Abstract
Background
Despite a wealth of data from high-income countries, there is limited information on the distinct epidemiological patterns observed in diverse, densely populated regions within Latin America. This retrospective analysis of COVID-19’s four major waves in Bogotá, Colombia, evaluates 1.77 million cases in detail.
Methods
A comprehensive suite of statistical methods was employed. Transmission dynamics were assessed by estimating the instantaneous reproduction number , while variant-specific transmission advantages were estimated using multinomial logistic regression models. Disease severity was assessed through a suite of indicators: Hospitalisation Case Ratio (HCR), intensive care unit case ratio (ICU-CR), case fatality ratio (CFR), hospitalisation fatality ratio (HFR), and ICU fatality ratio (ICU-FR). Additionally, we analysed the distribution of hospitalisations, ICU admissions, and fatalities by age group and wave. We employed a Bayesian hierarchical model to capture epidemiological delays—such as onset-to-death, hospitalisation, and ICU admission durations to estimate hospital and ICU stay durations.
Results
Our findings reveal substantial variation in , with peaks exceeding 2.5 during the ancestral and Omicron waves. Over the course of the pandemic, we observed a 78% reduction in CFR, underscoring shifts in clinical severity. The third wave, associated with the Mu variant, recorded the highest case and death counts, alongside a decreased CFR, an elevated HFR, and a shift in the most affected age group towards younger populations. In contrast, the fourth wave, driven by the Omicron variant, exhibited the highest reproduction number and the lowest overall severity. This wave was characterised by a significant increase in pediatric hospitalisations. The study reveals a continued decline in the mean durations of hospital and ICU stays across the four waves, with hospital stays decreasing from 10.84 to 7.85 days and ICU stays dropping from 16.2 to 12.4 days.
Conclusions
This study reveals significant shifts in transmission and severity metrics—including mortality, hospitalisation, and ICU rates and stays—across age groups during Bogotá’s four COVID-19 waves. These insights underscore the value of retrospective analyses to understand the pandemic’s varied impact and inform public health strategies in diverse urban settings.
Supplementary Information
The online version contains supplementary material available at 10.1186/s44263-024-00105-x.
Keywords: COVID-19, Dynamics, Severity, Delays, Colombia
Background
Understanding the statistical characteristics of an epidemic is crucial for modelling and managing public health emergencies. During the COVID-19 pandemic, early estimates based on preliminary reports [1, 2] were crucial for projecting spreading scenarios across different geographies. This pandemic exhibited marked geographical heterogeneity, influenced initially by variations in population demographics and health system capacities, and later by the diversity in interventions and contact patterns [3]. The evolving epidemiological scenario was further complicated by the advent of novel SARS-CoV-2 variants and unequal vaccination rates, presenting challenges in comprehending the regional disease dynamics [4].
In Latin America, especially in major urban centres like Bogotá, Colombia, distinct epidemiological patterns emerged during the COVID-19 pandemic. Some of these trends were initially identified in the early phases of the outbreak [5], yet a comprehensive retrospective characterisation remains lacking. While numerous studies have detailed epidemiological parameters across various high-income countries [6, 7], there is a notable deficit in holistic retrospective analyses that integrate epidemiological, clinical, and genomic data on a global scale, particularly in the Latin American context.
Addressing this void, our study presents a nuanced statistical analysis and comparative examination of the transmission and severity of the first four COVID-19 waves in Bogotá. Covering March 2020 to April 2022, our work distinguishes itself by synthesising diverse data sources -–such as the confirmed COVID-19 case database from the District Health Secretary of Bogotá and the surveillance data published by GISAID—to elucidate the pandemic’s multifaceted dynamics in a key Latin American urban setting.
Methods
Data
Confirmed cases
The confirmed cases database of the District Health Secretary of Bogotá stores individual information on dates: symptoms onset, admission to general hospitalisation and intensive care units (ICU), discharge from hospitalisation services, and death. It also contains information on the condition of patients, the level of severity of the infection, and demographic details such as age and sex [8]. This database is maintained and updated with the daily report of confirmed cases from polymerase chain reaction and antigen tests and the information on patients' status provided by the National Epidemiology Surveillance System.
To compute stays at general hospital beds we created end dates using the following hierarchy of available dates: ICU entrance, discharge, and death. Similarly, we calculated stays in ICU creating the end dates from discharge and death dates, following the same hierarchy. To validate the estimated end dates, we recreated daily occupancy curves from the confirmed cases report. We compared them with the official report of ICU occupancy for both services available on the open data websites: Datos Abiertos Bogotá [9] and Saludata Bogotá [10].
Genomic data
The genomic surveillance data was accessed through GISAID [11, 12]. It contains the genomic sequences for SARS-CoV-2, processed by different laboratories all across the country. We classified the viral lineages using the nomenclature presented in the literature [13] and counted the sequences grouped by epidemic week, starting from the 12th week of 2021, which is the earliest date available.
Statistical methods
Start and end dates of waves
We computed the start and end date of each wave using the first derivative criteria for change of convexity applied on the daily series of new cases. For that purpose, we differentiated this series and calculated its roots using simple linear interpolation. To avoid multiple roots generated by the typical series oscillations, we smoothed both series using the Gaussian smoothing method with a kernel width of 10 days.
Reproduction number
We estimated the time-varying using the epidemiological R package EpiEstim [14]. We used the daily report of new cases from the confirmed cases database, grouped by onset date, to distinguish imported cases from local cases. We assumed an incubation period of 5 days and a serial interval of 6.48 days with a standard deviation equal to 3.83 days [15, 16]. Additionally, we estimated the delay time of the database using the percentile 0.9 of the distribution of differences between reporting and onset dates of cases. We ran these estimations for the total confirmed population in Bogotá and for adults 60 years of age or older to compute the possible effects of changes in the focalisation of massive testing strategies in Bogotá.
Transmission advantage
We evaluated the transmission advantage using a multinomial logistic regression with a single explanatory variable given by
where is the intercept of the model and is the variant-specific parameter for the time covariate, which is computed with respect to a reference (or pivot) variant. For simplicity, we chose Alpha as the pivot variant, which is the first observation in time that we have.
In general terms, the coefficients can be used to calculate the transmission advantage of a variant with respect to the pivot variant (in our case Alpha) by means of the following relation [17].
Where is the generation time of Alpha [18], and the coefficient was divided by seven to normalise its value to daily scale. Thus, we can compute the transmission advantages between any two different variants and , using the transmission rates and as follows:
Severe outcomes
We computed the HCR, ICU-CR, CFR, HFR and ICU-FR disaggregated by sub-populations where is the number of wave and the age-group. For the case ratios we used the following:
Where is the cumulative number of hospitalised patients () and the cumulative number of patients at the ICU () in a subpopulation, and is the cumulative number of cases by sub-population. On the other hand, the fatality ratios were calculated as:
In this case, and is the cumulative number of deaths given that they belong to the population [19].
We also computed the percentages of Hospitalisation, ICU admission, and deaths per age group and wave:
where is the number of cases for each outcome per wave and age group and is the total number of cases for each outcome per wave. In all cases, we estimated 95% Confidence Intervals (95% CI) using binomial proportions.
Probability distributions of epidemiological delays
We fitted the probability density functions (PDFs) to the observed distributions for onset to death, general hospitalisation, and ICU entrance; and for stays at general hospital and ICU beds. We used a Bayesian hierarchical model20 to fit the parameters of each distribution. In this order, we assumed that the set of parameters of the th wave was normally distributed as follows:
Where runs over parameters of certain PDF, is the wave, is the value of the th parameter of the PDF estimated for Bogotá and is the standard deviation which is assumed to be distributed as a truncated normal distribution. For simplicity, we assumed normal truncated distributions as prior probabilities for the parameters at the district level. All the parameters were estimated within a 95% Bayesian Credible Interval (95% BCI).
We ran all the estimations of posterior samples using the Hamiltonian Monte Carlo (HMC) algorithm implemented in Stan, setting four chains of 2000 iterations (1000 for warming up and 1000 for sampling). To get the best fitting for each epidemiological distribution, we compared the models by pairs using the Bayes Factor (BF), as follows:
Here is the evidence of the model , computed using the Laplace approximation corrected with Thermodynamic Integration [20, 21].
We fitted the multinomial regressions and the Bayesian hierarchical models using the HMC algorithm implemented in Stan. In all the cases, we used a typical number of 2000 iterations (1000 for warming up and 1000 for sampling) and sampled over 4 chains.
All methods implemented in this paper are publicly available on GitHub [22].
Results
Overview
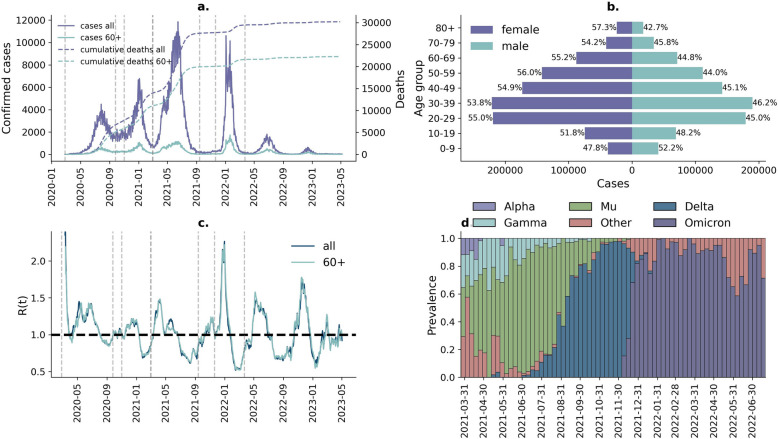
Bogotá comprises an estimated 8 million inhabitants. According to official data [23], 52% of the population is female and 48% is male. The age pyramid is composed of 13% adults older than 60 years, 53% between 18 and 59 years, and 24% younger than 18 years. The health system is composed of around 80% contributive affiliates and 20% subsidised individuals. During the COVID-19 pandemic, the city experienced about a third of the impact in terms of cases and deaths in the country. Between March 2020 and April 2022, Bogotá reported a total of around 1.77 million confirmed cases and 112,985 hospitalisations, including 38,088 ICU admissions, and 29,512 deaths associated with COVID-19. Across this period, there were four discrete “waves” of transmission (Fig. 1a, and details in Additional file 1: Table S1, also see Statistical methods section for how “waves” were defined). Across all waves, a larger number of COVID-19 cases were reported in females than in males (Fig. 1b). The highest R(t) values were registered at the beginning of the first wave and close to the peak of the fourth wave (Fig. 1c). The former is associated with the original virus lineage from Wuhan and the latter with the Omicron variant (Fig. 1d). Genomic sequencing data is only available from March 2021 onwards and shows the third wave (the largest and deadliest, with 781,276 cases and 13,188 deaths during this period) was dominated by the Mu variant. Although the Delta variant dominated between August and September 2021, this did not lead to the resurgence of transmission and caused another wave. The fourth wave was dominated by Omicron (Fig. 1d).
Fig. 1.
Overview of the COVID-19 pandemic in Bogotá: a confirmed cases (left) and cumulative deaths (right) during the pandemic; b distribution of cases by age group and sex; c instantaneous reproduction number during the pandemic; d prevalence of SARS-CoV-2 variants since March 2021. Dashed lines in panels a and c correspond to the start and end dates of the four waves
Transmission advantage
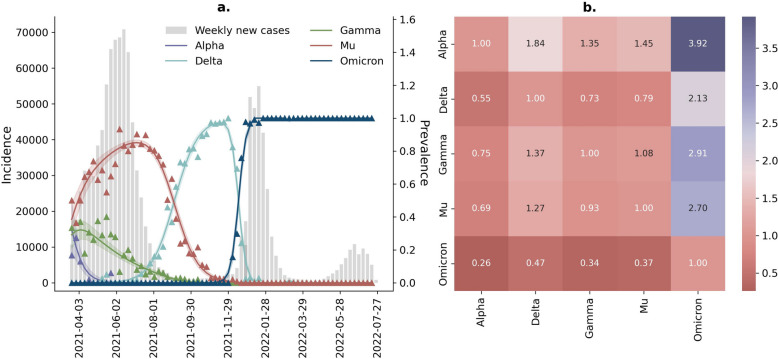
Analysis of genomic sequencing data collated from cases in Bogotá over the period from March 2021 onwards highlights a dynamic pattern of establishment and replacement of variant lineages (Fig. 2). During the initial period following the availability of sequencing data, we observed the replacement of the Gamma variant by the Mu variant (mainly associated with the third wave). The Mu variant was subsequently replaced by the Delta variant (though this establishment occurred without leading to a resurgence of transmission). Following this, Delta was replaced by the Omicron variant (which caused the fourth wave of transmission in Bogotá) (Fig. 2a).
Fig. 2.
Transmission advantage for variants detected in Bogotá since March 2021. a Multinomial regressions setting Alpha as the pivot variant (95% BCI). b Results for the transmission advantage between Alpha, Delta, Gamma, Mu, and Omicron. The first row of the heatmap contains the coefficient obtained directly for the multinomial regressions, which describes the advantage of a variant (Alpha, Delta, Gamma, Mu, and Omicron) with respect to the reference variant Alpha. Notice that the first matrix element in this diagram trivially equals zero, as well as the whole diagonal of the matrix, because there is no advantage of any variant with respect to itself
We applied a multinomial regression methodology to calculate the relative transmission of each variant (see the “Methods” section). Our analyses highlight that Delta, Gamma, Mu, and Omicron were more transmissible than Alpha, being 1.84 (95% BCI 1.63–2.13), 1.35 (95% BCI 1.19–1.56), 1.45 (95% BCI 1.28–1.68), and 3.92 (95% BCI 3.35–4.61) times more transmissible, respectively. Omicron exhibited the highest transmission advantage with respect to all the variants, being 2.13 (95% BCI 1.95–2.33), 2.91 (95% BCI 2.65–3.2), and 2.7 (95% BCI 2.46–2.96) times more transmissible than Delta, Gamma, and Mu, respectively.
Note that, after the third wave, Delta became dominant in Bogotá, leading to the extinction of Mu. This fact makes it necessarily more transmissible than Mu (1.3 times its transmission advantage). Despite this, Delta did not cause a new outbreak after the end of the third wave.
Severe outcomes
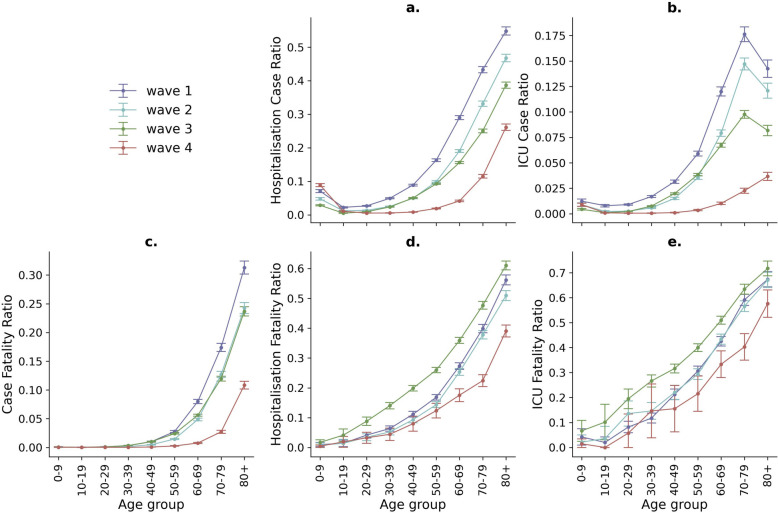
The HCR, the ICU-CR, and the CFR decreased dramatically across the four waves of the pandemic (see Fig. 3a–c).
Fig. 3.
Severity parameters per age group and wave of the COVID-19 pandemic in Bogotá (95% CI). a, b Hospitalisation case ratio (HCR) and ICU case ratio (ICU-CR); c Case fatality ratio (CFR); d, e hospitalisation fatality ratio (HFR) and ICU fatality ratio (ICU-FR)
During the first wave, the all-ages CFR was noted to be 2.70% (95% CI 2.60–2.80). In the second wave, there was a substantial decrease at 1.80% (95% CI 1.80–1.90). The third wave saw a further marginal reduction in the CFR, at 1.70% and a significant change was observed in the fourth wave, where the CFR dropped substantially to 0.60% (95% CI 0.50–0.60). It means a 78% reduction of CRF from the first to the fourth wave (See details in Additional file 1: Table S6).
Contrastingly, the HFR showed varying trends across the four waves of the pandemic. In the first wave, the HFR was 23.90% (95% CI 23.50–24.40), and slightly higher in the second wave, at 24.30% (95% CI 23.80–24.90). A notable increase was observed in the third wave, where the HFR rose to 31.00% (95% CI 30.50–31.40), while the fourth wave saw a significant decrease in the HFR, dropping to 18.90% (95% CI 18.00–19.70). Similar trends were observed for ICU-FR with the highest values during the third wave (see Fig. 3 and Additional file 1: Table S6).
The age group with the highest difference to the overall trend of the other three waves was 50–59, with an HFR and ICU-FR of 26.00% (95% CI 25.10–26.80) and 40.00% (95% CI 38.50–41.50), respectively. Interestingly, the values of the ICU-FR showed an increasing trend with age up to the 70–79 age group and then a decline for the 80 + age group during the first to the third waves (Fig. 3b).
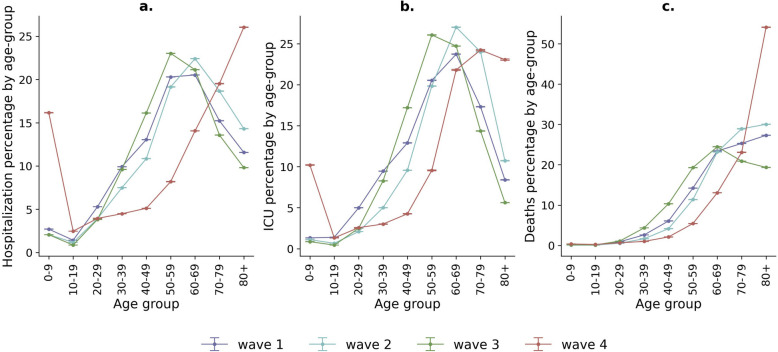
In addition to the severe outcome ratios, we calculated the percentages of the population in general hospital and ICU services, as well as the distribution of deaths by age group for each wave (Fig. 4). The overall behaviour of the first three waves (Fig. 4a–c) was similar, yet the fourth wave exhibited drastic changes. Despite a significant decrease in the number of the population presenting severe outcomes during the fourth wave (Additional file 1: Fig. S4), the percentage of children below 10 years of age in general hospitalisation and ICUs (Fig. 4a, b) increased in comparison to the other waves. Moreover, the percentage of deaths predominantly occurred in the population aged 80 years and older, reversing the decreasing trend observed in this age group during the first three waves.
Fig. 4.
Age percentage distribution of COVID-19 cases in hospitalisation, ICU services and deaths by wave (95% CI). The corresponding values for each point of the same colour sum up to 100%
It is noteworthy that, except for the fourth wave, the hospitalisation and ICU percentages per wave were highest among those aged between 50 and 69 years, and lowest for individuals under 20 years old. For example, in the third wave, the least hospitalised age group was 10–19 years, and the most hospitalised was 50–59 years, with 0.85% (95% CI 0.85–0.86) and 23.02% (95% CI 23.01–23.03) of the hospitalised cases in that wave, respectively (Additional file 1: Table S6).
Furthermore, during the fourth wave, which corresponded to a period when the Omicron variant was prevalent in the city (Fig. 1d), there was a marked increase in the percentage of hospitalised cases and ICU admissions for the age group 0–9 years. The percentages rose from 2.70% (95% CI 2.70–2.71) in the first wave to 16.17% (95% CI 16.15–16.19) in the fourth wave for general hospitalisation and from 1.32% (95% CI 1.31–1.33) in the first wave to 10.20% (95% CI 10.15–10.26) for ICU admissions. However, these drastic increases did not result in a corresponding rise in the percentage of deaths for this age group.
Conversely, the percentage of deaths among the elderly (80 + years) dramatically increased in the fourth wave to 54.11% (95% CI 54.03–54.19). This age group accounted for 928 of the 1715 deaths that occurred during the fourth wave (Additional file 1: Table S1). Still, this rise in the contribution of deaths from this age group did not reflect an increase in the total number of deaths compared to previous waves, due to the overall lower severity of the Omicron variant as indicated by the reported fatality ratios (Fig. 3).
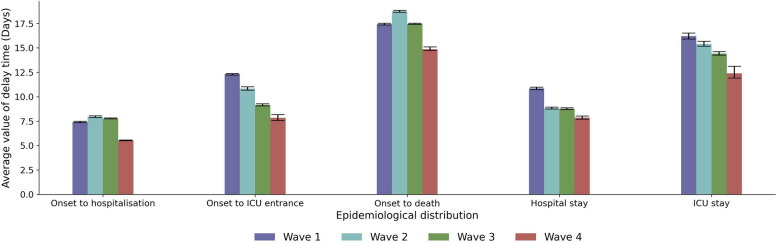
Epidemiological delay distributions
We utilised a previously developed hierarchical Bayesian framework [24] to fit different statistical distributions to data describing the delays between key epidemiological outcomes (e.g. COVID-19 symptom onset and death). Our results highlight marked variation in the different epidemiological distributions across the four waves, with a clear downward trend for the fourth wave compared to previous ones. The highest values were observed for the second wave for the parameters onset to hospitalisation, and onset to death. Also, narrower distributions were observed for the fourth wave (Fig. 5). In most cases, we found evidence in favour of the Generalised Log-Normal distribution as the best model, except for the onset-to-death distributions, for which the best model was the Gamma distribution (Additional file 1: Fig. S3 and Tables S4 and S5).
Fig. 5.
Epidemiological delay distributions. Average number of days for onset to hospitalisation, onset to ICU admission, onset to death, hospital stay, and ICU stay by wave (95% BCI)
For the time from symptoms onset to hospitalisation, mean values of around 7–8 days were observed for the first three waves, with a significant reduction for the fourth wave to 5.54 (95% BCI 5.49–5.57) days. For the time from symptoms onset to ICU admission, the mean values were reduced from the first to the fourth waves and went from 12.31 (95% BCI 12.22–12.38) days to 7.84 (95% BCI 7.55–8.17). For the time from symptoms onset to death, the mean value decreased from the first to the fourth waves, from 17.42 (95% BCI 17.33–17.5) to 14.87 (95% BCI 14.65–15.03) days.
For the duration of the hospital stay, a reduction of around 2 days, from 10.84 (95% BCI 10.72–10.98) to 8.83 (95% BCI 8.74–8.93) days, was observed from the first to the second wave. No further changes were observed for the third wave. However, a further reduction of almost 1 day, from 8.77 (95% BCI 8.69–8.86) to 7.85 (95% BCI 7.7–8.01) days, was observed for the fourth wave with respect to the third wave. Interestingly, for the duration of the ICU stay, a reduction of about 1 day was observed in each consecutive wave, going from 16.2 (95% BCI 15.91–16.52) to 15.4 (95% BCI 15.16–15.67) and to 14.41 (95% BCI 14.25–14.61), during the first, second, and third waves respectively. A further decrease of 2 days was observed for the fourth wave, for which the mean ICU stay time was 12.4 (95% BCI 11.9–13.11).
Discussion
This study constitutes the first detailed exploration of the COVID-19 pandemic’s progression in Colombia, employing an extensive dataset that includes 177 million cases, 105,831 deaths, 36,313 hospital admissions, and 28,274 ICU admissions from Bogotá between 2020 and 2022. It uniquely compares transmission and severity parameters across four distinct waves, offering critical insights into the dynamic nature of the pandemic.
The analysis of transmission, as indicated by the , revealed significant fluctuations. During the early days of the pandemic in March 2020, linked to the ancestral strain, the reached 2.8, denoting high transmission. A similar peak in transmission ( =2.7) occurred in the fourth wave, around November 2021, corresponding with the Omicron variant's emergence. Studies have reported that the attack rates for the first three waves in Bogotá were 30% (95% CI 27.3–32.8) [25], 53% (95% CI 45–62) [5], and 70–80% [26]. Genomic data available from the latter part of the second wave disclosed a sequence of variant displacement: Gamma initially overtook Alpha, followed by Mu’s dominance in the third wave, the most extensive of the pandemic. This prevalence of Mu between April and July 2021 is hypothesised to have led to a significant proportion of the local population contracting the virus. The transition from the third to the fourth wave saw the Delta variant ascend to dominance, causing the decline of Mu. This trend mirrors observations from India and Europe, underscoring Delta’s heightened transmission [27]. However, Delta's rise in Bogotá did not trigger a new outbreak, likely due to the reduced pool of susceptible individuals after the third wave. The emergence of Omicron towards the end of 2021 then swiftly replaced Delta, instigating a large wave of infections, attributable to its ability to evade immune responses.
Our findings show a transmission advantage of 2.13 (95% BCI 1.95–2.33) of Omicron over Delta, 2.91 (95% BCI 2.65–3.2) over Mu, and 2.7 (95% BCI 2.46–2.96) over Alpha. These results align with previous studies, which reported similar transmission advantages for emerging variants, such as the increased transmissibility of Alpha over the wild-type strain observed in England [28], Gamma over prior lineages in Brazil [29], Delta over previously circulating lineages in India [30], as well as a global analysis of variant transmission advantages across many countries [31]. Although our method cannot decompose the different contributing components for each variant considered here, the transmission advantage likely arose due to a combination of higher intrinsic transmissibility [32] and some immunity escape [33]. Evidence exists supporting the relevance of both features, though the comparative contributions of each to the transmission advantage likely vary depending on the variant being considered. We note that our method (which utilises information routinely collected by public health agencies conducting genomic sequencing) cannot decompose the relative contributions of transmissibility increase and immune evasion to the observed transmission advantage; this would represent a productive area of future research.
In examining severity parameters, a consistent reduction across the waves was noted in the likelihood of hospitalisation, ICU admission, or death among confirmed cases. This trend aligns with observations from other nations, such as the UK, where a shift in severity estimates from the first wave was noted, primarily due to advancements in medical care [34]. Nonetheless, some intriguing patterns in severity emerged during the third and fourth waves that are worth mentioning.
The third wave, though characterised by high case and mortality rates at the population level, exhibited a lower CFR but a higher risk of death post-hospitalisation or ICU admission. This pattern does not necessarily point to an inherently more severe Mu variant, but rather to a greater influx of severe cases into hospitals due to the largest infection surge. The strain on hospital capacity during this period reached an all-time high, as evidenced by peak hospital and ICU occupancy rates (Additional file 1: Fig. S2).
On the other hand, the fourth wave, predominantly driven by Omicron, presented the lowest values of all severity parameters during the pandemic: CFR, HCR, ICU-CR, HFR, and ICU-FR. The marked decrease in severity was accompanied by a notable reduction in delay times across various metrics (onset to hospitalisation, onset to death and hospitalisation stays), potentially reflecting the overall less severe nature of this wave. This reduction could be a consequence of both the substantial vaccination coverage (60% with at least the first dose) and the reduced severity of the Omicron variant. Intriguingly, despite lower severity metrics, this wave witnessed an increase in mortality among those over 80 and a rise in hospitalisations in the under-10 age group.
The vaccination strategy against COVID-19 in Bogotá likely played an important role in the decrease of severity parameters across waves, particularly on the fourth wave. The vaccination programme in the country only started after the second wave. The two-dose scheme in Bogotá started by age-order but covered less than 1% of the population by the beginning of the third wave and reached 30% by the end of that wave. Nonetheless, it surpassed 50% by the beginning of the fourth wave. A retrospective analysis estimated the vaccine effectiveness at 82.7% (95% CI 82.1–83.2) for preventing hospitalisation and 86.0% (95% CI 85.5–86.5) for preventing death in the country [35]. A waning in the protective effect of vaccines against symptomatic infection by Omicron was reported by a case–control study in Bogotá, although it was not reflected in the protection against hospitalisation [36]. These studies clearly show that vaccination contributed to the reduction of severity, at the population level, particularly in the fourth wave which correlates with our findings. Some studies have also demonstrated a lower severity of Omicron compared to Delta [28, 37] and a reduction of severity of subsequent infections after natural prior exposure. These factors together (vaccination, lower intrinsic severity, and prior immunity) may have contributed to a decrease in severity parameters in the fourth wave, despite an increase in variant transmission.
A high heterogeneity has been found in delay times across regions. A systematic review [38] found that South America had the longest average length of stay in hospitals at 20.85 days (95% CI 14.80–26.91), while Africa had the lowest value at 8.56 days (95% CI 1.00–22.76). Note that in our results, the length of stay in hospital was disaggregated by successive times in general hospital beds and ICU beds, with values between 7.85 and 10.84 days (95% BCI 7.7–10.98) and 12.4–16.2 days (95% BCI 11.9–16.52), respectively. The onset to death exhibited similar values during the first three waves: 17.42 days (95% BCI 17.33–17.5), 18.75 days (95% BCI 18.66–18.84), and 17.47 days (95% BCI 17.42–17.51), comparable to the average value reported in China, which was 17.8 days (95% CI 16.9–19.2) [39]. Highly heterogeneous results have been reported for this parameter in Brazil, with an average of 15.2 days (95% BCI 15.1–15.3) and highly variable across states [24]. Our estimates of the length of stay show an important decrease (of about 3 days) during the fourth wave (median 14.87, 14.65, 15.03), which was dominated by the Omicron variant and with most of the population vaccinated. Studies in Denmark have demonstrated that vaccination played a role in the reduction of this parameter [40]. In summary, the fourth wave exhibited the shortest epidemiological timelines of the pandemic. The duration from symptom onset to hospitalisation and death remained relatively stable throughout the first three waves. Meanwhile, other hospital stay durations consistently decreased from one wave to the next.
Conclusions
Overall, this study underscores the dynamic and evolving nature of the COVID-19 pandemic in Bogotá, marked by significant variations in transmission rates, severity, and clinical outcomes across the four waves. The data indicate a gradual reduction in disease severity, as evidenced by a substantial decrease in the Case Fatality Ratio (CFR) and shorter hospital and ICU stays over time. Each wave, driven by distinct SARS-CoV-2 variants, had unique impacts on different age groups and healthcare demands. The findings highlight the critical importance of ongoing surveillance and adaptive healthcare strategies to address the challenges posed by emerging variants, reinforcing the need for flexible, informed public health responses. Additionally, the study emphasises the role of collaboration between governmental and academic institutions, which has been essential for developing the technological infrastructure necessary to access comprehensive data and enable robust analysis. Such partnerships are vital for deepening our understanding of pandemic dynamics and informing future outbreak preparedness and response strategies.
Supplementary Information
Additional file 1: Table S1. Overview of confirmed cases, hospitalisations, ICUs, deaths, and maximum instantaneous reproduction number R(t) in each wave. Table S2. Relative transmission advantage between Alpha, Delta, Gamma, Mu, and Omicron (95% BCI). Table S3. Probability density functions for the estimation of epidemiological distributions. Table S4. Bayes factors for the models fitted to the epidemiological distributions. Table S5. Mean and variance of the epidemiological distributions estimated from the best model (95% BCI). Table S6. Hospitalisation Case Ratio (HCR), ICU Case Ratio (ICU-CR), Case Fatality Ratio (CFR), Hospitalisation Fatality Ratio (HFR), ICU Fatality Ratio (ICU-FR), Hospitalisation percentage (HOSP-%), ICU admission percentage (ICU-%) and Death percentage (DEATH-%) by age-group for each wave (95% CI). Figure S1. New daily confirmed cases and daily difference. In both cases, a Gaussian smoothing method was performed to avoid multiple roots around close dates. The roots selected as start and end dates of each waves are represented using black dashed lines. Figure S2. Comparison between daily occupancies reported by Saludata Bogotá [9, 10] and reconstruction made using Confirmed cases dataset a) General hospital bed occupancy. b) ICU occupancy. Figure S3. Observed delay times fitted to different probability density functions. a-d) Onset to hospitalisation, e–h) Onset to ICU, i-l) Onset to death, m-p) Stay at the general hospital, q-t) ICU stay. In every case, the cumulative distribution of the data compared to the best distribution, according to the BF, is shown on the left axis. The left axis gives the probability of occurrence of each event, while the right axis shows the cumulative probability of the best model. Figure S4. Population counts for severe outcomes, disaggregated by age. a) Hospitalisations, b) ICU, c) Deaths.
Acknowledgements
We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based. We also thank all the public health professionals who reported, cleaned, verified and submitted the information we used for this analysis.
Abbreviations
- GISAID
Global Initiative on Sharing All Influenza Data
- ICU
Intensive care unit
Instantaneous reproduction number
- CFR
Case fatality ratio
- HCR
Hospitalisation case ratio
- ICU-CR
Intensive care unit case ratio
- HFR
Hospitalisation fatality ratio
- ICU-FR
Intensive care unit fatality ratio
Probability density function
- 95% CI
95% Confidence interval
- 95% BCI
95% Bayesian credible interval
- BF
Bayes Factor
- HMC
Hamiltonian Monte Carlo
Authors’ contribution
DSQ and ZMC designed the study. DSQ and NTD prepared data for analysis and conducted data analysis and visualisations. DO, AC, FA and DM collated the data, cleaned, organised, and verified the quality of the datasets. CW advised and supervised the statistical analysis. ZMC: conceived project idea, revised analysis, administered and supervised the project. All authors read and approved the final manuscript.
Funding
DSQ, NT, and ZMC are funded in whole or in part by the TRACE-LAC project (Enhancing Tools for Response, Analytics and Control of Epidemics in Latin America and the Caribbean funded by the International Development Research Center (IDRC), grant number: 109848–002. C.W. was supported by Sir Henry Wellcome Postdoctoral Fellowship, reference 224190/Z/21/Z. This research was funded in whole, or in part, by the Wellcome Trust (reference 224190/Z/21/Z). This work was supported by the Ministerio de Ciencia, Tecnología e Innovación de Colombia, MinCiencias, AGORA Project: “Alianza para la Generación de evidencia sobre Covid-19, su Respuesta y lecciones Aprendidas para la postpandemia y futuras epidemias” (Contract No. 637–2022).
Data availability
The genomic dataset is available at https://gisaid.org (retrieval date: 2022-08-02).
GISAID Identifier: EPI_SET_240129mb
Doi: 10.55876/gis8.240129mb
All genome sequences and associated metadata in this dataset are published in GISAID’s EpiCoV database. To view the contributors of each individual sequence with details such as accession number, Virus name, Collection date, Originating Lab and Submitting Lab and the list of Authors, visit 10.55876/gis8.240129mb
EPI_SET_240129mb is composed of 5,395 individual genome sequences. The collection dates range from 2021-03-22 to 2022-07-21; Data were collected in one countries and territories;
All sequences in this dataset are compared relative to hCoV-19/Wuhan/WIV04/2019 (WIV04), the official reference sequence employed by GISAID (EPI_ISL_402124).Learn more at https://gisaid.org/WIV04
The confirmed cases dataset is available at https://datosabiertos.bogota.gov.co/ (retrieval date: 2023-05-15).
Codes for this paper are available at https://github.com/TRACE-LAC/covid19-waves-bogota/
Declarations
Ethics approval and consent to participate
This study used public, anonymised, and de-identified surveillance data from the public repository made available by the Secretary of Health of Bogotá during the COVID-19 pandemic. Ethics Approval was granted through the Research Ethics Committee of the District Secretary of Health, code SDSCTI20220004.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Linton N, et al. Incubation Period and Other Epidemiological Characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. 2020;9:538. 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker PGT, et al. The impact of COVID-19 and strategies for mitigation and suppression in low- and middle-income countries. Science. 2020;369:413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas LJ, et al. Spatial heterogeneity can lead to substantial local variations in COVID-19 timing and severity. Proc Natl Acad Sci U S A. 2020;117:24180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan WS, et al. Geographical prevalence of SARS-CoV-2 variants, August 2020 to July 2021. Sci Rep. 2022;12:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laajaj R, et al. COVID-19 spread, detection, and dynamics in Bogota, Colombia. Nat Commun. 2021;12:4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Carretero R, Vazquez-Gomez O, Gil-Prieto R, Gil-de-Miguel A. Hospitalization burden and epidemiology of the COVID-19 pandemic in Spain (2020–2021). BMC Infect Dis. 2023;23:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atherstone CJ, et al. COVID-19 Epidemiology during Delta variant dominance period in 45 high-income countries, 2020–2021. Emerg Infect Dis. 2023;29:1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Confirmed Cases Bogotá - Datos Abiertos Bogotá. https://datosabiertos.bogota.gov.co/dataset/numero-de-casos-confirmados-por-el-laboratorio-de-covid-19-bogota-d-c. Accessed 15 May 2023.
- 9.ICU occupancy Bogotá - Datos Abiertos Bogotá. https://datosabiertos.bogota.gov.co/dataset/ocupacion-de-camas-uci-covid-19-bogota-d-c. Accessed 3 Jan 2023.
- 10.General hospital occupancy Bogotá - SaluData Bogotá. https://saludata.saludcapital.gov.co/osb/wp-content/uploads/medios/Ocupacion-Hospitalizacion-COVID-19.csv. Accessed 3 Jan 2023.
- 11.GISAID. Global Initiative on Sharing All Influenza Data (GISAID). GISAID Identifier: EPI_SET_240129mb. 10.55876/gis8.240129mb. GISAIDhttps://gisaid.org/. Accessed 2 Aug 2022.
- 12.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SARS-CoV-2 variants of concern as of 14 October 2022. European Centre for Disease Prevention and Control https://www.ecdc.europa.eu/en/covid-19/variants-concern.
- 14.Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 2013;178:1505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson N, et al. Report 9: Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID19 Mortality and Healthcare Demand; 2020. https://spiral.imperial.ac.uk/handle/10044/1/77482. 10.25561/77482. [DOI] [PMC free article] [PubMed]
- 17.Davies NG, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart WS, et al. Generation time of the alpha and delta SARS-CoV-2 variants: an epidemiological analysis. Lancet Infect Dis. 2022;22:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghani AC, et al. Methods for estimating the case fatality ratio for a novel, emerging infectious disease. Am J Epidemiol. 2005;162:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X-L, Wong WH. Simulating ratios of normalizing constants via a simple identity: a theoretical exploration. Stat Sin. 1996;6:831–60. [Google Scholar]
- 21.Gelman A, Meng X-L. Simulating normalizing constants: from importance sampling to bridge sampling to path sampling. SSO Schweiz Monatsschr Zahnheilkd. 1998;13:163–85. [Google Scholar]
- 22.Four Waves of SARS-CoV-2 in Bogotá: a detailed retrospective statistical comparison. https://github.com/TRACE-LAC/covid19-waves-bogota. Accessed 14 July 2023.
- 23.Censo Nacional de Población y Vivienda - CNPV - 2018 – Datos Abiertos Colombia. https://www.datos.gov.co/Estad-sticas-Nacionales/Censo-Nacional-de-Poblaci-n-y-Vivienda-CNPV-2018/qzc6-q9qw/about_data. Accessed 2 Aug 2022.
- 24.Hawryluk I, et al. Inference of COVID-19 epidemiological distributions from Brazilian hospital data. J R Soc Interface. 2020;17:20200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercado-Reyes M, et al. Seroprevalence of anti-SARS-CoV-2 antibodies in Colombia, 2020: a population-based study. Lancet Reg Health Am. 2022;9:100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.España G, et al. The impact of vaccination strategies for COVID-19 in the context of emerging variants and increasing social mixing in Bogotá, Colombia: a mathematical modelling study. medRxiv 2021.08.06.21261734. 2021 10.1101/2021.08.06.21261734.
- 27.McCrone JT, et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature. 2022;610:154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia S, Wardle J, Nash RK, Nouvellet P, Cori A. Extending EpiEstim to estimate the transmission advantage of pathogen variants in real-time: SARS-CoV-2 as a case-study. Epidemics. 2023;44:100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faria NR, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mlcochova P, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell F, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King KL, et al. SARS-CoV-2 variants of concern Alpha and Delta show increased viral load in saliva. medRxiv. 2022 10.1101/2022.02.10.22270797. [DOI] [PMC free article] [PubMed]
- 33.Cele S, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapter 10: improvements in care of COVID-19. GOV.UK https://www.gov.uk/government/publications/technical-report-on-the-covid-19-pandemic-in-the-uk/chapter-10-improvements-in-care-of-covid-19. Accessed 8 Jan 2023.
- 35.Rojas-Botero ML, et al. Real-world effectiveness of COVID-19 vaccines among Colombian adults: a retrospective, population-based study of the ESPERANZA cohort. PLOS Glob Public Health. 2023;3:e0001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paternina-Caicedo A, et al. Comparative effectiveness and duration of protection of ChAdOx1, CoronaVac, BNT162b2, mRNA-1273, and Ad26.COV2.S COVID-19 vaccines for symptomatic and hospitalized Mu, Delta, and Omicron: a test-negative case-control study. Vaccine. 2023;41:6291–9. [DOI] [PubMed] [Google Scholar]
- 37.Hyams C, et al. Severity of Omicron (B.1.1.529) and Delta (B.1.617.2) SARS-CoV-2 infection among hospitalised adults: a prospective cohort study in Bristol, United Kingdom. Lancet Reg Health Eur. 2023;25:100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alimohamadi Y, et al. Hospital length of stay for COVID-19 patients: a systematic review and meta-analysis. Multidiscip Respir Med. 2022;17:856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verity R, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker R, et al. Length of hospital stay and risk of intensive care admission and in-hospital death among COVID-19 patients in Norway: a register-based cohort study comparing patients fully vaccinated with an mRNA vaccine to unvaccinated patients. Clin Microbiol Infect. 2022;28:871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Overview of confirmed cases, hospitalisations, ICUs, deaths, and maximum instantaneous reproduction number R(t) in each wave. Table S2. Relative transmission advantage between Alpha, Delta, Gamma, Mu, and Omicron (95% BCI). Table S3. Probability density functions for the estimation of epidemiological distributions. Table S4. Bayes factors for the models fitted to the epidemiological distributions. Table S5. Mean and variance of the epidemiological distributions estimated from the best model (95% BCI). Table S6. Hospitalisation Case Ratio (HCR), ICU Case Ratio (ICU-CR), Case Fatality Ratio (CFR), Hospitalisation Fatality Ratio (HFR), ICU Fatality Ratio (ICU-FR), Hospitalisation percentage (HOSP-%), ICU admission percentage (ICU-%) and Death percentage (DEATH-%) by age-group for each wave (95% CI). Figure S1. New daily confirmed cases and daily difference. In both cases, a Gaussian smoothing method was performed to avoid multiple roots around close dates. The roots selected as start and end dates of each waves are represented using black dashed lines. Figure S2. Comparison between daily occupancies reported by Saludata Bogotá [9, 10] and reconstruction made using Confirmed cases dataset a) General hospital bed occupancy. b) ICU occupancy. Figure S3. Observed delay times fitted to different probability density functions. a-d) Onset to hospitalisation, e–h) Onset to ICU, i-l) Onset to death, m-p) Stay at the general hospital, q-t) ICU stay. In every case, the cumulative distribution of the data compared to the best distribution, according to the BF, is shown on the left axis. The left axis gives the probability of occurrence of each event, while the right axis shows the cumulative probability of the best model. Figure S4. Population counts for severe outcomes, disaggregated by age. a) Hospitalisations, b) ICU, c) Deaths.
Data Availability Statement
The genomic dataset is available at https://gisaid.org (retrieval date: 2022-08-02).
GISAID Identifier: EPI_SET_240129mb
Doi: 10.55876/gis8.240129mb
All genome sequences and associated metadata in this dataset are published in GISAID’s EpiCoV database. To view the contributors of each individual sequence with details such as accession number, Virus name, Collection date, Originating Lab and Submitting Lab and the list of Authors, visit 10.55876/gis8.240129mb
EPI_SET_240129mb is composed of 5,395 individual genome sequences. The collection dates range from 2021-03-22 to 2022-07-21; Data were collected in one countries and territories;
All sequences in this dataset are compared relative to hCoV-19/Wuhan/WIV04/2019 (WIV04), the official reference sequence employed by GISAID (EPI_ISL_402124).Learn more at https://gisaid.org/WIV04
The confirmed cases dataset is available at https://datosabiertos.bogota.gov.co/ (retrieval date: 2023-05-15).
Codes for this paper are available at https://github.com/TRACE-LAC/covid19-waves-bogota/