Abstract
Background
Advances in our understanding of the molecular biology of meningiomas have led to significant gains in the ability to predict patient prognosis and tumor recurrence and to identify novel targets for therapeutic design. Specifically, classification of meningiomas based on DNA methylation has greatly improved our ability to risk stratify patients, however new questions have arisen in terms of the underlying impact these DNA-methylation signatures have on meningioma biology.
Methods
This study utilizes RNA-sequencing data from 486 meningioma samples corresponding to 3 meningioma DNA-methylation groups (merlin-intact, immune-enriched, and hypermitotic), followed by in vitro experiments utilizing human meningioma cell lines.
Results
We identify alterations in RNA splicing between meningioma DNA-methylation groups including individual splicing events that correlate with hypermitotic meningiomas and predict tumor recurrence and overall patient prognosis and compile a set of splicing events that can accurately predict DNA-methylation classification based on RNA-seq data. Furthermore, we validate these events using reverse transcription polymerase chain reaction (RT-PCR) in patient samples and meningioma cell lines. Additionally, we identify alterations in RNA-binding proteins and splicing factors that lie upstream of RNA splicing events, including upregulation of SRSF1 in hypermitotic meningiomas which we show drives alternative RNA splicing changes. Finally, we design splice-switching antisense oligonucleotides to target RNA splicing changes in NASP and MFF observed in hypermitotic meningiomas, providing a rationale for RNA-based therapeutic design.
Conclusions
RNA splicing is an important driver of meningioma phenotypes that can be useful in prognosticating patients and as a potential exploit for therapeutic vulnerabilities.
Keywords: meningioma, RNA splicing, RNA-based therapeutics, risk stratification, transcriptomics
For the podcast associated with this article, please visit ‘https://soc-neuro-onc.libsyn.com/alterations-in-rna-splicingbetween-meningioma-dna-methylation-groups’
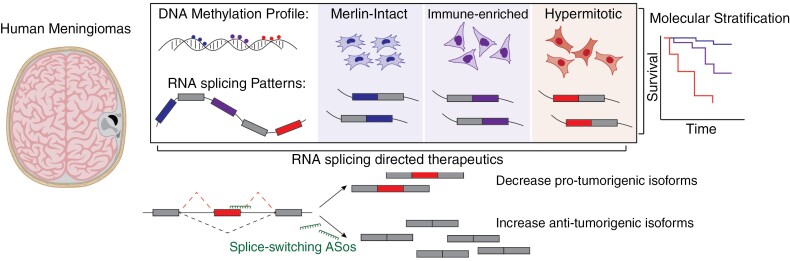
Graphical Abstract
Graphical Abstract.
Key Points.
Meningioma DNA-methylation groups harbor unique alternative RNA splicing changes that correlate with tumor recurrence and overall survival.
RNA splicing events associated with clinically high-risk meningiomas can be directly targeted with splice-switching antisense oligonucleotides.
Importance of the Study.
DNA-methylation profiling has revealed molecular groups of meningiomas that are associated with distinct gene expression programs, therapeutic vulnerabilities, and clinical outcomes. However, RNA processing events across meningioma DNA-methylation groups have not been assessed. Specifically, RNA splicing, a key step in gene expression that promotes transcriptomic and proteomic diversity, has yet to be systematically characterized in human meningiomas. This study identifies key RNA splicing events associated with high-risk (hypermitotic) meningioma groups. We further identify RNA-binding proteins that are differentially expressed and regulate these splicing events in meningiomas. Finally, we create splice-switching antisense oligonucleotides directed at oncogenic splicing events that are toxic to hypermitotic meningioma cell lines in vitro. Together, our results provide the first systematic identification of alternative splicing events across molecular groups of meningiomas with potential utility in clinical diagnostics, prognostication, and therapeutics.
Meningiomas are the most common intracranial tumor.1 While most are benign,1 a subset are aggressive with high rates of recurrence despite standard treatment.2,3 Recent studies have revised meningioma classifications based on tumor genomics,4–7 epigenetics,8–14 and gene expression signatures,15,16,17 and nominated novel therapeutic targets that are in preclinical or early clinical trials for aggressive meningiomas.8,9,18 Among these molecular approaches to meningioma classification, DNA methylation is a powerful tool for predicting patient outcomes.9–11,14 However, we lack a comprehensive understanding of the biological differences between DNA-methylation groups and the downstream impact of these changes on tumor growth or therapeutic vulnerability. Interestingly, DNA-methylation groups with more malignant tumors harbor dysregulated expression signatures across genes involved in RNA processing and splicing.9,11 Alternative RNA splicing (AS) is a key step in gene expression regulation that allows individual genes to encode multiple RNA isoforms, facilitating transcriptomic diversity that underlies cellular phenotypes.19,20 AS is dysregulated in cancers where the expression of RNA isoforms is skewed towards those that promote hallmark phenotypes of cancer.21,22 This shift in RNA isoforms is due to underlying defects in RNA processing machinery including splicing factors (SFs), a family of RNA-binding proteins (RBPs) that regulate AS in a dose-dependent manner. SFs are recurrently mutated in hematological malignancies but predominantly undergo copy number and expression level changes in solid tumors.21,22 Despite its importance in cancer biology, a systematic analysis of AS changes in meningiomas is lacking. A few studies examined the impact of individual AS isoforms on meningioma tumorigenesis, revealing important interactions with isoforms of CHEK2 and NF2 loss of function,23 alternative splicing in NF2 itself,24 and tumor dependency on RBPs.25 These findings suggest a role for AS in meningioma biology, and an unbiased high-throughput analysis might therefore discover key RNA isoforms that impact meningioma tumorigenesis.
Here, we systematically analyze 486 meningioma samples and reveal differences in AS patterns across DNA-methylation groups, identifying AS events that predict tumor recurrence and overall survival across independent patient cohorts. Additionally, we identify upstream RBPs that are differentially expressed between DNA-methylation groups, including those upregulated in hypermitotic meningiomas, which have the worst clinical outcomes, and demonstrate that depletion of these proteins impairs meningioma cell proliferation. Finally, we develop splice-switching antisense oligonucleotides (ASOs) that target hypermitotic-associated AS events in NASP and MFF, providing a proof-of-principle in rational therapeutic design against AS in meningiomas.
Methods
Human Cell Lines
HO1654, ID1654, NU02141, NU02171, IOMM-Lee, and BenMen cell lines are maintained in DMEM (Gibco) with 10% FBS, 1% penicillin streptomycin (Sigma), and 1× glutamax (Gibco). Cells are grown at 37 °C with 5% CO2. Cells are routinely tested negative for mycoplasma using the MycoAlert™ Mycoplasma Detection Kit (Lonza), and early passages aliquots are used.
Patient samples
Meningioma tissue and MRI with brief clinical history were provided as deidentified samples from the University of Connecticut Health Center biobank (IRB #IE-08-310-1). MRI images shown are contrast-enhanced T1-weighted series. For RT-PCR analysis, meningioma tissue was lysed into RLT buffer (Qiagen) and continued through RNA extraction and PCR as below.
RNA Extraction and Reverse Transcription
Cells are lysed using RLT buffer (Qiagen) supplemented with 1% β-mercaptoethanol. RNA is purified using an RNAeasy kit (Qiagen) with DNAse I. About 250 to 500 ng of RNA is reverse transcribed using Superscript III reverse transcriptase (Invitrogen).
Semiquantitative PCR for Splicing Detection
About 20 ng of cDNA are amplified with Phusion hot start II DNA polymerase (Thermo Fisher) and primers listed in Supplementary Table S3A. PCR products are separated in 1% to 2% agarose gel stained with SYBR Safe (Invitrogen) and imaged using ChemiDoc MP Imaging System (Bio-rad). PCR bands are quantified using ImageLab 6.0 (Bio-rad) and the percent spliced-in (PSI) ratio of each transcript is calculated as the exon-included band intensity divided by the intensity of included and skipped isoform bands. ΔPSI is calculated as PSIcase − PSIcontrol.
Cell Line Transfections
Cell lines are reverse transfected with siRNAs (Ambion Silencer Select siRNA) (Supplementary Table S3B) or uniformly modified 2ʹ-O-methoxyethyl (2ʹMOE) ASOs with phosphorothioate backbones (IDT) (Supplementary Table S3C) using Lipofectamine RNAiMAX (Invitrogen). siRNAs and ASOs are diluted to final concentration of 10 nM and 50 to 500 nM, respectively, in 100 μL Optimum media (Gibco), supplemented with 1.5 µL of lipofectamine RNAiMAX. Following incubation at room temperature, 2.5 × 105 cells/mL of resuspended cells in 500 µL media are added to the siRNA or ASO mix. About 125 µL or 500 µL of the mix are platted into 96- or 24-well plates for phenotyping or RNA and protein extraction. For everolimus co-treatment, cells were transfected with ASOs as above and plated in everolimus (Thermo) containing media to final concentration of 10 nM.
Western Blot Analysis
Cells are harvested in 2 mM EDTA in PBS and lysed in Laemmli buffer (50 mM Tris-HCl pH 6.2, 5% β-mercaptoethanol, 10% glycerol, 3% SDS). Protein lysates are ran on 8% to 16% gradient gels (Bio-rad), transferred onto nitrocellulose membranes (Millipore) and blocked with 5% milk in Tween-20-TBST (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% Tween-20). Blots are incubated with primary and secondary antibodies listed in Supplementary Table S3D, and imaged with a ChemiDoc MP Imaging System (Bio-rad). Protein expression is quantified using ImageLab 6.0 software (Bio-rad), normalized to loading control and expressed as fold change (FC) to controls.
Phenotypic Assays
ASO or siRNA-treated cells are seeded into 96 well imaging plates (Perkin Elmer) at 2.5 × 104 cells per well. For caspase activation: 48 h after transfection cells are incubated for 1 h with 5 µM Cell Event Caspase-3/7 detection reagent (Invitrogen) and 5 ng/mL Hoechst (Life Technologies). For cell proliferation: 48 h after transfection, cells are labeled with 10 µM EdU for 6 h, fixed in 4% paraformaldehyde, and permeabilized with 0.5% tritonX-100. EdU is detected using the Click-iT cell proliferation kit (Thermo Fisher) with alexa-647 azide, and counterstained with Hoechst (5 ng/mL). For all assays, 9 fields of view per replicate are imaged with a 10× objective on an Opera Phenix high-content imaging system (Perkin Elmer). Caspase+ or EdU+ cells and total Hoechst + nuclei are counted using the Columbus analysis software (Perkin Elmer) and presented as the percentage of Caspase+ or EdU+ cells.
Immunofluorescence
About 48 h following the transfection cells were washed with PBS, fixed with 4% paraformaldehyde, washed with IF buffer (7.6 g/L NaCl, 1.896 g/L Na2HPO4, 0.414 g/L NaH2PO4, 0.5 g/L NaN3, 1 g/L BSA, 0.2% Triton X-100, 0.05% Tween-20, pH 7.4), permeabilized with 0.5% TritonX-100, and blocked with 10% goat serum (Sigma). Primary and secondary antibodies are listed in Supplementary Table S3D. Cells were counterstained with Hoechst and phalloidin-647 (Thermo), and imaged with a 20× objective using an Opera Phenix high-content imaging system (Perkin Elmer).
Human Meningioma Cohorts
RNA-seq data from human meningioma samples was previously published (GSE183653, GSE212666).9,14,17 For each analysis, meningioma samples were split between a discovery (GSE212666 n = 302, 150 bp paired-ended reads) and validation (GSE183653, n = 184, 50 bp single-ended reads) cohort as described.
Differential Splicing And Survival Analysis in Human Meningioma Samples
Differential splicing analysis is carried out using an in-house computational pipeline that incorporates rMATS for event level splicing quantification26 (v2.0 https://github.com/TheJacksonLaboratory/splicing-pipelines-nf). To stratify patients based on splicing, PSI values for individual splicing events were extracted and samples were categorized as “high inclusion” (z-score > 0.5), “low inclusion” (z-score < −0.5), or “other” (−0.5 < z-score < 0.5). Survival analysis was performed using Survival and Survminer R packages.
Differential Gene Expression Analysis in Human Meningioma Samples
Differential gene expression is performed using DESeq227 in R with gene count matrices from STAR mapped fastq files filtered for read counts > 10, by comparing DNA-methylation groups (hypermitotic vs merlin-intact, hypermitotic vs immune-enriched, immune-enriched vs merlin-intact). Significant differential expression is assessed using padjusted-value < 0.05 (Benjamini-Hochberg). To compare splicing-factor expression, normalized gene counts were z-scored for each cohort and plotted with median gene expression compared using a Wilcoxon test.
Visualization of eCLIP and ChIP-Seq Data from ENCODE/ENCORE
ENCODE data for SRSF1 eCLIP-seq from HepG2 (ENCSR989VIY) and K562 (ENCSR432XUP) was visualized in the UCSC genome browser as peak call outputs from ENCODE analysis.28,29
Gene Ontology Analysis Using Enrichr
Gene lists from differential expression analysis were analyzed with Enrichr (https://maayanlab.cloud/Enrichr/). Results for GO Biological Processes 2023 were plotted using R.
Graphs and Figures
Plots were generated in R (v3.6.3) or excel (Microsoft) and then formatted using Illustrator (Adobe). Figures were generated using Illustrator (Adobe) in compliance with the Nature Publishing Group policy concerning image integrity. Figures were supplemented with images from BioRender.
Quantification and Statistical Analysis
Plots include mean ± stdev or median ± interquartile range, as well as individual replicates/samples where applicable. For RT-PCR, western blot, and immunofluorescence data is presented as the mean ± stdev and significant differences to a control are assessed using a 2-tailed unpaired t-test. For plots generated in R, statistics are done using the ggpubr package.
Results
Alternative RNA Splicing Patterns Distinguish Meningioma DNA-Methylation Groups and Predict Patient Outcomes
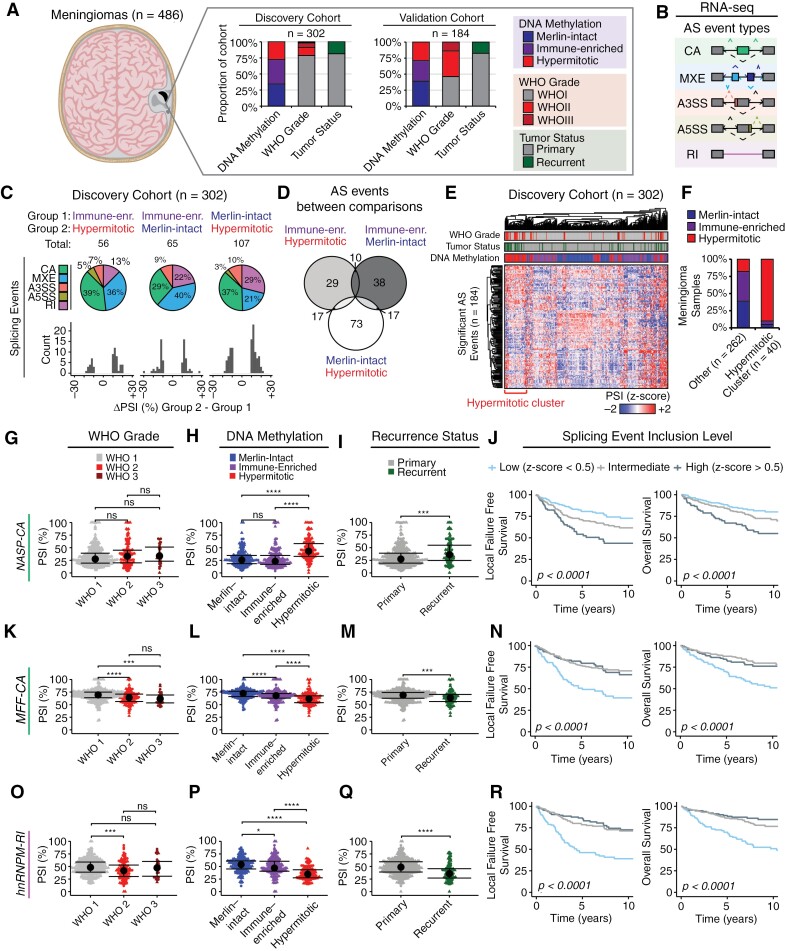
To understand AS differences between meningiomas we analyzed RNA-sequencing data from 486 meningiomas for which there was paired DNA-methylation classification based on the UCSF classifier (merlin-intact n = 176, immune-enriched n = 174, and hypermitotic n = 136),9,14 split into discovery (n = 302) and validation cohorts (n = 184) (Figure 1A). We focused on comparing these groups as several studies have described biological differences between DNA-methylation groups that provide important prognostic value.9,11,14 We utilized a computational pipeline that incorporates rMATS for predicting and quantifying AS events from RNA-seq data.26,30 AS events were classified into 5 types: cassette alternative exons (CA), mutually exclusive exons (MXE), alternative 3ʹ splice sites (A3SS), alternative 5ʹ splice sites (A5SS), and retained introns (RI), and for each AS event a “percent-spliced in” (PSI) score was calculated representing the ratio of isoforms including the event vs others (Figure 1B). Within the discovery cohort we identified 56 significant AS events (ΔPSI > 10%, FDR < 0.05, P < 0.05) between immune-enriched and hypermitotic meningiomas, 65 significant AS events between immune-enriched and merlin-intact meningiomas, and 107 significant AS events between merlin-intact and hypermitotic meningiomas, the majority of which were CA or MXE events across all DNA-methylation groups (Figure 1C, Supplementary Table S1). A total of 228 AS events were detected in these 3 comparisons, some of which were shared across multiple comparisons (Figure 1D). A total of 184 unique AS events were used to cluster all tumors, revealing a cluster enriched for hypermitotic meningiomas (Figure 1E and F). This subset of meningiomas were also enriched for high-risk subtypes from other DNA-methylation classifiers,10 gene expression signatures,15,17 molecularly integrated grades,12,31 and CDKN2A deletions (Supplementary Figure S1A). We also observed a non-significant trend towards higher proportions of meningiomas harboring TERT promoter (TERTp), BAP1, and SMARCB1 mutations (Supplementary Figure S1A). Additionally, we compared recurrent vs primary samples, identifying 13 significant AS events when considering all meningiomas regardless of DNA-methylation group, 506 significant AS events between recurrent vs primary merlin-intact meningiomas, 104 significant AS events between recurrent vs primary immune-enriched meningiomas, and 33 significant AS events between recurrent vs primary hypermitotic meningiomas (Supplementary Figure S1B and C, Supplementary Table S1D to G).
Figure 1.
RNA splicing differences underlie meningioma DNA-methylation groups. (A) Description of human meningioma samples (n = 486), split into discovery (n = 302) and validation (n = 184) cohorts. Meningioma samples were subjected to DNA-methylation profile and group assignment, as described previously, as well as RNA-sequencing. Histograms represent the relative proportion of meningiomas in each cohort by DNA-methylation group, conventional WHO grade, and recurrence status. (B) RNA-sequencing data was subjected to a computational pipeline that predicts 5 types of alternative RNA splicing events (CA: cassette alternative exon; MXE: mutually exclusive exon, A3SS/A5SS: alternative 3ʹ/5ʹ splice site, RI: retained intron) based on exonic, intronic, and junctional read counts. (C) Summary plots displaying significant differential AS events (Δpercent spliced-in (PSI) > 10%, FDR < 0.05, P < 0.05) between DNA-methylation groups in the discovery cohort. Pie charts demonstrate distribution of AS events by event type, histograms depict total count of AS events by ΔPSI. (D) Overlap of significant AS events detected across DNA-methylation group comparisons. (E) Significant AS events detected in the discovery cohort are plotted as a heatmap of PSI z-score across tumor samples. Hierarchical clustering based on Euclidean distance. (F) Proportion of tumors representative of each DNA-methylation group in the hypermitotic splicing signature cluster from (E) vs the remainder of meningioma samples in the discovery cohort. (G to J) NASP-CA inclusion was quantified for both the discovery (circle points) and validation cohorts (triangle points) and displayed as PSI across WHO grades (E), DNA-methylation groups (F), and recurrence status (G) (median ± IQR, Wilcoxon test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns—not significant). Tumors were stratified based on NASP-CA inclusion level as high (z-score > 0.5), low (z-score < −0.5), or other (−0.5 < z-score > 0.5). Patient outcomes were assessed with Kaplan–Meier estimates of local failure free recurrence (LFFR) (left) and overall survival (OS) (right) (H). (K to N) hnRNPM-RI inclusion across cohorts (K to M), and across patient outcomes (N), same as in (G to J). (O to R) MFF-CA inclusion across cohorts (O to Q), and across patient outcomes (R), same as in (G to J).
Among the AS events significantly enriched in hypermitotic meningiomas, we identified inclusion of a CA exon in NASP that generates a long NASP isoform (also known as testicular or tNASP32,33) (Figure 1G and H). NASP is an essential gene in mammals that regulates cell cycle progression through histone maintenance,34 and is dysregulated in multiple human tumor types.35–37 Depletion of tNASP can impair cancer cell proliferation in vitro.38,39 Interestingly, this event was not different across WHO grades but was enriched in recurrent meningioma samples (Figure 1I). Importantly, stratifying meningioma samples based on NASP splicing demonstrated significantly worse outcomes in local freedom from recurrence (LFFR) (5-y LFFR [95% confidence interval] = 54% [44% to 67%] NASP-CA high; 81% [74% to 87%] NASP-CA low; 71% [64% to 79%] NASP-CA intermediate) and overall survival (OS) for patients with high NASP-CA inclusion (5-y OS [95% confidence interval] = 67% [58% to 78%] NASP-CA high; 88% [83% to 94%] NASP-CA low; 84% [78% to 89%] NASP-CA intermediate) (Figure 1J). A second example of AS events distinguishing the 3 meningioma groups was an RI event in HNRNPM that was suppressed in higher WHO grade, the hypermitotic DNA-methylation group, and recurrent meningiomas (Figure 1K to M). hnRNPM is a SF whose expression is frequently dysregulated in human tumors.22 Patients with low HNRNPM-RI inclusion had lower rates of LFFR (5-y LFFR [95% confidence interval] = 81% [74% to 86%] HNRNPM-RI high; 51% [41%–62%] HNRNPM-RI low; 77% [71% to 83%] HNRNPM-RI intermediate) and OS (5-y OS [95% confidence interval] = 83% [76% to 90%] HNRNPM-RI high; 70% [62% to 79%] HNRNPM-RI low; 88% [83% to 92%] HNRNPM-RI intermediate) (Figure 1N). A third example of a gene impacted by AS was MFF which encodes a protein that regulates mitochondrial permeability and cell death pathways, and when inhibited decreases cancer cell viability.40–42 Multiple AS events were seen in MFF, including a CA event with decreased inclusion in WHO2, hypermitotic, and recurrent meningiomas (Figure 1O to Q). Low inclusion of MFF-CA was associated with decreased rates of LFFR and OS (5-y LFFR [95% confidence interval] = 86% [80% to 93%] MFF-CA high; 46% [38% to 57%] MFF-CA low; 79% [72% to 85%] MFF-CA intermediate) and OS (5-y OS [95% confidence interval] = 88% [83% to 94%] MFF-CA high; 70% [62% to 78%] MFF-CA low; 86% [81% to 92%] MFF-CA intermediate) (Figure 1R). We observed additional AS events in other transcripts whose splicing has previously been described to impact cancer phenotypes, including cancer cell metabolism (PFKM),43 cytoskeletal dynamics and angiogenesis (ADD3),44,45 and ribosome biogenesis and cell turnover (EIF4A2)46 (Supplementary Figure S1D to O).
Given the prognostic value of individual AS events on predicting patient survival as described above, we next aimed to classify these tumors based on AS event PSI values de novo (Supplementary Figure S2A). We utilized 1000 AS events with the highest standard deviation across samples in the discovery cohort after filtering for read count (nsupporting-reads > 10) and extremes of event inclusion or skipping (PSICohort-average > 90% or <10%) (Supplementary Figure S2B). Hierarchical clustering using Euclidean distance resulted in 6 clusters with 2 (clusters 4 and 6) containing a disproportionate amount of hypermitotic and immune-enriched meningiomas compared to merlin-intact meningiomas (Supplementary Figure S2C to E). Compared to other groupings based on AS profiles, these groups demonstrated lower rates of LFFR but no significant impact on OS (Supplementary Figure S2F and G). K-means clustering gave similar results with 2 clusters (clusters 2 and 3) showing higher rates of aggressive meningiomas and decreased LFFR without significant impact on OS (Supplementary Figure S2H to L). Taken together, these data suggest that AS can be utilized as a powerful tool for assessing meningioma recurrence and patient prognosis and identifies potential aggressive subgroups with specific AS alterations.
Alternative RNA Splicing Can Reliably Predict Meningioma DNA-Methylation Group
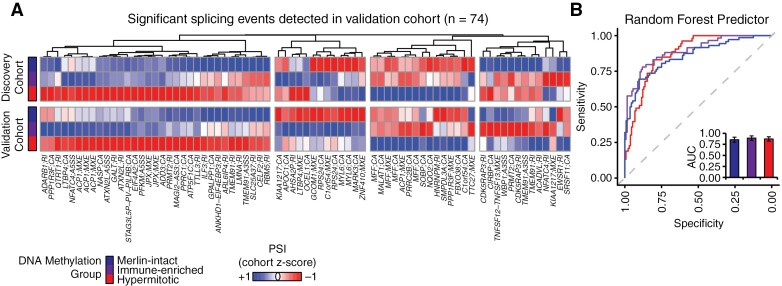
Since AS events differed between methylation groups, we next aimed to identify a set of AS events that could reliably predict DNA-methylation signatures across patient cohorts. We therefore filtered the 184 significant AS events from the discovery cohort (Figure 1E) for those that were also detected in the validation cohort, resulting in 74 AS events with very similiar patterns of PSI difference across meningioma DNA methylation groups in both cohorts (Figure 2A). We further filtered this set of events for those that could be identified across all tumor samples in both cohorts, resulting in 43 events that were utilized to train a random forest classifier with z-scaled PSI values from the discovery cohort, and used this model to predict DNA-methylation groups of the validation cohort (Figure 2B). This model had a high predictive value for each methylation group (AUCmerlin-intact = 0.82 [0.75 to 0.88], AUCimmune-enriched = 0.83 [0.77 to 0.89], AUChypermitotic = 0.86 [0.81 to 0.91]) (Figure 2B and Supplementary Figure S3). Together, suggesting that AS profiles can be used to classify tumor DNA-methylation group.
Figure 2.
Alternative RNA splicing patterns accurately predict meningioma DNA-methylation groups. (A) PSI values from 74 AS events that were significantly different between DNA-methylation groups in the discovery cohort and detected in the validation cohort are plotted as z-score of mean group PSI across both cohorts. Hierarchical clustering by Euclidean distance. (B) z-scaled PSI values (A) were used as input variables for a random forest algorithm trained on the discovery cohort and tested on the validation cohort. Receiver operating curves for this random forest classifier were applied to the validation cohort. Area under the curve (AUC) values are shown in the bar plots (AUC ± 95% CI).
RT-PCR Can Readily Detect Alternative RNA Splicing Changes in Human Meningioma Samples and Cell Lines
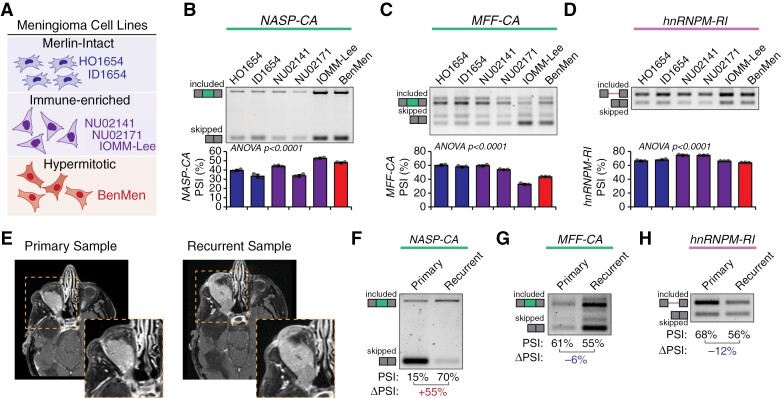
Current methods to capture DNA-methylation status from human tumors require sophisticated sequencing techniques with large upfront costs and time investment, which can restrict their use to large academic centers. Given that AS events can readily predict DNA-methylation groups (Figure 2) we aimed to develop RT-PCR based assays to both validate the RNA-seq data and provide potential testing strategies. We focused on 3 events described above (NASP-CA, MFF-CA, HNRNPM-RI) given their biological significance and ability to stratify patient outcomes (Figure 1). We first validated these AS events in 6 meningioma cell lines with DNA-methylation patterns consistent with either merlin-intact (HO1654, ID1654), immune-enriched (NU02141, NU02171, IOMM-Lee), or hypermitotic (BenMen) meningiomas (Figure 3A) using RT-PCR with primers simultaneously detecting both RNA isoforms. While we observed some inherent variability between cell lines, BenMen cells had higher inclusion of NASP-CA, lower inclusion of MFF-CA and hnRNPM-RI then most other cell lines, consistent with primary hypermitotic tumors (Figure 3B to D). Notably, while classified as an immune-enriched meningioma cell line, IOMM-Lee exhibited a highly proliferative growth rate and splicing patterns more consistent with hypermitotic meningiomas (Figure 3B to D).
Figure 3.
Classification of meningioma cell lines and patient samples using PCR-based detection of alternative RNA splicing events. (A) Cell lines derived from human meningioma tumors previously described to have DNA-methylation profiles consistent with merlin-intact (UCSF-HO, UCSF-ID), immune-enriched (NU02141, NU02171, IOMM-Lee), and hypermitotic meningiomas (BenMen). (B to D) Splicing of NASP-CA (F), MFF-CA (G), and hnRNPM-RI (H) was measured across meningioma cell lines using RT-PCR primers that amplify both included and skipped isoforms. Representative gels along with percent spliced (PSI) quantification from band intensity are shown (n = 3; mean ± SD; ANOVA). (E) Axial post-gadolinium contrast T1-wieghted MRI sequences from a patient with right orbital meningioma demonstrating the original tumor and recurrence. (F to H) Splicing of NASP-CA (F), MFF-CA (G), or hnRNPM-RI (H) was measured in primary and recurrent tumor samples from (E) using RT-PCR primers that amplify both included and skipped isoforms. Representative gels along with percent spliced (PSI) quantification from band intensity are shown (ΔPSI = PSIrecurrent − PSIprimary).
We sought to further validate these AS events in a clinical scenario of tumor recurrence in a patient with a meningioma extending into the right orbit that underwent initial resection followed by recurrence (Figure 3E). We observed in the recurrent vs the primary tumor sample: increased exon inclusion of NASP-CA (ΔPSI = +55%), decreased exon inclusion of MFF-CA (ΔPSI = −6%), and decreased intron retention of HNRNPM-RI (ΔPSI = −12%) (Figure 3F to H). Together, analysis of cell lines and an independent human sample supports the use of RT-PCR assays to detect splicing changes associated with aggressive meningioma behavior.
RNA-binding proteins and SFs are differentially expressed across meningioma groups
Since dysregulation of AS frequently occurs due to changes in RBP and SF expression,21,22 we aimed to assess differences in the splicing machinery across meningioma DNA-methylation groups. We performed differential gene expression analysis on the discovery cohort, which readily clustered samples by DNA-methylation group (Supplementary Figure S4). Overall, we observed 1805 upregulated and 1099 downregulated genes comparing hypermitotic to merlin-intact meningiomas; 1347 upregulated and 1887 downregulated genes comparing hypermitotic to immune-enriched meningiomas; and 2788 upregulated and 1347 downregulated genes comparing immune-enriched to merlin-intact meningiomas (upregulated: Log2FC > 1, padj < 0.05; downregulated: Log2FC < −1, padj < 0.05) (Supplementary Figure S4B to D, Supplementary Table S2). Consistent with previous studies,9 genes upregulated in hypermitotic meningiomas were associated with mitosis, those in immune-enriched meningiomas with immune cell activation and inflammatory responses, and those in merlin-intact meningiomas with cellular differentiation and epidermal development (Supplementary Figure S4E to G).
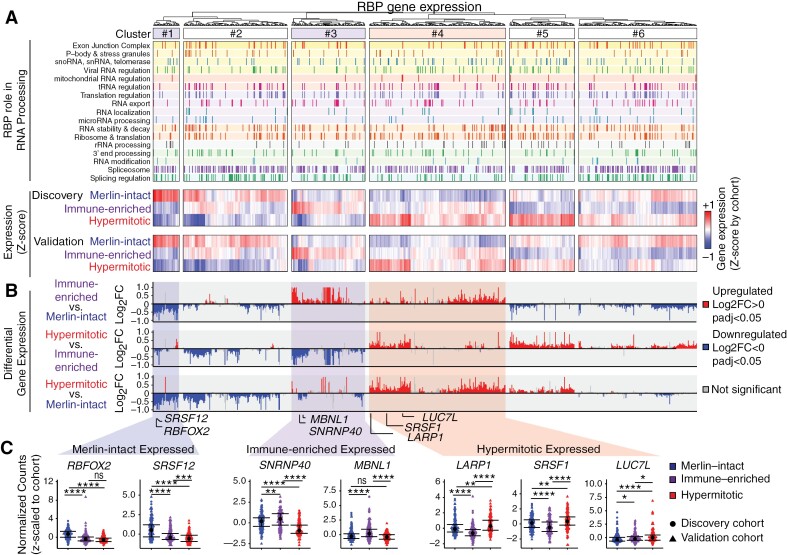
We next examined the expression of 770 annotated RBPs across DNA-methylation groups using normalized count values z-scaled by cohort to control for any absolute differences between the cohorts. Tumors were classified into 6 clusters according to RBP expression: RBPs differentially expressed and preferentially upregulated in merlin-intact (cluster 1: e.g., RBFOX2, SRSF12), immune-enriched (cluster 3: e.g., SNRNP40, MBNL1), and hypermitotic meningiomas (cluster 4: e.g., LARP1, SRSF1, LUC7L) (Figure 4A-C). The remaining clusters had more variable RBP expression. We further performed differential expression analysis of RBP genes annotated as directly associated with the spliceosome or as regulating AS, utilizing a lower FC cutoff (Log2FC>|0|) as even small changes in SF-levels occurring in human tumors can cause profound effects on cellular phenotypes.21,22,47 We identified 76 upregulated (Log2FC > 0, padj < 0.05) and 96 downregulated (Log2FC < 0, padj < 0.05) SFs when comparing immune-enriched vs merlin-intact, 103 upregulated and 69 downregulated SFs in hypermitotic vs immune-enriched, and 83 upregulated and 87 downregulated SFs in hypermitotic vs merlin-intact meningiomas (Supplementary Figure S4H). Together these data demonstrate differential expression of RBPs and SFs across meningioma DNA-methylation groups and suggests the importance of these difference in driving underlying splicing changes.
Figure 4.
Splicing-factor expression is dysregulated across meningioma DNA-methylation groups. (A) Gene expression of 770 RBPs across meningioma DNA-methylation groups using DESeq2. Normalized counts were extracted and z-scaled by cohort (discovery vs validation) and plotted as a heatmap with hierarchical clustering based on Euclidean distance (top). Metadata provided on the left highlights each RBPs role in known RNA processing steps. (B) Gene expression data from both cohorts was then aggregated and differential expression of RBPs was further compared between DNA-methylation groups and represented as Log2FC (bottom, plots scaled to Log2FC of ±1) with downregulated genes in blue (Log2FC < 0, padj < 0.05) and upregulated genes in red (Log2FC > 0, padj < 0.05). (C) Expression of RBPs with enrichment in merlin-intact (RBFOX2, SRSF12), immune-enriched (SNRNP40, MBNL1), and hypermitotic meningiomas (LARP1, SRSF1, LUC7L). Z-scaled normalized counts of each respective RBP across tumor samples is plotted by DNA-methylation group (n = 486; median ± IQR; Wilcoxon test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns—not significant).
DDX39A and SRSF1 Are Upregulated in Hypermitotic Meningiomas and Impact Proliferation
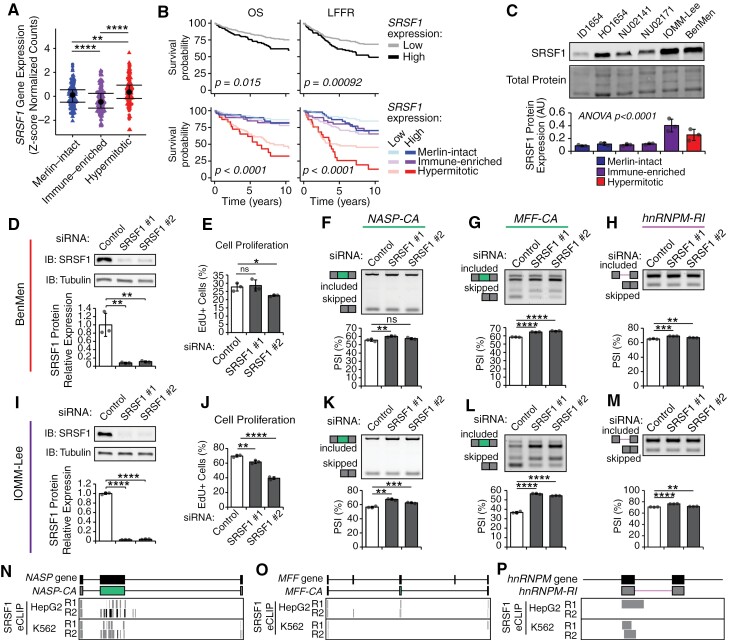
SFs can act as potent oncogenes when overexpressed in human cancers.21,22 We therefore focused on 2 RBPs, DDX39A, and SRSF1, upregulated in hypermitotic meningiomas (Figure 5, Supplementary Figure S5A) and previously implicated in other cancers. DDX39A is an RNA-helicase which functions in mRNA nuclear export48 and telomere maintenance,49 and has been associated with poor prognosis in pediatric neuroblastomas.50 We observed lower rates of OS and LFFR for patient samples with high DDX39A expression, both regardless of their DNA-methylation status and when accounting for their underlying DNA-methylation group (Supplementary Figure S5B). BenMen and IOMM-Lee, 2 highly proliferative meningioma cell lines, exhibit higher levels of DDX39A protein compared to other cell lines (Supplementary Figure S5C). Knockdown (KD) of DDX39A decreased proliferation in both IOMM-Lee and BenMen cells (Supplementary Figure S5D to G) and had a reproducible but mild impact on AS of NASP-CA (ΔPSI = −5%) in BenMen cells (Supplementary Figure S5H). However, DDX39A KD did not impact AS of MFF-CA and had inconsistent effects on HNRNPM-RI splicing across siRNAs (Supplementary Figure S5I and J), suggesting that DDX39A might impact meningioma cell phenotypes through alternative routes aside from its role in splicing. In comparison, SRSF1 is a well validated oncogenic SF in other human cancers where it promotes AS of pro-tumorigenic isoforms.21,22 We observed increased SRSF1 expression in hypermitotic vs merlin-intact and immune-enriched meningiomas (Figure 5A). Meningioma patient samples with high SRSF1 expression, regardless of DNA-methylation group, demonstrated lower OS and LFFR rates. Additionally, when incorporating DNA-methylation information, high SRSF1 expression identified a group of hypermitotic tumors that tended to have lower OS and LFFR (Figure 5B). Finally, we observed higher SRSF1 protein levels in hypermitotic BenMen cells and hyperproliferative immune-enriched IOMM-Lee cells (Figure 5C). Together, these data suggest high SRSF1 levels may contribute to meningioma aggressiveness.
Figure 5.
SRSF1 is upregulated in hypermitotic meningiomas and contributes to splicing changes and meningioma cell proliferation in vitro. (A) SRSF1 gene expression was quantified from RNA-seq across both discovery and validation cohorts. Z-scaled normalized counts across tumor samples are plotted for each DNA-methylation group (n = 486; median ± IQR; Wilcoxon test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns—not significant). (B) Relationship of SRSF1 expression on patient outcomes was assessed by first stratifying patients based on SRSF1 expression (top) or first by meningioma DNA-methylation and then by SRSF1 expression (bottom) as high (z-score > 0.5) or low (z-score < 0.5). Survival outcomes were assessed with Kaplan-Meier estimates of overall survival (OS) (left) and LFFR (right). (C) SRSF1 protein expression was assessed across meningioma cell lines, representative of different DNA-methylation groups, by western blotting with antibodies directed against SRSF1 and normalized to total protein input. Representative gels along with quantification are shown (n = 3, mean ± SD, ANOVA). (D and I) Expression of SRSF1 in BenMen (D) and IOMM-Lee (I) cells transfected with siRNAs targeting SRSF1 or a negative control assessed at 48 h post-transfection using western blotting and normalized to GAPDH as a loading control. Representative gels along with quantification are shown (n = 3; mean ± SD; t-test to control, **P < 0.01, ****P < 0.0001 ns—not significant). (E and J) Cell proliferation in BenMen (E) and IOMM-Lee (J) cells transfected with siRNAs targeting SRSF1 or a negative control assessed using an EdU incorporation assay, counterstained with Hoechst, and plotted as percent EdU+ to total Hoechst+ cells (n = 3; mean ± SD; t-test to control, *P < 0.05, **P < 0.01, ****P < 0.0001 ns—not significant). (F and M) Splicing of NASP-CA (F and K), MFF-CA (G and L), and hnRNPM-RI (H and M) in BenMen (F, G, and H) and IOMM-Lee (K, L, and M) cells transfected with siRNAs targeting SRSF1 or a negative control assessed with RT-PCR primers that amplify both included and skipping bands. Representative gels along with PSI quantification from band intensity are shown (n = 3; mean ± SD; t-test to control, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns—not significant). (N to P) Location of SRSF1 eCLIP peaks from ENCODE HepG2 and K562 cell lines in exonic and intronic regions nearby the NASP-CA (N), MFF-CA (O), and hnRNPM-RI (P) AS events. Individual boxes represent peak calls for each replicate (R1 and R2) for indicated cell lines. Upper diagram shows the genomic region, middle diagram shows the transcript structure of the specific AS event.
To examine how SRSF1 expression influences meningioma cell phenotypes we utilized siRNAs to deplete SRSF1 in BenMen and IOMM-Lee cells. In both cell lines we achieved >90% SRSF1 protein KD and observed decreased cell proliferation and mis-splicing of target transcripts (Figure 5D to M). Specifically, SRSF1 KD increased inclusion of MFF-CA and hnRNPM-RI (Figure 5G,L,H,M), suggesting that baseline SRSF1 expression promotes skipping of these events in hypermitotic meningioma samples. Conversely, SRSF1 KD increased NASP-CA inclusion (Figure 5F and K), suggesting the increased inclusion of NASP-CA observed in hypermitotic meningiomas is driven by other SFs, such as DDX39A, or context-specific splicing in these samples. Changes in splicing of target transcripts following perturbation of SF-levels can be explained by both direct effects of the SF on the target and indirect effects caused by secondary changes in other SFs or binding site occupancy. To address this, we utilized publicly available enhanced cross-linking and immunoprecipitation (eCLIP-seq) ENCODE data from HepG2 hepatocellular carcinoma and K562 leukemia cell lines28 to examine SRSF1 binding around the NASP-CA, MFF-CA, and hnRNPM-RI AS events (Figure 5N to P). SRSF1 eCLIP peaks were found, in at least one cell line, within the alternative exon and surrounding exonic sequences in NASP-CA and MFF-CA events (Figure 5N and O), as well as at the 5ʹ end of the hnRNPM-RI event (Figure 5P). Together, these data suggest that SRSF1 plays a direct role in regulating of these AS events observed in human meningiomas.
Therapeutic Targeting of Alternative Splicing Events in NASP and MFF
RNA splicing has been an attractive target for the design of targeted therapies, ranging from broad spectrum splicing inhibition to highly specific isoform-level targeting. Given the established role of NASP and MFF in tumorigenesis and their prognostic value in predicting meningioma patient outcomes (Figure 1), we aimed to develop splice-switching antisense oligonucleotides (ASOs) to reverse NASP-CA and MFF-CA splicing changes observed in hypermitotic meningiomas.
We designed 3 ASOs targeting an intronic splicing silencer and hnRNPA1 binding site upstream of MFF-CA to promote its inclusion (Supplementary Figure S6A), and reverse the low MFF-CA inclusion observed in hypermitotic meningiomas. All 3 ASOs promoted MFF-CA inclusion, with ASO-MFF#1 and ASO-MFF#2 promoting strong dose-dependent effects (Supplementary Figure S6B and C). While MFF-CA directed ASOs had very modest effects on cell viability in an initial screen in BenMen cells (Supplementary Figure S6G), we aimed to further explore the impact of MFF-CA splicing on meningioma cell phenotypes. Treatment of BenMen cells with ASO-MFF#1 or ASO-MFF#2 increased MFF-CA inclusion as well as relative levels of high-molecular weight (H-MW) to low-molecular weight (L-MW) MFF protein isoforms (Supplementary Figure S7A to C). ASO-MFF treatment in BenMen cells induced variable results on cell proliferation with ASO-MFF#1 increasing and ASO-MFF#2 decreasing proliferation (Supplementary Figure S7D). Additionally, we observed little to no impact of ASO-MFF#1 or ASO-MFF#2 on MFF localization in BenMen cells (Supplementary Figure S7E). We observed similar results in IOMM-Lee cells (Supplementary Figure S7F to H). Given the role of MFF and mTOR signaling in mitochondrial function,51 we co-treated BenMen and IOMM-Lee cells with ASO-MFF#1 or ASO-MFF#2 and the mTOR inhibitor everolimus (Supplementary Figure S7I to L). Everolimus treatment reduced levels of phosphorylated S6 kinase and S6 ribosome and decreased proliferation of ASO-CTL treated BenMen and IOMM-Lee cells (Supplementary Figure S7I to L). However, co-treatment of ASO-MFF and everolimus produced variable results on cell proliferation, with ASO-MFF#2 creating an additive reduction in proliferation of BenMen cells co-treated with everolimus (Supplementary Figure S7J), while both ASO-MFF#1 and ASO-MFF#2 produced mild but significant increases in proliferation of IOMM-Lee cells co-treated with everolimus (Supplementary Figure S7L). Together, these data suggest targeting MFF-CA with ASOs induces changes in MFF-CA splicing and MFF protein isoform expression. However, given its established role in mitochondrial metabolism and turnover,40,52,53 peroxisome biogenesis,54 and autophagy,52,55 the variable effects on cell proliferation may be related to cellular stress and metabolic pathways.
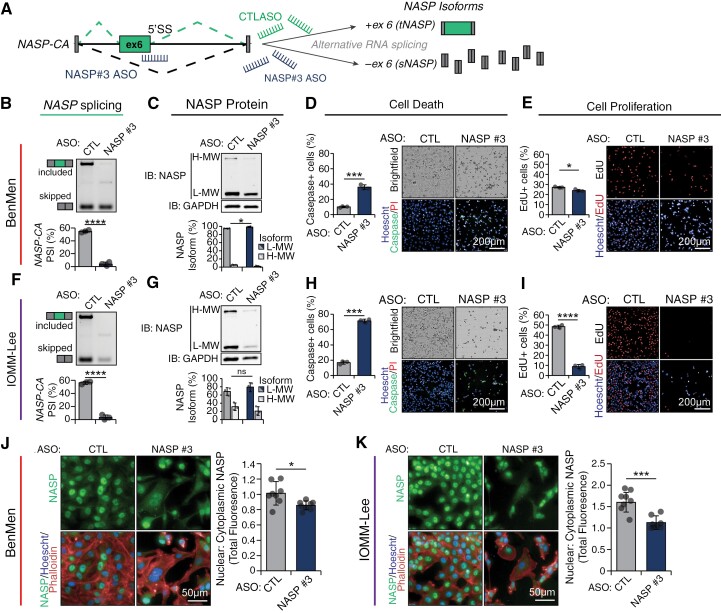
Similarly, we designed 3 splice-switching ASOs targeting the 5ʹ splice site (5ʹSS) of the NASP-CA event to block exon recognition and decrease NASP-CA inclusion, which is detected in hypermitotic meningiomas (Figure 6A, Supplementary Figure S6D). All ASOs significantly decreased NASP-CA inclusion in a dose-dependent manner in IOMM-Lee and BenMen cells, with ASO-NASP#3 promoting almost complete exon skipping (Supplementary Figure S6E and F). To explore the cytotoxic effects of targeting NASP splicing, we treated BenMen and IOMM-Lee cells with control ASO or ASO-NASP#3 which led to a significant reduction in NASP-CA inclusion (Figure 6B and F), and decreased ratio of tNASP (H-MW) to sNASP (L-MW) protein isoforms (Figure 6C and G) in both cell lines. ASO-NASP#3 treatment significantly increased cell death (FCBenMen = 3.5, FCIOMM-Lee = 4.2) (Figure 6D and H), and decreased cell proliferation (FCBenMen = 0.89, FCIOMM-Lee = 0.18) (Figure 6E and I) compared to ASO-CTL, consistent with its impact on overall cell viability (Supplementary Figure S6G). Cells treated with ASO-NASP#3 demonstrated redistribution of NASP protein from the nucleus to the cytoplasm (Figure 6J and K), consistent with previously reported nuclear predominance of tNASP vs sNASP,56 likely do to differential inclusion of a 339 amino acid sequence that regulates its association with histone variants and its nuclear vs cytoplasmic localization.57 Together, these data suggest that NASP-targeting ASOs induces cytotoxic effects through altering protein isoform abundance of tNASP/sNASP and its subcellular localization.
Figure 6.
Splice-switching ASO targeting the hypermitotic meningioma associated NASP-CA splicing event are toxic to meningioma cells in vitro. (A) ASO-NASP#3 targets the 5ʹ splice site (5ʹSS) of NASP-CA resulting in increased exon skipping. Schematic structure of the NASP-CA exon in green, the relative position of ASO-NASP#3, and predicted splicing consequences are shown. (B and F) NASP-CA splicing in BenMen (B) and IOMM-Lee (F) cells transfected with 200 nM of ASO-NASP#3 or ASO-CTL is measured 48 h post-transfection by RT-PCR using primers that amplify the skipped and included isoform. Representative gels along with percent spliced (PSI) quantification from band intensity are shown (n = 3, mean ± SD, t-test; ****P < 0.0001). (C and G) NASP protein isoform expression in BenMen (C) and IOMM-Lee (G) cells transfected with 200 nM of ASO-NASP#3 or ASO-CTL is measured 48 h post-transfection by western blotting using an antibody that recognizes both the high-molecular weight (H-MW; tNASP) and low-molecular weight (L-MW; sNASP) isoforms, or GAPDH for loading control. Protein isoforms are normalized to GAPDH and then represented as a percentage of total NASP protein. Representative gels along with quantification are shown (n = 3, mean ± SD, t-test; *P < 0.05, ns—not significant). (D and H) Cell death in BenMen (D) and IOMM-Lee (H) cells transfected with 200 nM of ASO-NASP#3 or ASO-CTL is measured 48 h post-transfection with a live stain caspase-3/7 activation detection reagent (cell event), counterstained with Hoechst and propidium iodide, and plotted as percent caspase+ to total Hoechst+ cells. Representative images (scale bar: 200 µm) along with quantification are shown (n = 3, mean ± SD, t-test; ***P < 0.001). (E and I) Cell proliferation in BenMen (E) and IOMM-Lee (I) cells transfected with 200 nM of ASO-NASP#3 or ASO-CTL is measured 48 h post-transfection using an EdU incorporation assay, counterstained with Hoechst, and plotted as percent EdU+ to total Hoechst+ cells. Representative images (scale bar: 200 μm) along with quantification are shown (n = 3, mean ± SD, t-test; *P < 0.05, ****P < 0.0001). (J and K) NASP protein localization in BenMen (J) and IOMM-Lee (K) cells transfected with 200 nM of ASO-NASP#3 or ASO-CTL is measured 48 h post-transfection using immunofluorescence and counterstained with Hoechst and phalloidin (F-actin). Relative nuclear to cytoplasmic distribution of NASP is quantified as the total nuclear to total cytoplasmic NASP fluorescence. Representative images (scale bar: 50 µm) along with quantification are shown (n = 8 individual fields; mean ± SD; t-test; ns—not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Discussion
Dysregulation of AS is a critical step in tumorigenesis observed across all solid tumor types. This study is the first to systematically identify differential AS events in meningiomas, particularly comparing DNA-methylation groups with distinct biological behaviors and clinical outcomes. In total, we uncover 184 differential AS events across meningioma DNA-methylation groups and identify several that scale with clinical outcomes. Together, these AS events provide novel opportunities for diagnostics, patient stratification, and targeted therapies.
Clinical genomics have revolutionized our approach to central nervous system tumors and have been readily adapted into clinical care.58–60 With advances in RNA-sequencing, stratification of meningioma patients based on underlying gene expression17 and now differential splicing has provided additional information to predict tumor recurrence and therapeutic response. However, these approaches come with financial burden and require significant time commitments for tissue preparation and downstream data analysis. PCR-based testing is a fast and inexpensive assay routinely employed in medical diagnostics. AS provides a unique opportunity, being internally normalized, to identify tumors with ‘high-risk’ isoform expression (e.g. NASP-CA, MFF-CA, HNRNPM-RI). Importantly, we validate that AS changes are readily detectable by standard PCR methods in patient samples and cell lines, and may aid diagnostics in resource-limited settings or nonacademic centers. Furthermore, analysis of gene expression or AS events in combination with clinical genomics and DNA-methylation profiling allows for enhanced discretion in meningioma patient prognosis.17
RBPs are a large family of proteins with multifunctional roles in splicing, transcription regulation, mRNA localization, mRNA stability and degradation, epitranscriptomics, and translation.61–63 Indeed, some of the most hypermitotic-enriched RBPs uncovered here not only impact RNA splicing but also regulate other aspects of RNA biology such as mRNA stability and degradation, transport, translation (e.g. DDX39A, MEX3A, IGF2BP1). SRSF1 is a well described SF that regulates splicing in many different human tumors, promoting isoforms that induce cellular transformation.21,22 We identified SRSF1 upregulation in hypermitotic meningiomas where high expression correlates with lower rates of OS and LFFR. Further we provide evidence that it directly interacts with and regulates AS events associated with hypermitotic meningiomas. Overall, this implicates SRSF1 as both a prognostic and therapeutic target for aggressive meningiomas, however the overall transcriptome changes observed are likely due to a combination of alterations in multiple RBPs/SFs.
AS is an important contributor to the immunogenicity of human tumors through the generation of cancer-specific epitopes and altered function of immune-related genes,22 and disrupting AS can synergize with immune-checkpoint blockade in preclinical models.64 While we do not identify a specific immune-related signature of AS events in immune-enriched meningiomas, we observe splicing changes in transcripts related to HLA alleles (HLA-DRB5), immune cell signaling (SE31CA65), and antitumor immune cell reactivity (ASAH166,67). Future directions are aimed at further investigating interactions between AS in human meningiomas and the immune system, including leveraging normal meninge tissue to predict AS-derived neoepitopes within human meningiomas.
Dysregulated AS can be exploited as a therapeutic option for human tumors, including aggressive meningiomas for which there is a paucity of reliable therapies. Broad spectrum splicing inhibition, while toxic to tumor cells, has significant systemic toxicities that need to be managed for these approaches to become clinically applicable.68,69 It remains to be tested whether more targeted deliveries using small devices or biopolymers could provide therapeutic efficacy while limiting toxicity. Therefore, novel approaches at targeting AS patterns by inhibiting underlying SFs or modulating AS events have arisen as promising therapies.21,70 In particular, ASOs targeting individual AS events have had tremendous clinical success in treating central nervous system diseases71 with stability and broad bioavailability in cerebrospinal fluid.72 Here, we designed ASOs targeting 2 AS events enriched in hypermitotic meningiomas, NASP-CA and MFF-CA. This approach reverses these AS events towards patterns seen in more benign meningiomas (merlin-intact and immune-enriched) and exhibit significant toxicity to hypermitotic meningioma cell lines in vitro. Further studies are needed to elucidate the exact mechanism of these ASOs, and to test their efficacy against patient-derived and in vivo meningioma models. Furthermore, potential synergism with radiation or other therapies warrants additional investigation. For example, given its importance as a histone shuttler, NASP-targeting ASOs could be combined with histone deactylase inhibitors currently tested in aggressive meningiomas (e.g. AR-42).73–75 Additionally, mTOR inhibitors (e.g. everolimus) are being explored in clinical trials for meningiomas refractory to conventional treatment,76–78 particularly as NF2 mutated tumors upregulate mTOR signaling.79,80 Given the relation between mTOR, mitochondrial dynamics, and autophagy,81 MFF-targeting ASOs may synergize with mTOR inhibitors. Together, this study provides an initial look at transcriptome diversity, and highlights the prognostic and therapeutic value of AS for meningiomas.
Supplementary material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Acknowledgments
We thank members of the Anczukow and Raleigh labs for helpful discussions. We acknowledge assistance from the Microscopy and Information Technology Shared Resources at The Jackson Laboratory (JAX), supported in part by the JAX Cancer Center (NCI P30CA034196), as well as The Northwestern Nervous System Tumor Bank supported by the P50CA221747 SPORE for Translational Approaches to Brain Cancer. The content is solely the responsibility of the authors and does not necessarily represent NIH official views.
Contributor Information
Nathan K Leclair, Graduate Program in Genetics and Development, UConn Health, Farmington, CT, USA; The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA.
Abrar Choudury, Department of Pathology, University of California San Francisco, San Francisco, CA, USA; Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA; Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA.
William C Chen, Department of Pathology, University of California San Francisco, San Francisco, CA, USA; Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA; Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA.
Stephen T Magill, Department of Neurological Surgery, Northwestern University, Chicago, IL, USA.
Kathleen McCortney, Department of Neurological Surgery, Northwestern University, Chicago, IL, USA.
Craig M Horbinski, Department of Pathology, Northwestern University, Chicago, IL, USA; Department of Neurological Surgery, Northwestern University, Chicago, IL, USA.
Zhenhong Chen, Department of Pathology, University of California San Francisco, San Francisco, CA, USA; Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA; Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA.
Ezequiel Goldschmidt, Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA.
Charlotte D Eaton, Department of Pathology, University of California San Francisco, San Francisco, CA, USA; Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA; Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA.
Ketan R Bulsara, Division of Neurosurgery, Department of Surgery, UConn Health, Farmington, CT, USA.
Wenya Linda Bi, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA; Department of Neurosurgery, Brigham and Women’s Hospital, Boston, MA, USA.
Akash J Patel, Department of Otolaryngology-Head and Neck Surgery, Baylor College of Medicine, Houston, TX, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX, USA; Department of Neurosurgery, Baylor College of Medicine, Houston, TX, USA.
Felix Sahm, CCU Neuropathology, German Consortium for Translational Cancer Research, German Cancer Research Center, Heidelberg, Germany; Department of Neuropathology, University Hospital Heidelberg, Heidelberg, Germany.
David Raleigh, Department of Pathology, University of California San Francisco, San Francisco, CA, USA; Department of Neurological Surgery, University of California San Francisco, San Francisco, CA, USA; Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA.
Olga Anczukow, Institute for Systems Genomics, UConn Health, Farmington, CT, USA; Department of Genetics and Genome Sciences, UConn Health, Farmington, CT, USA; The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA.
Funding
Funding for this work was provided by The Jackson Laboratory (OA, NKL), National Institutes of Health (P30CA034196 to OA, R01 CA262311 to DRR, R01NS118039 to CH, R01NS117104 to CH, P50CA221747 to CH), and the Lou and Jean Malnati Brain Tumor Institute (CH).
Conflict of interest statement None declared.
Authorship statement
NKL, DRR, and OA designed the study, analyzed data, and wrote the paper. NKL conducted experiments and performed bioinformatics analyses. DRR and OA supervised the study. AC, STM, and DRR established the dataset. KM, CMH, ZC, EG, and CDE established meningioma cell lines. KRB provided clinical tissue sample. WCC, WLB, AJP, and FS provided additionally metadata for tumor classification. All authors discussed the results and manuscript.
Data Availability
RNA-seq data from human meningioma samples were downloaded as fastq files from the NIH gene expression omnibus (GSE183653, GSE212666)
References
- 1. Ostrom QT, Price M, Neff C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015-2019. Neuro Oncol. 2022;24(Suppl 5):v1–v95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goldbrunner R, Stavrinou P, Jenkinson MD, et al. EANO guideline on the diagnosis and management of meningiomas. Neuro Oncol. 2021;23(11):1821–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen WC, Perlow HK, Choudhury A, et al. Radiotherapy for meningiomas. J Neurooncol. 2022;160(2):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spiegl-Kreinecker S, Lötsch D, Neumayer K, et al. TERT promoter mutations are associated with poor prognosis and cell immortalization in meningioma. Neuro Oncol. 2018;20(12):1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Youngblood MW, Miyagishima DF, Jin L, et al. Associations of meningioma molecular subgroup and tumor recurrence. Neuro Oncol. 2021;23(5):783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prager BC, Vasudevan HN, Dixit D, et al. The meningioma enhancer landscape delineates novel subgroups and drives druggable dependencies. Cancer Discov. 17412020;10(11):1722–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choudhury A, Magill ST, Eaton CD, et al. Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat Genet. 2022;54(5):649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 11. Nassiri F, Liu J, Patil V, et al. A clinically applicable integrative molecular classification of meningiomas. Nature. 2021;597(7874):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Driver J, Hoffman SE, Tavakol S, et al. A molecularly integrated grade for meningioma. Neuro Oncol. 2022;24(5):796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choudhury A, Chen WC, Lucas CG, et al. Hypermitotic meningiomas harbor DNA methylation subgroups with distinct biological and clinical features. Neuro Oncol. 2023;25(3):520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patel AJ, Wan Y-W, Al-Ouran R, et al. Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc Natl Acad Sci U S A. 2019;116(43):21715–21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen WC, Vasudevan HN, Choudhury A, et al. A prognostic gene-expression signature and risk score for meningioma recurrence after resection. Neurosurgery. 2020;88(1):202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen WC, et al. Targeted gene expression profiling predicts meningioma outcomes and radiotherapy responses. Nat Med. 2023;29(Dec):3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brastianos PK, Galanis E, Butowski N, et al. ; International Consortium on Meningiomas. Advances in multidisciplinary therapy for meningiomas. Neuro Oncol. 2019;21(Suppl 1):i18–i31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gehring NH, Roignant JY.. Anything but ordinary – emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet. 2021;37(4):355–372. [DOI] [PubMed] [Google Scholar]
- 20. Wright CJ, Smith CWJ, Jiggins CD.. Alternative splicing as a source of phenotypic diversity. Nat Rev Genet. 2022;23(11):697–710. [DOI] [PubMed] [Google Scholar]
- 21. Urbanski LM, Leclair N, Anczukow O.. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA. 2018;9(4):e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradley RK, Anczukow O.. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer. 2023;23(3):135–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang HW, Kim T-M, Song SS, et al. Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma. Neoplasia. 2012;14(1):20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufmann D, Leistner W, Kruse P, et al. Aberrant splicing in several human tumors in the tumor suppressor genes neurofibromatosis type 1, neurofibromatosis type 2, and tuberous sclerosis 2. Cancer Res. 2002;62(5):1503–1509. [PubMed] [Google Scholar]
- 25. Gauchotte G, Hergalant S, Vigouroux C, et al. Cytoplasmic overexpression of RNA-binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia. J Pathol. 2017;242(4):421–434. [DOI] [PubMed] [Google Scholar]
- 26. Urbanski L, Brugiolo M, Park SH, et al. MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. 2022;41(8):111704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Nostrand EL, Freese P, Pratt GA, et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583(7818):711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Consortium EP, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen S, Park JW, Lu Z-X, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111(51):E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maas SLN, Stichel D, Hielscher T, et al. ; German Consortium on Aggressive Meningiomas (KAM). Integrated molecular-morphologic meningioma classification: a multicenter retrospective analysis, retrospectively and prospectively validated. J Clin Oncol. 2021;39(34):3839–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richardson RT, Batova IN, Widgren EE, et al. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275(39):30378–30386. [DOI] [PubMed] [Google Scholar]
- 33. Liu W, Lu X, Zhao Z-H, et al. SRSF10 is essential for progenitor spermatogonia expansion by regulating alternative splicing. Elife. 2022;11:e78211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nabeel-Shah S, Ashraf K, Pearlman RE, Fillingham J.. Molecular evolution of NASP and conserved histone H3/H4 transport pathway. BMC Evol Biol. 2014;14:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang X, Feng Y, Gan Z, et al. NASP antagonize chromatin accessibility through maintaining histone H3K9me1 in hepatocellular carcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(10):3438–3448. [DOI] [PubMed] [Google Scholar]
- 36. Yu B, Chen X, Li J, et al. microRNA-29c inhibits cell proliferation by targeting NASP in human gastric cancer. BMC Cancer. 2017;17(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kong F, Li L, Wang C, Zhang Q, He S.. MiR-381-3p suppresses biological characteristics of cancer in head-neck squamous cell carcinoma cells by targeting nuclear autoantigenic sperm protein (NASP). Biosci Biotechnol Biochem. 2020;84(4):703–713. [DOI] [PubMed] [Google Scholar]
- 38. Alekseev OM, Richardson RT, Tsuruta JK, O’Rand MG.. Depletion of the histone chaperone tNASP inhibits proliferation and induces apoptosis in prostate cancer PC-3 cells. Reprod Biol Endocrinol. 2011;9:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fang J, Wang H, Xi W, et al. Downregulation of tNASP inhibits proliferation through regulating cell cycle-related proteins and inactive ERK/MAPK signal pathway in renal cell carcinoma cells. Tumour Biol. 2015;36(7):5209–5214. [DOI] [PubMed] [Google Scholar]
- 40. Seo JH, Chae YC, Kossenkov AV, et al. MFF regulation of mitochondrial cell death is a therapeutic target in cancer. Cancer Res. 2019;79(24):6215–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao VA. Targeting mitochondrial fission to trigger cancer cell death. Cancer Res. 2019;79(24):6074–6075. [DOI] [PubMed] [Google Scholar]
- 42. Seo JH, Agarwal E, Chae YC, et al. Mitochondrial fission factor is a novel Myc-dependent regulator of mitochondrial permeability in cancer. EBioMedicine. 2019;48:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jourdain AA, Begg BE, Mick E, et al. Loss of LUC7L2 and U1 snRNP subunits shifts energy metabolism from glycolysis to OXPHOS. Mol Cell. 2021;81(9):1905–1919.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang JZ, Fu X, Fang Z, et al. QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer. J Mol Cell Biol. 2021;13(5):347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiang KM, Zhang P, Li N, et al. Loss of cytoskeleton protein ADD3 promotes tumor growth and angiogenesis in glioblastoma multiforme. Cancer Lett. 2020;474:118–126. [DOI] [PubMed] [Google Scholar]
- 46. Liu L, Vujovic A, Deshpande NP, et al. The splicing factor RBM17 drives leukemic stem cell maintenance by evading nonsense-mediated decay of pro-leukemic factors. Nat Commun. 2022;13(1):3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park S, Brugiolo M, Akerman M, et al. Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Rep. 2019;29(9):2672–2688.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saleem I, Mirza S, Sarkar A, et al. The mammalian ecdysoneless protein interacts with RNA helicase ddx39a to regulate nuclear mRNA export. Mol Cell Biol. 2021;41(7):e0010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoo HH, Chung IK.. Requirement of DDX39 DEAD box RNA helicase for genome integrity and telomere protection. Aging Cell. 2011;10(4):557–571. [DOI] [PubMed] [Google Scholar]
- 50. Otake K, Uchida K, Ide S, et al. Identification of DDX39A as a potential biomarker for unfavorable neuroblastoma using a proteomic approach. Pediatr Blood Cancer. 2016;63(2):221–227. [DOI] [PubMed] [Google Scholar]
- 51. de la Cruz Lopez KG, Toledo Guzman ME, Sanchez EO, Garcia Carranca A.. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front Oncol. 2019;9:1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toyama EQ, Herzig S, Courchet J, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351(6270):275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanchez-Alvarez R, De Francesco EM, Fiorillo M, Sotgia F, Lisanti MP.. Mitochondrial fission factor (MFF) inhibits mitochondrial metabolism and reduces breast cancer stem cell (CSC) activity. Front Oncol. 2020;10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Passmore JB, Carmichael RE, Schrader TA, et al. Mitochondrial fission factor (MFF) is a critical regulator of peroxisome maturation. Biochim Biophys Acta Mol Cell Res. 2020;1867(7):118709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang CS, Lin SC.. AMPK promotes autophagy by facilitating mitochondrial fission. Cell Metab. 2016;23(3):399–401. [DOI] [PubMed] [Google Scholar]
- 56. Alekseev OM, Bencic DC, Richardson RT, Widgren EE, O'Rand MG.. Overexpression of the Linker histone-binding protein tNASP affects progression through the cell cycle. J Biol Chem. 2003;278(10):8846–8852. [DOI] [PubMed] [Google Scholar]
- 57. Kato D, Osakabe A, Tachiwana H, Tanaka H, Kurumizaka H.. Human tNASP promotes in vitro nucleosome assembly with histone H3.3. Biochemistry. 2015;54(5):1171–1179. [DOI] [PubMed] [Google Scholar]
- 58. Ji J, Kaneva K, Hiemenz MC, et al. Clinical utility of comprehensive genomic profiling in central nervous system tumors of children and young adults. Neurooncol. Adv.. 2021;3(1):vdab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pratt D, Sahm F, Aldape K.. DNA methylation profiling as a model for discovery and precision diagnostics in neuro-oncology. Neuro Oncol. 2021;23(23 Suppl 5):S16–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galbraith K, Vasudevaraja V, Serrano J, et al. Clinical utility of whole-genome DNA methylation profiling as a primary molecular diagnostic assay for central nervous system tumors—a prospective study and guidelines for clinical testing. Neurooncol. Adv.. 2023;5(1):vdad076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hentze MW, Castello A, Schwarzl T, Preiss T.. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327–341. [DOI] [PubMed] [Google Scholar]
- 62. Wahl MC, Will CL, Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. [DOI] [PubMed] [Google Scholar]
- 63. Wiener D, Schwartz S.. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22(2):119–131. [DOI] [PubMed] [Google Scholar]
- 64. Lu SX, De Neef E, Thomas JD, et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184(15):4032–4047.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Long L, Wei J, Lim SA, et al. CRISPR screens unveil signal hubs for nutrient licensing of T cell immunity. Nature. 2021;600(7888):308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rother N, Yanginlar C, Prévot G, et al. Acid ceramidase regulates innate immune memory. Cell Rep. 2023;42(12):113458. [DOI] [PubMed] [Google Scholar]
- 67. Vijayan Y, James S, Viswanathan A, et al. Targeting acid ceramidase enhances antitumor immune response in colorectal cancer. J Adv Res. 2023:S2090–1232(23)00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hong DS, Kurzrock R, Naing A, et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32(3):436–444. [DOI] [PubMed] [Google Scholar]
- 69. Schneider-Poetsch T, Chhipi-Shrestha JK, Yoshida M.. Splicing modulators: on the way from nature to clinic. J Antibiot (Tokyo). 2021;74(10):603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bashari A, Siegfried Z, Karni R.. Targeting splicing factors for cancer therapy. RNA. 2023;29(4):506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Finkel RS, Mercuri E, Darras BT, et al. ; ENDEAR Study Group. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377(18):1723–1732. [DOI] [PubMed] [Google Scholar]
- 72. Geary RS, Norris D, Yu R, Bennett CF.. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. [DOI] [PubMed] [Google Scholar]
- 73. Bush ML, Oblinger J, Brendel V, et al. AR42, a novel histone deacetylase inhibitor, as a potential therapy for vestibular schwannomas and meningiomas. Neuro Oncol. 2011;13(9):983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Collier KA, Valencia H, Newton H, et al. A phase 1 trial of the histone deacetylase inhibitor AR-42 in patients with neurofibromatosis type 2-associated tumors and advanced solid malignancies. Cancer Chemother Pharmacol. 2021;87(5):599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Welling DB, Collier KA, Burns SS, et al. Early phase clinical studies of AR-42, a histone deacetylase inhibitor, for neurofibromatosis type 2-associated vestibular schwannomas and meningiomas. Laryngoscope Investig Otolaryngol. 10192021;6(5):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shih KC, Chowdhary S, Rosenblatt P, et al. A phase II trial of bevacizumab and everolimus as treatment for patients with refractory, progressive intracranial meningioma. J Neurooncol. 2016;129(2):281–288. [DOI] [PubMed] [Google Scholar]
- 77. Mair MJ, Berghoff AS, Brastianos PK, Preusser M.. Emerging systemic treatment options in meningioma. J Neurooncol. 2023;161(2):245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pinker B, Barciszewska AM.. mTOR signaling and potential therapeutic targeting in meningioma. Int J Mol Sci. 2022;23(4):1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. James MF, Han S, Polizzano C, et al. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29(15):4250–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG.. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol Cell Biol. 2009;29(15):4235–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim J, Kundu M, Viollet B, Guan KL.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from human meningioma samples were downloaded as fastq files from the NIH gene expression omnibus (GSE183653, GSE212666)