Abstract
Aims
Cellular communication network factor 2 (CCN2) is a matricellular protein implicated in fibrotic diseases, with ongoing clinical trials evaluating anti-CCN2-based therapies. By uncovering CCN2 as abundantly expressed in non-diseased artery tissue, this study aimed to investigate the hypothesis that CCN2 plays a pivotal role in maintaining smooth muscle cell (SMC) phenotype and protection against atherosclerosis.
Methods and results
Global- and SMC-specific Ccn2 knockout mouse models were employed to demonstrate that Ccn2 deficiency leads to SMC de-differentiation, medial thickening, and aorta elongation under normolipidaemic conditions. Inducing hyperlipidaemia in both models resulted in severe aorta malformation and a 17-fold increase in atherosclerosis formation. Lipid-rich lesions developed at sites of the vasculature typically protected from atherosclerosis development by laminar blood flow, covering 90% of aortas and extending to other vessels, including coronary arteries. Evaluation at earlier time points revealed medial lipid accumulation as a lesion-initiating event. Fluorescently labelled LDL injection followed by confocal microscopy showed increased LDL retention in the medial layer of Ccn2 knockout aortas, likely attributed to marked proteoglycan enrichment of the medial extracellular matrix. Analyses leveraging data from the Athero-Express study cohort indicated the relevance of CCN2 in established human lesions, as CCN2 correlated with SMC marker transcripts across 654 transcriptomically profiled carotid plaques. These findings were substantiated through in situ hybridization showing CCN2 expression predominantly in the fibrous cap.
Conclusion
This study identifies CCN2 as a major constituent of the normal artery wall, critical in regulating SMC differentiation and aorta integrity and possessing a protective role against atherosclerosis development. These findings underscore the need for further investigation into the potential effects of anti-CCN2-based therapies on the vasculature.
Keywords: Cellular communication network factor 2 (CCN2), Aorta, Atherosclerosis, Smooth muscle cell
Graphical Abstract
Graphical Abstract.
Time of primary review: 30 days
1. Introduction
Atherosclerosis is a chronic inflammatory disease that develops in large- to medium-sized arteries near branching sites and on the inside of arterial curvatures.1 The disease progresses over decades by gradual accumulation of lipids, cells, and extracellular matrix within the arterial intimal layer at these pre-dilection sites.2 Advanced atherosclerotic plaques are characterized by the presence of a lipid-rich necrotic core, which is superimposed by a fibrous cap that protects the plaque from rupture and abrupt thrombus formation.3
Several cell types are involved in the development of atherosclerosis, including leukocytes, endothelial cells, and vascular smooth muscle cells (SMCs). In healthy arteries, SMCs primarily reside in the medial layer and are rich in myofilaments required for muscle contraction. The contractile SMC phenotype is maintained by myocardin (MYOCD) that regulates several genes encoding proteins of the contractile apparatus.4 During atherogenesis, inflammatory cytokines released from the developing plaque down-regulate MYOCD causing SMC de-differentiation and loss of their contractile phenotype.5 In a process involving migration and proliferation, de-differentiated SMCs then enter the intimal layer and contribute to lesion development. The conventional perception has been that SMC-derived cells only contribute to the formation of the fibrous cap as ‘myofibroblasts’. However, lineage-tracing experiments in mice have revealed that SMC-derived cells also express markers commonly associated with mesenchymal stem cells,6 chondrocytes,7 and macrophages.6,8–10
Cellular communication network factor 2 [CCN2, previously known as connective tissue growth factor (CTGF)11] is a matricellular protein established as a disease-promoting factor in the context of fibrosis and cancer.12 CCN2 is a promising target in various fibrotic conditions, and anti-CCN2-based therapies are currently in clinical trials for idiopathic pulmonary fibrosis, pancreatic cancer, and Duchenne muscular dystrophy (NCT03955146, NCT04229004, and NCT02606136).
CCN2 is not a growth factor as indicated by its original name, and no specific CCN2 receptor has been identified. Rather, CCN2 is a multi-domain protein capable of binding various integrins, non-integrin receptors, extracellular matrix components, cytokines, and growth factors, and the myriad of reported cellular effects attributed to CCN2 appears to be highly tissue and context dependent.12
While CCN2 is highly expressed in developing vasculature,13–15 its expression in healthy adult tissues, including arteries, has been reported to be largely absent16,17 and without physiological significance.18,19 In contrast, CCN2 has been shown to be highly expressed during vascular injury and disease, including atherosclerosis,16,18–22 but the role of CCN2 in atherosclerosis remains unknown.
In this study, we demonstrate that CCN2 is one of the most highly transcribed genes in healthy/non-lesioned artery tissue in both humans and mice, while its expression is relatively low in other tissues. Based on observations that SMCs are the primary source of CCN2 in arteries, we formulated two hypotheses. First, we proposed that CCN2 plays a critical role in maintaining artery tissue homoeostasis by preserving the contractile SMC phenotype. Second, we hypothesized that depletion of CCN2 would lead to increased formation of atherosclerosis under hyperlipidaemic conditions. To test these hypotheses, we employed in vitro experiments and in vivo investigations using human tissue samples and mouse models. Our data reveal a novel and crucial role of CCN2 as a regulator of SMC integrity, establishing it as a potent atheroprotective molecule.
2. Methods
2.1. In silico analysis
Bulk RNA analyses were performed using the GTEx Portal V823 and the STARNET database.24 Single-nucleus RNA-sequencing (RNA-seq) analysis of human thoracic aortas, coronary, and carotid plaques was based on GSE165824 from Pirruccello et al.,25 GSE131778 from Wirka et al.,26 and GSE155512 from Pan et al.,27 respectively, and analysed using Seurat v4 package28 in R and annotated according to original studies.
2.2. Mouse procedures
To generate global Ccn2 knockout mice, Ccn2fl/fl mice (C57BL/6J)29 (developed by Professor Andrew Leask, University of Western Ontario) were crossed with Rosa26-CreERT2 mice.30 Littermates (Ccn2fl/fl mice with or without the Rosa26-CreERT2 transgene) were used for experiments, and all mice were treated with tamoxifen at 8 weeks of age by intra-peritoneal injection with 1 mg tamoxifen (T5648, Sigma Aldrich) in 100 µL corn oil (C8267, Sigma Aldrich) each day for 4 consecutive days. Hereafter, global knockouts and littermate controls were referred to as Ccn2Δ/Δ and Ccn2fl/fl, respectively.
To generate SMC-specific Ccn2 knockout mice, Ccn2fl/fl mice were crossed with Myh11-CreERT2 mice (B6.FVB-Tg[SMMHC-Cre/ERT2]1Soff/J, kindly provided by Professor Stefan Offermanns, Max-Planck Institute for Heart and Lung Research, Germany, using the CreERT2 construct generated by Pierre Chambon, GIE-CERBM, France). Male Ccn2fl/fl/Myh11-CreERT2 mice and Ccn2wt/wt/Myh11-CreERT2 mice from synchronized breeding pairs were treated with tamoxifen as described for global Ccn2 knockout mice and referred to as Ccn2SMCΔ/Δ and Ccn2wt/wt, respectively. Of note, we previously reported excision of flCcn2 starting at 4 weeks of age, prior to tamoxifen administration.31 This premature excision would affect aortic phenotype starting prior to tamoxifen administration; however, this problem was not observed in Rosa26-CreERT2-based Ccn2 knockout mice and is premature partial Ccn2 deficiency is unlikely to significantly affect our findings.
Primers used for genotyping were as follows: Ccn2 locus: 5′AATACCAATGCACTTGCCTGGATGG and 5′-GAAACAGCAATTACTACAACGGGAGTGG; and Rosa26 locus: 5′AAAGTCGCTCTGAGTTGTTAT, 5′GGAGCGGGAGAAATGGATATG, and 5′CCTGATCCTGGCAATTTCG. The Myh11-CreERT2 transgene was genotyped using the standard protocol from Jackson Laboratories for this line.
Hyperlipidaemia was induced by a single intravenous injection of 1·1011 viral genomes of rAAV8-D377Y-mPcsk9 produced at the University of North Carolina Vector Core (Chapel Hill, NC, USA) from pAAV/ApoEHCR-hAAT-D377Y-mPcsk9-BGHpA32 in combination with Western diet (wd; 21% fat and 0.21% cholesterol) (D12079B, Research Diets Inc.). Blood was harvested from the facial vein to monitor plasma cholesterol (CHOL2, Roche Diagnostics).
Mice were either euthanized by CO2/O2 inhalation, and tissues were either snap frozen in liquid nitrogen or immersed in 4% formaldehyde in phosphate-buffered saline (PBS) for 24 h and then transferred to PBS supplemented with 0.05% sodium azide. In experiments requiring analysis of full-length aortas, mice were euthanized by intra-peritoneal injection of pentobarbital (250 mg/kg) and lidocaine (20 mg/kg) and then perfused at 100 mmHg with PBS for 30 s followed by 4% formaldehyde in PBS for 5 min through the left ventricle using the cut right atrium as a route of drainage. Mice were then immersed in 4% formaldehyde in PBS for 6 h and stored in PBS supplemented with 0.05% sodium azide at 4°C.
For all experiments, mouse genotype was unknown to investigators, and all experimental procedures and analyses were performed blinded. However, the severity of the phenotype was self-revealing in most morphometric and histological analyses. All mice from litters of all synchronized breeding were included in experiments, and no mice were excluded from experiments other than premature death.
All procedures were performed in accordance with protocols approved by the local authorities (the Danish Animal Experiments Inspectorate) and conform to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
2.3. Blood pressure measurements in conscious mice
Blood pressure was measured in conscious-free moving mice; the procedure was adopted from previously published studies.33–35 In brief, 2 weeks after tamoxifen treatment, two catheters were funnelled subcutaneously via small incision in the neck of the mice and inserted in the left femoral artery and vein. For this procedure, mice were given 100 mg/kg ketamine and 10 mg/kg xylazine intravenously and administered Temgesic (0.1 mg/kg) for pain relief subcutaneously during surgery and an intravenous bolus of 50 µL Temgesic the following day. Mice had a 4-day recovery period before continuous online measurement of diastolic, systolic, and mean arterial blood pressure and heart rate. The catheters contained isotonic glucose solution with 100 U/mL heparin, and infusion was given continuously at low rate (10 µL/h). Baseline measurements were performed for 6 consecutive days followed by induction of hypertension by intravenous Angiotensin II (ANGII) infusion (60 ng/kg/min) for 4 consecutive days of. Mice were euthanized by CO2/O2 inhalation.
2.4. Wire myography
Mice used for measuring blood pressure served a dual purpose, being utilized for wire myography after sacrifice. Myography was conducted as described previously.36 In brief, thoracic aortas were dissected, perivascular adipose tissue was removed, and aortas were placed in a physiological PSS solution (PSS; 115 mM NaCl, 25 mM NaHCO3, 2.5 mM K2HPO4, 1.2 mM MgSO4, 5.5 mM glucose, 10 mM HEPES, and 1.3 mM CaCl2). Aortas were divided into three 2 mm long pieces and mounted on 200 µm pins (Danish Myo Technology, Søften, Denmark). The chambers were continuously infused with 5% CO2 during the experiment. Blood vessels were allowed to rest for 20 min in PSS at 37°C. Subsequently, vessels were stretched by 200 µm every 3 min until internal diameter and measured wall tension corresponded to 100 mmHg according to the law of LaPlace. Once 100 mmHg was reached, the diameter was adjusted to 90% of the diameter at 100 mmHg, and measurements were recorded and used for experiments. Vessels were then allowed to rest for 30 min in PSS, and smooth muscle viability was tested by stimulation with 32 mM K+. The response of the SMCs to 10−6 M phenylephrine was tested, and relaxation in response to 10−5 M acetylcholine was used to evaluate endothelial function. Sensitivity and maximal response to PE and U46619 were finally evaluated through exposure to cumulative concentrations of each drug (PE 10−9 M to 10−5 M) and U46916 10−9 M to 10−5 M).
2.5. Evaluation of LDL permeability and retention in vivo
Blood for LDL isolation was collected in EDTA-coated tubes. Plasma was isolated by centrifugation at 4000 g for 10 min and loaded into a 39 mL Quick-Seal® Round-Top Polypropylene tube, which was then sealed and placed in a type 70 TI Fixed-Angle Titanium Rotor with a 13 mm Diameter Aluminum Tube Spacer. Ultracentrifugation was performed at 350 000 g for 18 h at 6°C in an OptimaTM XE-100 ultracentrifuge. Tubes were punctured, and the yellow phase was extracted. Potassium bromide (KBr) was used to adjust the extracted phase to a density of 1.063 g/mL. The adjusted solution was then ultracentrifuged as before, and the top yellow LDL-containing phase was extracted. LDL protein concentration was measured by the Pierce BCA Protein Assay Kit using a bovine serum albumin (BSA)-based standard. LDL was then conjugated at a 1:5 molar ratio to Atto 565 N-hydroxysuccinimide (NHS) ester fluorochrome (Sigma Aldrich, 72464-1MG-F) at room temperature for 18 h. Labelled LDL was then purified on a PD10 desalting column with PBS to remove excess fluorochrome. Each mouse received LDL corresponding to 500 μg protein via a 100 μL tail vein injection and was sacrificed either 1 h (endothelial permeability) or 24 h (LDL retention) after injection. Mice were euthanized by CO2/O2 inhalation, and aortas were immersed in 4% formaldehyde for 24 h and then transferred to PBS supplemented with 0.05% sodium azide. To quantify the amount of LDL retained in the aortas of mice, formalin-fixed aortic arch and descending thoracic aorta were cryo-embedded at a 5 μm thickness. Cross-sections were washed for 10 min in MilliQ-H2O and mounted with DAPI-containing slowfade and coverslips. Images were captured using an Olympus FV1000MPE upright confocal laser scanning confocal microscope using the FV-ASW (version 4.2C) software. The intensity of Atto-565 LDL in cryo-sections was quantified using automatic macros in Fiji (ImageJ).
2.6. Transcriptomics
RNA was extracted from snap-frozen tissue or cells using NORGEN Total RNA Purification Kit (37500, Norgen Biotek Corp.), and cDNA was made by High-Capacity cDNA Reverse Transcription Kit (4368814, Thermo Fisher Scientific). Quantitative PCR was performed using SYBR™ Green PCR Master Mix (4364344, Thermo Fisher Scientific) and the ViiA7 Real-Time PCR system (Thermo Fisher Scientific). Primer pairs used for mouse transcripts were as follows: Ccn2: 5′GAGGAAAACATTAAGAAGGGC and 5′CAGGCACAGGTCTTGATGAAC; Myocd: 5′CCTGCCTGCTGGGAGTTAAT and 5′GCCGTAACTGTAAGACTGATCG; Myh11: 5′ATCACGGGGGAGCTGGAAA and 5′CGGCGAGCAGGTAGTAGAAG; Acta2: 5′CCTTCGTGACTACTGCCGAG and 5′ATAGGTGGTTTCGTGGATGC; Lgals3: 5′CCCCCGCTTCAATGAGAACA and 5′CAAAGGGGAAGGCTGACTGT; and Gapdh: 5′GGCTGCCCAGAACATCAT and 5′CGGACACATTGGGGGTAG. Primer pairs used for human transcripts were as follows: CCN2: 5′CATCTTCGGTGGTACGGTGT and 5′TTCCAGTCGGTAAGCCGC; MYOCD: 5′AAAAAGCCCAAGGACCCCAA and 5′GTCCATAGGTGGAGGGGACT; MYH11: 5′TCCAAATACGCGGATGAGAGG and 5′TGAGCATTTTGTTGGTCCGC; ACTA2: 5′CCAGAGCCATTGTCACACAC and 5′CAGCCAAGCACTGTCAGG; LGALS3: 5′ACTGGTTGAACCTGACCACT and 5′AGCACTGGTGAGGTCTATGTC; and GAPDH: 5′CAGCGACACCCACTCCTC and 5′TGAGGTCCACCACCCTGT. Gapdh/GAPDH was used as reference gene.
For microarray analysis, background correction, normalization, and gene expression index calculation of probe intensities were done in Transcriptome Analysis Console (TAC) software (Thermo Fisher Scientific) using the robust multi-array average method. Probes representing non-expressed genes as defined by TAC were removed from further analysis. All subsequent calculations were performed using the open-source R-environment (R version 3.6.1). First, the normalized probe expression matrix was collapsed by gene symbol using maximal probe intensity, thus providing one gene expression value per sample per gene. Next, the experiment series associated with batch effects were removed/reduced by COMBAT batch removal using the ComBat function from the sva R-package. Differential gene expression analysis between Ccn2Δ/Δ and Ccn2fl/fl SMCs was conducted on the gene expression measured by the microarrays using an unpaired limma t-test embedded in the limma R-package.37
For RNA-seq analysis, sample preparation was performed using Illumina Stranded mRNA kit (Illumina, San Diego, CA, USA) and 300 ng total RNA, following the manufacturer’s instructions, and sequencing of 2 × 150 bp was performed using NovaSeq 6000 (Illumina, San Diego, CA, USA). Quality control of the raw sequencing data was performed using FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw reads were trimmed using Bbduk v38 to remove low-quality reads (mean phred score < 10), short reads (<25 bp), and adapter contamination (https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbduk-guide/). Both left trimming and right trimming were performed, and read pairs were trimmed to the same length. STAR v2.7.10a with default settings, for alignment of the trimmed, paired reads to the mus musculus reference genome and transcriptome (Ensembl Mus_musculus.GRCm39 v105). Next, uniquely aligned reads were extracted and indexed using SAMtools v1.10 before counting reads in genomic features. The counting was conducted using HTSeq v0.11.2 with reverse strand specificity and with the intersection-strict mode to handle reads overlapping more than one feature. All subsequent calculations and analyses were performed using the open-source R-environment. The count data were filtered based on counts-per-million (cpm), and only genes with cpm > 1 in a number of samples corresponding to the smallest group were included for further analysis. The library size was re-computed and normalized using the trimmed mean of M-values (TMM) normalization method. Differential gene expression including calculations of dispersion estimates and fitting of negative binomial generalized linear models (Ccn2SMCΔ/Δ aortas compared with Ccn2wt/wt aortas) was analysed using the edgeR R-package.38,39 Comparisons were adjusted for multiple testing using the Benjamin–Hochberg method to control the false discovery rate (fdr). Genes with fdr ≤ 0.05 were considered as significantly differentially expressed between the compared groups. Full lists of differentially regulated genes from microarray and RNA-seq experiments are provided in supplementary material online, Tables S1 and S2.
2.7. Mass spectrometry
Samples for mass spectrometry were prepared as previously described.36 In brief, snap-frozen tissues were homogenized using a TissueLyser system (Qiagen) with stainless steel beads (Qiagen) in a lysis buffer [100 mM DTT, 5% sodium deoxycholate, 1% β-octylglucoside, 20 mM Tris, pH 8.8, supplemented with complete, Mini, EDTA-free Protease Inhibitor Cocktail Tablets and PhosSTOP (Roche)]. Proteins were then denatured (99°C for 20 min, 80°C for 100 min), alkylated (150 mM iodoacetamide for 30 min at room temperature), precipitated (−20°C acetone for 1 h), re-solubilized (8 M urea for 30 min at room temperature), and trypsinized [0.5 µg/µL trypsin (Promega) in 50 mM ammonium bicarbonate and 1 M urea at 37°C overnight]. After acidification, tryptic peptides were purified on custom-made Poros R2/R3 (Thermo Scientific) micro-columns, dried, re-constituted, and finally re-suspended in 2% acetonitrile (ACN) and 0.1% formic acid. The concentration of tryptic peptides was determined (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific) and normalized across samples.
For targeted multiple-reaction-monitoring (MRM) analysis, heavy isotope-labelled standard peptides (JPT Peptide Technologies) were added to each sample aiming at a 0.1–10 peak area ratio between each endogenous peptide assayed and its corresponding heavy peptide. Each sample (1 µg endogenous tryptic peptides) was run on an Easy-nLC II NanoLC system using a C18 trapping column (length 2 cm; internal diameter 100 µm) for desalting and a C18 analytical column (length 10 cm; internal diameter 75 µm) for peptide separation (Thermo Scientific). Peptides were eluted with a three-step 45 min gradient of 0.1% formic acid in ACN at a flow rate of 300 nL/min. Peptides were ionized using a Nanospray Flex ion source (Thermo Scientific) and analysed on a TSQ Vantage triple quadruple mass spectrometer (Thermo Scientific) in MRM mode. MRM raw files were processed using Pinpoint 1.3 (Thermo Scientific). The peak area ratio between endogenous and heavy isotope-labelled spiked peptide was used for data analysis. The peak area ratio for Ccn2 (target peptide: TTTLPVEFK) was normalized to Gapdh (target peptides: GAAQNIIPASTGAAK and LISWYDNEYGYSNR) within the same sample.31
For explorative proteomics of mouse aortas, samples (4 μg tryptic peptides per sample) were randomly labelled with 10- or 18-plex tandem mass tags (TMT or TMTpro, Thermo Scientific), high-pH fractionated, and analysed by nano-LC-MS/MS virtually as previously described.40 All Eclipse raw data files were processed and quantified using Proteome Discoverer version 2.4 (Thermo Scientific) as previously described.40 Full list of differentially regulated proteins is provided in supplementary material online, Tables S3 and S4.
2.8. Pathway analysis
Gene set enrichment analysis (GSEA) of omic data was performed using clusterProfiler 4.0 package41 in R based on ranked log2-transformed fold changes. For both RNA-seq and mass spectrometry data, default gseGO settings were used for ‘biological process’ or ‘molecular function’ terms: ont = ‘BP’, keyType = ‘SYMBOL’, nPerm = 10000, minGSSize = 10, maxGSSize = 800, pvalueCutoff = 0.05, verbose = TRUE, OrgDb = ‘org.Mm.eg.db’, pAdjustMethod = ‘BH’. For the analysis of mass spectrometry data sets, no enrichment was found, and pAdjustMethod = ‘none’ was used to obtain the presented data. Redundant terms were removed using the simplify function with default settings (gse, cutoff = 0.7, by = ‘p.adjust’, select_fun = min). All enriched terms identified in the analyses are provided in supplementary material online, Tables S5 (microarray), S6 (RNA-seq), and S7 (mass spectrometry).
2.9. Oil Red O staining of en face-prepared aortas
Formalin-fixed aortas were dissected, cut open longitudinally, and stained with a 2:1 mix of 0.5% Oil Red O in isopropanol (O1391, Sigma Aldrich) and de-ionized water for 10 min at 37°C, briefly washed in isopropanol (I9516, Sigma Aldrich) and transferred to de-ionized water. Aortas were pinned to a black rubber material and immersed in PBS, and images were generated using a Zeiss AxioCam ERc 5 s camera attached to a Zeiss Stemi 2000-C microscope. Thoracic aortic area and percentage of thoracic aorta stained by Oil Red O were determined using an automated macro in Fiji (ImageJ).
2.10. Tissue preparation, histology, and immunohistochemistry
Human internal thoracic arteries were obtained from coronary artery bypass grafting procedures performed at Odense University Hospital. All donors had given written consent, and the study was performed in accordance with protocols approved by the local ethics committee (S-20100044) and conformed to the principles of the Declaration of Helsinki. Tissue was fixed in 4% formaldehyde in PBS for 24 h, embedded in paraffin, sectioned at 5 µm thickness, and deparaffinized prior to histological staining.
Formalin-fixed mouse aortas were cryo-protected in sucrose (25% in PBS for 24 h and then 50% in PBS for 24 h), embedded in optimal cutting temperature compound (Tissue-Tek, Sakura), and sectioned at 5 µm thickness.
For Masson trichome staining, sections were air-dried and exposed to Papaniculaus 1 (6 min), running tap water (8 min), 1.2% picric acid (5 min), tap water (1 min), 1% Biebrich scarlet (10 min), 1% phosphor wolfram acid (10 min), 2.5% methyl blue (2 min), acetic acid (30 s), 1% phosphor wolfram acid (5 min), and acetic acid (3 min). Slides were then dehydrated, and coverslips were mounted using Aquatex (Sigma Aldrich). For Oil Red O staining, sections were exposed to 7% ethanol (1 min), Oil Red O (2.5 min), 100% ethanol (1 min), tap water (1 min), Mayer’s haematoxylin (2 min), tap water (1 min), 0.3% sodium carbonate (10 s), and running tap water (1 min). Coverslips were then mounted using Aquatex (Sigma Aldrich).
For Alcian blue staining, sections were air-dried and exposed to 3% acetic acid (3 min), Alcian blue solution (ab150662, Abcam) (30 min), 3% acetic acid (10 s), running tap water (2 min), distilled water (2 min), nuclear fast red solution (ab150662, Abcam) (5 min), running tap water (2 min), and distilled water (2 min). Coverslips were then mounted using Aquatex (Sigma Aldrich).
ACTA2, MYH11, and CD68 immunohistochemistry of human tissues was performed at the Department of Pathology, Odense University Hospital, using primary antibodies BS66 (BSH-7459-100, Nordic Biosite, 1:1000), SMMS-1 (M3558, Dako, 1:100), and PG-M1 (M0876, Dako, 1:50), respectively, utilizing the full-automated OptiView DAB IHC Detection Kit (760–700). Images were recorded by a Hamamatsu model 2 OHT scanner.
2.11. Immunofluorescence
Sections were air-dried; washed in de-ionized water (1 min) and PBS (3 min); blocked in 5% BSA in PBS (30 min); incubated with rabbit anti-mouse Acta2 antibody (ab5694, Abcam) (1:300 in 5% NGS in PBS) and rat anti-mouse Lgals3 antibody (M3/38, Cedarlane) (1:600 in 5% NGS in PBS) (2 h); washed in PBS (3 × 3 min); and incubated with appropriate secondary antibody [Alexa Fluor 647-conjugated goat anti-rabbit IgG (A21244, Invitrogen) (1:300 in 5% BSA in PBS), Alexa Fluor 647-conjugated goat anti-rabbit IgG (A21244, Invitrogen) (1:300 in 5% BSA in PBS), or Alexa Fluor 555-conjugated goat anti-rat IgG (A21434, Invitrogen) (1:300 in 5% BSA in PBS)] for 1 h at room temperature.
When co-staining for lipid, BODIPY (493/503) (D3922, Invitrogen) (10 mg/mL in DMSO) was co-incubated with secondary antibodies (1:600 in 5% BSA in PBS). Slides were then washed in PBS (3 × 3 min), and coverslips were mounted with SlowFade Gold Antifade Mountant with DAPI (S36942, Thermo Fisher Scientific). Images were recorded by a Hamamatsu Model 2 OHT scanner, and immunofluorescence images were recorded with an Olympus FV1000MPE upright confocal laser scanning confocal microscope (excitation lasers 405, 488, 559, and 635 nm) using the FV-ASW (version 4.2C) software.
2.12. In situ hybridization
In situ hybridization (ISH) was performed with RNAscope® 2.5 VS Reagent Kit (323250, Advanced Cell Diagnostics, Newark, CA, USA) on a Ventana Discovery Ultra platform (Roche Diagnostics, Penzberg, Germany). Sections were exposed to 24-h antigen retrieval and 16-min protease treatment. The probes applied were specific for human CCN2 mRNA (cat no. 560589) and for dapB (4-hydroxy-tetrahydrodipicolinate reductase from Escherichia coli) as negative control (cat no. 312039). Signal was detected using Fast Red detection kit (8127166001, Roche Diagnostics, Penzberg, Germany).
2.13. SMC cultivation
Ccn2Δ/Δ and Ccn2fl/fl mice were euthanized by cervical dislocation 6 weeks after tamoxifen treatment. The right atrium was cut and mice were flushed via the left ventricle using sterile PBS (70011044, Thermo Fisher Scientific). The descending thoracic aortas were dissected free from perivascular tissue while in normal growth media [Dulbecco's modified Eagle's medium supplemented with 20% foetal bovine serum (FBS) (10270106, Life Technologies), 1% Penicillin-Streptomycin (15140122, Life Technologies), 1% L-glutamine (G7513, Sigma Aldrich) containing 0.01% amphotericin B (15290018, Thermo Fisher Scientific)]. Aortas were cut into 1 × 1 mm pieces, placed in a 96-well plate (6267170, Buch & Holm), and digested in normal growth media containing 1.42 mg/mL collagenase (C6885, Sigma Aldrich) for 6 h at 37°C. Cell suspensions were then centrifuged at 350 g for 5 min, re-suspended in normal growth media, and seeded in 24-well plates (6267168, Buch & Holm). Fourth passage cells were starved for 48 h in normal growth media (without FBS), trypsinated (25200072, Sigma Aldrich), centrifuged for 350 g for 5 min, and stored at −80°C until use.
Human aortic SMCs (HAoSMCs) (C-12533, PromoCell) were grown in Smooth Muscle Cell Growth Medium 2 (C-22062, PromoCell). At 80% confluency, third passage cells were starved for 48 h in Basal Medium 2, phenol red-free (C-22267, PromoCell), trypsinated (25200072, Sigma Aldrich), centrifuged at 220 g for 5 min, and seeded (6000 cells/cm2) in Smooth Muscle Cell Growth Medium 2 in 12-well plates (6267167, Buch & Holm). After 24 h, media were removed and cells were tranfected with 50 nM ON-TARGETplus Human CTGF (1490) siRNA—SMARTpool (L-012633-01-0005, Horizon) or ON-TARGETplus Non-targeting Pool (D-001810-10-05, Horizon) in Basal Medium 2, phenol red-free using Lipofectamine™ RNAiMAX Transfection Reagent (13778075, Thermo Fisher Scientific) according to the manufacturer’s instructions for 24 h. Transfection media were replaced by Smooth Muscle Cell Growth Medium 2, and after 24 h, media (centrifuged at 1500 g for 10 min) and cells (trypsinated (25200072, Sigma Aldrich) and centrifuged at 220 g for 5 min) were harvested and stored at −80°C until use.
Presented data are representative of at least three independent experiments.
2.14. oxLDL uptake assay
HAoSMCs were cultured and RNA silencing was conducted as described above. After 24 h, the medium was changed and the cells were stimulated with diI-conjugated oxLDL (diI-oxLDL) (10 μg/mL, L34358, Thermo Fisher) for 48 h, starting 48 h after transfection. The cells were washed three times with PBS at 37°C before trypsinization. The trypsin was inactivated with SMC growth medium, and the cells were centrifuged at 220 g for 5 min at 4°C. The supernatant was removed, and the cells were re-suspended in an ice-cold flow cytometry buffer [PBS containing 1% BSA (422661, VWR) and 0.1 mM EDTA (15578-038, Invitrogen)]. The cell suspension was filtered through a 22 μm syringe filter (15181499, Fisher) and a cell strainer (352235, Falcon) and kept on ice and in the dark until flow cytometry.
The samples were run on a BD LSR II flow cytometer, with diI-oxLDL data collected using the yellow laser (561 nm), the band-pass filter (585/15 nm), and the long-pass filter (570 nm).
For imaging, the cells were fixed with 4% PFA (9713, 5000, VWR) for 10 min at room temperature in the dark, after three washes with PBS. The nuclei were stained with DAPI diluted in PBS (1:10 000, D9542, Sigma Aldrich), and the wells were washed again with ice-cold PBS before imaging. Fluorescent images were captured using a BioTek Citation 1 Cell Imaging Multimode Reader with the DAPI and RFP channels.
To analyse the data, negative diI signal values were replaced with 0, and a pseudo-value of 1 was added to all measurements to enable log-transformation of the data. The resulting data were plotted as frequency polygons with a normalized area of 1 for each sample. Only cells with a log(signal + 1) value > 0 (corresponding to 35% of cells from each sample) are visible on the plots. To distinguish between autofluorescence and diI signal, a threshold was defined as the intersection between samples not treated with diI-oxLDL and samples treated with diI-oxLDL. The threshold was set at a log(signal + 1) value of 4.6. The percentage of cells above the threshold in each sample was defined as cells taking up diI-oxLDL, after subtracting the mean of the groups that were not treated with diI-oxLDL. The same method was used to calculate the mean signal intensity for cells taking up diI-oxLDL.
2.15. Western blotting
HAoSMCs were lysed with 1 × RIPA buffer (20-188, Merck Millipore) containing one PhosSTOP tablet (04 906 837 001, Roche) and one cOmplete ULTRA tablets, Mini, EDTA-free (05 892 791 001, Roche). Protein concentrations were quantified using the Pierce BCA Protein Assay Kit (23227, Thermo Fischer Scientific). Normalized samples were prepared with NuPAGE™ LDS sample buffer (4×) (NP0007, Thermo Fischer Scientific™) and NuPAGE™ Sample Reducing Agent (10×) (NP0009, Thermo Fischer Scientific™) and then loaded on Mini-Protean TGX Gels (4561086, Bio-Rad). Gels were run for 35 min in running buffer (1610732, Bio-Rad). Proteins were transferred to PVDF transfer membranes (88518, Thermo Fischer Scientific™) using transfer buffer (1610734, Bio-Rad). Membranes were blocked for 1 h in TBST with 5% milk and incubated overnight with primary antibody in either TBST with 5% BSA, TBST with 5% milk, or TBST [1:500 pSMAD2 (138D4, Cell Signaling TECHNOLOGY®), 1:1000 SMAD2/3 (8685, Cell Signaling TECHNOLOGY), 1:1000 TGFBR2 (66636-1-Ig, Proteintech), 1:1000 ACTA2 (ab5694, Abcam), and 1:5000 GAPDH (ab9485, Abcam)]. Following incubation for 1 h with secondary antibodies [anti-rabbit (P0448, Agilent Technologies) and anti-mouse (P0447, Agilent Technologies)], membranes were visualized with SuperSignal™ West Femto (34094, Thermo Fischer Scientific™) or Western Lightning® ECL pro (NEL122001EA, PerkinElmer).
2.16. Athero-Express study cohort analyses
The Athero-Express biobank includes patients undergoing carotid endarterectomy, of which the study design has been published before.42–45 The study is in line with the Declaration of Helsinki, and informed consent was provided by all study participants after the approval for this study by medical ethical committees of the different hospitals (the University Medical Center Utrecht NL and St. Antonius Hospital Nieuwegein NL) was obtained. The morphological grading was based on haematoxylin–eosin staining and staining of collagen and calcification, or by immunohistochemical staining of CD68 (Roche cat# 790-2931) (macrophages) and ACTA2 (Biosite BSH-7459-1, clone BS66).
2.17. High-sensitivity C-reactive peptide measurements
Analyses involved 314 patients who underwent carotid endarterectomy with available transcriptome data and high-sensitivity C-reactive peptide (hsCRP) data. Patients were categorized based on hsCRP levels measured in citrate plasma collected before surgery into two groups: those with hsCRP levels ≥ 2 mg/L and those with <2 mg/L, as outlined previously.46 Patients with hsCRP levels > 40 mg/L, indicative of acute infection, were excluded.
2.18. Interleukin 6 measurements
Analyses involved 352 patients who underwent carotid endarterectomy with available transcriptome data and plasma interleukin 6 (IL6) levels. IL6 levels were quantified using the OLINK proteomics platform in plasma samples collected pre-surgery, as detailed previously47
2.19. Statistical analysis
For two-group comparison of normal distributed data (as assessed by Shapiro–Wilk test), unpaired t-test with Welch’s correction was used and data were represented as mean and standard error of the mean (SEM). If data from at least one group were not normal distributed, Mann–Whitney U test was used, and data were represented as median with interquartile range. Whenever we observed a significant effect of genotype for both sexes in the global Ccn2 knockout mouse model, a two-group comparison between the knockout group of both sexes was conducted (after normalization of values to the respective wild-type groups) to assess differences in the relative effect of genotype between sexes. The results of these tests have been integrated into the figure legends. Comparison of groups over time was performed using two-way analysis of variance (ANOVA) and presented as mean and SEM. Differences in survival curves were assessed by log-rank test.
For analyses based on the Athero-Express study cohort, comparison of CCN2 expression (Figure 7A) and SMC markers (Figure 7B) between plaque clusters was performed by Kruskal–Wallis test followed by Dunn’s multiple comparisons test; correlation of CCN2 to each gene across all plaques (Figure 7C) was performed by Pearson correlation significance test and Bonferroni adjusted for multiple testing (data provided in supplementary material online, Table S8). GSEA was conducted as described under ‘pathway analysis’, and enriched terms identified in the analysis are provided in supplementary material online, Table S9. Comparison of groups in Figure 7E was performed using Wilcoxon rank-sum test. The association between plasma hsCRP levels and CCN2 expression was assessed using ANOVA to compare the groups. A linear regression model was utilized to assess the relationship between plasma IL6 levels and CCN2 expression in the patients.
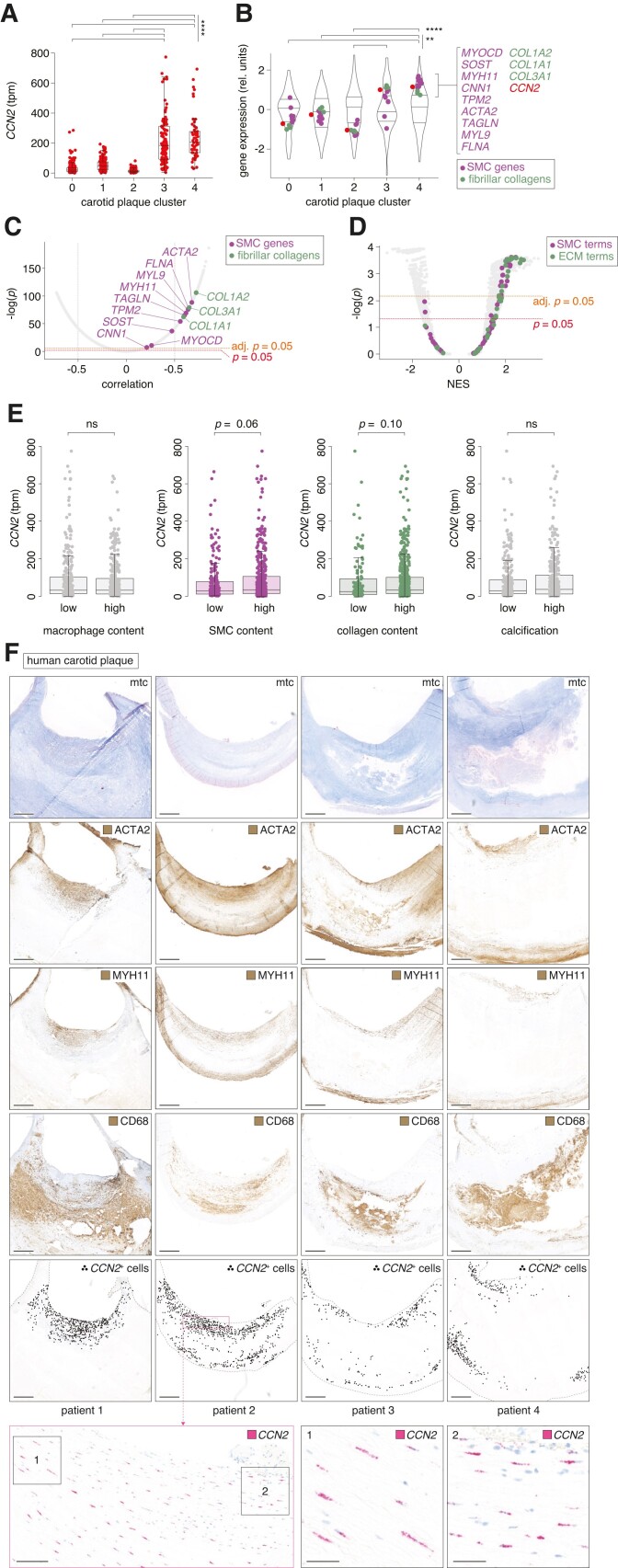
Figure 7.
CCN2 expression correlates with SMC markers in advanced human plaques. (A) CCN2 transcripts per million (tpm) in carotid plaques from the Athero-Express study cohort42–45 (n = 654) divided into five transcriptome-based plaque clusters as previously described.42 Data are shown as median and interquartile range. (B) Gene expression in the five transcriptome-based plaque clusters from the Athero-Express study cohort. MYOCD and genes encoding pre-specified markers for contractile SMCs are shown in magenta, while genes encoding fibrillar collagens are shown in green. CCN2 is shown in red. The distribution of gene expression for all measured transcripts is represented as violin plots. Expression values are scaled per cluster. (C) Correlation of CCN2 with any other gene across the 654 carotid plaques from the Athero-Express study cohort. MYOCD and genes encoding pre-specified markers for contractile SMCs are shown in magenta, while genes encoding fibrillar collagens are shown in green. (D) GSEA based on genes ranked by correlation to CCN2 (as shown in C). Terms related to SMCs are shown in magenta, while terms related to extracellular matrix (ECM) is shown in green. (E) Associations between CCN2 expression level and human carotid plaque content of macrophages, ACTA2+ SMCs, collagen, and calcification, respectively, semi-quantitatively divided into low and high levels of each histologically assessed parameter. The data are based on the Athero-Express study cohort (n = 654). (F) CCN2, ACTA2, MYH11, and CD68 expression in human carotid plaques (n = 4) from the Odense Artery Biobank by ISH and immunohistochemistry, respectively. CCN2-positive cells are marked with black dots to assist visualization of CCN2+ cell distribution at low magnification. Scale bars (overview) = 1 mm. Scale bars (magnification levels) = 100 and 25 µm, respectively. Mtc, Masson’s trichrome. Data in A and B were analysed by Kruskal–Wallis test followed by Dunn’s multiple comparisons test. Data in C were analysed by Pearson correlation significance test and Bonferroni adjusted for multiple testing. Data in D were analysed using clusterProfiler 4.0 package41 in R. Data in E were analysed by Wilcoxon rank-sum test.
All images shown in the figures are representative images/stainings for each treatment group.
In graphs where data values are not absolute, individual values are normalized to the mean of the control group. P ≤ 0.05 was the criterion for reliable differences between groups. In figures, asterisks indicate the level of significance: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and ns: non-significant.
3. Results
3.1. CCN2 is highly expressed in non-diseased artery tissues
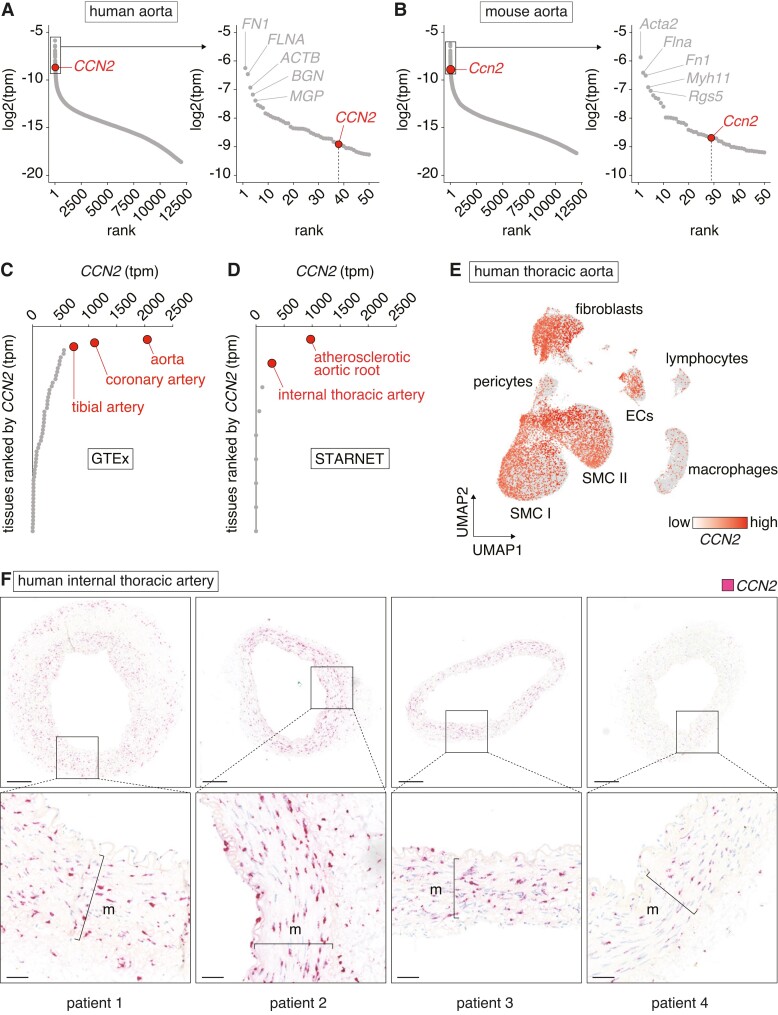
To evaluate relative CCN2 expression within the aorta transcriptome, we used RNA-seq data from human and mouse aorta samples and found CCN2 to be among the top 50 most abundant transcripts in both species (excluding mitochondrial genes) (Figure 1A and B and supplementary material online, Figure S1). Moreover, relative CCN2 expression was markedly elevated in human arteries compared with a range of non-vascular tissues (Figure 1C and D), implying a critical role for CCN2 in artery tissues.
Figure 1.
CCN2 is highly expressed in non-diseased artery tissues. (A, B) Genes ranked by expression level in aortas from humans (n = 432) (A) and mice (n = 4) (B). Dots represent median expression level for each gene. CCN2 is among the top 50 most abundant transcripts in both species (excluding mitochondrial genes). (C, D) CCN2 transcripts per million (tpm) across all tissues from the GTEx (C) and STARNET (D) databases. Dots represent median expression level in each tissue. Artery tissues are shown in red. Relative CCN2 expression was markedly elevated in human arteries compared with non-vascular tissues (E). UMAP plot based on single-nucleus RNA-seq data from non-lesioned human thoracic aortas (n = 6) overlaid with CCN2 expression. SMCs, fibroblasts, and endothelial cells express CCN2, while lymphocytes and macrophages displayed little to no CCN2 expression (F). CCN2 ISH of human internal thoracic arteries (n = 4). Most staining was observed in the medial layer. Refer to supplementary material online, Figure S2 for negative control ISH. Scale bars (overview) = 250 µm; scale bars (magnification) = 50 µm. m = media.
To gain insights into the potential cell sources of CCN2 in the aortic wall, single-nucleus RNA-seq data obtained from human thoracic aortas were analysed.25 This analysis showed that SMCs, fibroblasts, and endothelial cells, but not lymphocytes and macrophages, are the main cells expressing CCN2 (Figure 1E). ISH analysis of human internal thoracic arteries confirmed this expression pattern and showed the most intense labelling associated with SMCs in the medial layer (Figure 1F and supplementary material online, Figure S2).
3.2. Ccn2 deficiency causes de-differentiation of SMCs
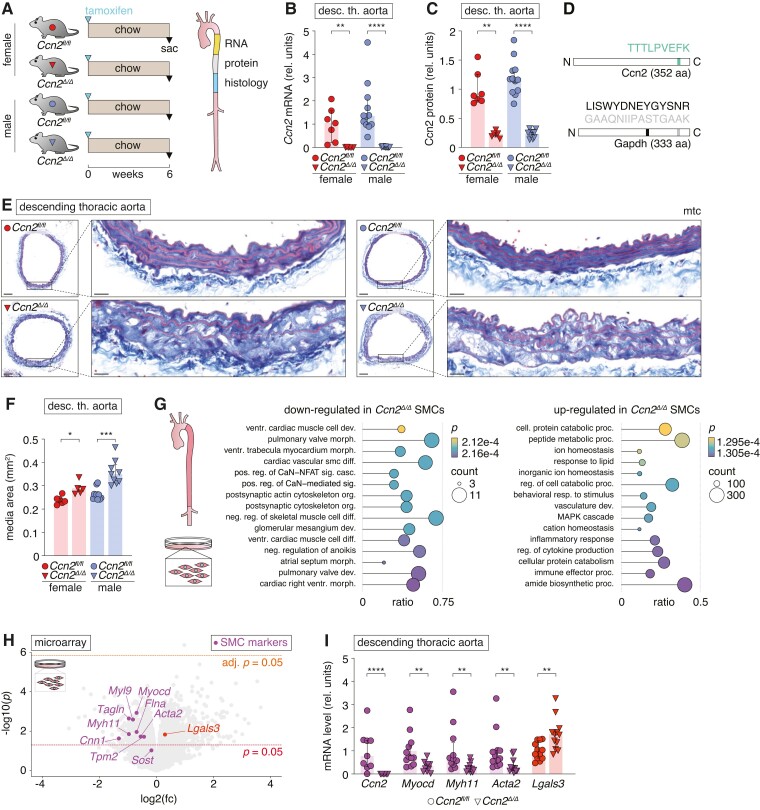
To investigate the role of CCN2 in the developed aorta, global Ccn2 knockout (Ccn2Δ/Δ) was induced by tamoxifen administration in adult mice of both sexes (Figure 2A). Six weeks after tamoxifen injection, Ccn2Δ/Δ mice presented undetectable levels of Ccn2 transcripts (Figure 2B) and an ∼75% reduction in Ccn2 protein levels in the aorta (Figure 2C and D). Histological assessment of the descending thoracic aortas from Ccn2Δ/Δ revealed a disorganized and expanded medial layer compared with the aortas of wild-type littermate controls (Ccn2fl/fl) (Figure 2E and F and supplementary material online, Figure S3A).
Figure 2.
Ccn2 deficiency causes de-differentiation of SMCs. (A) Experiment design and aorta segments used for RNA, protein, and histological analyses. (B, C) Ccn2 mRNA (B) and Ccn2 protein (C) level in descending thoracic aortas. (D) Peptides of Ccn2 and Gapdh quantified to obtain results in C. Peptide positions are indicated. (E) Cross-sections of descending thoracic aortas stained by Masson’s trichrome (mtc). Scale bars (overview) = 100 µm; scale bars (magnification) = 20 µm. (F) Medial cross-sectional area of descending thoracic aorta. We found no difference in the relative effect of genotype on this measure between female and male mice. (G) GSEA based on microarray data obtained from primary Ccn2Δ/Δ (n = 9) and Ccn2fl/fl (n = 6) SMCs. The 15 most down- and up-regulated gene ontology biological process terms (based on significance) are shown. Dot sizes indicate the number of genes regulated in Ccn2Δ/Δ SMCs for each term. The ratio shows coverage of a given term by genes regulated in Ccn2Δ/Δ SMCs, and dot colours indicate the level of significance. (H) Volcano plot showing regulation of genes in primary Ccn2Δ/Δ SMCs compared with Ccn2fl/fl SMCs. Two thousand one hundred seventy-five genes were up-regulated and 1366 genes were down-regulated (unadjusted P < 0.05) in Ccn2Δ/Δ SMCs. Only Hlx displayed significant up-regulation after correction for multiple testing (see supplementary material online, Table S1). Myocd and genes encoding pre-specified markers for contractile SMCs are shown in magenta, and Lgals3 is shown in red. (I) Validation of selected genes by qPCR of descending thoracic aortas. Data in B, C, F, and I were analysed by unpaired t-test with Welch’s correction or Mann–Whitney U test. Data in G were analysed using clusterProfiler 4.0 package41 in R.
To evaluate the molecular effects of Ccn2 deficiency specifically in SMCs, isolated SMCs from Ccn2Δ/Δ and Ccn2fl/fl aortas were next investigated by microarray analysis. Pathway enrichment analysis supported the transition of SMCs from the contractile phenotype (down-regulation of genes associated with contractile apparatus-related terms) to a pro-inflammatory phenotype (up-regulation of genes associated with the pro-inflammatory response, immune system, and cytokine production) (Figure 2G). Indeed, Myocd and 7/8 pre-defined markers of the contractile phenotype (Myh11, Acta2, Cnn1, Sost, Flna, Myl9, Tagln, and Tpm2)26 were reduced, while Galectin 3 (Lgals3) known to be associated with trans-differentiation of SMCs was up-regulated (Figure 2H). Although these transcripts were only nominally significant in the microarray analysis, their regulation was confirmed in vivo by qPCR using aortas from Ccn2Δ/Δ and Ccn2fl/fl mice (Figure 2I). Proteomic profiling of Ccn2Δ/Δ aortas supported an increase in Lgals3 expression (adjusted P < 0.05), and although effects on contractile SMC markers were less pronounced at the protein level, Myh11 was nominally reduced, and unbiased pathway enrichment analysis corroborated that down-regulation of terms related to the contractile apparatus was a central effect of Ccn2 deficiency (see supplementary material online, Figure S3B and C).
3.3. Ccn2 deficiency causes severe atherosclerosis not limited to typical pre-dilection sites in hyperlipidaemic mice
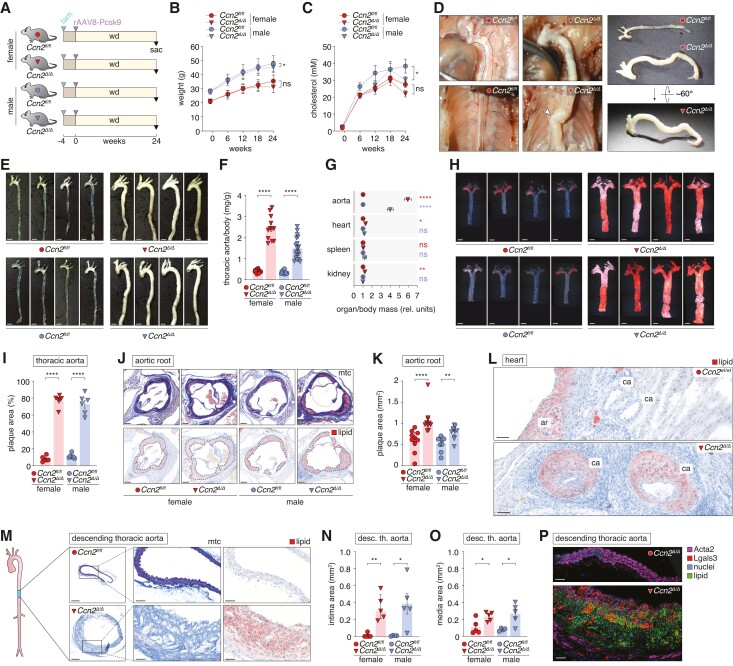
To investigate the consequence of Ccn2 deficiency for atherosclerosis development, mice of both sexes were exposed to 24 weeks of hyperlipidaemia. This was achieved using a standardized model32 combining wd with a single intravenous injection of rAAV8-D377Y-mPcsk9 to deplete LDL receptors 4 weeks after Ccn2 deletion (Figure 3A). Through the course of the experiment, Ccn2Δ/Δ males displayed a modest reduction in weight gain and plasma cholesterol with similar trends observed for Ccn2Δ/Δ females (Figure 3B and C). At sacrifice, Ccn2Δ/Δ mice presented a very distinct aorta phenotype as compared with Ccn2fl/fl controls. Ccn2Δ/Δ aortas were pallid and severely enlarged, which hindered natural alignment with the spine, and a resulting kink of the descending thoracic aortas was consistently observed (Figure 3D). Thoracic aortas had a 4- to 6-fold increase in mass relative to body weight (Figure 3E and F) that was limited to aorta/arteries as relative organ mass of the heart, spleen, and kidney was similar between genotypes (Figure 3G). The atherosclerosis phenotype extended to the abdominal aorta (see supplementary material online, Figure S4A). Remarkably, the severe aortic phenotype did not affect survival (see supplementary material online, Figure S4B).
Figure 3.
Ccn2 deficiency causes severe atherosclerosis not limited to typical pre-dilection sites in hyperlipidaemic mice. (A) Experiment design (n = 14/12/10/19 for Ccn2fl/fl females, Ccn2Δ/Δ females, Ccn2fl/fl males, Ccn2Δ/Δ males, respectively, after excluding three mice that met the pre-specified humane endpoints (see supplementary material online, Figure S4B). (B, C). Mouse weight (B) and plasma cholesterol (C) through the course of the experiment. (D) In situ and ex vivo images of Ccn2fl/fl and Ccn2Δ/Δ aortas. While aortas of Ccn2fl/fl mice were aligned with the spine, the enlargement of aortas in Ccn2Δ/Δ mice hindered this and resulted in a consistently observed kink (arrowhead) at the level of the descending thoracic aorta. (E) Thoracic aortas after 24 weeks of hyperlipidaemia. Scale bars = 2.5 mm. (F) Mass of thoracic aortas relative to body weight after 24 weeks of hyperlipidaemia. The relative effect of the genotype was significantly higher in female mice. (G) Mass of the thoracic aorta, heart, spleen, and kidney (mean of two kidneys) relative to body weight. Each organ/body mass ratio is normalized to the mean of the female Ccn2fl/fl group for each organ to enable comparison of effects across the organs. (H) Oil Red O staining of en face-prepared thoracic aortas after 24 weeks of hyperlipidaemia. Scale bars = 2.5 mm. (I) Percentage of en face-prepared thoracic aorta area stained positive for lipid. We found no difference in the relative effect of genotype on this measure between female and male mice. (J, K) Cross-sections of aortic roots stained by Masson’s trichrome (mtc) and Oil Red O (scale bars = 100 µm, J) and quantitation of aortic root plaque area (K). The relative effect of the genotype was significantly higher in female mice. (L) Oil Red O staining showing atherosclerosis in coronary arteries (ca) near aortic roots (ar) from Ccn2Δ/Δ mice, which was not observed in Ccn2fl/fl mice. Scale bars = 50 µm. (M) Cross-sections of descending thoracic aortas stained by Masson’s trichrome (mtc) or Oil Red O. Scale bars (overview) = 200 µm; Scale bars (magnification) = 50 µm. (N, O) Intimal (N) and medial (O) cross-sectional area of descending thoracic aorta. We found no difference in the relative effect of genotype on these measures between female and male mice. (P) Immunofluorescence co-staining of descending thoracic aorta (adjacent to sections shown in M) for Acta2 (staining was performed with an Alexa Fluor 647-conjugated secondary antibody but shown in magenta), Lgals3 (red), lipid (green), and nuclei (blue). Scale bars = 50 µm. Some analyses shown on the figure were performed on randomly selected subsets from each group. Data in B and C were analysed by two-way ANOVA. Data in F, G, I, and K and N and O were analysed by unpaired t-test with Welch’s correction or Mann–Whitney U test.
Oil Red O staining of en face-prepared thoracic aortas showed that 75% of the thoracic aortic area of Ccn2Δ/Δ mice stained positive for lipid in contrast to 10% in thoracic aortas from Ccn2fl/fl mice (Figure 3H and I). When considering that the thoracic aorta area was 1.75 times larger in Ccn2Δ/Δ mice (see supplementary material online, Figure S3C), there was a 13-fold increase in absolute lesion formation in Ccn2Δ/Δ mice.
At typical pre-dilection sites (aortic root and brachiocephalic artery), where atherosclerosis developed in Ccn2fl/fl mice, the extent of plaque formation was accentuated in Ccn2Δ/Δ mice (Figure 3J and K and supplementary material online, Figure S4D–F), but we found no significant effects on necrotic core area or content of collagen, Acta2, or Lgals3 in aortic root lesions (see supplementary material online, Figure S4G–K). Intriguingly, Ccn2 deficiency also caused severe atherosclerosis in coronary arteries, which are clinically the most relevant vessels, but rarely inflicted by lesions in mouse atherosclerosis models (Figure 3L). In general, Ccn2Δ/Δ mice displayed lesion formation throughout the aorta even in areas typically protected from atherosclerosis formation by laminar blood flow and high shear stress. Histological assessment of the descending thoracic aorta confirmed severe atherosclerotic lesion formation (Figure 3M–O) with the contribution of both Lgals3− (non-macrophage) and Lgals3+ (macrophages or transformed SMCs) lipid-uptaking cells (Figure 3P).
To evaluate the kinetics of the aortic phenotype, an independent cohort of mice were sacrificed after 4 and 12 weeks of hyperlipidaemia (see supplementary material online, Figure S5A). We noticed that already at this early time point, Ccn2Δ/Δ aortas were more flaccid upon dissection (see supplementary material online, Figure S5B), and despite no detectable effect on body weight, aortas were 10 and 17% longer at the 4- and 12-week time points, respectively (see supplementary material online, Figure S5C and D). Compared with the 24-week time point, the atherosclerosis phenotype after 12 weeks of hyperlipidaemia was generally less pronounced, although one Ccn2Δ/Δ mouse displayed a more advanced phenotype with 5-fold increase in relative whole-aorta mass compared with Ccn2f/fl mice (see supplementary material online, Figure S5E). Moderate lesions had formed after 4 weeks of hyperlipidaemia with more atherosclerosis formation at the 12-week time point (see supplementary material online, Figure S5F). At both time points, Ccn2Δ/Δ mice had accelerated lesion formation (see supplementary material online, Figure S5G and H). As observed for Ccn2Δ/Δ mice after 24 weeks of hyperlipidaemia, atherosclerosis development was not confined to typically prone areas at sites of disturbed laminar blood flow, as early lesions were scattered all over the thoracic aorta.
3.4. SMC-specific Ccn2 deletion recapitulates effects on SMC phenotype and medial expansion observed in global Ccn2 knockout mice
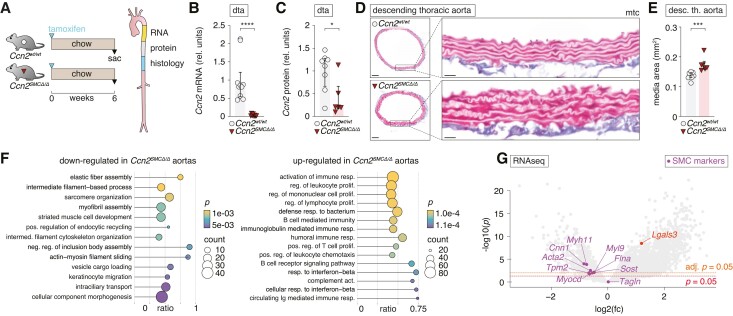
Hypothesizing that the vascular phenotype in global Ccn2-deficient mice was driven by SMC-dependent mechanisms, we next generated SMC-specific Ccn2 knockout mice (Ccn2SMCΔ/Δ). First, we analysed aortas from Ccn2SMCΔ/Δ and Ccn2wt/wt mice on regular chow 6 weeks after treatment with tamoxifen (Figure 4A). Ccn2 deletion was confirmed at the mRNA level (Figure 4B), which, similar to our observations from global Ccn2 mice, led to a 75% reduction in the level of Ccn2 protein in the aorta at this time point (Figure 4C). Notably, SMC-specific deletion of Ccn2 was sufficient to drive medial expansion (Figure 4D and E). Transcriptomic profiling of descending thoracic aortas from Ccn2SMCΔ/Δ mice showed down-regulation of terms related to the contractile apparatus, while terms related to the pro-inflammatory response and the immune response were up-regulated (Figure 4F). Myocd and 7/8 of the pre-defined contractile SMC phenotype markers were significantly down-regulated, while Lgals3 was up-regulated (Figure 4G) consistent with observations from global Ccn2 knockout SMCs.
Figure 4.
SMC-specific Ccn2 deletion recapitulates effects on SMC phenotype and medial expansion observed in global Ccn2 knockout mice. (A) Experiment design (resulting in data shown in B–G) and aorta segments used for RNA, protein, and histological analyses. (B, C) Ccn2 mRNA (B) and Ccn2 protein (C) level in descending thoracic aortas. dta, descending thoracic aorta. (D, E) Cross-sections of descending thoracic aortas stained by Masson’s trichrome (mtc) [scale bars (overview) = 100 µm; scale bars (magnification) = 20 µm] (D) and quantitation of the medial area (E). (F) GSEA based on RNA-seq of descending thoracic aorta from Ccn2SMCΔ/Δ (n = 7) and Ccn2wt/wt aortas (n = 8). The 15 most down- and up-regulated gene ontology biological process terms (based on significance) are shown. Dot sizes indicate the number of genes regulated in Ccn2SMCΔ/Δ aortas for each term. The ratio shows the coverage of a given term by genes regulated in Ccn2SMCΔ/Δ aortas, and dot colours indicate the level of significance. (G) Volcano plot showing regulation of all genes in Ccn2SMCΔ/Δ aortas. Two thousand four hundred eighty-seven (18.7%) genes were up-regulated and 139 (1%) genes were down-regulated (adjusted P < 0.05) in Ccn2SMCΔ/Δ aortas (see supplementary material online, Table S2). Myocd and genes encoding pre-specified markers for contractile SMCs are shown in magenta, and Lgals3 is shown in red. Data in B and C and E were analysed by unpaired t-test with Welch’s correction or Mann–Whitney U test. Data in F were analysed using clusterProfiler 4.0 package41 in R.
To ascertain if the effect of Ccn2 deficiency on SMCs affected the vasculature functionally, baseline blood pressure and ANGII-induced hypertension were recorded in conscious, freely moving, unstressed mice by indwelling chronic catheters for 12 days (see supplementary material online, Figure S6A). At baseline, there was a modest decrease in systolic and mean arterial blood pressure in Ccn2SMCΔ/Δ mice compared Ccn2wt/wt mice (see supplementary material online, Figure S6B, D, and G). ANGII infusion increased blood pressure consistently and stably, but the dynamic increase and stable plateau reached were the same for both genotypes (see supplementary material online, Figure S6C, E, F, and H). This indicates that the functionality of arterioles was preserved and that the significance of Ccn2 in arteriolar biology is less critical as compared with conduit arteries, in which Ccn2 deletion resulted in thoracic aortas less responsive to agonist-induced force development (α1-adrenoceptor and TP receptor) in isometric wire myography experiments (see supplementary material online, Figure S6I–P).
3.5. Atheroprotective effects of Ccn2 are exerted through SMC-dependent mechanisms
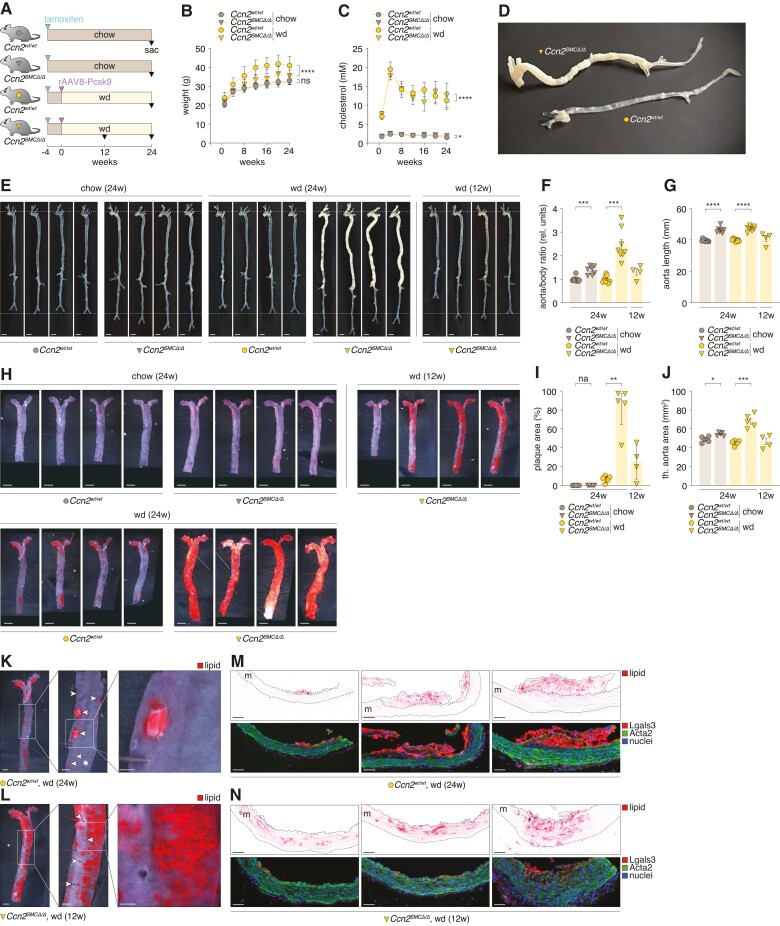
Next, to investigate the consequence of SMC-specific Ccn2 deficiency on atherosclerosis development, we induced hyperlipidaemia in Ccn2SMCΔ/Δ and Ccn2wt/wt mice as previously described by single intravenous injection of rAAV8-D377Y-mPcsk9 combined with wd-feeding for 24 weeks. Un-injected chow-fed Ccn2SMCΔ/Δ and Ccn2wt/wt mice were included as normolipidaemic controls (Figure 5A). A subset of Ccn2SMCΔ/Δ mice were sacrificed after 12 weeks to monitor phenotype progression at this time point. As for global Ccn2 knockout males, Ccn2SMCΔ/Δ mice gained less weight through the experiment (Figure 5B), and plasma cholesterol was significantly lower through most of the 24 weeks (Figure 5C).
Figure 5.
Atheroprotective effects of Ccn2 are exerted through SMC-dependent mechanisms. (A) Experiment design (n = 9/7/9/8 for chow-fed Ccn2wt/wt and Ccn2SMCΔ/Δ mice and wd-fed Ccn2wt/wt and Ccn2SMCΔ/Δ mice, respectively, after excluding five mice that met the pre-specified humane endpoints (see supplementary material online, Figure S7D). In addition, four Ccn2SMCΔ/Δ mice were sacrificed after 12 weeks of wd feeding to enable phenotype assessment at this time point. (B, C) Mouse weight (B) and plasma cholesterol (C) through the course of the experiment. (D, E) Ex vivo images of Ccn2wt/wt and Ccn2SMCΔ/Δ aortas. (F) Mass of aortas relative to body weight. (G) Aorta length (beginning of aortic arch to iliac bifurcation). (H) Oil Red O staining of en face-prepared thoracic aortas. Scale bars = 2.5 mm. (I) Percentage of en face-prepared thoracic aorta area stained positive for lipid. (J) Area of en face-prepared descending thoracic aortas. (K, L) Magnification of lipid-stained areas of descending thoracic aortas from Ccn2wt/wt mice after 24 weeks of hyperlipidaemia (K) and Ccn2SMCΔ/Δ mice after 12 weeks of hyperlipidaemia (L). Arrowheads in point to ostia of aortic branches. (M + N) Histological images of Oil Red O-stained lesions (top) and the same specimens stained for Lgals3 (red), Acta2 (green), and nuclei. m = media. Scale bars = 50 µm. Data in B and C were analysed by two-way ANOVA. Data in F and G and I and G were analysed by unpaired t-test with Welch’s correction or Mann–Whitney U test.
Analysis of aortas from hyper- and normolipidaemic Ccn2SMCΔ/Δ and Ccn2wt/wt mice revealed that SMC-specific Ccn2 deletion was sufficient to recapitulate the severe aorta phenotype observed in global Ccn2 knockout mice, including aorta elongation and atherosclerosis formation (Figure 5D–G and supplementary material online, Figure S7A–C). Ninety per cent of en face-prepared thoracic aortas from Ccn2SMCΔ/Δ mice stained positive for lipid, whereas only 8% of thoracic aortas from Ccn2fl/fl mice were stained. Considering that the thoracic aortic area was 1.5 times larger in Ccn2SMCΔ/Δ mice, there was a remarkable 17-fold increase in absolute lesion formation in Ccn2SMCΔ/Δ mice (Figure 5H–J). As for global Ccn2 knockout mice, SMC-specific Ccn2 deletion did not have a significant impact on survival of the mice (see supplementary material online, Figure S7D).
3.6. Initial lesion formation in SMC-specific Ccn2-deficient mice is preceded by medial lipid accumulation
To understand the mechanisms underlying the severity and atypical topological distribution of atherosclerosis in Ccn2SMCΔ/Δ mice, we focused our investigation on the descending thoracic aorta, which (except for aortic branching sites) is typically resistant to atherosclerosis in wild-type mice, even after 24 weeks of hyperlipidaemia (Figure 5K). We examined Ccn2SMCΔ/Δ mice sacrificed after 12 weeks of hyperlipidaemia to evaluate lesions at an earlier time point. While lipid staining was less pronounced after 12 weeks of hyperlipidaemia, the lipid distribution was similar to aortas from Ccn2SMCΔ/Δ mice after 24 weeks of hyperlipidaemia (Figure 5L) (as we also observed for global Ccn2 knockout mice after 12 weeks of hyperlipidaemia; see supplementary material online, Figure S5F). This indicated that lesion development occurs diffusely across the aorta instead of spreading from a few focal sites as observed in wild-type mice.
Lipid deposition in aortas from Ccn2SMCΔ/Δ mice was generally more diffuse than the well-defined lesions observed in Ccn2wt/wt mouse aortas. Interestingly, lipid deposition occurred at atypical sites at a time when lipid was not deposited at aortic branch ostia, suggesting that the mechanisms underlying lipid accumulation in the aortic wall in Ccn2SMCΔ/Δ mice are more potent atherosclerosis-promoting processes compared with effects of disturbed laminar blood flow.
To further elucidate the nature of lipid deposition, we next investigated en face-prepared aortas by histology. In Ccn2wt/wt mice, lesions were classical ‘fatty streaks’ composed of lipid-engorging macrophages/foam cells (Lgals3+) confined to the intimal layer, with largely unaffected underlying media (Figure 5M). In contrast, lipid staining of aortas from Ccn2SMCΔ/Δ mice at the 12-week time point was primarily attributed to lipid accumulation in the expanded medial layer with scarce or no participation of luminal macrophages (Figure 5N). Considering these results, we revisited aortas from global Ccn2 knockout mice after 12 weeks of hyperlipidaemia and found similar medial lipid accumulation (see supplementary material online, Figure S8).
3.7. Ccn2 deficiency augments the capacity of medial extracellular matrix to retain LDL
The accumulation of lipid in the aortic medial layer can result from various processes, including augmented lipoprotein uptake by modulated SMCs, increased endothelial permeability to lipoproteins, enhanced capacity of the arterial extracellular matrix to retain lipoproteins, or a combination of these factors.
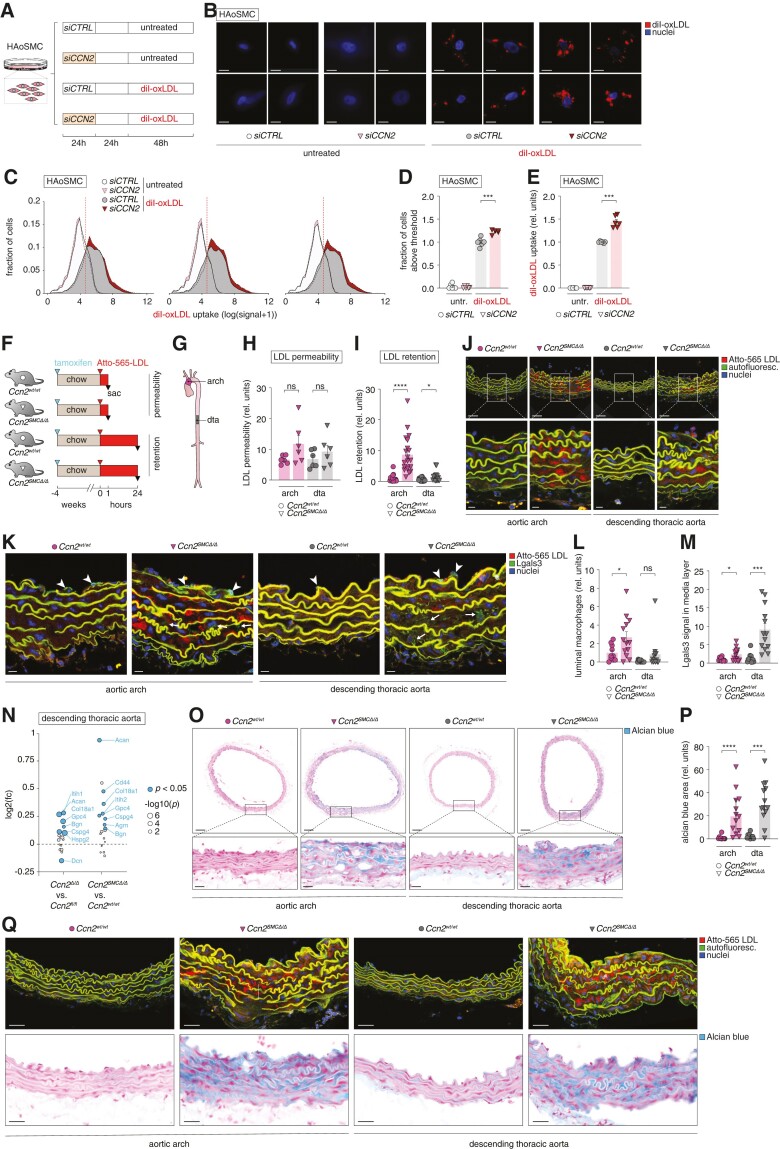
To investigate the impact of CCN2 deficiency on SMC lipoprotein uptake, we employed RNA interference to silence CCN2 in HAoSMCs. Successful knockdown of CCN2 led to down-regulation of MYOCD and ACTA2, alongside up-regulation of LGALS3, mimicking in vivo effects (see supplementary material online, Figure S9). Following treatment with diI-oxLDL for 48 h (Figure 6A), we observed a significant 20% increase in the proportion of cells taking up oxLDL and an overall 40% increase in the total uptake of oxLDL compared with control cells (Figure 6B–E).
Figure 6.
Ccn2 deficiency augments the capacity of medial extracellular matrix to retain LDL. (A) Experiment design (resulting in data shown in B–E). The experiment was replicated three independent times obtaining similar results. (B) Images of CCN2-silenced and control HAoSMCs either untreated or treated with diI-conjugated oxidized LDL (diI-oxLDL, red) and nuclei were stained with DAPI (blue). Scale bars = 5 µm. (C–E) Histograms showing three examples of flow cytometry data from the four experimental groups (C); quantitation of proportion of HAoSMCs with an oxLDL uptake above the defined cut-off (vertical red dotted line in C) (D) and the mean uptake for this population (E). (F, G) Experiment design (n = 6/6 for chow-fed Ccn2wt/wt and Ccn2SMCΔ/Δ mice used for investigation of endothelial permeability to LDL, and n = 15/20 for chow-fed Ccn2wt/wt and Ccn2SMCΔ/Δ mice used for investigation of LDL retention) (F) and aorta segments used for analyses in H–Q (G). dta, descending thoracic aorta. (H–J) Atto-565 LDL signal in the aortic arch and dta 1 h (H) and 24 h (I) after injection as quantified based on confocal fluorescence microscopy images shown for the 24-h time point in J. Scale bars (top) = 50 µm; scale bars (magnification) = 10 µm. (K) Sections adjacent to those analysed in H–J were stained for Lgals3 by immunofluorescence. Arrowheads point to endothelium-associated luminal macrophages (high level of dense Lgals3 signal), whereas arrows point to more diffuse and low levels of Lgals3 staining in the medial layer (presumed to be modulated SMCs). Scale bars = 10 µm. (L, M) Quantitation of endothelium-associated luminal Lgals3-positive macrophages (L) and medial Lgals3 signal (M) based on Lgals3 stainings represented in K. (N) Effect of global and SMC-specific Ccn2 knockout on the level of aortic proteoglycans (defined previously1) as assessed by mass spectrometry (full data sets are provided in supplementary material online, Tables S3 and S4). Blue dots show significantly regulated proteins, and dot size corresponds to the level of significance. Identities of all significantly regulated proteoglycans are shown. (O, P) Alcian blue staining of sections adjacent to those shown in J and K (O), and quantitation of Alcian blue-positive area (P). Scale bars (top) = 100 µm; scale bars (magnification) = 25 µm. (Q) Comparison of Atto-565 LDL signal (from analysis in H–J) and Alcian blue staining (from analysis in O and P). Scale bars = 25 µm. Data in D, E, H, I, L, M, and P were analysed by unpaired t-test with Welch’s correction or Mann–Whitney U test.
To assess lipoprotein permeability and retention, human LDL was labelled with Atto-565 NHS ester and injected intravenously into normolipidaemic Ccn2SMCΔ/Δ and Ccn2wt/wt mice. Mice were either sacrificed after 1 h to monitor initial influx of LDL as a proxy of endothelial permeability or after 24 h (once LDL had been cleared from the circulation) as a measure of LDL retention in the vessel wall as previously described48,49 (Figure 6F). For both analyses, we utilized the atherosclerosis-prone inner curvature of the aortic arch and the atherosclerosis-resistant descending thoracic aorta (Figure 6G). Although we observed a tendency towards increased permeability at both anatomical sites, this effect did not reach statistical significance (Figure 6H), and we did not observe differential expression of genes associated with endothelial tight junctions (see supplementary material online, Figure S10A). In contrast, we found a significant increase in retained LDL at both anatomical sites, with a 9-fold increase in LDL retention in the aortic arch and a 2.5-fold increase in the descending thoracic aorta (Figure 6I and J).
RNA-seq data from aortas of normolipidaemic Ccn2SMCΔ/Δ and Ccn2wt/wt mice unveiled heightened expression of macrophage markers Lgals3 and Cd68 and the adhesion molecule Vcam1 implying the involvement of macrophages in the Ccn2SMCΔ/Δ aorta phenotype even in a normolipidaemic setting (see supplementary material online, Figure S10B). By immunofluorescence staining of Lgals3 (Figure 6K), we found increased numbers of endothelium-associated luminal macrophages in the aortic arch of Ccn2SMCΔ/Δ mice (and a similar trend in descending thoracic aortas) (Figure 6L), but the LDL-retaining medial layer was most often devoid of macrophages suggesting that mechanisms driving LDL retention operate independently of macrophages. We did, however, observe more diffuse Lgals3 staining in some areas of the medial layer (Figure 6M), which was likely attributed to expression from modulated SMCs (corroborated by CCN2-silencing experiments; supplementary material online, Figure S9).
LDL retention can be mediated through electrostatic interactions between positively charged residues on apolipoprotein B and negatively charged proteoglycans within the extracellular matrix.1 By mass spectrometry of Ccn2SMCΔ/Δ aortas, we found most arterial proteoglycans50 to be significantly over-expressed in Ccn2SMCΔ/Δ aortas of normolipidaemic mice (and corroborated these findings by revisiting mass spectrometry data from the global Ccn2 knockout model) (Figure 6N). These observations were further validated by distinct Alcian blue staining of the medial layer of Ccn2SMCΔ/Δ aortas with negligible staining in Ccn2wt/wt aortas (Figure 6O and P). Notably, areas exhibiting high LDL retention in adjacent histological sections displayed dense Alcian blue staining (see Figure 6Q). The marked up-regulation of proteoglycans indicated SMC-modulation trajectory towards chondrocyte-like cells,5 as supported by marked increase in Runx2 expression, a key regulator of chondrocyte/osteoblast differentiation of SMCs51 (see supplementary material online, Figure S10C and D).
To gain insights into the signalling pathways influenced by Ccn2 deprivation, we specifically investigated the TGFβ-signalling pathway, which has previously been suggested to be down-regulated in conditions of Ccn2 deficiency.52,53 However, our investigations did not reveal down-regulation of TGFβR2 expression (see supplementary material online, Figure S11A–D and H) or reduced TGFβ signalling (see supplementary material online, Figure S11E–G and I) in CCN2-silenced HAoSMCs in baseline or oxLDL stimulation conditions. Nor did we detect regulation of Tgfβr2 in Ccn2SMCΔ/Δ mouse aortas (see supplementary material online, Figure S11J) or down-regulation of a panel of previously reported TGFβ-responsive genes54 in the aorta transcriptome (see supplementary material online, Figure S11K).
3.8. CCN2 expression correlates with SMC markers in advanced human plaques
To evaluate whether CCN2 could have similar roles in affecting SMC phenotype in advanced human atherosclerosis, we leveraged data from 654 advanced carotid plaques from the Athero-Express study cohort.42–45 In a recent investigation by Mokry et al.,42 bulk transcriptomics of these plaques led to the identification of five distinct clusters. CCN2 expression was markedly higher in plaque clusters 3 and 4 (Figure 7A), which are enriched in SMC markers and genes encoding collagens 1 and 3 (fibrillar collagens) alpha chains (Figure 7B). Across all 654 plaques (regardless of plaque clustering), CCN2 correlated significantly with the eight pre-defined SMC markers, MYOCD, and genes encoding fibrillar collagens (Figure 7C), and unbiased GSEAs, based on genes ranked by correlation to CCN2, revealed significant enrichment of multiple gene ontology terms pertaining to SMC and extracellular matrix (Figure 7D). Furthermore, histomorphological assessment of plaques from the Athero-Express study cohort, encompassing parameters such as macrophage content, SMC content, collagen content, and calcification, revealed a suggestive trend towards heightened CCN2 expression in plaques exhibiting elevated levels of ACTA2+ SMC content (P = 0.06), albeit not with other parameters (Figure 7E).
Complementary immunohistochemistry and ISH analyses of serial carotid plaque sections unveiled localization of CCN2 expression predominantly within the ACTA2+ and MYH11+ fibrous cap region, with minimal expression observed in CD68+ regions (Figure 7F). Moreover, corroborative evidence from previously published single-cell RNA-seq data from both coronary26 and carotid27 plaques showed CCN2 expression within clusters co-expressing SMC markers and genes encoding fibrillar collagens (see supplementary material online, Figure S12A and B).
Collectively, these findings indicate a potential involvement of CCN2 in the differentiation or maintenance of ACTA2+-expressing SMCs of the fibrous cap, reminiscent of its role in non-lesioned mouse aortas. However, we did not find significant correlations between CCN2 expression and plaque phenotype (as assessed by histomorphometry), plaque symptoms at the time of surgery, or major adverse cardiovascular events within 3 years after carotid endarterectomy in the Athero-Express study cohort (see supplementary material online, Table S10). In the analysis of 352 patients, linear regression models revealed a significant negative association between plasma IL6 levels and plaque CCN2 expression (P = 0.03), while hsCRP levels in 314 patients were not associated with plaque CCN2.
4. Discussion
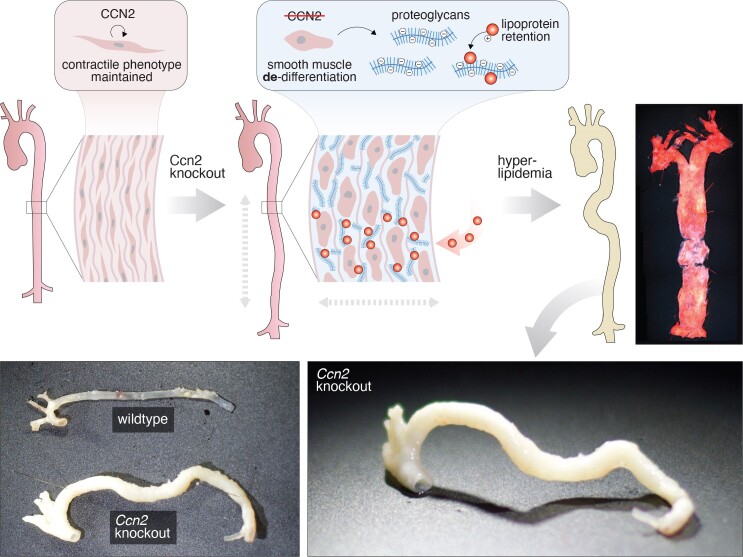
CCN2 has traditionally been regarded as a disease-promoting factor and thus a valuable target for therapeutic intervention.12 The present study challenges this perception by demonstrating that CCN2 plays an essential role in maintaining the contractile phenotype of vascular SMCs and that SMC deprivation of Ccn2 causes medial thickening, aorta malformation, and severe atherosclerosis, even at sites usually protected by laminar blood flow. Importantly, genotypic influences on plasma cholesterol or blood pressure were excluded as confounding factors, suggesting that mechanisms within the vascular wall are responsible for driving the atherosclerosis phenotype. We propose that the primary mechanism involves the transition of Ccn2-deprived SMCs into a chondrocyte-like state, resulting in a marked increase in proteoglycans within the arterial media, which enhances the ability of the artery wall to retain circulating LDL, which according to the response-to-retention hypothesis is the key initiating step of atherosclerosis, sufficient to drive subsequent pathological processes.1
Proteoglycans are negatively charged due to sulfate and carboxylic acid groups in their glycosaminoglycan side chains and entrap LDL in the vessel wall by electrostatically interacting with stretches of positively charged residues on APOB100, as experimentally demonstrated for biglycan55 and heparan-sulfate proteoglycan 2.56 The high-molecular weight proteoglycan aggrecan was the predominant proteoglycan enriched in the aorta of both mouse models employed in the study. In the extracellular matrix of cartilage, aggrecan–hyaluronic acid complexes are interspersed in a collagen meshwork and provide the osmotic properties required to resist de-swelling under compressive load.57 Osmotic swelling in cartilage tissue occurs due to interstitial hydration, driven by the high concentration of interstitial sodium ions (and other positively charged ions), which are attracted by the abundance of negatively charged aggrecan–hyaluronic acid complexes in the tissue.57 It is conceivable that interstitial swelling, caused by the accumulation of aggrecan and other proteoglycans, contributes to the medial thickening of aortas observed in both global and SMC-specific Ccn2 knockout mouse models.
Proteoglycans are enriched in the intimal layer of human arteries at atherosclerosis susceptibility sites.58,59 These areas often exhibit eccentric intimal thickenings, which are considered non-pathological responses to localized flow dynamics.58,59 Supporting the response-to-retention hypothesis, the initial deposition of lipids is observed in the proteoglycan-rich extracellular matrix at these sites prior to the accumulation of inflammatory cells.60 Early studies in rabbits proposed that susceptible and resistant artery segments differ primarily in lipoprotein efflux rather than lipoprotein influx, suggesting increased lipoprotein entrapment at these sites.61 Interestingly, interventions that override the natural topological distribution of atherosclerosis-susceptible sites, such as artificially inducing disturbed laminar blood flow49 or balloon injury,62 are associated with local enrichment of arterial proteoglycan expression and increased ability of the artery wall to retain LDL. Genetic interventions disrupting the topological distribution of atherosclerosis are rare but include genetic models of progeria, where features like medial thickening, increased LDL retention, and atherosclerosis formation are observed throughout the aorta,63 similar to, although less aggressive than, the Ccn2 knockout phenotype.
Although the increased LDL retention capacity is a prominent feature in Ccn2 knockout mice and likely the main contributor to the atherosclerosis phenotype, our data also indicate the involvement of other mechanisms, as we observed a trend towards increased endothelial permeability to LDL and augmented oxLDL uptake in CCN2-silenced SMCs. Moreover, we observe an increased recruitment of macrophages to the vessel wall, likely due to augmented secretion of chemokines and adhesion molecules, as indicated by RNA-seq data. This recruitment is likely a parallel or subsequent mechanism to LDL retention as macrophages do not appear to be directly involved at sites where LDL accumulates.
Our systematic evaluation of CCN2 expression across a range of human tissues revealed a markedly higher level of CCN2 expression in artery tissues relative to non-vascular tissues, with CCN2 being one of the most highly expressed genes in both human and mouse aortas. Intriguingly, among the 50 top-ranking genes in human aortas, 20 can be considered housekeeping genes abundantly expressed across most tissues, and another eight genes (MYH11, MGP, TPM2, COL6A2, FLNA, ACTA2, TAGLN, and MYL9) are abundant in all SMC-dense tissues. In comparison, CCN2 expression was confined to artery tissues suggesting a specific role for SMCs of the vasculature. The reason for previous studies’ assertion that CCN2 expression is primarily found in developing vasculature13–15 and minimally in adult non-diseased artery tissue16–19 remains unclear. Previous studies have relied on probes or antibodies, and it cannot be ruled out that limitations in the sensitivity of these reagents might have influenced the conclusions drawn. The present study is the first to investigate CCN2 expression in absolute terms (transcripts per million) and relative to other transcripts in human and mouse artery tissues, and we readily detect CCN2 by ISH and mass spectrometry. Moreover, our data show that Ccn2 is highly physiologically relevant in adult aortic tissue—at least in mice.
Ccn2 has recently been studied in the context of aortic aneurysm disease, in which Ccn2 deletion led to the abrupt formation of aortic aneurysms in experimental models of this disease.53,64 Although neither of these studies reported on aortic dimensions or atherosclerosis, they both support the impact of Ccn2 depletion on the modulation of SMCs in line with our findings. Both groups propose that CCN2 acts on SMCs through the promotion of TGFβ signalling either by directly interacting with TGFβ and its receptors,53 or by up-regulating TGFβR2 via activation of EGFR.52 Interestingly, we did not detect reduced TGFβR2 expression levels or down-regulation of TGFβ signalling in our study. The reason for this may underline context-dependent effects of CCN2 and the notion that recapitulating the role of full-length Ccn2 of the medial layer in vivo by in vitro experiments,52,53 or by treating mice with recombinant Ccn2 domain 4,52 can be problematic.
Our study involves both global and SMC-specific deletion of Ccn2. However, due to the location of the Myh11-CreERT2 transgene on the Y chromosome, our investigations using this model were limited to male mice. Additionally, the Myh11 promoter is a generic SMC promoter not exclusive to vascular SMCs. Therefore, we cannot rule out the possibility that phenotypic traits, such as reduced body weight or lower plasma cholesterol, may be influenced by factors like reduced intestinal cholesterol absorption due to Ccn2 deletion in intestinal SMCs. However, these effects would be expected to decrease atherosclerosis. Moreover, the high expression of CCN2 in arterial tissues compared with non-vascular tissues, along with the prominent SMC and aorta phenotypes observed under normolipidaemic conditions, strongly indicates that the mechanisms underlying atherosclerosis are inherent to the vascular wall.
It is intriguing to speculate whether the potent influence of CCN2 on SMC phenotype persists in advanced lesions. In other words, do SMCs within the plaque that continue to express CCN2 or are exposed to CCN2 in the local microenvironment tend to adopt characteristics similar to myofibroblast-like cells found in the fibrous cap? Supporting this notion, CCN2 expression is present in fibrous cap cells within human carotid lesions and CCN2 expression correlated with SMC markers and genes encoding fibrillar collagens across bulk transcriptomic profiled carotid plaques from the Athero-Express study cohort.42 These observations are particularly interesting considering previous research demonstrating that treatment of de-differentiated SMCs with full-length recombinant CCN2 in vitro promotes their transition towards a fibroblast-like phenotype.27 From other disease contexts, CCN2 has been shown to be required for inducing fibroblast differentiation into ACTA2+ myofibroblasts, the primary effector cells in fibrosis, and for sustaining this specific phenotype.12,65–67 Therefore, contrary to the established deleterious role of CCN2 in most fibrotic diseases, these same properties might be highly beneficial in the context of atherosclerosis, where a high lesion content of ACTA2+ cells and collagen production are associated with plaque stability and reduced risk of rupture,3 which is also supported by the inverse relationship between plaque CCN2 and circulating IL6, potentially reflecting that plaques low in CCN2 are more pro-inflammatory. On the contrary, prior studies have demonstrated elevated CCN2 protein levels in complicated plaques compared with fibrous cap plaques.19 Additionally, high CCN2 plasma levels have been associated with future myocardial infarction and mortality.68,69 Therefore, further investigations are warranted to directly examine the role of CCN2 in advanced plaques and to elucidate how CCN2 intervention may impact plaque stability.
Our findings suggest the need for further investigation into the potential effects of anti-CCN2-based therapies on the vasculature. Currently, pamrevlumab, which successfully attenuated the progression of idiopathic pulmonary fibrosis in a Phase II trial, has advanced to a Phase III clinical trial for efficacy and safety testing in these patients. Furthermore, pamrevlumab is being developed for pancreatic cancer and Duchenne muscular dystrophy. Based on our results, we recommend that future trials involving anti-CCN2 therapy should incorporate vascular endpoints such as ankle-brachial pressure index, carotid intima-media thickness, or coronary calcium score to monitor the potential adverse vascular effects.
In conclusion, our study identifies a new critical role for CCN2 in preserving artery integrity and in preventing atherosclerosis. Moreover, our findings highlight the importance of further research into the potential effects of anti-CCN2-based therapies on the vasculature.
Supplementary Material
Acknowledgements
We thank Gitte Kitlen, Mie Grønning Rytz Hansen, Nete Kronborg Vandel, Anja Toft, Camilla Enggaard, Niels Larsen, Amalie Bøgsted, Julie May Jensen, Inger Nissen, Christian Enggaard, Johan Holmberg, Katarzyna Kawka, Morten Ploug Kühlmann, Camilla Rasmussen, Lars Vitved, Søren Hansen, and Pia Søndergaard Jensen for the excellent technical assistance. We thank Professor Jacob Fog Bentzon (Aarhus University Hospital, Denmark) for providing pAAV/ApoEHCR-hAAT-D377Y-mPcsk9-BGHpA, Professor Pierre Chambon (GIE-CERBM, France) for generating the CreERT2 construct, Professor Stefan Offermanns (Max-Planck Institute for Heart and Lung Research, Germany) for providing Myh11-CreERT2 mice, and Professor Peter Bie for valuable discussions.
Contributor Information
Jannik H Larsen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark; Centre for Individualized Medicine in Arterial Diseases, Odense University Hospital, J. B. Winsløws Vej 4, DK-5000 Odense C, Denmark.
Julie S Hegelund, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Matilde K Pedersen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Cecilie M Andersson, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Caroline A Lindegaard, Department of Biochemistry and Molecular Biology, University of Southern Denmark, Odense, Denmark.
Didde R Hansen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Jane Stubbe, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Jes S Lindholt, Centre for Individualized Medicine in Arterial Diseases, Odense University Hospital, J. B. Winsløws Vej 4, DK-5000 Odense C, Denmark; Department of Cardiothoracic and Vascular Surgery, Odense University Hospital, Odense, Denmark.
Camilla S Hansen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Andrietta Grentzmann, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Maria Bloksgaard, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Boye L Jensen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Raúl R Rodriguez-Díez, Department of Cell Biology, Complutense University School of Medicine, Madrid, Spain.
Marta Ruiz-Ortega, Cellular and Molecular Biology in Renal and Vascular Pathology Laboratory, IIS-Fundación Jiménez Díaz-Universidad Autónoma Madrid, Madrid, Spain.
Sebastian Albinsson, Department of Experimental Medical Science, Vascular Physiology Environment, Lund University, Lund, Sweden.
Gerard Pasterkamp, Laboratory of Clinical Chemistry and Haematology, University Medical Center, Heidelberglaan 100, Utrecht, The Netherlands.
Michal Mokry, Laboratory of Clinical Chemistry and Haematology, University Medical Center, Heidelberglaan 100, Utrecht, The Netherlands; Department of Cardiology, University Medical Center Utrecht, Heidelberglaan 100, Utrecht, The Netherlands.
Andrew Leask, College of Dentistry, University of Saskatoon, Saskatoon, SK, Canada.
Roel Goldschmeding, Department of Pathology, University Medical Center Utrecht, Utrecht, The Netherlands.
Bartosz Pilecki, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Grith L Sorensen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Charles Pyke, Research and Early Development, Novo Nordisk A/S, Måløv, Denmark.
Martin Overgaard, Department of Clinical Biochemistry, Odense University Hospital, Odense, Denmark.
Hans C Beck, Department of Clinical Biochemistry, Odense University Hospital, Odense, Denmark.
Daniel F J Ketelhuth, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark.
Lars M Rasmussen, Centre for Individualized Medicine in Arterial Diseases, Odense University Hospital, J. B. Winsløws Vej 4, DK-5000 Odense C, Denmark; Department of Clinical Biochemistry, Odense University Hospital, Odense, Denmark.
Lasse B Steffensen, Department of Molecular Medicine, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark; Centre for Individualized Medicine in Arterial Diseases, Odense University Hospital, J. B. Winsløws Vej 4, DK-5000 Odense C, Denmark.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
J.H.L., L.M.R., and L.B.S. generated the hypothesis, initiated the project, and designed the experiments; J.H.L., J.S.H., M.K.P., C.M.A., C.A.L., D.R.H., C.S.H., A.G., B.L.J., S.A., B.P., C.P., M.O., and H.C.B. performed the experiments; J.S.L., G.P., M.M., C.P., M.O., H.C.B., and L.M.R. provided human data and human material and performed the related analyses; J.S., J.S.L., M.B., B.L.J., R.R.R.-D., M.R.O., S.A., G.P., M.M., A.L., R.G., G.L.S., D.F.J.K., and L.M.R. provided the intellectual input and techniques and edited the manuscript; J.H.L. and L.B.S. designed the figures; and J.H.L. and L.B.S. wrote the manuscript, which was approved by all authors.
Funding
This work was supported by the Centre for Individualized Medicine in Arterial Diseases, Odense University Hospital, Arvid Nilssons Fond, Direktør Emil C. Hertz og Hustru Inger Hertz’ Fond. D.F.J.K. is supported by grants from the Novo Nordisk Foundation (0064142 and 0075258), Danmarks Frie Forskningsfond (2034-00136B), Simon Fougner Hartmanns Familiefond (2023-0066), and the University of Southern Denmark. L.B.S. is supported by the Danish Diabetes Academy (funded by the Novo Nordisk Foundation).
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Borén J, Williams KJ. The central role of arterial retention of cholesterol-rich apolipoprotein-B-containing lipoproteins in the pathogenesis of atherosclerosis: a triumph of simplicity. Curr Opin Lipidol 2016;27:473–483. [DOI] [PubMed] [Google Scholar]
- 2. Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, Tokgözoğlu L, Lewis EF. Atherosclerosis. Nat Rev Dis Primers 2019;5:56. [DOI] [PubMed] [Google Scholar]
- 3. Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res 2014;114:1852–1866. [DOI] [PubMed] [Google Scholar]
- 4. Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 2006;20:1545–1556. [DOI] [PubMed] [Google Scholar]
- 5. Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol 2019;16:727–744. [DOI] [PubMed] [Google Scholar]
- 6. Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 2018;9:4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Albarrán-Juárez J, Kaur H, Grimm M, Offermanns S, Wettschureck N. Lineage tracing of cells involved in atherosclerosis. Atherosclerosis 2016;251:445–453. [DOI] [PubMed] [Google Scholar]
- 9. Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res 2014;115:662–667. [DOI] [PubMed] [Google Scholar]
- 10. Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jørgensen HF. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res 2016;119:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perbal B, Tweedie S, Bruford E. The official unified nomenclature adopted by the HGNC calls for the use of the acronyms, CCN1-6, and discontinuation in the use of CYR61, CTGF, NOV and WISP 1-3 respectively. J Cell Commun Signal 2018;12:625–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramazani Y, Knops N, Elmonem MA, Nguyen TQ, Arcolino FO, van den Heuvel L, Levtchenko E, Kuypers D, Goldschmeding R. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol 2018;68–69:44–66. [DOI] [PubMed] [Google Scholar]
- 13. Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 2003;130:2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chuva de Sousa Lopes SM, Feijen A, Korving J, Korchynskyi O, Larsson J, Karlsson S, ten Dijke P, Lyons KM, Goldschmeding R, Doevendans P, Mummery CL. Connective tissue growth factor expression and SMAD signaling during mouse heart development and myocardial infarction. Dev Dyn 2004;231:542–550. [DOI] [PubMed] [Google Scholar]
- 15. Hall-Glenn F, De Young RA, Huang B-L, van Handel B, Hofmann JJ, Chen TT, Choi A, Ong JR, Benya PD, Mikkola H, Iruela-Arispe ML, Lyons KM. CCN2/connective tissue growth factor is essential for pericyte adhesion and endothelial basement membrane formation during angiogenesis. PLoS One 2012;7:e30562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ponticos M. Connective tissue growth factor (CCN2) in blood vessels. Vascul Pharmacol 2013;58:189–193. [DOI] [PubMed] [Google Scholar]
- 17. Oemar BS, Lüscher TF. Connective tissue growth factor. Friend or foe? Arterioscler Thromb Vasc Biol 1997;17:1483–1489. [DOI] [PubMed] [Google Scholar]
- 18. Oemar BS, Werner A, Garnier JM, Do DD, Godoy N, Nauck M, Marz W, Rupp J, Pech M, Luscher TF. Human connective tissue growth factor is expressed in advanced atherosclerotic lesions. Circulation 1997;95:831–839. [DOI] [PubMed] [Google Scholar]
- 19. Cicha I, Yilmaz A, Klein M, Raithel D, Brigstock DR, Daniel WG, Goppelt-Struebe M, Garlichs CD. Connective tissue growth factor is overexpressed in complicated atherosclerotic plaques and induces mononuclear cell chemotaxis in vitro. Arterioscler Thromb Vasc Biol 2005;25:1008–1013. [DOI] [PubMed] [Google Scholar]
- 20. Rupérez M, Lorenzo O, Blanco-Colio LM, Esteban V, Egido J, Ruiz-Ortega M. Connective tissue growth factor is a mediator of Angiotensin II-induced fibrosis. Circulation 2003;108:1499–1505. [DOI] [PubMed] [Google Scholar]
- 21. Kanazawa S, Miyake T, Kakinuma T, Tanemoto K, Tsunoda T, Kikuchi K. The expression of platelet-derived growth factor and connective tissue growth factor in different types of abdominal aortic aneurysms. J Cardiovasc Surg (Torino) 2005;46:271–278. [PubMed] [Google Scholar]
- 22. Wang X, LeMaire SA, Chen L, Shen YH, Gan Y, Bartsch H, Carter SA, Utama B, Ou H, Coselli JS, Wang XL. Increased collagen deposition and elevated expression of connective tissue growth factor in human thoracic aortic dissection. Circulation 2006;114:I200–I205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration & Visualization—EBI; Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis & Coordinating Center (LDACC); NIH Program Management; Biospecimen Collection; Pathology; eQTL Manuscript Working Group; Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature 2017;550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koplev S, Seldin M, Sukhavasi K, Ermel R, Pang S, Zeng L, Bankier S, Di Narzo A, Cheng H, Meda V, Ma A, Talukdar H, Cohain A, Amadori L, Argmann C, Houten SM, Franzen O, Mocci G, Meelu OA, Ishikawa K, Whatling C, Jain A, Jain RK, Gan LM, Giannarelli C, Roussos P, Hao K, Schunkert H, Michoel T, Ruusalepp A, Schadt EE, Kovacic JC, Lusis AJ, Bjorkegren JLM. A mechanistic framework for cardiometabolic and coronary artery diseases. Nat Cardiovasc Res 2022;1:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pirruccello JP, Chaffin MD, Chou EL, Fleming SJ, Lin H, Nekoui M, Khurshid S, Friedman SF, Bick AG, Arduini A, Weng LC, Choi SH, Akkad AD, Batra P, Tucker NR, Hall AW, Roselli C, Benjamin EJ, Vellarikkal SK, Gupta RM, Stegmann CM, Juric D, Stone JR, Vasan RS, Ho JE, Hoffmann U, Lubitz SA, Philippakis AA, Lindsay ME, Ellinor PT. Deep learning enables genetic analysis of the human thoracic aorta. Nat Genet 2022;54:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, Ihuegbu CO, Bush EC, Worley J, Vlahos L, Laise P, Solomon RA, Connolly ES, Califano A, Sims PA, Zhang H, Li M, Reilly MP. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation 2020;142:2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hao Y, Hao S, Andersen-Nissen E, Mauck WM III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu S, Shi-wen X, Abraham DJ, Leask A. CCN2 is required for bleomycin-induced skin fibrosis in mice. Arthritis Rheum 2011;63:239–246. [DOI] [PubMed] [Google Scholar]
- 30. Fontes MS, Kessler EL, van Stuijvenberg L, Brans MA, Falke LL, Kok B, Leask A, van Rijen HV, Vos MA, Goldschmeding R, van Veen TA. CTGF knockout does not affect cardiac hypertrophy and fibrosis formation upon chronic pressure overload. J Mol Cell Cardiol 2015;88:82–90. [DOI] [PubMed] [Google Scholar]
- 31. Steffensen LB, Stubbe J, Overgaard M, Larsen JH. Tamoxifen-independent Cre-activity in SMMHC-CreERT2 mice. Atheroscler Plus 2022;48:8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bjorklund MM, Hollensen AK, Hagensen MK, Dagnaes-Hansen F, Christoffersen C, Mikkelsen JG, Bentzon JF. Induction of atherosclerosis in mice and hamsters without germline genetic engineering. Circ Res 2014;114:1684–1689. [DOI] [PubMed] [Google Scholar]
- 33. Mattson DL. Long-term measurement of arterial blood pressure in conscious mice. Am J Physiol 1998;274:R564–R570. [DOI] [PubMed] [Google Scholar]
- 34. Thuesen AD, Finsen SH, Rasmussen LL, Andersen DC, Jensen BL, Hansen PBL. Deficiency of T-type Ca(2+) channels Ca(v)3.1 and Ca(v)3.2 has no effect on Angiotensin II-induced hypertension but differential effect on plasma aldosterone in mice. Am J Physiol Renal Physiol 2019;317:F254–F263. [DOI] [PubMed] [Google Scholar]
- 35. Thangaraj SS, Enggaard C, Stubbe J, Palarasah Y, Hansen PBL, Svenningsen P, Jensen BL. Interleukin 17A infusion has no acute or long-term hypertensive action in conscious unrestrained male mice. Pflugers Arch 2022;474:709–719. [DOI] [PubMed] [Google Scholar]
- 36. Steffensen LB, Stubbe J, Lindholt JS, Beck HC, Overgaard M, Bloksgaard M, Genovese F, Holm Nielsen S, Tha MLT, Bang-Moeller SK, Hong Lin MKT, Larsen JH, Hansen DR, Jones GT, Bown MJ, Karsdal MA, Rasmussen LM. Basement membrane collagen IV deficiency promotes abdominal aortic aneurysm formation. Sci Rep 2021;11:12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mulorz J, Spin JM, Beck HC, Thi MLT, Wagenhauser MU, Rasmussen LM, Lindholt JS, Tsao PSC, Steffensen LB. Hyperlipidemia does not affect development of elastase-induced abdominal aortic aneurysm in mice. Atherosclerosis 2020;311:73–83. [DOI] [PubMed] [Google Scholar]
- 41. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mokry M, Boltjes A, Slenders L, Bel-Bordes G, Cui K, Brouwer E, Mekke JM, Depuydt MAC, Timmerman N, Waissi F, Verwer MC, Turner AW, Khan MD, Hodonsky CJ, Diez Benavente E, Hartman RJG, van den Dungen NAM, Lansu N, Nagyova E, Prange KHM, Kovacic JC, Björkegren JLM, Pavlos E, Andreakos E, Schunkert H, Owens GK, Monaco C, Finn AV, Virmani R, Leeper NJ, de Winther MPJ, Kuiper J, de Borst GJ, Stroes ESG, Civelek M, de Kleijn DPV, den Ruijter HM, Asselbergs FW, van der Laan SW, Miller CL, Pasterkamp G. Transcriptomic-based clustering of human atherosclerotic plaques identifies subgroups with different underlying biology and clinical presentation. Nat Cardiovasc Res 2022;1:1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hellings WE, Moll FL, De Vries JP, Ackerstaff RG, Seldenrijk KA, Met R, Velema E, Derksen WJ, De Kleijn DP, Pasterkamp G. Atherosclerotic plaque composition and occurrence of restenosis after carotid endarterectomy. JAMA 2008;299:547–554. [DOI] [PubMed] [Google Scholar]
- 44. Hellings WE, Peeters W, Moll FL, Piers SR, van Setten J, Van der Spek PJ, de Vries JP, Seldenrijk KA, De Bruin PC, Vink A, Velema E, de Kleijn DP, Pasterkamp G. Composition of carotid atherosclerotic plaque is associated with cardiovascular outcome: a prognostic study. Circulation 2010;121:1941–1950. [DOI] [PubMed] [Google Scholar]
- 45. Verhoeven BA, Velema E, Schoneveld AH, de Vries JP, de Bruin P, Seldenrijk CA, de Kleijn DP, Busser E, van der Graaf Y, Moll F, Pasterkamp G. Athero-Express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur J Epidemiol 2004;19:1127–1133. [DOI] [PubMed] [Google Scholar]
- 46. Elsenberg EH, Sels JE, Hillaert MA, Schoneveld AH, van den Dungen NA, van Holten TC, Roest M, Jukema JW, van Zonneveld AJ, de Groot PG, Pijls N, Pasterkamp G, Hoefer IE. Increased cytokine response after toll-like receptor stimulation in patients with stable coronary artery disease. Atherosclerosis 2013;231:346–351. [DOI] [PubMed] [Google Scholar]
- 47. Giral P. Targeted proteomics improves cardiovascular risk prediction in secondary prevention: the impact of statin treatment? Eur Heart J 2022;43:3811–3811. [DOI] [PubMed] [Google Scholar]
- 48. Lewis EA, Muniz-Anquela R, Redondo-Angulo I, Gonzalez-Cintado L, Labrador-Cantarero V, Bentzon JF. Capacity for LDL (low-density lipoprotein) retention predicts the course of atherogenesis in the murine aortic arch. Arterioscler Thromb Vasc Biol 2023;43:637–649. [DOI] [PubMed] [Google Scholar]
- 49. Steffensen LB, Mortensen MB, Kjolby M, Hagensen MK, Oxvig C, Bentzon JF. Disturbed laminar blood flow vastly augments lipoprotein retention in the artery wall: a key mechanism distinguishing susceptible from resistant sites. Arterioscler Thromb Vasc Biol 2015;35:1928–1935. [DOI] [PubMed] [Google Scholar]
- 50. Cikach FS, Koch CD, Mead TJ, Galatioto J, Willard BB, Emerton KB, Eagleton MJ, Blackstone EH, Ramirez F, Roselli EE, Apte SS. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight 2018;3:e97167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin ME, Chen TM, Wallingford MC, Nguyen NB, Yamada S, Sawangmake C, Zhang J, Speer MY, Giachelli CM. Runx2 deletion in smooth muscle cells inhibits vascular osteochondrogenesis and calcification but not atherosclerotic lesion formation. Cardiovasc Res 2016;112:606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tejera-Munoz A, Marquez-Exposito L, Tejedor-Santamaria L, Rayego-Mateos S, Orejudo M, Suarez-Alvarez B, Lopez-Larrea C, Ruiz-Ortega M, Rodrigues-Diez RR. CCN2 increases TGF-beta receptor type II expression in vascular smooth muscle cells: essential role of CCN2 in the TGF-beta pathway regulation. Int J Mol Sci 2021;23:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y, Liu X, Xu Q, Xu W, Zhou X, Lin Z. CCN2 deficiency in smooth muscle cells triggers cell reprogramming and aggravates aneurysm development. JCI Insight 2023;8:e162987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen PY, Qin L, Li G, Malagon-Lopez J, Wang Z, Bergaya S, Gujja S, Caulk AW, Murtada SI, Zhang X, Zhuang ZW, Rao DA, Wang G, Tobiasova Z, Jiang B, Montgomery RR, Sun L, Sun H, Fisher EA, Gulcher JR, Fernandez-Hernando C, Humphrey JD, Tellides G, Chittenden TW, Simons M. Smooth muscle cell reprogramming in aortic aneurysms. Cell Stem Cell 2020;26:542–557.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skalen K, Gustafsson M, Rydberg EK, Hulten LM, Wiklund O, Innerarity TL, Boren J. Subendothelial retention of atherogenic lipoproteins in early atherosclerosis. Nature 2002;417:750–754. [DOI] [PubMed] [Google Scholar]
- 56. Tran-Lundmark K, Tran PK, Paulsson-Berne G, Friden V, Soininen R, Tryggvason K, Wight TN, Kinsella MG, Boren J, Hedin U. Heparan sulfate in perlecan promotes mouse atherosclerosis: roles in lipid permeability, lipid retention, and smooth muscle cell proliferation. Circ Res 2008;103:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chandran PL, Horkay F. Aggrecan, an unusual polyelectrolyte: review of solution behavior and physiological implications. Acta Biomater 2012;8:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stary HC, Blankenhorn DH, Chandler AB, Glagov S, Insull W Jr, Richardson M, Rosenfeld ME, Schaffer SA, Schwartz CJ, Wagner WD, Wissler RW. A definition of the intima of human arteries and of its atherosclerosis-prone regions. A report from the committee on vascular lesions of the council on arteriosclerosis, American Heart Association. Arterioscler Thromb 1992;12:120–134. [DOI] [PubMed] [Google Scholar]
- 59. Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol 1995;15:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakashima Y, Fujii H, Sumiyoshi S, Wight TN, Sueishi K. Early human atherosclerosis: accumulation of lipid and proteoglycans in intimal thickenings followed by macrophage infiltration. Arterioscler Thromb Vasc Biol 2007;27:1159–1165. [DOI] [PubMed] [Google Scholar]
- 61. Schwenke DC, Carew TE. Quantification in vivo of increased LDL content and rate of LDL degradation in normal rabbit aorta occurring at sites susceptible to early atherosclerotic lesions. Circ Res 1988;62:699–710. [DOI] [PubMed] [Google Scholar]
- 62. Srinivasan SR, Xu JH, Vijayagopal P, Radhakrishnamurthy B, Berenson GS. Injury to the arterial wall of rabbits produces proteoglycan variants with enhanced low-density lipoprotein-binding property. Biochim Biophys Acta 1993;1168:158–166. [DOI] [PubMed] [Google Scholar]
- 63. Hamczyk MR, Villa-Bellosta R, Gonzalo P, Andres-Manzano MJ, Nogales P, Bentzon JF, Lopez-Otin C, Andres V. Vascular smooth muscle-specific progerin expression accelerates atherosclerosis and death in a mouse model of Hutchinson–Gilford progeria syndrome. Circulation 2018;138:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodrigues-Diez RR, Tejera-Munoz A, Esteban V, Steffensen LB, Rodrigues-Diez R, Orejudo M, Rayego-Mateos S, Falke LL, Cannata-Ortiz P, Ortiz A, Egido J, Mallat Z, Briones Ana M, Bajo MA, Goldschmeding R, Ruiz-Ortega M. CCN2 (cellular communication network factor 2) deletion alters vascular integrity and function predisposing to aneurysm formation. Hypertension 2022;79:e42–e55. [DOI] [PubMed] [Google Scholar]
- 65. Liu S, Herault Y, Pavlovic G, Leask A. Skin progenitor cells contribute to bleomycin-induced skin fibrosis. Arthritis Rheumatol 2014;66:707–713. [DOI] [PubMed] [Google Scholar]
- 66. Vanhove T, Goldschmeding R, Kuypers D. Kidney fibrosis: origins and interventions. Transplantation 2017;101:713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsang M, Quesnel K, Vincent K, Hutchenreuther J, Postovit LM, Leask A. Insights into fibroblast plasticity: cellular communication network 2 is required for activation of cancer-associated fibroblasts in a murine model of melanoma. Am J Pathol 2020;190:206–221. [DOI] [PubMed] [Google Scholar]
- 68. Hunt KJ, Jaffa MA, Garrett SM, Luttrell DK, Lipson KE, Lopes-Virella MF, Luttrell LM, Jaffa AA; VADT Investigators . Plasma connective tissue growth factor (CTGF/CCN2) levels predict myocardial infarction in the Veterans Affairs Diabetes Trial (VADT) cohort. Diabetes Care 2018;41:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gerritsen KG, Falke LL, van Vuuren SH, Leeuwis JW, Broekhuizen R, Nguyen TQ, de Borst GJ, Nathoe HM, Verhaar MC, Kok RJ, Goldschmeding R, Visseren FL, Group SS. Plasma CTGF is independently related to an increased risk of cardiovascular events and mortality in patients with atherosclerotic disease: the SMART study. Growth Factors 2016;34:149–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.