Abstract
G protein‐coupled receptors (GPCRs) are vital cell surface receptors that govern a myriad of physiological functions. Despite their crucial role in regulating antitumor immunity and tumorigenesis, therapeutic applications targeting GPCRs in oncology are currently limited. This review offers a focused examination of selected protumorigenic chemokine and metabolite‐sensing GPCRs. Specifically, the review highlights five GPCRs able to orchestrate tumor immunobiology at three main levels: tumor immunity, cancer cell expansion, and blood vessel development. The review culminates by illuminating emerging therapies and discussing innovative strategies to harness the full potential of GPCR‐targeted treatments, by applying a multireceptor and patient‐specific logic.
Keywords: Angiogenesis, Cancer, GPCRs, Immunity, Treatment
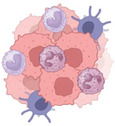
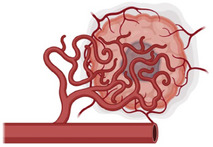
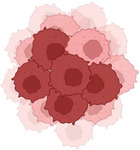
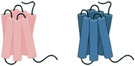
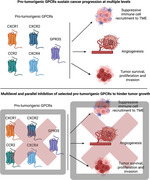
(Top). Representative protumorigenic GPCRs can regulate cancer at multiple levels, including immune cell recruitment to TME, angiogenesis, and tumor biology. (Bottom). Multiple and parallel targeting of selected protumorigenic GPCRs might overcome receptor redundancy and impede tumor growth on various fronts, thus increasing the therapeutic potential of GPCR‐targeted therapies.
Introduction
G protein‐coupled receptors (GPCRs) represent approximately 4% of the human genome and stand as the largest family of cell‐surface receptors [1, 2]. GPCRs are instrumental in orchestrating a plethora of physiological processes, ranging from cell migration, cell survival, and proliferation [1, 2]. Their dysregulation is linked to a spectrum of human diseases, including cancer [3]. GPCRs can control many aspects of cancer immunity and tumorigenesis, including immune cell recruitment, tumor‐cell proliferation, cancer invasion, and angiogenesis [4, 5, 6]. The complex interplay of GPCR–ligand interactions, characterized by their multifunctional redundancy and occasionally contradictory roles in tumor biology, has complicated our full understanding of its various dimensions and importance for cancer patients [5]. Indeed, despite their ability to regulate such a broad range of key functions in tumors, there are few approved compounds targeting GPCRs in the context of cancer [7]. This review provides a concise exploration of the biology of chemokine and metabolite sensing GPCRs, focusing on five protumorigenic receptors with a parallel influence on three main aspects of tumor immunobiology: immune cell recruitment; angiogenesis; cancer cell proliferation, survival, and migration. While other GPCRs can individually regulate additional aspects of tumor progression, such as resistance to cell death, genome instability, and mutations, these concepts have been extensively discussed [3, 5, 7–13] and will not be the focus of this review. Finally, we discuss recent findings that highlight the role of specific GPCRs as promising targets in anticancer immunotherapy and examine intriguing new potential approaches to boost the efficacy of GPCR‐targeted treatments.
Basics of GPCR biology
In the realm of cellular communication, GPCRs play a crucial role in bridging external signals to internal cellular responses. Each GPCR exhibits a distinctive molecular architecture, characterized by a seven‐transmembrane domain, flanked by an extracellular amino terminus and an intracellular carboxyl terminus [2]. GPCRs show versatility in interacting with a wide array of ligands, including chemokines, lipids, and metabolites. These interactions trigger conformational changes that lead to receptor activation. Upon ligand engagement, GPCRs reveal intracellular domains that facilitate coupling with the G‐protein heterotrimer composed of α, β, and γ subunits. This interaction prompts the exchange of GDP for GTP on the Gα subunit, provoking the separation of Gα from the Gβγ complex [14]. Subsequently, both the Gα‐GTP and Gβγ subunits activate a cascade of downstream effectors, propagating the signal elicited by the initial agonist binding. GPCRs can activate multiple Gα proteins, which fall into four main families: Gαs, Gαi, Gαq, and Gα12/13 [13]. These proteins initiate various signaling pathways, influencing processes such as cAMP production, phospholipase, and phosphodiesterase activation, and intracellular calcium levels [13] (Fig. 1). Gβγ dimers also play a significant role, regulating ion channels and activating enzymes like phospholipase C and PI3Ks (Fig. 1). In addition, β‐arrestins are key regulators of the receptor recycling upon activation and participate to intracellular signaling by fine‐tuning dynamic receptor responses and by engaging a scaffolding activity to activate ERK and JNK3 [13] (Fig. 1). Altogether, these signaling networks ultimately exert a profound influence on gene transcription and regulate cell migration, survival, and proliferation to maintain homeostasis and modulate antipathogen or antitumor responses [5, 15].
Figure 1.
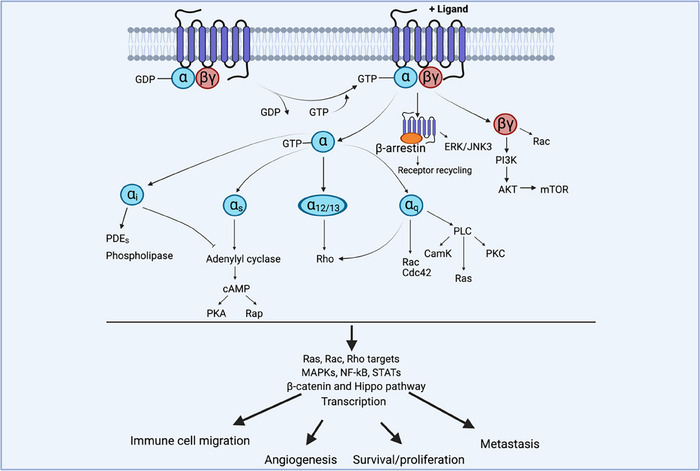
Basics of GPCR signaling. When an agonist binds to its cognate GPCR, it induces a conformational change that activates the receptor and enables its coupling with G proteins. This activation triggers the exchange of GDP for GTP on the Gα subunit, leading to its dissociation from the Gβγ dimers. Both components remain membrane‐bound but are now free to interact with downstream signaling proteins. GPCRs activate various Gα proteins (Gαs, Gαi, Gαq, Gα12/13), initiating signaling pathways that influence cAMP production, phospholipase activation, and calcium levels. Downstream effectors include second messenger systems, GEFs, and Rho/Ras GTPases, which activate signaling cascades that regulate key cellular functions through MAPK, AKT, mTOR, and other kinases and phosphatases to influence transcription, cell migration, cell proliferation, and survival.
CXCR1 and CXCR2
Immune cells
CXCR1 and CXCR2 are instrumental for the recruitment of myeloid cells to the tumor microenvironment (TME) [5]. These Gαi‐coupled receptors are expressed mainly in neutrophils and monocytes/macrophages [10, 16]. The chemokine ligands, CXCL8 for CXCR1 and CXCL1‐8 for CXCR2, can be produced by both cancer cells and other cells within the TME, including stromal cells and immune cells [17]. These chemokines create a gradient that directs the extravasation and migration of immune cells toward the tumor parenchyma. Nevertheless, this receptor‐ligand axis usually promotes the infiltration of protumorigenic neutrophils and myeloid‐derived suppressor cells (MDSCs), which contribute to immunosuppression and promote tumor growth by several distinct mechanisms, including the production of immune suppressive cytokines and proangiogenic factors [5, 18, 19, 20].
Angiogenesis
Within the repertoire of chemokine receptors, CXCR2 stands out as notably associated with angiogenesis [4]. CXCR2 activation triggers a characteristic proangiogenic cascade involving phosphorylation of ERK and PI3K (Fig. 1). Considering these notions, it is not surprising that CXCR2‐expressing endothelial cells significantly contribute to tumor angiogenesis [21, 22, 23]. Thus, CXCR2 ligands are potent angiogenic factors, and their signaling is crucial for the migration, proliferation, and tube formation of endothelial cells — all essential steps in the expansion of blood vessels within tumors [22, 24, 25]. In addition, CXCR2‐expressing tumor‐associated neutrophils can promote angiogenesis by the production of several factors, including metalloproteases (MMP9) and secreted proteins (Bv8) [25, 26, 27, 28].
Tumor cells
Beyond their roles in immune cell recruitment and angiogenesis, CXCR1 and CXCR2 can be expressed on tumor cells, including melanoma [29], ovarian cancer [23], esophageal squamous cell carcinoma [30], breast cancer [31], and pancreatic ductal adenocarcinoma (PDAC) [30, 32]. In this context, the activation of these receptors promotes oncogenic processes, such as tumor proliferation, migration, and metastasis [29, 31]. This can occur through the modulation of downstream signaling pathways like PI3K/Akt, MAPK, NF‐κB, and STAT3 (Fig. 1) in response to tumor‐associated macrophages (TAM)‐derived ligands [32, 33, 34]. Specifically, the phosphorylation of Akt and ERK1/2, the activation of STAT3, and the modulation of the NF‐kB pathway through SOX4 binding cooperate to boost cancer proliferation, survival, and invasion.
Thus, CXCR1 and CXCR2 promote tumor growth and proliferation by supporting the recruitment of immunosuppressive myeloid cells, by directly and indirectly promoting vessel expansion, and by regulating cancer cell survival and proliferation (Table 1). In accordance with this view, their expression and activation often correlate with increased angiogenesis, tumor growth, and a generally poor prognosis [23, 35, 36, 37]. While antibody neutralization of CXCL8 has shown therapeutic potential in a breast cancer xenograft model [38], this approach has still not induced a clear benefit in human cancer patients, probably due to ligand redundancy [5].
Table 1.
Comprehensive reference table detailing the impact of CXCR1/2, CCR2, CXCR4, and GPR35 on tumor immunobiology across immune cell recruitment, angiogenesis, and cancer cell dynamics within the TME.
| GPCRs |
Immune cell recruitment to TME
|
Angiogenesis
|
Cancer cell proliferation, survival, invasion
|
|
CXCR1 CXCR2
|
Acharyya et al. [20] Li et al. [18] Teijeira et al. [19] |
Keane et al. [22] Yang et al. [21] Ijichi et al. [23] |
Maeda et al. [32] Miyamoto et al. [35] Wang et al. [34] |
|
CCR2
|
Qian et al. [42] Li et al. [40] Nywening et al. [39] |
Salcedo et al. [49] Izhak et al. [48] Bartneck et al. [47] Pausch et al. [44] |
Fang et al. [53] Macanas‐Pirard et al. [52] Fein et al. [54] |
|
CXCR4
|
Arwert et al. [57] Chiodoni et al. [60] Steele et al. [58] |
Orimo et al. [66] Xu et al. [62] Du et al. [63] |
Muller et al. [68] Murakami et al. [71] Darash‐Yahana et al. [67] |
|
GPR35
|
Shu et al. [99] Yue et al. [92] |
McCallum et al. [101] Pagano et al. [98] Li et al. [100] |
Wang et al. [104] Schneditz et al. [105] |
Abbreviation: TME, tumor microenvironment.
CCR2
Immune cells
CCR2 preferentially couples to Gαi proteins (Fig. 1) and plays a pivotal role in the recruitment and trafficking of myeloid cells to the TME. It is primarily known for its interaction with its chemokine ligands, notably CCL2 and CCL7. CCL2 is produced by tumor cells, stromal cells, and infiltrating immune cells to boost CCR2‐expressing cell recruitment to the tumor site. Indeed, CCR2‐expressing myeloid cells, such as monocytes, MDSCs, and TAMs [39, 40, 41, 42], usually favor tumor progression by suppressing antitumor immune responses and by modulating angiogenesis. Nevertheless, despite CCL7 being a ligand for CCR2 — where its augmented levels would typically be presumed to boost suppressive monocyte recruitment — researchers have discovered a paradoxical effect. Indeed, the restoration of CCL7 expression has been shown to increase T‐cell infiltration and boost the recruitment of antitumor myeloid cells [43]. These data suggest that the outcome of CCR2‐ligand regulation may hinge on the subsequent differentiation of monocytes into either tumor‐promoting or inflammatory cells and that CCR2 regulation might be ligand‐dependent.
Angiogenesis
CCR2 and its ligands are involved in tumor angiogenesis in different types of cancers, including renal cell carcinoma, PDAC, and hepatocellular carcinoma (HCC) [44, 45, 46, 47]. Indeed, CCR2‐driven recruitment of myeloid cells contributes to vessel expansion, as these cells produce factors that support the growth of blood vessels within the tumor [44, 45, 46, 47, 48]. In addition, tumor‐associated endothelial cells express CCR2 and respond to CCL2 to support angiogenesis [49], highlighting both a direct and indirect role for this receptor in regulating the tumor vasculature.
Tumor cells
Cancer cells exhibit a high prevalence of CCR2 expression. For instance, in‐depth analysis of osteosarcoma cases has confirmed widespread CCR2 expression across the patients analyzed [50]. In renal cell carcinoma, around half of metastatic tumors express CCR2 [51]. Acute myeloid leukemia patients also show CCR2‐expressing cancer cells [52]. CCR2 expression in tumor cells can boost metastatization and invasion and promote tumor survival [52, 53]. For example, CCL2 can promote breast cancer cell survival by activating MAPK signaling (Fig. 1) and the SMAD pathway to enhance cancer cell invasiveness [53]. Furthermore, CCR2‐deficient breast cancer cells are also more sensitive to T‐cell‐mediated killing in vivo via CD103+ cross‐presenting dendritic cells [54].
In summary, CCR2 expression shapes TME by influencing the recruitment of myeloid cells, by directly and indirectly regulating tumor angiogenesis, and by promoting cancer cell proliferation and invasion (Table 1). Thus, CCR2 shows an overall tendency to promote tumor progression in different settings and at multiple levels. In accordance with this view, inhibition of CCR2 or its ligand CCL2 has been proposed as a therapeutic strategy in the context of HCC [40], prostate cancer [55], and breast cancer [42].
CXCR4
Immune cells
CXCR4 is a key GPCR that regulates immune cell recruitment to the TME. Upon activation, CXCR4 can couple to both Gαi and Gα12/13 proteins, with the latter also required for the induction of cell migration [56] (Fig. 1). The interaction of CXCR4 with its chemokine ligand CXCL12 guides the migration of both myeloid and adaptive immune cells [5]. This recruitment may foster antitumor immunity, but it can also sustain tumor progression by supporting the recruitment of protumorigenic cells or by inhibiting inflammatory immune cell retention within the TME. For instance, CXCL12 produced by perivascular fibroblasts attracts monocyte‐derived TAMs, which express CXCR4 upon exposure to cancer‐cell‐derived TGFβ [57]. These TAMs then facilitate cancer cell intravasation and metastasis formation in a murine breast cancer model [57]. More recently, CXCL12 expression by tumor‐associated lymphatic endothelial cells was shown to be key in guiding the egress of tumor‐infiltrating T cells, and especially of TCF1+ T cells to draining lymph nodes [58]. Intriguingly, tumors can also remotely regulate CXCL12 distribution in the bone marrow to boost the egress of MDSCs that accumulate within the TME and suppress anticancer immunity [59, 60].
Angiogenesis
CXCR4 and its ligand CXCL12 synergize with other GPCR–ligand axes to promote the recruitment of myeloid cells able to support new vessel formation [17]. In addition, this axis may directly regulate angiogenesis. In line with this view, previous work found a higher expression of CXCR4 in endothelial cells within HCC specimens compared with healthy liver tissue [61, 62]. In this context, CXCR4 expression by tumor endothelial cells can promote vessel sprouting and support tumor growth, representing a promising therapeutic target for combination therapies [62, 63]. Finally, cancer‐associated fibroblasts, which contribute to tumor angiogenesis, show increased CXCR4 expression [64, 65, 66].
Tumor cells
CXCR4 is upregulated in a wide array of malignancies, including but not limited to kidney, lung, brain, prostate, breast, pancreatic, ovarian, and skin cancers. This overexpression plays a role in promoting tumor proliferation, metastasis, and resistance to treatment [9, 67, 68, 69, 70]. Research utilizing mouse models demonstrated that CXCR4 is instrumental in directing cancer cells toward CXCL12‐abundant tissues like the lungs, liver, and bone marrow [68, 71]. Interestingly, hypoxia — a common condition within the TME — dramatically increases CXCR4 levels by stabilizing hypoxia‐inducible factor 1 subunit alpha, suggesting a role for CXCR4 in fostering metastatic colonization [72].
Overall, CXCR4 regulates immune cell trafficking to tumors, it can directly and indirectly influence angiogenesis and support the proliferation and invasion of cancer cells (Table 1). Accordingly, CXCR4 inhibition is predicted to have preferentially beneficial effects on tumor clearance. This conclusion is supported by data from a preclinical mouse model of HCC [73] and a human PDAC tumor explant model [74] in which the CXCR4 inhibitor AMD3100 synergized with anti‐PD1 therapy, as well as encouraging results observed for the CXCR4 inhibitor BL‐8040 in combination with anti‐PD1 in patients with PDAC [75].
GPR35
Immune cells
GPR35, a GPCR sensitive to metabolites, plays multiple roles in both immune and non‐immune cells, including the regulation of myeloid cell migration [76, 77]. GPR35 shows a context‐dependent G‐protein coupling ability, with both Gαi and Gα12/13 contributing to receptor intracellular signaling [78, 79, 80, 81, 82] (Fig. 1). GPR35 can be activated by endogenous tryptophan derivatives like kynurenic acid [83] and the serotonin metabolite 5‐hydroxyindole‐acetic acid [76, 84, 85, 86], as well as lysophosphatidic acid [87]. Recent studies have highlighted GPR35's role in myeloid cell recruitment across various inflammatory contexts [84, 85, 86, 87, 88, 89, 90, 91, 92]. Specifically, our recent work has shown how GPR35, stimulated by platelet and mast cell‐derived 5‐hydroxyindole‐acetic acid, can influence granulocyte migration to tissues under inflammatory conditions [84, 85]. Intriguingly, it was observed a correlation between high GPR35 expression and poorer prognosis across various cancer types [93, 94]. In addition, tumor‐associated myeloid cells express GPR35 across a spectrum of cancers [95, 96, 97, 98]. While emerging preliminary findings imply that GPR35 could influence immune cell accumulation and function in the TME [92, 99] (Table 1), the precise mechanisms by which this receptor contributes to cell migration and recruitment to tumors, as well as the specific ligands involved, are yet to be fully elucidated.
Angiogenesis
GPR35 can be expressed by endothelial cells and plays a role in their regulatory processes [100, 101, 102]. The impact of endothelial GPR35 expression in tumor angiogenesis, however, has not yet been fully determined. Recent studies have shed light on the significance of GPR35 expression in TAMs and its importance in the regulation of angiogenesis in colorectal cancer models [98]. These findings suggest that GPR35 may indirectly influence the formation of new blood vessels within tumors. Significantly, GPR35 exhibits marked upregulation during hypoxia, induced by hypoxia‐inducible factor 1 subunit alpha, which directly interacts with a hypoxia‐responsive element located within the GPR35 promoter region [103]. Consequently, GPR35 becomes integral to the transcriptional machinery governing adaptation to hypoxic conditions, known to be one of the most potent proangiogenic stimuli. This underscores the potential significance of GPR35 in promoting sprouting neo‐angiogenesis, particularly within hypoxic environments such as solid tumors. Therefore, the exploration of how GPR35 modulates angiogenesis in cancer through both direct and indirect pathways is an interesting field of ongoing research.
Tumor cells
GPR35 is upregulated in a wide range of primary tumors and cell lines, including colorectal cancer [93], gastric cancer [99], lung cancer [104], and PDAC [93]. This wide expression across several cancer cells suggests a key role for GPR35 in sustaining tumor growth and survival. In line with this view, GPR35 expression can promote glycolysis, proliferation, and oncogenic signaling in cancer cells by a ligand‐independent mechanism [105] and confer drug resistance [104].
This set of preliminary studies suggests an overall protumorigenic role for GPR35 expression through multiple mechanisms. Of note, inhibitors of the human GPR35 receptors are available [106], whether these drugs are useful in this context is unknown (Table 1). Tryptophan metabolism is often dysregulated in cancer [107], with increased levels of kynurenic acid [108] and serotonin [109] within the TME. In addition, platelets [110, 111] and mast cells [112, 113] may display an activated state in the TME, potentially representing an important source of GPR35‐activating metabolites. These observations shed light on the possibility of targeting tryptophan and serotonin metabolisms to dampen GPR35‐dependent tumorigenesis. In agreement with this view, recent work has provided evidence for the role of peripheral serotonin inhibition in the regulation of response to immunotherapy [114]. The role of other metabolite‐sensing GPCRs that do not encompass this multilevel modulation of cancer immunobiology, including GPR81, GPR40, LPARs, and others, is extensively reviewed elsewhere [115, 116, 117, 118] and will not be commented on in this review.
The power of many: could parallel targeting of specific GPCRs boost anticancer treatments?
GPCRs are emerging as potential targets in cancer treatment [115, 119]. Despite their role in regulating a wide array of critical functions in tumors, the therapeutic landscape for targeting GPCRs in cancer remains elusive. This is partially due to the intricate dynamics of GPCR–ligand interactions, with sometimes paradoxical roles in tumor biology. In some instances, however, clear therapeutic benefit was not achieved even when targeting a single GPCR–ligand axis with a clear tendency to promote tumor progression at multiple levels, such as in the case of the neutralizing human CCL2 antibody carlumab [5]. For instance, in the context of CCL2‐CCR2 neutralization, other GPCRs like CCR1, CXCR1, and CXCR2 may support suppressive myeloid cell recruitment to tumors [5, 16]. To address these challenges, we believe that a promising strategy may involve the simultaneous inhibition of multiple protumorigenic GPCR/ligand pathways within tumors. A collective inhibition of protumorigenic receptors might enhance antitumor immunity and impede tumor growth on various fronts. The concomitant neutralization of both receptors and cognate ligands should also be considered to avoid functional redundancy. Importantly, personalized medicine strategies should guide the receptor list selection for patient‐specific therapeutic targeting, leveraging advanced technologies such as spatial transcriptomics, spectral flow cytometry, and scRNA‐seq. These innovative tools enable the collection of vast biological data from tumor biopsies, facilitating tailored decisions on which receptor–ligand axes are more likely to drive tumor progression in each patient (Fig. 2).
Figure 2.
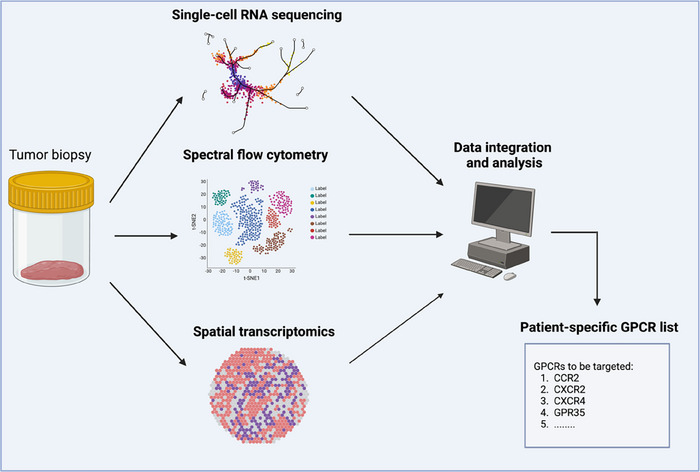
Personalized medicine approaches to develop tailored GPCR anticancer treatment. The figure represents a proposed personalized medicine pipeline to perform patient‐specific GPCR cancer treatments. Tumor biopsies are analyzed with cutting‐edge techniques such as scRNA‐seq, spectral flow cytometry, and spatial transcriptomics. Data are then integrated and analyzed using dedicated bioinformatic pipelines to compile a protumorigenic GPCR list to be targeted in each specific patient.
Adoptive cellular therapy involves the transfer of immune cells, mostly (CAR‐)T cells, that have been primed or engineered to respond to a tumor antigen. While these approaches have revolutionized cancer treatment, they provide few objective benefits for the treatment of solid tumors, mostly due to a lack of proper recruitment and intratumor positioning of the transferred cells [5]. To overcome these limitations, an innovative GPCR‐based method is emerging in the context of cellular immunotherapies. The overexpression of selective GPCRs in CAR‐T cells leads to increased cell recruitment within the tumor and increased capability to kill cancer cells. For instance, CCR2‐overexpressing CAR‐T cells are recruited more efficiently into the TME in preclinical mouse models [120]. Similarly, overexpression of other GPCRs can increase transferred cell recruitment to different solid tumors [5]. Nevertheless, to further enhance the effectiveness of adoptive cell transfer therapies, we believe it is necessary to move beyond the strategy of overexpressing a single GPCR. Indeed, simultaneous modulation of several receptors may substantially improve the homing of adoptively transferred cells by harnessing the full spectrum of receptor–ligand interactions critical to this process (Fig. 3A and B). Thus, it is plausible that endowing transferred cells with a palette of GPCRs akin to those found on cells proficiently recruited to tumor sites — such as MDSCs and TAMs — could significantly augment their infiltration and retention into the TME. These engineered cells may be named “Trojan Horse CAR‐T cells”, mimicking the GPCR profile of suppressive cells whose recruitment is usually favored by tumors (Fig. 3B). This way, it could be possible to “deceive” the tumor into facilitating the infiltration of therapeutic T cells and use the tumor's own tactics against it to enhance treatment efficacy. This approach may also offer a secondary benefit: indeed, to resist such treatment, the tumor would be forced to suppress essential pathways it relies on for recruiting suppressive myeloid cells, thus potentially cornering the tumor into a self‐defeating position.
Figure 3.
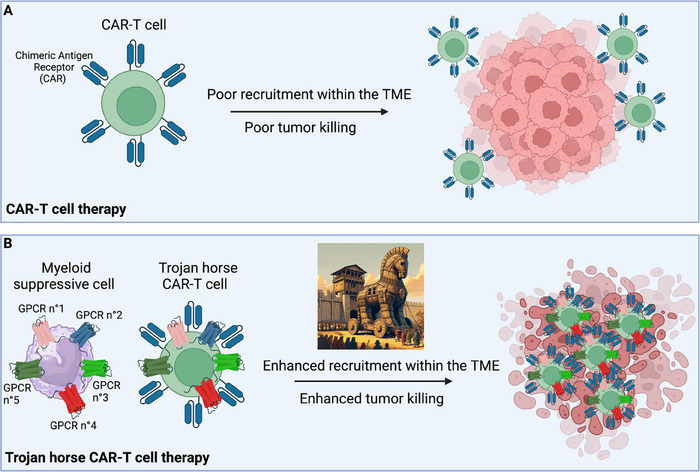
Enhancing CAR‐T cell recruitment to the TME: the Trojan horse trick. The figure depicts a potential approach to boost the efficacy of CAR‐T cell treatment for solid tumors. (A) CAR‐T are usually poorly recruited to the TME, which results in low tumor‐killing efficiency. (B) “Trojan horse” CAR‐T cells display a palette of GPCRs that mirrors those found in cells proficiently recruited to the TME, such as suppressive monocytes. This approach may potentiate the recruitment of the transferred CAR‐T cells, resulting in increased tumor‐killing efficiency. TME, tumor microenvironment.
Conclusion
Our understanding of how GPCR regulates cancer progression at multiple levels is rapidly advancing. This highlighted the complexity and redundancy of GPCR–ligand interactions in tumors, often hindering the success of therapies targeting single receptor–ligand axes.
To overcome these challenges, in this review, we discuss the role of five representative GPCRs in modulating cancer immunobiology at multiple levels and propose a novel therapeutic strategy based on the parallel targeting of key protumorigenic receptor/ligand axes. We also discuss how personalized medicine approaches could guide the selection of receptor–ligand targets implicated in tumor progression for each patient. Finally, we comment on the possibility of enhancing the efficacy of adoptive cellular therapies by overexpressing multiple GPCRs to improve engineered T‐cell recruitment to solid tumors. To this end, we propose a novel strategy named the “Trojan Horse trick”, which involves engineering therapeutic cells to mimic the GPCR profiles of myeloid cells that are efficiently recruited to the TME. In conclusion, we believe that parallel targeting of key protumorigenic GPCRs coupled with personalized strategies has the potential to significantly advance cancer treatments, offering more effective and tailored therapeutic options for cancer patients.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Author contributions
Marco De Giovanni conceptualized the study, prepared the figures, and wrote the manuscript. Donato Inverso, Carlotta Tacconi, and Serena Ranucci provided insightful inputs and revised the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202350870.
Abbreviations
- HCC
hepatocellular carcinoma
- MDSC
myeloid‐derived suppressor cells
- PDAC
pancreatic ductal adenocarcinoma
- TAM
tumor‐associated macrophages
- TME
tumor microenvironment
Acknowledgements
This work was supported by the Giovanni Armenise Harvard Foundation Career Development Award (to M.D.G.), the Italian Association for Cancer Research (AIRC) Start‐Up Grant 27564 (to M.D.G.), the ERC Starting Grant 101116224 (to M.D.G), the AIRC Start‐Up Grant 26183 (to D.I.), and Cariplo Grant 2021‐1542 (to C.T.). The authors thank all the Iannacone and Guidotti lab members (San Raffaele Scientific Institute, Milan, Italy) for helpful discussions. Figures were created with BioRender.com.
Open Access Funding provided by BIBLIOSAN.
Data availability statement
Data sharing is not applicable as no new data were generated.
References
- 1. Lu, E. and Cyster, J. G. , G‐protein coupled receptors and ligands that organize humoral immune responses. Immunol. Rev. 2019. 10.1111/imr.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosenbaum, D. M. , Rasmussen, S. G. F. and Kobilka, B. K. , The structure and function of G‐protein‐coupled receptors. Nature 2009. 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dorsam, R. T. and Gutkind, J. S. , G‐protein‐coupled receptors and cancer. Nat. Rev. Cancer 2007. 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 4. Mehrad, B. , Keane, M. P. and Strieter, R. M. , Chemokines as mediators of angiogenesis. 2007. 97: 755–762. [PMC free article] [PubMed] [Google Scholar]
- 5. Mempel, T. R. , Lill, J. K. and Altenburger, L. M. , How chemokines organize the tumour microenvironment. Nat. Rev. Cancer 2024. 10.1038/s41568-023-00635-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balkwill, F. , Cancer and the chemokine network. Nat. Rev. Cancer 2004. 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 7. Bar‐Shavit, R. , Maoz, M. , Kancharla, A. , Nag, J. K. , Agranovich, D. , Grisaru‐Granovsky, S. and Uziely, B. , G protein‐coupled receptors in cancer. Int. J. Mole. Sci. 2016. 10.3390/ijms17081320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu, Y. , An, S. , Ward, R. , Yang, Y. , Guo, X. X. , Li, W. and Xu, T. R. , G protein‐coupled receptors as promising cancer targets. Cancer Lett. 2016. 10.1016/j.canlet.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 9. Balkwill, F. , Cancer and the chemokine network. Nat. Rev. Cancer 2004. 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 10. Olson, T. S. and Ley, K. , invited review Chemokines and chemokine receptors in leukocyte trafficking. 2002. 10.1152/ajpregu.00738.2001.-Che. [DOI] [PubMed]
- 11. Arang, N. and Gutkind, J. S. , G protein‐coupled receptors and heterotrimeric G proteins as cancer drivers. Preprint at Wiley Blackwell 2020. 10.1002/1873-3468.14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dorsam, R. T. and Gutkind, J. S. , G‐protein‐coupled receptors and cancer. FEBS Lett. 2007. 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 13. Wu, V. , Yeerna, H. , Nohata, N. , Chiou, J. , Harismendy, O. , Raimondi, F. , Inoue, A. et al., Illuminating the onco‐GPCRome: Novel G protein‐coupled receptor‐driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019. 294: 11062–11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neves, S. R. , Ram, P. T. and Iyengar, R. , G protein pathways. Science 2002. 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 15. De Giovanni, M. and Iannacone, M. , In vivo imaging of adaptive immune responses to viruses. Curr. Opin. Virol. 2018. 10.1016/j.coviro.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geissmann, F. , Jung, S. and Littman, D. R. , Blood monocytes consist of two principal subsets with distinct migratory properties cytes. Immunity 2003. 10.1016/s1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- 17. Chao, T. , Furth, E. E. and Vonderheide, R. H. , CXCR2‐dependent accumulation of tumor‐associated neutrophils regulates T‐cell immunity in pancreatic ductal adenocarcinoma. Cancer Immunol. Res. 2016. 4: 968–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li, J. , Byrne, K. T. , Yan, F. , Yamazoe, T. , Chen, Z. , Baslan, T. , Richman, L. P. et al., Tumor cell‐intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 2018. 49. 10.1016/j.immuni.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teijeira, Á. , Garasa, S. , Gato, M. , Alfaro, C. , Migueliz, I. , Cirella, A. , de Andrea, C. et al., CXCR1 and CXCR2 chemokine receptor agonists produced by tumors induce neutrophil extracellular traps that interfere with immune cytotoxicity. Immunity 2020. 52. 10.1016/j.immuni.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 20. Acharyya, S. , Oskarsson, T. , Vanharanta, S. , Malladi, S. , Kim, J. , Morris, P. G. , Manova‐Todorova, K. et al., A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012. 150. 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang, G. , Rosen, D. G. , Liu, G. , Yang, F. , Guo, X. , Xiao, X. , Xue, F. et al., CXCR2 promotes ovarian cancer growth through dysregulated cell cycle, diminished apoptosis, and enhanced angiogenesis. Clin. Cancer Res. 2010. 16: 3875–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keane, M. P. , Belperio, J. A. , Xue, Y. Y. , Burdick, M. D. and Strieter, R. M. , Depletion of CXCR2 inhibits tumor growth and angiogenesis in a murine model of lung cancer. J. Immunol. 2004. 172: 2853–2860. [DOI] [PubMed] [Google Scholar]
- 23. Ijichi, H. , Chytil, A. , Gorska, A. E. , Aakre, M. E. , Bierie, B. , Tada, M. , Mohri, D. et al., Inhibiting Cxcr2 disrupts tumor‐stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Invest. 2011. 121: 4106–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang, D. , Wang, H. , Brown, J. , Daikoku, T. , Ning, W. , Shi, Q. , Richmond, A. et al., CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J. Exp. Med. 2006. 203. 10.1084/jem.20052124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith, D. R. , Polverini, P. J. , Kunkel, S. L. , Orringer, M. B. , Whyte, R. I. , Burdick, M. D. , Wilke, C. A. et al., Inhibition of interleukin 8 attenuates angiogenesis in bronchogenic carcinoma. J. Exp. Med. 1994. 179. 10.1084/jem.179.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raccosta, L. , Fontana, R. , Maggioni, D. , Lanterna, C. , Villablanca, E. J. , Paniccia, A. , Musumeci, A. et al., The oxysterol‐cxcr2 axis plays a key role in the recruitment of tumor‐promoting neutrophils. J. Exp. Med. 2013. 210. 10.1084/jem.20130440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh, H. , Wang, D. , Daikoku, T. , Sun, H. , Dey, S. K. and DuBois, R. N. , CXCR2‐expressing myeloid‐derived suppressor cells are essential to promote colitis‐associated tumorigenesis. Cancer Cell 2013. 24. 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nozawa, H. , Chiu, C. and Hanahan, D. , Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc. Natl. Acad. Sci. USA. 2006. 103. 10.1073/pnas.0601807103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dhawan, P. and Richmond, A. , Role of CXCL1 in tumorigenesis of melanoma. [PMC free article] [PubMed]
- 30. Hosono, M. , Koma, Y.‐I. , Takase, N. , Urakawa, N. , Higashino, N. , Suemune, K. , Kodaira, H. et al., CXCL8 derived from tumor‐associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression by promoting migration and invasion of cancer cells. 2017. 8: 106071–106088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mishra, A. , Suman, K. H. , Nair, N. , Majeed, J. and Tripathi, V. , An updated review on the role of the CXCL8‐CXCR1/2 axis in the progression and metastasis of breast cancer. Preprint at Springer Science and Business Media B.V. 2021. 10.1007/s11033-021-06648-8. [DOI] [PubMed] [Google Scholar]
- 32. Maeda, S. , Kuboki, S. , Nojima, H. , Shimizu, H. , Yoshitomi, H. , Furukawa, K. , Miyazaki, M. et al., Duffy antigen receptor for chemokines (DARC) expressing in cancer cells inhibits tumor progression by suppressing CXCR2 signaling in human pancreatic ductal adenocarcinoma. Cytokine 2017. 95. 10.1016/j.cyto.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 33. Meng, Z. W. , Zhang, L. , Cai, X. R. , Wang, X. , She, F. F. and Chen, Y. L. , IL‐8 is a novel prometastatic chemokine in intrahepatic cholangiocarcinoma that induces CXCR2‐PI3K/AKT signaling upon CD97 activation. Sci. Rep. 2024. 14: 8478 10.1038/s41598-024-58952-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang, N. , Liu, W. , Zheng, Y. , Wang, S. , Yang, B. , Li, M. , Song, J. et al., CXCL1 derived from tumor‐associated macrophages promotes breast cancer metastasis via activating NF‐κB/SOX4 signaling. Cell Death. Dis. 2018. 9. 10.1038/s41419-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyamoto, M. , Shimizu, Y. , Okada, K. , Kashii, Y. , Higuchi, K. and Watanabe, A. , Effect of interleukin‐8 on production of tumor‐associated substances and autocrine growth of human liver and pancreatic cancer cells. Cancer Immunol. Immunother. 1998. 47: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaffer, T. and Ma, D. , The emerging role of chemokine receptor CXCR2 in cancer progression. AME Publishing Company, 2016. 10.21037/tcr.2016.10.06. [DOI] [Google Scholar]
- 37. Nannuru, K. , Sharma, B. , Varney, M. and Singh, R. , Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis. J. Carcinog. 2011. 10. 10.4103/1477-3163.92308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dominguez, C. , McCampbell, K. K. , David, J. M. and Palena, C. , Neutralization of IL‐8 decreases tumor PMN‐MDSCs and reduces mesenchymalization of claudin‐low triple‐negative breast cancer. JCI Insight 2017. 2. 10.1172/jci.insight.94296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nywening, T. M. , Belt, B. A. , Cullinan, D. R. , Panni, R. Z. , Han, B. J. , Sanford, D. E. , Jacobs, R. C. et al., Targeting both tumour‐associated CXCR2+ neutrophils and CCR2+ macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut 2018. 67. 10.1136/gutjnl-2017-313738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li, X. , Yao, W. , Yuan, Y. , Chen, P. , Li, B. , Li, J. , Chu, R. et al., Targeting of tumour‐infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 2017. 66. 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 41. Mitchem, J. B. , Brennan, D. J. , Knolhoff, B. L. , Belt, B. A. , Zhu, Y. , Sanford, D. E. , Belaygorod, L. et al., Targeting tumor‐infiltrating macrophages decreases tumor‐initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013. 73. 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qian, B. Z. , Li, J. , Zhang, H. , Kitamura, T. , Zhang, J. , Campion, L. R. , Kaiser, E. A. et al., CCL2 recruits inflammatory monocytes to facilitate breast‐tumour metastasis. Nature 2011. 475. 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng, Q. , Saghafinia, S. , Chryplewicz, A. , Fournier, N. , Christe, L. , Xie, Y. Q. , Guillot, J. et al., Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science 2022. 378. 10.1126/science.abl7207. [DOI] [PubMed] [Google Scholar]
- 44. Pausch, T. M. , Aue, E. , Wirsik, N. M. , Freire Valls, A. , Shen, Y. , Radhakrishnan, P. , Hackert, T. et al., Metastasis‐associated fibroblasts promote angiogenesis in metastasized pancreatic cancer via the CXCL8 and the CCL2 axes. Sci. Rep. 2020. 10. 10.1038/s41598-020-62416-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deshmane, S. L. , Kremlev, S. , Amini, S. and Sawaya, B. E. , Monocyte chemoattractant protein‐1 (MCP‐1): An overview. Preprint, 2009. 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed]
- 46. Arakaki, R. , Yamasaki, T. , Kanno, T. , Shibasaki, N. , Sakamoto, H. , Utsunomiya, N. , Sumiyoshi, T. et al., CCL2 as a potential therapeutic target for clear cell renal cell carcinoma. Cancer Med. 2016. 5. 10.1002/cam4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bartneck, M. , Schrammen, P. L. , Möckel, D. , Govaere, O. , Liepelt, A. , Krenkel, O. , Ergen, C. et al., The CCR2 + macrophage subset promotes pathogenic angiogenesis for tumor vascularization in fibrotic livers. CMGH 2019. 7. 10.1016/j.jcmgh.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Izhak, L. , Wildbaum, G. , Jung, S. , Stein, A. , Shaked, Y. and Karin, N. , Dissecting the autocrine and paracrine roles of the CCR2‐CCL2 axis in tumor survival and angiogenesis. PLoS One 2012. 7. 10.1371/journal.pone.0028305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salcedo, R. , Ponce, M. L. , Young, H. A. , Wasserman, K. , Ward, J. M. , Kleinman, H. K. , Oppenheim, J. J. et al., Human endothelial cells express CCR2 and respond to MCP‐1: Direct role of MCP‐1 in angiogenesis and tumor progression. Blood 2000. 96. 10.1182/blood.v96.1.34.013a49_34_40. [DOI] [PubMed] [Google Scholar]
- 50. von Luettichau, I. , Segerer, S. , Wechselberger, A. , Notohamiprodjo, M. , Nathrath, M. , Kremer, M. , Henger, A. et al., A complex pattern of chemokine receptor expression is seen in osteosarcoma. BMC Cancer 2008. 8. 10.1186/1471-2407-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wyler, L. , Napoli, C. U. , Ingold, B. , Sulser, T. , Heikenwälder, M. , Schraml, P. and Moch, H. , Brain metastasis in renal cancer patients: Metastatic pattern, tumour‐associated macrophages and chemokine/chemoreceptor expression. Br. J. Cancer 2014. 110. 10.1038/bjc.2013.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Macanas‐Pirard, P. , Quezada, T. , Navarrete, L. , Broekhuizen, R. , Leisewitz, A. , Nervi, B. and Ramírez, P. A. , The CCL2/CCR2 axis affects transmigration and proliferation but not resistance to chemotherapy of acute myeloid leukemia cells. PLoS One 2017. 12. 10.1371/journal.pone.0168888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fang, W. B. , Jokar, I. , Zou, A. , Lambert, D. , Dendukuri, P. and Cheng, N. , CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein‐ and p42/44 mitogen‐activated protein kinase (MAPK)‐dependent mechanisms. J. Biol. Chem. 2012. 287. 10.1074/jbc.M112.365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fein, M. R. , He, X. Y. , Almeida, A. S. , Bruzas, E. , Pommier, A. , Yan, R. , Eberhardt, A. et al., Cancer cell CCR2 orchestrates suppression of the adaptive immune response. J. Exp. Med. 2020. 217. 10.1084/JEM.20181551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loberg, R. D. , Ying, C. , Craig, M. , Day, L. L. , Sargent, E. , Neeley, C. , Wojno, K. et al., Targeting CCL2 with systemic delivery of neutralizing antibodies induces prostate cancer tumor regression in vivo. Cancer Res. 2007. 67. 10.1158/0008-5472.CAN-07-1286. [DOI] [PubMed] [Google Scholar]
- 56. Tan, W. , Martin, D. and Gutkind, J. S. , The Gα13‐Rho signaling axis is required for SDF‐1‐induced migration through CXCR. J. Biol. Chem. 2006. 281: 39542–39549. [DOI] [PubMed] [Google Scholar]
- 57. Arwert, E. N. , Harney, A. S. , Entenberg, D. , Wang, Y. , Sahai, E. , Pollard, J. W. and Condeelis, J. S. , A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep. 2018. 23. 10.1016/j.celrep.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Steele, M. M. , Jaiswal, A. , Delclaux, I. , Dryg, I. D. , Murugan, D. , Femel, J. , Son, S. et al., T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control. Nat. Immunol. 2023. 10.1038/s41590-023-01443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ostrand‐Rosenberg, S. and Fenselau, C. , Myeloid‐derived suppressor cells: immune‐suppressive cells that impair antitumor immunity and are sculpted by their environment. J. Immunol. 2018. 200. 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chiodoni, C. , Cancila, V. , Renzi, T. A. , Perrone, M. , Tomirotti, A. M. , Sangaletti, S. , Botti, L. et al., Transcriptional profiles and stromal changes reveal bone marrow adaptation to early breast cancer in association with deregulated circulating microRNAs a C. Cancer Res. 2020. 80. 10.1158/0008-5472.CAN-19-1425. [DOI] [PubMed] [Google Scholar]
- 61. Li, W. , Gomez, E. and Zhang, Z. , Immunohistochemical expression of stromal cell‐derived factor‐1 (SDF‐1) and CXCR4 ligand receptor system in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2007. 26: 527–533. [PubMed] [Google Scholar]
- 62. Xu, J. , Liang, J. , Meng, Y. M. , Yan, J. , Yu, X. J. , Liu, C. Q. , Xu, L. et al., Vascular CXCR4 expression promotes vessel sprouting and sensitivity to sorafenib treatment in hepatocellular carcinoma. Clin. Cancer Res. 2017. 23. 10.1158/1078-0432.CCR-16-2131. [DOI] [PubMed] [Google Scholar]
- 63. Du, R. , Lu, K. V. , Petritsch, C. , Liu, P. , Ganss, R. , Passegué, E. , Song, H. et al., HIF1α induces the recruitment of bone marrow‐derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell 2008. 13: 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eck, S. M. , Côté, A. L. , Winkelman, W. D. and Brinckerhoff, C. E. , CXCR4 and matrix metalloproteinase‐1 are elevated in breast carcinoma‐associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol. Cancer Res. 2009. 7. 10.1158/1541-7786.MCR-09-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kojima, Y. , Acar, A. , Eaton, E. N. , Mellody, K. T. , Scheel, C. , Ben‐Porath, I. , Onder, T. T. et al., Autocrine TGF‐β and stromal cell‐derived factor‐1 (SDF‐1) signaling drives the evolution of tumor‐promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. USA. 2010. 107. 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Orimo, A. , Gupta, P. B. , Sgroi, D. C. , Arenzana‐Seisdedos, F. , Delaunay, T. , Naeem, R. , Carey, V. J. et al., Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF‐1/CXCL12 secretion. Cell 2005. 121. 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 67. Darash‐Yahana, M. , Pikarsky, E. , Abramovitch, R. , Zeira, E. , Pal, B. , Karplus, R. , Beider, K. et al., Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004. 18. 10.1096/fj.03-0935fje. [DOI] [PubMed] [Google Scholar]
- 68. Müller, A. , Homey, B. , Soto, H. , Ge, N. , Catron, D. , Buchanan, M. E. , McClanahan, T. et al., Involvement of chemokine receptors in breast cancer metastasis. Nature 2001. 410. 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 69. Vandercappellen, J. , Van Damme, J. and Struyf, S. , The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008. 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 70. Chatterjee, S. , Behnam Azad, B. and Nimmagadda, S. , The intricate role of CXCR4 in cancer. Adv. Cancer. Res. 2014. 10.1016/B978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murakami, T. , Maki, W. , Cardones, A. R. , Fang, H. , Tun Kyi, A. , Nestle, F. O. and Hwang, S. T. , Expression of CXC chemokine receptor‐4 enhances the pulmonary metastatic potential of murine B16 melanoma cells. Cancer Res. 2002. 62: 7328–7334. [PubMed] [Google Scholar]
- 72. Schioppa, T. , Uranchimeg, B. , Saccani, A. , Biswas, S. K. , Doni, A. , Rapisarda, A. Bernasconi, S. et al., Regulation of the Chemokine Receptor CXCR4 by Hypoxia. J. Exp. Med. 2003. 198. 10.1084/jem.20030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen, Y. , Ramjiawan, R. R. , Reiberger, T. , Ng, M. R. , Hato, T. , Huang, Y. , Ochiai, H. et al., CXCR4 inhibition in tumor microenvironment facilitates anti‐PD‐1 immunotherapy in sorafenib‐treated HCC in mice HHS public access. Hepatology 2015. 61: 1591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Seo, Y. D. , Jiang, X. , Sullivan, K. M. , Jalikis, F. G. , Smythe, K. S. , Abbasi, A. , Vignali, M. et al., Mobilization of CD8+ T cells via CXCR4 blockade facilitates PD‐1 checkpoint therapy in human pancreatic cancer. Clin. Cancer Res. 2019. 25. 10.1158/1078-0432.CCR-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bockorny, B. , Semenisty, V. , Macarulla, T. , Borazanci, E. , Wolpin, B. M. , Stemmer, S. M. , Golan, T. et al., BL‐8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: the COMBAT trial. Nat. Med. 2020. 26. 10.1038/s41591-020-0880-x. [DOI] [PubMed] [Google Scholar]
- 76. De Giovanni, M. , Chen, H. , Li, X. and Cyster, J. G. , GPR35 and mediators from platelets and mast cells in neutrophil migration and inflammation. Immunol. Rev. 2023. 10.1111/imr.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Milligan, G. , GPR35: from enigma to therapeutic target. Trends Pharmacol. Sci., 2023. 10.1016/j.tips.2023.03.001. [DOI] [PubMed] [Google Scholar]
- 78. Oka, S. , Ota, R. , Shima, M. , Yamashita, A. and Sugiura, T. , GPR35 is a novel lysophosphatidic acid receptor. Biochem. Biophys. Res. Commun. 2010. 395: 232–237. [DOI] [PubMed] [Google Scholar]
- 79. De Giovanni, M. , Tam, H. , Valet, C. , Xu, Y. , Looney, M. R. and Cyster, J. G. , GPR35 promotes neutrophil recruitment in response to serotonin metabolite 5‐HIAA. Cell 2022: 1–16. 10.1016/j.cell.2022.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaya, B. , Doñas, C. , Wuggenig, P. , Diaz, O. E. , Morales, R. A. , Melhem, H. , Hernández, P. P. et al., Lysophosphatidic acid‐mediated GPR35 signaling in CX3CR1+ macrophages regulates intestinal homeostasis. Cell Rep. 2020. 32. 10.1016/j.celrep.2020.107979. [DOI] [PubMed] [Google Scholar]
- 81. Wang, J. , Simonavicius, N. , Wu, X. , Swaminath, G. , Reagan, J. , Tian, H. and Ling, L. , Kynurenic acid as a ligand for orphan G protein‐coupled receptor GPR35. J. Biol. Chem. 2006. 281: 22021–22028. [DOI] [PubMed] [Google Scholar]
- 82. Agudelo, L. Z. , Ferreira, D. M. S. , Cervenka, I. , Bryzgalova, G. , Dadvar, S. , Jannig, P. R. , Pettersson‐Klein, A. T. et al., Kynurenic acid and Gpr35 regulate adipose tissue energy homeostasis and inflammation. Cell Metab. 2018. 27: 378–392.e5. [DOI] [PubMed] [Google Scholar]
- 83. Wang, J. , Simonavicius, N. , Wu, X. , Swaminath, G. , Reagan, J. , Tian, H. and Ling, L. , Kynurenic acid as a ligand for orphan G protein‐coupled receptor GPR35. J. Biol. Chem. 2006. 281. 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 84. De Giovanni, M. , Tam, H. , Valet, C. , Xu, Y. , Looney, M. R. and Cyster, J. G. , GPR35 promotes neutrophil recruitment in response to serotonin metabolite 5‐HIAA. Cell 2022. 185. 10.1016/j.cell.2022.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. De Giovanni, M. , Dang, E. V. , Chen, K. Y. , An, J. , Madhani, H. D. and Cyster, J. G. , Platelets and mast cells promote pathogenic eosinophil recruitment during invasive fungal infection via the 5‐HIAA‐GPR35 ligand‐receptor system. Immunity 2023. 56. 10.1016/j.immuni.2023.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. De Giovanni, M. , Vykunta, V. S. , Biram, A. , Chen, K. Y. , Taglinao, H. , An, J. , Sheppard, D. et al., Mast cells help organize the Peyer's patch niche for induction of IgA responses. 2024. 9: eadj7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kaya, B. , Doñas, C. , Wuggenig, P. , Diaz, O. E. , Morales, R. A. , Melhem, H. , Hernández, P. P. et al., Lysophosphatidic acid‐mediated GPR35 signaling in CX3CR1+ macrophages regulates intestinal homeostasis. Cell Rep. 2020. 32. 10.1016/j.celrep.2020.107979. [DOI] [PubMed] [Google Scholar]
- 88. Barth, M. C. , Ahluwalia, N. , Anderson, T. J. T. , Hardy, G. J. , Sinha, S. , Alvarez‐Cardona, J. A. , Pruitt, I. E. et al., Kynurenic acid triggers firm arrest of leukocytes to vascular endothelium under flow conditions. J. Biol. Chem. 2009. 284. 10.1074/jbc.M109.024042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Miyamoto, K. , Sujino, T. , Harada, Y. , Ashida, H. , Yoshimatsu, Y. , Yonemoto, Y. , Nemoto, Y. et al., The gut microbiota‐induced kynurenic acid recruits GPR35‐positive macrophages to promote experimental encephalitis. Cell Rep. 2023. 42. 10.1016/j.celrep.2023.113005. [DOI] [PubMed] [Google Scholar]
- 90. Sharmin, O. , Abir, A. H. , Potol, A. , Alam, M. , Banik, J. , Rahman, A. , Tarannum, N. et al., Activation of GPR35 protects against cerebral ischemia by recruiting monocyte‐derived macrophages. Sci. Rep. 2020. 10. 10.1038/s41598-020-66417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li, T. , Yue, Y. , Ma, Y. , Zhong, Z. , Guo, M. , Zhang, J. , Wang, Z. et al., Fasting‐mimicking diet alleviates inflammatory pain by inhibiting neutrophil extracellular traps formation and neuroinflammation in the spinal cord. Cell Commun. Signal. 2023. 21. 10.1186/s12964-023-01258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yue, J. , Guo, H. , Xu, P. , Ma, J. and Wu, Y. , Activation of the GPR35 on ILC2 drives immunosuppression to promote lung cancer progression. 2023. 13: 2426–2438. [PMC free article] [PubMed] [Google Scholar]
- 93. Mackiewicz, T. , Włodarczyk, J. , Zielińska, M. , Włodarczyk, M. , Durczyński, A. , Hogendorf, P. , Dziki, Ł. et al., Increased GPR35 expression in human colorectal and pancreatic cancer samples: a preliminary clinical validation of a new biomarker. Adv. Clin. Exp. Med. 2023. 32. 10.17219/acem/157291. [DOI] [PubMed] [Google Scholar]
- 94. Mackiewicz, T. , Jacenik, D. , Talar, M. and Fichna, J. , The GPR35 expression pattern is associated with overall survival in male patients with colorectal cancer. Pharmacol. Rep. 2022. 74. 10.1007/s43440-022-00371-2. [DOI] [PubMed] [Google Scholar]
- 95. Bi, K. , He, M. X. , Bakouny, Z. , Kanodia, A. , Napolitano, S. , Wu, J. , Grimaldi, G. et al., Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 2021. 39. 10.1016/j.ccell.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pelka, K. , Hofree, M. , Chen, J. H. , Sarkizova, S. , Pirl, J. D. , Jorgji, V. , Bejnood, A. et al., Spatially organized multicellular immune hubs in human colorectal cancer. Cell 2021. 184. 10.1016/j.cell.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sun, D. , Wang, J. , Han, Y. , Dong, X. , Ge, J. , Zheng, R. , Shi, X. et al., TISCH: a comprehensive web resource enabling interactive single‐cell transcriptome visualization of tumor microenvironment. Nucleic. Acids. Res. 2021. 49. 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pagano, E. , Elias, J. E. , Schneditz, G. , Saveljeva, S. , Holland, L. M. , Borrelli, F. , Karlsen, T. H. et al., Activation of the GPR35 pathway drives angiogenesis in the tumour microenvironment. Gut 2022. 71. 10.1136/gutjnl-2020-323363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shu, C. , Wang, C. , Chen, S. , Huang, X. , Cui, J. , Li, W. and Xu, B. , ERR‐activated GPR35 promotes immune infiltration level of macrophages in gastric cancer tissues. Cell Death Discov. 2022. 8. 10.1038/s41420-022-01238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li, H. , Nguyen, H. , Meda Venkata, S. P. , Koh, J. Y. , Kowluru, A. , Li, L. , Rossi, N. F. et al., Novel role of GPR35 (G‐protein‐coupled receptor 35) in the regulation of endothelial cell function and blood pressure. Hypertension 2021. 78. 10.1161/HYPERTENSIONAHA.120.15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McCallum, J. E. , Mackenzie, A. E. , Divorty, N. , Clarke, C. , Delles, C. , Milligan, G. and Nicklin, S. A. , G‐protein‐coupled receptor 35 mediates human saphenous vein vascular smooth muscle cell migration and endothelial cell proliferation. J. Vasc. Res. 2016. 52. 10.1159/000444754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Xiang, Q. , Zhou, D. , Xiang, X. , Le, X. , Deng, C. , Sun, R. , Li, C. et al., Neuroglobin plays as tumor suppressor by disrupting the stability of GPR35 in colorectal cancer. Clin. Epigenetics 2023. 15. 10.1186/s13148-023-01472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ronkainen, V. P. , Tuomainen, T. , Huusko, J. , Laidinen, S. , Malinen, M. , Palvimo, J. J. , Ylä‐Herttuala, S. et al., Hypoxia‐inducible factor 1‐induced G protein‐coupled receptor 35 expression is an early marker of progressive cardiac remodelling. Cardiovasc. Res. 2014. 101: 69–77. [DOI] [PubMed] [Google Scholar]
- 104. Wang, W. , Han, T. , Tong, W. , Zhao, J. and Qiu, X. , Overexpression of GPR35 confers drug resistance in NSCLC cells by β‐arrestin/Akt signaling. Onco. Targets Ther. 2018. 11. 10.2147/OTT.S175606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schneditz, G. , Elias, J. E. , Pagano, E. , Zaeem Cader, M. , Saveljeva, S. , Long, K. , Mukhopadhyay, S. et al., GPR35 promotes glycolysis, proliferation, and oncogenic signaling by engaging with the sodium potassium pump. Sci. Signal 2019. 12. 10.1126/scisignal.aau9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jenkins, L. , Harries, N. , Lappin, J. E. , MacKenzie, A. E. , Neetoo‐Isseljee, Z. , Southern, C. , McIver, E. G. et al., Antagonists of GPR35 display high species ortholog selectivity and varying modes of action. J. Pharmacol. Exp. Ther. 2012. 343. 10.1124/jpet.112.198945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Platten, M. , Nollen, E. A. A. , Röhrig, U. F. , Fallarino, F. and Opitz, C. A. , Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019. 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 108. Walczak, K. , Wnorowski, A. , Turski, W. A. and Plech, T. , Kynurenic acid and cancer: facts and controversies. Cell. Mole. Life Sci. 2020. 10.1007/s00018-019-03332-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Balakrishna, P. , George, S. , Hatoum, H. and Mukherjee, S. , Serotonin pathway in cancer. Int. J. Mole. Sci. 2021. 10.3390/ijms22031268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chen, L. , Zhu, C. , Pan, F. , Chen, Y. , Xiong, L. , Li, Y. , Chu, X. et al., Platelets in the tumor microenvironment and their biological effects on cancer hallmarks. Preprint, 2023. 10.3389/fonc.2023.1121401. [DOI] [PMC free article] [PubMed]
- 111. Bambace, N. M. and Holmes, C. E. , The platelet contribution to cancer progression. J. Thrombosis Heamostasis 2011. 10.1111/j.1538-7836.2010.04131.x. [DOI] [PubMed] [Google Scholar]
- 112. Komi, D. E. A. and Redegeld, F. A. , Role of mast cells in shaping the tumor microenvironment. Clin. Rev. Allergy Immunol. 2020. 10.1007/s12016-019-08753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ribatti, D. , Mast cells and resistance to immunotherapy in cancer. Preprint, 2023. 10.1007/s00005-023-00676-x. [DOI] [PMC free article] [PubMed]
- 114. Schneider, M. A. , Heeb, L. , Beffinger, M. M. , Pantelyushin, S. , Linecker, M. , Roth, L. , Lehmann, K. et al., Attenuation of peripheral serotonin inhibits tumor growth and enhances immune checkpoint blockade therapy in murine tumor models. Sci. Transl. Med. 2021. 13. 10.1126/scitranslmed.abc8188. [DOI] [PubMed] [Google Scholar]
- 115. Arora, C. , Matic, M. , Dichiaro, P. , De Oliveira Rosa, N. , Carli, F. , Clubb, L. , Amir, L. et al., The landscape of cancer rewired GPCR signaling axes. 10.1101/2023.03.13.532291 bioRxiv. [DOI] [PMC free article] [PubMed]
- 116. Cosín‐Roger, J. , Ortiz‐Masia, D. , Barrachina, M. D. and Calatayud, S. , Metabolite sensing GPCRs: promising therapeutic targets for cancer treatment? Cells 2020. 10.3390/cells9112345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mills, G. B. and Moolenaar, W. H. , The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer 2003. 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 118. Nakashima, C. , Shingo, K. , Fujiwara‐Tani, R. , Luo, Y. , Kawahara, I. , Goto, K. , Sasaki, T. et al., Expression of long‐chain fatty acid receptor GPR40 is associated with cancer progression in colorectal cancer: a retrospective study. Oncol. Lett. 2018. 15: 8641–8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wu, V. H. , Yung, B. S. , Faraji, F. , Saddawi‐Konefka, R. , Wang, Z. , Wenzel, A. T. , Song, M. J. et al., The GPCR–Gαs–PKA signaling axis promotes T cell dysfunction and cancer immunotherapy failure. Nat. Immunol. 2023. 24: 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Craddock, J. A. , Lu, A. , Bear, A. , Pule, M. , Brenner, M. K. , Rooney, C. M. and Foster, A. E. , Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J. Immunother. 2010. 33. 10.1097/CJI.0b013e3181ee6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as no new data were generated.