Abstract
Tuberculosis (TB) is one of the deadliest infectious diseases globally, ranking as 13th leading cause of mortality and morbidity. According to the Global Tuberculosis Report 2022, TB claimed the lives of 1.6 million people worldwide in 2021. Among the casualties, 1 870 000 individuals with HIV co-infections contributed to 6.7% of the total fatalities, accounting TB as the second most lethal infectious disease following COVID-19. In the quest to identify biomarkers for disease progression and anti-TB therapy, microRNAs (miRNAs) have gained attention due to their precise regulatory role in gene expression in disease stages and their ability to distinguish latent and active TB, enabling the development of early TB prognostic signatures. miRNAs are stable in biological fluids and therefore will be useful for non-invasive and broad sample collection. However, their inherent lack of specificity and experimental variations may lead to false-positive outcomes. These limitations can be overcome by integrating standard protocols with machine learning, presenting a novel tool for TB diagnostics and therapeutics. This review summarizes, discusses and highlights the potential of miRNAs as a biomarker, particularly their differential expression at disease stages. The review assesses the advantages and obstacles associated with miRNA-based diagnostic biomarkers in pulmonary TB and facilitates rapid, point-of-care testing.
Keywords: miRNA diagnostic marker, miRNA, Mycobacterium tuberculosis, pulmonary tuberculosis
Introduction
Mycobacterium tuberculosis (Mtb) is the causative agent of pulmonary tuberculosis (PTB), which primarily affects the lungs (Ref. 1). Mtb has evolved multiple mechanisms to evade the antimicrobial actions of macrophages and modulate the host defence system, enabling its survival and replication within the immune cells (Ref. 2). Countries where tuberculosis (TB) is endemic still rely on conventional diagnostic techniques, despite their inherent limitations (Refs 3, 4). TB confirmatory tests are not feasible for children under the age of six (Ref. 5). Notably, culturing Mtb is time-consuming and not optimal for diagnosing extrapulmonary TB. This approach does not allow distinguishing between latent and active TB infection (Ref. 6). To achieve the objectives and to enable effective disease management and control set forth by the World Health Organization (WHO) under the ‘End TB Strategy’ by 2035 necessitates the timely and accurate detection of TB, and also requires systemic screening of high-risk populations (Ref. 7). Despite recent advancements in diagnostic technologies, accurate and prompt diagnosis remains a challenge due to the lack of an ideal TB biomarker, which explicitly identifies Mtb infection through transcriptomic, metabolic and proteomic approaches, while also distinguishing latent and active TB infection (Refs 8, 9, 10, 11, 12).
Various studies suggest that microRNAs (miRNAs) regulate a broad spectrum of immune cells such as antimicrobial activity of macrophages, cytokine production, antigen presentation, dendritic cells, cytotoxicity of NK cells, B-cell and T-cell activation and differentiation (Refs 13, 14). The importance of miRNAs in regulating autophagy, tumour necrosis factor (TNF)-α secretion and stimulating interferon (IFN)-γ during Mtb infection has been further underscored by both in vitro and in vivo studies (Refs 15, 16, 17). A study by Agarwal et al. also proved the modulation of cell-mediated immune responses by miRNAs which is an essential approach to limit the transmission of infection (Ref. 18). These findings highlight the pivotal role of miRNA in shaping the immune response against Mtb infection and offering potential targets for therapeutic interventions. miRNA is a small single-stranded RNA (~18–24 nucleotides) molecule which is highly conserved and non-coding (Ref. 13). These miRNAs play a crucial role in the negative regulation of gene expression at the post-transcriptional level by interacting with the mRNA of their target genes (Ref. 19). Mature miRNAs exhibit a robust binding affinity to the 3′-untranslated regions of target mRNAs, resulting in either mRNA degradation or inhibition of translation. Consequently, miRNAs are recognized as key epigenetic modulators (Ref. 20). The interplay between miRNAs and epigenetic mechanisms has the potential to influence the regulation of various immunological responses, both in individuals with active and latent TB, as well as in healthy individuals (Ref. 4).
miRNAs as a diagnostic biomarker for pulmonary TB diagnosis
Potential and pitfall of miRNA as a biomarker
In recent years, miRNAs have gained substantial attention as potential biomarkers for prognostic, diagnostic and therapeutic implications in the field of PTB (Ref. 21). Their resilience, tissue-specific expression patterns, and association with diverse biological processes and disease pathways render them highly appealing as a biomarker (Ref. 22). miRNAs have demonstrated significant potential as biomarkers across various sample types, including peripheral blood mononuclear cell (PBMC), serum, urine, blood, plasma, pleural fluid and pulmonary epithelial lining fluid (Ref. 23). The ability to assess miRNAs in these non-invasive samples offers the potential for identifying individuals at risk of progressing to active TB. This holds immense value for effective patient management and timely interventions. Nevertheless, despite extensive research endeavours, their suitability for clinical applications still needs to be firmly established. In addition to their ability to differentiate between latent and active TB and to identify specific TB cases with HIV co-infection (Refs 4, 24), miRNAs exhibit extensive association with diverse conditions including cancer, diabetes, heart disease, pregnancy, psoriasis and infectious disorders (Ref. 23). The versatility and broad applicability of miRNAs as biomarkers imply their potential for diagnosing and treating a diverse spectrum of diseases and medical conditions.
Delivery of miRNAs using nanoparticles holds immense potential for treating various diseases including TB and addressing the unique challenges posed by TB. In addition to targeting miRNAs to inhibit their function, significant progress has been made in the direct delivery of miRNAs to the lungs. This approach strengthens the immune response and offers a potential innovative strategy for TB treatment by specifically targeting Mtb. Studies conducted on mouse models have demonstrated the successful modulation of TGF-β1 expression, a cytokine linked to TB pathogenesis (Ref. 25). Various delivery methods, such as lentiviral vectors, lipid conjugates and small exosome-like vesicles, have been investigated for in vivo miRNA delivery. These methods are used either to restore or inhibit miRNA expression (Ref. 26). A recent study showcased the effective delivery of specific anti-miRNA molecules into B cells and upon activation, they efficiently delivered the specific anti-miRNA to antigen-activated T cells, resulting in the successful silencing of miR-195 (Ref. 27). The critical factor in implementing direct delivery lies in the careful selection of miRNAs. One potential candidate for direct delivery is miR-155, given its role as a facilitator of TLR signalling (Ref. 28) and its ability to enhance autophagy, thereby promoting the clearance of Mtb (Ref. 29).
Similarly, delivering miR-125b holds the potential to suppress TNF biosynthesis during Mtb infection. Such delivery methods have also been successfully established in pancreatic cancer and lung cancer (Refs 30, 31). The inhibitory impact of miR-99b on pro-inflammatory cytokine release during Mtb infection provides a foundation for the development of miRNA-based therapies for TB management (Ref. 32). miR-23a-5p facilitates the survival of Mtb by influencing the host's innate immune response through the modulation of NF-κB pathway and inhibits autophagy. This underscores the importance of miR-23a-5p as a therapeutic target for host immune manipulation (Ref. 33). The list of differentially expressed miRNAs involved in the Mtb pathogenesis and survival is detailed in Table 1.
Table 1.
Differentially expressed miRNAs are involved in the pathogenesis and survival of Mtb
| Biological process | miRNA (up/downregulated) |
|---|---|
| Autophagy | miR-23a-5p (up), miR-27a-5p (up), miR-31 (up), miR-33a-5p (up), miR-33b-5p (up), miR-125a-5p (up), miR-144-5p (up), miR-423-5p (up), miR-889-5p (up), miR-17-5p (down), miR-26a-5p (down) |
| Inflammation | miR-99b-5p (up), miR-125b-5p (up), miR-146a-5p (up), miR-233-3p (up), Let-7 (up) |
| Apoptosis | miR-21-5p (up), miR-27b-3p (up), miR-29 (up), miR-155-5p (up), miR-325-3p (up), miR-582-5p (up), miR-20b-5p (down), miR-145 (down) |
The human genome encompasses more than 2000 functional miRNAs, with the potential to regulate approximately one-third of human genes. This broad regulatory capacity arises from the unique ability of a single miRNA to simultaneously target multiple mRNA molecules. However, it is worth noting that mRNAs lack strict specificity toward individual genes or diseases, therefore identifying a precise miRNA signature that offers high sensitivity and specificity in accurately detecting TB is challenging. This difficulty arises from the fact that miRNA expression level varies depending on the disease stage and the host immune response (Refs 34, 35). Also, the fact that a single gene can be regulated by multiple miRNAs suggests that the administration of exogenous miRNAs may potentially lead to off-target effects (Ref. 36). This limitation emphasizes the importance of careful evaluation and optimization of miRNA-based therapies and efficacy to minimize the risk of unintended impacts. Developing standard protocols for miRNA extraction, study design, involving larger population size and considering diagnostic odds ratio becomes crucial to ensure reproducibility across various laboratory and clinical settings (Ref. 37). Discrepancies may also arise due to several factors, including variation in sample types, storage and processing methods, validation techniques, inadequate statistical evaluation during candidate selection and challenges in the data normalization (Ref. 38). The diversity of the strains, which includes widespread and multi-drug resistance strains, makes it difficult to estimate how long an infection has lasted (Ref. 39). This emphasizes the need for comprehensive approaches to study the involvement of miRNA in TB. miRNA profiles are commonly observed in inflammatory diseases, including TB. Therefore, solely relying on miRNA signatures to distinguish active TB from other inflammatory diseases can pose a substantial challenge (Ref. 22). Given this limitation, it is imperative to establish rigorous criteria and validation processes when selecting miRNA as a reliable biomarker for comprehensive TB assessment.
The multifaceted role of miRNA could potentially regulate a vast number of biological processes, for example, transcription, miRNA processing and target interaction to fine-tune gene expression (Ref. 40). One such example is the miR-155 which is a pleiotropic miRNA that plays a crucial role in the modulation of inflammatory response (Ref. 41). The expression level of miR-155 is shown to increase in Mtb-infected macrophages as well as in non-tuberculous Mycobacterium avium-intracellulare complex (MAI/MAC) infected murine and human macrophages. Interestingly, miR-155 is the established regulator of inflammation which enhances COX-2 expression in both Mtb and non-tuberculous mycobacterium (NTM) infections (Refs 42, 43, 44). Moreover, four differentially expressed serum miRNAs (hsa-miR-484, hsa-miR-584-5p, hsa-miR-625-3p and hsa-miR-4732-5p) involved in host immune response have been identified as being expressed at higher levels among non-tuberculous mycobacteria-pulmonary disease patients than healthy controls, making these as potential biomarkers for diagnosis and intervention (Ref. 45). Kim et al. via miRNA expression assay as well as gene expression analysis on macrophage from Mycobacteroides abscessus (Mab)-infected patients has found that increased expression levels of miR-144-3p lead to upregulation of proinflammatory cytokines/chemokines and enhanced intracellular growth of Mab (Ref. 46). Also, miR-125a-5p enhances autophagy by targeting the inhibition of STAT3 expression and blocks the intracellular survival of Mtb (Ref. 47). These limitations have been observed in studies of miRNA–target interactions, where a single miRNA could bind to multiple gene targets associated with NTM infections, leading to potential false-positive results.
Stability of miRNAs in biofluids and ease of detection
The preservation of miRNAs in biological fluids is a critical consideration when assessing their potential as biomarkers in individuals with TB. miRNAs offer several advantages as potential biomarkers, primarily due to the presence of protective exosomes and their resilience to RNase degradation, and freeze–thaw cycles, making them stable across various biological samples. Their stability in serum, plasma and sputum, along with their reliable measurements and high sensitivity, suggests miRNAs as valuable molecular markers for infectious diseases (Refs 48, 49). This inherent stability renders miRNAs well-suited for non-invasive diagnostic tests and promising candidates for biomarker research.
Sputum is commonly utilized for diagnosing PTB (Ref. 50). While miRNAs in sputum are relatively stable, their preservation can be affected by various factors including sample handling and processing. To ensure stability, it is recommended to store the sputum samples at low temperatures (i.e. −80 °C) while employing RNA stabilizing agents and minimizing repeated freeze–thaw cycles. Urine is easy to collect non-invasively, making it a good specimen for biomarker studies. miRNAs in urine remain stable primarily due to the presence of protective vesicles and their resistance to RNase activity, though variations in urine composition and pH have major effects on stability. Even after storing urine samples at different temperatures for five days and subjecting them to 10 freeze–thaw cycles, a considerable amount of miRNA remained, allowing for quantitative analysis despite some degradation (Ref. 51). Thus, considerable effort and attention are required to standardize the sample collection and handling procedures while considering variations in urine composition and pH to ensure reliable miRNA analysis in urine-based biomarker studies. Plasma is a rich source of extracellular vesicles, including exosomes, which play a protective role in preserving miRNA from degradation (Ref. 52). Plasma can be processed to obtain cell-free miRNA, which remains relatively stable. A study demonstrated that miR-1, miR-21 and miR-29b levels remained stable for at least 24 h at room temperature in whole blood. Storage of samples at −80 °C significantly extended the stability of miRNA. However, in long-termed stored whole blood samples (9 months), miRNA levels were significantly altered, contrasting with plasma samples where miRNA levels remained stable. It was shown that disturbance of the plasma/serum sample and repeated freeze–thaw cycles led to reduced miRNA levels (Ref. 53). Other reasons for the stability of miRNAs in blood (serum or plasma) and urine samples include protection via association with other molecules (e.g. RNA–protein complex) or modifications of the miRNAs that render them resistant to RNase activity (Ref. 54). These characteristics further enhance the suitability of miRNAs in biological fluids for biomarker analysis.
Advantages of miRNAs over current diagnostic methods
Diagnostic techniques such as ultra-high-performance liquid chromatography-tandem mass spectrometry are often used to screen plasma lipids in TB patients, lung cancer patients, community-acquired pneumonia patients and normal healthy controls (Ref. 55). A recent study by Zou et al. has assessed the value of next-generation sequencing (NGS) in combination with Xpert MTB/RIF for the early precise diagnosis of PTB (Ref. 56). Additionally, a novel approach using exosomal DNA and droplet digital PCR platforms also being used to detect Mtb DNA in clinical samples (Ref. 57).
Molecular tests, including PCR, LAMP, Xpert MTB/RIF and line probe assays are being explored alongside culture, which is an invaluable contributor to the diagnosis of female genital TB (Ref. 58). A recent review summarized various studies on Mtb exo-molecules, including protein and miRNA, and the method employed to detect exosomes in biological fluids and cell culture supernatants (Ref. 59). Another study showed the use of auramine-O-staining using LED microscopy with bleach-treated sputum samples for the detection of PTB (Ref. 60). The study also demonstrated assessing the diagnostic performance of microscopy by Ziehl Neelsen (ZN) and Auramine Staining (AO) in conjunction with Gene Xpert/CBNAAT (cartridge-based nucleic acid amplification test) for the diagnosis of PTB (Ref. 61).
To identify robust biomarkers for PTB screening, researchers have utilized the ratio-based method focusing on mitochondria-derived small RNAs in human PBMCs (Ref. 62). miRNA-based diagnosis of PTB offers distinct advantages over current diagnostic approaches and contributes to improved TB control and management. Various studies summarize the role of exo-molecules, including proteins and miRNAs, and the method employed to detect exosomes in biological fluids and cell culture supernatants (Ref. 59). Circulating miRNAs are emerging as highly promising biomarkers for TB due to their exceptional specificity, accessibility and sensitivity (Ref. 63). In response to Mtb infection, miRNAs exhibit altered gene expression patterns, aiding in the differentiation of active TB from latent TB and healthy individuals (Refs 4, 64). Their detection at very low concentrations enhances their sensitivity as biomarkers. MiRNAs can be easily extracted from various sources such as blood, or other non-invasive liquid biopsies, during the early stage of TB infection, even prior to the manifestation of clinical symptoms, which is one of the major limitations of conventional diagnostic methods (Ref. 65). This non-invasive approach reduces patient discomfort and eliminates the need for invasive procedures like bronchoscopy or lung biopsies, which are common in current diagnostic methods (Refs 65, 66).
Other non-invasive tests involve collecting samples such as exhaled breath aerosol (XBA) to detect volatile organic compounds (VOCs) using gas chromatography-mass spectrometry. The detection of VOCs in exhaled breath and condensate, which is sensitive to external and internal variables, has long been a focus of breath-based TB testing (Refs 67, 68, 69, 70). However, due to the variability in test performance, the clinical use of breath-based TB testing has not yet reached its full potential (Ref. 69). Electronic nose test has been reported to have an estimated sensitivity and specificity of 92% (95% confidence interval (CI) 82–97%) and 93% (95% CI 88–96%), respectively as compared to other VOC-based breath tests which had a wide range of sensitivity (62–100%) and specificity (11–84%) (Ref. 71). XBA appears to be a sensitive and highly specific specimen containing Mtb pathogens that can be detected using molecular diagnostic methods (Refs 69, 72). Limitations include low concentration of pathogen nucleic acids, the need for intensive technical efforts, lengthy collection periods for sampling and high cost of instrumentation (Ref. 73).
Various conventional radiological tests such as chest radiograph (CXR), computed tomography (CT), magnetic resonance imaging (MRI), 18F-fluorodeoxyglucose positron emission tomography/CT (18F-FDG PET/CT) are employed for primary diagnosis of TB (Ref. 74). CXRs are rapid imaging tools used to detect lung abnormalities, including nodules, cavities and infiltrates, which can indicate active TB disease. They are highly sensitive to PTB diagnosis (87–98%), but their accuracy can be limited, especially in extrapulmonary TB cases (Ref. 75). Jaeger et al. demonstrated the potential for discriminating between drug-resistant (DR) and drug-sensitive TB using chest X-rays through image analysis and machine learning (ML) methods, employing an artificial neural network in combination with a set of shape and texture features (Ref. 76). CT is a gold-standard medical imaging technique for detecting morphological changes in lung parenchyma. High-resolution CT showed sensitivity and specificity of 90.9 and 96.4% respectively, in identifying active PTB in smear-positive patients (Ref. 77). Although CT techniques have significantly reduced radiation dose, the level of radiation exposure remains high. MRI has been established as a radiation-free alternative to CT for several lung diseases (Ref. 78). The challenges of MRI in TB diagnosis impose susceptibility artefacts caused by low-signal intensity in normal lung tissue, low proton density and wide air–tissue interfaces. These artefacts are exacerbated by the constant movement of chest part during imaging. 18F-FDG PET/CT is a non-invasive imaging technique often used to differentiate between active and inactive PTB, as active tuberculoma exhibit a much higher standardized uptake value (SUVmax) than inactive tuberculoma. Limitations of 18F-FDG-PET/CT include the non-specificity of elevated FDG uptake, difficulty in distinguishing between physiologic from pathologic FDG uptake, and time required for adequate patient preparation, including dietary precautions and FDG biodistribution (Ref. 79). The artificial intelligence community has developed software for patient triage and diagnosis support, focusing on targets such as CXR, ultrasound images, cough and lung sounds (Ref. 80). However, challenges such as the small size and low quality of algorithm training data, as well as regulatory barrier, have hindered the adaption of these tools in clinical settings (Ref. 81).
miRNAs show high specificity for tissue or cell types, for instance, miRNA-122-5p is notably enriched in the liver (Ref. 82), while miRNA-140 is exclusively expressed in the cartilage (Ref. 83). Monitoring changes in the expression level of the tissue-or-cell-specific miRNA during treatment can help to evaluate treatment response, detect treatment failure or relapses, and guide adjustments to the treatment regimen if needed (Ref. 63). In addition, miRNAs are also associated with drug-resistance strains in TB (Ref. 4), thus, will be useful for the identification of drug-resistance strains in TB patients and for providing insights into the treatment regimens to combat DR TB. The diverse methodologies, such as qPCR, microarray analysis and NGS, are progressively becoming more accessible, rapid and cost-effective. Consequently, they are being employed for the examination of miRNA-based diagnostic and markers in disease.
Validation of miRNAs as biomarkers for pulmonary TB diagnosis
Differentially expressed miRNAs in TB patients
Various studies have examined differential expressions of miRNAs during TB infection using diverse experimental models and sample types, such as human lung tissues, PBMC, serum and macrophages. A recent study demonstrated that individuals infected with TB showed varying expression patterns of 35 miRNAs, among these 12 miRNAs were upregulated, while 23 miRNAs exhibited downregulation (Ref. 84). Remarkably, miR-17/92 cluster members (miR-17, miR-19b and miR-20a), along with miR-20b, miR-30b and miR-30d, were found to be present at lower levels in TB patients compared to healthy controls, likely due to increased adaptive immune responses during TB infection. Conversely, there was an observed increase in the abundance of miR-20b, miR-212, miR-220b and miR-650 in serum, which might reflect immune reactivity during TB infection. Previous validations of these miRNA signatures affirmed their key role in modulating host immune response or metabolic processes (Ref. 84). Precisely, downregulation of miR-20b inhibits Mtb survival in macrophages by activating the NLRP3/caspase-1/IL-1β pathway, thus mitigating the inflammatory response through increased NLRP3 expression in a TB mice model (Ref. 85). Several studies have indicated conflicting findings regarding the expression of miR-20b in TB (Refs 23, 86). While some studies have reported upregulation of miR-20b in serum and whole blood samples from TB patients, others have shown downregulation of miR-20b in serum and macrophages of TB patients (Refs 22, 87). These discrepancies highlight the need for further investigation into the exact role of miR-20b and its potential as a diagnostic biomarker.
Another comprehensive study revealed 41 differentially expressed miRNAs in PTB patients as compared to healthy individuals (Ref. 88). Various studies have identified important miRNAs related to TB, for instance, hsa-miR-150-3p, hsa-miR-150-5p, hsa-miR-874-5p and hsa-miR-941 were detected to be significant miRNAs. In the context of childhood TB, circulating miR-150, miR-146a and miR-125b were recognized as potential diagnostic markers. Furthermore, in sputum samples of TB patients compared to controls, differential expression was observed for hsa-miR-3179 and hsa-miR-19b-2, in which hsa-miR-3179 showed the most pronounced increase and hsa-miR-19b-2 exhibited a significant decrease in expression (Refs 88, 89). Another study compared the miRNA profile of THP-1 cells infected with Mtb H37Rv and Mtb H37Ra strains. The study identified differentially expressed miRNAs such as miR-30a, miR-30e, miR-155, miR-1275, miR-3665, miR-3178, miR-4484, miR-4668-5p and miR-4497. These miRNAs could potentially serve as markers to distinguish between different Mtb strains (Ref. 90). In THP-1 macrophages infected with different clinical strains of Mtb, seven miRNAs (hsa-miR-16, hsa-miR-137, hsa-miR-140-3p, hsa-miR-193a-3p, hsa-miR-501-5p, hsa-miR-598 and hsa-miR-95) showed differential expression. Most miRNAs, except hsa-miR-95, were upregulated in active TB compared to healthy controls. Two miRNAs, hsa-miR-101 and hsa-miR-150 distinguish latent TB infection (LTBI) from active TB, while hsa-miR-146b-3p and hsa-miR-296-5p were specific to LTBI group (Ref. 91).
In active PTB, a study identified 95 differentially expressed miRNA profiles in sputum samples, with 43 miRNAs being overexpressed and 52 miRNAs under-expressed. Further validation confirmed miR-19b-2 was under-expressed, while miR-3179 and miR-147 were over-expressed in the TB group (Ref. 89). Another study detected 87 differentially regulated miRNAs in the plasma of TB patients compared to healthy controls, highlighting the potential of miRNAs signature as biomarkers for active TB (Ref. 92). In the comparison between active and latent TB, seven miRNAs exhibited differential expression, with six miRNAs (hsa-miR-21, hsa-miR-223, hsa-miR-302a, hsa-miR-424, hsa-miR-451 hsa-miR-486-5p) being upregulated in active TB patients (Ref. 93). The primary human macrophages infected with Mtb strains showed differential expression of miRNAs such as miR-155, miR-146a, miR-145, miR-222, miR-27a and miR-27b (Ref. 94). It is worth noting that the expression profile of miRNA can vary depending on the specific stage of TB infection, the type of sample analysed and the experimental methodology employed. Therefore, there may be some discrepancies among different studies, though identifying statistically significant differential expressed miRNA across the studies holds promise and would be informative for identifying potential diagnostic miRNA markers (Table 2).
Table 2.
Differentially expressed miRNAs found in biofluids of tuberculosis patients in different studies
| Clinical sample | miRNA (up/down) | Functions | Ref. |
|---|---|---|---|
| Serum | miR-29a (up) | miR-29a, regulator of Wnt signalling pathway genes and lipid metabolism | (Ref. 124) |
| miR-361-5p (up), miR-576-3p (up), miR-889 (up) | miR-361-5p, targets SP-1 transcription factor, a key signalling pathway for IL-10; miR-576-3p, involved in immune system development; miR-889, is associated with respiratory system development | (Ref. 125) | |
| miR-197 (up) | Work as a biological indicator for breast cancer screening | (Ref. 126) | |
| miR-16 (up), miR-29a (up), miR-125b (up), miR-155 (up) | miR-16, modulates inflammation; miR-125b, regulates EMT; miR-155, regulates the adaptive immune response | (Ref. 127) | |
| miR-17-5p (up), miR-20b-5p (up), miR-423-5p (up) |
miR-17-5p and miR-423-5p, regulate cell proliferation; miR-20b-5p, regulates wound healing process |
(Ref. 128) | |
| miR-125b-5p (up), miR-146a-5p (up) | miR-146a-5p, regulate cancer cell proliferation. miR-146a-5p, regulating iNOS expression and NO production in macrophages upon M. bovis BCG infection | (Ref. 17) | |
| Plasma | miR-22-3p (down), miR-320a (down), miR-769-5p (down) | miR-22-3p, involves in the DNA damage machinery; miR-320a, involves in the modulation of cytokine production, cell proliferation, migration and invasion; miR-769-5p, involves in the prognosis of pancreatic cancer and non-small cell lung cancer | (Ref. 129) |
| miR-29a-3p (up), miR-155-5p (up), miR-361-5p (up) | miR-29a-3p, regulated several hallmarks of cancer including cell growth, migration, and tumour formation; miR-155-5p, key regulatory factor in the innate immunity; miR-361-5p, suppresses lung cancer progression and decreases glycolytic metabolism, proliferation, and invasion of breast cancer cell | (Ref. 95) | |
| miR-31 (up), miR-155 (up), miR-146a (down) | miR-31, regulates lipid metabolism. miR-146a regulates IL-6 production and inflammation; miR-146a, regulates IL-6 production and inflammation | (Ref. 96) | |
| PBMC | miR-155 (up) | miR-155, acts as a positive regulator of cytokine production in macrophages | (Ref. 99) |
| miR-31 (down) | miR-31, inhibition of IFN γ-mediated autophagy | (Ref. 130) | |
| miR-199b-5p (up), miR-582–5p (up), miR-892b (down) | miR-199b-5p, promotes cell proliferation, migration and suppresses apoptosis in cervical cancer cells; miR-582–5p, inhibits apoptosis of monocytes; miR-892b, tumour-suppressor in breast cancer | (Ref. 131) | |
| Urine | miR-155 (up), miR-625-3p (up) | miR-625-3p, regulates cancer cell proliferation | (Ref. 132) |
| Sputum | miR-155 (up) | miR-155, inhibits apoptosis of monocytes by targeting FOXO3 | (Ref. 49) |
Assessing the impact of miRNAs on treatment outcomes
miRNA expression profiles have the potential to predict how individuals will respond to specific treatments. To evaluate the diagnostic usefulness of miRNA, receiver operating characteristic (ROC) curves are commonly employed which provide a visual representation of the test performance. A study by Ndzi et al. showed that plasma-expressed miR-29a RNA exhibited promising diagnostic performance (84.35%) in distinguishing between active and latent TB, as well as good performance (81.37%) in distinguishing between active TB from control groups or active-TB cases in children (Ref. 95). Another study by Kathirvel et al. found miR-31, miR-155 and miR-146a demonstrated excellent diagnostic area under the ROC curve (AUC) values of 0.978, 0.953 and 0.903 (95% CI) respectively, suggesting their effectiveness as diagnostic biomarkers for detecting active-TB in children (Ref. 96). Additional ROC analysis of miR-29a-3p indicated 86% sensitivity, 73% specificity and an AUC of 0.763 (95% CI 0.668–0.858), further supporting it as a diagnostic marker for PTB (Ref. 4). The well-established immune regulatory role of miRNA-155 in macrophages due to miRNA imbalance is widely acknowledged. A comprehensive meta-analysis comprising 122 studies focused on active TB indicated the high diagnostic accuracy of miRNA-155 with a specificity of 0.85 (95% CI 0.77–0.91) and sensitivity of 0.87 (95% CI 0.76–0.93). Interestingly, these diagnostic accuracy metrics were even more pronounced in children compared to adults, implying the potential of miRNA-155 as a diagnostic marker for active TB (Ref. 97).
miRNA-378 showed elevated expression in the active TB group in contrast to the latent TB group, with an impressive AUC of 0.849 for distinguishing the normal group from the active TB group with a sensitivity of 77.42% and specificity of 79.37%. Furthermore, when it came to distinguishing the normal group from the latent TB group, miR-378 displayed an AUC of 0.898, accompanied by a sensitivity of 80.66% and specificity of 82.54%. These findings suggest that miR-378 is a promising candidate for discerning different stages of TB (Ref. 98). The study conducted by Barry et al. (Ref. 92) showed five differentially expressed miRNAs (miR-29a, miR-99b, miR-21, miR-146a and miR-652) and demonstrated their remarkable diagnostic performance. These miRNAs showed a sensitivity of 94%, a specificity of 88% and an AUC of 0.976 in distinguishing PTB from healthy controls. However, among treated patients, significant alterations in the expression of miR-29a, miR-99b, miR-26a and miR-146a were observed, highlighting their potential as novel biomarkers for PTB, with implications for monitoring treatment outcomes.
The potential of miRNAs in distinguishing active TB, LTBI and healthy individual
Distinguishing between active and latent TB and a healthy individual's infection holds significant clinical importance, as it informs appropriate treatment strategies and aids in comprehending the progression of the disease. However, accurately discerning between these two states can be a challenging task due to the similarities in symptoms and the limitations of current diagnostic methods. Several miRNAs, including miR-29a, miR-155, miR-223 and miR-99b, have consistently exhibited upregulation in active TB compared to both latent TB and healthy controls (Refs 92, 99). However, their failure to upregulate in latent TB suggests their potential role as a discriminative marker between these two conditions. MiR-29a is known to target various genes involved in immune responses, and inflammation, and modulate the extracellular matrix, suggesting their key role in the pathogenesis of active TB. The studies also revealed that elevated levels of miR-29a are associated with severe disease manifestations, such as increased lung involvement, a higher bacterial burden and less favourable treatment outcomes (Ref. 23).
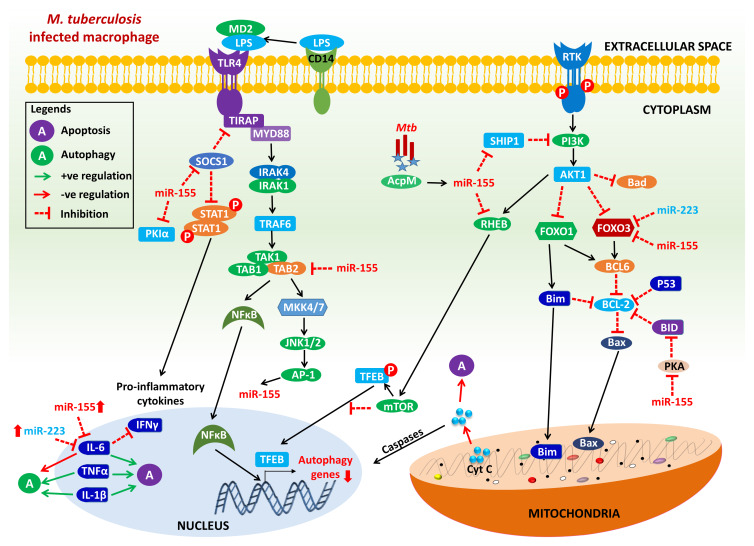
MiR-155 regulates cellular pathways within macrophages and is upregulated in active TB. This miRNA (miR-155) actively participates in the activation of macrophages and dendritic cells, fostering the release of pro-inflammatory cytokines such as TNF-α and interleukin-6, which play a pivotal role in host defence against Mtb infection (Fig. 1). Elevated levels of miR-155 have been correlated with extensive lung involvement and a higher bacterial burden (Ref. 44). Remarkably, miR-155 demonstrated superior diagnostic value for PTB and effectively distinguishes PTB from other non-PTB (Refs 44, 97). Similarly, miR-223 exhibits upregulation in active TB and is involved in regulating granulocyte differentiation, particularly in the development and maturation of neutrophils. Dysregulation of miR-223 in active TB may potentially influence neutrophil function, contributing to the altered immune response. Several studies have noted that a lower level of miR-223 is associated with severe disease manifestations (Fig. 1).
Figure 1.
Schematic representation of miRNA regulation of apoptosis and autophagy in Mtb-infected macrophage. MiR-155 and miR-223 are potential regulators of apoptosis and autophagy in M. tuberculosis. During Mtb infection, acyl carrier protein (AcpM; Rv2244) of Mtb promotes the expression of miR-155, which targets SH2 domain-containing inositol 5-phosphatase-1 (SHIP1) to activate the Akt1/mTOR pathway. The activated Akt1/mTOR signalling pathway inhibits the nuclear translocation of transcription factor EB (TFEB) and reduces the expression of autophagy and lysosomal genes, which is likely to induce antimicrobial defence and improves intracellular mycobacterial survival by inhibiting phagosome–lysosome fusion in macrophages (Ref. 133). MiR-155 can abolish expression of SHIP1, leading to activation of phosphatidylinositol 3-kinase (PI3 K) signalling-mediated inhibition of pro-apoptotic factors including Bad, FOXO-1 and FOXO-3. In addition, miR-155 and miR-223 can also directly target and suppress the expression of FOXO-3 which is not able to induce a variety of apoptotic stimuli and reducing the release of cytochrome c from mitochondria, which in turn inhibits a series of downstream biochemical reactions, mediates caspase inactivation and downregulate the apoptotic process (Ref. 134). MiR-155 is induced by TLR activation which inhibits suppressor of cytokine signaling-1 (SOCS1) and activate signal transducer and activator of transcription-1 (STAT1) to induce the production of pro-inflammatory cytokines such as IL-6, IL-1β and TNF-α (Refs 16, 135). miR-155 and miR-223 also inhibit IL-6 expression which in turn inhibits IFN-γ inducted autophagy in M. tuberculosis-infected macrophages (Refs 136, 137). MiR-155 also inhibits TAB2 which can abrogate the activation of NFκB to induce the activation of autophagy-related genes. Inhibition of TAB2 y miR-155 also leads to the suppression of JNK pathway-related proteins to regulate innate immune responses in macrophages (Refs 16, 137). MiR-155 also targets Ras homologue enriched in brain (Rheb) and mediates the positive regulation of autophagy during Mtb infection (Ref. 63). Adapted from references (Refs 16, 63, 133, 134, 135, 136, 137).
On the other hand, miR-150 is expressed in lymphocytes and holds a crucial function in the development and differentiation of both B and T cells. The reduced expression of miR-150 in active TB has an impact on lymphocyte differentiation, potentially influencing immune response against Mtb. This downregulation is associated with severe disease manifestation in active TB (Ref. 100). The expression of miRNA-146a is triggered by Mtb-induced inflammation, subsequently activating the pro-inflammatory TLR-TRAF6-IRAKI-NFκB signalling pathway. As a result, miRNA-146a plays a significant role in the pathogenesis of TB, suggesting its potential as a biomarker for detecting active PTB (Ref. 101). In children with active TB, as compared to healthy controls, there is notable overexpression of miR-21, miR-29a, miR-31 and miR-155, along with the downregulation of miR-146a. These findings suggest that these miRNAs could serve as effective diagnostic biomarkers for detecting active TB in children (Ref. 96). A study in a Chinese population has indicated that hsa-miR-451a has considerable potential for predicting active TB development from LTBI. The study also revealed that individuals with LTBI who had lower levels of hsa-miR-16-5p and hsa-miR-451a were at an increased risk of developing active TB disease (Ref. 102).
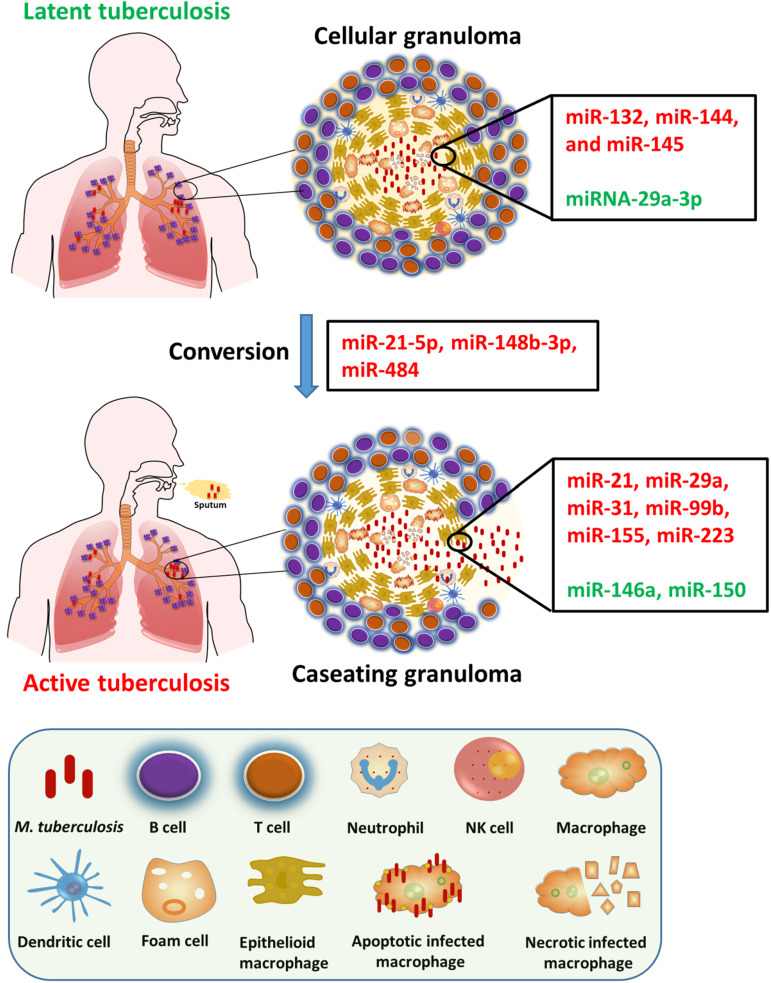
Specific miRNAs have been linked to LTBI. For instance, miR-132, miR-144 and miR-145 showed downregulation in latent TB compared to active TB and healthy control groups. In contrast, miRNA-29a-3p expression is elevated in latent TB. The increased expression of miRNA-29a-3p inhibits IFN-γ expression in T cells, dampens the immune response and may potentially increase the susceptibility to PTB. However, it is worth noting that studies have not found a correlation between miRNA-29a-3p and the IFN-γ expression (Refs 4, 103). The dysregulated expression of these miRNAs in latent TB is intricately linked to the modulation of cellular pathways involved in immune response and inflammation. These altered expressions may play a pivotal role in maintaining the latent state of TB infection. Similarly, miR-29a demonstrates a downregulation in latent TB compared to both active TB and healthy controls and plays a key role in regulating extracellular matrix remodelling and immune responses. The reduced expression miR-29a in latent TB may contribute to the maintenance of a quiescent state. Various studies have demonstrated that miR-21-5p and miR-148b-3p were upregulated in the serum of TB patients in comparison to healthy controls (Ref. 104) while miR-484 showed upregulation in serum exosomes of TB patients (Ref. 105) (Fig. 2).
Figure 2.
Dysregulated miRNA expression in active TB versus LTBI. The process of Mtb infection starts when bacilli from an active-TB patient are transmitted to a nearby host via aerosol droplets. Localized alveolar macrophages then ingest the Mtb allowing it to enter lung interstitial tissue. T and B cells, neutrophils and other immune system cells are recruited to the site of infection and converge with the infected macrophage to form a granuloma. To ensure its survival, Mtb prompt the release of both pro- and anti-inflammatory cytokines from granuloma, leading to the successful evasion of the host immune response and resulting in the state of latency or the development of active TB disease. Here, red indicates upregulated factors, while green signifies downregulated ones.
The potential of miRNAs as biomarkers for distinguishing between active and latent TB is an ongoing area of investigation, requiring further validation and refinement of these findings. Different clinical samples may have different expression of miR-155 and miR-20b, which could be due to a variety of factors that affect the results of miRNA analysis (Refs 37, 53, 106). These might consist of variations in sample collection timing, comorbidities and population variability. Other than this, the normalization techniques utilized by various reports vary, and it is still unknown how drug treatment affects miRNA responsiveness. As a result, different subjects may have various expression patterns. The results of the study may also be impacted using various data-analysis pipelines and acquisition systems for miRNA quantification. The differential expression patterns of miRNAs in these two disease states suggest their potential utility in enhancing TB diagnosis and gaining insights into disease progression.
Obstacles and progress in utilizing miRNAs for diagnostic purposes
Need for standardized protocols for miRNA-based diagnostics
The translation of miRNAs into effective diagnostic tool faces several challenges. First, the heterogeneity and specificity of miRNA expression profiles can vary significantly among different tissues and disease conditions (Ref. 22). Therefore, achieving a balance between sensitivity and specificity is crucial to ensure accurate diagnosis, considering the potential overlap of miRNA signatures between diseases. Secondly, the process of sample collection, handling and storage can introduce biasness that impact miRNA detection, potentially leading to erroneous results (Ref. 34). Notably, significant challenges arise from inconsistencies in sample collection, handling and processing such as the presence of blood cell contamination during sample preparation and the absence of consensus regarding data normalization (Ref. 107). It is essential to acknowledge that several diseases, including TB, can lead to the release of nucleic acids in blood circulation, resulting in elevated levels of circulating RNA in TB patients compared to healthy individuals (Ref. 108). Hence, when conducting analysis for the detection of circulating biomarkers, it may be more precise to use an equal volume input rather than an equivalent RNA quantity (Ref. 109). Third, the existing methods for miRNA detection, including microarray platforms and immunoassays exhibit limitations in terms of sensitivity, specificity and dynamic range due to the extremely short sequences and relatively low copy numbers of miRNAs (Ref. 110). Fourth, factors like miRNA homologues cause cross-reactivity and misidentification. The analysis of 169 specimens using culture resulted in 100% sensitivity and specificity for detecting Mtb. However, the MTB/RIF assay incorrectly assigned rifampicin resistance in 4/13 (31%) cases (Ref. 111). Hence, the establishment of standardized protocols for sample handling, rigorous validation studies and advanced bioinformatics tools becomes imperative to minimize preanalytical variations and ensure the reliability of outcomes. The integration of miRNA analysis with other diagnostic techniques, such as imaging or protein-based biomarkers, can enhance the accuracy and reliability of diagnosis (Ref. 112). This multi-modal approach may be advantageous in holding great promise for personalized medicine.
The development of innovative techniques, such as NGS and digital droplet PCR, can overcome several limitations, although challenges related to cost-effectiveness and standardization still persists. Additionally, computational challenges inherent in data analysis, and interpretation to extract meaningful insights from complex miRNA expression data demand advanced bioinformatics tools and algorithms (Ref. 113). Analysing extensive datasets to identify disease-specific miRNA signatures and distinguishing them from normal variations pose a substantial computational challenge. In recent years, various computational methods based on sequence complementarity between miRNA and the mRNAs have been developed. However, the outcomes generated by these computational methods in predicting interactions remain inconsistent and yield false-positive rates. These methods often forecast hundreds of thousands of target mRNAs for each miRNA (Ref. 114). According to studies, the estimated false-positive rate of these predictions ranges from 24 to 70% (Refs 115, 116). In the absence of high-throughput experimental techniques for miRNA target prediction, several computational approaches have emerged. Some of these methods combine expression data with sequence analysis, and the integration of miRNA and mRNA expression data has proven to be a valuable strategy for filtering sequence-based putative predictions.
Potential of miRNAs to improve accuracy and efficiency of TB diagnosis
Standardized techniques would enable the development of reliable diagnostic criteria and facilitate cross-laboratory comparisons, quality control and reproducibility while alleviating the technical constraints associated with the miRNA identification methods. Moreover, standardized procedures would streamline the integration of miRNA analysis with other diagnostic modalities, such as imaging or protein-based indicators, thereby enhancing the precision and comprehensiveness of diagnostic strategies (Ref. 112). These reliable diagnostic assays can aid in early disease identification and accurate prognosis and promote personalized medicine by understanding inter- and intra-individual drug metabolism potencies (Ref. 117). In one of the study protocols for a randomized controlled trial, the effectiveness of a standard anti-TB drug therapy regimen was evaluated for patients with multi-DR TB (STREAM). In Bangladesh, several cohort studies revealed a nine-month programme with excellent results (Ref. 118). A study published by Hoving et al., examined point-of-care ultrasound (PoCUS) performance in identifying HIV-associated TB in HIV-positive patients. They established independent PoCUS predictors of HIV-associated TB suitable for use by emergency centre practitioners, performed an external validation of the focused assessment with sonography for HIV/TB (FASH) protocol, and determined the diagnostic accuracy of individual PoCUS features (Ref. 119). For miRNA-based diagnostic testing, the FDA should demand precise procedures and thorough validation studies to enhance regulatory control. This would simplify approval processes and boost confidence in miRNA-based diagnostic methods. To encourage the integration of biomarkers into standard clinical practice, researchers must promote test utility, conduct thorough validation studies and meet specific clinical demands (Ref. 120). It is essential to use the right methods for sample collection, RNA isolation, detection and data analysis. By reducing the sample volume, it is possible to improve detection, reproducibility and cross-validation studies to harmonize the data. In a study conducted in Lima, Peru, Mtb was detected in oral swabs using Xpert MTB/RIF ULTRA (Xpert Ultra; Cepheid, CA, USA), with a sensitivity of 45% (95% CI 29–62%) and specificity of 100% (95% CI 89–100%) using liquid culture of sputum as reference test (Ref. 121).
Conclusions and future prospects
In summary, miRNAs have emerged as promising diagnostic biomarkers, despite the obstacles in heterogeneity, technological limitations and data analysis complexities. The implementation of standardized procedures is essential to tackle these challenges and create reliable and reproducible miRNA-based diagnostics. Through the adaptation of standardized approaches, we can unlock the potential of miRNAs and pave the way for their integration into routine clinical practice, ultimately enhancing patient care and outcomes. Despite the difficulties, miRNAs have the potential to revolutionize the field of TB diagnostics. Their distinctive features, such as stability, tissue specificity and presence in various biofluids, make them appealing prospects for early disease detection, monitoring treatment responses, and predicting patient outcomes. However, to fully harness the diagnostic capabilities of miRNAs, it is imperative to confront the challenges and establish standardized protocols.
ML techniques such as Support Vector Machine-Hidden Markov Model (SVM HMM), naïve Bayes and deep learning (DL) have initiated a new era of research in miRNA discovery and target prediction. They hold the promise of identifying potential disease biomarkers with high accuracy and lower false-positive rates. As a result, there is a pressing need for advancement in ML approaches within the field of miRNA research, particularly for novel identification, target prediction and functional annotation for clinical biomarkers (Ref. 122). However, many of these methods rely on predefined features that demand significant computational effort and resources, often yielding suboptimal miRNA target prediction. Recently, a novel hybrid DL-based approach has been developed to enhance miRNA target prediction accuracy. This approach integrates convolutional neural networks for capturing spatial features and recurrent neural networks for recognizing sequential features. It offers the advantage of learning both the intrinsic spatial and sequential characteristics of miRNA and its target. Notably, this represents the first study that compares various existing DL-based approaches for miRNA target prediction using the user-friendly tool, miTAR (Ref. 123). In conclusion, miRNA-based diagnostics for TB hold significant promise for the future, but their clinical implementation will necessitate further research and refinement.
Author contributions
R. A. and R. C.: conceptualization, investigation, writing – original draft, writing – review and editing; S. K. and J. M. V.: writing – review and editing; J. J. K., R. C.: supervision.
Funding statement
The authors declare that no funds, grants or other support were received during the preparation of this manuscript.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
References
- 1.Bagcchi S (2023) WHO's global tuberculosis report 2022. The Lancet Microbe 4, e20. [DOI] [PubMed] [Google Scholar]
- 2.Hmama Z et al. (2015) Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunological Reviews 264, 220–232. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins HE et al. (2017) Mortality in children diagnosed with tuberculosis: a systematic review and meta-analysis. The Lancet Infectious Diseases 17, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angria N et al. (2022) Expression of miRNA-29a-3p and IFN-γ as biomarkers in active and latent pulmonary tuberculosis. Annals of Medicine and Surgery 83, 104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joel DR et al. (2014) Diagnosis of paediatric tuberculosis using sputum induction in Botswana: programme description and findings. The International Journal of Tuberculosis and Lung Disease 18, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afsar I et al. (2018) Comparison of culture, microscopic smear and molecular methods in diagnosis of tuberculosis. Revista Espanola de Guimioterapia 31, 435. [PMC free article] [PubMed] [Google Scholar]
- 7.Floyd K et al. (2018) Global tuberculosis targets and milestones set for 2016–2035: definition and rationale. The International Journal of Tuberculosis and Lung Disease 22, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasmer RM, Nahid P and Hopewell PC (2002) Latent tuberculosis infection. New England Journal of Medicine 347, 1860–1866. [DOI] [PubMed] [Google Scholar]
- 9.Brodie D and Schluger NW (2005) The diagnosis of tuberculosis. Clinics in Chest Medicine 26, 247–271. [DOI] [PubMed] [Google Scholar]
- 10.Kunst H (2006) Diagnosis of latent tuberculosis infection: the potential role of new technologies. Respiratory Medicine 100, 2098–2106. [DOI] [PubMed] [Google Scholar]
- 11.Nahid P, Pai M and Hopewell PC (2006) Advances in the diagnosis and treatment of tuberculosis. Proceedings of the American Thoracic Society 3, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pai M, Kalantri S and Dheda K (2006) New tools and emerging technologies for the diagnosis of tuberculosis: part II. Active tuberculosis and drug resistance. Expert Review of Molecular Diagnostics 6, 423–432. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 14.Kundu M and Basu J (2021) The role of microRNAs and long non-coding RNAs in the regulation of the immune response to Mycobacterium tuberculosis infection. Frontiers in Immunology 12, 687962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L et al. (2022) MicroRNAs as immune regulators and biomarkers in tuberculosis. Frontiers in Immunology 13, 1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JK et al. (2017) MicroRNA in innate immunity and autophagy during mycobacterial infection. Cellular Microbiology 19, e12687. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarty S et al. (2019) Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis 116, 37–43. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal RG, Sharma P and Nyati KK (2019) microRNAs in mycobacterial infection: modulation of host immune response and apoptotic pathways. Immune Network 19, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anglicheau D, Muthukumar T and Suthanthiran M (2010) MicroRNAs: small RNAs with big effects. Transplantation 90, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao Q, Chen Y and Zhou X (2019) The roles of microRNAs in epigenetic regulation. Current Opinion in Chemical Biology 51, 11–17. [DOI] [PubMed] [Google Scholar]
- 21.Weiner J, Maertzdorf J and Kaufmann SHE (2013) The dual role of biomarkers for understanding basic principles and devising novel intervention strategies in tuberculosis. Annals of the New York Academy of Sciences 1283, 22–29. [DOI] [PubMed] [Google Scholar]
- 22.Sinigaglia A et al. (2020) Tuberculosis-associated microRNAs: from pathogenesis to disease biomarkers. Cells 9, 2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattnaik B et al. (2022) Micro RNAs as potential biomarkers in tuberculosis: a systematic review. Non-coding RNA Research 7, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsson O et al. (2022) Expression of microRNAs is dysregulated by HIV while Mycobacterium tuberculosis drives alterations of small nucleolar RNAs in HIV positive adults with active tuberculosis. Frontiers in Microbiology 12, 808250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosas-Taraco AG et al. (2011) Local pulmonary immunotherapy with siRNA targeting TGFβ1 enhances antimicrobial capacity in Mycobacterium tuberculosis infected mice. Tuberculosis 91, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryniarski K et al. (2013) Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. Journal of Allergy and Clinical Immunology 132, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almanza G et al. (2013) Synthesis and delivery of short, noncoding RNA by B lymphocytes. Proceedings of the National Academy of Sciences 110, 20182–20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J et al. (2014) MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Molecular Immunology 62, 29–36. [DOI] [PubMed] [Google Scholar]
- 29.Wang J et al. (2013) MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathogens 9, e1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su M-J, Aldawsari H and Amiji M (2016) Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Scientific Reports 6, 30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talekar M et al. (2016) Combination wt-p53 and microRNA-125b transfection in a genetically engineered lung cancer model using dual CD44/EGFR-targeting nanoparticles. Molecular Therapy 24, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh Y et al. (2013) Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. Journal of Biological Chemistry 288, 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu X et al. (2017) MiR-23a-5p modulates mycobacterial survival and autophagy during Mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Experimental Cell Research 354, 71–77. [DOI] [PubMed] [Google Scholar]
- 34.Condrat CE et al. (2020) miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells 9, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel EA et al. (2022) MicroRNAs as diagnostic biomarkers for tuberculosis: a systematic review and meta-analysis. Frontiers in Immunology 13, 954396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P et al. (2020) A systematic way to infer the regulation relations of miRNAs on target genes and critical miRNAs in cancers. Frontiers in Genetics 11, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moldovan L et al. (2014) Methodological challenges in utilizing miRNAs as circulating biomarkers. Journal of Cellular and Molecular Medicine 18, 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueberberg B et al. (2014) Are microRNAs suitable biomarkers of immunity to tuberculosis? Molecular and Cellular Pediatrics 1, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Espinal MA (2003) The global situation of MDR-TB. Tuberculosis 83, 44–51. [DOI] [PubMed] [Google Scholar]
- 40.Chevillet J et al. (2014) Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19, 6080–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigorito E et al. (2013) Mir-155: an ancient regulator of the immune system. Immunological Reviews 253, 146–157. [DOI] [PubMed] [Google Scholar]
- 42.Yuan Z et al. (2021) MicroRNA-155 modulates macrophages’ response to non-tuberculous mycobacteria through COX-2/PGE2 signaling. Pathogens 10, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu L et al. (2018) miR-155 modulates cockroach allergen- and oxidative stress-induced cyclooxygenase-2 in asthma. The Journal of Immunology 201, 916–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothchild AC et al. (2016) MiR-155–regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences 113, E6172–E6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han SA et al. (2020) miRNA expression profiles and potential as biomarkers in nontuberculous mycobacterial pulmonary disease. Scientific Reports 10, 3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HJ et al. (2021) MiR-144-3p is associated with pathological inflammation in patients infected with Mycobacteroides abscessus. Experimental & Molecular Medicine 53, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y et al. (2020) Levels of miR-125a-5p are altered in Mycobacterium avium-infected macrophages and associate with the triggering of an autophagic response. Microbes and Infection 22, 31–39. [DOI] [PubMed] [Google Scholar]
- 48.Ntoumou E et al. (2017) Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clinical Epigenetics 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying H et al. (2020) MicroRNA-155 from sputum as noninvasive biomarker for diagnosis of active pulmonary tuberculosis. Iranian Journal of Basic Medical Sciences 23, 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan L, Zhang Q and Xiao H (2017) Clinical diagnostic value of simultaneous amplification and testing for the diagnosis of sputum-scarce pulmonary tuberculosis. BMC Infectious Diseases 17, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mall C et al. (2013) Stability of miRNA in human urine supports its biomarker potential. Biomarkers in Medicine 7, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Q et al. (2023) A plasma 3-marker microRNA biosignature distinguishes spinal tuberculosis from other spinal destructive diseases and pulmonary tuberculosis. Frontiers in Cellular and Infection Microbiology 13, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glinge C et al. (2017) Stability of circulating blood-based microRNAs – pre-analytic methodological considerations. PLoS ONE 12, e0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell PS et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Y-S et al. (2021) Identification of potential lipid biomarkers for active pulmonary tuberculosis using ultra-high-performance liquid chromatography-tandem mass spectrometry. Experimental Biology and Medicine 246, 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou X et al. (2023) Value analysis of next-generation sequencing combined with Xpert in early precise diagnosis of pulmonary tuberculosis. Diagnostic Microbiology and Infectious Disease 107, 115921. [DOI] [PubMed] [Google Scholar]
- 57.Cho SM et al. (2020) A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clinical Microbiology and Infection 26, 942.e1–942.e5. [DOI] [PubMed] [Google Scholar]
- 58.Munne KR et al. (2020) Female genital tuberculosis in light of newer laboratory tests: a narrative review. Indian Journal of Tuberculosis 67, 112–120. [DOI] [PubMed] [Google Scholar]
- 59.Biadglegne F et al. (2021) Composition and clinical significance of exosomes in tuberculosis: a systematic literature review. Journal of Clinical Medicine 10, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gizaw N et al. (2020) The yield of Auramine O staining using led microscopy with bleach treated sputum samples for detection of pulmonary tuberculosis at St. Peter tuberculosis specialized hospital, Addis Ababa, Ethiopia. Journal of Clinical Tuberculosis and Other Mycobacterial Diseases 18, 100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma M et al. (2023) Comparison of conventional diagnostic methods with molecular method for the diagnosis of pulmonary tuberculosis. Indian Journal of Tuberculosis 70, 182–189. [DOI] [PubMed] [Google Scholar]
- 62.Ling Z et al. (2022) mtTB: a web-based R/shiny app for pulmonary tuberculosis screening. Frontiers in Cellular and Infection Microbiology 12, 850279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabir N et al. (2018) miRNAs in tuberculosis: new avenues for diagnosis and host-directed therapy. Frontiers in Microbiology 9, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chauhan D and Davuluri KS (2022) microRNAs associated with the pathogenesis and their role in regulating various signaling pathways during Mycobacterium tuberculosis infection. Frontiers in Cellular and Infection Microbiology 12, 1009901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gayosso-Gómez LV and Ortiz-Quintero B (2021) Circulating microRNAs in blood and other body fluids as biomarkers for diagnosis, prognosis, and therapy response in lung cancer. Diagnostics 11, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miotto P et al. (2013) miRNA signatures in sera of patients with active pulmonary tuberculosis. PLoS ONE 8, e80149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berna AZ and Odom John AR (2021) Breath metabolites to diagnose infection. Clinical Chemistry 68, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pham YL and Beauchamp J (2021) Breath biomarkers in diagnostic applications. Molecules 26, 5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallace MAG and Pleil JD (2018) Evolution of clinical and environmental health applications of exhaled breath research: review of methods and instrumentation for gas-phase, condensate, and aerosols. Analytica Chimica Acta 1024, 18–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh C et al. (2021) Breath-based diagnosis of infectious diseases. Clinics in Laboratory Medicine 41, 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saktiawati AMI et al. (2019) Diagnosis of tuberculosis through breath test: a systematic review. EBioMedicine 46, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duguid JP (1946) The size and the duration of air-carriage of respiratory droplets and droplet-nuclei. Epidemiology and Infection 44, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson B et al. (2020) Sensitivity optimisation of tuberculosis bioaerosol sampling. PLoS ONE 15, e0238193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arya R et al. (2024) Exploring the potential of exosomes as biomarkers in tuberculosis and other diseases. International Journal of Molecular Sciences 25, 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.World Health Organization (2013) Systematic Screening for Active Tuberculosis: Principles and Recommendations. Geneva: World Health Organization. [PubMed] [Google Scholar]
- 76.Jaeger S et al. (2018) Detecting drug-resistant tuberculosis in chest radiographs. International Journal of Computer Assisted Radiology and Surgery 13, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh JJ et al. (2012) High-resolution CT for identify patients with smear-positive, active pulmonary tuberculosis. European Journal of Radiology 81, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Busi Rizzi E et al. (2011) Detection of pulmonary tuberculosis: comparing MR imaging with HRCT. BMC Infectious Diseases 11, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pijl JP et al. (2021) Limitations and pitfalls of FDG-PET/CT in infection and inflammation. Seminars in Nuclear Medicine 51, 633–645. [DOI] [PubMed] [Google Scholar]
- 80.Mulrenan C, Rhode K and Fischer BM (2022) A literature review on the use of artificial intelligence for the diagnosis of COVID-19 on CT and chest X-ray. Diagnostics 12, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aristidou A, Jena R and Topol EJ (2022) Bridging the chasm between AI and clinical implementation. The Lancet 399, 620. [DOI] [PubMed] [Google Scholar]
- 82.Lagos-Quintana M et al. (2002) Identification of tissue-specific microRNAs from mouse. Current Biology 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 83.Miyaki S et al. (2010) MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes & Development 24, 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Z et al. (2015) Differential expression of miRNAs and their relation to active tuberculosis. Tuberculosis 95, 395–403. [DOI] [PubMed] [Google Scholar]
- 85.Lou J et al. (2017) MiR-20b inhibits Mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Experimental Cell Research 358, 120–128. [DOI] [PubMed] [Google Scholar]
- 86.Hu X et al. (2019) Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine 40, 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang D, Yi Z and Fu Y (2019) Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. Journal of Cellular Biochemistry 120, 5889–5896. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Zhu M and Hu X (2018) Integrated miRNA and mRNA expression profiling to identify mRNA targets of dysregulated miRNAs in pulmonary tuberculosis. Epigenomics 10, 1051–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi Z et al. (2012) Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS ONE 7, e43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Das K, Saikolappan S and Dhandayuthapani S (2013) Differential expression of miRNAs by macrophages infected with virulent and avirulent Mycobacterium tuberculosis. Tuberculosis 93, S47–S50. [DOI] [PubMed] [Google Scholar]
- 91.Zheng L et al. (2015) Differential microRNA expression in human macrophages with Mycobacterium tuberculosis infection of Beijing/W and non-Beijing/W strain types. PLoS ONE 10, e0126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barry SE et al. (2018) Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. Journal of Infection 77, 341–348. [DOI] [PubMed] [Google Scholar]
- 93.Wang C et al. (2011) Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS ONE 6, e25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Furci L et al. (2013) Alteration of human macrophages microRNA expression profile upon infection with Mycobacterium tuberculosis. International Journal of Mycobacteriology 2, 128–134. [DOI] [PubMed] [Google Scholar]
- 95.Ndzi EN et al. (2019) MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis 114, 69–76. [DOI] [PubMed] [Google Scholar]
- 96.Kathirvel M, Saranya S and Mahadevan S (2020) Expression levels of candidate circulating microRNAs in pediatric tuberculosis. Pathogens and Global Health 114, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X et al. (2021) Diagnostic value of microRNA-155 in active tuberculosis: a systematic review and meta-analysis. Medicine 100, e27869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun X et al. (2022) High miRNA-378 expression has high diagnostic values for pulmonary tuberculosis and predicts adverse outcomes. BMC Molecular and Cell Biology 23, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu J et al. (2012) Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Human Immunology 73, 31–37. [DOI] [PubMed] [Google Scholar]
- 100.Zhou B et al. (2007) miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proceedings of the National Academy of Sciences 104, 7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buha I et al. (2022) Association between active pulmonary tuberculosis and miRNA-146a: a preliminary study from Serbia. The Journal of Infection in Developing Countries 16, 1317–1322. [DOI] [PubMed] [Google Scholar]
- 102.Xin H et al. (2022) The association between circulating microRNAs and the risk of active disease development from latent tuberculosis infection: a nested case-control study. Microbiology Spectrum 10, e02625–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Afum-Adjei Awuah A et al. (2014) Dynamics of T-cell IFN-γ and miR-29a expression during active pulmonary tuberculosis. International immunology 26, 579–582. [DOI] [PubMed] [Google Scholar]
- 104.Wang C et al. (2018) Screening and identification of four serum miRNAs as novel potential biomarkers for cured pulmonary tuberculosis. Tuberculosis 108, 26–34. [DOI] [PubMed] [Google Scholar]
- 105.Alipoor SD et al. (2019) Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Disease Markers 2019, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mompeón A et al. (2020) Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Scientific Reports 10, 5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tiberio P et al. (2015) Challenges in using circulating miRNAs as cancer biomarkers. BioMed Research International 2015, 731479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang Z-K et al. (2018) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cellular Physiology and Biochemistry 45, 1230–1240. [DOI] [PubMed] [Google Scholar]
- 109.Zampetaki A and Mayr M (2012) Analytical challenges and technical limitations in assessing circulating miRNAs. Thrombosis and Haemostasis 108, 592–598. [DOI] [PubMed] [Google Scholar]
- 110.Kappel A and Keller A (2017) miRNA assays in the clinical laboratory: workflow, detection technologies and automation aspects. Clinical Chemistry and Laboratory Medicine (CCLM) 55, 636–647. [DOI] [PubMed] [Google Scholar]
- 111.Williamson DA et al. (2012) An evaluation of the Xpert MTB/RIF assay and detection of false-positive rifampicin resistance in Mycobacterium tuberculosis. Diagnostic Microbiology and Infectious Disease 74, 207–209. [DOI] [PubMed] [Google Scholar]
- 112.Sempere LF (2012) Recent advances in miRNA-based diagnostic applications. Expert Review of Molecular Diagnostics 12, 557–559. [DOI] [PubMed] [Google Scholar]
- 113.Shaker F et al. (2020) Web-based tools for miRNA studies analysis. Computers in Biology and Medicine 127, 104060. [DOI] [PubMed] [Google Scholar]
- 114.Alexiou P et al. (2009) Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 25, 3049–3055. [DOI] [PubMed] [Google Scholar]
- 115.Sethupathy P, Megraw M and Hatzigeorgiou AG (2006) A guide through present computational approaches for the identification of mammalian microRNA targets. Nature Methods 3, 881–886. [DOI] [PubMed] [Google Scholar]
- 116.Bentwich I (2005) Prediction and validation of microRNAs and their targets. FEBS Letters 579, 5904–5910. [DOI] [PubMed] [Google Scholar]
- 117.Nakano M and Nakajima M (2018) Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Expert Opinion on Drug Metabolism & Toxicology 14, 493–504. [DOI] [PubMed] [Google Scholar]
- 118.Nunn AJ et al. (2014) Evaluation of a standardized treatment regimen of anti-tuberculosis drugs for patients with multi-drug-resistant tuberculosis (STREAM): study protocol for a randomized controlled trial. Trials 15, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Van Hoving DJ et al. (2020) Point-of-care ultrasound predictors for the diagnosis of tuberculosis in HIV-positive patients presenting to an emergency center. Journal of Acquired Immune Deficiency Syndromes 83, 415–423. [DOI] [PubMed] [Google Scholar]
- 120.Wu AC et al. (2018) Current status and future opportunities in lung precision medicine research with a focus on biomarkers. an American Thoracic Society/National Heart, Lung, and Blood Institute Research Statement. American Journal of Respiratory and Critical Care Medicine 198, e116–e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mesman AW et al. (2019) Mycobacterium tuberculosis detection from oral swabs with Xpert MTB/RIF ULTRA: a pilot study. BMC Research Notes 12, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parveen A et al. (2020) Applications of machine learning in miRNA discovery and target prediction. Current Genomics 20, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gu T et al. (2021) miTAR: a hybrid deep learning-based approach for predicting miRNA targets. BMC Bioinformatics 22, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu Y et al. (2011) Circulating microRNAs in patients with active pulmonary tuberculosis. Journal of Clinical Microbiology 49, 4246–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qi Y et al. (2012) Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infectious Diseases 12, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abd-El-Fattah AA et al. (2013) Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochemistry and Biophysics 67, 875–884. [DOI] [PubMed] [Google Scholar]
- 127.Wagh V, Urhekar A and Modi D (2017) Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis 102, 24–30. [DOI] [PubMed] [Google Scholar]
- 128.Tu H et al. (2019) Elevated pulmonary tuberculosis biomarker miR-423-5p plays critical role in the occurrence of active TB by inhibiting autophagosome-lysosome fusion. Emerging Microbes & Infections 8, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cui J-Y et al. (2017) Characterization of a novel panel of plasma microRNAs that discriminates between Mycobacterium tuberculosis infection and healthy individuals. PLoS ONE 12, e0184113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang JX et al. (2015) Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genetics and Molecular Research 14, 17235–17243. [DOI] [PubMed] [Google Scholar]
- 131.Zhang Y et al. (2019) Integrated bioinformatics analysis and validation revealed potential immune-regulatory miR-892b, miR-199b-5p and miR-582-5p as diagnostic biomarkers in active tuberculosis. Microbial Pathogenesis 134, 103563. [DOI] [PubMed] [Google Scholar]
- 132.Wang J et al. (2018) Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerging Microbes & Infections 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paik S et al. (2022) Mycobacterial acyl carrier protein suppresses TFEB activation and upregulates miR-155 to inhibit host defense. Frontiers in Immunology 13, 946929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abdalla AE et al. (2020) Intelligent mechanisms of macrophage apoptosis subversion by Mycobacterium. Pathogens 9, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Behrouzi A et al. (2019) The role of host miRNAs on Mycobacterium tuberculosis. ExRNA 1, 40. [Google Scholar]
- 136.Dutta RK et al. (2012) IL-6 inhibits IFN-γ induced autophagy in Mycobacterium tuberculosis H37Rv infected macrophages. The International Journal of Biochemistry & Cell Biology 44, 942–954. [DOI] [PubMed] [Google Scholar]
- 137.Zhou X, Li X and Wu M (2018) miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduction and Targeted Therapy 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]