Abstract
Colorectal cancer (CRC) is a prevalent malignant tumor often leading to liver metastasis and mortality. Despite some success with PD-1/PD-L1 immunotherapy, the response rate for colon cancer patients remains relatively low. This is closely related to the immunosuppressive tumor microenvironment mediated by tumor-associated macrophages (TAMs). Our previous work identified that a phosphoglycerate mutase 1 (PGAM1) allosteric inhibitor, HKB99, exerts a range of anti-tumor activities in lung cancer. Here, we found that upregulation of PGAM1 correlates with increased levels of M2-like tumor-associated macrophages (TAMs) in human colon cancer samples, particularly in liver metastatic tissues. HKB99 suppressed tumor growth and metastasis in cell culture and syngeneic tumor models. M2-polarization, induced by colon cancer cell co-culture, was reversed by HKB99. Conversely, the increased migration of colon cancer cells by M2-TAMs was remarkably restrained by HKB99. Notably, a decrease in TAM infiltration was required for the HKB99-mediated anti-tumor effect, along with an increase in CD8+ T cell infiltration. Moreover, HKB99 improved the efficacy of anti-PD-1 treatment in syngeneic tumors. Overall, this study highlights HKB99's inhibitory activity in TAM-mediated colon cancer progression. Targeting PGAM1 could lead to novel therapeutic strategies and enhance the effectiveness of existing immunotherapies for colon cancer.
Key words: Phosphoglycerate mutase 1, Allosteric inhibitor, Tumor-associated macrophages, Macrophage polarization, Anti-PD-1, Immunotherapy, Liver metastasis, Colon cancer
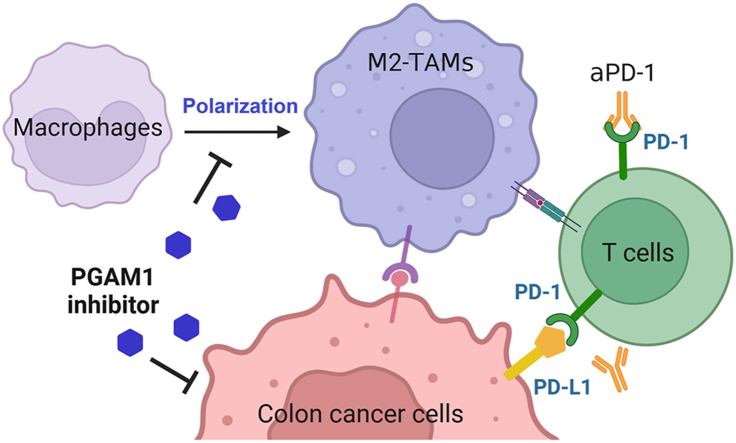
Graphical abstract
This study uncovers HKB99's inhibitory activity in TAM-mediated colon cancer progression, and PGAM1 could be considered as a potential therapeutic target for colon cancer therapy.
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy and second leading cause of cancer-related deaths1, 2, 3. 70% of patients with CRC develop liver metastasis, which is a major cause of death4. Immune checkpoint blockade (ICB) targeting programmed cell death protein/programmed death ligand 1 (PD-1/PD-L1) has led to remarkable advances in the treatment of various cancers5,6. However, the vast majority of CRC patients fail to respond to ICB, except for those with DNA mismatch repair deficiency (dMMR) and microsatellite instability (MSI)7. Therefore, it is necessary to uncover novel therapeutic vulnerabilities and inhibitors to increase the ICB response rate.
Tumor-associated macrophages (TAMs), the most abundant tumor-infiltrating immune cells in the tumor microenvironment (TME), are functionally categorized into tumor-supportive macrophages (M2-TAMs) and tumor-suppressive macrophages (M1-TAMs)8, 9, 10. M2-TAMs, with high levels of CD163, CD206, and anti-inflammatory cytokines, including transforming growth factor β (TGF-β), interleukin-6 (IL-6), and interleukin-10 (IL-10), contribute to immune escape for tumor growth and metastasis11, 12, 13, 14. M2-TAMs are enriched in liver metastatic CRCs and diminish ICB efficacy15, 16, 17. Accumulating evidence suggests that the interleukin-33 (IL-33)/ST2 pathway plays a critical role in recruiting macrophages into the tumor microenvironment and promoting colon cancer progression18,19.
Metabolic reprogramming is a crucial hallmark of cancer20. Targeting distinctive metabolic pathways has emerged as a promising strategy for reshaping the TME and reversing tumor immune evasion21, 22, 23. Phosphoglycerate mutase 1 (PGAM1) is a pivotal enzyme in aerobic glycolysis that catalyzes the reversible conversion of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG). Emerging evidence indicates that PGAM1 plays a multifunctional role in tumorigenesis and tumor progression via its non-canonical protein–protein interaction function beyond its metabolic activity24, 25, 26. Our previous work has identified HKB99 as a novel PGAM1 allosteric inhibitor to block conformational change of PGAM1. HKB99 exerts multiple anti-tumor effects in lung cancer, including the suppression of tumor growth and metastasis27. Moreover, HKB99 counteracts resistance to multiple generations of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), such as erlotinib and osimertinib, by blocking the interaction of PGAM1 with JAK2 and STAT3, subsequently inactivating the IL-6/JAK2/STAT3 signaling pathway28,29. However, whether pharmacological inhibition of PGAM1 by HKB99 affects the TME and tumor progression in CRC remains unclear.
In this study, clinical data showed that the upregulation of PGAM1 positively correlates with the level of the M2-TAMs signature in colon cancer. PGAM1 inhibition by HKB99 inhibits tumor growth and liver metastasis in syngeneic CRC models. Moreover, HKB99 affected the crosstalk between M2-TAMs and colon cancer cells. Therefore, HKB99 improves the efficacy of anti-PD-1 treatment in colon cancer models. Thus, targeting PGAM1 represents a promising strategy for enhancing the efficacy of ICB in colon cancer therapy.
2. Materials and methods
2.1. Cell lines and cell culture
Human colon cancer cell lines (HCT8 and HCT116), murine colon cancer cell lines (MC38 and CT26), human THP-1 monocyte cell line, and murine Raw 264.7 macrophage cell line were obtained from the American Type Culture Collection (ATCC). CT26-luc cells were gifted by Prof. Chao Fang (Shanghai Jiao Tong University of Medicine, Shanghai, China). HCT8, HCT116, CT26, and CT26-luc cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS (Gemini), MC38 and Raw 264.7 cells were cultured in DMEM supplemented with 10% FBS. THP-1 cells were cultured in RPMI-1640 medium supplemented with 10 mmol/L HEPES and 10% heat-inactivated fetal bovine serum (FBS). All the cells were incubated in a humidified incubator at 37 °C with 5% CO2. Cell identification was performed using short tandem repeat (STR) analysis, and mycoplasma testing was performed every six months to ensure the authenticity and purity of the cell lines used in the study.
2.2. Cell viability assay
Cells were seeded in 96-well plates at a density of 3000 cells/well and allowed to reach 30% confluence the next day. The cells were then treated with the drug of interest at various concentrations and incubated for 72 h. Cell viability was measured using a cell counting kit-8 (Dojindo) according to the manufacturer's instructions. Optical density (OD) at 450 nm was measured using a 550 microplate reader (Bio-Rad). The cell growth inhibition rate was calculated using Eq. (1):
| Cell growth inhibition rate (%) = [1 − (OD drug – OD blank) / (OD control – OD blank)] × 100 | (1) |
2.3. Clonogenic assay
Colon cancer cells were seeded into a 6-well plate at a density of 1000 cells/well and treated with different concentrations of the drugs. The cells were cultured for 7–10 days and stained with crystal violet before imaging. The number of colonies was calculated using the ImageJ software (Bethesda).
2.4. M2-like macrophages polarization
Cell co-culture models were established to detect the polarization macrophages induced by colon cancer cells30. Human THP-1 monocytes were polarized into M2 macrophages by HCT8 conditioned media or co-cultured with HCT8 cells. Firstly, THP-1 cells were seeded into the lower chamber of the trans-well device (Corning) and treated with 200 nmol/L phorbol 12-myristate 13-acetate (PMA, Sigma) for 24 h to induce M0 macrophages. After PBS wash, M0 macrophages were then incubated with HCT8 conditioned media for 24 h or co-cultured with HCT8 cells for 48 h using a cell culture insert (0.4-μm pore, Corning). Similarly, mouse Raw 264.7 macrophages were induced to M2-like polarization when co-cultured with MC38 cells. In detail, 5 × 105 cells/mL cancer cells were seeded in the upper chamber with M0 macrophages in the lower chamber of six-well plates containing 2.0 mL of RPMI-1640 or DMEM. The two cell lines were co-cultured in a humidified atmosphere consisting of 95% air and 5% CO2 at 37 °C.
2.5. Trans-well migration assay
A Corning chamber was used, with serum-free basic media placed in the inner chamber and media containing 20% serum in the lower chamber. MC38, CT26, and HCT8 cells were cultured for 24 h, whereas HCT116 cells were cultured for 48 h. After incubation, the chambers were fixed with 4% paraformaldehyde and stained with crystal violet. Finally, an upright Leica DM6 microscope was used to capture images, and ImageJ software was used for analysis and counting.
2.6. Wound healing assay
Colon cancer cells were seeded in a 96-well plate, and cell confluence reached approximately 90% the following day. A scratch tool (Essen Bioscience) was used to create uniform scratches. The cells were washed with media 1–2 times and treated with HKB99 or M2-TAMs conditioned media. Wound confluence was detected using an IncuCyte live-cell analysis system (Essen Bioscience).
2.7. Quantitative real-time PCR (qRT-PCR)
Total cellular RNA was extracted according to the manufacturer's instructions (Takara), and the RNA concentration was determined using a Nanodrop microspectrophotometer (Thermo Fisher Scientific). cDNA was prepared from 1 μg RNA using the PrimeScript™ RT reagent kit (Takara). Real-time quantitative detection was performed using a TB Green® Premix Ex Taq™ kit (Takara). The PCR primers used are listed in Supporting Information Table S1.
2.8. Animal studies
Female BALB/c or C57BL/6 mice aged eight weeks were provided by the Animal Science Department of Shanghai Jiao Tong University School of Medicine. All experimental procedures involving mice were approved by the Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University School of Medicine (JUMC2023-143-A) and were conducted in accordance with IACUC regulations. Mice were housed under specific pathogen-free (SPF) conditions on a 12-h light/dark cycle with chow and water provided ad libitum.
2.9. Subcutaneous tumor models
To establish subcutaneous colon cancer models, 5 × 105 CT26 or MC38 cells were injected into the right flank of the mice. When subcutaneous tumors reached a volume of 50–100 mm3, the mice were randomly divided into different treatment groups. HKB99 (50 mg/kg body weight, mpk) was intraperitoneally (i.p.) administered once a day. For macrophage depletion models as described previously31, clodronate liposomes (Yeasen) were administered as 50 mpk every 3 days starting 2 days before HKB99 treatment. For the anti-PD-1 combination model, the mice were intraperitoneally injected with 5 mg/kg of anti-PD-1 on Days 8, 11, and 15 after cell inoculation. The tumor volume was measured using a caliper and calculated according to Eq. (2):
| Volume = (Length × Width2)/2 | (2) |
The initial tumor volume (TVinitial) and tumor volume at time T (TVT) were recorded.
| Relative tumor volume (RTV, %) = TVT/TVinitial × 100 | (3) |
RTVT and RTVC indicate the RTV of the drug treatment and control groups, respectively. The anti-tumor effect of the drug was calculated as tumor growth inhibition (TGI) value according to Eq. (4):
| TGI (%) = (1–RTVT/RTVC) × 100 | (4) |
2.10. Intrasplenic liver metastasis model of colon cancer
Mouse CT26-luc colon cancer cells intrasplenic injection model was established in BALB/c syngenic immunocompetent mice as described previously32,33. Mice aged 7–8 weeks were anesthetized with pentobarbital sodium and a left lateral flank incision was made to expose the spleen. 5 × 105 CT26-luc cells (50 μL) were slowly injected into the spleen, and the surgical incision was closed using a surgical thread. Luciferin-based bioluminescence signal was measured to validate viable metastasis foci throughout the mice via IVIS Spectrum In Vivo Imaging System (PerkinElmer) after the mice were injected with potassium luciferin (150 mg/kg). The bioluminescence image and signals were analyzed by Living Image software (PerkinElmer). On Day 8 after cell injection, the mice were randomly divided into two groups and treated with vehicle and HKB99 (50 mpk, once a day, i.p.) until the end of the study. Mice were euthanized and dissected on Day 19. Metastatic foci in the dissected liver were quantified by morphometry and metastatic surface area compared with total surface area, as described previously34.
2.11. Flow cytometry
Macrophages were washed with PBS and digested with ACCUTASE (Sigma) for 15 min to prepare single-cell suspensions. The isolated cells were stained with anti-CD206 (BioLegend) at 4 °C for 30 min. Finally, each sample was resuspended in a staining buffer and analyzed using a Attune NxT2 flow cytometer (Thermo Fisher Scientific). The data were analyzed using FlowJo software (TreeStar).
Tumors were cut into pieces and incubated with a digestion solution containing collagenase I and DNase I at 37.5 °C for 30 min. The suspension was filtered through a 70 μm cell strainer and centrifuged at 1200 × g for 6 min to remove the supernatant. The cell pellet was resuspended in staining buffer to obtain a single-cell suspension. Single-cell suspensions were stained with LIVE/DEAD Fixable Near IR dye (Thermo Fisher Scientific) in PBS at room temperature for 5 min before staining. For surface marker staining, cells were blocked for 15 min with anti-CD16/32 and stained with the following antibodies for 30 min at 4 °C: BV605-CD45, BV650-CD3, AF647-CD8a, Perp-cy5.5-CD4, FITC-CD11b, and Perp-cy5.5-F4/80 (BioLegend). For CD206 staining, cells were fixed in CytoFix/CytoPerm buffer (BD Biosciences) for 20 min and washed using 1 × Perm/Wash buffer (BD Biosciences). Finally, the samples were resuspended in staining buffer and analyzed using a CytoFLEX LX flow cytometer (Beckman Coulter). The data were analyzed using FlowJo software (TreeStar).
2.12. Hematoxylin–eosin (H&E) and immunohistochemistry (IHC) staining
H&E and IHC staining was performed as previously described27. Briefly, tumor and liver tissues were formalin-fixed, paraffin-embedded, and cut into 4 μm-thick sections. For H&E staining, deparaffinized slides were baked, passed through graded alcohols, and then incubated for 5 min in Harris hematoxylin before staining with 0.5% eosin for 1 min. For IHC staining, the F4/80, CD206, and CD8 antibodies were obtained from Invitrogen and Abcam, respectively. Semi-quantitative analysis of the average of CD8 positive numbers and CD206 positive areas was conducted using ImageJ software, as previously described35,36.
2.13. Statistical analysis
All assays were repeated at least three times, and group comparisons were performed using two-tailed unpaired or paired Student's t-tests using GraphPad Prism 8.4 software. P value < 0.05 was considered significant (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ∗∗∗∗P < 0.0001, respectively).
2.14. Data availability
Relative PGAM1 mRNA level analysis was performed using data obtained from The Cancer Genome Atlas (TCGA) colon adenocarcinoma database and the Genotype-Tissue Expression (GTEx) database via the UCSC Xena database37. Data analyzed in colon cancer liver metastasis samples were obtained from the GEO database GSE41258. The proportion of macrophages analyzed in colon cancer and liver metastasis samples using the website of Single Cell RNA-seq Data Visualization and Analysis (scDVA) and data were downloaded from GSE16452238. The data of correlation analysis between PGAM1 and immune cells were downloaded from the TCGA colon adenocarcinoma database, and then statistically and visually analyzed using R version 3.6.3, and the GSVA package version 1.34.039,40. Graphical abstract was created with BioRender.com.
3. Results
3.1. Aberrant up-regulation of PGAM1 positively correlates with M2-TAMs signature in human colon cancer
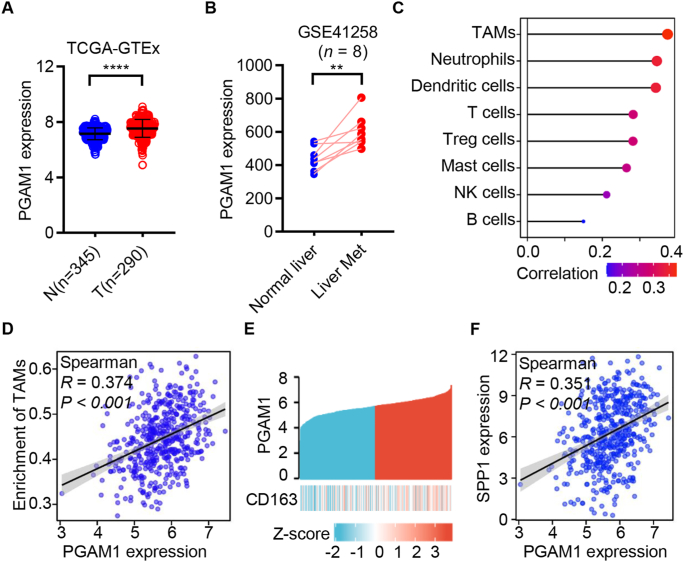
To investigate whether PGAM1 is involved in the TME in colon cancer, the expression of PGAM1 in colon cancer and normal tissues in TCGA and the GTEx databases was analyzed. As shown in Fig. 1A, the mRNA level of PGAM1 was significantly upregulated in colon cancer patients compared with normal colon tissues. Moreover, the mRNA level of PGAM1 was much higher in liver metastatic colon cancer tissues than in the corresponding adjacent normal liver tissues (Fig. 1B).
Figure 1.
Phosphoglycerate mutase 1 (PGAM1) up-regulation correlates with M2 tumor-associated macrophages (M2-TAMs) levels in human colon cancer. (A) The expression of PGAM1 was analyzed in normal (n = 345) and tumor (n = 290) tissues of colon cancer using the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) database. (B) The expression of PGAM1 was analyzed in normal liver tissue and liver metastasis lesions of colon cancer patients using the GSE41258 (n = 8). (C) The correlation analysis between PGAM1 and enrichment of immune cells in TCGA colon adenocarcinoma database (n = 480). (D) The correlation analysis between PGAM1 and M2-TAMs in the TCGA colon adenocarcinoma database (n = 480). (E) The TCGA colon adenocarcinoma database was stratified into high- and low-PGAM1 expression groups, and CD163 expression in two groups was presented by heatmap (n = 480). (F) The association between PGAM1 and SPP1 in TCGA colon adenocarcinoma database (n = 480). ∗∗P < 0.01; ∗∗∗∗P < 0.0001.
Second, to investigate the correlation between PGAM1 and tumor-infiltrating immune cell status in colon specimens, we analyzed the mRNA levels of PGAM1 and the enrichment of immune cells in the TCGA database. The data revealed that PGAM1 levels were most closely correlated with TAMs (Fig. 1C and D, Supporting Information Fig. S1A–S1G). We also observed that M2-TAMs were enriched in liver metastatic tissues relative to primary colon cancer tissues in the GSE164522 dataset (Fig. S1H)38. Moreover, heatmap analysis indicated that high PGAM1 expression positively correlated with CD163, a marker of M2-TAMs (Fig. 1E). Consistently, SPP141, another M2-TAMs marker, was also positively associated with PGAM1 (Fig. 1F). Taken together, these clinical data support the hypothesis that the upregulation of PGAM1 is positively associated with the level of M2-TAMs in human colon cancer.
3.2. HKB99 inhibits cell proliferation and migration in colon cancer cells
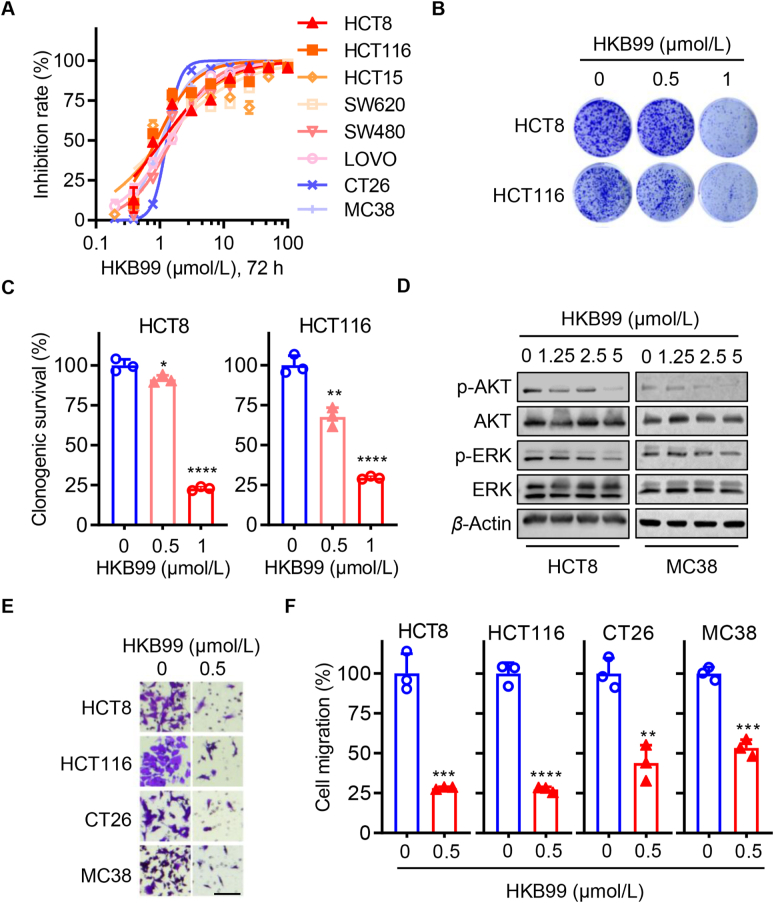
Since our previous work indicated that HKB99 inhibits cell proliferation and migration in lung cancer, we continuously explored whether HKB99 exerts a similar anti-tumor effect in colon cancer cells. As expected, the results of the CCK8 assays showed that HKB99 inhibited the proliferation of multiple human and murine colon cancer cells with an IC50 of approximately 1 μmol/L (Fig. 2A). The data of clonogenic assays showed that 1 μmol/L HKB99 significantly suppressed colony formation of HCT8 and HCT116 cells, while 0.5 μmol/L HKB99 had a slight inhibitory effect (Fig. 2B and C). Immunoblotting was performed and confirmed that HKB99 inhibited the level of p-AKT and p-ERK in a concentration-dependent manner in both human HCT8 and MC38 cells (Fig. 2D), which is consistent with our previous findings27. To exclude the possibility of off-target effects, PGAM1 inhibition by siPGAM1#1 and siPGAM1#2 at 25 nmol/L for 72 h of incubation also inhibited the p-AKT level in MC38 cells (Fig. S1I). Meanwhile, the sensitivity to HKB99 treatment was significantly decreased by siPGAM1#1 and siPGAM1#2 transfection in MC38, which demonstrates HKB99's on-target-based effect in colon cancer cells (Fig. S1J). Moreover, trans-well data showed that 0.5 μmol/L HKB99 efficiently inhibited cell migration in human HCT8 and HCT116, murine CT26, and MC38 colon cancer cells (Fig. 2E and F). Overall, these results suggest that HKB99 perturbs the proliferation and migration of colon cancer cells.
Figure 2.
Cell proliferation and migration are suppressed by HKB99 treatment in colon cancer cells. (A) Cell viability was detected by CCK8 assays after being treated with HKB99 for 72 h in multiple colon cancer cells as indicated including human HCT116, HCT15, SW620, HCT8, SW480, LOVO, and murine MC38 and CT26 cells. (B) Clonogenic assays were performed after the HCT8 and HCT116 cells were treated with 0.5 and 1.0 μmol/L HKB99 respectively. (C) Quantitative statistical results of clonogenic assays in (B). (D) The proteins as indicated were immunoblotted with β-actin as a loading control after HCT8 and MC38 cells were treated with 1.25, 2.5, and 5 μmol/L HKB99 for 1 h. (E) Trans-well assays were performed in HCT8, CT26, and MC38 incubated with 0.5 μmol/L HKB99 for 24 h and HCT116 for 48 h respectively. Scale bars, 100 μm. (F) The statistical result of (E). Data are shown as mean ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
3.3. HKB99 restrains M2-like polarization of TAMs and antagonizes the cell migration induced by M2-TAMs in colon cancer cells
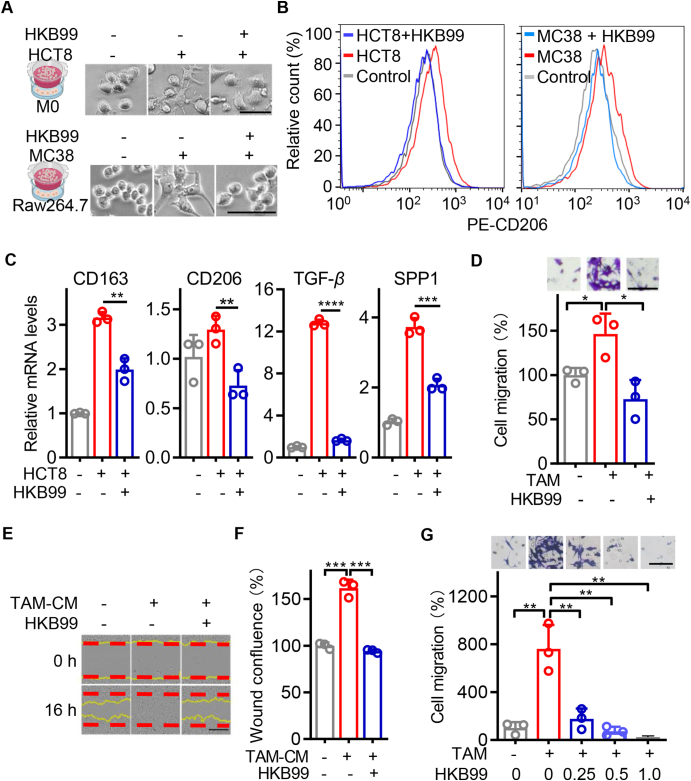
Regarding the aforementioned correlation of PGAM1 and TAMs in colon cancer, we investigated the cell-extrinsic role of HKB99 in colon cancer cells. Human THP-1 monocytes were differentiated into M0 macrophages by PMA pretreatment. When co-cultured with HCT8 colon cancer cells, M2-like macrophages exhibited a flat, amoeboid shape, and branching morphology. However, with incubation of 0.5 μmol/L HKB99, they transitioned back to a M0-like round shape (Fig. 3A). Consistently, flow cytometry using the typical M2 marker CD206, as well as qRT-PCR of M2 biomarkers, including CD163, CD206, TGF-β, and SPP1, confirmed that HKB99 decreased the M2-like polarization of the TAMs induced by HCT8 cell co-culture (Fig. 3B and C). Similar results were observed in murine Raw 264.7 macrophages co-cultured with MC38 colon cancer cells (Fig. 3A and B).
Figure 3.
HKB99 abrogates M2-TAMs induced cell migration in colon cancer cells. (A) Upper, the representative typical images of THP-1-derived macrophages co-cultured with HCT8 cells for 48 h in the presence of 0.5 μmol/L HKB99 treatment as indicated. Scale bars, 100 μm. Lower, the representative images of Raw 264.7 macrophages co-cultured with MC38 cells for 24 h in the presence of 1.0 μmol/L HKB99 treatment as indicated. Scale bars, 50 μm. (B) Flow cytometry assays determined the CD206 positive fraction of macrophages after cells were treated as described in (A). The x-axis (divided in channel log100 to 104) represents the relative fluorescence intensity of CD20642. (C) The expression of M2 markers including CD163, CD206, TGF-β, and SPP1 were detected by qRT-PCR after macrophages were treated as described in (A). (D) HCT8 cells were co-cultured with M2-TAMs in the presence of 0.5 μmol/L HKB99 treatment for 48 h. Cell migration was detected by trans-well assay. Scale bar, 100 μm. (E) HCT8 cells were incubated with TAM-conditioned medium (CM) in the presence of 0.5 μmol/L HKB99 treatment for 16 h. Cell migration was detected by wound healing assays. Scale bar, 400 μm. (F) The statistical result of (E). (G) MC38 cells were co-cultured with M2-TAMs in the presence of 0.25, 0.5, and 1.0 μmol/L HKB99 treatment for 24 h. Cell migration was detected by trans-well assay. Scale bar, 100 μm. Data are shown as mean ± SD. ∗P < 0.05; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Next, the immunoblotting data showed that 1 μmol/L HKB99 treatment effectively downregulated the level of IL-33 in MC38 cells, while it had little effect on IL-33 in Raw 264.7 macrophages (Supporting Information Fig. S2A). Interestingly, the increase in IL-33 receptor, ST2 caused by MC38 co-culture was constitutively suppressed by a 1 μmol/L HKB99 treatment in Raw 264.7 cells, which was not reversed with a 12 h-pretreatment of 30 ng/mL exogenous recombinant murine IL-33 (rmIL-33) (Fig. S2B). Accordingly, flow cytometry and cell morphology data revealed that HKB99 significantly reduced M2-like polarization of Raw264.7 macrophages induced by MC38 co-culture, which was also not reversed by rmIL33 rescue even at a high concentration of 100 ng/mL (Fig. S2C–S2E). Taken together, these results suggested that HKB99 primarily inhibits the expression of ST2 in TAMs, thereby suppressing IL-33/ST2 mediated M2-like polarization of TAMs induced by colon cancer cells.
Finally, trans-well assays were performed to evaluate the role of HKB99 in crosstalk between TAMs and colon cancer cells. M0 macrophages derived from THP-1 monocytes after PMA pretreatment were incubated with conditioned media (CM) from HCT8 cells for 24 h. Flow cytometry data confirmed that the number of CD206-positive macrophages was enhanced by HCT8-CM, which echoed the results in Fig. 3B (Fig. S2F). Subsequently, HCT8 cells were co-cultured with the induced M2-TAMs and trans-well assays were performed (Fig. S2G). The data showed that migration of HCT8 cells was markedly promoted by M2-TAMs co-culture, which was reversed in the presence of HKB99 treatment (Fig. 3D). Consistently, the results of wound-healing assays also showed that HKB99 significantly abrogated the enhanced cell migration induced by TAM-conditioned medium (TAM-CM) in HCT8 cells (Fig. 3E and F). Meanwhile, similar findings were observed that HKB99 effectively counteracted the increased cell migration in MC38 cells, which was stimulated by Raw 264.7 M2-TAMs (Fig. 3G and Fig. S2H). Collectively, all the above results supported that HKB99 restrains M2-like polarization of TAMs, thereby counteracts the promotion of M2-TAMs on the migration of colon cancer cells.
3.4. HKB99 inhibits tumor growth and diminishes M2-TAMs infiltration in vivo
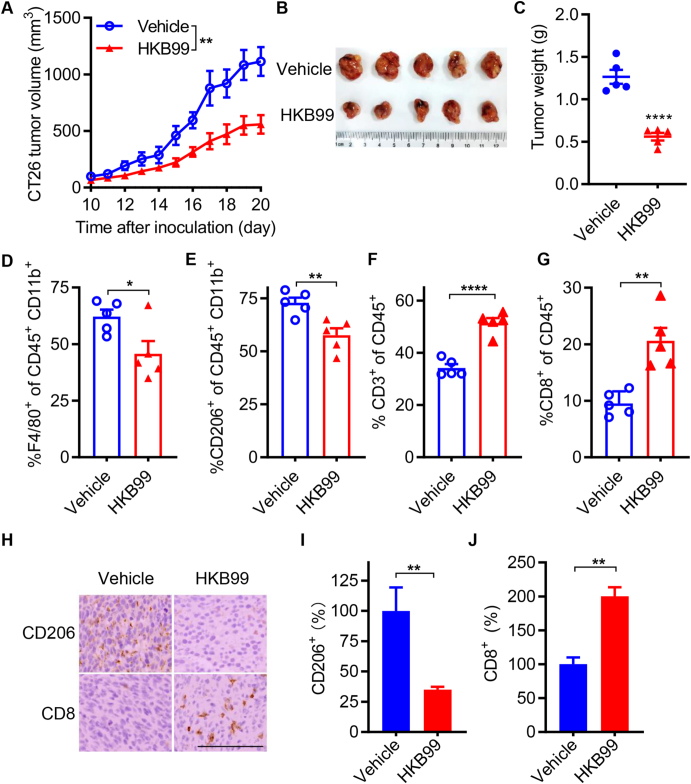
To further investigate the role of HKB99 in tumor growth and TAMs reprogramming in vivo, immune-competent models with murine CT26 colon cancer cells were established, and HKB99 was administered at a dose of 50 mg/kg per day when tumors reached 50–100 mm3. Our results indicated that HKB99 treatment significantly inhibited CT26 tumor growth (Fig. 4A and B). The average weight of subcutaneous syngeneic tumors in the vehicle group was approximately 1.27 g, while the average weight of tumors in the HKB99-treated group was about 0.56 g (Fig. 4C).
Figure 4.
HKB99 alters the infiltration of CD206+ M2-TAMs and CD8+ T cells in mouse CT26 tumors. (A) Tumor growth curve of CT26 tumor in BALB/c syngeneic immune-competent mice and HKB99 were injected at a dose of 50 mg/kg/day. (B) The image of CT26 tumors. (C) The tumor weight is as indicated. (D–G) The proportion of F4/80+ macrophages, CD206+ M2-TAMs, CD3+ T cells and CD8+ T cells were determined as indicated by flow cytometry. (H) IHC staining of CD206 and CD8 in tumors. Scale bars, 100 μm. (I, J) The statistical result of (H). Data, mean ± SEM (n = 5 for each group). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗∗P < 0.0001.
Flow cytometry was performed, and lineage markers were analyzed to identify specific immune cell populations. The results showed that HKB99 remarkably reduced CD45+ CD11b+ F4/80+ macrophages and the infiltration of CD45+ CD11b+ CD206+ M2-TAMs (Fig. 4D and E). HKB99 enhanced the infiltration of T cells, including CD45+ CD3+ T cells and cytotoxic CD45+ CD8+ T cells (Fig. 4F and G). Immunohistochemistry staining (IHC) also revealed that the CD206 level was decreased and the CD8 level was increased by HKB99 treatment (Fig. 4H–J). Thus, our findings suggest that HKB99 remodels the proportion of immune cells and exerts anti-tumor effects in colon cancer.
3.5. HKB99 significantly diminishes liver metastases and consistently alters the CD206+ TAMs and CD8+ T cells infiltration in vivo
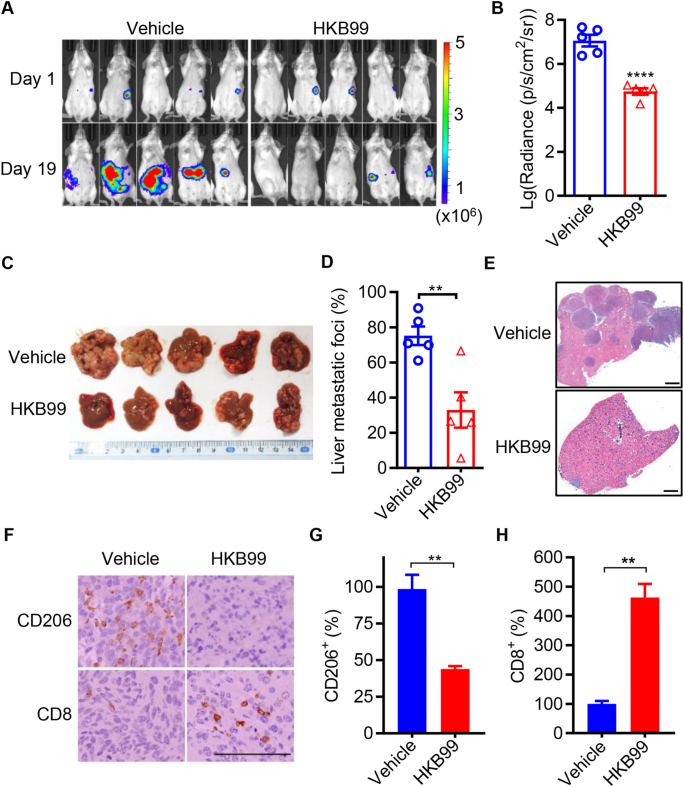
As HKB99 suppressed the TAMs promoted cell migration of colon cancer cells in vitro, we then explored the effect of HKB99 on tumor metastasis and TAMs infiltration in vivo. To mimic the liver metastasis occurring in colon cancer patients, a mouse CT26-luc colon cancer cells intrasplenic injection model was established in BALB/c syngenic mice. Bioluminescence signal intensity in the whole body was determined and showed that HKB99 decreased the signal of CT26-luc tumors compared with the vehicle control in mice (Fig. 5A and B). Notably, a significant reduction in the liver metastatic foci was observed in the dissected livers and H&E-stained tissue sections by HKB99 treatment on Day 19 (Fig. 5C–E). Consistently, the bioluminescence signal and weight of dissected liver tissues also revealed that HKB99 administration attenuated liver metastasis of colon cancer (Supporting Information Fig. S3A–S3C).
Figure 5.
HKB99 perturbs liver metastases and reduces M2-TAMs in CT26-luc tumors. (A) The luciferin-based bioluminescence signal was measured via IVIS spectrum in the murine intrasplenic liver metastasis model on Day 1 and Day 19 after CT26-luc cells injection. HKB99 was injected at a dose of 50 (mg/kg body weight, mpk) daily starting on Day 8 after the CT26-luc injection. (B) The statistical results of (A). (C) The image of CT26-luc liver metastases. (D) The quantification of liver metastatic foci for (C). (E) The typical tiled image of the whole liver section of H&E staining showed the colon cancer liver metastases as indicated. Scale bars, 1 mm. (F) The IHC staining of CD8 and CD206. Scale bars, 100 μm. (G, H) The statistical result of (F). Data are presented as mean ± SEM (n = 5 for each group). ∗∗P < 0.01; ∗∗∗∗P < 0.0001.
Similar to previous CT26 subcutaneous models, IHC data showed that HKB99 treatment led to a significant decrease in CD206 levels and a concurrent increase in CD8 levels (Fig. 5F–H). In summary, HKB99 reshapes the proportion of immune cells and suppresses liver metastasis in colon cancer.
3.6. HKB99 exerts an anti-tumor effect partially dependent on macrophages
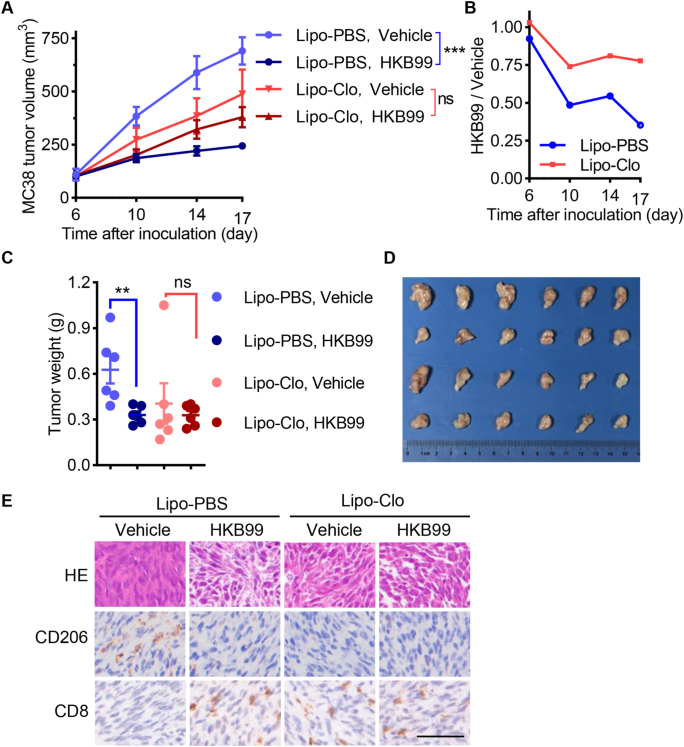
To further decipher whether macrophages were involved in the anti-tumor effect of HKB99, macrophage depletion models were established as shown in Fig. S3D. As expected, macrophages were effectively depleted by clodronate liposomes (Lipo-Clo) treatment in MC38 tumor-bearing mice (Fig. S3E). Lipo-Clo suppressed the tumor growth compared with PBS control liposomes (Lipo-PBS) treatment group in vehicle groups of MC38 tumor-bearing mice, which supported that macrophage infiltration facilitates tumor growth (Fig. 6A). Consequently, a reduced anti-tumor effect of HKB99 was observed in Lipo-Clo treated mice, whereas the significant growth inhibition of MC38 tumors was obtained with HKB99 administration in Lipo-PBS treated mice (Fig. 6A–D). Correspondingly, the CD8+ T cells increase along with CD206+ M2-TAMs reduction by HKB99 treatment was diminished after macrophage depletion (Fig. 6E). Interestingly, increased CD8+ T infiltration in MC38 tumor was observed in Lipo-Clo treated mice (Fig. 6E), which is in consistent with previous finds that TAMs impede CD8+ T cells from reaching tumor cells43. Meanwhile, no significant difference in mice body weight indicated that the mice were tolerant to Lipo-Clo and HKB99 treatment during the study period (Fig. S3F). Collectively, these data suggested that HKB99 exerts anti-tumor effect at least partially dependent on inhibiting M2-TAMs infiltration.
Figure 6.
HKB99 exerts an anti-tumor effect partially dependent on inhibiting M2-TAMs infiltration. (A) Tumor growth curve of MC38 tumor in C57BL/6 mice. Lipo-Clo was administered at 50 mpk every 3 days starting 2 days before HKB99 treatment. HKB99 was injected at a dose of 50 mpk daily. Lipo-PBS and Vehicle were used as Lipo-Clo and HKB99's control respectively. (B) the relative size of HKB99-treated tumors compared with the vehicle-treated group with Lipo-PBS or Lipo-Clo administration respectively. (C) Tumor weight as indicated. (D) The image of MC38 tumors. (E) The H&E and IHC staining of the MC38 tumor section with CD206 and CD8 was indicated. Scale bars, 50 μm. Data, mean ± SEM (n = 6 for each group). ∗∗P < 0.01; ∗∗∗∗P < 0.0001, ns, non-significant.
3.7. HKB99 augments the tumoricidal efficacy of PD-1 blockade in vivo
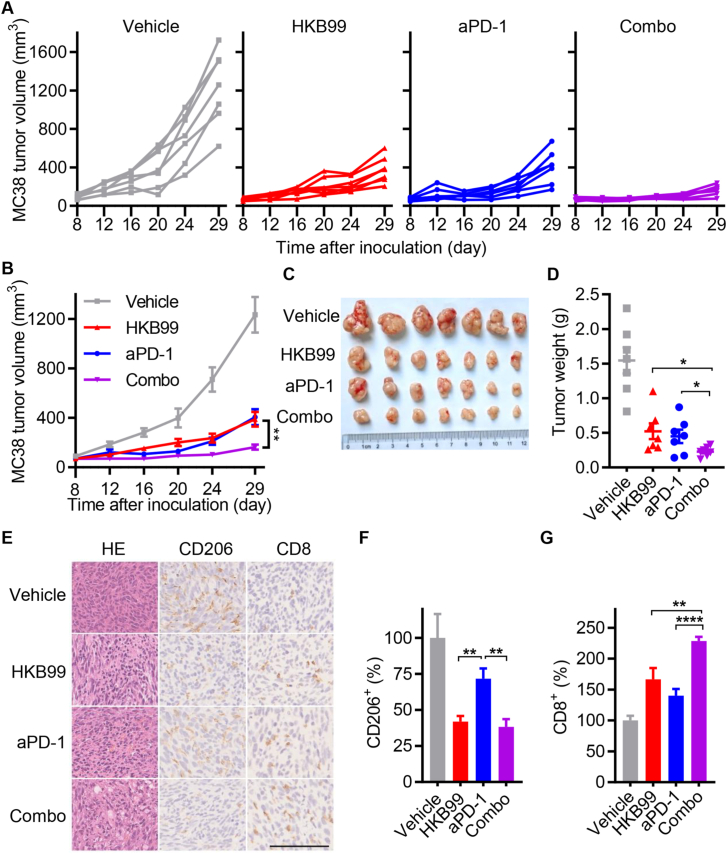
The above results intrigued us to explore whether HKB99 could enhance the tumoricidal effect of anti-PD-1 treatment in colon cancer. In support of this possibility, mice were injected with MC38 cells (0.5 × 106) and randomized according to tumor size when reaching 50–100 mm3. We treated MC38 tumor-bearing mice with HKB99, either alone or in combination with an anti-PD-1 antibody (Fig. S3G). As shown in Fig. 7A–C, HKB99 or anti-PD-1 antibody treatment alone moderately inhibited MC38 tumor growth with a TGI value of 60.9% and 62.4% on Day 29, while combination treatment almost ablated the tumors with a TGI value of 82.3%. The tumor weight similarly showed the regression of MC38 tumors in the combination group with 0.23 g average versus 1.54 g, 0.52 g, and 0.45 g in the control, HKB99, and anti-PD-1 antibody group respectively (Fig. 7D). Meanwhile, the mice exhibited steady weight gain, indicating the favorable biosafety of the combination therapy (Fig. S3H).
Figure 7.
HKB99 improves the efficacy of anti-PD-1 treatment in colon cancer. (A, B) The tumor growth curve of MC38 tumor in C57BL/6 immune-competent mice as indicated. (C) The image of the MC38 dissected tumors. (D) The tumor weight of MC38 tumors. (E) The H&E and IHC staining of the MC38 tumor section with CD206 and CD8 was indicated. Scale bars, 100 μm. (F, G) The statistical result of (E). Data, mean ± SEM (n = 7 for each group). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗∗P < 0.0001.
Moreover, as shown in Fig. 7E–G, H&E staining revealed that HKB99 and anti-PD-1 antibody combination treatment induced significant tumor necrosis in MC38 tumors. Notably, IHC staining indicated that HKB99 alone or in combination with anti-PD-1 antibody obviously reduced CD206 levels, while anti-PD-1 antibody treatment alone did not alter CD206 levels relative to the vehicle control. Importantly, CD8 levels were prominently enhanced in the combination group compared with the levels in the HKB99 or anti-PD-1 antibody alone group. Taken together, our data indicate that HKB99 alters M2-TAMs and cytotoxic CD8+ T cell infiltration, and synergistically augments the tumoricidal efficacy of PD-1 blockade in vivo.
4. Discussion
Recent studies have revealed that TAMs exhibit aberrant glucose metabolism to facilitate malignant tumor growth and metastasis. Targeting key enzymes in glucose metabolism has emerged as a promising strategy for reshaping the TME and reversing tumor immune evasion44. Herein, we found that the aberrant upregulation of PGAM1 positively correlates with the M2-TAMs signature in human colon cancer. Functionally, HKB99 inhibits tumor growth and metastasis in colon cancer cells and syngeneic models. Further analysis demonstrated that treatment with HKB99 increased cytotoxic CD8+ T cells and decreased M2-TAMs in colon cancer models, whereas macrophage depletion attenuated the anti-tumor role of HKB99. Mechanistically, HKB99 alleviated the polarization of M2-TAMs in colon cancer cells. Conversely, HKB99 suppressed the enhanced cell migration of colon cancer cells induced by M2-TAMs co-culture. Importantly, HKB99 treatment effectively augmented the tumoricidal activity of anti-PD-1 antibodies against colon cancer in vivo. Overall, our study uncovers HKB99's inhibitory activity in TAM-mediated colon cancer progression, and PGAM1 could be considered a potential therapeutic target for colon cancer immunotherapy.
Emerging evidence has shown that aberrant glucose metabolism results in the accumulation of a variety of oncometabolites, including lactate and succinate, in the TME to suppress anti-tumor immunity and assist tumor progression45. M2-TAMs have recently been highlighted as the major cell population for increased glucose uptake in the TME, which fuels the O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) of lysosomal cathepsin B to promote cancer metastasis and chemoresistance46. Therefore, targeting glucose metabolism represents a promising strategy for switching TAMs from pro-tumor to anti-tumor functions for cancer therapy.
A recent study showed that PGAM1 is involved in the process of N6-methyladenosine-modified circular RNA QSOX1, which regulates glycolysis and promotes immune escape in colorectal cancer cells47. Elevated levels of PGAM1 are associated with immune cell infiltration, specifically macrophages, which indicates that PGAM1 could serve as a prognostic biomarker and an immunotherapeutic target in clear cell renal cell carcinoma48. Our previous work identified that PGAM1 inhibition by HKB99 and siPGAM1s leads to inactivation of AKT and ERK, the key effectors of the PI3K and MAPK pathways, respectively, in lung adenocarcinoma27. Notably, another study identified that PGAM1 inhibition promotes ferroptosis and downregulates PD-L1 levels through energy stress and ROS-dependent AKT inactivation in hepatocellular carcinoma cells, which synergistically enhances the efficacy of anti-PD-1 immunotherapy49.
The present study highlights the role of HKB99 in the crosstalk between colon cancer cells and M2-TAMs. HKB99 counteracted M2 polarization of TAMs induced by colon cancer cell co-culture, whereas the IL33/ST2 pathway was suppressed by HKB99 under these co-culture circumstances. Additionally, macrophage depletion attenuated the anti-tumor effect of HKB99 in mouse syngeneic models, which underscores HKB99's inhibitory activity in TAM-mediated colon cancer progression. However, the deeper molecular mechanism deserved further investigation.
Notably, PD-1/PD-L1 blockade therapy has shown unsatisfactory response rates in colon cancer. Several studies have suggested that immunosuppressive macrophages (M2-TAMs) in the TME play a crucial role in mediating anti-PD-1 resistance in various cancers50, 51, 52, 53. Our findings indicated that HKB99 treatment decreased M2-TAMs while increasing CD8+ T cell infiltration, which synergistically improved the anti-tumor effects of PD-1 blockade in mouse colon tumors.
5. Conclusions
Our study illustrates that the allosteric inhibitor of PGAM1, HKB99, exerts an immunostimulatory effect by modulating M2-TAMs polarization and CD8+ T cell infiltration, and thereby suppresses tumor growth and liver metastasis in colon cancer. Additionally, HKB99 effectively augmented the anti-tumor immune effects of anti-PD-1 treatment. Overall, targeting PGAM1 has potential therapeutic value for improving the currently available immunotherapies against colon cancer.
Author contributions
Cheng Wang: Writing – original draft, Visualization, Methodology, Formal analysis, Data curation. Minghao Zhang: Writing – original draft, Visualization, Methodology, Formal analysis, Data curation. Shunyao Li: Visualization, Methodology, Data curation. Miaomiao Gong: Writing – review & editing, Visualization, Methodology, Formal analysis, Data curation. Ming-yu Luo: Visualization, Methodology, Investigation, Formal analysis. Mo-cong Zhang: Visualization, Methodology, Formal analysis. Jing-Hua Zou: Methodology, Formal analysis. Ningxiang Shen: Visualization, Methodology. Lu Xu: Visualization, Investigation, Formal analysis. Hui-min Lei: Visualization, Methodology. Ling Bi: Visualization, Formal analysis. Liang Zhu: Validation, Resources. Zhengting Wang: Validation, Resources. Hong-zhuan Chen: Writing – review & editing, Supervision, Resources, Conceptualization. Lu Zhou: Writing – review & editing, Resources, Investigation, Conceptualization. Ying Shen: Writing – review & editing, Visualization, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Conflicts of interest
Ying Shen, Lu Zhou, Hong-zhuan Chen, Liang Zhu, and Mingyu Luo are named inventors of authorized patents (ZL201910284333.4 and ZL201810273938.9) to the Chinese Patent Office related to the work.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82073868), the Science and Technology Commission of Shanghai Municipality (20S11900100 and 21TQ016, China), and China Postdoctoral Science Foundation (2022M722138). We are grateful for support from the Core Facility of Basic Medical Sciences, Shanghai Jiao Tong University School of Medicine.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.09.007.
Contributor Information
Miaomiao Gong, Email: gongmiaomiao1029@163.com.
Hong-zhuan Chen, Email: hongzhuan_chen@hotmail.com.
Lu Zhou, Email: zhoulu@fudan.edu.cn.
Ying Shen, Email: yshen0510@sjtu.edu.cn.
Appendix A. Supporting information
The following is the Supporting Information to this article:
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Wagle N.S., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233–254. doi: 10.3322/caac.21772. [DOI] [PubMed] [Google Scholar]
- 3.Cheng C., Yao H., Li H., Liu J., Liu Z., Wu Y., et al. Blockade of the deubiquitinating enzyme USP48 degrades oncogenic HMGA2 and inhibits colorectal cancer invasion and metastasis. Acta Pharm Sin B. 2024;14:1624–1643. doi: 10.1016/j.apsb.2024.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z., Kim S.Y., Tu W., Kim J., Xu A., Yang Y.M., et al. Extracellular vesicles in fatty liver promote a metastatic tumor microenvironment. Cell Metab. 2023;35 doi: 10.1016/j.cmet.2023.04.013. 1209–26.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagchi S., Yuan R., Engleman E.G. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y., Wang X., He S., Duan Z., Zhang Y., Sun X., et al. Immunostimulatory gene therapy combined with checkpoint blockade reshapes tumor microenvironment and enhances ovarian cancer immunotherapy. Acta Pharm Sin B. 2024;14:854–868. doi: 10.1016/j.apsb.2023.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas E.M., Wright J.A., Blake S.J., Page A.J., Worthley D.L., Woods S.L. Advancing translational research for colorectal immuno-oncology. Br J Cancer. 2023;129:1442–1450. doi: 10.1038/s41416-023-02392-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Visser K.E., Joyce J.A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403. doi: 10.1016/j.ccell.2023.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Cassetta L., Pollard J.W. A timeline of tumour-associated macrophage biology. Nat Rev Cancer. 2023;23:238–257. doi: 10.1038/s41568-022-00547-1. [DOI] [PubMed] [Google Scholar]
- 10.Ostuni R., Kratochvill F., Murray P.J., Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 11.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christofides A., Strauss L., Yeo A., Cao C., Charest A., Boussiotis V.A. The complex role of tumor-infiltrating macrophages. Nat Immunol. 2022;23:1148–1156. doi: 10.1038/s41590-022-01267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A., Allavena P., Marchesi F., Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21:799–820. doi: 10.1038/s41573-022-00520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H., Li N., Wei X., Li Q., Guo J., Yang G., et al. Biomimetic "Gemini nanoimmunoregulators" orchestrated for boosted photoimmunotherapy by spatiotemporally modulating PD-L1 and tumor-associated macrophages. Acta Pharm Sin B. 2024;14:1345–1361. doi: 10.1016/j.apsb.2023.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y.C., Yang S.X., Ma J.Q., Chen Z.C., Song G.H., Rao D.N., et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–153. doi: 10.1158/2159-8290.CD-21-0316. [DOI] [PubMed] [Google Scholar]
- 16.Yang S., Qian L., Li Z.X., Li Y., Bai J., Zheng B., et al. Integrated multi-omics landscape of liver metastases. Gastroenterology. 2023;164:407–423. doi: 10.1053/j.gastro.2022.11.029. [DOI] [PubMed] [Google Scholar]
- 17.Yu J., Green M.D., Li S., Sun Y., Journey S.N., Choi J.E., et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med. 2021;27:152–164. doi: 10.1038/s41591-020-1131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi S., Elhance A., Van Duzer A., Kumar S., Leitenberger J.J., Oshimori N. Tumor-initiating cells establish an IL-33-TGF-β niche signaling loop to promote cancer progression. Science. 2020;369 doi: 10.1126/science.aay1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang M., Li Y., Huang K., Qi S., Zhang J., Zgodzinski W., et al. IL33 promotes colon cancer cell stemness via JNK activation and macrophage recruitment. Cancer Res. 2017;77:2735–2745. doi: 10.1158/0008-5472.CAN-16-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faubert B., Solmonson A., DeBerardinis R.J. Metabolic reprogramming and cancer progression. Science. 2020;368 doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Reyes I., Chandel N.S. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 22.Bader J.E., Voss K., Rathmell J.C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cel. 2020;78:1019–1033. doi: 10.1016/j.molcel.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone R.D., Powell J.D. Metabolism of immune cells in cancer. Nat Rev Cancer. 2020;20:516–531. doi: 10.1038/s41568-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitosugi T., Zhou L., Elf S., Fan J., Kang H.B., Seo J.H., et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitosugi T., Zhou L., Fan J., Elf S., Zhang L., Xie J., et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D., Jin N., Sun W., Li X., Liu B., Xie Z., et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2016;36:2900–2909. doi: 10.1038/onc.2016.446. [DOI] [PubMed] [Google Scholar]
- 27.Huang K., Liang Q., Zhou Y., Jiang L.L., Gu W.M., Luo M.Y., et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cel Metab. 2019;30 doi: 10.1016/j.cmet.2019.09.014. 1107–19.e8. [DOI] [PubMed] [Google Scholar]
- 28.Liang Q., Gu W.M., Huang K., Luo M.Y., Zou J.H., Zhuang G.L., et al. HKB99, an allosteric inhibitor of phosphoglycerate mutase 1, suppresses invasive pseudopodia formation and upregulates plasminogen activator inhibitor-2 in erlotinib-resistant non-small cell lung cancer cells. Acta Pharmacol Sin. 2020;42:115–119. doi: 10.1038/s41401-020-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Q., Gong M., Zou J.H., Luo M.Y., Jiang L.L., Wang C., et al. A phosphoglycerate mutase 1 allosteric inhibitor overcomes drug resistance to EGFR-targeted therapy via disrupting IL-6/JAK2/STAT3 signaling pathway in lung adenocarcinoma. Drug Resist Updat. 2023;68 doi: 10.1016/j.drup.2023.100957. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y., Yang J., Xu D., Gao X.M., Zhang Z., Hsu J.L., et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68:1653–1666. doi: 10.1136/gutjnl-2019-318419. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J., Naulaerts S., Boudhan L., Martin M., Gatto L., Van den Eynde B.J. Tumour immune rejection triggered by activation of α2-adrenergic receptors. Nature. 2023;618:607–615. doi: 10.1038/s41586-023-06110-8. [DOI] [PubMed] [Google Scholar]
- 32.Dafflon C., Santamaría-Martínez A., Ordóñez-Morán P. An intrasplenic injection model for the study of cancer stem cell seeding capacity. Methods Mol Biol. 2020;2171:293–302. doi: 10.1007/978-1-0716-0747-3_20. [DOI] [PubMed] [Google Scholar]
- 33.Dupaul-Chicoine J., Arabzadeh A., Dagenais M., Douglas T., Champagne C., Morizot A., et al. The Nlrp3 inflammasome suppresses colorectal cancer metastatic growth in the liver by promoting natural killer cell tumoricidal activity. Immunity. 2015;43:751–763. doi: 10.1016/j.immuni.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Caino M.C., Seo J.H., Aguinaldo A., Wait E., Bryant K.G., Kossenkov A.V., et al. A neuronal network of mitochondrial dynamics regulates metastasis. Nat Commun. 2016;7 doi: 10.1038/ncomms13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strasser K., Birnleitner H., Beer A., Pils D., Gerner M.C., Schmetterer K.G., et al. Immunological differences between colorectal cancer and normal mucosa uncover a prognostically relevant immune cell profile. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2018.1537693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng S., Zou Y., Xie X., Liang J.Y., Yang A., Yu K., et al. Development and validation of a stromal immune phenotype classifier for predicting immune activity and prognosis in triple-negative breast cancer. Int J Cancer. 2020;147:542–553. doi: 10.1002/ijc.33009. [DOI] [PubMed] [Google Scholar]
- 37.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Zhang Q., Xing B., Luo N., Gao R., Yu K., et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell. 2022;40 doi: 10.1016/j.ccell.2022.02.013. 424–37.e5. [DOI] [PubMed] [Google Scholar]
- 39.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozato Y., Kojima Y., Kobayashi Y., Hisamatsu Y., Toshima T., Yonemura Y., et al. Spatial and single-cell transcriptomics decipher the cellular environment containing HLA-G+ cancer cells and SPP1+ macrophages in colorectal cancer. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2022.111929. [DOI] [PubMed] [Google Scholar]
- 42.Adan A., Alizada G., Kiraz Y., Baran Y., Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- 43.Peranzoni E., Lemoine J., Vimeux L., Feuillet V., Barrin S., Kantari-Mimoun C., et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Nat Acad Sci U S A. 2018;115:E4041–E4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., Cao X. Glucose metabolism of TAMs in tumor chemoresistance and metastasis. Trends Cel Biol. 2023;33:967–978. doi: 10.1016/j.tcb.2023.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Baryła M., Semeniuk-Wojtaś A., Róg L., Kraj L., Małyszko M., Stec R. Oncometabolites-A link between cancer cells and tumor microenvironment. Biology. 2022;11:270. doi: 10.3390/biology11020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Q., Shen Q., Liu Y., Shi Y., Huang W., Wang X., et al. Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal cathepsin B to promote cancer metastasis and chemoresistance. Cancer Cell. 2022;40 doi: 10.1016/j.ccell.2022.08.012. 1207–22.e10. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z., Zheng N., Li J., Li C., Zheng D., Jiang X., et al. N6-Methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat. 2022;65 doi: 10.1016/j.drup.2022.100886. [DOI] [PubMed] [Google Scholar]
- 48.Wen C.Y., Hsiao J.H., Tzeng Y.T., Chang R., Tsang Y.L., Kuo C.H., et al. Single-cell landscape and spatial transcriptomic analysis reveals macrophage infiltration and glycolytic metabolism in kidney renal clear cell carcinoma. Aging (Albany NY) 2023;15:11298–11312. doi: 10.18632/aging.205128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y., Wang Y., Lu Z., Wan J., Jiang L., Song D., et al. PGAM1 inhibition promotes HCC ferroptosis and synergizes with anti-PD-1 immunotherapy. Adv Sci. 2023;10 doi: 10.1002/advs.202301928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez-Usatorre A., Kadioglu E., Boivin G., Cianciaruso C., Guichard A., Torchia B., et al. Overcoming microenvironmental resistance to PD-1 blockade in genetically engineered lung cancer models. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abd1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M., Huang L., Ding G., Huang H., Cao G., Sun X., et al. Interferon gamma inhibits CXCL8–CXCR2 axis mediated tumor-associated macrophages tumor trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2019-000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi T., Zhang Y.P., Wang Y., Song X.R., Wang H.B., Zhou X.Y., et al. DKK1 promotes tumor immune evasion and impedes anti-PD-1 treatment by inducing immunosuppressive macrophages in gastric cancer. Cancer Immunol Res. 2022;10:1506–1524. doi: 10.1158/2326-6066.CIR-22-0218. [DOI] [PubMed] [Google Scholar]
- 53.Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F., et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relative PGAM1 mRNA level analysis was performed using data obtained from The Cancer Genome Atlas (TCGA) colon adenocarcinoma database and the Genotype-Tissue Expression (GTEx) database via the UCSC Xena database37. Data analyzed in colon cancer liver metastasis samples were obtained from the GEO database GSE41258. The proportion of macrophages analyzed in colon cancer and liver metastasis samples using the website of Single Cell RNA-seq Data Visualization and Analysis (scDVA) and data were downloaded from GSE16452238. The data of correlation analysis between PGAM1 and immune cells were downloaded from the TCGA colon adenocarcinoma database, and then statistically and visually analyzed using R version 3.6.3, and the GSVA package version 1.34.039,40. Graphical abstract was created with BioRender.com.