Abstract
Cytopharmaceutical based on macrophages is a breakthrough in the field of targeted drug delivery. However, it remains a challenge to localize and control drug release while retaining macrophage activity and exerting its immunotherapeutic effect. Herein, a localized light-triggered release macrophage cytopharmaceutical (USIP@M) was proposed, which could utilize the tumor targeting and immunotherapy effects of macrophages to reverse the immune suppression of tumor microenvironment (TME). Amphiphilic block copolymers with ultraviolet (UV)-responsive o-nitrobenzyl groups were synthesized and co-loaded with sorafenib (SF), IMD-0354 (IMD), and upconverting nanoparticles (UCNPs), which were then taken up by macrophages, and the targeted delivery of drugs was realized by using the tumor tropism of macrophages. UCNPs converted near-infrared light with strong penetrability and high safety into UV light, which promoted the photoresponsive depolymerization of block copolymers and production of exosomes from USIP@M, accelerated drug efflux and maintained the activity of macrophages. IMD simultaneously polarized carrier macrophages and tumor-associated macrophages to exert the antitumor effect of macrophages, enhance T cell immunity, and alleviate the immunosuppressive state of TME. Synergistically with the chemotherapeutic effect of SF, it could effectively kill tumors. In conclusion, based on the localized light-triggered release strategy, this study constructed a novel macrophage cytopharmaceutical that could localize and control drug release while retaining the activity of macrophages and exerting its immunotherapeutic effect, which could effectively treat solid tumors.
Key words: Cytopharmaceutical, Macrophage, Light-triggered release, Upconverting nanoparticle, O-Nitrobenzyl, Exosome, Solid tumor, Cell-chemotherapy
Graphical abstract
A localized light-triggered release macrophage cytopharmaceutical was proposed, utilizing the tumor targeting and immunotherapy effects of macrophages to reverse the immune suppression of tumor microenvironment.
1. Introduction
Cytopharmaceuticals have attracted extensive attention in recent years, in which drugs or drug-loaded nanoparticles are loaded with living cells ex vivo1,2, displaying great promise in targeted drug delivery in terms of natural targeting, superior biocompatibility, prolonged circulation time, and flexible morphology3. Natural cells including red blood cells, immune cells (T cells, macrophages, neutrophils, natural killer cells, etc.), and stem cells have all been used in carrier cells of cytopharmaceuticals4, which could effectively deliver the drugs to corresponding pathological sites with ultimately maximize treatment efficacy and minimize side effects5. However, the limited integration of physiological functions of living cells and their ability to deliver drugs in existing studies has slowed down their development rate.
The macrophages comprise up to 30%–50% of cells in a tumor mass, and show great advantages as carrier cells of cytopharmaceuticals due to the tumor tropism, unique phagocytic ability, and immunotherapeutic effects compared with other cells6. Firstly, compelling evidence has shown that inflammatory chemokines such as CC-chemokine ligand 2 (CCL2) in tumor sites possess strong chemotactic capability to recruit macrophages, leading to higher recruitment and infiltration of macrophages in tumor tissue7. Secondly, macrophages also facilitate the internalization of drugs or drug-loaded nanoparticles as professional phagocytic cells. Moreover, proinflammatory M1-type macrophages can phagocytose tumor cells and have the potential to participate in antitumor immunity8. Thus, the use of living macrophages as appealing carrier cells of cytopharmaceuticals has become a research hotspot in recent years. Our research group9 used M1-type macrophages to co-incubate with sorafenib (SF)-loaded lipid nanoparticles to design a cytopharmaceutical and drug delivery system (M1/SLNP). M1-type macrophages could not only provide immunotherapy as therapeutic tools but also deliver drugs to tumor tissues for drug delivery. The system made full use of the tumor tropism and antitumor immunity of M1-type macrophages to inhibit tumorigenesis and progression effectively. Zhang et al.10 prepared silica nanocomplex-loaded macrophages to achieve tumor-tropic delivery of antitumor drugs doxorubicin (DOX), which had a two-phase drug release profile in macrophages. Despite the good progress of these studies, most macrophage cytopharmaceuticals still lack designs for triggered drug release during delivery. Although some studies have used endogenous stimulation11 or photothermal therapy12 to achieve drug release at tumor sites or exert therapeutic effects, these methods also lead to the death of macrophages, making them unable to participate in antitumor immunity. Therefore, how to control drug release while exerting its therapeutic effect as immune cells is still a challenge for cytopharmaceuticals.
To this end, we proposed a localized light-triggered release strategy. As a commonly used external stimulus, light can precisely control drug release in terms of irradiation time, site and wavelength, etc13. Most photoresponsive depolymerization materials are sensitive to ultraviolet (UV) light, and a certain degree of UV irradiation can promote the increase of exosome secretion of macrophages, thereby promoting the efflux of drugs14. The amphiphilic block copolymer formed by hydrophobic polymethacrylate with an O-nitrobenzyl group and hydrophilic poly (ethylene oxide) can be depolymerized under the irradiation of UV light to realize the light-triggered release of drugs15,16. The application of UV light-controlled photoresponsive depolymerization nanoparticles in macrophage cytopharmaceuticals can promote the production of exosomes and control drug release. However, one practical roadblock that hampers the in vivo application is that the tissue penetration depth of UV is very low. Compared with UV light, near-infrared (NIR) light is better suited to biomedical applications17. Longer-wavelength NIR light has less attenuation in blood and tissues, leading to deeper tissue penetration and higher safety, without causing significant damage to healthy cells. Recently, a strategy to trigger photoreactions by NIR light using upconverting nanoparticles (UCNPs) has been extensively studied18. UCNPs can absorb two or more long-wavelength and low-energy photons under the excitation of NIR irradiation, and convert them to longer-wavelength and higher-energy photons. Based on this principle, UCNPs can realize the conversion from NIR light to UV light19. It is shown that UCNPs can effectively combine the drug-controlled release ability of UV light with the deep tissue penetration of NIR light. Here, the above strategy was used for localized drug light-triggered release from macrophages. On the one hand, the UV light response of sensitive materials can be realized to trigger the release of intracellular drugs. On the other hand, it can stimulate the increase of exosome secretion of macrophages and promote the efflux of drugs.
Another challenge is that TME stays in an immunosuppressive state. After macrophages reach the TME, they are often polarized into M2-type macrophages, which secrete cytokines and chemokines that support the recruitment of immunosuppressive regulatory T cells (Tregs) and favor a remodeling of the extracellular matrix that restrains tumor infiltration by effector T cells7,20. The tumor immunosuppressive microenvironment limits the application of macrophage cytopharmaceuticals. Based on this, in order to control the macrophages that reach the TME with the M1 phenotype, and effectively subvert the endogenous tumor-associated macrophages (TAMs) into the M1 phenotype, thereby reversing the immunosuppressive TME, we encapsulated the drug IMD-0354 (IMD), which could polarize macrophages to M1 phenotype, into nanoparticles within macrophages21. Upon UV light converted from NIR light stimulation, the released IMD both polarized carrier macrophages to M1 phenotype and repolarized TAMs in the TME. At the same time, in order to further improve the therapeutic effect, we co-loaded SF, a first-line chemotherapy drug for hepatocarcinoma22 and melanoma23, into the photocleavage nanoparticles, which not only exerted cell therapy but also exerted the dual antitumor effect of SF to jointly inhibit tumor growth.
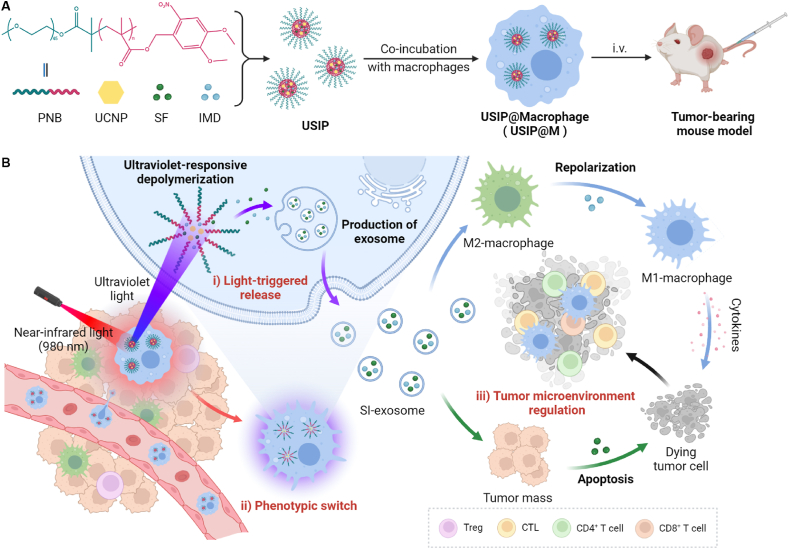
In conclusion, we proposed a localized light-triggered release strategy based on macrophages (Fig. 1). The photoresponsive depolymerization micelles loaded with UCNPs, SF, and IMD (USIPs) were co-incubated with macrophages to construct a localized light-triggered release macrophage cytopharmaceutical USIP@M. Utilizing the tumor tropism of macrophages, USIP@Ms were actively targeted to tumor sites. Using NIR light as an exogenous stimulus, the UCNPs were used to convert it into UV light in the cell to make the micelles photocleavage, promote the production of exosomes from USIP@M, accelerate drug efflux, and maintain the activity of macrophages. This strategy not only utilized the tumor tropism ability of macrophages but also realized the triggered release of drugs in macrophages through the transformation of NIR and UV light, thereby inducing the polarization of macrophage carriers and TAMs to M1 phenotype and exerting chemoimmunity effect, which provided a new idea for the research of macrophage cytopharmaceuticals.
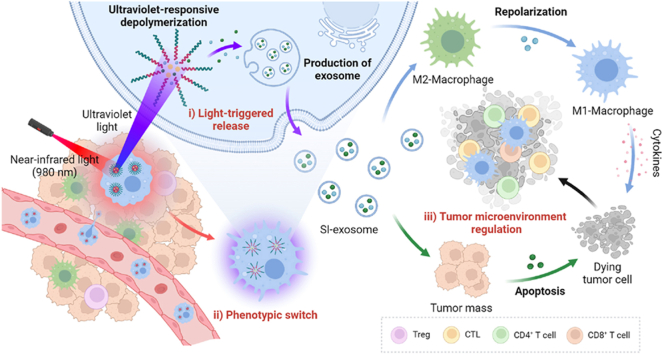
Figure 1.
Schematic illustration of localized light-controlled USIP@M. (A) Preparation of USIP@M by co-incubating USIP and macrophage, and tumor-bearing mouse model was treated by intravenous (i.v.) administration with USIP@M. (B) The delivery process of USIP@M in vivo. USIP@M accumulated at the tumor site utilizing the tumor tropism of macrophages. Under the action of NIR light, the UCNPs emitted UV light in the cell to photoresponsive depolymerize the micelles and promote the release of drugs in exosome form (SI-exosome). SI-exosome induced the polarization of macrophage carriers and TAMs to M1 phenotype, and the apoptosis of tumor cells to jointly achieve the regulation of TME. This figure was created with BioRender.com.
2. Materials and methods
2.1. Materials
SF was purchased from Shanghai Biochempartner Co., Ltd. (Shanghai, China). IMD was provided by Selleckchem Co., Ltd. (Houston, TX, USA). UCNPs were provided by Ruixi Biological Technology Co., Ltd. (Xi'an, China). Obtain DiR and coumarin-6 (C6) through Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). Purchase thiazolyl blue tetrazolium bromide (MTT) and percoll cell isolation solution through Solarbio Science & Technology Co., Ltd. (Beijing, China). Purchase DMEM (high glucose) from Sperikon Life Science & Biotechnology (Sichuan, China). FBS was purchased from Inner Mongolia Opcel Biotechnology (Inner Mongolia, China). Fast cell freeze solution was purchased from Shanghai Life-iLab Biotech Co., Ltd. (Shanghai, China). Obtain estern blot antibodies (anti-GAPDH, anti-CD81, anti-TSG101, anti-β-Tubulin, and anti-CCR2) through Affinity Biosciences Co., Ltd. (Liyang, China). Cytokine ELISA kits for IL-12, IL-10, TGF-β, and TNF-α were purchased from MultiSciences Biotech Co., Ltd. (Hangzhou, China). Cytokine ELISA kits for IL-6 were purchased from Dakewe Biotech Co., Ltd. (Nanjing, China). Purchase serum-free cryopreservation medium without phenol red from MeilunBio Co., Ltd. (Dalian, China). Purchase flow cytometry antibodies (PerCP/Cy5.5 anti-mouse F4/80, PE anti-mouse CD80, APC anti-mouse CD206, APC anti-mouse CD3, Alexa Fluor® 488 anti-mouse CD86, FITC anti-mouse CD4, PE anti-mouse CD4, PE anti-mouse CD8a, Alexa Fluor® 488 anti-mouse IFN-γ and Alexa Fluor® 647 anti-mouse FOXP3) through BioLegend (San Diego, CA, USA). All the reagents used were of analytical reagent grade.
2.2. Cell lines and animals
Murine-derived hepatocarcinoma cell line H22, murine-derived malignant melanoma cell strains B16F10, and murine-derived macrophage cell line RAW264.7 were purchased from Cell Bank/Stem Cell Bank (Shanghai, China). Murine-derived macrophage cell line RAW264.7 was cultured in DMEM (high glucose) containing 10% FBS. Using RPMI 1640 medium contained 10% FBS, H22 cells and B16F10 cells were cultured. Obtain female BALB/c mice and female C57BL/6 mice through SPF Biotechnology Co., Ltd. (Beijing, China). All experimental procedures were executed according to the protocols approved by the Shandong University Animal Care and Use Committee (approval number: 230013).
2.3. Synthesis of the monomer 4,5-dimethoxy-2-nitrobenzyl methacrylate (NBMA)
4, 5-Dimethoxyl-2-nitrobenzyl alcohol (260 mg) was first dissolved in 2 mL of dry DMF under stirring and then adding 255 μL triethylamine into the solution. Add magnets to it and fill it with N2. Then the methacryloyl chloride (130 μL diluted with 200 μL dry DMF) was added slowly to the mixed solution in an ice bath. After stirring for 1 h, the whole system was kept at 25 °C overnight. After the DMF was removed by washing with water, the crude product was dissolved in chloroform, purified by washing through 1 mol/L hydrochloric acid and 1 mol/L potassium chloride and dried with the help of anhydrous sodium sulfate. Then filter the solution, remove the chloroform with a rotary evaporator, and finally dry under vacuum overnight. The structure of NBMA was verified by proton nuclear magnetic resonance (1H NMR, Bruker, Avance DPX-300, Germany).
2.4. Synthesis of macroinitiator (PEO2K-Br)
Monmethoxy-polyethylene oxide (PEO2K, Mw = 2000) (500 mg) and triethylamine (174 μL) were dissolved in 3 mL of dry dichloromethane and stirred in an ice bath. In the mixed solution, 2-bromoisobutyryl bromide (154.5 μL diluted in 1 mL of dichloromethane) was added dropwise and then stirred overnight at 25 °C. Using a rotary evaporator, the resulting solution was dried. The obtained dry solid was re-dissolved in dichloromethane and washed three times with NaCl solution (v/v, 1:1). The organic phase was added dropwise to ice ether, and a white flocculent precipitate was produced. Centrifuge at 5000 rpm for 2 min (Eppendorf, 5804R, Germany), discard the supernatant and dry in a vacuum. The structure of PEO2k-Br was verified by 1H NMR.
2.5. Synthesis of amphiphilic block copolymer (PNB)
Cu(I)Br (5.3 mg), PEO2k-Br (37 mg), NBMA (500 mg), N,N,N′,N″,N″-pentamethyl diethylenetriamine (6.5 mg) was dissolved in 1 mL DMSO, placed in a 5 mL Schlenk bottle, degassed three times and sealed under vacuum. After stirring for 10 min at 25 °C, the resulting solution was bathed in oil (90 °C) for 2 h, precipitated in methanol, filtered, and dried. After the crude product had been dissolved and THF used as the eluent, the excess catalyst was removed through a neutral Al2O3 column. The resulting yellow filtrate was concentrated under reduced pressure and precipitated in ether. Then it was collected by filtration and vacuum drying. The structure of PNB was verified by 1H NMR and gel permeation chromatography (GPC).
2.6. Preparation and characterization of USIP
The preparation of light-responsive micelles loaded with UCNPs, SF, and IMD. 50 mg PNB, 50 mg UCNPs, 10 mg SF, and 2.3 mg IMD were first dissolved in 5 mL THF. A system of 20 mL pure water was prepared, and then the above-mixed solution was added dropwise. At 25 °C, the whole solution was stirred for 4 h. Subsequently, the organic solvent was removed by dialysis. After centrifugal collection and resuspension, the obtained nanoparticles USIP were stored in the dark. According to the actual need to reduce the preparation system in equal proportions. The light-responsive micelles loaded with UCNPs and C6 (UCP), light-responsive micelles loaded with DiR (DP), light-responsive micelles loaded with UCNPs and SF (USP), and light-responsive micelles loaded with UCNPs and IMD (UIP) were prepared as described above.
Using transmission electron microscopy (TEM, Hitachi, HT7700, Japan) to characterize the morphology of USIP. The intensity size and zeta potential of USIP were determined by a Zetasizer (Malvern, Nano-ZS90, Britain). Using high-performance liquid chromatography (HPLC) to quantitatively analyze the content of SF and IMD. The drug loading (DL) of SF and IMD were calculated.
2.7. Photoresponsive characteristics of the USIP
Take an appropriate amount of USIP and irradiate it with NIR light (980 nm, 1 W, 10 min). After dilution, the particle size changes before and after NIR light irradiation were measured with a Zetasizer. The morphology of USIP irradiated by NIR light was characterized by TEM.
2.8. Preparation of USIP@M
USIP@M was obtained by co-incubation of USIP and RAW 264.7 cells. Based on the single-factor study, we determined the optimal formulations of USIP@M by investigating the concentration of SF (50, 100, 200, 300, and 400 μg/mL) and incubation time (1, 2, and 4 h). RAW 264.7 cells were seeded into 96-well plates (1 × 104 cells/well) for cell viability detection. A series of doses of USIP were incubated at different times by adding them into the wells. Then, the supernatant was replaced. When 48 h had passed, MTT and DMSO were introduced. The cell viability was measured according to the results of 570 nm. RAW 264.7 cells were seeded into 12-well plates (1 × 106 cells/well) for DL indicators. After being cultured in a conventional culture condition for a period of time, USIP with the different concentrations of SF (50, 100, 200 μg/mL) was added and the cells were incubated for different times (1, 2, and 4 h), and the supernate was taken. The drug concentration in the supernatant before and after incubating was determined by HPLC, and the DL for USIP@M was calculated. UCP@M, DP@M, USP@M, and UIP@M were prepared as described above.
2.9. Characterization of USIP@M
Formation of USIP@M by laser confocal microscopy (LCM). The hydrophobic fluorescent substance C6 was used to replace the hydrophobic drugs SF and IMD to prepare UCP for subsequent experiments. At conventional culture conditions, use a confocal special cell culture dish to seed RAW264.7 cells (1 × 105 cells/dish). After the cells adhered, UCP was added at a final concentration of C6 in 20 μg/mL. The cells were incubated for another 2 h to obtain UCP@M. Wash twice, add rat serum to block, add Alexa Fluor® 647 anti-mouse F4/80 for 1 h, wash again with PBS, and then finally observe the formation of UCP@M under an LCM (Zeiss, LSM 900, Germany).
In order to obtain the cumulative release profiles, SI@M, USIP@M and USIP@M with NIR irradiation (980 nm) of 1 W/cm2 for 10 min (USIP@M+L) were incubated with serum-free DMEM for 0.5, 1, 2, 4, 8, 12, 24, 48 or 72 h. At the corresponding point in time, the supernatant of different preparations was collected, and fresh medium was supplemented until 72 h. The drug in the supernatant was quantified using HPLC.
2.10. Investigation of photoresponsive release form of USIP@M
USIP@M was irradiated with NIR light (980 nm, 1 W, 10 min) and incubated at 37 °C under 5% CO2 conditions. After 24 h, extract the supernatant, and perform a series of centrifugation (3000×g for 10 min, 10,000×g for 30 min, 140,000×g for 90 min) to obtain exosomes. The exosomes released within 24 h after preparation of USIP@M were obtained as described above. The exosome protein content was evaluated by the bicinchoninic acid assay, and the exosome key markers (CD81 and TSG101) were detected by western blotting analysis.
In order to evaluate the ability of NIR light-triggered drug release in vivo, IVISense 680 fluorescent dye and C6 were selected to mark injected macrophages and drug, respectively. H22 tumor-bearing BALB/c mice were i.v. injected with fluorescently labeled macrophage or UCP@M at 24 h before harvesting the tumors. The tumors were placed in 4% paraformaldehyde, paraffin-embedded, and sliced to be imaged. The UCP@M group was set into an NIR irradiation group (1 W/cm2 for 10 min) and without an NIR irradiation group (UCP@M+L and UCP@M). For groups requiring NIR irradiation, the mice were irradiated at 4 h before harvesting the tumors. For the UCP@M+L+skin group, the tumor was covered with 5 mm pigskin during NIR irradiation.
2.11. Phenotype investigation
Phenotype analysis of USIP@M 24 h after irradiation with or without NIR light was evaluated. The prepared USIP@M and USIP@M+L groups preparations were cultured with fresh culture medium for 24 h. RAW264.7 cells incubated in 10% FBS-containing DMEM and RAW264.7 cells incubated with IMD were used as the control groups. For flow cytometry analysis, the cells were stained with PerCP/Cy5.5 anti-mouse F4/80, APC anti-mouse CD206, and Alexa Fluor® 488 anti-mouse CD86. For ELISA analysis, the culture supernatant of the cells for 24 h was collected, and the level of cytokines (IL-10, TGF-β, and TNF-α) was measured using ELISA kits.
2.12. TAMs phenotype regulation
RAW264.7 cells were seeded into 12-well plates (1 × 106 cells/well) containing IMD, USIP, and USIP with NIR irradiation (980 nm) at a power density of 1 W/cm2 for 10 min (USIP+L) and released medium from USIP@M+L, and the cells were further incubated for 24 h. The tumor-conditioned medium (TCM) group consisted of TAMs cultured in TCM without any formulations. Following 24 h of incubation. The RAW264.7 cells were stained with APC anti-mouse CD206 (MMR) antibody, PE anti-mouse CD80 antibody, and Brilliant Violet 421™ anti-mouse F4/80 antibody for flow cytometry analysis.
For in vivo phenotype regulation, the H22 tumor-bearing mouse model was used. The fluorescent labeling dye IVISense 680 was selected to label injected macrophage from macrophage, USIP@M, and USIP@M+L groups. The mice were injected with macrophage, USIP@M, and USIP@M+L every 4 days for 2 times (at a dosage of 3.0 × 106 cells per mouse). For the USIP@M+L group, 1 day after each administration, the mice were irradiated at a power density of 1 W/cm2 for 10 min. 48 h after the second injection, mice were euthanized, and the single-cell suspensions from tumors were stained for flow cytometry analysis. The injected macrophages could be distinguished by flow cytometry.
2.13. The apoptosis analysis of H22 cells
To investigate the effect of different formulations on tumor cell apoptosis, H22 cells were seeded into 12-well plates (2 × 105 cells/well) containing NS, Free SF, USIP, USIP+L, and released medium from USIP@M+L, and the cells were further incubated for 24 h. The cells were then collected, incubated with Annexin V-FITC at room temperature in the dark for 10 min and PI for 5 min in turn, and analyzed using flow cytometry.
2.14. The cell viability of H22 cells
The CCK-8 assay was used to evaluate the cell viability of different formulations on H22 cells. H22 cells were seeded in 96-well plates (1 × 104 cells/well) containing NS, Free SF, USIP, USIP+L and released medium from USIP@M+L with different concentrations of SF (0.1, 1, 5, 10, 25 μg/mL) for 24 h. Subsequently, the CCK-8 reagent was added to the wells, and 2 h later the absorbance of each well was measured at 450 nm to calculate the cell survival rate.
2.15. Evaluation of cellular uptake in vitro
Free C6, UCP, released medium from UCP@M and UCP@M + L were introduced into 12-well plates containing H22 cells (1 × 105 cells/well) and further incubated for 2 or 4 h. Finally, the cells from each experimental group were imaged. In addition, the above cells were collected and measured using flow cytometric to quantify the cellular uptake.
2.16. The migration capacity of USIP@M in vitro
The in vitro migration ability of USIP@M was investigated by Transwell assay. The macrophages and USIP@Ms were seeded in upper chambers (8.0 μm pore size, polycarbonate membrane) with 3 × 105 cells per chamber, and 600 μL of DMEM media or H22-cell condition media was added to the lower chambers. After incubating for 6 h, the cells in the lower chambers were observed and counted.
2.17. Tumor targeting capability in vivo
For the investigation of tumor targeting capabilities, free DiR, DP, and DP@M were prepared by using the hydrophobic dye DiR instead of the hydrophobic drug SF and IMD. The hepatocarcinoma-bearing mice were used, which were established at the right axilla by inoculating subcutaneously 1 × 106 H22 cells. The mice were intravenous (i.v.) administration with free DiR, DP, or DP@M (at a dosage of 3.0 × 106 cells per injection, equal to 1.5 mg/kg DiR). After 1, 2, 4, 8, 12, and 24 h of injection, the mice were anesthetized. Using the IVIS spectrum in vivo imaging system, the mice were observed. For further ex vivo evaluation, tumors or main organs were obtained from sacrificed mice at 24 h.
2.18. Evaluation of targeting protein
The targeting proteins CCR2 from different preparations were verified by Western blotting experiments. The proteins of the macrophage, USIP@M, USIP@M+L, USIP, and PNB groups were extracted respectively, and the protein samples were denatured and then added to the gel. The electrophoresis and membrane transfer were performed sequentially. The anti-β-Tubulin and anti-CCR2 were incubated and washed. Finally, the signal was observed by the gel imaging system.
2.19. Evaluation of deep penetration
H22 tumor-bearing BALB/c mice were used as animal models, and the hydrophobic fluorescent dye C6 was selected to replace the hydrophobic drugs SF and IMD to prepare UCP and UCP@M. When the tumor volume was appropriate, hepatocarcinoma-bearing mice were i.v. injected with free C6, UCP, and UCP@M respectively. The UCP@M group was set into an NIR irradiation group (1 W/cm2 for 10 min) and without an NIR irradiation group (UCP@M+L and UCP@M). For groups requiring NIR irradiation, the mice were irradiated at 4 h before harvesting the tumors. 24 h after injection, the mice were sacrificed and the tumor tissues were dissected out. The tumors were placed in 4% paraformaldehyde, paraffin-embedded and sliced to observe the deep penetration of different preparations in the tumor site.
2.20. The antitumor efficacy evaluation of USIP@M in hepatocarcinoma-bearing mouse model
Using the hepatocarcinoma-bearing mouse model, the antitumor effect of USIP@M was evaluated. The mice were randomly assigned and i.v. injected with (1) NS, (2) macrophage, (3) SF, (4) IMD, (5) IMD+SF (SF and IMD mixed solution), (6) USIP+L, (7) USP@M+L, (8) UIP@M+L, (9) USIP@M and (10) USIP@M+L every 4 days for 5 times. For groups requiring NIR irradiation (980 nm), 1 day after administration, the mice were irradiated at a power density of 1 W/cm2 for 10 min, once every 4 days, for a total of 5 times. Mice that were injected with macrophage, USP@M+L, UIP@M+L, USIP@M, or USIP@M+L each received 3.0 × 106 cells per injection, equal to 5 mg/kg SF and 1.17 mg/kg IMD. Mice requiring an injection of SF, IMD, IMD+SF, or USIP+L each received the equivalent dose of medicine (5 mg/kg SF and 1.17 mg/kg IMD). The tumor volume and body weight were measured every other day. The mice were sacrificed, and tumors were excised and weighed on Day 20 after the first administration. Based on tumor volume, the suppression rates for tumors were calculated.
2.21. Immunohistochemistry evaluation
After the in vivo antitumor efficacy study, the major organs (heart, liver, spleen, lung, and kidney) and tumors were obtained, fixed in 4% paraformaldehyde, and embedded in paraffin for histological analysis. Sections of tumors were stained with H&E and TUNEL. Sections of major organs were stained with H&E.
2.22. The preliminary safety evaluation
For the hemolysis test, the 2% erythrocyte suspension was prepared based on erythrocytes from Wistar rats. After incubation with a series of doses (5, 10, 20, 40, and 80 μg/mL according to the amount of SF) of USIP@M for 3 h, the mixed solutions were centrifuged and photographed. The supernatants of mixed solutions were detected to obtain the absorbance at 540 nm. The hemolysis rate (%) was calculated.
For serum cytokine assessment, serum samples from H22 tumor-bearing BALB/c mice at 48 h after the first treatment according to the dosage mentioned in part of “2.20 The antitumor efficacy evaluation of USIP@M in a hepatocarcinoma-bearing mouse model” were collected, and analyzed for concentrations of IL-6 and TNF-α using the ELISA Kit.
2.23. In vivo immunological evaluation
In order to analyze the proportion of immune cells in tumors, tumors were isolated, and single-cell suspensions were obtained. The lymphocytes were isolated by 40% and 60% Percoll solution in turn, and treated with rat serum to block nonspecific binding. The macrophage was analyzed by labeling with anti-F4/80-PerCP/Cyanine5.5, anti-CD80-PE, and anti-CD206-APC antibodies. For T cells, cells were analyzed by labeling with anti-CD3-APC, anti-CD4-FITC, and anti-CD8-PE antibodies. For CD8+IFN-γ+ T cell, monensin was added to inhibit the protein transport, and then the cell was labeled with anti-CD3-APC, anti-CD8-PE and anti-IFN-γ-Alexa Fluor® 488 antibodies. For Treg, cells were analyzed by labeling with anti-CD25-Alexa Fluor® 488, anti-CD4-PE, and anti-FOXP3-Alexa Fluor® 647 antibodies. Using flow cytometry, cells were detected. Using FlowJo (V10), the results were analyzed.
Peripheral blood was collected from mice in each group on Day 20, and the serum was collected by centrifugation. The levels of cytokines (IFN-γ, IL-12, TNF-α, IL-10, and TGF-β) were investigated by ELISA kits.
2.24. The antitumor efficacy evaluation of USIP@M in B16F10-bearing mouse model
Using the B16F10-bearing mouse model, the antitumor effect was evaluated. The mice were randomly assigned and i.v. injected with (1) NS, (2) macrophage, (3) USIP+L, (4) USIP@M, and (5) USIP@M+L every 4 days for 4 times. For groups requiring NIR irradiation (980 nm), 1 day after administration, the mice were irradiated at a power density of 1 W/cm2 for 10 min, once every 4 days, for a total of 4 times. Mice that were injected with macrophage, USIP@M, or USIP@M+L each received 3.0 × 106 cells per injection, equal to 5 mg/kg SF and 1.17 mg/kg IMD. The tumor volume and body weight were measured every other day. The mice were sacrificed, and tumors were excised and weighed on Day 16 after the first administration. Based on tumor volume, the suppression rates for tumors were calculated. On Day 16, the tumors were prepared single-cell suspensions for flow cytometry as described in the “In vivo immunological evaluation” section.
2.25. Statistical analysis
The data analysis was performed using GraphPad Prism 8 for Student's t-test and one-way analysis of variance (ANOVA). The differences were considered to be statistically significant when ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001. All results were reported as the mean ± standard deviation (SD).
3. Results and discussion
3.1. USIP had the capability of photoresponsive depolymerization
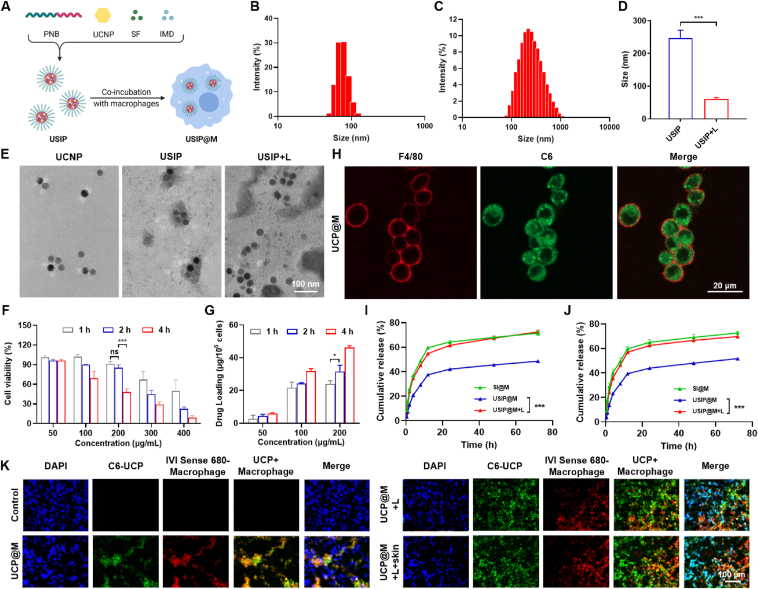
The monomer NBMA and macroinitiator PEO2K-Br were synthesized and used to prepare amphiphilic block copolymer PNB. The structures were first verified by 1H NMR and then further confirmed by GPC (Supporting Information Fig. S1). The light-responsive micelle (USIP) was prepared by encapsulating UCNPs, SF, and IMD, and then the USIP@M was prepared by co-incubating macrophages with USIP (Fig. 2A). The particle sizes of UCNP and USIP were 77.00 ± 2.34 and 218.50 ± 4.20 nm, respectively (Fig. 2B and C). The corresponding polydispersity indexes were 0.136 ± 0.007 and 0.196 ± 0.016. SF and IMD were efficiently loaded into USIP with drug loadings (DL, %) of 5.42 ± 0.91% and 1.27 ± 0.12%, respectively. Overall, these results demonstrate the successful preparation of USIP. The emission spectra of the USIP and UCNP were investigated (Supporting Information Fig. S2). Compared with UCNP, the emission spectra of UV light of the USIP were much weaker around the 350 nm wavelength, indicating that the group of O-nitrobenzyl within PNB in USIP could absorb the emitted UV photons when UCNP was irradiation. The hydrophilic-hydrophobic balance of USIP was broken by hydrophilic polymethacrylic acid transforming from the polymethacrylate due to the photocleavage of o-nitrobenzyl groups, which resulted in the depolymerization of USIP. Therefore, the capability of photoresponsive depolymerization was studied. After the irradiation with NIR light (980 nm, 1 W, 10 min), the UCNPs emitted UV light in the cell, the particle sizes became smaller because of the UV-triggered depolymerizing (246.93 ± 24.55 nm vs 60.77 ± 4.68 nm, P < 0.001, Fig. 2D). And the complete depolymerization of USIP was visible according to TEM images (Fig. 2E).
Figure 2.
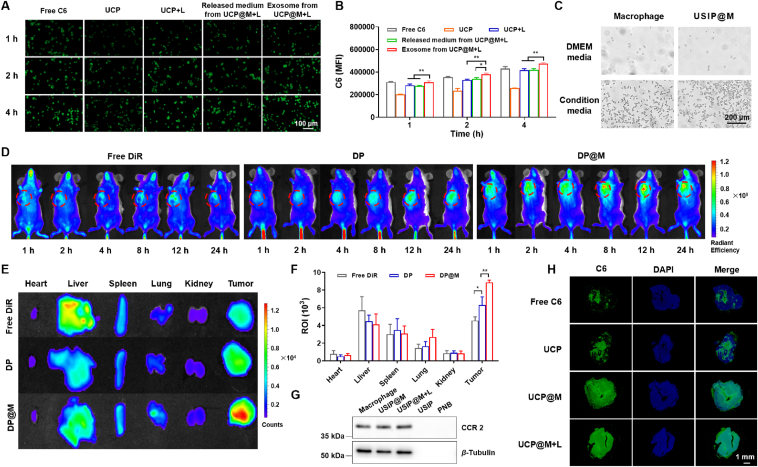
The characterizations of photoresponsive depolymerization for USIP and the feasibility of photoresponsive drug release for USIP@M. (A) Schematic illustration of the preparation process of USIP and USIP@M, this figure was created with BioRender.com. Hydrodynamic size of (B) UCNP and (C) USIP. (D) Changes of hydrodynamic size before (USIP) and after (USIP+L) NIR irradiation. (E) TEM images of UCNP, USPI, and USIP+L group, scale bar = 100 nm. (F) The cell viability of USIP in macrophages at various concentrations (according to the amount of SF) and incubation time. (G) The drug loading of macrophages for USIP at various concentrations (according to the amount of SF) and incubation time. (H) CLSM images of UCP@M, scale bar = 20 μm. The cell membrane was stained with anti-F4/80 antibody (red) and the drug was replaced by C6. Release profiles of (I) SF and (J) IMD from SI@M, USIP@M, and USIP@M with NIR irradiation (USIP@M+L). (K) CLSM images of tumor sections after administration at 24 h. IVISense 680 fluorescent dye and C6 were selected to mark injected macrophages (red) and drug (green), respectively, scale bar = 100 μm. Data are presented as mean ± SD (n = 3). ∗P < 0.05 and ∗∗∗P < 0.001.
3.2. USIP@M could achieve photoresponsive drug release
USIP@M was successfully prepared by co-incubating macrophages with USIP in vitro. The incubation concentration (according to the amount of SF) and incubation time in the co-incubating of macrophages and USIP were optimized by single-factor assays (Fig. 2F and G). The results showed that 2 h was the optimal incubation time and 200 μg/mL was the optimal incubation concentration (according to the amount of SF) with the higher DL and no obvious cytotoxicity to macrophages. Under this condition, the DL efficiency of USIP@M was 15.73 ± 1.97%, and USIP@M had an SF content at 31.46 ± 3.94 μg/106 cells and IMD content at 7.39 ± 0.22 μg/106 cells. Visualized under confocal laser scanning microscopy (CLSM), hydrophobic dye C6 was chosen to make the drug visible, which could display a green fluorescence signal. Red fluorescence signal showed the cell membrane with the help of Alexa Fluor® 647 anti-mouse F4/80. The CLSM images of UCP@M showed that macrophages contained a large number of green fluorescence signals of UCP, indicating that UCP was effectively loaded into macrophages to form UCP@M (Fig. 2H). The above results showed that USIP could be successfully loaded into macrophages.
The cell viability at the optimal incubation time and concentration of USIP@M was also evaluated by the calcein-AM/PI staining kit, in which green and red fluorescence signals represented living and dead cells, respectively. As shown in the fluorescence microscopy images, large amounts of green fluorescence were detected in the macrophage group and USIP@M group, which suggested the living state with high cell viability of the macrophage and USIP@M. Then, the USIP@M was irradiated with NIR light (980 nm, 1 W, 10 min), showing no difference in green fluorescence compared with macrophage and USIP@M groups (Supporting Information Fig. S3). The results indicated that the irradiation with NIR light had no obvious phototoxicity to USIP@M itself.
In order to obtain the release profiles, SF and IMD from SI@M, USIP@M, and USIP@M with NIR irradiation (980 nm, 1 W) for 10 min (USIP@M+L) were quantified using HPLC over 72 h (Fig. 2I and J). The proportion of cumulative released SF from the USIP@M+L group at 72 h was 72.50 ± 1.62% (Fig. 2I), which was significantly higher than that from the USIP@M group (48.59 ± 0.43%, P < 0.001). The analysis based on IMD quantitative data in the USIP@M+L group at 72 h was also significantly higher than that from the USIP@M group (69.75 ± 0.54% 51.66 ± 0.97%, P < 0.001, Fig. 2J). Furtherly, we evaluated the ability of NIR light-triggered drug release in vivo (Fig. 2K). IVISense 680 fluorescent dye and C6 were selected to mark injected macrophages (red) and drug (green), respectively. The strong fluorescence colocalization of green and red (yellow) was observed in the UCP@M group, which indicated that the drug remained inside injected macrophages in the absence of NIR irradiation. After NIR irradiation (UCP@M+L group), the green fluorescence appeared outside the colocalization region, which meant that the drug was released from injected UCP@M. Surprisingly, even with 5 mm pigskin blocking, NIR light-triggered drug release was still possible.
3.3. USIP@M generated drug exosomes and aggravated phenotypic polarization by UV light converted from NIR light
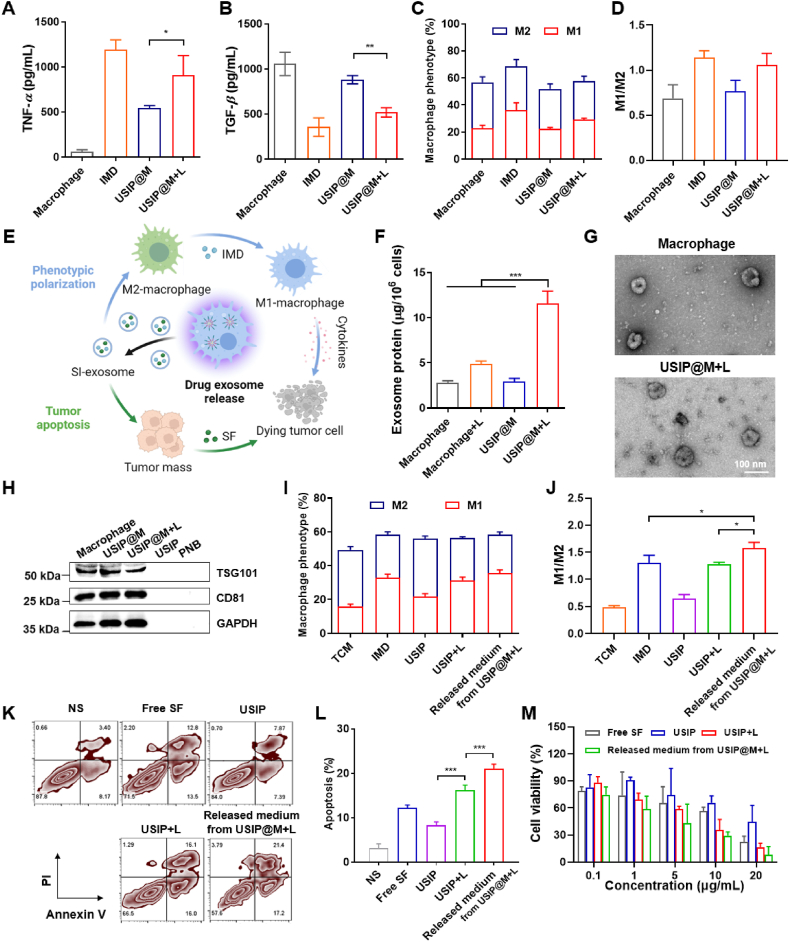
To evaluate the effect of USIP-loaded and NIR-irradiated drug release on macrophage self-phenotype, we assessed the expression of several factors, including immunosuppressed cytokines (TGF-β and IL-10), pro-inflammatory cytokines (TNF-α) and the percentage of M1 or M2-type macrophages. After loading USIP, the levels of pro-inflammatory and anti-inflammatory factors secreted by USIP@M showed comparable secretion relative to the control group. However, after IMD treatment and NIR irradiation of USIP@M, TNF-α showed upregulated secretion, while immunosuppressed cytokines TGF-β and IL-10 were decreased because IMD could promote the phenotypic polarization of macrophages (Fig. 3A and B, Supporting Information Fig. S4). USIP@M+L group showed higher TNF-α levels than that in the group of USIP@M (P < 0.05). On the contrary, the levels of TGF-β were decreased in the USIP@M + L group, which was significantly lower than that in the USIP@M group (P < 0.01). The analysis based on flow cytometry data showed the same conclusion (Fig. 3C and D).
Figure 3.
UV light converted from NIR light promoted the drug exosomes release with therapeutic effects from USIP@M. (A–D) Phenotype analysis of USIP@M 24 h after irradiation with or without NIR light, including the secretion of (A) TNF-α and (B) TGF-β, (C) the percentage of M1 and M2-type macrophages, (D) the ratio of M1 to M2-type macrophages. The macrophage group (untreated RAW264.7 cells) served as controls. The IMD group (RAW264.7 cells incubated with IMD) contained the same concentration of IMD as USIP@M and USIP@M+L group. (E) Schematic diagram of drug exosome release and its therapeutic effect, this figure was created with BioRender.com. Stimulated by the converted UV light, USIP@M could release SI-exosomes containing SF and IMD, which induced the polarization of macrophage carriers to the M1 phenotype and the apoptosis of tumor cells. (F) The exosome protein content was released within 24 h after preparation of USIP@M and USIP@M+L. The macrophage group (untreated RAW264.7 cells) served as controls. (G) TEM images of exosomes from macrophage and USIP@M+L groups, scale bar = 100 nm. (H) Western blotting analysis of exosome markers (CD81 and TSG101) of released exosomes from different preparations. (I, J) Phenotype analysis of TAMs after coculture with IMD, USIP, USIP+L, and released medium from USIP@M+L in TCM for 24 h. (K, L) The representative flow cytometry plots and flow cytometric analysis of H22 cells apoptosis. (M) The cell viability of H22 cells after coculture with free SF, USIP, USIP+L, and released medium from USIP@M+L for 24 h by CCK-8 assay at various concentrations (according to the amount of SF). Data are presented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
Due to the ability of UV irradiation to promote the production of exosomes by living cells13 and the UCNPs within USIP@M could convert NIR light into UV light in the cell, the produced exosome was evaluated by total protein quantification using the bicinchoninic acid assay (Fig. 3E). The results further confirmed that UV light converted from NIR light increased the generation of exosomes from USIP@M (P < 0.001, Fig. 3F). TEM images of exosomes from macrophage and USIP@M+L groups showed no differences in the appearance of exosomes (Fig. 3G). In the condition that USIP and PNB groups were negative controls, exosome markers (CD81 and TSG101) were detected by Western blotting analysis generated by macrophage, USIP@M, and USIP@M+L groups (Fig. 3H). The exosomes from macrophage, USIP@M and USIP@M+L groups showed no differences in the characterization of exosomes, the corresponding particle sizes were 106.93 ± 3.39, 117.87 ± 6.36 and 112.80 ± 3.35 nm, and the corresponding zeta potentials were −13.13 ± 1.07, −13.00 ± 1.14 and −13.37 ± 1.42 mV (Supporting Information Table S1).
3.4. The effect of regulating TAMs phenotype and inducing tumor apoptosis for USIP@M after NIR irradiation triggering
The effect of regulating TAMs phenotype from M2-type macrophages to M1-type macrophage reversion was evaluated. TAMs were prepared by culturing RAW264.7 cells in TCM for 24 h. We explored the phenotype analysis of TAMs after coculture with IMD, USIP, and USIP+L and released medium from USIP@M+L in TCM for 24 h. The results showed that the ratio of M1 to M2-type macrophages for the released medium from the USIP@M+L group was 1.58 ± 0.10, while that of the USIP+L group was only 1.27 ± 0.04 (P < 0.05), indicating that the use of macrophage cytopharmaceutical could promote TAMs phenotype polarization more effectively (Fig. 3I and J). For in vivo phenotype regulation, we used IVISense 680 fluorescent dye labeling injected macrophage from macrophage, USIP@M, and USIP@M+L groups to distinguish adoptively transferred macrophage and TAM (Supporting Information Fig. S5). The experiment results showed that USIP@M could transform into and maintain the M1 phenotype after NIR irradiation in vivo. Compared with the USIP@M group, the endogenous TAM from the USIP@M+L group was more likely to be polarized to M1 phenotype (P < 0.001), which indicated the action of USIP@M after NIR irradiation could convert TAM to play an anti-tumour role.
The effects on H22 cells apoptosis and viability were evaluated. According to flow cytometry analysis, the results showed that the apoptosis ratio of the USIP+L group was 16.20 ± 1.15%, while that of the USIP group was only 8.35 ± 0.72% (P < 0.001), indicating that USIP had a better role in inducing apoptosis only after photoresponsive depolymerization. The released medium from the USIP@M+L group had an apoptosis ratio of 21.00 ± 1.06%, which was significantly higher than the USIP+L group (P < 0.001), indicating that the use of macrophage cytopharmaceutical could induce cell apoptosis more effectively (Fig. 3K and L). By using the CCK-8 assay, the in vitro cytotoxicity of the released medium from the USIP@M+L group on H22 cells was obtained. The IC50 of the released medium from the USIP@M+L group was 0.76 ± 0.34 μg/mL, while the IC50 value of free SF, USIP, and USIP+L groups were 5.56 ± 2.22 (P < 0.001), 20.20 ± 8.01 (P < 0.001), and 3.77 ± 0.47 (P < 0.001) μg/mL, respectively (Fig. 3M and Supporting Information Table S2). The above results demonstrate that the USIP@M+L group could effectively promote tumor cell apoptosis and kill tumor cells in vitro.
3.5. USIP@M had the capability of tumor-targeting and tumor-penetrating
The cell uptake of USIP, USIP+L, the released medium from USIP@M+L, and the exosome from USIP@M+L were observed using hepatocarcinoma cell line H22. C6 was selected as a tracer agent to prepare UCP and UCP@M. According to the results of fluorescence images (Fig. 4A) and flow cytometric (Fig. 4B), the fluorescence intensity in each group gradually increased from 1 to 4 h. Moreover, the fluorescence intensity of exosomes from the UCP@M+L group was significantly higher than that of the UCP+L group (P < 0.01) and released medium from the UCP@M+L group (P < 0.01) at 4 h. The above results showed that the exosome from UCP@M after NIR irradiation could be efficiently ingested into H22 cells, which may be related to the fact that exosomes stimulated by the converted UV light themselves could promote cell uptake.
Figure 4.
Tumor targeting and deep penetration ability enhancement of macrophage cytopharmaceuticals. (A) Fluorescence images and (B) flow cytometric results of cellular uptake in H22 cells, including free C6, UCP, UCP+L, the released medium from UCP@M+L and the exosome from UCP@M+L group at the same concentration (400 ng/mL) of C6, scale bar = 100 μm. (C) Micrographs for evaluating migration capacity of macrophage and USIP@M in vitro, scale bar = 200 μm. (D) In vivo imaging of the H22 tumor-bearing mice at different times after treatment with free DiR, DP, and DP@M. (E) Ex vivo imaging and (F) the average fluorescent intensity after the H22 tumor-bearing mice were dissected at 24 h post-administration. DiR was a near-infrared tracer in D-F. (G) Western blotting analysis of targeting protein CCR2 from different preparations. (H) Tumor sections in a hepatocarcinoma-bearing mouse model of 24 h after injection of free C6, UCP, UCP@M, and UCP@M+L, respectively, scale bar = 1 mm. Data are presented as mean ± SD (n = 3). ∗P < 0.05 and ∗∗P < 0.01.
The effect of loading USIP on the chemotactic migration ability of macrophage toward H22 cells was verified by Transwell assay. H22-cell condition media was seeded into the lower chambers, and macrophage or USIP@M was loaded into the upper chambers. We found that USIP@M could efficiently transfer into lower chambers, with migration percentages comparable to the macrophage group. Meanwhile, when H22-cell condition media was absent, there was a low migration percentage (Fig. 4C and Supporting Information Fig. S6). Next, the effect of USIP loading on tumor targeting of macrophage was investigated by NIR fluorescence imaging on H22 tumor-bearing mice. Using tracer agent DiR, DP, and DP@M were prepared. In the real-time imaging in vivo, the fluorescence signal intensity of the DP@M group was the strongest at 12 h, indicating that the tumor site high accumulation of DP@M at 12 h (Fig. 4D). The tumor homing ability of macrophages contributed to the tumor-targeting ability of DP@M significantly stronger than that of DP group (P < 0.01) or free DiR group (P < 0.001, Fig. 4E and F). For the free DiR group, the DP group had a higher tumor site fluorescence signal (P < 0.05), indicating that DP was more concentrated in the tumor site due to enhanced personality and retention effect compared with free DiR. As shown in Fig. 4G, the targeting protein CCR2 on different preparations was further determined by western blotting. In macrophage, USIP@M, and USIP@M+L groups, we could observe the targeting protein CCR2, but not in USIP or PNB groups, which confirmed the existence of the targeting protein. These results indicated that USIP@M inherited the tumor-targeting ability of macrophages.
We also explored the tumor-penetrating ability of free C6, UCP, UCP@M, and UCP@M+L groups on H22 tumor-bearing mice by constructing C6-loaded formulations (Fig. 4H). Tumor sections were stained with DAPI (blue) to locate tumor cells and C6 (green) could track UCP and UCP@M inside tumor tissue. The C6 signals in the UCP@M and UCP@M+L groups covered almost the entire tumor section, especially penetrating deep into the tumor. On top of that, the fluorescence intensity of the UCP@M+L group was stronger than the UCP@M group in the entire tumor section. However, only a small fraction of tumor sections in the free C6 group showed green fluorescence signals, indicating poor tumor penetration ability. The results showed that UCP@M and UCP@M+L had good deep tumor penetration ability, and the exosomes of macrophage could be stimulatively produced after UCNP converted NIR into UV, resulting in stronger deep tumor-penetrating ability in vivo.
3.6. The preliminary safety evaluation of USIP@M was excellent
The safety of USIP@M was preliminarily evaluated through a hemolysis test, histopathological analysis of the major organs, and serum cytokine assessment. There was no significant tissue damage was observed, indicating that USIP@M was safe for major organs (Supporting Information Fig. S7). The safety of injection was investigated by hemolysis test, and the results showed no obvious hemolysis phenomenon in the experimental concentration range (5, 10, 20, 40, and 80 μg/mL), which demonstrated that USIP@M had less interaction with erythrocytes and was suitable for i.v. administration (Supporting Information Fig. S8). Based on the serum IL-6 and TNF-α levels, cytokine storms did not occur after the initial administration of USIP@M (Supporting Information Fig. S9). Additionally, the body weight changes of mice during the treatment cycle provided further evidence of safety (Fig. 5B and 7D). Overall, these results indicated that USIP@M had excellent preliminary safety.
Figure 5.
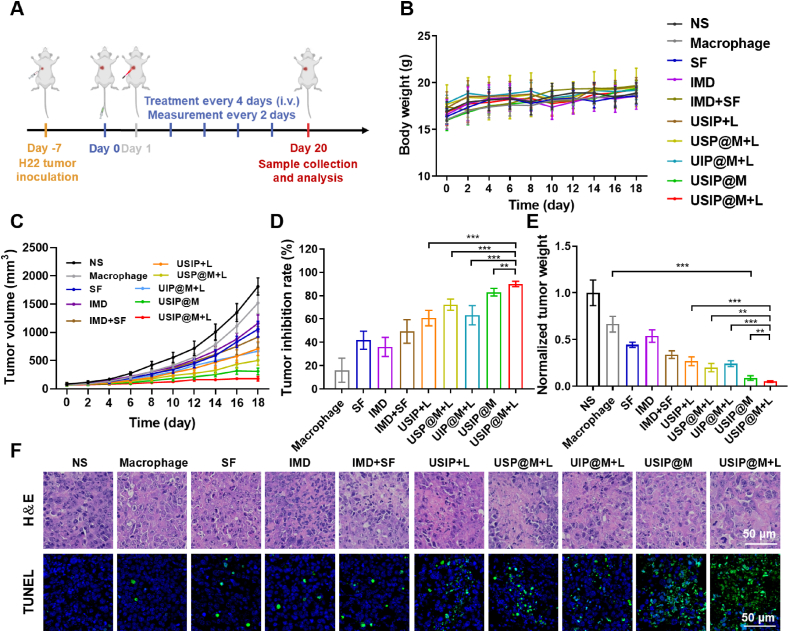
USIP@M showed a powerful antitumor efficiency under NIR light in hepatocarcinoma-bearing mouse models. (A) Schedule of in vivo administration approach. Different formulations were used to treat mice, and some of the groups were irradiated with NIR light (980 nm) of 1 W/cm2 for 10 min (at a dosage of 3.0 × 106 cells per injection, equal to 5 mg/kg SF and 1.17 mg/kg IMD). When the tumor volumes reached about 2000 mm3, the mice were sacrificed. This figure was created with BioRender.com. (B) Body weight changes, (C) tumor volume changes, (D) tumor inhibition rate, and (E) normalized tumor weight after i.v. injection with NS, macrophage, SF, IMD, IMD + SF (SF and IMD mixed solution), USIP+L, USP@M+L, UIP@M+L, USIP@M and USIP@M+L. (F) Immunohistochemical analysis of H&E (scale bar = 50 μm) and TUNEL (scale bar = 50 μm) in H22 tumors. Data are presented as mean ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
Figure 7.
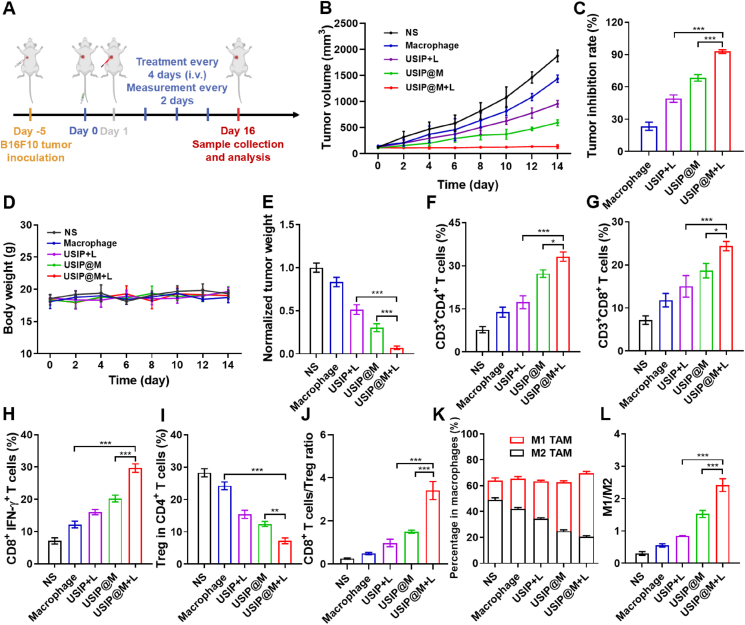
USIP@M exhibited excellent antitumor immune effect under NIR light in the B16F10-bearing mouse model. (A) Schedule of in vivo administration approach. Different formulations were used to treat mice, and some of the groups were irradiated with NIR light (980 nm) of 1 W/cm2 for 10 min (at a dosage of 3.0 × 106 cells per injection, equal to 5 mg/kg SF and 1.17 mg/kg IMD). When the tumor volumes reached about 2000 mm3, the mice were sacrificed. This figure was created with BioRender.com. (B) Tumor volume changes, (C) tumor inhibition rate, (D) body weight changes, and (E) normalized tumor weight after i.v. injection with NS, macrophage, USIP+L, USIP@M and USIP@M+L. The proportion of intratumor infiltration (F) CD4+ T cells, (G) CD8+ T cells, (H) CD8+IFN-γ+ T cells (I) Treg in CD4+ T cells. (J) The ratio of CD8+ T cells to Treg in tumor tissue. (K) The ratio of M1 to M2-type macrophages and (L) the percentage of M1 or M2-type macrophages in all macrophages. Data are presented as mean ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
3.7. The antitumor effect of USIP@M after NIR irradiation in the H22 tumor-bearing mouse model
The in vivo antitumor efficacy of USIP@M was evaluated in the H22 tumor-bearing mouse model. Mice were treated with the formulations of NS, Macrophage, SF, IMD, IMD+SF, USIP+L, USP@M+L, UIP@M+L, USIP@M, and USIP@M+L groups by i.v. injection. The formulations were injected every 4 days 5 times, in which the doses of 3.0 × 106 cells per injection or equal to 5 mg/kg SF and 1.17 mg/kg IMD (Fig. 5A). The tumor volume of the SF group, IMD group, and IMD+SF group decreased slightly with poor therapeutic effect when compared with NS group, which might be related to the lack of targeting of free drugs (Fig. 5C, Supporting Information Figs. S10 and S11). The tumor inhibition rates of Macrophage, SF, IMD, IMD+SF, USIP+L, USP@M+L, UIP@M+L, USIP@M, and USIP@M+L groups were 16.09 ± 10.35%, 41.83 ± 7.77%, 36.01 ± 8.20%, 49.37 ± 10.10%, 60.74 ± 6.62%, 72.25 ± 4.77%, 63.17 ± 8.27%, 82.97 ± 3.24%, and 90.01 ± 2.32%, respectively (Fig. 5D). Although the antitumor effect of USIP+L group with passive targeting effect was better than that of free drugs (SF group, IMD group, and IMD+SF group), its tumor inhibition rate was much lower than USIP@M+L group (P < 0.001), due to the better tumor targeting effect of carrier macrophage and the immunotherapeutic antitumor efficacy of macrophage which were polarized to M1 phenotype after drug photoresponsive release. Unsurprisingly, the suppression rates for tumors of the USP@M+L group (P < 0.001) and UIP@M+L group (P < 0.001) were not as good as that of the USIP@M+L group, which may have something to do with the absence of IMD immunotherapy effect in the USP@M+L group, while the tumor-killing effect of SF was absent in the UIP@M+L group. Even when compared with the USIP@M group, the USIP@M+L group was still able to significantly inhibit tumor growth (P < 0.01), showing the best antitumor effect, which indicated that UV light converted from NIR light could promote the production of exosome and drug-triggered release, and improve the antitumor efficacy of USIP@M. The excised tumors were weighed, and the significance analysis based on normalized tumor weight was in accordance with the conclusion obtained from the above volume (Fig. 5E).
After euthanized, the mice were dissected to collect tumor tissues after treatment for 20 days. The proliferation and apoptosis of tumor tissues were assessed by H&E and TUNEL staining (Fig. 5F). As shown by TUNEL staining, the USIP@M+L group showed a lot of green fluorescence, which meant the highest level of cell apoptosis. These results suggested that USIP@M after photoresponsing drug release could effectively promote apoptosis and inhibit proliferation for tumors, exhibiting a powerful antitumor efficiency.
3.8. USIP@M repolarized TAM and enhanced T cell immunity to remodel TME under NIR light
T cells are important effectors in the immune system for fighting against tumors and are mainly divided into CD8+ T cells and CD4+ T cells. The immune status of TME regulated by USIP@M was evaluated by measuring the frequencies of intratumoral CD3+CD4+ T cells, CD3+CD8+ T cells, CD8+IFN-γ+ T cells, and Tregs. After administration with different formulations, the lymphocytes within the tumor were collected and analyzed by flow cytometry to evaluate the T cell content and the ability of USIP@M to improve the TME. The flow cytometry analysis results of the USIP@M+L group showed that the percentage of intratumoral CD3+CD4+ and CD3+CD8+ T cells infiltration were significantly increased (Fig. 6A, B, and I). Furthermore, the levels of CD8+IFN-γ+ T cells and Tregs in the tumor tissue were analyzed. As shown in the flow cytometry analysis, intratumoral CD8+IFN-γ+ T cell infiltration had a similar trend to CD3+CD8+ T cells. The content of intratumoral CD8+IFN-γ+ T cells in the USP@M+L group was signally lower than that in the USIP@M+L group (P < 0.001) because the lack of IMD weakened its immune stimulation (Fig. 6C and J). The content of CD8+IFN-γ+ T cells in the USIP@M+L group was 16.20 ± 0.89%, significantly higher than that in the macrophage group (P < 0.001), and the content of Tregs in the USIP@M+L group was 6.51 ± 0.60%, significantly lower than that in the Macrophage group (P < 0.001), indicating that USIP loading and localized light-triggered release could promote the infiltration of T cells in the tumor (Fig. 6D and K). The content of CD8+IFN-γ+ T cells in the USIP+L group was 10.21 ± 1.69%, which was significantly lower than that in the USIP@M+L group (P < 0.001). At the same time, the content of Tregs in the USIP+L group was 13.33 ± 0.67%, which was double higher when compared to 6.51 ± 0.60% in the USIP@M+L group (P < 0.001). Moreover, the ratio of CD8+ T cells to Tregs in different groups was calculated (Fig. 6E). The ratio of CD8+ T cells to Tregs in the USIP@M+L group was 3.66 ± 0.45, significantly better than that in the USIP@M group with a ratio of 2.62 ± 0.13 (P < 0.001), indicating that localized light-triggered release strategy was crucial to the change of T cells. The above results showed effective intratumor infiltration of T cells and improvement of immunosuppression.
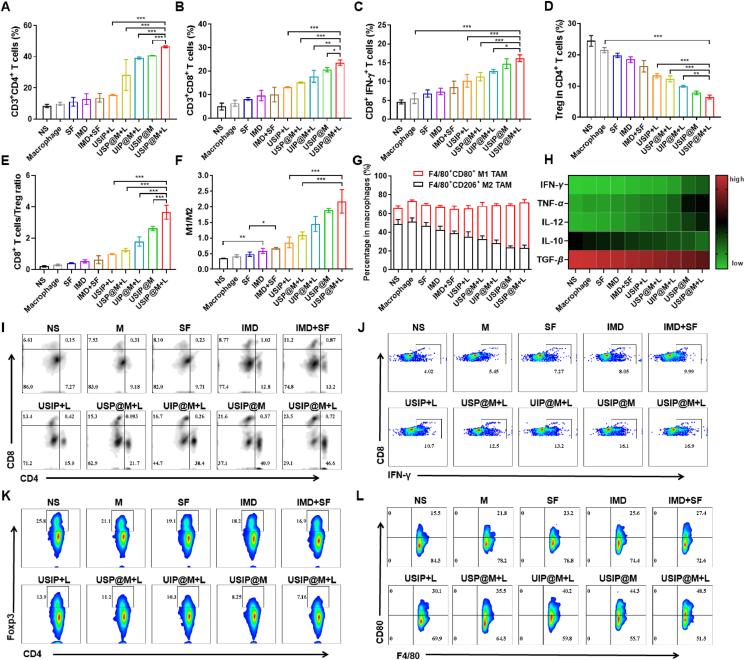
Figure 6.
USIP@M initiated the antitumor immune response mediated by T cells and macrophages under NIR light in a hepatocarcinoma-bearing mouse model. The proportion of intratumor infiltration (A) CD4+ T cells, (B) CD8+ T cells, (C) CD8+IFN-γ+ T cells, (D) Treg in CD4+ T cells. (E) The ratio of CD8+ T cells to Treg in tumor tissue. (F) The ratio of M1 to M2-type macrophages and (G) the percentage of M1 or M2-type macrophages in all macrophages. (H) The levels of cytokines in peripheral blood, including IFN-γ, TNF-α, IL-12, IL-10, and TGF-β, were quantified using ELISA. The representative flow cytometry plots of (I) CD4+ and CD8+ T cells, (J) CD8+IFN-γ+ T cells, (K) Treg in CD4+ T cells, and (L) M1-type macrophages. Data are presented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
TAMs affect tumor progression in a manner that is dependent on their polarization. Macrophages possessing the M1 phenotype have been shown to improve outcomes in tumor therapy. After proving that the carrier macrophages could polarize towards the M1 phenotype after the UV light converted from NIR light responsive release of drugs in vitro, we sought to evaluate the phenotype changes of TAMs in vivo after the photoresponsive release of drugs by USIP@M. The macrophages in tumor tissues treated with different formulations were analyzed by flow cytometry (Fig. 6F, G, and L). When compared with the NS group based on flow cytometry analysis results, the phenotypic ratio of TAMs changed little for the macrophage group, while M1-type macrophages increased significantly and M2-type macrophages decreased dramatically in the IMD group (P < 0.01), UIP@M+L group (P < 0.001) and USIP@M+L group (P < 0.001). Among them, the percentage of M1-type macrophages in the USIP@M+L group was the highest, and the percentage of M1-type macrophages in the USP@M+L group (without containing IMD) was slightly lower than that in the USIP@M+L group (P < 0.001), while the percentage of M2-type macrophages was the opposite, which suggested that IMD was necessary for phenotypic changes in TAMs. Furtherly, a higher ratio of M1 to M2-type macrophages in the USIP@M+L group was calculated compared with 1.88 ± 0.06 in the USIP@M group, which again proved that photoresponsing drug release was extremely important for efficacy. These results suggested that the intratumoral percentage of M1-type macrophages increased due to the photoresponsive release of IMD, and furtherly changed the ratio of M1 to M2 in TME through the immunomodulation of M1-type macrophages.
In addition, the pro-inflammatory cytokines (IFN-γ, TNF-α, and IL-12) and immunosuppressed cytokines (IL-10 and TGF-β) were also studied. Based on cytokine heat map results, the level of pro-inflammatory cytokines (IFN-γ, TNF-α, and IL-12) in the USIP@M+L group was the highest of all the groups. Along with that, the level of immunosuppressed cytokines (IL-10 and TGF-β) in the USIP@M+L group was the lowest of all the groups (Fig. 6H). The changes in cytokine levels were consistent with the results of flow cytometry analysis results, which together supported that USIP@M under NIR irradiation could improve immune suppression and activate antitumor immunity.
3.9. The antitumor effect of USIP@M after NIR irradiation in B16F10 tumor-bearing mouse model
We used B16F10 tumor-bearing mouse models to further evaluate the antitumor effect of USIP@M (Fig. 7A). Mice were treated with the formulations of NS, Macrophage, USIP+L, USIP@M, and USIP@M+L groups by i.v. injection. Compared with the USIP@M group, the USIP@M+L group was able to significantly inhibit tumor growth (P < 0.001), which proved that irradiation of NIR light improved the antitumor effect of USIP@M, which might be due to the effect of UV light converted from NIR light because UV light could stimulate the production of exosomes from USIP@M, accelerate drug efflux while maintaining the activity of macrophages (Fig. 7B and C, Supporting Information Fig. S12). The tumor inhibition rates of Macrophage, USIP+L, USIP@M, and USIP@M+L groups were 23.44 ± 3.79%, 49.15 ± 3.43%, 68.38 ± 3.06%, and 92.82 ± 1.75%, respectively (Fig. 7C). And there are no significant changes for the body weight of mice in these groups during treatment cycle (Fig. 7D). The excised tumors were weighed, and the significance analysis based on normalized tumor weight was in accordance with the conclusion obtained from the above volume (Fig. 7E). Similar to the hepatocarcinoma-bearing mouse model, the USIP@M+L group significantly improved CD3+CD4+ T cells (Fig. 7F), CD3+CD8+ T cells (Fig. 7G), CD8+IFN-γ+ T cells (Fig. 7H), Tregs (Fig. 7I), CD8+ T cells/Treg ratio (Fig. 7J) and M1-type macrophages (Fig. 7K and L, Supporting Information Fig. S13) infiltration, indicating that the TME reversing capacity of USIP@M under NIR light in B16F10-bearing mouse model.
4. Conclusions
In summary, a macrophage cytopharmaceutical based on localized light-triggered release was constructed, which made full use of the tumor tropism of macrophages and enhanced the targeting delivery and accumulation of drugs at tumor sites. UCNPs were used to convert NIR light with strong penetrability and high safety into UV light, which could promote the depolymerization of USIP and stimulate the production of exosomes from USIP@M, accelerating drug efflux while maintaining the activity of macrophages. Due to the increase of exosomes containing drugs, the uptake of drugs by tumor cells was significantly enhanced and the polarization of macrophage carriers as well as TAMs to M1-type with antitumor effect was higher proportion, which further enhanced T cell immunity. Combined with the chemotherapeutic effect of SF, USIP@M had a remarkable antitumor effect, which provided a new idea for the research of macrophage cytopharmaceuticals.
Author contributions
Jinhu Liu: Writing – original draft, Validation. Han Yang: Writing – original draft. Xiao Sang: Validation. Tong Gao: Validation. Zipeng Zhang: Validation. Shunli Fu: Validation. Huizhen Yang: Validation. Lili Chang: Validation. Xiaoqing Liu: Validation. Shuang Liang: Validation. Shijun Yuan: Validation. Suyun Wei: Validation. Yuxin Yang: Validation. Xiaoxin Yan: Validation. Xinke Zhang: Validation. Weiwei Mu: Validation. Yongjun Liu: Writing – review & editing. Na Zhang: Writing – review & editing.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
Jinhu Liu and Han Yang contributed equally to this work. This work was supported by the National Natural Science Foundation of China (82373809, 82173756, 82173757), and the Shandong Excellent Youth Fund (ZR2022YQ76, China). The authors appreciate the pharmaceutical biology sharing platform of Shandong University for supporting the cell-related experiments. The authors thank the Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work. We thank the support provided by BioRender.com in creating Figure 1, Figure 2, Figure 3, Figure 5, Figure 7A (Agreement number: IX2754RE87).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.08.033.
Appendix A. Supporting information
The following is the Supporting Information to this article:
References
- 1.Yang L.X., Yang Y., Chen Y., Xu Y.H., Peng J.L. Cell-based drug delivery systems and their in vivo fate. Adv Drug Deliver Rev. 2022;187 doi: 10.1016/j.addr.2022.114394. [DOI] [PubMed] [Google Scholar]
- 2.Li W.S., Su Z.G., Hao M.X., Ju C.Y., Zhang C. Cytopharmaceuticals: an emerging paradigm for drug delivery. J Control Release. 2020;328:313–324. doi: 10.1016/j.jconrel.2020.08.063. [DOI] [PubMed] [Google Scholar]
- 3.Luo Z.Y., Lu Y.C., Shi Y.Y., Jiang M.S., Shan X.Y., Li X., et al. Neutrophil hitchhiking for drug delivery to the bone marrow. Nat Nanotechnol. 2023;18:647–656. doi: 10.1038/s41565-023-01374-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y.X., Qin D.T., Zou J.H., Li X.B., Guo X.D., Tang Y., et al. Living leukocyte-based drug delivery systems. Adv Mater. 2023;35 doi: 10.1002/adma.202207787. [DOI] [PubMed] [Google Scholar]
- 5.Chen C., Zhang Y.Q., Chen Z.W., Yang H.H., Gu Z. Cellular transformers for targeted therapy. Adv Drug Deliver Rev. 2021;179 doi: 10.1016/j.addr.2021.114032. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C.Y., Cheng Y.Y., Huang P., Wang C.R., Wang W.P., Wang M.J., et al. X-ray-guided in situ genetic engineering of macrophages for sustained cancer immunotherapy. Adv Mater. 2023;35 doi: 10.1002/adma.202208059. [DOI] [PubMed] [Google Scholar]
- 7.Christofides A., Strauss L., Yeo A., Cao C., Charest A., Boussiotis V.A. The complex role of tumor-infiltrating macrophages. Nat Immunol. 2022;23:1148–1156. doi: 10.1038/s41590-022-01267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Z.H., Davidson D., Li R., Zhong M.C., Qian J., Chen J., et al. Inflammatory macrophages exploit unconventional pro-phagocytic integrins for phagocytosis and anti-tumor immunity. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.110111. [DOI] [PubMed] [Google Scholar]
- 9.Hou T., Wang T.Q., Mu W.W., Yang R., Liang S., Zhang Z.P., et al. Nanoparticle-loaded polarized-macrophages for enhanced tumor targeting and cell-chemotherapy. Nano-micro Lett. 2021;13:6. doi: 10.1007/s40820-020-00531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W.Z., Wang M.Z., Tang W., Wen R., Zhou S.Y., Lee C., et al. Nanoparticle-laden macrophages for tumor-tropic drug delivery. Adv Mater. 2022;34 doi: 10.1002/adma.202109925. [DOI] [PubMed] [Google Scholar]
- 11.Qiu Y., Ren K.B., Zhao W., Yu Q.W., Guo R., He J., et al. A "dual-guide" bioinspired drug delivery strategy of a macrophage-based carrier against postoperative triple-negative breast cancer recurrence. J Control Release. 2021;329:191–204. doi: 10.1016/j.jconrel.2020.11.039. [DOI] [PubMed] [Google Scholar]
- 12.An L., Wang Y.Y., Lin J.M., Tian Q.W., Xie Y.X., Hu J.Q., et al. Macrophages-mediated delivery of small gold nanorods for tumor hypoxia photoacoustic imaging and enhanced photothermal therapy. Acs Appl Mater Inter. 2019;11:15251–15261. doi: 10.1021/acsami.9b00495. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y., Ye H., Chen Y.B., Zhu R.Y., Yin L.C. Photoresponsive drug/gene delivery systems. Biomacromolecules. 2018;19:1840–1857. doi: 10.1021/acs.biomac.8b00422. [DOI] [PubMed] [Google Scholar]
- 14.Ruan S.B., Erwin N., He M. Light-induced high-efficient cellular production of immune functional extracellular vesicles. J Extracell Vesicles. 2022;11 doi: 10.1002/jev2.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G.J., Jaskula-Sztul R., Esquibel C.R., Lou I., Zheng Q.F., Dammalapati A., et al. Neuroendocrine tumor-targeted upconversion nanoparticle-based micelles for simultaneous NIR-controlled combination chemotherapy and photodynamic therapy, and fluorescence imaging. Adv Funct Mater. 2017;27 doi: 10.1002/adfm.201604671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan B., Boyer J.C., Branda N.R., Zhao Y. Near-infrared light-triggered dissociation of block copolymer micelles using upconverting nanoparticles. J Am Chem Soc. 2011;133:19714–19717. doi: 10.1021/ja209793b. [DOI] [PubMed] [Google Scholar]
- 17.Thangudu S., Kaur N., Korupalli C., Sharma V., Kalluru P., Vankayala R. Recent advances in near infrared light responsive multi-functional nanostructures for phototheranostic applications. Biomater Sci-uk. 2021;9:5432–5443. doi: 10.1039/d1bm00631b. [DOI] [PubMed] [Google Scholar]
- 18.Chu H.Q., Zhao J., Mi Y.S., Di Z.H., Li L.L. NIR-light-mediated spatially selective triggering of anti-tumor immunity via upconversion nanoparticle-based immunodevices. Nat Commun. 2019;10:2839. doi: 10.1038/s41467-019-10847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S., Butt H.J. Near-infrared-sensitive materials based on upconverting nanoparticles. Adv Mater. 2016;28:1208–1226. doi: 10.1002/adma.201502843. [DOI] [PubMed] [Google Scholar]
- 20.Kloosterman D.J., Akkari L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell. 2023;186:1627–1651. doi: 10.1016/j.cell.2023.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., Liang S., Jiang D.D., Gao T., Fang Y.X., Fu S.L., et al. Manipulation of TAMs functions to facilitate the immune therapy effects of immune checkpoint antibodies. J Control Release. 2021;336:621–634. doi: 10.1016/j.jconrel.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Tang W.W., Chen Z.Y., Zhang W.L., Cheng Y., Zhang B., Wu F., et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5:87. doi: 10.1038/s41392-020-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda T., Tsubaki M., Kato N., Genno S., Ichimura E., Enomoto A., et al. Sorafenib treatment of metastatic melanoma with c-Kit aberration reduces tumor growth and promotes survival. Oncol Lett. 2021;22:827. doi: 10.3892/ol.2021.13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.