Abstract
Objective
Educational offerings to fill the bioinformatics knowledge gap are a key component to enhancing access and use of health data from the All of Us Research Program. We developed a Train the Trainer-based, innovative training series including project-based learning, modular on-demand demonstrations, and unstructured tutorial time as a model for educational engagement in the All of Us community.
Materials and Methods
We highlight our training modules and content, with training survey data informing cycles of development in the creation of a 6-module training series with modular demonstrations.
Results
We have conducted 2 public iterations of the Train the Trainer (Tx3) Series based on survey feedback while training over 300 registered researchers to access and analyze data on the All of Us Researcher Workbench.
Discussion and Conclusion
Future directions of the Tx3 Series include enhanced focus on project-based learning and learner requests for modularity and asynchronous materials access.
Keywords: community-based participatory research, computational biology, training programs
Introduction
Training researchers to utilize cloud-based bioinformatics platforms is critical to ensuring broad uptake and enhanced use of resources such as the All of Us Research Program, an NIH-funded effort to provide the research community with distributed access to large-scale genomics and health data. Mirroring its goals to build a population diverse human health database, the All of Us Research Program also aims to ensure equity of researcher access to the data, so all researchers, including those from underrepresented communities, have access and can perform research addressing both broad medical and specific community needs. To this end, the All of Us Researcher Workbench is an online cloud-based platform consisting of genomics, health records, survey data, and a collection of analysis tools.1 Launched as a beta release on May 27, 2020, the Researcher Workbench provides a hub for users to perform analyses, store code and results, as well as share workspaces with selected users. However, novice users to the All of Us Researcher Workbench, even if otherwise expert healthcare researchers, often lack skills in programming/cloud-computing or in the fundamentals of large-scale genomics data analysis.2–5 Indeed, in a study of over 400 healthcare professionals, Dolezel and McLeod6 suggest that the “theory-practice” gap in genomics education can be overcome by “real world big-data” experience and training. To help close the knowledge gap and promote researcher engagement, the Baylor College of Medicine’s All of Us Evenings with Genetics team developed a distributed learning, modular, virtual educational training series entitled “Train the Trainer” (Tx3).
Together with the goal of recruiting at least 1 million participants, the All of Us Research Program also seeks to attract at least 10 000 investigators to register (completed by April 2024) and productively use the Researcher Workbench to further human healthcare. To this end, we adopted a Train-the-Trainer approach to this learning challenge to accelerate the generation of both subject matter experts and teachers of the material.7–10 We acknowledge other efforts in this arena (RTI International’s Researcher Academy for example), but contend that Tx3 is unique in depth, scope, and methods. We note that all authors participated in the on-going development of materials and represent various backgrounds in the fields of genetics, genomics, bioinformatics, and education, with combined decades of experience.
The Tx3 Series has thus far served over 300 pre-registered Workbench learners through 2 iterations of training in late 2023 and early 2024. The goal for attendees is familiarity with available Workbench All of Us data and use policies, cohort and dataset selection, introduction to the Jupyter notebook, data manipulation and statistical analyses, and an introduction to utilizing genomic data. Here, we describe the Tx3 Series, our iterative development, learner feedback, lessons learned, and future developments in educational training and outreach. We offer this as a model to others seeking to develop educational offerings for the All of Us Research Program, leveraging our foundation, experience, and materials. Our PowerPoint slide decks are available as PDF documents (https://doi.org/10.5281/zenodo.11453503).
Materials and methods
Development of the Tx3 Series
The Tx3 Series is based heavily on the work of Coleman and colleagues,11 which consisted of a 4-class training plus educational materials “boot camp” developed for the annual All of Us Evenings with Genetics Biomedical Researcher Summit (May 2022).12 Our initial (pre-Tx3) efforts retained the 4-module format, launched in early 2023 and consisted of 2, 2-hour sessions per week. Surveys as well as interviews with NIH All of Us Community Engagement Partners (https://allofus.nih.gov/funding-and-program-partners/communications-and-engagement-partners), including representatives of the American Association on Health and Disability, the Delta Research and Education Foundation, and the National Alliance for Hispanic Health, emphasized the need for additional computer language and statistical analysis training. This input, together with experience to date, demonstrated the breadth of user experience and expectations. Therefore, in July and August 2023, the Tx3 curriculum was restructured and expanded as a 6-module series (see Table 1) to provide a slower pace with more depth per subject while simultaneously researching and adopting the Train-the-Trainer approach (Tx3).7–10
Table 1.
The 6 modules comprising the All of Us Tx3 Series.
| Modules | Main content | Pre-lab |
|---|---|---|
|
The All of Us Research Program and its data | Familiarize yourself with the User Support Hub and its “Getting Started” materials |
| The All of Us User Support Hub | ||
| The All of Us Researcher Workbench | ||
| Featured Workspaces on the Researcher Workbench | ||
| What you can expect from this series | ||
|
Visualizing and statistically comparing All of Us data: (A) Comparing normal distributions. (B) Example python code to plot histogram. (C) Example python code to perform a T-test. | Familiarize yourself with the Cohort Builder |
| Steps to create a project and data selection on the Researcher Workbench: (A) Create/copy a workspace. (B) Select a cohort and data to compare. (C) View selected data in the Jupyter notebook. | ||
| Plot histograms and assess height differences using python codes: (A) Identify the correct data in DataFrames. (B) Plot and save histograms. (C) Compare distributions with a T-test. (D) Celebrate your first AoU data analysis. | ||
|
Brief Review: (A) Workbench Components. (B) Create or Duplicate a Workspace. | Creating an All of Us dataset by setting your phenotype correctly |
| Introduction to phenotypes: (A) Terminology in phenotype study. (B) Tutorial workspace for phenotype selection. | ||
| Create an All of Us dataset: (A) Cohort Builder. (B) Concept Sets. (C) Create a Dataset. (D) Tutorial workspace examples. | ||
| Jupyter Notebook Introduction: (A) Background on Jupyter Notebook. (B) Access the All of Us data through Jupyter Notebook. | ||
|
Brief Review: (A) Define your phenotype. (B) Create cohorts, concept sets, and datasets. | Create a test educational workspace and duplicate a workspace |
| Jupyter Notebooks on the Researcher Workbench: (A) Exporting a Dataset. (B) Computing environments. (C) File storage options. | Create an Analysis Environment by exporting a workspace dataset | |
| Getting started with Jupyter Notebooks: (A) Introduction to the Jupyter Notebook. (B) Introduction to code snippets. (C) Using code snippets to save and retrieve data. (D) Backing up your Jupyter Notebook. (E) Other helpful tips. | Jupyter Notebook Features and Code Snippets | |
| Using code snippets to interact with Workspace Bucket | ||
| Back Up Notebooks—Save HTML or HTML Snapshots | ||
|
Getting to the Support Hub While Logged In | Review the Featured Workspace: Data Wrangling |
| Getting Started Resources | ||
| Data Wrangling Examples | ||
| Further Data Checking and Cleaning | ||
| Statistical analysis resources | ||
|
Brief review of previous modules | Tutorial workspace—How to work with All of Us Genomic Data |
| Significance of the All of Us genomic data: (A) Inclusive genomics improves everyone’s health. (B) Genomics data available. |
|
|
| Background on a Genome-Wide Association Study (GWAS): (A) The missing diversity in human genetic studies. (B) What is a GWAS? (C) Simplest Regression Model of Association. | Demo—Siloed Analysis of All of Us and UK Biobank Genomic data | |
| Steps to a GWAS project on the All of Us Researcher Workbench: (A) An Introduction to GWAS using Hail. | Demo—PheWas smoking | |
| Phenotype—Type 2 diabetes |
The Tx3 Series is sequential, with each module building upon the others while providing training videos, separate demonstration videos, and PowerPoint slide decks for both review and downstream lesson building. Programmatically, we emphasized a project-based approach, encouraging participants to bring analysis projects and develop research questions. Weekly sessions were also restructured: (1) a 2-hour session divided into didactic presentations and active-learning live demonstrations and (2) an unstructured 2-hour tutorial time for introductory/beginner tutoring on the All of Us platform as well as research and project-based questions. The Tx3 Series is open to participants with diverse backgrounds regardless of experience in programming, statistical analysis, and genomics research, while requiring attendees to be registered users of the All of Us Researcher Workbench. Weekly participant surveys were conducted in an iterative fashion by providing attendees with a link to an online survey questionnaire at the end of each session. Survey questions were designed specifically to capture the feedback of potential trainers. For example, we asked attendees to indicate their reason for attending the training session, as trainers or researchers. They were also asked to assess their confidence and provide suggestions on how we can improve their confidence in training others. We reviewed surveys in-depth during team meetings and responsively incorporated feedback into course development (see Table 2 and Figures S1-S3).
Table 2.
Selected comments from surveys conducted after each session.
| Positive | Constructive | |
|---|---|---|
|
I am totally new to workbench, but the information was very useful and engaging. Looking forward to learning a lot. | It will be better for me when I catch up with everyone and use Chrome instead |
| I am excited to do the demonstration work and to get unstuck with analysis! | This learning level is intermediate, it's not for beginners. | |
| Conducive learning environment and supportive facilitators. | The presenter tried to explain it but the material was high level for a novice so I will need to review again. | |
|
Thank you for this opportunity. I greatly appreciate it! | Live demo using the RW in addition to the slide show will be better |
| This was great! One of the best ones I have attended. Jun you are really great at explaining what is happening and how to navigate the workbench and why. | The pacing was pretty wonky for this lesson—I think it would make sense to ask participants to be prepared in certain ways before starting the class (eg, make sure everyone has the demo workspace duplicated so that we can all build the dataset together and spin up the computing environment together). It was also a bit confusing to have a set of slides that laid out the lesson but then the slides are too complicated/detailed to properly understand as participants, so we have to go through the slides and then go back to then have the presenter or various participants share their screens to walk back through the steps. | |
|
Homework exercises would be a good way to proceed | (comment from session 5) Putting the R and python codes side by side would have been more helpful to me. Like the way the first session was conducted |
| Very informative and helpful. I thought the level of detail and pace were great! The slides were very detailed and easy to follow | Thank you. Overall, the resources provided will be very helpful for getting going with Workbench but attending the sessions alone was not sufficient. Will take a great deal more work reviewing all the session videos and resources on my won. | |
| These sessions are slowly improving my understanding of the WB—thanks for organizing |
For example, in response to the timing of subject matter, we developed and distributed a course syllabus (Figure S4). In response to requests for pre-session materials, we produced pre-session slide decks and suggested readings. In response to the pacing of a session, we used a slower pace and more attendee check-ins.
Results
Tx3 Series enhances All of Us access and outreach
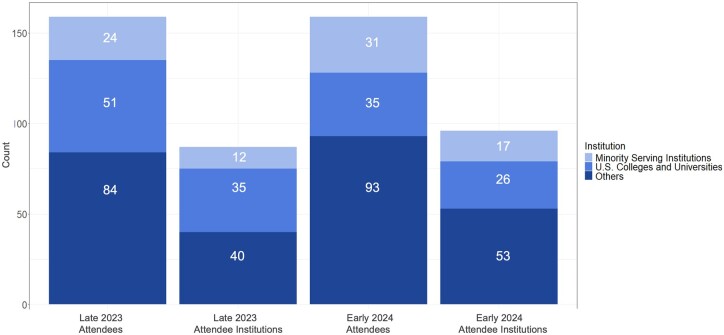
The 6-module Tx3 Series was deployed internally as a beta test in September-October 2023 with a cohort of 15 researchers. The next 2 iterations were publicly available in November-December 2023 and February-March 2024, with >150 attendees for each. One aim of the Baylor College of Medicine All of Us Evenings with Genetics grant is to increase access and outreach engaging Researcher Workbench users across multiple demographic sectors. To advertise the Tx3 Series, we used flyers (Figure S5) email, listservs, the Workbench User Support Hub Calendar, X (Twitter), Facebook, LinkedIn, and targeted outreach to related training programs, especially at Minority Serving Institutions (MSIs). We derived institutions from registrant email addresses where possible (15 registrants used Gmail or similar) and as shown in Figure 1, we have served 52 learners from colleges and universities identified as MSIs (17% of total, n = 305).13 In addition, the Tx3 Series drew participants from 141 unique institutions (including colleges, universities, research institutes, medical schools, hospitals, healthcare systems/networks, and government agencies in the United States and Puerto Rico), with 34 of these entities deemed as minority-serving (24% of total, n = 141).
Figure 1.
Attendance numbers for the 2 public Tx3 iterations held to date. Attendance is presented by maximum attendees per session together with their respective institutions and categorized by those from colleges and universities and whether those institutions are minority serving together with other institutions including medical schools, hospitals and healthcare systems, government agencies, etc.
Discussion
Challenges and lessons learned
Developing a virtual bioinformatics training series for a broad audience creates both opportunities and challenges. Here, we address 2 challenges we identified and resolved, so that other programs interested in similar efforts could start from an awareness of these issues.
Addressing a broad range of learners through unstructured tutorials and peer training
Participants in the All of Us Tx3 Series have a range of expertise from significant experience in computer programming and data analysis (and thus mainly just needed All of Us Researcher Workbench training) to data science and computer programming novices (often similarly needing All of Us Researcher Workbench training). Traditionally, this is resolved through stratifying training modules into “beginner” or “advanced” topics. However, that increases the overhead to offer training and reduces the availability for each type of participant. We sought to overcome this through unstructured tutorial time, informally called “Workbench Wednesdays,” which are 2-hour virtual “drop-in help desk” sessions. Participants bring questions ranging from technical coding approaches to research project design questions such as identifying specific rare disease variants in the All of Us dataset. Participants share questions, are paired with All of Us instructors, and split into 1 to 5 Zoom breakout rooms for focused assistance. Many attendees returned for multiple weeks as they developed projects and competence in the Researcher Workbench. Beginners could get extra help in these sessions, while advanced learners could progress and even help others. The small-group sessions were anecdotally highly valuable. We plan to explore the impact of these unstructured sessions in future iterations and develop surveys to gauge advanced participant interest in leading sessions.
Scaling class size and technical demonstrations
The All of Us Researcher Workbench is designed for team-science; however, there remain challenges when scaled to a large (50+) participant group of. Challenges we encountered centered around sharing Workbench workspaces with hundreds of registered participants, demonstration lag times (in starting Jupyter Notebook or saving/retrieving files), pacing of demonstrations with advanced and beginner learners, and accommodating All of Us data privacy by not showing row-level data during live demonstrations, even to our registered participants as we cannot control materials custody downstream. Ultimately, we developed several small “workarounds” such as pre-recorded video clips, pre-loading shared workspaces, and asking participants to sign-in and start their notebooks prior to the start of the session. The All of Us Data and Research Center1 developed an internal synthetic dataset that we intend to explore in further iterations but with the caveat that it does not contain genomic data.
Conclusions and future directions
As our Tx3 Series proved highly popular over the last year, we intend to continue iterating for on-going development, informed by participant feedback while expanding offerings such as enhanced modularity and the use of inverted learning models wherein advanced learners lead demonstrations at an instructor-level. Because many participants experienced challenges with computer programming and statistical analysis, we plan to develop and publish workspace modules that can be easily swapped based on some commonly requested applications such as pharmacogenomics, genomics association analysis and PheWAS analysis. This will reduce the training needed to run algorithms on the Researcher Workbench and will still ensure interaction with code by modifying only the key components (applying a “code-builder” approach similar in process to the cohort builder or by using code-snippets and libraries). In addition, many requests from participants focused on cohort building and the data cleaning or “wrangling” needed to convert a query in the dataset builder to a useful, non-redundant, harmonized, analysis-ready dataset. We will focus more content on these areas in future iterations, so that learners have code modules ready and available to apply in managing cohorts and datasets beyond the creation stage and into the analysis stage. We have recently seen multiple groups making use of combined analysis of large-scale datasets, such as comparing All of Us results with Genomics England results, and we intend to create a module on how to bring in external datasets to All of Us analysis. In our efforts to ensure that learners not only use project-based learning but also communicate their work to larger audiences, we will trial synchronizing at least 1 Tx3 session with the American Society of Human Genetics (ASHG) annual meeting abstract deadline, so that trainees with active All of Us projects can engage the Tx3 Series as they develop their projects into ASHG abstract submissions. Lastly, we are looking forward to developing additional modules based on application of Artificial Intelligence (AI) features to the All of Us Researcher Workbench, so that cohort building, data wrangling, and even the coding currently necessary for analysis can be minimized, and the questions a researcher can ask of the data can be prioritized. Since 2022, our work in developing the Tx3 Series has trained more than 300 All of Us Researcher Workbench users, and we anticipate seeing All of Us research publications from both our trainees and their trainees in the future.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the All of Us participants, without which, this work would not be possible.
Contributor Information
Deborah I Ritter, Department of Pediatrics, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX 77030, United States.
Jinyoung Byun, Section of Epidemiology and Population Sciences, Department of Medicine Institute for Clinical and Translational Research, Baylor College of Medicine, Houston, TX 77030, United States.
Jun Wang, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Stephen Richards, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States; Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, United States.
Pamela N Luna, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
LaTerrica Williams, Department of Pediatrics-Psychology, Baylor College of Medicine, Houston, TX 77030, United States.
Julie R Coleman, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Jasmine N Baker, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Shamika Ketkar, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Ashley M Butler, Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030, United States.
Latanya Hammonds-Odie, Department of Biological Sciences, School of Science and Technology, Georgia Gwinnett College, Lawrenceville, GA 30043, United States.
Elizabeth G Atkinson, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States; Jan and Dan Duncan Neurological Research Institute, Baylor College of Medicine, Houston, TX 77030, United States.
Kim C Worley, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Debra D Murray, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Brendan Lee, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Steven E Scherer, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Author contributions
Deborah Ritter (Conceptualization, Methodology, Writing—original draft, Writing—review & editing), Jinyoung Byun (Conceptualization, Methodology, Writing—original draft, Writing—review & editing), Jun Wang (Conceptualization, Methodology, Writing—original draft, Writing—review & editing), Stephen Richards (Conceptualization, Methodology, Writing—original draft, Writing—review & editing), Pamela Luna (Conceptualization, Methodology, Writing—original draft, Writing—review & editing), LaTerrica Williams (Conceptualization, Formal Analysis, Writing—original draft, Writing—review & editing), Julie Coleman (Conceptualization, Methodology, Resources, Writing—original draft, Writing—review & editing), Jasmine Baker (Conceptualization, Resources, Writing—original draft, Writing—review & editing), Shamika Ketkar (Conceptualization, Resources, Writing—original draft, Writing—review & editing), Ashley Butler (Conceptualization, Formal analysis, Methodology, Writing—original draft, Writing—review & editing), Latanya Hammonds-Odie (Conceptualization, Project administration, Writing—original draft, Writing—review & editing), Elizabeth Atkinson (Conceptualization, Resources, Writing—original draft, Writing—review & editing), Kim Worley (Conceptualization, Methodology, Resources, Supervision, Writing—original draft, Writing—review & editing), Debra Murray (Conceptualization, Funding acquisition, Project administration, Supervision, Writing—original draft, Writing—review & editing), Brendan Lee (Conceptualization, Funding acquisition, Writing—original draft, Writing—review & editing), and Steven Scherer (Conceptualization, Investigation, Methodology, Project administration, Supervision, Writing—original draft, Writing—review & editing)
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work was funded by the National Institutes of Health Award #: OT2 OD031932 awarded to B.L. and D.D.M.
Conflicts of interest
The authors declare no competing interests.
Data availability
The lesson slide deck pdf files underlying this article are available in Zenodo (https://doi.org/10.5281/zenodo.11453503). Further materials underlying this article may be available upon reasonable request to the corresponding author.
References
- 1. Mayo KR, Basford MA, Carroll RJ, et al. The All of Us Data and Research Center: creating a secure, scalable, and sustainable ecosystem for biomedical research. Annu Rev Biomed Data Sci. 2023;6:443-464. 10.1146/annurev-biodatasci-122120-104825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Işık EB, Brazas MD, Schwartz R, et al. Grand challenges in bioinformatics education and training. Nat Biotechnol. 2023;41(8):1171-1174. 10.1038/s41587-023-01891-9 [DOI] [PubMed] [Google Scholar]
- 3. National Academies of Sciences, Engineering, and Medicine; Policy and Global Affairs; Committee on Women in Science, Engineering, and Medicine; Committee on Increasing the Number of Women in Science, Technology, Engineering, Mathematics, and Medicine (STEMM); Helman A, Bear A, Colwell R, eds. Promising Practices for Addressing the Underrepresentation of Women in Science, Engineering, and Medicine: Opening Doors. National Academies Press (US; ); 2020. [PubMed] [Google Scholar]
- 4. Pevzner P, Shamir R. Computing has changed biology—biology education must catch up. Science. 2009;325(5940):541-542. 10.1126/science.1173876 [DOI] [PubMed] [Google Scholar]
- 5. Wolff A, Gooch D, Montaner JJC, et al. Creating an understanding of data literacy for a data-driven society. JoCI. 2016;12(3):9-26. [Google Scholar]
- 6. Dolezel D, McLeod A. Big-data skills: bridging the data science theory-practice gap in healthcare. Perspect Health Inf Manag. 2021;18(Winter):1j. [PMC free article] [PubMed] [Google Scholar]
- 7. Pearce J, Mann MK, Jones C, et al. The most effective way of delivering a train-the-trainers program: a systematic review. J Contin Educ Health Prof. 2012;32(3):215-226. 10.1002/chp.21148 [DOI] [PubMed] [Google Scholar]
- 8. Triplett NS, Sedlar G, Berliner L, et al. Evaluating a train-the-trainer approach for increasing EBP training capacity in community mental health. J Behav Health Serv Res. 2020;47(2):189-200. 10.1007/s11414-019-09676-2 [DOI] [PubMed] [Google Scholar]
- 9. Orfaly RA, Frances JC, Campbell P, et al. Train-the-trainer as an educational model in public health preparedness. J Public Health Manag Pract. 2005;Suppl:S123-S127. 10.1097/00124784-200511001-00021 [DOI] [PubMed] [Google Scholar]
- 10. Pancucci S. Train the Trainer: the bricks in the learning community scaffold of professional development. Int J Educ Pedagog Sci. 2007;1(11):597-604. [Google Scholar]
- 11. Coleman JR, , BakerJN, , Kekar S. Development and evaluation of a training curriculum to engage researchers on accessing and analyzing the All of Us data. J Am Med Inform Assoc. 2024;31(12):2857-2868. 10.1093/jamia/ocae240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coleman JR, Baker JN, Ketkar S. . All of Us evenings with genetics data trainings for the 2024 Biomedical Researchers Faculty Summit. Zenodo. 2024. https://doi.org/10.5281/zenodo.12009041 [Google Scholar]
- 13.The Rutgers Center for Minority Serving Institutions. Rutgers-New Brunswick Graduate School of Education. Accessed March 23, 2024. https://cmsi.gse.rutgers.edu/msi-directory
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lesson slide deck pdf files underlying this article are available in Zenodo (https://doi.org/10.5281/zenodo.11453503). Further materials underlying this article may be available upon reasonable request to the corresponding author.