Abstract
Background
COVID-19 disease has spread rapidly worldwide, causing high mortality. Accessible biomarkers capable of early identification of patients at risk of severe form are needed in clinical practice. The aim of the study was to determine the biological markers that predict a critical condition.
Methods
Retrospective study including patients with confirmed COVID-19 hospitalized between September 2020 and June 2021. The primary endpoint was progression to critical status within 7 days from admission. We defined two groups:
Critical group: Patients who developed a critical condition or died or transferred to the ICU before or at 7 th day.
Non-critical group: Patients who remained in non-critical respiratory status until 7 th day or discharged before or at 7 th day.
Results
Our study included 456 patients, with a sex ratio of 1.32 and an average age of 62 years. At the 7 th day of hospitalization, 115 (25.2%) patients were in the critical group and 341 (74.8%) patients were in the non-critical group. The univariate logistic regression indicated that laboratory findings between non-critical and critical groups showed that C-reactive protein (CRP) (p=0.047), D-Dimer (p=0.011), creatinine (0.026), creatine kinase (p=0.039), lactate dehydrogenase (p=0.04), and troponin (p=0.001) were all higher among patients in critical group. However, lymphocyte (p<0.001) and platelet (p<0.001) counts were significantly lower among the critical group. Multivariate logistic regression model, identified four independent risk factors: lymphopenia (OR=2.771, 95%CI=1.482-5.181, p=0.001), Neutrophil to Lymphocyte Ratio (OR=2.286, 95%CI=1.461-3.578, p<0.001), thrombocytopenia (OR=1.944, 95%CI=1.092-3.459, p=0.024), and CRP>71.5 (OR=1.598, 95% CI=1.042-2.45, p=0.032) were associated to critical group.
Conclusions
Our results show the predictive value of some biological markers to evaluate the prognosis of COVID-19 pneumonia. A prognostic score could be proposed for guiding clinical care and improving patient outcomes.
Keywords: COVID-19, Viral pneumonia, Critical illness, Adult respiratory distress syndrome, Biomarkers, Prognosis
Introduction
In December 2019, a novel coronavirus caused viral pneumonia cases in Wuhan, China, spreading globally. Termed SARS-CoV-2, it was declared a “public health emergency of international concern” in January 2020, impacting various levels. Tunisia reported imported COVID-19 cases in March 2020, with a milder initial wave due to strict measures. The second wave strained care facilities in July 2020 due to eased preventive measures. 1 Tunisia’s limited healthcare resources posed challenges, necessitating hospital reorganization and resource optimization. Intensive care units expanded, and departments like pulmonology were dedicated to COVID-19 care. Identifying high-risk patients and predictors of severe outcomes has become of paramount importance. Severe SARS-CoV-2 cases exhibited vascular processes caused by inflammation, leading to complications such as acute respiratory distress syndrome (ARDS) and cardiovascular issues. 2 , 3 Urgent identification of predictors guided risk assessment and intervention studies. This contributed to the effective allocation of resources and informed clinical decision-making. Moreover, distinguishing severe cases using parameters like haematological and inflammatory markers improved clinical management. 4 , 5 This is a study performed at University Hospital Center Mongi Slim, focused on COVID-19 patients hospitalized between October 2020 and June 2021 in the COVID-19 unit comprising Pulmonology and Rheumatology departments. The objectives of this study was to describe the clinical and evolutionary characteristics of hospitalized patients with COVID-19 and to determine predictive biological markers of a critical condition on the 7th day of admission.
Methods
This was a retrospective study, conducted at University Hospital Center Mongi Slim, including patients hospitalized for COVID-19 pneumonia between October 2020 and June 2021 in the COVID-19 unit: Pulmonology Department and Rheumatology Department, which were jointly managed in collaboration between the two teams.
Study design
Inclusion and exclusion criteria :
Eligible patients were those admitted to the COVID-19 unit between October 2020 and June 2021 with moderate to severe COVID-19 pneumonia at admission, confirmed for SARS-CoV-2 via Reverse Transcription-Polymerase Chain Reaction (RT-PCR), Rapid Antigen Test (RAT), or positive SARS-CoV-2 serology (IgM or IgG) for vaccine-naive patients.
Patients were not included if they had a mild COVID-19 infection throughout their hospitalization and another reason for hospitalization, if they were patients admitted to the COVID-19 unit after an initial stay in the intensive care unit, or if they remained in the emergency department for more than 7 days before being admitted to the COVID-19 unit.
We excluded patients who were admitted to the COVID-19 unit with hypoxemic pneumonia and suggestive findings of COVID-19 pneumonia on thoracic CT scan but with negative virological results (RT-PCR, RAT, and SARS-CoV-2 serologies), and an alternative cause was identified during the course of the disease. Additionally, patients were excluded if virological documentation data could not be located.
Outcome measure :
We defined the development of a critical state before or on the 7th day of hospitalization (D7) as the outcome measure. Patients were categorized into two groups:
-
1)Critical Group: Event group including:
-
•Patients who developed a critical state by D7. The definition of a critical state will be detailed in the following paragraph.
-
•Patients who died before or on D7.
-
•Patients transferred to the intensive care unit before or on D7.
-
•
-
2)Non-Critical Group: Non-event group including:
-
•Patients who maintained a stable and non-critical respiratory state until D7.
-
•Patients discharged from the hospital before or on D7.
-
•Patients transferred to the intensive care unit before or on D7.
-
•
Definition of a Critical State : We defined a critical state as:
-
•
The occurrence of Acute Respiratory Distress Syndrome (ARDS) requiring invasive or non-invasive respiratory support. For this criterion, we defined the oxygen requirement level of 10 liters with a high-concentration mask as the threshold for evolving into ARDS.
-
•
Vital distress or shock, sepsis, and/or organ failure. In all cases, intensive care unit admission is indicated. We relied on the definitions from the World Health Organization (WHO) version of November 2021 6 and the National Evaluation and Accreditation Agency for Health (INEAS) version of April 2021 7 to establish the definition of a critical state.
Definition of ARDS : We defined the oxygen requirement level of 10 liters with a high-concentration mask as the threshold for evolving into ARDS. This threshold was considered throughout the COVID unit’s activity based on the medical team’s experience as an indicator of a critical state and warranting alerting the intensive care units.
Data collection
Data were retrospectively and anonymously collected between June 2023 and January 2024. All demographic, clinical, biological, radiological, therapeutic, and progression-related data were extracted from the medical records. The medical records followed a standardized format with predefined items consistently used in both departments of COVID-19 unit. The collected information included demographic data, clinical details recorded at admission and D7, and biological data obtained at admission. For the latter, all patients underwent a routine standard biological assessment at admission complete blood count (CBC) including leukocytes, lymphocytes, neutrophils, platelets, hemostasis profile (Prothrombin Time, Activated Partial Thromboplastin Time, D-Dimers), renal profile (Urea, Creatinine, Blood Ionogram), liver profile (Aspartate Aminotransferase (AST), Alanine Aminotransferase (ALT)), markers of muscle injury (Creatine Kinase (CK), Lactate Dehydrogenase (LDH)), and C-Reactive Protein (CRP). Some parameters were unavailable due to reagent shortages, and others were optional based on clinical presentation, such as Troponin and N-terminal prohormone of brain natriuretic peptide (NT-ProBNP). Chest computed tomography (CT) data were recorded at admission and upon aggravation during evolution. Therapeutic data included oxygen therapy, antibiotic treatment, corticosteroids, anticoagulation, and other measures, adhering to a uniform protocol guided by INEAS recommendations. Evolvement data covered hospital discharge, transfers to intensive care, COVID-19 unit deaths, and complication occurrences.
Statistical analysis
For the analysis of the association between two qualitative variables, we used Pearson’s Chi-squared test to compare two frequencies when the conditions for its application were met, and the Fisher’s test otherwise. For the analysis of the association between a qualitative variable and a quantitative variable, we employed the Student’s t-test to compare two means in the case of a normal distribution, and the non-parametric Mann Whitney test otherwise. Correlations between two quantitative variables were calculated using the Pearson correlation coefficient, with significance tested bilaterally. In the multivariate study, a binary logistic regression model was followed, and the risk was computed as the Odds Ratio (OR) with a retained 95% confidence interval (CI 95%). To assess performance, we utilized parameters related to the Receiver Operating Characteristic (ROC) Curve, as well as sensitivity and specificity. Pre- and post-test probabilities were determined through likelihood ratios represented on the Fagan nomogram. We set the significance threshold at p ≤ 5%.
Ethical considerations
This retrospective study received approval from the Ethics Committee of University Hospital Center Mongi Slim, under approval number 57/2023 on Friday, 22 December 2023.
We followed strict ethical committee guidelines that allowed for exemption of consent, due to the non-intrusive nature of the study and the use of non-identified data to ensure confidentiality and anonymity of participants. All personally identifiable data were anonymized prior to analysis to protect individuals’ privacy. We also carefully assessed the risks and benefits of our research, ensuring to minimize the former and maximize the latter for participants and the scientific community. No financial or other conflicts of interest were identified by the authors of this study. Policies regarding the future use of data and their potential sharing with other researchers were strictly established in accordance with ethical guidelines.
Results
Among 626 patients hospitalized in the COVID-19 unit of University Hospital Center Mongi Slim for a period of 9 months (October 2020 - June 2021), we selected 456 patients after applying the inclusion and exclusion criteria. The number of included, not included, and excluded patients is summarized in the flowchart below ( Figure 1).
-
1)
Patient characteristics
On the 7th day of hospitalization, 115 (25.2%) evolved to a critical condition. Among the patients in the study, 260 (57%) were males; 196 (43%) were females. The gender of the patients was not statistically significant between critical and non-critical groups (p=0.106). As shown in Table 1, Chi-square outcome indicated that the age differed significantly between the critical and non-critical patients (p=0.004). In fact, the mean age for the non-critically ill patients was 61.5 ± 13.2 years while the critically ill patients had a mean age of 65.7 ± 14. Chronic concomitant diseases were found among 322 patients (70.6%), with no significant difference between both groups. The mean time from onset of symptoms to admission into the COVID-19 unit was 8.5 ± 4.12 days. This lapse was significantly shorter for the critically ill patients: 7.9 vs 8.8 days (p=0.022). Among the total cases, 31.4% had an extent of lesions greater than 50% on chest CT, the critical group had a higher proportion of cases with an extent exceeding 50% (37% versus 25.6%, p=0.021).
-
2)
Laboratory test outcomes
The CBC findings shown in Table 2 indicate that concentration of the lymphocytes were statistically lower among critically ill patients (0.85 ± 0.24) compared to non-critically ill patients (1.10 ± 0.33, p<0.001). Lymphopenia also occurred among 89.5% of patients in critical group (p=0.002) ( Table 3). Similarly, the platelet count was lower among the critical group (203 vs 235, p<0.001). The level of WBC and neutrophil did not statistically differ between both groups.
The mean Neutrophil-to-Lymphocyte ratio (NLR) was 7.52. The NLR at admission was higher in the critical group with a marginally significant difference (p=0.073). There was also a significant correlation between the NLR and the extent of lesions on the chest CT (r=0.213, p<10 −3).
The CRP concentration outcome is the other biochemistry test that was statistically significantly different between critically ill (122 ± 78) and non-critically ill patients (106 ± 72, p=0.047). Upon admission, data regarding levels of D-dimer (full data available for 136 patients), hypersensitive troponin (full data available for 117 patients), B-type natriuretic peptide (full data available for 67 patients) were obtained. As shown in Table 2, in comparison to the non-critically ill patients, patients with critical illness had significantly higher levels of D-Dimer, hypersensitive and NT-pro-BNP. Besides, patients with severe pneumonia had higher rates of rhabdomyolysis markers than patients exhibiting less critical forms of the disease; (CK: 127 vs 83, p=0.039), (LDH: 831 vs 701, p=0.04). However, the findings presented in Table 2 indicate that the concentration of AST and ALT did not vary between both groups.
Figure 1. Flow chart illustrating the patient selection process.
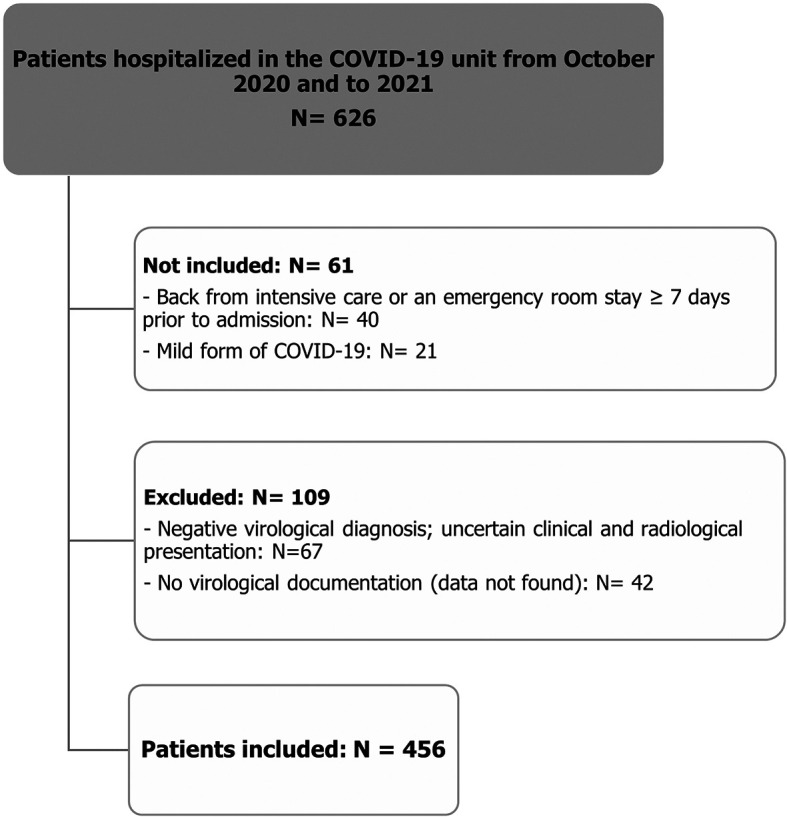
Table 1. Baseline characteristics of 456 patients infected with COVID-19 pneumonia on admission.
| Characteristics | Total number (N=456) | Non-critical group (N=341) | Critical group (N=115) | p-value |
|---|---|---|---|---|
| Gender | 0.106 | |||
| Male N, (%) | 260 (57) | 187 (55) | 73 (64) | |
| Female N, (%) | 196 (43) | 154 (45) | 42 (36) | |
| Age, mean, SD (years) | 62.56 ± 13.5 | 61,5 ± 13.2 | 65,7 ± 14 | 0.004 |
| Onset of symptoms to admission, mean, SD ( days) | 8.5 ± 4.12 | 8.8 ± 4.12 | 7.9 ± 4.11 | 0.022 |
| BMI, kg/m 2 , SD | 29.3 ± 5 | 29.33 ± 5.45 | 29.06 ± 5.88 | 0.672 |
| Chronic concomitant diseases, N (%) | 322 (70.8) | 236 (69.2) | 86 (75.4) | 0.256 |
| Hypertension, N (%) | 198 (43.4) | 146 (42.8) | 52 (45.6) | 0.653 |
| Diabetes, N (%) | 171 (37.5) | 126 (37) | 45 (39.5) | 0.676 |
| Dyslipidemia, N (%) | 72 (15.8) | 51 (15) | 21 (18.4) | 0.401 |
| Coronary artery disease, N (%) | 45 (9.9) | 35 (10.3) | 10 (8.8) | 0.626 |
| Obesity, N (%) | 203 (40) | 157 (46) | 46 (40) | 0.298 |
| COPD, N (%) | 23 (5) | 16 (4.7) | 7 (6.1) | 0.554 |
| Symptoms | ||||
| Asthenia, N (%) | 324 (71) | 238 (70.8) | 86 (74.8) | 0.308 |
| Fever, N (%) | 286 (63) | 211 (62.6) | 75 (65.2) | 0.522 |
| Myalgia, N (%) | 221 (48.5) | 179 (53.3) | 42 (36.5) | 0.003 |
| Dyspnea, N (%) | 305 (67) | 226 (67.3) | 79 (68.7) | 0.633 |
| Cough, N (%) | 229 (50) | 169 (50.3) | 60 (52.2) | 0.628 |
| Peripheral oxygen saturation in room air, %, SD | 88 ± 4.24 | 88.8 ± 4 | 87.1 ± 4.5 | <0.001 |
| CT findings | ||||
| Extent of lesions > 50%, N (%) | 129 (31.4) | 86 (25,6) | 43 (37) | 0.021 |
| Bilateral distribution, N (%) | 386 (98.2) | 287 (98.6) | 99 (97) | 0.42 |
| Ground-glass opacity, N (%) | 385 (96.7) | 283 (93) | 102 (95) | 0.568 |
| Consolidations, N (%) | 276 (70.6) | 202 (69.7) | 74 (73.3) | 0.662 |
| Pleural effusion, N (%) | 27 (6.9) | 17 (6) | 10 (9) | 0.183 |
| Pericardial effusion, N (%) | 40 (10.2) | 23 (8) | 17 (16) | 0.013 |
| Outcome | ||||
| Transfer to intensive care unit, N (%) | 81 (16.6) | 11 (3.2) | 70 (60.9) | <0.001 |
| Deaths in the COVID-19 unit, N (%) | 22 (4.8) | 1 (0.3) | 21 (1.3) | <0.001 |
Table 2. Laboratory findings among critical and non-critical COVID-19 patients on admission to hospital.
| Variables | Normal range | Total number (N=456) | Non-critical group (N=341) | Critical group (N=115) | p-value |
|---|---|---|---|---|---|
| Leukocyte, × 10 9/L | 4-10 | 8.1 ± 3.8 | 7.4 ± 2.4 | 7 ± 2 | 0.4 |
| Lymphocyte, × 10 9/L | 1.5-4 | 1.1 ± 0.6 | 1.1 ± 0.33 | 0.8 ± 0.24 | <0.001 |
| Neutrophil, × 10 9/L | 1.5-7 | 6.5 ± 3.7 | 5.7 ± 0.19 | 5.8 ± 0.38 | 0.926 |
| Platelet, × 10 9/L | 150-400 | 252 ± 114 | 235 ± 6.53 | 203 ± 8.76 | <0.001 |
| NLR (ratio) | - | 7.52 ± 8.05 | 7.12 ± 0.43 | 8.69 ± 0.76 | 0.073 |
| C-reactive protein, mg/L | <8 | 110 ± 73 | 106 ± 71.7 | 122 ± 78.2 | 0.047 |
| D-dimer (N=136), μg/L | 0-500 if < 50 years 0-(age*10) if > 50 years | 1214 ± 1311 | 712 ± 1401 (N=99) | 1027 ± 1035 (N=37) | 0.011 |
| Creatinine, μmol/L | 50-101 | 86.6 ± 62.5 | 85 ± 64.3 | 91.6 ± 56 | 0.026 |
| Blood urea nitrogen, mmol/L | 2.5-7 | 7.8 ± 6 | 6.2 ± 6.3 | 7 ± 4.4 | 0.061 |
| Creatine Kinase, U/L | <168 | 146.7 ± 168 | 83 ± 153 | 127 ± 199 | 0.039 |
| Lactate Dehydrogenase, U/L | 125-222 | 784.7 ± 34 | 701 ± 342 | 831 ± 334 | 0.04 |
| Aspartate aminotransferase, U/L | 5-40 | 40.5 ± 31 | 39 ± 26.25 | 45 ± 40.6 | 0.172 |
| Alanine aminotransferase, U/L | 5-45 | 34.7 ± 29 | 34 ± 26.8 | 36 ± 34.5 | 0.740 |
| Natremia, mmol/L | 135-145 | 134.7 ± 4.5 | 135 ± 4.1 | 133 ± 5.5 | 0.011 |
| Kalemia, mmol/L | 3.5-5 | 4.09 ± 0.6 | 4.12 ± 0.56 | 3.98 ± 0.63 | 0.029 |
| Hypersensitive troponin (N=117), ng/L | <19 | 180.2 ± 1570 | 15.5 ± 57.2 (N=86) | 637 ± 3036 (N=31) | 0.001 |
| NT-Pro-BNP (N=67), pg/mL | <125 | 989.7 ± 1149 | 779 ± 1315 (N=41) | 1322 ± 1608 (N=26) | 0.011 |
Table 3. Comparison of biological abnormalities between critically and non-critically ill COVID-19 pneumonia patient groups.
| Total number (N=456) | Non-critical group (N=341) | Critical group (N=115) | p-value | |
|---|---|---|---|---|
| Hyperleucocytosis, N (%) | 110 (24.1) | 80 (23.5) | 30 (26.1) | 0.569 |
| Lymphopenia, N (%) | 355 (77.9) | 253 (76) | 102 (89.5) | 0.002 |
| Neutrophilia, N (%) | 155 (34.6) | 113 (34) | 42 (36.8) | 0.573 |
| Thrombocytopenia, N (%) | 59 (12.9) | 37 (11.1) | 22 (19.5) | 0.023 |
| Elevated CRP, N (%) | 424 (93.5) | 315 (96) | 109 (98.2) | 0.375 |
| D-Dimer > 1000 ng/ml, N (%) | 50 (40.8) | 31 (31.3) | 19 (51.4) | 0.031 |
| Elevated blood urea nitrogen, N (%) | 186 (40.8) | 135 (39.6) | 51 (44.3) | 0.369 |
| Elevated aminotransferase, N (%) | 157 (39.5) | 116 (39.7) | 41 (39) | 1 |
| Elevated Creatine Kinase, N (%) | 55 (24.2) | 31 (18.6) | 24 (40) | 0.001 |
| Elevated Lactate dehydrogenase, N (%) | 190 (41.6) | 135 (39.5) | 55 (47.8) | 1 |
| Elevated tropnin, N (%) | 21 (18) | 9 (10.5) | 12 (38.7) | <0.001 |
Univariate analysis
The univariate logistic regression indicated that laboratory findings between non-critical and critical groups showed numerous differences including C-reactive protein (p=0.047), D-Dimer (p=0.011) and creatinine (0.026) as well as, creatine kinase (p=0.039), lactate dehydrogenase (p=0.04), troponin (p=0.001) and NT-pro-BNP level (p=0.011) which were all higher among patients in critical condition. On the other hand, lymphocyte (p<0.001) and platelet (p<0.001) counts, natremia (p=0.011) and kalemia (p=0.029) were significantly lower among the critical group, whereas liver enzymes showed no significant differences. Table 3 summarizes the biological abnormalities on admission.
Multivariate analysis
Based on the multivariate logistic regression model, four variables were demonstrated as independent risk factors. As shown in Table 4, the results indicated that lymphopenia (OR=2.771, 95%CI=1.482-5.181, p=0.001), NLR (OR=2.286, 95%CI=1.461-3.578, p<0.001), thrombocytopenia (OR=1.944, 95%CI=1.092-3.459, p=0.024), and CRP>71.5 (OR=1.598, 95% CI=1.042-2.45, p=0.032) were associated with critical outcome.
Table 4. Multivariate analysis of biological markers predicting critical condition.
| Parameters | OR | 95%CI | p-value |
|---|---|---|---|
| Lymphopenia | 2.771 | 1.482-5.181 | 0.001 |
| NLR > 5 | 2.286 | 1.461-3.578 | <0.001 |
| Thrombocytopenia | 1.944 | 1.092-3.459 | 0.024 |
| CRP > 71.5 mg/L | 1.598 | 1.042-2.45 | 0.032 |
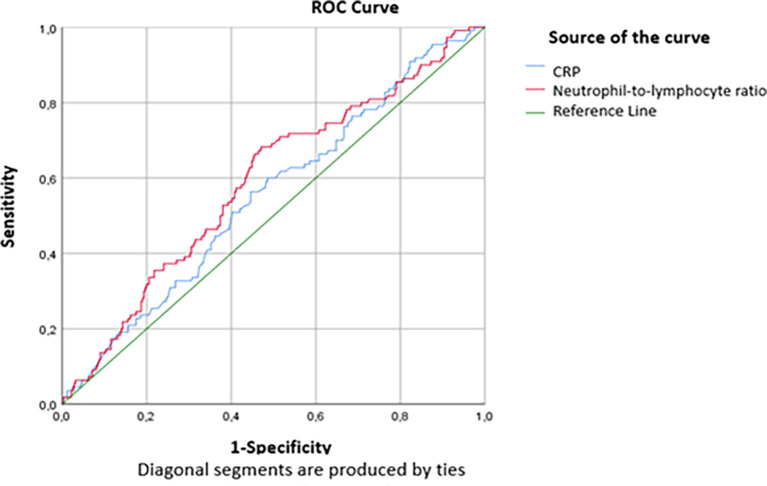
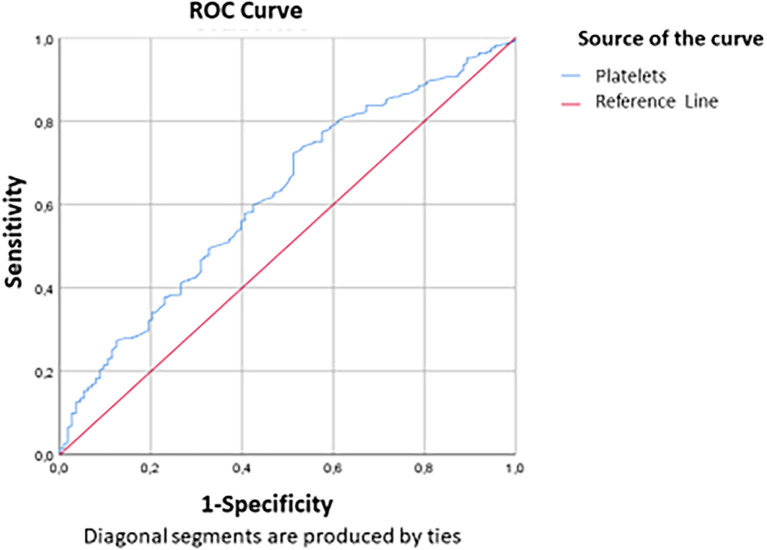
The threshold values of 71.5mg/L for the CRP level and 5 for the RNL were defined according to the study of the ROC curve according to the critical and non-critical groups. The findings in Table 4 show that statistically significant AUCs were obtained for CRP (AUC=0.533, p=0.032) and NLR (AUC=0.589, P<0.001). Thus, the performance of the NLR was slightly superior to that of the CRP in predicting a critical condition on day 7 ( Figure 2). For a threshold equal to 71.5 mg/L, the sensitivity and specificity of CRP to predict critical illness were respectively equal to 66.1% and 38.1%, while for a threshold equal to 5, the sensitivity and specificity of the NLR to predict the critical illness were respectively equal to 68.7% and 51% ( Table 5). Figure 2 and Figure 3 show the area under the ROC curve of CRP and NLR ( Figure 2) and platelet count ( Figure 3). The platelet count was negatively correlated with the occurrence of a critical condition and it was quite efficient in predicting non-critical cases; (AUC=0.62; CI95%=0.56-0.679) ( Figure 2). The best threshold was estimated at 187.5*10 3 elements/mm 3 corresponds to a sensitivity and specificity equal to 72.4% and 48.7% respectively with a quite good accuracy ( Table 6).
Figure 2. ROC curve indicating the performance of CRP and NLR in detecting the critical cases.
Table 5. Performance parameters of CRP and NLR.
| Cut-off | AUC (95%CI) | Sensitivity | Specificity | |
|---|---|---|---|---|
| CRP (mg/L) | 71.5 | 0.533 (0.492-0.614) | 66.1% | 38.1% |
| NLR | 5 | 0.589 (0.528-651) | 68.7% | 51% |
AUC: Area under ROC curve.
Figure 3. ROC curve indicating platelet count performance in predicting non-critical cases.
Table 6. Performance parameters of platelet count.
| Cut-off | AUC (95%CI) | Sensitivity | Specificity | |
|---|---|---|---|---|
| Platelets (10 9/L) | 187.5 | 0.62 (0.56-0.679) | 72.4% | 48.7% |
AUC: Area under ROC curve.
Discussion
Based on the univariate analysis of our study, some biomarkers were predictive of the progression to a critical state on the 7th day of hospitalization for COVID-19 pneumonia.
A significant difference in biochemical and CBC test parameters was found between critically and non-critically ill patients, with elevated CRP, D-dimer, creatinine, CK, LDH, troponin and NT-Pro-BNP among the critical group compared to the non-critical group. Added to that, lymphocytes and platelet count were higher among the non-critical group. Our findings showed that the parameters with good specificity and sensitivity in predicting a critical outcome among COVID-19 patients are: CRP>71.5 mg/L and NLR>5.
Analysis of demographic characteristics revealed that critically ill patients were older than non-critically-ill ones which aligns with previous studies’ results. 8 However, this age difference was smaller compared to that reported previously, noted as 20 years. 9
We underline the fact that the study sample of COVID-19 patients cared during the study involved more males than females, as well among both groups (critical and non-critical), which supports previous observations that indicate gender as a potential determinant of likelihood to develop serious complications and unfavorable outcome. In fact, a multicenter study conducted in China, including 1099 patients hospitalized for COVID-19 pneumonia in 552 hospitals, 10 had shown a clear male predominance (58%). This male susceptibility has been clearly elucidated in an Italian study, 11 in which, two significant factors incriminated in the initiation of viral infection were emphasized, namely angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), both of which are influenced by gender.
Moreover, this observed difference could also be attributed to behavioral differences between the sexes, notably that smoking and alcohol consumption are more often seen among men 12 heightening the risk of chronic comorbidities such as cardiovascular and chronic pulmonary diseases, which have been associated with a greater likelihood of developing severe illness with COVID-19. 13
Hematological biomarkers
Our univariate analysis findings regarding the CBC parameters were different to previous findings. 14 , 15 Previous studies concluded that increased neutrophils count is characteristic of patients with severe cases. 16 Evidence suggests that severe COVID-19 is associated with elevated neutrophil levels, which increase inflammation via cytokine storm, and hemorrhages especially in the lungs, which occur as a result of neutrophil-induced tissue damage.
According to our data, lymphopenia occurred in 78% of cases. The lymphocyte count ranged between 500 and 1000/mm 3 among 36.6% of our patients while 8.8% had deep lymphopenia (< 500/mm 3). Furthermore, in the critical group of patients, lymphopenia was more common (89%). Our results also showed, using the multivariate logistic regression model, that lymphopenia was an independent risk factor for a severe illness. These findings are in line with the literature. Indeed, several studies have assessed the impact of lymphopenia on disease severity. For example, Zhang and al. 17 reported an average blood lymphocyte count of 1200/mm 3, and Jiang and al. 9 showed that 41% of patients had a lymphocyte count of less than 1000/mm 3. This count was significantly lower in patients with an unfavorable outcome. Additionally, Bellan and al. 18 demonstrated that a low lymphocyte count at admission was correlated with the risk of mortality due to SARS-CoV-2 infection. Two Chinese meta-analyses 19 , 20 have concluded that patients with severe COVID-19 pneumonia had lower blood lymphocyte counts compared to non-severe forms, and lymphopenia was associated with a three-fold higher risk of severe infection, particularly ARDS. They, therefore, suggest that lymphocyte count and lymphopenia could be an effective indicator for quickly identifying patients at risk of severe pneumonia. Indeed, lymphocytes play a fundamental role in maintaining adaptive immune responses against viral infections. When viral particles resulting from the replication of SARS-CoV-2 spread through the respiratory mucosa, it leads to a cytokine storm, generating a cascade of immune responses primarily involving CD4 and CD8 T lymphocytes. 21 Four main mechanisms have been recognized as responsible for lymphopenia 22 : Sequestration of lymphocytes in target organs and their direct infection by the virus, as these cells express the ACE2 receptor; Damage of lymphatic organs such as the thymus and spleen by the virus; Apoptosis of lymphocytes due to the increase of pro-inflammatory cytokines (TNF-α, IL-6, etc.) and Inhibition of lymphocytes by substances produced during certain metabolic disorders.
In our study, thrombocytopenia was present among 13% of cases and was correlated with the progression to a critical condition (p=0.023). We demonstrated that thrombocytopenia was associated with an odds ratio (OR) of 1.94 of developing a critical condition. Our findings are consistent with those of the literature. Indeed, a meta-analysis of 21 studies including 3,377 SARS-CoV-2 positive patients 4 found that platelet count was negatively correlated with the severity of the infection as well as mortality. Another meta-analysis involving 1,779 COVID-19 patients 23 revealed that thrombocytopenia was associated with a threefold higher risk of severe disease, and mortality. Moreover, the platelet count was also correlated with disease severity scores and the risk of mortality in intensive care units. 24 , 25 Platelet count was considered as an independent risk factor for COVID-19 mortality. This count was significantly lower in COVID-19 patients who died compared to survivors. 18 , 26 Platelet kinetics could also help to identify patients at risk of an unfavourable outcome. 27 In COVID-19 patients, thrombocytopenia appears to be multifactorial. In the case of ARDS, there is a significant platelet consumption resulting from the combination of viral infection and mechanical ventilation, leading to endothelial damage and platelet activation, aggregation, and thrombus formation in the lungs. 23 , 27 Moreover, the lung can be a site for platelet release from mature megakaryocytes, and alteration of the pulmonary capillary bed can affect platelet degranulation and pulmonary megakaryocytopoiesis. 23 , 27
In our study, the mean NLR was 7.52. This ratio was higher in the critical group patients (p=0.073). As we previously mentioned, lymphopenia was more common among patients in a critical condition. It appears logical to observe a trend of increasing NLR in severe forms. This finding was consistent with studies of both Yang and al. 28 and Yan and al., 29 where a higher NLR was correlated with severe forms and mortality, respectively (p<10 −3). NLR is a biomarker that integrates two subtypes of leukocytes representing two inversely related immune pathways. It provides information about systemic inflammation and is a useful biomarker for predicting bacterial infection, including pneumonia, better than absolute leukocyte, lymphocyte, or neutrophil counts. 29 A high NLR reflects a discrepancy in the inflammatory response. 29 Numerous studies and meta-analyses 30 – 32 have proved that an increased NLR is associated with a poor prognosis in various diseases, such as cardiovascular diseases, solid cancers, and infections.
Increased NLR during COVID-19 could be a result of the expression of inflammatory cytokines, an unusual increase in pathological low-density neutrophils, and the upregulation of genes involved in the lymphocyte cell death pathway caused by SARS-CoV-2 infection. 33 Furthermore, two studies 34 , 35 have proposed integrating the NLR into nomograms to prove the prognostic value of this biomarker. In these studies, the authors demonstrated that NLR values at admission were higher in severe or critical cases 34 and that patients with an NLR greater than 4.87 were 8 times more likely to develop a severe or critical form of the disease. 35 The NLR threshold varies according to studies. An NLR threshold >3.7 on Day 7 was a major predictor of progression to severe illness, while an NLR>11.75 was significantly correlated with in-hospital mortality from all causes. 29
Our study showed that for a threshold ≥ 5, the sensitivity and specificity of NLR in predicting critical status on Day 7 were 68.7% and 51%, respectively. This sensitivity was 56.52% in the study by Liu and al. 35 Moreover, we further demonstrated that patients with an increased NLR (>5) were as much as 2.286 times (95% CI: 1.461-3.578) more likely to develop the severe/critical type than those with a low NLR (≤5). Therefore, NLR is an objective, simple, and inexpensive parameter that could be used in routine practice as a prognostic biomarker.
Thrombotic complications and coagulopathy represent a major event during SARS-CoV-2 infection. An increase in D-dimer and fibrinogen levels and a decrease in prothrombin time indicate a state of hypercoagulability. Many studies have shown that high levels of D-dimer are associated with severe 36 , 37 and critical 8 forms, the need for intensive care management, 2 , 15 and in-hospital mortality due to COVID-19 pneumonia. 26 , 38 This hypercoagulability in COVID-19 patients could result from several mechanisms 39 : in viral infections, there is often an unbalance between pro-inflammatory response and anti-inflammatory response; this can lead to endothelial cell dysfunction and excessive thrombin production; hypoxia contributes to thrombosis by increasing blood viscosity. In addition to that, hospitalized patients, typically have risk factors for hypercoagulability due to age, comorbidities, extended hospital stay, and invasive treatments.
Increased D-dimer levels indicate that the fibrinolytic system is activated in COVID-19 patients. Furthermore, the increased release of cytokines during viral infections stimulates coagulation cascade. 40 Moreover, D-dimer >2.0 mg/L at admission were an independent predictor of death, and dynamic changes in serum D-dimers were closely associated with disease severity. A reduction in D-dimer levels was observed in recovered patients, independently of anticoagulant treatment, while a continuous increase in D-dimer levels was predictive of a higher risk of thrombosis and unfavorable outcomes. 39 , 41 Thus, D-dimers are an early and reliable marker for predicting a poor prognosis in COVID-19 hospitalized patients. 39
In our study, thromboembolic events occurred among 11 patients. The incidence of pulmonary embolism (PE) in COVID-19 patients is still unknown and likely underestimated. 42 Bilaloglu and al. reported a prevalence of PE of 3.2% in a study involving 3,334 COVID-19 patients in New York, 43 while according to a French study, 44 the incidence of PE was 23.7%. This difference could be explained by differences in disease severity, patient characteristics, and the limited use of CT angiography among patients.
Biochemical biomarkers
Biochemical tests findings, showed that the critical group of patients had elevated level of CRP, D-Dimer, creatinine, lactate dehydrogenase, creatine kinase, troponin and NT-Pro-BNP, which corroborates the conclusion made by Feng and al. 37
The CRP was elevated (>8 mg/L) in 93.5% of patients with an average of 110 ± 73 mg/L. We noted that the CRP on admission was significantly higher in the critical group (p=0.047). In multivariate analysis, an elevated CRP level was correlated with an increased risk of progressing to a critical condition on the 7th day of admission (OR: 1.598; 95% CI: 1.042-2.45; p=0.032), and for a threshold of 71.5 mg/L, the sensitivity and specificity of CRP in predicting a critical condition were 66.1% and 38.1% respectively. Many studies have shown that elevated CRP levels are correlated with critical forms, 8 , 36 disease progression, 45 and mortality from COVID-19 pneumonia. 18 As known, CRP is a non-specific marker of inflammation induced by interleukin-6 secretion. In clinical practice, it is used as a biomarker for various inflammatory and infectious conditions. High CRP levels have been directly correlated with the inflammation’s degree and disease severity. 46 Moreover, CRP levels among dead COVID-19 patients were decuple higher than survivors. 47
A meta-analysis of 20 studies including 4,843 COVID-19 patients and focusing on the clinical utility of CRP, 48 emphasized that high CRP level was associated with a fourfold higher risk of an unfavorable outcome (p<10 −3). In fact, at the early stages of COVID-19 infection, an increase in CRP was directly associated with the development of lung lesions, reflecting the severity of the disease. 48 , 49 Furthermore, Ali and al. 50 demonstrated that CRP level could predict disease worsening among non-severe cases, indicating 5% risk of progressing to a severe form for each unit increase in the CRP rate. Added to that, the CRP level has also been reported as a reliable biomarker for treatment responses in COVID-19 patients 16 ; in fact, this marker could be used to select patients who would benefit from treatment with tocilizumab, another IL-6 receptor inhibitor similar to sarilumab. 51 , 52
Our findings indicated that NLR and CRP are good predictors of unfavorable outcome. The reported excellent accuracy of these parameters in the prediction of COVID-19 patients’ outcome corroborates findings of the studies of Yang and al. 28 and. Liu and al. 35
We found that blood creatinine (p=0.026) and urea levels (p=0.061) were higher in the critical group. Furthermore, urea levels were significantly higher in elderly patients (> 70 years old) (p<10 −3) and in patients with high blood pressure (p=0.012). Elevated blood urea (≥ 7 mmol/L) was noted in 46.6%. These findings align with the literature, where it has been demonstrated that elevated blood creatinine and urea values are associated with severe disease, unfavorable prognosis, and significant mortality. 38 , 53 , 54 The mechanism of renal involvement in COVID-19 is likely to be multifactorial. 53 It involves direct cytopathic effects on kidney tissue by the virus leading to renal cell necrosis as well as indirect damage by cytokines and metabolites induced by hypoxia, shock, or rhabdomyolysis.
Increased liver enzymes level was found in 39.5%, especially in male patients (p<10 −3), while AST and ALT rates did not vary between both groups. Numerous studies have demonstrated the association between high transaminase levels and the severity of the disease, 36 , 37 , 54 , 55 transfer to the intensive care unit 2 , 15 , 56 , 57 and death 26 , 38 due to COVID-19. Moreover, the good accuracy of AST and ALT as a predictor of ICU admission have been clearly shown with AUC>0.7. 58 Added to that, Malik and al. demonstrated in their meta-analysis 48 that high levels of AST and ALT (> 40 IU/L) were associated with a threefold higher risk of a poor prognosis. Some studies have shown that COVID-19 only transiently increases transaminases. Cytolysis is rather due to liver damage secondary to systemic inflammatory processes or to the use of hepatotoxic drugs, especially antivirals such as Lopinavir and Ritonavir during patient management. 48 , 59 However, viral RNA has been detected in the liver at high titers, exceeding viremia, during autopsies, suggesting that SARS-CoV-2 hepatic infection can contribute to elevation of transaminase levels in patients with severe forms of COVID-19. 41 , 60 In addition, hypoxia observed in COVID-19 patients induces hepatocellular necrosis through the production of free radicals that increase the release of hepatotoxic pro-inflammatory factors. 48
Regarding rhabdomyolysis markers, we found an elevation of LDH and CK levels among 98% and 24% of our patients, respectively. We also demonstrated that muscle lysis enzymes were significantly higher in critical group patients. Our data were consistent with the literature, where it has been shown that high levels of LDH and CK were associated with the severity 5 , 61 and progression 62 of the disease, transfer to intensive care units, and mortality 54 from COVID-19. In a meta-analysis assessing the prognostic value of LDH levels, 38 it was found that elevated LDH levels were associated with a risk of mortality (OR: 16) and severe disease (OR: 6). Furthermore, a study conducted on COVID-19 patients 63 showed that increased LDH levels at the early stage of the disease can predict lung damage and severe cases of COVID-19. These high LDH levels may result from decreased tissue oxygenation leading to stimulation of the glycolytic pathway or from damage of multiple organs in case of multi-organ failure. 48 Additionally, severe infections can induce cytokine-mediated tissue damage and LDH release. Since LDH is present in alveoli, it is common to observe elevated LDH levels in patients with severe forms of COVID-19. 64 Moreover, CK is a marker of muscle injury. Thus, an acute elevation of CK indicates rhabdomyolysis. The mechanism of viral myositis is unclear, but some authors suggest that myocyte damage is immune-mediated through the deposition of immune complexes in muscles. 48 Therefore, LDH and CK levels are considered as important biomarkers in the prognosis of patients with severe and critical COVID-19. 41
Concerning cardiac injury markers, we found that elevated levels of cardiac troponins were correlated with critical conditions (p=0.001). Indeed, troponins have a prognostic value in sepsis 65 and have been proposed as severity markers 15 of the disease and predictors of COVID-19 mortality. 26 , 38 These disturbances in cardiac enzymes can result from viral myocarditis, which is more frequent during the cytokine storm, myocardial damage caused by cytokines or microangiopathies and coronary spasms secondary to hypoxia. 66
Furthermore, according to our data, we observed an increase in NT-Pro-BNP levels among 25 patients (5.5%), indicating a left heart failure. Additionally, the mean NT-Pro-BNP value was higher in the critical group of patients (p=0.011). NT-Pro-BNP is a hormone secreted by the left ventricular cardiomyocytes in response to increased stretching of myocardial fibers. This biomarker helps diagnose and estimate the severity of heart failure. The mechanisms of heart failure can be attributed to an imbalance between increased cardiac output and reduced oxygen supply, with the possibility for type 2 myocardial infarction. 67 According to Li and al., 36 NT-Pro-BNP>500pg/L were significantly associated with severe forms of COVID-19, and Hong and al., 8 demonstrated that critically ill patients had higher NT-Pro-BNP levels (p=0.002). Therefore, cardiac biomarkers, including troponins and NT-Pro-BNP, can reflect cardiovascular involvement in COVID-19, since they are independent risk factors for poor prognosis and mortality. 67 , 68
Limitation
Our study has shown that some biochemical and CBC tests are important in predicting COVID-19 patients’ need for ICU care. Specifically, laboratory tests that should be prioritized to determine patient risk of developing severe COVID-19 pneumonia, include lymphopenia, NLR, thrombocytopenia, CRP, D-Dimer, creatinine, LDH, CK, troponin and NT-Pro-BNP. NLR is most preferred as it was noted to be a very good test.
However, limitations should be considered in the interpretation of our findings. First, our study is retrospective and monocentric. Second, some biological parameters were missing in the medical records and other specialized tests are not commonly performed in our hospital such as Interleukine-6 and Procalcitonin given their cost. Also, the lack of systematic data on blood gas analysis on Day 7, did not allow us to evaluate PaO2/FiO2 ratio, to define ARDS. Besides, the study did not consider the influence of pre-existing health conditions. Despite its limitations, this study has provided insights into laboratory parameters that can be used to predict the severity of COVID-19 cases allowing prediction of severe illness at the time of admission.
It is therefore recommended that healthcare providers consider these parameters in making evidence-based decisions regarding patient management especially where there are limited ICU facilities.
Conclusion
In conclusion, we have identified certain biological markers that can be used to assess the risk of COVID-19 pneumonia progressing to a critical state. In patients hospitalized with moderate to severe forms, we recommend close monitoring of leukocyte count, lymphocyte count, platelet count and CRP.
Since the beginning of the pandemic, it has been scientifically important to analyze the discriminatory capacity of hematological, biochemical, inflammatory and immunological biomarkers in patients with COVID-19, with or without a severe or critical form. Determining risk categories after the diagnosis of COVID-19 is essential for better resource allocation, improved clinical management and prevention of serious complications.
Ethical approval and consent
This retrospective study received approval from the Ethics Committee of University Hospital Center Mongi Slim, under approval number 57/2023 on Friday, 22 December 2023.
We followed strict ethical committee guidelines that allowed for exemption of consent (institute policy), due to the non-intrusive nature of the study and the use of non-identified data to ensure confidentiality and anonymity of participants. All personally identifiable data were anonymized prior to analysis to protect individuals’ privacy. We also carefully assessed the risks and benefits of our research, ensuring to minimize the former and maximize the latter for participants and the scientific community Policies regarding the future use of data and their potential sharing with other researchers were strictly established in accordance with ethical guidelines.
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; peer review: 2 approved with reservations]
Data availability
The data supporting the findings of this study are not publicly available due to ethical concerns. However, fully de-identified data will be made available upon request from reviewers and readers. Interested parties should contact the corresponding author at donia.belkhir@fmt.utm.tn to request access to the data. Data requests will be reviewed in accordance with the local ethical committee.
References
- 1. Chouikha A, Fares W, Laamari A, et al. : Molecular Epidemiology of SARS-CoV-2 in Tunisia (North Africa) through Several Successive Waves of COVID-19. Viruses. 2022 Mar 17;14(3):624. 10.3390/v14030624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. : Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 [cited 2022 Dec 17];395(10223):497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia - PMC. [cited 2023 Nov 1]. Reference Source
- 4. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. [cited 2023 Mar 7]. 10.1515/cclm-2020-0369/html [DOI] [PubMed]
- 5. Danwang C, Endomba FT, Nkeck JR, et al. : A meta-analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID-19). Biomark. Res. 2020 Aug 31 [cited 2023 Mar 24];8:37. 10.1186/s40364-020-00217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. gps_covid.pdf:[cited 2023 Nov 1]. Reference Source
- 7. Living guidance for clinical management of COVID-19:[cited 2023 Nov 1]. Reference Source
- 8. Hong W, Chen Q, Qian S, et al. : Critically Ill vs. Non-Critically Ill Patients With COVID-19 Pneumonia: Clinical Features, Laboratory Findings, and Prediction. Frontiers in Cellular and Infection. Microbiology. 2021 [cited 2022 Dec 17];11. 10.3389/fcimb.2021.550456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ning J, Yan-Nan L, Jing B, et al. : Clinical features and risk factors associated with severe COVID-19 patients in China. Chin. Med. J. 2021 Apr 20 [cited 2022 Dec 17];134(08):944–953. 10.1097/CM9.0000000000001466 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jie GW, Yi NZ, Hu Y, et al. : Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 Apr 30 [cited 2022 Dec 7];382(18):1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gemmati D, Bramanti B, Serino ML, et al. : COVID-19 and Individual Genetic Susceptibility/Receptivity: Role of ACE1/ACE2 Genes, Immunity, Inflammation and Coagulation. Might the Double X-chromosome in Females Be Protective against SARS-CoV-2 Compared to the Single X-Chromosome in Males? Int. J. Mol. Sci. 2020 May 14;21(10):3474. 10.3390/ijms21103474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reitsma MB, Fullman N, Ng M, et al. : Smoking prevalence and attributable disease burden in 195 countries and territories, 1990–2015: a systematic analysis from the Global Burden of Disease Study 2015. Lancet. 2017 May 13 [cited 2023 Oct 31];389(10082):1885–1906. 10.1016/S0140-6736(17)30819-X Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guan WJ, Liang WH, Zhao Y, et al. : Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020 May;55(5):2000547. 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frater JL, Zini G, d’Onofrio G, et al. : COVID-19 and the clinical hematology laboratory. Int. J. Lab. Hematol. 2020 [cited 2023 Oct 27];42(S1):11–18. 10.1111/ijlh.13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D, Hu B, Hu C, et al. : Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. 2020 Mar 17 [cited 2022 Oct 5];323(11):1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bivona G, Agnello L, Ciaccio M: Biomarkers for Prognosis and Treatment Response in COVID-19 Patients. Ann. Lab. Med. 2021 Nov 1;41(6):540–548. 10.3343/alm.2021.41.6.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J, Yu M, Tong S, et al. : Predictive factors for disease progression in hospitalized patients with coronavirus disease 2019 in Wuhan, China. J. Clin. Virol. 2020 Jun [cited 2023 Feb 28];127:104392. 10.1016/j.jcv.2020.104392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellan M, Patti G, Hayden E, et al. : Fatality rate and predictors of mortality in an Italian cohort of hospitalized COVID-19 patients. Sci. Rep. 2020 Nov 26 [cited 2023 Jan 20];10:20731. 10.1038/s41598-020-77698-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang I, Pranata R: Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J. Intensive Care. 2020 May 24 [cited 2023 Mar 12];8(1):36. 10.1186/s40560-020-00453-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Q, Meng M, Kumar R, et al. : Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A systemic review and meta-analysis. Int. J. Infect. Dis. 2020 Jul [cited 2022 Dec 17];96:131–135. 10.1016/j.ijid.2020.04.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sette A, Crotty S: Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 Feb 18 [cited 2023 Mar 7];184(4):861–880. 10.1016/j.cell.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan L, Wang Q, Zhang D, et al. : Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target. Ther. 2020 Mar 27;5(1):33. 10.1038/s41392-020-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippi G, Plebani M, Henry BM: Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin. Chim. Acta. 2020 Jul 1 [cited 2022 Dec 11];506:145–148. 10.1016/j.cca.2020.03.022 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vanderschueren S, De Weerdt A, Malbrain M, et al. : Thrombocytopenia and prognosis in intensive care. Crit. Care Med. 2000 Jun [cited 2023 Mar 7];28(6):1871–1876. 10.1097/00003246-200006000-00031 Reference Source [DOI] [PubMed] [Google Scholar]
- 25. Khurana D, Deoke SA: Thrombocytopenia in critically ill patients: Clinical and laboratorial behavior and its correlation with short-term outcome during hospitalization. Indian. J. Crit. Care Med. 2017 Dec [cited 2023 Mar 13];21(12):861–864. 10.4103/ijccm.IJCCM_279_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou F, Yu T, Du R, et al. : Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar 28 [cited 2022 Dec 7];395(10229):1054–1062. 10.1016/S0140-6736(20)30566-3 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Sun W, Guo Y, et al. : Association between platelet parameters and mortality in coronavirus disease 2019: Retrospective cohort study. Platelets. 2020 May 18;31(4):490–496. 10.1080/09537104.2020.1754383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang AP, Ping LJ, Qiang TW, et al. : The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020 Jul 1 [cited 2022 Dec 11];84:106504. 10.1016/j.intimp.2020.106504 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan X, Li F, Wang X, et al. : Neutrophil to lymphocyte ratio as prognostic and predictive factor in patients with coronavirus disease 2019: A retrospective cross-sectional study. J. Med. Virol. 2020 [cited 2023 Feb 28];92(11):2573–2581. 10.1002/jmv.26061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhat T, Teli S, Rijal J, et al. : Neutrophil to lymphocyte ratio and cardiovascular diseases: a review. Expert. Rev. Cardiovasc. Ther. 2013 Jan 1 [cited 2023 Mar 15];11(1):55–59. 10.1586/erc.12.159 [DOI] [PubMed] [Google Scholar]
- 31. Guthrie GJK, Charles KA, Roxburgh CSD, et al. : The systemic inflammation-based neutrophil–lymphocyte ratio: Experience in patients with cancer. Crit. Rev. Oncol. Hematol. 2013 Oct 1 [cited 2023 Mar 15];88(1):218–230. 10.1016/j.critrevonc.2013.03.010 Reference Source [DOI] [PubMed] [Google Scholar]
- 32. Azab B, Jaglall N, Atallah JP, et al. : Neutrophil-Lymphocyte Ratio as a Predictor of Adverse outcomes of Acute Pancreatitis. Pancreatology. 2011 Aug 1 [cited 2023 Mar 15];11(4):445–452. 10.1159/000331494 Reference Source [DOI] [PubMed] [Google Scholar]
- 33. Yan Q, Li P, Ye X, et al. : Longitudinal Peripheral Blood Transcriptional Analysis Reveals Molecular Signatures of Disease Progression in COVID-19 Patients. J. Immunol. 2021 May 1 [cited 2023 Mar 15];206(9):2146–2159. 10.4049/jimmunol.2001325 [DOI] [PubMed] [Google Scholar]
- 34. Liu J, Liu Y, Xiang P, et al. : Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020 May 20 [cited 2023 Jan 22];18:206. 10.1186/s12967-020-02374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu YP, Li GM, He J, et al. : Combined use of the neutrophil-to-lymphocyte ratio and CRP to predict 7-day disease severity in 84 hospitalized patients with COVID-19 pneumonia: a retrospective cohort study. Ann. Transl. Med. 2020 May [cited 2022 Aug 19];8(10):635–635. 10.21037/atm-20-2372 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Xu S, Yu M, et al. : Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020 Jul [cited 2023 Feb 28];146(1):110–118. 10.1016/j.jaci.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng Y, Ling Y, Bai T, et al. : COVID-19 with Different Severities: A Multicenter Study of Clinical Features. Am. J. Respir. Crit. Care Med. 2020 Jun 1 [cited 2022 Dec 17];201(11):1380–1388. 10.1164/rccm.202002-0445OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang D, Yin Y, Hu C, et al. : Clinical course and outcome of 107 patients infected with the novel coronavirus, SARS-CoV-2, discharged from two hospitals in Wuhan, China. Crit. Care. 2020 Apr 30 [cited 2023 Feb 28];24:188. 10.1186/s13054-020-02895-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang L, Yan X, Fan Q, et al. : D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemost. 2020 Jun;18(6):1324–1329. 10.1111/jth.14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, Yang X, Jiao H, et al. : Coagulopathy in patients with COVID-19: a systematic review and meta-analysis. Aging (Albany NY). 2020 Nov 24 [cited 2023 Mar 24];12(24):24535–24551. 10.18632/aging.104138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong GY, Ding M, Dong X, et al. : Risk factors for severe and critically ill COVID-19 patients: A review. Allergy. 2021 [cited 2023 Mar 13];76(2):428–455. 10.1111/all.14657 [DOI] [PubMed] [Google Scholar]
- 42. Price LC, McCabe C, Garfield B, et al. : Thrombosis and COVID-19 pneumonia: the clot thickens!. Eur. Respir. J. 2020 Jul 30 [cited 2023 Mar 25];56(1):2001608. 10.1183/13993003.01608-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bilaloglu S, Aphinyanaphongs Y, Jones S, et al. : Thrombosis in Hospitalized Patients With COVID-19 in a New York City Health System. JAMA. 2020 Aug 25 [cited 2023 Mar 25];324(8):799–801. 10.1001/jama.2020.13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bompard F, Monnier H, Saab I, et al. : Pulmonary embolism in patients with COVID-19 pneumonia. Eur. Respir. J. 2020 Jul 30 [cited 2023 Mar 26];56(1):2001365. 10.1183/13993003.01365-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu W, Tao ZW, Wang L, et al. : Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin. Med. J. 2020 May 5 [cited 2023 Jan 13];133(9):1032–1038. 10.1097/CM9.0000000000000775 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chalmers S, Khawaja A, Wieruszewski PM, et al. : Diagnosis and treatment of acute pulmonary inflammation in critically ill patients: The role of inflammatory biomarkers. World. J. Crit. Care Med. 2019 Sep 11 [cited 2023 Mar 22];8(5):74–96. 10.5492/wjccm.v8.i5.74 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prognostic value of C-reactive protein in patients with COVID-19 - PMC. [cited 2023 Mar 21]. Reference Source
- 48. Malik P, Patel U, Mehta D, et al. : Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid. Based Med. 2021 Jun 1 [cited 2022 Dec 17];26(3):107–108. 10.1136/bmjebm-2020-111536 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L: C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020 Jun 1 [cited 2023 Mar 21];50(4):332–334. 10.1016/j.medmal.2020.03.007 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ali N: Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J. Med. Virol. 2020 Nov [cited 2023 Mar 23];92(11):2409–2411. 10.1002/jmv.26097 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ponti G, Maccaferri M, Ruini C, et al. : Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020 Sep;57(6):389–399. 10.1080/10408363.2020.1770685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang C, Wu Z, Li JW, et al. : Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020 May [cited 2023 Mar 23];55(5):105954. 10.1016/j.ijantimicag.2020.105954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng Y, Luo R, Wang K, et al. : Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020 May;97(5):829–838. 10.1016/j.kint.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hu L, Chen S, Fu Y, et al. : Risk Factors Associated With Clinical Outcomes in 323 Coronavirus Disease 2019 (COVID-19) Hospitalized Patients in Wuhan, China. Clin. Infect. Dis. 2020 Nov 19 [cited 2023 Jan 8];71(16):2089–2098. 10.1093/cid/ciaa539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang D, Li R, Wang J, et al. : Correlation analysis between disease severity and clinical and biochemical characteristics of 143 cases of COVID-19 in Wuhan, China: a descriptive study. BMC Infect. Dis. 2020 Jul 16;20(1):519. 10.1186/s12879-020-05242-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gopaul CD, Ventour D, Thomas D: Laboratory predictors for COVID-19 Intensive Care Unit admissions in Trinidad and Tobago. Dialogues Health. 2022 Dec [cited 2023 Mar 8];1:100022. 10.1016/j.dialog.2022.100022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang G, Zhang J, Wang B, et al. : Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Respir. Res. 2020 [cited 2023 Mar 8];21:74. 10.1186/s12931-020-01338-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mardani R, Ahmadi Vasmehjani A, Zali F, et al. : Laboratory Parameters in Detection of COVID-19 Patients with Positive RT-PCR; a Diagnostic Accuracy Study. Arch. Acad. Emerg. Med. 2020 Apr 4 [cited 2023 Oct 28];8(1):e43. Reference Source [PMC free article] [PubMed] [Google Scholar]
- 59. Cai Q, Huang D, Yu H, et al. : COVID-19: Abnormal liver function tests. J. Hepatol. 2020 Sep;73(3):566–574. 10.1016/j.jhep.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fan Z, Chen L, Li J, et al. : Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin. Gastroenterol. Hepatol. 2020 Jun 1 [cited 2023 Mar 29];18(7):1561–1566. 10.1016/j.cgh.2020.04.002 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang G, Hu C, Luo L, et al. : Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J. Clin. Virol. 2020 Jun;127:104364. 10.1016/j.jcv.2020.104364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ji D, Zhang D, Xu J, et al. : Prediction for Progression Risk in Patients With COVID-19 Pneumonia: The CALL Score. Clin. Infect. Dis. 2020 Sep 15 [cited 2023 Jan 8];71(6):1393–1399. 10.1093/cid/ciaa414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang J, Meng G, Li W, et al. : Relationship of chest CT score with clinical characteristics of 108 patients hospitalized with COVID-19 in Wuhan, China. Respir. Res. 2020 [cited 2023 Oct 28];21:180. 10.1186/s12931-020-01440-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han Y, Zhang H, Mu S, et al. : Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging (Albany NY). 2020 Jun 24 [cited 2023 Mar 31];12(12):11245–11258. 10.18632/aging.103372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bessière F, Khenifer S, Dubourg J, et al. : Prognostic value of troponins in sepsis: a meta-analysis. Intensive Care Med. 2013 Jul 1 [cited 2023 Apr 1];39(7):1181–1189. 10.1007/s00134-013-2902-3 [DOI] [PubMed] [Google Scholar]
- 66. Tersalvi G, Vicenzi M, Calabretta D, et al. : Elevated Troponin in Patients With Coronavirus Disease 2019: Possible Mechanisms. J. Card. Fail. 2020 Jun;26(6):470–475. 10.1016/j.cardfail.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Battaglini D, Lopes-Pacheco M, Castro-Faria-Neto HC, et al. : Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Front. Immunol. 2022 Apr 27 [cited 2023 Mar 30];13:857573. 10.3389/fimmu.2022.857573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen T, Wu D, Chen H, et al. : Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020 Mar 26 [cited 2023 Jan 13];368:m1091. 10.1136/bmj.m1091 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]