Summary
The utilization of high amounts of nitrate fertilizers for crop yield leads to nitrate pollution of ground and surface waters. In this study, we report the assimilation and utilization of nitrate luxuriant levels, 20 times more than the highest N fertilizer application in Europe, by transgenic poplars overexpressing a cytosolic glutamine synthetase (GS1). In comparison with the wild‐type controls, transgenic plants grown under high N levels exhibited increased biomass (171.6%) and accumulated higher levels of proteins, chlorophylls and total sugars such as glucose, fructose and sucrose. These plants also exhibited greater nitrogen‐use efficiency particularly in young leaves, suggesting that they are able to translocate most of the resources to the above‐ground part of the plant to produce biomass. The transgenic poplar transcriptome was greatly affected in response to N availability with 1237 genes differentially regulated in high N, while only 632 genes were differentially expressed in untransformed plants. Many of these genes are essential in the adaptation and response against N excess and include those involved in photosynthesis, cell wall formation and phenylpropanoid biosynthesis. Cellulose production in the transgenic plants was fivefold higher than in control plants, indicating that transgenic poplars represent a potential feedstock for applications in bioenergy. In conclusion, our results show that GS transgenic poplars can be used not only for improving growth and biomass production but also as an important resource for potential phytoremediation of nitrate pollution.
Keywords: Populus, glutamine synthetase, transgenic trees, nitrate pollution, biomass, bioenergy
Introduction
Nitrogen (N) is a key structural component of many plant biomolecules and consequently an essential macronutrient required for plant growth and development. In spite that this element is highly abundant in nature, it is frequently a limiting factor for plant growth. Most part of N is present in the atmosphere as dinitrogen, a form of N that cannot be assimilated by plants except for those associated with N‐fixing microorganisms. Nitrate and ammonium are usually the major forms of inorganic N available to be assimilated by plants but their relative abundance in natural soils is quite low. For that reason, plants have evolved efficient and highly regulated mechanisms for N acquisition and assimilation.
Intensive N fertilization in agriculture has increased food production worldwide in the last decades, but it has also provided an excess of inorganic N that is not used by crop production. The excess reduced N is rapidly converted to nitrate by nitrification and, under anaerobic conditions, it may be denitrified to gaseous N compounds such as nitrous oxides (NOx) and emitted to the atmosphere (Schlesinger, 2009). Therefore, the massive utilization of N fertilizers has led to environmental problems with harmful effects including contamination of ground and surface waters and N‐induced eutrophication of terrestrial and aquatic systems (Galloway et al., 2008). In Europe, where nitrate fertilization dominates, nitrate pollution is a common problem in many areas with extensive agriculture. Poplar species growing in riparian zones are frequently exposed to high levels of N nutrition, mainly as nitrate, coming from streams draining agricultural lands (Rennenberg et al., 2010). The ability of poplar trees to filter nitrate and achieve substantial reductions in the concentration of nitrate in contaminated waters have been reported (O'Neill and Gordon, 1994). In fact, poplars are well adapted to nitrate acquisition (Min et al., 1998) through high‐affinity and low‐affinity nitrate transporters which encoded by a large gene family (Bai et al., 2013).
Nitrate is a major macronutrient but also acts as a signal molecule to regulate plant metabolism and development (Miller et al., 2007). Nitrate transporters are rapidly down‐regulated when plants become N replete reaching the appropriate N status (Miller et al., 2007). This is particularly important for woody perennials such as poplars, which integrate acquisition of external N with the endogenous metabolic processes of seasonal N recycling to maintain the N economy (Cantón et al., 2005; Rennenberg et al., 2010).
Once taken up and transported, nitrate is reduced to ammonium and incorporated to the pool of organic molecules in the reaction catalysed by the glutamine synthetase (GS, EC 6.3.1.2). GS plays a central role in the complex matrix of plant N metabolism as the enzyme catalyses the ATP‐dependent condensation of ammonium and glutamate to form glutamine, a precursor of all nitrogenous compounds required for plant growth (Lea and Ireland, 1999). In addition, ammonium assimilated by GS can come from various metabolic activities of N recycling such as photorespiration, deamination of phenylalanine, and the mobilization of proteins and nucleic acids during senescence or in response to pathogen attack (Cren and Hirel, 1999). A small family of nuclear genes expressed in photosynthetic and nonphotosynthetic tissues encodes GS polypeptides which are assembled into oligomeric isoenzymes located either in the cytosol or in the plastids (Bernard and Habash, 2009). In poplar, the GS family is organized in four groups of duplicated genes, three of which code for cytosolic GS isoforms (GS1.1, GS1.2 and GS1.3) and one that codes for the plastidic GS isoform (GS2) (Castro‐Rodríguez et al., 2011). Recent results support that GS gene redundancy may contribute to the homoeostasis of N metabolism in functions associated with changes in glutamine use in multiple metabolic pathways (Castro‐Rodriguez et al., 2015).
Consistent with the central role of GS in N metabolism, increased growth was observed in transgenic poplars overexpressing constitutively a pine GS gene (Fu et al., 2003; Gallardo et al., 1999; Jing et al., 2004). Furthermore, enhanced GS expression in poplar resulted in enhanced efficiency in N assimilation (Man et al., 2005) and altered wood chemistry (Coleman et al., 2012).
In this study, we report assimilation and utilization of luxuriant levels of N by transgenic poplar overexpressing a pine cytosolic GS. Furthermore, transgenic poplars accumulate enhanced levels of cellulose in the aerial part of the plant. Poplar species are well suited for use in phytoremediation of a variety of environmental pollutants such as heavy metals, pesticides and waste products (Yadav et al., 2010). Our results strongly support that GS poplars have improved potential for environmental remediation of polluted areas and at the same time they can be used as feedstocks for the biofuels or fibre markets. Field trials are necessary to assess the extent of this potential.
Results
Vegetative growth of transgenic poplars under high nitrate levels
Transgenic hybrid poplar (P. tremula × P. alba) plants exhibited higher growth than untransformed controls (WT) under adequate (10 mm) and luxuriant (50 mm) levels of nitrate, especially in the aerial part (Figure 1; Table 1). A total of 12 plants were analysed per treatment. During the 3‐month growth period, plants grown under high nitrate (50 mm) showed greater vegetative growth; the fresh weight in the aerial part increased by 85.5% in controls and 193% in transgenic plants; meanwhile, dry weight (DW) increased by 53.5% in controls and 171.6% in the transgenics (Table 1). Transgenic plants exhibited root biomass increases under high nitrate supplementation of 44.5% in fresh weight and 27.9% increases in DW; in WT plants, main roots showed an 80% increase in fresh weight when were grown under high nitrate, whereas root fresh weight decreased (approximately 20%) in response to adequate 10 mm nitrate supply (Table 1).
Figure 1.
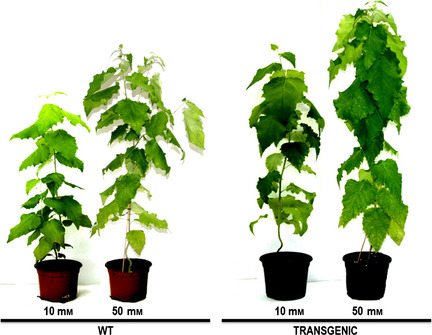
Phenotypes of 3‐month‐old untransformed controls (WT) and transgenic poplars irrigated with a solution containing adequate (10 mm) or high (50 mm) nitrate concentration.
Table 1.
Biomass accumulation in control and transgenic poplars under nitrate nutrition
| WT | Transgenic | ||||||
|---|---|---|---|---|---|---|---|
| Sample | Nitrate (mm) | Length (cm) | FW (g) | DW (g) | Length (cm) | FW (g) | DW (g) |
| Aerial Plant | 10 | 31.84 ± 0.70 | 5.40 ± 0.12cde | 2.28 ± 0.07cd | 39.16 ± 1.03 | 7.61 ± 0.19bc | 3.10 ± 0.10bc |
| Main Roots | 2.10 ± 0.28 | 0.78 ± 0.10 | 2.36 ± 0.98 | 1.04 ± 0.43 | |||
| Lateral Roots | 6.32 ± 1.47cd | 1.33 ± 0.42 | 4.72 ± 0.97 | 1.50 ± 0.59 | |||
| Aerial Plant | 50 | 51.62 ± 1.09 | 10.02 ± 0.30b | 3.51 ± 0.16b | 63.63 ± 1.20a | 22. 35 ± 1.12a | 8.42 ± 0.27a |
| Main Roots | 3.78 ± 0.65 | 0.82 ± 0.30 | 4.00 ± 0.82 | 1.71 ± 0.50 | |||
| Lateral Roots | 5.31 ± 0.44cde | 1.12 ± 0.50 | 6.23 ± 1.44cde | 1.54 ± 0.46 | |||
The parameters length, fresh weight (FW) and dry weight (DW) were compared between WT and transgenics grown under different concentrations of nitrate. Means with different lowercase letters are significantly different, P < 0.01 (ANOVA and Tukey's test).
Total N and C levels in transgenic plants reflect enhanced ability to assimilate nitrate
To further study the increased growth of transgenic plants under high nitrate, total N and C contents, nitrogen‐utilization efficiency (NUtE) and nitrogen‐uptake efficiency (NUpE) were examined in the same samples in which the biomass analyses were performed. Analyses were performed for three regions of the aerial part of the plants (L1/S1, L2/S2 and L3/S3) and two regions of the roots (R1 and R2) (Figure 2). Similar N and C contents were observed in WT and transgenic plants growing under 10 mm nitrate. In contrast, higher levels were observed in the transgenic plants at 50 mm nitrate. These increases were mainly observed in young leaves. Increases of eightfold in N content and sixfold in C content were observed in young leaves of transgenic poplar at 50 mm nitrate.
Figure 2.
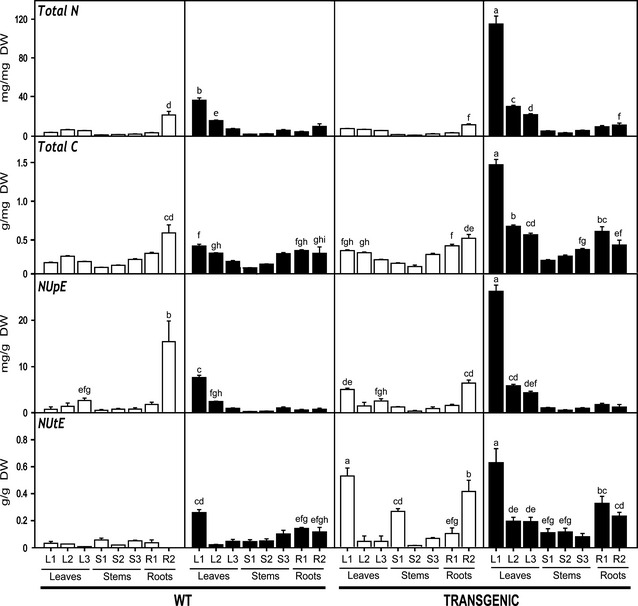
Spatial distribution of total N, total C, NutE and NupE in WT and transgenic poplars growing at different nitrate levels. Open bars correspond to poplars irrigated with a solution containing 10 mm of nitrate. Closed bars correspond to poplars irrigated with a solution containing 50 mm of nitrate. Values are means ± SD of three independent plant samples. Different letters indicate significant differences between samples at P < 0.001. The same notation is applicable to legends of Figures 3, 7 and 8.
The parameter NUtE illustrates the use of N to produce biomass during growth under different N regimes (Good et al., 2004). Thus, N availability differentially affected biomass accumulation in WT and transgenic plants. Transgenic plants accumulated more biomass than the WT in nearly all tissues, especially in young leaves. Significant differences between WT and transgenic plants under 10 mm nitrate were observed in young leaves, stems and secondary roots. In WT plants, increased NUtE at high nitrate was only observed in young leaves, whereas values remained unaltered in other plant regions.
The parameter NUpE showed the differences in N uptake and N metabolism between WT and transgenic plants under adequate and high N levels (Figure 2). Transgenic plants exhibited greater ability to remove N excess from soil and move it to leaves, particularly to young leaves. Enhanced N allocation was also observed in L2 (intermediate leaves) and L3 (mature leaves). WT plants under high nitrate showed limited ability to remove N excess when compared to the transgenics. Interestingly, enhanced N‐uptake ability was observed in secondary roots of WT plants under adequate N availability.
Changes in soluble protein and chlorophyll contents in response to N excess
To further characterize the response of transgenic poplar to nitrate excess, protein and chlorophyll contents were determined (Figure 3). Compared with 10 mm nitrate conditions, both WT and transgenic plants grown on 50 mm nitrate exhibited greater levels of protein per fresh weight with the higher values exclusively found in young leaves and stems. Interestingly, young leaves of transgenic plants grown under nitrate excess accumulated significantly higher protein levels than WT plants grown under the same conditions. When compared to untransformed controls, transgenic plants also accumulated greater contents of chlorophylls a and b in their leaves under high nitrate. The accumulation of chlorophylls was significantly higher in young leaves of transgenic plants.
Figure 3.
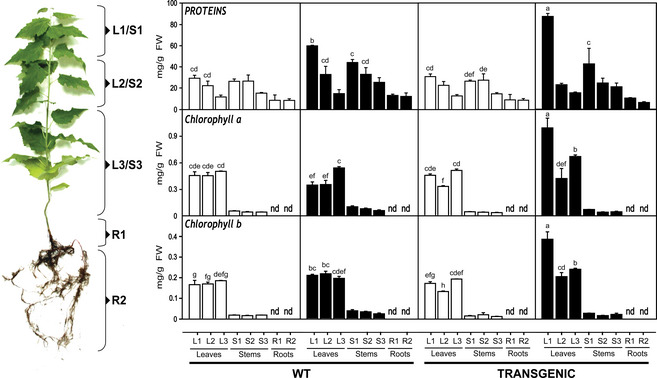
Spatial distribution of protein and chlorophyll contents in WT and transgenic poplars growing at different nitrate levels.
Changes in the transcriptome in response to N availability
The above data showed that the most significant changes that might explain the observed growth differences between WT and transgenic trees occurred in the young leaves under high N. To understand the molecular basis for these differences, transcriptomic studies in young leaves of plants grown at 10 and 50 mm nitrate were performed. Changes in the leaf transcriptome were more pronounced in the transgenics than in the WT (Figure 4, compare 50 mm WT vs 50 mm Transgenics and 10 mm WT vs 10 mm Transgenics; Table S2). Figure 4 also shows that 213 genes are in common between WT and transgenic plants in their response at high nitrate levels. Table 2 shows that a total of 1237 genes were a minimum of twofold differentially expressed in the transgenic leaves, 826 up‐regulated and 411 down‐regulated. In contrast, only 632 genes were differentially regulated in response to nitrate excess in untransformed controls, although a similar proportion of up‐regulated and down‐regulated genes were observed. Changes in the leaf transcriptome were also observed between untransformed and transgenic plants grown at high nitrate (Figure 4, 50 mm WT vs 50 mm Transgenics; Table S2). However, lower differences in the number of genes differentially expressed were observed when untransformed and transgenic plants were grown at adequate levels of nitrate (Figure 4, 10 mm WT vs 10 mm Transgenics; Table S2). Table 2 shows that 1346 genes were differentially expressed in the leaves of transgenics with respect to WT at nitrate excess. The proportion of up‐regulated and down‐regulated genes was similar to that of observed in the transgenics when grown at high nitrate. However, only 631 genes were differentially expressed in the transgenic plants when grown at adequate levels of nitrate with 391 up‐regulated and 240 down‐regulated genes (Table 2).
Figure 4.
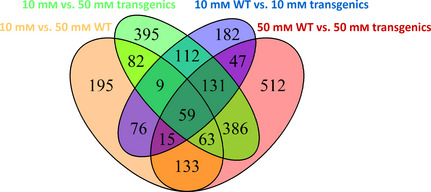
Changes in the transcriptome of WT and transgenic poplars grown at different nitrate levels. Venn diagram depicting the overlap between genes differentially expressed in 10 mm vs 50 mm WT, 10 mm vs 50 mm Transgenics, 50 mm WT vs 50 mm Transgenics and 10 mm WT vs 10 mm Transgenics.
Table 2.
Differentially expressed genes in control and transgenic poplar leaves in response to high nitrate nutrition
| Condition | Total | Up‐regulated | Down‐regulated |
|---|---|---|---|
| 10 mm vs 50 mm WT | 632 | 428 | 204 |
| 10 mm vs 50 mm transgenics | 1237 | 826 | 411 |
| 50 mm WT vs 50 mm transgenics | 1346 | 856 | 490 |
| 10 mm WT vs 10 mm transgenics | 631 | 391 | 240 |
Effect of increased nitrate levels in WT and transgenic plants (10 mm vs 50 mm WT and 10 mm vs 50 mm Transgenics) and comparison of WT and transgenic plants under the same concentration of nitrate supply (50 mm WT vs 50 mm Transgenics and 10 mm WT vs 10 mm Transgenics).
Validation of transcriptome analysis
To validate the transcriptome profiles in response to nutritional changes, we selected a number of representative genes for accurate determination of their expression levels using qPCR (Figure S1, Table S5). Overall, the expression analysis of the selected genes confirmed the microarray results. PtPIP (plasma membrane intrinsic protein), PtNAC (NAC domain protein), PtMYB (MYB transcription factor) and PtFBA (fructose‐biphosphate aldolase) had enhanced expression in the transgenic plants under high N levels (Figure S1, panel 10 vs 50 mm Transgenics). In contrast, PtFBox (flavin‐binding domain F‐box protein), PtWR (wound‐responsive protein‐related), PtPTR (proton‐dependent oligopeptide transport protein) and PtTIP (water channel tonoplast intrinsic protein) had lower expression in the same samples. PtFBox, PtPIP and PtWR were expressed differently in WT plants under high nitrate (Figure S1, panel 10 vs 50 mm WT).
Functional enrichment analysis of the leaf transcriptome in response to N availability
In regard to the identity of the genes differentially regulated in response to nitrate excess, we first analysed those related to primary and secondary metabolism. Among the metabolic pathways that were activated in transgenic poplar in response to high nitrate were the biosynthesis of phenylpropanoids, terpenoids and flavonoids, the biosynthesis of the cell wall and the pathways involved in photosynthesis and carbon metabolism (Figures 5 and S2; Tables S1, S3 and S4). Genes involved in biosynthesis and transport of amino acids, aromatic compound synthesis, carboxylic and organic acid transport, glucosinolate, glucans and cellulose showed up‐regulation (Tables S1, S3 and S4). However, transgenic plants grown under adequate nitrate levels had higher expression of ammonium transporters and TIP aquaporins. The regulation of transcription was altered in the transgenic plants growing at 50 mm nitrate with increased expression of a huge number of transcription factors (Figures S2 and S3; Tables S1, S3 and S4). There were a number of changes in genes involved in signalling, protein degradation and pathogenesis related (PR) triggering changes in the expression profiles (Figure 6) but not function enrichment in response to nitrate nutrition.
Figure 5.
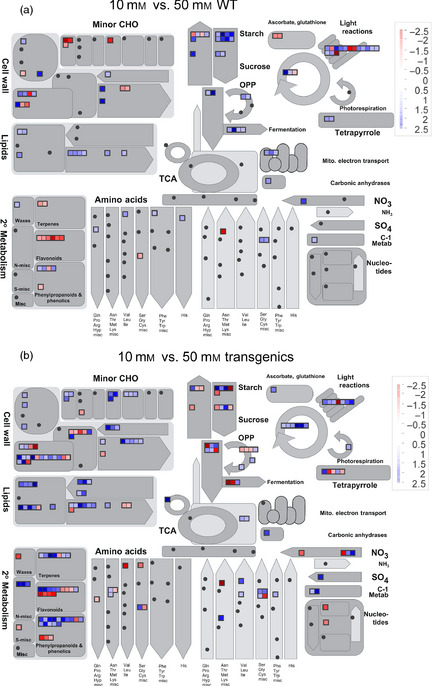
Mapman representation of a metabolism response overview. (a) Comparison of WT plants growing at 10 and 50 mm nitrate. (b) Comparison of transgenic plants growing at 10 and 50 mm nitrate. Blue scale indicates up‐regulated genes at 50 mm. Red scale indicates down‐regulated genes at 50 mm.
Figure 6.
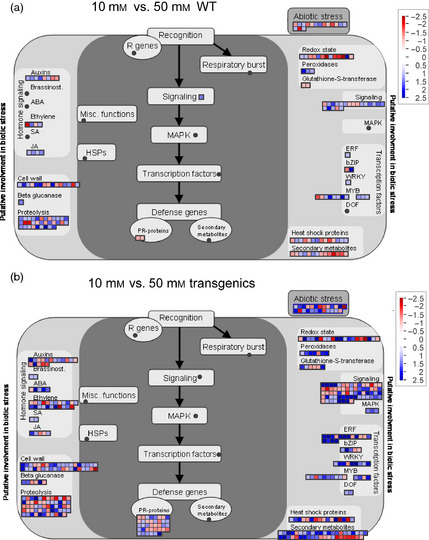
Mapman representation of a stress response overview. (a) Comparison of WT plants growing at 10 and 50 mm nitrate. (b) Comparison of transgenic plants growing at 10 and 50 mm nitrate. Blue scale indicates up‐regulated genes at 50 mm. Red scale indicates down‐regulated genes at 50 mm.
WT plants under adequate N displayed differential expression of genes involved in secondary metabolism mainly in flavonoid synthesis (Figure 5). In contrast, under high nitrate, WT plants enhanced expression of genes involved in regulation of carbon metabolism (as for example starch degradation) and in transcription factors (Figures 6, S2 and S3, and Tables S3 and S4). We identified changes in gene expression related to biotic and abiotic stresses, signalling and regulatory factors. Most of the signalling genes differentially expressed involved in the response to stress were up‐regulated in high nitrate (Figure 6; Tables S1 and S2 and S4). Finally, there were a higher number of genes involved in proteolysis that were also up‐regulated (Figure 6; Tables S1, S3 and S4).
Increased expression levels of secondary and minor CHO metabolism genes were found in transgenic plants when the WT and transgenic plants were compared (Figures S2 and S4; Tables S1, S3 and S4). In all cases, WT plants had higher expression levels of genes involved in central carbon metabolism (glycolysis and starch and sucrose metabolism) (Figures S2 and S4; Tables S1, S3 and S4). Nevertheless, in transgenic plants under 50 mm nitrate, photosynthesis‐related genes were up‐regulated (Figure S4). Nitrate nutrition had huge effects over the expression of genes involved in transcription, signalling and PR (Figures S2, S5 and S6; Tables S1, S3 and S4). At 10 mm nitrate, there were more transcription factors, signalling and PR genes up‐regulated in WT plants, while at 50 mm this tendency was inverted being up‐regulated this group of genes in the transgenic plants (Figures S2, S5 and S6; Tables S1, S3 and S4). The histones were a particular case. All differentially expressed histone genes were down‐regulated at adequate nitrate levels whereas up‐regulated in transgenic plants at high nitrate (Figure S6 and Table S4).
Transcript levels of poplar GS genes and GS1a in response to high nitrate
As N excess affected morphological and physiological traits of WT and transgenic plants, we were interested in determining to what extent the expression of endogenous GS genes and the transgene were also affected (Figure 7). In WT plants, higher levels of PtGS1.1 transcripts were observed under high N, especially in leaves and young stems. In transgenic plants, high expression levels of PtGS1.1 were observed in intermediate leaves (L2), older leaves (L3), and lateral roots (R2) independently of N availability (Figure 7). PtGS1.3 expression was preferentially detected in the stems S1 of WT and transgenic plants, but transcript abundance was clearly enhanced in the stems of transgenic plants with a significant increase in S2 of transgenics under high nitrate (Figure 7). PtGS2 was mainly expressed in young leaves of WT and transgenics, and the expression levels were not affected by the N availability (Figure 7). The pine GS1a transgene was expressed in all regions of the plants examined independent of N availability, but higher levels of GS1a were observed in leaves in comparison with other tissues analysed (Figure 7).
Figure 7.
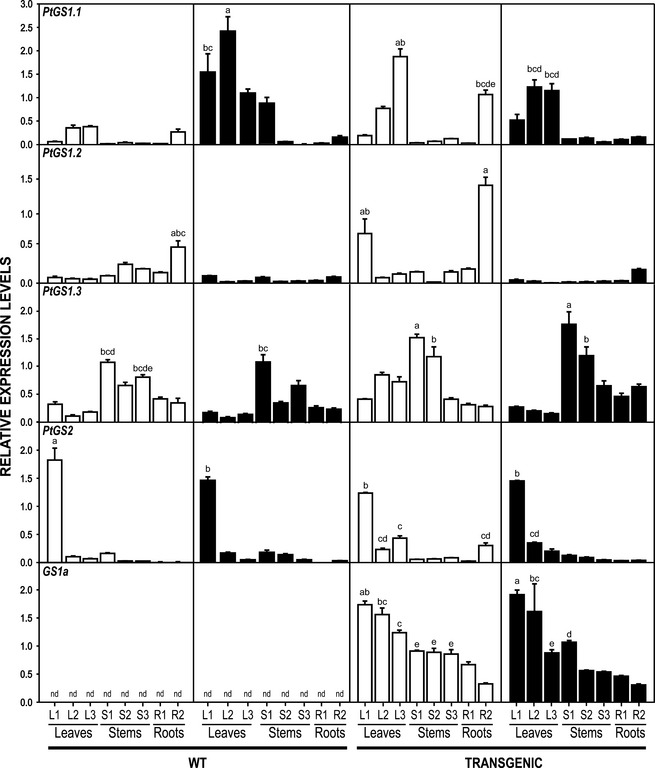
Spatial distribution of GS transcripts in WT and transgenic poplars growing at different nitrate levels.
Changes in carbohydrates and lignin content in response to N excess
To understand the balance between N and C utilization, levels of carbohydrates and total lignin were examined in WT and transgenic plants (Figure 8). In general, glucose, fructose and sucrose profiles were similar for transgenic plants under all conditions. Glucose, fructose and sucrose profiles in WT plants grown under 10 mm nitrate were clearly lower than in plants grown under high nitrate nutrition. In transgenic plants, free sugars accumulated preferentially in leaves, and especially in young leaves under high N. This was particularly evident for glucose. Higher levels of starch were found in lateral roots of transgenic plants grown under high nitrate nutrition followed by WT plants grown under the same conditions. In fact, starch amounts were significantly higher in lateral roots grown under 50 mm nitrate than in roots grown under 10 mm nitrate. Maximum cellulose contents were observed in response to high N availability in transgenic plants; there was no significant effect on the WT plants grown under high nitrate nutrition. Cellulose contents showed significant increases in young leaves (L1 and L2) and in young stems (S1 and S2) mainly in S1. Total lignin showed similar profiles in all the plants increasing from the young leaves to the lateral roots (from L1 to R2). However, lignin contents were higher in the roots of plants grown at 50 mm nitrate. Furthermore, the transgenic plants at 50 mm nitrate exhibited the higher lignin content in the roots (R1 and R2) and in the middle stem (S2).
Figure 8.
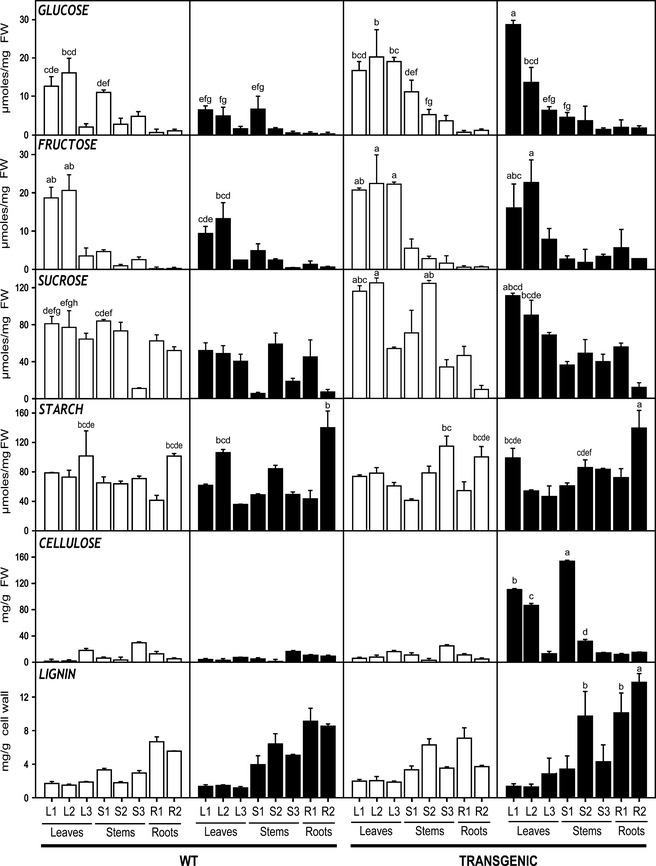
Spatial distribution of carbohydrates and lignin in WT and transgenic poplars growing at different nitrate levels.
Discussion
The utilization of high amounts of nitrate fertilizers for crop yield leads to nitrate pollution of ground and surface waters. In Europe, application rates of N fertilizers vary from the lowest level of 42 kg/ha for agricultural land in Portugal to 243 kg/ha for grassland in the Netherlands (Erisman et al., 2011). In this study, we report the assimilation and utilization of luxuriant levels of N by transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a) (equivalent to 5.3 t/ha N, much higher than the average N fertilizer application in Europe). The assimilation of these high levels of nitrate resulted in increased poplar biomass, growth and nitrogen‐use efficiency (NUE). These results are consistent with previous reports of enhanced growth in young transgenic plants expressing the pine GS1a (Fu et al., 2003; Gallardo et al., 1999) and in a field trial of GS1a transgenic poplar (Coleman et al., 2012; Jing et al., 2004), and improved NUE of these plants (Man et al., 2005).
Our data indicate that GS transgenic poplars are able to maintain balanced C and N assimilation even under luxuriant N levels. We have shown that GS1a transgenic plants provided with high N levels (50 mm nitrate) exhibit increased biomass (171.6%) compared with transgenic and WT plants grown under adequate N levels (10 mm nitrate). Total soluble proteins and contents of chlorophylls a and b also increased significantly in young leaves under luxuriant N levels. Previous analyses of GS1a transgenic poplars also observed reduced free ammonium amounts and increased glutamine levels in response to nitrate (Man et al., 2005). We hypothesized that increased availability of organic nitrogen in the form of glutamine confers metabolic advantages to transgenic plants, resulting in improved growth and biomass (Cánovas et al., 2006). Furthermore, it has been suggested that overexpression of cytosolic GS results in enhanced primary assimilation of ammonium and reassimilation of N released in photorespiration, protein remobilization and lignin biosynthesis (Cánovas et al., 2007; Man et al., 2005; Oliveira et al., 2002). N assimilation and biomass production have a relationship with efficient N uptake and allocation into the plant. Previous studies suggest that leaf area and biomass are principal sinks under increased N availability in poplar trees (Cooke et al., 2005).
Total sugars (glucose, fructose and sucrose) decreased in WT plants under luxuriant N levels, suggesting a shortage of carbon skeletons for N assimilation. However, their levels increased in the transgenics under these nutritional conditions, suggesting that GS‐overexpressing plants have greater capacity to assimilate N. This likely reflects increased photosynthetic rates and carbon fixation capacity (Cooke et al., 2005; Man et al., 2005). In fact, it was previously determined that net photosynthetic rates were higher in transgenic than in nontransgenic plants (El‐Khatib et al., 2004). GS transgenic poplar growing in field trials grew faster and produced more biomass in shorter time (Jing et al., 2004), and composition of cell wall carbohydrates, including glucose, galactose, xylose and mannose in wood was increased (Coleman et al., 2012). Starch accumulated in the poplar roots under luxuriant N levels, and some studies have proposed a negative correlation between starch concentration and plant N status (Wetzel et al., 1995). In contrast, we have found starch accumulation in secondary roots of both transgenic and WT plants under high N, suggesting that the production of C skeletons (e.g. sucrose) in the aerial part of the plant is sufficient for N assimilation, and assimilated C excess could likely be stored in the roots. In fact, the transcriptomic data indicate that a β‐amylase similar to PtBAMY1 (pt_40322) was up‐regulated in young leaves of transgenic poplar under N luxuriant levels. These findings suggest that starch breakdown was activated providing C skeletons for N assimilation (Geisler‐Lee et al., 2006).
Differences in C and N allocation in poplar are correlated with different growth strategies during biomass formation and N usage (Li et al., 2012a,b; Novaes et al., 2009). Our data indicate that transgenic plants have greater capacity for N uptake and use even under luxuriant N levels. NUtE in transgenic roots was higher than in WT roots under adequate and luxuriant levels, and this is can be explained considering that in comparison with the WT, transgenic plants have a greater capacity to carry out primary N assimilation (from the soil) and, therefore, greater capacity to exploit mineral resources. Transgenics consequently display enhanced tolerance to high N levels. The highest NUtE and NUpE values were found in young leaves of transgenic plants under luxuriant N levels, the major organ where ammonium is assimilated by the glutamine synthetase/glutamate synthase (GS‐GOGAT) cycle and then transferred to amino acids and other N compounds (Bernard and Habash, 2009). As previously suggested, poplar trees translocate most of resources to above‐ground tissues (Cooke et al., 2005). Interestingly, the NUpE was five times higher in the young leaves of transgenic plants under high N reflecting that N allocation and metabolism in transgenic plants are more efficient than in WT plants, confirming previous studies (Man et al., 2005). Consequently, GS1 overexpression provides advantages for N assimilation in the leaves and improves the assimilation of carbon skeletons providing more biomass in the aerial part. NUpE involves different metabolic pathways, and N compounds allocated in the leaves could enhance the metabolic status of the plant and plant N use (Hirose, 2012). The high NUpE and low NUtE in the roots of WT plants under adequate N likely reflect that these plants are as efficient in N uptake but not in N use when compared to transgenics. During vegetative growth, leaves and roots are the principal sinks for inorganic N and the synthesis of amino acids for transport (Kant et al., 2011).
The poplar transcriptome is altered during growth under high nitrate levels, resulting in changes in the expression of a range of genes involved in different processes (this paper, Cooke et al., 2003, 2005). The number of genes differentially expressed in the leaves of GS transgenic plants was significantly higher than in the WT. In fact, 1346 genes were differentially regulated between transgenic and WT plants (minimum twofold) when plants were grown under luxuriant N compared with only 631 differentially expressed under N adequate levels. In transgenic plants, genes involved in phenylpropanoid, terpenoid and flavonoid biosynthesis were up‐regulated under N luxuriant levels, consistent with enhanced carbon metabolism and N recycling. For example, the cellulose synthase like gene PtCslD4 (pt_28112) was overexpressed only in transgenic plants under high N. This gene is mainly expressed in the shoot tips and is involved in poplar cell wall formation and modification (Suzuki et al., 2006).
Lignin biosynthesis has been reported to decrease under high N availability in poplar (Novaes et al., 2009; Pitre et al., 2007), and our data show that there are lower lignin contents in the apical regions (young leaves and stems) likely reflecting the redirection of C skeletons for biosynthesis of other carbohydrates such as cellulose. This is consistent with the overexpression of PtCslD4 and cellulose accumulation in young leaves. Under N luxuriant levels, young poplars increased shoot biomass and wood cellulose contents (Cooke et al., 2005). N fertilization stimulates carbon allocation in shoots over roots and alters wood chemistry traits with increases in cellulose and hemicellulose levels and reduction in lignin content (Novaes et al., 2009).
Furthermore, expression of genes for numerous physiological processes was modified in the transgenics including genes for biosynthesis and catabolism of amino acids and aromatic compounds and nutrient transport. Expression of genes for membrane transport proteins was also activated; for example, PtAAP11 (pt_05342) has an important role during N transfer during xylem differentiation (Couturier et al., 2010), and we have found that this amino acid transporter was up‐regulated in transgenic and WT plants under high N, suggesting that this gene responds to N availability and could modify wood formation. A high‐affinity nitrate transporter similar to Arabidopsis AtNRT2.7 (pt_00777) was up‐regulated in transgenic plants under high nitrate. In contrast, PtAMT3.1 (pt_32332), highly expressed in senescent leaves (Couturier et al., 2007), was down‐regulated under high N, suggesting that N availability repressed its expression.
The gene for win‐like VSP 425 protein (pt_ pt_34191) was highly expressed under N luxuriant levels in transgenic plants, suggesting that these vegetative storage proteins promote purine degradation and increase NUE through N remobilization from sink to source (Werner and Witte, 2011). These results are consistent with previous reports describing that win4 and bsp respond to high N concentration levels (Coleman et al., 1994; Cooke et al., 2005).
Specific AP2 domain‐containing transcription factors were up‐regulated only in transgenic plants under high N. These genes are induced by external stimuli (Zhuang et al., 2008), suggesting that these families of transcriptional regulators may play a crucial role in plant growth. Other transcription factors overexpressed in the GS transgenics under high N are NAC‐ (Li et al., 2012a,b; Zhao et al., 2014) and MYB‐related proteins involved in wood formation in Populus (McCarthy et al., 2010; Wang et al., 2014; Zhong et al., 2011). WRKY proteins implicated in abiotic stress responses (He et al., 2012) showed similar profiles, and all these results are consistent with the observed tolerance of GS transgenics to different types of stress (Cánovas et al., 2006). Overall, transcription factors were up‐regulated in transgenic plants, particularly at high nitrate levels.
Expansins associated with woody tissues have an important role during xylogenesis (Gray‐Mitsumune et al., 2004). PtEXLA1.1 (pt_03200), a gene mainly associated with developing wood tissues, was up‐regulated under high N levels in transgenic poplar. When PttEXPA1 was overexpressed in poplar, the transgenic plants increased expansion activity on cellulose–xyloglucan composites, resulting in stem elongation, leaf expansion and enhanced cell wall expansion (Gray‐Mitsumune et al., 2008). Genes implicated in cell wall growth, including expansin and xyloglucan, for example xyloglucosil transferase (pt_12600), were activated in response to N, as were genes involved in photosynthesis and Calvin cycle (Scheible et al., 2004). This expression pattern confirms that for enhanced vegetative growth in transgenic plants under high N, it is necessary to have a preferential flux of carbon to the shoot and young leaves (Geisler‐Lee et al., 2006).
Transcripts for various genes implicated in DNA structure, repair and general process were up‐regulated in WT plants under high N but down‐regulated under adequate N nutrition. Rapid changes in histones produce specific DNA/chromatin modifications that play a crucial role in the regulation of responses to plant abiotic and biotic stress (Boycheva et al., 2014). These findings together with the observed up‐regulation of transcription factors suggest a general reorganization of the regulation of gene expression induced by N nutrition, which affects differently to transgenic and WT plants.
Endogenous GS transcripts in Populus display organ‐specific expression profiles (Figure 7) confirming previous studies (Castro‐Rodríguez et al., 2011). Interestingly, the expression of PtGS1.1 increased more than fourfold in the leaves of WT plants under luxuriant N levels. PtGS1.1 codes for a high‐affinity ammonium enzyme that seems to play a crucial role in ammonium recycling in mature leaves (Castro‐Rodríguez et al., 2015). The ectopically expressed pine GS1a has an expression profile independent of N availability. The GS1a sequence is a putative target gene for miR‐3630‐3p miRNA. It has been reported that miR‐3630‐3p targeted nucleotide‐binding leucine‐rich repeat (NB‐LRR) and F‐box genes (Barozai et al., 2012) are induced during abiotic stress (Li et al., 2011). MicroRNAs are potential regulators of transgene targets throughout the plant, especially when their expression is associated with enhanced vegetative growth (Fu et al., 2012; Zhang and Li, 2013). Considering these previous reports, miR‐3630‐3p could be important in the regulation of the pine transgene under high N.
Our results highlight the accumulation of biomass in aerial parts of GS1a transgenic plants in response to high N availability. Cellulose content in the leaves of transgenics increased fivefold in response to luxuriant N levels, whereas lignin content increased in basal stems and roots of plants grown under similar conditions. Previous studies have shown that increased biomass in trees is concomitant with reduced levels of lignin, reflecting a strong correlation between the balance in lignin biosynthesis and biomass accumulation in woody tissues (Novaes et al., 2010). However, more lignin provides more structural support to growing plants with increased biomass (Donaldson, 2001). Our results indicate that wood formation in stems was regulated by increasing lignin content in the middle and basal section to support the biomass accumulation in apical regions. Analysis of wood chemistry of field‐grown GS1a transgenics has shown enhanced levels of hemicellulose‐associated polymers (galactomannans) and decreased lignin contents (Coleman et al., 2012). Similar results were reported for poplar exposed to enhanced N fertilizer treatments (Pitre et al., 2007). These data suggest that GS1a transgenic poplars grown under high nitrate levels may be of significant value in pulp and biofuels applications because they have improved pulping traits (high S/G ratios) and enhanced fibre characteristics (Coleman et al., 2012). Taken together, our results strongly support the use of GS1a transgenic poplars as potential phytoremediation tools for environmental control of polluted areas and as feedstocks for the biofuels or fibre markets. Field trials will be required to assess these potential applications.
Experimental procedures
Plant material, culture conditions and sampling
Untransformed (WT) hybrid poplar (Populus tremula × P. alba, clone INRA 7171‐B, INRA) and a transgenic line (line 4‐29) overexpressing a glutamine synthetase cytosolic pine (GS1a) under the control of cauliflower mosaic virus 35S promoter were produced and maintained in vitro as previously described (Fu et al., 2003; Gallardo et al., 1999). The line 4‐29 was identified as one of the superior performing lines in the growth studies performed in the greenhouse and under natural conditions (Fu et al., 2003; Jing et al., 2004). Rooted shoots of transformed and WT plants were maintained in plant growth chambers with a photoperiod of 16 h light with light intensity of 295 μmol m2/s, a constant temperature of 24 °C and 80% humidity. Plants were cultivated in plastic pots containing a potting mix and vermiculite in proportions 1:1 and watered with distilled water supplemented with macro‐ and micronutrients for optimal growth (Gallardo et al., 1999; Jing et al., 2004). After 8 weeks, plants with similar height (approximately 60 cm) were divided by genotype: WT and transgenic poplars (24 individuals per genotype) and each group of plants were then subdivided randomly into two groups. One of the groups was irrigated with a nutrient solution enriched with 10 mm potassium nitrate, considered as adequate for poplar vegetative growth according to the previous data reported by Man et al. (2005). The remaining group of plants was supplied with 50 mm nitrate, a luxuriant concentration of nitrate for poplar according to Cooke et al. (2003). The plants were watered with the nutrient solution once a week by flooding, for a total of 4 weeks. Three‐month‐old P. tremula × P. alba plants were harvested and samples taken from eight different sections from the shoot apex to the root tip.
Growth measurements were carried out taking into account differences in height growth of the plants (12 plants were analysed per treatment). The aerial regions of the plants were divided into three parts: the 1st, 2nd, 3rd, 4th and 5th apical leaves (L1) and the corresponding stem (S1); the intermediate region with the 6th, 7th, 8th, 9th and 10th leaves (L2) and the corresponding stem (S2); and the more basal region including 11th, 12th, 13th, 14th and 15th leaves (L3) and the corresponding stem (S3). The root was sectioned into R1, main root and R2, secondary root. Harvested leaves, stems and roots were immediately frozen in liquid nitrogen. The samples were ground into fine powder in liquid nitrogen with a mortar and pestle and stored at −80 °C until processing. Frozen powder (100 mg) from the samples of each plant was dried at 70 °C for 48 h to determine the fresh to dry mass. For further studies, a fine powder was prepared from leaves, stems or roots of plants subjected to the two nutritional treatments. Extractions were performed from each plant per triplicate.
Determination of proteins and chlorophylls
Soluble proteins were extracted from poplar samples with a mortar and pestle according to the following procedure: 1 g of tissue, 1 g of fine sand and 1 mL of extraction buffer [0.175 m Tris pH 8.8, 0.1% (w/v) SDS, 15% (v/v) glycerol, 0.3 m mercaptoethanol] were homogenized (Castro‐Rodríguez et al., 2011) and the resulting extract was centrifuged at 10 000 g 4 °C for 30 min. The supernatant was used for the determination of soluble proteins using the assay described by Bradford (1976).
The extraction of total chlorophylls was performed from 100 mg of frozen samples on liquid nitrogen and 80% (v/v) acetone, and the content was determined according to Graan and Ort (1984).
Determination of soluble sugars
Sugars were extracted from frozen powder (100 mg) of leaves, stems and roots following the procedure described by de la Torre et al. (2014). Sucrose, glucose and fructose levels were measured enzymatically following the reduction of NADP+ at 340 nm after successive addition of the coupling enzymes: glucose‐6‐P‐dehydrogenase (4 units/mL), hexokinase (10 units/mL), phosphoglucose isomerase (5 units/mL) and invertase (Sekin, 1978). Starch was measured as described by Smith and Zeeman (2006). All enzymes were obtained from Roche Diagnostics, Mannheim (Germany).
Determination of cellulose and lignin
Cellulose content was determined by the anthrone method (Updegraff, 1969) following exactly the protocol previously described (de la Torre et al., 2014). Total lignin quantification was performed following the thioacidolysis method described by Lange et al. (1995) according to the modifications specified by de la Torre et al. (2014).
Determination of total C and N content, NUtE and NUpE
Fine powder (100 mg) was dried in an oven at 70 °C for 48 h. The percentages of N (%) and C (%) of the triplicate samples were determined by an elemental macro‐analyser Leco truSpec CHNS (Leco Corporation, St. Joseph, MI) at the Atomic Spectrometry Unit, University of Málaga. The NUtE was calculated using the increase in biomass DW of leaves, stems and roots divided by the total N supplied (g per plant) for each plant in the treatment (10 and 50 mm) (Good et al., 2004). The NUpE was calculated using the observed increase in total N content of leaves, stems and roots divided by the N supplied (g per plant) (Good et al., 2004).
Expression analysis of GS genes
RNA was isolated from leaves, stems and roots as described by Canales et al. (2010), and all traces of genomic DNA were removed by digestion with Dnase I (Promega Corporation, Madison, WI). RNA concentration and purity were quantified using a NanoDrop© ND‐1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, DE). cDNA synthesis was performed as described previously (Canales et al., 2010). The qPCR was carried out in a thermal cycler CFX384 (Bio‐Rad, Hercules, CA) under the following conditions: 3 min at 95 °C (1 cycle), 1 s at 95 °C and 5 s at 60 °C (40 cycles) and a melting curve from 60 to 95 °C to verify the reaction specificity. Primers for PtGS1.1, PtGS1.2, PtGS1.3, PtGS2, previously described in Castro‐Rodríguez et al. (2011), and PpGS1a (Forward: 5′‐AATTACAAAGTGGAGGCCAG; Reverse: 5′‐AGCCCTCGCCATATTACAAGT) were used. Actin2 and Ubiquitin were used as reference genes (Brunner et al., 2004). PCR reactions were performed in triplicate and relative expression levels were calculated using the R package qPCR (Ritz and Spiess, 2008). The raw fluorescence data from each reaction were fitted to the MAK2 model, which requires no assumptions about the amplification efficiency of the qPCR assay (Boggy and Woolf, 2010). The initial target concentrations (DO parameter) for each gene were deduced from the MAK2 model using the qPCR package and normalized to the geometric mean of the reference genes.
Microarray analysis
RNA was isolated as previously described, and RNA quality was assessed using the RNA Pico Assay for the 2100 Bioanalyzer (Agilent, Santa Clara, CA). Gene expression analyses were performed with the Agilent Populus whole genome array (4 × 44 k) (Tsai et al., 2011). Microarray labelling, hybridization and washing were performed according to Agilent Low Input Quick Amp Labeling Kit‐two color protocol (Agilent Technologies; cat# 5190–2306). For each amplification and labelling reaction, 200 ng of total RNA was employed. cRNAs with dye‐specific activities (pmol Cy3 or Cy5 per μg cRNA) higher than 6 were used for the hybridizations. For each field of hybridization, 825 ng of a specific Cy3‐labelled cRNA and 825 ng of a specific Cy5‐labelled cRNA were used. Two technical and two biological replicates were performed. Microarray slides were scanned using GenePix 4100A scanner with a resolution of 5 μm/pixel. Data were extracted from the scanned images using the GenePix v6.0 software (Molecular Devices, Sunnyvale, CA).
Normalization and differential expression analysis of the data were performed using limma package in the R environment (Smyth, 2004). Normalization was carried out using quantile method and eBayes statistics to determine the differential expression between the comparisons (Smyth et al., 2005). Statistical significance was corrected for multiple testing using the Benjamini–Hochberg procedure. The differentially expressed genes were selected with a significance P‐value <0.05 and log2FC >1 and <−1.
Overall changes of the genes involved in metabolic pathways of plants, or in response to stress have been represented by Mapman tool and analysed by the functional Mapman categories (Thimm et al., 2004). Mapman functional categories for differentially expressed genes are presented in Table S4. An overview of the Mapman functional categories including the differentially expressed genes is shown in Figures S7 and S8.
The microarray data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series Accession Number GSE61801 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62449).
Microarray validation
The differential expression of selected genes in the microarrays was tested using qPCR in the same samples as described above. Sequences of specific primers are listed in Table S5; all primers used the same annealing temperature and time (60 °C, 5 s).
Statistical analysis
Statistical variables were analysed by two‐way ANOVA test and Tukey's multiple comparisons with a significance of P < 0.001. The identification of homogenous groups significantly different, and ANOVAs were performed in the R environment with the agricolae package using the HSD.test command.
Supporting information
Figure S1 Comparison between qPCR and microarray expression data to validate the microarray hybridizations.
Figure S2 Function enrichment analysis of the Mapman categories in the four analysed comparisons.
Figure S3 Mapman representation of the transcription related genes including differentially expressed genes.
Figure S4 Mapman representation of the metabolism response overview, including differentially expressed genes.
Figure S5 Mapman representation of the stress response overview, including differentially expressed genes.
Figure S6 Mapman representation of the transcription related genes including differentially expressed genes.
Figure S7 Overview of the Mapman functional categories including the differentially expressed genes.
Figure S8 Overview of the Mapman functional categories including the differentially expressed genes.
Table S1 Differentially expressed genes table.
Table S2 Limma analysis data for all differentially genes.
Table S3 Enrichment analysis of the Mapman functional categories.
Table S4 Table including all the Mapman Bin annotations for differentially expressed genes in each sample comparison.
Table S5 Genes primers for microarray validation.
Acknowledgements
We would like to thank the anonymous reviewers for their thorough evaluation and constructive recommendations that helped to improve this manuscript. We are indebted to CJ Tsai (University of Georgia) for giving us access to the Agilent poplar microarray. This work was supported by grants from the Spanish Ministerio de Economía y Competitividad (BIO2012‐33797) and Junta de Andalucía (BIO2012‐0474).
Private reviewer link to microarray data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wpgfeumwddkna=GSE62449
References
- Bai, H. , Euring, D. , Volmer, K. , Janz, D. and Polle, A. (2013) The nitrate transporter (NRT) gene family in poplar. PLoS One, 8, e72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozai, M.Y. , Baloch, I.A. and Din, M. (2012) Identification of MicroRNAs and their targets in Helianthus . Mol. Biol. Rep. 39, 2523–2532. [DOI] [PubMed] [Google Scholar]
- Bernard, S.M. and Habash, D.Z. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620. [DOI] [PubMed] [Google Scholar]
- Boggy, G.J. and Woolf, P.J. (2010) A mechanistic model of PCR for accurate quantification of quantitative PCR data. PLoS One, 5, e12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva, I. , Vassileva, V. and Lantcheva, A. (2014) Histone acetyltransferases in plant development and plasticity. Curr. Genomics, 15, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brunner, A.M. , Yakovlev, I.A. and Strauss, S.H. (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales, J. , Flores‐Monterrosso, A. , Rueda‐Lopez, M. , Avila, C. and Cánovas, F.M. (2010) Identification of genes regulated by ammonium availability in the roots of maritime pine trees. Amino Acids, 39, 991–1001. [DOI] [PubMed] [Google Scholar]
- Cánovas, F.M. , Gallardo, F. , Jing, Z.P. and Pascual, B. (2006) Transgenic approaches to engineer nitrogen metabolism. In Tree Transgenesis. Recent Developments ( Fladung, M. and Ewald, S. , eds), pp. 157–178. Heildelberg: Springer Verlag. [Google Scholar]
- Cánovas, F.M. , Avila, C. , Cantón, F.R. , Cañas, R.A. and de la Torre, F. (2007) Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 58, 2307–2318. [DOI] [PubMed] [Google Scholar]
- Cantón, F.R. , Suárez, M.F. and Cánovas, F.M. (2005) Molecular aspects of nitrogen mobilisation and recycling in trees. Photosynth. Res. 83, 265–278. [DOI] [PubMed] [Google Scholar]
- Castro‐Rodríguez, V. , García‐Gutiérrez, A. , Canales, J. , Avila, C. , Kirby, E.G. and Cánovas, F.M. (2011) The glutamine synthetase gene family in Populus . BMC Plant Biol. 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro‐Rodríguez, V. , García‐Gutiérrez, A. , Cañas, R.A. , Pascual, M.B. , Avila, C. and Cánovas, F.M. (2015) Redundancy and metabolic function of the glutamine synthetase gene family in poplar. BMC Plant Biol. 15, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, G.D. , Bañados, M.P. and Chen, T.H.H. (1994) Poplar bark storage protein and a related wound‐induced gene are differentially induced by nitrogen. Plant Physiol. 106, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, H.D. , Cánovas, F.M. , Man, H. , Kirby, E.G. and Mansfield, S.D. (2012) Enhanced expression of glutamine synthetase (GS1a) confers altered fibre and wood chemistry in field grown hybrid poplar (Populus tremula X alba) (717‐1B4). Plant Biotech. J. 10, 883–889. [DOI] [PubMed] [Google Scholar]
- Cooke, J.E.K. , Brown, K.A. , Wu, R. and Davis, J.M. (2003) Gene expression associated with N‐induced shifts in resource allocation in poplar. Plant Cell Environ. 26, 757–770. [Google Scholar]
- Cooke, J.E.K. , Martin, T.A. and Davis, J.M. (2005) Short‐term physiological and developmental responses to nitrogen availability in hybrid poplar. New Phytol. 167, 41–52. [DOI] [PubMed] [Google Scholar]
- Couturier, J. , Montanini, B. , Martin, F. , Brun, A. , Blaudez, D. and Chalot, M. (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150. [DOI] [PubMed] [Google Scholar]
- Couturier, J. , de Fay, E. , Fitz, M. , Wipf, D. , Blaudez, D. and Chalot, M. (2010) PtAAP11, a high affinity amino acid transporter specifically expressed in differentiating xylem cells of poplar. J. Exp. Bot. 61, 1671–1680. [DOI] [PubMed] [Google Scholar]
- Cren, M. and Hirel, B. (1999) Glutamine synthetase in higher plants regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 40, 1187–1193. [Google Scholar]
- Donaldson, L.A. (2001) Lignification and lignin topochemistry—an ultrastructural view. Phytochemistry, 57, 859–873. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Khatib, R.T. , Hamerlynck, E.R. , Gallardo, F. and Kirby, E.G. (2004) Transgenic poplar characterized by ectopic expression of a pine cytosolic glutamine synthetase gene exhibits enhanced tolerance to water stress. Tree Physiol. 21, 729–736. [DOI] [PubMed] [Google Scholar]
- Erisman, J.W. , van Grinsven, H. , Grizzetti, B. , Bouraoui, F. , Powlson, D. , Sutton, M.A. , Bleeker, A. and Reis, S. (2011) The European nitrogen problem in a global perspective. In The European Nitrogen Assessment ( Sutton, M.A. , Howard, C.M. , Erisman, J.W. , Billen, G. , Bleeker, A. , Grennfelt, P. , van Grinsven, H. and Grizzetti, B. , eds), pp. 9–31. Cambridge: Cambridge University Press. [Google Scholar]
- Fu, J. , Sampalo, R. , Gallardo, F. , Cánovas, F.M. and Kirby, E.G. (2003) Assembly of a cytosolic pine glutamine synthetase holoenzyme in leaves of transgenic poplar leads to enhanced vegetative growth in young plants. Plant Cell Environ. 26, 411–418. [Google Scholar]
- Fu, C. , Sunkar, R. , Zhou, C. , Shen, H. , Zhang, J.‐Y. , Matts, J. , Wolf, J. , Mann, D.G.J. , Stewart, C.N. Jr , Tang, Y. and Wang, Z.‐Y. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotech. J. 10, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, F. , Fu, J. , Cantón, F.R. , García‐Gutiérrez, A. , Cánovas, F.M. and Kirby, E.G. (1999) Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta, 210, 19–26. [DOI] [PubMed] [Google Scholar]
- Galloway, J.N. , Townsend, A.R. , Erisman, J.W. , Bekunda, M. , Cai, Z. , Freney, J.R. , Martinelli, L.A. , Seitzinger, S.P. and Sutton, M.A. (2008) Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science, 320, 889–892. [DOI] [PubMed] [Google Scholar]
- Geisler‐Lee, J. , Geisler, M. , Coutinho, P.M. , Segerman, B. , Nishikubo, N. , Takahashi, J. , Aspeborg, H. , Djerbi, S. , Master, E. , Andersson‐Gunnerås, S. , Sundberg, B. , Karpinski, S. , Teeri, T.T. , Kleczkowski, L.A. , Henrissat, B. and Mellerowicz, E.J. (2006) Poplar carbohydrate‐active enzymes. Gene identification and expression analyses. Plant Physiol. 140, 946–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, A.G. , Shrawat, A.K. and Muench, D.G. (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605. [DOI] [PubMed] [Google Scholar]
- Graan, T. and Ort, D.R. (1984) Quantitation of the rapid electron donors to P700, the functional plastoquinone pool, and the ratio of the photosystems in spinach chloroplasts. J. Biol. Chem. 259, 14003–14010. [PubMed] [Google Scholar]
- Gray‐Mitsumune, M. , Mellerowicz, E.J. , Abe, H. , Schrader, J. , Winzéll, A. , Sterky, F. , Blomquist, K. , McQueen‐Mason, S. , Teeri, T.T. and Sundberg, B. (2004) Expansins abundant in secondary xylem belong to subgroup A of the alpha‐expansin gene family. Plant Physiol. 135, 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray‐Mitsumune, M. , Blomquist, K. , McQueen‐Mason, S. , Teeri, T.T. , Sundberg, B. and Mellerowicz, E.J. (2008) Ectopic expression of a wood‐abundant expansin PttEXPA1 promotes cell expansion in primary and secondary tissues in aspen. Plant Biotech. J. 6, 62–72. [DOI] [PubMed] [Google Scholar]
- He, H.S. , Dong, Q. , Shao, Y.H. , Jiang, H.Y. , Zhu, S.W. , Cheng, B.J. and Xiang, Y. (2012) Genome‐wide survey and characterization of the WRKY gene family in Populus trichocarpa . Plant Cell Rep. 31, 1199–1217. [DOI] [PubMed] [Google Scholar]
- Hirose, T. (2012) Leaf‐level nitrogen use efficiency: definition and importance. Oecologia, 169, 591–597. [DOI] [PubMed] [Google Scholar]
- Jing, Z.P. , Gallardo, F. , Pascual, M.B. , Sampalo, R. , Romero, J. , de Navarra, A.T. and Cánovas, F.M. (2004) Improved growth in a field trial of transgenic hybrid poplar overexpressing glutamine synthetase. New Phytol. 164, 137–145. [DOI] [PubMed] [Google Scholar]
- Kant, S. , Bi, Y.‐M. and Rothstein, S.J. (2011) Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62, 1499–1509. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Lapierre, C. and Sanderman, H. (1995) Elicitor‐induced spruce stress lignin. Structural similarity to early developmental lignin. Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, P.J. and Ireland, R.J. (1999) Nitrogen metabolism in higher plants. In Plant Amino Acids, Biochemistry and Biotechnology ( Singh, B.K. , ed.), pp. 1–47. New York: Marcel Dekker. [Google Scholar]
- Li, H. , Dong, Y. , Yin, H. , Wang, N. , Yang, J. , Liu, X. , Wang, Y. , Wu, J. and Li, X. (2011) Characterization of the stress associated microRNA in Glycine max by deep sequencing. BMC Plant Biol. 11, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, M. , Luo, X.C. , Qu, L. , Gai, Y. , Jiang, X. , Liu, T. , Bai, H. , Janz, D. , Polle, A. , Peng, C. and Luo, Z.‐B. (2012a) N‐fertilization has different effects on the growth, carbon and nitrogen physiology, and wood properties of slow‐ and fast‐growing Populus species. J. Exp. Bot. 63, 6173–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Lin, Y.C. , Sun, Y.H. , Song, J. , Chen, H. , Zhang, X.H. , Sederoff, R.R. and Chiang, V.L. (2012b) Splice variant of the SND1 transcription factor is a dominant negative of SND1 member and their regulation in Populus trichocarpa . Proc. Natl Acad. Sci. USA, 109, 14699–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, H.M. , Boriel, R. , El‐Khatib, R. and Kirby, E.G. (2005) Characterization of transgenic poplar with ectopic expression of pine cytosolic glutamine synthetase under conditions of varying nitrogen availability. New Phytol. 167, 31–39. [DOI] [PubMed] [Google Scholar]
- McCarthy, R.L. , Zhong, R. , Fowler, S. , Lyskowski, D. , Piyasena, H. , Carleton, K. , Spicer, C. and Ye, Z.H. (2010) The poplar MYB transcription factors, PtrMYB3 and Ptr MYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Miller, A.J. , Fan, X. , Orsel, M. , Smith, S.J. and Wells, D. (2007) Nitrate transport and signaling. J. Exp. Bot. 58, 2297–2306. [DOI] [PubMed] [Google Scholar]
- Min, X. , Siddiqi, Q. , Guy, R.D. , Glass, A.D.M. and Kronzucker, H.J. (1998) Induction of nitrate uptake and nitrate reductase activity in trembling aspen and lodgepole pine. Plant Cell Environ. 21, 1039–1046. [Google Scholar]
- Novaes, E. , Osorio, L. , Drost, D.R. , Mile, B.L. , Boaventura‐Novaes, C.R.D. , Benedict, C. , Dervinis, C. , Yu, Q. , Syke, R. , Davis, M. , Martin, T.A. , Peter, G.F. and Kirst, M. (2009) Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytol. 182, 878–890. [DOI] [PubMed] [Google Scholar]
- Novaes, E. , Kirst, M. , Chiang, V. , Winter‐Sederoff, H. and Sederoff, R. (2010) Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol. 154, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, I.C. , Brears, T. , Knight, T.J. , Clark, A. and Coruzzi, G.M. (2002) Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, G.J. and Gordon, A.M. (1994) The nitrogen filtering capability of Carolina poplar in an artificial riparian zone. J. Environ. Qual. 23, 1218–1223. [Google Scholar]
- Pitre, F.E. , Cooke, J.E.K. and Mackay, J.J. (2007) Short‐term effects of nitrogen availability on wood formation and fibre properties in hybrid poplar. Trees‐Struct. Funct. 21, 249–259. [Google Scholar]
- Rennenberg, H. , Wilhagen, H. and Ehlting, B. (2010) Nitrogen nutrition of poplar trees. Plant Biol. 12, 275–291. [DOI] [PubMed] [Google Scholar]
- Ritz, C. and Spiess, A.N. (2008) qpcR: an R package for sigmoidal model selection in quantitative real‐time polymerase chain reaction analysis. Bioinformatics, 24, 1549–1551. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R. , Morcuende, R. , Czechowski, T. , Fritz, C. , Osuna, D. , Palacios‐Rojas, N. , Schindelasch, D. , Thimm, O. , Udvardi, M.K. and Stitt, M. (2004) Genome‐wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger, W.H. (2009) On the fate of anthropogenic nitrogen. Proc. Natl Acad. Sci. USA, 106, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekin, S. (1978) Enzymatic determination of glucose, fructose and sucrose in Tobacco. Tobacco Sci. 23, 75–77. [Google Scholar]
- Smith, A.M. and Zeeman, S.C. (2006) Quantification of starch in plant tissues. Nat. Protoc. 1, 1342–1345. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. , Michaud, J. and Scott, H. (2005) The use of within‐array replicate spots for assessing differential expression in microarray experiments. Bioinformatics, 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- Suzuki, S. , Li, L. , Sun, Y.‐H. and Chiang, V.L. (2006) The cellulose synthase gene superfamily and biochemical functions of xylem specific cellulose synthase like genes in Populus trichocarpa . Plant Physiol. 142, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O. , Bläsing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Krüger, P. , Selbig, J. , Müller, L.A. , Rhee, S.Y. and Stitt, M. (2004) MAPMAN: a user‐drive tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- de la Torre, F. , El‐Azaz, J. , Ávila, C. and Cánovas, F.M. (2014) Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiol. 164, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.J. , Ranjan, P. , DiFazio, S.P. , Tuskan, G.A. and Johnson, V. (2011) Poplar genome microarrays. In Genetics, Genomics and Breeding of Poplar ( Joshi, C.P. , DiFazio, S.P. and Kole, C. , eds), pp. 112–127. Enfield: Science Publishers. [Google Scholar]
- Updegraff, D.M. (1969) Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Li, E. , Porth, I. , Chen, J.‐G. , Mansfield, S.D. and Douglas, C.J. (2014) Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis . Sci. Rep. 4, 5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, A.K. and Witte, C.‐P. (2011) The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci. 13, 381–387. [DOI] [PubMed] [Google Scholar]
- Wetzel, S. , Sennerby‐Forsse, L. and Burgess, D. (1995) Storage compounds in Populus cuttings in response to two different nitrogen regimen in nutrient uptake and cycling in forest ecosystems. In Nutrient Uptake and Cycling in Forest Ecosystems ( Nilsson, L.O. , Hüttl, R.F. and Johansson, U.T. , eds), pp. 677–685. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Yadav, R. , Arora, P. , Kumar, S. and Chaudhury, A. (2010) Perspectives for genetic engineering of poplars for enhanced phytoremediation abilities. Ecotoxicology, 19, 1754–1588. [DOI] [PubMed] [Google Scholar]
- Zhang, H. and Li, L. (2013) SQUAMOSA promoter binding protein‐like7 regulated microRNA408 is required for vegetative development in Arabidopsis . Plant J. 74, 98–109. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Sun, J. , Xu, P. , Zhang, R. and Li, L. (2014) Intron‐Mediated alternative splicing of Wood‐associated NAC transcription factor1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164, 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , McCarthy, R.L. , Lee, C. and Ye, Z.H. (2011) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, J. , Cai, B. , Peng, R.‐H. , Zhu, B. , Jin, X.‐F. , Xue, Y. , Gao, F. , Fu, X.‐Y. , Tian, Y.‐S. , Zhao, W. , Qiao, Y.‐S. , Zhang, Z. , Xiong, A.‐S. and Yao, Q.‐H. (2008) Genome‐wide analysis of the AP2/ERF gene family in Populus trichocarpa . Biochem. Biophys. Res. Commun. 371, 468–474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison between qPCR and microarray expression data to validate the microarray hybridizations.
Figure S2 Function enrichment analysis of the Mapman categories in the four analysed comparisons.
Figure S3 Mapman representation of the transcription related genes including differentially expressed genes.
Figure S4 Mapman representation of the metabolism response overview, including differentially expressed genes.
Figure S5 Mapman representation of the stress response overview, including differentially expressed genes.
Figure S6 Mapman representation of the transcription related genes including differentially expressed genes.
Figure S7 Overview of the Mapman functional categories including the differentially expressed genes.
Figure S8 Overview of the Mapman functional categories including the differentially expressed genes.
Table S1 Differentially expressed genes table.
Table S2 Limma analysis data for all differentially genes.
Table S3 Enrichment analysis of the Mapman functional categories.
Table S4 Table including all the Mapman Bin annotations for differentially expressed genes in each sample comparison.
Table S5 Genes primers for microarray validation.


