Abstract
Adeno-associated virus (AAV) is a safe and efficient gene delivery vehicle for gene therapies. However, its relatively small packaging capacity limits its use as a gene transfer vector. Here, we describe a strategy to deliver large genes that exceed the AAV’s packaging capacity using up to four AAV vectors and the CRE-lox DNA recombination system. We devised novel lox sites by combining non-compatible and reaction equilibrium-modifying lox site variants. These lox sites facilitate sequence-specific and near-unidirectional recombination of AAV vector genomes, enabling efficient reconstitution of up to 16 kb of therapeutic genes in a pre-determined configuration. Using this strategy, we have developed AAV gene therapy vectors to deliver IFT140, PCDH15, CEP290, and CDH23 and demonstrate efficient production of full-length proteins in cultured mammalian cells and mouse retinas. Notably, AAV-IFT140 gene therapy vectors ameliorated retinal degeneration and preserved visual functions in an IFT140-associated retinitis pigmentosa mouse model. The CRE-lox approach described here provides a simple, flexible, and effective platform for generating AAV gene therapy vectors beyond AAV’s packaging capacity.
Keywords: AAV, CRE, gene therapy, reconstitution, large gene delivery
Graphical Abstract
Graphical Abstract.
Delivery of large genes using multiple AAV vectors and the CRE-lox DNA recombination system.
Introduction
Adeno-associated virus (AAV) is one of the most broadly used viral vectors for gene therapy. With diverse natural and engineered serotypes, AAVs can transduce a wide range of cell types, while eliciting minimal immune responses [1–4]. Furthermore, AAV-mediated gene delivery appears to result in long-term transgene expression, which is an important qualification for a cure for a genetic disease. The safety, broad tropism, and long-term transgene expression have made AAV the preferred vector for gene therapy in numerous clinical trials.
However, AAV has a major disadvantage as a gene transfer vector. AAVs can accommodate up to 5 kb of viral genomes [5], and this packaging capacity is insufficient to carry full-length coding sequences of many disease-causing genes. For instance, the full-length coding sequence of ABCA4, which is the most frequently mutated gene among all Mendelian retinal degeneration genes [6], spans 6822 bp. A ciliopathy gene CEP290 is associated with multiple genetic diseases ranging from isolated early-onset retinal degeneration to oculorenal dysplasia and lethal Meckel-Gruber syndrome [7–11]. Mutations in CEP290 are one of the most frequent causes of Leber congenital amaurosis (LCA), severe vision loss at birth or in infancy, and its coding sequence is 7440 bp-long [6, 8, 12]. The coding sequence of CDH23, mutations of which cause Usher syndrome affecting both vision and hearing, spans 10 065 bp [13, 14]. Since viral packaging signals (i.e. inverted terminal repeats (ITRs)) and other essential regulatory elements (e.g. a promoter and a transcription termination signal) must be included in viral vectors, the practical limit of a therapeutic gene that may be delivered via AAVs is ~4 kb. This limited packaging capacity of AAV precludes its use as a gene transfer vector when large gene delivery is needed.
Various strategies have been developed to deliver genes larger than the packaging capacity of AAV vectors [15, 16]. In these strategies, cargo genes are split and packaged in two (or three) AAV vectors for delivery to target cells. The split genes can be reconstituted at the DNA, mRNA, or protein level. The DNA-level reconstitution approach relies on occasional DNA recombination events that occur between AAV genomes after internalization. This strategy includes dual (or triple) AAV trans-splicing and hybrid methods [17–20]. A new method to combine two AAV vector genomes using the CRE-lox DNA recombination system has been recently reported [21]. In these approaches, the reconstituted therapeutic cassette produces mRNAs encoding full-length proteins. Additionally, a novel strategy, REVeRT (reconstitution via mRNA trans-splicing), has been reported to facilitate the reconstitution of large genes at the mRNA level [22]. The protein-level reconstitution approach employs split inteins and protein trans-splicing [23–25]. Protein splicing is an auto-catalytic process where an intervening protein segment (intein) excises itself from a precursor protein and attaches its two flanking regions (exteins) through a peptide bond. Some inteins are naturally split, originating from two separate genes [24, 25], and the protein trans-splicing approach utilizes these naturally split inteins. In this approach, the N- and C-terminal halves of a protein are separately delivered and produced from corresponding AAV vectors, and reconstitution is achieved via protein trans-splicing mediated by split inteins.
Although these approaches have produced encouraging outcomes in animal models [18, 20, 22, 26–30], the need for more efficient and versatile delivery methods remains. For instance, AAV trans-splicing methods require high-dose AAV injections to compensate for their low efficiency and the random configuration of recombination events. This problem becomes more pronounced when more than two AAV vectors are needed. Similarly, the reconstitution efficiency of REVeRT appears to decrease significantly when the cargo gene is split into three AAV vectors. The split intein-based reconstitution approach entails the production of truncated proteins, which could potentially have dominant negative effects. Furthermore, the efficiency of protein splicing is heavily affected by the amino acids immediately adjacent to split inteins [25, 31, 32], and the impact of truncation on the folding and stability of both truncated protein products and reconstituted proteins is hard to predict. Due to these constraints, determining optimal splitting positions requires a screening process, and suitable positions may not always be available.
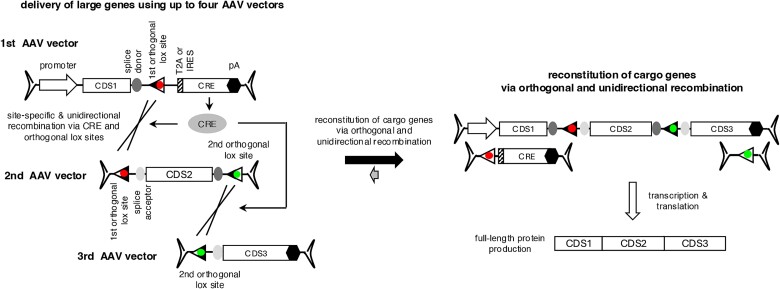
In the present work, we describe a novel strategy to deliver large genes using up to four AAV vectors. Cargo genes are split into 2-4 AAV vectors and reconstituted by using the CRE-lox DNA recombination system. The use of novel lox sites, which were generated by combining non-compatible and reaction equilibrium-modifying lox site variants, enables efficient reconstitution of a therapeutic cassette in a pre-determined configuration.
Results
Development of novel lox site variants for sequence-specific, unidirectional recombination
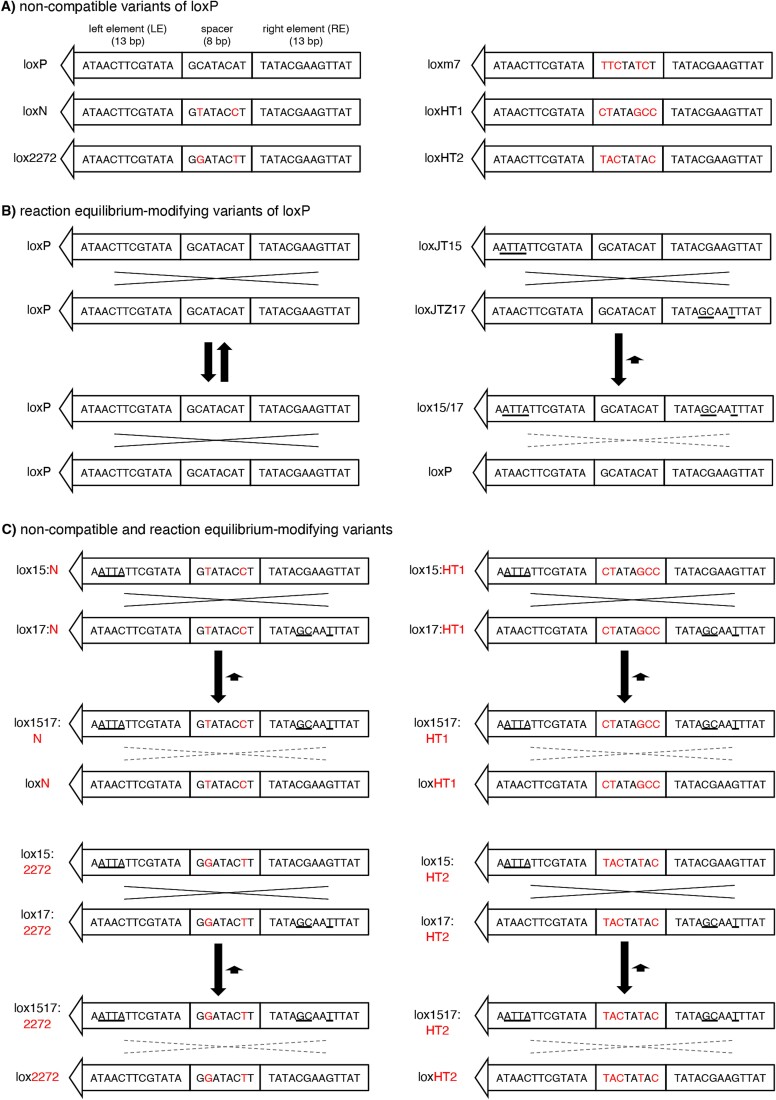
The canonical loxP site consists of two 13-bp inverted repeats (left and right elements; LE and RE, respectively) separated by an asymmetric 8-bp spacer/core sequence (Fig. 1A). While the left and right elements serve as the binding sites for the CRE recombinase, the spacer participates in the strand exchange reaction and determines the compatibility between lox sites (i.e. whether two lox sites can recombine or not) [33–35]. The asymmetry of the spacer imparts directionality to the loxP site.
Figure 1.
Lox site variants that enable CRE-dependent splicing of multiple AAV vector genomes. (A) Non-compatible mutant variants of loxP. Sequence differences (red) in the spacer region prevent recombination between non-compatible lox sites. Left and right elements (LE and RE, respectively) are palindromic. (B) Reaction equilibrium-modifying variants of loxP. LoxJT15 and loxJTZ17 sites have mutations (underlined) in either LE or RE but not in both. Recombination between two loxP sites (left) is fully reversible because substrates and products are identical. In contrast, recombination between loxJT15 and loxJTZ17 produces an LE/RE double mutant (lox15/17) and a loxP sequence. LE/RE double mutants are poor substrates for CRE, and consequently, the reverse reaction is significantly slower than the forward reaction. (C) Lox site variants that prevent recombination between non-compatible lox sites and inhibit reverse reactions. The spacer sequences of loxJT15 and loxJTZ17, which are from loxP, are replaced with those of the non-compatible lox sites (loxN, lox2272, loxm7 (not shown), loxHT1, and loxHT2).
There are two classes of lox site variants. One is non-compatible mutant variants, which include loxm7, loxN, and lox2272 [36–38] (Fig. 1A). These variants have mutations within the spacer sequence, and these mutations prevent strand exchange (and consequently recombination) between non-compatible lox sites while allowing recombination between homologous (or compatible) sites. A high-throughput screen identified fully non-compatible and promiscuous lox sites [39]. The second group comprises reaction equilibrium-modifying variants (Fig. 1B). These variants have mutations in either LE or RE, but not in both (e.g. loxJT15, loxJTZ17, lox71, and lox66) [40, 41]. Single-element mutations do not affect the binding of CRE to the lox site, and recombination between these mutant lox sites occurs as efficiently as between canonical loxP sites. However, recombination between LE and RE single mutants produces an LE/RE double mutant and a canonical loxP site. The presence of mutations in both LE and RE significantly reduces the affinity of the LE/RE double mutant to CRE, making it a poor substrate. While the recombination between canonical loxP sites is fully reversible as the initial substrates and the products have the same lox sites, the reaction equilibrium is drastically shifted toward the forward direction when LE and RE single mutants are used as substrates because the reverse reaction is much slower than the forward reaction. This causes the CRE-mediated recombination nearly unidirectional when the reaction equilibrium-modifying lox sites are used.
Although the CRE-lox system is highly efficient, canonical loxP sites (or any single species of lox sites) cannot be used to combine more than two DNA molecules. When there are two or more loxP sites in a single DNA molecule, the intervening “floxed” sequence is rapidly excised (Fig. S1A). The reverse reaction (i.e. insertion) is much slower than the forward reaction. Furthermore, if all DNA fragments have the same lox sites, recombination can occur in any combination (Fig. S1B), preventing the specification of the order of DNA segments in end products, leading to the production of unintended, non-functional products. Lastly, since the CRE-lox recombination is fully reversible, the reconstituted DNA constantly goes through the assembly-disassembly cycle, limiting the yield of the reconstituted DNAs (Fig. S1C). In principle, the yields of reconstituted products are 50%, 25%, and 12.5% when 2, 3, and 4 fragments are used as substrates, respectively, even when the excision and the order of DNA segment problems are disregarded.
To enable the reconstitution of large genes using multiple AAV vectors and the CRE-lox DNA recombination system, we devised novel lox sites by combining non-compatible and reaction equilibrium-modifying lox site variants (Fig. 1C). We chose the loxJT15-loxJTZ17 pair for the reaction equilibrium-modifying mutants because this pair was the most effective in inhibiting reverse-direction recombination [40]. For the non-compatible lox sites, we selected loxN, lox2272, loxm7, and two additional lox sites identified by a high-throughput screen with spacer sequences CTATAGCC (named loxHT1 herein) and TACTATAC (loxHT2) [39]. The loxN-based pair, for example, was generated by replacing the spacer sequence of loxJT15 and loxJTZ17 (GCATACAT) with that of loxN (GTATACCT). If these hybrid lox sites are fully non-compatible with one another, they should prevent the excision of intervening sequences and unintended recombinations (Fig. S1D and E). At the same time, they should significantly increase the yield of reconstituted DNA products by inhibiting reverse reactions, particularly when using 3 or more AAV vectors (Fig. S1F).
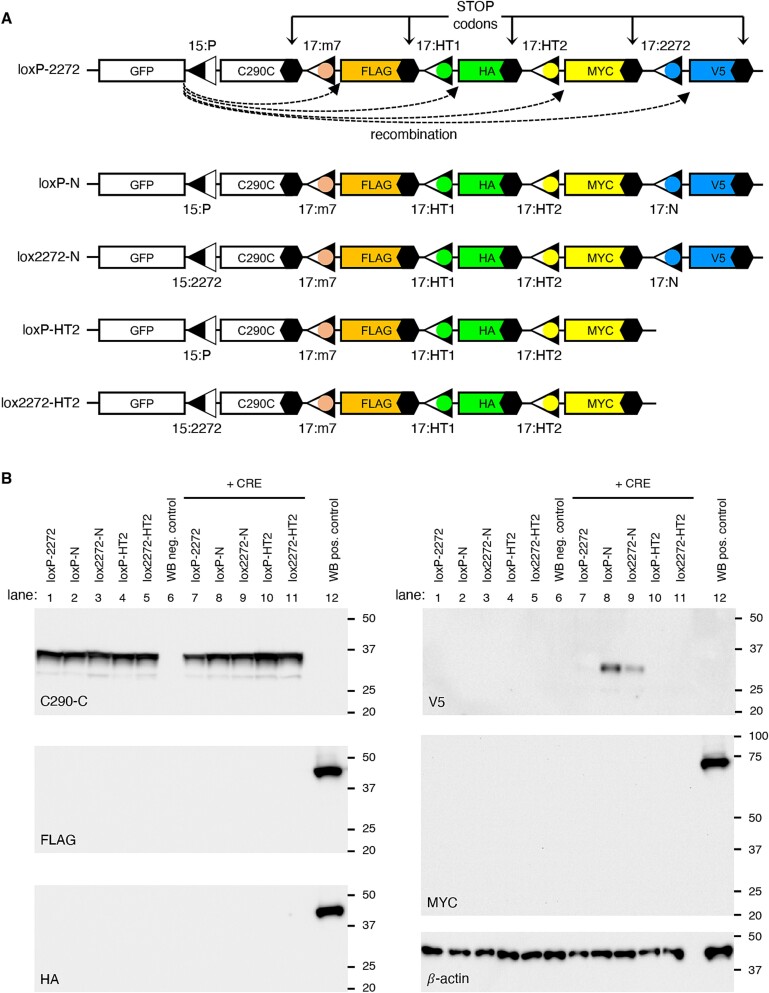
To assess the compatibility of the developed hybrid lox sites in mammalian cells, we designed GFP expression cassettes capable of tracking recombination events between different lox sites (Fig. 2 and Fig. S2; see Supplementary Materials for sequences). The first reporter construct, loxP-2272, was designed to survey the compatibility of loxJT15 with four hybrid lox sites (Fig. 2A). The reporter is composed of a CMV promoter, a GFP coding sequence (CDS), a 156-bp segment from the human CEP290 C-terminus (C290C; amino acids 2428–2479; in frame with GFP), and a stop codon. A loxJT15 site (15:P) was inserted between GFP and C290C such that recombination events between loxJT15 and any downstream lox sites would result in the excision of C290C and the production of new GFP fusion proteins with an identification tag (Fig. S2). FLAG, HA, MYC, and V5 tags were used to report the recombination of loxJT15 with loxJTZ17:m7 hybrid (hereafter denoted as 17:m7 for brevity), 17:HT1, 17:HT2, and 17:2272 sites, respectively. Recombination between loxJT15 (15:P) and 17:HT1, for instance, results in the production of GFP + HA fusion proteins (~30 kDa). Of note, recombination events not involving the 15:P site, such as between 17:m7 and 17:2272, do not lead to the fusion of associated tags with GFP and therefore go unreported. Four additional reporter constructs (loxP-N, lox2272-N, loxP-HT2, and lox2272-HT2) were generated to examine the compatibility among the hybrid lox sites that we have developed (Fig. 2A).
Figure 2.
Assessment of incompatibility among hybrid lox sites. (A) Schematics of reporter constructs to detect recombination events between loxJT15 (15:P), loxJTZ17:m7 (17:m7), loxJTZ17:HT1 (17:HT1), loxJTZ17:HT2 (17:HT2), loxJTZ17:2272 (17:2272), and loxJTZ17:N (17:N). The names of the reporter constructs (loxP-2272, loxP-N, lox2272-N, loxP-HT2, and lox2272-HT2) are shown on the left. Black hexagons denote stop codons. C290C: A 156-bp fragment from human CEP290 C-terminus (aa 2428–2479). (B) The spacers of loxP and lox2272 are fully incompatible with each other and with those of loxm7, loxHT1, and loxHT2. Reporter constructs shown in (A) were transfected to HEK293T cells with and without a CRE expression vector, and cell lysates were subjected to SDS-PAGE and immunoblotting. C290-C, FLAG, HA, V5, MYC, and β-actin antibodies were used for immunoblotting. A lysate derived from untransfected cells served as the negative control (lane 6), while lysates obtained from cells transfected with MYC-BBS1, FLAG-LZTFL1, and HA-LZTFL1 expression vectors were used as the positive control (lane 12). β-actin was used as a loading control. Numbers on the right of each panel indicate the locations of protein size markers.
When transfected into HEK293T cells, these reporters produced 35-kDa GFP + C290C fusion proteins, which could be detected by our CEP290 antibody (Fig. 2B and Fig. S2). However, when co-transfected with a CRE expression plasmid (pAAV-EF1α-CRE), reporters containing the 17:N site (i.e. loxP-N and lox2272-N) expressed GFP + V5 fusion proteins. In contrast, no new GFP fusion proteins were detected in cells transfected with loxP-2272, loxP-HT2, and lox2272-HT2. These results suggest that the spacer of loxN is partially compatible with those of loxP and lox2272, while loxP and lox2272 are fully incompatible with each other and with loxm7, loxHT1, and loxHT2. Since these reporters are designed to disclose only recombination events that involve the first lox site, which is linked to GFP, the compatibility among downstream lox sites (i.e. loxm7, loxHT1, and loxHT2) was not tested in this assay. As a positive control for Western blotting (lane 12), lysates from cells transfected with FLAG-LZTFL1, HA-LZTFL1, and MYC-BBS1 expression vectors were included to rule out the possibility of Western blotting failure, with β-actin serving as a loading control. Based on these results, we selected the spacer sequences of loxP, lox2272, and loxHT1 for the assembly of up to four AAV vector genomes. The spacers of loxm7 and loxHT2 may be used as alternatives to loxHT1 in this assembly strategy.
CRE-lox-mediated reconstitution of large genes delivered by tripartite AAV vectors
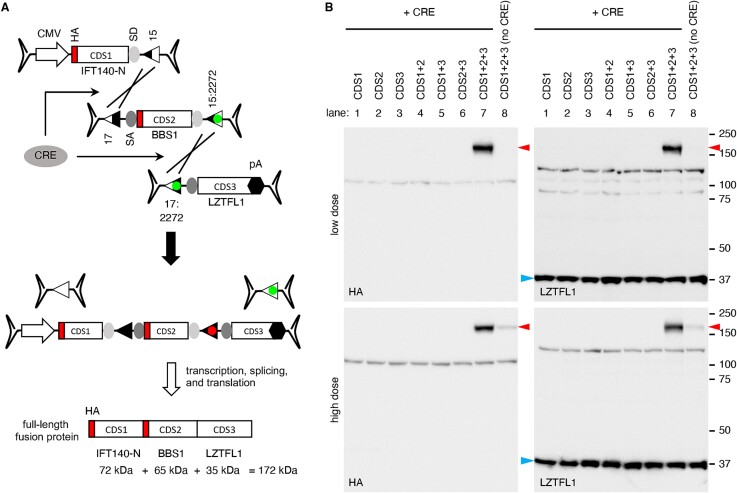
To evaluate the feasibility of our approach in delivering large genes using tripartite AAV vectors, we designed a set of three AAV vectors containing the coding sequences of three human genes: 5′ 1923 bp of IFT140 (IFT140-N), full-length BBS1, and full-length LZTFL1 (Fig. 3A). The first vector comprises a CMV promoter, 5′ 1923 bp of the IFT140 CDS, a splice donor (SD) site, and a loxJT15 site. An HA tag sequence was added to the 5′ end of IFT140 to facilitate the detection of expressed proteins. The second vector contains a loxJTZ17 site, a splice acceptor (SA) site, the BBS1 CDS (1775 bp; including an HA tag and two linker sequences), an SD site, and a lox15:2272 site. The third vector is composed of a lox17:2272 site, a SA site, the LZTFL1 CDS (988 bp; including a linker sequence), and a bovine growth hormone (BGH) transcription termination signal. Recombination between these three AAV vector genomes in the correct arrangement will lead to the reconstitution of an expression cassette encoding IFT140N + BBS1 + LZTFL1 fusion proteins.
Figure 3.
CRE-lox-mediated reconstitution of large genes delivered by tripartite AAV vectors: Proof-of-concept. (A) Schematic representation of large gene reconstitution by CRE-lox mediated recombination of three AAV vector genomes. A gene-of-interest is split into three fragments (CDS1, 2, and 3) and delivered to target cells via three separate AAV vectors. For a proof-of-concept demonstration, IFT140-N (5′ 1923 bp of IFT140), BBS1, and LZTFL1 coding sequences were used as CDS1, 2, and 3, respectively. HA tag sequences were added to the 5′ ends of IFT140-N and BBS1. CRE recombinase was delivered via a separate AAV vector. SD: Splice donor site, SA: Splice acceptor site, 15: loxJT15, 17: loxJTZ17, and pA: polyA signal. (B) Production of IFT140N + BBS1 + LZTFL1 fusion proteins (red arrowheads) from a tripartite AAV vector set. AAV vectors depicted in panel (A) were transduced to 293T cells, and the expression of IFT140N + BBS1 + LZTFL1 fusion proteins was examined by SDS-PAGE and immunoblotting using HA and LZTFL1 antibodies. Numbers on the right mark the locations of protein standards. A separate AAV vector, AAV-EF1α-CRE, was co-transduced to express CRE (lanes 1–7). Lane 1: CDS1 (IFT140-N) only, lane 2: CDS2 (BBS1) only, lane 3: CDS3 (LZTFL1) only, lane 4: CDS1 + CDS2, lane 5: CDS1 + CDS3, lane 6: CDS2 + CDS3, lane 7: CDS1 + CDS2 + CDS3, lane 8: CDS1 + CDS2 + CDS3 (without CRE). Endogenous LZTFL1 (blue arrowheads) was used as a loading control.
These AAV vectors, all utilizing serotype 2, were transduced individually or in various combinations to HEK293T cells at two different doses. The “low” dose involved transduction at a multiplicity of infection (MOI) of 1.5 × 104 genome copies (GC) per cell of each vector, while the “high” dose utilized an MOI of 6.0 × 104 GC/cell of each vector. CRE recombinase was delivered via a separate AAV vector (AAV2/2-EF1α-CRE) with an MOI of 0.3 × 104 GC/cell for the low dose and 1.2 × 104 GC/cell for the high dose. As shown in Fig. 3B, robust expression of the IFT140N + BBS1 + LZTFL1 fusion protein (red arrowheads) was observed using both HA and LZTFL1 antibodies at both low and high doses (upper and lower panels, respectively). The migration rate of the protein was consistent with the predicted molecular weight of the full-length fusion protein, approximately 172 kDa. Notably, no protein production was detected when the three AAV vectors were transduced individually or any of the three was omitted. Although the expression of IFT140N + BBS1 + LZTFL1 fusion proteins was not detected in the absence of CRE (lane 8) at the low dose, a small amount was detectable at the high dose (averaging 7% of CRE co-transduced cells; n = 3). These proteins are likely attributed to spontaneous and fortuitous recombination of the three AAV vector genomes in the correct configuration. Endogenous LZTFL1, marked by blue arrowheads, served as a loading control. These data indicate that the CRE- and hybrid lox site-mediated reconstitution is significantly more efficient than the trans-splicing approach for reconstituting large genes.
CRE-lox-mediated reconstitution of large genes delivered by quadripartite AAV vectors
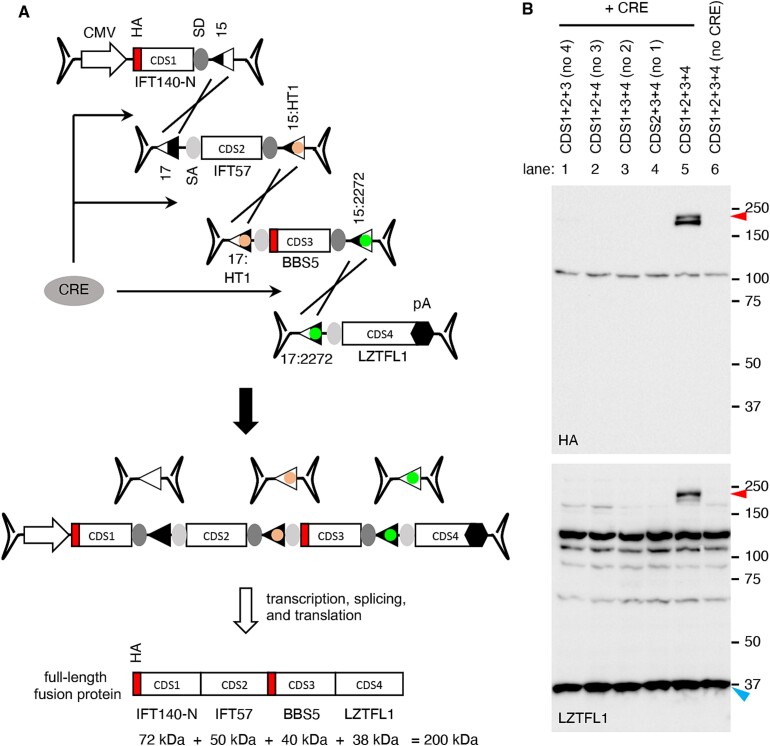
We expanded our approach to utilize quadripartite AAV vectors. For a proof-of-concept demonstration, we designed AAV vectors containing the CDSs of the 5′ 1923 bp of IFT140, IFT57, BBS5, and LZTFL1 (Fig. 4A). The first and fourth AAV vectors that contained the IFT140-N and LZTFL1 CDSs were the same ones used in the tripartite set above. The second vector was constructed with a loxJTZ17 site, a SA site, the IFT57 CDS (1330 bp; including two linker sequences), a SD site, and a lox15:HT1 site. The third vector was composed of a lox17:HT1 site, a SA site, the BBS5 CDS (1090 bp; including an HA-tag and two linker sequences), a SD site, and a lox15:2272 site. If recombination occurs as intended, it will result in the reconstitution of an expression cassette encoding IFT140N + IFT57 + BBS5 + LZTFL1 fusion proteins with a predicted molecular weight of ~200 kDa.
Figure 4.
CRE-lox-mediated reconstitution of large genes delivered by quadripartite AAV vectors: Proof-of-concept. (A) Schematic representation of large gene reconstitution by CRE-lox mediated recombination of four AAV vector genomes. A gene-of-interest is split into four fragments (CDS1, 2, 3, and 4) and delivered to target cells via four separate AAV vectors. For a proof-of-concept demonstration, IFT140-N (5′ 1923 bp of IFT140), IFT57, BBS5, and LZTFL1 coding sequences were used as CDS1, 2, 3, and 4, respectively. HA tag sequences were added to the 5′ ends of IFT140-N and BBS5. Others are the same as in Fig. 3. (B) Production of IFT140N + IFT57 + BBS5 + LZTFL1 fusion proteins (red arrowheads) from a quadripartite AAV vector set. AAV vectors depicted in panel (A) were transduced to 293 T cells, and the expression of IFT140N + IFT57 + BBS5 + LZTFL1 fusion proteins was examined by SDS-PAGE and immunoblotting using HA and LZTFL1 antibodies. Endogenous LZTFL1 (blue arrowheads) was used as a loading control. Others are the same as in Fig. 3.
These AAV vectors were transduced into 293T cells in various combinations at an MOI of 2.5 × 104 GC/cell of each vector. The AAV-EF1α-CRE vector was transduced at an MOI of 0.5 × 104 GC/cell. Upon transduction of the quadripartite AAV vectors and AAV-EF1α-CRE, the production of IFT140N + IFT57 + BBS5 + LZTFL1 fusion proteins was readily detected by immunoblotting using HA and LZTFL1 antibodies (Fig. 4B; lane 5). The fusion protein appeared to be unstable, with some instances of truncation near the C-terminus, resulting in doublets. When any of the four AAV vectors was omitted, the full-length fusion protein was not produced (lanes 1–4). Furthermore, in the absence of CRE, the fusion protein was not detected (lane 6). These data validate the successful reconstitution of a gene expression cassette delivered by quadripartite AAV vectors and underscore the efficacy of our CRE-lox-mediated recombination approach.
CRE-lox-mediated reconstitution of IFT140 delivered by bipartite AAV vectors
We applied the CRE-lox-mediated DNA recombination approach to IFT140, a gene associated with retinitis pigmentosa (RP) and short-rib thoracic dysplasia [42–44]. Although the full-length CDS of IFT140 (4389 bp) is small enough to be accommodated within a single AAV vector, additional regulatory sequences such as a promoter, a transcription termination signal, and two inverted terminal repeats (ITRs) must be included in the gene therapy vector, and the addition of such sequences makes the IFT140 expression cassette to exceed the AAV’s packaging capacity. Therefore, at least two AAV vectors are required to deliver the IFT140 gene. We constructed dual AAV-IFT140 vectors using the CRE-lox-mediated DNA recombination approach and examined whether full-length IFT140 could be successfully delivered by this method.
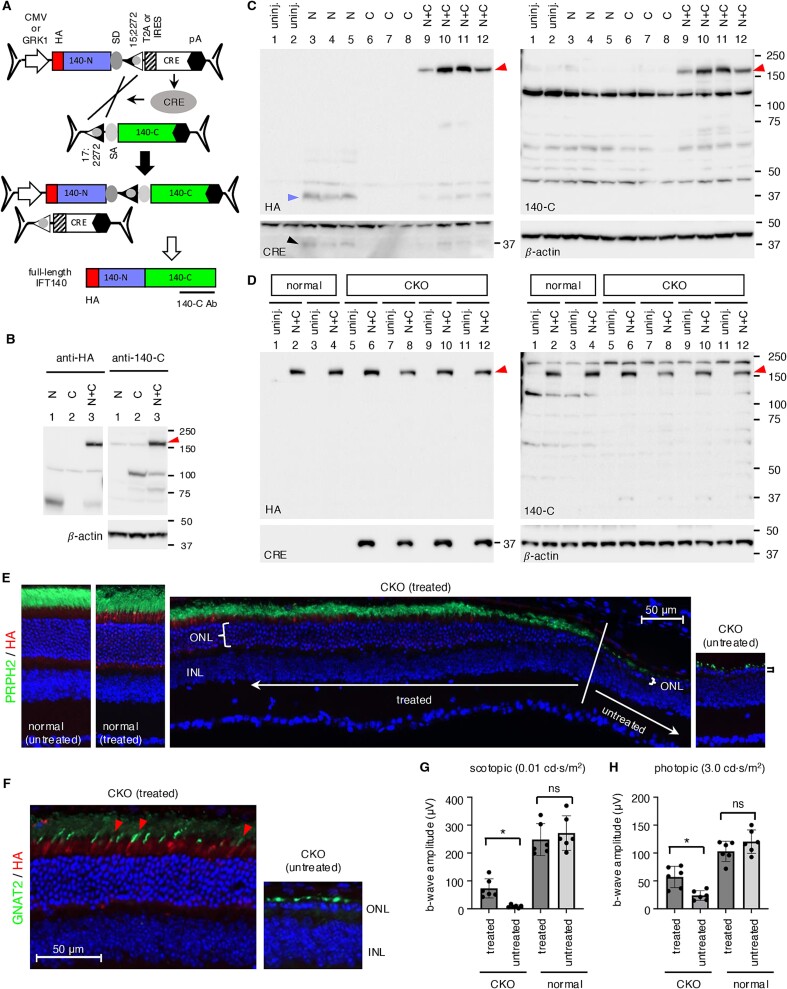
The 5′ vector was composed of a CMV promoter, 5′ 1923 bp of the IFT140 CDS, an SD site, and a lox15:2272 site, and the 3′ vector contained a lox17:2272 site, an SA site, the rest of the IFT140 CDS (2466 bp), and a BGH transcription termination signal (Fig. 5A). Since the combined payload capacity of two AAV vectors is ~9 kb and the size of the IFT140 expression cassette is 6–6.5 kb, there is space to accommodate the CRE gene within the dual AAV-IFT140 vectors. We incorporated the CRE CDS, along with an N-terminal T2A “self-cleaving” peptide, after the lox15:2272 site in the 5′ vector. A BGH polyA signal was also added following the CRE gene. The inclusion of CRE in this configuration not only eliminates the need for a separate AAV vector for CRE delivery but also offers an additional benefit of CRE “self-inactivation” by causing the separation of the CRE CDS from its promoter as recombination progresses. To facilitate protein detection, an HA tag was introduced at the N-terminus of IFT140. Additionally, we used an IFT140 antibody (140-C Ab), which was raised against human IFT140 aa1114-1462, to detect the C-terminal portion of the protein.
Figure 5.
Reconstitution of IFT140 by CRE-lox-mediated recombination. (A) Schematic representation of IFT140 reconstitution by CRE-lox-mediated recombination of bipartite AAV vector genomes. T2A: T2A “self-cleaving” peptide, IRES: Internal ribosome entry site. Others are the same as in Fig. 3. (B) Production of full-length IFT140 proteins (red arrowheads) through CRE-lox-mediated recombination in HEK293T cells. HEK293T cells were transduced with dual AAV vectors depicted in panel A (with a CMV promoter), and cell lysates were subjected to SDS-PAGE followed by immunoblotting with HA and IFT140-C antibodies. Lane 1: AAV-IFT140_N only, lane 2: AAV-IFT140_C only, lane 3: Dual AAV-IFT140_N+C. β-actin was used as a loading control. (C) Expression of full-length IFT140 in normal mouse retinas using the dual AAV-IFT140 CRE/lox set. Dual AAV-GRK1p-IFT140 vectors were administered via subretinal injection into mouse eyes (serotype: Anc80L65, dose: 8 × 108 GC per vector), and eyes were collected 2 weeks post-injection for immunoblotting with HA, IFT140-C, and CRE antibodies. Lanes 1–2: Uninjected (n = 2), lanes 3–5: AAV-IFT140_N only (n = 3), lanes 6–8: AAV-IFT140_C only (n = 3), and lanes 9–12: AAV-IFT140_N+C (n = 4). Each lane corresponds to an individual eye. The blue and black arrowheads indicate the IFT140-N product and CRE, respectively. (D) Reconstitution of IFT140 expression in Ift140 CKO mice using dual AAV-GRK1p-IFT140 vectors. Dual AAV-GRK1p-IFT140 CRE/lox vectors were delivered to the subretinal space of normal (Ift140fl/fl;iCre75−) and Ift140 CKO (Ift140fl/fl;iCre75+) mice at P16, and IFT140 expression was analyzed by immunoblotting at P55. Both injected and uninjected eyes were collected, and their protein extracts were loaded side-by-side (indicated by black lines). Each lane corresponds to an individual eye. (E and F) Preservation of photoreceptor cells in Ift140 CKO mice by dual AAV-GRK1p-IFT140 CRE/lox vectors. Dual AAV-GRK1p-IFT140 CRE/lox vectors were delivered to the subretinal space of normal and Ift140 CKO mice at P16, and retinal histology was examined by immunohistochemistry using HA, PRPH2, and GNAT2 antibodies at P52. Nuclei were counterstained with DAPI. Brackets delineate the outer nuclear layer (ONL). Scale bar: 50 μm. INL: Inner nuclear layer. (G and H) Preservation of retinal function in Ift140 CKO mice by IFT140 subretinal gene therapy. Normal (n = 6) and Ift140 CKO (n = 6) mice were administered dual AAV-GRK1p-IFT140 vectors in one eye at P16, and rod (G) and cone (H) functions were evaluated by scotopic and photopic ERG at P45. Uninjected contralateral eyes were used as controls. Rod function was measured using 0.01 cd⋅s/m2 dim flashes after dark adaptation, while cone function was measured using 3.0 cd⋅s/m2 bright flashes after light adaptation. ERG b-wave amplitudes (mean ± SD) are shown in the graphs. Asterisks indicate statistical significance (two-tailed Student’s t-test; P < 0.05). ns: not significant.
Dual AAV-CMV-IFT140 vectors (with serotype 2) were transduced into 293T cells at an MOI of 3 × 104 GC/cell for each vector. Upon transduction, the 5′ vector produced a ~70 kDa protein detectable by the HA antibody (Fig. 5B; lane 1). The 3′ vector produced a ~100 kDa protein (lane 2), which is presumably due to the intrinsic promoter activity of the ITRs [45]. When both the 5′ and 3′ vectors were co-transduced, robust expression of full-length IFT140 was observed (lane 3; red arrowhead), demonstrating efficient reconstitution of the IFT140 expression cassette in 293T cells.
Dual AAV-IFT140 CRE-lox vectors prevent or delay retinal degeneration in a mouse model of IFT140-associated retinitis pigmentosa
We further investigated whether the CRE/lox-based dual AAV-IFT140 vectors could produce full-length IFT140 proteins in vivo and prevent retinal degeneration caused by the loss of IFT140 function. Since IFT140 is a relatively low-abundance protein, we adopted the rhodopsin kinase (GRK1) promoter, which is active in both rods and cones [46], instead of the CMV promoter (Fig. 5A). Additionally, to minimize the potential impact of producing N-terminal truncated proteins, we reduced the size of the IFT140-N fragment to 810 bp and placed the remaining 3576 bp in the 3′ vector, which lacked a promoter. These AAV vectors were prepared with the Anc80L65 serotype [4] and delivered to mouse retinas via subretinal injections.
We first administered the dual AAV-GRK1p-IFT140 vectors into the eyes of 1- to 2-month-old wild-type mice and examined IFT140 protein production by immunoblotting (Fig. 5C). To this end, animals received the dual AAV-GRK1p-IFT140 vectors either individually (n = 3) or in combination (n = 4) at a dose of 8 × 108 GC of each vector. Treated eyes were collected two weeks post-injection, and protein extracts were analyzed by SDS-PAGE followed by immunoblotting. When injected individually, the 5′ vector produced ~37 kDa truncated protein products (lanes 3–5; blue arrowhead), while no protein production was observed with the 3′ vector (lanes 6–8). However, when the 5′ and 3′ vectors were co-delivered, full-length IFT140 proteins were readily detected with both HA and IFT140-C antibodies in all four injected animals (lanes 9–12). These data underscore the reliability of the CRE/lox-mediated DNA reconstitution approach for delivering large genes using AAV.
We then evaluated the therapeutic efficacy of the dual AAV-GRK1p-IFT140 vectors in Ift140 mutant mice. Because constitutive inactivation of Ift140 is embryonic lethal in mice, we used Ift140 conditional knockout (CKO) mice, in which exon 7 is floxed [47, 48]. For retina-specific Ift140 ablation, we employed the iCre75 driver (also known as rhodopsin-iCRE; [49]) to selectively ablate Ift140 expression in rod photoreceptors. Of note, iCre75-driven CRE activity is detectable as early as post-natal day (P) 7 [49]. It also should be noted that the incompatibility between loxP (in the Ift140 floxed allele) and lox2272 (in the AAV-IFT140 vectors) sites prevents recombination between the Ift140fl alleles and the AAV vector genomes. Since retinal degeneration is already noticeable at P24 and progresses rapidly in Ift140fl/fl;iCre75+ mice (Fig. S3; also see below), we delivered the AAV gene therapy vectors at an early stage of degeneration (P16).
To confirm the production of full-length IFT140 proteins in Ift140 CKO (Ift140fl/fl;iCre75+) mice, treated and untreated contralateral eyes were collected at P55 (39 days post-injection), and protein extracts were subjected to SDS-PAGE followed by immunoblotting. As shown in Fig. 5D, full-length IFT140 expression was observed in all CKO mice injected (n = 4). Notably, the 37-kDa IFT140 N-terminal truncated protein product was no longer detected in either CKO or normal (Ift140fl/fl;iCre75−) mouse eyes, suggesting that unassembled 5′ vectors may have been depleted by this time. Consistent with this, CRE expression was not detected in treated normal mouse eyes. The CRE proteins present in injected CKO mouse eyes are likely (or mostly) derived from the iCre75 transgene rather than the 5′ AAV-IFT140 vector. The absence of CRE in uninjected eyes is due to the complete loss of rod photoreceptors in CKO mice by this age (see below).
To assess whether the dual AAV-GRK1p-IFT140 vectors could prevent retinal degeneration, we examined retinal sections from treated and untreated contralateral eyes of normal (n = 2) and Ift140 CKO (n = 3) mice using immunohistochemistry at P52 (Fig. 5E and F). HA, PRPH2, and GNAT2 antibodies were used to visualize IFT140, photoreceptor outer segments, and cone outer segments, respectively. In untreated CKO mouse eyes, the vast majority of photoreceptor cells had been lost by this age, with only 1–2 rows of photoreceptor cell nuclei remaining. In contrast, 5–8 rows of photoreceptor nuclei were preserved within the central two-thirds of the treated regions (n = 3 mice). HA-IFT140 expression was also observed in cones (Fig. 5F; red arrowheads). In normal mice, the administration of AAV-GRK1p-IFT140 vectors did not appear to cause obvious toxic effects, at least at the dose used (8 × 108 GC of each vector).
Lastly, we evaluated the light responsiveness of the treated eyes by electroretinography (ERG) at P45. To assess the rod function, mice were dark-adapted and subjected to dim light flashes (0.01 cd⋅s/m2) (Fig. 5G; see Fig. S4 for ERG waveforms). In Ift140 CKO mice, untreated contralateral eyes exhibited minimal ERG responses, with an average b-wave amplitude ± standard deviation (SD) of 8.7 ± 5.2 μV (n = 6). In contrast, significant ERG responses were detected in treated eyes, with an average amplitude of 73.2 ± 34.3 μV (n = 6). Statistical analysis using a two-tailed Student’s t-test revealed a significant difference between the two groups (P = 0.005). No significant differences were observed between treated and untreated normal eyes (248.2 ± 57.2 μV vs. 271.4 ± 61.7 μV; n = 6; P = 0.516). Cone function was evaluated by stimulating eyes with bright light flashes (3.0 cd⋅s/m2) after light adaptation (Fig. 5H and Fig. S4). Although iCre75 was only expressed in rods, cone function was also reduced in Ift140 CKO mice at P45. However, eyes treated with the AAV-GRK1p-IFT140 gene therapy vectors exhibited significantly higher ERG responses, with an average b-wave amplitude of 57.0 ± 19.1 μV, (n = 6) compared to untreated contralateral eyes (23.8 ± 8.6 μV, n = 6; Student’s t-test P = 0.006). Similar to the scotopic ERG results, no significant differences were observed in photopic ERG between treated and untreated eyes (102.9 ± 18.3 μV vs. 112.1 ± 30.7 μV, n = 6, P = 0.549) in normal mice. A longer-term study is currently underway. Overall, these findings indicate that the dual AAV-GRK1p-IFT140 vectors can effectively prevent or delay retinal degeneration in Ift140 CKO mice.
Reconstitution of PCDH15 by CRE-lox-mediated recombination
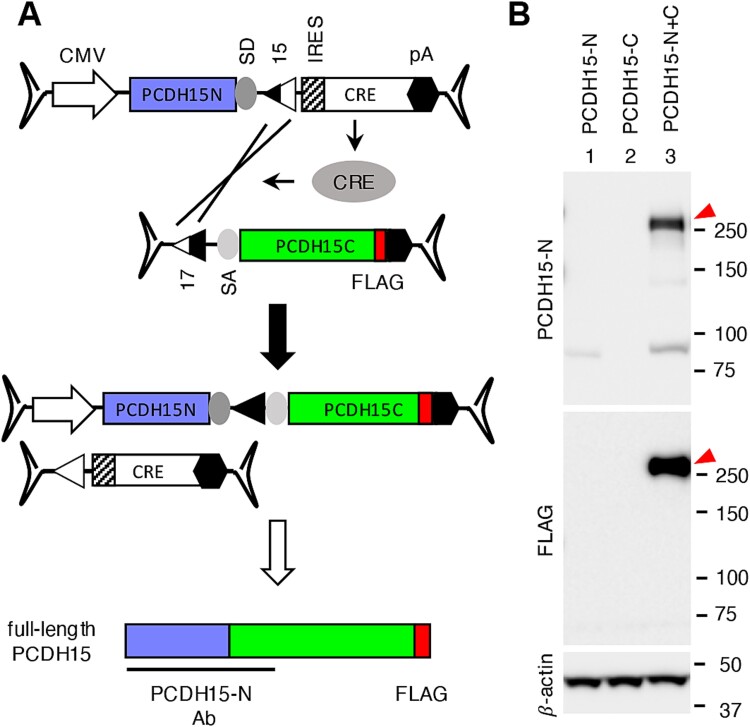
Mutations in PCDH15 cause Usher syndrome type 1F (USH1F), which is characterized by profound congenital hearing impairment and progressive vision loss [50–52]. The full-length human PCDH15 CDS spans 5865 bp, necessitating two AAV vectors for delivery. We applied the CRE-lox approach to PCDH15 and created dual AAV vectors (Fig. 6).
Figure 6.
Reconstitution of PCDH15 using the CRE-lox recombination approach. (A) Strategy for the PCDH15 reconstitution using dual AAV-PCDH15 vectors. PCDH15 CDS was divided into two segments (1932 bp and 3933 bp) for delivery, and a FLAG tag (red) sequence was added to the 3′ end of the gene. PCDH15-N antibody was raised against the N-terminal half of human PCDH15 (aaQ27-A1376). Others are the same as in Fig. 3. (B) Reconstitution of PCDH15 by dual AAV-PCHD15 CRE-lox vectors. HEK293T cells were transduced with dual AAV2/2-CMV-PCHD15 CRE-lox vectors as indicated, and cell lysates were subjected to SDS-PAGE and immunoblotting with PCDH15-N and FLAG antibodies. Red arrowheads mark the reconstituted full-length PCDH15 proteins.
The full-length human PCDH15 CDS was divided into two segments (1932 bp and 3933 bp) and inserted into the 5′ and 3′ vectors, respectively (Fig. 6A). Similar to the IFT140 gene, the CRE CDS was included within the 5′ vector, but in this case, an internal ribosome entry site (IRES) was used instead of the T2A self-cleaving peptide. Additionally, we introduced a FLAG tag to the C-terminus of PCDH15 to facilitate protein detection. For the detection of the N-terminal portion of PCDH15, a polyclonal antibody (PCDH15-N Ab) raised against recombinant human PCDH15 protein (aa Q27-A1376) was used.
When transduced to HEK293T cells (at an MOI of 3 × 104 GC/cell for each vector), the 5′ vector produced an ~85 kDa protein detected by the PCDH15-N antibody, while no protein production was observed from the 3′ vector (Fig. 6B, lanes 1 and 2). However, when both vectors were transduced together, robust expression of full-length PCDH15 was observed (lane 3; red arrowheads). These results demonstrate that the CRE-lox-based dual AAV-PCDH15 vectors are an effective method for delivering full-length PCDH15 gene.
Reconstitution of CEP290 by CRE-lox-mediated recombination
CEP290 mutations are associated with various ciliopathies, ranging from isolated retinal dystrophy to syndromic conditions such as Bardet-Biedl syndrome, Joubert syndrome, and Meckel-Gruber syndrome [7–9, 53–55]. Although the full-length human CEP290 CDS (7440 bp) can be accommodated in two AAV vectors, a better outcome was obtained when CEP290 was split into three AAV vectors in a prior study to develop CEP290 gene therapy vectors using the split intein strategy [30]. However, even with the optimized set, the yield of full-length CEP290 was very low, and a large amount of truncated protein products remained unspliced.
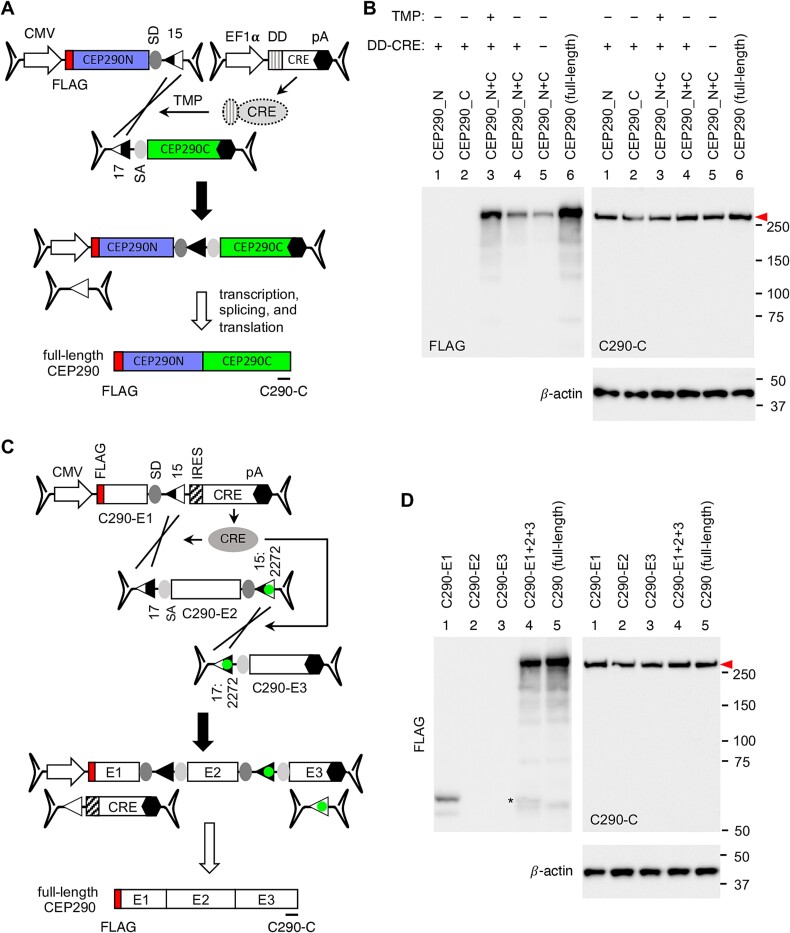
We applied the CRE-lox approach to CEP290 and developed bipartite and tripartite AAV-CEP290 vectors. For the bipartite set (Fig. 7A), the CEP290 CDS was divided into two fragments (3527 bp and 3913 bp) and inserted into the 5′ and 3′ vectors, respectively. CRE was delivered through a separate AAV vector (AAV-EF1α-DD-CRE). To prevent constitutive overexpression of CRE, we adopted dihydrofolate reductase destabilizing domain (DD)-fused CRE [56, 57], which undergoes rapid degradation through the proteasomal pathway but can be temporarily stabilized by the addition of trimethoprim (TMP) (Fig. S5). For the tripartite set (Fig. 7C), CEP290 was split into 3 segments (5′ (E1): 1065 bp, middle (E2): 2913 bp, and 3′ (E3): 3462 bp), and the CRE gene was included within the 5′ vector. The loxJT15 and loxJTZ17 pair was used to join the 5′ and middle vectors and the lox15:2272 and lox17:2272 pair was used for the middle and 3′ vectors. A FLAG tag sequence was introduced at the 5′ end of CEP290 to facilitate the detection of the N-terminal portion of CEP290 in both sets, and the CEP290-C antibody was used to detect the C-terminal portion of CEP290.
Figure 7.
CRE-lox mediated reconstitution of CEP290. (A) Schematic representation of the bipartite AAV-CEP290 vectors. The 5′ vector is composed of a CMV promoter, 5′ 3527 bp of human CEP290, an SD site, and a loxJT15 site. The 3′ vector consists of a loxJTZ17 site, an SA site, 3′ 3913 bp of CEP290, and a BGH polyA signal. A FLAG tag was added to the 5′ end of CEP290. Destabilizing domain (DD)-fused CRE was delivered via a separate AAV vector (AAV-EF1α-DD-CRE). The location of the C290-C antibody epitope was marked by a black line at the bottom of the schematic. (B) Production of full-length CEP290 proteins (red arrowhead) by CRE-lox-mediated recombination in 293T cells. HEK293T cells were transduced with dual AAV2/2-CEP290 (MOI: 3 × 104 GC/cell per vector) and AAV2/2-EF1α-DD-CRE (MOI: 1 × 104 GC/cell) vectors. After transduction, cells were treated with 10 μM trimethoprim (TMP) for 48 h to stabilize DD-CRE, and cell lysates were subjected to SDS-PAGE followed by immunoblotting with FLAG and C290-C antibodies. Lane 1: AAV-CEP290_N only, lane 2: AAV-CEP290_C only, lanes 3–4: Dual AAV-CEP290_N+C, and lane 5: pSS-FS-hCEP290 plasmid transfected (full-length; positive control). Cells in lanes 1–3 were co-transduced with AAV2/2-EF1α-DD-CRE. Numbers on the right denote the location of protein standards. β-actin was used as a loading control. (C) Schematic representation of the tripartite AAV-CEP290 vectors. The 5′ vector (E1) is composed of a CMV promoter, 5′ 1065 bp of human CEP290, an SD site, a loxJT15 site, IRES, CRE CDS, and a BGH polyA signal. The middle vector (E2) is composed of a loxJTZ17 site, an SA site, 2913 bp of CEP290 CDS, an SD site, and a lox15:2272 site. The 3′ vector (E3) consists of a lox17:2272 site, an SA site, 3′ 3462 bp of CEP290, and a BGH polyA signal. Others are the same as in panel A. (D) Production of full-length CEP290 proteins (red arrowhead) by CRE-lox-mediated recombination in 293T cells. Others are the same as in panel B.
To evaluate the reconstitution and expression of CEP290 from the bipartite AAV-CEP290 vectors, HEK293T cells were transduced with the dual AAV-CEP290 vectors at an MOI of 3 × 104 GC/cell of each vector and AAV-EF1α-DD-CRE at an MOI of 1 × 104 GC/cell. After transduction, cells were treated with 10 μM TMP for 48 h to stabilize DD-CRE, and CEP290 expression was assessed by immunoblotting (Fig. 7B). FLAG-tagged, full-length CEP290 expression was detectable in cells transduced with all three AAV vectors and treated with TMP (lane 3). FLAG-CEP290 expression was also detectable without TMP treatment (lane 4), but at a significantly lower level (average 24% of TMP-treated cells; n = 3). A small amount of FLAG-CEP290 was observed in the absence of CRE (lane 5; average 9% of TMP-treated cells; n = 3), representing CEP290 gene reconstitution via spontaneous recombination between the dual AAV-CEP290 vectors. Plasmid DNAs encoding full-length CEP290 were transfected and used as a positive control (lane 6). For some unknown reason, over-expressed CEP290 appears to be unstable and degraded, and consequently, we were not able to observe increased CEP290 protein levels beyond endogenous expression using the CEP290-C antibody. β-actin was used as a loading control.
The tripartite AAV-CEP290 vectors were transduced into 293T cells at an MOI of 3 × 104 GC/cell of each vector, and FLAG-CEP290 expression was examined as above. As shown in Fig. 7D, full-length CEP290 expression was detectable only when all three AAV vectors were transduced. These data demonstrate that both bipartite (with a separate AAV-CRE vector) and tripartite AAV-CEP290 vectors efficiently deliver the full-length CEP290 gene to target cells.
Reconstitution of CDH23 by CRE-lox mediated recombination
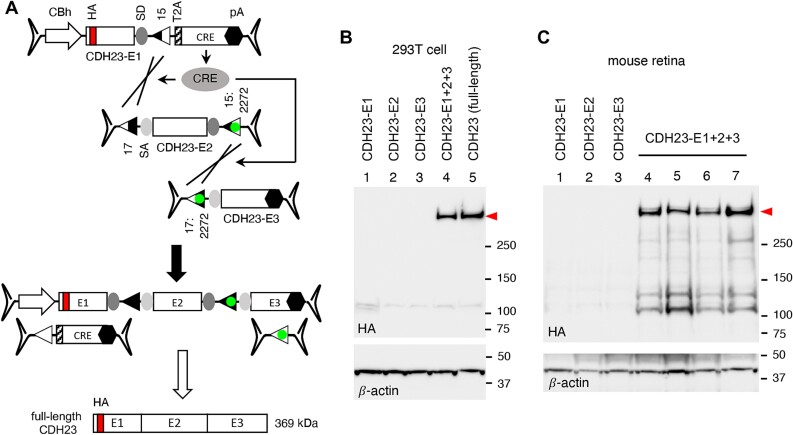
We then applied the CRE-lox-mediated DNA recombination approach to CDH23 gene therapy vectors. Inactivating mutations of CDH23 cause Usher syndrome type 1D (USH1D) [13, 14]. The full-length human CDH23 CDS spans 10 065 bp, requiring three AAV vectors for delivery. Since the total payload capacity of three AAV vectors is ~14 kb (excluding ITRs), we split the CDH23 CDS into 3 fragments (5′ vector (E1): 2176 bp, middle vector (E2): 4077 bp, and 3′ vector (E3): 3812 bp) and included the CRE gene (with a T2A peptide) in the 5′ vector (Fig. 8A). The loxJT15 and loxJTZ17 pair was used to join the 5′ and the middle vectors and the lox15:2272 and lox17:2272 pair was used for the middle and the 3′ vectors. CDH23 is a type-I single transmembrane protein with an N-terminal signal peptide, and an HA tag was inserted after the signal peptide for protein detection. A CBh promoter and a BGH polyA signal were used as a promoter and a transcription termination signal, respectively.
Figure 8.
CRE-lox-mediated reconstitution of CDH23 delivered by tripartite AAV vectors. (A) Schematic of the CDH23 reconstitution using tripartite AAV-CDH23 vectors. The CDH23 CDS (10 065 bp) was split into three pieces (E1: 2176 bp, E2: 4077 bp, and E3: 3812 bp), and the CRE gene was included in the 5′ (E1) vector for self-inactivation after recombination. A T2A “self-cleaving” peptide was used for CRE expression. An HA tag (red) was added to the N-terminus of CDH23 for detection (after the signal peptide). (B) Production of full-length CDH23 proteins (red arrowhead) by CRE-lox-mediated recombination in 293T cells. HEK293T cells were transduced with tripartite AAV2/2-CDH23 vectors (MOI: 3 × 104 GC/cell of each vector), and cell lysates were subjected to SDS-PAGE followed by immunoblotting with HA antibodies. Lane 1: AAV-CDH23-E1 only, lane 2: AAV-CDH23-E2 only, lane 3: AAV-CDH23-E3 only, lane 4: AAV-CDH23-E1, E2, and E3 co-transduced, and lane 5: pSS-HA-CDH23-SF plasmid transfected (full-length; positive control). β-actin was used as a loading control. (C) Expression of CDH23 from the tripartite AAV-CDH23 vectors in mouse retinas. Tripartite AAV-CDH23 vectors were subretinally administered to wild-type mice as indicated (lane 1: E1 vector alone, lane 2: E2 vector alone, lane 3: E3 vector alone, lanes 4–7: E1, E2, and E3 co-injected) at the dose of 3 × 109 GC per vector (n = 3–4 per vector or vector set). Treated eyes were collected 3 weeks post-injection and retinal protein extracts were subjected to SDS-PAGE and immunoblotting. Each lane represents an individual eye.
To test whether the tripartite AAV-CDH23 vectors could deliver full-length CDH23, we transduced HEK293T cells with these vectors at an MOI of 3 × 104 GC/cell of each vector (serotype AAV2) and examined the production of full-length CDH23 proteins by immunoblotting. When all three vectors were co-transduced, robust expression of CDH23 was observed (Fig. 8B; red arrowhead). A plasmid containing a full-length CDH23 expression cassette was used as a positive control (lane 5), and β-actin was used as a loading control. To test whether these vectors could be used to deliver CDH23 in vivo, we administered the tripartite AAV-CDH23 vectors to wild-type mouse eyes by subretinal injections (serotype: AAV5, dose: 3 × 109 GC per vector). Consistent with the results in 293T cells, full-length CDH23 proteins were readily detected in all eyes injected with the tripartite AAV vectors (Fig. 8C, lanes 4–7).
Discussion
Reconstitution of therapeutic genes via the CRE-lox-mediated DNA recombination offers several advantages compared to other approaches that have been reported thus far.
First, compared to spontaneous recombination-dependent approaches such as trans-splicing, overlapping, and hybrid dual or triple AAV approaches [17–20], the CRE-lox-mediated recombination drastically increases the recombination efficiency and the yield of correctly reconstituted genes. This is especially true when tripartite or quadripartite AAV vectors are used. The use of non-compatible, hybrid lox sites prevents the excision of floxed sequences, ensures recombination in a pre-determined configuration, and inhibits the disassembly of reconstituted genes (Fig. S1D-F). The enhanced efficiency and yield reduce the number of AAV particles needed to transduce target cells, and the use of fewer AAV vectors reduces the potential risks of viral vector-derived toxicity and inflammation.
Second, the CRE-lox-mediated DNA recombination approach provides more flexibility regarding splitting positions compared to the protein trans-splicing approach. The efficiency of protein trans-splicing is influenced by the amino acid residues adjacent to split inteins [25, 31, 32]. The first residue within the C-extein is particularly important, and Cys, Ser, and Thr residues are strongly preferred. This constraint limits the number of possible locations where a protein may be split. Moreover, protein truncations can affect a protein’s structure, stability, and localization, and these factors also influence the overall efficiency and yield of the protein reconstitution. If the target gene encodes a transmembrane or secreted protein (e.g. PCDH15 and CDH23), the topology and secretion of each protein fragment should be considered when determining splitting positions. Ideal splitting positions for the split intein approach should support the protein trans-splicing process and have minimal or no effect on the folding and stability of each truncated protein fragment and reconstituted proteins. Additionally, the truncated protein products should be localized to the same compartment or close locations to increase the likelihood of engagement, while the size of each fragment should be small enough to fit into single AAV vectors. Therefore, identifying optimal splitting positions could be challenging and usually involves comparing multiple candidate sites empirically. And the complexity increases if the target gene requires 3 or 4 AAV vectors. In contrast, when using the CRE-lox approach, protein structure, stability, localization, and topology are not factors to consider since the reconstitution occurs at the DNA level. Cargo capacity expansion can be achieved by simply adding additional sets of non-compatible lox sites to AAV vectors.
Another significant advantage of the CRE-lox approach over protein trans-splicing is the lack or minimal production of truncated proteins. Protein trans-splicing requires the production of “half” proteins before reconstitution, which can have dominant negative or harmful effects if continuously expressed. In contrast, truncated protein production is either absent or low when the CRE-lox strategy is used because the AAV vectors lack a promoter or a polyA signal, which stabilizes mRNA. Although ITRs have some intrinsic promoter activities [45], they are weak in most cells. When the CRE gene is included in the 5′ vector, a truncated protein and CRE are initially produced. However, as recombination progresses and the 5′ vector is converted to a full-length therapeutic cassette, the production of truncated proteins and CRE diminishes. The lack or minimal production of truncated proteins may be crucial for certain genes if such protein products are toxic to cells, and the ‘self-inactivation’ feature of the CRE-containing 5′ vector provides an additional layer of safety to the CRE-lox approach.
Lastly, although the CRE-lox approach requires the delivery of CRE in addition to therapeutic genes, the practical payload capacity of AAV vectors with the CRE-lox approach is either comparable with or larger than that of the split intein-based approach. The protein trans-splicing approach requires each AAV vector to have its own promoter and transcription termination signal to produce therapeutic gene products and split inteins. The repeated inclusion of transcriptional regulatory elements erodes the AAV vector’s combined payload capacity. In contrast, the CRE-lox approach only requires one promoter and one transcriptional termination signal for the entire vector set, which becomes more beneficial as more AAV vectors are needed.
One notable concern of the CRE-lox approach is the prolonged expression of CRE recombinase in transduced cells. Prolonged expression of CRE could lead to unintended recombination in the human genome, potentially resulting in unwanted mutations or genomic instability. Moreover, prolonged CRE expression could also lead to immune responses, which may limit the effectiveness of AAV gene therapies. This could potentially result in the destruction of cells expressing the therapeutic gene or a reduction in the efficacy of the AAV gene therapy over time. In this regard, the inclusion of CRE in 5′ vectors and being “self-inactivated” by recombination significantly reduces these risks. Alternatively, an inducible promoter or destabilizing domain-fused CRE may be used to control CRE expression. Other site-specific DNA recombinases that display higher specificity than CRE may be used as well. It is noteworthy that while numerous transgenic rodent lines expressing CRE have been created and some phenotypes have been reported [58], constitutive expression of CRE does not appear to cause serious health concerns in rodents. However, a thorough investigation of genes or sites in the human genome that could be modified by CRE will be necessary. As with any medical treatment, it is crucial to carefully consider the potential risks and benefits associated with CRE-lox-dependent AAV gene therapies.
In summary, the CRE-lox approach described in this study offers a simple, versatile, and efficient platform for producing AAV-based, generic gene replacement therapy vectors capable of delivering large genes. As this approach delivers full-length genes, the gene therapy vectors developed using this method have the potential to be generally applicable to all patients with loss-of-function mutations.
Materials and methods
Plasmid DNAs
To generate the GFP expression reporter loxP-N (Fig. 2), a double-stranded DNA fragment corresponding to the loxJT15 + C290C + 17:m7 + FLAG+17:HT1 + HA + 17:HT2 + MYC + 17:N + V5 portion was synthesized (Integrated DNA Technologies) and inserted at the C-terminus of GFP within the pEGFP-C3 plasmid (Clontech) using a GenBuilder Cloning kit (GenScript). Reporter loxP-2272 was generated by substituting the lox17:N site (ATAACTTCGTATAGTATACCTTATAGCAATTTAT) within loxP-N with the lox17:2272 site (ATAACTTCGTATAGGATACTTTATAGCAATTTAT) using a Q5 Site-Directed Mutagenesis kit (New England Biolabs) and PCR. Reporter lox2272-N was produced by replacing the loxJT15 site (AATTATTCGTATAGCATACATTATACGAAGTTAT) in the loxP-N reporter with the lox15:2272 site (AATTATTCGTATAGGATACTTTATACGAAGTTAT). Reporters loxP-HT2 and lox2272-HT2 were derived by eliminating the lox17:N sites in loxP-N and lox2272-N using a Q5 Site-Directed Mutagenesis kit and PCR. The DNA sequences downstream of GFP are shown in the Supplementary Material.
The AAV shuttle plasmids used in this study are listed in the Supplementary Material, and their sequences are available upon request. Briefly, the pFBAAV-related plasmids were generated by modifying the pFBAAVCMVmcsBGHpA plasmid (from the Viral Vector Core Facility, University of Iowa). Plasmid pAAV-CBh-JT1522 was custom-synthesized and procured from VectorBuilder. Other pAAV-related plasmids were developed by modifying these plasmids using standard molecular biology techniques and the GenBuilder Cloning kit (GenScript). PCR primers and oligonucleotides used were obtained from Integrated DNA Technologies (IDT). Plasmids pcDNA3-MYC-hBBS1, pCS2FLAG-hLZTFL1, and pCS2HA-hLZTFL1 were previously described [59, 60]. Plasmid DNAs containing human IFT140 (BC035577) and PCDH15 (NM_033056.4) CDSs were acquired from transOMIC and GenScript, respectively. These plasmids were used as PCR templates to produce pFBAAV-CMV-IFT140N-15, pAAV-CMV-IFT140N-1522-CRE, pFBAAV-1722-IFT140C-pA, pAAV-GRK1p-IFT140N-1522-CRE, pFBAAV-1722-IFT140C-pAv3, pAAV-CMV-PCDH15N-15-IRES-CRE, and pFBAAV-17-PCDH15C-pA. A plasmid containing the full-length human CEP290 CDS (NM_025114.4) was reported earlier [61] and used as a PCR template for bipartite (pFBAAV-CMV-CEP290N-JT15 and pFBAAV-17-CEP290C-pA) and tripartite AAV-CEP290 vectors (pAAV-CBh-CEP290-E1-15-CRE, pAAV-17-CEP290-E2-1522, and pFBAAV-1722-CEP290-E3-pA). The human CDH23 CDS was divided into 5 segments and synthesized by IDT according to the NM_022124.6 sequence. An HA tag was introduced after CDH23’s signal peptide (N-terminal 28 residues). These 5 segments were PCR-amplified using Q5 High-Fidelity DNA polymerase and inserted into the multiple cloning site (MCS) of pAAV-CBh-JT15-CREv2 (first segment), pAAV-JTZ17-mid-1522 (second and third segments), and pFBAAV-1722-pA (fourth and fifth segments) using the GenBuilder Cloning kit to produce pAAV-CBh-CDH23-E1-15-CRE, pAAV-17-CDH23-E2-1522, and pFBAAV-1722-CDH23-E3-pA, respectively. A full-length CDH23 expression vector, pSS-HA-hCDH23-SF, was generated by PCR amplification of the CDH23-E1, E2, and E3 fragments and insertion of these 3 fragments into the MCS of the pSS-SF plasmid [62] using the GenBuilder Cloning kit (sequence available upon request).
Transfection and AAV transduction in HEK293T/17 cells
HEK293T/17 cells were acquired from American Type Culture Collection (ATCC; #CRL-11268) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco) and 100 units/ml penicillin/streptomycin (Gibco) at 37°C in a humidified 5% CO2 incubator. Plasmid DNAs were transfected in 12-well plates (Sarstedt) using FuGENE HD Transfection Reagent (Promega) following the manufacturer’s instructions. Twenty-four hours post-transfection, cells were transferred to 6-well plates and cultured for 48 h before harvesting.
AAV vectors with serotype 2 were produced by VectorBuilder and used for transducing HEK293T/17 cells. Approximately 5 × 104 cells/well were seeded in a 24-well plate (Sarstedt) 24 h prior to transduction. On the day of transduction, AAV vectors were thawed on ice, and appropriate volumes were taken to achieve the desired MOI and added to 250 μl of DMEM with 2% FBS. After removing the existing culture medium, the AAV-containing medium was added to the cells. The cells were then incubated for 16–17 h in the cell culture incubator. Transduced cells were expanded in 6-well plates with complete culture medium and further incubated for 72 h before harvesting. For trimethoprim (TMP) treatment, transduced cells were expanded in 6-well plates with complete medium containing 10 μM TMP (Sigma), cultured for 48 h, and then switched to a normal culture medium for an additional 24 h before harvesting.
Mouse, animal study approval, and AAV subretinal injection
Wild-type C57BL/6J mice (strain #: 000664) were acquired from the Jackson laboratory. Ift140fl and iCre75 mice were generously provided by Dr Gregory J. Pazour (University of Massachusetts) and Dr Ching-Kang Chen (University of Texas Health Science Center at San Antonio), respectively [47–49]. Genotyping was performed as previously described [47, 48, 61, 63].
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Iowa and conducted in accordance with the recommendations outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animals were maintained in 12-hour light–dark cycles and fed standard mouse chow ad libitum.
For subretinal injections, AAV/Anc80-GRK1p-IFT140 vectors (L65 variant) were prepared by the Viral Vector Core Facility at the University of Iowa. AAV-CDH23 vectors were packaged in the AAV5 serotype and produced by VectorBuilder. On the day of injection, AAV vectors were thawed on ice and mixed/diluted to desired titers (8 × 108 GC/μl per vector for AAV-IFT140 and 3 × 109 GC/μl per vector for AAV-CDH23) in PBS supplemented with 0.001% Poloxamer 188 (Sigma Aldrich).
For the gene expression analysis in normal mice, 1-2-month-old wild-type C57BL/6J mice (both males and females) were used and at least 3 mice were injected per vector or vector set. For IFT140 subretinal gene therapy in Ift140 CKO mice, injections were administered at P16 in both males and females. Each animal received a subretinal injection in one eye, with the contralateral eye serving as an uninjected control. Mice were anesthetized with a ketamine/xylazine mixture (87.5 mg/kg ketamine, 12.5 mg/kg xylazine), and 10% povidone-iodine and 1% tropicamide solutions were applied. Under a Zeiss OPMI VISU 150 surgical microscope, eyes were gently rotated using forceps, and a conjunctival peritomy was made with Vannas scissors, followed by a sclerotomy made posterior to the limbus using a 30-gauge needle. Transscleral subretinal injections were performed with a Hamilton syringe attached to a blunt-end 32-gauge needle inserted subretinally at an oblique angle, delivering 1 μl of vector solution. After injections, an antibiotic/steroid ophthalmic ointment (neomycin and dexamethasone) was applied. Animals were excluded from follow-up analysis if no obvious blebs were observed or if significant hemorrhage occurred at the time of injection.
Protein extraction, SDS-PAGE, and immunoblotting
To extract proteins from HEK293T/17 cells, cells in 6-well plates were briefly rinsed with ice-cold PBS and lysed with 300 μl of ice-cold lysis buffer (50 mM HEPES pH 7.0, 150 mM NaCl, 2 mM EGTA, 2 mM MgCl2, 1% Triton X-100) supplemented with Protease Inhibitor Cocktail (Bimake). Lysates were centrifuged at 20 000 × g for 15 min at 4°C to precipitate insoluble materials.
To extract proteins from mouse retinas, mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Eyes were enucleated and anterior segments were removed using micro-dissecting scissors. Collected tissues were homogenized with a pestle in a lysis buffer (50 mM HEPES pH 7.0, 150 mM NaCl, 2 mM EGTA, 2 mM MgCl2, 1% Triton X-100) supplemented with the Protease Inhibitor Cocktail. Homogenates were centrifuged at 20 000 × g for 15 min at 4°C to precipitate insoluble materials.
Supernatants were mixed with NuPAGE LDS Sample Buffer (Invitrogen) and Reducing Agent (Invitrogen), and 40–50 μg of proteins were loaded per lane on NuPAGE 4%–12% (wt/vol) Bis-Tris gels (Invitrogen; for all genes except CDH23) or 3%–8% Tris-Acetate gels (Invitrogen; for CDH23). Proteins were transferred onto nitrocellulose membranes (BioRad) and immunoblotting was performed following standard protocols. Antibodies used are listed in Table S1. Proteins were detected by using horse radish peroxidase (HRP)-conjugated secondary antibodies and SuperSignal West Dura Extended Duration Substrate (Thermo Scientific). Images were taken with a ChemiDoc Imaging system (Bio-Rad).
Immunohistochemistry
Mouse eyes were collected, fixed, and embedded in Neg-50 Frozen Section Medium (Richard-Allan Scientific) as described previously [61, 63]. The frozen eye cups were mounted on a cryostat chuck and oriented to obtain sections along the superior–inferior axis. Retinal sections (8-μm thick) from the middle half of the eye cups were collected at intervals of every 5–6 sections using a CryoStar NX70 cryostat (Epredia) and used for immunohistochemistry. The immunostaining and imaging procedures were described previously [61].
Electroretinography (ERG)
The ERG recording procedures were described previously [64]. Scotopic and photopic ERG responses were measured with 15 flashes of stimulus light at 0.01 cd⋅s/m2 and 3.0 cd⋅s/m2, respectively (color temperature: 6500 K). Statistical analyses were performed using two-tailed, two-sample t-tests with unequal variances. A P-value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Gregory J. Pazour (University of Massachusetts) and Ching-Kang Chen (University of Texas Health Science Center at San Antonio) for generously providing Ift140fl and iCre75 transgenic mice.
Contributor Information
Poppy Datta, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Kun-Do Rhee, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Rylee J Staudt, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Jacob M Thompson, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Ying Hsu, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Salma Hassan, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Arlene V Drack, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Department of Pediatrics, The University of Iowa Carver College of Medicine, 200 Hawkins Drive, Iowa City, IA 52242, United States.
Seongjin Seo, Department of Ophthalmology and Visual Sciences, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States; Institute for Vision Research, The University of Iowa Carver College of Medicine, 375 Newton Road, Iowa City, IA 52242, United States.
Author contributions
Conception and study design: S.S., vector design: S.S., materials: P.D., K.D.R., R.J.S., subretinal injection: P.D., K.D.R., J.M.T., Y.H., S.H., A.V.D., data collection: P.D., K.D.R., S.S., data analysis: S.S., funding acquisition: S.S., A.V.D., manuscript drafting: S.S., manuscript revision: K.D.R., Y.H., and A.V.D.
Conflict of interest statement: S.S. and P.D. are co-inventors on a patent application (PCT/US23/77973, filed by the University of Iowa) titled “Method to deliver large genes using virus and a DNA recombination system.” The remaining authors declare no competing interests.
Funding
This work was supported by National Institutes of Health grant R01-EY034176 (to S.S.), Retina Research Foundation Pilot grant (to S.S.), Flagella Vision Foundation (to S.S. and A.V.D.), and the Ronald Keech professorship (to A.V.D.). This work was also supported in part by National Institutes of Health grant P30-EY025580 to the University of Iowa.
References
- 1. Bennett J. Immune response following intraocular delivery of recombinant viral vectors. Gene Ther 2003;10:977–982. [DOI] [PubMed] [Google Scholar]
- 2. Lugin ML, Lee RT, Kwon YJ. Synthetically engineered adeno-associated virus for efficient, safe, and versatile gene therapy applications. ACS Nano 2020;14:14262–14283. [DOI] [PubMed] [Google Scholar]
- 3. Asokan A, Schaffer DV, Samulski RJ. The AAV vector toolkit: poised at the clinical crossroads. Mol Ther 2012;20:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zinn E, Pacouret S, Khaychuk V. et al. In Silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep 2015;12:1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther 2010;18:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stone EM, Andorf JL, Whitmore SS. et al. Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology 2017;124:1314–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. den Hollander AI, Koenekoop RK, Yzer S. et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006;79:556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perrault I, Delphin N, Hanein S. et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat 2007;28:416. [DOI] [PubMed] [Google Scholar]
- 9. Baala L, Audollent S, Martinovic J. et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet 2007;81:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valente EM, Silhavy JL, Brancati F. et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet 2006;38:623–625. [DOI] [PubMed] [Google Scholar]
- 11. Sayer JA, Otto EA, O'Toole JF. et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 2006;38:674–681. [DOI] [PubMed] [Google Scholar]
- 12. Coppieters F, Lefever S, Leroy BP. et al. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat 2010;31:1097–1108. [DOI] [PubMed] [Google Scholar]
- 13. Bolz H, von Brederlow B, Ramirez A. et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes usher syndrome type 1D. Nat Genet 2001;27:108–112. [DOI] [PubMed] [Google Scholar]
- 14. Di Palma F, Holme RH, Bryda EC. et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for usher syndrome type 1D. Nat Genet 2001;27:103–107. [DOI] [PubMed] [Google Scholar]
- 15. Trapani I, Puppo A, Auricchio A. Vector platforms for gene therapy of inherited retinopathies. Prog Retin Eye Res 2014;43:108–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trapani I, Tornabene P, Auricchio A. Large gene delivery to the retina with AAV vectors: are we there yet? Gene Ther 2021;28:220–222. [DOI] [PubMed] [Google Scholar]
- 17. Ghosh A, Yue Y, Lai Y. et al. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol Ther 2008;16:124–130. [DOI] [PubMed] [Google Scholar]
- 18. Dyka FM, Boye SL, Chiodo VA. et al. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum Gene Ther Methods 2014;25:166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carvalho LS, Turunen HT, Wassmer SJ. et al. Evaluating efficiencies of dual AAV approaches for retinal targeting. Front Neurosci 2017;11:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dyka FM, Molday LL, Chiodo VA. et al. Dual ABCA4-AAV vector treatment reduces pathogenic retinal A2E accumulation in a mouse model of autosomal recessive Stargardt disease. Hum Gene Ther 2019;30:1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barrett B, Kerns S, Kevany B. et al. Dual AAV vector strategy for expression of large genes targeted for Stargardt disease gene therapy development. Invest Ophthalmol Vis Sci 2021;62:1484. [Google Scholar]
- 22. Riedmayr LM, Hinrichsmeyer KS, Thalhammer SB. et al. mRNA trans-splicing dual AAV vectors for (epi)genome editing and gene therapy. Nat Commun 2023;14:6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah NH, Muir TW. Inteins: Nature's gift to protein chemists. Chem Sci 2014;5:446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 1998;95:9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iwai H, Zuger S, Jin J. et al. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett 2006;580:1853–1858. [DOI] [PubMed] [Google Scholar]
- 26. Trapani I, Colella P, Sommella A. et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol Med 2014;6:194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapani I, Toriello E, de Simone S. et al. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum Mol Genet 2015;24:6811–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McClements ME, Barnard AR, Singh MS. et al. An AAV dual vector strategy ameliorates the Stargardt phenotype in adult Abca4(−/−) mice. Hum Gene Ther 2019;30:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Villiger L, Grisch-Chan HM, Lindsay H. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med 2018;24:1519–1525. [DOI] [PubMed] [Google Scholar]
- 30. Tornabene P, Trapani I, Minopoli R. et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci Transl Med 2019;11. 10.1126/scitranslmed.aav4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chong S, Williams KS, Wotkowicz C. et al. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J Biol Chem 1998;273:10567–10577. [DOI] [PubMed] [Google Scholar]
- 32. Shah NH, Eryilmaz E, Cowburn D. et al. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J Am Chem Soc 2013;135:5839–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoess RH, Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA 1984;81:1026–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res 1986;14:2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoess RH, Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 1985;181:351–362. [DOI] [PubMed] [Google Scholar]
- 36. Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 1998;216:55–65. [DOI] [PubMed] [Google Scholar]
- 37. Siegel RW, Jain R, Bradbury A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett 2001;505:467–473. [DOI] [PubMed] [Google Scholar]
- 38. Livet J, Weissman TA, Kang H. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007;450:56–62. [DOI] [PubMed] [Google Scholar]
- 39. Missirlis PI, Smailus DE, Holt RA. A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination. BMC Genomics 2006;7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomson JG, Rucker EB 3rd, Piedrahita JA. Mutational analysis of loxP sites for efficient Cre-mediated insertion into genomic DNA. Genesis 2003;36:162–167. [DOI] [PubMed] [Google Scholar]
- 41. Albert H, Dale EC, Lee E. et al. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 1995;7:649–659. [DOI] [PubMed] [Google Scholar]
- 42. Xu M, Yang L, Wang F. et al. Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum Genet 2015;134:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hull S, Owen N, Islam F. et al. Nonsyndromic retinal dystrophy due to Bi-allelic mutations in the ciliary transport gene IFT140. Invest Ophthalmol Vis Sci 2016;57:1053–1062. [DOI] [PubMed] [Google Scholar]
- 44. Perrault I, Saunier S, Hanein S. et al. Mainzer-Saldino syndrome is a ciliopathy caused by IFT140 mutations. Am J Hum Genet 2012;90:864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Earley LF, Conatser LM, Lue VM. et al. Adeno-associated virus serotype-specific inverted terminal repeat sequence role in vector transgene expression. Hum Gene Ther 2020;31:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khani SC, Pawlyk BS, Bulgakov OV. et al. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest Ophthalmol Vis Sci 2007;48:3954–3961. [DOI] [PubMed] [Google Scholar]
- 47. Jonassen JA, SanAgustin J, Baker SP. et al. Disruption of IFT complex a causes cystic kidneys without mitotic spindle misorientation. J Am Soc Nephrol 2012;23:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crouse JA, Lopes VS, Sanagustin JT. et al. Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton (Hoboken) 2014;71:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S, Chen D, Sauve Y. et al. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 2005;41:73–80. [DOI] [PubMed] [Google Scholar]
- 50. Ahmed ZM, Riazuddin S, Bernstein SL. et al. Mutations of the protocadherin gene PCDH15 cause usher syndrome type 1F. Am J Hum Genet 2001;69:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alagramam KN, Murcia CL, Kwon HY. et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet 2001;27:99–102. [DOI] [PubMed] [Google Scholar]
- 52. Alagramam KN, Yuan H, Kuehn MH. et al. Mutations in the novel protocadherin PCDH15 cause usher syndrome type 1F. Hum Mol Genet 2001;10:1709–1718. [DOI] [PubMed] [Google Scholar]
- 53. Chang B, Khanna H, Hawes N. et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet 2006;15:1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cideciyan AV, Aleman TS, Jacobson SG. et al. Centrosomal-ciliary gene CEP290/NPHP6 mutations result in blindness with unexpected sparing of photoreceptors and visual brain: implications for therapy of Leber congenital amaurosis. Hum Mutat 2007;28:1074–1083. [DOI] [PubMed] [Google Scholar]
- 55. Frank V, den Hollander AI, Bruchle NO. et al. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum Mutat 2008;29:45–52. [DOI] [PubMed] [Google Scholar]
- 56. Iwamoto M, Bjorklund T, Lundberg C. et al. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol 2010;17:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sando R 3rd, Baumgaertel K, Pieraut S. et al. Inducible control of gene expression with destabilized Cre. Nat Methods 2013;10:1085–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmidt EE, Taylor DS, Prigge JR. et al. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA 2000;97:13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seo S, Guo DF, Bugge K. et al. Requirement of Bardet-Biedl syndrome proteins for leptin receptor Signaling. Hum Mol Genet 2009;18:1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seo S, Zhang Q, Bugge K. et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and smoothened. PLoS Genet 2011;7:e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Datta P, Hendrickson B, Brendalen S. et al. The myosin-tail homology domain of centrosomal protein 290 is essential for protein confinement between the inner and outer segments in photoreceptors. J Biol Chem 2019;294:19119–19136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Humbert MC, Weihbrecht K, Searby CC. et al. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA 2012;109:19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Datta P, Ruffcorn A, Seo S. Limited time window for retinal gene therapy in a preclinical model of ciliopathy. Hum Mol Genet 2020;29:2337–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Datta P, Cribbs JT, Seo S. Differential requirement of NPHP1 for compartmentalized protein localization during photoreceptor outer segment development and maintenance. PLoS One 2021;16:e0246358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.