Abstract
Objectives
The All of Us Research Program is a precision medicine initiative aimed at establishing a vast, diverse biomedical database accessible through a cloud-based data analysis platform, the Researcher Workbench (RW). Our goal was to empower the research community by co-designing the implementation of SAS in the RW alongside researchers to enable broader use of All of Us data.
Materials and Methods
Researchers from various fields and with different SAS experience levels participated in co-designing the SAS implementation through user experience interviews.
Results
Feedback and lessons learned from user testing informed the final design of the SAS application.
Discussion
The co-design approach is critical for reducing technical barriers, broadening All of Us data use, and enhancing the user experience for data analysis on the RW.
Conclusion
Our co-design approach successfully tailored the implementation of the SAS application to researchers' needs. This approach may inform future software implementations on the RW.
Keywords: data analysis, software, data ecosystem, diversity, precision medicine
Background and significance
The All of Us Research Program, funded by the National Institutes of Health (NIH), is a precision medicine initiative seeking to enroll at least one million individuals across the United States into a longitudinal cohort study.1 Nearly 45% of participants belong to racial and ethnic minority groups, and over 75% are classified as underrepresented in biomedical research, making the All of Us dataset the most diverse of its kind.2,3 The program aims to accelerate health research discoveries and enable innovative, personalized healthcare solutions. To support these objectives, the All of Us Data and Research Center (DRC) developed the Researcher Workbench (RW), a secure cloud-based platform for analyzing All of Us data.4,5 The RW is equipped with custom software and tools that facilitate powerful data analysis and collaborative research. As of March 2024, there were over 10 000 registered users on the RW, which contains participant-level data including surveys (N = 413 360), electronic health records (EHRs; N = 255 640), genomics (N = 245 400), physical measurements (N = 337 540), and wearable device data (N = 15 620). The platform is constantly evolving, with the addition of new features, tools, and data types to support researchers from diverse backgrounds.
A core component of the RW is its suite of tools for data analysis. At its initial launch in May 2020, it featured Jupyter notebooks for Python and R, with a plan for expanding tool offerings as the researcher base grew. Among data analysis tools, SAS has become one of the most widely used by researchers in health science, including epidemiologists, statisticians, and social scientists, among others. It was one of the top 3 statistical analysis tools requested by researchers after the RW’s launch, with 35% of user requests for additional tools referencing SAS as their preferred software for data analysis.6 Specifically, SAS appeals to users across a broad spectrum of programming proficiency by providing a sophisticated programming environment in addition to a point-and-click interface to allow all users to create code quickly and easily. Providing additional tools based on researcher feedback and domain interest is critical to reducing technical barriers and enabling broad use of the All of Us data on the RW.
Therefore, the goal of this work was to co-design the implementation of SAS on the RW with biomedical researchers. Our goal consists of 2 objectives. The first was to integrate the cloud-based version of SAS Studio with existing tools on the RW, including the Cohort Builder and Dataset Selector, which feature a user-friendly, point-and-click interface for creating, reviewing, and annotating cohorts without requiring programming knowledge.7,8 In addition to the existing capabilities of SAS, including descriptive statistics, data visualization, and data management, our implementation aims to support cloud-based information storage and retrieval on the RW, respect the built-in security controls of the RW, and facilitate end-to-end usage of All of Us data for researchers. The second objective was to develop a comprehensive collection of user support articles, analytic guides, and tutorials regarding best practices on using SAS Studio to reduce technical barriers for users. Our work returns value to communities by improving the utility of the RW for researchers, and our multidisciplinary co-design approach may inform future software implementations on the RW to enable broader use of the All of Us data.
Materials and methods
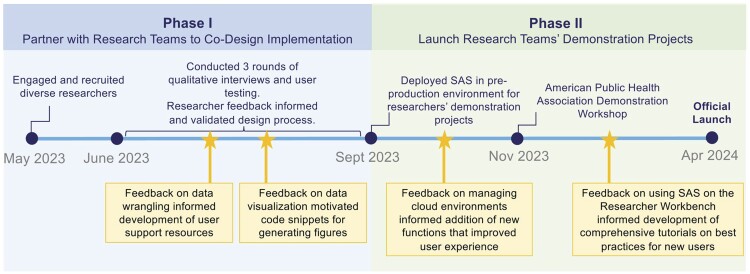
Implementing SAS Studio on the RW was a collaborative effort involving a multidisciplinary team of project managers, cloud architects, software engineers, data scientists, and user experience (UX) designers across multiple research institutions and industry partners, including the NIH, Vanderbilt University Medical Center, Verily Life Sciences, the Broad Institute of MIT and Harvard, and SAS Institute. To aid in the user-driven implementation of SAS, the NIH Program Office partnered with 6 researchers experienced in using both SAS and the RW to co-design the implementation. The researchers, whose training and backgrounds span the fields of nursing, medicine, public health, social and behavioral sciences, biostatistics, and epidemiology, were recruited from the All of Us Researcher Ambassador network (Table 1). They formed 3 teams of 2 based on prior collaborations and shared research interests. Team 1 consisted of experts in public health and biomedical research, with a focus on social and behavioral health, nursing, and health disparities. The researchers in Team 2 specialized in epidemiology and research on health disparities. Team 3 combined expertise in social and behavioral science, biostatistics, epidemiology, and health disparities research. From June to September 2023, researchers participated in a total of 3 interviews, including 2 rounds of qualitative interviews and 1 user testing interview (Figure 1). From September 2023 to March 2024, they conducted research projects using SAS Studio on the RW showcasing best practices for analyzing a variety of different data types, including EHRs and surveys (Figure 1). Use cases included characterizing social determinants of health and sociodemographic barriers to healthcare across various diseases, such as type 2 diabetes, depression, heart attack, and stroke (Table 1).
Table 1.
Summary of research teams and their demonstration projects.
| Researcher expertise | Affiliations | Prior experience with SAS | Project objective | Condition(s) of interest | Data types utilized | Variables of interest |
|---|---|---|---|---|---|---|
| Team 1 (2 researchers) | ||||||
| Social and behavioral health, nursing, health disparities | Yale University, Seattle University, United States | 10+ years | Characterize the relationship between social determinants of health and life-threatening medical conditions | Heart attack, stroke, severe allergic reaction, asthma attack, traumatic injuries | Electronic health records, surveys | Access to healthcare, education, gender identity, race/ethnicity, occurrence of life-threatening conditions |
| Team 2 (2 researchers) | ||||||
| Epidemiology, medicine, health disparities | The University of Texas Rio Grande Valley, United States | 3+ years | Explore and understand potential disparities in prescriptions of newer generation diabetes medication | Diabetes | Electronic health records, surveys | Drug exposure, lab measurements, sociodemographic information, health insurance information |
| Team 3 (2 researchers) | ||||||
| Social and behavioral science, biostatistics, epidemiology, health disparities | New York University, United States | 10+ years | Examine sociodemographic patterns, including health disparities, in diagnosis and treatment of mental health conditions | Depression, anxiety, eating disorders, autism spectrum disorder | Electronic health records, surveys | Timing of diagnosis and treatment, gender identity, socioeconomic information, race/ethnicity |
Figure 1.
Overview of project timeline.
Each team participated in 3 interviews aimed at aligning research goals with UX needs, informing and validating user interface (UI) design, and provide insight on the researchers’ end-to-end journey with SAS Studio on the RW, including data collection, exploratory data analysis, statistical analysis, and result interpretation. The first interview aimed to understand the research teams’ experience with SAS Studio and their goals for using it to analyze All of Us data on the RW. The second interview was designed to understand researchers’ data analysis workflow with a focus on their specific use cases. For the third interview, research teams participated in UX testing during which they were guided through a user journey of the beta version of SAS Studio on the RW to identify necessary adjustments for integrating it into the RW, including optimizing program launch and pause functions in a cloud computing environment and improving user interaction with the landing page. Representatives from each research team participated in the interviews virtually. Each interview was conducted with the consent of the researchers and recorded. The interview questions were developed by a team of experienced product managers, cloud architects, and UI designers who have extensive experience with the RW. The questions were adapted from similar frameworks used to inform the implementation of other software and tools on the RW and further refined by an expert from the SAS Institute, ensuring that they were tailored for the SAS environment. During each interview, members of the DRC transcribed researchers’ responses in real time. After the interviews were completed, the DRC team reviewed both the recordings and transcriptions to consolidate feedback, which was analyzed by identifying common themes and patterns in the responses (Table 2). To ensure the integrity of our qualitative analysis and avoid bias, we used a thematic analysis approach where 2 team members independently reviewed the feedback to identify common themes. They then met to discuss their findings, and any discrepancies were resolved by a third team member. These team members held diverse roles, including project managers, biostatisticians, and healthcare professionals familiar with the practical applications of the software, allowing us to cross validate themes from multiple perspectives to ensure a comprehensive and unbiased interpretation of researcher feedback. For further details, see the interview guides and protocol for the user testing in the Supplementary Material of this article. Our study focused on user testing for software implementation on the RW and was not designed to contribute to or develop generalizable knowledge. Therefore, institutional review board approval was not required.
Table 2.
Summary of user experience interviews and results.
| Number of participants | Feedback | Analysis of results | Changes implemented |
|---|---|---|---|
| Interview 1 | |||
| 5 interviewees, 2 interviewers, 5 observers | Interviewees reported that they have previous experience with Base SAS, SAS Viya, and SAS University Edition. | On average, interviewees had 4.6 years of prior experience with SAS. None were familiar with SAS Studio. Therefore, it was necessary to develop new user support materials to lower the technical barrier of using SAS Studio on the Researcher Workbench. | The All of Us Data and Research Center (DRC) collaborated with the SAS Institute to develop a variety of educational materials and training events, enabling users to maximize the utilization of SAS Studio on the Researcher Workbench. |
| All interviewees expressed strong interest in using SAS for performing descriptive statistical analysis of the All of Us data. | The interviewees’ feedback highlighted a common trend among users, emphasizing the importance of SAS as a tool for conducting descriptive statistical analysis. This feedback helped inform areas for enhancing user support and training. | The DRC developed analytic guides and Featured Workspaces that provide users with hands-on tutorials for using SAS Studio to perform a wide range of descriptive statistics procedures. Specifically, these tutorials provide step-by-step instructions focused on optimizing descriptive statistical functions in SAS Studio on the Researcher Workbench. | |
| All interviewees indicated that data cleaning and wrangling can be challenging, especially for a large-scale, cloud-based dataset like All of Us. | The interviewees’ feedback highlights the challenge of analyzing large-scale datasets on a cloud computing platform. This underscores the need for additional support and training in data cleaning and wrangling in SAS Studio, particularly for handling complex datasets in cloud environments. | A variety of user support materials, including articles, office hours presentations, and a Featured Workspace dedicated to best practices and tips for data wrangling and data cleaning. Furthermore, the All of Us Research Support Team is available to answer further questions. | |
| Interview 2 | |||
| 4 interviewees, 2 interviewers, 5 observers | All interviewees expressed interest in exporting their analysis results (e.g., summary statistics) from SAS as a .csv file. In addition, 2 interviewees highlighted that generating graphs and visualizations with other tools, such as spreadsheet software or other data analysis programs, is a critical component of their routine workflow when performing statistical procedures not available in SAS. | The interviewees’ feedback suggests a strong preference for exporting analysis files from SAS Studio. As such, interoperability with other tools was crucial for their data analysis tasks. The essential role of external tools for graphing and visualization highlights a potential gap in SAS’s graphical functionalities. | The DRC implemented a process for users to export summary statistics and aggregate counts from the Researcher Workbench in accordance with the All of Us Research Program’s Data and Statistics Dissemination Policy.11 Users can use these files to create visualizations in external applications. The DRC also published user support resources and hosted education and training events to familiarize users with SAS Studio’s functionalities. These include pre-programmed SAS Snippets for generating graphs and visualizations and SAS Tasks for executing a variety of statistical, artificial intelligence, and machine learning procedures. |
| Interview 3 | |||
| 4 interviewees, 1 interviewer, 6 observers | All interviewees expressed confusion about how to launch the SAS Studio application, specifically regarding the purpose of the lightning bolt icon and whether it is necessary to click the SAS icon to start the application. | It is necessary to improve the design of the SAS Studio’s start page so it is more intuitive for users to launch the application. | A function to allow users to click on the SAS logo to launch directly instead of going to the cloud icon was implemented. |
| Interviewees expressed uncertainty about the implications of the “delete” feature for a cloud-computing environment. | The interviewees’ feedback indicates a lack of clarity about the process of deleting a cloud environment on the Researcher Workbench. | The DRC developed user support articles that thoroughly explain the process for pausing and deleting cloud environments. The DRC also implemented an auto-delete function that helps users save costs by automatically deleting cloud environments after a specified period of inactivity (i.e., 7 or more days). In addition, the DRC introduced a “Cloud Environments” page, which provides a clear overview of the status of all initiated applications, allowing users to easily manage, pause, or delete their cloud environments across all applications on the Researcher Workbench. | |
| Interviewees expressed uncertainty about technical terms like “paused,” “persistent disk,” “compute resource,” “cloud compute,” etc | Clarity and explanations of technical terms are needed and must be accessible on the application launch page. | A banner titled “How to use SAS on Workbench” was added to the SAS Studio launch page with links to 3 user support resources for the following topics: (1) how to launch and use the SAS application, (2) how to use persistent disk storage, and (3) how to understand cloud environment costs. | |
| Interviewees expressed that they want clear and timely notifications about the progress of cloud environment provisioning, including estimated wait times to better plan their tasks. They further emphasized the importance of visual and pop-up notifications to alert them when the environment is ready. | It is important to clearly communicate and visualize the status of the application provisioning process, ensuring that users are informed that the process is underway and providing them with an expected timeline for when the SAS Studio application will be ready to use. | Similar to the other applications in the Researcher Workbench, the application start page and sidebar panel display color-changing indicators for environment provisioning status (ie, yellow when the app is paused, green when the app is actively running, and a spinning wait cursor when the application is still provisioning) so users are aware of the status and are free to navigate away from the start page to complete other tasks in the Researcher Workbench while still tracking the progress of application provisioning in the sidebar panel. | |
Based results from the thematic analysis, the DRC developed user support articles with step-by-step instructions for using SAS Studio on the RW and a tutorial Featured Workspace, which included markdown SAS code and recommended best practices for using the application to explore and analyze All of Us data.9,10 In addition, the UI was enhanced to include clarifying text and informational pop-up banners to help optimize UX.
Results
From the interviews, we identified 5 common points of critical feedback: (1) data wrangling and cleaning for a large, cloud-based dataset, such as All of Us, can be challenging regardless of the statistical software used; (2) some researchers prefer to export data from SAS to a spreadsheet tool to create data visualizations; (3) researchers are reluctant to delete their SAS application on the UI due to a lack of clarity on how the action may affect their ability to save and share their work; (4) researchers need options to pause the application to prevent incurring cloud storage and compute costs associated with a running application; and (5) researchers want additional guidance on navigating SAS Studio’s cloud-based capabilities, which are not available in the more commonly used desktop version of Base SAS software (Table 2).
Feedback from users was used to inform improvements to the features and UI of the RW (Figure 2), including (1) the All of Us Research Support Team at the DRC has published articles and tutorials with recommendations on best practices for data wrangling, data cleaning, and managing computational cost; (2) SAS code snippets to automatically create figures were included to help researchers easily visualize data; (3) refining the process of deleting applications, including adding clarity to the language to improve understanding to help users save their data and analysis while managing costs by avoiding continuously running an application on the cloud; (4) users can now pause applications to save costs; and (5) the DRC and SAS Institute collaborated on developing user support articles, analytic guides, and tutorials regarding best practices for using SAS Studio in the RW, including how to save work and easily share SAS files with colleagues (Table 2).
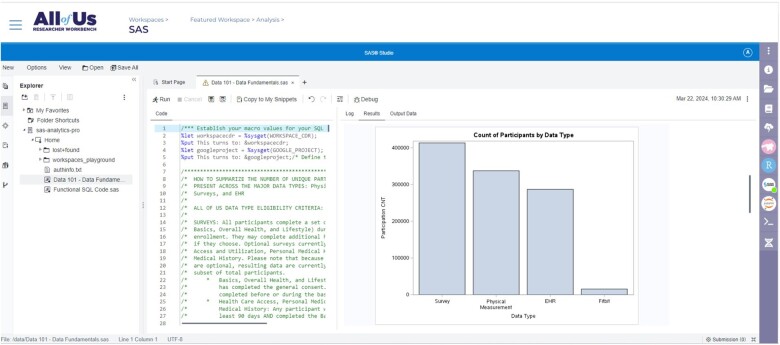
Figure 2.
Image of the SAS Studio application in the Researcher Workbench.
Furthermore, throughout the duration of their research projects, the research teams were invited to participate in weekly office hours hosted by the All of Us Research Support Team, which consisted of at least one software developer who was responsible for deploying SAS to the RW and at least 2 data scientists with extensive expertise in analyzing All of Us data and more than 20 years of experience using SAS. During these office hours, the research teams were given the opportunity to ask questions or report any potential issues or barriers they encountered while using the software. These office hours sessions revealed 2 main points of feedback: (1) the default settings for the random access memory (RAM) and central processing units (CPUs) in SAS Studio were insufficient to fully execute analysis for the size of the datasets which the researchers were attempting to build and analyze, and (2) researchers need clarity on the storage options for SAS files when they want to save their work to their RW accounts. In response to this feedback, SAS Studio UI was modified to give users the ability to customize the RAM and CPUs for the application in order to maximize the computational performance of SAS on large datasets. Additionally, one of the user support articles which was published in tandem with the launch of SAS Studio in the RW provides users with step-by-step instructions regarding their file storage options for SAS files.10 In addition to these readily available online resources, user support is available via email at support@researchallofus.org or bi-weekly office hours hosted by the All of Us Research Support Team.
Discussion
Introducing SAS on the RW will return value to communities by equipping researchers with an additional tool to analyze and maximize the utility of the All of Us data. Our approach of co-designing technical implementations in collaboration with researchers has provided valuable insight into their needs and priorities to ensure an ideal UX with SAS Studio on the RW. Building a multidisciplinary team with diverse researchers, experienced product and project managers, cloud architects/engineers, data scientists, and SAS consultants was key to ensuring that our approach is sustainable for future work. Continued engagement with research teams throughout the co-design process offered opportunities to build trust, identify and prioritize research goals, and tailor the implementation to meet their needs. As such, this work resulted in an effective integration of SAS on the RW and sustainable data products, including user support materials, which will provide the broader research community with additional resources for working with All of Us data.
A potential limitation of our study is the relatively small sample of researchers who participated in the interviews. While they had diverse training and backgrounds, the researchers’ use cases primarily focused on survey and EHRs data. Other data types, including genomic and wearables data, typically require distinct methodologies and assumptions for analysis. As a result, the generalizability of the researchers’ feedback on data analysis may be limited. In addition, all researchers were well-versed with both SAS and the RW, so our results may not fully represent the experiences and potential challenges of diverse users. Therefore, future directions include surveying RW users and collecting their feedback on long-term usability, including how well the current implementation of SAS Studio integrates into their daily workflows. This could provide insight on how researcher feedback from the current study has been addressed and the impact of subsequent updates to the software on the RW.
Continuing to refine the tools on the RW and broaden our understanding of diverse user needs is critical to empower a wider range of researchers. This can help researchers optimize the use of the All of Us dataset, providing opportunities to advance personalized medicine and improve public health outcomes.
Supplementary Material
Acknowledgments
In addition to the funded partners, the All of Us Research Program would not be possible without the contributions made by its participants.
Contributor Information
Izabelle Humes, Vanderbilt Institute of Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Cathy Shyr, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Moira Dillon, Verily Life Sciences, San Francisco, CA 94080, United States.
Zhongjie Liu, Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142, United States.
Jennifer Peterson, SAS Institute, Cary, NC 27513, United States.
Chris St Jeor, Zencos, Cary, NC 27518, United States.
Jacqueline Malkes, SAS Institute, Cary, NC 27513, United States.
Hiral Master, Vanderbilt Institute of Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Brandy Mapes, Vanderbilt Institute of Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Romuladus Azuine, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Nakia Mack, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Bassent Abdelbary, College of Health Professions, The University of Texas Rio Grande Valley, Edinburg, TX 78539, United States.
Joyonna Gamble-George, School of Public Health, Yale University, New Haven, CT 06510, United States.
Emily Goldmann, School of Global Public Health, New York University, New York, NY 10003, United States.
Stephanie Cook, School of Global Public Health, New York University, New York, NY 10003, United States.
Fatemeh Choupani, College of Nursing, Seattle University, Seattle, WA 98122, United States.
Rubin Baskir, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Sydney McMaster, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Chris Lunt, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Karriem Watson, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Minnkyong Lee, All of Us Research Program Office, National Institutes of Health, Bethesda, MD 20892, United States.
Sophie Schwartz, Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142, United States.
Ruchi Munshi, Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142, United States.
David Glazer, Verily Life Sciences, San Francisco, CA 94080, United States.
Eric Banks, Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142, United States.
Anthony Philippakis, Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142, United States.
Melissa Basford, Vanderbilt Institute of Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Dan Roden, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Paul A Harris, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, TN 37203, United States.
Author contributions
Izabelle Humes and Cathy Shyr had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors reviewed the results and approved the final version of the manuscript. Drafting of the manuscript: Izabelle Humes and Cathy Shyr. Critical revision of the manuscript for important intellectual content: Izabelle Humes, Cathy Shyr, Hiral Master, Brandy Mapes, Melissa Basford, Paul A. Harris, David Glazer, and Jennifer Peterson. Conceptualization and methodology: Romuladus Azuine, Nakia Mack, Brandy Mapes, Melissa Basford, and Paul A. Harris. Investigation: Izabelle Humes, Cathy Shyr, Moira Dillon, Zhongjie Liu, Bassent Abdelbary, Joyonna Gamble-George, Fatemeh Choupani, Stephanie Cook, and Emily Goldmann. Data collection and data analysis: Izabelle Humes, Cathy Shyr, Moira Dillon, and Zhongjie Liu. Obtained funding: Romuladus Azuine, Nakia Mack, Melissa Basford, Paul A. Harris, and Brandy Mapes. Subject recruitment: Romuladus Azuine, Nakia Mack, Sydney McMaster, Minnkyong Lee, Rubin Baskir, and Karriem Watson. Resources: Chris Lunt, Rubin Baskir, Karriem Watson, Romuladus Azuine, Nakia Mack, Sydney McMaster, and Minnkyong Lee. Project administration: Izabelle Humes, Moira Dillon, Sophie Schwartz, Ruchi Munshi, Hiral Master, and Brandy Mapes. Technical or material support: Chris St Jeor, Jennifer Peterson, and Jacqueline Malkes. Supervision: Romuladus Azuine, Melissa Basford, Paul A. Harris, Brandy Mapes, Dan Roden, Anthony Philippakis, and Eric Banks.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
The All of Us Research Program is supported by the National Institutes of Health, Office of the Director. For a list of participating members of the program, see https://allofus.nih.gov/funding-and-program-partners. The work by the authors of this article are funded by 3OT2OD035404 from the National Institutes of Health and 1K99LM014429-01 from the National Library of Medicine.
Conflicts of interest
All of the authors listed declare that they have no conflicts of interest to disclose. The authors affiliated with the SAS Institute did not participate in the data collection, data analysis, or data interpretation for this project.
Data availability
The data underlying this article are available in the article and in its online supplementary material. Further data from the user experience testing and details about the testing protocol will be shared on reasonable request to the corresponding author.
Attributions
The Researcher Workbench described in this paper uses SAS Viya Analytics Pro software. Copyright ©2023 SAS Institute Inc.
References
- 1. Denny JC, Rutter JL, Goldstein DB, et al. ; All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med. 2019;381(7):668-676. https://www.nejm.org/doi/10.1056/NEJMsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Master H, Kouame A, Hollis H, Marginean K, Rodriguez K. 2022. Q4R9 v7 data characterization report. 2023. Accessed January 15, 2024. https://support.researchallofus.org/hc/en-us/articles/14558858196628-2022Q4R9-v7-Data-Characterization-Report
- 3. Mapes B, Foster CS, Kusnoor SV, et al. ; All of Us Research Program. Diversity and inclusion for the All of Us Research Program: a scoping review. PLos One. 2020;15(7):e0234962. 10.1371/journal.pone.0234962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayo K, Basford M, Carroll R, et al. The All of Us Data and Research Center: creating a secure, scalable, and sustainable ecosystem for biomedical research. Annu Rev Biomed Data Sci. 2023;6(6):443-464. 10.1146/annurev-biodatasci-122120-104825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. All of Us Research Program. Workbench. Accessed December 12, 2023. https://www.researchallofus.org/data-tools/workbench
- 6. Masuadi E, Mohamud M, Almutairi M, Alsunaidi A, Alswayed AK, Aldhafeeri OF. Trends in the usage of statistical software and their associated study designs in health sciences research: a bibliometric analysis. Cureus. 2021;13(1):e12639. 10.7759/cureus.12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. All of Us Research Program. Selecting participants using the Cohort Builder tool. Accessed December 13, 2023. https://support.researchallofus.org/hc/en-us/articles/360039585591-Selecting-participants-using-the-Cohort-Builder-tool
- 8. All of Us Research Program. Using the Concept Set Selector and Dataset Builder tools to build your dataset. Accessed December 13, 2023. https://support.researchallofus.org/hc/en-us/articles/4556645124244-Using-the-Concept-Set-Selector-and-Dataset-Builder-tools-to-build-your-dataset
- 9. All of Us Research Program. Featured workspaces—table of contents. Accessed July 16, 2024. https://support.researchallofus.org/hc/en-us/articles/360059633052-Featured-Workspaces-Table-of-Contents
- 10. All of Us Research Program. Exploring All of Us data using SAS Studio. Accessed July 16, 2024. Https://support.researchallofus.org/hc/en-us/articles/24413326641684-Exploring-All-of-Us-data-using-SAS-Studio#01HTZE5Y0NAQZFRHZVDGYEMD34
- 11. All of Us Research Program. Data and Statistics Dissemination Policy. Accessed May 19, 2024. Https://www.researchallofus.org/faq/data-and-statistics-dissemination-policy/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. Further data from the user experience testing and details about the testing protocol will be shared on reasonable request to the corresponding author.