Abstract
Background
Previous studies have indicated that human flora may affect the development of scabies, however, no studies have proven a causal relationship between human flora and scabies, which would be detrimental to future in-depth studies on human flora and scabies.
Methods
Mendelian randomization (MR) was used to analyze the causal effect between human microbiota and scabies, with data on intestinal flora and skin flora from two large published studies and data on scabies from the FinnGen database. Five MR analysis methods were used to increase the reliability of the results, and sensitivity analyses were conducted to increase the robustness of the results.
Results
Our results suggest that 13 intestinal flora as well as 7 skin flora can have a causal effect on scabies.
Conclusion
Overall, our results demonstrate a causal relationship between intestinal and skin flora and scabies and are consistent with previous observational findings. This will contribute to the future development of probiotic agents for the prevention or treatment of scabies.
Keywords: gut microbiota, skin microbiota, scabies, FinnGen, Mendelian randomization
Introduction
Scabies is a globally prevalent skin condition usually thought to be caused by an infection with a mite called Sarcoptes scabiei, with itchy skin (worse at night) being the main clinical manifestation.1 It is now suggested that the scabies mite may affect the skin barrier function by the following mechanism: the mite produces large quantities of saliva when digging holes in the skin, which stimulates the body to develop an immune and inflammatory response, compromising the skin integrity and possibly promoting secondary bacterial infections.2,3 Although scabies can be contracted through contact with contaminated items such as the clothing of a person with scabies, direct skin contact is still the primary route of mite infection.4 It has been reported that patients with poor hygiene, use of immunosuppressive medications (such as steroid hormone use, immunosuppression), chronic diseases (such as hypertension, and diabetes), or AIDS are more likely to suffer from scabies.5 Since physical contact is unavoidable in communal living areas, immune-compromised populations are more susceptible to mite attacks, and how to target susceptible populations for effective protection is a major public health How to protect susceptible populations effectively is a major public health challenge.6
With the in-depth study of human microecology in recent years, people have gradually realized the role played by microorganisms in the human body. Microorganisms are distributed in various parts of the body, such as the skin and intestines, and they interact with each other and play a very important role in maintaining the homeostasis of the internal environment of the human body.7–10 The human flora plays a crucial role in immunity regulation. It has been reported that intestinal flora can enhance the body’s immunity by maintaining the integrity of the intestinal mucosa and signaling pathways of the host, and influence the progression of diseases such as irritable bowel syndrome and inflammatory bowel disease.11–13 In addition, the skin flora also plays an unimportant role in maintaining the skin barrier against external pathogens.14 Several studies have been carried out to demonstrate the importance of the relationship between the human flora and skin diseases (such as psoriasis, and atopic dermatitis).15,16 However, there is a lack of studies on the association between scabies and gut and skin flora, and no study has demonstrated a causal effect between scabies and gut or skin flora.
Mendelian randomization (MR) is a research tool to explain the causal relationship between disease and exposure from a genetic point of view, using genetic information about exposure, such as single nucleotide polymorphisms (SNPs), as instrumental variables (IVs), which are derived from published articles on human genetics research. According to Mendel’s laws of inheritance, parental alleles segregate randomly when gametes are formed and each gamete carries one allele, a process similar to randomized grouping in a randomized controlled trial that ensures that the distribution of genetic variation in a population is random. Because genetic variants are randomly assigned before the onset of disease, confounding factors are avoided, providing strong evidence to explain the causal relationship between exposure and outcome.17 This study aimed to utilize MR to clarify the causal relationship between the skin, intestinal flora, and scabies. It provides a theoretical reference for future studies related to scabies control.
Material and Methods
Data Sources
In this study, all genetic data were obtained from the European population. Scabies data were obtained from the FinnGen database, which combines genetic data with health record data and is designed to study genetic variants associated with the disease.18 The total number of cases in the scabies cohort was 411,729, comprising a total of 1244 cases and 410,485 controls.
Data on intestinal flora were obtained from a study involving 5959 individuals.19 The data included 473 flora-related data. Data for skin flora were obtained from a study involving 597 individuals. Data for this study were obtained from the PopGen database and the Cooperative Health Research in the Region Augsburg (KORA) platform, with the PopGen cohort comprising 324 individuals and the KORA FF4 cohort comprising a total of 273 individuals, involving a total of 150 flora-related data.20
IVs Selection
The main purpose of using IVs in MR analyses is to address endogeneity. That is, when exposure factors are associated with potential confounders, traditional observational research methods are unable to estimate the causal relationship between exposure factors and outcomes accurately. IVs provide a way to assess the causal relationship between exposure factors and outcomes by using genetic variants strongly associated with exposure factors as instrumental variables. In this study, SNPs were used as IVs in MR analyses.MR analyses were based on three main principles: (1) there is a strong correlation between genes and exposures (correlation assumption). (2) SNPs are not related to confounders (independence assumption). (3) SNPs affect outcomes only through exposures (exclusivity assumption).21 To fulfill these three core assumptions, choosing the right IVs is critical. Firstly, to fulfill the correlation assumption, we selected SNPs that were strongly correlated with intestinal and skin flora, respectively, and used these SNPs as IVs. At the beginning, we set the threshold for the P-value to 5 × 10−8, but this would result in too few SNPs to allow further MR analysis. We therefore set the threshold for the P value to 1×10−5.22 In addition, to further ensure that there was a strong correlation between IVs and exposure, we retained only those IVs with an F statistic greater than 10 to prevent MR results from being affected by weak IVs.23
Subsequently, since genetic variants that are located too close to each other in the genome are usually more inclined to be co-inherited to offspring, this may lead to a weakening of the effect of IVs, also known as the phenomenon of Linkage Disequilibrium (LD).24 Therefore, we removed SNPs in the presence of LD (R2 < 0.01 and clumping distance = 10,000 kb).
MR Analysis
This study was statistically analyzed using R software (version 4.2.1), and R packages such as “TwoSampleMR” and “MendelianRandomization” were used. We performed MR analysis using skin and intestinal flora as exposures and scabies as an outcome. To increase the accuracy of MR results, we used five methods to perform MR analysis (including Inverse variance weighted (IVW), MR-Egger, Weighted median, Simple mode, and Weighted mode), of which we mainly used the IVW method because it was considered the most reliable.25
Subsequently, we performed sensitivity analyses to ensure that the exclusivity assumption was valid. First, Cochran’s Q statistic was employed in this study to evaluate the presence of heterogeneity, which was considered to exist if P<0.05. Subsequently, the MR-Egger intercept test, as well as the Mendelian Randomized Polytropic Residuals and Outliers (MR-PRESSO) test, were used to detect polytropy, which was considered to exist if P<0.05. Subsequently, a “leave-one-out” analysis was performed to ensure that there were no SNPs that did not overly influence the overall MR effect.
Reverse MR Analysis
To clarify whether scabies have a causal effect on intestinal and skin flora, we performed reverse MR analysis, designating scabies as the exposure and intestinal and skin flora as the outcome. Due to the limitation of the number of SNPs, we similarly set the threshold of P-value to 1×10−5 when screening for SNPs associated with scabies, and these SNPs were similarly required to have F-statistic values greater than 10, as well as the absence of an LD effect.
Results
The Causal Effect of Intestinal Flora on Scabies
In this study, all analyzed SNPs were strongly correlated with exposure (P < 1×10−5), with F values > 10 and no LD effect.
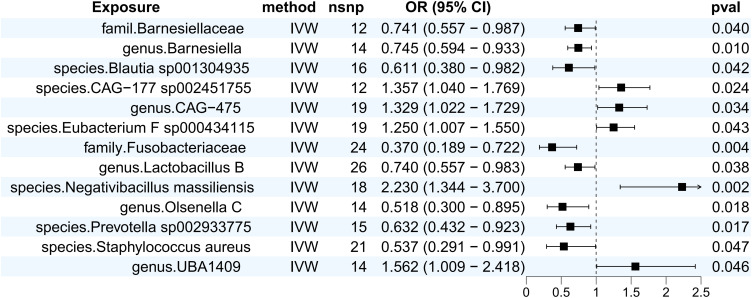
We performed MR analyses of intestinal flora as exposure and scabies as outcome, and identified a total of 13 flora that were causally associated with scabies.IVW results suggested that family.Barnesiellaceae (OR (95% CI) = 0.741 (0.557–0.987), p = 0.040), genus.Barnesiella (OR (95% CI) = 0.745 (0.594–0.933), p = 0.010), species.blautia sp001304935 (OR (95% CI) = 0.611 (0.380–0.982), p = 0.042), family. Fusobacteriaceae (OR (95% CI) = 0.370 (0.189–0.722), p = 0.004), genus.Lactobacillus B (OR (95% CI) = 0.740 (0.557–0.983), p = 0.038), species.Prevotella sp002933775 (OR (95% CI) = 0.632 (0.432–0.923), p = 0.017), genus.Olsenella C (OR (95% CI) = 0.518 (0.300–0.895), p = 0.018), species.Staphylococcus aureus (OR (95% CI) = 0.537 (0.291–0.991), p = 0.047) was able to reduce the risk of scabies, species.CAG-177 sp002451755 (OR (95% CI) = 1.357 (1.040–1.769), p = 0.024), genus.CAG-475 (OR (95% CI) = 1.329 (1.022–1.729), p = 0.034), species.Eubacterium_F sp000434115 (OR (95% CI) = 1.250 (1.007 −1.550), p = 0.043), species.Negativibacillus massiliensis (OR (95% CI) = 2.230 (1.344–3.670), p = 0.002), genus.UBA1409 (OR (95% CI) = 1.562 (1.009–2.418), p = 0.046) increased the risk of scabies. Meanwhile, we plotted the corresponding forest plot (Figure 1) as well as the scatterplot (Figure 2). It is noteworthy that on the scatterplot, all five statistical methods showed a consistent direction, which increased the reliability of the findings.
Figure 1.
Forest plot of the results of MR Analysis of gut microbiota and scabies.
Abbreviations: IVW, inverse variance weighted; nsnp, number of single nucleotide polymorphism; OR, odds ratio; CI: confidence interval.
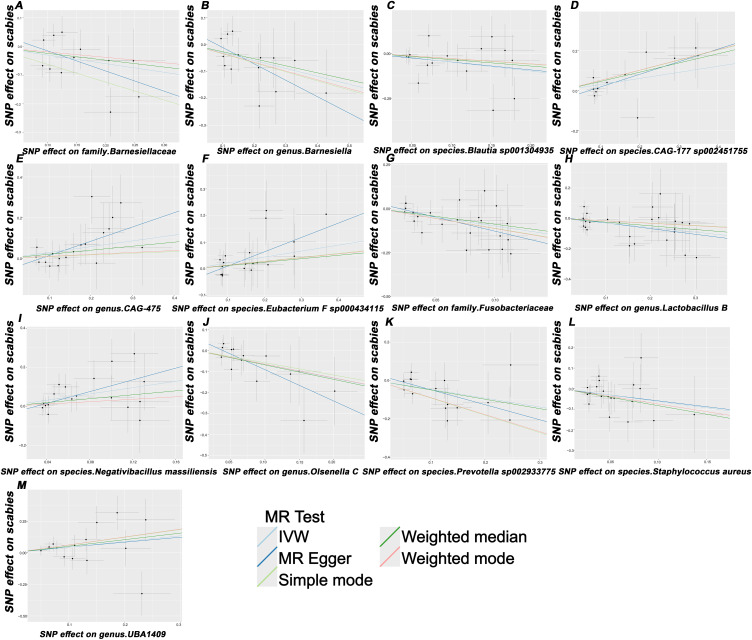
Figure 2.
Scatterplot of the causal effect of (A) family Barnesiellaceae, (B) genus Barnesiella, (C) species Blautia sp001304935, (D) species CAG-177 sp002451755, (E) genus CAG-475, (F) species Eubacterium F sp000434115, (G) family Fusobacteriaceae, (H) genus Lactobacillus B, (I) species Negativibacillus massiliensis, (J) genus.Olsenella C, (K) species.Prevotella sp002933775, (L) species.Staphylococcus aureus, and (M) genus.UBA1409 on scabies risk. Each black dot represents a SNP, with the effect of the SNP on exposure plotted on the x-axis and the effect on outcome plotted on the y-axis. These dots are displayed on a scatterplot, where the slope of each line indicates causality. If the slope is positive, it indicates a positive causal relationship between exposure and outcome.
Abbreviations: IVW, inverse variance weighted; SNP, single nucleotide polymorphism.
The results of the sensitivity analysis indicated no heterogeneity or pleiotropy, and the leave-one-out method analysis suggested that there were no SNPs that had an excessive effect on the total effect (Figure S1). Specific MR analysis results as well as sensitivity results are detailed in Supplementary Table 1.
The Causal Effect of Scabies on Intestinal Flora
Subsequently, we conducted a reverse MR analysis using scabies as exposure and gut flora as outcome, and the results suggested that scabies was not reverse causally associated with the 13 flora mentioned above, and that it had a causal effect on 14 gut flora. The IVW results suggested that scabies had a causal effect on species.Alistipes (OR (95% CI) = 0.956 (0.917–0.997), p = 0.034), species.Blautia A sp900066355 (OR (95% CI) = 0.972 (0.947–0.998), p = 0.034), species.Blautia A sp900066355 (OR (95% CI) = 0.972 (0.947–0.998), p = 0.037), species.Coprobacillus cateniformis (OR (95% CI) = 0.968 (0.938–0.998), p = 0.037), genus.Coprobacillus (OR (95% CI) = 0.963 (0.931–0.996), p = 0.030), species.Eisenbergiella sp900066775 (OR (95% CI) = 0.956 (0.925–0.989), p = 0.008), species.QALR01 sp003150035 (OR (95% CI) = 0.974 (0.949–0.999), p = 0.038), species.RUG420 sp900317985 (OR (95% CI) = 0.984 (0.970–0.999), p = 0.034), family.Thioalkalivibrionaceae (OR (95% CI) = 0.980 (0.967–0.993), p = 0.003) produce an CAG-273 sp003507395 (OR (95% CI) = 1.073 (1.021–0.998), p = 1.129), genus.Enterococcus (OR (95% CI) = 1.019 (1.004–1.035), p = 0.013), family.Fervidobacteriaceae (OR (95% CI) = 1.021 (1.007–1.036), p = 0.004), genus.Gillisia (OR (95% CI) = 1.019 (1.001–1.037), p = 0.041), species.Kandleria vitulina (OR (95% CI) = 1.038 (1.009–1.068), p = 0.009), species.UBA5394 sp002409725 (OR (95% CI) = 1.024 (1.000–1.049), p = 0.049) produced a promoting effect. The sensitivity analysis results suggested there was no heterogeneity or pleiotropy, and the leave-one-out results suggested the absence of SNPs that disproportionately affected the results. Specific MR analysis results as well as sensitivity results are detailed in Supplementary Table 2.
The Causal Effect of Skin Flora on Scabies
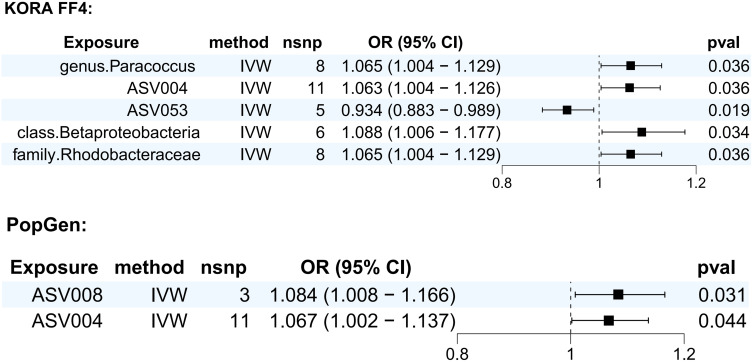
We performed MR analysis for the KORA FF4 cohort as well as the PopGen cohort, respectively. In the KORA FF4 cohort, five skin flora had a causal effect on scabies. IVW results showed that genus. Paracoccus (OR (95% CI) = 1.065 (1.004–1.129), p = 0.036), ASV004 (OR (95% CI) = 1.063 (1.004–1.126), p = 0.036), class. Betaproteobacteria (OR (95% CI) = 1.088 (1.006–1.177), p = 0.034), family. Rhodobacteraceae (OR (95% CI) = 1.065 (1.004–1.129), p = 0.036) were positively associated with the risk of scabies, and ASV053 (OR (95% CI) = 0.934 (0.883–0.989), p = 0.019) was negatively associated with the risk of scabies.
In the PopGen cohort, 2 skin flora had a causal effect on scabies, with IVW results suggesting that ASV008 (OR (95% CI) = 1.084 (1.008–1.166), p = 0.031), ASV005 (OR (95% CI) = 1.067 (1.002–1.137), p = 0.044) was positively associated with the risk of developing scabies.
We then plotted a forest plot (Figure 3) and a scatterplot (Figure 4) of the MR results, with all five statistics having the same orientation in the scatterplot. The sensitivity analysis results suggested there was no heterogeneity or pleiotropy. Specific results of the MR analyses and sensitivity analyses are shown in Supplementary Table 3. The leave-one-out results did not show any SNPs that had an excessive effect on the total (Figure S2).
Figure 3.
Forest plot of the results of MR Analysis of skin microbiota and scabies.
Abbreviations: IVW, inverse variance weighted; OR, odds ratio; CI, confidence interval; nsnp, number of single nucleotide polymorphism.
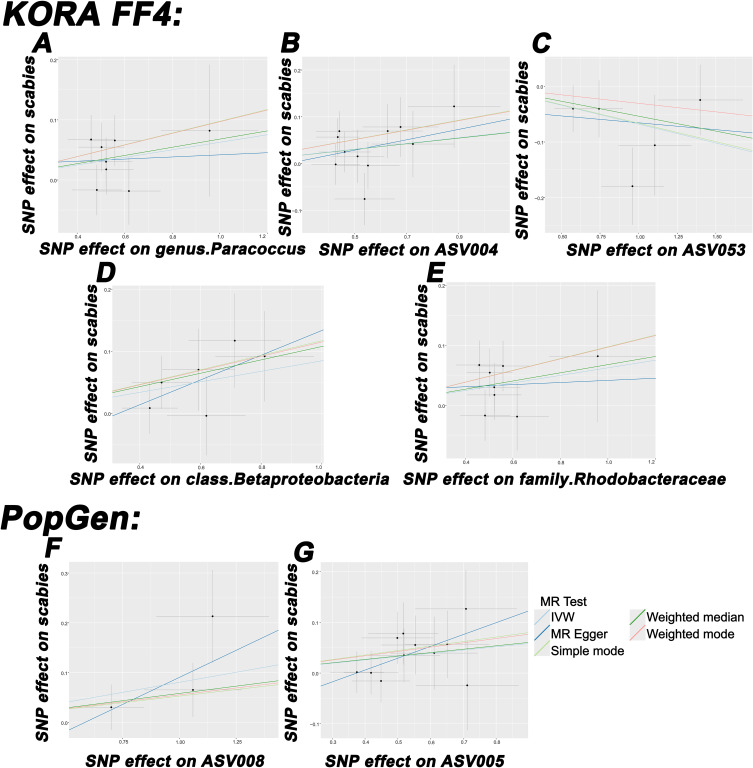
Figure 4.
Scatterplot of the causal effect of (A) genus Paracoccus, (B) ASV004, (C) ASV053, (D) class Betaproteobacteria, (E) family Rhodobacteraceae, (F) ASV008, and (G) ASV005 on scabies risk. Each black dot represents a SNP, with the effect of the SNP on exposure plotted on the x-axis and the effect on outcome plotted on the y-axis. These dots are displayed on a scatterplot, where the slope of each line indicates causality. If the slope is positive, it indicates a positive causal relationship between exposure and outcome.
Abbreviations: IVW, inverse variance weighted; SNP, single nucleotide polymorphism; IVW, inverse variance weighted; SNP, single nucleotide polymorphism.
The Causal Effect of Scabies on Skin Flora
Reverse MR results suggested that scabies was not reverse causally associated with the seven skin flora of the appeal. In the KORA FF4 cohort, scabies inhibited ASV026 (OR (95% CI) = 0.602 (0.408–0.889), p = 0.011).
In the PopGen cohort, scabies had an inhibitory effect on family.Clostridiales (OR (95% CI) = 0.663 (0.452–0.974), p = 0.036), genus.Streptococcus (OR (95% CI) = 0.647 (0.442–0.948), p = 0.026), ASV007 (OR (95% CI) = 0.550 (0.315–0.959), p = 0.035), ASV035 (OR (95% CI) = 0.619 (0.402–0.954), p = 0.030), and ASV045 (OR (95% CI) = 1.712 (1.077–2.719), p = 0.023), ASV057 (OR (95% CI) = 1.906 (1.159–3.135), p = 0.011) and growth-promoting effect.
Sensitivity analysis results suggested no heterogeneity or pleiotropy. Besides, the leave-one-out results did not show SNPs that had an excessive effect on the total effect. The specific results of the MR analysis as well as the sensitivity analysis are shown in Supplementary Table 4.
Discussion
In this study, we identified a total of 13 intestinal flora and 7 skin flora that exert a causal effect on scabies. Seven of the gut flora and one of the skin flora had a protective effect, and six of the gut flora and six of the skin flora increased the risk of developing scabies.
The gut microbiome refers to the microbial communities present in the human intestine, including bacteria, fungi, viruses, and archaea. Its composition is primarily influenced by environmental factors, particularly diet, while genetic factors play a relatively minor role.26 Gut microbes are responsible for processing and digesting nutrients and significantly impact overall health by regulating the immune system.27,28 Recently, the “gut-skin axis” theory has gained attention, revealing a close relationship between gut health and skin health. Gut microbiota and their metabolites can affect skin conditions through various pathways.29
The skin, the largest organ of the human body, constantly interacts with the external environment and employs various defense mechanisms to protect the host from infections.30 The skin microbiome, which includes bacteria, fungi, and viruses, plays an essential role in immune regulation, inflammatory responses, protective functions, and nutrient metabolism. Under normal conditions, the skin microbiome establishes a symbiotic relationship with host tissues through both innate and adaptive immune systems. However, an imbalance in the skin microbiome can compromise the skin barrier, increasing susceptibility to external pathogens such as scabies mites.31
It is noteworthy that, although previous studies have established a correlation between gut and skin microbiota and conditions such as psoriasis and atopic dermatitis, this research is the initial one to report a causal connection between gut and skin microbiota and scabies. Most prior studies have primarily focused on the diagnosis and pharmacological treatment of scabies. This study offers valuable insights into the potential role of probiotics in adjunctive therapy.
In the gut microbiome, the abundance of the Barnesiellaceae family has been found to be reduced in patients with Behçet’s disease. One possible reason for this is that Barnesiellaceae may exert anti-inflammatory effects by lowering TNF-α levels.32 And individuals with scabies often exhibit elevated TNF-α levels,33 which may explain why our results indicate a negative correlation between Barnesiellaceae and the risk of scabies.
The Lactobacillus genus is a common probiotic in the digestive tract, playing an indispensable role in regulating immune responses and suppressing inflammation. Additionally, Lactobacillus directly combats skin pathogens by producing antimicrobial metabolites that influence the metabolism of these pathogens. It has also been reported that Lactobacillus can be formulated into oral probiotics, demonstrating effectiveness in the prevention and treatment of skin conditions such as atopic dermatitis.34–36
Our results align with previous studies. The genus Eubacterium has been extensively researched and is believed to be associated with the development of skin diseases such as psoriasis and atopic dermatitis.37 In an animal study, researchers found that the levels of Fusobacteriaceae were lower than normal in dogs with dust mite-induced atopic dermatitis.38 Similarly, a reduced abundance of Fusobacteriaceae has been observed in patients with immune disorders like Crohn’s disease.39 Although the specific mechanisms remain unclear, there may be a connection to the body’s immune response, warranting further investigation to clarify these mechanisms.
Our results suggest that the genus Paracoccus may promote the occurrence of scabies within the skin microbiome. Previous studies have established a strong association between Paracoccus and skin diseases. Research has consistently shown a high abundance of Paracoccus near skin pustules, in patients with Darier disease (a genetic skin disorder), and individuals with atopic dermatitis.40–42 Overall, Paracoccus may increase host susceptibility to pathogens and negatively impact the maintenance of the skin mucosal barrier.
However, most current studies are observational and do not establish a causal relationship between gut and skin microbiota and skin diseases such as scabies. This research offers a significant advantage by analyzing genetic factors, thereby eliminating confounding influences and demonstrating the causal effect of microbiota on scabies. Additionally, this study conducts a multi-site microbiota analysis rather than focusing solely on gut or skin microbiota, which will enhance our understanding of the relationship between the overall human microbiota and disease.
This study has some limitations. First, due to insufficient genetic data, we only analyzed the microbiomes of the skin and gut. It is important to acknowledge that microbiomes in different parts of the body interact and influence one another, highlighting the need for future analyses of additional body sites. Second, our sample population consisted solely of individuals of European descent, which may limit the applicability of our findings to other ethnic groups. Third, the number of SNPs analyzed was limited, leading us to set the P-value threshold for selected SNPs at 1 × 10−5, which is higher than the conventional threshold of 5×10−8. This may affect the strength of the association between the selected SNPs and exposure.
Conclusion
In summary, this study identifies the gut and skin microbiota that are causally linked to scabies, which will aid in the future development of probiotic formulations to support scabies treatment. Additionally, it paves the way for further research into the interactions between human microbiota and scabies, clarifying how human microbiota influences the onset and progression of the condition.
Acknowledgments
Thanks to the participants and investigators of the FinnGen study.
Funding Statement
This study was supported by the Fundamental Research Funds for the Central Universities (2022ZFJH003), Shandong Provincial Laboratory Project (SYS202202) and the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022001A).
Data Sharing Statement
Data on scabies are available in the Finnish database (https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_AB1_SCABIES.gz), and data on intestinal flora and skin flora are available in the GWAS catalog (https://www.ebi.ac.uk/gwas/).
Ethics Statements
According to Article 32 of the Ethical Review Measures for Life Science and Medical Research Involving Human Beings of the People’s Republic of China, the data used in this study will not cause any form of harm to human beings, nor will it touch sensitive personal privacy or trade secrets, so the ethical review can be exempted. In addition, the database used in this study was publicly available and legally available.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Walton SF, Currie BJ. Problems in diagnosing scabies, a global disease in human and animal populations. Clin Microbiol Rev. 2007;20(2):268–279. doi: 10.1128/cmr.00042-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan MS, Arlian LG, Markey MP. Sarcoptes scabiei mites modulate gene expression in human skin equivalents. PLoS One. 2013;8(8):e71143. doi: 10.1371/journal.pone.0071143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer K, Holt D, Currie B, Kemp D. Scabies: important clinical consequences explained by new molecular studies. Adv Parasitol. 2012;79:339–373. doi: 10.1016/b978-0-12-398457-9.00005-6 [DOI] [PubMed] [Google Scholar]
- 4.Thomas C, Coates SJ, Engelman D, Chosidow O, Chang AY. Ectoparasites: scabies. J Am Acad Dermatol. 2020;82(3):533–548. doi: 10.1016/j.jaad.2019.05.109 [DOI] [PubMed] [Google Scholar]
- 5.Bergamin G, Hudson J, Currie BJ, Mounsey KE. A systematic review of immunosuppressive risk factors and comorbidities associated with the development of crusted scabies. Int J Infect Dis. 2024;143:107036. doi: 10.1016/j.ijid.2024.107036 [DOI] [PubMed] [Google Scholar]
- 6.Sunderkötter C, Wohlrab J, Hamm H. Scabies: epidemiology, Diagnosis, and Treatment. Dtsch Arztebl Int. 2021;118(41):695–704. doi: 10.3238/arztebl.m2021.0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Shen X, Johnson JS, et al. Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease. Cell Host Microbe. 2024;32(4):506–526.e9. doi: 10.1016/j.chom.2024.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bian F, Yan D, Wu X, Yang C. A Biological Perspective of TLR8 Signaling in Host Defense and Inflammation. Infect Microb Dis. 2023;5(2):44–55. doi: 10.1097/im9.0000000000000119 [DOI] [Google Scholar]
- 9.Zheng Q, Li Y, Ni J, et al. Causality Between Gut Microbiota and Inflammatory Bowel Disease: a Bidirectional Mendelian Randomization Study. Infect Microb Dis. 2024;6(2):93–99. DOI: 10.1097/im9.0000000000000147 [DOI] [Google Scholar]
- 10.Pan J, Zhang X, Shi D, et al. Correlation of Oral Microbiota With Different Immune Responses to Antiretroviral Therapy in People Living With HIV. Infect Microb Dis. 2024;6(2):85–92. DOI: 10.1097/im9.0000000000000148 [DOI] [Google Scholar]
- 11.Chen Y, Feng S, Li Y, Zhang C, Chao G, Zhang S. Gut microbiota and intestinal immunity-A crosstalk in irritable bowel syndrome. Immunology. 2024;172(1):1–20. doi: 10.1111/imm.13749 [DOI] [PubMed] [Google Scholar]
- 12.Xie Y, Liu F. The role of the gut microbiota in tumor, immunity, and immunotherapy. Front Immunol. 2024;15:1410928. doi: 10.3389/fimmu.2024.1410928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito Y, Amagai M. Dissecting skin microbiota and microenvironment for the development of therapeutic strategies. Curr Opin Microbiol. 2023;74:102311. doi: 10.1016/j.mib.2023.102311 [DOI] [PubMed] [Google Scholar]
- 15.Wrześniewska M, Wołoszczak J, Świrkosz G, Szyller H, Gomułka K. The Role of the Microbiota in the Pathogenesis and Treatment of Atopic Dermatitis-A Literature Review. Int J Mol Sci. 2024;25(12):6539. doi: 10.3390/ijms25126539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Qu L, Mijakovic I, Wei Y. Advances in the human skin microbiota and its roles in cutaneous diseases. Microb Cell Fact. 2022;21(1):176. doi: 10.1186/s12934-022-01901-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson SC, Butterworth AS, Burgess S. Mendelian randomization for cardiovascular diseases: principles and applications. Eur Heart J. 2023;44(47):4913–4924. doi: 10.1093/eurheartj/ehad736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518. doi: 10.1038/s41586-022-05473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y, Havulinna AS, Liu Y, et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat Genet. 2022;54(2):134–142. doi: 10.1038/s41588-021-00991-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moitinho-Silva L, Degenhardt F, Rodriguez E, et al. Host genetic factors related to innate immunity, environmental sensing and cellular functions are associated with human skin microbiota. Nat Commun. 2022;13(1):6204. doi: 10.1038/s41467-022-33906-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Q, Liu JH, Ma WY, Cheng ZL, Hao PS, Luo NN. Genomics-Microbiome Based Assessment of Bidirectional Causality Between Gut Microbiota and Psoriasis. Clin Cosmet Invest Dermatol. 2024;17:435–445. doi: 10.2147/ccid.S450227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang K, Wang P, Xu Z, et al. Causal Effects of Gut Microbiome on Systemic Lupus Erythematosus: a Two-Sample Mendelian Randomization Study. Front Immunol. 2021;12:667097. doi: 10.3389/fimmu.2021.667097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan T, Hu J, Liu W, et al. Causal Association Between Anemia and Cardiovascular Disease: a 2-Sample Bidirectional Mendelian Randomization Study. J Am Heart Assoc. 2023;12(12):e029689. doi: 10.1161/jaha.123.029689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess S, Davey Smith G, Davies NM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2019;4:186. doi: 10.12688/wellcomeopenres.15555.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji X, Wu S, Zhao D, et al. Revealing the Impact of Gut Microbiota on Acne Through Mendelian Randomization Analysis. Clin Cosmet Invest Dermatol. 2024;17:383–393. doi: 10.2147/ccid.S451104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 27.Abdul Rahim MBH, Chilloux J, Martinez-Gili L, et al. Diet-induced metabolic changes of the human gut microbiome: importance of short-chain fatty acids, methylamines and indoles. Acta Diabetol. 2019;56(5):493–500. doi: 10.1007/s00592-019-01312-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 29.Mahmud MR, Akter S, Tamanna SK, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. 2022;14(1):2096995. doi: 10.1080/19490976.2022.2096995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen AV, Soulika AM. The Dynamics of the Skin’s Immune System. Int J Mol Sci. 2019;20(8):1811. doi: 10.3390/ijms20081811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 32.van der Houwen TB, van Laar JAM, Kappen JH, et al. Behçet’s Disease Under Microbiotic Surveillance? A Combined Analysis of Two Cohorts of Behçet’s Disease Patients. Front Immunol. 2020;11:1192. doi: 10.3389/fimmu.2020.01192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Aal AA A, Hassan MA, Gawdat HI, Ali MA, Barakat M. Immunomodulatory impression of anti and pro-inflammatory cytokines in relation to humoral immunity in human scabies. Int J Immunopathol Pharmacol. 2016;29(2):188–194. doi: 10.1177/0394632015627464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie A, Chen A, Chen Y, et al. Lactobacillus for the treatment and prevention of atopic dermatitis: clinical and experimental evidence. Front Cell Infect Microbiol. 2023;13:1137275. doi: 10.3389/fcimb.2023.1137275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delanghe L, Spacova I, Van Malderen J, Oerlemans E, Claes I, Lebeer S. The role of lactobacilli in inhibiting skin pathogens. Biochem Soc Trans. 2021;49(2):617–627. doi: 10.1042/bst20200329 [DOI] [PubMed] [Google Scholar]
- 36.Kanda N, Hoashi T, Saeki H. Nutrition and Atopic Dermatitis. J Nippon Med Sch. 2021;88(3):171–177. doi: 10.1272/jnms.JNMS.2021_88-317 [DOI] [PubMed] [Google Scholar]
- 37.Mao R, Yu Q, Li J. The causal relationship between gut microbiota and inflammatory dermatoses: a Mendelian randomization study. Front Immunol. 2023;14:1231848. doi: 10.3389/fimmu.2023.1231848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierezan F, Olivry T, Paps JS, et al. The skin microbiome in allergen-induced canine atopic dermatitis. Vet Dermatol. 2016;27(5):332–e82. doi: 10.1111/vde.12366 [DOI] [PubMed] [Google Scholar]
- 39.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rensburg JJ, Lin H, Gao X, et al. The Human Skin Microbiome Associates with the Outcome of and Is Influenced by Bacterial Infection. mBio. 2015;6(5):e01315. doi: 10.1128/mBio.01315-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reiter O, Leshem A, Alexander-Shani R, et al. Bacterial Skin Dysbiosis in Darier Disease. Dermatology. 2024;240(3):443–452. doi: 10.1159/000537714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XP, Liu YY, Zhang CY, et al. An Observational Study: association Between Atopic Dermatitis and Bacterial Colony of the Skin Based on 16S rRNA Gene Sequencing. Clin Cosmet Invest Dermatol. 2024;17:1649–1659. doi: 10.2147/ccid.S464431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data on scabies are available in the Finnish database (https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_AB1_SCABIES.gz), and data on intestinal flora and skin flora are available in the GWAS catalog (https://www.ebi.ac.uk/gwas/).