cancer cells are assumed to form a branching tree of cells, neither of which is actually necessary for a system to evolve. We will analyze both assumptions in detail, searching for limits and critiques to the model and distinguishing those that are real limitations to how much evolutionary theories can explain cancer (factual extent), and those that are invitations to improve the current clonal evolution model (theoretical limitations).
Assumption #1: fitness relies largely on genotype
The clonal evolution model draws from the modern evolutionary synthesis, which is a quantitative genetic theory of Darwinian evolution defining evolution as changes in the frequencies of genetic variants in a population through space and time. As a mathematical and theoretical framework, it requires some simplifying assumptions, in particular the organism is reduced to its genetics. In oncology, specific genetic alterations have been identified and characterized for their key functional roles in the development and evolution of the disease, which led to the success of the notion of oncogenes and tumor suppressor genes in the 1980s.116,117 However, it is well known from developmental biology that the same genotype can produce widely different phenotypes in normal cells116,117 and this also applies to cancer cells.118 The assumption that the fitness of cancer cells largely relies on their genetics faces a number of challenges.
Identifying key clonal evolution genes is challenging
Large consortiums have invested a lot of effort in sequencing tumors to identify cancer genes and several confounding factors have been identified, such as the size of the genes and their location in fragile sites.64,119–121 The statistical significance of the presence of some mutations needs to be evaluated according to some background factors such as the mutation rate of each nucleotide, structural variations, and purity of the sample. Given that mutations in typical cancer driver genes are present in somatic healthy tissues, it also appears crucial to compare the frequency of mutations in cancer to their frequency in non-cancerous tissues. For example, NOTCH1 mutations are common in esophageal cancer and skin cancer, leading to the assumption that it is a driver gene in those cancers. However, as NOTCH1 mutations are more common in normal skin and esophageal epithelium than in the cancers of those tissues, NOTCH1 mutations may actually play a protective role against both skin and esophageal cancer.122,123 Thus, identifying driver mutations is not trivial.
There is also a conceptual problem in identifying cancer genes.124 The same mutation can be neutral or even deleterious in one context and become a driver in another, and vice versa. The concept of the cell of origin illustrates this phenomenon: the same mutation may have varying impact depending on which cell it occurs in.125 Similarly, therapies change the environment of the cells which can select for clones with specific resistant mutations that expand during treatment and regress at treatment withdrawal.126 Treatments induce drastic and rapid selective pressures, but slower changes are also possible, with the same consequences. Aging, for example, changes the fitness landscape of the cells, so that a mutation could be neutral or even deleterious in a young person but be advantageous in the context of aging.101–103,127 Fitness is by definition always relative to a particular environment. The environment of cancer cells changes, throughout the natural development of the disease but also due to aging, interventions and other changes in exposures. The difficulties in identifying driver mutations complicates efforts to define clones based on driver mutations (see Box 1).
Box 1 |. What is a clone?
The model of clonal evolution is based on the analysis, in time and space, of cells belonging to different clones. The concept of the clone is therefore central, but is rarely explicitly defined. There is an implicit consensus to see cancer clones as populations of cells that share a common identity inherited from a common ancestor.274 But what alterations people use to identify a clone differs from study to study.
Traditionally, the identity of a clone is based on the genotype, or rather parts of it. A typical definition of the clone is for example: "a set of cells that descend from a common ancestor and thus share genetic features".275 For practical reasons, clonal evolution studies are often based on driver mutations. Some studies use techniques allowing broader analysis of the genome such as whole exome-sequencing, or even whole genome sequencing, which does not rely on a priori knowledge of which mutations are involved in cancer. Finally, some studies favor the use of neutral mutations as an unbiased way to track clonal evolution.276
There is also a more fundamental ontological and epistemic challenge in the identification of a clone. The clonal evolution model relies on reconstruction of clones from incomplete information: we neither have access to all the cancer cells, nor to all the heritable properties of each single cell. This can lead to ćlonal illusioń, when a subclonal mutation appears to be clonal because a biopsy was only taken within the subclone.277 It also relies on choices made by investigators as to which alterations are relevant for defining a clone and which can be ignored.124 Changing the list of mutations that are deemed relevant and thus used to reconstruct clonal evolution affects that reconstruction. The number of clones identified, for example, will depend on the number of genes studied as well as the depth of sequencing.95 As an extreme illustration of this issue, one may define a clone as the set of cells that are genomically identical. However, whole genome sequencing at the single-cell level would likely reveal that each individual cancer cell is unique, and therefore count as a new clone, making the concept of a clone useless.259 The history of the clonal evolution model shows that these choices of what alterations should be used to define a clone have changed through time. This historical contingency highlights the arbitrariness in definitions of clones. In a pluralistic approach, one could argue that which clone delineation is the right or best one will depend on the perspectives and aims one adopts. Whether, and how much, clone delineation is an issue largely depends on what is expected from the clonal evolution model. It is for example an issue for any study that quantifies intratumoral heterogeneity by counting clones. It is much less of an issue for those who may want to reconstruct phylogenetic relationships between various cancer cells (for example between metastases at different locations).
There are some alternative views of clonal identity: several studies have now been using epigenetics in their reconstruction of cancer evolution.27,37,129,143,144,147,278 A few have supported a phenotypic or functional clonal identity. For example, a clone was defined as "a group of cells with the same phenotype, which have expressed that phenotype consistently since their most recent common ancestor".63 That definition might call a set of stem cells a clone if they shared the same phenotype and had a recent common ancestor, but would exclude all the non-stem cells that were part of the same branch on the cell lineage tree. In contrast, in developmental biology, a clone has been defined as “the in vivo descendants of a single ancestral cell", equivalent to a monophyletic clade in a phylogeny.279
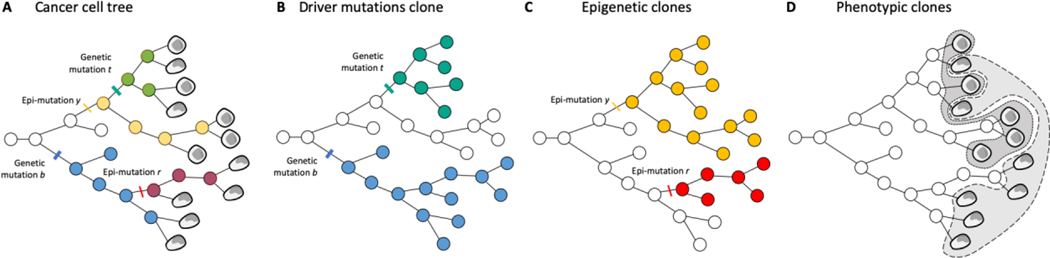
Under every cancer is a cell lineage tree describing the relationships and history of the cells within the cancer.74,168 Defining clones is a matter of dividing up that evolutionary tree (Figure 2).
Genes are not the only inherited material
It is now well appreciated that many epigenetic properties are heritable at cell division and can contribute to clonal evolution,128–137 including the evolution of metastasis138,139 and resistance to drugs.133,140,141 However, epigenetics encompasses a broad range of phenomena, some of which are random and heritable, while other are under the control of the cell and may change in response to some form of signaling, for example during differentiation. The heritable alterations could be integrated in clonal evolution as epimutations.142 Studies have found a concordance between genetic and epigenetic clonal evolution, which makes sense given that there is one unifying lineage of the cancer cells.143–145Additionally, there can be multiple epigenetic subclones within one genetic clone, or vice versa.146,147 This again raises the question what information to use to delineate and count clones (Box 1, Figure 2). Epigenetic alterations challenge the theoretical accuracy of the clonal evolution model when the model relies only on genetic mutations, but it does not challenge its factual extent.
Figure 2|. Defining clones is a matter of dividing up the cancer cells tree.
a, The tree of cancer cells, with genetic, epigenetic, and phenotypic information overlaid. b, The cancer cells tree with clones identified through driver mutations. 3 clones are identified (white with whatever mutations the transformed cell had, blue with the occurrence of the blue mutation b, turquoise with the turquoise mutation t). c, The cancer cells tree with clones identified through epigenetic alterations. 3 clones are identified (white with the epigenetic state of the first transformed cell, red with epigenetic mutation r, yellow with epigenetic mutation y). They are not the same clones as in the genetic clones. d, Phenotypic clones. There are 2 cell types. The turquoise genetic clone is mixed and contains both cell types, while the white and blue genetic clones contain a different cell type. The yellow epigenetic clone is mixed, while the red and white epigenetic clones contain the same cell type. Are there 3 clones, 5 clones, or more?
Heritable properties may extend beyond the genetic material and its epigenetic alteration. As cell reproduction happens through cell division, daughter cells inherit a portion of the cytoplasm of the mother cell, including organelles, RNA, proteins. They also inherit the microenvironment that the mother cell may have modified.213–215 This may generate short-term heritability of the cells’ phenotype with time scales depending on the fluctuations and turnover rate of those molecules and the microenvironment. In bacteria, the use of śister machineś that separate the two daughter cells in independent growth channels, revealed that different non-genetic traits present different ´memorý patterns under different time-scales, from 2 to 10 generations.148 In a melanoma cell line, 227 genes that show transient heritable high expression (around 40h, sometimes reaching 5 days) were identified,149 of which 162 were previously associated with resistance to therapy.150 Cells sorted for high expression of two of these transiently heritable genes, specifically EGFR and nerve growth factor receptor (NGFR) showed a much higher level of resistance to the MEK inhibitor trametinib than unsorted cells.
The inheritance of properties at different time-scales remains largely ignored, however given the short duration of therapeutic interventions, these alternative forms of inheritance may be of clinical relevance. As discussed below, improving our understanding of inheritance might improve the theoretical performance of clonal evolution.
One genotype can take on many phenotypes
Normal somatic cells in multicellular organisms are phenotypically plastic. Phenotypic plasticity has long been studied in the field of development and is increasingly recognized as important in cancer as well. From a single genotype, cells can take on different phenotypes as a result of at least three non-exclusive processes: differentiation, response to other extrinsic signals, and stochastic fluctuations (of gene products and epigenetic modifications). The latter two can include dedifferentiation, transdifferentiation, or other changes in cell states such as metabolic changes. Cancer cells inherit and modify the phenotypic plasticity of the normal cells from which they evolve.
Currently, phenotypic variations that are independent of the genotype are disregarded by the clonal evolution model. Thus, plasticity is taken as a challenge to the factual extent of the clonal evolution model: the more non-heritable plasticity is causally involved in cancer, the less evolutionary processes are. For example, a study of dormancy and relapse in estrogen receptor–positive breast cancer treated with adjuvant endocrine therapies showed that the treatment stochastically induces a dormant state, which was unstable, in a fraction of cancer cells from a random diversity of clones.133 In such cases, evolution by natural selection does not play much role in cancer cells ability to survive treatment. However, conceptual fuzziness about phenotypic plasticity obscures the debate.151 Some cases of plasticity might heritable and in fact only be a theoretical challenge which may be incorporated to improve the clonal evolution model.
First, the phenotype of cancer cells often relies on non-genetic, yet heritable, properties, such as epimutations. Such epigenetic-driven phenotypic diversity can easily be integrated in the clonal evolution model by incorporating epimutations. The same should apply to any other non-genetic heritable property that contributes to the cell phenotype.
Second, the degree of phenotypic plasticity can be heritable, encoded in the genome or epigenome (reviewed in ref151). Certain cell lineages may be more plastic than others, providing them an advantage in changing microenvironments. Other cells have phenotypes that are remarkably stable, such as fully differentiated cells. Some (epi)mutations may increase cell plasticity, and selection on those (epi)mutations will flow indirectly through direct selection on the phenotypes they produce. There is relevant literature on the evolution of mutation rates152–155 and the evolution of phenotypic plasticity156–158 that might be used to integrate phenotypic plasticity into the clonal evolution model.
Third, phenotypic plasticity, includes a variety of phenomena such as differentiation (a robust, channeled, predictable process in multicellular organisms), response to extrinsic signals (also partly predictable, although conditional on microenvironmental signals), and stochastic fluctuations (unpredictable). The differences between these processes matters if one wants to address the empirical limitations of evolutionary accounts of cancers. Why these types of plasticity need to be conceptually distinguished, can be illustrated by two hypothetical contrasting examples. First, when a treatment selects stem cells due to their upregulation of efflux pumps, selection for efflux pumps will maintain the clonal composition. This is because all genetic clones contain cancer stem cells, although there may be changes in clone size based on the proportions of cancer stem cells in the different clones.81 Second, when a cell cycle-specific treatment selects for a slow-cycling or non-proliferating cells, and if quiescence is induced by a particular microenvironment, then clones that happen to be in that microenvironment will survive. These two cases present different challenges to the clonal evolution model. In the first case, the maintenance of all genetic clones may hide the selection of stem cells. In the second case, resistance might wrongly be attributed to the genetics of the clone when it is its location that caused the resistance. Other scenarios involving plasticity may also challenge the clonal evolution model. For example, the clonal evolution model takes the cancer stem cells to be the units of selection,79 but dedifferentiation may occur159,160 allowing non-stem cells to contribute to the evolutionary dynamics.81,161
While phenotypic plasticity in response to external stimuli represents a major limitation to the factual involvement of clonal evolution, differentiation-based and stochastic-based types of plasticity represent theoretical challenges. As far as differentiation remains robust and predictable, this type of plasticity could be included in mathematical models of clonal evolution.81 Similarly, stochastic plasticity is amenable to mathematical modeling that can be introduced in evolutionary models of cancer.162
Many genotypes can produce the same phenotype
Such phenomena can be cases of convergent evolution, phenotypic plasticity, or non-heritable factors. Selective pressures may select for the same phenotype in independent lineages, which is well accounted for by the evolutionary models of cancer. We have argued that the hallmarks of cancer are cases of convergent evolution that occur due to natural selection acting on unrelated cells.58 This challenge to the genetic evolutionary model of cancer led to the hypothesis that we gain more clarity and control by focusing on the evolution of phenotypes.163 As we have argued above, differentiation processes and responses to environmental signals can result in similar phenotypes regardless of the underlying genetics. There are other non-heritable determinants of phenotype that may be independent of the clones genetics. For example, cancer cells can transfer proteins, lipids and nucleic acids via extracellular vesicles which can contribute to the malignant phenotype.164 In a mouse model of gliomas, it was shown that cancer cells with mutated EGFR can transfer the truncated oncogenic form EGFRvIII to cancer cells lacking the mutation, leading to the acquisition of the mutated phenotype by these cells.165 Distinguishing between heritable and non-heritable determinants of phenotypes is key, as the second is a direct challenge to the factual extent of the clonal evolution model, although evolutionary theory can account for non-heritable determinants of phenotype as a form of noise, by including measures of the heritability or evolvability of traits in models of phenotypic evolution.166,167
Assumption #2: A cancer cell lineage is a tree
The clonal evolution model assumes a tree shape of the cell lineages,168 where cell lineages divide and die but do not fuse. Clones are defined by descent from a single common ancestor cell (Box 1). Inheritance is vertical, from the parental cell. Moreover, basing the clonal evolution model on cell lineages implicitly assumes that cells are the only unit of selection. Several observations question these assumptions.
Cell fusion and cancer cell modes of reproduction
Cancer cells can fuse into viable, reproducing cells that can give rise to clonal populations of cells.169,170 Cell fusion in cancer is considered rare and has been largely neglected. Yet, several studies reported fusions between cancer cells and non-cancer cells (hybridomas being a famous case of such fusion), or between different cancer cells. In patients, fusions have been reported with hematopoietic cells in the context of bone marrow transplantations (BMT) that offer markers to track cell fusions.169,171,172 As an example, a mother who underwent BMT from her son, later developed a renal carcinoma with a fraction of the cancer cells containing both chromosomal alterations of the cancer cells, and a Y chromosome from her son’s cells.171 Another study analyzed tumor biopsies from 7 patients with various cancers (pancreatic ductal adenocarcinoma, renal cell carcinoma, head and neck squamous cell carcinoma, and lung adenocarcinoma), and found evidence of fusions between cancer cells and leukocytes in all of them.169 Following up on this result, they searched for circulating hybrid cells in patients with pancreatic cancer and found that the quantity of circulating hybrid cells correlated with advanced disease and was associated with poor prognosis, which was not the case of non-fused circulating tumor cells, raising the possibility that fusions may increase the risk of cancer progression.
The role of cell fusion in cancer cell evolution has been studied in vitro and in animal models. Hybrids of cancer cell lines and macrophages, once transplanted into mice, had a shorter doubling time than the maternal cancer cell line, suggesting that cell fusion can increase the fitness of the cells.169 Cell fusion can also provide cancer cells with new properties. For example, fusion with hematopoietic cells such as macrophages can endow the fused cancer cells with migrating abilities.169,173–177 Several studies discuss fusion as a possible alternative mechanism to (but not mutually exclusive with) the Darwinian clonal evolution explanation of metastasis according to which the ability to metastasize is gained through a process of evolution by natural selection.176,177 Other studies have shown that fusion between cancer cell lines can generate new cell lines that are more malignant and therapeutically resistant than the original cell lines.169,170 Cell fusion has also been discussed as a possible mechanism to produce cancer stem cells.178,179 Last but not least, cell fusion is a mechanism that can generate diversity through subsequent stochastic loss of genetic material.180
Cell fusions do not undermine the factual extent of clonal evolution but add a level of complexity regarding processes involved in their evolution that is not currently taken into account by the clonal evolution model.
Horizontal gene transfer
The clonal evolution model assumes that inheritance is vertical with cell division. But several mechanisms of horizontal gene transfer have been described such as exchange of mitochondria and DNA. Co-culture of a lung cancer cell line depleted of mtDNA with bone marrow non-hematopoietic cells or skin fibroblast,181 or transplantation of breast cancer cell lines depleted of mtDNA182 showed rescue of mitochondrial function through mitochondrial transfer. Introduction of mitochondria from mesenchymal stromal cells (MSCs) into cancer cells led to an increase in oxidative phosphorylation, ATP production, migration and proliferation of the cancer cells.183 Horizontal transfer of mitochondria through tunneling nanotubes from endothelial cells to MCF7 breast cancer cells was shown to improve chemoresistance to doxorubicin in vitro.184 Similar results were also obtained through co-culture with bone marrow stromal cells in vitro or after engraftment of acute myeloid leukemia blast cells from patients or cell lines in mice.185 Coculture of glioblastoma stem cells (GSCs) and MSCs also led to metabolic rewiring of GSCs following mitochondria transfer from MSCs to GSCs, increased proliferation and resistance to temozolomide in GSCs.186
Horizontal gene transfer can also occur in cancer due to the uptake of DNA from extracellular vesicles, including apoptotic bodies from dying cancer cells.187 This has been shown in vitro187,188 180,181 as well as in mouse experiments,189 and can be prevented by treatment with DNAses.189 Microvesicles have been shown to carry oncogenes such as MYC or HRASV12 ref188,190,191 that can be taken up by other cells. The extent and importance of horizontal gene transfer in cancer through extracellular vesicles remains an open question.
Horizontal gene transfer does not contradict an evolutionary model of cancer. Rather it adds complexity to the model. Adaptation through natural selection can occur through horizontal transfer in addition to vertical inheritance.
Trogocytosis and horizontal transfer
Beside gene transfer, phenotypic properties can also be horizontally transferred through trogocytosis, a mechanism of membrane fragment transfer between cells. Trogocytosis is a relevant phenomenon for cancer cell evolution as it allows cells to acquire new phenotypic properties independently of their genetics and epigenetics, leading to a mismatch between the genotype and phenotype. For example, in a colon cancer mouse model using patient-derived xenografts, cancer cells were able to acquire lymphocyte membrane proteins including lymphocyte cellular markers, as well as immune regulatory surface proteins, which suppress activation of immune cells.192 Similar results were obtained using a mouse model of leukemia,193 although in this study, Natural Killer (NK) cells and CD8+ T cells acquire the checkpoint receptor programmed cell death protein 1 (PD1) from leukemia cells, which results in the suppression of NK cell antitumor immunity. Trogocytosis has been implicated in chimeric antigen receptor-T cell (CAR-T) cell and CAR-NK cell escape.194,195 Trogocytosis can only be meaningful to clonal evolution if the phenotype it induces can be transmitted through at least one cell division. Cell surface materials are partly maintained through cell division, but the symmetry of inheritance to daughter cells is unknown, as well as the number of divisions after which the phenotype is lost.
Cell cannibalism is another process that can provide properties to cells that change their fitness under particular selective pressure. For example, metastatic melanoma cells can cannibalize T lymphocytes, which allow them to survive under serum deprivation.196 Similarly, a breast cancer cell line was shown to cannibalize mesenchymal stromal cells, which also enhanced their survival in the context of starvation, through the induction of a dormant state.197 Again, its contribution to clonal evolution depends on the heritability of those phenotypes.
Horizontal transmission of phenotypes due to trogocytosis or cell cannibalism reduces the heritability of those phenotypes and so reduces the factual extent of the clonal evolution model.
Intracellular microbes
Although clonal evolution refers to the evolution of cancer cells, presence of microbes (such as viruses, bacteria and fungi) inside cancer cells may call for a multispecies view of clonal evolution.198 Viruses have been implicated in carcinogenesis for a long time, and in cases where they directly cause carcinogenesis and even integrate into the cancer genomes, they become part of the genetic material that is evolving in cancers.199,200 Recent studies have observed that bacteria and fungi are not just around some cancer cells, but that they can also reside inside them201–205 and it was even suggested that most intratumoral bacteria are intracellular, residing in both cancer cells and immune cells.202
Intracellular microbes can change their host cell functional properties and fitness, thereby contributing to clonal evolution. They can increase the fitness of cancer cells, for example, in colorectal cancers, Fusobacterium nucleatum have been associated with increased tumorigenicity.206,207 Fusobacterium-positive tumors engrafted in patient-derived xenografts models while Fusobacterium-negative ones did not, a difference that was reversed by antibiotic treatment.201 Bacteria can inhibit or elicit immune responses, for example through peptide presentation203,208 and can also mediate cancer cells resistance to therapies, for example through metabolization of chemotherapies into an inactive form.209 Human Papilloma Virus (HPV) interferes with multiple tumor suppressor mechanisms and increases the fitness of the neoplastic cells that they infect.210
Thus, intracellular microbes may contribute to clonal evolution in predictable ways that could be implemented in the clonal evolution model. The heterogeneous and focal distribution of intracellular bacteria lead to the notion of ´microbial intraclonal diversitý whereby different fitness of cancer cells of the same genetic clone is dependent on the presence of intracellular bacteria.198 There are, however, theoretical challenges to the integration of the role of microbes in clonal evolution. One of them is the mode of transmission of these microbes. They may be transmitted vertically, and thus simply represent a subclone of infected cancer cells, as well as horizontally by transiting from one cell to another. In the second case, they question the tree-shape of inheritance patterns among cancer cells. Moreover, bacteria themselves may undergo their own evolutionary dynamics. Bioinformatic analysis of the metabolic activity of intratumoral bacteria suggested that bacteria are under their own selective pressures shaped by the tumor cells and their microenvironment.202 Thus various selective processes may act on cancer cell lineages and microbial lineages.198
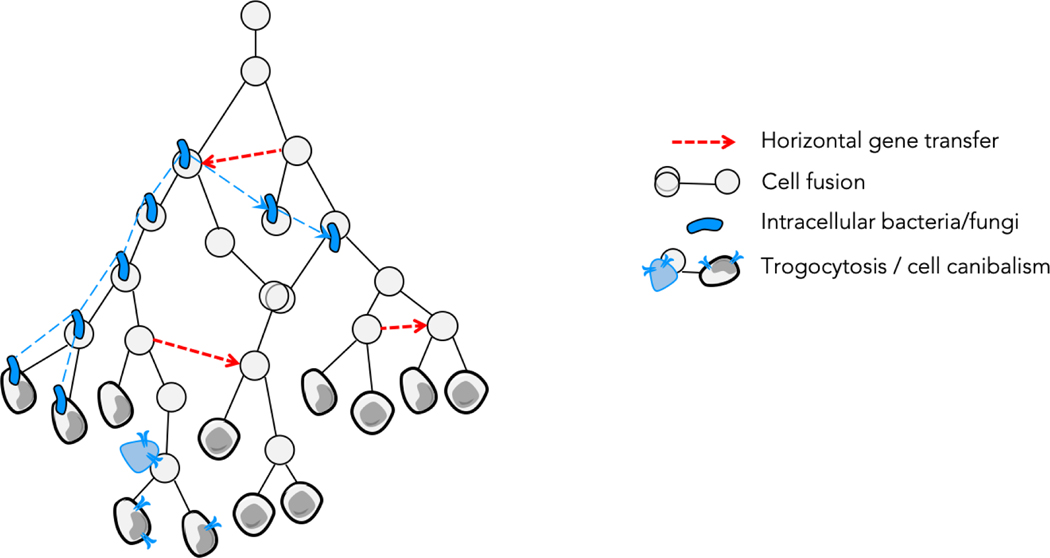
The evolution of cancer cells may thus not be a simple branching tree of cells, but might involve fusion and transmission between branches (Figure 3). Genetic and non-genetic inheritance may not just be vertical but may also sometimes be horizontal. Additionally, clonal evolution might not only concern cancer cells but also intracellular bacteria and fungi that may change the fitness of cancer cells. These mechanisms challenge the theoretical efficacy of the current clonal evolution model rather than the factual extent of the involvement of cancer cell evolution. They thus open avenues to improve the theoretical power of the clonal evolution model.
Figure 3|. Clonal evolution is more complex than a bifurcating tree of cells.
Clones can use horizontal gene transfer (dotted arrows) to transfer heritable information, thereby breaking the assumption of vertical inheritance. The purple bean-shaped cells represent intracellular microorganisms, which evolve and have their own lineages indicated by the purple dotted lines. Elements of the cells can also be transmitted horizontally from cells, including non-malignant cells, to cancer cells, through absorption of extracellular vesicles, tunneling nanotubes, trogocytosis or cell cannibalism, which is represented by the transfer of cell surface receptors from the blue cell (for example a lymphocyte) to the gray cancer cell.
Improving the theory of cancer
The above challenges and limitations of the clonal evolution model suggest opportunities for improving the model. We will focus on four potential extensions to the clonal evolution model: the diversity and time scale of inheritance beyond genetics, phenotypic plasticity, reticulating modes of evolution, and the concept of clone.
Inheritance and time scale
Inheritance is a central concept for any evolutionary theory of cancer. There cannot be evolution without some sort of inheritance. But inheritance is loosely defined, and its definition is subject to debate.
Several classifications of forms of inheritance have been proposed.211 This includes the distinction of internal from external channels of inheritance.212 Internal inheritance comprises factors that are transmitted through cell division (or cell fusion) such as genetic and epigenetic inheritance. External inheritance contains every transmission that is not passed through cell division. Internal inheritance can be traced through cell lineages while external inheritance cannot.
It is relatively easy to extend genetic models of inheritance to epigenetic models, at least for randomly mutating epigenetic states.143,213,214 But internal inheritance can also extend beyond genetics and epigenetics. One approach to studying non-genetic and non-epigenetic heritable phenotypes could be to quantify the heritability of those phenotypes using the breeders’ equation.215 By measuring change in cancer cell phenotype (in vitro or in vivo) over different timescales and selective pressures, cancer biologists could quantify the degree of heritability of a phenotype of interest. This would account for both internal and external modes of inheritance, including non-(epi)genetic inheritance of the cytoplasm and niche construction. Note that these measures of heritability depend on the degree of heritable variation in the population. A phenotype could be heritable but would appear to be not if the population did not include variation in the substrates that encode that phenotype.
There is external inheritance in cancer, at least through niche construction.216 There is a form of co-evolution between organisms and the niches that they alter, that can be represented in formal models of evolution.217 When neoplastic cells engage in niche construction, by activating fibroblasts,218,219 recruiting immune cells,220–222 inducing angiogenesis,223 and otherwise altering their microenvironment,224–227 they generate a form of external inheritance that changes the selection pressures on themselves, and thereby change their own evolutionary trajectories.217 The timescale of such external inheritance can be long. For example, activated fibroblasts can maintain their pro-tumor phenotype even in the absence of cancer cells, through autocrine loops, epigenetic alterations, and even genetic alterations.228 Efforts have been made to start integrating ecology in the clonal evolution model,94 but accounting for niche construction remains challenging.
There have also been debates about which kind of inheritance matters more.229–231 One argument is that there is a causal asymmetry between factors transmitted over many generations and those transmitted over one generation, the former playing a more substantial role in evolution because they can accumulate over long periods of time by natural selection.232 In cancer, however, selective pressures change at very different time scales. Aging slowly reshapes the selective landscape over decades.101–103 The hallmarks of cancer, that are necessary for transformation, must last for the time span of neoplastic progression, which can take decades.147 Short-term forms of inheritance cannot meaningfully contribute to these long processes of evolution by natural selection.232 However, there are other barriers to neoplastic progression, such as survival in circulation during metastasis, which likely only needs to be overcome for a matter of hours or days. In that case, inheriting some key proteins from a parental cell, or a niche, like a cluster of neoplastic cells that may safely travel through the bloodstream, may last long enough to deliver the neoplastic cell to a new metastatic microenvironment. Similarly, treatments like radiotherapy and chemotherapies, which impose intense selective pressures on cancer cells, are often applied for the duration of only a few cell generations, similar to the time-scale of cytoplasmic inheritance.
The focus on genetic clonal evolution can only partially capture the short time scale events of evolution and several avenues could help improve the model in this regard. One approach involves reconstructing clonal evolution using a variety of molecular clocks. DNA, copy number variants (CNVs), and single nucleotide variants (SNVs) mostly change over years.233,234 Methylation of CpG sites have faster clock rates than SNVs, although those rates may vary from site to site in the genome.162 Other epigenetic modifications to histones235,236 or changes to cell state through signal transduction and transitions to new attractor states in the genetic regulatory network237,238 can occur rapidly and may require their own molecular clocks in clonal evolution models.
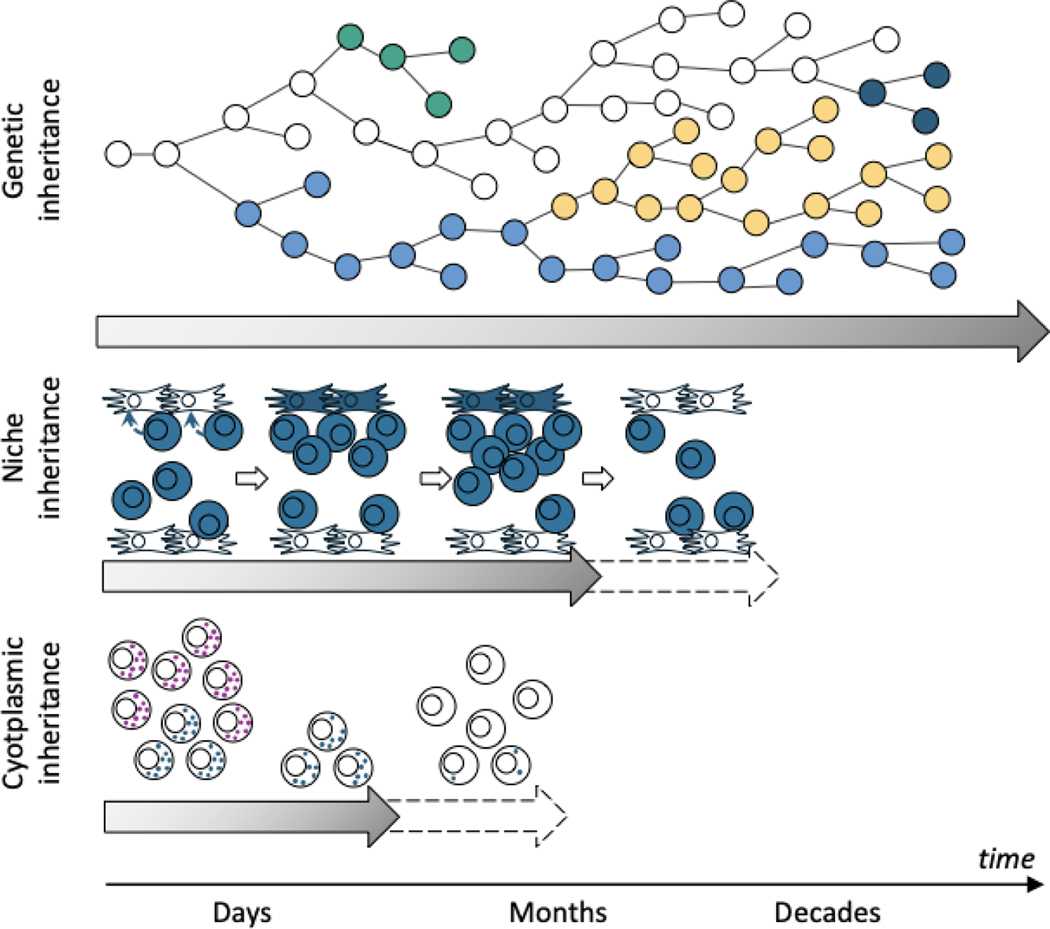
Future research should characterize the time scales of different mechanisms of inheritance (Figure 4). Any phenotypic change that lasts less than the cell lifespan cannot be a meaningful heritable property for clonal evolution. Any phenotypic change that lasts less than the time scale of a selective pressure will have little effect on the cell population’s evolution in response to that pressure. In contrast, mechanisms of inheritance that produce phenotypic changes that last as long or longer than selective pressures, will be important to a population’s response to these pressures, as long as there is relevant variation in the cell population. There are mathematical methods for disentangling the different forms of inheritance, including genetic, epigenetic and ecological inheritance.239
Figure 4|. Types of inheritance over various timescales.
Genetic inheritance may contribute at any time scale and persist throughout life. Niche construction by cancer cells may have selective effects lasting for days to months, though the exact timescale is unknown. Some entities such as cytoplasmic and cell surface proteins may be inherited for a very short period of time, likely just days. They may be important contributors to evolution by natural selection in case of abrupt and short changes in selective pressures such as cancer therapies.
Phenotypic plasticity
Phenotypic plasticity can weaken the relationship between genotypes and phenotypes, reducing the heritability of the phenotype, and thereby dampening the effects of natural selection. However, phenotypic plasticity itself can be an adaptive phenotype and has evolved multiple times by natural selection.156–158 At least one historical theoretical framework supports a contribution of phenotypic plasticity to genetic evolution: the Baldwin effect, a process by which an initial phenotypic accommodation is later reinforced by genetic adaptation.
The Baldwin effect, in its simplified form,240,241 refers to a three-step process: first, some individuals (in our case, cancer cells) accommodate environmental changes through phenotypic plasticity. This initial phase of phenotypic plasticity allows some individuals to avoid extinction. Second, some genetic (and thus heritable) changes occur in the surviving population that allow them to survive in the new environment without having to pay the cost of phenotypic plasticity. Third, these genetically determined phenotypes are favored by natural selection and finally spread in the population because they achieve the same or more fit phenotypes as the phenotypically plastic individuals without having to pay the cost of plasticity, which may be a metabolically demanding process. Hence, the Baldwin effect is a particular case where an adaptation is initially individual and non-heritable and becomes hereditary and selected.
Several studies suggest mechanisms of initial phenotypic adaptation that are secondarily genetically selected.133,242,243 For example, when exposed to EGFR inhibitors, most EGFR mutated lung cancer cells die, but a few can survive through a phenotypic accommodation called ´persisteŕ state. Sequencing and drug screening of relapsed clones developing from persistent cells of a single initial EGFR mutated clone revealed secondary selection of heterogeneous resistance mechanisms,242 suggesting a case of Baldwin effect. Similarly, it was observed that the EGFRT790M resistance mutation is not present at the time of therapy in some cases and can evolve from persister cells.243 Persister cells need not be quiescent. Brief exposure to vincristine can activate efflux pumps which protect cells from the drug, allowing them to persist in an active state. That activation is stably inherited by epigenetic modifications5 which may later be stabilized by genetic alterations.
The Baldwin effect has long been neglected in evolutionary biology as a minor phenomenon that simply ´buys timé,244 and thus is of interest only in cases where selective pressures change too fast for the traditional variation and selection process to produce adaptation226 Cancer is precisely one of those situations where selective pressure may change quickly (for example the onset of treatment) and buying time might be a crucial evolutionary mechanism that could be incorporated in the clonal evolution model.
Reticulation, introgression and open lineages
Cell lineages are usually assumed to form a tree, but as discussed above, cancer cell fusion can occur, and inheritance can also be horizontal, with possible transfer of mitochondria, extrachromosomal DNA, pieces of cells such as membranes, or intracellular bacteria and fungi. Those processes all violate the tree assumption and impact phylogenetic reconstruction, as the cell tree not necessarily matches the gene trees (Figure 3). It also impacts clonal deconvolution of bulk assays,245,246 as horizontal gene transfer and cell fusion violate many of the assumptions those algorithms are based upon. Multiple discordant gene trees may exist within a cell lineage, reflecting the diverse flows of genes through all these independent phenomena. Similar lineage violations are common in prokaryotes, giving rise to the concept of reticulated evolution. Reticulation has led many to argue that the tree of life is a misrepresentation.233,234,239 It may be more appropriate to represent evolution as a network or web of interlaced branches.247–249 Some suggest replacing ćlades ´ by ćlans ´ when there is no clear unique common ancestor, to avoid unjustified assumptions coming with the traditional terminology of phylogenetics.250 This could apply to clones. Several measures have been developed and used such as the concordance factors,251 diversity indices for unrooted trees;252 and new computational methods and statistics have been developed to identify lateral transfer and analyze phylogenetic networks.253,254 In general, the revised view of evolution that developed around reticulated evolution calls for a multi-level perspective, sometimes also referred to as integrative evolution,252,255 where multiple genetic worlds contribute to the observed diversity.256
These conceptual and computational tools represent opportunities to improve our understanding of clonal evolution. Given that horizontal genetic transfers can provide evolutionary short-cuts to acquire functional adaptations such as the ability to metastasize or multi-drug resistance, it is important to evaluate the implications of such reticulation events in cancer progression, and to adopt an integrative clonal evolution model.
Quantifying Clonal Diversity
Counting clones as a proxy of intratumor heterogeneity and cancer cell evolution is based on two implicit assumptions: that the cells within a clone are homogeneous (intra-clonal homogeneity) and distinct from other clones (inter-clonal heterogeneity), so that their number reflects the diversity of the tumor cell population. But as discussed above, these assumptions might be disputable (Figure 2). If we use mutations to delineate clones (Figure 2a), there are 3 clones and thus 3 relevant populations of cancer cells with distinctive characteristics. However, there is a different delineation of clones with epigenetics (Figure 2b), and their integration (Figure 2d) suggests that there are more than 3 relevant populations of cancer cells.
One can measure the degree of intra-clonal homogeneity and inter-clonal heterogeneity. This could be done by measuring the distance between any 2 cells inside and between clones for any phenotypic or genotypic measure. For example, a measure of transcriptomic diversity inside barcoded lineages in three mouse models of acute myeloid leukemia257 observed that clones are clearly not homogeneous, as single cell RNA sequencing often reveals smears of cells across phenotype space, rather than tight clusters. In contrast, another study observed high intra-clonal homogeneity inside barcoded lineages and inter-clonal heterogeneity between barcoded lineages in a human melanoma cell line.258 Single cell genomics analysis can also be used to measure clonal diversity.168,259–264 One analysis found evidence for the existence of just a few clones, with little variation within clones and a large degree of differences between clones based on copy number profiling of two breast cancer cases.168 In contrast, the same team found massive genomic diversity in point mutations in two other breast cancer cases, leading the authors to question the concept of a clone as no two tumor cells were found to be genetically identical.259
It is possible to abandon the counting of clones altogether. As they are a proxy for measuring the structure and degree of genetic diversity in a neoplasm, we may replace the counting of clones with other methods of characterizing that diversity. For example, if the evolutionary relationships between samples can be reconstructed into a tree, then there are a variety of tree statistics that may actually be more sensitive and representative of the genetic diversity than the number of clones.265 In fact, some of these measures provide information on the population dynamics and selection within the neoplasm.265
Conclusions
There is no doubt that somatic cells undergo evolutionary processes, including both genetic drift and natural selection. They acquire heritable alterations, some of which affect their reproduction and survival. The fact that cancer cells evolve was realized in the 1950s and mostly investigated since the 21st century. However, how much evolutionary processes explain cancer remains open to debate. The success of the evolutionary theory of cancer for explaining many of the phenomena of cancer, and for developing better interventions, clearly shows the utility of the theory. However, it is also clear that non-evolutionary mechanisms contribute to cancer. The field is currently gauging the respective role of evolutionary and non-evolutionary mechanisms in cancer. But this cannot be done if we lack clarity on the boundaries between evolutionary and non-evolutionary mechanisms. There are many theories and models of cancer today266 whose integration is highly challenging.267 It’s unclear which ones are compatible or not with evolutionary theories. In general, other theories of cancer, like the cancer stem cell theory,268 the atavistic theory,269,270 and the wound that will not heal,271–273 describe the biological constraints and affordances within which somatic cells evolve. To address this issue, we focused on the boundaries of evolutionary processes, distinguishing two kinds of boundaries: theoretical and factual (Figure 5). The factual boundaries of the evolutionary dynamics of cancer extend beyond the current theory and the evolutionary theory of cancer needs to be expanded to account for phenotypic plasticity, alternative modes of inheritance, horizontal gene transfer, cell lineage fusions, and better measures of clonal diversity. Much more remains to be discovered about those phenomena and their relative importance in cancer. An improved evolutionary theory of cancer should lead to improvements in our prediction, prognosis, and treatment of cancer.
Figure 5|. Explanatory power of clonal evolution.
When aiming to explain cancer in its entirety, evolutionary processes can explain only some, but not all cancer phenomena. Other theories, such as the cancer stem cell theory or some versions of the persister cell models provide complementary explanations for various phenomena in cancer. The clonal evolution model only captures parts of the causal role of evolutionary processes in cancer. There are thus opportunities to increase our current ability to provide evolutionary accounts of cancer, by integrating heredity, plasticity, reticulate evolution and clone diversity into the clonal evolution model. The proportion of cancer phenomena that can be explained by evolution is unknown.
Acknowledgments
This work has been funded by CNRS MITI through the 80|Prime program, Cancéropôle IDF (n°2021–1-EMERG-54-CNRS DR 5–1), SIRIC (INCa-DGOS-Inserm-ITMO Cancer_18002), McDonnell Foundation, Ligue Nationale Contre le Cancer EL2020 (to LL), as well as by NIH grants U54 CA217376, U2C CA233254, R21 CA257980 and R01 CA140657 (to CCM) as well as CDMRP Breast Cancer Research Program Award BC132057 and the Arizona Biomedical Research Commission grant ADHS18–198847 (to CCM). The findings, opinions and recommendations expressed here are those of the authors and not necessarily those of the universities where the research was performed or the National Institutes of Health.
Glossary terms
- chronic myelomonocytic leukemia
A type of chronic leukemia affecting the myeloid cells and defined by a persisting monocytosis.
- stasis
long periods during which no or few evolutionary changes occur.
- convergent evolution
independent evolution of a similar phenotype in different lineages.
- breeder’s equation
ΔZ=h2S describes the change in the mean phenotype (Z) in the population as a function of its heritability (h) and the selection pressure (S) placed on that phenotype.
- clades
A monophyletic group, i.e. a group of all the individuals that derive from a common ancestor.
- Clans
an unrooted analogue of monophyletic group or clade, coming from the Gaelic for family. This definition comes from reference250.
- epistemic challenge
Challenges that relate to our knowledge.
- intratumor heterogeneity
Variation between cells within a neoplasm.
- Hybridomas
Cell lines coming from the fusion of B cells with immortal myeloma cell lines that are used to produced monoclonal antibodies
- karyotype
The set of chromosomes of a cell, including any large scale abnormalities visible in a mitotic spread.
- clonal mutation
A mutation that derives from a single ancestral cell and is shared among all its descendants.
- subclonal mutation
A mutation that is present in only a subset of the cells within a clone.
- clonal expansion
An increase in the number of cells deriving from a common ancestor that defines the clone.
- fitness
The ability of a cell to survive and proliferate in its current microenvironment.
- neutral evolution
Changes in allele frequency in a population due to chance rather than fitness.
- negative selection
The removal of mutations from a population due to their negative effect on the fitness of the organism or cell.
- gradualism
Evolution through slow continuous small changes in the phenotypes of organisms over time
- punctuation
Evolution through sudden important changes in the phenotypes of organisms
- molecular clocks
Molecular processes that change proportional to time and can so be used to infer time since past events.
- linear evolution
Evolution characterized by a sequence of fixation events where one genotype is replaced by another in the population, such that change over time can be described by a single sequence of genotypes.
- branched evolution
Evolution in which lineages split such that multiple lineages coexist, and can be represented as a tree.
- epithelial proliferative unit
An organizational sub-structure of epithelial tissues consisting of one or a few stem cells along with their partially and fully differentiated progeny.
- phylogenetic reconstruction
The process of inferring the ancestral relationships between organisms or cells.
- monophyletic clade
The set of all the species that descended from a common ancestor.
- driver gene
A gene in which particular genetic alterations increase the fitness of a cell, causing a clonal expansion.
- deleterious mutation
A mutation that decreases the fitness of an organism or cell.
- advantageous mutation
A mutation that increases the fitness of an organism or cell, also known as a driver mutation.
- epimutation
A non-genetic heritable epigenetic change in DNA methylation or the chromatin.
- phenotypic plasticity
The ability of a cell to adopt different phenotypes without changing its genotype.
- lineage tree
A type of branching graph diagramming ancestral relationships with only a single path between any two given nodes in the tree.
- hybrid cells
Cells that directly derive from more than one ancestor.
- vertical inheritance
The passage of properties from parents to offspring.
- horizontal gene transfer
The transmission of genetic material from organisms or cells that are not direct ancestors of the recipient.
- microvesicles
Lipid bilayer-delimited particles that are released from the cell membrane.
- trogocytosis
Transfer of plasma membrane fragments from one cell to another.
- reticulated evolution
Transmission of heritable properties from one lineage to another.
- niche construction
Modification of the environment by an organism, generally to the benefit of that organism.
- transformation
The change from a normal cell state to an malignant state.
- neoplastic progression
The process of change from normal tissue to cancer and on to metastatic disease.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Marusyk A, Janiszewska M & Polyak K Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer cell 37, 471–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goyette M-A, Lipsyc-Sharf M & Polyak K Clinical and translational relevance of intratumor heterogeneity. Trends in Cancer 9, 726–737 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams MJ et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nature Genetics 50, 895–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams MJ, Werner B, Barnes CP, Graham TA & Sottoriva A Identification of neutral tumor evolution across cancer types. Nature Genetics 48, 238–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisco AO et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nature Communications 4, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Househam J et al. Phenotypic plasticity and genetic control in colorectal cancer evolution. Nature 611, 744–753 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowell P The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 8.Foulds L The experimental study of tumor progression: a review. Cancer Res 14, 327–339 (1954). [PubMed] [Google Scholar]
- 9.Foulds L Tumor progression. Cancer Res 17, 355–356 (1957). [PubMed] [Google Scholar]
- 10.Morange M What history tells us XXVIII. What is really new in the current evolutionary theory of cancer? Journal of Biosciences 37, 609–612 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Boveri T Concerning the Origin of Malignant Tumours by Theodor Boveri. Translated and annotated by Henry Harris. Journal of Cell Science 121, 1–84 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Makino S & Kano K Cytological studies of tumors. IX. Characteristic chromosome individuality in tumor strain-cells in ascites tumors of rats. J Natl Cancer Inst 13, 1213–35 (1953). [PubMed] [Google Scholar]
- 13.Sato H On the chromosomes of Yoshida sarcoma; studies with tumor cells proliferated in the peritoneal cavity of the rat transplanted with a single cell. Gan 43, 1–16 (1952). [PubMed] [Google Scholar]
- 14.Makino S Further evidence favoring the concept of the stem cell in ascites tumors of rats. Ann N Y Acad Sci 63, 818–30 (1956). [DOI] [PubMed] [Google Scholar]
- 15.Hauschka TS The chromosomes in ontogeny and oncogeny. Cancer Res 21, 957–974 (1961). [PubMed] [Google Scholar]
- 16.Nowell PC & Hungerford DA Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst 25, 85–109 (1960). [PubMed] [Google Scholar]
- 17.Linder D & Gartler SM Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas. Science 150, 67–9 (1965). [DOI] [PubMed] [Google Scholar]
- 18.Linder D & Gartler SM Problem of single cell versus multicell origin of a tumor. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability 4:, 625–633 (1967). [Google Scholar]
- 19.Fialkow PJ, Klein E, Klein G, Clifford P & Singh S Immunoglobulin and glucose-6-phosphate dehydrogenase as markers of cellular origin in Burkitt lymphoma. J Exp Med 138, 89–102 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiel E, Rodt H, Huhn D & Thierfelder S Decrease and altered distribution of human T antigen on chronic lymphatic leukemia cells of T type, suggesting a clonal origin. Blood 47, 723–36 (1976). [PubMed] [Google Scholar]
- 21.Ford CE Selection Pressure in Mammalian Cell Populations. in Cytogenetics of Cells in Culture (ed. Harris R) 27–45 (Academic Press, New York and London, 1964). doi: 10.1016/B978-1-4832-3076-4.50009-9. [DOI] [Google Scholar]
- 22.Lejeune J Aberrations Chromosomiques et Cancer. in Proceedings of the 9th International Cancer Congress (ed. Harris RJC) 71–85 (Springer, Berlin, Heidelberg, 1967). doi: 10.1007/978-3-662-41838-3_5. [DOI] [Google Scholar]
- 23.Ionov Y, Peinado MA, Malkhosyan S, Shibata D & Perucho M Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363, 558–561 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Shibata D, Navidi W, Salovaara R, Li Z-H & Aaltonen LA Somatic microsatellite mutations as molecular tumor clocks. Nat Med 2, 676–681 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Tsao J-L et al. Colorectal Adenoma and Cancer Divergence: Evidence of Multilineage Progression. The American Journal of Pathology 154, 1815–1824 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsao J-L et al. Genetic reconstruction of individual colorectal tumor histories. Proceedings of the National Academy of Sciences 97, 1236–1241 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata D & Tavaré S Counting divisions in a human somatic cell tree: how, what and why? Cell Cycle 5, 610–614 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Barrett MT et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 22, 106–109 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marusyk A & Polyak K Tumor heterogeneity: Causes and consequences. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 1805, 105–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maley CC et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38, 468–473 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine 366, 883–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burrell RA, McGranahan N, Bartek J & Swanton C The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–45 (2013). [DOI] [PubMed] [Google Scholar]
- 33.McGranahan N & Swanton C Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell 27, 15–26 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Stanta G & Bonin S Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Frontiers in Medicine 5, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maley CC et al. The Combination of Genetic Instability and Clonal Expansion Predicts Progression to Esophageal Adenocarcinoma. Cancer Research 64, 7629–7633 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Wong DJ et al. p16(INK4a) lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res 61, 8284–8289 (2001). [PubMed] [Google Scholar]
- 37.Siegmund KD, Marjoram P, Woo Y-J, Tavaré S & Shibata D Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America 106, 4828–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasa Y, Nowak MA & Michor F Evolution of resistance during clonal expansion. Genetics 172, 2557–2566 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhang HC et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nature Medicine 21, 440–448 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Morrissy AS et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature (2016) doi: 10.1038/nature16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ercan D et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 29, 2346–2356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt MW, Loeb LA & Salk JJ The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol 13, 335–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X & Zhang Z Understanding the Genetic Mechanisms of Cancer Drug Resistance Using Genomic Approaches. Trends in Genetics 32, 127–137 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Salgia R & Kulkarni P The Genetic/Non-genetic Duality of Drug ‘Resistance’ in Cancer. Trends in Cancer 4, 110–118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Cunningham JJ, Brown JS & Gatenby RA Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nature Communications 8, 1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Cunningham J, Brown J & Gatenby R Evolution-based mathematical models significantly prolong response to abiraterone in metastatic castrate-resistant prostate cancer and identify strategies to further improve outcomes. eLife 11, e76284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enriquez-Navas PM et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Science translational medicine 8, 327ra24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gatenby RA, Silva AS, Gillies RJ & Frieden BR Adaptive therapy. Cancer research 69, 4894–903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gatenby RA, Zhang J & Brown JS First Strike–Second Strike Strategies in Metastatic Cancer: Lessons from the Evolutionary Dynamics of Extinction. Cancer Research 79, 3174–3177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maley CC, Reid BJ & Forrest S Cancer prevention strategies that address the evolutionary dynamics of neoplastic cells: simulating benign cell boosters and selection for chemosensitivity. Cancer Epidemiol Biomarkers Prev 13, 1375–1384 (2004). [PubMed] [Google Scholar]
- 51.Haas L et al. Acquired resistance to anti-MAPK targeted therapy confers an immune-evasive tumor microenvironment and cross-resistance to immunotherapy in melanoma. Nat Cancer 2, 693–708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merlo LM, Pepper JW, Reid BJ & Maley CC Cancer as an evolutionary and ecological process. Nat Rev Cancer 6, 924–935 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Greaves M & Maley CC Clonal evolution in cancer. Nature 481, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crespi B & Summers K Evolutionary biology of cancer. Trends in ecology & evolution 20, 545–52 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Gerlinger M et al. Cancer: Evolution Within a Lifetime. Annual Review of Genetics 48, 215–236 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Lewontin RC Adaptation. Scientific American 239, 212–8, 220, 222 passim (1978). [DOI] [PubMed] [Google Scholar]
- 57.Germain P-L Cancer cells and adaptive explanations. Biology & Philosophy 27, 785–810 (2012). [Google Scholar]
- 58.Fortunato A et al. Natural Selection in Cancer Biology: From Molecular Snowflakes to Trait Hallmarks. Cold Spring Harbor perspectives in medicine 7, a029652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lean C & Plutynski A The evolution of failure: explaining cancer as an evolutionary process. Biology and Philosophy 31, 39–57 (2016). [Google Scholar]
- 60.Germain PL & Laplane L Metastasis as supra-cellular selection? A reply to Lean and Plutynski. Biology and Philosophy 32, 281–287 (2017). [Google Scholar]
- 61.Ling S et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proceedings of the National Academy of Sciences 112, E6496–6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sottoriva A et al. A Big Bang model of human colorectal tumor growth. Nature Genetics 47, 209–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sottoriva A, Barnes CP & Graham TA Catch my drift? Making sense of genomic intra-tumour heterogeneity. Biochimica et Biophysica Acta - Reviews on Cancer 1867, 95–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martincorena I et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell vol. 171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson DC et al. Neutral tumor evolution in myeloma is associated with poor prognosis. Blood 130, 1639–1643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyne A-M, Laplane L & Perié L To portray clonal evolution in blood cancer, count your stem cells. Blood 137, 1862–1870 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Baca SC et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephens PJ et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rausch T et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao R et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature genetics 48, 1119–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cross WC, Graham TA & Wright NA New paradigms in clonal evolution: punctuated equilibrium in cancer. The Journal of Pathology 240, 126–136 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Davis A, Gao R & Navin N Tumor evolution: Linear, branching, neutral or punctuated? Biochimica et Biophysica Acta - Reviews on Cancer 1867, 151–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anderson K et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Frankell AM et al. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature 1–9 (2023) doi: 10.1038/s41586-023-05783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abbosh C et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carreira S et al. Tumor clone dynamics in lethal prostate cancer. Science Translational Medicine 6, 254ra125–254ra125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weissman IL Stem cells are units of natural selection for tissue formation, for germline development, and in cancer development. Proceedings of the National Academy of Sciences 112, 8922–8928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greaves M Cancer stem cells as ‘units of selection’. Evol Appl 6, 102–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schiffman JD, White RM, Graham TA, Huang Q & Aktipis A The Darwinian dynamics of motility and metastasis. in Frontiers in Cancer Research: Evolutionary Foundations, Revolutionary Directions 135–176 (Springer; New York, 2016). doi: 10.1007/978-1-4939-6460-4. [DOI] [Google Scholar]
- 81.Sprouffske K et al. An evolutionary explanation for the presence of cancer nonstem cells in neoplasms. Evolutionary applications 6, 92–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Birtwell D, Luebeck G & Maley CC The evolution of metapopulation dynamics and the number of stem cells in intestinal crypts and other tissue structures in multicellular bodies. Evolutionary Applications 13, 1771–1783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burns KH Transposable elements in cancer. Nat Rev Cancer 17, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Wu S, Bafna V & Mischel PS Extrachromosomal DNA (ecDNA) in cancer pathogenesis. Current Opinion in Genetics & Development 66, 78–82 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Di Bona M & Bakhoum SF Micronuclei and Cancer. Cancer Discovery 14, 214–226 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maynard Smith J & Szathmary E The Major Transitions in Evolution. (Oxford University Press, Oxford, England, 1995). [Google Scholar]
- 87.Okasha S Evolution and the Levels of Selection. (Oxford University Press, Oxford, 2006). [Google Scholar]
- 88.Vendramin R, Litchfield K & Swanton C Cancer evolution: Darwin and beyond. The EMBO Journal 40, e108389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Letouzé E et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 8, 1315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Z-M et al. Early and multiple origins of metastatic lineages within primary tumors. Proceedings of the National Academy of Sciences 113, 2140–2145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu Z et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 51, 1113–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Al Bakir M et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature 616, 534–542 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bolan PO et al. Genotype-Fitness Maps of EGFR-Mutant Lung Adenocarcinoma Chart the Evolutionary Landscape of Resistance for Combination Therapy Optimization. Cell Systems 10, 52–65.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maley CC et al. Classifying the evolutionary and ecological features of neoplasms. Nature Reviews Cancer 17, 605–619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andor N et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nature Medicine 22, 105–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Black JRM & McGranahan N Genetic and non-genetic clonal diversity in cancer evolution. Nature Reviews Cancer 1–14 (2021) doi: 10.1038/s41568-021-00336-2. [DOI] [PubMed] [Google Scholar]
- 97.Florez MA et al. Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem Cell 29, 882–904 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fabre MA et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 606, 335–342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitchell E et al. Clonal dynamics of haematopoiesis across the human lifespan. Nature 606, 343–350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Solary E & Laplane L The role of host environment in cancer evolution. Evolutionary Applications 13, 1756–1770 (2020). [Google Scholar]
- 101.Rozhok A & DeGregori J A generalized theory of age-dependent carcinogenesis. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DeGregori J Evolved tumor suppression: why are we so good at not getting cancer? Cancer Res 71, 3739–3744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marongiu F & DeGregori J The sculpting of somatic mutational landscapes by evolutionary forces and their impacts on aging-related disease. Molecular Oncology 16, 3238–3258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berenblum I The Cocarcinogenic Action of Croton Resin. Cancer Research 1, 44–48 (1941). [Google Scholar]
- 105.Balmain A The critical roles of somatic mutations and environmental tumor-promoting agents in cancer risk. Nature Genetics 52, 1139–1143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Illmensee K & Mintz B Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proceedings of the National Academy of Sciences of the United States of America 73, 549–53 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mintz B & Illmensee K Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci U S A 72, 3585–9 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weaver VM et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. The Journal of cell biology 137, 231–45 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bissell MJ & Radisky D Putting tumours in context. Nature Reviews. Cancer 1, 46–54 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCullough KD, Coleman WB, Smith GJ & Grisham JW Age-dependent induction of hepatic tumor regression by the tissue microenvironment after transplantation of neoplastically transformed rat liver epithelial cells into the liver. Cancer Res 57, 1807–1813 (1997). [PubMed] [Google Scholar]
- 111.Maffini MV, Calabro JM, Soto AM & Sonnenschein C Stromal regulation of neoplastic development: age-dependent normalization of neoplastic mammary cells by mammary stroma. Am J Pathol 167, 1405–1410 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boumahdi S & de Sauvage FJ The great escape: tumour cell plasticity in resistance to targeted therapy. Nature Reviews Drug Discovery 19, 39–56 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Marine J-C, Dawson S-J & Dawson MA Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 20, 743–756 (2020). [DOI] [PubMed] [Google Scholar]
- 114.Shen S, Vagner S & Robert C Persistent Cancer Cells: The Deadly Survivors. Cell 183, 860–874 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Feinberg AP & Levchenko A Epigenetics as a mediator of plasticity in cancer. Science 379, eaaw3835 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morange M The discovery of cellular oncogenes. History and philosophy of the life sciences 15, 45–58 (1993). [PubMed] [Google Scholar]
- 117.Morange M From the regulatory vision of cancer to the oncogene paradigm, 1975–1985. Journal of the history of biology 30, 1–29 (1997). [DOI] [PubMed] [Google Scholar]
- 118.Pérez-González A, Bévant K & Blanpain C Cancer cell plasticity during tumor progression, metastasis and response to therapy. Nat Cancer 4, 1063–1082 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cannataro VL, Gaffney SG & Townsend JP Effect Sizes of Somatic Mutations in Cancer. JNCI: Journal of the National Cancer Institute 110, 1171–1177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Plutynski A The Cancer Genome Atlas Project: Data-driven, Hypothesis-driven or Something In-Between? in Perspectives on the Human Genome Project and Genomics (eds. Donohue C & Love AC) (Minnesota Studies in the Philosophy of Science, Minneapolis, 2021). [Google Scholar]
- 122.Martincorena I et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pradeu T et al. Reuniting philosophy and science to advance cancer research. Biological Reviews 98, 1668–1686 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Visvader JE Cells of origin in cancer. Nature 469, 314–322 (2011). [DOI] [PubMed] [Google Scholar]
- 126.Siravegna G et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nature Medicine 21, 795–801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Henry CJ et al. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. The Journal of clinical investigation 125, 4666–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pastore A et al. Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL. Nature Communications 10, 1874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heide T et al. The co-evolution of the genome and epigenome in colorectal cancer. Nature 611, 733–743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ushijima T, Clark SJ & Tan P Mapping genomic and epigenomic evolution in cancer ecosystems. Science 373, 1474–1479 (2021). [DOI] [PubMed] [Google Scholar]
- 131.Gaiti F et al. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature 569, 576–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meir Z, Mukamel Z, Chomsky E, Lifshitz A & Tanay A Single-cell analysis of clonal maintenance of transcriptional and epigenetic states in cancer cells. Nat Genet 52, 709–718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rosano D et al. Long-term Multimodal Recording Reveals Epigenetic Adaptation Routes in Dormant Breast Cancer Cells. Cancer Discovery 14, 866–889 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ciriello G et al. Cancer Evolution: A Multifaceted Affair. Cancer Discovery 14, 36–48 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ciriello G & Magnani L The many faces of cancer evolution. iScience 24, 102403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Patten DK et al. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nature Medicine 24, 1469–1480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Michealraj KA et al. Metabolic Regulation of the Epigenome Drives Lethal Infantile Ependymoma. Cell 181, 1329–1345.e24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chatterjee A, Rodger EJ & Eccles MR Epigenetic drivers of tumourigenesis and cancer metastasis. Seminars in Cancer Biology 51, 149–159 (2018). [DOI] [PubMed] [Google Scholar]
- 139.McDonald OG et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 49, 367–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS & Borley J Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer 14, 747–753 (2014). [DOI] [PubMed] [Google Scholar]
- 141.Sharma SV et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 141, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oey H & Whitelaw E On the meaning of the word ‘epimutation’. Trends in Genetics 30, 519–520 (2014). [DOI] [PubMed] [Google Scholar]
- 143.Brocks D et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell reports 8, 798–806 (2014). [DOI] [PubMed] [Google Scholar]
- 144.Mazor T et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell 28, 307–317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oakes CC et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer discovery 4, 348–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sasca D & Huntly BJP Independence of epigenetic and genetic diversity in AML. Nature Medicine 22, 708–709 (2016). [DOI] [PubMed] [Google Scholar]
- 147.Li S et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nature Medicine 22, 792–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vashistha H, Kohram M & Salman H Non-genetic inheritance restraint of cell-to-cell variation. eLife 10, e64779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shaffer SM et al. Memory Sequencing Reveals Heritable Single-Cell Gene Expression Programs Associated with Distinct Cellular Behaviors. Cell 182, 947–959.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shaffer SM et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Whiting FJH, Househam J, Baker A-M, Sottoriva A & Graham TA Phenotypic noise and plasticity in cancer evolution. Trends in Cell Biology 0, (2023). [DOI] [PubMed] [Google Scholar]
- 152.Lynch M et al. Genetic drift, selection and the evolution of the mutation rate. Nature Reviews Genetics 17, 704–714 (2016). [DOI] [PubMed] [Google Scholar]
- 153.Sniegowski PD, Gerrish PJ, Johnson T & Shaver A The evolution of mutation rates: separating causes from consequences. BioEssays 22, 1057–1066 (2000). [DOI] [PubMed] [Google Scholar]
- 154.Metzgar D & Wills C Evidence for the Adaptive Evolution of Mutation Rates. Cell 101, 581–584 (2000). [DOI] [PubMed] [Google Scholar]
- 155.Good BH & Desai MM Evolution of Mutation Rates in Rapidly Adapting Asexual Populations. Genetics 204, 1249–1266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pigliucci M Evolution of phenotypic plasticity: where are we going now? Trends in Ecology & Evolution 20, 481–486 (2005). [DOI] [PubMed] [Google Scholar]
- 157.Scheiner SM Genetics and Evolution of Phenotypic Plasticity. Annual Review of Ecology and Systematics 24, 35–68 (1993). [Google Scholar]
- 158.Berrigan D & Scheiner SM Modeling the Evolution of Phenotypic Plasticity. in Phenotypic Plasticity: Functional and Conceptual Approaches (eds. DeWitt TJ & Scheiner SM) 0 (Oxford University Press, 2004). doi: 10.1093/oso/9780195138962.003.0006. [DOI] [Google Scholar]
- 159.Chaffer CL et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A 108, 7950–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chaffer CL et al. Poised Chromatin at the ZEB1 Promoter Enables Breast Cancer Cell Plasticity and Enhances Tumorigenicity. Cell 154, 61–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Laplane L Cancer stem cells modulate patterns and processes of evolution in cancers. Biology and Philosophy 33, (2018). [Google Scholar]
- 162.Gabbutt C et al. Fluctuating methylation clocks for cell lineage tracing at high temporal resolution in human tissues. Nat Biotechnol 1–11 (2022) doi: 10.1038/s41587-021-01109-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gatenby RA, Gillies RJ & Brown JS Of cancer and cave fish. Nat Rev Cancer 11, 237–238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kalluri R & McAndrews KM The role of extracellular vesicles in cancer. Cell 186, 1610–1626 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Al-Nedawi K et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10, 619–624 (2008). [DOI] [PubMed] [Google Scholar]
- 166.Bourrat P Unifying heritability in evolutionary theory. Studies in History and Philosophy of Science 91, 201–210 (2022). [DOI] [PubMed] [Google Scholar]
- 167.Hansen TF, Pélabon C & Houle D Heritability is not Evolvability. Evol Biol 38, 258–277 (2011). [Google Scholar]
- 168.Navin N et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gast CE et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv 4, eaat7828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Miller FR, Mohamed AN & McEachern D Production of a More Aggressive Tumor Cell Variant by Spontaneous Fusion of Two Mouse Tumor Subpopulations1. Cancer Research 49, 4316–4321 (1989). [PubMed] [Google Scholar]
- 171.Yilmaz Y, Lazova R, Qumsiyeh M, Cooper D & Pawelek J Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant 35, 1021–1024 (2005). [DOI] [PubMed] [Google Scholar]
- 172.Chakraborty A et al. Donor DNA in a renal cell carcinoma metastasis from a bone marrow transplant recipient. Bone Marrow Transplant 34, 183–186 (2004). [DOI] [PubMed] [Google Scholar]
- 173.Goldenberg DM, Pavia RA & Tsao MC In vivo hybridisation of human tumour and normal hamster cells. Nature 250, 649–651 (1974). [DOI] [PubMed] [Google Scholar]
- 174.Kerbel RS, Lagarde AE, Dennis JW & Donaghue TP Spontaneous fusion in vivo between normal host and tumor cells: possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol Cell Biol 3, 523–538 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miller FR, McInerney D, Rogers C & Miller BE Spontaneous fusion between metastatic mammary tumor subpopulations. J Cell Biochem 36, 129–136 (1988). [DOI] [PubMed] [Google Scholar]
- 176.Rachkovsky M et al. Melanoma × macrophage hybrids with enhanced metastatic potential. Clin Exp Metastasis 16, 299–312 (1998). [DOI] [PubMed] [Google Scholar]
- 177.Pawelek JM & Chakraborty AK Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer 8, 377–386 (2008). [DOI] [PubMed] [Google Scholar]
- 178.Lu X & Kang Y Cell fusion hypothesis of the cancer stem cell. Adv Exp Med Biol 714, 129–140 (2011). [DOI] [PubMed] [Google Scholar]
- 179.Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J & Terzis AJA The origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer 5, 899–904 (2005). [DOI] [PubMed] [Google Scholar]
- 180.Miroshnychenko D et al. Spontaneous cell fusions as a mechanism of parasexual recombination in tumour cell populations. Nat Ecol Evol 5, 379–391 (2021). [DOI] [PubMed] [Google Scholar]
- 181.Spees JL, Olson SD, Whitney MJ & Prockop DJ Mitochondrial transfer between cells can rescue aerobic respiration. Proceedings of the National Academy of Sciences 103, 1283–1288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tan AS et al. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell Metabolism 21, 81–94 (2015). [DOI] [PubMed] [Google Scholar]
- 183.Caicedo A et al. MitoCeption as a new tool to assess the effects of mesenchymal stem/stromal cell mitochondria on cancer cell metabolism and function. Sci Rep 5, 9073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pasquier J et al. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med 11, 1–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moschoi R et al. Protective mitochondrial transfer from bone marrow stromal cells to acute myeloid leukemic cells during chemotherapy. Blood 128, 253–264 (2016). [DOI] [PubMed] [Google Scholar]
- 186.Nakhle J et al. Mitochondria Transfer from Mesenchymal Stem Cells Confers Chemoresistance to Glioblastoma Stem Cells through Metabolic Rewiring. Cancer Research Communications 3, 1041–1056 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Holmgren L Horizontal gene transfer: you are what you eat. Biochem Biophys Res Commun 396, 147–151 (2010). [DOI] [PubMed] [Google Scholar]
- 188.Bergsmedh A et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proceedings of the National Academy of Sciences 98, 6407–6411 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Trejo-Becerril C et al. Cancer Progression Mediated by Horizontal Gene Transfer in an In Vivo Model. PLOS ONE 7, e52754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Balaj L et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2, 180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Elzanowska J, Semira C & Costa-Silva B DNA in extracellular vesicles: biological and clinical aspects. Mol Oncol 15, 1701–1714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shin JH et al. Colon cancer cells acquire immune regulatory molecules from tumor-infiltrating lymphocytes by trogocytosis. Proc Natl Acad Sci U S A 118, e2110241118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hasim MS et al. When killers become thieves: Trogocytosed PD-1 inhibits NK cells in cancer. Sci Adv 8, eabj3286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hamieh M et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568, 112–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li Y et al. KIR-based inhibitory CARs overcome CAR-NK cell trogocytosis-mediated fratricide and tumor escape. Nat Med 28, 2133–2144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lugini L et al. Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res 66, 3629–3638 (2006). [DOI] [PubMed] [Google Scholar]
- 197.Bartosh TJ, Ullah M, Zeitouni S, Beaver J & Prockop DJ Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc Natl Acad Sci U S A 113, E6447–E6456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sepich‐Poore GD et al. Cancer’s second genome: Microbial cancer diagnostics and redefining clonal evolution as a multispecies process. BioEssays 44, 2100252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cantalupo PG, Katz JP & Pipas JM Viral sequences in human cancer. Virology 513, 208–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Moore PS & Chang Y Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 10, 878–889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bullman S et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nejman D et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 368, 973–980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kalaora S et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Narunsky-Haziza L et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 185, 3789–3806.e17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Casasanta MA et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Science Signaling 13, eaba9157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rubinstein MR et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kostic AD et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gur C et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 42, 344–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Geller LT et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kranjec C & Doorbar J Human papillomavirus infection and induction of neoplasia: a matter of fitness. Curr Opin Virol 20, 129–136 (2016). [DOI] [PubMed] [Google Scholar]
- 211.Lamm E Inheritance Systems. The Stanford Encyclopedia of Philosophy (2021). [Google Scholar]
- 212.Odling-Smee J Niche Inheritance. in Evolution: The extended synthesis (eds. Pigliucci M & Müller GB) (2010). [Google Scholar]
- 213.Capra JA & Kostka D Modeling DNA methylation dynamics with approaches from phylogenetics. Bioinformatics 30, i408–i414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hong YJ, Marjoram P, Shibata D & Siegmund KD Using DNA Methylation Patterns to Infer Tumor Ancestry. PLOS ONE 5, e12002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Heywood JS AN EXACT FORM OF THE BREEDER’S EQUATION FOR THE EVOLUTION OF A QUANTITATIVE TRAIT UNDER NATURAL SELECTION. Evolution 59, 2287–2298 (2005). [PubMed] [Google Scholar]
- 216.Verschuren EW Niche Reconstruction to Revert or Transcend the Cancer State. in Rethinking Cancer (eds. Strauss B, Bertolaso M, Ernberg I & Bissell MJ) 353–390 (The MIT Press, 2021). doi: 10.7551/mitpress/12111.003.0021. [DOI] [Google Scholar]
- 217.Odling-Smee J, Erwin DH, Palkovacs EP, Feldman MW & Laland KN Niche Construction Theory: A Practical Guide for Ecologists. The Quarterly Review of Biology 88, 3–28 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Fang Z et al. Signaling pathways in cancer-associated fibroblasts: recent advances and future perspectives. Cancer Commun (Lond) 43, 3–41 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Alkasalias T, Moyano-Galceran L, Arsenian-Henriksson M & Lehti K Fibroblasts in the Tumor Microenvironment: Shield or Spear? International Journal of Molecular Sciences 19, 1532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Galdiero MR et al. Tumor associated macrophages and neutrophils in cancer. Immunobiology 218, 1402–1410 (2013). [DOI] [PubMed] [Google Scholar]
- 221.Kumagai S et al. An Oncogenic Alteration Creates a Microenvironment that Promotes Tumor Progression by Conferring a Metabolic Advantage to Regulatory T Cells. Immunity 53, 187–203.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 222.Wang X et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3+Treg cells in pancreatic ductal adenocarcinoma. Oncogene 36, 3048–3058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Carmeliet P & Jain RK Angiogenesis in cancer and other diseases. Nature 407, 249–257 (2000). [DOI] [PubMed] [Google Scholar]
- 224.Hanahan D & Coussens LM Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 21, 309–322 (2012). [DOI] [PubMed] [Google Scholar]
- 225.Naito Y, Yoshioka Y, Yamamoto Y & Ochiya T How cancer cells dictate their microenvironment: present roles of extracellular vesicles. Cell. Mol. Life Sci. 74, 697–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.de Visser KE & Joyce JA The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 41, 374–403 (2023). [DOI] [PubMed] [Google Scholar]
- 227.Obenauf AC et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520, 368–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mezawa Y & Orimo A Phenotypic heterogeneity, stability and plasticity in tumor-promoting carcinoma-associated fibroblasts. The FEBS Journal 289, 2429–2447 (2022). [DOI] [PubMed] [Google Scholar]
- 229.Griffiths PE & Gray RD Darwinism and Developmental Systems. in Cycles of Contingency: Developmental Systems and Evolution (eds. Oyama S, Griffiths P & Gray RD) 195–218 (MIT Press, 2001). [Google Scholar]
- 230.Sterelny K Niche Construction, Developmental Systems and the Extended Replicator. in Cycles of Contingency: Developmental Systems and Evolution (eds. Oyama S, Griffiths P & Gray RD) (MIT Press, 2001). [Google Scholar]
- 231.Laland K et al. Does evolutionary theory need a rethink? Nature 514, 161–164 (2014). [DOI] [PubMed] [Google Scholar]
- 232.Merlin F On Griffiths and Gray’s Concept of Expanded and Diffused Inheritance. Biol Theory 5, 206–215 (2010). [Google Scholar]
- 233.Kostadinov RL et al. NSAIDs modulate clonal evolution in Barrett’s esophagus. PLoS Genet 9, e1003553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Martinez P et al. Evolution of Barrett’s esophagus through space and time at single-crypt and whole-biopsy levels. Nat Commun 9, 794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Anink-Groenen LCM, Maarleveld TR, Verschure PJ & Bruggeman FJ Mechanistic stochastic model of histone modification pattern formation. Epigenetics & Chromatin 7, 30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bruno S, Williams RJ & Vecchio DD Epigenetic cell memory: The gene’s inner chromatin modification circuit. PLOS Computational Biology 18, e1009961 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lugagne J-B & Dunlop MJ Cell-machine interfaces for characterizing gene regulatory network dynamics. Curr Opin Syst Biol 14, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Luscombe NM et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 (2004). [DOI] [PubMed] [Google Scholar]
- 239.Danchin É et al. Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat Rev Genet 12, 475–486 (2011). [DOI] [PubMed] [Google Scholar]
- 240.Simpson GG The Baldwin Effect. Evolution 7, 110–117 (1953). [Google Scholar]
- 241.Loison L Epigenetic inheritance and evolution: a historian’s perspective. Philosophical Transactions of the Royal Society B: Biological Sciences 376, 20200120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ramirez M et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nature Communications 7, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hata AN et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nature Medicine 22, 262–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pennisi E Buying time. Science 362, 988–991 (2018). [DOI] [PubMed] [Google Scholar]
- 245.Tanner G, Westhead DR, Droop A & Stead LF Benchmarking pipelines for subclonal deconvolution of bulk tumour sequencing data. Nat Commun 12, 6396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Caravagna G et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nature Genetics 52, 898–907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Quammen D The Tangled Tree: A Radical New History of Life. (Simon & Schuster, 2019). [DOI] [PubMed] [Google Scholar]
- 248.Beauregard-Racine J et al. Of woods and webs: possible alternatives to the tree of life for studying genomic fluidity in E. coli. Biology Direct 6, 39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dagan T & Martin W Getting a better picture of microbial evolution en route to a network of genomes. Philosophical Transactions of the Royal Society B: Biological Sciences 364, 2187–2196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wilkinson M, McInerney JO, Hirt RP, Foster PG & Embley TM Of clades and clans: terms for phylogenetic relationships in unrooted trees. Trends in Ecology & Evolution 22, 114–115 (2007). [DOI] [PubMed] [Google Scholar]
- 251.Baum DA Concordance trees, concordance factors, and the exploration of reticulate genealogy. TAXON 56, 417–426 (2007). [Google Scholar]
- 252.Lapointe F-J, Lopez P, Boucher Y, Koenig J & Bapteste E Clanistics: a multi-level perspective for harvesting unrooted gene trees. Trends in Microbiology 18, 341–347 (2010). [DOI] [PubMed] [Google Scholar]
- 253.Huson DH & Bryant D Application of Phylogenetic Networks in Evolutionary Studies. Molecular Biology and Evolution 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 254.Beiko RG & Ragan MA Detecting Lateral Genetic Transfer. in Bioinformatics: Data, Sequence Analysis and Evolution (ed. Keith JM) 457–469 (Humana Press, Totowa, NJ, 2008). doi: 10.1007/978-1-60327-159-2_21. [DOI] [Google Scholar]
- 255.Bapteste E & Burian RM On the need for integrative phylogenomics, and some steps toward its creation. Biol Philos 25, 711–736 (2010). [Google Scholar]
- 256.Boucher Y & Bapteste E Revisiting the concept of lineage in prokaryotes: a phylogenetic perspective. Bioessays 31, 526–536 (2009). [DOI] [PubMed] [Google Scholar]
- 257.Fennell KA et al. Non-genetic determinants of malignant clonal fitness at single-cell resolution. Nature 601, 125–131 (2022). [DOI] [PubMed] [Google Scholar]
- 258.Schaff DL, Fasse AJ, White PE, Vander Velde RJ & Shaffer SM Clonal differences underlie variable responses to sequential and prolonged treatment. Cell Syst 15, 213–226.e9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang Y et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Funnell T et al. Single-cell genomic variation induced by mutational processes in cancer. Nature 612, 106–115 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Voet T et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Research 41, 6119–6138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Andor N et al. Joint single cell DNA-seq and RNA-seq of gastric cancer cell lines reveals rules of in vitro evolution. NAR Genomics and Bioinformatics 2, lqaa016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Nam AS et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571, 355–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Rodriguez-Meira A et al. Single-cell multi-omics identifies chronic inflammation as a driver of TP53-mutant leukemic evolution. Nat Genet 55, 1531–1541 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lemant J, Le Sueur C, Manojlović V & Noble R Robust, Universal Tree Balance Indices. Systematic Biology 71, 1210–1224 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bertolaso M Philosophy of Cancer: A Dynamic and Relational View. (Springer, Dordrecht, 2016). doi: 10.1007/978-94-024-0865-2. [DOI] [Google Scholar]
- 267.Plutynski A Cancer and the goals of integration. Studies in History and Philosophy of Science Part C :Studies in History and Philosophy of Biological and Biomedical Sciences 44, 466–476 (2013). [DOI] [PubMed] [Google Scholar]
- 268.Reya T, Morrison SJ, Clarke MF & Weissman IL Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001). [DOI] [PubMed] [Google Scholar]
- 269.Lineweaver CH, Bussey KJ, Blackburn AC & Davies PCW Cancer progression as a sequence of atavistic reversions. Bioessays 43, e2000305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Davies PCW & Lineweaver CH Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys Biol 8, 015001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Schäfer M & Werner S Cancer as an overhealing wound: an old hypothesis revisited. Nature Reviews Molecular Cell Biology 9, 628–638 (2008). [DOI] [PubMed] [Google Scholar]
- 272.Dvorak HF Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 315, 1650–1659 (1986). [DOI] [PubMed] [Google Scholar]
- 273.Dvorak HF Tumors: Wounds That Do Not Heal–Redux. Cancer Immunology Research 3, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Jourdain Antoine, et al. Evolution clonale : qu’est-ce qu’un clone ? in La biologie au défi de l’histoire. Mélanges offerts à Michel Morange (eds. Loison L & Pradeu T) 222 (Editions Matériologiques, 2021). [Google Scholar]
- 275.Pogrebniak KL & Curtis C Harnessing Tumor Evolution to Circumvent Resistance. Trends in Genetics 34, 639–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Merlo LMF et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer prevention research (Philadelphia, Pa.) 3, 1388–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Huebner A, Dietzen M & McGranahan N SnapShot: Tumor evolution. Cell 184, 1650–1650.e1 (2021). [DOI] [PubMed] [Google Scholar]
- 278.Kim JY, Beart RW & Shibata D Stability of colon stem cell methylation after neo-adjuvant therapy in a patient with attenuated familial adenomatous polyposis. BMC Gastroenterol 5, 19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]