Summary
Leaf senescence is a complex process, which has dramatic consequences on crop yield. In sunflower, gap between potential and actual yields reveals the economic impact of senescence. Indeed, sunflower plants are incapable of maintaining their green leaf area over sustained periods. This study characterizes the leaf senescence process in sunflower through a systems biology approach integrating transcriptomic and metabolomic analyses: plants being grown under both glasshouse and field conditions. Our results revealed a correspondence between profile changes detected at the molecular, biochemical and physiological level throughout the progression of leaf senescence measured at different plant developmental stages. Early metabolic changes were detected prior to anthesis and before the onset of the first senescence symptoms, with more pronounced changes observed when physiological and molecular variables were assessed under field conditions. During leaf development, photosynthetic activity and cell growth processes decreased, whereas sucrose, fatty acid, nucleotide and amino acid metabolisms increased. Pathways related to nutrient recycling processes were also up‐regulated. Members of the NAC, AP2‐EREBP, HB, bZIP and MYB transcription factor families showed high expression levels, and their expression level was highly correlated, suggesting their involvement in sunflower senescence. The results of this study thus contribute to the elucidation of the molecular mechanisms involved in the onset and progression of leaf senescence in sunflower leaves as well as to the identification of candidate genes involved in this process.
Keywords: sunflower, leaf senescence, transcriptomics, metabolomics, data integration, candidate genes
Introduction
Leaf senescence is a complex process, controlled by multiple genetic and environmental variables, which has strong impact on crop yield (Gregersen et al., 2013). In sunflower, the fourth most important oil crop worldwide, the senescence process reduces the capacity of plants to maintain their green leaf area for longer periods, thus leading to economic losses (Aguirrezábal et al., 2003; Dosio et al., 2000). In different crops, including sunflower, a delay in leaf senescence thus has an important impact on yield, by maintaining photosynthetic leaf area especially during the reproductive stage (DelaVega et al., 2011; Gregersen et al., 2013; Kusaba et al., 2013; Sadras et al., 2000a,b). During leaf senescence, a massive degradation of photosynthetic proteins takes place. In addition, Martínez et al. (2008) and Carrión et al. (2013) demonstrated that the accumulation of small ‘senescence‐associated vacuoles’ (SAVs) with intense proteolytic activity in senescing leaves of soya bean and Arabidopsis thaliana is associated with chloroplast protein breakdown. This finding suggests autophagy is involved in the process. Considering the complexity and dynamic nature of these processes, embarking on an experimental systems biology approach is crucial in understanding the leaf senescence process (Guo, 2013). At the transcriptional level, the regulation of senescence has been well studied in model species such as A. thaliana (Gepstein et al., 2003; Lim et al., 2003; Lin and Wu, 2004). Furthermore, this process has been partially assessed in other species such as rice (Lee et al., 2001), tobacco (Pageau et al., 2006), pea (Pic et al., 2002), wheat (Uauy et al., 2006), cotton (Kong et al., 2013), maize (Sekhon et al., 2012; Zhang et al., 2014), turnip (Gombert et al., 2006), barley (Hollmann et al., 2014; Jukanti et al., 2008) and sunflower (Cabello et al., 2006; Fernandez et al., 2012a; Moschen et al., 2014).
Transcription factors (TFs) are key proteins in the regulation of gene expression and signal transduction networks regulating different biological processes. Several TF families have been associated with leaf senescence in many species, particularly the NAC (Balazadeh et al., 2010, 2011; Guo and Gan, 2006; He et al., 2005; Hu et al., 2010; Kim et al., 2009, 2013b; Matallana‐Ramirez et al., 2013; Nuruzzaman et al., 2010; Wang and Dane, 2013), MYB (Jaradat et al., 2013; Zhang et al., 2011), AP2 (Chen et al., 2012a,b; Koyama et al., 2013) and WRKY (Besseau et al., 2012; Miao et al., 2008; Rushton et al., 2012; Ulker et al., 2007; Zhou et al., 2011) families.
The advances in genome sequencing and gene expression profiling tools allowed transcriptome analyses during natural and induced senescence (Breeze et al., 2011; Buchanan‐Wollaston et al., 2005; Guo and Gan, 2012; Guo et al., 2004). These transcriptomic analyses increased the understanding of senescence; senescence regulation at the metabolic level, however, remains limited.
Recently, Watanabe et al. (2013) performed a detailed spatio‐temporal dissection of metabolic changes throughout senescence in A. thaliana. This studied covered approximately 260 metabolites, including different pigments, lipids, sugars, amino acids, organic acids, secondary metabolites and ionic nutrients.
An integrative study of different omics approaches for the study of leaf senescence was recently conducted in maize (Sekhon et al., 2012) and A. thaliana (Balazadeh et al., 2014).
The aim of the present study was to characterize transcriptional and metabolic pathways related to the triggering and progression of leaf senescence in sunflower, through the integration of transcriptomic, metabolomic and physiological profiles evaluated at different developmental stages. Moreover, considering the increasing concern about whether glasshouse experiments could reproduce the natural growth conditions of field experiments (Gregersen et al., 2013), we conducted the assays simultaneously under both conditions.
Results
Physiological measurement of leaf senescence
Plant development and flowering time (100 °Cd later) were delayed under glasshouse conditions compared to field conditions (Table 1). The plants grown in the glasshouse lacked the natural leaf senescence phenotype observed in field experiments. Thus, in glasshouse experiments, leaves remained green throughout plant development until an abrupt senescence concomitant with general plant senescence.
Table 1.
Leaf sampling for transcriptomic and metabolomic analyses at three thermal time points
| Condition | T‐0 | T‐1 | Flowering | T‐2 |
|---|---|---|---|---|
| Field (°CDAE) | 564 | 768 | 810 | 861 |
| Glasshouse (°CDAE) | 621 | 871 | 910 | 1089 |
We also assessed the progression of leaf senescence throughout leaf development by measuring changes in chlorophyll content (glasshouse and field experiments) as well as nitrogen and soluble carbohydrates (field experiment). Under field conditions, maximum chlorophyll content reached 0.03 mg/cm2, more than 200 °Cd ahead from flowering time. From 700 °Cd after emergence, chlorophyll began to decline up to undetectable levels close to 900 °Cd (Figure 1a). The chlorophyll content under glasshouse conditions remained apparently unaltered along the evaluated time points (Figure 1a).
Figure 1.
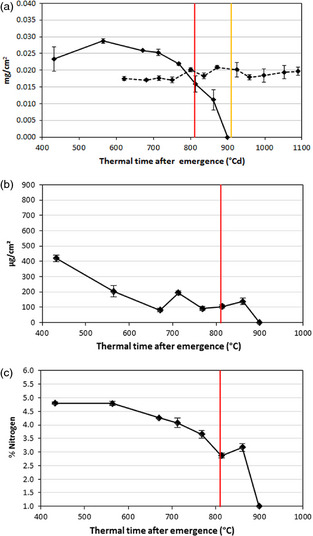
Physiological measurements during leaf senescence. (a) Chlorophyll content in mg/cm2 under field (solid line) and glasshouse conditions (dotted line); (b) total soluble carbohydrates in μg/cm² under field conditions and (c) total nitrogen percentage under field conditions. The red line indicates anthesis time under field conditions and the orange line indicates anthesis time under glasshouse conditions. Error bars correspond to standard errors.
After an initial high value above 400 μg/cm², total soluble carbohydrates (TSC) decreased to values between 100 and 200 μg/cm² from 600 up to 850 °Cd just after flowering. Then, TSC dropped abruptly until leaf death around 900 °Cd (Figure 1b). Similarly, nitrogen content (%) started to decline at 600 °Cd after emergence (Figure 1c) with a similar rate to that of the chlorophyll degradation and before flowering time; at that time, the N content had already been reduced to 60% of its maximum content.
Transcriptomic analysis
Microarray data analysis allowed the detection of 10 173 and 7517 probes in the field and glasshouse experiments, respectively; these probes represent differentially expressed genes between the different time points throughout leaf development (T‐1 vs. T‐0 and T‐2 vs. T‐0). Values were considered significant with P‐values lower than 0.05 and with a log2 fold change higher or lower than 2. Microarray validation through qPCR analysis yielded similar expression profiles in nine of the ten evaluated transcripts in comparison with the microarray data from the field experiment (Figure S1). Principal component analysis (PCA) of transcriptomic data showed a lower variability among biological replicates in the field experiment over the glasshouse condition. This result could, however, reflect the sample unit definition in each condition, as each biological replicate consisted of three randomly selected plants from each plot in the field experiment and one plant per plot in the glasshouse experiment (Figure 2a). Hereafter, elevated mRNA levels will be described as up‐regulated and decreased mRNA levels as down‐regulated.
Figure 2.
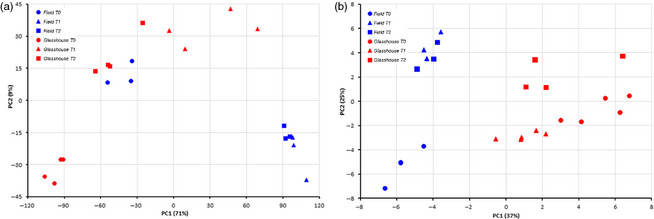
Principal component analyses. (a) Transcript analysis. (b) Metabolite analysis. The blue and red dots correspond to the biological replicates of field and glasshouse experiments, respectively, during leaf development.
Functional analysis
A gene set analysis allowed the detection of the differentially enriched functional categories represented on the microarray (Figure 3). This analysis revealed that most GO categories of biological processes and molecular function were down‐regulated rather than up‐regulated during senescence under both conditions (Figure 3). Furthermore, the list of differentially regulated functional categories was invariant between glasshouse and field conditions at the pre‐anthesis time (T1‐T0). Among the up‐regulated genes, these GO categories contained genes mainly related to transcriptional regulation, TFs and ligase activity, especially involved in the carbon–nitrogen and carbon–oxygen link formation. Furthermore, among these genes, there were genes related to transferase activity, specifically nitrate and phosphate group transferases (Figure S2a and c). Among the down‐regulated genes, we found high enrichment of oxido‐reductase, transferase and isomerase catalytic activities, especially those related to glucosyltransferase activities involved in cell wall synthesis, as well as of genes related to carbon and sulphide metabolite groups (Figure S2b and d).
Figure 3.
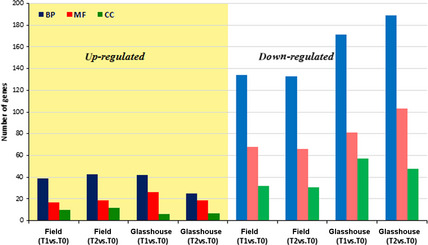
Distribution of significant GO terms. BP: Biological process, MF, molecular function; CC, cellular component. The bars on the left indicate the number of up‐regulated GO and the bars on the right indicate the number of down‐regulated GOs in both field and glasshouse experiments.
Senescence‐associated genes (SAGs) analysis
To assess the behaviour of previously reported SAGs, we performed a BLAST search of approximately 1200 A. thaliana SAG sequences reported on the leaf senescence database (http://psd.cbi.pku.edu.cn/) (Li et al., 2014) against the sunflower database (SUR v1.0) (Fernandez et al., 2012b). The output of this search was that 369 candidate unigenes had high sequence similarity. Of these candidates, 167 and 103 putative SAGs were differentially expressed during leaf senescence in the field (Figure 4a) and glasshouse (Figure 4b) experiments, respectively. Analysis of the most relevant molecular functions revealed that most of them were up‐regulated and corresponded to genes associated either with nutrient recycling, lipid and/or carbohydrate metabolism, transcription regulation, protein degradation or redox regulation.
Figure 4.
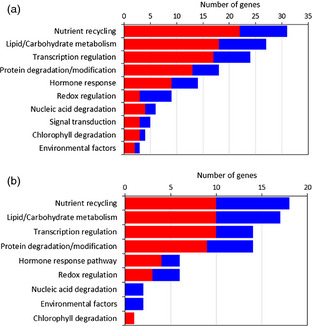
Differentially expressed SAGs assessed by microarray analysis. Differentially expressed genes during leaf development (T0 vs. T1 and/or T2 vs. T0) in the (a) field and (b) glasshouse experiments. The red and blue bars indicate the number of up‐regulated and down‐regulated genes, respectively.
As mentioned before, a higher number of differentially expressed SAGs were detected under field conditions. For this reason, we selected the detected up‐ and down‐regulated SAGs from this experiment for further analysis. The selected SAGs were those with a log2 fold change higher or lower than 2 (Table S2). Among this subset, the most significantly expressed categories were enzymes related to nutrient recycling such as glyoxylase I and lysine‐ketoglutarate reductase enzymes. SAG12, a cysteine protease, which is normally used as a senescence marker gene (Lohman et al., 1994), was also up‐regulated during leaf senescence. Its senescence‐associated up‐regulation was previously demonstrated in several species including agronomic crops such as barley (Jukanti et al., 2008; Parrott et al., 2010) and wheat (Gregersen and Holm, 2007; Ruuska et al., 2008). Furthermore, TFs previously reported as SAGs, such as representatives of the NAC, MYB, MBF1, HSF and AP2‐EREBP families, also exhibited high expression levels during leaf senescence. Similarly, genes related to lipid and carbohydrate metabolisms and protein degradation displayed high expression levels throughout this process.
Analysis of transcription factors
We identified sunflower TFs by comparing approximately 23 000 sequences of TFs from Arabidopsis lyrata, A. thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera and Zea mays. These sequences are available at the plant TF database (http://plntfdb.bio.uni-potsdam.de/v3.0/) (Pérez‐Rodríguez et al., 2010) and were compared with SUR v1.0 database (Fernandez et al., 2012b). A total of 687 candidate unigenes with high sequence similarity to TFs were identified. The expression analysis of these candidate TFs revealed that 140 and 123 were differentially expressed along leaf senescence under field (Figure 5a) and glasshouse conditions, respectively (Figure 5b). The most represented families correspond to the AP2‐EREBP, MYB, NAC, bZIP and HB TF families. Several members of these TF families have been previously reported to be associated with leaf senescence as well as with different biotic and abiotic stress responses in model species. Furthermore, most of these TF families showed a high ratio of up‐/down‐regulated genes throughout the senescence process.
Figure 5.
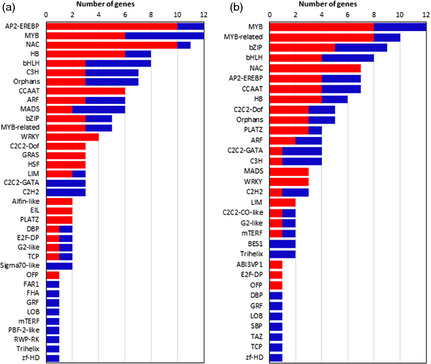
Differentially expressed putative transcription factors derived from the microarray analysis. Differentially expressed genes during leaf development (T0 vs. T1 and/or T2 vs. T0) in the (a) field and (b) glasshouse experiments. The red and blue bars indicate the number of up‐regulated and down‐regulated genes, respectively.
Similarly to the results of the SAG analysis, we detected several putative TFs with higher expression levels during leaf senescence in the field experimental condition. Moreover, when we selected the top up‐ and down‐regulated TFs with a log2 fold change higher or lower than 2, we observed that the most represented TF families not only exhibited a large number of differentially expressed genes, but also showed higher expression levels in relation to other gene families (Table S3).
Spearman correlation analysis was performed to assess possible co‐expression and coregulation patterns during natural leaf senescence. This analysis was performed between the differentially expressed TFs of the field experiment (Figure 6). It revealed many positive correlations mainly among the up‐regulated genes. The NAC TF family showed a large number of inter‐relationships with other candidate TFs including those of the NAC, AP2‐EREBP, MYB, MYB‐related and ARF families. Among the NAC TF family, we found high sequence identity with genes associated with leaf senescence and different stresses in A. thaliana: NAC19, NAC02, NAC47, NAC55 and NAC81.
Figure 6.
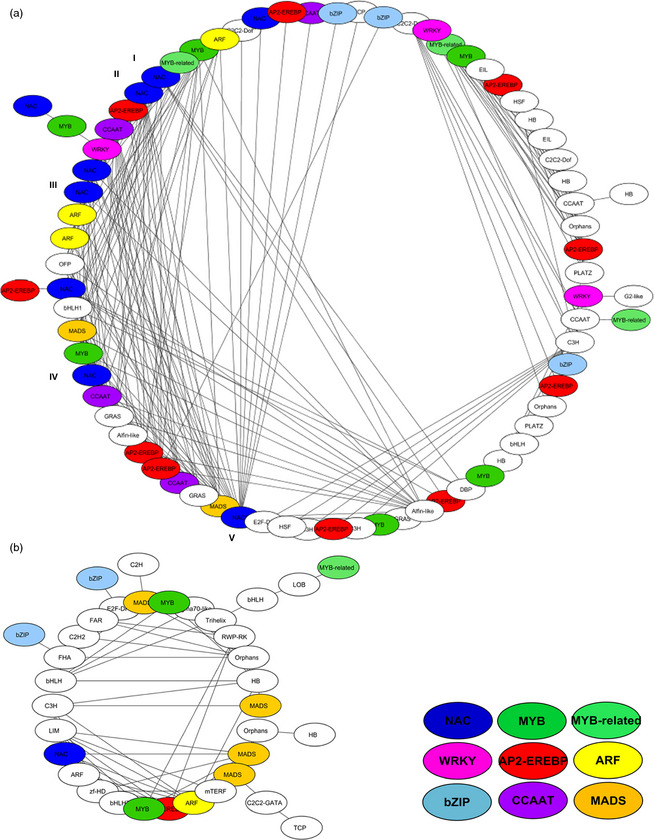
Transcription factor correlation analysis. Spearman correlations between differentially expressed TFs during leaf senescence progress under natural conditions, the field experiment (T0 vs. T1 and/or T2 vs. T0). (a) Up‐regulated and (b) down‐regulated TFs during leaf senescence. Correlation P‐value <0.01. Roman numerals indicate high sequence similarity to Arabidopsis thaliana genes: I: NAC19, II: NAC02, III: NAC47, IV: NAC55 and V: NAC81.
Metabolic analysis
Primary metabolite analysis
By GC‐TOF‐MS analysis, we detected around 60 primary metabolites during leaf development in both conditions, including different amino acids, organic acids, sugars and sugar alcohols (Figure 7).
Figure 7.
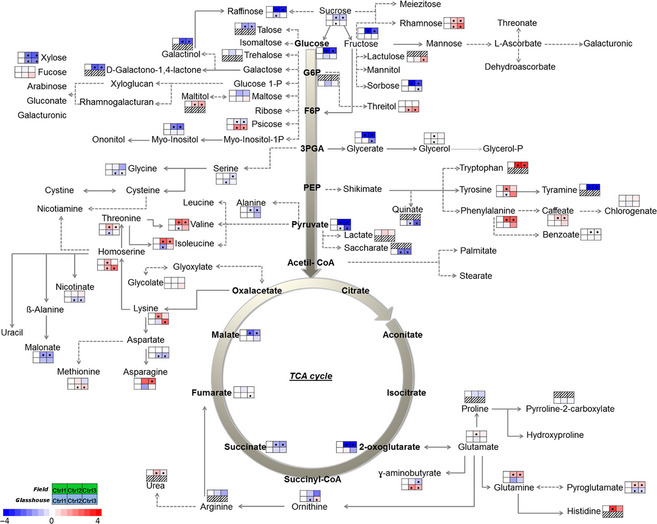
Metabolic profile analysed by GC‐TOF‐MS. Each graph represents the fold change considering the time 0 as a control; columns represent the three evaluated times for each metabolite. The first row corresponds to the field experiment, whereas the second row corresponds to the glasshouse experiment. The points within each graph indicate significant differences (P‐value <0.05).
The metabolic profiles of field‐ and glasshouse‐grown materials showed similar patterns. In both experiments, carbohydrate levels along with metabolites of the tricarboxylic acid (TCA) cycle decreased during leaf senescence. By contrast, many other metabolites involved in the amino acid metabolism increased their level, in particular aromatic amino acids such as tryptophan, tyrosine and phenylalanine, and branched‐chain amino acids such as isoleucine and valine. We also observed high levels of asparagine and glutamine, amino acids involved in the nutrient recycling process.
As in the transcriptomic analysis, these changes were considerably stronger in the field experiment, with higher fold changes during the progress of leaf development. The different growth conditions could explain the observed differences. For instance, in the field experiment, plants receive higher light intensity, which could enhance the differences in metabolite levels.
Similarly to the transcriptomic results, PCA of metabolite data revealed a closer clustering among biological replicates of the field experiment compared to the glasshouse condition (Figure 2b). With these results in mind, we focused the integrated data analysis on the field experiment to assess leaf senescence under natural environmental conditions.
Ionic nutrient analysis
Four anionic nutrients (chloride, nitrate, sulphate and phosphate) and five cationic nutrients (sodium, ammonium, potassium, magnesium and calcium) were detected and quantified by ion chromatography in samples from both field and glasshouse experiments (Figure 8). Among the anionic nutrients, nitrate levels increased during leaf development under both conditions, whereas sulphate content decreased throughout leaf senescence. Phosphate contents slightly decreased under field conditions, whereas remained apparently unaltered under glasshouse conditions. In contrast, cationic nutrients did not display well‐defined patterns during leaf senescence.
Figure 8.
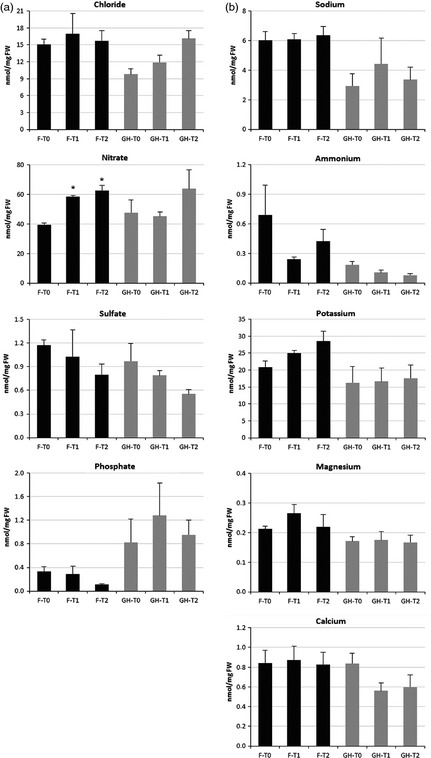
Ion chromatography analysis. (a) anion and (b) cation analysis. Ionic nutrients in nanomole per milligram fresh weight. F, field; GH, glasshouse. T0, T1 and T2 correspond to the three leaf samplings. The asterisk indicates significant differences (P‐value <0.05).
Data integration
With the aim of characterizing the progression of leaf senescence in sunflower, we performed a transcriptomic and metabolic data integration using MapMan (Thimm et al., 2004). Field (Figure 9) and glasshouse (Figure S3) experiments showed similar up‐ and down‐regulated patterns, with a higher fold change observed in the field experiment. For this reason, we focused our analysis in the field experiment.
Figure 9.
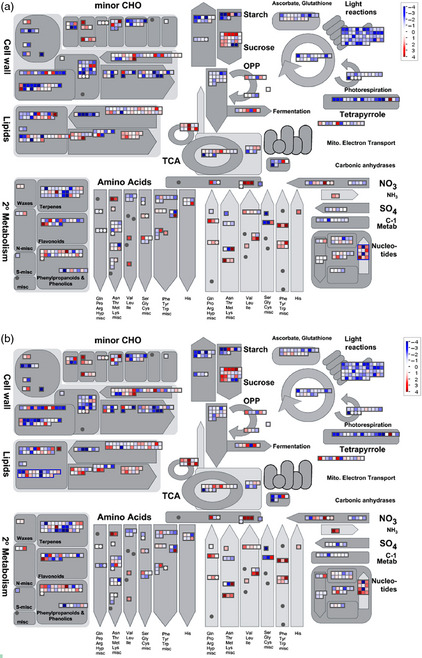
Metabolism overview in the field experiment. (a) Pre‐anthesis time T1 vs. T0 and (b) postanthesis time T2 vs. T0. Genes and metabolites are represented by squares and circles, respectively. Colour intensity corresponds to the expression ratio at logarithmic scale (red: up‐regulated, blue: down‐regulated).
Data integration allowed the detection of the most important biological processes during leaf senescence in sunflower plants. Under the evaluated conditions, we observed a decrease in the expression levels of genes and in metabolite contents related to photosynthesis and photorespiration as well as to the ascorbate–glutathione cycle involved in antioxidant defence mechanisms. Similarly, glycolysis‐associated genes were also down‐regulated, and as expected, the metabolites involved in these pathways were showed lower levels. On the other hand, sucrose and starch synthesis genes had low expression levels throughout leaf senescence with the exception of sucrose degradation genes whose expression increased through the process. In addition, we found high expression of genes related to lipid degradation pathways, the glyoxylate cycle, nucleotide and amino acid degradation, as well as genes associated with nutrient recycling.
At the pre‐anthesis stage, prior to the appearance of first visual symptoms of leaf senescence, we detected high expression levels of genes and contents of metabolites related to nutrient recycling. Beta‐oxidation of fatty acids and the glyoxylate cycle‐related genes showed particularly high expression levels whilst the expression of malate synthase and isocitrate lyase enzymes was also considerably higher. Furthermore, genes related to amino acid and nucleic acid degradation were up‐regulated during leaf development. For instance, glutamate dehydrogenase was up‐regulated and thus could be anticipated to lead to an enhanced production of ammonia. Ammonia in turn is substrate of the glutamine synthase enzyme, which was also up‐regulated, and thus could be anticipated to generate glutamine as a transport amino acid. The asparagine synthetase gene was also up‐regulated at an early stage during senescence, thus potentially increasing asparagine production, another important process of mobilization during nutrient recycling.
Discussion
Transcriptomic analysis of sunflower leaf senescence
Different genes are up‐ or down‐regulated during senescence, and these genes might be involved in multiple signalling pathways interconnected in a network that controls leaf senescence. In the present study, a transcription profile analysis of leaf senescence in sunflower plants grown under field or glasshouse conditions was performed using Agilent microarray technology (Fernandez et al., 2012b).
A functional enrichment analysis via gene set analysis methodology (Montaner and Dopazo, 2010) demonstrated a decrease in mitochondrial activity and cell growth during senescence (Figure S2b and d). In addition, an enrichment of genes related to transcriptional regulation and nutrient recycling was detected (Figure S2a and c). Similar results were obtained in A. thaliana, in which the transcriptional profile was assessed throughout senescence (Breeze et al., 2011). This process was further described in other nonmodel plants such as barley (Parrott et al., 2007) and wheat (Gregersen and Holm, 2007).
Genes related to recycling processes, such as kinases, phosphatases, proteases and hydrolases, were not only found to be differentially expressed throughout senescence, but most of them were specifically up‐regulated during this process (Table S2). In this analysis, we identified differentially expressed TFs mainly from the NAC, MYB, AP2‐EREBP, HB and bZIP families (Figure 5). Members of these TF families have been widely studied and reported as candidate genes activating the leaf senescence programme in different species (Balazadeh et al., 2008; Breeze et al., 2008). Genes with high sequence similarity to ANAC055, ANAC019 and ANAC029 from A. thaliana were highly expressed during sunflower senescence (AT3G15500, AT1G52890 and AT1G69490, respectively) (Table S3). In addition, two genes with high similarity to ANAC055 and ANAC019, which are involved in jasmonic acid and/or ethylene signalling pathways particularly in response to pathogen attack (Bu et al., 2008), were also detected as highly correlated. ANAC019 was further postulated as a triggering factor of senescence with a role in the flavonoid and anthocyanin biosynthesis; ANAC055, on the other hand, could accelerate senescence and seems to be involved in the chitin response (Hickman et al., 2013). Additionally, ANAC029, also called NAC‐LIKE ACTIVATED BY AP3/PI (AtNAP), has previously been reported as a senescence activator (Guo and Gan, 2006).
Two MYB TFs, which displayed high expression levels, exhibit high sequence identity to the A. thaliana MYB44 and MYB73 (AT5G67300 and AT4G37260, respectively) (Table S3). MYB44 is expressed during plant development especially in senescent leaves and behaves as a negative regulator of ABA, stress, wounding and senescence, through an homoeostatic function, by maintaining the growth (Jaradat et al., 2013). This TF may be induced by ethylene (Liu et al., 2010). MYB73 is induced by salt stress and is a negative regulator of salt overly sensitive genes (SOS) (Kim et al., 2013a).
Regarding AP2‐EREBP TF family, three members with high sequence similarity to ERF104, ERF3 and ERF72 (AT5G61600, AT1G50640 and AT3G16770, respectively) showed high expression levels during senescence in sunflower (Table S3). These TFs have not yet been directly associated with senescence process; however, this family is part of a regulatory network involved in hormonal regulation by sugars and redox signalling in response to several abiotic stresses (Dietz et al., 2010).
HAHB4 (AF339748) is a sunflower HD‐Zip TF family that has been extensively studied in last years in response to biotic and abiotic stress (Dezar et al., 2005a,b; Gago et al., 2002; Manavella et al., 2006, 2008a,b). HAHB4 displayed high levels during leaf senescence in sunflower (Table S3). This TF is positively regulated by ethylene and, once induced, it itself negatively regulates ethylene biosynthesis‐ and signalling‐related genes (Manavella et al., 2006). HAHB4 is also involved in response to wounding and abiotic stress, positively regulating ethylene and jasmonic acid production and negatively controlling ethylene sensitivity and abscisic acid accumulation (Manavella et al., 2008b).
High levels of correlation between these TFs families were detected (Figure 6), especially among members of the NAC family in agreement with previous results described for A. thaliana (Balazadeh et al., 2008). In this analysis, we also found a transcript with high sequence identity to ANAC02 or ATAF1. This transcript plays an important role in the adaptation processes to different environmental stresses and in plant development regulating ABA synthesis (Jensen et al., 2013). It also responds to different pathogens (Wang et al., 2009) and is involved in ethylene signalling pathways (Shan et al., 2012). Furthermore, ATAF1 might be associated with an upstream regulation of the signalling pathway ORE1 and EIN2 (Balazadeh et al., 2013), thus activating their expression and inhibiting the expression of Golden2‐like (GLKs) genes, which are important for chloroplast development and maintenance (Rauf et al., 2013). ORE1 also acts as an antagonist of GLK protein and in this way adds more complexity to this regulation pathway (Rauf et al., 2013). In A. thaliana, the ORE1 TF can induce leaf senescence (Balazadeh et al., 2010). In addition, the micro‐RNA miR164 may suppress ORE1 transcript levels; miR164 and ORE1 may be regulated in a loop that would also involve EIN2, where EIN2 would promote the expression of ORE1 and would inhibit miR164 (Kim et al., 2009). In a previous work conducted in sunflower, the expression profiles of candidate genes Ha‐EIN2 and Ha‐NAC01 (with high sequence identity to ORE1) were evaluated together with miR164 levels (Moschen et al., 2014). In that study, the candidate genes and miR164 displayed high expression levels prior to anthesis and ahead of the first senescence symptoms. This finding was in line with the increase in the nutrient remobilization rate. In addition, Kim et al. (2014) demonstrated that ANAC019, ANAC047 and ANAC055 together with ORE1 are preferentially under the control of EIN2 during leaf ageing. These three NAC TFs are within the highly correlated group. Therefore, this TF family is a putative regulator of leaf senescence in sunflower.
Transcriptomic analysis plays an important role in the study of leaf senescence. These results suggest a complex regulatory network underlying the senescence process; thus, the identification of regulatory networks based on expression profiling is an important starting point for the detection of new key genes involved in the triggering of the senescence process.
Metabolic analysis of sunflower leaf senescence
Senescence development in plants is a physiologically coordinated process that is critical for nutrient recycling from older leaves to growing organs. Metabolite remobilization from mature and senescent leaves to the different sinks, particularly into seed development, affects their quality and quantity and is one of the most important aspects in crop improvement (Gregersen et al., 2013). In this study, we performed a metabolic profile analysis of leaf senescence in sunflower in the same leaves assessed through transcriptomic profile.
Carbon metabolism
The role of sugars in senescence has been widely discussed in recent years. Sugars are central elements of the source–sink relationships (Balibrea Lara et al., 2004; Roitsch and González, 2004) and have been reported as growth (Smeekens et al., 2010) and photosynthetic rate regulators (Wingler et al., 1998). However, the effect of sugars on senescence is controversial and differs between different species (Nooden, 1988; Wingler et al., 2009; Yoshida, 2003). In A. thaliana, for example, sugar accumulation occurs during leaf development, whereas exogenous application of sugars induces an early senescence (Diaz et al., 2005; van Doorn, 2008; Masclaux et al., 2000; Masclaux‐Daubresse et al., 2005; Quirino et al., 2001; Watanabe et al., 2013; Wingler et al., 2006, 2012). In sunflower, on the other hand, sugar content decreases during leaf development in field and glasshouse experimental conditions (Figure 7). This finding is in line with previous studies, in which the photosynthetic rate decreased together with sugar levels in a mature leaf (Quirino et al., 2000). Furthermore, sunflower is a plant with a strong demand for nutrient, especially sugars as substrate for oil synthesis, during the grain‐filling phase. Likewise, low levels of sugars may increase production and/or ethylene sensitivity, which acts as senescence enhancer (Grbic and Bleecker, 1995; Hoeberichts et al., 2007). Several studies have described the role of sugars in the senescence programme. However, the results are variable and depend on the species, treatments, leaf and leaf section assessed; therefore, general conclusions concerning their effect are ambiguous (van Doorn, 2008).
Amino acid metabolism
Aromatic amino acids, which are derivatives of shikimate pathway, such as tryptophan, tyrosine and phenylalanine showed high levels throughout senescence (Figure 7). These amino acids have an important role as precursors of the secondary metabolite synthesis such as flavonoids. Specifically, flavonoids act as cellular protectants during senescence (Pichersky et al., 2006; Radwanski and Last, 1995; Watanabe et al., 2013). Particularly, tyrosine is a precursor of vitamin E or α‐tocopherol biosynthesis, which acts as a powerful antioxidant in tissues (Almeida et al., 2011; Collakova and DellaPenna, 2003; Falk and Munné‐Bosch, 2010; Quadrana et al., 2013). Indeed, vitamin E accumulation occurs in senescent leaf tissue (Abbasi et al., 2009; Molina‐Torres and Martinez, 1991; Rise et al., 1989).
Furthermore, branched amino acids such as isoleucine and valine also displayed high levels throughout leaf development. These amino acids have a role in respiration as alternative substrates under stress and increase their level under dark‐induced senescence (Araújo et al., 2010, 2011). Glutamine and asparagine levels also increased through leaf senescence. These amino acids are involved in nitrogen and carbon transport between the different organs and are the most abundant amino acids in the xylem and phloem, which indicates that they are actively involved in nutrient remobilization (Lea and Miflin, 1980; Urquhart and Joy, 1981).
GABA highly increased during senescence under glasshouse conditions (Figure 7). GABA is a product of glutamic acid decarboxylation, and its accumulation is associated with several stresses (Kinnersley and Turano, 2000; Obata and Fernie, 2012; Roberts, 2007) and also to senescence leaves of legumes and tobacco (Lahdesmaki, 1968; Masclaux et al., 2000).
Different roles have been proposed for GABA accumulation. For instance, it could be an amplifier of stress signals through the increase of ethylene synthesis; GABA could also be involved in temporary storage of nitrogen and could be an anaplerotic compound by providing TCA cycle intermediates during stress response (Kinnersley and Turano, 2000).
Ionic nutrients
Among the different ionic nutrients detected in this analysis, nitrate represents one of the most relevant groups during senescence. The concentration of this element increased during senescence in both evaluated conditions (Figure 8). This fining differs from a study in A. thaliana. In A. thaliana, the nitrate content decreased during senescence in the whole plant with an accumulation gradient from the tip to the base of the leaves (Watanabe et al., 2013). However, an increase in nitrate content has been detected in older leaves of tobacco plants, whereas the opposite effect was observed in young leaves (Masclaux et al., 2000). This increase could be related to the role of nitrates as osmoprotectants in senescent vacuoles (McIntyre, 1997), which would explain the elevated cell turgor observed in senescent cells (Brugière et al., 2000).
The sulphate content decreased progressively during leaf development in sunflower and this decrease negatively correlates with the increase in nitrate content (Blom‐Zandstra and Lampe, 1983; Diaz et al., 2005). However, the mechanisms by which sulphates are remobilized and exported during leaf senescence have not yet been clearly defined. Oilseed species such as canola (Brassica napus L.) are particularly sulphur demanding and highly sensitive to the lack of this compound with considerable consequences for seed yield and quality (Dubousset et al., 2009; Janzen and Bettany, 1983; Scherer, 2001).
Altogether, these results suggest high nutrient mobility throughout the leaf senescence process in sunflower plants. Effective nutrient remobilization from leaf to seed during senescence is very important for weight increase and seed quality, especially in agronomically important crops such as sunflower. The transition between sink and source that takes place in leaves is important and critical for a proper development of the plant (Watanabe et al., 2013).
Integrative analysis of sunflower leaf senescence
Throughout the study of complex cellular processes such as leaf senescence, the experimental systems biology approach attempts to assess the dynamics and integration of the different molecular components associated with physiological responses.
In annual crops, senescence is typically induced in the whole plant after flowering. Foliar nutrients are remobilized and translocated into the fruit, and this leads ultimately to cell death. Therefore, this is a process tightly controlled by the development of reproductive structures (Davies and Gan, 2012; Gregersen et al., 2013). This highlights the strong pressure of the source–sink relationships that control the onset of senescence in monocarpic plant species.
In the present study, we focused leaf sampling at early stages of plant development in both field and glasshouse experiments, to find candidate genes involved in triggering of the senescence process. In natural field growth conditions, chlorophyll and nitrogen content sharply decreased after flowering, whereas in glasshouse condition, chlorophyll content remains relative stable throughout the sampling period likely due to radiation limitation (Figure 1). In rice, it was reported that low irradiance strongly retarded the decline in chlorophyll content which remained almost constant until late senescence when an abrupt decreased took place (Hidema et al., 1991). The relationship between productivity and senescence is complex. Productivity would benefit from a delay in senescence, and this delay has to be coordinated with the early stages of development, particularly at the beginning of the reproductive phase growth (Gregersen et al., 2013).
Studies at transcriptional and metabolic levels in sunflower leaves allowed us to detect significant changes in the metabolism at an early stage of development (T1 vs. T0), prior to anthesis and before the onset of the first senescence symptoms assessed by physiological measurements especially in the field experiment (Figure 9a and Figure S3a). In the postanthesis period (T2 vs. T0), the detected changes were similar to those observed at an early stage. However, at postanthesis, we detected the highest values of gene expression and metabolite levels associated with different cell metabolism (Figure 9 and Figure S3a).
The genes and metabolites related to the photosynthetic processes were down‐regulated during leaf development. A decrease in photosynthetic activity directly impacts sugar metabolism. In this study, we detected a decrease in sugar levels during senescence, in contrast to what happens in A. thaliana, where sugar accumulation occurs (Watanabe et al., 2013). The genes related to sucrose degradation, such as different cell wall, vacuolar and neutral invertases and fructokinases, displayed high levels of expression.
Lipid metabolism‐associated genes were highly expressed, especially those related to the beta‐oxidation process. The beta‐oxidation process culminates in acetyl‐CoA production, which is substrate of both the TCA cycle and the enzyme malate synthase. Acetyl‐CoA also displayed high expression levels. Glyoxylate cycle enzymes such as isocitrate lyase and malate synthase increased their activity at early stages of senescence in barley (Gut and Matile, 1988) and cucumber (Graham et al., 1992). These findings indicate a role of carbon recycling during senescence. However, the glyoxylate cycle in A. thaliana does not play a major role in nutrient recycling, and the malate synthase expression is specifically restricted to young seedlings and root tips of older plants; in senescent leaves, both malate synthase and isocitrate lyase were undetectable (Charlton et al., 2005).
A critical component associated with the senescence process is the protein degradation (Roberts et al., 2012). Genes associated with protein processing, ubiquitination and degradation such as kinases, phosphatases, cysteine protease (SAG12), F‐box protein, and heat‐shock proteins (Hsp70 and Hsp90) also displayed high expression levels during sunflower senescence. Proteases also show high expression levels in senescent leaves in different species (Desclos et al., 2009; Hollmann et al., 2014; Schiltz et al., 2004) and many of them are reported as SAGs. Throughout protein renovation and degradation, ammonium release occurs. Indeed, ammonium release is accelerated during senescence (Gregersen, 2011). This ammonium group could be channelled back into amino acid structures by the action of glutamine synthetase, which plays a central role in nitrogen remobilization (Bernard and Habash, 2009; Buchanan‐Wollaston et al., 2003; Martin et al., 2006; Tabuchi et al., 2007). In support of this theory, enzymes involved in recycling, such as asparagine synthetase and glutamine synthase transcripts, as well as the associated metabolites, asparagine and glutamine, showed high levels during sunflower leaf development. This finding indicates a strong recycling activity at early stages of leaf development, prior to the appearance of the first senescence symptoms.
In summary, in sunflower many biological processes were up‐ and down‐regulated at an early stage of plant development, prior to anthesis and before the onset of the first visual leaf senescence symptoms. We identified candidate genes associated with the senescence process, especially NAC TFs that could act as triggers for senescence. Furthermore, we demonstrated the importance of conducting experiments under field conditions that reproduce their natural growing environments. Even though the use of larger pots in glasshouse help to simulate field conditions, the radiation limitations are difficult to overcome. This limitation leads to a marked delay in leaf senescence and, thus, to a temporal shift of the gene expression pattern and metabolomic profiles in glasshouse compared to field experiments. Therefore, a better characterization of the molecular processes that underlie leaf senescence in sunflower was assessed in field experiments.
This is the first study that uses an integrated approach related to the senescence process in sunflower. Thus this study provides an important starting point for future analysis and opens new insights to explore alternative strategies and possibilities.
Understanding the onset of the process will in turn impact on the development of different senescence management strategies and could help controlling the grain‐filling process. This could eventually improve crop yields, which represent an important challenge for the future of agriculture attending to the increase in both, world population and climate risks that affect productivity.
Experimental procedures
Plant material and experimental conditions
The sunflower leaf senescence assays were conducted under field and glasshouse conditions. The field experiment was carried out at the INTA Balcarce Experimental Station (37°45′ S, 58°18′ W) as previously described (Moschen et al., 2014). The glasshouse experiment was performed at the Biotechnology Institute, INTA Castelar (34°36′ S, 58°40′ W). Both experiments were sowed simultaneously during the 2010/11 growing season to maintain similar photoperiod. The sunflower hybrid VDH 487 (Advanta Seeds, Balcarce, Argentina) was sown at a 7.2 plants/m2 with three and five biological replicates (plots) in the field and glasshouse experiments, respectively; each biological replicate consisted of three randomly selected plants from each plot in the field experiment. In the glasshouse experiment, one plant per plot was evaluated.
Under glasshouse conditions, 20 kg pots containing a mixture of soil, sand and peat (5 : 3 : 1) as substrate were used (one plant per pot). In both experiments, temperature, diseases, weeds and insects were adequately controlled. Soil fertility assured maximum yields under nonlimiting water conditions.
Transcriptomic and metabolic profiles were performed using the leaf 10 (numbered from the bottom to the top of the plant) at three different development stages in both experiments, labelled as T‐0 (young leaf), T‐1 (pre‐anthesis leaf) and T‐2 (postanthesis leaf, with senescence symptoms) (Table 1). Time was expressed on a thermal time basis by daily integration of air temperature with a threshold temperature of 6 °C and considering plant emergence as thermal time origin (°CDAE: °C days after emergence) (Kiniry et al., 1992).
Physiological measurements
The physiological measurements were assessed as previously described (Moschen et al., 2014) including chlorophyll, TSC and nitrogen determinations.
Transcriptomic analysis
RNA isolation, quantification and quality controls
The sampled leaves were immediately frozen in liquid nitrogen upon collection and saved at −80 °C until processing. High‐quality total RNA was isolated from 100 mg of frozen tissue using TriPure, according to the manufacturer's instructions (Roche, Buenos Aires, Argentina). The genomic DNA was eliminated after treatment with DNase I for 20 min at room temperature using DNase I (Invitrogen, Buenos Aires, Argentina). The RNA concentration was measured using a Nanodrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). The purity and integrity of total RNA was determined by 260/280 nm ratio and by NanoBioanalyzer RNA‐6000 analysis (Agilent Technologies, Palo Alto, CA).
Microarray analysis
The transcriptomic profile was performed using a previously developed custom sunflower microarray, Agilent 4x44K (Fernandez et al., 2012b). Data preprocessing was performed using the Limma library (Smyth, 2005) from the R‐package (R Core Team, 2012). The background correction was performed using the rma algorithm from the backgroundCorrect function. Between‐array normalization was performed using the quantile method from the normalizeBetweenArray function. Finally, fold changes were transformed to log2 scale and the information from technical replicates was incorporated by calculating the median parameter.
Differential gene expression analysis
Statistical analysis was performed using in‐house routines to fit, gene by gene, a linear mixed‐effects model. The Sunflower Custom Oligo Microarray includes four arrays per chip; therefore, the chip effect (incomplete block) was included as a random effect. The set of routines mentioned above were based on the lme function of the nlme library of R (Pinheiro et al., 2012) implemented in InfoStat statistical software (Di Rienzo et al., 2013). The differential gene expression analysis was carried out using the limma package. Gene set analysis was conducted according to the gene ontology terms (Ashburner et al., 2000) using FatiScan (Al‐Shahrour et al., 2007) integrated in the Babelomics suite (Al‐Shahrour et al., 2005). The correction of P‐values in a multiple test context was performed using the Benjamini–Hochberg method, and the respective adjusted P‐values (Benjamini and Hochberg, 1995) were obtained.
Microarray validation: qPCR analysis
The expression of 10 genes was evaluated in relation to elongation factor‐1α (EF‐1α), which has been previously selected as a reference gene for expression senescence experiments (Fernandez et al., 2011) (Figure S1). Specific primer pairs were designed using Primer3 software (Rozen and Skaletsky, 2000) with default parameters (Table S1).
For each sample, 500 ng DNase‐treated RNA was reverse‐transcribed using Superscript III first strand synthesis system (Invitrogen) and random hexamer primers according to the manufacturer's instructions (Moschen et al., 2014).
Metabolic analysis
GC‐TOF‐MS analysis
The metabolite extraction was performed promoting the extraction of lipophilic and polar compounds according to recently published protocols (Roessner‐Tunali et al., 2003) and further adaptations to sunflower tissue samples (Peluffo et al., 2010). The samples were derivatized and injected (1 μL) into the GC‐TOF‐MS system (LECO Corporation, St. Joseph, MI). Chromatography was performed on a 30 m SPB‐50 column with 0.25 mm inner diameter and 0.25 lm film thickness (Supelco, Belfonte, CA). The injection temperature was 230 °C, the interface was set to 250 °C, and the ion source was adjusted to 200 °C. The carrier gas was He at a constant flow ratio of 1 mL/m. The chromatograms and spectra were evaluated using the ChromaTOF (LECO Corporation) and TagFinder (Luedemann et al., 2008). The ion spectra were compared to the Golm metabolome database (http://gmd.mpimp-golm.mpg.de/). Metabolite levels were normalized to fresh weight and the internal control ribitol. Changes in metabolite levels along leaf development were calculated as the fold change relative to the first sampled time (T‐0) as control.
Ion chromatography
Ionic nutrients were assessed by ion chromatography. A total of 500 μL of 4 °C ULC water (ultra liquid chromatography) was added into each metabolite sample extraction. The samples were shaken in vortex 10 s followed by centrifugation 30 min at 16000 × g at 4 °C. A total of 480 μL of supernatant was transferred into glass vials. Serial dilutions of MgCl2, KNO3, KH2PO4, MgSO4, NH4SO4 and CaCl2: 100, 50, 25, 12.5, 6.25 and 3.125 μm were used as standard solutions. An ionic nutrient analysis was carried out using Ion Chromatograph Dionex IC3000 (Thermo Scientific, Fitchburg, WI).
Transcriptomic and metabolic integration
MapMan analysis
MapMan software (Thimm et al., 2004) was used to integrate transcriptomic and metabolic profiles. For mapping generation, fasta sequences of Helianthus annuus were downloaded from Sunflower Unigene Repository (SUR v1.0) (Fernandez et al., 2012b). Annotation was processed through Mercator annotation pipeline (Lohse et al., 2014).
The log2 ratio of fold change (T1/T0 and T2/T0) was calculated for each experimental condition. The expression cut‐off was log2 fold change higher or lower than 2 and with a P‐value lower than 0.05. The resulting data table was used for MapMan analysis.
Supporting information
Figure S1 Microarray validation.
Figure S2 Significant GO distribution for molecular function at pre‐anthesis time (T1 vs. T0).
Figure S3 Metabolism overview under glasshouse conditions.
Table S1 Primer sequences for qPCR validation analysis.
Table S2 Up‐ and down‐regulated SAGs with a log2 fold change higher or lower than 2 under field conditions.
Table S3 Putative transcription factors up‐ and down‐regulated with a log2 fold change higher or lower than 2 under field conditions.
Supplementary Legends
Acknowledgements
We want to thank Luis Mendez, Carlos Antonelli, Silvio Giuliano, Guillermo Dosio and Luis Aguirrezabal for support in field experiments at INTA Balcarce and Martin Fernandez, Agustin Montenegro, Matias Rodriguez and Juan Ignacio Tevez for their support in glasshouse experiments. Dr. Julia Sabio y Garcia is gratefully acknowledged for critical reading of this manuscript.
References
- Abbasi, A.‐R. , Saur, A. , Hennig, P. , Tschiersch, H. , Hajirezaei, M. , Hofius, D. , Sonnewald, U. and Voll, L.M. (2009) Tocopherol deficiency in transgenic tobacco (Nicotiana tabacum L.) plants leads to accelerated senescence. Plant Cell Environ. 32, 144–157. [DOI] [PubMed] [Google Scholar]
- Aguirrezábal, L.A.N. , Lavaud, Y. , Dosio, G.A.A. , Izquierdo, N.G. , Andrade, F.H. and González, L.M. (2003) Weight per seed and oil concentration in a sunflower hybrid are accounted for by intercepted solar radiation during a definite period of seed filling. Crop Sci. 43, 152–161. [Google Scholar]
- Almeida, J. , Quadrana, L. , Asís, R. , Setta, N. , de Godoy, F. , Bermúdez, L. , Otaiza, S.N. , Corrêa da Silva, J.V. , Fernie, A.R. , Carrari, F. and Rossi, M. (2011) Genetic dissection of vitamin E biosynthesis in tomato. J. Exp. Bot. 62, 3781–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Shahrour, F. , Minguez, P. , Vaquerizas, J.M. , Conde, L. and Dopazo, J. (2005) BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high‐throughput experiments. Nucleic Acids Res. 33, W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Shahrour, F. , Arbiza, L. , Dopazo, H. , Huerta‐Cepas, J. , Minguez, P. , Montaner, D. and Dopazo, J. (2007) From genes to functional classes in the study of biological systems. BMC Bioinformatics, 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, W.L. , Ishizaki, K. , Nunes‐Nesi, A. , Larson, T.R. , Tohge, T. , Krahnert, I. , Witt, S. , Obata, T. , Schauer, N. , Graham, I.A. , Leaver, C.J. and Fernie, A.R. (2010) Identification of the 2‐hydroxyglutarate and isovaleryl‐CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell, 22, 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, W.L. , Tohge, T. , Ishizaki, K. , Leaver, C.J. and Fernie, A.R. (2011) Protein degradation – an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498. [DOI] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C.A. , Blake, J.A. , Botstein, D. , Butler, H. , Cherry, J.M. , Davis, A.P. , Dolinski, K. , Dwight, S.S. , Eppig, J.T. , Harris, M.A. , Hill, D.P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J.C. , Richardson, J.E. , Ringwald, M. , Rubin, G.M. and Sherlock, G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Riaño‐Pachón, D.M. and Mueller‐Roeber, B. (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana . Plant Biol. 10, 63–75. [DOI] [PubMed] [Google Scholar]
- Balazadeh, S. , Wu, A. and Mueller‐Roeber, B. (2010) Salt‐triggered expression of the ANAC092‐dependent senescence regulon in Arabidopsis thaliana . Plant Signal. Behav. 5, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Kwasniewski, M. , Caldana, C. , Mehrnia, M. , Zanor, M.I. , Xue, G.‐P.P. and Mueller‐Roeber, B. (2011) ORS1, an H2O2‐responsive NAC transcription factor, controls senescence in Arabidopsis thaliana . Mol Plant, 4, 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Garapati, P. , Xue, G. and Mueller‐Roeber, B. (2013) A transcription factor upstream of ORE1 and GLK1 integrates ABA signalling with drought‐induced senescence. In 6th European Workshop on Leaf Senescence, 14–18 October. INRA, Versailles, France. [Google Scholar]
- Balazadeh, S. , Schildhauer, J. , Araújo, W.L. , Munné‐Bosch, S. , Fernie, A.R. , Proost, S. , Humbeck, K. and Mueller‐Roeber, B. (2014) Reversal of senescence by N resupply to N‐starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. J. Exp. Bot. 000, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balibrea Lara, M.E. , Gonzalez Garcia, M.‐C. , Fatima, T. , Ehness, R. , Lee, T.K. , Proels, R. , Tanner, W. and Roitsch, T. (2004) Extracellular invertase is an essential component of cytokinin‐mediated delay of senescence. Plant Cell, 16, 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 57, 289–300. [Google Scholar]
- Bernard, S. and Habash, D. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620. [DOI] [PubMed] [Google Scholar]
- Besseau, S. , Li, J. and Palva, E.T. (2012) WRKY54 and WRKY70 co‐operate as negative regulators of leaf senescence in Arabidopsis thaliana . J. Exp. Bot. 63, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom‐Zandstra, G. and Lampe, J.E.M. (1983) The effect of chloride and sulphate salts on the nitrate content in lettuce plants (Lactuca sativa L.). J. Plant Nutr. 6, 611–628. [Google Scholar]
- Breeze, E. , Harrison, E. , Page, T. , Warner, N. , Shen, C. , Zhang, C. and Buchanan‐Wollaston, V. (2008) Transcriptional regulation of plant senescence: from functional genomics to systems biology. Plant Biol (Stuttg), 10, 99–109. [DOI] [PubMed] [Google Scholar]
- Breeze, E. , Harrisona, E. , McHattiea, S. , Hughesa, L. , Hickmana, R. , Hilla, C. , Kiddle, S. , Kim, Y. , Penfold, C.A. , Jenkins, D. , Zhang, C. , Morris, K. , Jenner, C. , Jackson, S. , Thomas, B. , Tabrett, A. , Legaie, R. , Moore, J.D. , Wild, D.L. , Ott, S. , Rand, D. , Beynon, J. , Denby, K. , Mead, A. and Buchanan‐wollaston, V. (2011) High‐resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell, 23, 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière, N. , Dubois, F. , Masclaux, C. , Sangwan, R.S. and Hirel, B. (2000) Immunolocalization of glutamine synthetase in senescing tobacco (Nicotiana tabacum L.) leaves suggests that ammonia assimilation is progressively shifted to the mesophyll cytosol. Planta, 211, 519–527. [DOI] [PubMed] [Google Scholar]
- Bu, Q. , Jiang, H. , Li, C.‐B. , Zhai, Q. , Zhang, J. , Wu, X. , Sun, J. , Xie, Q. and Li, C. (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid‐signaled defense responses. Cell Res. 18, 756–767. [DOI] [PubMed] [Google Scholar]
- Buchanan‐Wollaston, V. , Earl, S. , Harrison, E. , Mathas, E. , Navabpour, S. , Page, T. and Pink, D. (2003) The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnol. J. 1, 3–22. [DOI] [PubMed] [Google Scholar]
- Buchanan‐Wollaston, V. , Page, T. , Harrison, E. , Breeze, E. , Lim, P.O. , Nam, H.G. , Lin, J.‐F.F. , Wu, S.‐H.H. , Swidzinski, J. , Ishizaki, K. and Leaver, C.J. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation‐induced senescence in Arabidopsis. Plant J. 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Cabello, P. , Agüera, E. and la Haba, P.de. (2006) Metabolic changes during natural ageing in sunflower (Helianthus annuus) leaves: expression and activity of glutamine synthetase isoforms are regulated differently during senescence. Physiol. Plant. 128, 175–185. [Google Scholar]
- Carrión, C.A. , Costa, M.L. , Martínez, D.E. , Mohr, C. , Humbeck, K. and Guiamet, J.J. (2013) In vivo inhibition of cysteine proteases provides evidence for the involvement of “senescence‐associated vacuoles” in chloroplast protein degradation during dark‐induced senescence of tobacco leaves. J. Exp. Bot. 64, 4967–4980. [DOI] [PubMed] [Google Scholar]
- Charlton, W.L. , Johnson, B. , Graham, I.A. and Baker, A. (2005) Non‐coordinate expression of peroxisome biogenesis, beta‐oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 23, 647–653. [DOI] [PubMed] [Google Scholar]
- Chen, G.‐H. , Chan, Y.‐L. , Liu, C.‐P. and Wang, L.‐C. (2012a) Ethylene response pathway is essential for ARABIDOPSIS A‐FIFTEEN function in floral induction and leaf senescence. Plant Signal. Behav. 7, 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.‐H. , Liu, C.‐P. , Chen, S.‐C.G. and Wang, L.‐C. (2012b) Role of ARABIDOPSIS A‐FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2. J. Exp. Bot. 63, 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova, E. and DellaPenna, D. (2003) The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol. 133, 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J. and Gan, S. (2012) Towards an integrated view of monocarpic plant senescence. Russ. J. Plant Physiol. 59, 467–478. [Google Scholar]
- DelaVega, A.J. , Cantore, M.A. , Sposaro, M.M. , Trápani, N. , López Pereira, M. and Hall, A.J. (2011) Canopy stay‐green and yield in non‐stressed sunflower. Field. Crop. Res. 121, 175–185. [Google Scholar]
- Desclos, M. , Etienne, P. , Coquet, L. , Jouenne, T. , Bonnefoy, J. , Segura, R. , Reze, S. , Ourry, A. and Avice, J.‐C. (2009) A combined 15N tracing/proteomics study in Brassica napus reveals the chronology of proteomics events associated with N remobilisation during leaf senescence induced by nitrate limitation or starvation. Proteomics, 9, 3580–3608. [DOI] [PubMed] [Google Scholar]
- Dezar, C.A. , Fedrigo, G.V. and Chan, R.L. (2005a) The promoter of the sunflower HD‐Zip protein gene Hahb4 directs tissue‐specific expression and is inducible by water stress, high salt concentrations and ABA. Plant Sci. 169, 447–456. [Google Scholar]
- Dezar, C.A. , Gago, G.M. , Gonzalez, D.H. and Chan, R.L. (2005b) Hahb‐4, a sunflower homeobox‐leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 14, 429–440. [DOI] [PubMed] [Google Scholar]
- Di Rienzo, J.A. , Casanoves, F. , Balzarini, M.G. , Gonzalez, L. , Tablada, M. , Robledo, C.W. InfoStat versión 2014 . Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. URL http://www.infostat.com.ar (2013) InfoStat.
- Diaz, C. , Purdy, S. , Christ, A. , Morot‐Gaudry, J.‐F. , Wingler, A. and Masclaux‐Daubresse, C. (2005) Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. A metabolic profiling approach. Plant Physiol. 1, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, K.‐J. , Vogel, M.O. and Viehhauser, A. (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma, 245, 3–14. [DOI] [PubMed] [Google Scholar]
- van Doorn, W.G. (2008) Is the onset of senescence in leaf cells of intact plants due to low or high sugar levels? J. Exp. Bot. 59, 1963–1972. [DOI] [PubMed] [Google Scholar]
- Dosio, G.A.A. , Aguirreza, L.A.N. , Andrade, F.H. , Pereyra, V.R. and Aguirrezábal, L.A.N. (2000) Solar radiation intercepted during seed filling and oil production in two sunflower hybrids. Crop Sci. 1644, 1637–1644. [Google Scholar]
- Dubousset, L. , Abdallah, M. , Desfeux, A.S. , Etienne, P. , Meuriot, F. , Hawkesford, M.J. , Gombert, J. , Ségura, R. , Bataillé, M.‐P. , Rezé, S. , Bonnefoy, J. , Ameline, A.F. , Ourry, A. , Le Dily, F. and Avice, J.C. (2009) Remobilization of leaf S compounds and senescence in response to restricted sulphate supply during the vegetative stage of oilseed rape are affected by mineral N availability. J. Exp. Bot. 60, 3239–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, J. and Munné‐Bosch, S. (2010) Tocochromanol functions in plants: antioxidation and beyond. J. Exp. Bot. 61, 1549–1566. [DOI] [PubMed] [Google Scholar]
- Fernandez, P. , Di Rienzo, J.A. , Moschen, S. , Dosio, G.A. , Aguirrezabal, L.A. , Hopp, H.E. , Paniego, N. and Heinz, R.A. (2011) Comparison of predictive methods and biological validation for qPCR reference genes in sunflower leaf senescence transcript analysis. Plant Cell Rep. 30, 63–74. [DOI] [PubMed] [Google Scholar]
- Fernandez, P. , Moschen, S. , Paniego, N. and Heinz, R.A. (2012a) Functional approaches to study leaf senescence in sunflower. In Senescence ( Nagata, T. , ed.), pp. 69–88. Croatia: InTech Open Access Publisher. [Google Scholar]
- Fernandez, P. , Soria, M. , Blesa, D. , Di Rienzo, J. , Moschen, S. , Rivarola, M. , Clavijo, B.J. , Gonzalez, S. , Peluffo, L. , Príncipi, D. , Dosio, G. , Aguirrezabal, L. , García‐García, F. , Conesa, A. , Hopp, E. , Dopazo, J. , Heinz, R.A. and Paniego, N. (2012b) Development, characterization and experimental validation of a cultivated sunflower (Helianthus annuus L.) gene expression oligonucleotide microarray. PLoS One, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago, G.M. , Almoguera, C. , Jordano, J. , Gonzalez, D.H. and Chan, R.L. (2002) Hahb‐4, a homeobox‐leucine zipper gene potentially involved in abscisic acid‐dependent responses to water stress in sunflower*. Plant Cell Environ. 25, 633–640. [Google Scholar]
- Gepstein, S. , Sabehi, G. , Carp, M.‐J.M.‐J. , Hajouj, T. , Nesher, M.F.O. , Yariv, I. , Dor, C. and Bassani, M. (2003) Large‐scale identification of leaf senescence‐associated genes. Plant J. 36, 629–642. [DOI] [PubMed] [Google Scholar]
- Gombert, J. , Etienne, P. , Ourry, A. and Le Dily, F. . (2006) The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J. Exp. Bot. 57, 1949–1956. [DOI] [PubMed] [Google Scholar]
- Graham, I.A. , Leaver, C.J. and Smith, S.M. (1992) Induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell, 4, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic, V. and Bleecker, A.B. (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602. [Google Scholar]
- Gregersen, P.L. (2011) Senescence and nutrient remobilization in crop plants. In The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops ( Hawkesford, M.J. and Barraclough, P. , eds), pp. 83–102. Oxford, UK: Wiley‐Blackwell. [Google Scholar]
- Gregersen, P.L. and Holm, P.B. (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 5, 192–206. [DOI] [PubMed] [Google Scholar]
- Gregersen, P.L. , Culetic, A. , Boschian, L. and Krupinska, K. (2013) Plant senescence and crop productivity. Plant Mol. Biol. 82, 603–622. [DOI] [PubMed] [Google Scholar]
- Guo, Y. (2013) Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 82, 519–528. [DOI] [PubMed] [Google Scholar]
- Guo, Y. and Gan, S. (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Guo, Y. and Gan, S.‐S. (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence‐promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35, 644–655. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Cai, Z. and Gan, S. (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. [Google Scholar]
- Gut, H. and Matile, P. (1988) Apparent induction of key enzymes of the glyoxylic acid cycle in senescent barley leaves. Planta, 176, 548–550. [DOI] [PubMed] [Google Scholar]
- He, X.J. , Mu, R.L. , Cao, W.H. , Zhang, Z.G. , Zhang, J.S. and Chen, S.Y. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. [DOI] [PubMed] [Google Scholar]
- Hickman, R. , Hill, C. , Penfold, C.A. , Breeze, E. , Bowden, L. , Moore, J.D. , Zhang, P. , Jackson, A. , Cooke, E. , Bewicke‐Copley, F. , Mead, A. , Beynon, J. , Wild, D.L. , Denby, K.J. , Ott, S. and Buchanan‐Wollaston, V. (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidema, J. , Makino, A. , Mae, T. and Ojima, K. (1991) Photosynthetic characteristics of rice leaves aged under different irradiances from full expansion through senescence. Plant Physiol. 97, 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts, F.A. , van Doorn, W.G. , Vorst, O. , Hall, R.D. and van Wordragen, M.F. (2007) Sucrose prevents up‐regulation of senescence‐associated genes in carnation petals. J. Exp. Bot. 58, 2873–2885. [DOI] [PubMed] [Google Scholar]
- Hollmann, J. , Gregersen, P.L. and Krupinska, K. (2014) Identification of predominant genes involved in regulation and execution of senescence‐associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 65, 3963–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, R. , Qi, G. , Kong, Y. , Kong, D. , Gao, Q. and Zhou, G. (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa . BMC Plant Biol. 10, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen, H.H. and Bettany, J.R. (1983) Sulfur nutrition of rapeseed: I. Influence of fertilizer nitrogen and sulfur rates. Soil Sci. Soc. Am. J. 48, 100–107. [Google Scholar]
- Jaradat, M.R. , Feurtado, J.A. , Huang, D. , Lu, Y. and Cutler, A.J. (2013) Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M.K. , Lindemose, S. , de Masi, F. , Reimer, J.J. , Nielsen, M. , Perera, V. , Workman, C.T. , Turck, F. , Grant, M.R. , Mundy, J. , Petersen, M. and Skriver, K. (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana . FEBS Open Bio. 3, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukanti, A.K. , Heidlebaugh, N.M. , Parrott, D.L. , Fischer, I.A. , McInnerney, K. and Fischer, A.M. (2008) Comparative transcriptome profiling of near‐isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol. 177, 333–349. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Woo, H.R. , Kim, J. , Lim, P.O. , Lee, I.C. , Choi, S.H. , Hwang, D. and Nam, H.G. (2009) Trifurcate feed‐forward regulation of age‐dependent cell death involving miR164 in Arabidopsis. Science, 323, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Nguyen, N.H. , Jeong, C.Y. , Nguyen, N.T. , Hong, S.‐W. and Lee, H. (2013a) Loss of the R2R3 MYB, AtMyb73, causes hyper‐induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170, 1461–1465. [DOI] [PubMed] [Google Scholar]
- Kim, Y.‐S. , Sakuraba, Y. , Han, S.‐H. , Yoo, S.‐C. and Paek, N.‐C. (2013b) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54, 1660–1672. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Hong, S.H. , Kim, Y.W. , Lee, I.H. , Jun, J.H. , Phee, B.‐K. , Rupak, T. , Jeong, H. , Lee, Y. , Hong, B.S. , Nam, H.G. , Woo, H.R. and Lim, P.O. (2014) Gene regulatory cascade of senescence‐associated NAC transcription factors activated by ETHYLENE‐INSENSITIVE2‐mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiniry, J.R. , Blanchet, R. , Williams, J.R. , Texier, V. , Jones, K. and Cabelguenne, M. (1992) Sunflower simulation using the EPIC and ALMANAC models. Field. Crop. Res. 30, 403–423. [Google Scholar]
- Kinnersley, A.M. and Turano, F.J. (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 19, 479–509. [Google Scholar]
- Kong, X. , Luo, Z. , Dong, H. , Eneji, A.E. , Li, W. and Lu, H. (2013) Gene expression profiles deciphering leaf senescence variation between early‐ and late‐senescence cotton lines. PLoS One, 8, e69847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T. , Nii, H. , Mitsuda, N. , Ohta, M. , Kitajima, S. , Ohme‐Takagi, M. and Sato, F. (2013) A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 162, 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M. , Tanaka, A. and Tanaka, R. (2013) Stay‐green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117, 221–234. [DOI] [PubMed] [Google Scholar]
- Lahdesmaki, P. (1968) The amount of gama‐aminobutyric acid and the activity of glutamic carboxylase in ageing leaves. Physiol. Plant. 21, 1322–1327. [Google Scholar]
- Lea, P. and Miflin, B. (1980) Transport and metabolism of asparagine and other nitrogen compounds within the plant. In The Biochemistry of Plants: a Comprehensive Treatise ( Miflin, B. , ed.), pp. 569–608. New York, NY; London: Academic Press Inc. LTD. [Google Scholar]
- Lee, R.H. , Wang, C.H. , Huang, L.T. and Chen, S.C. (2001) Leaf senescence in rice plants: cloning and characterization of senescence up‐regulated genes. J. Exp. Bot. 52, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Zhao, Y. , Liu, X. , Peng, J. , Guo, H. and Luo, J. (2014) LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res. 42, D1200–D1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, P.O. , Woo, H.R. and Nam, H.G. (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 8, 272–278. [DOI] [PubMed] [Google Scholar]
- Lin, J.‐F.F. and Wu, S.‐H.H. (2004) Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Lü, B. , Wang, X. , Zhang, C. , Zhang, S. , Qian, J. , Chen, L. , Shi, H. and Dong, H. (2010) Thirty‐seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. J. Biosci. 35, 435–450. [DOI] [PubMed] [Google Scholar]
- Lohman, K.N. , Gan, S. , John, M.C. and Amasino, R.M. (1994) Molecular analysis of natural leaf senescence in Arabidopsis thaliana . Physiol. Plant. 92, 322–328. [Google Scholar]
- Lohse, M. , Nagel, A. , Herter, T. , May, P. , Schroda, M. , Zrenner, R. , Tohge, T. , Fernie, A.R. , Stitt, M. and Usadel, B. (2014) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37, 1250–1258. [DOI] [PubMed] [Google Scholar]
- Luedemann, A. , Strassburg, K. , Erban, A. and Kopka, J. (2008) TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC‐MS)‐based metabolite profiling experiments. Bioinformatics, 24, 732–737. [DOI] [PubMed] [Google Scholar]
- Manavella, P.A. , Arce, A.L. , Dezar, C.A. , Bitton, F. , Renou, J.‐P.P. , Crespi, M. and Chan, R.L. (2006) Cross‐talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb‐4 transcription factor. Plant J. 48, 125–137. [DOI] [PubMed] [Google Scholar]
- Manavella, P.A. , Dezar, C.A. , Ariel, F.D. , Drincovich, M.F. and Chan, R.L. (2008a) The sunflower HD‐Zip transcription factor HAHB4 is up‐regulated in darkness, reducing the transcription of photosynthesis‐related genes. J. Exp. Bot. 59, 3143–3155. [DOI] [PubMed] [Google Scholar]
- Manavella, P.A. , Dezar, C.A. , Bonaventure, G. , Baldwin, I.T. and Chan, R.L. (2008b) HAHB4, a sunflower HD‐Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 56, 376–388. [DOI] [PubMed] [Google Scholar]
- Martin, A. , Lee, J. , Kichey, T. , Gerentes, D. , Zivy, M. , Tatout, C. , Dubois, F. , Balliau, T. , Valot, B. , Davanture, M. , Terce‐Laforgue, T. , Quillere, I. , Coque, M. , Gallais, A. , Gonzalez‐Moro, M.‐B. , Bethencourt, L. , Habash, D.Z. , Lea, P.J. , Charcosset, A. , Perez, P. , Murigneux, A. , Sakakibara, H. , Edwards, K.J. and Hirel, B. (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell, 18, 3252–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, D.E. , Costa, M.L. , Gomez, F.M. , Otegui, M.S. and Guiamet, J.J. (2008) “Senescence‐associated vacuoles” are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J. 56, 196–206. [DOI] [PubMed] [Google Scholar]
- Masclaux, C. , Valadier, M.H. , Brugiere, N. , Morot‐Gaudry, J.F. and Hirel, B. (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta, 211, 510–518. [DOI] [PubMed] [Google Scholar]
- Masclaux‐Daubresse, C. , Carrayol, E. and Valadier, M.‐H. (2005) The two nitrogen mobilisation‐ and senescence‐associated GS1 and GDH genes are controlled by C and N metabolites. Planta, 221, 580–588. [DOI] [PubMed] [Google Scholar]
- Matallana‐Ramirez, L.P. , Rauf, M. , Farage‐Barhom, S. , Dortay, H. , Xue, G.‐P.P. , Dröge‐Laser, W. , Lers, A. , Balazadeh, S. , Mueller‐ Roeber, B. , Droge‐Laser, W. and Mueller‐Roeber, B. (2013) NAC transcription factor ORE1 and senescence‐induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant, 1, 1–34. [DOI] [PubMed] [Google Scholar]
- McIntyre, G.I. (1997) The role of nitrate in the osmotic and nutritional control of plant development. Aust. J. Plant Physiol. 24, 103. [Google Scholar]
- Miao, Y. , Smykowski, A. and Zentgraf, U. (2008) A novel upstream regulator of WRKY53 transcription during leaf senescence in Arabidopsis thaliana . Plant Biol (Stuttg), 10(Suppl 1), 110–120. [DOI] [PubMed] [Google Scholar]
- Molina‐Torres, J. and Martinez, M.L. (1991) Tocopherols and leaf age in Xanthium strumarium L. New Phytol. 118, 95–99. [Google Scholar]
- Montaner, D. and Dopazo, J. (2010) Multidimensional gene set analysis of genomic data. PLoS One, 5, e10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschen, S. , Bengoa Luoni, S. , Paniego, N.B. , Hopp, H.E. , Dosio, G.A.A. , Fernandez, P. and Heinz, R.A. (2014) Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS One, 9, e104379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooden, L.D. (1988) Whole plant senescence. In Senescence and Aging in Plants ( L. D, Nooden and A. C., Leopold , eds), pp. 392–439. San Diego, CA: Academic Press. [Google Scholar]
- Nuruzzaman, M. , Manimekalai, R. , Sharoni, A.M. , Satoh, K. , Kondoh, H. , Ooka, H. and Kikuchi, S. (2010) Genome‐wide analysis of NAC transcription factor family in rice. Gene, 465, 30–44. [DOI] [PubMed] [Google Scholar]
- Obata, T. and Fernie, A.R. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 69, 3225–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageau, K. , Reisdorf‐Cren, M. , Morot‐Gaudry, J.‐F. and Masclaux‐Daubresse, C. (2006) The two senescence‐related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicoti. J. Exp. Bot. 57, 547–557. [DOI] [PubMed] [Google Scholar]
- Parrott, D.L. , McInnerney, K. , Feller, U. and Fischer, A.M. (2007) Steam‐girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence‐associated genes. New Phytol. 176, 56–69. [DOI] [PubMed] [Google Scholar]
- Parrott, D.L. , Martin, J.M. and Fischer, A.M. (2010) Analysis of barley (Hordeum vulgare) leaf senescence and protease gene expression: a family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytol. 187, 313–331. [DOI] [PubMed] [Google Scholar]
- Peluffo, L. , Lia, V. , Troglia, C. , Maringolo, C. , Norma, P. , Escande, A. , Esteban Hopp, H. , Lytovchenko, A. , Fernie, A.R. , Heinz, R. and Carrari, F. (2010) Metabolic profiles of sunflower genotypes with contrasting response to Sclerotinia sclerotiorum infection. Phytochemistry, 71, 70–80. [DOI] [PubMed] [Google Scholar]
- Pérez‐Rodríguez, P. , Riaño‐Pachón, D.M. , Corrêa, L.G.G. , Rensing, S.A. , Kersten, B. and Mueller‐Roeber, B. (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38, D822–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic, E. , de la Serve, T.B. , Tardieu, F. and Turc, O. (2002) Leaf senescence induced by mild water deficit follows the same sequence of macroscopic, biochemical, and molecular events as monocarpic senescence in pea. Plant Physiol. 128, 236–246. [PMC free article] [PubMed] [Google Scholar]
- Pichersky, E. , Noel, J.P. and Dudareva, N. (2006) Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science, 311, 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. and Sarkar, D. (2012) nlme: linear and nonlinear mixed effects models. R package.
- Quadrana, L. , Almeida, J. , Otaiza, S.N. , Duffy, T. , Corrêa da Silva, J.V. , de Godoy, F. , Asís, R. , Bermúdez, L. , Fernie, A.R. , Carrari, F. and Rossi, M. (2013) Transcriptional regulation of tocopherol biosynthesis in tomato. Plant Mol. Biol. 81, 309–325. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F. , Noh, Y.S. , Himelblau, E. and Amasino, R.M. (2000) Molecular aspects of leaf senescence. Trends Plant Sci. 5, 278–282. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F. , Reiter, W.‐D. and Amasino, R.D. (2001) One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence‐associated. Plant Mol. Biol. 46, 447–457. [DOI] [PubMed] [Google Scholar]
- R Core Team (2012) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, ISBN 3‐900051‐07‐0. [Google Scholar]
- Radwanski, E.R. and Last, R.L. (1995) Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell, 7, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf, M. , Arif, M. , Dortay, H. , Matallana‐Ramírez, L.P. , Waters, M.T. , Gil Nam, H. , Lim, P.‐O. , Mueller‐Roeber, B. and Balazadeh, S. (2013) ORE1 balances leaf senescence against maintenance by antagonizing G2‐like‐mediated transcription. EMBO Rep. 14, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rise, M. , Cojocaru, M. , Gottlieb, H.E. and Goldschmidt, E.E. (1989) Accumulation of alpha‐Tocopherol in Senescing Organs as Related to Chlorophyll Degradation. Plant Physiol. 89, 1028–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M.R. (2007) Does GABA act as a signal in plants?: hints from molecular studies. Plant Signal. Behav. 2, 408–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, I.N. , Caputo, C. , Criado, M.V. and Funk, C. (2012) Senescence‐associated proteases in plants. Physiol. Plant. 145, 130–139. [DOI] [PubMed] [Google Scholar]
- Roessner‐Tunali, U. , Hegemann, B. , Lytovchenko, A. , Carrari, F. , Bruedigam, C. , Granot, D. and Fernie, A.R. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133, 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch, T. and González, M.‐C. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Rushton, D.L. , Tripathi, P. , Rabara, R.C. , Lin, J. , Ringler, P. , Boken, A.K. , Langum, T.J. , Smidt, L. , Boomsma, D.D. , Emme, N.J. , Chen, X. , Finer, J.J. , Shen, Q.J. and Rushton, P.J. (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. [DOI] [PubMed] [Google Scholar]
- Ruuska, S.A. , Lewis, D.C. , Kennedy, G. , Furbank, R.T. , Jenkins, C.L.D. and Tabe, L.M. (2008) Large scale transcriptome analysis of the effects of nitrogen nutrition on accumulation of stem carbohydrate reserves in reproductive stage wheat. Plant Mol. Biol. 66, 15–32. [DOI] [PubMed] [Google Scholar]
- Sadras, V.O. , Echarte, L. and Andrade, F.H. (2000a) Profiles of leaf senescence during reproductive growth of sunflower and maize. Ann. Bot. 85, 187–195. [Google Scholar]
- Sadras, V.O. , Quiroz, F. , Echarte, L. , Escande, A. and Pereyra, V.R. (2000b) Effect of Verticillium dahliae on photosynthesis, leaf expansion and senescence of field‐grown sunflower. Ann. Bot. 86, 1007–1015. [Google Scholar]
- Scherer, H.W. (2001) Sulphur in crop production—invited paper. Eur. J. Agron. 14, 81–111. [Google Scholar]
- Schiltz, S. , Gallardo, K. , Huart, M. , Negroni, L. , Sommerer, N. and Burstin, J. (2004) Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling. Plant Physiol. 135, 2241–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon, R.S. , Childs, K.L. , Santoro, N. , Foster, C.E. , Buell, C.R. , de Leon, N. and Kaeppler, S.M. (2012) Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 159, 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, W. , Kuang, J. , Chen, L. , Xie, H. , Peng, H. , Xiao, Y. , Li, X. , Chen, W. , He, Q. , Chen, J. and Lu, W. (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 63, 5171–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens, S. , Ma, J. , Hanson, J. and Rolland, F. (2010) Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 274–279. [DOI] [PubMed] [Google Scholar]
- Smyth, G. (2005) Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor ( Gentleman, R. , Carey, V. , Dudoit, S. , Irizarry, R. and Huber, W. , eds), pp. 397–420. New York: Springer. [Google Scholar]
- Tabuchi, M. , Abiko, T. and Yamaya, T. (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 58, 2319–2327. [DOI] [PubMed] [Google Scholar]
- Thimm, O. , Blasing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Kruger, P. , Selbig, J. , Muller, L.A. , Rhee, S.Y. , Stitt, M. , Bläsing, O. , Krüger, P. and Müller, L.A. (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Uauy, C. , Distelfeld, A. , Fahima, T. , Blechl, A. and Dubcovsky, J. (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science, 314, 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker, B. , Shahid Mukhtar, M. and Somssich, I.E. (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta, 226, 125–137. [DOI] [PubMed] [Google Scholar]
- Urquhart, A.A. and Joy, K.W. (1981) Use of Phloem exudate technique in the study of amino Acid transport in pea plants. Plant Physiol. 68, 750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. and Dane, F. (2013) NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiologiae Plantarum, 35(5), 1397–1408. [Google Scholar]
- Wang, X. , Basnayake, B.M.V.S. , Zhang, H. , Li, G. , Li, W. , Virk, N. , Mengiste, T. and Song, F. (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 22, 1227–1238. [DOI] [PubMed] [Google Scholar]
- Watanabe, M. , Balazadeh, S. , Tohge, T. , Erban, A. , Giavalisco, P. , Kopka, J. , Mueller‐Roeber, B. , Fernie, A.R. and Hoefgen, R. (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 162, 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler, A. , Von Schaewen, A. , Leegood, R.C. , Lea, P.L. and Quick, P.W. (1998) Regulation of leaf senescence by cytokinin, sugars, and light. Plant Physiol. 116, 329–335. [Google Scholar]
- Wingler, A. , Purdy, S. , MacLean, J.A. and Pourtau, N. (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57, 391–399. [DOI] [PubMed] [Google Scholar]
- Wingler, A. , Masclaux‐Daubresse, C. and Fischer, A.M. (2009) Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 60, 1063–1066. [DOI] [PubMed] [Google Scholar]
- Wingler, A. , Delatte, T. , O'Hara, L. , Primavesi, L. , Jhurreea, D. , Paul, M. and Schluepmann, H. (2012) Trehalose 6‐Phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 158, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. (2003) Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 6(1), 79–84. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Ju, H.‐W. , Chung, M.‐S. , Huang, P. , Ahn, S.‐J. and Kim, C.S. (2011) The R‐R‐type MYB‐like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 52, 138–148. [DOI] [PubMed] [Google Scholar]
- Zhang, W.Y. , Xu, Y.C. , Li, W.L. , Yang, L. , Yue, X. , Zhang, X.S. and Zhao, X.Y. (2014) Transcriptional analyses of natural leaf senescence in maize. PLoS One, 9, e115617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Jiang, Y. and Yu, D. (2011) WRKY22 transcription factor mediates dark‐induced leaf senescence in Arabidopsis. Mol. Cells, 31, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Microarray validation.
Figure S2 Significant GO distribution for molecular function at pre‐anthesis time (T1 vs. T0).
Figure S3 Metabolism overview under glasshouse conditions.
Table S1 Primer sequences for qPCR validation analysis.
Table S2 Up‐ and down‐regulated SAGs with a log2 fold change higher or lower than 2 under field conditions.
Table S3 Putative transcription factors up‐ and down‐regulated with a log2 fold change higher or lower than 2 under field conditions.
Supplementary Legends


