Abstract
Background and Aims
Deep learning applied to electrocardiograms (ECG-AI) is an emerging approach for predicting atrial fibrillation or flutter (AF). This study introduces an ECG-AI model developed and tested at a tertiary cardiac centre, comparing its performance with clinical models and AF polygenic score (PGS).
Methods
Electrocardiograms in sinus rhythm from the Montreal Heart Institute were analysed, excluding those from patients with pre-existing AF. The primary outcome was incident AF at 5 years. An ECG-AI model was developed by splitting patients into non-overlapping data sets: 70% for training, 10% for validation, and 20% for testing. The performance of ECG-AI, clinical models, and PGS was assessed in the test data set. The ECG-AI model was externally validated in the Medical Information Mart for Intensive Care-IV (MIMIC-IV) hospital data set.
Results
A total of 669 782 ECGs from 145 323 patients were included. Mean age was 61 ± 15 years, and 58% were male. The primary outcome was observed in 15% of patients, and the ECG-AI model showed an area under the receiver operating characteristic (AUC-ROC) curve of .78. In time-to-event analysis including the first ECG, ECG-AI inference of high risk identified 26% of the population with a 4.3-fold increased risk of incident AF (95% confidence interval: 4.02–4.57). In a subgroup analysis of 2301 patients, ECG-AI outperformed CHARGE-AF (AUC-ROC = .62) and PGS (AUC-ROC = .59). Adding PGS and CHARGE-AF to ECG-AI improved goodness of fit (likelihood ratio test P < .001), with minimal changes to the AUC-ROC (.76–.77). In the external validation cohort (mean age 59 ± 18 years, 47% male, median follow-up 1.1 year), ECG-AI model performance remained consistent (AUC-ROC = .77).
Conclusions
ECG-AI provides an accurate tool to predict new-onset AF in a tertiary cardiac centre, surpassing clinical and PGS.
Keywords: Atrial fibrillation, Deep learning, Electrocardiogram, Polygenic scores
Structured Graphical Abstract
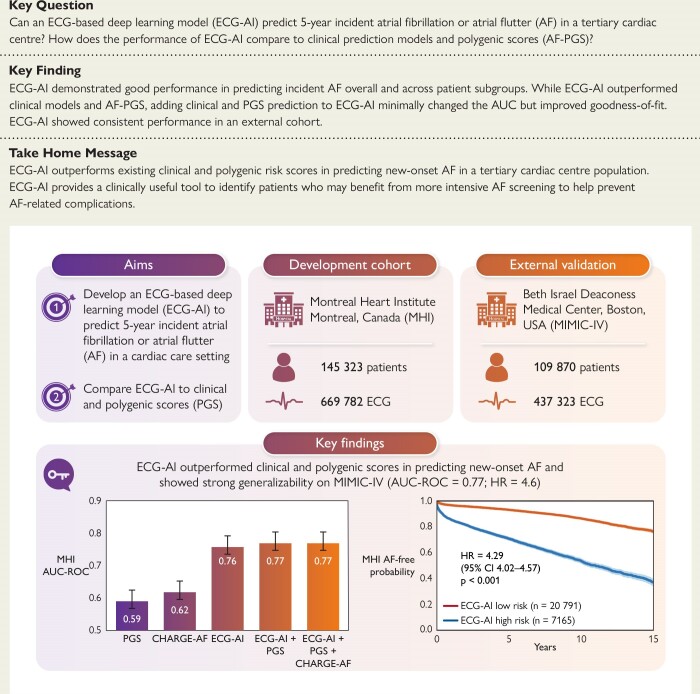
Structured Graphical Abstract.
An ECG-AI model trained at the MHI (a tertiary cardiac centre) predicts 5-year incident atrial fibrillation or flutter (AF) in an internal independent test data set (MHI; AUC-ROC .78) and an external population (MIMIC-IV; AUC-ROC .77). The ECG-AI outperforms existing clinical (CHARGE-AF) and polygenic scores (PGS). Adding PGS and CHARGE-AF to ECG-AI improved goodness of fit (likelihood ratio test P < .001), with minimal changes to the AUC-ROC (.76–.77). Created with Biorender.com. HR, hazard ratio; MIMIC-IV, Medical Information Mart for Intensive Care-IV; AUC-ROC, area under the receiver operating characteristic curve.
See the editorial comment for this article ‘Another piece in the puzzle of atrial fibrillation risk: clinical, genetic, and electrocardiogram-based artificial intelligence', by S. Kany et al., https://doi.org/10.1093/eurheartj/ehae691.
Introduction
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia in adults1 and is associated with an increased risk of stroke, heart failure, cognitive decline, hospitalizations, and death.2–6 Oral anticoagulation (OAC) significantly reduces the risk of stroke in patients with AF.7,8 Early rhythm-control strategies have also been shown to be associated with a lower risk of adverse cardiovascular outcomes compared with initial rate-control strategies.9 Importantly, it has been estimated that in about one-third of patients, AF is asymptomatic, contributing to under-detection in a large proportion of patients.10 Early detection of AF provides an opportunity to implement measures aimed at the primary prevention of AF-related morbidity and mortality with an early initiation of appropriate therapy, including OAC when indicated, and risk factor modification. Improving the ability to identify individuals at high risk of developing AF can help define populations in whom more intensive AF screening would be cost-effective.11
Simple predictive scores for new-onset AF, such as CHARGE-AF (Cohorts for Aging and Research in Genomic Epidemiology–Atrial Fibrillation) and HATCH, have been explored for that purpose.12,13 Moreover, the CHA2DS2-VASc score, which was developed to predict the risk of stroke in AF patients, has also been shown to predict incident AF.14 Recent technological advances in artificial intelligence (AI) have enabled the development of novel AF prediction models, which have shown promising performances in predicting new-onset AF using single or multiple lead sinus rhythm electrocardiograms (ECGs) in primary care settings or across diverse hospital network populations.15–20 Furthermore, recent studies suggest a good predictive ability of AF polygenic scores (AF-PGS) in a primarily general population, with a possible additive value to clinical AF prediction models.21–25
In this study, an open-weight ECG-AI model is introduced to predict incident AF at the Montreal Heart Institute (MHI), Canada. The analysis plan and reporting adhere to key quality criteria recently developed for clinical AI prediction modelling studies.26–28 This study stands out by exploring innovative risk markers of AF (ECG-AI and AF-PGS) within a tertiary cardiac care institution with a high prevalence of heart failure (HF) and coronary artery disease (CAD), where AF is more prevalent, is associated with increased risk for complications, and may be mediated by distinct risk markers compared with the general population.29,30 Moreover, unlike most previously published literature on ECG-AI for incident AF prediction, the presented ECG-AI model is open-weight (i.e. publicly available) to spur further innovation and improve accessibility in ECG-AI technology, potentially accelerating advancements in AF prediction and management strategies. Furthermore, the study evaluates the ECG-AI model across diverse populations, including variations in socio-economic status, age, and sex, thereby addressing the important issue of bias in AI applications.31 A comprehensive assessment of the ECG-AI performance using discrimination, calibration, decision curve, and time-to-event analyses and confirming its generalizability in an external cohort was conducted. Finally, a novel approach of combining ECG-AI, AF-PGS, and traditional clinical prediction in AF risk stratification is assessed.
Methods
Study population
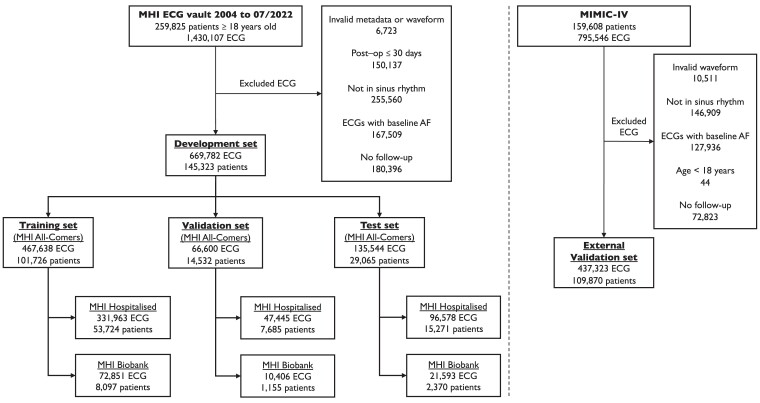
In a retrospective cohort, all ECGs in the MHI database acquired between 2004 and 2022 were considered. Electrocardiograms were excluded if they had invalid metadata or waveform (i.e. missing derivations or erroneous signals defined by a maximum voltage > 10 mV) or showed no sinus rhythm (see Supplementary data online, Figure S1). Electrocardiograms were also excluded if performed within 30 days of cardiac surgery, were acquired in patients with pre-existing AF or atrial flutter (using the same definition as the outcome, see next section), or were acquired in patients without subsequent follow-up at MHI (Figure 1). The remaining ECGs were then randomly split by distributing patients into non-overlapping training (70%), validation (10%), and test (20%) sets, stratified according to age, sex, and outcome ensuring balanced distribution of these variables among data sets (see Supplementary data online, Tables S1–S3). In each set, three groups were defined (Table 1). The ‘MHI All-Comers’ group included all the ECGs in each set. Two subgroups of the ‘MHI All-Comers’ are defined: the ‘MHI Hospitalized’ group included the ECGs of patients who were hospitalized at the MHI and the ‘MHI Biobank’ included the subset of patients in the MHI hospital biobank, a prospective hospital-based cohort of >20 000 participants of which 16 876 have available genotypic data. Detailed comorbidities are only reported for the ‘MHI Hospitalized’ group, in whom clinical diagnoses were ascertained using International Classification of Diseases (ICD) codes (see Supplementary data online, Table S4).
Figure 1.
Electrocardiogram and patient flowchart for the Montreal Heart Institute cohort and the external validation cohort Medical Information Mart for Intensive Care-IV (MIMIC-IV). A single ResNet-50 model initialized with random weights was trained using the training set. Hyperparameter tuning was performed using the validation set. The best performing model in the validation set was selected based on the lowest loss, and then, this model performance was reported on three subgroups within the test set, i.e. ‘MHI All-Comers’, ‘MHI Hospitalized’, and ‘MHI Biobank’. For the latter group, after removing patients with missing data, CHARGE-AF and AF-PGS scores were available for 2301 out of the 2370 patients. External validation was performed in the MIMIC-IV data set from the Beth Israel Deaconess Medical Center in Boston, USA
Table 1.
Demographic and clinical characteristics of the development and external validation data sets
| MHI All-Comers | MHI Hospitalized | MHI Biobank | MIMIC-IV | |
|---|---|---|---|---|
| ECG | 669 782 | 475 986 | 104 850 | 437 323 |
| Patients | 145 323 | 76 680 | 11 622 | 109 870 |
| ECG per patient | 2.0 (Q1: 1.0, Q3: 5.0) | 3.0 (Q1: 2.0, Q3: 8.0) | 6.0 (Q1: 3.0, Q3: 12.0) | 2.0 (Q1: 1.0, Q3: 4.0) |
| Patient-level data | ||||
| Age (years) | 61.3 (± 15.2) | 64.3 (± 13.7) | 63.2 (± 11.6) | 59.2 (± 17.9) |
| Male | 84 087 (57.9%) | 50 114 (65.4%) | 7326 (63.0%) | 51 627 (47%) |
| CIMD | 3.2 (± 1.4) | 3.2 (± 1.3) | 3.2 (± 1.4) | |
| MHI Hospitalized | 76 680 (52.8%) | 76 680 (100.0%) | 7919 (68.1%) | |
| Follow-up (years) | 3.2 (Q1: .3, Q3: 8.6) | 4.0 (Q1: .3, Q3: 9.7) | 9.6 (Q1: 4.6, Q3: 13.6) | 1.1 (Q1: .03, Q3: 4.7) |
| 5-year incident AF | 22 695 (15.6%) | 18 492 (24.1%) | 2846 (24.5%) | 16 610 (15.1%) |
| ECG-level data | ||||
| Age (years) | 62.8 (± 14.8) | 64.2 (± 14.1) | 64.0 (± 12.0) | 61.2 (± 16.5) |
| Follow-up (years) | 4.2 (Q1: 1.2, Q3: 8.3) | 4.3 (Q1: 1.1, Q3: 8.6) | 6.6 (Q1: 3.2, Q3: 10.5) | 1.7 (Q1: .2, Q3: 4.5) |
| 5-year incident AF | 80 183 (12.0%) | 70 230 (14.8%) | 13 772 (13.1%) | 65 301 (14.9%) |
| Years to incident AF | 2.0 (Q1: .1, Q3: 5.6) | 1.9 (Q1: .1, Q3: 5.5) | 3.3 (Q1: .7, Q3: 7.0) | 1.1 (Q1: .1, Q3: 3.3) |
‘MHI All-Comers’ group includes all the development set electrocardiograms. ‘MHI Hospitalized’ group includes the electrocardiograms of patients who have been hospitalized at the MHI. ‘MHI Biobank’ includes the subset of patients in the Montreal Heart Institute hospital biobank. ‘MIMIC-IV’ includes all eligible patients and electrocardiograms of the external validation data set. The number of electrocardiograms per patient, follow-up duration, and time to incident atrial fibrillation are provided in quartiles format, indicating the median, 25th percentile (Q1), and 75th percentile (Q3). Age and Canadian Index for Multiple Deprivation (CIMD) are presented as mean ± standard deviation.
Outcome
The primary outcome, termed ‘incident AF’, included new-onset AF or atrial flutter. Incident AF at 5 years was modelled as a binary outcome and was determined based on available outpatient and inpatient clinical and medico-administrative databases and ECG diagnoses, which incorporated ECG acquisitions, hospitalization records, emergency room visits, AF clinic visits, and electrophysiology procedures (see Supplementary data online, Figure S2). The sensitivity and specificity of this definition of incident AF were assessed in 200 randomly selected patients using manual chart reviews as a gold standard. The same clinical and administrative databases were used as eligible follow-up encounters to establish maximum follow-up time, with censoring at the date of last follow-up at MHI, heart transplantation, or death.
Electrocardiogram acquisition
Electrocardiograms were retrieved in XML format using the MUSE Cardiology Information System (GE Healthcare, Chicago, IL). Each XML file contains data for 12 ECG derivations, with each derivation capturing voltage readings over a 10 s period sampled at 250 Hz. Each voltage was standardized by removing the mean and scaling to unit variance of the training set population voltages. Since this scaling method is inherently sensitive to outliers, ECGs with extreme voltage values (>10 mV) were considered to be outliers and discarded from the data set.
Electrocardiogram-based deep learning model
A single ResNet-50 model32 initialized with random weights was trained in the training set using four A6000 GPUs (NVIDIA, Santa Clara, CA, USA). The model receives a single 12-lead ECG as input, with a duration of 10 s per lead at a sampling rate of 250 Hz. Multiple ECG recordings from the same patient were independently fed into the training model. Hyperparameters were optimized on the validation set using a Bayesian grid-search approach. The best performing model in the validation set was selected based on the lowest loss, and then, this model performance was reported on three subgroups within the internal MHI test set, i.e. ‘MHI All-Comers’, ‘MHI Hospitalized’, and ‘MHI Biobank’. Using TensorFlow’s GradientTape (version 2.9.1), the gradient of the model’s prediction was computed with respect to the input ECG sample, resulting in a saliency map that highlights the most influential parts of the ECG signal, thereby providing explainability.33,34 The ECG-AI development details are provided in the Supplementary data online, Note S1.
Clinical risk models
Four different clinical risk models were tested, including ‘Age & Sex’, HATCH, CHA2DS2-VASc, and CHARGE-AF (details in Supplementary data online, Tables S5 and S6).12–14 Clinical risk scores were incorporated into logistic regression (LR) models, which were fitted using the training and validation sets. The CHARGE-AF score was only calculated for patients in the MHI biobank cohort at the time of inclusion in the biobank, since some components were not available in the other patient subgroups. The ECG-AI prediction based on the single ECG closest to inclusion in the MHI biobank (and CHARGE-AF calculation) was used to compare the predictions of ECG-AI with those of CHARGE-AF.
Polygenic score calculation in the Montreal Heart Institute biobank
The predictive ability of AF-PGS, both alone and in combination with ECG-AI and CHARGE-AF, was assessed in the MHI biobank cohort. The previously published AF-PGS from Khera et al.35 (PGS catalogue ID PGS000016) was converted to GRCh38 genomic build. The MHI biobank cohort previously underwent array genotyping on the Illumina Global Screening Array followed by standard genotypic quality control and genome-wide imputation on the TOPMed reference panel. The AF-PGS was computed using weights from PGS000016, including 6 502 964 single-nucleotide polymorphisms out of 6 730 541 in the original score (97%). The raw AF-PGS was standardized using a standard scaler, followed by a logistic transformation to convert the values to a range between 0 and 1. The single ECG closest to enrolment in the MHI biobank was used to compare ECG-AI predictions with AF-PGS predictions.
Statistical analysis, performance metrics, and reporting
After ECG-AI training, a LR model was used to integrate ECG-AI probability predictions with clinical and polygenic scores. The LR model was fitted on the training and validation sets and tested on the test set. Model performance was assessed using several metrics in the test set. Discrimination performance is reported using the area under the receiver operating characteristic (AUC-ROC) curve, the area under the precision–recall curve (PRC), and the diagnostic odds ratio (DOR). The DOR is the ratio of the odds of disease in test positives relative to the odds of disease in test negatives.36
Calibration was assessed using calibration curves by fitting a spline to the calibration data using the UnivariateSpline function from the SciPy Python library (version 1.10.1) with a smoothing factor of 1. To quantify the calibration performance, the estimated calibration index (ECI) was computed as the root mean squared difference between the mean predicted probabilities and the spline-fitted calibration curve.37
Decision analysis curves (DCAs) were constructed by plotting net benefit (NB) against various threshold probabilities, considering both discrimination and calibration.38 The NB was computed at different decision thresholds as follows, where N is the total number of samples and t is the threshold probability:
In a deployment scenario, a predictive model would be used to guide downstream intensive AF screening in high-risk populations. Therefore, the NB was compared with the default policies of ‘Screen None’ or ‘Screen All’ for AF. The ‘Screen None’ NB is 0 given that TP and FP are 0. ‘Screen All’ implies no perceived downsides of over-screening. To calculate the ‘Screen All’ NB, (TP/N) was replaced by the prevalence and (FP/N) by (1 − prevalence). For instance, with a 10% AF event rate, ‘Screen All’ implies 10% correct and 90% incorrect classification at a given threshold. Sensitivity, specificity, and DOR were calculated at the event rate threshold in each group that maximizes the NB and considered to be an optimal threshold.
Finally, time-to-event analyses were conducted in which time 0 was defined as the date of each patient’s first ECG. For the MHI Biobank subset, the ECG closest to biobank enrolment was used. This approach was chosen to simulate a prospective deployment scenario with the longest possible follow-up and to provide a fair comparison with CHARGE-AF, calculated at biobank enrolment. An exploratory analysis was also conducted by choosing the ECG generating the highest predicted AF probability as time 0. Stratification into high-risk and low-risk groups was determined using predictions from the ECG-AI model at the chosen optimal classification threshold. Survival curves were estimated using the Kaplan–Meier (KM) method. The survival distributions between high-risk and low-risk groups were compared using the log-rank test. Hazard ratios (HR) between the two groups were calculated by fitting a Cox proportional hazards model after verifying proportionality assumptions.
All results are reported on the test set which excludes patients included in the training and validation sets. The ECG-AI prediction was reported at the ECG level, whereby multiple ECG recordings from the same patient were independently fed into the training model. The ECG-AI prediction was also reported at the patient level by averaging the model’s probability outputs for ECGs grouped according to both their 5-year AF outcome and the patient’s identity (see Supplementary data online, Figure S3).
Confidence intervals (CIs) are reported using bootstrapping with 1000 iterations. For normally distributed data, results are presented as mean ± standard deviation. For non-normally distributed data, results are presented using quartiles. The DeLong method was used to statistically compare the ROC curves of different predictive models.39 To evaluate the improvement in the model’s goodness of fit with the addition of new variables to ECG-AI, a log-likelihood ratio test (LRT) was conducted.
Data analysis and visualization were performed using Python (version 3.8) with the following libraries: scikit-learn (version 1.3.2), lifelines (version 0.27.8), matplotlib (version 3.7.5), and seaborn (version 0.13.2).
Subgroup analyses
The study aimed to ensure that ECG-AI performance remained consistent across diverse patient populations. For this purpose, pre-defined subgroup analyses were performed by stratifying the testing data set by sex (male and female), age (<65 and ≥65 years), and socio-economic status. The latter was assessed using the Canadian Index for Multiple Deprivation (CIMD), a measure of socio-economic conditions based on the 2021 Canadian Census of Population microdata and derived using patient postal codes.40 The composite summary score ranges from 1 to 5, with 1 representing the least deprived and 5 representing the most deprived. Furthermore, the performance of ECG-AI was tested in subgroups with and without the two most common cardiac conditions, namely, HF and CAD, defined using ICD codes in the ‘MHI Hospitalized’ subgroup of the test data set (see Supplementary data online, Table S4).
External validation
To investigate the generalizability of the ECG-AI model outside the MHI, an external validation analysis was performed using the Medical Information Mart for Intensive Care (MIMIC)-IV, a large de-identified data set of patients admitted to the emergency department or an intensive care unit at the Beth Israel Deaconess Medical Center in Boston, USA.41–43 Similar ECG inclusion/exclusion criteria from the MHI data set were used. Electrocardiogram voltages were standardized using a standard scaler, adjusting for the external validation set by removing the mean and scaling to unit variance. Incident AF at 5 years (the primary outcome) was modelled as a binary outcome and determined based on ECG and hospitalization diagnoses. Performance of ECG-AI in this external data set was also assessed using AUC-ROC, PRC, calibration, DCA, and time-to-event analyses as described above.
Results
Description of the Montreal Heart Institute study population
A total of 669 782 ECGs (47% of the screened ECGs) acquired from 145 323 patients met the inclusion criteria (Figure 1). In the ‘MHI All-Comers’ group, the mean age was 61 ± 15 years and 58% of the patients were male. Each patient had a median of two ECGs [first quartile (Q1): 1, third quartile (Q3): 5]. The 5-year incident AF outcome was observed in 12.0% of ECGs and 15.6% of patients. In a validation study where all available medical records were manually retrieved and reviewed for a randomly selected subset of 200 patients, the specificity and sensitivity for classifying the primary AF outcome were 100% and 91% (95% CI: 83.9–98.6), respectively (see Supplementary data online, Table S7). The median time to incident AF was 2 years (Q1: .1, Q3: 5.6). The ‘MHI Hospitalized’ group included 71% of the ECGs and 53% of the patients from the ‘MHI All-Comers’ group (Table 1). Clinical characteristics for the ‘MHI Hospitalized’ group were comparable among the training, validation, and test sets (Table 2). The MHI cohort had a prevalence of CAD of 71.4% and HF of 13.4%.
Table 2.
Detailed patient comorbidities for MHI Hospitalized group (overall, training, validation, and test subgroups) and the external validation cohort (MIMIC-IV)
| Overall | Training | Validation | Test | MIMIC-IV | |
|---|---|---|---|---|---|
| Number of patients | 76 680 | 53 724 | 7685 | 15 271 | 109 870 |
| Heart failure | 10 260 (13.4%) |
7214 (13.4%) |
999 (13.0%) |
2047 (13.4%) |
15 273 (13.9%) |
| Coronary artery disease | 54 725 (71.4%) |
38 362 (71.4%) |
5455 (71.0%) |
10 908 (71.4%) |
25 643 (23.3%) |
| Chronic obstructive pulmonary disease | 8952 (11.7%) |
6285 (11.7%) |
892 (11.6%) |
1775 (11.6%) |
10 479 (9.5%) |
| Hypertension | 48 285 (63.0%) |
33 777 (62.9%) |
4805 (62.5%) |
9703 (63.5%) |
60 705 (55.3%) |
| Diabetes | 20 682 (27.0%) |
14 507 (27.0%) |
2086 (27.1%) |
4089 (26.8%) |
25 630 (23.3%) |
| Stroke | 847 (1.1%) |
594 (1.1%) |
82 (1.1%) |
171 (1.1%) |
4589 (4.2%) |
| Dyslipidaemia | 50 978 (66.5%) |
35 688 (66.4%) |
5167 (67.2%) |
10 123 (66.3%) |
44 440 (40.4%) |
| Obesity | 20 621 (26.9%) |
14 520 (27.0%) |
2059 (26.8%) |
4042 (26.5%) |
15 003 (13.7%) |
| Chronic kidney disease | 10 360 (13.5%) |
7290 (13.6%) |
1049 (13.6%) |
2021 (13.2%) |
15 750 (14.3%) |
| Sleep apnoea | 4482 (5.8%) |
3122 (5.8%) |
458 (6.0%) |
902 (5.9%) |
11 101 (10.1%) |
| Hyperthyroidism | 358 (.5%) |
255 (.5%) |
32 (.4%) |
71 (.5%) |
1064 (1.0%) |
| Vascular disease | 8169 (10.7%) |
5694 (10.6%) |
829 (10.8%) |
1646 (10.8%) |
8021 (7.3%) |
MIMIC-IV, Medical Information Mart for Intensive Care-IV.
Performance results for each of the pre-defined prediction models in the test set are summarized in Supplementary data online, Table S8, and further described below.
Atrial fibrillation prediction in MHI All-Comers using ECG-based deep learning, age, and sex
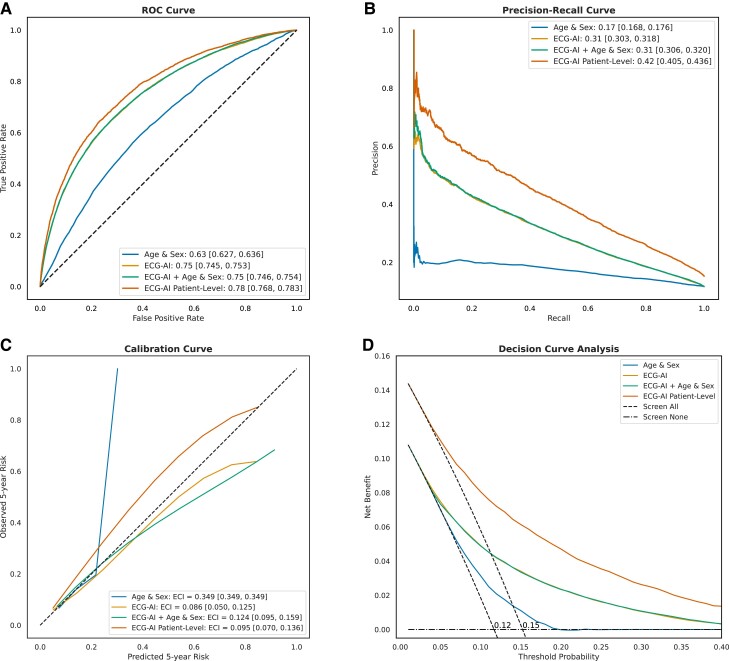
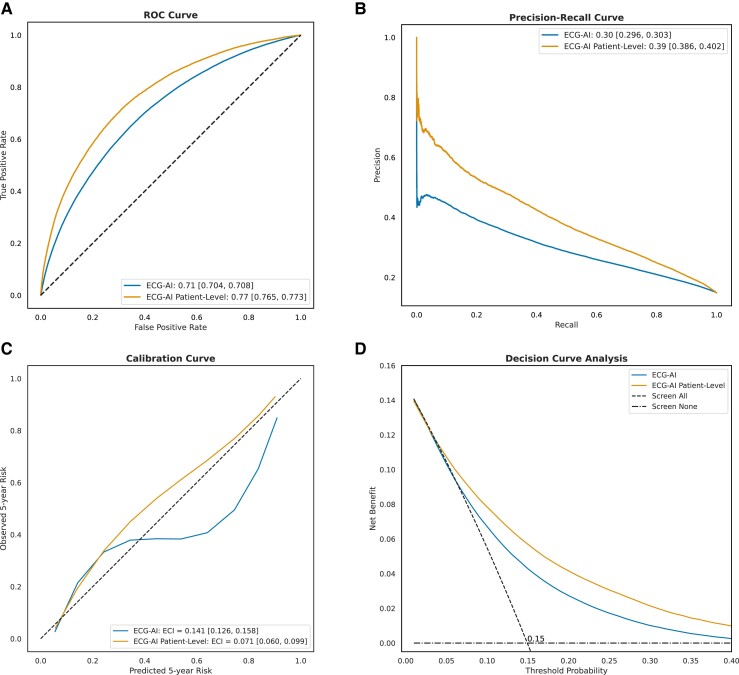
The ECG-AI model demonstrated good discriminative ability in the test set to identify incident AF with significantly better performance compared with the Age & Sex LR model, with a higher AUC-ROC curve of .75 (95% CI: .745–.753) vs. .63 (95% CI: .627–.636; P < .001) and improved precision–recall area of .31 (95% CI: .30–.32) vs. .17 (95% CI: .168–.176) (Figure 2). The ECG-AI model also exhibited the best calibration with an ECI of .086. Adding age and sex to the ECG-AI model as a post-training LR model yielded a similar AUC-ROC (.75) and did not provide an overall better fit (LRT statistic < 0, P = 1). At the patient level, the ECG-AI model showed the highest AUC-ROC of .78 (95% CI: .768–.783) and precision–recall area of .42 (95% CI: .41–.44).
Figure 2.
MHI All-Comers test set (29 065 patients, 135 544 ECG) performance assessment of the four models: (i) Age & Sex logistic regression, (ii)Electrocardiogram-based deep learning (ECG-AI), (iii) ECG-AI + Age & Sex, and (iv) ECG-AI patient level. ECG-AI and ECG-AI + Age & Sex overlap in A, B, and D. (A) The receiver operating characteristic curve, plotting the true positive rate against the false positive rate for each model, with the area under the curve indicating discriminatory power and reported in the legend. (B) The precision–recall curve, plotting precision against recall, with the area under the curve reported in the legend. (C) The calibration curve, showing the relationship between predicted and observed 5-year AF risk; the slope and intercept are calculated using linear regression, and the curve is plotted using a univariate spline with smoothing factor of 1. The estimated calibration index (ECI, reported in the legend) is the root mean squared difference between the mean predicted probabilities and the spline-fitted calibration curve. (D) The decision curve analysis, plotting net benefit against threshold probability. The ‘Screen All’ line is different for patient-level and ECG-level curve.
By means of the DCA, ECG-AI model consistently showed the highest NB across a range of threshold probabilities, with significant improvement over the Age & Sex model, with the highest separation observed at a probability threshold corresponding to the event rate (i.e. 12% at the ECG level and 15% at the patient level). Using a classification threshold of 12%, the ECG-AI model showed a sensitivity of 66%, a specificity of 75%, and a negative predictive value of 93% at the patient level. Supplementary data online, Table S9, provides the classification metrics at the patient level for various thresholds.
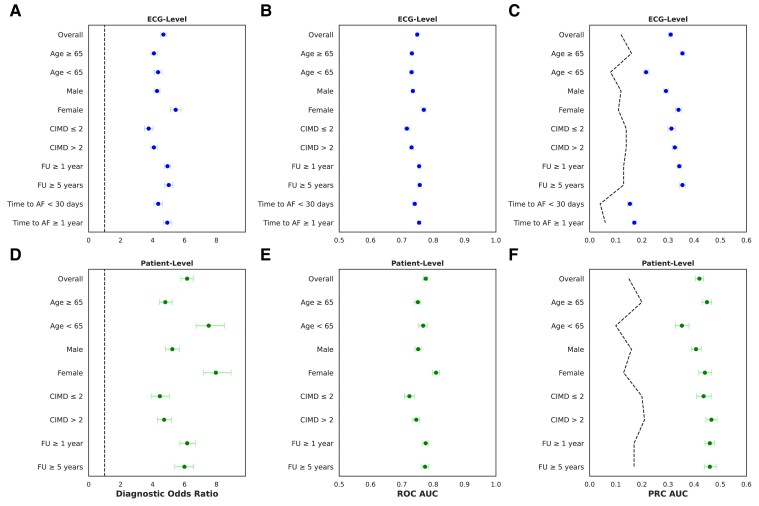
Subgroup analyses are shown in Figure 3. The ECG-AI demonstrated better discrimination performance in female patients (AUC-ROC .77), compared with male patients (AUC-ROC .735; DeLong P < .001). The model did not show significant differences in discrimination performance across other subgroups, including age, CIMD, follow-up duration, and time interval to AF diagnosis (Figure 3).
Figure 3.
Electrocardiogram-based deep learning (ECG-AI) discrimination performance metrics overall and in subgroups of the MHI All-Comers test set (29 065 patients, 135 544 ECG) at the ECG level (A–C) and patient level (D–F). (A and D) The diagnostic odds ratio which is calculated as (sensitivity/(1 − sensitivity))/(specificity/(1 − specificity)) at an optimal threshold of 12% for ECG level and 15% for patient level. (B and E) The receiver operating characteristic area under the curve (ROC AUC). (C and F) The precision–recall curve area under the curve (PRC AUC). The dashed lines represent prevalence, indicating the proportion of true positive cases within the population, important for interpreting precision–recall curve which is sensitive to class imbalance. Confidence intervals for all metrics were derived from 1000 bootstrap iterations. CIMD, Canadian Index for Multiple Deprivation; FU, follow-up)
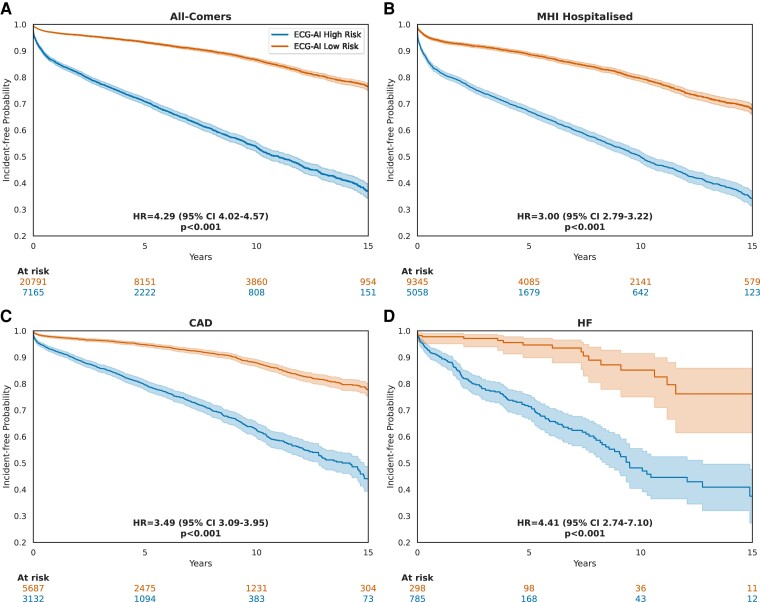
Time-to-event analysis results showed that patients with a high-risk ECG, as predicted by ECG-AI with a threshold probability of ≥12%, had significantly lower incident-free probabilities compared with those having a low-risk ECG with a HR of 4.29 (95% CI: 4.02–4.57; P < .001) at an extended follow-up of up to 15 years (Figure 4). In sensitivity analyses, KM curves were plotted after removing cases where AF was diagnosed within 30 days (see Supplementary data online, Figure S4) and within 1 year (see Supplementary data online, Figure S5) from the index ECG. The model also demonstrated consistent discrimination performance after excluding ECGs with a time to AF diagnosis of <1 year (Figure 3), which could represent pre-existing paroxysmal AF rather than true incident AF. Time-to-event subgroup analyses showed consistent results when stratifying for age and sex (see Supplementary data online, Figure S6).
Figure 4.
Incident atrial fibrillation–free probability: Kaplan-Meier curves using electrocardiogram-based deep learning (ECG-AI) to stratify patients at classification threshold of 12%. Index electrocardiograms with calculated time to atrial fibrillation diagnosis of 0 were removed. Hazard ratios (HR) were calculated by fitting a Cox proportional hazards model. P-values are calculated using the log-rank test. (A) KM curves of patients in the ‘MHI All-Comers’ group. Only the first electrocardiogram of each patient was used. (B) KM curves of patients in the ‘MHI Hospitalized’ group. Only the first electrocardiogram of each patient was used. (C) KM curves of patients with a prior history of CAD. Only the first electrocardiogram acquired after the earliest record of coronary artery disease diagnosis was used. (D) KM curves of patients with a prior history of heart failure. Only the first electrocardiogram acquired after the earliest record of heart failure diagnosis was used
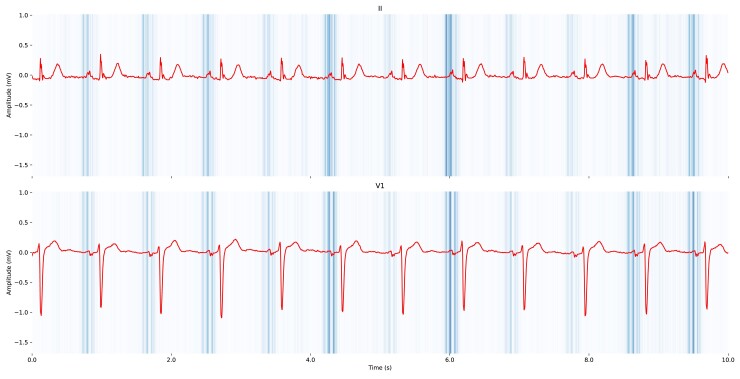
Saliency maps highlighted the P-wave area as having the highest influence on the model’s prediction (Figure 5). Signal artefacts and ectopic beats appeared to contribute less to the model’s prediction (see Supplementary data online, Figures S7–S9).
Figure 5.
Saliency maps for two electrocardiogram derivations, II and V1, which visualize the importance of different segments of the electrocardiogram signals in predicting atrial fibrillation using electrocardiogram-based deep learning (ECG-AI). The saliency maps were generated using TensorFlow’s GradientTape to compute the gradient of the model’s prediction with respect to the input electrocardiogram sample, providing explainability. The maps show regions of low to high saliency, indicated by the colour gradient from light (low saliency) to dark (high saliency). The derivations II and V1 are shown, with notable high saliency around the P-wave that the model found most relevant for predicting atrial fibrillation
Atrial fibrillation prediction in MHI Hospitalized patients using ECG-based deep learning, HATCH, and CHA2DS2-VASc
The ECG-AI model was then compared with the traditional clinical risk scores to predict incident AF in the MHI Hospitalized cohort, where clinical data for each patient were adjudicated. The ECG-AI model achieved an AUC-ROC of .73 (95% CI: .725–.735), indicating superior discrimination compared with the CHA2DS2-VASc (AUC-ROC = .55, 95% CI: .548–.558) and HATCH (AUC-ROC = .52, 95% CI: .515–.524) models (see Supplementary data online, Figure S10). Similarly, the PRC revealed that the ECG-AI model had the highest precision–recall area of .34 (95% CI: .33–.35), outperforming the clinical models. Adding CHA2DS2-VASc or HATCH clinical scores to ECG-AI after training did not provide an overall better fit on the test set (LRT statistic < 0, P = 1). A sensitivity analysis was also performed, restricting cases to those with a follow-up duration of over 1 year, which did not significantly impact the discrimination performance of clinical risk models and ECG-AI (see Supplementary data online, Table S10). The ECG-AI model performance was similar in different subgroups including in cases with actionable AF, i.e. where OAC would be recommended based on the CHA2DS2-VASc score, with an ECG-level AUC-ROC of .728 (95% CI: .722–.734) (see Supplementary data online, Figure S11). Discrimination performance was reduced in patients with a history of HF (AUC-ROC of .69, 95% CI: .68–.70) compared with the overall results, while performance was better in patients with a history of CAD, with an AUC-ROC of .75 (95% CI: .746–.758) (see Supplementary data online, Figure S11). The time-to-event analysis also showed that ECG-AI prediction was associated with a long-term hazard of developing AF in the ‘MHI Hospitalized’ group (HR 3.0, 95% CI: 2.79–3.22; P < .001), in patients with history of CAD (HR 3.49, 95% CI: 3.09–3.95; P < .001) and HF (HR 4.41, 95% CI: 2.74–7.10; P < .001) (Figure 4A–D), and in patients without documented diagnoses of HF or CAD (see Supplementary data online, Figure S12).
Atrial fibrillation prediction in MHI Biobank patients using ECG-based deep learning, AF-PGS, and CHARGE-AF
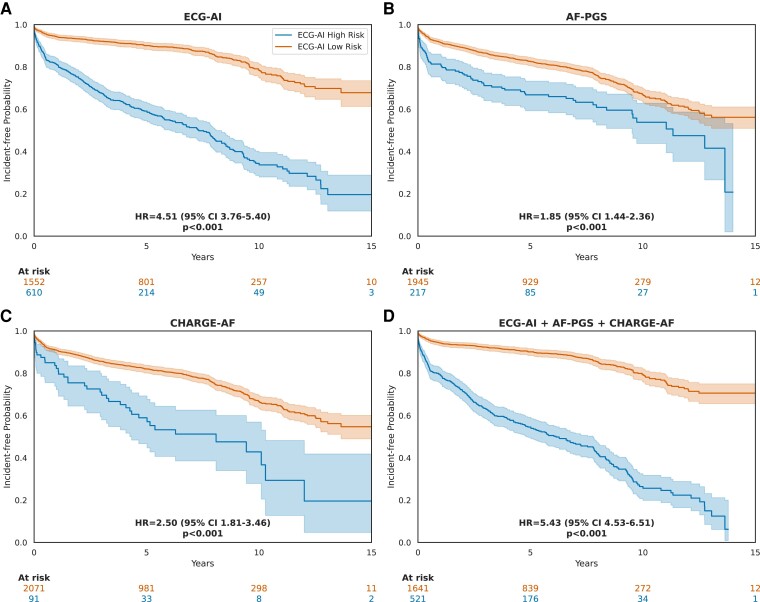
A single ECG per patient, acquired closest to patient enrolment in the MHI Biobank, was used to compare ECG-AI predictions with AF-PGS and CHARGE-AF predictions in the MHI Biobank group. A total of 2301 patients with complete AF-PGS data and CHARGE-AF score were included from the test set. The AF-PGS and CHARGE-AF models showed poorer discrimination performances, with respective AUC-ROC of .59 (95% CI: .57–.63; DeLong P < .001) and .62 (95% CI: .60–.65; DeLong P < .001) compared with ECG-AI (AUC-ROC of .76, 95% CI: .74–.79) (see Supplementary data online, Figure S13). While the addition of AF-PGS and/or CHARGE-AF to ECG-AI as a post-training set yielded similar AUC-ROC compared with ECG-AI, this addition improved the calibration performance with a reduction of ECI from .157 (95% CI: .125–.198) using ‘ECG-AI alone’ to .095 (95% CI: .052–.147) using ‘ECG-AI + AF-PGS’ and .079 (95% CI: .046–.116) using ‘ECG-AI + AF-PGS + CHARGE-AF’ (see Supplementary data online, Figure S13). The LRT further confirmed that the more complex models provided a significantly better overall fit (P = .0002 for ‘ECG-AI + AF-PGS’; P < .0001 for ‘ECG-AI + AF-PGS + CHARGE-AF’), compared to ‘ECG-AI alone’. The DCA, which is influenced by both discrimination and calibration, also showed an improved NB in models that add AF-PGS and/or CHARGE-AF to ECG-AI (see Supplementary data online, Figure S13). Time-to-event analyses in the MHI Biobank group showed superior HR when ECG-AI was used to stratify patients into high-risk and low-risk groups [HR 4.51 (95% CI: 3.76–5.40); P < .001] compared with AF-PGS [HR 1.85 (95% CI: 1.44–2.36); P < .001] and CHARGE-AF [HR 2.50 (95% CI: 1.81–3.46); P < .001] (Figure 6).
Figure 6.
Incident atrial fibrillation–free probability: Kaplan-Meier curves using different models to stratify patients in the MHI Biobank group. Index electrocardiograms with calculated time to AF diagnosis equal to 0 days were removed. Hazard ratios were calculated by fitting a Cox proportional hazards model. P-values are calculated using the log-rank test. (A) Electrocardiogram-based deep learning (ECG-AI) model. Classification threshold = 12%. (B) AF-polygenic score (AF-PGS) model. Classification threshold = top decile (10%) of PGS. (C) CHARGE-AF score. Classification threshold = 21% based on the decision curve analysis. (D) ECG-AI + AF-PGS + CHARGE-AF model. AF-PGS and CHARGE-AF are added to ECG-AI post-training using a logistic regression. Classification threshold = 21% based on the decision curve analysis
External validation of the ECG-based deep learning model in the Medical Information Mart for Intensive Care-IV data set
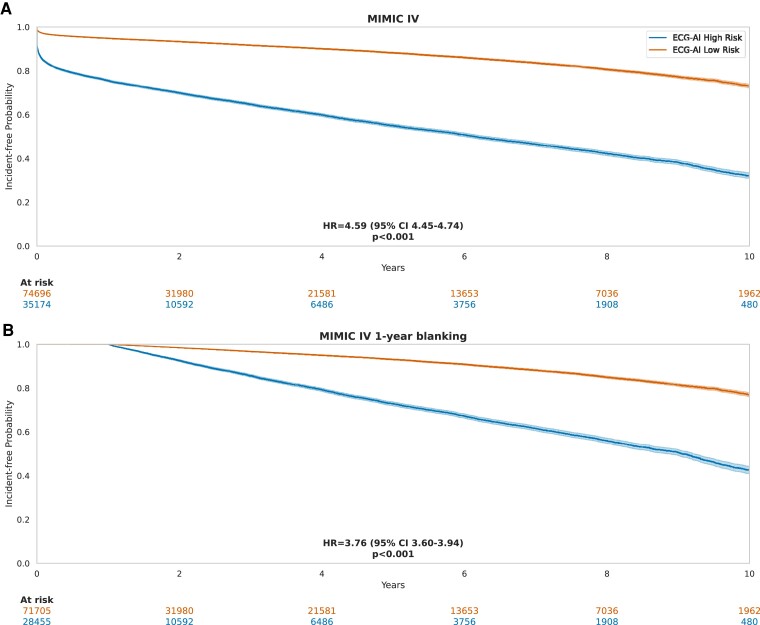
A total of 437 323 ECGs recorded in 109 870 patients from the MIMIC-IV data set were used to externally validate the ECG-AI model. Patients were aged 59 ± 18 years, 47% were males, and median follow-up was 1.1 years (Q1: .03, Q3: 4.7) (Table 1). The 5-year incidence of AF was 15.1%. As shown in Figure 7, ECG-AI demonstrated good discrimination, calibration, and net clinical benefit. Time-to-event analyses demonstrated that the 32% of patients with ECG-AI predicted high AF risk had a 4.6-fold increased hazard (95% CI 4.45–4.74) of developing AF during long-term follow-up (P < .001), with consistent results when excluding ECGs with time to AF < 1 year (Figure 8).
Figure 7.
Performance assessment of electrocardiogram-based deep learning (ECG-AI) in the Medical Information Mart for Intensive Care-IV (MIMIC-IV) external validation data set (109 870 patients, 437 323 ECG). (A) The receiver operating characteristic curve, plotting the true positive rate against the false positive rate for each model, with the area under the curve indicating discriminatory power (reported in legend). (B) The precision–recall curve, plotting precision against recall with the area under the curve reported in legend. (C) The calibration curve, showing the relationship between predicted and observed 5-year atrial fibrillation risk; the slope and intercept are calculated using linear regression, and the curve is plotted using a univariate spline with smoothing factor of 1. The estimated calibration index (ECI, reported in legend) is the root mean squared difference between the mean predicted probabilities and the spline-fitted calibration curve. (D) The decision curve analysis, plotting net benefit against threshold probability.
Figure 8.
Incident-free atrial fibrillation probability: Kaplan–Meier curves using electrocardiogram-based deep learning (ECG-AI) to stratify patients in the Medical Information Mart for Intensive Care-IV (MIMIC-IV) external validation cohort. Electrocardiograms with calculated time to atrial fibrillation diagnosis equal to 0 days were removed. Hazard ratios (HR) were calculated by fitting a Cox proportional hazards model. P-values are calculated using the log-rank test. (A) ECG-AI model. Classification threshold = 12%. (B) ECG-AI model when excluding ECGs with time to AF < 1 year.
Discussion
The development of an ECG-based deep learning model (ECG-AI) designed to predict 5-year incident AF risk at an academic cardiac centre was presented. The study aimed to adhere to key quality criteria recently developed for clinical AI prediction modelling studies.26–28 The ECG-AI demonstrated superior discrimination and calibration performance compared with both clinical and polygenic scores (Structured Graphical Abstract). Recent studies reported on the development of incident AF prediction models using 12-lead ECG during normal sinus rhythm in a diverse hospital network population.15–19 To our knowledge, this study represents the first attempt to predict incident AF in a cardiac care centre using an ECG-AI model and comparing it to both clinical and polygenic scores. A multifaceted performance assessment was conducted, evaluating discrimination, calibration, and NB using decision curve analysis to comprehensively assess the predictive capabilities of the models.27 This is crucial since solely reporting AUC-ROC can be overly optimistic in the setting of imbalanced data with a relatively small event rate,44 and the NB better reflects the impact of clinical implementation. Reassuringly, the ECG-AI model demonstrated good performance in an independent external validation cohort (MIMIC-IV), supporting the generalizability of the model across different clinical settings. Importantly, while most published ECG-AI models remain proprietary, this study not only shares the open weights of the model but also provides the complete code for replicating the validation using the MIMIC-IV data set. This comprehensive approach promotes transparency and reproducibility, allowing researchers to independently validate our findings. Furthermore, our open-source code enables other investigators to retrain and fine-tune the model on their own data sets, facilitating adaptation to diverse clinical contexts. By establishing this robust, accessible baseline, the aim is to accelerate future research and improvement in ECG-based AF risk prediction across various healthcare settings.
Electrocardiogram-based deep learning outperforms clinical models in predicting 5-year incident atrial fibrillation
Atrial fibrillation clinical risk scores are a combination of established AF risk factors. In a recent meta-analysis, CHARGE-AF achieved a C-statistic of .71 (95% CI: .66–.76), HATCH .67 (95% CI: .61–.73), and CHA2DS2-VASc .69 (95% CI: .64–.74).45 Further, the CHARGE-AF score performance has been consistently reported in the .7–.8 range across both ambulatory and general healthcare populations.12,16,46–48 In our study, the performance of these three clinical models was lower than previously reported. This discrepancy could be attributed to several factors. First, classification bias may arise from inconsistencies in how AF is recorded across different settings, potentially leading to underreporting or misclassification of AF cases. The 91% sensitivity of our AF outcome adjudication validation study indicated a modest risk of classification bias (see Supplementary data online, Table S7), which could have also negatively impacted the performance of ECG-AI. There was no significant change in the discrimination performance of clinical risk models in the sensitivity analysis restricting cases to those with a follow-up duration of over 1 year, suggesting that short-term follow-up bias had a negligible impact on our findings (see Supplementary data online, Table S10). Second, the population in our study, drawn from a cardiac care centre, is significantly different from the primarily general population included in the meta-analysis.45 Clinical risk models may perform less well in a cardiac centre patient population owing to the higher prevalence of cardiovascular comorbidities. Consistent with our findings, a recent study by Marston et al.49 reported that the CHARGE-AF clinical score achieved a C-index of .65 in predicting incident AF among patients with cardiovascular conditions.
Our findings indicate that the ECG-AI model outperforms traditional clinical risk models, CHA2DS2-VASc, HATCH, and CHARGE-AF, across various performance metrics. Saliency maps revealed that the P-wave area had the most significant impact on ECG-AI’s prediction of AF risk, which is similar to the finding of Khurshid et al.16 The pathophysiological plausibility of the predictive ability of ECG-AI can thus be attributed to the assumption of an underlying ECG signature indicative of significant atrial myopathy, which represents a vulnerable substrate for AF. In fact, Verbrugge et al.50 recently found that a higher deep learning probability of paroxysmal AF is associated with greater atrial myopathy on echocardiography and invasive haemodynamic testing.
The ECG-AI model maintained good performance across different demographic groups, including age, sex, and socio-economic status (CIMD). This confirmation is important since current deep learning models have limited explainability and, therefore, carry the potential to reflect and perpetuate health disparities.31,51 Of interest, ECG-AI showed significantly better discrimination performance in females compared with males. This better performance in females was also observed in a prior study for clinical AF prediction.48 Accurate prediction of AF in females is of clinical interest since female AF patients are at increased risk for adverse outcomes including stroke and death compared with males.52 The ECG-AI also maintained consistent performance in survival analysis when selecting the single ECG with the highest AF probability per patient (see Supplementary data online, Figure S14).
Electrocardiogram-based deep learning outperforms AF-PGS in predicting 5-year incident atrial fibrillation
Genetic variation has been shown to play an important role in determining long-term AF risk. In the MHI Biobank cohort, AF-PGS showed poor discrimination performances compared with ECG-AI. The lower performance of AF-PGS in our study compared with prior AF-PGS studies can be attributed to differences in the clinical setting, where our study involved patients seen in a tertiary cardiac institute while prior studies have mostly considered application of AF-PGS in the general population. In this study, the first attempt to combine ECG-AI with AF-PGS to predict AF risk is reported. Interestingly, while this combination did not significantly enhance discrimination performance, it did improve calibration performance, resulting in an overall better fit and NB for the combined model compared with ECG-AI and with greater benefit than when only the CHARGE-AF was added. This finding suggests that AF-PGS potentially provides additional predictive value beyond ECG-AI in assessing AF risk in a cardiac care centre population.
Electrocardiogram-based deep learning prediction is associated with long-term atrial fibrillation risk
The time-to-event analysis demonstrated a sustained separation of the KM incidence-free survival curves for up to 15 years after the index ECG. This finding further supports the potential of ECG-AI to stratify AF risk. Additionally, these results are consistent with those of a previous study evaluating AF prediction using ECG-AI, thereby strengthening the overall body of evidence.17 The ECG-AI model maintained its ability to discriminate between high- and low-risk populations for new-onset AF after further stratifying the time-to-event analysis by age and sex groups, two established risk factors for AF. The model’s superiority over age and sex alone was most evident in younger age groups and female patients, aligning with the findings of Raghunath et al.17 Finally, stratifying patient using ECG-AI was associated with a higher HR of incident AF compared with AF-PGS and CHARGE-AF further affirming the advantage of ECG-AI (Figure 6).
Limitations and future directions
The results should be interpreted in the context of our study design, which was retrospective and confined to a single tertiary cardiac referral centre. In Canada, coronary angiography is largely performed in academic centres like MHI, while HF care is more widely distributed, contributing to the higher relative prevalence of CAD in the MHI population. External validation was conducted with the MIMIC-IV cohort, which includes patients admitted to the emergency department or an intensive care unit, a population that differs from the MHI cohort and has a short median follow-up of 1.1 years. Despite these differences, the consistent performance of ECG-AI in patients without CAD and in the MIMIC-IV cohort is reassuring for its generalizability. Although the AF classification bias was mitigated by using multiple sources and conducting a validation study against manual adjudication, a residual classification bias may be present and may have negatively impacted ECG-AI performance. The 5-year AF incidence in our cohort is higher than in general healthcare-related data sets.16,48 This discrepancy can be attributed to our study population consisting of cardiac patients, who have a higher risk of developing AF and who may have undergone more intensive monitoring leading to more frequent detection of AF compared with general healthcare-related data sets. Furthermore, in a retrospective analysis, it is challenging to distinguish between the detection of pre-existing paroxysmal AF and the prediction of truly incident AF, despite purposefully excluding patients with known AF at baseline. It cannot be excluded that some sinus rhythm ECG recorded after AF diagnosis may have been included. Reassuringly, the sensitivity analysis excluding ECG with a time to AF diagnosis of <1 year (to minimize inclusion of patients with pre-existing paroxysmal AF) showed consistent results with the overall All-Comers cohort (Figure 3; Supplementary data online, Figure S5). Furthermore, this retrospective study did not account for right censoring when modelling AF as a binary outcome. Despite consistent results from sensitivity analyses with follow-up restrictions, future studies could incorporate time-to-event analysis in prospective cohorts where right censoring can be properly accounted for. The study did not assess performance bias related to ethnicity, as such data were not available in the entire population and the MHI Biobank subgroup (where genetic ancestry could have been inferred) is predominantly of European ancestry. Another limitation of this study is that new-onset AF and atrial flutter were analysed in a combined manner, which may affect the granularity and specificity of the results. Future work could consider separating these conditions, which can be beneficial in specific clinical scenarios, such as predicting new-onset AF in patients referred for atrial flutter ablation procedure.
Our study leverages a ResNet-50 architecture for ECG-AI prediction of incident AF, building upon previous seminal works that utilized less complex convolutional neural network structures.15–17 Importantly, while most published ECG-AI models remain proprietary, the open weights of the MHI ECG-AI model were shared. This open-weight approach promotes transparency and establishes a robust baseline for future research. As the field of AI rapidly evolves, with recent advancements suggesting that more complex models could enhance predictive performance, our open-weight ResNet-50 model serves as a valuable benchmark. Researchers can now use this model to validate its performance in diverse populations and directly compare new, potentially more sophisticated architectures against our model, accelerating progress in the field.
While adding clinical and genetic prediction to ECG-AI improved performance in the MHI Biobank subgroup, further analysis is warranted to ensure the generalizability of this observation in diverse populations. Future developments also hold the promise of training a deep learning model end to end with all these modalities combined.53 Furthermore, the added clinical gain of ECG-AI remains uncertain beyond net clinical benefit modelling, and the cost-effectiveness remains unexplored. Finally, inherent challenges with ECG-AI persist, such as workflow integration and the acceptance of AI by both clinicians and patients in the medical field.54
Conclusion
Our study contributes to the growing body of evidence demonstrating that deep learning applied to a resting 12-lead ECG during sinus rhythm can effectively predict the risk of new-onset AF with high performance in a tertiary cardiac care centre population. This prediction outperforms existing clinical and polygenic scores. Our study demonstrates the potential of ECG-AI models to significantly enhance AF prediction in a cardiac care setting, paving the way for tailored strategies for early detection of AF to prevent adverse outcomes.
Supplementary Material
Acknowledgements
We wish to thank Julie Todd for her assistance in big data extraction and Sewanou Hermann Honfo and Luca Scimeca for the valuable discussions on statistical and deep learning modelling, as well as Amélie Jeuken, Léanie Moreau, Breanna Chen, Agnès Hage-Chehine, Louise-Sabine Louis-Aimé, and Samuel Moussa for their assistance in data collection. The Structured Graphical Abstract was created with Biorender.com.
Contributor Information
Gilbert Jabbour, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Alexis Nolin-Lapalme, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Quebec Artificial Intelligence Institute (MILA), Montreal, Quebec, Canada.
Olivier Tastet, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Denis Corbin, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Paloma Jordà, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada.
Achille Sowa, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Jacques Delfrate, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
David Busseuil, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Julie G Hussin, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; Quebec Artificial Intelligence Institute (MILA), Montreal, Quebec, Canada; Université de Montréal Beaulieu-Saucier Pharmacogenomics Center, Montreal, Quebec H1T 1C8, Canada.
Marie-Pierre Dubé, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; Université de Montréal Beaulieu-Saucier Pharmacogenomics Center, Montreal, Quebec H1T 1C8, Canada.
Jean-Claude Tardif, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; Université de Montréal Beaulieu-Saucier Pharmacogenomics Center, Montreal, Quebec H1T 1C8, Canada; Montreal Health Innovations Coordinating Center, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Léna Rivard, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada.
Laurent Macle, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada.
Julia Cadrin-Tourigny, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada.
Paul Khairy, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; Montreal Health Innovations Coordinating Center, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Robert Avram, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada; HeartWise.Ai, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada.
Rafik Tadros, Montreal Heart Institute Research Centre, 5000 Belanger St, Montreal, Quebec H1T 1C8, Canada; Faculty of Medicine, Université de Montréal, 2900 Edouard Montpetit Blvd, Montreal, Quebec H3T 1J4, Canada.
Supplementary data
Supplementary data are available at European Heart Journal online.
Declarations
Disclosure of Interest
M.-P.D. holds a minor equity interest in DalCor Pharmaceuticals. J.-C.T. reports research grants from AstraZeneca, Boehringer Ingelheim, Ceapro, DalCor Pharmaceuticals, Esperion, Merck, Novartis, Novo Nordisk, and Pfizer; honoraria from DalCor Pharmaceuticals, Pendopharm, and Pfizer; and minor equity interest in DalCor Pharmaceuticals.
Data Availability
The associated code required to run the trained ResNet-50 model used in this study is available on GitHub. The ECG-AI model is also made publicly available. Please visit our repository at https://github.com/HeartWise-AI/ecg-ai-af-mhi.
Funding
P.J. is supported by a fellowship grant from Takeda Canada and CIHR Institute of Genetics (TAK401439). J.H., L.R., J.C.-T., and R.A. are supported by the Fonds de Recherche du Québec. J.H. is funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2022-04262). M.-P.D., J.-C.T., and R.T. are supported by the Canada Research Chairs programme. J.C.-T.and R.T. are supported by the Philippa and Marvin Carsley Chair. P.K. is supported by the André Chagnon Research Chair.
Ethical Approval
The study complies with the Declaration of Helsinki. The protocols were approved by the MHI ethics committee (submission IDs 2023-3160, 2023-1756, and 2021-2928). All MHI Biobank participants signed an informed consent. For other MHI patients, waiver of informed consent was obtained from the ethics committee.
Pre-registered Clinical Trial Number
None supplied.
References
- 1. Tsao CW, Aday AW, Almarzooq ZI, Anderson CAM, Arora P, Avery CL, et al. Heart Disease and Stroke Statistics-2023 Update: a report from the American Heart Association. Circulation 2023;147:e93–621. 10.1161/CIR.0000000000001123 [DOI] [PubMed] [Google Scholar]
- 2. Goette A, Kalman JM, Aguinaga L, Akar J, Cabrera JA, Chen SA, et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterization, and clinical implication. Europace 2016;18:1455–90. 10.1093/europace/euw161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin D, Mansour MC, Ruskin JN, Heist EK. Atrial fibrillation-mediated cardiomyopathy. Circ Arrhythm Electrophysiol 2019;12:e007809. 10.1161/CIRCEP.119.007809 [DOI] [PubMed] [Google Scholar]
- 4. Santhanakrishnan R, Wang N, Larson MG, Magnani JW, McManus DD, Lubitz SA, et al. Atrial fibrillation begets heart failure and vice versa: temporal associations and differences in preserved versus reduced ejection fraction. Circulation 2016;133:484–92. 10.1161/CIRCULATIONAHA.115.018614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koh YH, Lew LZW, Franke KB, Elliott AD, Lau DH, Thiyagarajah A, et al. Predictive role of atrial fibrillation in cognitive decline: a systematic review and meta-analysis of 2.8 million individuals. Europace 2022;24:1229–39. 10.1093/europace/euac003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papanastasiou CA, Theochari CA, Zareifopoulos N, Arfaras-Melainis A, Giannakoulas G, Karamitsos TD, et al. Atrial fibrillation is associated with cognitive impairment, all-cause dementia, vascular dementia, and Alzheimer’s disease: a systematic review and meta-analysis. J Gen Intern Med 2021;36:3122–35. 10.1007/s11606-021-06954-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hart RG, Pearce LA, Aguilar MI. Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med 2007;146:857–67. 10.7326/0003-4819-146-12-200706190-00007 [DOI] [PubMed] [Google Scholar]
- 8. Ruff CT, Giugliano RP, Braunwald E, Hoffman EB, Deenadayalu N, Ezekowitz MD, et al. Comparison of the efficacy and safety of new oral anticoagulants with warfarin in patients with atrial fibrillation: a meta-analysis of randomised trials. Lancet 2014;383:955–62. 10.1016/S0140-6736(13)62343-0 [DOI] [PubMed] [Google Scholar]
- 9. Kirchhof P, Camm AJ, Goette A, Brandes A, Eckardt L, Elvan A, et al. Early rhythm-control therapy in patients with atrial fibrillation. N Engl J Med 2020;383:1305–16. 10.1056/NEJMoa2019422 [DOI] [PubMed] [Google Scholar]
- 10. Dilaveris PE, Kennedy HL. Silent atrial fibrillation: epidemiology, diagnosis, and clinical impact. Clin Cardiol 2017;40:413–8. 10.1002/clc.22667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petzl AM, Jabbour G, Cadrin-Tourigny J, Purerfellner H, Macle L, Khairy P, et al. Innovative approaches to atrial fibrillation prediction: should polygenic scores and machine learning be implemented in clinical practice? Europace 2024;26:euae201. 10.1093/europace/euae201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J Am Heart Assoc 2013;2:e000102. 10.1161/JAHA.112.000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suenari K, Chao T-F, Liu C-J, Kihara Y, Chen T-J, Chen S-A. Usefulness of HATCH score in the prediction of new-onset atrial fibrillation for Asians. Medicine (Baltimore) 2017;96:e5597. 10.1097/MD.0000000000005597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zuo M-L, Liu S, Chan K-H, Lau K-K, Chong B-H, Lam K-F, et al. The CHADS2 and CHA 2DS 2-VASc scores predict new occurrence of atrial fibrillation and ischemic stroke. J Interv Card Electrophysiol 2013;37:47–54. 10.1007/s10840-012-9776-0 [DOI] [PubMed] [Google Scholar]
- 15. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–7. 10.1016/S0140-6736(19)31721-0 [DOI] [PubMed] [Google Scholar]
- 16. Khurshid S, Friedman S, Reeder C, Di Achille P, Diamant N, Singh P, et al. ECG-based deep learning and clinical risk factors to predict atrial fibrillation. Circulation 2022;145:122–33. 10.1161/CIRCULATIONAHA.121.057480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raghunath S, Pfeifer JM, Ulloa-Cerna AE, Nemani A, Carbonati T, Jing L, et al. Deep neural networks can predict new-onset atrial fibrillation from the 12-lead ECG and help identify those at risk of atrial fibrillation-related stroke. Circulation 2021;143:1287–98. 10.1161/CIRCULATIONAHA.120.047829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan N, Duffy G, Dhruva SS, Oesterle A, Pellegrini CN, Theurer J, et al. Deep learning of electrocardiograms in sinus rhythm from US veterans to predict atrial fibrillation. JAMA Cardiol 2023;8:1131–9. 10.1001/jamacardio.2023.3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hygrell T, Viberg F, Dahlberg E, Charlton PH, Kemp Gudmundsdottir K, Mant J, et al. An artificial intelligence-based model for prediction of atrial fibrillation from single-lead sinus rhythm electrocardiograms facilitating screening. Europace 2023;25:1332–8. 10.1093/europace/euad036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Attia ZI, Harmon DM, Behr ER, Friedman PA. Application of artificial intelligence to the electrocardiogram. Eur Heart J 2021;42:4717–30. 10.1093/eurheartj/ehab649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weng L-C, Preis SR, Hulme OL, Larson MG, Choi SH, Wang B, et al. Genetic predisposition, clinical risk factor burden, and lifetime risk of atrial fibrillation. Circulation 2018;137:1027–38. 10.1161/CIRCULATIONAHA.117.031431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muse ED, Wineinger NE, Spencer EG, Peters M, Henderson R, Zhang Y, et al. Validation of a genetic risk score for atrial fibrillation: a prospective multicenter cohort study. PLoS Med 2018;15:e1002525. 10.1371/journal.pmed.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lazarte J, Dron JS, McIntyre AD, Skanes AC, Gula LJ, Tang AS, et al. Evaluating polygenic risk scores in “lone” atrial fibrillation. CJC Open 2021;3:751–7. 10.1016/j.cjco.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borschel CS, Ohlrogge AH, Geelhoed B, Niiranen T, Havulinna AS, Palosaari T, et al. Risk prediction of atrial fibrillation in the community combining biomarkers and genetics. Europace 2021;23:674–81. 10.1093/europace/euaa334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khurshid S, Mars N, Haggerty CM, Huang Q, Weng L-C, Hartzel DN, et al. Predictive accuracy of a clinical and genetic risk model for atrial fibrillation. Circ Genom Precis Med 2021;14:e003355. 10.1161/CIRCGEN.121.003355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Royen FS, Asselbergs FW, Alfonso F, Vardas P, van Smeden M. Five critical quality criteria for artificial intelligence-based prediction models. Eur Heart J 2023;44:4831–4. 10.1093/eurheartj/ehad727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Smeden M, Heinze G, Van Calster B, Asselbergs FW, Vardas PE, Bruining N, et al. Critical appraisal of artificial intelligence-based prediction models for cardiovascular disease. Eur Heart J 2022;43:2921–30. 10.1093/eurheartj/ehac238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Collins GS, Moons KGM, Dhiman P, Riley RD, Beam AL, Van Calster B, et al. TRIPOD + AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024;385:e078378. 10.1136/bmj-2023-078378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zafrir B, Lund LH, Laroche C, Ruschitzka F, Crespo-Leiro MG, Coats AJS, et al. Prognostic implications of atrial fibrillation in heart failure with reduced, mid-range, and preserved ejection fraction: a report from 14 964 patients in the European Society of Cardiology Heart Failure Long-Term Registry. Eur Heart J 2018;39:4277–84. 10.1093/eurheartj/ehy626 [DOI] [PubMed] [Google Scholar]
- 30. Fauchier L, Bisson A, Bodin A, Herbert J, Angoulvant D, Danchin N, et al. Outcomes in patients with acute myocardial infarction and new atrial fibrillation: a nationwide analysis. Clin Res Cardiol 2021;110:1431–8. 10.1007/s00392-021-01805-2 [DOI] [PubMed] [Google Scholar]
- 31. Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019;366:447–53. 10.1126/science.aax2342 [DOI] [PubMed] [Google Scholar]
- 32. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. New York City: IEEE, 2016, 770–778. [Google Scholar]
- 33. Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. 2016. Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv, arXiv:160304467, preprint: not peer reviewed. [Google Scholar]
- 34. Simonyan K, Vedaldi A, Zisserman A. 2013. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv, arXiv:13126034 , preprint: not peer reviewed. [Google Scholar]
- 35. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–24. 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 2003;56:1129–35. 10.1016/S0895-4356(03)00177-X [DOI] [PubMed] [Google Scholar]
- 37. Van Hoorde K, Van Huffel S, Timmerman D, Bourne T, Van Calster B. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. J Biomed Inform 2015;54:283–93. 10.1016/j.jbi.2014.12.016 [DOI] [PubMed] [Google Scholar]
- 38. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565–74. 10.1177/0272989X06295361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 40.Statistics Canada. 2023. The Canadian Index of Multiple Deprivation, 2021. https://www150.statcan.gc.ca/n1/pub/45-20-0001/452000012023002-eng.htm.
- 41. Johnson A, Bulgarelli L, Pollard T, Gow B, Moody B, Horng S, et al. MIMIC-IV version 3.0. PhysioNet 2024. 10.13026/hxp0-hg59 (15 August 2024, date last accessed). [DOI]
- 42. Johnson AEW, Bulgarelli L, Shen L, Gayles A, Shammout A, Horng S, et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci Data 2023;10:1. 10.1038/s41597-022-01899-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 2000;101:E215–20. 10.1161/01.CIR.101.23.e215 [DOI] [PubMed] [Google Scholar]
- 44. Avram R, Olgin JE, Tison GH. The rise of open-sourced machine learning in small and imbalanced datasets: predicting in-stent restenosis. Can J Cardiol 2020;36:1574–6. 10.1016/j.cjca.2020.02.002 [DOI] [PubMed] [Google Scholar]
- 45. Himmelreich JCL, Veelers L, Lucassen WAM, Schnabel RB, Rienstra M, van Weert H, et al. Prediction models for atrial fibrillation applicable in the community: a systematic review and meta-analysis. Europace 2020;22:684–94. 10.1093/europace/euaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christophersen IE, Yin X, Larson MG, Lubitz SA, Magnani JW, McManus DD, et al. A comparison of the CHARGE-AF and the CHA2DS2-VASc risk scores for prediction of atrial fibrillation in the Framingham Heart Study. Am Heart J 2016;178:45–54. 10.1016/j.ahj.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hulme OL, Khurshid S, Weng L-C, Anderson CD, Wang EY, Ashburner JM, et al. Development and validation of a prediction model for atrial fibrillation using electronic health records. JACC Clin Electrophysiol 2019;5:1331–41. 10.1016/j.jacep.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khurshid S, Kartoun U, Ashburner JM, Trinquart L, Philippakis A, Khera AV, et al. Performance of atrial fibrillation risk prediction models in over 4 million individuals. Circ Arrhythm Electrophysiol 2021;14:e008997. 10.1161/CIRCEP.120.008997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marston NA, Garfinkel AC, Kamanu FK, Melloni GM, Roselli C, Jarolim P, et al. A polygenic risk score predicts atrial fibrillation in cardiovascular disease. Eur Heart J 2023;44:221–31. 10.1093/eurheartj/ehac460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verbrugge FH, Reddy YNV, Attia ZI, Friedman PA, Noseworthy PA, Lopez-Jimenez F, et al. Detection of left atrial myopathy using artificial intelligence-enabled electrocardiography. Circ Heart Fail 2022;15:e008176. 10.1161/CIRCHEARTFAILURE.120.008176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa S, et al. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circ Arrhythm Electrophysiol 2020;13:e007988. 10.1161/CIRCEP.119.007988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tadros R, Ton AT, Fiset C, Nattel S. Sex differences in cardiac electrophysiology and clinical arrhythmias: epidemiology, therapeutics, and mechanisms. Can J Cardiol 2014;30:783–92. 10.1016/j.cjca.2014.03.032 [DOI] [PubMed] [Google Scholar]
- 53. Boonstra MJ, Weissenbacher D, Moore JH, Gonzalez-Hernandez G, Asselbergs FW. Artificial intelligence: revolutionizing cardiology with large language models. Eur Heart J 2024;45:332–45. 10.1093/eurheartj/ehad838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leclercq C, Witt H, Hindricks G, Katra RP, Albert D, Belliger A, et al. Wearables, telemedicine, and artificial intelligence in arrhythmias and heart failure: proceedings of the European Society of Cardiology Cardiovascular Round Table. Europace 2022;24:1372–83. 10.1093/europace/euac052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The associated code required to run the trained ResNet-50 model used in this study is available on GitHub. The ECG-AI model is also made publicly available. Please visit our repository at https://github.com/HeartWise-AI/ecg-ai-af-mhi.